Abstract
Plant growth responds to various environmental and developmental cues via signaling cascades that influence gene expression at the level of transcription and pre-mRNA splicing. Alternative splicing of pre-mRNA increases the coding potential of the genome from multiexon genes and regulates gene expression through multiple mechanisms. Serine/arginine-rich (SR) proteins, a conserved family of splicing factors, are the key players of alternative splicing and regulate pre-mRNA splicing under stress conditions. The rice (Oryza sativa) genome encodes 22 SR proteins categorized into six subfamilies. Three of the subfamilies are plant-specific with no mammalian orthologues, and the functions of these SR proteins are not well known. The clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) system is a genome engineering tool that cleaves the target DNA at specific locations directed by a guide RNA (gRNA). Recent advances in CRISPR/Cas9-mediated plant genome engineering make it possible to generate single and multiple functional knockout mutants in diverse plant species. In this study, we targeted each rice SR locus and produced single knockouts. To overcome the functional redundancy within each subfamily of SR genes, we utilized a polycistronic tRNA-gRNA multiplex targeting system and targeted all loci of each subfamily. Sanger sequencing results indicated that most of the targeted loci had knockout mutations. This study provides useful resource materials for understanding the molecular role of SR proteins in plant development and biotic and abiotic stress responses.
1. Introduction
Plants are constantly exposed to diverse and changing environmental conditions. Changes in their habitat can adversely affect their growth and development [1,2]. Therefore, plants have evolved numerous molecular mechanisms to survive under environmental fluctuations. Among these, regulation of gene expression is a major form of adaptation in response to biotic and abiotic stresses [3,4]. Pre-mRNA splicing is primarily a co-transcriptional process conserved among eukaryotes. It is a key step in gene regulation and is performed by the spliceosome, a large macromolecular complex. Spliceosomes comprise of five small nuclear ribonucleoproteins (snRNPs: U1, U2, U4/U6, and U5) and are assisted by ~200 accessory associated proteins [5,6]. Spliceosome assembly occurs independently on each intron, beginning with the binding of the U1 snRNP and U2 auxiliary factor (U2AF) to the 5′ and 3′ splice sites, respectively. The resulting scaffold, termed the E complex, recruits the U2 snRNP to the branch-point sequence, forming pre-spliceosome complex A. Subsequently, the U5-U4/U6 tri-snRNP is recruited to the assembling spliceosome to form the B complex. Spliceosome assembly reaches completion upon subsequent conformational rearrangements that yield the catalytically active spliceosomal C complex, which catalyzes the transesterification reactions that lead to the excision of the intervening intron and ligation of adjoining exons [7,8].
One of the important aspects of alternative splicing (AS) of pre-mRNA is that it enhances the coding capacity of the genome from multiexon genes. Regions encoding different protein domains can be selectively retained or excised from the mature mRNA so that a single gene can generate multiple distinct proteins that may vary greatly in structure, function, and other properties [9,10]. AS occurs when splice sites are differentially selected and multiple isoforms are produced from the same transcript. AS is fine-tuned by a group of RNA-binding proteins that bind the sequence signals in the pre-mRNA and guide the spliceosome complex for splicing regulation. Serine/arginine-rich proteins are important RNA-binding proteins that function as splicing factors, determine splice sites (SSs), and regulate pre-mRNA splicing under normal and stress conditions [11,12]. Various environmental stresses influence widespread changes in the AS of pre-mRNAs, especially from stress-responsive genes [13,14,15,16]. Members of the SR protein family, such as SRSF1 and SRSF2 in animals, were examined for their crucial functions in constitutive splicing as they were discovered to recruit U1 snRNP to the 5′ SS and U2 snRNP binding to the 3′ SS. Moreover, they bridge the interaction between these initial SS recognition occurrences in the pre-spliceosome and the mature spliceosome [17,18]. Several studies have corroborated the role of SR proteins in stress responses. The loss-of-function mutants of rs40 and rs41, scl30a, and sr45 showed hypersensitivity to salt and abscisic acid (ABA) stress in Arabidopsis [19,20,21]. Moreover, the sr34b mutant showed increased sensitivity to cadmium (Cd) stress and expression of SR34b increased under Cd stress [22]. The SR proteins also play important roles in mineral nutrient homeostasis in rice (O. sativa) [23]. The biological roles of SR proteins in animals are further extended to mRNA nuclear export, mRNA stability, genome maintenance, and translation [24,25,26].
The clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated protein (Cas) adaptive immunity system has been harnessed for genome editing applications across eukaryotic species [27,28,29]. For targeted genome engineering, CRISPR/Cas9 is modified into two components: A single-guide RNA (sgRNA) and a Cas9 endonuclease, which is guided by the sgRNA to a specified genomic locus. The Cas9 endonuclease cleaves a specific genomic DNA sequence by introducing double-strand breaks (DSBs) at the target site. These DSBs can be repaired by one of two endogenous mechanisms: Nonhomologous end joining (NHEJ) or homology-directed repair (HDR). The repair of DSBs via NHEJ often leads to the formation of small insertion/deletion (InDel) mutations. These InDel mutations can disrupt coding or regulatory sequences of the target gene, resulting in loss-of-function mutations. Repair of double-strand breaks by HDR requires the simultaneous delivery of a DNA repair template that carries the desired modification to be incorporated into the repaired locus. This powerful molecular tool is used for basic research [30,31], medical applications [31,32,33], and crop improvement [34,35].
In this study, we utilized the CRISPR/Cas9 technology to target all members of the SR gene family in rice. We produced single knockout mutants and multigene mutants to overcome the functional redundancy within each subgroup. Sanger sequencing analysis of the T0 generation indicated that most of the single targeted genes had knockout mutations.
2. Material and Methods
2.1. Plant Materials and Vector Construction
O. sativa L. ssp. japonica cv. Nipponbare was used for all experiments. The expression of Cas9 was driven by OsUBIQUITIN. The pRGEB32 vector was used for callus transformations. The sgRNAs were designed to target the 1st or 2nd exon of each locus. The sgRNA was synthesized as a polycistronic tRNA-gRNA (PTG) fragment. For multiplex targeting, synthetic PTG fragments contained more than one sgRNA that were separated by tRNA. Each fragment had BsaI overhangs for cloning under the OsU3 promoter in pRGEB32 [36]. The sequences of the PTG fragments are given in Table S1.
2.2. Agrobacterium tumefaciens-Mediated Transformation of Rice
Agrobacterium tumefaciens-mediated rice transformation was performed using the strain EHA105 as described previously [30,37]. Briefly, callus induction is done at 2N6 media. The Agrobacterium cells set to a density of OD600 = 0.3 incubated with calli for 5 min. After co-cultivation under dark at 25 °C for three days, rice calli were selected on media supplemented with Hygromycin (50 mg/L) and Timentin (200 mg/L). After the screening stage, actively growing calli were sub-cultured onto a regenerative medium for regeneration under continuous light. After 2–3 weeks, transgenic seedlings were transferred to sterile plastic containers containing fresh rooting medium and grown for 2–3 weeks before being transferred into soil. Transgenic rice plants were grown in a growth chamber at 28 °C with a 16 h light/8 h dark cycle.
2.3. Genotyping of the SR Mutant Plants
After one week, when plants were established on soil, DNA was extracted from leaf samples frozen in liquid nitrogen after collection. Frozen plant material was homogenized to a fine powder. Afterwards, 300 µL of the extraction buffer (200 mM Tris-Cl (pH 7.5), 250 mM NaCl, 25 mM EDTA (Ethylenediaminetetraacetic acid) and 0.5% SDS (Sodium dodecyl sulfate)) were added to the sample, vortexed to mix the sample with the buffer. 300 µL of PCI (Phenol:Chloroform:IsoAmylalcohol 25:24:1) were added, vigorously vortexed and spun down for 10 min at 13,000 rpm and room temperature (RT). The aqueous phase (upper) was precipitated with isopropanol (80% of total volume) and 3M sodium acetate (10% of total volume) incubated for 10 min at RT and then centrifuged at 13,000 rpm for 10 min at RT. The supernatant was removed and the pellet was washed with 500 µL of 70% ethanol and successive centrifugation for 5 min at 13,000 rpm. The ethanol was completely removed and the pellet was air-dried for 15–20 min. The pellet was dissolved in 50 µL TE (Tris-EDTA) buffer. 1–2 µL were used for further PCR. PCR was conducted using gene-specific primers. Purified PCR products were cloned using the CloneJET PCR Cloning Kit (K1231, Thermo Fisher Scientific, Waltham, MA, USA). The ligation mixture was directly used for transformation into Escherichia coli competent cells. Recombinant clones were selected and DNA was purified and subjected to Sanger sequencing. The oligo sequences used for genotyping are listed in Table S2.
3. Results
3.1. Targeted Multiplex Genome Engineering of the SR Gene Family in Rice
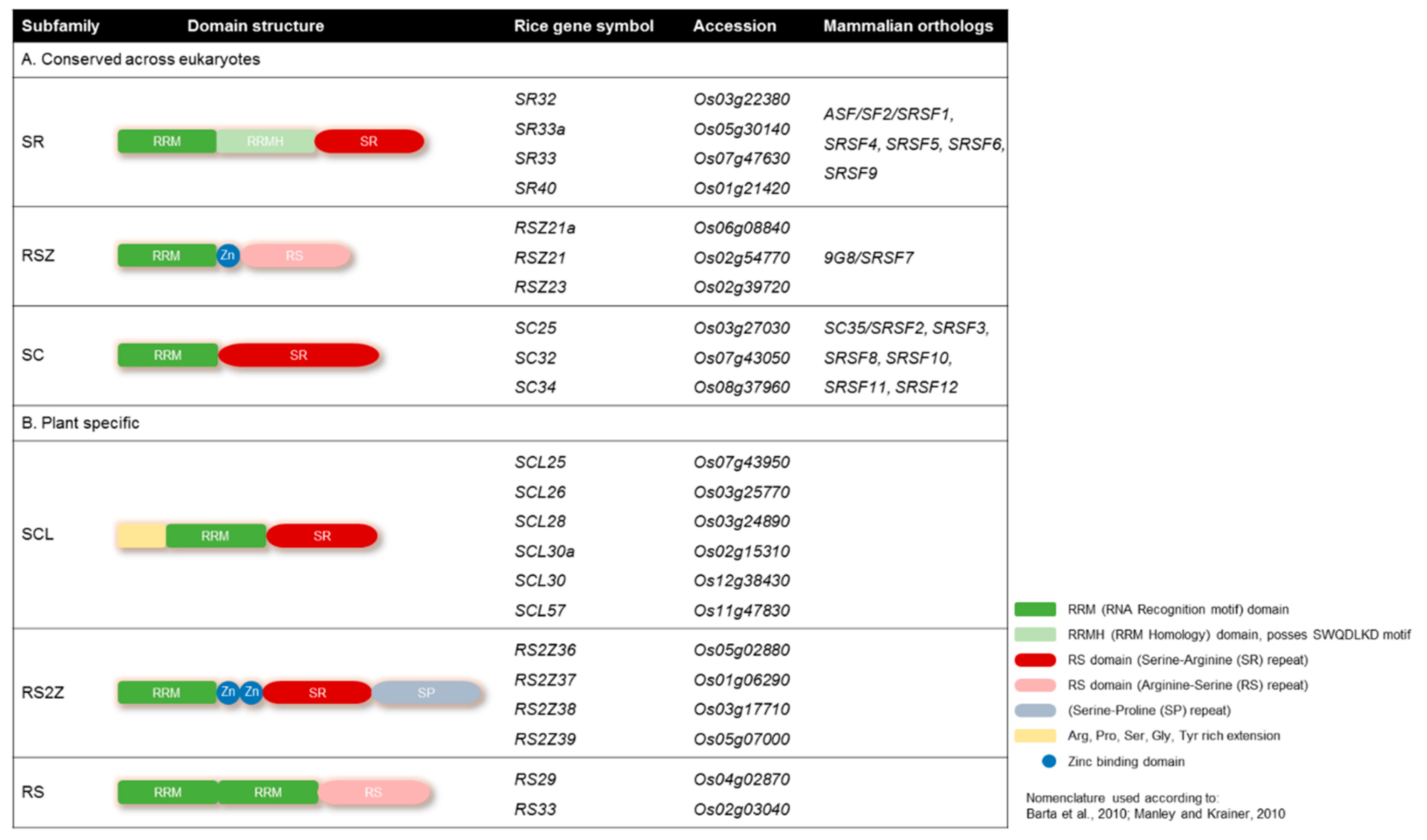
Plant SR proteins are defined as having one or two N-terminal RNA binding domains (RBDs), also called RNA recognition motifs (RRMs), and a C-terminal arginine/serine-rich (RS) domain with SR or RS dipeptide content [38,39] (Figure 1). The N-terminal domains are responsible for binding with the mRNA transcript, and the C-terminal region is responsible for protein-protein interaction during the splicing process. Rice has 22 members of SR proteins, the most among all known species, which can be categorized into six subfamilies. Three of these subfamilies (SR, RSZ, and SC) are conserved among eukaryotes, while the rest (SCL, RS2Z, and RS) are plant-specific (Figure 1). The plant-specific subfamilies have unique structural features; for example, the SCL subfamily is similar to the SC subfamily but possesses a unique N-terminal domain rich in charged amino acids (Figure 1). The RS2Z subfamily has two zinc knuckles instead of one in RSZ members. RS2Z members also have an additional serine- and proline-rich C-terminal domain. The second RRM domain of RS subfamily members is deficient in the SWQDLKD motif and possess RS dipeptides in the C-terminus (Figure 1).
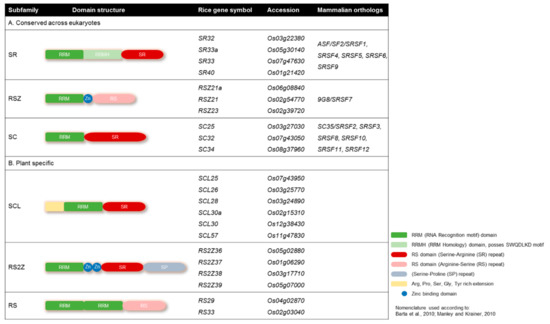
Figure 1.
Domain structures of the serine/arginine-rich (SR) proteins. Domain architecture of the SR protein subfamilies with their mammalian orthologs. (A) Highly conserved eukaryotic SR protein subfamilies; (B) plant-specific SR protein subfamilies. Rice has 22 SR proteins, 10 of which are conserved among eukaryotes, while the other 12 are plant-specific. The nomenclature adopted from [38,39].
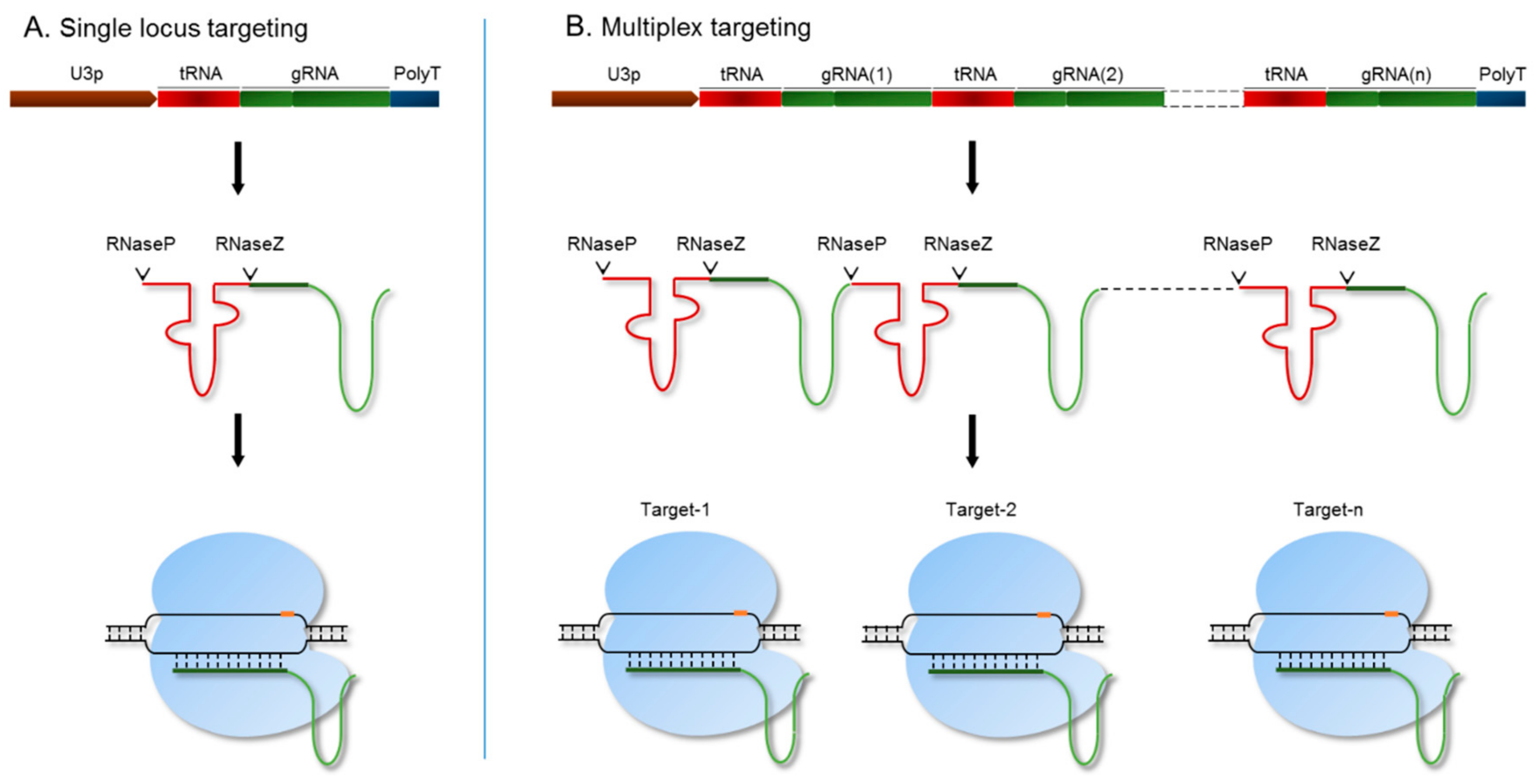
For targeted engineering of the SR genomic regions, we utilized a polycistronic tRNA-gRNA (PTG)/Cas9 system, which allows for precise processing of functional gRNA [36] (Figure 2). This system is expressed under the rice OsU3 promoter with a poly(T) terminator. The endogenous machinery of RNaseP and RNaseZ cleaves the PTG transcript by recognizing transfer RNA (tRNA) sequences [36]. As a result, the gRNA is released as a single molecule and loaded onto the Cas9 endonuclease to target a specific genomic region. To target a single locus, we used a synthetic fragment of the tRNA-gRNA-terminator with BsaI overhangs to clone under the OsU3 promoter (Table S1). We designed a PTG fragment for each SR member and cloned each into the binary vector pRGEB32 [36]. We then performed A. tumefaciens-mediated rice callus transformation for each of the individual targets to produce single-gene knockout mutants. Genes within each subfamily of the SR family have high sequence similarity and probably have redundant functions. Therefore, we designed and synthesized a single PTG fragment with gRNAs to target all the members of a subfamily (Figure 2B). Each gRNA targeting a single locus is released after processing from the PTG transcript and forms a complex with the Cas9 endonuclease to simultaneously target all the loci of an SR subfamily [36] (Figure 2).
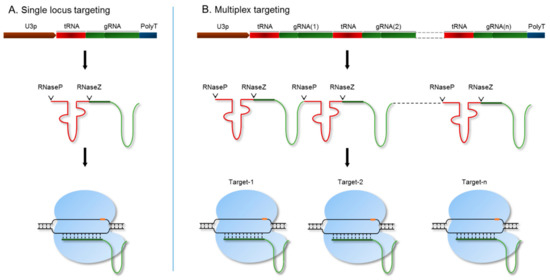
Figure 2.
The polycistronic tRNA-gRNA (PTG)/ CRISPR/CRISPR-associated protein 9 (Cas9) system for targeting single or multiple genes. The synthetic PTG molecule consists of a tRNA-gRNA unit. (A) For targeting a single locus, PTG was expressed under the OsU3 promoter. The PTG fragment is spliced by RNaseP and RNaseZ, and the sgRNA is released to further guide the Cas9 endonuclease to its genomic target; (B) multiple loci were targeted using the PTG system. Mature sgRNAs are released by the cleavage activity of RNaseP and RNaseZ. Each sgRNA directs the Cas9 endonuclease to its target genomic region.
The gRNAs were specifically designed in the first or second exon to perturb the function of the protein. After the rice callus transformation, the plants were transferred to soil. Once plants were established on soil for 10 days the DNA was extracted from the leaves and genotyped by Sanger sequencing.
3.2. The SR Subfamily
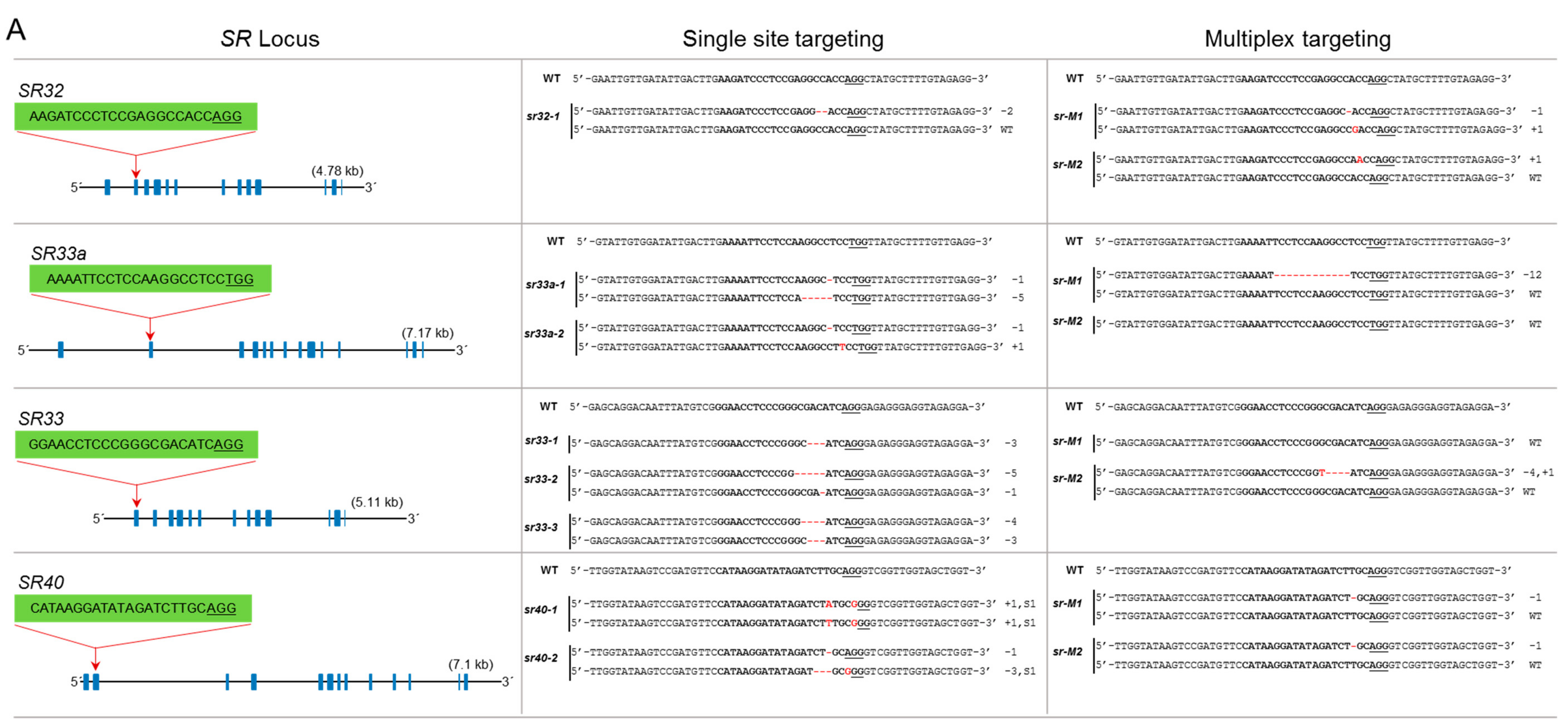
The SR subfamily has four members: SR32, SR33a, SR33, and SR40. These proteins have the SWQDLKD motif and have the mammalian orthologs ASF/SF2/SRSF1. SR32 has 13 exons, and we targeted the 2nd exon. We genotyped 10 lines but did not recover a mutant for this locus (Figure 3A). SR33a has 14 exons. The gRNA was designed to target the 2nd exon, and we successfully recovered knockout biallelic mutants for this gene. SR33 has a high sequence similarity to SR33a and contains 13 exons. We used a gRNA targeting the 1st exon and produced three mutants. sr33-1 is an in-frame mutant and sr33-3 is a biallelic mutant with in-frame and knockout mutations. sr33-2 is a complete knockout mutant (Figure 3A). SR-40 has 13 exons, and we targeted the 2nd exon; only the sr40-1 mutant is a complete knockout mutant. In the multiplex targeting experiment, we did not recover a complete knockout mutant for all four loci. The sr-M1 mutant has a complete knockout mutation for SR32 but was heterozygous for a mutated and wild-type allele for SR33a and SR40, while no mutation was generated for SR33. The sr-M2 mutant is heterozygous for a mutated and wild-type allele for SR32, SR33, and SR-40 but has no mutation for SR33a (Figure 3A).
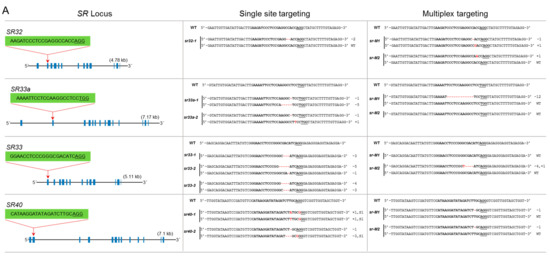

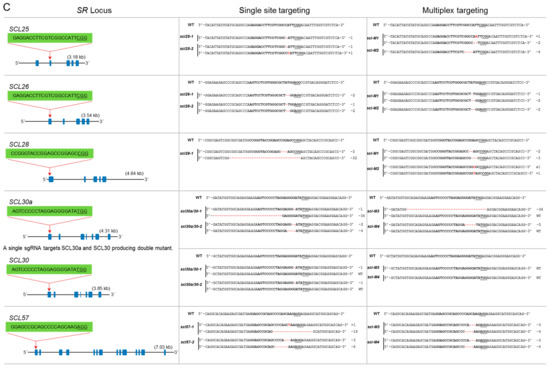

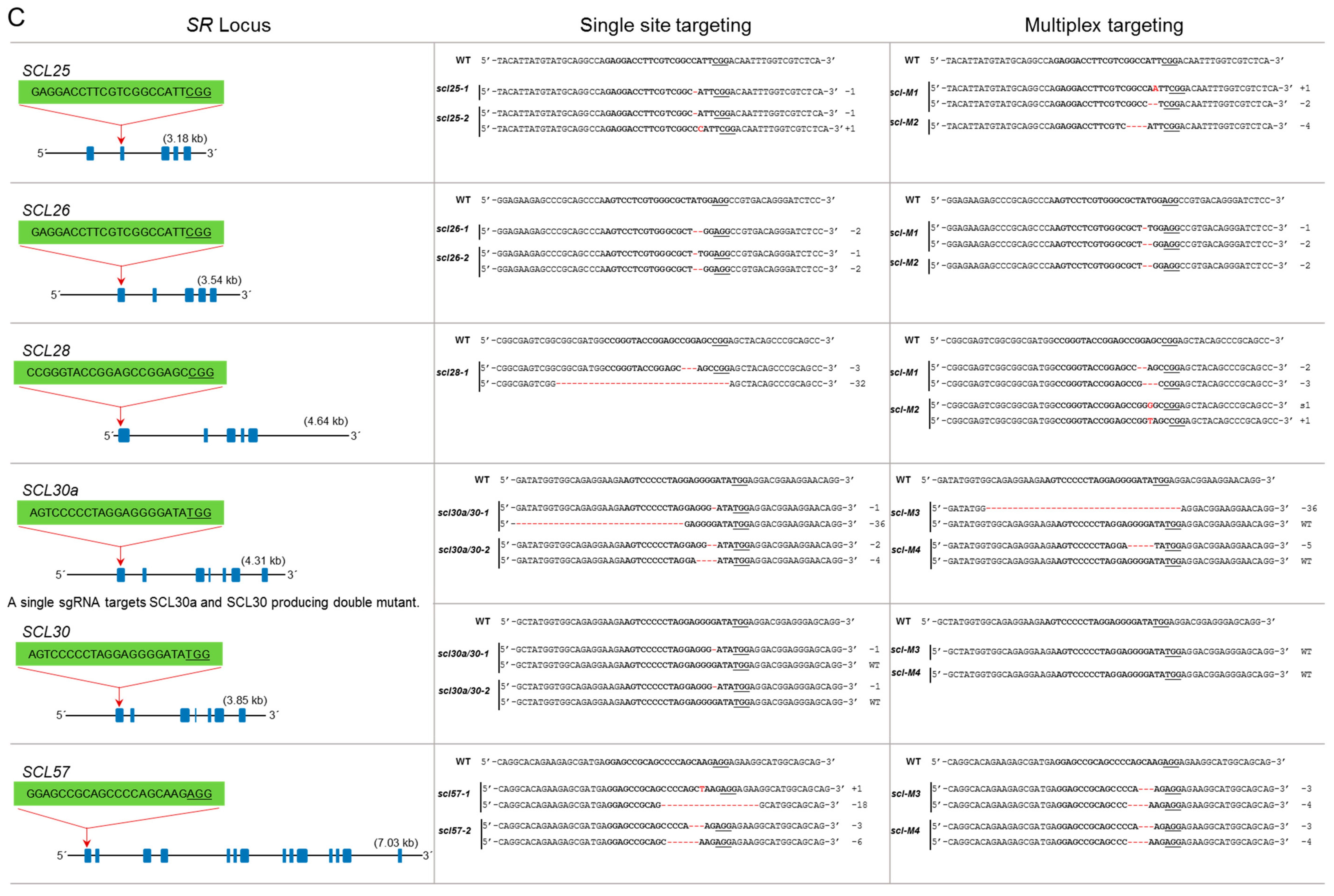
Figure 3.
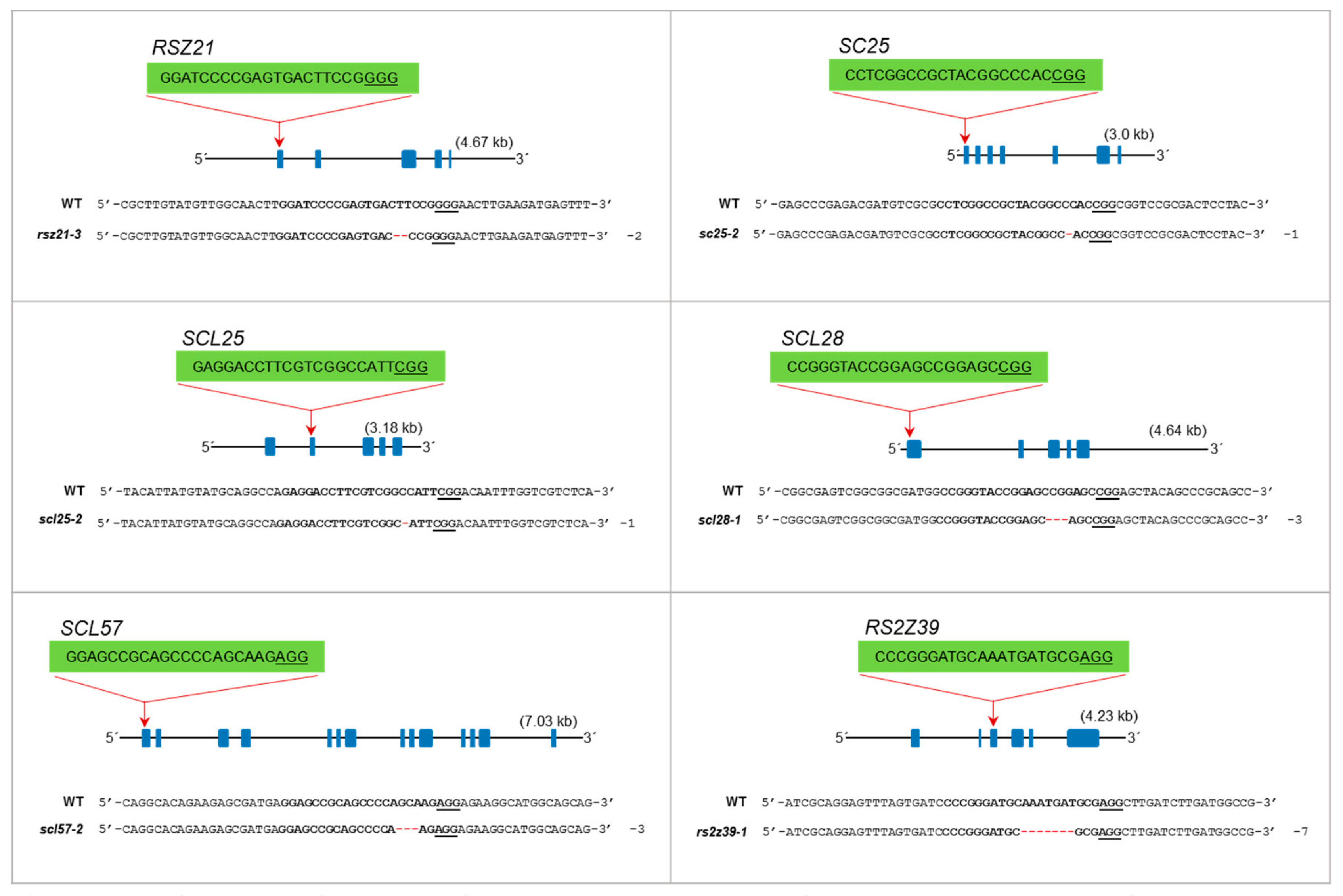
Single and multiplex mutagenesis of SR proteins using the PTG system. sgRNAs were designed to target each SR locus mostly in the first or second exons. The sgRNA target sequence is indicated in bold. The protospacer adjacent motif (PAM) sequence is underlined. Blue boxes indicate exons, and lines represent introns. Multiplex targeting was done for each subgroup of the SR family, in which all loci were targeted simultaneously. The multiplex mutants were named according to their subfamily name preceded by “M” for “multiplex.” (A) Single and multiplex mutants are identified for the SR subfamily; (B) single and multiplex mutants for RSZ and SC subfamilies; (C) the SCL subfamily has six members. Multiplex targeting was done for SCL25/26/28 and SCL30a/30/57. SCL30a and SCL30 are highly conserved; a single sgRNA was used to target both loci; (D) mutagenesis of RS2Z and RS subfamilies. RS2Z36 and RS2Z38 are highly conserved, and a single sgRNA was used to target both loci.
3.3. The RSZ Subfamily
The RSZ subfamily has three members: RSZ21a, RSZ21, and RSZ23. This subfamily is characterized by one zinc knuckle and has an SRSF7/9G8 motif found in mammalian counterparts. RSZ21a has five exons, and we targeted the 1st exon. The mutant line is biallelic with one allele containing an in-frame mutation. RSZ21 was targeted at its 1st exon, and two monoallelic and one biallelic mutants were generated. rsz21-1 is a monoallelic in-frame mutant, while the other two mutants are complete knockouts. RSZ23 contains only four exons, and the gRNA targeted the 1st exon. We did not recover a single knockout mutant for this gene; both mutants for this locus are heterozygous and have a wild-type allele (Figure 3B). In multiplex targeting, we did not recover a complete triple knockout mutant. The rsz-M1 and rsz-M2 mutants are biallelic mutants for RSZ21a and RSZ23 but heterozygous for RSZ21 containing a mutated allele and the wild-type allele (Figure 3B).
3.4. The SC Subfamily
The SC subfamily has three members: SC25, SC32, and SC34. This group has one RRM domain and an SR domain. The mammalian orthologs are SRSF2/SC35. SC25 has seven exons, and the gRNA was designed to target the 1st exon. We generated two single knockout biallelic mutants for this locus. SC32 has eight exons, and we targeted the 1st exon. The mutants for this locus are all complete knockouts and one is a biallelic mutant. SC34 contains eight exons, and the gRNA was designed to target the 1st exon. The single mutants for this locus are complete knockouts except for the sc34-2 mutant, which has one allele with an in-frame deletion (Figure 3B). During multiplex targeting, we produced the sc-M1 mutant, which is a complete knockout for SC25 and SC34 but has no mutation for SC32. The sc-M2 mutant has a knockout mutation for SC34 and a biallelic mutation for SC32 with one in-frame deletion of 21 amino acids and is heterozygous for SC25 containing a mutated allele and the wild-type allele (Figure 3B).
3.5. The SCL Subfamily
This is the largest subfamily of SR genes with six members: SCL25, SCL26, SCL28, SCL30a, SCL30, and SCL57. This is a plant-specific subfamily but is similar to SC35; hence, it is named SC35-lik3 (SCL). This group has an N-terminal extension of charged amino acids. SCL25 has five exons, and we targeted the 2nd exon. We generated single knockout mutants for this locus. Similarly, SCL26 has five exons, and the gRNA targeted the 1st exon. The mutants for this locus have one- and two-nucleotide deletions. SCL28 has five exons, and the gRNA target site was in the 1st exon. The only recovered mutant is biallelic and has a deletion of 32 nucleotides and an in-frame deletion of three nucleotides (Figure 3C) SCL30a and SCL30 have highly similar nucleotide sequences; a single gRNA was designed to target both loci at their 1st exons. The scl30a/30-1 mutant is biallelic for a mutation in SCL30a but heterozygous for SCL30 with a mutated and a wild-type allele at this locus. Similarly, scl30a/30-2 is a knockout for SCL30a but is heterozygous for SCL30 (Figure 3C). SCL57 contains 14 exons, and the gRNA targeted the 1st exon. The mutants produced are biallelic with different in-frame deletions (Figure 3C). For efficient multiplex mutagenesis, we targeted only three loci of this subfamily for each multiplex reaction. scl-M1 is a triple mutant for SCL25/26/28 and has only one in-frame deletion for SCL28. scl-M2 is a triple knockout mutant for all three loci, SCL25/26/28 (Figure 3C). The scl-M3 mutant is heterozygous with an in-frame mutation for SCL30a and a wild-type allele, a biallelic mutation for SCL57, and no mutation for SCL30. The scl-M4 mutant is heterozygous for SCL30a, has no mutation for SCL30, and a biallelic mutation for SCL57 (Figure 3C).
3.6. The RS2Z Subfamily
Proteins of this plant-specific family have two zinc knuckles and a characteristic serine/proline-rich region at their C termini. This subfamily has four members: RS2Z36, RS2Z37, RS2Z38, and RS2Z39. RS2Z36 and RS2Z38 have high sequence similarity, and a single gRNA was designed to target both loci at their 4th exons. rs2z36/38-1 is a double mutant for RS2Z36 and RS2Z38, while rs2z36/38-2 is a knockout mutant for RS2Z36 but was heterozygous for RS2Z38 with a mutated allele and a wild-type allele. RS2Z37 has six exons, and the gRNA targeted the 1st exon. rs2z37-1 is a monoallelic in-frame mutant, but rs2z37-2 is a complete knockout biallelic mutant. RS2Z39 has six exons, and the gRNA targeted the 3rd exon. The mutants recovered for this locus are homozygous, but only the rs2z39-2 mutant has an in-frame mutant allele (Figure 3D). During multiplex targeting, we recovered triple mutants for RS2Z36, RS2Z37, and RS2Z39, but the RS2Z38 locus was not mutated (rs2z-M1) and the other was heterozygous, containing a wild-type allele and a mutated allele (rs2z-M2) (Figure 3D).
3.7. The RS Subfamily
The RS subfamily is plant-specific and has only two members in rice. RS29 has five exons, and the gRNA targeted the 2nd exon. We recovered two mutants, a monoallelic mutant with an in-frame deletion of 18 nucleotides and a heterozygous mutant with a mutated allele and the wild-type allele. RS33 has five exons, and the target site for the gRNA was in the 1st exon. The mutants for this locus are complete knockouts (Figure 3D). We did not recover a double knockout mutant for these loci during multiplex targeting. The rs-M1 mutant is heterozygous for RS29 and has an in-frame deletion for RS33. Similarly, the rs-M2 mutant has in-frame deletions for RS29 as well as for RS33 (Figure 3D).
3.8. Heritability in the Seed Progeny of SR Family Mutants
Recovery of homozygous mutants in the progeny is essential to use these mutants for phenotypic analysis to understand the responses of these mutant to growth, development, abiotic and biotic stress cues. To confirm the heritability of these mutations in seed progeny, we conducted a genotypic analysis on their progeny plants (Figure 4). We selected the T0 heterozygous mutants to analyze the segregation of mutations in their seed progeny including rsz21-3, sc25-2, scl25-2, scl28-1, scl57-2, and rs2z39-1. The genotyping of the T1 generation identified homozygous monoallelic lines for these mutants. Our genotyping analysis data reveal that the single mutants are heritable in seed progeny and we can recover homozygous mutants. These data indicate that our collection of mutants of the SR gene family will be useful for the community to use these mutants to understand the molecular underpinnings of the SR regulation in response to stress and growth cues.
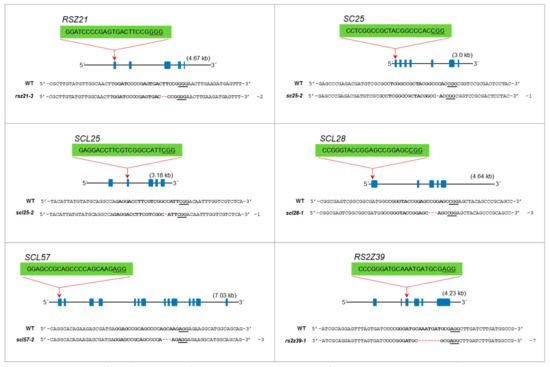
Figure 4.
Analysis of seed progeny of SR mutants. Genotyping of SR gene mutations in seed progeny. The heterozygous parent plants were segregating to homozygous monoallelic mutants for RSZ21, SC25, SCL25, SCL28, SCL57 and RS2Z39.
4. Discussion
Our study provides excellent resource materials to further analyze the biological function of a family of splicing regulators in rice growth, development, and stress responses. Understanding the role of SR proteins in AS in response to environmental stresses will have implications in crop improvement [10,12,40,41]. Pre-mRNA splicing is an important regulatory mechanism during which the noncoding sequences from eukaryotic genes are excised with precision. SR proteins are RNA-binding proteins that assist the spliceosome mainly in splice site selection and are an important component of constitutive and alternative splicing. Most of our knowledge of SR proteins came from animal systems, where, in addition to mRNA splicing, these proteins are involved in mRNA export, stability, and translation; RNA metabolism; gene regulation; subcellular localization of transcripts; genome stability; chromatin binding; and miRNA processing. However, whether plant SR proteins are involved in these processes is still unknown and needs further investigation. Since plants have more SR proteins that are structurally different from mammalian SR proteins further points to some additional roles of these proteins in the plant life cycle. In plants, only Arabidopsis SR proteins have been extensively studied, in which most SR mutants are hypersensitive to abiotic stresses like salt, ABA, drought, and heavy metals [19,21,22,42]. In rice, only a couple of studies have been reported, in which overexpression of RSZ36 and SRp33b changed the splicing patterns of RSZ36 and SRp32, respectively [43]. In another study, Zheng et al. showed that AS plays a critical role in mineral homeostasis and SR proteins are important regulators of the Zn, Mn, and P nutrition uptake and remobilization [23]. The SR pre-mRNAs themselves are alternatively spliced under different environmental stresses and may affect the downstream AS targets [42]. This high number of SR genes in rice probably enables them to adapt to adverse environmental conditions.
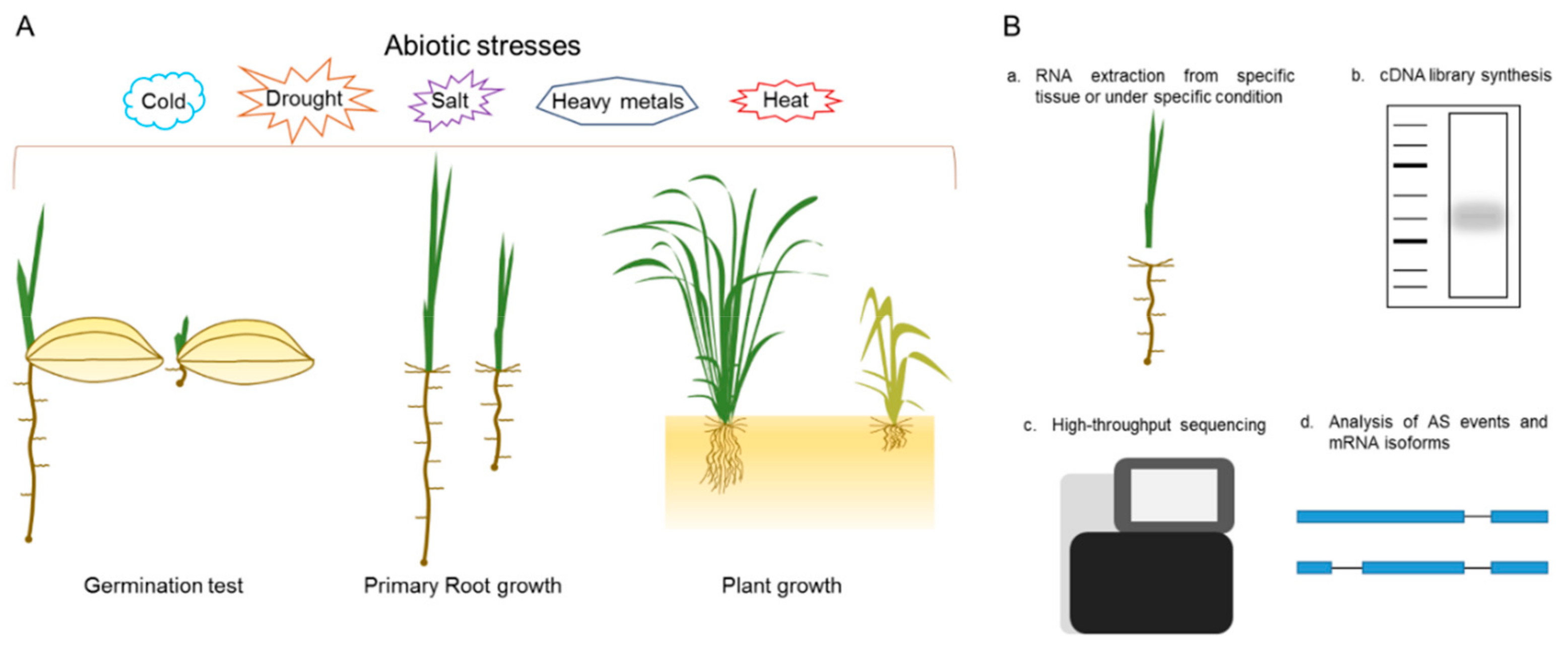
Here, we proposed a platform for physiological and molecular analysis of our SR family mutants (Figure 5). For phenotypic analysis, the mutants’ response to different abiotic stresses, such as drought, salt, extreme temperatures, and heavy metals, can be investigated. The stress treatments can be conducted at various growth stages, such as seed germination, primary root inhibition, or plant growth, in soil or in hydroponic conditions (Figure 5A). These stress treatments will be applied to single or multiplex SR family mutants and will help to determine their molecular role during plant growth, development and in response to stress cues. The multiplex mutants were not homozygous for all targeted loci and produced combinations of single, double and triple mutants. We hypothesized that these are a good starting material for comprehensive genetic and phenotypic analysis and their segregating progeny will serve as a useful resource for the plant splicing and alternative research community. For molecular analysis, RNA-seq may be the best approach. RNA can be extracted from specific tissues (roots, shoots, and reproductive organs) or under specific conditions and used for the cDNA library synthesis. The library will be used for high-throughput sequencing and, after read alignment and splice junction predictions, can help to determine the mRNA isoforms in specific tissues or under specific conditions (Figure 5B). This approach is useful for the determination of general and specific mRNA targets of each SR family member.
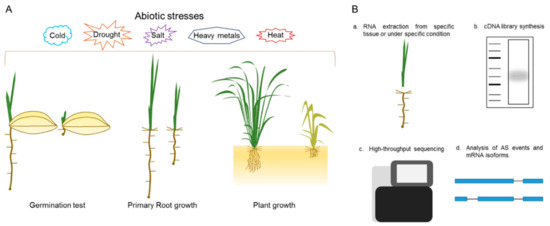
Figure 5.
Platform for the analysis of SR mutants. (A) Molecular and physiological phenotyping can be done at different growth stages of SR mutants. This can be achieved by providing different abiotic stresses like salt, drought, and extreme temperatures, etc; (B) a general pipeline for the global analysis of alternative splicing (AS) in SR mutants.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4425/10/8/596/s1, Table S1: Synthetic fragments used as sgRNAs for targeting single and multiple SR loci, Table S2: List of oligos used for genotyping, Table S3: Summary for genotyping of SR mutants.
Author Contributions
Conceptualization, M.M.M.; Data curation, H.B., A.P. and L.L.; Formal analysis, H.B. and L.L.; Investigation, H.B. and A.P.; Methodology, M.M.M.; Project administration, M.M.M.; Supervision, M.M.M.; Writing—original draft, H.B.; Writing—review & editing, A.S.N.R. and M.M.M.
Funding
This study is supported by King Abdullah University of Science and Technology.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Alexander, J.M.; Diez, J.M.; Levine, J.M. Novel competitors shape species’ responses to climate change. Nature 2015, 525, 515–518. [Google Scholar] [CrossRef] [PubMed]
- Becklin, K.M.; Anderson, J.T.; Gerhart, L.M.; Wadgymar, S.M.; Wessinger, C.A.; Ward, J.K. Examining Plant Physiological Responses to Climate Change through an Evolutionary Lens. Plant. Physiol. 2016, 172, 635–649. [Google Scholar] [CrossRef] [PubMed]
- Mastrangelo, A.M.; Marone, D.; Laido, G.; De Leonardis, A.M.; De Vita, P. Alternative splicing: Enhancing ability to cope with stress via transcriptome plasticity. Plant. Sci. 2012, 185, 40–49. [Google Scholar] [CrossRef] [PubMed]
- Guerra, D.; Crosatti, C.; Khoshro, H.H.; Mastrangelo, A.M.; Mica, E.; Mazzucotelli, E. Post-transcriptional and post-translational regulations of drought and heat response in plants: A spider’s web of mechanisms. Front. Plant. Sci. 2015, 6, 57. [Google Scholar] [CrossRef] [PubMed]
- Wahl, M.C.; Will, C.L.; Luhrmann, R. The Spliceosome: Design Principles of a Dynamic RNP Machine. Cell 2009, 136, 701–718. [Google Scholar] [CrossRef]
- Fica, S.M.; Nagai, K. Cryo-electron microscopy snapshots of the spliceosome: Structural insights into a dynamic ribonucleoprotein machine. Nat. Struct. Mol. Biol. 2017, 24, 791–799. [Google Scholar] [CrossRef] [PubMed]
- Will, C.L.; Luhrmann, R. Spliceosome structure and function. Cold Spring Harb Perspect. Biol. 2011, 3, a003707. [Google Scholar] [CrossRef]
- Koncz, C.; Dejong, F.; Villacorta, N.; Szakonyi, D.; Koncz, Z. The spliceosome-activating complex: Molecular mechanisms underlying the function of a pleiotropic regulator. Front. Plant. Sci. 2012, 3, 9. [Google Scholar] [CrossRef]
- Syed, N.H.; Kalyna, M.; Marquez, Y.; Barta, A.; Brown, J.W.S. Alternative splicing in plants—Coming of age. Trends Plant. Sci. 2012, 17, 616–623. [Google Scholar] [CrossRef]
- Staiger, D.; Brown, J.W. Alternative splicing at the intersection of biological timing, development, and stress responses. Plant. Cell I. 2013, 25, 3640–3656. [Google Scholar] [CrossRef]
- Zhou, Z.; Fu, X.D. Regulation of splicing by SR proteins and SR protein-specific kinases. Chromosoma 2013, 122, 191–207. [Google Scholar] [CrossRef] [PubMed]
- Morton, M.; AlTamimi, N.; Butt, H.; Reddy, A.S.N.; Mahfouz, M. Serine/Arginine-rich protein family of splicing regulators: New approaches to study splice isoform functions. Plant. Sci. 2019, 283, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Yan, K.; Liu, P.; Wu, C.A.; Yang, G.D.; Xu, R.; Guo, Q.H.; Huang, J.G.; Zheng, C.C. Stress-induced alternative splicing provides a mechanism for the regulation of microRNA processing in Arabidopsis thaliana. Mol. Cell 2012, 48, 521–531. [Google Scholar] [CrossRef] [PubMed]
- Ling, Y.; Alshareef, S.; Butt, H.; Lozano-Juste, J.; Li, L.; Galal, A.A.; Moustafa, A.; Momin, A.A.; Tashkandi, M.; Richardson, D.N.; et al. Pre-mRNA splicing repression triggers abiotic stress signaling in plants. Plant. J. 2017, 89, 291–309. [Google Scholar] [CrossRef]
- Laloum, T.; Martin, G.; Duque, P. Alternative Splicing Control of Abiotic Stress Responses. Trends Plant. Sci. 2018, 23, 140–150. [Google Scholar] [CrossRef] [PubMed]
- AlShareef, S.; Ling, Y.; Butt, H.; Mariappan, K.G.; Benhamed, M.; Mahfouz, M.M. Herboxidiene triggers splicing repression and abiotic stress responses in plants. BMC Genom. 2017, 18, 260. [Google Scholar] [CrossRef]
- Fu, X.D.; Maniatis, T. The 35-kDa mammalian splicing factor SC35 mediates specific interactions between U1 and U2 small nuclear ribonucleoprotein particles at the 3′ splice site. Proc. Natl. Acad. Sci. USA 1992, 89, 1725–1729. [Google Scholar] [CrossRef]
- Kohtz, J.D.; Jamison, S.F.; Will, C.L.; Zuo, P.; Luhrmann, R.; Garcia-Blanco, M.A.; Manley, J.L. Protein-protein interactions and 5′-splice-site recognition in mammalian mRNA precursors. Nature 1994, 368, 119–124. [Google Scholar] [CrossRef]
- Carvalho, R.F.; Carvalho, S.D.; Duque, P. The plant-specific SR45 protein negatively regulates glucose and ABA signaling during early seedling development in Arabidopsis. Plant. Physiol. 2010, 154, 772–783. [Google Scholar] [CrossRef]
- Chen, T.; Cui, P.; Chen, H.; Ali, S.; Zhang, S.; Xiong, L. A KH-domain RNA-binding protein interacts with FIERY2/CTD phosphatase-like 1 and splicing factors and is important for pre-mRNA splicing in Arabidopsis. PLoS Genet. 2013, 9, e1003875. [Google Scholar] [CrossRef]
- Cruz, T.M.; Carvalho, R.F.; Richardson, D.N.; Duque, P. Abscisic acid (ABA) regulation of Arabidopsis SR protein gene expression. Int J. Mol. Sci. 2014, 15, 17541–17564. [Google Scholar] [CrossRef]
- Zhang, W.; Du, B.; Liu, D.; Qi, X. Splicing factor SR34b mutation reduces cadmium tolerance in Arabidopsis by regulating iron-regulated transporter 1 gene. Biochem. Biophys. Res. Commun. 2014, 455, 312–317. [Google Scholar] [CrossRef]
- Dong, C.; He, F.; Berkowitz, O.; Liu, J.; Cao, P.; Tang, M.; Shi, H.; Wang, W.; Li, Q.; Shen, Z.; et al. Alternative Splicing Plays a Critical Role in Maintaining Mineral Nutrient Homeostasis in Rice (Oryza sativa). Plant. Cell 2018, 30, 2267–2285. [Google Scholar] [CrossRef]
- Huang, Y.; Steitz, J.A. SRprises along a messenger’s journey. Mol. Cell 2005, 17, 613–615. [Google Scholar] [CrossRef]
- Reed, R.; Cheng, H. TREX, SR proteins and export of mRNA. Curr. Opin. Cell Biol. 2005, 17, 269–273. [Google Scholar] [CrossRef]
- Barta, A.; Kalyna, M.; Lorković, Z.J. Plant SR proteins and their functions. Curr. Top. Microbiol. Immunol. 2008, 326, 83–102. [Google Scholar]
- Li, J.F.; Norville, J.E.; Aach, J.; McCormack, M.; Zhang, D.; Bush, J.; Church, G.M.; Sheen, J. Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat. Biotechnol. 2013, 31, 688–691. [Google Scholar] [CrossRef]
- Mali, P.; Yang, L.; Esvelt, K.M.; Aach, J.; Guell, M.; DiCarlo, J.E.; Norville, J.E.; Church, G.M. RNA-guided human genome engineering via Cas9. Science 2013, 339, 823–826. [Google Scholar] [CrossRef]
- Aman, R.; Ali, Z.; Butt, H.; Mahas, A.; Aljedaani, F.; Khan, M.Z.; Ding, S.; Mahfouz, M. RNA virus interference via CRISPR/Cas13a system in plants. Genome Biol. 2018, 19, 1. [Google Scholar] [CrossRef]
- Butt, H.; Eid, A.; Ali, Z.; Atia, M.A.M.; Mokhtar, M.M.; Hassan, N.; Lee, C.M.; Bao, G.; Mahfouz, M.M. Efficient CRISPR/Cas9-Mediated Genome Editing Using a Chimeric Single-Guide RNA Molecule. Front. Plant. Sci. 2017, 8, 1441. [Google Scholar] [CrossRef]
- Butt, H.; Eid, A.; Momin, A.A.; Bazin, J.; Crespi, M.; Arold, S.T.; Mahfouz, M.M. CRISPR directed evolution of the spliceosome for resistance to splicing inhibitors. Genome Biol. 2019, 20, 73. [Google Scholar] [CrossRef]
- Aouida, M.; Eid, A.; Mahfouz, M.M. CRISPR/Cas9-mediated target validation of the splicing inhibitor Pladienolide, B. Biochim. Open 2016, 3, 72–75. [Google Scholar] [CrossRef]
- Mahas, A.; Neal Stewart, C., Jr.; Mahfouz, M.M. Harnessing CRISPR/Cas systems for programmable transcriptional and post-transcriptional regulation. Biotechnol. Adv. 2018, 36, 295–310. [Google Scholar] [CrossRef]
- Butt, H.; Jamil, M.; Wang, J.Y.; Al-Babili, S.; Mahfouz, M. Engineering plant architecture via CRISPR/Cas9-mediated alteration of strigolactone biosynthesis. BMC Plant. Biol. 2018, 18, 174. [Google Scholar] [CrossRef]
- Gao, C. The future of CRISPR technologies in agriculture. Nat. Rev. Mol. Cell Biol. 2018, 19, 275–276. [Google Scholar] [CrossRef]
- Xie, K.; Minkenberg, B.; Yang, Y. Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc. Natl. Acad. Sci. USA 2015, 112, 3570–3575. [Google Scholar] [CrossRef]
- Hiei, Y.; Komari, T. Agrobacterium-mediated transformation of rice using immature embryos or calli induced from mature seed. Nat. Protoc. 2008, 3, 824–834. [Google Scholar] [CrossRef]
- Barta, A.; Kalyna, M.; Reddy, A.S.N. Implementing a rational and consistent nomenclature for serine/arginine-rich protein splicing factors (SR proteins) in plants. Plant. Cell 2010, 22, 2926–2929. [Google Scholar] [CrossRef]
- Manley, J.L.; Krainer, A.R. A rational nomenclature for serine/arginine-rich protein splicing factors (SR proteins). Genes Dev. 2010, 24, 1073–1074. [Google Scholar] [CrossRef]
- Reddy, A.S.; Marquez, Y.; Kalyna, M.; Barta, A. Complexity of the alternative splicing landscape in plants. Plant. Cell. 2013, 25, 3657–3683. [Google Scholar] [CrossRef]
- Ling, Y.; Serrano, N.; Gao, G.; Atia, M.; Mokhtar, M.; Woo, Y.H.; Bazin, J.; Veluchamy, A.; Benhamed, M.; Crespi, M.; et al. Thermopriming triggers splicing memory in Arabidopsis. J. Exp. Bot. 2018, 69, 2659–2675. [Google Scholar] [CrossRef]
- Palusa, S.G.; Ali, G.S.; Reddy, A.S.N. Alternative splicing of pre-mRNAs of Arabidopsis serine/arginine-rich proteins: Regulation by hormones and stresses. Plant. J. 2007, 49, 1091–1107. [Google Scholar] [CrossRef]
- Isshiki, M.; Tsumoto, A.; Shimamoto, K. The serine/arginine-rich protein family in rice plays important roles in constitutive and alternative splicing of pre-mRNA. Plant. Cell 2006, 18, 146–158. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).