Abstract
Treatment decisions for breast cancer are based on staging and hormone receptor expression and include chemotherapies and endocrine therapy. While effective in many cases, some breast cancers are resistant to therapy, metastasize and recur, leading to eventual death. Higher percentages of tumor-initiating cancer stem cells (CSCs) may contribute to the increased aggressiveness, chemoresistance, and worse outcomes among breast cancer. This may be particularly true in triple-negative breast cancers (TNBCs) which have higher percentages of CSCs and are associated with worse outcomes. In recent years, increasing numbers of long non-coding RNAs (lncRNAs) have been identified as playing an important role in breast cancer progression and some of these have been specifically associated within the CSC populations of breast cancers. LncRNAs are non-protein-coding transcripts greater than 200 nucleotides which can have critical functions in gene expression regulation. The preclinical evidence regarding lncRNA antagonists for the treatment of cancer is promising and therefore, presents a potential novel approach for treating breast cancer and targeting therapy-resistant CSCs within these tumors. Herein, we summarize the lncRNAs that have been identified as functionally relevant in breast CSCs. Furthermore, our review of the literature and analysis of patient datasets has revealed that many of these breast CSC-associated lncRNAs are also enriched in TNBC. Together, this suggests that these lncRNAs may be playing a particularly important role in TNBC. Thus, certain breast cancer-promoting/CSC-associated lncRNAs could be targeted in the treatment of TNBCs and the CSCs within these tumors should be susceptible to anti-lncRNA therapy.
1. TNBCs and CSCs – in Need of New Targets
Breast cancer is a heterogeneous disease, categorized clinically based on expression of hormone receptors, estrogen receptor (ER), progesterone receptor (PR), or human epidermal growth factor 2 receptor (HER2) [1]. The presence of hormone receptors in breast tumors allows for treatment with hormone receptor antagonists in addition to chemotherapies [2]. Breast tumors lacking expression of these receptors, termed triple-negative breast cancers (TNBCs), are treated primarily with chemotherapies and are associated with worse outcomes, at least in part due to therapy resistance [3]. Transcriptome analyses have sub-classified breast cancers into at least four major subtypes (luminal A, luminal B, HER2, and basal-like) [4]. The basal-like subtype is comparatively aggressive and is enriched with TNBCs, with at least 75% of basal-like breast cancers being triple-negative [5].
The aggressiveness of TNBC/basal-like breast cancers has been attributed to the enrichment of cancer stem cell (CSC) populations within these subtypes [6,7,8,9,10,11,12,13,14]. Within tumors, CSCs are the most tumorigenic cells, initiating new tumors with high efficiency [15]. Most concerning in terms of mitigating the risk of metastasis and recurrence in the treatment of TNBC/basal-like breast cancers is the resistance of CSCs to chemotherapies [16].
Breast CSCs are typically identified by the expression of cell surface markers (CD44highCD24low or CD133high), or high aldehyde dehydrogenase (ALDHhigh) activity quantified by the Aldefluor assay [15,17,18]. In addition, pluripotent stem cell markers SRY box 2 (SOX2), octamer-binding transcription factor 4 (OCT4), and NANOG are often overexpressed in the CSCs of different cancers [19]. In further connection with TNBC, stemness factors such as SOX2 and Myc are enriched in TNBC [20,21]. In fact, the gene expression signatures of TNBCs exhibited striking similarities to those of mammary stem cells [22]. The developmental Wnt/β-catenin, Notch, and Hedgehog (Hh) pathways are also more active in both CSCs and TNBC [23,24,25].
A further shared characteristic of TNBCs and breast CSCs is epithelial-mesenchymal transition (EMT), a phenotypic change in cells and process that contributes to invasion and distant metastasis [26]. EMT-inducing transcription factors are enriched in TNBCs and mesenchymal proteins are upregulated, while epithelial proteins are down-regulated [22]. EMT confers tumor cells with CSC-like characteristics [27,28].
Novel targeted therapies based on the unique molecular characteristics and mutations associated with TNBCs are being tested, including inhibitors for poly-ADP ribose polymerase (PARP) [29], phosphoinositide 3-kinase (PI3K) [30], mitogen-activated protein kinase (MEK) [31,32,33,34], heat shock protein-90 (Hsp90) [35], histone deacetylases [36], and androgens [37]. Moreover, some evidence suggests immunotherapies may work on TNBCs [38].
Given the high abundance of CSCs within TNBC/basal-like breast cancer, novel therapies that also target CSCs may better reduce the risk of relapse and improve the outcomes of TNBC patients. Components of dysregulated CSC-enriched molecular pathways (e.g., Notch, Wnt, and Hh) may exemplify novel druggable targets for signaling antagonists [16,39,40]. Additionally, increasing evidence suggests that it may be possible to target CSCs via CSC-specific or associated non-coding RNAs [41,42,43,44].
2. A Role for lncRNAs in Breast Cancer
Genomic and transcriptomic analyses have demonstrated that as much as 85% of the human genome is transcribed [45]. Hence, most of the genome encodes non-coding RNA transcripts, which necessitate an expanded appreciation for their diverse biological functions [46]. Long non-coding RNAs (lncRNAs) are independently transcribed RNA species greater than 200 nucleotides that lack open reading frames (ORFs) [47]. Functionally characterized lncRNAs have been shown to act as transcriptional enhancers, transcription factor decoys, transcriptional guides, scaffolds for molecular interactions, and competitive endogenous RNAs (ceRNAs) that sponge miRNAs, proteins, and other molecules [47].
Many lncRNAs promote cancer development, metastasis, and drug resistance, and exhibit elevated tissue-specific expression in many human cancers, supporting their potential clinical relevance [48]. Expression profiles of numerous lncRNAs are associated with poor prognosis, survival, and disease recurrence in breast cancer [49,50] and may provide insight into disease management strategies. Importantly, tissue-specific lncRNA expression in breast cancer may serve as a clinically useful biomarker to differentiate between normal and tumor tissue or breast cancer subtype [51,52,53,54]. Expression of lncRNAs in breast cancer may also serve as markers, which can contribute to earlier breast cancer diagnosis and enhanced efficacy in disease treatment [52,54,55,56,57,58,59]. Targeting lncRNAs may also antagonize cancer progression, improving clinical outlook. Pre-clinical evidence has supported the potential use of lncRNA antagonists such as GapmeRs (modified antisense oligonucleotides) in breast cancer treatment [60]. Hence, there are plausible therapeutic strategies that could target breast CSC-associated lncRNAs if these lncRNAs are indeed shown to be critical for promoting CSC properties and disease progression.
3. Breast CSC-Associated lncRNAs
A literature review identified 18 lncRNAs reported to be associated with breast CSC populations and/or promoting features associated with CSCs. Some of these lncRNAs are well-studied (e.g., HOTAIR and H19), while others are more recently characterized (e.g., NRAD1). We describe the associations of these lncRNAs with breast CSC populations and summarize the findings in Table 1 and Figure 1.

Table 1.
Summary of long non-coding RNA (lncRNA) functions in breast cancer stem cells (CSCs).
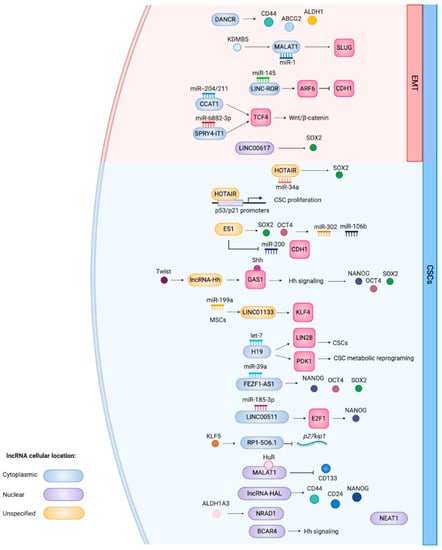
Figure 1.
Schematic overview of the CSC and EMT-related functions of lncRNAs that have been associated with breast CSCs. This is a representation of functions summarized in Table 1. LncRNAs implicated with EMT genes and the processes are located in the red area of the cell. If known, the cellular localization of the lncRNA is indicated by the color coding. The figure was created with Biorender.com.
3.1. HOTAIR
LncRNA HOTAIR (HOX Antisense Intergenic RNA) is developmentally essential by regulating expression of genes in the HOX family [83] and has been functionally linked to cancer progression [84,85]. HOTAIR’s pro-oncogenic activity is mediated in part by its interaction with the polycomb repressive complex 2 (PRC2). Specifically, for breast CSCs, HOTAIR regulates proliferation, self-renewal, and the tumor-forming capacity of CSCs in TNBC MDA-MB-231 cells and ER+ MCF7 cells, where it acts as a ceRNA to sponge miR-34a, allowing the upregulation of its target stemness gene, SOX2 [61]. Furthermore, by binding the promoters of tumor suppressors p53 and p21, HOTAIR affects the proliferation of the CSC populations within the breast cancer cell lines [61].
3.2. H19
Another developmentally important lncRNA, H19, promotes cancer progression in several cancers including breast [52,86,87,88] and is enriched in the ALDHhigh breast CSC populations of TNBCs [63]. H19 sponges miRNA tumor suppressor, let-7, facilitating a concomitant increase in the breast CSC-enriched pluripotency factor LIN28. The sponging of let-7 by H19 also leads to the increased expression of the glycolytic enzyme pyruvate dehydrogenase kinase 1 (PDK1), which promotes the metabolic reprogramming of breast CSCs [62].
3.3. NEAT1
The lncRNA nuclear paraspeckle assembly transcript 1 (NEAT1) has been identified in several human cancers and has been intimately linked to breast cancer progression by stimulating cell proliferation, EMT, and metastasis [89,90]. NEAT1 is reportedly elevated in TNBC and exerts an oncogenic function (regulation of apoptosis and cell cycle progression) in the subtype by promoting tumor growth, chemoresistance and the maintenance of breast CSCs [64]. NEAT1 knockdown in TNBC cells reduced CD44highCD24low, ALDHhigh, and SOX2high CSC populations.
3.4. MALAT1
Metastasis associated lung adenocarcinoma transcript 1 (MALAT1) was first shown to contribute to metastasis and poor patient survival in non-small cell lung cancer (NSCLC) [91] and later in the progression of TNBC [65]. MALAT1 promotes TNBC progression and aggressiveness through its regulation by the oncogenic histone demethylase KDM5B (Lysine-specific demethylase 5B), which plays a critical role in the formation and maintenance of breast CSCs [66]. Conversely, MALAT1 may act as a suppressor of breast cancer metastasis. MALAT1 and RNA-binding protein HuR form a repressive complex that regulates expression of CSC marker CD133 [67]. This complex was absent from metastatic TNBC tumor cells and present in non-metastatic cells. The absence of the MALAT1/HuR complex promotes EMT in a CD133-dependent manner.
3.5. BCAR4
LncRNA breast cancer anti-estrogen resistance 4 (BCAR4) was first identified as playing an oncogenic role in breast cancer [92]. Xing and colleagues [68] illustrated the potential role of BCAR4 in breast CSCs through its role in Hh signaling.
3.6. DANCR
Differentiation antagonizing non-protein coding RNA (DANCR) knockdown resulted in the downregulation of stemness factors CD44, ABCG2, and ALDH1 in MDA-MB-231 cells [69]. DANCR expression was correlated with TNM stages, histological grade and lymph node metastasis, and decreased survival in TNBC patients. DANCR knockdown in MDA-MB-231 and MDA-MB-468 cells reduced EMT, stemness, and inflammation [93]. Furthermore, in the normal breast epithelial cell line MCF10A, overexpression of DANCR led to acquisition of EMT, cancer stemness and inflammation properties in the normal cells [93].
3.7. NRAD1/LINC00284
Non-coding RNA in the aldehyde dehydrogenase 1A pathway (NRAD1), formerly referred to as LINC00284, is enriched in the ALDHhigh CSC populations of TNBCs [70]. Its expression is regulated by CSC marker ALDH1A3. The tumorigenic NRAD1 increased mammosphere formation and contributes to gene expression regulation by ALDH1A3 through chromatin interactions.
3.8. LINC-ROR
Long intergenic non-coding RNA, Regulator of Reprogramming (LINC-ROR), contributes to re-programming of differentiated cells and maintenance of embryonic stem cells [94]. LINC-ROR regulates the expression of stemness factors SOX2, OCT4, and NANOG (targets of miR-145), by sponging miR-145 [95]. LINC-ROR induced EMT and invasion/migration in MCF10A cells and LINC-ROR knockdown reduced the EMT phenotype in TNBC cells [71]. Importantly, this LINC-ROR-induced EMT program produced stem-like breast cells, evidenced by the increase of the breast CSC phenotype (CD44highCD24low) in LINC-ROR-expressing MCF10A cells. Moreover, LINC-ROR-expressing MCF10A cells demonstrated enhanced mammosphere forming ability. Together, these results suggest that LINC-ROR-induced EMT promotes the formation of breast CSCs [71].
3.9. LINC01133
LINC01133 has been associated with the progression and suppression of multiple cancers, including breast cancer [72,96,97,98]. LINC01133 is reportedly highly enriched in TNBC relative to HER2-enriched and luminal cell lines and it is associated with poor patient survival in TNBC [72]. Mesenchymal stem cells induce expression of LINC01133 in TNBC cells, which are associated with the generation of breast CSC-like cells and modulation of the miR-199a-FOXP2 pathway [72]. LINC01133 also regulates the pluripotency factor, Kruppel-Like Factor 4 (KLF4), which promotes stemness in breast cancer [99].
3.10. LINC00617
LINC00617 is the human ortholog of the evolutionary conserved lncRNA TUNA, which was shown to regulate pluripotency and neural differentiation of mouse embryonic stem cells [100]. LINC00617 expression is significantly higher in breast cancer tissues relative to non-cancerous tissues, promotes the invasion/migration of MCF7 breast cancer cells and induces EMT in the TNBC MDA-MB-468 cells [73]. The LINC00617-induced EMT program stimulated an increase in the CD44highCD24low cells, SOX2 expression, mammosphere formation potential, and metastasis. The LINC00617-mediated oncogenic activity was induced through its regulation of SOX2.
3.11. CCAT1
Colon cancer-associated transcript-1 (CCAT1) was first studied in colon cancer and subsequently in other cancers, including breast cancer [75,101]. In spheroid culture, breast cancer cells had a higher expression of CCAT1 [74]. CCAT1 increased expression of the stemness markers NANOG, SOX2, OCT4 and ALDH1A1, and promoted proliferation, invasion, and migration in the breast cancer cells [74]. CCAT1 is a cytoplasmic lncRNA and mediates its effects by sponging miR-204/211. Downstream targets of miR-204/211 include TCF4 (a key transcription factor in the Wnt/β-catenin signaling pathway).
3.12. SPRY4-IT1
SPRY4-IT1 is an intronic lncRNA transcribed from one of the introns of the SPRY4 gene, which encodes a tumor suppressor [102]. SPRY4-IT1 is upregulated in CD44highCD24low ER+ MCF-7 cells and expression of stemness factors OCT4, c-Myc, NANOG and SOX2 are positively correlated with SPRY4-IT1 expression levels [76]. Further analysis revealed that SPRY4-IT1 mediates its stemness promoting effects by acting as a ceRNA to sequester miR-6882-3p [76], which targets TCF7L2/TCF4.Thus, SPRY4-IT1 promotes stemness by promoting the Wnt/β-catenin signaling pathway.
3.13. LncRNA-Hh
Thus far, Twist-induced lncRNA-Hh has been characterized only in breast cancer where it directly targets growth arrest-specific 1 (GAS1) to initiate Hh signaling in breast cancer, promoting SOX2 and OCT4 expression, EMT, tumorigenesis, and cells with CSC properties [77].
3.14. RP1-5O6.5
LncRNA RP1-5O6.5 is regulated by the oncogenic protein KLF5 and induced breast cancer growth and metastasis by inhibiting translation of cell cycle inhibitor p27kip1 [78]. Through its regulation of p27kip1, RP1-5O6.5 also increases stemness in breast cancer cells [78]. The proportion of CD44highCD24low cells in TNBC cell lines with RP1-506.5 knockdown was significantly diminished.
3.15. LINC00511
LINC00511 has been implicated in breast cancer tumorigenesis and stemness through modulation of the miR-185-3p/E2F1/NANOG axis [79]. LINC00511 was also among a shortlist of lncRNAs identified as enriched in ALDHhigh CSC populations of TNBCs [70]. Cytoplasmic LINC00511 functions as a ceRNA, sequestering miR-185-3p from gaining access to its target transcription factor E2F1. Increased E2F1 upregulates NANOG, contributing to stemness in breast cancer [79]. Silencing LINC00511 reduced mammosphere formation TNBCs [79].
3.16. FEZF1-AS1
Known to be highly expressed in various cancers, FEZF1-AS1 is involved in cell proliferation, migration, invasion and the Warburg Effect [103]. FEZF1-AS1 increases the expression of stem cell markers NANOG, OCT4 and SOX2 in MDA-MD-231 TNBC cells by acting as ceRNA sponging miR-39a [80].
3.17. LncRNA ES1/LINC01108
The role of lncRNA ES1 (also called LINC01108 or ENST01108) has been studied in human embryonic stem cells [104]. Knockdown of the lncRNA in TNBC MDA-MD-231 and HER2+ SKBR3 cell lines reduced stemness factors SOX2 and OCT4 and their targets, miR-302 and miR-106b, were downregulated [81]. Further, the upregulation of lncRNA ES1 increased expression of Snail/Zeb1 and miR-106b and suppressed the expression of E-cadherin and miR-200, thereby implicating ES1 as a driver of EMT.
3.18. LncRNA-HAL
LncRNA-HAL is located in the intronic region in the MNT gene and its expression is correlated with expression of stemness factors such as CD44, CD24 and NANOG in breast cancer [82]. lncRNA-HAL interacts with histone proteins H2B and H1.2, suggesting that lncRNA-HAL may mediate its effects by binding and altering chromatin, leading to altered gene expression [82].
4. Breast CSC-Associated lncRNA Correlations with CSC Markers and Signaling Pathways in TNBC and All Breast Cancer Patient Tumors
Most of the above described associations between lncRNAs and breast CSC populations were made in cell lines. To assess the clinical relevance of the associations in breast cancer, we analyzed breast cancer patient tumor gene expression correlations using RNA seq data provided in The Cancer Genome Atlas (TCGA) in the Cell 2015 dataset via cBioPortal [105,106]. We extracted Spearman’s correlation values between the individual lncRNAs and the various CSC markers and the key players in CSC-associated signaling pathways (Table 2). Notably, there were extractable expression data for only seven of the 18 lncRNAs described above (HOTAIR, H19, NEAT1, MALAT1, BCAR4, DANCR, NRAD1/LINC00284), which unfortunately prevented us from assessing the entire panel of lncRNAs. We evaluated the correlations and the direction and strength in the CSC-enriched subtype (i.e., TNBC) compared to all breast cancer subtypes. Our analyses revealed relationships between the seven lncRNAs and some of the CSC/stemness genes. Often, the expression of different lncRNAs was correlated with different genes. Additionally, the strength of significant correlations between lncRNAs and the stemness-related genes was often amplified in TNBC patient tumor data compared to the complete dataset containing the unfiltered data for all breast cancer subtypes. This result is consistent with the hypothesized importance of CSC-enriched lncRNAs in TNBC and their role in maintaining and enhancing a stemness phenotype in this breast cancer subtype.

Table 2.
LncRNA co-expression with CSC markers, stemness factors, EMT genes, and players in CSC-associated signaling pathways.
Some interesting correlations include the strong correlation between CSC marker ALDH1A1 and MALAT1 (Table 2). This may be due to the regulation of MALAT1 by ALDH1A1, as MALAT1 was reported to be regulated by ALDH1A1 in lung cancer [107]. Similarly, NRAD1(LINC00284) was highly correlated with ALDH1A3, and we previously reported that the lncRNA is regulated by the CSC marker [70]. Furthermore, we noted a strong relationship between NRAD1 and CD133, and NRAD1 and CD24 expression (which is counterintuitive as CD24 is downregulated in CSC populations defined by CD44highCD24low, Table 2). Notably, CD24 overexpression is involved with TNBC metastasis, advanced tumor staging and decreased survival [108]. Additionally, CD133 and CD24 overexpression have been reported to promote EMT [67,108], and while our analysis did not reveal any correlation between NRAD1 (LINC00284) and EMT transcription factor expression, this finding may suggest an unreported putative function of NRAD1 in EMT.
CD49f is a marker associated with chemoresistant and tumor-initiating cell populations in TNBC [109]. We identified that NEAT1 expression is correlated with CD49f (Table 2). While the relationship between NEAT1 and CD49f has not been studied previously, this correlation may provide insight into the mechanism underlying the chemoresistance found in NEAT1-enriched tumors [64].
In patient data filtered by TNBC subtype, we observed that DANCR is positively correlated with the expression of oncogenic transcription factor c-Myc. DANCR is regulated by c-Myc in lymphoma [110], but to our knowledge, this has not been shown in breast cancer. This would be interesting to investigate given the strong correlation in patient tumors.
We also noted that H19 is highly correlated with EMT-mediating transcription factors Slug and Twist. This observation supports the role of H19 as an EMT mediator in breast cancer [111] and suggests that this function is also relevant in the TNBC subtype. The strong co-expression between MALAT1 and EMT-mediating transcription factor zinc finger E-box-binding homeobox 1 (Zeb1) further supports the clinical relevance of MALAT1 as an EMT driver through Zeb1-mediated functions. Notably, MALAT1 regulates Zeb1 expression through miRNA sponging in hepatocellular carcinoma [112]. While DANCR was found to play a role in EMT [93], our analysis of the breast cancer patient tumor data does not reflect that finding. In fact, Zeb1 and Zeb2 are negatively correlated with DANCR expression.
LncRNAs HOTAIR, H19, and MALAT1 are involved in Wnt signaling [113,114,115]. Our analysis of the breast cancer clinical data is consistent with this, as all three lncRNAs were positively correlated with Wnt signaling transcription factor TCF4.
5. Are Breast CSC-Associated lncRNAs Enriched in TNBCs/Basal-Like Breast Cancers?
Numerous lncRNAs are reportedly upregulated in TNBCs and contribute to their aggressiveness [49,116,117,118,119,120]. Zhang et al. performed a detailed analysis of patient tumor RNA-seq data (TCGA) and revealed more than 50 lncRNAs that are upregulated in TNBCs/basal-like breast cancers; most of which are uncharacterized and represent potentially untapped targets in TNBC [121]. Of these, approximately a fourth were later identified to be enriched in the ALDHhigh CSC populations of TNBCs (e.g., LINC00511, NRAD1/LINC00284) [70]. Conversely, we noted that of the 18 lncRNAs thus far described to be associated with breast CSCs, 11 have been reported to be enriched in TNBCs (Table 1). This proportion is higher than expected, suggesting that breast CSC-associated lncRNAs tend to be enriched in TNBC, and this is possibly due to the enrichment of CSCs within this subtype [6,7,8,9,10,11,12,13,14].
We performed our own subtype enrichment analysis of the seven breast CSC-associated lncRNAs for which we could extract expression data from the 2015 Cell breast cancer dataset accessible at TCGA via cbioportal (Table 3). This revealed that two of the seven CSC-lncRNAs, NRAD1/LINC00284 and DANCR, are significantly enriched in TNBC/basal-like breast cancers. The striking enrichment of NRAD1 in basal-like breast cancer and TNBC suggests that NRAD1 may serve as an attractive diagnostic or prognostic target. In agreement with previous reports [122,123], our analysis also showed that NEAT1 and MALAT1 were enriched in ER+ breast cancers. This conflicts with other studies that have shown that these lncRNAs are TNBC-enriched [64,67].

Table 3.
LncRNA and protein target fold-enrichment by breast cancer subtype.
For a comparison with potential protein-coding targets, we extended our analysis to include oncogene c-Myc, the hormone receptors, CSC markers, and key CSC-associated signaling pathway players (Table 3). As expected, the hormone receptors are enriched in the corresponding subtypes, and some of the CSC markers and c-Myc are enriched in TNBC/basal-like breast cancers. This data supports further study into the potential clinical relevance of at least a couple of these breast CSC-associated lncRNAs (i.e., NRAD/LINC00284 and DANCR) alongside potential protein targets, in the treatment of TNBC and basal-like breast cancers.
6. Clinical Value of Breast CSC-Associated lncRNAs in TNBC
Major barriers to effective remediation of TNBCs include their lack of therapeutic targets and their enrichment in tumor-initiating CSCs which confer therapeutic resistance and disease recurrence. Thus, including adjuvant therapies that target CSCs in TNBC may result in significant improvements in patient outcomes. Below, we discuss the evidence for targeting breast CSC-associated lncRNAs in the treatment of TNBC.
6.1. Therapeutic Targets
The investigation of lncRNAs as novel therapeutic targets provokes thoughtful questions regarding their effectiveness over protein targets. Besides the enrichment of TNBC/basal-like breast cancers (Table 3), another indicator of potential clinical value is correlation with patient outcomes. We therefore utilized KMPlotter and compared the hazard ratios of breast cancer patients with tumors with high expression of the breast CSC-associated lncRNAs or protein targets versus those with tumors that had low expression (Table 4). For the lncRNAs, we could only compare the six lncRNAs for which there was expression data. Among the lncRNAs, NRAD1 and DANCR were significantly associated with higher hazard ratios in basal-like breast cancers but not TNBC patients. This could be due to the smaller number of patients within the TNBC cohort (i.e., 40 patients). In comparison to some protein targets, we noted that ALDH1A3 was significantly associated with worse outcomes in basal-like breast cancers, as has been reported previously [124]. Notably, DANCR and NRAD1 were associated with higher hazard ratios than other potential protein targets assessed (e.g., CSC markers, c-Myc). Together, these analyses further flag NRAD1 and DANCR as candidates for further investigation as therapeutic targets.

Table 4.
LncRNA hazard ratio by breast cancer subtype.
Several pre-clinical methods have been described for targeting ncRNAs including antisense oligonucleotides (ASOs), CRISPR-Cas9 and RNAi-based methods. ASOs are DNA oligos that suppress RNA molecules by binding to them in an antisense fashion, generating a DNA-RNA duplex that can be recognized and degraded by RNase H, thereby suppressing or modifying protein expression [60,126]. ASOs have demonstrated success in both animal models and human clinical trials, and strikingly, ASOs were recently FDA-approved for clinical use for neurodegenerative diseases [60]. Synthetic and highly stable ASOs called locked nucleic acid (LNA) GapmeRs have also demonstrated some success inhibiting lncRNAs in vivo [127,128,129].
Targeting ncRNAs with CRISPR/Cas9 has been reported in cancer [130]. Specifically, lncRNA UCA1 (urothelial carcinoma-associated 1), upregulated in bladder cancer, could be inhibited by transcript-specific CRISPR/Cas9-associated gRNAs (guide RNAs) in vitro and in vivo, demonstrating its potential therapeutic value [131]. These findings suggest that there are strategies in development that will enable the clinical targeting of functionally characterized oncogenic lncRNAs that are specific to certain breast cancer subtypes (e.g., TNBC/basal-like breast cancer) and the CSCs within these tumors.
Vaidya et al. describe an effective TNBC therapy that uses nanoparticle-mediated transfer of RNAi molecules targeting the TNBC/CSC enriched lncRNA DANCR [132]. This therapy led to reduced tumor progression and was associated with no side effects in a murine xenograft model of TNBC. These results prompt the further investigation of nanoparticle-mediated targeting of oncogenic lncRNAs as a precision approach to TNBC therapy.
6.2. Challenges
While the lncRNA silencing methods are promising therapeutic strategies, a thorough assessment of off-target effects, potential toxicity, and drug delivery/precision targeting is required. Additionally, the multiple transcript variants of lncRNAs is more of a potential confounding factor than when considering strategies for targeting proteins. Depending on the specifics of the lncRNA silencing strategy, not all transcript variants will be targeted, possibly reducing the efficacy of the treatment if that variant is functional. In addition, the functions of most lncRNAs remain uncharacterized, further complicating the design of effective treatments and delivery strategies. Finally, whether lncRNAs represent more suitable targets over protein-coding genes is unknown and would require comparative analyses in clinical trials.
6.3. Biomarkers
Several studies have supported the use of lncRNAs as clinically relevant biomarkers in breast cancer. The expression of these lncRNAs may serve as valuable prognostic or diagnostic indicators in addition to markers of chemoresistance or response to treatment. Thus, relevant lncRNA expression may inform patient stratification efforts to identify those individuals who will respond best to a given treatment. The characterization of subtype-specific lncRNAs is particularly important in the context of breast cancer, where the analysis of expression levels may signify a specific subtype and aid treatment decisions.
For example, Jiang and colleagues developed an mRNA-lncRNA signature that was capable of TNBC patient classification into groups with low or high risks of disease recurrence [133]. In addition, the signature could predict response to taxane-based chemotherapy in TNBC patients. Moreover, Lv and colleagues identified a four-lncRNA signature (RP11-434D9.1, LINC00052, BC016831, and IGKV) to differentiate TNBC from non-TNBC [54]. Some other studies also identified lncRNA signatures indicative of survival [57], response to chemotherapy [59], and risk of recurrence [134].
Cancer detection through liquid biopsy via analysis of the molecules in the plasma, serum, and urine would allow for real-time, non-invasive detection of disease and promote early management strategies. Thus far, the majority of ncRNAs implicated as cancer biomarkers in liquid biopsies are miRNAs. Indeed, circulating exosomal miRNAs have been implicated as cancer biomarkers in TNBC, particularly in the blood [135].
Several lncRNAs have been detected via liquid biopsy and may serve as prognostic or diagnostic biomarkers in numerous malignancies. Of the lncRNAs discussed here, MALAT1 (in prostate cancer and multiple myeloma), HOTAIR (in NSCLC and multiple myeloma), and SPRY4-IT1 (in esophageal squamous-cell carcinoma) have been implicated as relevant biomarkers in liquid biopsies [136]. LncRNAs as liquid biopsy biomarkers have yet to be discovered for breast cancer, although this will likely come in the future.
7. Conclusions
Here, we have reviewed a series of oncogenic breast CSC-associated lncRNAs involved in breast cancer progression. We have also provided a review of the literature and analysis of patient data to determine the correlations of these lncRNAs with CSC markers and key signaling pathway players, subtype and patient survival. Intriguingly, our analyses unveiled correlations that have not been previously identified in the literature and may provide a starting point for further investigation of the molecular mechanisms underlying clinically pertinent links between lncRNAs and stemness-related gene expression.
LncRNAs greatly outnumber protein-coding genes in the human genome. Recently, a far-reaching analysis of lncRNAs in cancer uncovered 7941 lncRNAs that were cancer and/or lineage specific [137], further supporting the potential relevance of lncRNAs in cancer management. Due to their tissue-specific expression and upregulated expression in cancers, lncRNAs represent attractive targets and biomarkers for breast cancer diagnosis/prognosis, response to therapy, disease recurrence, and differentiation between tumor and non-tumor tissue. In addition, subtype-specific lncRNAs can help inform diagnoses and subsequent treatment routes, aiding in patient stratification efforts.
LncRNAs are involved in several aspects of breast CSCs; thus, the targeting of CSC-associated lncRNAs may represent a novel Achilles heel in the treatment of CSC-enriched TNBCs. Clinical trials for TNBCare focused on protein targets, including immune checkpoint inhibitors. Non-coding RNAs are under investigation but this is limited to miRNA. As we have demonstrated in this manuscript, numerous lncRNAs exert their regulatory oncogenic functions through interactions with miRNAs, and therefore, lncRNAs may represent equally valuable clinical targets. The tissue-specificity and enrichment of some oncogenic lncRNAs in breast CSCs and TNBCs (e.g., DANCR and NRAD1) make potential clinical investigation a possibility. A thorough analysis of the function of these lncRNAs, and characterization of the transcript variants and the potential off-target effects of the targeting strategy are first required before clinical trials are even considered.
Funding
This research was funded by the Canadian Institutes of Health Research (CIHR, PJT 162313) and the APC was funded by the Dalhousie Medical Research Foundation, Breakthrough Accelerator Award (no specific grant number). JMB is also partly supported by a Cancer Research Training Program (CRTP) scholarship from the Beatrice Hunter Cancer Research Institute (BHCRI) and the Terry Fox Research Institute. MCW is also partly supported by a Genomics in Medicine scholarship from the Dalhousie Medical Research Foundation and CRTP scholarship funded through the BHCRI.
Acknowledgments
The results presented in Table 2 and Table 3 were derived in part from data generated by The Cancer Genome Atlas (TCGA) research network. https://www.cancer.gov/tcga.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Sørlie, T.; Perou, C.M.; Tibshirani, R.; Aas, T.; Geisler, S.; Johnsen, H.; Hastie, T.; Eisen, M.B.; Van De Rijn, M.; Jeffrey, S.S.; et al. Gene Expression Patterns of Breast Carcinomas Distinguish Tumor Subclasses with Clinical Implications. Proc. Natl. Acad. Sci. USA 2001, 98, 10869–10874. [Google Scholar] [CrossRef] [PubMed]
- Carey, L.A.; Perou, C.M.; Livasy, C.A.; Dressler, L.G.; Cowan, D.; Conway, K.; Karaca, G.; Troester, M.A.; Chiu, K.T.; Edmiston, S.; et al. Race, Breast Cancer Subtypes, and Survival in the Carolina Breast Cancer Study. J. Am. Med. Assoc. 2006, 295, 2492–2502. [Google Scholar] [CrossRef] [PubMed]
- Carey, L.A.; Dees, E.C.; Sawyer, L.; Gatti, L.; Moore, D.T.; Collichio, F.; Ollila, D.W.; Sartor, C.I.; Graham, M.L.; Perou, C.M. The Triple Negative Paradox: Primary Tumor Chemosensitivity of Breast Cancer Subtypes. Clin. Cancer Res. 2007, 13, 2329–2334. [Google Scholar] [CrossRef] [PubMed]
- Perou, C.M.; Sørile, T.; Eisen, M.B.; Van De Rijn, M.; Jeffrey, S.S.; Ress, C.A.; Pollack, J.R.; Ross, D.T.; Johnsen, H.; Akslen, L.A.; et al. Molecular Portraits of Human Breast Tumours. Nature 2000, 406, 747–752. [Google Scholar] [CrossRef]
- Bertucci, F.; Finetti, P.; Cervera, N.; Esterni, B.; Hermitte, F.; Viens, P.; Birnbaum, D. How Basal Are Triple-Negative Breast Cancers? Int. J. Cancer 2008, 123, 236–240. [Google Scholar] [CrossRef]
- Li, H.; Ma, F.; Wang, H.; Lin, C.; Fan, Y.; Zhang, X.; Qian, H.; Xu, B. Stem Cell Marker Aldehyde Dehydrogenase 1 (ALDH1)-Expressing Cells Are Enriched in Triple-Negative Breast Cancer. Int. J. Biol. Markers 2013, 28. [Google Scholar] [CrossRef]
- Ma, F.; Li, H.; Li, Y.; Ding, X.; Wang, H.; Fan, Y.; Lin, C.; Qian, H.; Xu, B. Aldehyde Dehydrogenase 1 (ALDH1) Expression Is an Independent Prognostic Factor in Triple Negative Breast Cancer (TNBC). Medicine (U.S.) 2017, 96. [Google Scholar] [CrossRef]
- Wang, H.; Wang, L.; Song, Y.; Wang, S.; Huang, X.; Xuan, Q.; Kang, X.; Zhang, Q. CD44+/CD24- Phenotype Predicts a Poor Prognosis in Triple-Negative Breast Cancer. Oncol. Lett. 2017, 14, 5890–5898. [Google Scholar] [CrossRef]
- Honeth, G.; Bendahl, P.O.; Ringnér, M.; Saal, L.H.; Gruvberger-Saal, S.K.; Lövgren, K.; Grabau, D.; Fernö, M.; Borg, Å.; Hegardt, C. The CD44+/CD24-Phenotype Is Enriched in Basal-like Breast Tumors. Breast Cancer Res. 2008, 10. [Google Scholar] [CrossRef]
- Ricardo, S.; Vieira, A.F.; Gerhard, R.; Leitão, D.; Pinto, R.; Cameselle-Teijeiro, J.F.; Milanezi, F.; Schmitt, F.; Paredes, J. Breast Cancer Stem Cell Markers CD44, CD24 and ALDH1: Expression Distribution within Intrinsic Molecular Subtype. J. Clin. Pathol. 2011, 64, 937–944. [Google Scholar] [CrossRef]
- Giatromanolaki, A.; Sivridis, E.; Fiska, A.; Koukourakis, M.I. The CD44+/CD24- Phenotype Relates to “triple-Negative” State and Unfavorable Prognosis in Breast Cancer Patients. Med. Oncol. 2011, 28, 745–752. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Sarkissyan, M.; Elshimali, Y.; Vadgama, J.V. Triple Negative Breast Tumors in African-American and Hispanic/Latina Women Are High in CD44+, Low in CD24+, and Have Loss of PTEN. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Tsang, J.Y.S.; Huang, Y.H.; Luo, M.H.; Ni, Y.B.; Chan, S.K.; Lui, P.C.W.; Yu, A.M.C.; Tan, P.H.; Tse, G.M. Cancer Stem Cell Markers Are Associated with Adverse Biomarker Profiles and Molecular Subtypes of Breast Cancer. Breast Cancer Res. Treat. 2012, 136, 407–417. [Google Scholar] [CrossRef] [PubMed]
- Idowu, M.O.; Kmieciak, M.; Dumur, C.; Burton, R.S.; Grimes, M.M.; Powers, C.N.; Manjili, M.H. CD44 +/CD24 -/Low Cancer Stem/Progenitor Cells Are More Abundant in Triple-Negative Invasive Breast Carcinoma Phenotype and Are Associated with Poor Outcome. Hum. Pathol. 2012, 43, 364–373. [Google Scholar] [CrossRef]
- Al-Hajj, M.; Wicha, M.S.; Benito-Hernandez, A.; Morrison, S.J.; Clarke, M.F. Prospective Identification of Tumorigenic Breast Cancer Cells. Proc. Natl. Acad. Sci. USA 2003, 100, 3983–3988. [Google Scholar] [CrossRef]
- Thomas, M.L.; Coyle, K.M.; Sultan, M.; Vaghar-Kashani, A.; Marcato, P. Chemoresistance in Cancer Stem Cells and Strategies to Overcome Resistance. Chemotherapy 2014, 3, 2. [Google Scholar] [CrossRef]
- Wright, M.H.; Calcagno, A.M.; Salcido, C.D.; Carlson, M.D.; Ambudkar, S.V.; Varticovski, L. Brca1 Breast Tumors Contain Distinct CD44+/CD24-and CD133+cells with Cancer Stem Cell Characteristics. Breast Cancer Res. 2008, 10. [Google Scholar] [CrossRef]
- Ginestier, C.; Hur, M.H.; Charafe-Jauffret, E.; Monville, F.; Dutcher, J.; Brown, M.; Jacquemier, J.; Viens, P.; Kleer, C.G.; Liu, S.; et al. ALDH1 Is a Marker of Normal and Malignant Human Mammary Stem Cells and a Predictor of Poor Clinical Outcome. Cell Stem Cell 2007, 1, 555–567. [Google Scholar] [CrossRef]
- Seymour, T.; Twigger, A.-J.; Kakulas, F. Pluripotency Genes and Their Functions in the Normal and Aberrant Breast and Brain. Int. J. Mol. Sci. 2015, 16, 27288–27301. [Google Scholar] [CrossRef]
- Liu, P.; Tang, H.; Song, C.; Wang, J.; Chen, B.; Huang, X.; Pei, X.; Liu, L. SOX2 Promotes Cell Proliferation and Metastasis in Triple Negative Breast Cancer. Front. Pharmacol. 2018, 9, 942. [Google Scholar] [CrossRef]
- Horiuchi, D.; Kusdra, L.; Huskey, N.E.; Chandriani, S.; Lenburg, M.E.; Gonzalez-Angulo, A.M.; Creasman, K.J.; Bazarov, A.V.; Smyth, J.W.; Davis, S.E.; et al. MYC Pathway Activation in Triple-Negative Breast Cancer Is Synthetic Lethal with CDK Inhibition. J. Exp. Med. 2012, 209, 679–696. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Choi, J.H.; Nam, J.S. Targeting Cancer Stem Cells in Triple-Negative Breast Cancer. Cancers 2019, 11, 965. [Google Scholar] [CrossRef] [PubMed]
- Koike, Y.; Ohta, Y.; Saitoh, W.; Yamashita, T.; Kanomata, N.; Moriya, T.; Kurebayashi, J. Anti-Cell Growth and Anti-Cancer Stem Cell Activities of the Non-Canonical Hedgehog Inhibitor GANT61 in Triple-Negative Breast Cancer Cells. Breast Cancer 2017, 24, 683–693. [Google Scholar] [CrossRef] [PubMed]
- Satriyo, P.; Bamodu, O.; Chen, J.-H.; Aryandono, T.; Haryana, S.; Yeh, C.-T.; Chao, T.-Y. Cadherin 11 Inhibition Downregulates β-Catenin, Deactivates the Canonical WNT Signalling Pathway and Suppresses the Cancer Stem Cell-Like Phenotype of Triple Negative Breast Cancer. J. Clin. Med. 2019, 8, 148. [Google Scholar] [CrossRef]
- Bhola, N.E.; Jansen, V.M.; Koch, J.P.; Li, H.; Formisano, L.; Williams, J.A.; Grandis, J.R.; Arteaga, C.L. Treatment of Triple-Negative Breast Cancer with TORC1/2 Inhibitors Sustains a Drug-Resistant and Notch-Dependent Cancer Stem Cell Population. Cancer Res. 2016, 76, 440–452. [Google Scholar] [CrossRef]
- Felipe Lima, J.; Nofech-Mozes, S.; Bayani, J.; Bartlett, J. EMT in Breast Carcinoma—A Review. J. Clin. Med. 2016, 5, 65. [Google Scholar] [CrossRef]
- Liu, S.; Cong, Y.; Wang, D.; Sun, Y.; Deng, L.; Liu, Y.; Martin-Trevino, R.; Shang, L.; McDermott, S.P.; Landis, M.D.; et al. Breast Cancer Stem Cells Transition between Epithelial and Mesenchymal States Reflective of Their Normal Counterparts. Stem Cell Rep. 2014, 2, 78–91. [Google Scholar] [CrossRef]
- Mani, S.A.; Guo, W.; Liao, M.J.; Eaton, E.N.; Ayyanan, A.; Zhou, A.Y.; Brooks, M.; Reinhard, F.; Zhang, C.C.; Shipitsin, M.; et al. The Epithelial-Mesenchymal Transition Generates Cells with Properties of Stem Cells. Cell 2008, 133, 704–715. [Google Scholar] [CrossRef]
- Gelmon, K.A.; Tischkowitz, M.; Mackay, H.; Swenerton, K.; Robidoux, A.; Tonkin, K.; Hirte, H.; Huntsman, D.; Clemons, M.; Gilks, B.; et al. Olaparib in Patients with Recurrent High-Grade Serous or Poorly Differentiated Ovarian Carcinoma or Triple-Negative Breast Cancer: A Phase 2, Multicentre, Open-Label, Non-Randomised Study. Lancet Oncol. 2011, 12, 852–861. [Google Scholar] [CrossRef]
- Costa, R.L.B.; Han, H.S.; Gradishar, W.J. Targeting the PI3K/AKT/MTOR Pathway in Triple-Negative Breast Cancer: A Review. Breast Cancer Res. Treat. 2018, 169, 397–406. [Google Scholar] [CrossRef]
- Duncan, J.S.; Whittle, M.C.; Nakamura, K.; Abell, A.N.; Midland, A.A.; Zawistowski, J.S.; Johnson, N.L.; Granger, D.A.; Jordan, N.V.; Darr, D.B.; et al. Dynamic Reprogramming of the Kinome in Response to Targeted MEK Inhibition in Triple-Negative Breast Cancer. Cell 2012, 149, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Loi, S.; Dushyanthen, S.; Beavis, P.A.; Salgado, R.; Denkert, C.; Savas, P.; Combs, S.; Rimm, D.L.; Giltnane, J.M.; Estrada, M.V.; et al. RAS/MAPK Activation Is Associated with Reduced Tumor-Infiltrating Lymphocytes in Triple-Negative Breast Cancer: Therapeutic Cooperation between MEK and PD-1/PD-L1 Immune Checkpoint Inhibitors. Clin. Cancer Res. 2016, 22, 1499–1509. [Google Scholar] [CrossRef] [PubMed]
- Bartholomeusz, C.; Xie, X.; Pitner, M.K.; Kondo, K.; Dadbin, A.; Lee, J.; Saso, H.; Smith, P.D.; Dalby, K.N.; Ueno, N.T. MEK Inhibitor Selumetinib (AZD6244; ARRY-142886) Prevents Lung Metastasis in a Triple-Negative Breast Cancer Xenograft Model. Mol. Cancer Ther. 2015, 14, 2773–2781. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Cui, K.; Nie, F.; Wang, L.; Brandl, M.B.; Jin, G.; Li, F.; Mao, Y.; Xue, Z.; Rodriguez, A.; et al. The Effect of MTOR Inhibition Alone or Combined with MEK Inhibitors on Brain Metastasis: An In Vivo Analysis in Triple-Negative Breast Cancer Models. Breast Cancer Res. Treat. 2012, 131, 425–436. [Google Scholar] [CrossRef] [PubMed]
- Proia, D.A.; Bates, R.C. Ganetespib and HSP90: Translating Preclinical Hypotheses into Clinical Promise. Cancer Res. 2014, 74, 1294–1300. [Google Scholar] [CrossRef] [PubMed]
- Schech, A.; Kazi, A.; Yu, S.; Shah, P.; Sabnis, G. Histone Deacetylase Inhibitor Entinostat Inhibits Tumor-Initiating Cells in Triple-Negative Breast Cancer Cells. Mol. Cancer Ther. 2015, 14, 1848–1857. [Google Scholar] [CrossRef] [PubMed]
- Caiazza, F.; Murray, A.; Madden, S.F.; Synnott, N.C.; Ryan, E.J.; O’Donovan, N.; Crown, J.; Duffy, M.J. Preclinical Evaluation of the AR Inhibitor Enzalutamide in Triple-Negative Breast Cancer Cells. Endocr. Relat. Cancer 2016, 23, 323–334. [Google Scholar] [CrossRef]
- Li, Z.; Qiu, Y.; Lu, W.; Jiang, Y.; Wang, J. Immunotherapeutic Interventions of Triple Negative Breast Cancer. J. Transl. Med. 2018, 16, 147. [Google Scholar] [CrossRef]
- Takebe, N.; Harris, P.J.; Warren, R.Q.; Ivy, S.P. Targeting Cancer Stem Cells by Inhibiting Wnt, Notch, and Hedgehog Pathways. Nat. Rev. Clin. Oncol. 2011, 8, 97–106. [Google Scholar] [CrossRef]
- Takebe, N.; Miele, L.; Harris, P.J.; Jeong, W.; Bando, H.; Kahn, M.; Yang, S.X.; Ivy, S.P. Targeting Notch, Hedgehog, and Wnt Pathways in Cancer Stem Cells: Clinical Update. Nat. Rev. Clin. Oncol. 2015, 12, 445–464. [Google Scholar] [CrossRef]
- Huang, X.; Xiao, R.; Pan, S.; Yang, X.; Yuan, W.; Tu, Z.; Xu, M.; Zhu, Y.; Yin, Q.; Wu, Y.; et al. Uncovering the Roles of Long Non-Coding RNAs in Cancer Stem Cells. J. Hematol. Oncol. 2017, 10. [Google Scholar] [CrossRef] [PubMed]
- Eades, G.; Zhang, Y.S.; Li, Q.L.; Xia, J.X.; Yao, Y.; Zhou, Q. Long Non-Coding RNAs in Stem Cells and Cancer. World J. Clin. Oncol. 2014, 5, 134–141. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhu, J.; Wang, F.; Guan, Z.; Ge, Y.; Yang, X.; Cai, J. LncRNAs and Their Role in Cancer Stem Cells. Oncotarget 2017, 8, 110685–110692. [Google Scholar] [CrossRef] [PubMed]
- Heery, R.; Finn, S.; Cuffe, S.; Gray, S. Long Non-Coding RNAs: Key Regulators of Epithelial-Mesenchymal Transition, Tumour Drug Resistance and Cancer Stem Cells. Cancers (Basel) 2017, 9, 38. [Google Scholar] [CrossRef]
- Djebali, S.; Davis, C.A.; Merkel, A.; Dobin, A.; Lassmann, T.; Mortazavi, A.; Tanzer, A.; Lagarde, J.; Lin, W.; Schlesinger, F.; et al. Landscape of Transcription in Human Cells. Nature 2012, 489, 101–108. [Google Scholar] [CrossRef]
- Fang, Y.; Fullwood, M.J. Roles, Functions, and Mechanisms of Long Non-Coding RNAs in Cancer. Genom. Proteom. Bioinform. 2016, 14, 42–54. [Google Scholar] [CrossRef]
- Rinn, J.L.; Chang, H.Y. Genome Regulation by Long Noncoding RNAs. Annu. Rev. Biochem. 2012, 81, 145–166. [Google Scholar] [CrossRef]
- Prensner, J.R.; Chinnaiyan, A.M. The Emergence of LncRNAs in Cancer Biology. Cancer Discov. 2011, 1, 391–407. [Google Scholar] [CrossRef]
- Shen, X.; Xie, B.; Ma, Z.; Yu, W.; Wang, W.; Xu, D.; Yan, X.; Chen, B.; Yu, L.; Li, J.; et al. Identification of Novel Long Non-Coding RNAs in Triple-Negative Breast Cancer. Oncotarget 2015, 6, 21730–21739. [Google Scholar] [CrossRef]
- Zhou, S.; He, Y.; Yang, S.; Hu, J.; Zhang, Q.; Chen, W.; Xu, H.; Zhang, H.; Zhong, S.; Zhao, J.; et al. The Regulatory Roles of LncRNAs in the Process of Breast Cancer Invasion and Metastasis. Biosci. Rep. 2018. [Google Scholar] [CrossRef]
- Malih, S.; Saidijam, M.; Malih, N. A Brief Review on Long Noncoding RNAs: A New Paradigm in Breast Cancer Pathogenesis, Diagnosis and Therapy. Tumor Biol. 2016, 37, 1479–1485. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Luo, Z.; Zhang, Y.; Zhang, L.; Wu, L.; Liu, L.; Yang, J.; Song, X.; Liu, J. Circulating LncRNA H19 in Plasma as a Novel Biomarker for Breast Cancer. Cancer Biomark. 2016, 17, 187–194. [Google Scholar] [CrossRef] [PubMed]
- Pecero, M.L.; Salvador-Bofill, J.; Molina-Pinelo, S. Long Non-Coding RNAs as Monitoring Tools and Therapeutic Targets in Breast Cancer. Cell. Oncol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Lv, M.; Xu, P.; Wu, Y.; Huang, L.; Li, W.; Lv, S.; Wu, X.; Zeng, X.; Shen, R.; Jia, X.; et al. LncRNAs as New Biomarkers to Differentiate Triple Negative Breast Cancer from Non-Triple Negative Breast Cancer. Oncotarget 2016, 7, 13047–13059. [Google Scholar] [CrossRef]
- Meng, J.; Li, P.; Zhang, Q.; Yang, Z.; Fu, S. A Four-Long Non-Coding RNA Signature in Predicting Breast Cancer Survival. J. Exp. Clin. Cancer Res. 2014, 33, 84. [Google Scholar] [CrossRef]
- Zhou, M.; Zhong, L.; Xu, W.; Sun, Y.; Zhang, Z.; Zhao, H.; Yang, L.; Sun, J. Discovery of Potential Prognostic Long Non-Coding RNA Biomarkers for Predicting the Risk of Tumor Recurrence of Breast Cancer Patients. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef]
- Sun, M.; Wu, D.; Zhou, K.; Li, H.; Gong, X.; Wei, Q.; Du, M.; Lei, P.; Zha, J.; Zhu, H.; et al. An Eight-LncRNA Signature Predicts Survival of Breast Cancer Patients: A Comprehensive Study Based on Weighted Gene Co-Expression Network Analysis and Competing Endogenous RNA Network. Breast Cancer Res. Treat. 2019, 175, 59–75. [Google Scholar] [CrossRef]
- Li, J.; Wang, W.; Xia, P.; Wan, L.; Zhang, L.; Yu, L.; Wang, L.; Chen, X.; Xiao, Y.; Xu, C. Identification of a Five-LncRNA Signature for Predicting the Risk of Tumor Recurrence in Patients with Breast Cancer. Int. J. Cancer 2018, 143, 2150–2160. [Google Scholar] [CrossRef]
- Zeng, Y.; Wang, G.; Zhou, C.-F.; Zhang, H.-B.; Sun, H.; Zhang, W.; Zhou, H.-H.; Liu, R.; Zhu, Y.-S. LncRNA Profile Study Reveals a Three-LncRNA Signature Associated with the Pathological Complete Response Following Neoadjuvant Chemotherapy in Breast Cancer. Front. Pharmacol. 2019, 10. [Google Scholar] [CrossRef]
- Schoch, K.M.; Miller, T.M. Antisense Oligonucleotides: Translation from Mouse Models to Human Neurodegenerative Diseases. Neuron 2017, 94, 1056–1070. [Google Scholar] [CrossRef]
- Deng, J.; Yang, M.; Jiang, R.; An, N.; Wang, X.; Liu, B. Long Non-Coding RNA HOTAIR Regulates the Proliferation, Self-Renewal Capacity, Tumor Formation and Migration of the Cancer Stem- like Cell (CSC) Subpopulation Enriched from Breast Cancer Cells. PLoS ONE 2017, 12. [Google Scholar] [CrossRef] [PubMed]
- Peng, F.; Wang, J.H.; Fan, W.J.; Meng, Y.T.; Li, M.M.; Li, T.T.; Cui, B.; Wang, H.F.; Zhao, Y.; An, F.; et al. Glycolysis Gatekeeper PDK1 Reprograms Breast Cancer Stem Cells under Hypoxia. Oncogene 2018, 37, 1062–1074. [Google Scholar] [CrossRef] [PubMed]
- Shima, H.; Kida, K.; Adachi, S.; Yamada, A.; Sugae, S.; Narui, K.; Miyagi, Y.; Nishi, M.; Ryo, A.; Murata, S.; et al. Lnc RNA H19 Is Associated with Poor Prognosis in Breast Cancer Patients and Promotes Cancer Stemness. Breast Cancer Res. Treat. 2018, 170, 507–516. [Google Scholar] [CrossRef] [PubMed]
- Shin, V.Y.; Chen, J.; Cheuk, I.W.Y.; Siu, M.T.; Ho, C.W.; Wang, X.; Jin, H.; Kwong, A. Long Non-Coding RNA NEAT1 Confers Oncogenic Role in Triple-Negative Breast Cancer through Modulating Chemoresistance and Cancer Stemness. Cell Death Dis. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Jin, C.; Yan, B.; Lu, Q.; Lin, Y.; Ma, L. Reciprocal Regulation of Hsa-MiR-1 and Long Noncoding RNA MALAT1 Promotes Triple-Negative Breast Cancer Development. Tumor Biol. 2016, 37, 7383–7394. [Google Scholar] [CrossRef] [PubMed]
- Bamodu, O.A.; Huang, W.-C.; Lee, W.-H.; Wu, A.; Wang, L.S.; Hsiao, M.; Yeh, C.-T.; Chao, T.-Y. Aberrant KDM5B Expression Promotes Aggressive Breast Cancer through MALAT1 Overexpression and Downregulation of Hsa-MiR-448. BMC Cancer 2016, 16, 160. [Google Scholar] [CrossRef]
- Latorre, E.; Carelli, S.; Raimondi, I.; D’Agostino, V.; Castiglioni, I.; Zucal, C.; Moro, G.; Luciani, A.; Ghilardi, G.; Monti, E.; et al. The Ribonucleic Complex HuR-MALAT1 Represses CD133 Expression and Suppresses Epithelial-Mesenchymal Transition in Breast Cancer. Cancer Res. 2016, 76, 2626–2636. [Google Scholar] [CrossRef]
- Xing, Z.; Park, P.K.; Lin, C.; Yang, L. LncRNA BCAR4 Wires up Signaling Transduction in Breast Cancer. Rna Biol. 2015, 12, 681–689. [Google Scholar] [CrossRef]
- Sha, S.; Yuan, D.; Liu, Y.; Han, B.; Zhong, N. Targeting Long Non-Coding RNA DANCR Inhibits Triple Negative Breast Cancer Progression. Biol. Open 2017. [Google Scholar] [CrossRef]
- Vidovic, D.; Huynh, T.T.; Konda, P.; Dean, C.; Cruickshank, B.M.; Sultan, M.; Coyle, K.M.; Gujar, S.; Marcato, P. ALDH1A3-Regulated Long Non-Coding RNA NRAD1 Is a Potential Novel Target for Triple-Negative Breast Tumors and Cancer Stem Cells. Cell Death Differ. 2019. [Google Scholar] [CrossRef]
- Hou, P.; Zhao, Y.; Li, Z.; Yao, R.; Ma, M.; Gao, Y.; Zhao, L.; Zhang, Y.; Huang, B.; Lu, J. LincRNA-ROR Induces Epithelial-to-Mesenchymal Transition and Contributes to Breast Cancer Tumorigenesis and Metastasis. Cell Death Dis. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Tu, Z.; Schmöllerl, J.; Cuiffo, B.G.; Karnoub, A.E. Microenvironmental Regulation of Long Noncoding RNA LINC01133 Promotes Cancer Stem Cell-Like Phenotypic Traits in Triple-Negative Breast Cancers. Stem Cells 2019, 37, 1281–1292. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhu, L.; Xu, L.; Qin, K.; Liu, C.; Yu, Y.; Su, D.; Wu, K.; Sheng, Y. Long Noncoding RNA Linc00617 Exhibits Oncogenic Activity in Breast Cancer. Mol. Carcinog. 2017, 56, 3–17. [Google Scholar] [CrossRef] [PubMed]
- Tang, T.; Guo, C.; Xia, T.; Zhang, R.; Zen, K.; Pan, Y.; Jin, L. LncCCAT1 Promotes Breast Cancer Stem Cell Function through Activating WNT/β-Catenin Signaling. Theranostics 2019, 9, 7384–7402. [Google Scholar] [CrossRef]
- Han, C.; Li, X.; Fan, Q.; Liu, G.; Yin, J. CCAT1 Promotes Triple-Negative Breast Cancer Progression by Suppressing MiR-218/ZFX Signaling. Aging (Albany NY) 2019, 11, 4858–4875. [Google Scholar] [CrossRef]
- Song, X.; Zhang, X.; Wang, X.; Chen, L.; Jiang, L.; Zheng, A.; Zhang, M.; Zhao, L.; Wei, M. LncRNA SPRY4-IT1 Regulates Breast Cancer Cell Stemness through Competitively Binding MiR-6882-3p with TCF7L2. J. Cell. Mol. Med. 2019. [Google Scholar] [CrossRef]
- Zhou, M.; Hou, Y.; Yang, G.; Zhang, H.; Tu, G.; Du, Y.E.; Wen, S.; Xu, L.; Tang, X.; Tang, S.; et al. LncRNA-Hh Strengthen Cancer Stem Cells Generation in Twist-Positive Breast Cancer via Activation of Hedgehog Signaling Pathway. Stem Cells 2016, 34, 55–66. [Google Scholar] [CrossRef]
- Jia, X.; Shi, L.; Wang, X.; Luo, L.; Ling, L.; Yin, J.; Song, Y.; Zhang, Z.; Qiu, N.; Liu, H.; et al. KLF5 Regulated LncRNA RP1 Promotes the Growth and Metastasis of Breast Cancer via Repressing P27kip1 Translation. Cell Death Dis. 2019, 10. [Google Scholar] [CrossRef]
- Lu, G.; Li, Y.; Ma, Y.; Lu, J.; Chen, Y.; Jiang, Q.; Qin, Q.; Zhao, L.; Huang, Q.; Luo, Z.; et al. Long Noncoding RNA LINC00511 Contributes to Breast Cancer Tumourigenesis and Stemness by Inducing the MiR-185-3p/E2F1/Nanog Axis. J. Exp. Clin. Cancer Res. 2018, 37. [Google Scholar] [CrossRef]
- Zhang, Z.; Sun, L.; Zhang, Y.; Lu, G.; Li, Y.; Wei, Z. Long Non-Coding RNA FEZF1-AS1 Promotes Breast Cancer Stemness and Tumorigenesis via Targeting MiR-30a/Nanog Axis. J. Cell. Physiol. 2018, 233, 8630–8638. [Google Scholar] [CrossRef]
- Keshavarz, M.; Asadi, M.H. Long Non-Coding RNA ES1 Controls the Proliferation of Breast Cancer Cells by Regulating the Oct4/Sox2/MiR-302 Axis. Febs J. 2019, 286, 2611–2623. [Google Scholar] [CrossRef]
- García-Venzor, A.; Mandujano-Tinoco, E.A.; Lizarraga, F.; Zampedri, C.; Krötzsch, E.; Salgado, R.M.; Dávila-Borja, V.M.; Encarnación-Guevara, S.; Melendez-Zajgla, J.; Maldonado, V. Microenvironment-Regulated LncRNA-HAL Is Able to Promote Stemness in Breast Cancer Cells. Biochim. Biophys. Acta Mol. Cell Res. 2019, 1866. [Google Scholar] [CrossRef] [PubMed]
- Rinn, J.L.; Kertesz, M.; Wang, J.K.; Squazzo, S.L.; Xu, X.; Brugmann, S.A.; Goodnough, L.H.; Helms, J.A.; Farnham, P.J.; Segal, E.; et al. Functional Demarcation of Active and Silent Chromatin Domains in Human HOX Loci by Noncoding RNAs. Cell 2007, 129, 1311–1323. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Jutooru, I.; Chadalapaka, G.; Johnson, G.; Frank, J.; Burghardt, R.; Kim, S.; Safe, S. HOTAIR Is a Negative Prognostic Factor and Exhibits Pro-Oncogenic Activity in Pancreatic Cancer. Oncogene 2013, 32, 1616–1625. [Google Scholar] [CrossRef] [PubMed]
- Loewen, G.; Jayawickramarajah, J.; Zhuo, Y.; Shan, B. Functions of LncRNA HOTAIR in Lung Cancer. J. Hematol. Oncol. 2014, 7, 90. [Google Scholar] [CrossRef]
- Wang, J.; Xie, S.; Yang, J.; Xiong, H.; Jia, Y.; Zhou, Y.; Chen, Y.; Ying, X.; Chen, C.; Ye, C.; et al. The Long Noncoding RNA H19 Promotes Tamoxifen Resistance in Breast Cancer via Autophagy. J. Hematol. Oncol. 2019, 12, 81. [Google Scholar] [CrossRef]
- Collette, J.; Le Bourhis, X.; Adriaenssens, E. Regulation of Human Breast Cancer by the Long Non-Coding RNA H19. Int. J. Mol. Sci. 2017, 18, 2319. [Google Scholar] [CrossRef]
- Zhu, Q.N.; Wang, G.; Guo, Y.; Peng, Y.; Zhang, R.; Deng, J.L.; Li, Z.X.; Zhu, Y.S. LncRNA H19 Is a Major Mediator of Doxorubicin Chemoresistance in Breast Cancer Cells through a Cullin4A-MDR1 Pathway. Oncotarget 2017, 8, 91990–92003. [Google Scholar] [CrossRef]
- Choudhry, H.; Albukhari, A.; Morotti, M.; Haider, S.; Moralli, D.; Smythies, J.; Schödel, J.; Green, C.M.; Camps, C.; Buffa, F.; et al. Tumor Hypoxia Induces Nuclear Paraspeckle Formation through HIF-2α Dependent Transcriptional Activation of NEAT1 Leading to Cancer Cell Survival. Oncogene 2015, 34, 4482–4490. [Google Scholar] [CrossRef]
- Zhang, M.; Wu, W.B.; Wang, Z.W.; Wang, X.H. LncRNA NEAT1 Is Closely Related with Progression of Breast Cancer via Promoting Proliferation and EMT. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 1020–1026. [Google Scholar]
- Ji, P.; Diederichs, S.; Wang, W.; Böing, S.; Metzger, R.; Schneider, P.M.; Tidow, N.; Brandt, B.; Buerger, H.; Bulk, E.; et al. MALAT-1, a Novel Noncoding RNA, and Thymosin Β4 Predict Metastasis and Survival in Early-Stage Non-Small Cell Lung Cancer. Oncogene 2003, 22, 8031–8041. [Google Scholar] [CrossRef]
- Meijer, D.; Van Agthoven, T.; Bosma, P.T.; Nooter, K.; Dorssers, L.C.J. Functional Screen for Genes Responsible for Tamoxifen Resistance in Human Breast Cancer Cells. Mol. Cancer Res. 2006, 4, 379–386. [Google Scholar] [CrossRef]
- Zhang, K.; Tan, X.; Guo, L. The Long Non-coding RNA DANCR Regulates the Inflammatory Phenotype of Breast Cancer Cells and Promotes Breast Cancer Progression via EZH2-dependent Suppression of SOCS3 Transcription. Mol. Oncol. 2019. [Google Scholar] [CrossRef]
- Loewer, S.; Cabili, M.N.; Guttman, M.; Loh, Y.H.; Thomas, K.; Park, I.H.; Garber, M.; Curran, M.; Onder, T.; Agarwal, S.; et al. Large Intergenic Non-Coding RNA-RoR Modulates Reprogramming of Human Induced Pluripotent Stem Cells. Nat. Genet. 2010, 42, 1113–1117. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xu, Z.; Jiang, J.; Xu, C.; Kang, J.; Xiao, L.; Wu, M.; Xiong, J.; Guo, X.; Liu, H. Endogenous MiRNA Sponge LincRNA-RoR Regulates Oct4, Nanog, and Sox2 in Human Embryonic Stem Cell Self-Renewal. Dev. Cell 2013, 25, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Kong, J.; Sun, W.; Li, C.; Wan, L.; Wang, S.; Wu, Y.; Xu, E.; Zhang, H.; Lai, M. Long Non-Coding RNA LINC01133 Inhibits Epithelial–Mesenchymal Transition and Metastasis in Colorectal Cancer by Interacting with SRSF6. Cancer Lett. 2016, 380, 476–484. [Google Scholar] [CrossRef]
- Zhang, J.; Zhu, N.; Chen, X. A Novel Long Noncoding RNA LINC01133 Is Upregulated in Lung Squamous Cell Cancer and Predicts Survival. Tumor Biol. 2015, 36, 7465–7471. [Google Scholar] [CrossRef] [PubMed]
- Zang, C.; Nie, F.Q.; Wang, Q.; Sun, M.; Li, W.; He, J.; Zhang, M.; Lu, K.H. Long Non-Coding RNA LINC01133 Represses KLF2, P21 and E-Cadherin Transcription through Binding with EZH2, LSD1 in Non Small Cell Lung Cancer. Oncotarget 2016, 7, 11696–11707. [Google Scholar] [CrossRef]
- Yu, F.; Li, J.; Chen, H.; Fu, J.; Ray, S.; Huang, S.; Zheng, H.; Ai, W. Kruppel-like Factor 4 (KLF4) Is Required for Maintenance of Breast Cancer Stem Cells and for Cell Migration and Invasion. Oncogene 2011, 30, 2161–2172. [Google Scholar] [CrossRef]
- Lin, N.; Chang, K.Y.; Li, Z.; Gates, K.; Rana, Z.A.; Dang, J.; Zhang, D.; Han, T.; Yang, C.S.; Cunningham, T.J.; et al. An Evolutionarily Conserved Long Noncoding RNA TUNA Controls Pluripotency and Neural Lineage Commitment. Mol. Cell 2014, 53, 1005–1019. [Google Scholar] [CrossRef]
- Liu, Z.; Chen, Q.J.; Hann, S.S. The Functions and Oncogenic Roles of CCAT1 in Human Cancer. Biomed. Pharmacother. 2019. [Google Scholar] [CrossRef] [PubMed]
- Tennis, M.A.; Van Scoyk, M.M.; Freeman, S.V.; Vandervest, K.M.; Nemenoff, R.A.; Winn, R.A. Sprouty-4 Inhibits Transformed Cell Growth, Migration and Invasion, and Epithelial-Mesenchymal Transition, and Is Regulated by Wnt7a through PPARγ in Non-Small Cell Lung Cancer. Mol. Cancer Res. 2010, 8, 833–843. [Google Scholar] [CrossRef]
- Shi, C.; Sun, L.; Song, Y. FEZF1-AS1: A Novel Vital Oncogenic LncRNA in Multiple Human Malignancies. Biosci. Rep. 2019. [Google Scholar] [CrossRef] [PubMed]
- Ng, S.-Y.; Johnson, R.; Stanton, L.W. Human Long Non-Coding RNAs Promote Pluripotency and Neuronal Differentiation by Association with Chromatin Modifiers and Transcription Factors. Embo J. 2012, 31, 522–533. [Google Scholar] [CrossRef] [PubMed]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The CBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the CBioPortal. Sci. Signal. 2013, 6. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Lu, R.; Gu, J.; Chen, Y.; Zhang, X.; Zhang, L.; Wu, H.; Hua, W.; Zeng, J. Aldehyde Dehydrogenase 1A1 Up-Regulates Stem Cell Markers in Benzo[a]Pyrene-Induced Malignant Transformation of BEAS-2B Cells. Environ. Toxicol. Pharmacol. 2016, 45, 241–250. [Google Scholar] [CrossRef]
- Kwon, M.J.; Han, J.; Seo, J.H.; Song, K.; Jeong, H.M.; Choi, J.-S.; Kim, Y.J.; Lee, S.-H.; Choi, Y.-L.; Shin, Y.K. CD24 Overexpression Is Associated with Poor Prognosis in Luminal A and Triple-Negative Breast Cancer. PLoS ONE 2015, 10, e0139112. [Google Scholar] [CrossRef]
- Gómez-Miragaya, J.; Palafox, M.; Paré, L.; Yoldi, G.; Ferrer, I.; Vila, S.; Galván, P.; Pellegrini, P.; Pérez-Montoyo, H.; Igea, A.; et al. Resistance to Taxanes in Triple-Negative Breast Cancer Associates with the Dynamics of a CD49f+ Tumor-Initiating Population. Stem Cell Rep. 2017, 8, 1392–1407. [Google Scholar] [CrossRef]
- Lu, Y.; Hu, Z.; Mangala, L.S.; Stine, Z.E.; Hu, X.; Jiang, D.; Xiang, Y.; Zhang, Y.; Pradeep, S.; Rodriguez-Aguayo, C.; et al. MYC Targeted Long Noncoding RNA DANCR Promotes Cancer in Part by Reducing P21 Levels. Cancer Res. 2018, 78, 64–74. [Google Scholar] [CrossRef]
- Zhou, W.; Ye, X.L.; Xu, J.; Cao, M.G.; Fang, Z.Y.; Li, L.Y.; Guan, G.H.; Liu, Q.; Qian, Y.H.; Xie, D. The LncRNA H19 Mediates Breast Cancer Cell Plasticity during EMT and MET Plasticity by Differentially Sponging MiR-200b/c and Let-7b. Sci. Signal. 2017, 10. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Yao, H.; Wang, K.; Liu, X. Long Non-Coding RNA MALAT1 Regulates ZEB1 Expression by Sponging MiR-143-3p and Promotes Hepatocellular Carcinoma Progression. J. Cell. Biochem. 2017, 118, 4836–4843. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Yang, S.; Su, N.; Wang, Y.; Yu, J.; Qiu, H.; He, X. Overexpression of Long Non-Coding RNA HOTAIR Leads to Chemoresistance by Activating the Wnt/β-Catenin Pathway in Human Ovarian Cancer. Tumor Biol. 2016, 37, 2057–2065. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Hao, G.; Sun, Y.; Li, L.; Wang, Y. Long Noncoding RNA H19 Mediated the Chemosensitivity of Breast Cancer Cells via Wnt Pathway and EMT Process. Onco Targets Ther. 2018, 11, 8001–8012. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Zhao, W.; Zhang, L.; Wang, L.; Wang, J.; Wan, Z.; Hong, Y.; Yu, L. MALAT1-MiR-101-SOX9 Feedback Loop Modulates the Chemoresistance of Lung Cancer Cell to DDP via Wnt Signaling Pathway. Oncotarget 2017, 8, 94317–94329. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Gao, S.; Li, H.; Lv, M.; Lu, C. Long Noncoding RNAs (LncRNAs) in Triple Negative Breast Cancer. J. Cell. Physiol. 2017, 232, 3226–3233. [Google Scholar] [CrossRef]
- Chen, C.; Li, Z.; Yang, Y.; Xiang, T.; Song, W.; Liu, S. Microarray Expression Profiling of Dysregulated Long Non-Coding RNAs in Triple-Negative Breast Cancer. Cancer Biol. Ther. 2015, 16, 856–865. [Google Scholar] [CrossRef]
- Liu, Y.-R.; Jiang, Y.-Z.; Xu, X.-E.; Yu, K.-D.; Jin, X.; Hu, X.; Zuo, W.-J.; Hao, S.; Wu, J.; Liu, G.-Y.; et al. Comprehensive Transcriptome Analysis Identifies Novel Molecular Subtypes and Subtype-Specific RNAs of Triple-Negative Breast Cancer. Breast Cancer Res. 2016, 18, 33. [Google Scholar] [CrossRef]
- Tian, T.; Gong, Z.; Wang, M.; Hao, R.; Lin, S.; Liu, K.; Guan, F.; Xu, P.; Deng, Y.; Song, D.; et al. Identification of Long Non-Coding RNA Signatures in Triple-Negative Breast Cancer. Cancer Cell Int. 2018, 18, 103. [Google Scholar] [CrossRef]
- Rodríguez Bautista, R.; Ortega Gómez, A.; Hidalgo Miranda, A.; Zentella Dehesa, A.; Villarreal-Garza, C.; Ávila-Moreno, F.; Arrieta, O. Long Non-Coding RNAs: Implications in Targeted Diagnoses, Prognosis, and Improved Therapeutic Strategies in Human Non- and Triple-Negative Breast Cancer. Clin. Epigenetics 2018. [Google Scholar] [CrossRef]
- Zhang, Y.; He, Q.; Hu, Z.; Feng, Y.; Fan, L.; Tang, Z.; Yuan, J.; Shan, W.; Li, C.; Hu, X.; et al. Long Noncoding RNA LINP1 Regulates Repair of DNA Double-Strand Breaks in Triple-Negative Breast Cancer. Nat. Struct. Mol. Biol. 2016, 23, 522–530. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Chen, D.; Gao, X.; Li, X.; Shi, G. LncRNA NEAT1 Regulates Cell Viability and Invasion in Esophageal Squamous Cell Carcinoma through the MiR-129/CTBP2 Axis. Dis. Markers 2017, 2017. [Google Scholar] [CrossRef] [PubMed]
- Huang, N.S.; Chi, Y.Y.; Xue, J.Y.; Liu, M.Y.; Huang, S.; Mo, M.; Zhou, S.L.; Wu, J. Long Non-Coding RNA Metastasis Associated in Lung Adenocarcinoma Transcript 1 (MALAT1) Interacts with Estrogen Receptor and Predicted Poor Survival in Breast Cancer. Oncotarget 2016, 7, 37957–37965. [Google Scholar] [CrossRef] [PubMed]
- Tamori, S.; Nozaki, Y.; Motomura, H.; Nakane, H.; Katayama, R.; Onaga, C.; Kikuchi, E.; Shimada, N.; Suzuki, Y.; Noike, M.; et al. Glyoxalase 1 Gene Is Highly Expressed in Basal-like Human Breast Cancers and Contributes to Survival of ALDH1-Positive Breast Cancer Stem Cells. Oncotarget 2018, 9, 36515–36529. [Google Scholar] [CrossRef]
- Györffy, B.; Lanczky, A.; Eklund, A.C.; Denkert, C.; Budczies, J.; Li, Q.; Szallasi, Z. An Online Survival Analysis Tool to Rapidly Assess the Effect of 22,277 Genes on Breast Cancer Prognosis Using Microarray Data of 1,809 Patients. Breast Cancer Res. Treat. 2010, 123, 725–731. [Google Scholar] [CrossRef]
- Lennox, K.A.; Behlke, M.A. Cellular Localization of Long Non-Coding RNAs Affects Silencing by RNAi More than by Antisense Oligonucleotides. Nucleic Acids Res. 2016, 44, 863–877. [Google Scholar] [CrossRef]
- Leucci, E.; Vendramin, R.; Spinazzi, M.; Laurette, P.; Fiers, M.; Wouters, J.; Radaelli, E.; Eyckerman, S.; Leonelli, C.; Vanderheyden, K.; et al. Melanoma Addiction to the Long Non-Coding RNA SAMMSON. Nature 2016, 531, 518–522. [Google Scholar] [CrossRef]
- Xing, Z.; Lin, A.; Li, C.; Liang, K.; Wang, S.; Liu, Y.; Park, P.K.; Qin, L.; Wei, Y.; Hawke, D.H.; et al. LncRNA Directs Cooperative Epigenetic Regulation Downstream of Chemokine Signals. Cell 2014, 159, 1110–1125. [Google Scholar] [CrossRef]
- Gutschner, T.; Hämmerle, M.; Eißmann, M.; Hsu, J.; Kim, Y.; Hung, G.; Revenko, A.; Arun, G.; Stentrup, M.; Groß, M.; et al. The Noncoding RNA MALAT1 Is a Critical Regulator of the Metastasis Phenotype of Lung Cancer Cells. Cancer Res. 2013, 73, 1180–1189. [Google Scholar] [CrossRef]
- Yang, J.; Meng, X.; Pan, J.; Jiang, N.; Zhou, C.; Wu, Z.; Gong, Z. CRISPR/Cas9-Mediated Noncoding RNA Editing in Human Cancers. RNA Biol. 2018, 15, 35–43. [Google Scholar] [CrossRef]
- Zhen, S.; Hua, L.; Liu, Y.H.; Sun, X.M.; Jiang, M.M.; Chen, W.; Zhao, L.; Li, X. Inhibition of Long Non-Coding RNA UCA1 by CRISPR/Cas9 Attenuated Malignant Phenotypes of Bladder Cancer. Oncotarget 2017, 8, 9634–9646. [Google Scholar] [CrossRef] [PubMed]
- Vaidya, A.M.; Sun, Z.; Ayat, N.; Schilb, A.; Liu, X.; Jiang, H.; Sun, D.; Scheidt, J.; Qian, V.; He, S.; et al. Systemic Delivery of Tumor-Targeting SiRNA Nanoparticles against an Oncogenic LncRNA Facilitates Effective Triple-Negative Breast Cancer Therapy. Bioconjug. Chem. 2019, 30, 907–919. [Google Scholar] [CrossRef]
- Jiang, Y.Z.; Liu, Y.R.; Xu, X.E.; Jin, X.; Hu, X.; Yu, K.D.; Shao, Z.M. Transcriptome Analysis of Triple-Negative Breast Cancer Reveals an Integrated MRNA-LncRNA Signature with Predictive and Prognostic Value. Cancer Res. 2016, 76, 2105–2114. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Ren, J.; Cui, Q.; Zhang, D.; Kong, D.; Liao, X.; Lu, M.; Gong, Y.; Wu, G. A Prognostic 10-LncRNA Expression Signature for Predicting the Risk of Tumour Recurrence in Breast Cancer Patients. J. Cell. Mol. Med. 2019, 23, 6775–6784. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Peng, R.; Wang, J.; Qin, Z.; Xue, L. Circulating MicroRNAs as Potential Cancer Biomarkers: The Advantage and Disadvantage. Clin. Epigenetics 2018, 59. [Google Scholar] [CrossRef] [PubMed]
- Sole, C.; Arnaiz, E.; Manterola, L.; Otaegui, D.; Lawrie, C.H. The Circulating Transcriptome as a Source of Cancer Liquid Biopsy Biomarkers. Semin. Cancer Biol. 2019, 58, 100–108. [Google Scholar] [CrossRef]
- Iyer, M.K.; Niknafs, Y.S.; Malik, R.; Singhal, U.; Sahu, A.; Hosono, Y.; Barrette, T.R.; Prensner, J.R.; Evans, J.R.; Zhao, S.; et al. The Landscape of Long Noncoding RNAs in the Human Transcriptome. Nat. Genet. 2015, 47, 199–208. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).