Abstract
Protein ubiquitination belongs to the best characterized pathways of protein degradation in the cell; however, our current knowledge on its physiological consequences is just the tip of an iceberg. The divergence of enzymatic executors of ubiquitination led to some 600–700 E3 ubiquitin ligases embedded in the human genome. Notably, mutations in around 13% of these genes are causative of severe neurological diseases. Despite this, molecular and cellular context of ubiquitination remains poorly characterized, especially in the developing brain. In this review article, we summarize recent findings on brain-expressed HECT-type E3 UBE3 ligases and their murine orthologues, comprising Angelman syndrome UBE3A, Kaufman oculocerebrofacial syndrome UBE3B and autism spectrum disorder-associated UBE3C. We summarize evolutionary emergence of three UBE3 genes, the biochemistry of UBE3 enzymes, their biology and clinical relevance in brain disorders. Particularly, we highlight that uninterrupted action of UBE3 ligases is a sine qua non for cortical circuit assembly and higher cognitive functions of the neocortex.
1. Development of the Neocortical Circuits
Uninterrupted communication between executive brain centers, on a molecular and single neuron level manifested as circuit activity, allows for integration of external and internal sensory stimuli and generation of appropriate efferent responses. The neocortex, paramount achievement of mammalian brain evolution, constitutes a biological substrate for higher cognitive abilities in Primata. Spatiotemporally synchronized patterns of activity in and between neuronal circuits are enabled by tightly controlled transcriptional–translational and morphoregulatory programs in developing neocortex [1].
Assembly of a neuronal circuits in the mammalian neocortex starts during early neurogenesis, where at the ventricular zone (VZ) of the neural tube, radial glial cells (RGCs), give rise to postmitotic cells that differentiate into glutamatergic excitatory neurons. One of the most prominent features of cortical neuron development is that the immature nerve cells born close to the VZ do not stay in situ but migrate radially to distribute horizontally into their target niches to establish neocortical layers. These cortical niches contain neurons originating from spatiotemporally defined RGC populations. Neurons of each layer share transcriptional signatures and display similar physiological properties and patterns of connectivity. Molecular signaling orchestrating the postmitotic differentiation is conserved between neurons of different layers. The initial transcriptional ground states in RGCs, often display very subtle differences, but represent spatiotemporally restricted developmental trajectories that seem to be the source of the excitatory neuronal subtype variations in the cerebral cortex [2,3]. Formation of cortical layers, which requires orthodox supervision of neurogenesis, neuronal migration, specification of a single axon and its guidance, formation and elaborate growth of dendritic arbor and synaptogenesis, is therefore fundamental for establishment of afferent inputs and efferent connectivity of the cortical circuits. Specification of different neuronal types and the intricate geometry of the neuron dictates its contribution to the activity of the neuronal circuit [4]. Assembled circuits in and outside of the cortex underlie higher cognitive functions. A well characterized physiological manifestation of such is long-term potentiation (LTP), the most studied form of synaptic plasticity associated with memory storage, that leads to long-term strengthening of synaptic transmission between neurons driven by a specific pattern of activity [5]. Formation and retrieval of memories represents another aspect of circuit activity, as it requires directional coupling between two brain regions, neocortex and hippocampus. On a circuit physiology level, these processes are thought to be supported by neocortical desynchronization of alpha/beta rhythm and synchronization of hippocampal theta/gamma oscillations [6,7].
2. Neuronal Circuit Assembly and Neurodevelopmental Disorders
Aberrant assembly of neuronal circuits represents the primary cause of higher cognitive dysfunction [4]. Malfunctioning of neuronal circuits associated with structural changes in neuronal morphology is a physiological underpinning of neurodevelopmental disorders. These include autism spectrum disorders (ASD), or intellectual disability (ID), both extremely heterogenous and multifactorial groups of diseases, with their psychiatric and cognitive comorbidities and ever-increasing prevalence [4]. Genetic predisposition combined with an environmental impact are the core ASD/ID etiology [8].
3. Protein Ubiquitination as a Potent Regulatory Principle
Neuronal transcriptional signatures are the effect of a discrete regulation of gene expression. Recent reports derive the sources of cortical neuron complexity using state-of-the-art single cell transcriptomics to cluster the types of cellular populations present in the developing brain. These elegant but somewhat reductive approaches turn a blind eye to the involvement of post-transcriptional modes of gene expression regulation in the brain development. Recent accumulating evidence reveals critical roles of post-translational modification of ubiquitination in the formation of neuronal networks [9,10].
4. Protein Ubiquitination Is a Reversible Post-Translational Modification
Since its discovery in 1975 [11], ubiquitin (Ub) has been linked to regulation of the broad range of cellular processes, from DNA repair to intracellular trafficking. A breakthrough discovery that protein ubiquitination results in 26S proteasomal targeting and substrate degradation was awarded a Nobel Prize in Chemistry in 2004 and shed light onto the complex biochemistry of the modification [12]. Ubiquitination involves conjugation of 8.6 kDa globular Ub to a substrate protein in a cascade of enzymatic reactions catalyzed by an E1 Ub activating enzyme, an E2 Ub conjugating enzyme, and an E3 Ub ligase. Of note, apart from the canonical successive sequential mode of Ub shuttle, additional biochemical scenarios of ubiquitination have emerged plausible as well [13]. Eventually, Ub is shuttled onto the substrate protein, a reaction catalyzed by the E3 ligases, which determine the specificity of the ubiquitome [14]. Canonically, Ub is linked to the substrate via an isopeptide bond between C-terminus glycine residue of Ub and a lysine (K) of the target protein. Non-canonical ubiquitination includes linkages employing the N-terminal methionine, serine, or threonine of the substrate protein [15]. Potential functional consequences of ubiquitination via the non-canonical sites remain yet to be described. K residues on substrate proteins can be conjugated with a single Ub or with polyUb chains (polyubiquitination). Homotypic polyUb chain can be formed through one of the seven K residues of Ub (K6, K11, K27, K29, K33, K48 and K63). The type of the polyUb chain specifies the functional consequence of ubiquitination. In the mouse brain, approximately 60% of Ub remains a free monomer, 35% is detected as a monoUb on substrates and 5% in polyUb chains [16].
Protein ubiquitylation is reversible and catalyzed by ubiquitin proteases, enzymes called deubiquitylases (DUBs). Human genome harbors some 90 loci encoding for DUBs, far less than for E3 ligases, indicating a rather broad biochemical promiscuity. However, some DUBs display a strong ubiquitin chain type affinity, i.e., they catalyze proteolytic cleavage of certain polyUb chains, rather than the ubiquitylated substrate per se [17].
5. Uninterrupted Function of E3 Ligases Is Fundamental for Brain Development
Given a distinct mode of catalysis, E3 ligases fall into two, or three main classes. Homologous to E6-AP (HECT) type E3 Ub ligases act as direct Ub acceptors, conjugating it to their C-terminal cysteine residue, before shuttling the Ub onto their substrates. HECT is a group of biochemically distinct enzymes with 28 members in humans identified to date. Our previous works signify the involvement of HECT-type E3 ligases in the most fundamental neurodevelopmental processes including establishment of dendritic arbor [18], neurite extension [19], and acquisition of axon and neuronal migration [20]. Of all E3 ligase genes, approximately 13% are mutated in a neurological disorder, with ASD/ID-associated mutations in E3 ligase genes representing the biggest group [21]. Among these are the brain expressed UBE3 enzymes: the founder of the subclass Ubiquitin Ligase E3A (UBE3A), with its gene loss-of-function resulting in Angelman syndrome (AS) and Ubiquitin Ligase E3B (UBE3B), linked to Kaufman oculocerebrofacial syndrome (KOS) as well as ASD-associated Ubiquitin Ligase E3C (UBE3C). In this review, we will focus on highlighting their association with severe neurodevelopmental disorders and their function in the assembly of neuronal circuits. We will discuss the fact that despite a high degree of homology at the level of peptide sequence, there is strikingly little functional redundancy between these three ligases. Our previous review covers the neuronal roles of UBE3 ligases [9]; we will therefore focus on the most recent published works here.
6. UBE3 Ligases Are Able to Assemble Different polyUb Chain Types
Classically, ubiquitination of a substrate protein has been linked to its degradation. Loss-of-function mutations in E3 ligases are associated with neurological diseases, implicating the pivotal role of specific degradation in fundamental neurodevelopmental pathways ergo cortical circuit assembly. Recent data sheds lights onto non-degradative ubiquitination, encoded by polyUb chain type conjugated to the substrate, indicative of these types of modifications at play during neuronal development. The K48- and K63-linked polyUb chains constitute the major types of polyUb chains in the mouse brain and human frontal cortex and K11-linked chains are three- to four-fold less abundant [16]. Canonical ubiquitination, involving conjugation of K48-linked Ub chain destines substrate proteins for proteasomal degradation, whereas functional consequences of K63-linked chain conjugation include non-proteasomal effects, such as trafficking of transmembrane proteins, lysosomal degradation, or in their mono form, modification of histones to organize chromatin structure [10,22]. Although there is sufficient evidence for physiological consequences of canonical K48- and K63-linked ubiquitylation, roles of other polyUb chain types remain elusive, especially in terms of the development of the nervous system. Of note, K6-, K27-, K29-, and K33-polyUb chains together represent less than 1% of total Ub found in the mouse brains and human frontal cortex [16].
Whereas monoUb has been linked to modulating enzymatic activity or protein–protein interactions [18], different polyUb linkages display fundamentally opposite biochemical conformations, which may explain the various substrate protein fates that they predestine towards [23]. Structurally, molecular interfaces between hydrophobic patches on adjacent Ubs in a dimer can be formed by K6, K11, K29, K33, and K48, generating a conformational landscape for interactions. Predominantly, K63-linked chains adopt an open conformation with two Ub molecules only contacting at the linkage site [24].
The determinants of chain type formed by a given E3 Ub ligase have been an area of debate. Regarding the HECT ligases, these are the function of the catalytic HECT domain and do not seem to depend on the E2 conjugating enzyme, but rather are located within the 60 amino acids of the C-terminus of the HECT domain C-lobe [25]. Within the HECT family, UBE3A preferentially forms K48 polyUb chain [25], Ube3b is able to form both K48- and K63-linked chain [26] and UBE3C catalyzes K48- and less preponderant K29- and K11-linked chains [27]. K29 chains exist in cells within heterotypic and branched polyUb chains, among other Ub species [28]. Such divergent biochemical properties of these homologs are somewhat surprising, given a high degree of amino acid sequence similarity between their HECT domains. Indispensable roles of each UBE3 ligase in a neuron are associated with inherently specific biochemical modes of action (Figure 1).

Figure 1.
Sequence alignment of the C-termini of Ube3 ligases (Uniprot accession numbers on the left). Numbers on the left and right indicate the extreme amino acid positions. Highlighted in red is the active site catalytic cysteine. Symbols “*”, “:”, “.”, denote the degree of single residue homology.
7. Functional and Pathophysiological Implications of the HECT Domain Organization of UBE3 Ligases
Understanding of neuronal roles of UBE3s requires an in-depth analysis of the biochemistry of their action. Transient binding of Ub (autoubiquitination) prior to its sequential transfer onto the substrates, is a distinct feature of HECT-type ligases. Evolutionarily conserved organization of the catalytic C-terminal HECT domain highlights its E2 conjugating enzyme-interacting N-lobe, a short flexible linker and a C-lobe, containing an Ub acceptor cysteine [29]. Conformational state of the HECT domain determines Ub binding and allows for proper orientation of the growing polyUb chain [30]. Unsurprisingly, amino acid substitutions in this domain engender dramatic effects for the enzymatic activity of a given E3.
We know a fair amount regarding the organization of UBE3A HECT domain and its mode of action [31]. Biochemical studies show a two-step mechanism, with a site for initial E2 binding and another for Ub chain elongation [32]. Moreover, fully catalytically active UBE3A is a trimer with the oligomerization interface using a HECT domain localized residue. Destabilization of the trimer formation also seems to be a feature of a specific loss-of-function Angelman syndrome mutation [33]. An AS-associated transversion mutation found in the HECT domain of UBE3A I827K destabilizes protein folding and causes C-lobe aggregation, which hinders UBE3A enzymatic activity [34]. A specific c-Abl-mediated tyrosine residue phosphorylation in UBE3A HECT domain has also been linked to its oligomerization and activity [35]. Until now, no indications for oligomerization of UBE3B or UBE3C exist.
KOS-associated point mutations in UBE3B leading to single amino acid substitutions in the HECT domain appear to affect the enzymatic activity of the ligase implicating conformational restrictions. Theoretical modeling of Q727P substitution predicts it renders the HECT domain unfit for substrate binding and positioning towards catalytic cysteine, required for efficient ubiquitination [36]. HECT-localized KOS-linked substitutions G779R and R997P fail to restore deficient dendritic arbors of Ube3b conditional forebrain-specific knockout (cKO) neurons, and the latter one exhibits altered subneuronal localization, when compared to the native UBE3B [26]. It is possible, that perinuclear localization of R997P UBE3B represents its ER-trapped folding-deficient form, similar to the case of another postsynapse-associated ASD-linked protein, neuroligin-4 [37]. The scenario of ER-localized processing of UBE3B should be verified experimentally.
Recent studies have shed light on the crystal structure of UBE3C [38]. Aside from the catalytic cysteine, an additional K903 site is also a major site of autoubiquitination in its HECT domain, whereas Q961, S1049, and three C-terminal amino acids are necessary for UBE3C activity [38]. Additionally, the N-terminal region just preceding the HECT domain is essential for stabilization and activity of UBE3C. Of note, ASD-associated point substitutions in UBE3C localize to its HECT domain. It is tempting to speculate the conformational bias of these mutants and inefficient UBE3C-mediated ubiquitination (Table 1).

Table 1.
Amino acid residues necessary for UBE3C enzymatic activity.
8. Developmental Expression and Molecular Evolution of UBE3 Ligases
There has been a slight confusion about the functional homology between UBE3s, as observed in introductions to multiple publications. All three UBE3s share a gross domain organization plan including the catalytic HECT domain and long stretches of low complexity, possibly accounting for the high interactivity of ligases with binding partners [41]. UBE3B and UBE3C display an N-terminal IQ motif. Apart from this, evolutionary, biochemical, and physiological comparison of these three ligases indicates that each of them represents a distinct biochemical entity with specific functions in a neuron.
Single cell RNA sequencing databases provide insights into spatiotemporal expression dynamics of Ube3 genes. These expression trajectories indicate that each mRNA is present in progenitors and postmitotic neurons in the developing murine neocortex [3]. Ube3a mRNA shows elevated levels in the latter cell type, Ube3b mRNA exhibits a slight decrease in its levels in postmitotic cells and Ube3c mRNA displays an enrichment in E12 multipotent progenitors, able to give rise to neurons of both upper and deeper layers of the neocortex (Figure 2). From the evolutionary biology point of view, UBE3A is more closely related to members of the small HERC family of ligases than to UBE3B and UBE3C [42].
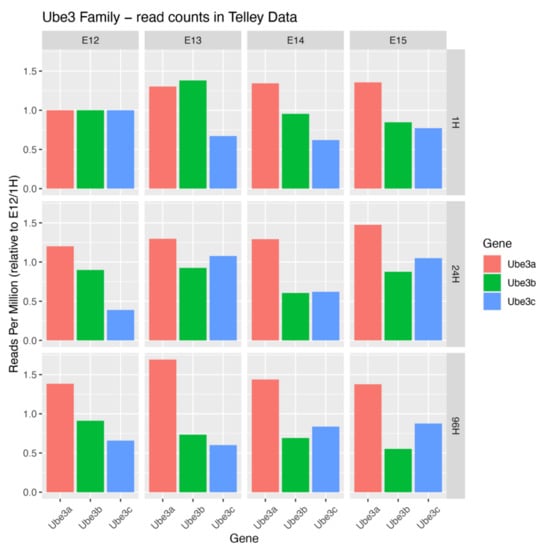
Figure 2.
Expression levels of Ube3 ligase genes in developing cortex. To plot expression levels of Ube genes, we downloaded the digital gene expression matrix (DGE) provided by Telley et al. [3], in which cells are annotated by the time of collection, and time since FlashTag labelling. We grouped all cells with the same annotation to counts per-gene, and by summing all such values we calculated a “library size” combination of variables, normalizing the counts to control for the number of cells of each type collected. For each gene, we then normalized the count values to the value at the earliest time of collection and “flashtagging”, in order to plot relative change in expression over time.
Phylogenetic analyses show that HECT E3s appear before animals or very early in their evolution and appearance of them precede the emergence of the cellular function that they control [43]. That said, evolutionary emergence of Ube3s dates long before the origin of the central nervous system. The history of Ube3a seems more tumultuous than that of Ube3b and Ube3c. Orthologues of Ube3a are identified in bilaterians, including vertebrates and fruit fly, but not in C. elegans indicating Ube3a loss in some nematodes [43]. Additionally, Ube3a shows secondary loss in choanoflagellate lineage. On top of this, the origin of Ube3a imprinting seems to be acquired in the mammalian radiation. Ube3b appeared relatively early in evolution and is present in fungi and other opisthokonta. Of the three Ube3s, Ube3c seems to be the youngest with its orthologues found in Bilateria and other Metazoa [44] (Figure 3). The phylogenetic searches for the ancestors of postsynaptic proteins demonstrate the presence of core protein orthologues in simple metazoans and choanoflagellates devoid of or with a primitive nervous system. This indicates an ancient origin of genes, including Ube3s and their orthologues, implicated in human neurodevelopmental disorders, which seem to regulate key cellular housekeeping processes in primitive organisms [42]. This might explain the critical role of UBE3s outside of the nervous system, as well.
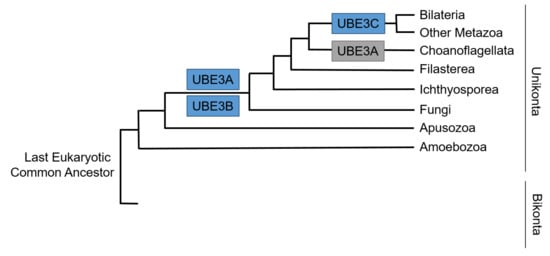
Figure 3.
Simplified pattern of acquisition of Ubiquitin Ligases E3s (UBE3s) across the tree of life of Eucaryota based on [44]. Gains are colored in blue and losses in grey.
9. The Founding Member of HECT Family, E6AP/UBE3A, Angelman Syndrome, and Its Role in the Developing Nervous System
The gene encoding for the founding member of HECT type ligase family, E6-associated protein, E6AP, or UBE3A, is imprinted in neurons in mice and humans resulting in maternal expression of UBE3A allele. Inactivation, deletions or mutation of maternal UBE3A allele are causative of Angelman syndrome (AS) with, among other symptoms, severe intellectual disability, speech impairment, behavioral disturbances, and developmental delay [45]. Duplications of 15q11.2–q13.3 locus, including UBE3A, is a genetic cause of the ASD [46]. We summarized some of the less recent findings on neuronal role of UBE3A in our previous review [9]. In human cortex, UBE3A is expressed in glutamatergic and GABAergic neurons and to the lower extent in glia [47]. Maternal Ube3a KO in mice (Ube3a m−/p+) recapitulates features of AS patients. Loss of Ube3a leads to ineffective LTP in the hippocampus, indicative of defects in neuronal circuitry associated with learning and memory [48].
10. Mother Allele-Specific Neuronal Expression of UBE3A
While UBE3A is located on chromosome 15, the mode of AS inheritance is not typical autosomal dominant or recessive [49,50]. Paternal alleles of human UBE3A and mouse Ube3a are silenced by large non-cording antisense RNA, expressed from the nearby loci [49]. The murine non-coding RNA called Ube3a antisense transcript (Ube3a-ATS) is transcribed by an atypical RNA polymerase type II and covers the entire Ube3a locus. Deletion of maternal Ube3a causes the complete lack of Ube3a protein expression, restored by additional targeting of paternal Ube3a-ATS [51]. Ube3a-ATS is a critical suppressor of the Ube3a expression from the paternal allele, which is likely the case in humans as well. Interestingly, Ube3a-ATS is expressed solely in neurons, without affecting expression of Ube3a in glial cells [52,53]. Of note, accumulating evidence suggests that the disinhibition of paternal allele of Ube3a expression offers a potential therapeutic approach for AS [54,55].
11. Molecular Biology of UBE3A, a Proteasome Regulator and Modulator of Cellular Signaling
Other reviews [56] highlight the mechanistic function of UBE3A, describing its association with several proteasome receptors, like DDI1 [57,58] or PSMD4 but also with proteasome itself, which is thought to control the proteolysis [59]. AS-associated point mutations in UBE3A N-terminus show impaired PSMD4 binding [60]. Further, AS-linked UBE3AT485A reduces proteasome activity and leads to a stabilization of β-catenin, indicative of its enhanced ability activate Wnt signaling [61]. The T485 residue is phosphorylated by protein kinase A (PKA) to (auto)-inhibit UBE3A, so that T485A engenders enhanced turnover of substrates in patient-derived cells and intensifies development of dendritic spines [62]. UBE3A has also been found to ubiquitinate proteasome related protein Rpn10, implicating UBE3A in a robust regulation of proteostasis [63]. As is the case for UBE3B, UBE3A also seems to play pivotal role in maintenance of mitochondrial function [64]. Quite intriguing is association of UBE3A with another HECT E3, HERC2 and formation of a high molecular weight complex of yet not understood function [65]. Upon interaction, HERC2 stimulates UBE3A enzymatic activity in a ubiquitination-independent manner [66]. There is a significant body of work on UBE3A in immune response to viral infection, cancer, and neurodegeneration. Unfortunately, we cannot mention all published works on UBE3A here, solely due to space limitations. Further on, we will focus on the roles of UBE3A regarding the assembly of cortical circuits for the sake of its functional comparison with UBE3B and UBE3C.
12. Isoform-Specific Neuromorphoregulatory Roles of UBE3A
Isoform- and tissue context-specific morphoregulatory roles of UBE3A have been fundamental for our understanding of its contribution to the development and functions of neuronal circuits. Five isoforms of UBE3A mRNAs were identified with three of the transcripts coding for protein isoforms [67]. Ube3a downregulation leads to less complex dendritic arbors in the cortex and hippocampus and abrogates growth of apical dendrites associated with loss of polarization of Golgi apparatus. Such phenotypes are reversible by re-expression of isoform 2, but not of isoform 1 or 3 of Ube3a [68]. In contrast to AS neurons, UBE3A-overexpressing neurons do not exhibit polarization problems [69]. Another report provides evidence that isoform 1 accounts for the majority of UBE3A in neurons [70]. Moreover, knockdown of alternatively spliced Ube3a1 devoid of its catalytic activity prompts robust dendritic growth, reduces the volume of spines and leads to decreased synaptic transmission. This function of Ube3a1 is encoded in its 3′ untranslated region, which acts as a competitive scavenger of microRNAs, specifically of miR-134 [71].
Expression of different isoforms of UBE3A has its relevance in the pathophysiology of AS. Developmental well-being of AS patients seem to rely on the isoforms of UBE3A affected in the genome. For instance, patients with Met1Thr mutation eliminating isoform 1 of maternal UBE3A exhibit far more advanced developmental skills as compared to other non-mosaic AS patients [72].
13. UBE3A Acts Locally at the Spine and Globally in the Nucleus to Regulate Synaptic Transmission
Ube3a localizes at the axon terminals and at the head and neck of dendritic spines [47] to regulate their density and length, glutamatergic and activity-dependent transmission [73]. Its ubiquitination targets degraded at the proteasome include Arc [48], responsible for endosomal recycling of α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptors (AMPARs), and phosphorylated ephexin-5 [74], a negative regulator of excitatory synapse development. Recently, Protein Phosphatase 2 Phosphatase Activator (PTPA) has joined the group of bona fide UBE3A substrates. Ube3a m−/p+ mice exhibit increased PTPA level and protein phosphatase 2A (PP2A) activity. Normalizing PTPA levels, as well as pharmacological inhibition of PP2A reversed developmental defects of excitatory synapses in AS mice [75]. Regarding other cellular pathways, Ube3a-mediated p18 targeting negatively regulates mTORC1 signaling and normalizing p18 overexpression in AS mouse model restores dendritic spine maturation, LTP and learning [76]. BMP repression is also involved in Ube3a-regulated synaptic growth in Drosophila [77].
Given UBE3A involvement in a developmental syndrome presenting a severe ID, a significant body of literature addresses synaptic roles of the ligase. Recently, extensive local synaptic actions of Small Ub-Like Modifier 1 (SUMO1), have been rebutted since in neurons, SUMO1 was found mainly in the nucleus [78]. Recent research on UBE3A also identifies its cytoplasmic and nuclear isoforms, with nuclear UBE3A as the predominant form, indicative of its nuclear roles, including regulation of transcription. Loss of UBE3A nuclear isoform, but not the cytoplasmic one, recapitulates behavioral and physiological phenotypes described for maternal Ube3a KO mouse [79]. Mimicking of the ASD-linked 15q11–13 chromosomal triplications by increasing nuclear UBE3A leads to diminished expression of excitatory synapse organizer cerebellin precursor 1 (Cbln1), associated with sociability in mice. Restoration of Cbln1 in midbrain ventral tegmental glutamatergic neurons alleviates Ube3a loss-linked sociability deficits [80]. It is possible that UBE3A acts to maintain proteostatic surveillance locally at single synapses but also globally regulate neuronal physiology by operating at the euchromatin-rich domains [56].
In line with this, UBE3A suppresses neuronal hyperexcitability by degradative ubiquitination of Ca2+- and voltage-dependent big potassium (BK) channels in human neurons and brain organoids. Inhibition of BK channels in AS mice alleviates the seizure susceptibility demonstrating the channelopathy as underlying cause of AS network dysfunction [81]. Moreover, UBE3A-mediated endocytosis of small conductance potassium channels (SKs), is critical for NMDAR activation ergo hippocampal LTP. Learning and memory deficits in AS mice are reversible by blocking SK2. This mechanism elucidates that SK channelopathy also contributes to cognitive aberrances and network dysfunction in AS [82].
14. Critical Developmental Window of UBE3A Expression
Neuron-restricted imprinting of UBE3A offers ways of intervention in AS mouse models, which have been useful to determine that pharmacological un-silencing of paternal allele in the adulthood is possible [54,55], yet proves ineffective in rehabilitating the cognitive abilities. Strikingly, rescuing anxiety, stereotypies and epilepsy in AS mice requires reinstatement of UBE3A during early development, but not juvenile or adult stages. This is in line with the findings that early embryonic deletion of Ube3a, but not its juvenile or adult loss, recapitulates all behavioral deficits of AS mice [83]. There seems to be no requirement of the developmental window for UBE3A reinstatement for the restoration of hippocampal LTP [84], or excitation/inhibition balance identified in mouse prefrontal cortex in AS mouse model [85]. GABAergic but not glutamatergic loss of Ube3a is responsible for perturbed excitation/inhibition imbalance with enhanced seizure susceptibility, signifying inhibitory neuron-specific loss of Ube3a as a principal reason for hyperactive neuronal circuits in AS mouse model [86].
15. Neuronal Phenotypes and Physiological Relevance of UBE3A Gain-of-Function
As mentioned before, maternal copy number gains of 15q11.2–q13.3 lead to Dup15q syndrome, characterized by intellectual disability, developmental delay, muscle hypotonia and speech impairment. Dup15q represents the most penetrant chromosomal anomaly in ASD patients accounting for 1–3% cases worldwide. UBE3A belongs to approximately 40 genes in that chromosomal region and its gain-of-function is thought to represent the molecular cause in Dup15q patients [87]. Indeed, transgenic mice, overexpressing isoform 2 of UBE3A in the glutamatergic neurons of the forebrain exhibited behavioral traits relevant to ASD [88]. Increased dosage of UBE3A in primary neuronal cultures or in the Ube3a ASD mouse model abrogates dendritic growth and synapse number in a mechanism involving caspase-3 activation and tubulin cleavage [89]. UBE3A has also been identified as a negative regulator of retinoic acid synthesis. Importantly, in animals with hyperactivation of UBE3A in the prefrontal cortex, retinoic acid supplements alleviated ASD-like phenotypes [90].
16. The Case of Forward Genetics: Kaufman Oculocerebrofacial Syndrome (KOS) and UBE3B Gene Discovery
In 1971, Robert Kaufman described four of seven siblings, who exhibited severe mental retardation with absent speech, characteristic facial dysmorphisms with blepharophimosis, aberrances of the eye, mongoloid slant of the palpebral fissures and small mandible. Already then, an autosomal recessive mode of disease inheritance seemed the most likely [91], later confirmed formally. Since then, cases of KOS, previously also known as blepharophimosis-ptosis-intellectual-disability syndrome, have been reported worldwide (OMIM #244450). Distinct features of KOS include face and eye malformations, microcephaly, severe psychomotor retardation, and growth arrest. Additionally, patients often display serum hypocholesterolemia, indicative of downregulation of cholesterol synthesis. Neurologically, hypoplastic or aplastic corpus callosum and anterior commissure, Chiari type I malformation, smaller pituitary gland, and seizures are found in some cases. KOS patients suffer from severe ID with absent speech [36,92].
Initially, two consecutive reports have revealed loss-of-function mutations in UBE3B, which inherited in autosomal recessive manner, are causative for KOS [36,93]. Since then, more KOS cases and novel mutations have been described, some of them resulting in frame shift and premature translation stop, but some of them leading to a single amino acid substitutions [92,94].
UBE3B gene was discovered as a gene upregulated after acoustic trauma in the chick basal papilla. Its expression dramatically increased in the lesion in chick inner ear, but not in undamaged regions [95]. Human UBE3B generates two splice isoforms, one devoid of the HECT domain, presumptively non-functional [96]. Notably, UBE3B has also been identified in a locus for a delayed-onset, progressive, high-frequency, nonsyndromic sensorineural hearing loss [97]. Some of the KOS patients also display hearing impairment [92]. Further research on UBE3B should address its possible molecular roles in the inner ear.
17. UBE3B in Non-KOS Diseases
Overall, UBE3B is highly expressed in the central nervous system, digestive tract, respiratory system, as well as in multiple cell lineages of skin and other soft tissues. Severe developmental delay in KOS patients and multi-organ defects signify the involvement of UBE3B in systemic homeostasis. Similarly to KOS, G>A polymorphism in cattle resulting in exon skipping and in-frame deletion of a portion of catalytic HECT domain is associated with ptosis, intellectual disability, growth retardation and increased mortality [98]. Besides KOS, UBE3B mutations have been identified is ASD patients [99]. Additionally, possible involvement in pathophysiology of schizophrenia onset further supports the pivotal role of UBE3B in formation of cortical circuits [100].
18. Molecular Context of UBE3B Actions in Neurons
Soon after UBE3B loss-of-function was discovered causative of KOS [36,93], the quest for unveiling its cellular and molecular roles, particularly in the central nervous system, began. Initial report on UBE3B explores its functions as an enzyme in the cell and proves its E3 ubiquitin ligase activity. Point mutation of the C-terminal catalytic cysteine or loss of the catalytic HECT domain leads to abrogation of the ubiquitin binding, indicative of the loss of enzymatic activity. As predicted from the domain organization, N-terminal IQ motif of UBE3B also interacts with calmodulin, and this interaction results in reduction of the ligase ubiquitination activity. Accordingly, deleting the IQ domain follows with an increase in the enzymatic activity of UBE3B, possibly in a mechanism involving a removal of self-inhibitory state due to the N-terminus sequestration of the HECT domain in a steady autoinhibited state, characteristic for some HECT-type ligases [101]. Contrary to the described role of the IQ domain as a calcium-independent calmodulin interaction site [102], variation of intracellular calcium seems to modulate calmodulin-UBE3B interaction, ergo UBE3B activity. How exactly calcium contributes to regulation of UBE3B function remains to be investigated. Regarding cellular physiology, knock-down (KD) of UBE3B in cells engenders mitotic arrest of cells and reduced cell survival associated aberrant punctate mitochondrial morphology and altered physiology [103].
The findings on UBE3B-mediated mitochondrial function and intracellular Ca2+ are especially relevant in the light of UBE3B implications in the stress response and cellular viability. In the first paper identifying mutations in UBE3B as a cause of KOS, researchers demonstrate that C. elegans ortholog of UBE3B, oxi-1 is a player in cellular protection under oxidative stress [36]. Moreover, siRNA-mediated UBE3B KD in human glioblastoma cells results in their sensitization to reactive oxygen species inducing agent, temozolomide [104]. Additionally, expression of IQ domain-devoid UBE3B, which displays higher enzymatic activity, in human cells induces apoptosis [103]. Given a reported higher enzymatic activity of this deletion mutant of UBE3B elsewhere, it would be worthwhile to test the link between its ubiquitination hyperactivity and cell death.
19. UBE3B as a Master Regulator of Metabolic Homeostasis
The next report on UBE3B function focuses on its role in the metabolic pathways and links it to the development of the central nervous system. In their previous report, this group of researchers first identified UBE3B as a candidate ASD gene in a whole-exome sequencing and homozygosity analysis of 16 probands. Additionally, Ube3b mRNA level increased in E16.5 mouse cortical neurons at DIV6 (day-in-vitro 6) after KCl-induced hyperpolarization, again linking UBE3B, neuronal excitability and Ca2+ [99]. Next, the authors follow up on the ligase and its role in metabolic homeostasis and nervous system using a full KO mouse model. Loss of Ube3b in mice leads to severe developmental defects including anatomical, behavioral, and metabolic disturbances, including growth retardation, hypoplasia of corpus callosum, enlarged ventricles and decreased cortical thickness. Using Golgi–Cox labeling, authors show that cortical neurons in Ube3b−/− mice display reduced dendritic branching and spine density. Using a cell line stably expressing tagged UBE3B, affinity chromatography and mass spectrometry, authors identify branched-chain α-ketoacid dehydrogenase kinase (BCKDK) as a putative UBE3B target, further validated in ubiquitination assays. BCKDK level was also increased in Ube3b−/− cortex, liver and skeletal muscle, indicative of a specific pathway for UBE3B-mediated BCKDK degradation. Consequently, authors demonstrate that metabolite profiles of Ube3b−/− mice display signs of severe metabolic perturbations, with high levels of, among others, plasma spermidine, and S-adenosylhomocysteine, and cortical urea. Authors also show that patients bearing compound heterozygous mutations in UBE3B display similar metabolic perturbations, as identified in mice. Mitochondrial physiology, as demonstrated before in a cell line, is affected in skeletal muscle of Ube3b−/−, but not in the liver. That is particularly interesting, given some indications of UBE3B indispensability for skeletal muscle function [105]. Behavioral phenotypes in the Ube3b−/− mice include loss of vocalizations, decreased grip strength and decreased grooming and nesting. The interpretation of behavioral changes is, however, not straightforward, as severe developmental and anatomical defects, likely account for poor sensorimotor performance of the Ube3b−/− animals and occlude the behavioral readouts of Ube3b-associated neuronal deficit-linked changes. Despite these limitations, full Ube3b KO mice are a model of human systemic KOS, a metabolic disease strongly affecting the skeletal and nervous system [106].
20. Neuronal Roles of UBE3B
Our most recent work on the murine ortholog of UBE3B addresses specifically its cell-autonomous role in developing nervous system and involvement in formation of neuronal circuits. To circumvent the syndromic lethality and the overt non-neuronal phenotypes in the Ube3b−/− mice, Emx1-driven, forebrain neuron and glia-specific mouse model was established, given a high expression of Ube3b in the cortex and the hippocampus. Ube3b associates with postsynaptic density fractions and regulates dendritic branching in a cell-autonomous manner. Conditional deletion of Ube3b in the forebrain is associated with decreased cortical volume, due to reduction of dendritic trees, and thinning of corpus callosum. Neurons and glia-restricted loss of Ube3b leads to increased spine density, a phenotype reversible by re-introducing Ube3b by in vivo transfection. Increased density of spines is in line with increased frequency of miniature excitatory postsynaptic potentials (mEPSPs). Regarding neuronal physiology, Ube3b maintains the ratio of NMDA to AMPA at the synapse and abundance of NR2A. Forebrain-restricted loss of Ube3b leads to altered activity of hippocampal circuit and induces complex behavioral changes. These include specific hippocampal circuitry-associated loss of spatial memory and increased social memory. Moreover, Ube3b cKO male mice exhibit a behavioral switch towards self-directed actions (like grooming and eating) at the expense of exteroceptive behaviors, such as exploration. On a molecular level, with regard to its synaptogenic function, Ube3b ubiquitinates γ-subunit of calcineurin, Ppp3cc and regulates its levels at the synapse [26]. Among the putative UBE3B interactors identified by affinity chromatography were regulatory subunits of protein phosphatases, as well [106]. Future research should determine how Ube3b regulates the activity of the phosphatases during neuronal development. On a biochemical level, Ube3b catalyzes formation of K48- as well as of K63-linked polyUb chains.
21. Ube3b Conditional and Conventional KO Mouse Models: Lessons Learned
The case of Ube3b and the abovementioned two reports reopens a discussion on the applicability of genetic mouse models and conclusions drawn based on neurodevelopmental phenotypes. While the Ube3b−/− mouse model serves to study human KOS, it is not clear if the brain phenotypes described for the full KO animals are due to a loss of Ube3b or constitute secondary effects of systemic dyshomeostasis and/or other endophenotypes. The forebrain-specific Ube3b cKO does not serve as a comprehensive model of KOS, since it restricts the loss of the gene to excitatory neurons and glia, but offers a possibility to determine the cell autonomous roles of Ube3b in the cerebral cortex and the hippocampus. Notably, of the most significantly increased metabolites in Ube3b−/− was urea, whose elevated levels are associated with brain inflammation, simplified dendritic arbors and reduced spine density in a mouse model of kidney failure [107]. Further research should determine the kidney function in Ube3b−/− mice and address the kidney-brain axis regarding the systemic influence of uremic metabolites on the developing neurons. It is tempting to speculate that loss of spines and synapses in Ube3b−/− mice is due to a non-cell autonomous mechanism. This should be verified in experiments addressing the effect of neuronal exposure to metabolites identified in plasma and cortex of Ube3b−/− mice on spine density. Additionally, future experiments should address the effects of calcineurin and/or other protein phosphatase inhibition on synapse number both in Ube3b conditional and conventional mouse models. This is particularly thought-provoking regarding the UBE3A-driven regulation of PP2A activity and small molecule-mediated rescue of AS-like phenotypes in Ube3am−/p+ mice.
According to the published works, Ube3b mRNA is depolarization-induced and calmodulin, a Ca2+-binding messenger, inhibits the enzymatic activity of Ube3b. On the other hand, Ube3b ubiquitinates and/ergo regulates the level of the γ-subunit of calcineurin, Ca2+-, and calmodulin-dependent serine/threonine protein phosphatase. Taken together, neuronal depolarization and Ca2+ are important regulators of Ube3b enzymatic activity in the developing neocortex, and in turn, the enzymatic activity of Ube3b controls the cellular responders to Ca2+ (Figure 4, Table 2).
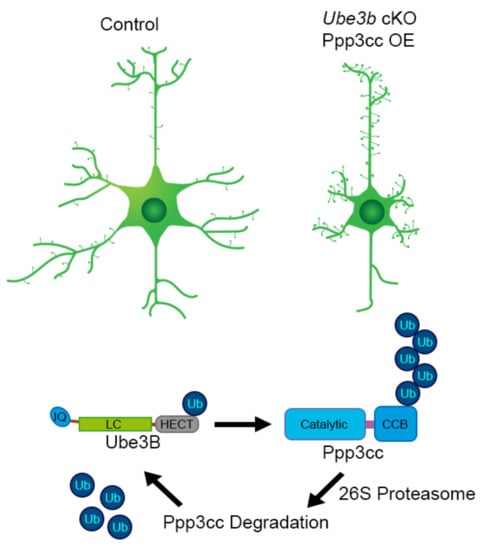
Figure 4.
Ube3b-mediated Ppp3cc regulation. In the forebrain-specific Ube3b cKO mouse, cortical and hippocampal neurons exhibit simpler, less arborized dendritic trees. This phenotype is mimicked, once neurons overexpress gamma-subunit of calcineurin, Ppp3cc, ubiquitination substrate of Ube3b. LC, low complexity; CBB, calcineurin B and calmodulin binding. PolyUb chain on position on Ppp3cc is randomly placed on the scheme to depict Ube3b-mediated ubiquitination.

Table 2.
Ubiquitination substrates of Ube3b and Ube3c.
22. Biological Functions and Clinical Importance of UBE3C: More Is Yet to Come
Of the three UBE3 ligases, neuronal roles of UBE3C remain uncharted. In the literature, UBE3C also exists as RTA-associated ubiquitin ligase (RAUL), or KIAA10. Since its identification through cDNA sequencing in myeloid cells [114], relatively little research concerning its role and function has been conducted. However, transcriptomic and proteomic approaches show that UBE3C is expressed throughout the body and enriched in skeletal muscles and the brain [115], where it interacts with human postsynaptic density [116] and such interaction undergoes dynamic scaling during sleep-wake cycle [117]. These findings are particularly relevant in the light of point mutations in UBE3C, resulting in single amino acid substitution in the catalytic domain region, described in ASD patients [39,40] (Table 1). Future studies should explore potential local UBE3C-mediated ubiquitination at the excitatory synapse. As is the case for UBE3A, UBE3C has been found in the nucleus [118]. Future research should explore a potential role of UBE3C in transcriptional control.
23. UBE3C in Human Disease
The involvement of UBE3C in various types of cancer has been most commonly reported. In many cases, upregulated UBE3C promotes the progression of the disease by tumorigenic and metastatic effects, associated with an increase in cellular migration and proliferation [110,119,120,121,122]. Additional populational genetic studies have associated UBE3C with disorders such as asthma [123], stroke [124], major depressive disorder [125,126], cocaine addiction [125], Parkinson’s [127], and Down’s syndrome [128].
24. UBE3C as a Proteasomal Regulator
UBE3C seems to have a distinct biological role as an E3 ligase that directly influences the cellular proteolytic machinery. Firstly, UBE3C and a DUB Usp14 continuously cycle on and off 26S proteasomes in ubiquitinated substrates-dependent manner and regulate cellular proteostasis [129]. UBE3C has also been described as a positive regulator of proteasomal processivity by propelling efficient proteolysis of accumulating partially degraded protein fragments [130]. Additionally, UBE3C-mediated ubiquitination of a proteasome subunit Rpn13 constitutes an autoinhibitory mechanism to prevent the attachment of Ub conjugates with stalled proteasomes [112]. Interaction with proteasomes and regulation of its physiology seems to be a common feature of UBE3A and UBE3C. Among other canonical degradation substrates of UBE3C are mostly transcriptional regulators (Table 2).
25. UBE3C Generates K29-Linked Ub Chains
Aside from its well-established role in proteolysis involving its inherent ability to conjugate K48-linked polyUb [131], UBE3C forms non-canonical K11- and K29-linked chains [27]. The role of these modification types has been poorly understood. It seems that K29-linked Ubs are found in heterotypical branched chains [28], implicating UBE3C as an important regulator of Ub chain morphology. Research on non-canonical Ub linkages is hampered by sparsity of available tools, however, K29 ubiquitination serves as a negative regulator of Wnt/β-Catenin [132], mediates viral infection, innate antiviral response [133,134], and is required for ER-mediated degradation. All of these processes are also associated with the function of UBE3C [113,122,135]. Considering a rather strong evidence of UBE3C involvement in the pathology of several human neurological diseases, future research should elucidate the cellular and molecular outcomes of UBE3C enzymatic activity and K29 ubiquitination in developing cortex.
26. Novel Concepts and Concluding Remarks
Despite classical grouping into one family, UBE3 ligases display fundamentally different characteristics, supported by the phylogenetic, biochemical, physiological, and clinical evidence. Each of UBE3 constitutes a separate biochemical entity, with specific ubiquitination pathways that they supervise, necessary for neuronal development. Based on the recent evidence, UBE3A has been shown to operate in the nucleus in an isoform-specific manner, where it globally regulates neuronal homeostasis. Other isoforms of UBE3A seem to have opposing effects in terms of their morphoregulatory and synaptogenic potential, and might operate locally in neurons, some of them not necessarily as enzymes. On top of this, UBE3A seems to modulate cellular proteolytic machinery. UBE3B associates with synaptic fractions, possibly forming a bigger protein complex, involving calcineurin. It would be interesting to test if its metabolic role is achieved in a global manner, by operating in different cellular compartments, similarly to UBE3A. Proteasome function depends on the association with ASD-linked UBE3C, able to assemble non-canonical polyUb chains. It would be extremely important to address developmental roles of not solely the ligase, but also of proteasome, its activity and non-degradative ubiquitination in ASD mouse models.
Future research will bring answers not only in terms of detailed molecular context of their action, but also regarding the biochemistry of non-conventional ubiquitination and its cellular and physiological consequences. This will aid clinical trial design for neurological and neurodegenerative diseases and cancer and shed light onto the complex machinery at play in the development and homeostasis of neuronal circuits.
Author Contributions
M.C.A. and K.J.C. wrote the manuscript, D.H. analyzed the expression dynamics of Ube3s and H.K., V.T. edited the draft. All authors have read and agreed to the published version of the manuscript.
Funding
The work in the lab is supported by the Russian Scientific Foundation (grant number 19-14-00345, VT), JSPS KAKENHI (grant number JP20K07334, HK), Ohsumi Frontier Science Foundation (HK), The Uehara Memorial Foundation (HK), and Takeda Science Foundation (HK).
Acknowledgments
We thank the authors of the publication 3 for the supplementary material that we used to plot the expression trajectories of Ube3 ligase genes. We would like to thank Nils Brose for his guidance during the work on ubiquitin and for his unyielding support throughout. We thank Theres Schaub for her helpful comments during editing.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Rakic, P. Evolution of the neocortex: A perspective from developmental biology. Nat. Rev. Neurosci. 2009, 10, 724–735. [Google Scholar] [CrossRef] [PubMed]
- Nowakowski, T.J.; Bhaduri, A.; Pollen, A.A.; Alvarado, B.; Mostajo-Radji, M.A.; Di Lullo, E.; Haeussler, M.; Sandoval-Espinosa, C.; Liu, S.J.; Velmeshev, D.; et al. Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science 2017, 358, 1318–1323. [Google Scholar] [CrossRef] [PubMed]
- Telley, L.; Agirman, G.; Prados, J.; Amberg, N.; Fièvre, S.; Oberst, P.; Bartolini, G.; Vitali, I.; Cadilhac, C.; Hippenmeyer, S.; et al. Temporal patterning of apical progenitors and their daughter neurons in the developing neocortex. Science 2019, 364. [Google Scholar] [CrossRef] [PubMed]
- Dierssen, M.; Ramakers, G.J.A. Dendritic pathology in mental retardation: From molecular genetics to neurobiology. Genes Brain Behav. 2006, 5 (Suppl. 2), 48–60. [Google Scholar] [CrossRef] [PubMed]
- Bliss, T.V.; Lomo, T. Long-lasting potentiation of synaptic transmission in the dentate area of the anaesthetized rabbit following stimulation of the perforant path. J. Physiol. (Lond.) 1973, 232, 331–356. [Google Scholar] [CrossRef]
- Griffiths, B.J.; Parish, G.; Roux, F.; Michelmann, S.; van der Plas, M.; Kolibius, L.D.; Chelvarajah, R.; Rollings, D.T.; Sawlani, V.; Hamer, H.; et al. Directional coupling of slow and fast hippocampal gamma with neocortical alpha/beta oscillations in human episodic memory. Proc. Natl. Acad. Sci. USA 2019, 116, 21834–21842. [Google Scholar] [CrossRef] [PubMed]
- Buzsáki, G.; Draguhn, A. Neuronal oscillations in cortical networks. Science 2004, 304, 1926–1929. [Google Scholar] [CrossRef]
- Masi, A.; DeMayo, M.M.; Glozier, N.; Guastella, A.J. An Overview of Autism Spectrum Disorder, Heterogeneity and Treatment Options. Neurosci Bull. 2017, 33, 183–193. [Google Scholar] [CrossRef]
- Ambrozkiewicz, M.C.; Kawabe, H. HECT-type E3 ubiquitin ligases in nerve cell development and synapse physiology. Febs Lett. 2015, 589, 1635–1643. [Google Scholar] [CrossRef]
- Kawabe, H.; Brose, N. The role of ubiquitylation in nerve cell development. Nat. Rev. Neurosci. 2011, 12, 251–268. [Google Scholar] [CrossRef]
- Goldstein, G.; Scheid, M.; Hammerling, U.; Schlesinger, D.H.; Niall, H.D.; Boyse, E.A. Isolation of a polypeptide that has lymphocyte-differentiating properties and is probably represented universally in living cells. Proc. Natl. Acad. Sci. USA 1975, 72, 11–15. [Google Scholar] [CrossRef] [PubMed]
- Hershko, A.; Ciechanover, A. THE UBIQUITIN SYSTEM. Annu. Rev. Biochem. 1998, 67, 425–479. [Google Scholar] [CrossRef] [PubMed]
- Hochstrasser, M. Lingering Mysteries of Ubiquitin-Chain Assembly. Cell 2006, 124, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Hershko, A.; Heller, H.; Elias, S.; Ciechanover, A. Components of ubiquitin-protein ligase system. Resolution, affinity purification, and role in protein breakdown. J. Biol. Chem. 1983, 258, 8206–8214. [Google Scholar]
- McDowell, G.S.; Philpott, A. Non-canonical ubiquitylation: Mechanisms and consequences. Int. J. Biochem. Cell Biol. 2013, 45, 1833–1842. [Google Scholar] [CrossRef]
- Kaiser, S.E.; Riley, B.E.; Shaler, T.A.; Trevino, R.S.; Becker, C.H.; Schulman, H.; Kopito, R.R. Protein standard absolute quantification (PSAQ) method for the measurement of cellular ubiquitin pools. Nat. Methods 2011, 8, 691–696. [Google Scholar] [CrossRef]
- McGouran, J.F.; Gaertner, S.R.; Altun, M.; Kramer, H.B.; Kessler, B.M. Deubiquitinating enzyme specificity for ubiquitin chain topology profiled by di-ubiquitin activity probes. Chem. Biol. 2013, 20, 1447–1455. [Google Scholar] [CrossRef]
- Kawabe, H.; Neeb, A.; Dimova, K.; Young, S.M.; Takeda, M.; Katsurabayashi, S.; Mitkovski, M.; Malakhova, O.A.; Zhang, D.-E.; Umikawa, M.; et al. Regulation of Rap2A by the ubiquitin ligase Nedd4-1 controls neurite development. Neuron 2010, 65, 358–372. [Google Scholar] [CrossRef]
- Hsia, H.-E.; Kumar, R.; Luca, R.; Takeda, M.; Courchet, J.; Nakashima, J.; Wu, S.; Goebbels, S.; An, W.; Eickholt, B.J.; et al. Ubiquitin E3 ligase Nedd4-1 acts as a downstream target of PI3K/PTEN-mTORC1 signaling to promote neurite growth. Proc. Natl. Acad. Sci. USA 2014, 111, 13205–13210. [Google Scholar] [CrossRef]
- Ambrozkiewicz, M.C.; Schwark, M.; Kishimoto-Suga, M.; Borisova, E.; Hori, K.; Salazar-Lázaro, A.; Rusanova, A.; Altas, B.; Piepkorn, L.; Bessa, P.; et al. Polarity Acquisition in Cortical Neurons Is Driven by Synergistic Action of Sox9-Regulated Wwp1 and Wwp2 E3 Ubiquitin Ligases and Intronic miR-140. Neuron 2018, 100, 1097–1115.e15. [Google Scholar] [CrossRef]
- George, A.J.; Hoffiz, Y.C.; Charles, A.J.; Zhu, Y.; Mabb, A.M. A Comprehensive Atlas of E3 Ubiquitin Ligase Mutations in Neurological Disorders. Front. Genet. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Hicke, L.; Riezman, H. Ubiquitination of a yeast plasma membrane receptor signals its ligand-stimulated endocytosis. Cell 1996, 84, 277–287. [Google Scholar] [CrossRef]
- Akutsu, M.; Dikic, I.; Bremm, A. Ubiquitin chain diversity at a glance. J. Cell Sci. 2016, 129, 875–880. [Google Scholar] [CrossRef] [PubMed]
- Komander, D.; Reyes-Turcu, F.; Licchesi, J.D.F.; Odenwaelder, P.; Wilkinson, K.D.; Barford, D. Molecular discrimination of structurally equivalent Lys 63-linked and linear polyubiquitin chains. Embo Rep. 2009, 10, 466–473. [Google Scholar] [CrossRef]
- Kim, H.C.; Huibregtse, J.M. Polyubiquitination by HECT E3s and the Determinants of Chain Type Specificity. Mol. Cell. Biol. 2009, 29, 3307–3318. [Google Scholar] [CrossRef]
- Ambrozkiewicz, M.C.; Borisova, E.; Schwark, M.; Ripamonti, S.; Schaub, T.; Smorodchenko, A.; Weber, A.I.; Rhee, H.J.; Altas, B.; Yilmaz, R.; et al. The murine ortholog of Kaufman oculocerebrofacial syndrome protein Ube3b regulates synapse number by ubiquitinating Ppp3cc. Mol. Psychiatry 2020. [Google Scholar] [CrossRef]
- Michel, M.A.; Elliott, P.R.; Swatek, K.N.; Simicek, M.; Pruneda, J.N.; Wagstaff, J.L.; Freund, S.M.V.; Komander, D. Assembly and specific recognition of k29- and k33-linked polyubiquitin. Mol. Cell 2015, 58, 95–109. [Google Scholar] [CrossRef]
- Kristariyanto, Y.A.; Abdul Rehman, S.A.; Campbell, D.G.; Morrice, N.A.; Johnson, C.; Toth, R.; Kulathu, Y. K29-Selective Ubiquitin Binding Domain Reveals Structural Basis of Specificity and Heterotypic Nature of K29 Polyubiquitin. Mol. Cell 2015, 58, 83–94. [Google Scholar] [CrossRef]
- Huang, L.; Kinnucan, E.; Wang, G.; Beaudenon, S.; Howley, P.M.; Huibregtse, J.M.; Pavletich, N.P. Structure of an E6AP-UbcH7 complex: Insights into ubiquitination by the E2-E3 enzyme cascade. Science (N. Y.) 1999, 286, 1321–1326. [Google Scholar] [CrossRef]
- Kim, H.C.; Steffen, A.M.; Oldham, M.L.; Chen, J.; Huibregtse, J.M. Structure and function of a HECT domain ubiquitin-binding site. Embo Rep. 2011, 12, 334–341. [Google Scholar] [CrossRef]
- Ries, L.K.; Liess, A.K.L.; Feiler, C.G.; Spratt, D.E.; Lowe, E.D.; Lorenz, S. Crystal structure of the catalytic C-lobe of the HECT-type ubiquitin ligase E6AP. Protein Sci. A Publ. Protein Soc. 2020, 29, 1550–1554. [Google Scholar] [CrossRef]
- Ronchi, V.P.; Klein, J.M.; Haas, A.L. E6AP/UBE3A ubiquitin ligase harbors two E2~ubiquitin binding sites. J. Biol. Chem. 2013, 288, 10349–10360. [Google Scholar] [CrossRef]
- Ronchi, V.P.; Klein, J.M.; Edwards, D.J.; Haas, A.L. The active form of E6-associated protein (E6AP)/UBE3A ubiquitin ligase is an oligomer. J. Biol. Chem. 2014, 289, 1033–1048. [Google Scholar] [CrossRef]
- Beasley, S.A.; Kellum, C.E.; Orlomoski, R.J.; Idrizi, F.; Spratt, D.E. An Angelman syndrome substitution in the HECT E3 ubiquitin ligase C-terminal Lobe of E6AP affects protein stability and activity. PLoS ONE 2020, 15, e0235925. [Google Scholar] [CrossRef]
- Chan, A.-L.; Grossman, T.; Zuckerman, V.; Campigli Di Giammartino, D.; Moshel, O.; Scheffner, M.; Monahan, B.; Pilling, P.; Jiang, Y.-H.; Haupt, S.; et al. c-Abl phosphorylates E6AP and regulates its E3 ubiquitin ligase activity. Biochemistry 2013, 52, 3119–3129. [Google Scholar] [CrossRef]
- Basel-Vanagaite, L.; Dallapiccola, B.; Ramirez-Solis, R.; Segref, A.; Thiele, H.; Edwards, A.; Arends, M.J.; Miró, X.; White, J.K.; Désir, J.; et al. Deficiency for the Ubiquitin Ligase UBE3B in a Blepharophimosis-Ptosis-Intellectual-Disability Syndrome. Am. J. Hum. Genet. 2012, 91, 998–1010. [Google Scholar] [CrossRef]
- Zhang, C.; Milunsky, J.M.; Newton, S.; Ko, J.; Zhao, G.; Maher, T.A.; Tager-Flusberg, H.; Bolliger, M.F.; Carter, A.S.; Boucard, A.A.; et al. A Neuroligin-4 Missense Mutation Associated with Autism Impairs Neuroligin-4 Folding and Endoplasmic Reticulum Export. J. Neurosci. 2009, 29, 10843–10854. [Google Scholar] [CrossRef]
- Singh, S.; Sivaraman, J. Crystal Structure of HECT Domain of UBE3C E3 Ligase and its Ubiquitination Activity. Biochem. J. 2020, 477, 905–923. [Google Scholar] [CrossRef]
- O’Roak, B.J.; Vives, L.; Girirajan, S.; Karakoc, E.; Krummm, N.; Coe, B.P.; Levy, R.; Ko, A.; Lee, C.; Smith, J.D.; et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature 2012, 485, 246–250. [Google Scholar] [CrossRef]
- Yuen, R.K.C.; Merico, D.; Cao, H.; Pellecchia, G.; Alipanahi, B.; Thiruvahindrapuram, B.; Tong, X.; Sun, Y.; Cao, D.; Zhang, T.; et al. Genome-wide characteristics of de novo mutations in autism. NPJ Genom. Med. 2016, 1. [Google Scholar] [CrossRef]
- Haynes, C.; Oldfield, C.J.; Ji, F.; Klitgord, N.; Cusick, M.E.; Radivojac, P.; Uversky, V.N.; Vidal, M.; Iakoucheva, L.M. Intrinsic disorder is a common feature of hub proteins from four eukaryotic interactomes. PLoS Comput. Biol. 2006, 2, e100. [Google Scholar] [CrossRef]
- Sato, M. Early origin and evolution of the angelman syndrome ubiquitin ligase gene Ube3a. Front. Cell. Neurosci. 2017, 11, 1–8. [Google Scholar] [CrossRef]
- Marín, I. Animal HECT ubiquitin ligases: Evolution and functional implications. BMC Evol. Biol. 2010, 10, 56. [Google Scholar] [CrossRef]
- Grau-Bové, X.; Sebé-Pedrós, A.; Ruiz-Trillo, I. A genomic survey of HECT ubiquitin ligases in eukaryotes reveals independent expansions of the HECT system in several lineages. Genome Biol. Evol. 2013, 5, 833–847. [Google Scholar] [CrossRef]
- Sadikovic, B.; Fernandes, P.; Zhang, V.W.; Ward, P.A.; Miloslavskaya, I.; Rhead, W.; Rosenbaum, R.; Gin, R.; Roa, B.; Fang, P. Mutation Update for UBE3A variants in Angelman syndrome. Hum. Mutat. 2014, 35, 1407–1417. [Google Scholar] [CrossRef]
- LaSalle, J.M.; Reiter, L.T.; Chamberlain, S.J. Epigenetic regulation of UBE3A and roles in human neurodevelopmental disorders. Epigenomics 2015, 7, 1213–1228. [Google Scholar] [CrossRef]
- Burette, A.C.; Judson, M.C.; Li, A.N.; Chang, E.F.; Seeley, W.W.; Philpot, B.D.; Weinberg, R.J. Subcellular organization of UBE3A in human cerebral cortex. Mol. Autism 2018, 9, 54. [Google Scholar] [CrossRef]
- Greer, P.L.; Hanayama, R.; Bloodgood, B.L.; Mardinly, A.R.; Lipton, D.M.; Flavell, S.W.; Kim, T.-K.; Griffith, E.C.; Waldon, Z.; Maehr, R.; et al. The Angelman Syndrome Protein Ube3A Regulates Synapse Development by Ubiquitinating Arc. Cell 2010, 140, 704–716. [Google Scholar] [CrossRef]
- Rougeulle, C.; Glatt, H.; Lalande, M. The Angelman syndrome candidate gene, UBE3A/E6-AP, is imprinted in brain. Nat. Genet. 1997, 17, 14–15. [Google Scholar] [CrossRef]
- Vu, T.H.; Hoffman, A.R. Imprinting of the Angelman syndrome gene, UBE3A, is restricted to brain. Nat. Genet. 1997, 17, 12–13. [Google Scholar] [CrossRef]
- Meng, L.; Person, R.E.; Beaudet, A.L. Ube3a-ATS is an atypical RNA polymerase II transcript that represses the paternal expression of Ube3a. Hum. Mol. Genet. 2012, 21, 3001–3012. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, K.; Joh, K.; Ohta, T.; Masuzaki, H.; Ishimaru, T.; Mukai, T.; Niikawa, N.; Ogawa, M.; Wagstaff, J.; Kishino, T. Neurons but not glial cells show reciprocal imprinting of sense and antisense transcripts of Ube3a. Hum. Mol. Genet. 2003, 12, 837–847. [Google Scholar] [CrossRef] [PubMed]
- Hsiao, J.S.; Germain, N.D.; Wilderman, A.; Stoddard, C.; Wojenski, L.A.; Villafano, G.J.; Core, L.; Cotney, J.; Chamberlain, S.J. A bipartite boundary element restricts UBE3A imprinting to mature neurons. Proc. Natl. Acad. Sci. USA 2019, 116, 2181–2186. [Google Scholar] [CrossRef]
- Huang, H.-S.; Allen, J.A.; Mabb, A.M.; King, I.F.; Miriyala, J.; Taylor-Blake, B.; Sciaky, N.; Dutton, J.W.; Lee, H.-M.; Chen, X.; et al. Topoisomerase inhibitors unsilence the dormant allele of Ube3a in neurons. Nature 2011, 481, 185–189. [Google Scholar] [CrossRef]
- Meng, L.; Ward, A.J.; Chun, S.; Bennett, C.F.; Beaudet, A.L.; Rigo, F. Towards a therapy for Angelman syndrome by targeting a long non-coding RNA. Nature 2015, 518, 409–412. [Google Scholar] [CrossRef]
- Lopez, S.J.; Segal, D.J.; LaSalle, J.M. UBE3A: An E3 Ubiquitin Ligase With Genome-Wide Impact in Neurodevelopmental Disease. Front. Mol. Neurosci. 2019, 11. [Google Scholar] [CrossRef]
- Ramirez, J.; Lectez, B.; Osinalde, N.; Sivá, M.; Elu, N.; Aloria, K.; Procházková, M.; Perez, C.; Martínez-Hernández, J.; Barrio, R.; et al. Quantitative proteomics reveals neuronal ubiquitination of Rngo/Ddi1 and several proteasomal subunits by Ube3a, accounting for the complexity of Angelman syndrome. Hum. Mol. Genet. 2018, 27, 1955–1971. [Google Scholar] [CrossRef]
- Elu, N.; Osinalde, N.; Beaskoetxea, J.; Ramirez, J.; Lectez, B.; Aloria, K.; Rodriguez, J.A.; Arizmendi, J.M.; Mayor, U. Detailed Dissection of UBE3A-Mediated DDI1 Ubiquitination. Front. Physiol. 2019, 10, 534. [Google Scholar] [CrossRef]
- Tomaic, V.; Pim, D.; Thomas, M.; Massimi, P.; Myers, M.P.; Banks, L. Regulation of the Human Papillomavirus Type 18 E6/E6AP Ubiquitin Ligase Complex by the HECT Domain-Containing Protein EDD. J. Virol. 2011, 85, 3120–3127. [Google Scholar] [CrossRef]
- Kühnle, S.; Martínez-Noël, G.; Leclere, F.; Hayes, S.D.; Harper, J.W.; Howley, P.M. Angelman syndrome-associated point mutations in the Zn2+-binding N-terminal (AZUL) domain of UBE3A ubiquitin ligase inhibit binding to the proteasome. J. Biol. Chem. 2018, 293, 18387–18399. [Google Scholar] [CrossRef]
- Yi, J.J.; Paranjape, S.R.; Walker, M.P.; Choudhury, R.; Wolter, J.M.; Fragola, G.; Emanuele, M.J.; Major, M.B.; Zylka, M.J. The autism-linked UBE3A T485A mutant E3 ubiquitin ligase activates the Wnt/β-catenin pathway by inhibiting the proteasome. J. Biol. Chem. 2017, 292, 12503–12515. [Google Scholar] [CrossRef] [PubMed]
- Yi, J.J.; Berrios, J.; Newbern, J.M.; Snider, W.D.; Philpot, B.D.; Hahn, K.M.; Zylka, M.J. An Autism-Linked Mutation Disables Phosphorylation Control of UBE3A. Cell 2015, 162, 795–807. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.Y.; Ramirez, J.; Franco, M.; Lectez, B.; Gonzalez, M.; Barrio, R.; Mayor, U. Ube3a, the E3 ubiquitin ligase causing Angelman syndrome and linked to autism, regulates protein homeostasis through the proteasomal shuttle Rpn10. Cell Mol. Life Sci. 2014, 71, 2747–2758. [Google Scholar] [CrossRef] [PubMed]
- Llewellyn, K.J.; Nalbandian, A.; Gomez, A.; Wei, D.; Walker, N.; Kimonis, V.E. Administration of CoQ10 analogue ameliorates dysfunction of the mitochondrial respiratory chain in a mouse model of Angelman syndrome. Neurobiol. Dis. 2015, 76, 77–86. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Noël, G.; Luck, K.; Kühnle, S.; Desbuleux, A.; Szajner, P.; Galligan, J.T.; Rodriguez, D.; Zheng, L.; Boyland, K.; Leclere, F.; et al. Network Analysis of UBE3A/E6AP-Associated Proteins Provides Connections to Several Distinct Cellular Processes. J. Mol. Biol. 2018, 430, 1024–1050. [Google Scholar] [CrossRef]
- Kühnle, S.; Kogel, U.; Glockzin, S.; Marquardt, A.; Ciechanover, A.; Matentzoglu, K.; Scheffner, M. Physical and functional interaction of the HECT ubiquitin-protein ligases E6AP and HERC2. J. Biol. Chem. 2011, 286, 19410–19416. [Google Scholar] [CrossRef]
- Yamamoto, Y.; Huibregtse, J.M.; Howley, P.M. The human E6-AP gene (UBE3A) encodes three potential protein isoforms generated by differential splicing. Genomics 1997, 41, 263–266. [Google Scholar] [CrossRef]
- Miao, S.; Chen, R.; Ye, J.; Tan, G.-H.; Li, S.; Zhang, J.; Jiang, Y.; Xiong, Z.-Q. The Angelman syndrome protein Ube3a is required for polarized dendrite morphogenesis in pyramidal neurons. J. Neurosci. 2013, 33, 327–333. [Google Scholar] [CrossRef]
- Tonazzini, I.; Van Woerden, G.M.; Masciullo, C.; Mientjes, E.J.; Elgersma, Y.; Cecchini, M. The role of ubiquitin ligase E3A in polarized contact guidance and rescue strategies in UBE3A-deficient hippocampal neurons. Mol. Autism 2019, 10, 41. [Google Scholar] [CrossRef]
- Sirois, C.L.; Bloom, J.E.; Fink, J.J.; Gorka, D.; Keller, S.; Germain, N.D.; Levine, E.S.; Chamberlain, S.J. Abundance and localization of human UBE3A protein isoforms. Hum. Mol. Genet. 2020. [Google Scholar] [CrossRef]
- Valluy, J.; Bicker, S.; Aksoy-Aksel, A.; Lackinger, M.; Sumer, S.; Fiore, R.; Wüst, T.; Seffer, D.; Metge, F.; Dieterich, C.; et al. A coding-independent function of an alternative Ube3a transcript during neuronal development. Nat. Neurosci. 2015, 18, 666–673. [Google Scholar] [CrossRef] [PubMed]
- Sadhwani, A.; Sanjana, N.E.; Willen, J.M.; Calculator, S.N.; Black, E.D.; Bean, L.J.H.; Li, H.; Tan, W.-H. Two Angelman families with unusually advanced neurodevelopment carry a start codon variant in the most highly expressed UBE3A isoform. Am. J. Med Genet. A 2018, 176, 1641–1647. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Kunz, P.A.; Mooney, R.; Philpot, B.D.; Smith, S.L. Maternal Loss of Ube3a Impairs Experience-Driven Dendritic Spine Maintenance in the Developing Visual Cortex. J. Neurosci. Off. J. Soc. Neurosci. 2016, 36, 4888–4894. [Google Scholar] [CrossRef] [PubMed]
- Margolis, S.S.; Salogiannis, J.; Lipton, D.M.; Mandel-Brehm, C.; Wills, Z.P.; Mardinly, A.R.; Hu, L.; Greer, P.L.; Bikoff, J.B.; Ho, H.-Y.H.; et al. EphB-mediated degradation of the RhoA GEF Ephexin5 relieves a developmental brake on excitatory synapse formation. Cell 2010, 143, 442–455. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Lou, S.-S.; Wang, T.; Wu, R.-J.; Li, G.; Zhao, M.; Lu, B.; Li, Y.-Y.; Zhang, J.; Cheng, X.; et al. UBE3A-mediated PTPA ubiquitination and degradation regulate PP2A activity and dendritic spine morphology. Proc. Natl. Acad. Sci. USA 2019, 116, 12500–12505. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Liu, Y.; Jia, Y.; Hao, X.; Lin, W.J.; Tran, J.; Lynch, G.; Baudry, M.; Bi, X. UBE3A-mediated p18/LAMTOR1 ubiquitination and degradation regulate mTORC1 activity and synaptic plasticity. eLife 2018, 7. [Google Scholar] [CrossRef]
- Li, W.; Yao, A.; Zhi, H.; Kaur, K.; Zhu, Y.-C.; Jia, M.; Zhao, H.; Wang, Q.; Jin, S.; Zhao, G.; et al. Angelman Syndrome Protein Ube3a Regulates Synaptic Growth and Endocytosis by Inhibiting BMP Signaling in Drosophila. PLoS Genet. 2016, 12, e1006062. [Google Scholar] [CrossRef]
- Daniel, J.A.; Cooper, B.H.; Palvimo, J.J.; Zhang, F.-P.; Brose, N.; Tirard, M. Analysis of SUMO1-conjugation at synapses. eLife 2017, 6. [Google Scholar] [CrossRef]
- Avagliano Trezza, R.; Sonzogni, M.; Bossuyt, S.N.V.; Zampeta, F.I.; Punt, A.M.; van den Berg, M.; Rotaru, D.C.; Koene, L.M.C.; Munshi, S.T.; Stedehouder, J.; et al. Loss of nuclear UBE3A causes electrophysiological and behavioral deficits in mice and is associated with Angelman syndrome. Nat. Neurosci. 2019, 22, 1235–1247. [Google Scholar] [CrossRef]
- Krishnan, V.; Stoppel, D.C.; Nong, Y.; Johnson, M.A.; Nadler, M.J.S.; Ozkaynak, E.; Teng, B.L.; Nagakura, I.; Mohammad, F.; Silva, M.A.; et al. Autism gene Ube3a and seizures impair sociability by repressing VTA Cbln1. Nature 2017, 543, 507–512. [Google Scholar] [CrossRef]
- Sun, A.X.; Yuan, Q.; Fukuda, M.; Yu, W.; Yan, H.; Lim, G.G.Y.; Nai, M.H.; D’Agostino, G.A.; Tran, H.-D.; Itahana, Y.; et al. Potassium channel dysfunction in human neuronal models of Angelman syndrome. Science (N. Y.) 2019, 366, 1486–1492. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Zhu, G.; Liu, Y.; Standley, S.; Ji, A.; Tunuguntla, R.; Wang, Y.; Claus, C.; Luo, Y.; Baudry, M.; et al. UBE3A Regulates Synaptic Plasticity and Learning and Memory by Controlling SK2 Channel Endocytosis. Cell Rep. 2015, 12, 449–461. [Google Scholar] [CrossRef] [PubMed]
- Sonzogni, M.; Hakonen, J.; Bernabé Kleijn, M.; Silva-Santos, S.; Judson, M.C.; Philpot, B.D.; van Woerden, G.M.; Elgersma, Y. Delayed loss of UBE3A reduces the expression of Angelman syndrome-associated phenotypes. Mol. Autism 2019, 10, 23. [Google Scholar] [CrossRef] [PubMed]
- Silva-Santos, S.; van Woerden, G.M.; Bruinsma, C.F.; Mientjes, E.; Jolfaei, M.A.; Distel, B.; Kushner, S.A.; Elgersma, Y. Ube3a reinstatement identifies distinct developmental windows in a murine Angelman syndrome model. J. Clin. Investig. 2015, 125, 2069–2076. [Google Scholar] [CrossRef] [PubMed]
- Rotaru, D.C.; van Woerden, G.M.; Wallaard, I.; Elgersma, Y. Adult Ube3a Gene Reinstatement Restores the Electrophysiological Deficits of Prefrontal Cortex Layer 5 Neurons in a Mouse Model of Angelman Syndrome. J. Neurosci. 2018, 38, 8011–8030. [Google Scholar] [CrossRef]
- Judson, M.C.; Wallace, M.L.; Sidorov, M.S.; Burette, A.C.; Gu, B.; van Woerden, G.M.; King, I.F.; Han, J.E.; Zylka, M.J.; Elgersma, Y.; et al. GABAergic Neuron-Specific Loss of Ube3a Causes Angelman Syndrome-Like EEG Abnormalities and Enhances Seizure Susceptibility. Neuron 2016, 90, 56–69. [Google Scholar] [CrossRef]
- Noor, A.; Dupuis, L.; Mittal, K.; Lionel, A.C.; Marshall, C.R.; Scherer, S.W.; Stockley, T.; Vincent, J.B.; Mendoza-Londono, R.; Stavropoulos, D.J. 15q11.2 Duplication Encompassing Only the UBE3A Gene Is Associated with Developmental Delay and Neuropsychiatric Phenotypes. Hum. Mutat 2015, 36, 689–693. [Google Scholar] [CrossRef]
- Copping, N.A.; Christian, S.G.B.; Ritter, D.J.; Islam, M.S.; Buscher, N.; Zolkowska, D.; Pride, M.C.; Berg, E.L.; LaSalle, J.M.; Ellegood, J.; et al. Neuronal overexpression of Ube3a isoform 2 causes behavioral impairments and neuroanatomical pathology relevant to 15q11.2-q13.3 duplication syndrome. Hum. Mol. Genet. 2017, 26, 3995–4010. [Google Scholar] [CrossRef]
- Khatri, N.; Gilbert, J.P.; Huo, Y.; Sharaflari, R.; Nee, M.; Qiao, H.; Man, H.-Y. The Autism Protein Ube3A/E6AP Remodels Neuronal Dendritic Arborization via Caspase-Dependent Microtubule Destabilization. J. Neurosci. 2018, 38, 363–378. [Google Scholar] [CrossRef]
- Xu, X.; Li, C.; Gao, X.; Xia, K.; Guo, H.; Li, Y.; Hao, Z.; Zhang, L.; Gao, D.; Xu, C.; et al. Excessive UBE3A dosage impairs retinoic acid signaling and synaptic plasticity in autism spectrum disorders. Cell Res. 2018, 28, 48–68. [Google Scholar] [CrossRef]
- Kaufman, R.L.; Rimoin, D.L.; Prensky, A.L.; Sly, W.S. An oculocerebrofacial syndrome. Birth Defects Orig. Artic. Ser. 1971, 7, 135–138. [Google Scholar] [PubMed]
- Basel-Vanagaite, L.; Yilmaz, R.; Tang, S.; Reuter, M.S.; Rahner, N.; Grange, D.K.; Mortenson, M.; Koty, P.; Feenstra, H.; Farwell Gonzalez, K.D.; et al. Expanding the clinical and mutational spectrum of Kaufman oculocerebrofacial syndrome with biallelic UBE3B mutations. Hum. Genet. 2014, 133, 939–949. [Google Scholar] [CrossRef] [PubMed]
- Flex, E.; Ciolfi, A.; Caputo, V.; Fodale, V.; Leoni, C.; Melis, D.; Bedeschi, M.F.; Mazzanti, L.; Pizzuti, A.; Tartaglia, M.; et al. Loss of function of the E3 ubiquitin-protein ligase UBE3B causes Kaufman oculocerebrofacial syndrome. J. Med. Genet. 2013, 50, 493–499. [Google Scholar] [CrossRef] [PubMed]
- Zaki, M.S.; Otaify, G.A.; Ismail, S.; Issa, M.Y.; El-Ruby, M.O.; Sadek, A.A.; Ashaat, E.A.; El Saeidi, S.A.; Aglan, M.S.; Temtamy, S.; et al. Blepharophimosis-ptosis-intellectual disability syndrome: A report of nine Egyptian patients with further expansion of phenotypic and mutational spectrum. Am. J. Med. Genet. A 2020. [Google Scholar] [CrossRef] [PubMed]
- Lomax, M.I.; Gong, T.-W.; Cho, Y.; Huang, L.; Oh, S.-H.; Adler, H.J.; Raphael, Y.; Altschuler, R.A. Differential Gene Expression Following Noise Trauma in Birds and Mammals. Noise Health 2001, 3, 19–35. [Google Scholar] [PubMed]
- Gong, T.-W.L.; Huang, L.; Warner, S.J.; Lomax, M.I. Characterization of the human UBE3B gene: Structure, expression, evolution, and alternative splicing. Genomics 2003, 82, 143–152. [Google Scholar] [CrossRef]
- Greene, C.C.; McMillan, P.M.; Barker, S.E.; Kurnool, P.; Lomax, M.I.; Burmeister, M.; Lesperance, M.M. DFNA25, a novel locus for dominant nonsyndromic hereditary hearing impairment, maps to 12q21-24. Am. J. Hum. Genet. 2001, 68, 254–260. [Google Scholar] [CrossRef]
- Venhoranta, H.; Pausch, H.; Flisikowski, K.; Wurmser, C.; Taponen, J.; Rautala, H.; Kind, A.; Schnieke, A.; Fries, R.; Lohi, H.; et al. In frame exon skipping in UBE3B is associated with developmental disorders and increased mortality in cattle. BMC Genom. 2014, 15, 890. [Google Scholar] [CrossRef]
- Chahrour, M.H.; Yu, T.W.; Lim, E.T.; Ataman, B.; Coulter, M.E.; Hill, R.S.; Stevens, C.R.; Schubert, C.R.; Greenberg, M.E.; ARRA Autism Sequencing Collaboration; et al. Whole-Exome Sequencing and Homozygosity Analysis Implicate Depolarization-Regulated Neuronal Genes in Autism. PLoS Genet. 2012, 8, e1002635. [Google Scholar] [CrossRef]
- Kohlbrenner, E.A.; Shaskan, N.; Pietersen, C.Y.; Sonntag, K.-C.; Woo, T.-U.W. Gene expression profile associated with postnatal development of pyramidal neurons in the human prefrontal cortex implicates ubiquitin ligase E3 in the pathophysiology of schizophrenia onset. J. Psychiatr Res. 2018, 102, 110–117. [Google Scholar] [CrossRef]
- Wang, Z.; Liu, Z.; Chen, X.; Li, J.; Yao, W.; Huang, S.; Gu, A.; Lei, Q.-Y.; Mao, Y.; Wen, W. A multi-lock inhibitory mechanism for fine-tuning enzyme activities of the HECT family E3 ligases. Nat. Commun. 2019, 10, 3162. [Google Scholar] [CrossRef] [PubMed]
- Alexander, K.A.; Cimler, B.M.; Meier, K.E.; Storm, D.R. Regulation of calmodulin binding to P-57. A neurospecific calmodulin binding protein. J. Biol. Chem. 1987, 262, 6108–6113. [Google Scholar] [PubMed]
- Braganza, A.; Li, J.; Zeng, X.; Yates, N.A.; Dey, N.B.; Andrews, J.; Clark, J.; Zamani, L.; Wang, X.-H.; St Croix, C.; et al. UBE3B Is a Calmodulin-regulated, Mitochondrion-associated E3 Ubiquitin Ligase. J. Biol. Chem. 2017, 292, 2470–2484. [Google Scholar] [CrossRef] [PubMed]
- Svilar, D.; Dyavaiah, M.; Brown, A.R.; Tang, J.; Li, J.; McDonald, P.R.; Shun, T.Y.; Braganza, A.; Wang, X.; Maniar, S.; et al. Alkylation sensitivity screens reveal a conserved cross-species functionome. Mol. Cancer Res. 2012, 10, 1580–1596. [Google Scholar] [CrossRef] [PubMed]
- Parkes, J.E.; Rothwell, S.; Day, P.J.; McHugh, N.J.; Betteridge, Z.E.; Cooper, R.G.; Ollier, W.E.; Chinoy, H.; Lamb, J.A. Myositis Genetics Consortium (MYOGEN) Systematic protein-protein interaction and pathway analyses in the idiopathic inflammatory myopathies. Arthritis Res. Ther. 2016, 18, 156. [Google Scholar] [CrossRef]
- Cheon, S.; Kaur, K.; Nijem, N.; Tuncay, I.O.; Kumar, P.; Dean, M.; Juusola, J.; Guillen-Sacoto, M.J.; Bedoukian, E.; Ierardi-Curto, L.; et al. The ubiquitin ligase UBE3B, disrupted in intellectual disability and absent speech, regulates metabolic pathways by targeting BCKDK. Proc. Natl. Acad. Sci. USA 2019, 116, 3662–3667. [Google Scholar] [CrossRef]
- Mazumder, M.K.; Paul, R.; Bhattacharya, P.; Borah, A. Neurological sequel of chronic kidney disease: From diminished Acetylcholinesterase activity to mitochondrial dysfunctions, oxidative stress and inflammation in mice brain. Sci. Rep. 2019, 9, 3097. [Google Scholar] [CrossRef]
- You, J.; Wang, M.; Aoki, T.; Tamura, T.; Pickart, C.M. Proteolytic Targeting of Transcriptional Regulator TIP120B by a HECT Domain E3 Ligase. J. Biol. Chem. 2003, 278, 23369–23375. [Google Scholar] [CrossRef]
- Gu, J.; Mao, W.; Ren, W.; Xu, F.; Zhu, Q.; Lu, C.; Lin, Z.; Zhang, Z.; Chu, Y.; Liu, R.; et al. Ubiquitin-protein ligase E3C maintains non-small-cell lung cancer stemness by targeting AHNAK-p53 complex. Cancer Lett. 2019, 443, 125–134. [Google Scholar] [CrossRef]
- Pan, S.-J.; Zhan, S.-K.; Ji, W.-Z.; Pan, Y.-X.; Liu, W.; Li, D.-Y.; Huang, P.; Zhang, X.-X.; Cao, C.-Y.; Zhang, J.; et al. Ubiquitin-protein ligase E3C promotes glioma progression by mediating the ubiquitination and degrading of Annexin A7. Sci. Rep. 2015, 5, 11066. [Google Scholar] [CrossRef]
- Gottlieb, C.D.; Thompson, A.C.S.; Ordureau, A.; Harper, J.W.; Kopito, R.R. Acute unfolding of a single protein immediately stimulates recruitment of ubiquitin protein ligase E3C (UBE3C) to 26S proteasomes. J. Biol. Chem. 2019, 294, 16511–16524. [Google Scholar] [CrossRef] [PubMed]
- Besche, H.C.; Sha, Z.; Kukushkin, N.V.; Peth, A.; Hock, E.-M.; Kim, W.; Gygi, S.; Gutierrez, J.A.; Liao, H.; Dick, L.; et al. Autoubiquitination of the 26S proteasome on Rpn13 regulates breakdown of ubiquitin conjugates. Embo J. 2014, 33, 1159–1176. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Hayward, G.S. The ubiquitin E3 ligase RAUL negatively regulates type i interferon through ubiquitination of the transcription factors IRF7 and IRF3. Immunity 2010, 33, 863–877. [Google Scholar] [CrossRef] [PubMed]
- Nomura, N.; Nagase, T.; Miyajima, N.; Sazuka, T.; Tanaka, A.; Sato, S.; Seki, N.; Kawarabayasi, Y.; Ishikawa, K.I.; Tabata, S. Prediction of the coding sequences of unidentified human genes. II. The coding sequences of 40 new genes (KIAA0041-KIAA0080) deduced by analysis of cDNA clones from human cell line KG-1. DNA Res. 1994, 1, 223–229. [Google Scholar] [CrossRef] [PubMed]
- Fagerberg, L.; Hallstrom, B.M.; Oksvold, P.; Kampf, C.; Djureinovic, D.; Odeberg, J.; Habuka, M.; Tahmasebpoor, S.; Danielsson, A.; Edlund, K.; et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell. Proteom. 2014, 13, 397–406. [Google Scholar] [CrossRef]
- Bayés, A.; van de Lagemaat, L.N.; Collins, M.O.; Croning, M.D.R.; Whittle, I.R.; Choudhary, J.S.; Grant, S.G.N. Characterization of the proteome, diseases and evolution of the human postsynaptic density. Nat. Neurosci. 2011, 14, 19–21. [Google Scholar] [CrossRef]
- Diering, G.H.; Nirujogi, R.S.; Roth, R.H.; Worley, P.F.; Pandey, A.; Huganir, R.L. Homer1a drives homeostatic scaling-down of excitatory synapses during sleep. Science 2017, 355, 511–515. [Google Scholar] [CrossRef]
- Okada, M.; Ohtake, F.; Nishikawa, H.; Wu, W.; Saeki, Y.; Takana, K.; Ohta, T. Liganded ERα Stimulates the E3 Ubiquitin Ligase Activity of UBE3C to Facilitate Cell Proliferation. Mol. Endocrinol. (Baltim. Md.) 2015, 29, 1646–1657. [Google Scholar] [CrossRef]
- Jiang, J.-H.; Liu, Y.-F.; Ke, A.-W.; Gu, F.-M.; Yu, Y.; Dai, Z.; Gao, Q.; Shi, G.-M.; Liao, B.-Y.; Xie, Y.-H.; et al. Clinical significance of the ubiquitin ligase UBE3C in hepatocellular carcinoma revealed by exome sequencing. Hepatology 2014, 59, 2216–2227. [Google Scholar] [CrossRef]
- Nara, M.; Teshima, K.; Watanabe, A.; Ito, M.; Iwamoto, K.; Kitabayashi, A.; Kume, M.; Hatano, Y.; Takahashi, N.; Iida, S.; et al. Bortezomib reduces the tumorigenicity of multiple myeloma via downregulation of upregulated targets in clonogenic side population cells. PLoS ONE 2013, 8, e56954. [Google Scholar] [CrossRef]
- Tang, L.; Yi, X.-M.; Chen, J.; Chen, F.-J.; Lou, W.; Gao, Y.-L.; Zhou, J.; Su, L.-N.; Xu, X.; Lu, J.-Q.; et al. Ubiquitin ligase UBE3C promotes melanoma progression by increasing epithelial-mesenchymal transition in melanoma cells. Oncotarget 2016, 7, 15738–15746. [Google Scholar] [CrossRef] [PubMed]
- Wen, J.L.; Wen, X.F.; Li, R.B.; Jin, Y.C.; Wang, X.L.; Zhou, L.; Chen, H.X. UBE3C promotes growth and metastasis of renal cell carcinoma via activating Wnt/β-catenin pathway. PLoS ONE 2015, 10, e0115622. [Google Scholar] [CrossRef] [PubMed]
- Pasaje, C.F.A.; Kim, J.H.; Park, B.L.; Park, J.S.; Uh, S.T.; Kim, M.K.; Park, C.S.; Shin, H.D. UBE3C genetic variations as potent markers of nasal polyps in Korean asthma patients. J. Hum. Genet. 2011, 56, 797–800. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chen, L.T.; Jiang, C.Y. MicroRNA Expression Profiles Identify Biomarker for Differentiating the Embolic Stroke from Thrombotic Stroke. BioMed Res. Int. 2018. [Google Scholar] [CrossRef]
- Yang, B.-Z.; Han, S.; Kranzler, H.R.; Farrer, L.A.; Gelernter, J. A genomewide linkage scan of cocaine dependence and major depressive episode in two populations. Neuropsychopharmacology 2011, 36, 2422–2430. [Google Scholar] [CrossRef]
- Garriock, H.A.; Kraft, J.B.; Shyn, S.I.; Peters, E.J.; Yokoyama, J.S.; Jenkins, G.D.; Reinalda, M.S.; Slager, S.L.; McGrath, P.J.; Hamilton, S.P. A Genomewide Association Study of Citalopram Response in Major Depressive Disorder. Biol. Psychiatry 2010, 67, 133–138. [Google Scholar] [CrossRef]
- Filatova, E.V.; Shadrina, M.I.; Alieva, A.K.; Kolacheva, A.A.; Slominsky, P.A.; Ugrumov, M.V. Expression analysis of genes of ubiquitin-proteasome protein degradation system in MPTP-induced mice models of early stages of Parkinson’s disease. Dokl. Biochem. Biophys. 2014, 456, 116–118. [Google Scholar] [CrossRef]
- Papoulidis, I.; Papageorgiou, E.; Siomou, E.; Oikonomidou, E.; Thomaidis, L.; Vetro, A.; Zuffardi, O.; Liehr, T.; Manolakos, E.; Vassilis, P. A patient with partial trisomy 21 and 7q deletion expresses mild Down syndrome phenotype. Gene 2014, 536, 441–443. [Google Scholar] [CrossRef]
- Kuo, C.-L.; Goldberg, A.L. Ubiquitinated proteins promote the association of proteasomes with the deubiquitinating enzyme Usp14 and the ubiquitin ligase Ube3c. Proc. Natl. Acad. Sci. USA 2017, 114, E3404–E3413. [Google Scholar] [CrossRef]
- Chu, B.W.; Kovary, K.M.; Guillaume, J.; Chen, L.; Teruel, M.N.; Wandless, T.J. The E3 ubiquitin ligase UBE3C enhances proteasome processivity by ubiquitinating partially proteolyzed substrates. J. Biol. Chem. 2013, 288, 34575–34587. [Google Scholar] [CrossRef]
- You, J.; Pickart, C.M. A HECT Domain E3 Enzyme Assembles Novel Polyubiquitin Chains. J. Biol. Chem. 2001, 276, 19871–19878. [Google Scholar] [CrossRef] [PubMed]
- Fei, C.; Li, Z.; Li, C.; Chen, Y.; Chen, Z.; He, X.; Mao, L.; Wang, X.; Zeng, R.; Li, L. Smurf1-Mediated Lys29-Linked Nonproteolytic Polyubiquitination of Axin Negatively Regulates Wnt/β-Catenin Signaling. Mol. Cell. Biol. 2013, 33, 4095–4105. [Google Scholar] [CrossRef] [PubMed]
- Karim, M.; Biquand, E.; Declercq, M.; Jacob, Y.; van der Werf, S.; Demeret, C. Nonproteolytic K29-Linked Ubiquitination of the PB2 Replication Protein of Influenza A Viruses by Proviral Cullin 4-Based E3 Ligases. mBio 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Chen, T.; Li, X.; Yang, M.; Tang, S.; Zhu, X.; Gu, Y.; Su, X.; Xia, M.; Li, W.; et al. Lys29-linkage of ASK1 by Skp1−Cullin 1−Fbxo21 ubiquitin ligase complex is required for antiviral innate response. eLife 2016, 5. [Google Scholar] [CrossRef]
- Van de Weijer, M.L.; Krshnan, L.; Liberatori, S.; Guerrero, E.N.; Robson-Tull, J.; Hahn, L.; Lebbink, R.J.; Wiertz, E.J.H.J.; Fischer, R.; Ebner, D.; et al. Quality Control of ER Membrane Proteins by the RNF185/Membralin Ubiquitin Ligase Complex. Mol. Cell 2020, 79, 768–781.e7. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).