Genome-Wide Identification of WRKY Transcription Factors in Chinese jujube (Ziziphus jujuba Mill.) and Their Involvement in Fruit Developing, Ripening, and Abiotic Stress
Abstract
:1. Introduction
2. Materials and Methods
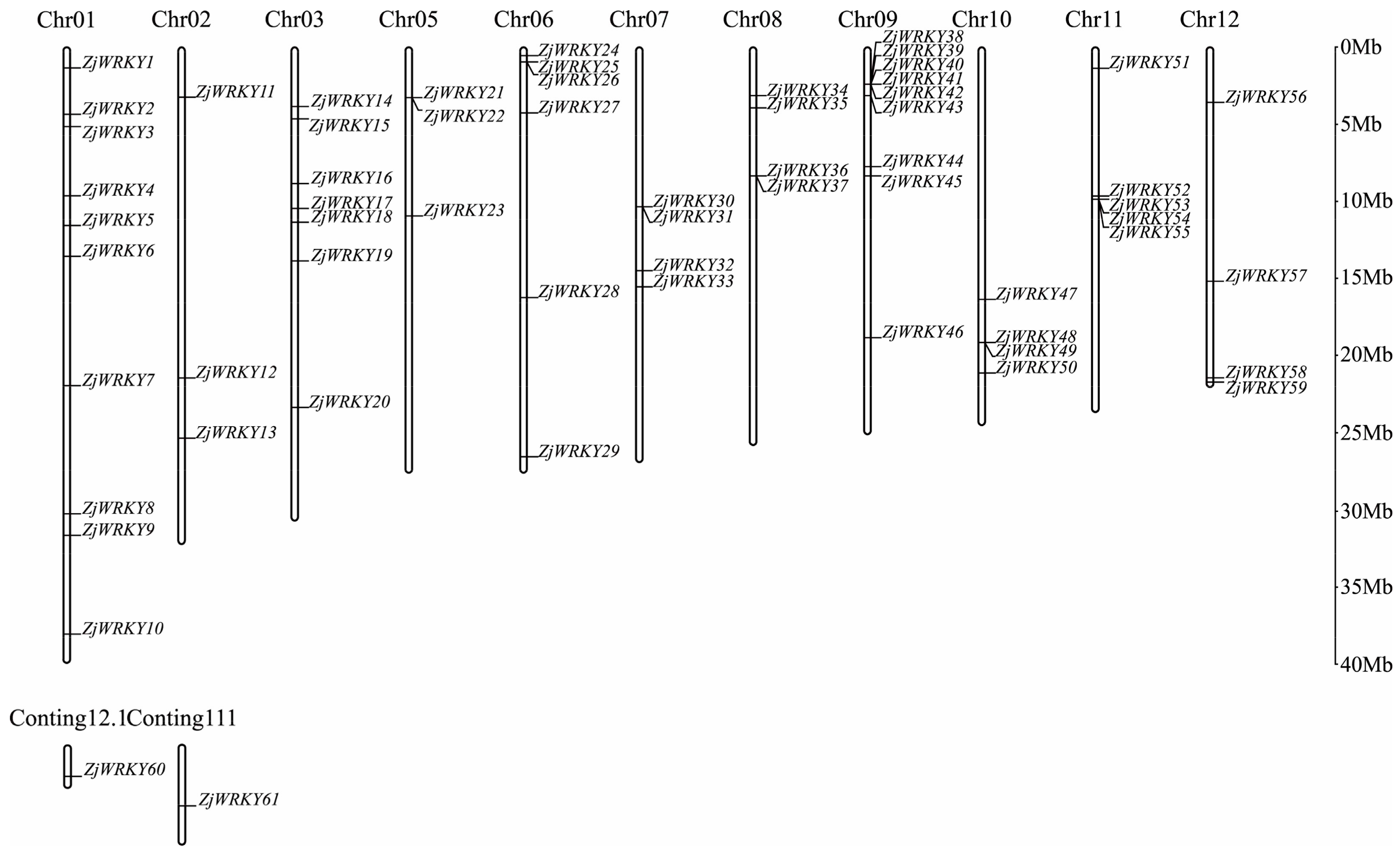
2.1. Identification of the WRKY Family Members in Chinese Jujube and Their Chromosomal Locations
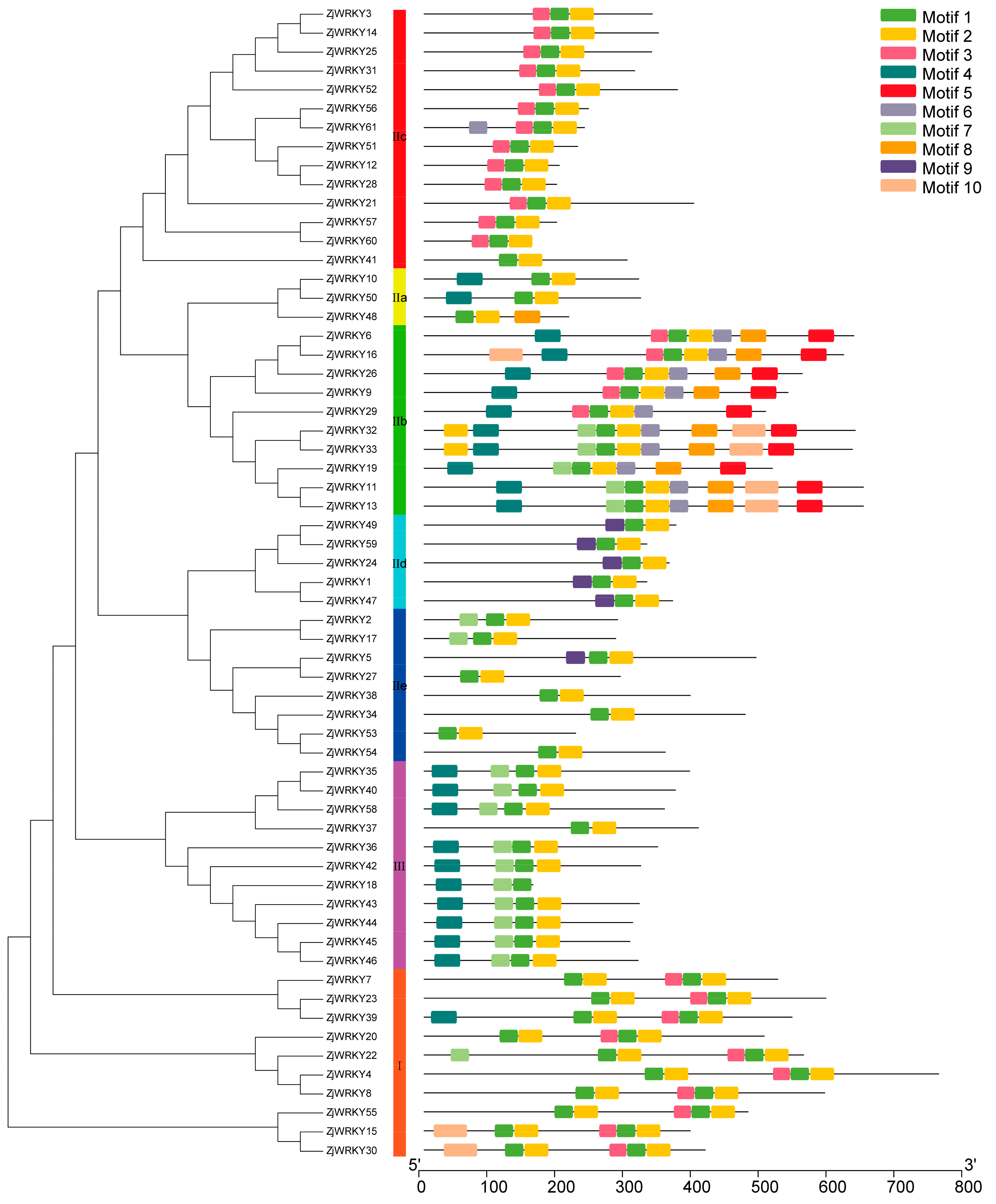
2.2. Sequence Alignment of WRKY Family Members and Construction of the Phylogenetic Tree
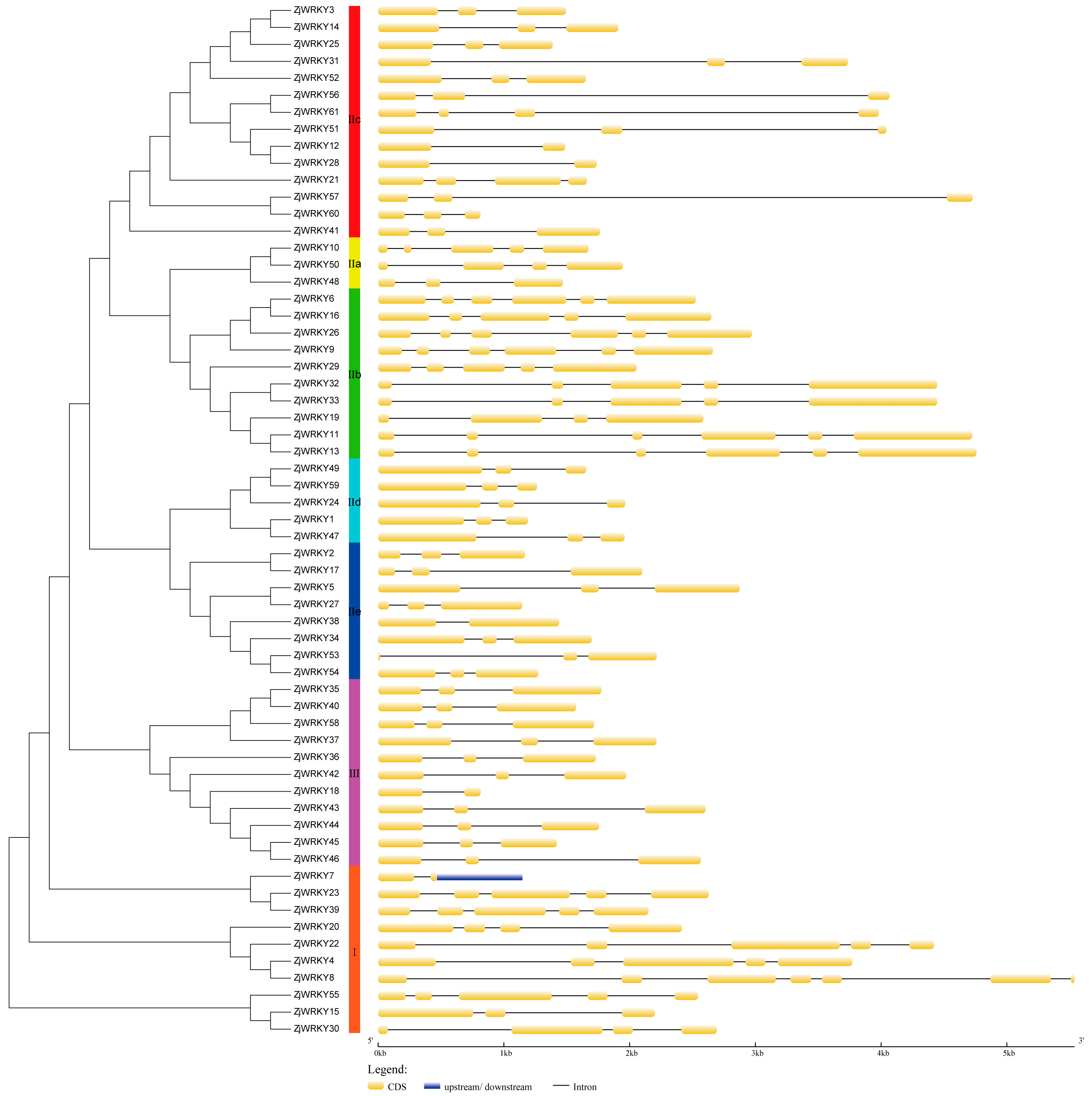
2.3. Structural and Analysis of WRKY Genes
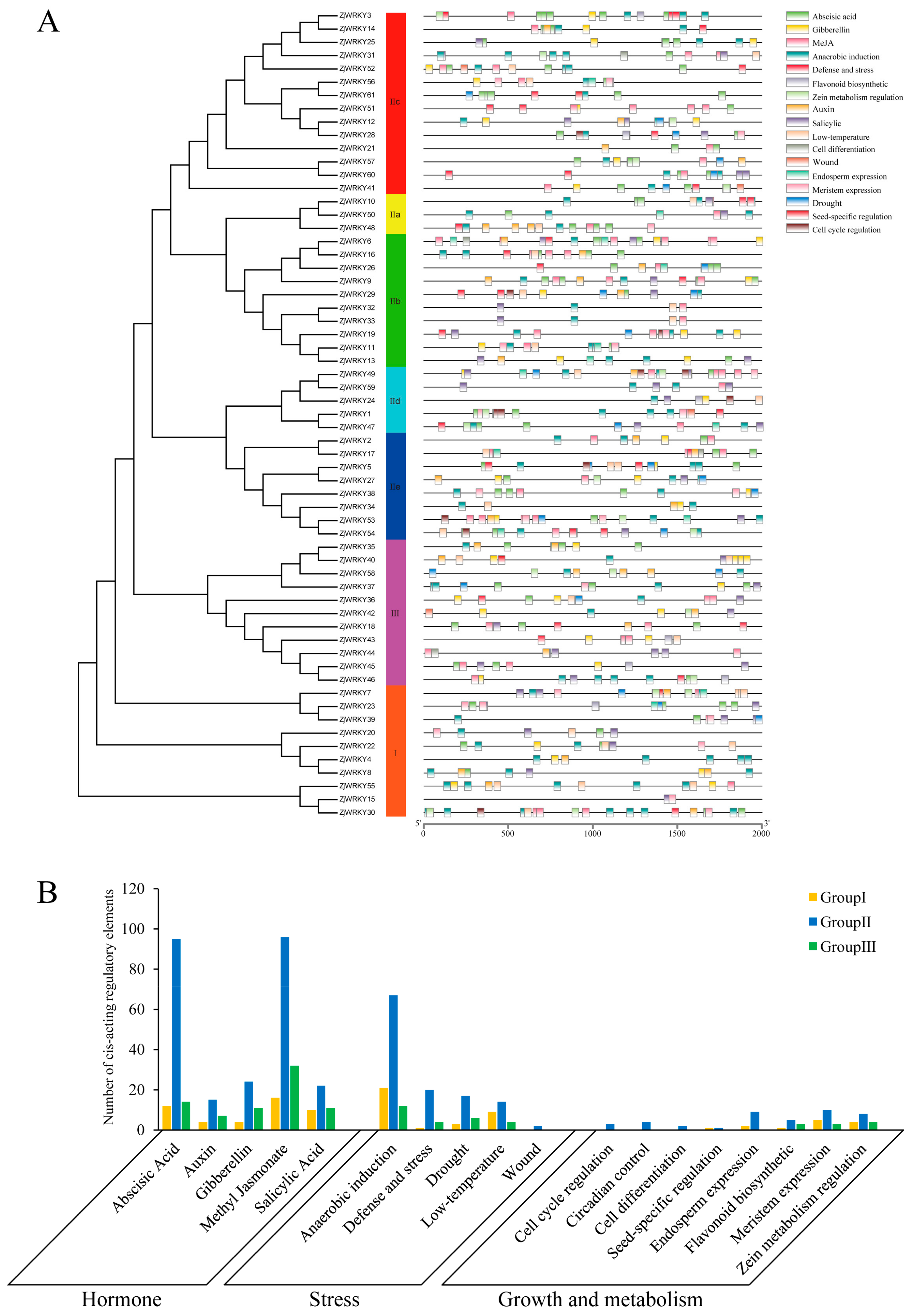
2.4. Analysis of Cis-Acting Regulatory Elements in the Promoter Regions of ZjWRKY Genes
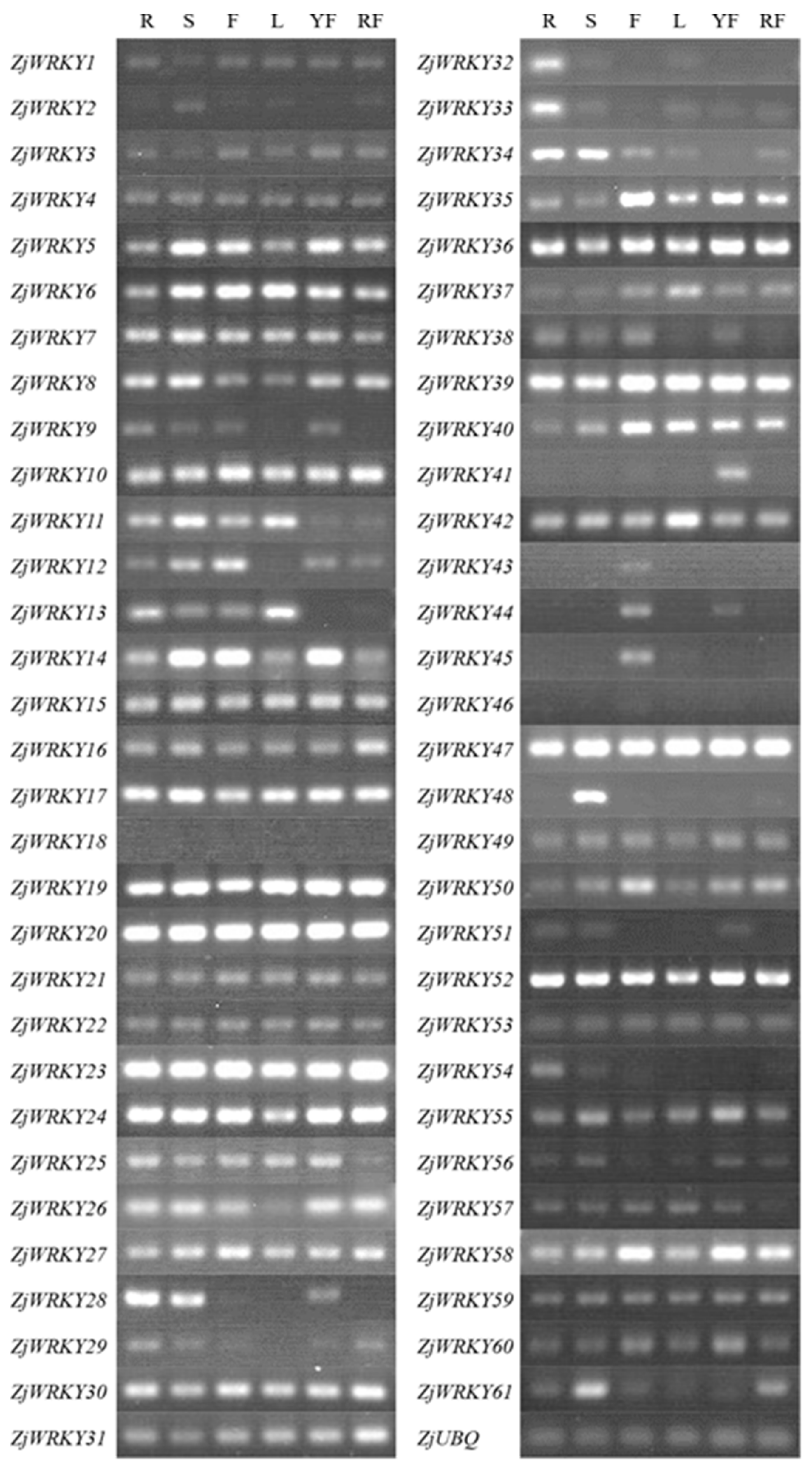
2.5. Tissue-Specific Expression Analysis of ZjWRKY Genes by Reverse Transcription Polymerase Chain Reaction (RT-PCR)
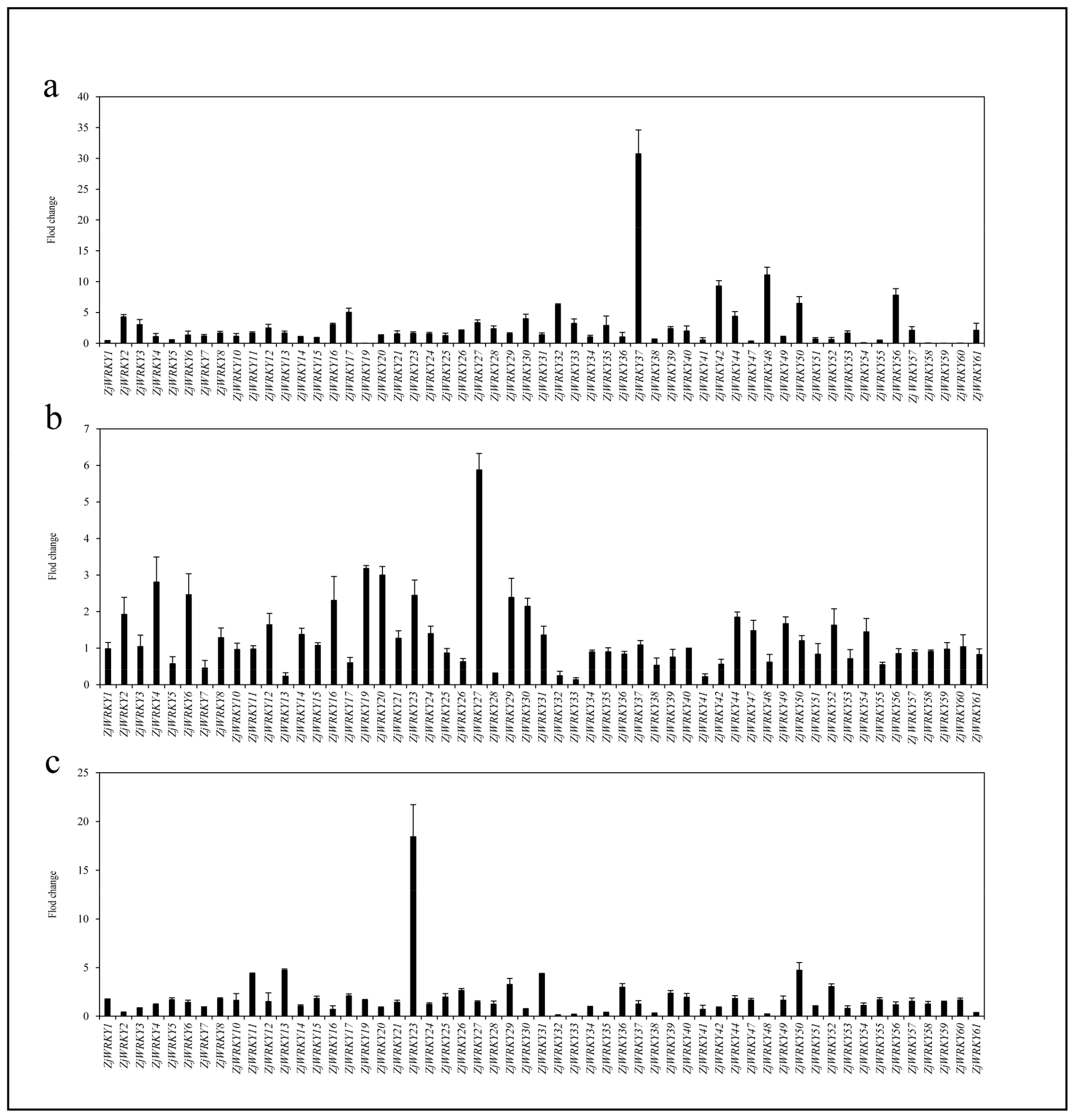
2.6. Transcriptome Analysis of ZjWRKY Genes During Jujube and Wild Jujube Fruit Development and Ripening
2.7. Salt Stress, Drought Stress, and ABA Treatment Experiments
2.7.1. Seedling Preparation
2.7.2. Drought Stress Treatment
2.7.3. NaCl Treatment
2.7.4. ABA Treatment
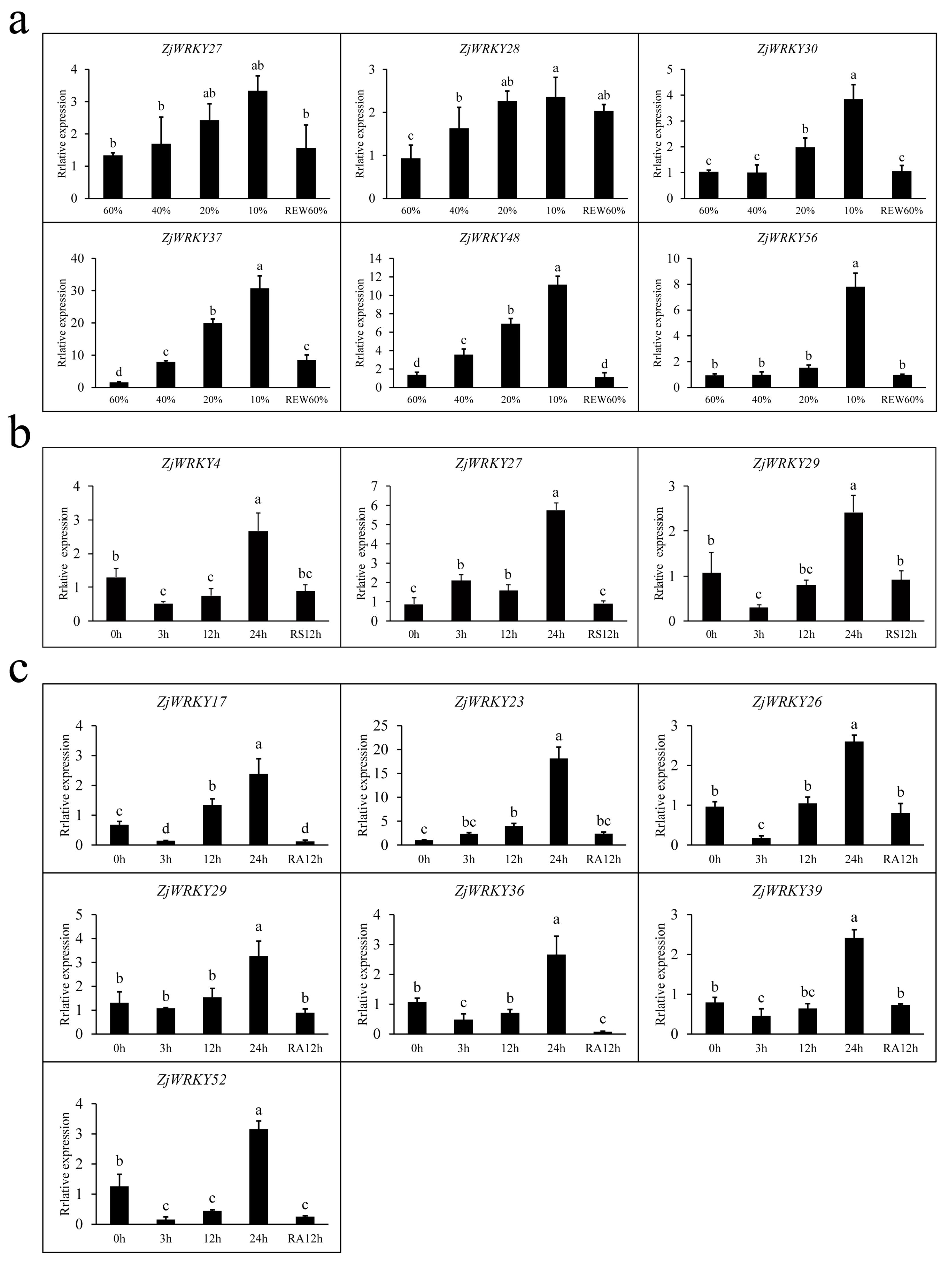
2.8. RT-qPCR Analyses of ZjWRKY Genes in Response to Abiotic Stresses
3. Results
3.1. Identification and Chromosomal Locations of ZjWRKY Family in Jujube
3.2. Phylogenetic Classification of ZjWRKY Genes
3.3. Conserved Motifs of the ZjWRKY and Structure of Their Genes
3.4. Cis-Acting Regulatory Elements of the ZjWRKY Promoters
3.5. Tissue-Specific Expression Profile of ZjWRKY Genes
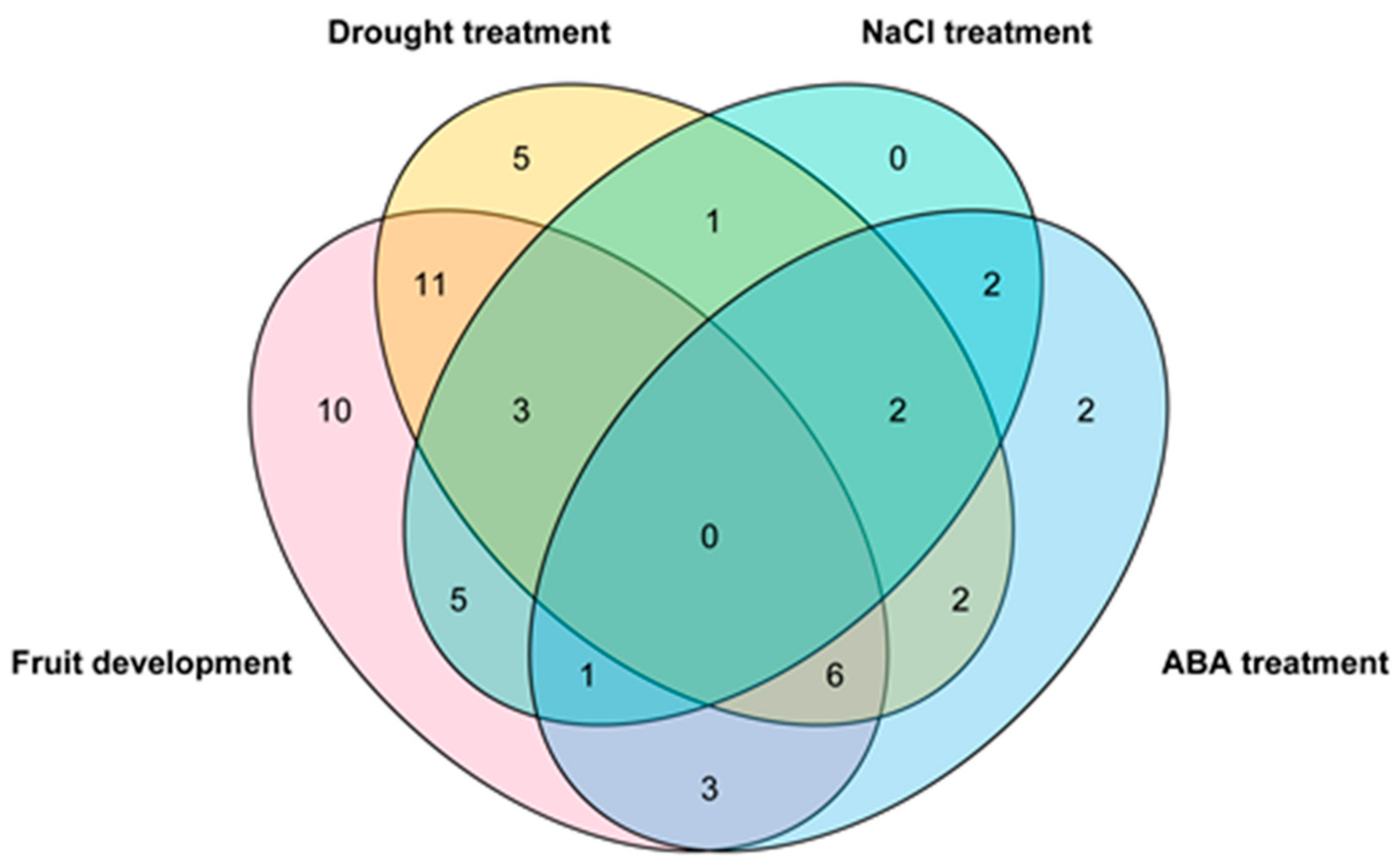
3.6. Transcriptional Dynamics of ZjWRKY Genes During Fruit Development and Ripening
3.7. Transcriptional Responses of ZjWRKY Genes to Salt, and Drought Stresses and ABA Treatment
4. Discussion
4.1. Identification of the WRKY Family Members in Chinese Jujube
4.2. ZjWRKY Expression Profile in Different Tissues
4.3. ZjWRKY Genes Associated with Jujube Fruit Development and Ripening
4.4. ZjWRKY Genes Involved in Response to Abiotic Stresses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Liu, M.J.; Wang, J.R.; Liu, P.; Zhao, J.; Zhao, Z.H.; Dai, L.; Li, X.S.; Liu, Z.G. Historical achievements and frontier advances in the production and research of Chinese jujube (Ziziphus jujuba) in China. Acta Hortic. Sin. 2015, 42, 1683–1698. [Google Scholar] [CrossRef]
- Song, S.; Zhou, H.; Sheng, S.; Cao, M.; Li, Y.; Pang, X. Genome-wide organization and expression profiling of the SBP-box gene family in Chinese jujube (Ziziphus jujuba Mill.). Int. J. Mol. Sci. 2017, 18, 1734. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J. China Forestry Statistical Yearbook; China Forestry Publishing House: Beijing, China, 2014; p. 474. [Google Scholar]
- Zhang, P.; Shi, Y.J.; Song, F.H.; Zhuo, R.T.; Wu, Z.B. Investigation on variation and correlation of the main nutrition quality traits of Ziziphus jujuba cv. Huizao from south of Xinjiang. J. Fruit Sci. 2011, 28, 77–81. [Google Scholar]
- Liu, M.J.; Zhao, J.; Cai, Q.L.; Liu, G.C.; Wang, J.R.; Zhao, Z.H.; Liu, P.; Dai, L.; Yan, G.; Wang, W.J.; et al. The complex jujube genome provides insights into fruit tree biology. Nat. Commun. 2014, 5, 5315. [Google Scholar] [CrossRef] [PubMed]
- Golldack, D.; Luking, I.; Yang, O. Plant tolerance to drought and salinity: Stress regulating transcription factors and their functional significance in the cellular transcriptional network. Plant. Cell Rep. 2011, 30, 1383–1391. [Google Scholar] [CrossRef]
- Zhu, J.K. Abiotic stress signaling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, D.; Yang, C.; Kong, N.; Shi, Z.; Zhao, P.; Nan, Y.; Nie, T.; Wang, R.; Ma, H.; et al. Genome-wide identification of the potato WRKY transcription factor family. PLoS ONE 2017, 12, e0181573. [Google Scholar] [CrossRef]
- Silva Monteiro de Almeida, D.; Oliveira Jordao do Amaral, D.; Del-Bem, L.E.; Bronze Dos Santos, E.; Santana Silva, R.J.; Peres Gramacho, K.; Vincentz, M.; Micheli, F. Genome-wide identification and characterization of cacao WRKY transcription factors and analysis of their expression in response to witches’ broom disease. PLoS ONE 2017, 12, e0187346. [Google Scholar] [CrossRef]
- Wang, L.; Zhu, W.; Fang, L.; Sun, X.; Su, L.; Liang, Z.; Wang, N.; Londo, J.P.; Li, S.; Xin, H. Genome-wide identification of WRKY family genes and their response to cold stress in Vitis vinifera. BMC Plant Biol. 2014, 14, 103. [Google Scholar] [CrossRef]
- Chu, X.; Wang, C.; Chen, X.; Lu, W.; Li, H.; Wang, X.; Hao, L.; Guo, X. The cotton WRKY gene GhWRKY41 positively regulates salt and drought stress tolerance in transgenic Nicotiana benthamiana. PLoS ONE 2015, 10, e0157026. [Google Scholar] [CrossRef]
- Duan, G.F.; Li, L.J.; Liu, Q.L. A WRKY transcription factor from Malus domestica negatively regulates dehydration stress in transgenic Arabidopsis. Acta Physiol. Plant 2014, 36, 541–548. [Google Scholar] [CrossRef]
- Zhang, Y.; Yu, H.; Yang, X.; Li, Q.; Ling, J.; Wang, H.; Gu, X.; Huang, S.; Jiang, W. CsWRKY46, a WRKY transcription factor from cucumber, confers cold resistance in transgenic-plant by regulating a set of cold-stress responsive genes in an ABA-dependent manner. Plant Physiol. Biochem. 2016, 108, 478–487. [Google Scholar] [CrossRef]
- Meng, D.; Li, Y.; Bai, Y.; Li, M.; Cheng, L. Genome-wide identification and characterization of WRKY transcriptional factor family in apple and analysis of their responses to waterlogging and drought stress. Plant Physiol. Biochem. 2016, 103, 71–83. [Google Scholar] [CrossRef]
- Vives-Peris, V.; Marmaneu, D.; Gómez-Cadenas, A.; Perez-Clemente, R.M. Characterization of Citrus WRKY transcription factors and their responses to phytohormones and abiotic stresses. Biol. Plantarum. 2017, 62, 1–12. [Google Scholar] [CrossRef]
- Dai, W.; Wang, M.; Gong, X.; Liu, J.H. The transcription factor FcWRKY40 of Fortunella crassifolia functions positively in salt tolerance through modulation of ion homeostasis and proline biosynthesis by directly regulating SOS2 and P5CS1 homologs. New Phytol. 2018, 219, 972–989. [Google Scholar] [CrossRef]
- Zhu, D.; Che, Y.; Xiao, P.; Hou, L.; Guo, Y.; Liu, X. Functional analysis of a grape WRKY30 gene in drought resistance. Plant Cell Tiss. Org. 2018, 132, 449–459. [Google Scholar] [CrossRef]
- Miao, Y.; Laun, T.; Zimmermann, P.; Zentgraf, U. Targets of the WRKY53 transcription factor and its role during leaf senescence in Arabidopsis. Plant Mol. Biol. 2004, 55, 853–867. [Google Scholar] [CrossRef]
- Guan, Y.; Meng, X.; Khanna, R.; LaMontagne, E.; Liu, Y.; Zhang, S. Phosphorylation of a WRKY transcription factor by MAPKs is required for pollen development and function in Arabidopsis. PLoS Genet. 2014, 10, e1004384. [Google Scholar] [CrossRef]
- Johnson, C.S.; Kolevski, B.; Smyth, D.R. TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell 2002, 14, 1359–1375. [Google Scholar] [CrossRef]
- Suttipanta, N.; Pattanaik, S.; Kulshrestha, M.; Patra, B.; Singh, S.K.; Yuan, L. The transcription factor CrWRKY1 positively regulates the terpenoid indole alkaloid biosynthesis in Catharanthus roseus. Plant Physiol. 2011, 157, 2081–2093. [Google Scholar] [CrossRef]
- Goel, R.; Pandey, A.; Trivedi, P.K.; Asif, M.H. Genome-wide analysis of the Musa WRKY gene family: Evolution and differential expression during development and stress. Front. Plant Sci. 2016, 7, 299. [Google Scholar] [CrossRef]
- Yuan, C.; Ahammed, G.J.; Yu, J.; Yao, Z.; Ruan, M.; Ye, Q.; Li, Z.; Wang, R.; Feng, K.; Zhou, G.J.S.R. Putative WRKYs associated with regulation of fruit ripening revealed by detailed expression analysis of the WRKY gene family in pepper. Sci. Rep. 2016, 6, 39000. [Google Scholar]
- Huang, J.; Zhang, C.; Zhao, X.; Fei, Z.; Wan, K.; Zhang, Z.; Pang, X.; Yin, X.; Bai, Y.; Sun, X. The jujube genome provides insights into genome evolution and the domestication of sweetness/acidity taste in fruit trees. PLoS Genet. 2016, 12, e1006433. [Google Scholar] [CrossRef]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador Vegas, A.; et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef]
- Johnson, L.S.; Eddy, S.R.; Portugaly, E. Hidden Markov model speed heuristic and iterative HMM search procedure. BMC Bioinform. 2010, 11, 431. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME Suite: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Postel, D.; Vanlemmens, P.; Gode, P.; Ronco, G.; Villa, P. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 2002, 31, 325–327. [Google Scholar]
- Zhang, C.; Huang, J.; Li, X. Identification of appropriate reference genes for RT-qPCR analysis in Ziziphus jujuba Mill. Sci. Hortic. Amst. 2015, 197, 166–169. [Google Scholar] [CrossRef]
- Abràmoff, M.D.; Magalhães, P.J.; Ram, S.J. Image processing with ImageJ. Biophotonics Int. 2004, 11, 36–42. [Google Scholar]
- Li, D.; Liu, P.; Yu, J.; Wang, L.; Dossa, K.; Zhang, Y.; Zhou, R.; Wei, X.; Zhang, X. Genome-wide analysis of WRKY gene family in the sesame genome and identification of the WRKY genes involved in responses to abiotic stresses. BMC Plant Biol. 2017, 17, 152. [Google Scholar] [CrossRef]
- Matsuura, H.; Ishibashi, Y.; Shinmyo, A.; Kanaya, S.; Kato, K.J.P. Genome-wide analyses of early translational responses to elevated temperature and high salinity in Arabidopsis thaliana. Plant Cell Physiol. 2010, 51, 448–462. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Chen, C.J.; Xia, R.; Chen, H.; He, Y.H. TBtools, a Toolkit for Biologists integrating various biological data handling tools with a user-friendly interface. BioRxiv 2018, 289660. [Google Scholar]
- Ramamoorthy, R.; Jiang, S.Y.; Kumar, N.; Venkatesh, P.N.; Ramachandran, S. A comprehensive transcriptional profiling of the WRKY gene family in rice under various abiotic and phytohormone treatments. Plant Cell Physiol. 2008, 49, 865–879. [Google Scholar] [CrossRef]
- Birnbaum, K.; Shasha, D.E.; Wang, J.Y.; Jung, J.W.; Lambert, G.M.; Galbraith, D.W.; Benfey, P.N.J.S. A gene expression map of the Arabidopsis root. Science 2003, 302, 1956–1960. [Google Scholar] [CrossRef]
- Bhattarai, K.K.; Atamian, H.S.; Kaloshian, I.; Eulgem, T. WRKY72-type transcription factors contribute to basal immunity in tomato and Arabidopsis as well as gene-for-gene resistance mediated by the tomato R gene Mi-1. Plant J. 2010, 63, 229–240. [Google Scholar] [CrossRef]
- Fujisawa, M.; Nakano, T.; Shima, Y.; Ito, Y. A large-scale identification of direct targets of the tomato MADS box transcription factor RIPENING INHIBITOR reveals the regulation of fruit ripening. Plant Cell 2013, 25, 371–386. [Google Scholar] [CrossRef]
- Lee, S.; Chung, E.J.; Joung, Y.H.; Choi, D. Non-climacteric fruit ripening in pepper: Increased transcription of EIL-like genes normally regulated by ethylene. Funct. Integr. Genomic 2010, 10, 135–146. [Google Scholar] [CrossRef]
- Marchive, C.; Mzid, R.; Deluc, L.; Barrieu, F.; Pirrello, J.; Gauthier, A.; Corio-Costet, M.-F.; Regad, F.; Cailleteau, B.; Hamdi, S.J.J.o.E.B. Isolation and characterization of a Vitis vinifera transcription factor, VvWRKY1, and its effect on responses to fungal pathogens in transgenic tobacco plants. J. Exp. Bot. 2007, 58, 1999–2010. [Google Scholar] [CrossRef]
- Wang, J.-N.; Kuang, J.-F.; Shan, W.; Chen, J.; Xie, H.; Lu, W.-J.; Chen, J.-W.; Chen, J.-Y. Expression profiles of a banana fruit linker histone H1 gene MaHIS1 and its interaction with a WRKY transcription factor. Plant Cell Rep. 2012, 31, 1485–1494. [Google Scholar] [CrossRef]
- Qi, Y.; Liu, M. Change of endogenous hormone in cultivars of Chinese jujube with different type of embryo abortion. Acta Hortic. Sin. 2004, 6, 800–802. [Google Scholar]
- Jiang, Y.; Deyholos, M.K. Comprehensive transcriptional profiling of NaCl-stressed Arabidopsis roots reveals novel classes of responsive genes. BMC Plant Biol. 2006, 6, 25. [Google Scholar] [CrossRef]
- Ding, Z.J.; Yan, J.Y.; Li, C.X.; Li, G.X.; Wu, Y.R.; Zheng, S.J. Transcription factor WRKY46 modulates the development of Arabidopsis lateral roots in osmotic/salt stress conditions via regulation of ABA signaling and auxin homeostasis. Plant J. 2015, 84, 56–69. [Google Scholar] [CrossRef]
- Chen, L.; Song, Y.; Li, S.; Zhang, L.; Zou, C.; Yu, D. The role of WRKY transcription factors in plant abiotic stresses. BBA-Gene Regul. Mech. 2012, 1819, 120–128. [Google Scholar] [CrossRef]
- Chen, H.; Lai, Z.; Shi, J.; Xiao, Y.; Chen, Z.; Xu, X. Roles of Arabidopsis WRKY18, WRKY40 and WRKY60 transcription factors in plant responses to abscisic acid and abiotic stress. BMC Plant Biol. 2010, 10, 281. [Google Scholar] [CrossRef]
- Liao, C.J.; Lai, Z.; Lee, S.; Yun, D.J.; Mengiste, T. Arabidopsis HOOKLESS1 regulates responses to pathogens and abscisic acid through interaction with MED18 and acetylation of WRKY33 and ABI5 chromatin. Plant Cell 2016, 28, 1662. [Google Scholar] [CrossRef]
- Jiang, Y.; Deyholos, M.K. Functional characterization of Arabidopsis NaCl-inducible WRKY25 and WRKY33 transcription factors in abiotic stresses. Plant Mol. Biol. 2009, 69, 91–105. [Google Scholar] [CrossRef]


| Name Symbol | Chromosome | Peptide Length | pI | MW | CDS Number | Group | Conserved Heptapeptide | Zinc-Finger Type |
|---|---|---|---|---|---|---|---|---|
| ZjWRKY1 | Chr1 | 329 | 9.57 | 35 | 3 | II d | WRKYGQK | CX5CX23HXH |
| ZjWRKY2 | Chr1 | 286 | 5.62 | 31 | 3 | II e | WRKYGQK | CX5CX23HXH |
| ZjWRKY3 | Chr1 | 337 | 6.37 | 37 | 3 | II c | WRKYGQK | CX4CX23HXH |
| ZjWRKY4 | Chr1 | 759 | 5.57 | 82 | 5 | I | WRKYGQK WRKYGQK | CX4CX22HXH CX4CX23HXH |
| ZjWRKY5 | Chr1 | 490 | 5.70 | 52 | 3 | II e | WRKYGQK | CX5CX23HXH |
| ZjWRKY6 | Chr1 | 634 | 6.12 | 68 | 6 | II b | WRKYGQK | CX5CX23HXH |
| ZjWRKY7 | Chr1 | 522 | 5.12 | 57 | 2 | I | WRKYGQK WRKYGQK | CX4CX22HXH CX4CX23HXH |
| ZjWRKY8 | Chr1 | 591 | 6.12 | 65 | 7 | I | WRKYGQK WRKYGQK | CX4CX22HXH CX4CX23HXH |
| ZjWRKY9 | Chr1 | 536 | 6.26 | 58 | 6 | II b | WRKYGQK | CX5CX19HXH |
| ZjWRKY10 | Chr1 | 317 | 8.68 | 35 | 5 | II a | WRKYGQK | CX5CX23HXH |
| ZjWRKY11 | Chr2 | 648 | 6.74 | 70 | 6 | II b | WRKYGQK | CX5CX23HXH |
| ZjWRKY12 | Chr2 | 200 | 9.21 | 23 | 2 | II c | WRKYGQK | CX4CX23HXH |
| ZjWRKY13 | Chr2 | 648 | 6.74 | 70 | 6 | II b | WRKYGQK | CX5CX23HXH |
| ZjWRKY14 | Chr3 | 346 | 6.76 | 39 | 3 | II c | WRKYGQK | CX4CX23HXH |
| ZjWRKY15 | Chr3 | 393 | 8.60 | 43 | 3 | I | WRKYGQK WRKYGQK | CX4CX22HXH CX4CX23HXH |
| ZjWRKY16 | Chr3 | 619 | 6.21 | 67 | 5 | II b | WRKYGQK | CX5CX23HXH |
| ZjWRKY17 | Chr3 | 283 | 5.46 | 31 | 3 | II e | WRKYGQK | CX5CX23HXH |
| ZjWRKY18 | Chr3 | 161 | 8.79 | 18 | 2 | III | WRKYGQK | N |
| ZjWRKY19 | Chr3 | 514 | 7.63 | 56 | 4 | II b | WRKYGQK | CX5CX23HXH |
| ZjWRKY20 | Chr3 | 502 | 6.52 | 55 | 4 | I | WRKYGQK WRKYGQK | CX4CX22HXH CX4CX23HXH |
| ZjWRKY21 | Chr5 | 398 | 7.08 | 44 | 4 | II c | WRKYGQK | CX4CX23HXH |
| ZjWRKY22 | Chr5 | 560 | 5.81 | 61 | 5 | I | WRKYGQK WRKYGQK | CX4CX22HXH CX4CX23HXH |
| ZjWRKY23 | Chr5 | 593 | 7.21 | 65 | 5 | I | WRKYGQK WRKYGQK | CX4CX22HXH CX4CX23HXH |
| ZjWRKY24 | Chr6 | 362 | 9.65 | 41 | 3 | II d | WRKYGQK | CX5CX23HXH |
| ZjWRKY25 | Chr6 | 336 | 6.43 | 37 | 3 | II c | WRKYGQK | CX4CX23HXH |
| ZjWRKY26 | Chr6 | 558 | 7.22 | 61 | 6 | II b | WRKYGQK | CX5CX23HXH |
| ZjWRKY27 | Chr6 | 290 | 5.27 | 32 | 3 | II e | WRKYGQK | CX5CX23HXH |
| ZjWRKY28 | Chr6 | 196 | 9.34 | 22 | 2 | II c | WRKYGQK | CX4CX23HXH |
| ZjWRKY29 | Chr6 | 504 | 5.50 | 56 | 5 | II b | WRKYGQK | CX5CX23HXH |
| ZjWRKY30 | Chr7 | 415 | 6.99 | 46 | 4 | I | WRKYGQK WRKYGQK | CX4CX22HXH CX4CX23HXH |
| ZjWRKY31 | Chr7 | 311 | 5.65 | 34 | 3 | II c | WRKYGQK | CX4CX23HXH |
| ZjWRKY32 | Chr7 | 636 | 6.62 | 69 | 5 | II b | WRKYGQK | CX5CX23HXH |
| ZjWRKY33 | Chr7 | 632 | 6.45 | 68 | 5 | II b | WRKYGQK | CX5CX23HXH |
| ZjWRKY34 | Chr8 | 474 | 5.19 | 52 | 3 | II e | WRKYGQK | CX5CX23HXH |
| ZjWRKY35 | Chr8 | 392 | 5.90 | 44 | 3 | III | WRKYGQK | CX7CX23HXC |
| ZjWRKY36 | Chr8 | 345 | 5.39 | 39 | 3 | III | WRKYGQK | CX7CX23HXC |
| ZjWRKY37 | Chr8 | 405 | 6.64 | 45 | 3 | III | WRKYGQK | CX7CX23HXC |
| ZjWRKY38 | Chr9 | 393 | 4.93 | 45 | 2 | II e | WRKYGQK | CX5CX23HXH |
| ZjWRKY39 | Chr9 | 543 | 7.09 | 60 | 5 | I | WRKYGQK WRKYGQK | CX4CX22HXH CX4CX23HXH |
| ZjWRKY40 | Chr9 | 371 | 5.26 | 42 | 3 | III | WRKYGQK | CX7CX23HXC |
| ZjWRKY41 | Chr9 | 300 | 5.35 | 33 | 3 | II c | WRKYGQK | CX4CX23HXH |
| ZjWRKY42 | Chr9 | 320 | 5.25 | 36 | 3 | III | WRKYGQK | CX7CX23HXC |
| ZjWRKY43 | Chr9 | 318 | 8.08 | 36 | 3 | III | WRKYGQK | CX7CX23HXC |
| ZjWRKY44 | Chr9 | 308 | 5.84 | 35 | 3 | III | WRKYGQK | CX7CX23HXC |
| ZjWRKY45 | Chr9 | 304 | 5.25 | 34 | 3 | III | WRQYGQK | CX7CX23HXC |
| ZjWRKY46 | Chr9 | 316 | 6.46 | 36 | 3 | III | WRKYGQK | CX7CX23HXC |
| ZjWRKY47 | Chr10 | 367 | 9.64 | 39 | 3 | II d | WRKYGQK | CX5CX23HXH |
| ZjWRKY48 | Chr10 | 214 | 9.32 | 23 | 3 | II a | WRKYGQK | CX5CX23HXH |
| ZjWRKY49 | Chr10 | 372 | 9.57 | 40 | 3 | II d | WRKYGQK | CX5CX23HXH |
| ZjWRKY50 | Chr10 | 320 | 8.65 | 35 | 4 | II a | WRKYGQK | CX5CX23HXH |
| ZjWRKY51 | Chr11 | 227 | 9.11 | 25 | 3 | II c | WRKYGQK | CX4CX23HXH |
| ZjWRKY52 | Chr11 | 374 | 5.16 | 41 | 3 | II c | WRKYGQK | CX4CX23HXH |
| ZjWRKY53 | Chr11 | 224 | 5.37 | 25 | 3 | II e | WRKYGQK | CX5CX23NXH |
| ZjWRKY54 | Chr11 | 356 | 5.92 | 39 | 3 | II e | WRKYGQK | CX5CX23HXH |
| ZjWRKY55 | Chr11 | 478 | 9.09 | 52 | 5 | I | WRKYGQK WRKYGQK | CX4CX22HXH CX4CX23HXH |
| ZjWRKY56 | Chr12 | 243 | 8.66 | 28 | 3 | II c | WRKYGQK | CX4CX23HXH |
| ZjWRKY57 | Chr12 | 196 | 6.73 | 22 | 3 | II c | WRKYGKK | CX4CX23HXH |
| ZjWRKY58 | Chr12 | 355 | 5.23 | 41 | 3 | III | WRKYGQK | CX7CX23HXC |
| ZjWRKY59 | Chr12 | 329 | 9.53 | 36 | 3 | II d | WRKYGQK | CX5CX23HXH |
| ZjWRKY60 | Conting12.1 | 159 | 5.46 | 18 | 3 | II c | WRKYGKK | CX4CX23HXH |
| ZjWRKY61 | Conting111 | 237 | 7.65 | 26 | 4 | II c | WRKYGQK | CX4CX23HXH |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, X.; Chen, R.; Wang, Y.; Wu, C.; Huang, J. Genome-Wide Identification of WRKY Transcription Factors in Chinese jujube (Ziziphus jujuba Mill.) and Their Involvement in Fruit Developing, Ripening, and Abiotic Stress. Genes 2019, 10, 360. https://doi.org/10.3390/genes10050360
Chen X, Chen R, Wang Y, Wu C, Huang J. Genome-Wide Identification of WRKY Transcription Factors in Chinese jujube (Ziziphus jujuba Mill.) and Their Involvement in Fruit Developing, Ripening, and Abiotic Stress. Genes. 2019; 10(5):360. https://doi.org/10.3390/genes10050360
Chicago/Turabian StyleChen, Xin, Ruihong Chen, Yanfeng Wang, Cuiyun Wu, and Jian Huang. 2019. "Genome-Wide Identification of WRKY Transcription Factors in Chinese jujube (Ziziphus jujuba Mill.) and Their Involvement in Fruit Developing, Ripening, and Abiotic Stress" Genes 10, no. 5: 360. https://doi.org/10.3390/genes10050360
APA StyleChen, X., Chen, R., Wang, Y., Wu, C., & Huang, J. (2019). Genome-Wide Identification of WRKY Transcription Factors in Chinese jujube (Ziziphus jujuba Mill.) and Their Involvement in Fruit Developing, Ripening, and Abiotic Stress. Genes, 10(5), 360. https://doi.org/10.3390/genes10050360




