Abstract
The sewage sludge isolate Pseudomonas nitroreducens HBP-1 was the first bacterium known to completely degrade the fungicide 2-hydroxybiphenyl. PacBio and Illumina whole-genome sequencing revealed three circular DNA replicons: a chromosome and two plasmids. Plasmids were shown to code for putative adaptive functions such as heavy metal resistance, but with unclarified ability for self-transfer. About one-tenth of strain HBP-1′s chromosomal genes are likely of recent horizontal influx, being part of genomic islands, prophages and integrative and conjugative elements (ICEs). P. nitroreducens carries two large ICEs with different functional specialization, but with homologous core structures to the well-known ICEclc of Pseudomonas knackmussii B13. The variable regions of ICEPni1 (96 kb) code for, among others, heavy metal resistances and formaldehyde detoxification, whereas those of ICEPni2 (171 kb) encodes complete meta-cleavage pathways for catabolism of 2-hydroxybiphenyl and salicylate, a protocatechuate pathway and peripheral enzymes for 4-hydroxybenzoate, ferulate, vanillin and vanillate transformation. Both ICEs transferred at frequencies of 10−6–10−8 per P. nitroreducens HBP-1 donor into Pseudomonas putida, where they integrated site specifically into tRNAGly-gene targets, as expected. Our study highlights the underlying determinants and mechanisms driving dissemination of adaptive properties allowing bacterial strains to cope with polluted environments.
1. Introduction
The recent industrial revolution has driven the intensive exploitation of natural resources and heavy use of chemicals, leading to massive environmental pollution. Besides its potential harmful effects on human health, pollution also strongly impacts the functionality of ecosystems. Even at the microbial level, polluting substances pose a strong selection, leading to the disappearance of sensitive strains, and the enrichment of adapted and resistant strains [1]. In some instances of organic pollution, strains have evolved to metabolize and transform the compounds under scrutiny [1]. Pollutant-degrading bacteria have attracted interest, because they may be applied to directly or indirectly remediate pollution and restore ecosystem health [1,2]. The inoculation of specific strains, such as Pseudomonas sp. JS150, Pseudomonas knackmussii B13, or Pseudomonas veronii 1YdBTEX2, has been shown to significantly enhance biodegradation of pollutants such as phenol, chlorobenzoate, benzene, toluene or xylene [3,4,5].
Pseudomonas nitroreducens HBP-1 (DSM 8897, previously named Pseudomonas azelaica HBP-1), was isolated in 1988 from sewage sludge in California [6], and is one of the rare bacteria able to completely degrade and use the biocide 2-hydroxybiphenyl (2HBP) as the sole carbon and energy source [6]. The metabolic pathway for 2HBP conversion in P. nitroreducens HBP-1 has been biochemically and genetically characterized in detail [6,7,8,9,10,11]. The enzymes responsible for the first steps of 2HBP metabolism are encoded by the hbpCAD locus. A broad-spectrum monooxygenase (HbpA) catalyzes the hydroxylation of 2HBP and a wide range of other 2-substituted phenols to their corresponding catechols [10]. Subsequent action of a dioxygenase (HbpC) leads to the cleavage of the hydroxylated ring, which is then converted to benzoate and 2-hydroxy-2,4-pentadienoic acid by means of the HbpD hydrolase. Both compounds are further metabolized via meta-cleavage degradation pathways. The expression of the hbpCAD genes is regulated at the transcriptional level by the sigma-54-dependent activator HbpR in the presence of 2HBP (or analogous molecules) [12,13,14].
The genomes of P. nitroreducens HBP-1 as well as its close relative P. azelaica strain Aramco J were recently released as drafts, consisting of 212 and 91 contigs, respectively [15,16]. Unfortunately, although the draft genomes provided insights into the gene content and functional capacities of P. nitroreducens, their incompleteness hampered comparative genomics analyses and evaluations of the potential role of mobile genetic elements in their evolution. The main goal of this study was, therefore, to produce a complete gapless genome of P. nitroreducens HBP-1 and to study the nature and functionalities of its potential mobile elements. We used a combination of Illumina short-read sequencing as well as PacBio long-read sequencing to assemble the HBP-1 genome. Regions of genome plasticity in HBP-1 were further analyzed for characteristics of genomic islands, putative prophages, and integrative and conjugative elements (ICEs). Mating assays between P. nitroreducens HBP-1 as donor and P. putida UWC1 as recipient were conducted to examine self-transfer of deduced ICEs. We found that P. nitroreducens contains two active ICEs that can transfer at low rates. The ICEs carry extensive sets of genes involved in aromatic compound metabolism and resistance to heavy metals, likely determining the ability of strain HBP-1 to cope with numerous toxic compounds. The capacity to spread adaptive functions via the transfer of ICEs could favor adaptation of further bacterial members in natural communities to withstand pollutant stress [17].
2. Materials and Methods
2.1. Culture Conditions
P. nitroreducens HBP-1 (formerly P. azelaica HBP-1) [6] and P. putida UWC1 miniTn7::Ptac-mcherry (UWCGC) (Gmr) [18] were routinely grown in lysogeny broth (LB Miller, Difco) at 30 °C and 180 rpm in an orbital shaker/incubator. The strains were preserved at −80 °C in LB containing 15% (vol/vol) glycerol. As a defined medium, 21C minimal medium [19] was used, which was supplemented with one of the following carbon sources (final concentrations): sodium succinate (20 mM), 2-HBP (2.5 mM, dosed 1:100 from a stock in dimethylsulfoxide), salicylate (2.5 mM), benzoate (2.5 mM), 4-hydroxybenzoate (2.5 mM), vanillate (2.5 mM), phthalate (2.5 mM), phenol (2.5 mM), or catechol (2.5 mM). Whenever necessary for the selection of genetic constructs, gentamicin (10 or 20 µg mL−1), rifampicin (25 µg mL−1), mercuric chloride (4 µM) or sodium arsenite (5 mM) were amended to the growth media.
2.2. DNA Isolation, Sequencing and Assembly
The P. nitroreducens HBP-1 genome was sequenced with both long-read (PacBio, Menlo Park, CA, USA) and short-read (Illumina paired-end) technology. For PacBio sequencing, a single clone grown from a glycerol stock of P. nitroreducens HBP-1 on a minimal medium–succinate plate, was inoculated into the same liquid medium and was harvested in mid-exponential growth phase (ca. 0.5 optical density units at 600 nm). Four aliquots of ca. 109 cells were pelleted by centrifugation at 14,000× g for 5 min, and DNA was extracted in parallel using a PowerSoil DNA extraction kit (MoBio Laboratories, Carlsbad, CA, USA). The resulting DNA aliquots were pooled together, precipitated with ethanol-sodium acetate, washed with 75% ethanol, briefly dried and dissolved in 5 mM Tri-HCl pH 8. DNA quality was analyzed on a 2100 Electrophoresis Bioanalyzer Instrument (Agilent Technologies, Santa Clara, CA, USA), and quantified with an Invitrogen™ Qubit™ 3 Fluorometer (Thermo Fisher Scientific Inc, Waltham, MA, USA). Subsequently, 7.4 μg of DNA were fragmented at 4100 rpm for 1 min with a Covaris g-TUBE device (Covaris Ltd., Brighton, UK) and 5 μg was used for preparing sequencing libraries (SMRTbell template prep kit 1.0, Pacific Biosciences, Menlo Park, CA, USA). DNA sequencing was performed on a PacBio RSII instrument (Pacific Biosciences, Menlo Park, CA, USA) at the Lausanne Genomic Technologies Facility using a single v3 SMRT™ Cell, P6-C4 chemistry and 4-h movies time. A total of 52,338 reads with a mean length of 13,833 nt were de novo assembled and circularized using the Hierarchical Genome Assembly Process (HGAP) version 3.0 [20], yielding three circular replicon assemblies with 83-fold average coverage. The assembly was validated using Illumina 50-nt paired-end sequence reads, obtained on a library prepared and sequenced essentially as described by Miyazaki et al. [4] in a single flow lane using the Illumina Genome Analyzer II sequencing platform, resulting in 8,000,273 reads. Subsequently QC-trimmed reads were mapped to the HGAP assembly using Bowtie [21], the output was converted to SAM/BAM format with SAMtools [22] and the assembly was validated with QualiMap version 2.2.1 [23,24].
In order to quantify and compare mean coverages of the three P. nitroreducens HBP-1 replicons, we resequenced another two DNA samples prepared from two succinate grown cultures inoculated either with a single or with five independent P. nitroreducens HBP-1 colonies. DNA was extracted from 109 cells by bead beating and magnetic bead purification (CleanNGS beads, LABGENE Scientific SA) following the manufacturer′s protocol. Indexed libraries were constructed by using the Vazyme TruePrep DNA Library Prep kit V2 for Illumina (Vazyme) starting from 1 ng DNA input. Libraries were paired-end sequenced on a Illumina HiSeq2500 with HiSeq Rapid SBS Kit V2 in a rapid 2 × 150 mode.
2.3. Annotation
The finished assembly of P. nitroreducens strain HBP-1 was annotated by the NCBI Prokaryotic Genome Annotation Pipeline (PGAP) [25,26] and deposited to GenBank (see below for accession numbers). In addition, Rapid Annotation using Subsystems Technology (RAST) [27] and Prokka [28] annotations were explored for maximum details about gene content. The functional gene content of the reported mobile genetic elements was inspected manually and, whenever needed, used to improve the automatic annotations based on analyses by BLAST to GenBank and UniProt Knowledge Base (UniProtKB) with the emphasis given to expert annotations. KEGG and MetaCyc [29], or specific literature were used as reference for metabolic pathways.
2.4. In Silico Analyses of Genomic Islands, ICEs and Prophages
IslandViewer [30] and PHASTER [31] were used with default parameters to search for genomic islands and phages, respectively. The data were visualized using ACT Artemis [32], DNA plotter [33], and Adobe Illustrator (Adobe Inc, San Jose, CA, USA, vs. 2020).
2.5. Characterization of ICE Attachment Sites
PCR primers were designed to amplify putative attL and attR recombination sites of ICEPni1 and ICEPni2 in P. nitroreducens HBP-1 and amplicons were verified by amplicon sequencing (Table 1). The primer sets used to amplify the specific attachment sites in P. nitroreducens and P. putida are listed in Table 2. PCRs were performed using 1 × GoTaq® G2 Green Master Mix (Promega), with 100 pM of each primer and 20 ng of purified DNA as a template with the following thermal cycler conditions: (i) 3 min at 94 °C; (ii) 30 cycles of 30 sec at 94 °C, 30 sec at the appropriate annealing temperature, and 1 min per kb at 72 °C; and (iii) 5 min at 72 °C. After agarose gel analyses, the intended PCR products were purified using a NucleoSpin® Gel and PCR Clean-up kit (Macherey-Nagel, Düren, Germany), and Sanger-sequenced at GATC Services (Eurofins Genomics, Luxembourg, Luxembourg). Nucleotide sequences were aligned and compared using DNASTAR Lasergene v.15 package and BLASTn.

Table 1.
Primers used in this study.

Table 2.
List of primers used to amplify integrative and conjugative element (ICE) recombination sites.
2.6. ICE Transfer Assays
The ability of the discovered ICEs to self-transfer was examined in conjugation assays between P. nitroreducens HBP-1 (donor) and P. putida UWC1 miniTn7::Ptac-mcherry (recipient) [18]. For ICEPni1 transfer, the donor was grown in liquid minimal medium with 20 mM succinate supplemented with either 4 mM sodium arsenite or 8 µM mercury chloride. For ICEPni2, the donor was grown in liquid minimal medium with 5 mM 2-HBP. The recipient was grown in succinate medium amended with 20 µg mL−1 Gm and 25 µg mL−1 Rif. Both cultures were harvested at OD600 0.5–1.0 by centrifugation for 5 min at 5000× g, washed once in 1 mL minimal medium, combined at 1:1 donor:recipient ratio, pelleted by centrifugation as above, resuspended and then deposited on a 0.2–µm cellulose acetate filter (Sartorius) placed on 1 mM succinate minimal media agar. Plates with filters were incubated for 48 h at 30 °C. The number of donor and recipient cells per mating ranged between 4.1 × 107 and 7.5 × 108 (per filter). After the incubation, the resulting mating mixes were resuspended in 0.5 mL of minimal medium, serially diluted and plated on selective media as follows. The number of donor cells (P. nitroreducens HBP-1) was enumerated on minimal medium agar plates supplemented with 2.5 mM 2HBP. Colonies of the UWCGC recipient (P. putida UWC1 miniTn7::Ptac-mcherry) were counted on minimal media agar amended with 20 mM succinate, 20 µg mL−1 gentamicin and 25 µg mL−1 Rif. P. putida ICEPni1-exconjugants were selected on minimal medium agar plates supplemented with 20 mM succinate, 20 µg mL−1 gentamicin and 4 µM mercury chloride, whereas, for ICEPni2 exconjugants, the minimal medium was amended with 20 µg mL−1 gentamicin and either 5 mM 2HBP or 2.5 mM salicylate. The exconjugant colonies were purified by streaking on the respective selective media and the presence of ICEs was confirmed by PCR as follows. The ICEPni1 marker merA was amplified with primer pair pPazICE1_merA_fw and pPazICE1_merA_rev, whereas ICEPni2 marker hbpA was amplified with pPazICE2_hbpA_fw and pPazICE2_hbpA_rev (Table 1). In addition, the P. putida background of putative exconjugants was confirmed by mCherry fluorescence detected with a Zeiss Axioplan II microscope equipped with a 100× Plan Achromat oil objective lens (Carl Zeiss, Oberkochen, Germany), a SOLA SE light engine (Lumencor, Beaverton, OR, USA), and a SPOT Xplorer 1.4 Mp Cooled CCD Camera (SPOT Imaging Solutions, a division of Diagnostic Instruments, Inc., Sterling Heights, MI, USA).
2.7. Database Submission
The complete gapless P. nitroreducens HBP-1 genome is available from Genbank under accession numbers CP049140, CP049142, CP049141 for the chromosome, and the two plasmids pPniHBP1_1 and pPniHBP1_2, respectively.
3. Results and Discussion
3.1. Genome of Pseudomonas Nitroreducens HBP-1
A complete genome of P. nitroreducens HBP-1 was sequenced using PacBio long read technology, assembled with HGAP and annotated by the NCBI Prokaryotic Genome Annotation Pipeline (PGAP) [25,26]. The final gapless P. nitroreducens genome is composed of three replicons: a circular chromosome of 6874,118 bp (CP049140), and two plasmids pPniHBP1_1 (CP049142, 425,042 bp) and pPniHBP1_2 (CP049141, 128,388 bp, Figure 1 and Table S1, sheet GenomeFeatures). Remapping of Illumina sequence reads on the complete assembled genome showed that the three replicons have roughly the same coverage, suggesting a single copy of each replicon per cell (Figure S1). Comparison with a previously published draft genome of HBP-1 [15] notably highlighted the presence of a third replicon (pPniHBP1_2), whereas only two repicons (a chromosome and a megaplasmid) had been proposed previously [15]. Further analysis indicated that the sequences of all three replicons (Figure 1) are found among the 212 contigs of the draft genome but had remained fragmented. Some of the previous contigs overlap the (arbitrary) starts and ends of the two newly assembled replicons, confirming that P. nitroreducens plasmids are closed circular DNA molecules (Figure 1A,B). The chromosome of P. nitroreducens aligned well with those of a number of other Pseudomonas species but presented a variety of regions of genome plasticity that likely correspond to genomic islands (Figure 1A, green and purple circles; discussed further below).
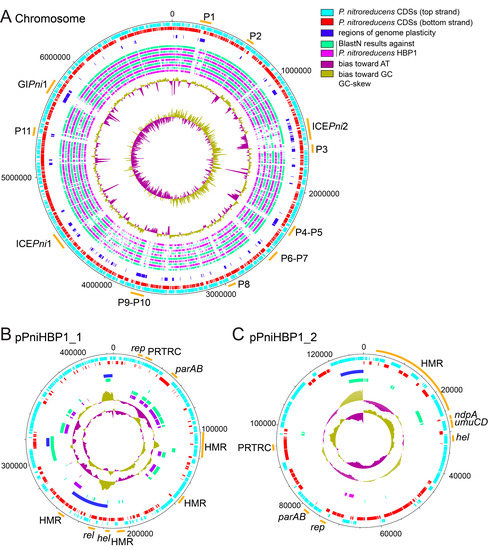
Figure 1.
Circular maps of replicons of the P. nitroreducens HBP-1 genome. (A). Chromosome with predictions of the main genomic island (GIPni1), prophages (P) and ICEs (ICEPni1 and ICEPni2). The outermost circles show the location of predicted ORFs on the top strand (light blue) and bottom strand (red), followed by IslandViewer and PHASTER predictions showing regions of genome plasticity (dark blue). The inner circles represent BlastN comparisons with genomes of other Pseudomonas species (alternating green and purple circles), from the outside to the inside: Pseudomonas sp. AK6U (Acc. No., NZ_CP025229.1), Pseudomonas knackmussii B13 (NZ_HG322950.1), Pseudomonas putida KT2440 (NC_002947.4), Pseudomonas aeruginosa PAO1 (NC_002516.2), Pseudomonas citronellolis SJTE-3 (NZ_CP015878.1), Pseudomonas oryzihabitans USDA-ARS-USMARC-56511 (NZ_CP013987.1), Pseudomonas pseudoalcaligenes CECT 5344 (NZ_HG916826.1), Pseudomonas resinovorans NBRC 106553 (NC_021499.1), Pseudomonas stutzeri 28a24 (NZ_CP007441.1), and Pseudomonas veronii 1YdBTEX2 (LT599583.1). The two innermost circles depict the GC plot (penultimate circle), and the GC skew (most central circle). Numbers around the black circle indicate the size in base pairs. (B). Plasmid pPniHBP1_1 with the indication of heavy metal resistance loci (HMR), active partition system (parAB or PRTRC), initiation replication protein (rep), relaxase (rel), helicase (hel). Organization is the same as that described for the map of the chromosome except for the BlastN comparisons, which were performed with (from the outside to the inside) Pseudomonas putida KT2440 (NC_002947.4), Pseudomonas aeruginosa genomic island PAGI-5 (EF611301.1), Pseudomonas aeruginosa strain PA298 plasmid pBM908 (CP040126.1), and Pseudomonas aeruginosa strain T2101 plasmid pBT2101 (CP039991.1). (C). Plasmid pPniHBP1_2 with the same indications as for pPniHBP1_1, and other features of interest (umuCD, ndpA). Organization is the same as that described for the map of the chromosome except for the BlastN comparisons, which were performed with (from the outside to the inside). Pseudomonas sp. SCB32 chromosome (CP045118.1), Pseudomonas aeruginosa strain AR_0356 plasmid (pAR0356, CP027167.1).
3.2. Genomic Insight into Plasmids of P. nitroreducens
P. nitroreducens HBP-1 carries two plasmids that were named pPniHBP1_1 and pPniHBP1_2 (Figure 1B,C). pPniHBP1_1 is circa 425 kb and codes for a putative RepB replication initiation protein G5B91_33395 [35]. At least two putative partition systems are encoded: a parA/parB locus (G5B91_33565/G5B91_33570), a gene coding for a PRTRC system ParB protein (G5B91_33435) and two orphan ParB-encoding genes (G5B91_35220 and G5B91_33555, Figure 1B and Table S1 pPniHBP1_1). pPniHBP1_1 also codes for a putative relaxase VirD2 (Figure 1B), but no other conjugative transfer components, suggesting that it could be a mobilizable plasmid relying on the T4SS of another autonomous conjugative element for its transfer [36]. pPniHBP1_1 carries three loci coding for resistance to heavy metals: copper, tellurite, mercury and arsenate/arsenite (Figure 1 and Table S1, sheet pPniHBP1_1).
Plasmid pPniHBP1_2 is 128,388 bp in length and encodes a RepB-type replication initiation protein, a ParA family protein, a ParB/RepB/Spo0J family partition protein and a replication terminus site-binding protein. All share about 50–65% aa identity with homologs from other Pseudomonas plasmids. Other plasmid-related functions include six ParB-homologs, a ThiF-Related Cassette (PRTRC), an abortive phage infection protein, a type-I restriction-modification system DNA methylase and a type IV toxin-antitoxin gene pair (AbiEi/AbiEii) (Table S1, sheet pPniHBP1_2). In addition, pPniHBP1_2 carries a 12-gene cluster that is well conserved among other Pseudomonas plasmids (notably, the P. resinovorans plasmid pCAR1). It seems unlikely that pPniHBP1_2 encodes its own conjugal transfer system since no hallmark type IV secretion system (T4SS) structural genes were identified. We did find genes for the putative conjugal transfer mating pair stabilization proteins TraN, the PilB T4P pilus assembly pathway ATPase-like protein, a helicase and an endonuclease (Table S1 pPniHBP1_2). This might indicate that pPniHBP1_2 is not self-transferable, but would depend on other mobile genetic elements for transfer [36]. The functional gene cargo on pPniHBP1_2 is predicted to code for proteins involved in sensing and defense by the efflux of heavy metal cations such as Co2+, Zn2+, Cd2+, Cu2+, Ag1+, Hg2+, Pb2+ (Table S1, sheet pPniHBP1_2) [37,38,39]. The plasmid is also predicted to code for the metabolism of diverse (amino)aromatic compounds (Table S1, sheet pPniHBP1_2).
3.3. Genomic Islands in the Genome of P. nitroreducens
The analysis of regions of genome plasticity in the P. nitroreducens chromosome by Island Viewer [30] and PHASTER [31] indicated several potential genomic islands (GIs) and prophages (Figure 1A). Moreover, the two plasmids might contain GIs (Figure 1B,C). The P. nitroreducens genome likely contains eight intact prophages (P1, P2, P6, P7, P8, P9, P10, P11), whereas a further seven regions (P3, P4, P5, P12, P13, P14, and P15) may encode satellite prophages, phage remnants or tailocins [40,41] (Figure 1A and Table 3). None of the phage-related GIs appeared to code for obvious adaptive functions.

Table 3.
PHASTER predictions of prophage locations in P. nitroreducens HBP-1.
Two GIs (Figure 1A) contained homologs to the hallmark gene VirB4 from the T4SS and, therefore, encompassed potentially transferable elements [42]. The product of both VirB4 homologs had a high similarity to the VirB4 protein of the prototypical element ICEclc of P. knackmussii B13, suggesting they belong to a mating-pair formation type MPFG that is characteristic for ICE T4SS [42]. BlastP comparison with VirB4ICEclc revealed 95% of amino acid (aa) identity over 98% of the protein sequence for VirB4ICEPni1, and 79% of aa identity over 99% for VirB4ICEPni2. The GIs that encompassed the virB4 homolog genes were thus renamed as putative ICEs ICEPni1 and ICEPni2 (integrative conjugative element of P. nitroreducens 1 and 2) for reasons outlined further below. Although ICEPni2 had been detected before and named ICEbhp [15], it was renamed here to comply with the suggestions put forward by Burrus and collaborators [43].
The third GI encompassed a 123-kb region between tRNAGln and tRNAMet (G5B91_27170 and 27745) on the P. nitroreducens HBP-1 genome (Figure 1A, designated as genomic island P. nitroreducens 1, GIPni1). GIPni1 did not contain any genes coding for horizontal transfer, thus preventing a more specific classification (Figure 1A and Table S1, sheet GIPni1). Sequence analysis revealed that GIPni1 bears two hallmark proteins shared by prophages and ICEs: a tyrosine type recombinase/integrase (G5B91_27195) and an AlpA-related transcriptional regulator/excisionase (G5B91_27730). Further resemblance to prophages was limited to three proteins: an inovirus Gp2 family protein, and two DUF3732 and DUF932 domain-containing proteins (Table S1, sheet GIPni1). There was little conservation in gene content between GIPni1 and other putative GIs occupying the same genome plasticity region in other Pseudomonas genomes, except for about a dozen genes surrounding the integrase gene and few other hypotheticals scattered throughout the GI (Table S1, sheet GIPni1). The large majority of the functions encoded on GIPni1 are enzymes associated with lipid metabolism. This concerns mainly fatty acid beta-oxidation pathway enzymes, many of which are represented by multiple alleles, for example, (long- and medium-chain) fatty acid-CoA ligases (4 alleles), acyl-CoA dehydrogenases (12), enoyl-CoA hydratases (8), thiolases (2), oxidoreductases (14), (acyl) CoA-transferases (3 alleles, Table S1, sheet GIPni1). Their corresponding genes are organized in operon-like structures and are associated with transcriptional regulators, transporters and electron-transfer components, likely forming complete and functional metabolic systems. In addition, the GIPni1 encodes a dozen different hydrolases and amidohydrolases, a glycerol-dehydrogenase and a glycolate oxidase, further extending its metabolic and adaptive potential for the host. Like in the case of the ICEPni2 (described further below), we hypothesize that the lipid- and fatty-acid-rich environment of the wastewater treatment plant has been selective to fix the acquired lipid metabolism-encoding GIPni1 in the genome of P. nitroreducens HBP1.
3.4. Defining the Borders of ICEPni1 and ICEPni2
Both putative ICE regions in the P. nitroreducens HBP-1 genome contained an integrase gene. The ICEPni1 integrase gene (G5B91_21265) shared 91% aa identity over 94% of the protein sequence with that of the ICEclc and was oriented adjacent to and in the same orientation as a tRNA operon encompassing three GCC tRNAGly and one TTC tRNAGlu gene (G5B91_21285-G5B91_21270). This strongly resembled the ICEclc “right end” structure (Figure 2) [34]. A repeat of the 18-bp 3′-end sequence of the tRNAGly (5′-G(A/T)CTCGTTTCCCGCTCCA-3′) was found about 95 kb upstream, suggesting that these repeats form attR and attL recombination sites (Figure 1, Figure 2A, and Table 4). The size of ICEPni1 in between the two 18-bp repeats was 95,926 bp (chromosome coordinates 4,376,070–4,471,995). The ICEPni1 attachment sites were almost identical to those of ICEclc, suggesting that they share a similar integration specificity [34].
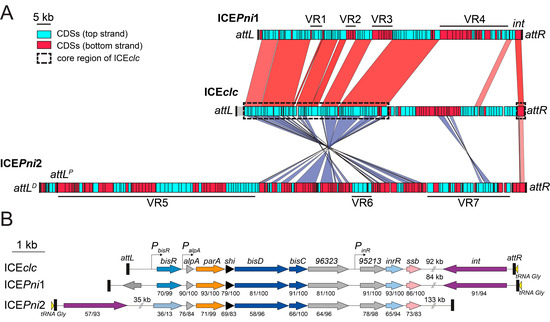
Figure 2.
Comparison of ICEPni1 and ICEPni2 from P. nitroreducens, and ICEclc from P. knackmussi B13 (A). Linear map of ICEPni1, ICEPni2, and ICEclc with indication of the location of predicted ORFs on the top strand (light blue boxes) and bottom strand (red boxes), the integrase-encoding gene (int), the attachment sites (att, black boxes), and variable regions (VR). The core region of ICEclc was framed with dashed black rectangles. The integrase gene of ICEclc on the rightmost end of the element was used as a reference point for the alignments. Comparisons were performed using BlastN and are displayed by colored areas linking related regions in the same (red) and inverted (purple) orientation. The intensity of the colored area reflects the percentage of nucleotide identity (minimum 65%) between the sequences. (B). Schematic representation (drawn to scale) of the genetic organization of ICEclc, ICEPni1 and ICEPni2 regulation loci. Genes are represented by arrowed boxes, and color-coded according to bioinformatic prediction or experimental demonstration of their function: purple, integration/excision; orange, active partition; light to dark blue, transcriptional activators; black, toxic effect; pink, single-stranded DNA protection; light yellow, tRNA; gray, unknown function. Promoters are represented by angled arrows pointing towards the transcription orientation. Numbers under genes (x/y) indicate the percentage of amino acid identity (x) and the coverage (y) of corresponding gene product in ICEclc.

Table 4.
Sequenced attachment sites of ICEPni1 and ICEPni2 in P. nitroreducens HBP-1 and in P. putida UWCGC.
The ICEPni2 integrase (G5B91_07495) showed 57% aa identity to IntB13 over 93% of the predicted protein sequence. The gene was located on the minus strand and was preceded by a CCC tRNAGly gene (G5B91_07500). Two sequences identical to the tRNAGly 18-bp 3′-end (5′-TTCCCTTCGCCCGCTCCA-3′) were found about 170 kb upstream, in two direct copies separated by ca. 6 kb from each other. We designated these as candidate proximal and distal attL recombination sites (attLPICEPni2 and attLDICEPni2 respectively) (Figure 2A and Table 4). As we show below, we only detected excision from the proximal recombination site attLPICEPni2 and identified its recombination from post-transfer integration in the new host. This indicated, therefore, that ICEPni2 would encompass the region 1410,790–1582,233 of the P. nitroreducens HBP-1 chromosome, and its size, including both repeats, would amount to 171,444 bp (Figure 2A).
3.5. ICEPni1 and ICEPni2 Are Related to ICEclc
Both ICEPni1 and ICEPni2 showed a region with extensive similarity to the core gene region of ICEclc, which codes for functions essential to the ICE lifestyle such as integration, excision, DNA processing, mating pore formation, and regulation (Figure 2A, dashed black rectangles) [44]. The analogous core region of ICEPni2 appears to be ‘inverted’ relative to the location of the integrase gene on ICEclc and ICEPni1 (Figure 2A). Homologs of the main regulatory genes for ICEclc activation were present on ICEPni1 and ICEPni2 (Figure 2B) [45]. This notably included a BisR activator homolog on ICEPni1 with 70% aa identity over 99% of the sequence related to BisR of ICEclc, and further, homologs of alpA, parA, shi, bisD and bisC. The strong gene synteny between regulatory loci suggests that ICEPni1 may be activated in a similar manner as ICEclc, which would thus entail a bistable formation of transfer competence in a specific small subpopulation of cells [45,46,47]. Gene homologs on ICEPni2 were more distantly related to the ICEclc regulatory module with around 70% nucleotide and 59–76% amino acid sequence identity, but still conserved synteny of alpA throughout bisC. In contrast, ICEPni2 did not appear to encode a BisR regulator homolog upstream of alpA, suggesting that its activation may proceed differently than for ICEPni1 and ICEclc (Figure 2B).
Comparison of the ICEs of P. nitroreducens with ICEclc further showed how conserved stretches are interspersed with unique variable regions (VRs), highlighting the modular pattern of their evolution (e.g., VR-regions indicated in Figure 2A). Some of the variable regions may have been maintained because they conferred selective advantages to P. nitroreducens, as will be discussed further below.
3.6. ICEPni1 Encodes Mercury, Arsenic and Formaldehyde Detoxification and a Bacteriophage Defense System
Manually curated annotations (Table S1, sheet ICEPni1) suggested that ICEPni1 carries three sets of genes for defense against toxic compounds. The first set (on VR1, G5B91_20860 to G5B91_20875) encodes the archetypal mercury resistance determinants. This includes mercury(II) uptake and reduction by a complex of mercuric transport protein MerT, mercuric transport protein periplasmic component MerP, mercuric reductase MerA under control of Hg(II)-responsive transcriptional regulator MerR. The second set (i.e., VR2, G5B91_20945 to G5B91_20965) codes for arsenate/arsenite resistance, with a classical transcriptional repressor ArsR, which controls an operon of arsenate reductase ArsC and arsenite efflux transporter ArsB. These, together, facilitate reduction of As(V) to As(III) and its efflux, followed by a trivalent organoarsenical oxidase ArsH, which oxidizes methyl-arsenite As(III) and other highly toxic trivalent organoarsenicals (e.g., herbicides MSMA, DSMA or CAMA) to less toxic pentavalent species [48,49]. The group of genes within VR4 (Figure 2A) likely codes for formaldehyde oxidation via glutathione-dependent and –independent pathways, and is represented by homologues of S-(hydroxymethyl) glutathione dehydrogenase FrmA (G5B91_21165) and S-formylglutathione hydrolase FrmB (G5B91_21180) on one hand, and formaldehyde dehydrogenase FdhA (G5B91_21240) on the other. VR4 also carried a mosaic of genes and remnants thereof belonging to different functional categories, e.g., carbon metabolism (notably, dye decolorizing peroxidase, nitrilase/amidase and oxidoreductase), DNA recombination and repair (replication-associated recombination protein A, excinuclease components UvrA and UvrB, two transposases), and a variety of transcriptional regulators (Table S1, sheet ICEPni1). In accordance with these predictions, P. nitroreducens was able to grow in the presence of up to 8 µM of mercury chloride and 10 mM sodium arsenite, and it transferred the mercury resistance trait via conjugation (see below). Finally, another feature of adaptive significance is encoded on VR3; an operon spanning loci G5B91_21005 to 21040 (Table S1, sheet ICEPni1), which encodes proteins resembling the recently described cyclic oligonucleotide-based anti-phage signaling system (CBASS) [50]. Taken together, the ICEPni1 cargo is densely packed with functions of detoxification and defense, which may have been advantageous for the survival of P. nitroreducens in the polluted (wastewater) environment from whence it was isolated.
3.7. ICEPni2 Encodes Metabolism of Aromatic Compounds and Fatty Acids
ICEPni2 contains genes for two complete central pathways for aromatic compound metabolism: a catechol meta-cleavage pathway (G5B91_07410 to G5B91_07475, 14 genes) and a protocatechuate ortho-cleavage pathway (G5B91_06945 to G5B91_07000, 12 genes) (Table S1, sheet ICEPni2). In addition, genes for so-called peripheral pathways encode the enzymes to channel mono- and di-aromatic substrates into the central pathways. Among these, the previously characterized hbpRCAD genes (G5B91_07355 to G5B91_07370) are responsible for transforming 2-hydroxy- or 2,2’-dihydroxy-biphenyl to 2-hydroxy-2,4-pentadienoic acid and either benzoate, or salicylate, respectively [6,14]. 2-hydroxy-2,4-pentadienoic acid enters the meta-pathway directly, whereas benzoate is first converted to catechol by action of a benzoate 1,2-dioxygenase and 1,6-dihydroxycyclohexa-2,4-diene-1-carboxylate dehydrogenase encoded by the host genome loci G5B91_12935-G5B91_12945 (not on ICEPni1). Salicylate may be decarboxylated to catechol by the ICEPni2-encoded salicylate hydroxylase (G5B91_07395). Further peripheral pathways encoded on ICEPni2 consist of a ferulate-CoA synthase FerA G5B91_06740, vanillin synthase/trans-feruloyl-CoA hydratase FerB G5B91_06745, vanillin dehydrogenase G5B91_06735, vanillate O-demethylase VanAB G5B91_06885-G5B91_06890 and 4-hydroxybenzoate 3-monooxygenase PobA G5B91_06935. These may catalyze the channeling of ferulate, vanillin, vanillate and 4-hydroxybenzoate into the protocatechuate pathway (Table S1, sheet ICEPni2). Finally, genes for a two-component aromatic hydroxylase G5B91_06770 and G5B91_06795, an FMN-dependent NADH-azoreductase (G5B91_06765), and for carbazole 1,9a-dioxygenase components G5B91_07470 and G5B91_07475, may imply the potential of P. nitroreducens to metabolize other undefined aromatic substrates. Interestingly, the genes for a major MexAB-OprM efflux system that is crucial to grow on 2HBP are not present on ICEPni2, but elsewhere on the chromosome (G5B91_02490–02510) [51].
A second large group of ICEPni2 catabolic functions is represented by around 30 genes organized in several transcriptional units (Figure 2A, VR5), which may encode transport and metabolism of fatty acids. Notably, all genes are present for the enzymes (some redundant) required for fatty acid beta-oxidation: the long-chain-fatty-acid-CoA ligase FadD (two ORFs), acyl-CoA dehydrogenase FadE (four ORFs), enoyl-CoA hydratase/isomerases FadB (two ORFs) and acetyl-CoA C-acyltransferases/thiolases FadA (two ORFs). Furthermore, ICEPni2 also encodes a 3-oxoacyl-ACP reductase FabG and enoyl-[acyl-carrier-protein] reductase FabI, which are involved in fatty acid synthesis and acyl-carrier-protein metabolism (Table S1, sheet ICEPni2). These genes were intermingled with ORFs encoding NAD- and FAD-dependent oxidoreductases, electron transfer proteins, hydrolases, transporters, transcriptional regulators, chemotactic and DUF-proteins (Table S1, sheet ICEPni2), which often co-occur with genes for fatty acid metabolism in other reference genomes, and on GIPni1 (data not shown). The presence of these functions on ICEPni2 may have provided a selective advantage in the fatty acid-rich sewage environment (as we discussed above for GIPni1) and, possibly, also to connect the acetyl-CoA-producing aromatic catabolic pathways to the central cell metabolism. Finally, close to the left end proximity of ICEPni2, we identified homologues of the imuA-imuB-imuC (dnaE2) gene cassette (G5B91_06695-G5B91_06705), which may encode an error-prone translesion DNA synthesis polymerase complex [52]. ImuABC polymerases are regulated by the SOS-response and facilitate replication bypass of damaged DNA, concomitantly enhancing spontaneous mutagenesis [53,54,55]. These are properties that might be advantageous for both resilience and for accelerated adaptive evolution to the strongly selective environments in which P. nitroreducens has prevailed. The variable gene content of ICEPni2 seemed “patchy”, i.e., indicative of their recent acquisition via multiple recombination events and from different donor genomes. The number of recombination-promoting sequences on ICEPni2 is limited to only one intact (istAB) and two mutated transposase genes (Table S1, sheet ICEPni2), suggesting that other recombination mechanisms may have prevailed in the acquisition of the variable gene content.
3.8. ICEPni1 Can Excise and Transfer to P. putida, and Integrates into One of Four tRNAGly Gene Targets
In order to test whether the identified ICEs were functional, we first examined their possible excision from the chromosome as a circular element carrying an attP attachment site [56]. A weak but reproducible signal for an amplified DNA fragment covering both extremities of ICEPni1 and containing its predicted attPICEPni1 site was obtained by PCR on DNA extracted from P. nitroreducens grown in MM with 20 mM succinate with or without 5 mM sodium arsenite or 4 µM mercury chloride or in MM with 5 mM 2HBP. The PCR amplicon was visible in DNA from three independent cultures that were sampled in exponential, early and late stationary growth phase (data not shown). This showed that ICEPni1 excision did occur, albeit at a low frequency. Subsequent amplicon sequencing confirmed that the attP site was formed by recombination at the inferred 18-bp repeats (Table 2 and Table 4).
To test the potential of ICEPni1 to transfer, we filter-mated P. nitroreducens with P. putida UWCGC as a recipient while selecting for mercury resistance (at 4 µM). P. putida UWCGC transconjugants resistant to mercury appeared at a frequency of 1.16 (±0.37) × 10−6 per donor CFU. The transconjugants were further Gm resistant and mCherry fluorescent, indicating they were genuine UWCGC derivatives. Among fifty colonies tested with PCR, nine (18%) amplified a fragment of the merA gene specific for ICEPni1 (primers pPazICE1_merA_fw/pPazICE1_merA_rev, Table 1), which confirmed transfer of ICEPni1 (with merA). The large proportion of mercury-resistant colonies without amplifiable merA of ICEPni1 suggested transfer of (an)other DNA element(s) from P. nitroreducens HBP-1 conferring mercury resistance. Our attempt to specifically amplify the merA allele carried by plasmid pPniHBP1_1 yielded no product, thus ruling out the transfer of this plasmid. Possibly, the second plasmid pPniHBP1_2 (Figure 1C) was transferred, which carries genes for ZntA-like P-type ATPase and a CzcCBA-like tripartite cation efflux system that might confer mercury resistance, but this was not tested.
In seven P. putida UWCGC transconjugants carrying merA of ICEPni1, we further analyzed the potential integration events. ICEPni1 had integrated into any of the four GCC tRNAGly genes on P. putida (Table 4, locus tags PP_t21, _t23, _t24, and _t59), exactly as was previously demonstrated for its sibling ICEclc [34]. Sequencing of the recombination site boundaries showed that attL retained the 18 bp sequence of the ICEPni1 attP-site (GACTCGTTTCCCGCTCCA, Table 4). This sequence has one mismatch with respect to attB in P. putida (GTCTCGTTTCCCGCTCCA), and to attR of ICEPni1 in P. nitroreducens HBP-1 (Table 4). All four sequenced attL sites of transconjugants contained this same nucleotide difference. This indicates that both excision and integration catalyzed by the ICEPni1 integrase can tolerate (some) mismatches in the recombination sites, thus likely allowing ICE establishment in broader range of hosts.
3.9. ICEPni2 Is also a Functional ICE
Excision of ICEPni2 was confirmed by PCR amplification of the attP junction in DNA extracted from cultures of P. nitroreducens. Interestingly, we could amplify a DNA fragment predicted to contain the junction between the proximal 18-bp repeat and attL (see above), but not the distal repeat. The junction was confirmed by sequencing (Table 2 and Table 4). Clear PCR amplification was detected on a gel only in two out of nineteen DNA samples, suggesting a generally low incidence of excision (data not shown).
Selection of P. putida UWCGC transconjugants from matings with P. nitroreducens HBP-1 on minimal medium agar with 5 mM 2-HBP and Gm yielded no colonies, possibly because of the toxicity of 2-HBP [51]. In contrast, selection on 2.5 mM salicylate and Gm yielded eight colonies among 8.6 × 108 donor cells, which would be equivalent to a transfer rate of 10−8 per donor CFU. From the DNA of all eight colonies, we could amplify a fragment of the hbpA gene, which is specific to ICEPni2. All colonies were also mCherry fluorescent, confirming that they are P. putida recipients. This also confirmed that ICEPni2 is transferable from P. nitroreducens. All recovered transconjugants had integrated ICEPni2 at the 3′-end of the CCC tRNAGly gene (locus_tag = PP_t64), suggesting a high specificity for this integration site. Interestingly, the presumed 18-bp attP site of ICEPni2 and the targeted attB site in P. putida had three mismatches, which resulted in different combinations of attL/attR sites flanking the integrated element in P. putida (Table 4). Recombination involving two non-identical sequences is known to result in the formation of a heteroduplex that is resolved in a variety of combinations of att sites in the integrated form of ICEPni2, which is not uncommon for tyrosine-type recombinases [57]. The low transfer rate may thus stem from a lower incidence of excision, absence of an efficient efflux detoxification system to allow productive growth on 2-HBP and salicylate [51], and from mismatch between the ICEPni2 attP site and the P. putida attB site that limits efficient recombination.
To verify the functionality of ICEPni2-encoded aromatic catabolism genes, we tested the ability of P. nitroreducens HBP-1 (ICE donor), P. putida UWCGC (recipient) and two P. putida UWCGC (ICEPni2) transconjugants to grow aerobically in minimal medium supplemented with 2.5 mM 2-HBP, salicylate, 4-hydroxybenzoate and vanillate as the sole carbon source. As expected, P. nitroreducens HBP-1 and P. putida UWCGC ICEPni2-transconjugants, but not P. putida UWCGC, were able to grow on 2-HBP and salicylate. This supported our hypothesis that ICEPni2 indeed contains all necessary genetic information for these two metabolic pathways. The above-mentioned inability to select for ICEPni2 transconjugants using 2-HBP most likely reflected the need for the P. putida recipient cells to integrate ICEPni2 catabolism into its own metabolic network before being able to grow on 2-HBP. In contrast to P. nitroreducens HBP-1, P. putida UWCGC ICEPni2-transconjugants did not grow on 4-hydroxybenzoate. This would indicate that the ICEPni2 transferred protocatechuate pathway may not have been functionally expressed in P. putida.
4. Concluding Remarks
The gapless sequence of the P. nitroreducens HBP-1 genome gives a clear view of the numerous gene acquisitions and adaptations that may have provided HBP-1 with selective advantages to cope with the polluted environment from whence it was isolated.
The exploration of HBP-1′s genomic plasticity indicated a variety of genetic elements that were most likely acquired by horizontal gene transfer, some of which (both ICEs) were capable of conjugative transfer from HBP-1 as donor. In contrast to previous reports [15], HBP-1 carries not only a single large plasmid but a second plasmid with a variety of potential adaptive functions (Figure 1B,C and Table S1). It will be interesting to learn more about the capability and mode of dissemination of these plasmids, and the importance of their adaptive gene functions to the host.
The P. nitroreducens HBP-1 genome is scattered with putative prophages, genomic islands and ICEs, most of which with unknown properties, origins and functionalities. A 126 kb long genomic island (GIPni1) was identified with unclear motility properties, as well as several putative prophages, or derivative elements whose mobility is compromised or dependent on helper element(s) (Figure 1 and Table 3) [36,58]. The two discovered ICEs, ICEPni1 and ICEPni2, code for many obvious adaptive functions, and confer P. nitroreducens with the capability to resist to heavy metals and to metabolize aromatic compounds, respectively (Figure 2A). Interestingly, both elements are related in their core structure to the prototypical element ICEclc (Figure 2), indicating the importance and wide distribution of this type of element among Gammaproteobacteria. In contrast to ICEclc, ICEPni1 and ICEPni2 transfer at much lower frequencies (10−6–10−8 per donor, compared to 10−2 for ICEclc), but we demonstrated their excision, transfer and integration in the chromosome of P. putida. They are therefore fully functional ICEs. Given the bistable development of transfer competence imposed by ICEclc on its host, it will be interesting to see if ICEPni1 and ICEPni2 act similarly. Such a mechanism is pivotal for the lifecycle of ICEclc [46,47,59], and differences and/or similarities in the regulation of ICEPni1 and ICEPni2 would strengthen our understanding of bistable behavior [45].
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4425/11/8/930/s1, Table S1: P. nitroreducens HBP-1 genome annotation, Figure S1: Read coverage plot of the P. nitroreducens HBP-1 genome. Graphical representation of the moving-average coverage per 100-bp window obtained from Illumina sequencing of P. nitroreducens HBP-1 replicons: chromosome (in blue), pPniHBP1_1 (in green) and pPniHBP1_2 (in red) using 1 colony (A. PNI1) or 5 colonies (B. PNI5) to inoculate an overnight culture. The mean and standard deviation of coverage are indicated for each replicon.
Author Contributions
Conceptualization, V.S. and J.R.v.d.M.; methodology, L.P.; validation, N.C., V.S. and C.B.; formal analysis, N.C., V.S., L.P. and J.R.v.d.M.; investigation, N.C., V.S., L.P., C.B.; data curation, V.S.; writing—original draft preparation, N.C.; writing—review and editing, N.C., V.S., L.P., C.B. and J.R.v.d.M.; visualization, C.B.; supervision, V.S. and J.R.v.d.M.; project administration, J.R.v.d.M.; funding acquisition, J.R.v.d.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Swiss National Science Foundation grant number 31003A_175638 to J.R.v.d.M.
Acknowledgments
Sequencing of P. nitroreducens strain HBP-1 was initiated as part of a Sequence a Genome Master class (SAGE2011) at the University of Lausanne, and we thank all assistants for their contribution: Laurent Falquet, Marc Robinson-Rechavy, Gilbert Greub, Ryo Miyazaki, Carlo Rivolta, Grégory Resch, Sandra Calderon, Walid Gharib, Shantanu Roy and Paola Benaglio. We thank Sudip Das for preparing and Illumina sequencing two libraries, and further the team of the Lausanne Genome Technology Facilities: Keith Harshman, Johann Weber, Emmanuel Beaudoing, and Mélanie Dupasquier. We also thank Burkhard Linke and Alexander Goessmann (Department of Biology, Bielefeld University, Bielefeld, Germany) for annotating the first draft of P. nitroreducens HBP-1 genome using GenDB [60].
Conflicts of Interest
The authors declare no conflict of interest.
References
- Atashgahi, S.; Sánchez-Andrea, I.; Heipieper, H.J.; van der Meer, J.R.; Stams, A.J.M.; Smidt, H. Prospects for harnessing biocide resistance for bioremediation and detoxification. Science 2018, 360, 743–746. [Google Scholar] [CrossRef] [PubMed]
- Megharaj, M.; Ramakrishnan, B.; Venkateswarlu, K.; Sethunathan, N.; Naidu, R. Bioremediation approaches for organic pollutants: A critical perspective. Environ. Int. 2011, 37, 1362–1375. [Google Scholar] [CrossRef] [PubMed]
- Mrozik, A.; Miga, S.; Piotrowska-Seget, Z. Enhancement of phenol degradation by soil bioaugmentation with Pseudomonas sp. JS150. J. Appl. Microbiol. 2011, 111, 1357–1370. [Google Scholar] [CrossRef] [PubMed]
- Miyazaki, R.; Bertelli, C.; Benaglio, P.; Canton, J.; De Coi, N.; Gharib, W.H.; Gjoksi, B.; Goesmann, A.; Greub, G.; Harshman, K.; et al. Comparative genome analysis of Pseudomonas knackmussii B13, the first bacterium known to degrade chloroaromatic compounds. Environ. Microbiol. 2015, 17, 91–104. [Google Scholar] [CrossRef]
- Morales, M.; Sentchilo, V.; Bertelli, C.; Komljenovic, A.; Kryuchkova-Mostacci, N.; Bourdilloud, A.; Linke, B.; Goesmann, A.; Harshman, K.; Segers, F.; et al. The genome of the toluene-degrading Pseudomonas veronii strain 1YdBTEX2 and its differential gene expression in contaminated sand. PLoS ONE 2016, 11, e0165850. [Google Scholar] [CrossRef]
- Kohler, H.P.; Kohler-Staub, D.; Focht, D.D. Degradation of 2-hydroxybiphenyl and 2,2′-dihydroxybiphenyl by Pseudomonas sp. strain HBP1. Appl. Environ. Microbiol. 1988, 54, 2683–2688. [Google Scholar] [CrossRef]
- Kohler, H.P.; Schmid, A.; van der Maarel, M. Metabolism of 2,2′-dihydroxybiphenyl by Pseudomonas sp. strain HBP1: Production and consumption of 2,2′,3-trihydroxybiphenyl. J. Bacteriol. 1993, 175, 1621–1628. [Google Scholar] [CrossRef]
- Kohler, H.P.; van der Maarel, M.J.; Kohler-Staub, D. Selection of Pseudomonas sp. strain HBP1 Prp for metabolism of 2-propylphenol and elucidation of the degradative pathway. Appl. Environ. Microbiol. 1993, 59, 860–866. [Google Scholar] [CrossRef]
- Reichlin, F.; Kohler, H.P. Pseudomonas sp. strain HBP1 Prp degrades 2-isopropylphenol (ortho-cumenol) via meta cleavage. Appl. Environ. Microbiol. 1994, 60, 4587–4591. [Google Scholar] [CrossRef]
- Suske, W.A.; Held, M.; Schmid, A.; Fleischmann, T.; Wubbolts, M.G.; Kohler, H.P. Purification and characterization of 2-hydroxybiphenyl 3-monooxygenase, a novel NADH-dependent, FAD-containing aromatic hydroxylase from Pseudomonas azelaica HBP1. J. Biol. Chem. 1997, 272, 24257–24265. [Google Scholar] [CrossRef][Green Version]
- Suske, W.A.; van Berkel, W.J.; Kohler, H.P. Catalytic mechanism of 2-hydroxybiphenyl 3-monooxygenase, a flavoprotein from Pseudomonas azelaica HBP1. J. Biol. Chem. 1999, 274, 33355–33365. [Google Scholar] [CrossRef] [PubMed]
- Jaspers, M.C.; Suske, W.A.; Schmid, A.; Goslings, D.A.; Kohler, H.P.; van der Meer, J.R. HbpR, a new member of the XylR/DmpR subclass within the NtrC family of bacterial transcriptional activators, regulates expression of 2-hydroxybiphenyl metabolism in Pseudomonas azelaica HBP1. J. Bacteriol. 2000, 182, 405–417. [Google Scholar] [CrossRef] [PubMed]
- Jaspers, M.C.; Sturme, M.; van der Meer, J.R. Unusual location of two nearby pairs of upstream activating sequences for HbpR, the main regulatory protein for the 2-hydroxybiphenyl degradation pathway of Pseudomonas azelaica HBP1. Microbiol. Read. Engl. 2001, 147, 2183–2194. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jaspers, M.C.; Schmid, A.; Sturme, M.H.; Goslings, D.A.; Kohler, H.P.; Roelof Van Der Meer, J. Transcriptional organization and dynamic expression of the hbpCAD genes, which encode the first three enzymes for 2-hydroxybiphenyl degradation in Pseudomonas azelaica HBP1. J. Bacteriol. 2001, 183, 270–279. [Google Scholar] [CrossRef]
- García, J.L.; Rozas, D.; Del Cerro, C.; Nogales, J.; El-Said Mohamed, M.; Díaz, E. Genome sequence of Pseudomonas azelaica HBP1, which catabolizes 2-hydroxybiphenyl fungicide. Genome Announc. 2014, 2. [Google Scholar] [CrossRef]
- El-Said Mohamed, M.; García, J.L.; Martínez, I.; Del Cerro, C.; Nogales, J.; Díaz, E. Genome sequence of Pseudomonas azelaica strain Aramco J. Genome Announc. 2015, 3. [Google Scholar] [CrossRef]
- Garbisu, C.; Garaiyurrebaso, O.; Epelde, L.; Grohmann, E.; Alkorta, I. Plasmid-mediated bioaugmentation for the bioremediation of contaminated soils. Front. Microbiol. 2017, 8, 1966. [Google Scholar] [CrossRef]
- Miyazaki, R.; van der Meer, J.R. A dual functional origin of transfer in the ICEclc genomic island of Pseudomonas knackmussii B13. Mol. Microbiol. 2011, 79, 743–758. [Google Scholar] [CrossRef]
- Gerhardt, P.; Murray, R.G.E.; Costilow, R.N.; Nester, E.W.; Wood, W.A.; Kreig, N.R.; Briggs Phillips, G. Manual of methods for general bacteriology. J. Clin. Pathol. 1981, 34, 1069. [Google Scholar]
- Chin, C.-S.; Alexander, D.H.; Marks, P.; Klammer, A.A.; Drake, J.; Heiner, C.; Clum, A.; Copeland, A.; Huddleston, J.; Eichler, E.E.; et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 2013, 10, 563–569. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- García-Alcalde, F.; Okonechnikov, K.; Carbonell, J.; Cruz, L.M.; Götz, S.; Tarazona, S.; Dopazo, J.; Meyer, T.F.; Conesa, A. Qualimap: Evaluating next-generation sequencing alignment data. Bioinformatics 2012, 28, 2678–2679. [Google Scholar] [CrossRef] [PubMed]
- Okonechnikov, K.; Conesa, A.; García-Alcalde, F. Qualimap 2: Advanced multi-sample quality control for high-throughput sequencing data. Bioinformatics 2016, 32, 292–294. [Google Scholar] [CrossRef] [PubMed]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef] [PubMed]
- Haft, D.H.; DiCuccio, M.; Badretdin, A.; Brover, V.; Chetvernin, V.; O’Neill, K.; Li, W.; Chitsaz, F.; Derbyshire, M.K.; Gonzales, N.R.; et al. RefSeq: An update on prokaryotic genome annotation and curation. Nucleic Acids Res. 2018, 46, D851–D860. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Caspi, R.; Billington, R.; Keseler, I.M.; Kothari, A.; Krummenacker, M.; Midford, P.E.; Ong, W.K.; Paley, S.; Subhraveti, P.; Karp, P.D. The MetaCyc database of metabolic pathways and enzymes—A 2019 update. Nucleic Acids Res. 2020, 48, D445–D453. [Google Scholar] [CrossRef]
- Bertelli, C.; Laird, M.R.; Williams, K.P.; Simon Fraser University Research Computing Group; Lau, B.Y.; Hoad, G.; Winsor, G.L.; Brinkman, F.S.L. IslandViewer 4: Expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 2017, 45, W30–W35. [Google Scholar] [CrossRef]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef] [PubMed]
- Carver, T.J.; Rutherford, K.M.; Berriman, M.; Rajandream, M.-A.; Barrell, B.G.; Parkhill, J. ACT: The Artemis Comparison Tool. Bioinformatics 2005, 21, 3422–3423. [Google Scholar] [CrossRef]
- Carver, T.; Thomson, N.; Bleasby, A.; Berriman, M.; Parkhill, J. DNAPlotter: Circular and linear interactive genome visualization. Bioinformatics 2009, 25, 119–120. [Google Scholar] [CrossRef] [PubMed]
- Sentchilo, V.; Czechowska, K.; Pradervand, N.; Minoia, M.; Miyazaki, R.; van der Meer, J.R. Intracellular excision and reintegration dynamics of the ICEclc genomic island of Pseudomonas knackmussii sp. strain B13. Mol. Microbiol. 2009, 72, 1293–1306. [Google Scholar] [CrossRef] [PubMed]
- Moscoso, M.; Eritja, R.; Espinosa, M. Initiation of replication of plasmid pMV158: Mechanisms of DNA strand-transfer reactions mediated by the initiator RepB protein. J. Mol. Biol. 1997, 268, 840–856. [Google Scholar] [CrossRef]
- Carraro, N.; Rivard, N.; Burrus, V.; Ceccarelli, D. Mobilizable genomic islands, different strategies for the dissemination of multidrug resistance and other adaptive traits. Mob. Genet. Elem. 2017, 7, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Nies, D.H. Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol. Rev. 2003, 27, 313–339. [Google Scholar] [CrossRef]
- Chevalier, S.; Bouffartigues, E.; Bodilis, J.; Maillot, O.; Lesouhaitier, O.; Feuilloley, M.G.J.; Orange, N.; Dufour, A.; Cornelis, P. Structure, function and regulation of Pseudomonas aeruginosa porins. FEMS Microbiol. Rev. 2017, 41, 698–722. [Google Scholar] [CrossRef]
- Perron, K.; Caille, O.; Rossier, C.; Van Delden, C.; Dumas, J.-L.; Köhler, T. CzcR-CzcS, a two-component system involved in heavy metal and carbapenem resistance in Pseudomonas aeruginosa. J. Biol. Chem. 2004, 279, 8761–8768. [Google Scholar] [CrossRef]
- Christie, G.E.; Calendar, R. Interactions between satellite bacteriophage P4 and its helpers. Annu. Rev. Genet. 1990, 24, 465–490. [Google Scholar] [CrossRef]
- Taylor, N.M.I.; van Raaij, M.J.; Leiman, P.G. Contractile injection systems of bacteriophages and related systems. Mol. Microbiol. 2018, 108, 6–15. [Google Scholar] [CrossRef] [PubMed]
- Guglielmini, J.; Néron, B.; Abby, S.S.; Garcillán-Barcia, M.P.; de la Cruz, F.; Rocha, E.P.C. Key components of the eight classes of type IV secretion systems involved in bacterial conjugation or protein secretion. Nucleic Acids Res. 2014, 42, 5715–5727. [Google Scholar] [CrossRef] [PubMed]
- Burrus, V.; Roussel, Y.; Decaris, B.; Guédon, G. Characterization of a novel integrative element, ICESt1, in the lactic acid bacterium Streptococcus thermophilus. Appl. Environ. Microbiol. 2000, 66, 1749–1753. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Toussaint, A.; Merlin, C. Mobile elements as a combination of functional modules. Plasmid 2002, 47, 26–35. [Google Scholar] [CrossRef]
- Carraro, N.; Richard, X.; Sulser, S.; Delavat, F.; Mazza, C.; van der Meer, J.R. An analog to digital converter controls bistable transfer competence of a widespread integrative and conjugative element. eLife 2020, 9. [Google Scholar] [CrossRef] [PubMed]
- Delavat, F.; Miyazaki, R.; Carraro, N.; Pradervand, N.; van der Meer, J.R. The hidden life of integrative and conjugative elements. FEMS Microbiol. Rev. 2017. [Google Scholar] [CrossRef]
- Minoia, M.; Gaillard, M.; Reinhard, F.; Stojanov, M.; Sentchilo, V.; van der Meer, J.R. Stochasticity and bistability in horizontal transfer control of a genomic island in Pseudomonas. Proc. Natl. Acad. Sci. USA 2008, 105, 20792–20797. [Google Scholar] [CrossRef]
- Chen, J.; Bhattacharjee, H.; Rosen, B.P. ArsH is an organoarsenical oxidase that confers resistance to trivalent forms of the herbicide monosodium methylarsenate and the poultry growth promoter roxarsone. Mol. Microbiol. 2015, 96, 1042–1052. [Google Scholar] [CrossRef]
- Yang, H.-C.; Rosen, B.P. New mechanisms of bacterial arsenic resistance. Biomed. J. 2016, 39, 5–13. [Google Scholar] [CrossRef]
- Lau, R.K.; Ye, Q.; Birkholz, E.A.; Berg, K.R.; Patel, L.; Mathews, I.T.; Watrous, J.D.; Ego, K.; Whiteley, A.T.; Lowey, B.; et al. Structure and mechanism of a cyclic trinucleotide-activated bacterial endonuclease mediating bacteriophage immunity. Mol. Cell 2020, 77, 723–733.e6. [Google Scholar] [CrossRef]
- Czechowska, K.; Reimmann, C.; van der Meer, J.R. Characterization of a MexAB-OprM efflux system necessary for productive metabolism of Pseudomonas azelaica HBP1 on 2-hydroxybiphenyl. Front. Microbiol. 2013, 4, 203. [Google Scholar] [CrossRef] [PubMed]
- Ippoliti, P.J.; Delateur, N.A.; Jones, K.M.; Beuning, P.J. Multiple strategies for translesion synthesis in bacteria. Cells 2012, 1, 799–831. [Google Scholar] [CrossRef] [PubMed]
- Erill, I.; Campoy, S.; Mazon, G.; Barbé, J. Dispersal and regulation of an adaptive mutagenesis cassette in the bacteria domain. Nucleic Acids Res. 2006, 34, 66–77. [Google Scholar] [CrossRef] [PubMed]
- Warner, D.F.; Ndwandwe, D.E.; Abrahams, G.L.; Kana, B.D.; Machowski, E.E.; Venclovas, C.; Mizrahi, V. Essential roles for imuA’- and imuB-encoded accessory factors in DnaE2-dependent mutagenesis in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 2010, 107, 13093–13098. [Google Scholar] [CrossRef]
- Remigi, P.; Capela, D.; Clerissi, C.; Tasse, L.; Torchet, R.; Bouchez, O.; Batut, J.; Cruveiller, S.; Rocha, E.P.C.; Masson-Boivin, C. Transient hypermutagenesis accelerates the evolution of legume endosymbionts following horizontal gene transfer. PLoS Biol. 2014, 12, e1001942. [Google Scholar] [CrossRef]
- Carraro, N.; Burrus, V. The dualistic nature of integrative and conjugative elements. Mob. Genet. Elem. 2015, 5, 98–102. [Google Scholar] [CrossRef]
- Jayaram, M.; Ma, C.-H.; Kachroo, A.H.; Rowley, P.A.; Guga, P.; Fan, H.-F.; Voziyanov, Y. An overview of tyrosine site-specific recombination: From an Flp perspective. Microbiol. Spectr. 2015, 3. [Google Scholar] [CrossRef]
- Novick, R.P.; Christie, G.E.; Penadés, J.R. The phage-related chromosomal islands of Gram-positive bacteria. Nat. Rev. Microbiol. 2010, 8, 541–551. [Google Scholar] [CrossRef]
- Delavat, F.; Mitri, S.; Pelet, S.; van der Meer, J.R. Highly variable individual donor cell fates characterize robust horizontal gene transfer of an integrative and conjugative element. Proc. Natl. Acad. Sci. USA 2016, 113, E3375–E3383. [Google Scholar] [CrossRef]
- Meyer, F.; Goesmann, A.; McHardy, A.C.; Bartels, D.; Bekel, T.; Clausen, J.; Kalinowski, J.; Linke, B.; Rupp, O.; Giegerich, R.; et al. GenDB—An open source genome annotation system for prokaryote genomes. Nucleic Acids Res. 2003, 31, 2187–2195. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).