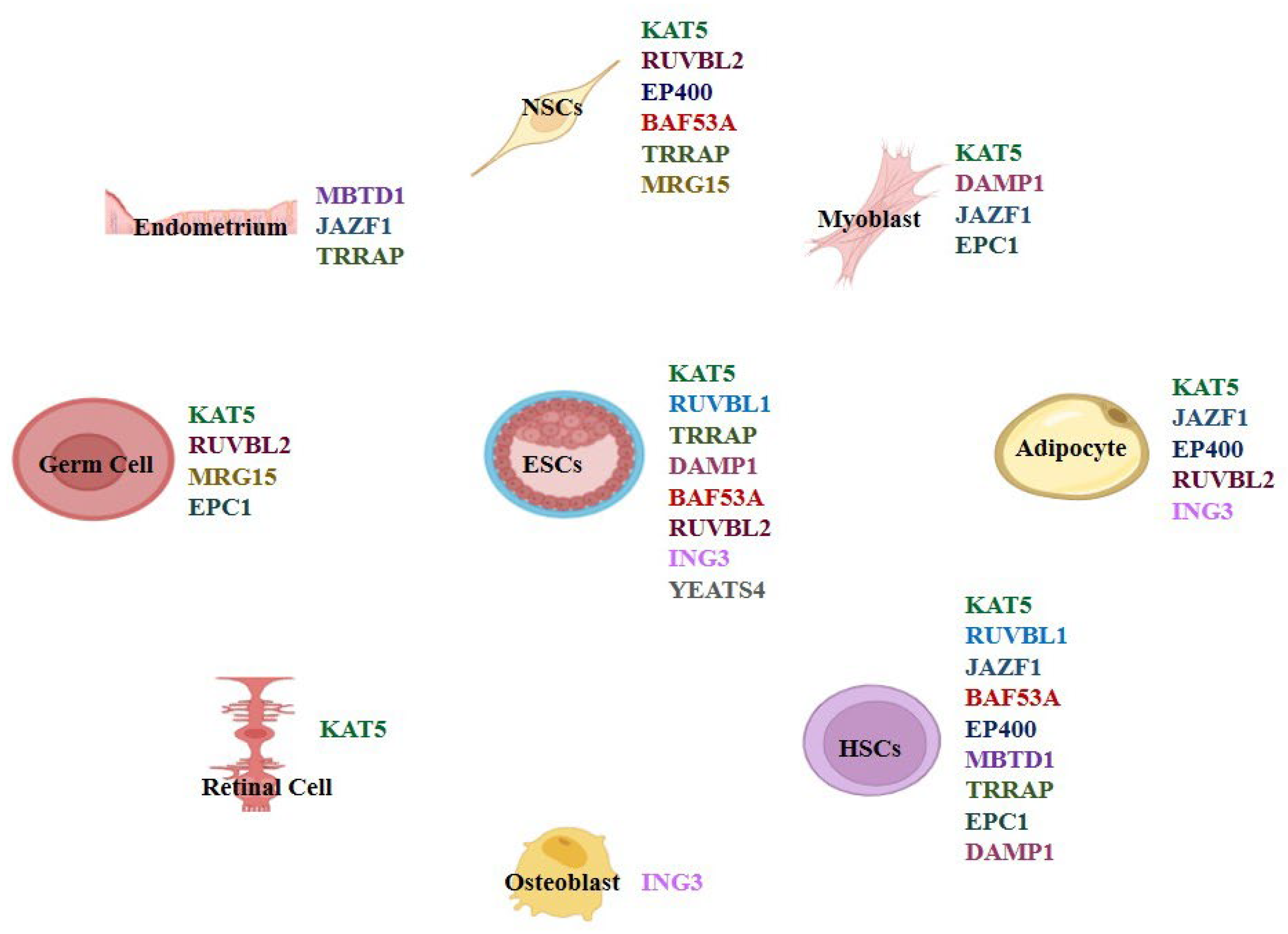
Functions of TIP60/NuA4 Complex Subunits in Cell Differentiation
Highlights
- TIP60/NuA4 functions as an integrated complex—catalytic (TIP60/KAT5), scaffold/ATPase (EP400/EPC1/TRRAP), and reader subunits (ING3, YEATS4, MBTD1, BAF53A/MRG15) cooperate to remodel chromatin during lineage-specific differentiation.
- Individual subunits have distinct yet convergent roles across neurogenesis, myogenesis, adipogenesis, hematopoiesis, and germ-cell development; viewing the whole complex explains overlapping differentiation phenotypes.
- A whole-complex perspective clarifies why disruption of different subunits yields similar outcomes and helps separate complex-dependent from subunit-specific effects.
- TIP60/NuA4 emerges as a central epigenetic hub with translational relevance for disorders of development, regeneration, and disease.
Abstract
1. Introduction
2. Functions of TIP60/NuA4 Subunits in Differentiation
2.1. TIP60/KAT5 and Differentiation
2.2. ING3 and Differentiation
2.3. MRG15 and Differentiation
2.4. MBTD1 and Differentiation
2.5. EPC1 and Differentiation
2.6. RUVBL1 and Differentiation
2.7. RUVBL2 and Differentiation
2.8. JAZF1 and Differentiation
2.9. TRRAP and Differentiation
2.10. DMAP1 and Differentiation
2.11. YEATS Family and Differentiation
2.12. YEATS Family and Neuronal Differentiation
2.13. BAF53A/ACTL6A and Differentiation
2.14. EP400 and Differentiation
3. Whole-Complex Perspective of TIP60/NuA4 in Differentiation
3.1. Therapeutic Outlook
3.2. Neurodegeneration
3.3. Oncology
3.4. Cardiac Injury
3.5. Immune Regulation
3.6. Translational Outlook
4. Gaps and Controversies
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Srinageshwar, B.; Maiti, P.; Dunbar, G.L.; Rossignol, J. Role of Epigenetics in Stem Cell Proliferation and Differentiation: Implications for Treating Neurodegenerative Diseases. Int. J. Mol. Sci. 2016, 17, 199. [Google Scholar] [CrossRef]
- Wu, H.; Sun, Y.E. Epigenetic Regulation of Stem Cell Differentiation. Pediatr. Res. 2006, 59, 21–25. [Google Scholar] [CrossRef]
- Zakrzewski, W.; Dobrzyński, M.; Szymonowicz, M.; Rybak, Z. Stem Cells: Past, Present, and Future. Stem Cell Res. Ther. 2019, 10, 68. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Wu, J.; Guo, H.; Yao, W.; Li, S.; Lu, Y.; Jia, Y.; Liang, X.; Tang, J.; Zhang, H. Post-Translational Modifications of Histones: Mechanisms, Biological Functions, and Therapeutic Targets. MedComm 2023, 4, e292. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.-J.; Man, N.; Tan, Y.; Nimer, S.D.; Wang, L. The Role of Histone Acetyltransferases in Normal and Malignant Hematopoiesis. Front. Oncol. 2015, 5, 108. [Google Scholar] [CrossRef]
- He, R.; Dantas, A.; Riabowol, K. Histone Acetyltransferases and Stem Cell Identity. Cancers 2021, 13, 2407. [Google Scholar] [CrossRef]
- Chen, K.; Wang, L.; Yu, Z.; Yu, J.; Ren, Y.; Wang, Q.; Xu, Y. Structure of the Human TIP60 Complex. Nat. Commun. 2024, 15, 7092. [Google Scholar] [CrossRef]
- Barry, R.M.; Sacco, O.; Mameri, A.; Stojaspal, M.; Kartsonis, W.; Shah, P.; De Ioannes, P.; Hofr, C.; Côté, J.; Sfeir, A. Rap1 Regulates TIP60 Function during Fate Transition between Two-Cell-like and Pluripotent States. Genes Dev. 2022, 36, 313–330. [Google Scholar] [CrossRef]
- Cai, Y.; Jin, J.; Tomomori-Sato, C.; Sato, S.; Sorokina, I.; Parmely, T.J.; Conaway, R.C.; Conaway, J.W. Identification of New Subunits of the Multiprotein Mammalian TRRAP/TIP60-Containing Histone Acetyltransferase Complex. J. Biol. Chem. 2003, 278, 42733–42736. [Google Scholar] [CrossRef] [PubMed]
- Chesi, A.; Wagley, Y.; Johnson, M.E.; Manduchi, E.; Su, C.; Lu, S.; Leonard, M.E.; Hodge, K.M.; Pippin, J.A.; Hankenson, K.D.; et al. Genome-Scale Capture C Promoter Interactions Implicate Effector Genes at GWAS Loci for Bone Mineral Density. Nat. Commun. 2019, 10, 1260. [Google Scholar] [CrossRef]
- Awe, J.P.; Byrne, J.A. Identifying Candidate Oocyte Reprogramming Factors Using Cross-Species Global Transcriptional Analysis. Cell. Reprogram. 2013, 15, 126–133. [Google Scholar] [CrossRef]
- Song, Y.; Hou, G.; Diep, J.; Ooi, Y.S.; Akopyants, N.S.; Beverley, S.M.; Carette, J.E.; Greenberg, H.B.; Ding, S. Inhibitor of Growth Protein 3 Epigenetically Silences Endogenous Retroviral Elements and Prevents Innate Immune Activation. Nucleic Acids Res. 2021, 49, 12706–12715. [Google Scholar] [CrossRef]
- Tominaga, K.; Sakashita, E.; Kasashima, K.; Kuroiwa, K.; Nagao, Y.; Iwamori, N.; Endo, H. Tip60/KAT5 Histone Acetyltransferase Is Required for Maintenance and Neurogenesis of Embryonic Neural Stem Cells. Int. J. Mol. Sci. 2023, 24, 2113. [Google Scholar] [CrossRef]
- Kim, J.-W.; Jang, S.-M.; Kim, C.-H.; An, J.-H.; Kang, E.-J.; Choi, K.-H. New Molecular Bridge between RelA/P65 and NF-ΚB Target Genes via Histone Acetyltransferase TIP60 Cofactor. J. Biol. Chem. 2012, 287, 7780–7791. [Google Scholar] [CrossRef]
- Janas, J.A.; Zhang, L.; Luu, J.H.; Demeter, J.; Meng, L.; Marro, S.G.; Mall, M.; Mooney, N.A.; Schaukowitch, K.; Ng, Y.H.; et al. Tip60-Mediated H2A.Z Acetylation Promotes Neuronal Fate Specification and Bivalent Gene Activation. Mol. Cell 2022, 82, 4627–4646.e14. [Google Scholar] [CrossRef]
- Ooi, S.K.T.; Qiu, C.; Bernstein, E.; Li, K.; Jia, D.; Yang, Z.; Erdjument-Bromage, H.; Tempst, P.; Lin, S.-P.; Allis, C.D.; et al. DNMT3L Connects Unmethylated Lysine 4 of Histone H3 to de Novo Methylation of DNA. Nature 2007, 448, 714–717. [Google Scholar] [CrossRef]
- Humbert, J.; Salian, S.; Makrythanasis, P.; Lemire, G.; Rousseau, J.; Ehresmann, S.; Garcia, T.; Alasiri, R.; Bottani, A.; Hanquinet, S.; et al. De Novo KAT5 Variants Cause a Syndrome with Recognizable Facial Dysmorphisms, Cerebellar Atrophy, Sleep Disturbance, and Epilepsy. Am. J. Hum. Genet. 2020, 107, 564–574. [Google Scholar] [CrossRef] [PubMed]
- Shen, E.; Shulha, H.; Weng, Z.; Akbarian, S. Regulation of Histone H3K4 Methylation in Brain Development and Disease. Philos. Trans. R. Soc. London Ser. B Biol. Sci. 2014, 369, 20130514. [Google Scholar] [CrossRef] [PubMed]
- Urban, I.; Kerimoglu, C.; Sakib, M.S.; Wang, H.; Benito, E.; Thaller, C.; Zhou, X.; Yan, J.; Fischer, A.; Eichele, G. TIP60/KAT5 Is Required for Neuronal Viability in Hippocampal CA1. Sci. Rep. 2019, 9, 16173. [Google Scholar] [CrossRef]
- Beaver, M.; Bhatnagar, A.; Panikker, P.; Zhang, H.; Snook, R.; Parmar, V.; Vijayakumar, G.; Betini, N.; Akhter, S.; Elefant, F. Disruption of Tip60 HAT Mediated Neural Histone Acetylation Homeostasis Is an Early Common Event in Neurodegenerative Diseases. Sci. Rep. 2020, 10, 18265. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Galbo, P.M.J.; Gong, W.; Storey, A.J.; Tsai, Y.-H.; Yu, X.; Ahn, J.H.; Guo, Y.; Mackintosh, S.G.; Edmondson, R.D.; et al. ZMYND11-MBTD1 Induces Leukemogenesis through Hijacking NuA4/TIP60 Acetyltransferase Complex and a PWWP-Mediated Chromatin Association Mechanism. Nat. Commun. 2021, 12, 1045. [Google Scholar] [CrossRef] [PubMed]
- Numata, A.; Kwok, H.S.; Zhou, Q.-L.; Li, J.; Tirado-Magallanes, R.; Angarica, V.E.; Hannah, R.; Park, J.; Wang, C.Q.; Krishnan, V.; et al. Lysine Acetyltransferase Tip60 Is Required for Hematopoietic Stem Cell Maintenance. Blood 2020, 136, 1735–1747. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, A.; Deiulio, A.; Martin, E.T.; Upadhyay, M.; Rangan, P. Tip60 Complex Promotes Expression of a Differentiation Factor to Regulate Germline Differentiation in Female Drosophila. Mol. Biol. Cell 2018, 29, 2933–2945. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Isono, K.-I.; Ohbo, K.; Endo, T.A.; Ohara, O.; Maekawa, M.; Toyama, Y.; Ito, C.; Toshimori, K.; Helin, K.; et al. EPC1/TIP60-Mediated Histone Acetylation Facilitates Spermiogenesis in Mice. Mol. Cell. Biol. 2017, 37, e00082-17. [Google Scholar] [CrossRef]
- Chen, P.B.; Hung, J.-H.; Hickman, T.L.; Coles, A.H.; Carey, J.F.; Weng, Z.; Chu, F.; Fazzio, T.G. Hdac6 Regulates Tip60-P400 Function in Stem Cells. Elife 2013, 2, e01557. [Google Scholar] [CrossRef]
- Hu, Y.; Fisher, J.B.; Koprowski, S.; McAllister, D.; Kim, M.-S.; Lough, J. Homozygous Disruption of the Tip60 Gene Causes Early Embryonic Lethality. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 2009, 238, 2912–2921. [Google Scholar] [CrossRef]
- Acharya, D.; Hainer, S.J.; Yoon, Y.; Wang, F.; Bach, I.; Rivera-Pérez, J.A.; Fazzio, T.G. KAT-Independent Gene Regulation by Tip60 Promotes ESC Self-Renewal but Not Pluripotency. Cell Rep. 2017, 19, 671–679. [Google Scholar] [CrossRef]
- Kim, J.-W.; Jang, S.-M.; Kim, C.-H.; An, J.-H.; Kang, E.-J.; Choi, K.-H. Tip60 Regulates Myoblast Differentiation by Enhancing the Transcriptional Activity of MyoD via Their Physical Interactions. FEBS J. 2011, 278, 4394–4404. [Google Scholar] [CrossRef]
- Jang, S.-M.; Kim, J.-W.; Kim, C.-H.; An, J.-H.; Johnson, A.; Song, P.I.; Rhee, S.; Choi, K.-H. KAT5-Mediated SOX4 Acetylation Orchestrates Chromatin Remodeling during Myoblast Differentiation. Cell Death Dis. 2015, 6, e1857. [Google Scholar] [CrossRef]
- Ma, X.; Wang, D.; Zhao, W.; Xu, L. Deciphering the Roles of PPARγ in Adipocytes via Dynamic Change of Transcription Complex. Front. Endocrinol. 2018, 9, 473. [Google Scholar] [CrossRef]
- van Beekum, O.; Brenkman, A.B.; Grøntved, L.; Hamers, N.; van den Broek, N.J.F.; Berger, R.; Mandrup, S.; Kalkhoven, E. The Adipogenic Acetyltransferase Tip60 Targets Activation Function 1 of Peroxisome Proliferator-Activated Receptor Gamma. Endocrinology 2008, 149, 1840–1849. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Koppen, A.; Rakhshandehroo, M.; Tasdelen, I.; van de Graaf, S.F.; van Loosdregt, J.; van Beekum, O.; Hamers, N.; van Leenen, D.; Berkers, C.R.; et al. Early Adipogenesis Is Regulated through USP7-Mediated Deubiquitination of the Histone Acetyltransferase TIP60. Nat. Commun. 2013, 4, 2656. [Google Scholar] [CrossRef]
- Kim, J.-W.; Jang, S.-M.; Kim, C.-H.; An, J.-H.; Choi, K.-H. Transcriptional Activity of Neural Retina Leucine Zipper (Nrl) Is Regulated by c-Jun N-Terminal Kinase and Tip60 during Retina Development. Mol. Cell. Biol. 2012, 32, 1720–1732. [Google Scholar] [CrossRef] [PubMed]
- Dantas, A.; Al Shueili, B.; Yang, Y.; Nabbi, A.; Fink, D.; Riabowol, K. Biological Functions of the ING Proteins. Cancers 2019, 11, 1817. [Google Scholar] [CrossRef]
- Bua, D.J.; Martin, G.M.; Binda, O.; Gozani, O. Nuclear Phosphatidylinositol-5-Phosphate Regulates ING2 Stability at Discrete Chromatin Targets in Response to DNA Damage. Sci. Rep. 2013, 3, 2137. [Google Scholar] [CrossRef] [PubMed]
- Gou, W.-F.; Yang, X.-F.; Shen, D.-F.; Zhao, S.; Sun, H.-Z.; Luo, J.-S.; Zheng, H.-C. Immunohistochemical Profile of ING3 Protein in Normal and Cancerous Tissues. Oncol. Lett. 2017, 13, 1631–1636. [Google Scholar] [CrossRef]
- Feng, X.; Hara, Y.; Riabowol, K. Different HATS of the ING1 Gene Family. Trends Cell Biol. 2002, 12, 532–538. [Google Scholar] [CrossRef]
- Thalappilly, S.; Feng, X.; Pastyryeva, S.; Suzuki, K.; Muruve, D.; Larocque, D.; Richard, S.; Truss, M.; von Deimling, A.; Riabowol, K.; et al. The P53 Tumor Suppressor Is Stabilized by Inhibitor of Growth 1 (ING1) by Blocking Polyubiquitination. PLoS ONE 2011, 6, e21065. [Google Scholar] [CrossRef][Green Version]
- Nagashima, M.; Shiseki, M.; Pedeux, R.M.; Okamura, S.; Kitahama-Shiseki, M.; Miura, K.; Yokota, J.; Harris, C.C. A Novel PHD-Finger Motif Protein, P47ING3, Modulates P53-Mediated Transcription, Cell Cycle Control, and Apoptosis. Oncogene 2003, 22, 343–350. [Google Scholar] [CrossRef]
- Nabbi, A.; Almami, A.; Thakur, S.; Suzuki, K.; Boland, D.; Bismar, T.A.; Riabowol, K. ING3 Protein Expression Profiling in Normal Human Tissues Suggest Its Role in Cellular Growth and Self-Renewal. Eur. J. Cell Biol. 2015, 94, 214–222. [Google Scholar] [CrossRef]
- Fink, D.; Yau, T.; Nabbi, A.; Wagner, B.; Wagner, C.; Hu, S.M.; Lang, V.; Handschuh, S.; Riabowol, K.; Rülicke, T. Loss of Ing3 Expression Results in Growth Retardation and Embryonic Death. Cancers 2019, 12, 80. [Google Scholar] [CrossRef]
- Suzuki, S.; Nozawa, Y.; Tsukamoto, S.; Kaneko, T.; Imai, H.; Minami, N. ING3 Is Essential for Asymmetric Cell Division during Mouse Oocyte Maturation. PLoS ONE 2013, 8, e74749. [Google Scholar] [CrossRef]
- Zhang, P.; Du, J.; Sun, B.; Dong, X.; Xu, G.; Zhou, J.; Huang, Q.; Liu, Q.; Hao, Q.; Ding, J. Structure of Human MRG15 Chromo Domain and Its Binding to Lys36-Methylated Histone H3. Nucleic Acids Res. 2006, 34, 6621–6628. [Google Scholar] [CrossRef]
- Tominaga, K.; Kirtane, B.; Jackson, J.G.; Ikeno, Y.; Ikeda, T.; Hawks, C.; Smith, J.R.; Matzuk, M.M.; Pereira-Smith, O.M. MRG15 Regulates Embryonic Development and Cell Proliferation. Mol. Cell. Biol. 2005, 25, 2924–2937. [Google Scholar] [CrossRef]
- Chen, M.; Takano-Maruyama, M.; Pereira-Smith, O.M.; Gaufo, G.O.; Tominaga, K. MRG15, a Component of HAT and HDAC Complexes, Is Essential for Proliferation and Differentiation of Neural Precursor Cells. J. Neurosci. Res. 2009, 87, 1522–1531. [Google Scholar] [CrossRef]
- Chen, M.; Pereira-Smith, O.M.; Tominaga, K. Loss of the Chromatin Regulator MRG15 Limits Neural Stem/Progenitor Cell Proliferation via Increased Expression of the P21 Cdk Inhibitor. Stem Cell Res. 2011, 7, 75–88. [Google Scholar] [CrossRef]
- Gupta, P.; Leahul, L.; Wang, X.; Wang, C.; Bakos, B.; Jasper, K.; Hansen, D. Proteasome Regulation of the Chromodomain Protein MRG-1 Controls the Balance between Proliferative Fate and Differentiation in the C. Elegans Germ Line. Development 2015, 142, 291–302. [Google Scholar] [CrossRef] [PubMed]
- Iwamori, N.; Tominaga, K.; Sato, T.; Riehle, K.; Iwamori, T.; Ohkawa, Y.; Coarfa, C.; Ono, E.; Matzuk, M.M. MRG15 Is Required for Pre-MRNA Splicing and Spermatogenesis. Proc. Natl. Acad. Sci. USA 2016, 113, E5408–E5415. [Google Scholar] [CrossRef] [PubMed]
- Takubo, K.; Htun, P.W.; Ueda, T.; Sera, Y.; Iwasaki, M.; Koizumi, M.; Shiroshita, K.; Kobayashi, H.; Haraguchi, M.; Watanuki, S.; et al. MBTD1 Preserves Adult Hematopoietic Stem Cell Pool Size and Function. Proc. Natl. Acad. Sci. USA 2023, 120, e2206860120. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Devoucoux, M.; Song, X.; Li, L.; Ayaz, G.; Cheng, H.; Tempel, W.; Dong, C.; Loppnau, P.; Côté, J.; et al. Structural Basis for EPC1-Mediated Recruitment of MBTD1 into the NuA4/TIP60 Acetyltransferase Complex. Cell Rep. 2020, 30, 3996–4002.e4. [Google Scholar] [CrossRef]
- Chadchan, S.B.; Maurya, V.K.; Krekeler, G.L.; Jungheim, E.S.; Kommagani, R. A Role for Malignant Brain Tumor Domain-Containing Protein 1 in Human Endometrial Stromal Cell Decidualization. Front. Cell Dev. Biol. 2020, 8, 745. [Google Scholar] [CrossRef]
- Honda, H.; Takubo, K.; Oda, H.; Kosaki, K.; Tazaki, T.; Yamasaki, N.; Miyazaki, K.; Moore, K.A.; Honda, Z.; Suda, T.; et al. Hemp, an Mbt Domain-Containing Protein, Plays Essential Roles in Hematopoietic Stem Cell Function and Skeletal Formation. Proc. Natl. Acad. Sci. USA 2011, 108, 2468–2473. [Google Scholar] [CrossRef]
- Searle, N.E.; Torres-Machorro, A.L.; Pillus, L. Chromatin Regulation by the NuA4 Acetyltransferase Complex Is Mediated by Essential Interactions Between Enhancer of Polycomb (Epl1) and Esa1. Genetics 2017, 205, 1125–1137. [Google Scholar] [CrossRef]
- Wang, Y.; Alla, V.; Goody, D.; Gupta, S.K.; Spitschak, A.; Wolkenhauer, O.; Pützer, B.M.; Engelmann, D. Epigenetic Factor EPC1 Is a Master Regulator of DNA Damage Response by Interacting with E2F1 to Silence Death and Activate Metastasis-Related Gene Signatures. Nucleic Acids Res. 2015, 44, 117–133. [Google Scholar] [CrossRef]
- Searle, N.E.; Pillus, L. Critical Genomic Regulation Mediated by Enhancer of Polycomb. Curr. Genet. 2018, 64, 147–154. [Google Scholar] [CrossRef]
- Liu, W.; Liu, X.; Li, L.; Tai, Z.; Li, G.; Liu, J.-X. EPC1/2 Regulate Hematopoietic Stem and Progenitor Cell Proliferation by Modulating H3 Acetylation and DLST. iScience 2024, 27, 109263. [Google Scholar] [CrossRef]
- Kee, H.J.; Kim, J.-R.; Nam, K.-I.; Park, H.Y.; Shin, S.; Kim, J.C.; Shimono, Y.; Takahashi, M.; Jeong, M.H.; Kim, N.; et al. Enhancer of Polycomb1, a Novel Homeodomain Only Protein-Binding Partner, Induces Skeletal Muscle Differentiation. J. Biol. Chem. 2007, 282, 7700–7709. [Google Scholar] [CrossRef]
- Kim, J.-R.; Kee, H.J.; Kim, J.-Y.; Joung, H.; Nam, K.-I.; Eom, G.H.; Choe, N.; Kim, H.-S.; Kim, J.C.; Kook, H.; et al. Enhancer of Polycomb1 Acts on Serum Response Factor to Regulate Skeletal Muscle Differentiation. J. Biol. Chem. 2009, 284, 16308–16316. [Google Scholar] [CrossRef] [PubMed]
- Tang, D.; Zhang, Z.; Zboril, E.; Wetzel, M.D.; Xu, X.; Zhang, W.; Chen, L.; Liu, Z. Pontin Functions as A Transcriptional Co-Activator for Retinoic Acid-Induced HOX Gene Expression. J. Mol. Biol. 2021, 433, 166928. [Google Scholar] [CrossRef] [PubMed]
- Boo, K.; Bhin, J.; Jeon, Y.; Kim, J.; Shin, H.-J.R.; Park, J.-E.; Kim, K.; Kim, C.R.; Jang, H.; Kim, I.-H.; et al. Pontin Functions as an Essential Coactivator for Oct4-Dependent LincRNA Expression in Mouse Embryonic Stem Cells. Nat. Commun. 2015, 6, 6810. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bereshchenko, O.; Mancini, E.; Luciani, L.; Gambardella, A.; Riccardi, C.; Nerlov, C. Pontin Is Essential for Murine Hematopoietic Stem Cell Survival. Haematologica 2012, 97, 1291–1294. [Google Scholar] [CrossRef]
- Dafinger, C.; Benzing, T.; Dötsch, J.; Schermer, B.; Liebau, M.C. Targeted Deletion of Ruvbl1 Results in Severe Defects of Epidermal Development and Perinatal Mortality. Mol. Cell. Pediatr. 2021, 8, 1. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Li, W.; Liu, H.; Yan, L.; Jiao, W.; Li, D.; Tang, Y.; Gu, G.; Xu, Z. Overexpression of Reptin in Renal Cell Carcinoma Contributes to Tumor Malignancies and Its Inhibition Triggers Senescence of Cancer Cells. Urol. Oncol. 2013, 31, 1358–1366. [Google Scholar] [CrossRef] [PubMed]
- Bauer, A.; Chauvet, S.; Huber, O.; Usseglio, F.; Rothbächer, U.; Aragnol, D.; Kemler, R.; Pradel, J. Pontin52 and Reptin52 Function as Antagonistic Regulators of Beta-Catenin Signalling Activity. EMBO J. 2000, 19, 6121–6130. [Google Scholar] [CrossRef]
- Jaishankar, A.; Barthelery, M.; Freeman, W.M.; Salli, U.; Ritty, T.M.; Vrana, K.E. Human Embryonic and Mesenchymal Stem Cells Express Different Nuclear Proteomes. Stem Cells Dev. 2009, 18, 793–802. [Google Scholar] [CrossRef]
- Hong, S.; Jo, J.; Kim, H.J.; Lee, J.E.; Shin, D.H.; Lee, S.-G.; Baek, A.; Shim, S.H.; Lee, D.R. RuvB-Like Protein 2 (Ruvbl2) Has a Role in Directing the Neuroectodermal Differentiation of Mouse Embryonic Stem Cells. Stem Cells Dev. 2016, 25, 1376–1385. [Google Scholar] [CrossRef]
- Barthéléry, M.; Jaishankar, A.; Salli, U.; Vrana, K.E. Reptin52 Expression during in Vitro Neural Differentiation of Human Embryonic Stem Cells. Neurosci. Lett. 2009, 452, 47–51. [Google Scholar] [CrossRef] [PubMed]
- Chapman, K.M.; Powell, H.M.; Chaudhary, J.; Shelton, J.M.; Richardson, J.A.; Richardson, T.E.; Hamra, F.K. Linking Spermatid Ribonucleic Acid (RNA) Binding Protein and Retrogene Diversity to Reproductive Success. Mol. Cell. Proteomics 2013, 12, 3221–3236. [Google Scholar] [CrossRef]
- Zhu, D.; Xu, L.; Wei, X.; Xia, B.; Gong, Y.; Li, Q.; Chen, X. PPARγ Enhanced Adiponectin Polymerization and Trafficking by Promoting RUVBL2 Expression during Adipogenic Differentiation. Gene 2021, 764, 145100. [Google Scholar] [CrossRef]
- Xie, X.; Chen, Y.; Xue, P.; Fan, Y.; Deng, Y.; Peng, G.; Yang, F.; Xu, T. RUVBL2, a Novel AS160-Binding Protein, Regulates Insulin-Stimulated GLUT4 Translocation. Cell Res. 2009, 19, 1090–1097. [Google Scholar] [CrossRef][Green Version]
- Fu, H.; Zhou, F.; Yuan, Q.; Zhang, W.; Qiu, Q.; Yu, X.; He, Z. MiRNA-31-5p Mediates the Proliferation and Apoptosis of Human Spermatogonial Stem Cells via Targeting JAZF1 and Cyclin A2. Mol. Ther. Nucleic Acids 2019, 14, 90–100. [Google Scholar] [CrossRef]
- Mameri, A.; Côté, J. JAZF1: A Metabolic Actor Subunit of the NuA4/TIP60 Chromatin Modifying Complex. Front. Cell Dev. Biol. 2023, 11, 1134268. [Google Scholar] [CrossRef]
- Procida, T.; Friedrich, T.; Jack, A.P.M.; Peritore, M.; Bönisch, C.; Eberl, H.C.; Daus, N.; Kletenkov, K.; Nist, A.; Stiewe, T.; et al. JAZF1, A Novel P400/TIP60/NuA4 Complex Member, Regulates H2A.Z Acetylation at Regulatory Regions. Int. J. Mol. Sci. 2021, 22, 678. [Google Scholar] [CrossRef]
- Ming, G.; Xiao, D.; Gong, W.; Liu, H.; Liu, J.; Zhou, H.; Liu, Z. JAZF1 Can Regulate the Expression of Lipid Metabolic Genes and Inhibit Lipid Accumulation in Adipocytes. Biochem. Biophys. Res. Commun. 2014, 445, 673–680. [Google Scholar] [CrossRef]
- Ming, G.; Li, X.; Yin, J.; Ai, Y.; Xu, D.; Ma, X.; Liu, Z.; Liu, H.; Zhou, H.; Liu, Z. JAZF1 Regulates Visfatin Expression in Adipocytes via PPARα and PPARβ/δ Signaling. Metabolism 2014, 63, 1012–1021. [Google Scholar] [CrossRef]
- Jeong, J.; Jang, S.; Park, S.; Kwon, W.; Kim, S.-Y.; Jang, S.; Ko, J.; Park, S.J.; Lim, S.-G.; Yoon, D.; et al. JAZF1 Heterozygous Knockout Mice Show Altered Adipose Development and Metabolism. Cell Biosci. 2021, 11, 161. [Google Scholar] [CrossRef] [PubMed]
- Yuasa, K.; Aoki, N.; Hijikata, T. JAZF1 Promotes Proliferation of C2C12 Cells, but Retards Their Myogenic Differentiation through Transcriptional Repression of MEF2C and MRF4-Implications for the Role of Jazf1 Variants in Oncogenesis and Type 2 Diabetes. Exp. Cell Res. 2015, 336, 287–297. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.-A.; Watson, J.K.; Nikolić, M.Z.; Rawlins, E.L. Fank1 and Jazf1 Promote Multiciliated Cell Differentiation in the Mouse Airway Epithelium. Biol. Open 2018, 7, bio033944. [Google Scholar] [CrossRef] [PubMed]
- Park, S.J.; Kwon, W.; Park, S.; Jeong, J.; Kim, D.; Jang, S.; Kim, S.-Y.; Sung, Y.; Kim, M.O.; Choi, S.-K.; et al. Jazf1 Acts as a Regulator of Insulin-Producing β-Cell Differentiation in Induced Pluripotent Stem Cells and Glucose Homeostasis in Mice. FEBS J. 2021, 288, 4412–4427. [Google Scholar] [CrossRef]
- Tavares, M.; Khandelwal, G.; Muter, J.; Viiri, K.; Beltran, M.; Brosens, J.J.; Jenner, R.G. JAZF1-SUZ12 Dysregulates PRC2 Function and Gene Expression during Cell Differentiation. Cell Rep. 2022, 39, 110889. [Google Scholar] [CrossRef]
- Liang, Y.; Lai, S.; Huang, L.; Li, Y.; Zeng, S.; Zhang, S.; Chen, J.; Deng, W.; Liu, Y.; Liang, J.; et al. JAZF1 Safeguards Human Endometrial Stromal Cells Survival and Decidualization by Repressing the Transcription of G0S2. Commun. Biol. 2023, 6, 568. [Google Scholar] [CrossRef] [PubMed]
- Meng, F.; Hao, P.; Du, H. Regulatory T Cells Differentiation in Visceral Adipose Tissues Contributes to Insulin Resistance by Regulating JAZF-1/PPAR-γ Pathway. J. Cell. Mol. Med. 2023, 27, 553–562. [Google Scholar] [CrossRef]
- Yin, B.-K.; Wang, Z.-Q. Beyond HAT Adaptor: TRRAP Liaisons with Sp1-Mediated Transcription. Int. J. Mol. Sci. 2021, 22, 12445. [Google Scholar] [CrossRef]
- Detilleux, D.; Raynaud, P.; Pradet-Balade, B.; Helmlinger, D. The TRRAP Transcription Cofactor Represses Interferon-Stimulated Genes in Colorectal Cancer Cells. Elife 2022, 11, e69705. [Google Scholar] [CrossRef]
- Elías-Villalobos, A.; Fort, P.; Helmlinger, D. New Insights into the Evolutionary Conservation of the Sole PIKK Pseudokinase Tra1/TRRAP. Biochem. Soc. Trans. 2019, 47, 1597–1608. [Google Scholar] [CrossRef]
- Fusi, L.; Paudel, R.; Meder, K.; Schlosser, A.; Schrama, D.; Goebeler, M.; Schmidt, M. Interaction of Transcription Factor FoxO3 with Histone Acetyltransferase Complex Subunit TRRAP Modulates Gene Expression and Apoptosis. J. Biol. Chem. 2022, 298, 101714. [Google Scholar] [CrossRef]
- Loizou, J.I.; Oser, G.; Shukla, V.; Sawan, C.; Murr, R.; Wang, Z.-Q.; Trumpp, A.; Herceg, Z. Histone Acetyltransferase Cofactor Trrap Is Essential for Maintaining the Hematopoietic Stem/Progenitor Cell Pool. J. Immunol. 2009, 183, 6422–6431. [Google Scholar] [CrossRef]
- Kang, K.-T.; Shin, M.-J.; Moon, H.-J.; Choi, K.-U.; Suh, D.-S.; Kim, J.-H. TRRAP Enhances Cancer Stem Cell Characteristics by Regulating NANOG Protein Stability in Colon Cancer Cells. Int. J. Mol. Sci. 2023, 24, 6260. [Google Scholar] [CrossRef]
- Finkbeiner, M.G.; Sawan, C.; Ouzounova, M.; Murr, R.; Herceg, Z. HAT Cofactor TRRAP Mediates Beta-Catenin Ubiquitination on the Chromatin and the Regulation of the Canonical Wnt Pathway. Cell Cycle 2008, 7, 3908–3914. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tapias, A.; Lázaro, D.; Yin, B.-K.; Rasa, S.M.M.; Krepelova, A.; Kelmer Sacramento, E.; Grigaravicius, P.; Koch, P.; Kirkpatrick, J.; Ori, A.; et al. HAT Cofactor TRRAP Modulates Microtubule Dynamics via SP1 Signaling to Prevent Neurodegeneration. Elife 2021, 10, e61531. [Google Scholar] [CrossRef] [PubMed]
- Yin, B.-K.; Lázaro, D.; Wang, Z.-Q. TRRAP-Mediated Acetylation on Sp1 Regulates Adult Neurogenesis. Comput. Struct. Biotechnol. J. 2023, 21, 472–484. [Google Scholar] [CrossRef]
- Wang, Z.; Plasschaert, L.W.; Aryal, S.; Renaud, N.A.; Yang, Z.; Choo-Wing, R.; Pessotti, A.D.; Kirkpatrick, N.D.; Cochran, N.R.; Carbone, W.; et al. TRRAP Is a Central Regulator of Human Multiciliated Cell Formation. J. Cell Biol. 2018, 217, 1941–1955. [Google Scholar] [CrossRef]
- Charles, N.A.; Holland, E.C. TRRAP and the Maintenance of Stemness in Gliomas. Cell Stem Cell 2010, 6, 6–7. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jiang, G.; Bi, K.; Tang, T.; Wang, J.; Zhang, Y.; Zhang, W.; Ren, H.; Bai, H.; Wang, Y. Down-Regulation of TRRAP-Dependent HTERT and TRRAP-Independent CAD Activation by Myc/Max Contributes to the Differentiation of HL60 Cells after Exposure to DMSO. Int. Immunopharmacol. 2006, 6, 1204–1213. [Google Scholar] [CrossRef]
- Rountree, M.R.; Bachman, K.E.; Baylin, S.B. DNMT1 Binds HDAC2 and a New Co-Repressor, DMAP1, to Form a Complex at Replication Foci. Nat. Genet. 2000, 25, 269–277. [Google Scholar] [CrossRef]
- Negishi, M.; Chiba, T.; Saraya, A.; Miyagi, S.; Iwama, A. Dmap1 Plays an Essential Role in the Maintenance of Genome Integrity through the DNA Repair Process. Genes Cells 2009, 14, 1347–1357. [Google Scholar] [CrossRef]
- Koizumi, T.; Negishi, M.; Nakamura, S.; Oguro, H.; Satoh, K.; Ichinose, M.; Iwama, A. Depletion of Dnmt1-Associated Protein 1 Triggers DNA Damage and Compromises the Proliferative Capacity of Hematopoietic Stem Cells. Int. J. Hematol. 2010, 91, 611–619. [Google Scholar] [CrossRef]
- Lee, Y.J.; Son, S.H.; Lim, C.S.; Kim, M.Y.; Lee, S.W.; Lee, S.; Jeon, J.; Ha, D.H.; Jung, N.R.; Han, S.Y.; et al. MMTR/Dmap1 Sets the Stage for Early Lineage Commitment of Embryonic Stem Cells by Crosstalk with PcG Proteins. Cells 2020, 9, 1190. [Google Scholar] [CrossRef]
- Mohan, K.N.; Ding, F.; Chaillet, J.R. Distinct Roles of DMAP1 in Mouse Development. Mol. Cell. Biol. 2011, 31, 1861–1869. [Google Scholar] [CrossRef] [PubMed]
- Fazzio, T.G.; Huff, J.T.; Panning, B. An RNAi Screen of Chromatin Proteins Identifies Tip60-P400 as a Regulator of Embryonic Stem Cell Identity. Cell 2008, 134, 162–174. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.S.L.; Palano, G.; Lim, C.; Moggio, A.; Drowley, L.; Plowright, A.T.; Bohlooly-Y, M.; Rosen, B.S.; Hansson, E.M.; Wang, Q.-D.; et al. CRISPR-Knockout Screen Identifies Dmap1 as a Regulator of Chemically Induced Reprogramming and Differentiation of Cardiac Progenitors. Stem Cells 2019, 37, 958–972. [Google Scholar] [CrossRef]
- Schulze, J.M.; Wang, A.Y.; Kobor, M.S. YEATS Domain Proteins: A Diverse Family with Many Links to Chromatin Modification and Transcription. Biochem. Cell Biol. 2009, 87, 65–75. [Google Scholar] [CrossRef]
- Pramparo, T.; Grosso, S.; Messa, J.; Zatterale, A.; Bonaglia, M.C.; Chessa, L.; Balestri, P.; Rocchi, M.; Zuffardi, O.; Giorda, R. Loss-of-Function Mutation of the AF9/MLLT3 Gene in a Girl with Neuromotor Development Delay, Cerebellar Ataxia, and Epilepsy. Hum. Genet. 2005, 118, 76–81. [Google Scholar] [CrossRef]
- Striano, P.; Elia, M.; Castiglia, L.; Galesi, O.; Pelligra, S.; Striano, S. A t(4;9)(Q34;P22) Translocation Associated with Partial Epilepsy, Mental Retardation, and Dysmorphism. Epilepsia 2005, 46, 1322–1324. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wen, H.; Xi, Y.; Tanaka, K.; Wang, H.; Peng, D.; Ren, Y.; Jin, Q.; Dent, S.Y.R.; Li, W.; et al. AF9 YEATS Domain Links Histone Acetylation to DOT1L-Mediated H3K79 Methylation. Cell 2014, 159, 558–571. [Google Scholar] [CrossRef]
- Qiao, Y.; Wang, X.; Wang, R.; Li, Y.; Yu, F.; Yang, X.; Song, L.; Xu, G.; Chin, Y.E.; Jing, N. AF9 Promotes HESC Neural Differentiation through Recruiting TET2 to Neurodevelopmental Gene Loci for Methylcytosine Hydroxylation. Cell Discov. 2015, 1, 15017. [Google Scholar] [CrossRef]
- Collins, E.C.; Appert, A.; Ariza-McNaughton, L.; Pannell, R.; Yamada, Y.; Rabbitts, T.H. Mouse Af9 Is a Controller of Embryo Patterning, like Mll, Whose Human Homologue Fuses with Af9 after Chromosomal Translocation in Leukemia. Mol. Cell. Biol. 2002, 22, 7313–7324. [Google Scholar] [CrossRef]
- Büttner, N.; Johnsen, S.A.; Kügler, S.; Vogel, T. Af9/Mllt3 Interferes with Tbr1 Expression through Epigenetic Modification of Histone H3K79 during Development of the Cerebral Cortex. Proc. Natl. Acad. Sci. USA 2010, 107, 7042–7047. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.-C.; Zhao, D.; Shi, J.; Peng, D.; Guan, H.; Li, Y.; Huang, Y.; Wen, H.; Li, W.; Li, H.; et al. Gas41 Links Histone Acetylation to H2A.Z Deposition and Maintenance of Embryonic Stem Cell Identity. Cell Discov. 2018, 4, 28. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Liu, S.; Li, X.; Li, X.D. YEATS Domains as Novel Epigenetic Readers: Structures, Functions, and Inhibitor Development. ACS Chem. Biol. 2023, 18, 994–1013. [Google Scholar] [CrossRef]
- Zhang, X.; Li, B.; Li, W.; Ma, L.; Zheng, D.; Li, L.; Yang, W.; Chu, M.; Chen, W.; Mailman, R.B.; et al. Transcriptional Repression by the BRG1-SWI/SNF Complex Affects the Pluripotency of Human Embryonic Stem Cells. Stem Cell Rep. 2014, 3, 460–474. [Google Scholar] [CrossRef]
- Zhu, B.; Ueda, A.; Song, X.; Horike, S.-I.; Yokota, T.; Akagi, T. Baf53a Is Involved in Survival of Mouse ES Cells, Which Can Be Compensated by Baf53b. Sci. Rep. 2017, 7, 14059. [Google Scholar] [CrossRef]
- Bao, X.; Tang, J.; Lopez-Pajares, V.; Tao, S.; Qu, K.; Crabtree, G.R.; Khavari, P.A. ACTL6a Enforces the Epidermal Progenitor State by Suppressing SWI/SNF-Dependent Induction of KLF4. Cell Stem Cell 2013, 12, 193–203. [Google Scholar] [CrossRef]
- Lu, W.; Fang, L.; Ouyang, B.; Zhang, X.; Zhan, S.; Feng, X.; Bai, Y.; Han, X.; Kim, H.; He, Q.; et al. Actl6a Protects Embryonic Stem Cells from Differentiating into Primitive Endoderm. Stem Cells 2015, 33, 1782–1793. [Google Scholar] [CrossRef] [PubMed]
- Cui, P.; Zhang, P.; Zhang, Y.; Sun, L.; Cui, G.; Guo, X.; Wang, H.; Zhang, X.; Shi, Y.; Yu, Z. HIF-1α/Actl6a/H3K9ac Axis Is Critical for Pluripotency and Lineage Differentiation of Human Induced Pluripotent Stem Cells. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2020, 34, 5740–5753. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Cui, P.; Li, Y.; Feng, G.; Tong, M.; Guo, L.; Li, T.; Liu, L.; Li, W.; Zhou, Q. Mitochondrially Produced ATP Affects Stem Cell Pluripotency via Actl6a-Mediated Histone Acetylation. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2018, 32, 1891–1902. [Google Scholar] [CrossRef]
- Lessard, J.; Wu, J.I.; Ranish, J.A.; Wan, M.; Winslow, M.M.; Staahl, B.T.; Wu, H.; Aebersold, R.; Graef, I.A.; Crabtree, G.R. An Essential Switch in Subunit Composition of a Chromatin Remodeling Complex during Neural Development. Neuron 2007, 55, 201–215. [Google Scholar] [CrossRef] [PubMed]
- Yoo, A.S.; Staahl, B.T.; Chen, L.; Crabtree, G.R. MicroRNA-Mediated Switching of Chromatin-Remodelling Complexes in Neural Development. Nature 2009, 460, 642–646. [Google Scholar] [CrossRef]
- Park, H.-J.; Tsai, E.; Huang, D.; Weaver, M.; Frick, L.; Alcantara, A.; Moran, J.J.; Patzig, J.; Melendez-Vasquez, C.V.; Crabtree, G.R.; et al. ACTL6a Coordinates Axonal Caliber Recognition and Myelination in the Peripheral Nerve. iScience 2022, 25, 104132. [Google Scholar] [CrossRef]
- Krasteva, V.; Buscarlet, M.; Diaz-Tellez, A.; Bernard, M.-A.; Crabtree, G.R.; Lessard, J.A. The BAF53a Subunit of SWI/SNF-like BAF Complexes Is Essential for Hemopoietic Stem Cell Function. Blood 2012, 120, 4720–4732. [Google Scholar] [CrossRef]
- Panatta, E.; Lena, A.M.; Mancini, M.; Smirnov, A.; Marini, A.; Delli Ponti, R.; Botta-Orfila, T.; Tartaglia, G.G.; Mauriello, A.; Zhang, X.; et al. Long Non-Coding RNA Uc.291 Controls Epithelial Differentiation by Interfering with the ACTL6A/BAF Complex. EMBO Rep. 2020, 21, e46734. [Google Scholar] [CrossRef]
- Fujii, T.; Ueda, T.; Nagata, S.; Fukunaga, R. Essential Role of P400/MDomino Chromatin-Remodeling ATPase in Bone Marrow Hematopoiesis and Cell-Cycle Progression. J. Biol. Chem. 2010, 285, 30214–30223. [Google Scholar] [CrossRef]
- Elsesser, O.; Fröb, F.; Küspert, M.; Tamm, E.R.; Fujii, T.; Fukunaga, R.; Wegner, M. Chromatin Remodeler Ep400 Ensures Oligodendrocyte Survival and Is Required for Myelination in the Vertebrate Central Nervous System. Nucleic Acids Res. 2019, 47, 6208–6224. [Google Scholar] [CrossRef]
- Fröb, F.; Sock, E.; Tamm, E.R.; Saur, A.-L.; Hillgärtner, S.; Williams, T.J.; Fujii, T.; Fukunaga, R.; Wegner, M. Ep400 Deficiency in Schwann Cells Causes Persistent Expression of Early Developmental Regulators and Peripheral Neuropathy. Nat. Commun. 2019, 10, 2361. [Google Scholar] [CrossRef]
- Ueda, T.; Watanabe-Fukunaga, R.; Ogawa, H.; Fukuyama, H.; Higashi, Y.; Nagata, S.; Fukunaga, R. Critical Role of the P400/MDomino Chromatin-Remodeling ATPase in Embryonic Hematopoiesis. Genes Cells 2007, 12, 581–592. [Google Scholar] [CrossRef] [PubMed]
- Couture, J.-P.; Nolet, G.; Beaulieu, E.; Blouin, R.; Gévry, N. The P400/Brd8 Chromatin Remodeling Complex Promotes Adipogenesis by Incorporating Histone Variant H2A.Z at PPARγ Target Genes. Endocrinology 2012, 153, 5796–5808. [Google Scholar] [CrossRef]
- Santopietro, M.V.; Ferreri, D.; Prozzillo, Y.; Dimitri, P.; Messina, G. The Multitalented TIP60 Chromatin Remodeling Complex: Wearing Many Hats in Epigenetic Regulation, Cell Division and Diseases. Epigenetics Chromatin 2025, 18, 40. [Google Scholar] [CrossRef] [PubMed]
- Bhatnagar, A.; Thomas, C.M.; Nge, G.G.; Zaya, A.; Dasari, R.; Chongtham, N.; Manandhar, B.; Kortagere, S.; Elefant, F. Tip60 HAT Activators as Therapeutic Modulators for Alzheimer’s Disease. Nat. Commun. 2025, 16, 3347. [Google Scholar] [CrossRef] [PubMed]
- Zohourian, N.; Coll, E.; Dever, M.; Sheahan, A.; Burns-Lane, P.; Brown, J.A.L. Evaluating the Cellular Roles of the Lysine Acetyltransferase Tip60 in Cancer: A Multi-Action Molecular Target for Precision Oncology. Cancers 2024, 16, 2677. [Google Scholar] [CrossRef]
- Wang, X.; Wan, T.C.; Kulik, K.R.; Lauth, A.; Smith, B.C.; Lough, J.W.; Auchampach, J.A. Pharmacological Inhibition of the Acetyltransferase Tip60 Mitigates Myocardial Infarction Injury. Dis. Model. Mech. 2023, 16, dmm049786. [Google Scholar] [CrossRef]
- Xiao, Y.; Nagai, Y.; Deng, G.; Ohtani, T.; Zhu, Z.; Zhou, Z.; Zhang, H.; Ji, M.Q.; Lough, J.W.; Samanta, A.; et al. Dynamic Interactions between TIP60 and P300 Regulate FOXP3 Function through a Structural Switch Defined by a Single Lysine on TIP60. Cell Rep. 2014, 7, 1471–1480. [Google Scholar] [CrossRef] [PubMed]
- Kumar, D.; Cinghu, S.; Oldfield, A.J.; Yang, P.; Jothi, R. Decoding the Function of Bivalent Chromatin in Development and Cancer. Genome Res. 2021, 31, 2170–2184. [Google Scholar] [CrossRef] [PubMed]
- Jacquet, K.; Fradet-Turcotte, A.; Avvakumov, N.; Lambert, J.-P.; Roques, C.; Pandita, R.K.; Paquet, E.; Herst, P.; Gingras, A.-C.; Pandita, T.K.; et al. The TIP60 Complex Regulates Bivalent Chromatin Recognition by 53BP1 through Direct H4K20me Binding and H2AK15 Acetylation. Mol. Cell 2016, 62, 409–421. [Google Scholar] [CrossRef]
- Gupta, H.; Gupta, A. Post-Translational Modifications of Epigenetic Modifier TIP60: Their Role in Cellular Functions and Cancer. Epigenetics Chromatin 2025, 18, 18. [Google Scholar] [CrossRef] [PubMed]

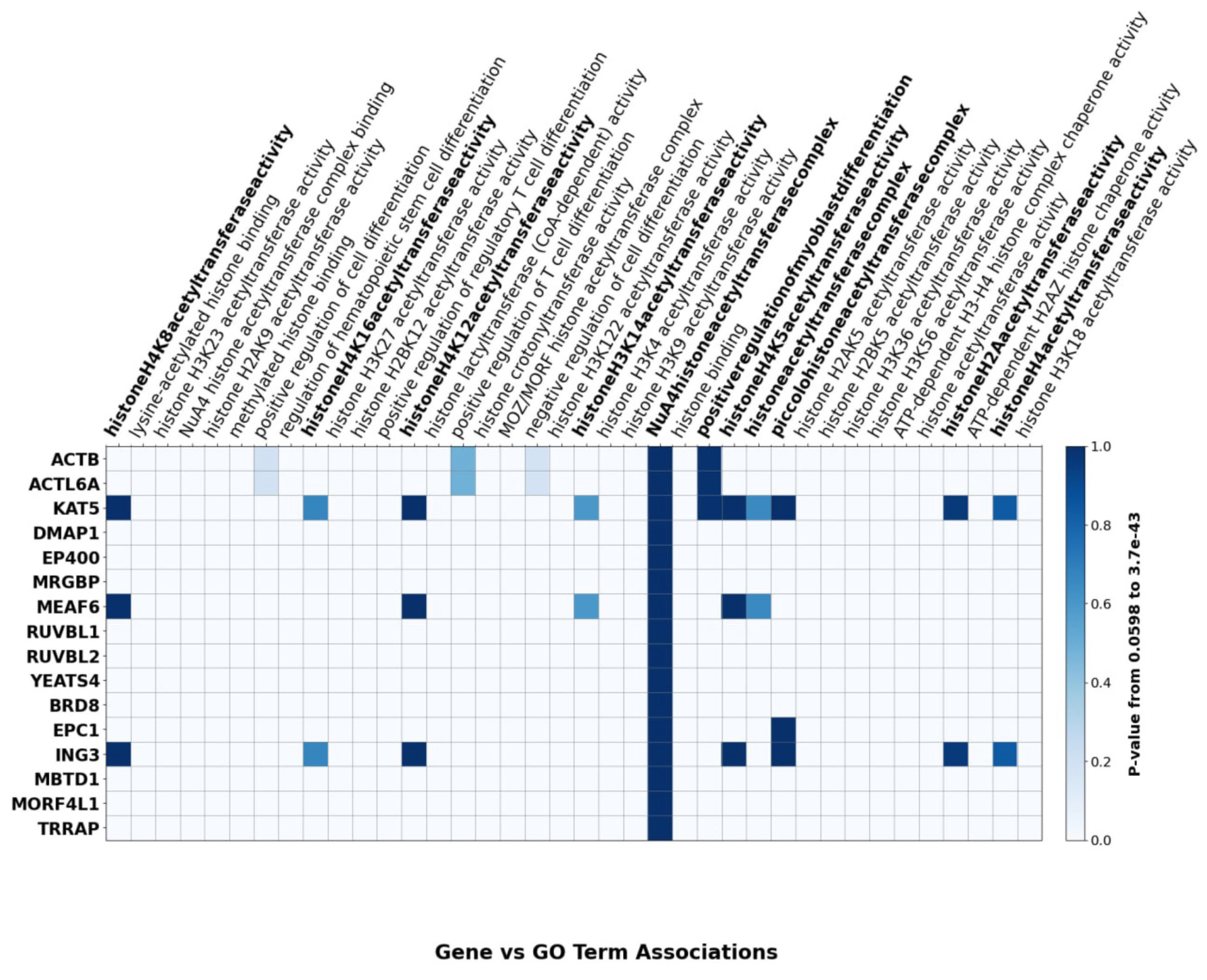
| Subunit | Full Name | Aliases | General Function/Category |
|---|---|---|---|
| KAT5 | Lysine Acetyltransferase 5 | TIP60 | Catalytic subunit; histone acetyltransferase (HAT) |
| EP400 | E1A Binding Protein p400 | p400 | ATP-dependent remodeler and central scaffold; histone variant exchange |
| EPC1 | Enhancer of Polycomb Homolog 1 | Epl1 (yeast homolog) | Scaffold; organizes/links modules and anchors HAT components |
| RUVBL1 | RuvB-Like AAA + ATPase 1 | Pontin, TIP49 | AAA+ ATPase; structural/regulatory core with RUVBL2 |
| RUVBL2 | RuvB-Like AAA + ATPase 2 | Reptin, TIP48 | AAA+ ATPase; structural/regulatory core with RUVBL1 |
| ING3 | Inhibitor of Growth Family Member 3 | — | PHD-finger, reader-type subunit; HAT module component |
| MRG15 | MORF4-Related Gene 15 | MORF4L1 | Chromodomain protein; reader-type subunit |
| MBTD1 | Malignant Brain Tumor Domain-Containing 1 | — | MBT-repeat reader; binds H4K20me |
| YEATS4 | YEATS Domain-Containing 4 | GAS41, YAF9 (yeast homolog) | YEATS-domain protein; reader-type subunit |
| TRRAP | Transformation/Transcription Domain-Associated Protein | — | Large scaffold; activator-binding hub (associates with EP400 SANT) |
| BRD8 | Bromodomain-Containing Protein 8 | — | Bromodomain subunit; links auxiliary components to EP400 N-terminus |
| DMAP1 | DNMT1-Associated Protein 1 | — | Auxiliary scaffold; stabilizes EP400 HSA/actin-related interfaces |
| ACTL6A | Actin-Like Protein 6A | BAF53A; ARP4 (yeast) | Actin-related subunit(s); two copies bound along EP400 HSA |
| VPS72 | Vacuolar Protein Sorting-Associated Protein 72 | YL1 | Histone chaperone; contacts RUVBL1/2 and EP400 |
| MEAF6 | MYST/Esa1-Associated Factor 6 | EAF6 (yeast homolog) | Small HAT subunit; stabilizer |
| MRGBP | MRG-Binding Protein | C20orf20 | Accessory protein; MRG15 partner |
| ACTB | β-Actin | Beta-actin | Actin subunit; forms heterodimer with ACTL6A |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hashemi, F.; Nourozi, A.; Loushab, M.S.; Riabowol, K. Functions of TIP60/NuA4 Complex Subunits in Cell Differentiation. Cells 2025, 14, 1720. https://doi.org/10.3390/cells14211720
Hashemi F, Nourozi A, Loushab MS, Riabowol K. Functions of TIP60/NuA4 Complex Subunits in Cell Differentiation. Cells. 2025; 14(21):1720. https://doi.org/10.3390/cells14211720
Chicago/Turabian StyleHashemi, Fatemeh, Aida Nourozi, Mojtaba Shaban Loushab, and Karl Riabowol. 2025. "Functions of TIP60/NuA4 Complex Subunits in Cell Differentiation" Cells 14, no. 21: 1720. https://doi.org/10.3390/cells14211720
APA StyleHashemi, F., Nourozi, A., Loushab, M. S., & Riabowol, K. (2025). Functions of TIP60/NuA4 Complex Subunits in Cell Differentiation. Cells, 14(21), 1720. https://doi.org/10.3390/cells14211720







