Abstract
The prevalence of Alzheimer’s disease (AD), the most common cause of age-associated dementia, is estimated to increase over the next decades. Evidence suggests neuro-immune signaling deregulation and risk genes beyond the amyloid-β (Aβ) deposition in AD pathology. We examined the temporal profile of inflammatory mediators and microglia deactivation/activation in the brain cortex and hippocampus of 3xTg-AD mice at 3- and 9-month-old. We found upregulated APP processing, decreased expression of CD11b, CX3CR1, MFG-E8, TNF-α, IL-1β, MHC-II and C/EBP-α and increased miR-146a in both brain regions in 3-month-old 3xTG-AD mice, suggestive of a restrictive regulation. Enhanced TNF-α, IL-1β, IL-6, iNOS, SOCS1 and Arginase 1 were only present in the hippocampus of 9-month-old animals, though elevation of HMGB1 and reduction of miR-146a and miR-124 were common features in the hippocampus and cortex regions. miR-155 increased early in the cortex and later in both regions, supporting its potential as a biomarker. Candidate downregulated target genes by cortical miR-155 included Foxo3, Runx2 and CEBPβ at 3 months and Foxo3, Runx2 and Socs1 at 9 months, which are implicated in cell survival, but also in Aβ pathology and microglia/astrocyte dysfunction. Data provide new insights across AD state trajectory, with divergent microglia phenotypes and inflammatory-associated features, and identify critical targets for drug discovery and combinatorial therapies.
1. Introduction
Alzheimer’s disease (AD) is, nowadays, the most common cause of dementia in elderly people, and, due to their complexity, no drugs have proven efficacy. However, the testing of some new treatments, now in the late stage of clinical development, may originate novel therapeutic approaches. A better understanding of the causes behind AD and an early diagnosis can open the door for future care and therapeutics. To achieve this goal, more than 150 AD animal models were developed according to AlzForum. Several mouse models have been generated to recapitulate behavior and neuropathology like that observed in AD patients. In this context, mouse models with mutations in more than one gene are considered to better mimic AD pathogenesis than those only reflecting single aspects [1].
To date, commonly used animal models are transgenic mice that overexpress human genes associated with familial AD, leading to the formation of senile plaques of amyloid-β (Aβ) peptide (e.g., overexpression of human amyloid precursor protein (APP)) alone or with presenilin-1 (PSEN1) and neurofibrillary tangles of hyperphosphorylated tau protein (e.g., overexpression of human microtubule associated protein tau (MAPT)), two hallmarks of AD [2]. Recently, the validity of such models has been questioned by the high failure rate of clinical trials of AD therapeutics (>99%) that were successful in preclinical trials using these models [3]. Thus, to enhance translational predictability, new models should be developed to incorporate genetics with multiple factors to further advance the molecular and cellular markers that trigger familial and sporadic AD-related cognitive decline [1,4]. Nevertheless, the data generated by experimental AD animal models can bring new highlights about specific aspects of AD pathogenesis if researchers consider the limitations of each model.
Although the introduction of wild-type forms of APP and PSEN1, as found in normal and non-familial AD cases, does not elicit AD pathology in mice, the transgenic mice overexpressing pathological human mutant proteins easily show Aβ aggregation, cognitive deficits, and some abnormal tau phosphorylation [5,6,7]. The mouse model APP/PS1ΔE9 that expresses APP with the Swedish mutation and mutant human PSEN1 with a deletion of exon 9 shows formation of amyloid plaques prior to typical cognitive impairments [8,9], is used to examine pathophysiological events associated to preclinical AD [10]. Another useful model of intraneuronal Aβ-induced neurodegeneration is the 5xFAD [11]. This mouse model expresses five familiar AD mutations, two for PSEN1 and three for APP, that induce Aβ overproduction to accelerate plaque development [12]. Consequently, 5xFAD mice develop intraneuronal Aβ at 1.5 months of age, amyloid plaques at 2 months and significant neuron loss at 9 months [12]. However, these models do not exhibit the development of the neurofibrillary tangles characteristic of the human disease [13]. On the other hand, the triple-transgenic mice model (3xTg-AD) progressively develops both Aβ plaques and neurofibrillary tangles with a temporal and spatial distribution that recapitulates the disease in humans [14,15]. These animals contain three major mutations associated with AD (APPSwe, PSEN1M146V and tauP301L), leading to intraneuronal Aβ accumulation first in the cortical region that later spreads into hippocampus, followed by the emergence of neurofibrillary tangles in the hippocampus that disseminate into cortical regions [14,15]. In previous studies conducted in the Center for Neuroscience and Cell Biology (CNC) at Coimbra, Portugal, the development of amyloid and tau pathologies in the 3xTg-AD mouse males was previously identified [16,17]. No differences in the number and phenotype of microglia and astrocytes, using the ionized calcium-binding adapter molecule 1 (IBA-1) and the glial fibrillary acidic protein (GFAP) assessment by immunohistochemistry, respectively, were identified in these animals at 3-month-old [16]. However, extensive microglia and astrocyte proliferation in the hippocampus and cortex of the 3xTg-AD mice were observed at 12 months. It should be noted that Aβ intraneuronal immunostaining was already found to be significantly enhanced in the hippocampus and prefrontal cortex in the mice at 3-month-old.
In these animals, behavioral alterations were identified as early as 2.5 months [18,19] and cognitive decline was detected at 4 months, when intraneuronal Aβ accumulated in the cortex, hippocampus and amygdala [20], which is consistent with the amyloid cascade hypothesis. Interestingly, the 3xTg-AD animals also showed extensive astrogliosis and microgliosis with upregulation of inflammatory molecules after 6 months [21,22], which occurs prior to neurofibrillary tangle detection.
An elevated number of activated astrocytes and microglia are usually found close to neurons and Aβ plaques [23]. Microglia, the resident immunocompetent and phagocytic cells in the central nervous system (CNS) [24], can be either neuroprotective or neurotoxic, thus promoting a decrease or worsening of disease progression, respectively [25]. In fact, while microglia phagocytic capacity contributes to CNS neuroprotection from excessive Aβ, microglial activation by Aβ soluble oligomers promotes excitotoxicity and neurodegeneration by the release of several inflammatory cytokines, such as tumor necrosis factor (TNF)-α and interleukin (IL)-1β, contributing to the onset and progression of AD [26,27]. Most curiously, single cell RNAseq studies using the 5xFAD mice and AD patient samples identified a novel microglia type associated with neurodegenerative diseases (DAM), the activation of which involved an initial downregulation of microglia checkpoints [28]. In addition, a dystrophic microglia phenotype was also identified in AD patient autopsied samples [29].
We have previously addressed changes in microglia reactivity with age and Aβ challenge using an in vitro aging model developed in our laboratory [30]. We showed that several neuroprotective functions, including phagocytosis, migration abilities and autophagy, are impaired by in vitro aging [31], while the Aβ-induced inflammatory response is also reduced in aged cells with the compromised expression of miRNAs associated with inflammatory responses (inflamma-miRNAs) [30]. miRNAs, a class of small, non-coding RNAS, are important players in controlling inflammation and have a key regulatory role in microglia polarization [32]. Most attractively, we demonstrated that exposure of young/reactive microglia to Aβ promotes cellular senescence [30], in accordance with the link between AD and immunosenescence [33]. Changes in several microglia inflammatory mediators, including inducible nitric oxide synthase (iNOS), TNF-α, IL-1β, high mobility group box 1 (HMGB1), suppressor of cytokine signaling 1 (SOCS1) and Arginase 1 gene expression levels, were found to be deregulated in our previous in vitro AD studies and aged-like microglia model [30,34].
In the present study, we decided to explore microglia reactivity and inflammatory mediators in the cortex and hippocampus of the 3xTg-AD at 3-, 6- and 9-month-old mice, which are temporal stages also used in other studies [35]. For that, we evaluated the mRNA expression of specific markers of microglia reactivity, namely functional genes, inflammatory cytokines, phenotype-related molecules, and inflamma-miRNAs. We report a reduction of CD11b, C-X3-C motif chemokine receptor 1 (CX3CR1) and milk fat globule-EGF factor 8 (MFG-E8) gene expression levels in the cortex and hippocampus of 3-month-old 3xTg-AD animals, as well as a decrease of inflammatory cytokines (TNF-α and IL-1β in both regions and IL-6, IL-18 and HMGB1 only in the brain cortex). Downregulation of the major histocompatibility complex class II (MHC-II) and CCAAT enhancer binding protein (C/EBP)-α pro-inflammatory markers was additionally observed in both brain regions, together with the anti-inflammatory SOCS1, transforming growth factor (TGF)-β and Arginase 1 genes, as well as with deregulated anti-inflammatory cytokines (IL-10 and IL-4) in the brain cortex. Increased levels of HMGB1 (both regions), of CX3CR1 (brain cortex) and of iNOS, TNF-α, IL-1β, IL-6, SOCS1 and Arginase 1 (hippocampus) were present in 9-month-old 3xTg-AD animals. We also provide evidence that miR-155, miR-124 and miR-146a were upregulated, mainly in the cortex of 3-month-old 3xTg-AD mice. Collectively, our data show an early dysfunctional/constrained microglia immune response that may represent a preventive intervention to hold the onset of the disease in the younger transgenic mice. However, with the appearance of homeostatic imbalance at 9 months, microglia in the 3xTg-AD mice develop an inflammatory response and lose their immune neuroprotective properties. In summary, according to our results, the 3xTg-AD mice at 3- month-old are a good model to explore the pathomechanisms that precede AD onset and to test novel therapeutic strategies towards balance and homeostasis.
2. Materials and Methods
2.1. Animals
Animal care followed the recommendations of the European Convention for the Protection of Vertebrate Animals Used for Experimental and Other Scientific Purposes (Council Directive 86/609/EEC) and National Law 1005/92 (rules for the protection of experimental animals). All animal procedures were approved by the Institutional Animal Care and Use Committee. Every effort was made to minimize suffering and the number of animals used in this study.
Breeding pairs of homozygous triple-transgenic AD (3xTg-AD) mice harboring PSEN1M146V, APPswe and tauP301L transgenes and age-matched non-transgenic mice with the same genetic background (C57BL/6 × 129 s), denominated as wild-type (WT) animals and used as controls, were kindly provided by Dr. Frank LaFerla from the University of California, Irvine (Irvine, CA, USA). Animals were genetically engineered as previously described [14,15]. Mice were bred and maintained at the CNC Faculty of Medicine animal house (license nº 520.000.000.2006, from the Portuguese animal welfare authorities), University of Coimbra, Portugal. The animals were maintained under controlled light and environmental conditions (12 h dark/light cycle, 23 ± 1 °C, 55 ± 5% relative humidity), having access to food and water, and breeding pairs were continuously genotyped for the detection of PSEN1M146V, APPswe and tauP301L transgenes, as shown in Figure 1. These 3xTg-AD mice did not show changes in IBA-1 or GFAP immunostaining at 3 months but revealed intraneuronal Aβ increase in the hippocampus and prefrontal cortex, as well as extensive microglia and astrocyte proliferation, at 12-month-old [16]. Based on such data and given our interest to identify early biomarkers of neuro-immune deregulation and homeostatic imbalance in the 3xTg-AD mouse model, we decided to assess potential glia-associated pathological drivers in male animals at 3-, 6- and 9-month-old. The animals were euthanized, and the brains were removed following transcardial perfusion with 20 mL of an ice-cold 0.9% NaCl solution. The brain hemispheres were separated and used for protein and mRNA extraction. For this purpose, each hemisphere was placed on an acrylic matrix and a 4 mm coronal section was cut with a stainless-steel razor. The hippocampal and cortical regions from this section were dissected and kept at −80 °C until protein and mRNA extraction.
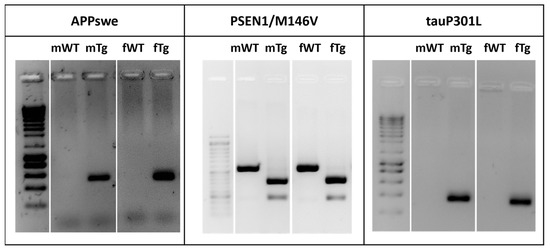
Figure 1.
3xTg-AD mice harbor APPswe, PSEN1M146V and tauP301L transgenes. Genotypic analysis for identification of 3xTg-AD mice. Representative gels comparing the human APPswe, PS1/M146V and tauP301L transgenes from tail DNA of breeding pairs (m—male and f—female) of WT and 3xTg-AD mice.
2.2. Evaluation of APP/Aβ Peptide and HMGB1 Expression
Detection of APP/Aβ protein and HMGB1 expression levels was processed by Western blot as usual in our laboratory [34]. Total protein extracts were obtained from brain extracts collected from the cortex and hippocampus of 3xTg-AD mice and their WT littermates. Briefly, tissues samples were lysed using TRIzol® (Life Technologies, Carlsbad, CA, USA), according to manufacturer’s instructions. Protein extracts were obtained as previously described [36] with minor alterations and stored at −80 °C. Protein concentrations were determined using BioRad protein assay (Bio-Rad, Hercules, CA, USA). Cell extracts containing equal amounts of protein (100–150 µg) were separated on a 10 to 15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) system and transferred to a nitrocellulose membrane. The membranes were blocked with 5% non-fat milk and incubated with the primary antibody mouse anti-Aβ (clone W0-2) antibody (1:500, SIGMA-Aldrich, St. Louis, MO, USA) that detects both APP and Aβ species, mouse HMGB1 (1:200, BioLegend, San Diego, CA, USA) and mouse β-actin (1:5000, SIGMA-Aldrich) overnight at 4 °C and then with horseradish peroxidase-labelled secondary antibodies for 1 h at room temperature. After extensive washes with saline buffer, immunoreactive bands were detected by LumiGLO® (Cell Signaling, Beverly, MA, USA) and visualized by chemiluminescence with ChemiDoc (Bio-Rad). Expression of HMGB1 expression was quantified by computerized image analysis using the Quantity One 1-D Analysis Software (Bio-Rad). Results were normalized to β-actin expression and expressed as fold vs. WT brain cortex from 3-month-old mice.
2.3. Gene and miRNA Expression Profiling
Determination of mRNA expression was performed by RealTime quantitative PCR (RT-qPCR) as usual in our laboratory [34]. Total RNA was extracted from hippocampal and cortical tissues of 3xTg-AD and WT animals using TRIzol® (LifeTechnologies), according to manufacturer’s instructions. Total RNA was quantified using Nanodrop ND-100 Spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). Aliquots of 1 µg of total RNA were treated with DNase I and then reverse transcribed into cDNA using oligo-dT primers and SuperScript II Reverse Transcriptase under the recommended conditions. RT-qPCR was performed using β-actin as an endogenous control to normalize the expression levels of transcription factors. The sequences used as primers are represented in Table 1. RT-qPCR was accomplished on a RT-PCR detection system (Applied Biosystems 7300 Fast Real-Time PCR System, Applied Biosystems, Madrid, Spain) using the SensiFAST SYBR® High-ROX kit (Bioline, Toronto, ON, Canada). The PCR was carried out in 96-well plates with each sample performed in triplicate, and no-template control was included for each amplification product. The RT-qPCR protocol was carried out under optimized conditions: 50 °C for 2 min followed by 95 °C for 2 min and, finally, 40 cycles at 95 °C for 0.05 min and 62 °C for 0.30 min. To verify the specificity of the amplification, a melt-curve analysis was performed immediately after the amplification protocol. Non-specific products of PCR were not found in any case. The results were normalized to β-actin in the same sample and relative mRNA concentrations calculated by the formula 2−ΔΔCT, considering 100% efficiency for each gene. ΔCT was the value obtained for each sample by assessing the difference between the mean CT value of each gene and the mean CT value of β-actin. ΔΔCT of one sample was the difference between its ΔCT value and the ΔCT of the sample chosen as reference, in our case the cortex of 3-month-old WT mice.

Table 1.
List of pairs of primers used to determine gene expression by RT-qPCR assays.
For miRNA analysis, cDNA conversion was performed with the universal cDNA Synthesis Kit (Qiagen, Hilden, Germany), as usual in our lab [34], using 5 ng total RNA according to the following protocol: 60 min at 42 °C followed by heat inactivation of the reverse transcriptase for 5 min at 95 °C. RT-qPCR was performed as previously indicated. For miRNA quantification, the miRCURY LNA™ Universal RT microRNA PCR system (Qiagen) was used in combination with pre-designed primers (Qiagen) for miR-155, miR-124, miR-146a and SNORD110 (reference gene) (Table 2). The reaction conditions consisted of polymerase activation/denaturation and well-factor determination at 95 °C for 10 min, followed by 50 amplification cycles at 95 °C for 10 s and 60 °C for 1 min (ramp rate 1.6°/s). The miRNA fold change with respect to the cortex of 3-month-old WT mouse samples was determined by the Pfaffl method, considering different amplification efficiencies of miRNAs in all experiments. The amplification efficiency for each target or reference RNA was determined according to the formula: E = 10(−1/S) – 1, where S is the slope of the obtained standard curve.

Table 2.
List of target sequences used to determine miRNA expression in RT-qPCR.
2.4. Assessment of miR-155 Target Expression
To assess the expression of genes regulated by miR-155, total RNA was extracted from cortical tissue of 3xTg-AD and WT animals and quantified as described above. A 500-ng amount of total RNA of each animal was pooled and mixed according to age (3 and 9 months) and disease condition (WT vs. 3xTg-AD). Aliquots of 500 ng of total mixed RNA were reverse transcribed into cDNA using the RT2 First Strand Kit (Qiagen, Hilden, Germany), according to manufacturer’s instructions. RT-qPCR was performed on a RT-PCR detection system (QuantStudio™ 7 Flex Real-Time PCR System, Applied Biosystems, Madrid, Spain) using the RT2 Profiler PCR Array 384-Well (4 × 96) Format (Qiagen) and a RT2 SYBR® Green qPCR Mastermix (Qiagen). The PCR was carried out in 384-well plates, and reverse transcription control (RTC) and positive PCR control (PPC) were included to determine the reverse transcription efficiency and PCR reaction, respectively. The results were normalized to a standard set of reference genes. The threshold cycle (CT) values were analyzed with the RT2 Profiler PCR Array Data Analysis Webportal and geometrically averaged and used for ΔΔCT calculations. Fold change was calculated by using ΔΔCT method [37], corresponding to the ratio of gene expression between the reference or control group (WT) and test group. Fold regulation was used to better read and interpret data. If the fold change values are greater than 1, the fold regulation and fold change values are the same and indicate upregulated or increased gene expression. Fold change values less than 1 imply that fold regulation is the negative inverse of the fold change and indicate downregulated or decreased gene expression. The results were presented in a heat map and further detailed in a scatter plot comparing the normalized expression of each gene between two groups (WT vs. 3xTg-AD). Gene-specific 2–ΔCT value in reference group was plotted on one axis against the corresponding value in test group on the other axis on a log base 10 scale to observe gene expression changes. Boundary lines were used to allow better visualization of upregulated and downregulated genes above and below a selected fold change value. The central line indicates unchanged gene expression. The section of the scatter plot above the fold change boundary lines contains genes upregulated in the y-axis group as compared to the x-axis group, and the section of the scatter plot below the fold change boundary lines contains genes downregulated in the y-axis group as compared to the x-axis group.
2.5. Statistical Analysis
Results of at least four different animals per experimental group are expressed as mean ± SEM. Significant differences between the assessed parameters were determined by the two-tailed Student’s t-test performed based on equal and unequal variance, as appropriate, using GraphPad Prism® 8.0 (GraphPad Software Inc., San Diego, CA, USA). p value less than 0.05 was considered statistically significant.
3. Results
3.1. The 3xTg-AD Animals Express Upregulated APP from 3 Months Forward
The 3xTg-AD mouse model, developed in LaFerla’s laboratory, presents cortical intraneuronal Aβ as early as 3 months, followed by cortical extracellular deposits and hippocampal intraneuronal Aβ at 6 months and diffuse plaque formation at 15 months [14]. So, first, we decided to evaluate APP/Aβ protein expression in 3-, 6- and 9-month-old animals before further analysis to be sure that the present cohort of 3xTg-AD mice expressed increased levels of these proteins. Using the Aβ (clone W0-2) antibody that detects both APP and Aβ, we were able to detect the APP species, as well as Aβ oligomers and Aβ monomers, as shown in Figure 2. As expected, we observed a clear expression of APP and Aβ monomers only in the 3xTg-AD mice at 3, 6 and 9 months, but no differences in Aβ oligomers when compared to age-matched WT animals. Although APP expression was not altered by age, Aβ monomers accumulation was age dependent. Since tau phosphorylation was only described in these 3xTg-AD mice at 12 months [14,38], a later time point than those evaluated, we did not assess such protein modifications.
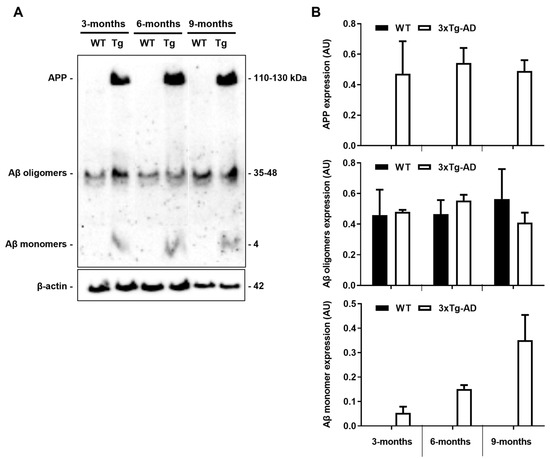
Figure 2.
Amyloid precursor protein (APP) and Aβ monomer expression levels are increased in the 3xTg-AD mouse model. Cortical samples from wild-type (WT) and 3xTg-AD animals were collected at 3-, 6- and 9-month-old and analyzed for APP/Aβ expression by Western blot using the Aβ (clone W0-2) antibody. (A) Representative image showing APP/Aβ bands. (B) Graph bars of the quantitative evaluation of APP/Aβ expression levels (arbitrary units, AU) are mean ± SEM representative of n = 3 animals per experimental group.
3.2. Expression of Microglia Homeostatic/Reactive Genes Is Decreased in the 3xTg-AD Mice at the Early Stage
Microglia were described to behave differently along AD progression in distinct animal models of the disease associated with Aβ pathology [39,40,41]. Our previous studies showed that microglia aged in vitro become irresponsive even when challenged with Aβ1-42 oligomers and fibrils [30,31]. In this section we decided to assess the temporal expression of microglia common markers in the 3xTg-AD animals. To do that, we evaluated the CD11b, CX3CR1 and MFG-E8 gene expression levels in the 3-, 6- and 9-month-old 3xTg-AD animals and age-matched WT mice. While CD11b, a β-integrin marker of microglia, is associated with microglial activation during neuroinflammation [42], CX3CR1 is involved in microglia reactivity [43] with still controversial effects in AD. Relative to MFG-E8, it was reported to be essential for microglia phagocytosis of affected neurons in the presence of Aβ peptide [44].
We observed a marked reduction of CD11b gene expression levels in the 3-month-old 3xTg-AD animals (~55% in the cortex and ~65% in the hippocampus, p < 0.01), together with that of CX3CR1 (~56% cortex and ~55% hippocampus, p < 0.05) and MFG-E8 (~89% cortex and ~80% hippocampus, p < 0.01) that disappeared in older animals (Figure 3). These results are in line with a restrictive microglia phenotype in the early-stage AD mice, suggesting an attempt to hold the inflammatory response to the initial increase of Aβ expression levels in the 3-month-old transgenic mice.
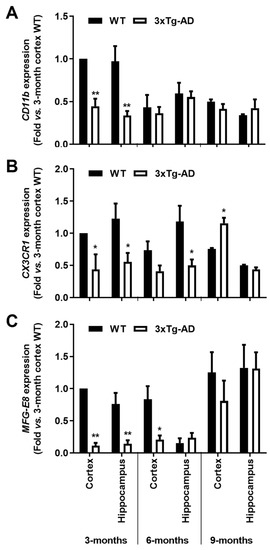
Figure 3.
Microglial typical markers are decreased in the brain cortex and hippocampus of the 3-month-old 3xTg-AD mice. Samples from the wild-type (WT) and 3xTg-AD animals were collected at 3-, 6- and 9-month-old and analyzed for (A) CD11b, (B) CX3CR1 and (C) MFG-E8 mRNAs by RT-qPCR. Results are mean ± SEM, representative of n = 4 animals per experimental group, and are expressed as fold change vs. 3-month-old cortex WT mice. * p < 0.05 and ** p < 0.01 vs. respective WT, two-tailed Student’s t-test.
3.3. Expression of Inflammatory Genes in the 3xTg-AD Mice Switch from Downregulated at 3 Months to Upregulated at 9 Months
Considering that microglia homeostatic/reactive genes were found depressed based on the expression of the CD11b, CX3CR1 and MFG-E8 markers, and since inflammatory genes were found downregulated in the SOD1G93A mouse model of amyotrophic lateral sclerosis before disease onset [45], we next assessed a set of inflammatory genes associated with neuroinflammation in neurodegenerative diseases. Actually, only a few studies using AD animal models have looked at the early stages of the disease, and, in those cases, pro-inflammatory cytokine expression was found to be most similar to WT animals [38,40] or sometimes even reduced [16,46].
Though we assessed samples from 3-, 6- and 9-month-old mice, the 6-month-old period did not reflect gross alterations, probably due to the altered dynamics of intra- and extracellular Aβ deposition at this temporal period [20], and it was excluded from subsequent data presentation.
As depicted in Figure 4, all the evaluated pro-inflammatory cytokines showed a generalized reduction of mRNA expression levels in the 3-month-old 3xTg-AD mice when compared to WT animals. In the cortex, TNF-α, IL- 6, IL-1β and IL-18 expression levels were decreased in the 3-month-old 3xTg-AD animals by more than 40% (p < 0.05). From these, TNF-α and IL-6 were still reduced in 9-month-old 3xTg-AD animals, while IL-18 and HMGB1 were upregulated. (1.4-fold, p < 0.01 and 1.3-fold, p < 0.05, respectively). When looking at the hippocampus, TNF-α, IL-1β and HMGB1 expression levels were found similarly depressed (p < 0.05) in the 3-month-old 3xTg-AD mice, but all were upregulated at 9 months (TNF-α, 2-fold, p < 0.01; IL-1β, 2.4-fold, p < 0.01; HMGB1, 2.1-fold, p < 0.05; and IL-6, 1.5-fold; p < 0.01). HMGB1 protein expression was found reduced in the 3-month-old 3xTg-AD mice, namely in the cortex (0.3-fold, p < 0.05) with no changes in samples from the 9-month-old animals (Figure 4H). Curiously, when evaluating the expression of anti-inflammatory cytokines (Figure 4E,F), while IL-10 was reduced in the cortex of 3-month-old 3xTg-AD mice (0.7-fold, p < 0.05), IL-4 expression was enhanced in samples from the same region and age (1.7-fold, p < 0.01) but markedly downregulated in the cortex of 9-month-old 3xTg-AD mice (1.3-fold in 3xTg-AD vs. 4.6-fold in WT mice, p < 0.05). Data show, for the first time, that the expression levels of inflammatory genes are downregulated early during AD disease in the 3xTg-AD mice, which is in accordance with the low expression of homeostatic and reactive markers demonstrated above.
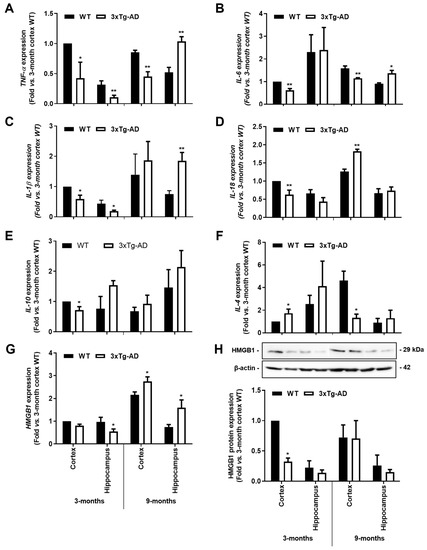
Figure 4.
Gene expression levels of pro-inflammatory cytokines are reduced in the 3xTg-AD animal model at the early stage, but some of them increase with disease progression. Brain samples from wild-type (WT) and 3xTg-AD animals were collected at 3 months and analyzed for (A) TNF-α, (B) IL-6, (C) IL-1β, (D) IL-18, (E) IL-10, (F) IL-4 and (G) HMGB1 mRNA expression by RT-qPCR, as well as for (H) representative Western blot for HMGB1 protein expression levels and respective quantification. Results are mean ± SEM, representative of n = 4 animals per experimental group, and are expressed as fold change vs. WT mouse cortex at 3-month-old. * p < 0.05 and ** p < 0.01 vs. respective WT, two-tailed Student’s t-test.
3.4. Microglia Genes Associated with Inflammatory Programs Differ with Age and Brain Regions in the 3xTg-AD Mice
It has been suggested that microglia pro-inflammatory polarization is lost in the early stages of AD and precede the occurrence of neuroinflammation. This later effect was demonstrated in AD models, including the APP/PS1, the APP/PS1ΔE9 and the 5xFAD mice [39,40]. Here, we assessed common pro- and anti-inflammatory markers of microglia-polarized subtypes in the cortex and the hippocampus of 3- and 9-month-old 3xTg-AD mice to better define the regional and age-dependent cell phenotypes. For that, we evaluated the mRNA expression of MHC-II, iNOS and C/EBP-α, which are usually considered pro-inflammatory microglial markers and SOCS1, TGF-β and Arginase 1, associated with anti-inflammatory properties.
At this point, we observed that both MHC-II and C/EBP-α were reduced by >55% and >35%, respectively, in both cortical and hippocampal tissues of the transgenic mice at 3-month-old (p < 0.05), as depicted in Figure 5A,B. Curiously, MHC-II was still reduced at 9 months in the 3xTg-AD hippocampus (~60%, p < 0.05, Figure 5A), while a slight upward trend was found for C/EBP-α (Figure 5B). For the expression of iNOS, mostly involved in the innate immune response of the conventional activated microglia [47], its predominant increase was only noticed at 9 months in the hippocampus (~2.9-fold, p < 0.05, Figure 5C). These data are in line with a restricted microglial phenotype early on and a more iNOS-associated cytotoxic profile later during AD progression.
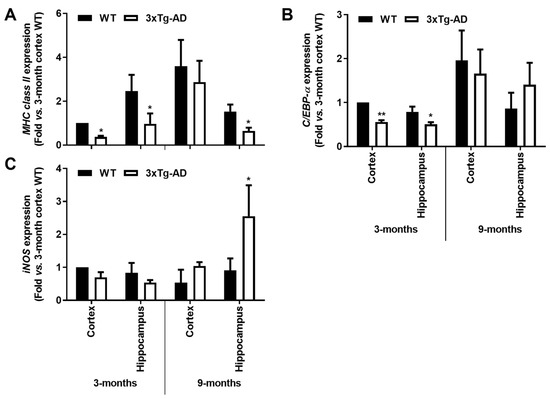
Figure 5.
Microglia-associated MHC-II and C/EBP-α pro-inflammatory markers are downregulated in the hippocampus and cortex from the 3-month-old 3xTg-AD mice, while iNOS induction is apparent in 9-month-old hippocampal mouse samples. Brain samples were collected from wild-type (WT) and 3xTg-AD animals at 3 and 9 months and analyzed for (A) MHC-II, (B) C/EBP-α and (C) iNOS mRNA expression by RT-qPCR. Results are mean ± SEM, representative of n = 4 animals per experimental group, and are expressed as fold change vs. 3-month-old cortical tissues from the WT mice. * p < 0.05 and ** p < 0.01 vs. respective WT, two-tailed Student’s t-test.
Reinforcing the depressed microglia polarization in the transgenic mice at 3-month- old, a marked reduction of anti-inflammatory markers was also noticed, namely in cortical tissues. Indeed, reduced gene expression of SOCS1 (<45%, p < 0.05), Arginase 1 (<35%, p < 0.05) and TGF-β (<65%, p < 0.05) was present in cortical samples from 3xTg-AD mice at 3-month-old. In the 9-month-old animals, only SOCS1 reduction was maintained in the cortical region. Contrasting data were obtained in the hippocampal region at this animal age, where not only the SOCS1 was found increased (>1.7-fold, p < 0.05, Figure 6A), but also Arginase 1 (>2-fold, p < 0.05, Figure 6B). Hippocampal increase of Arginase 1 in the transgenic animals at 9 months may indicate a compensatory mechanism against iNOS induction. Overall, both microglia pro- and anti-inflammatory gene-associated markers were found reduced in the early-stage disease of 3xTg-AD mice, suggesting deactivated microglia.
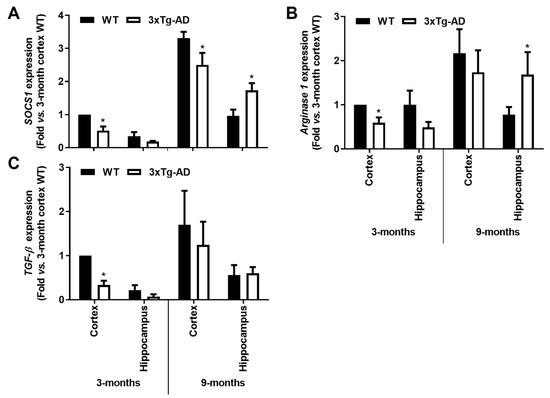
Figure 6.
Microglia-associated SOCS1 and Arginase 1 anti-inflammatory markers are downregulated in the cortex from the 3-month-old 3xTg-AD mice but upregulated in the 9-month hippocampal samples, with a further TGF-β decrease in the first. Brain samples were collected from wild-type (WT) and 3xTg-AD animals at 3 and 9 months and analyzed for (A) SOCS1, (B) Arginase 1 and (C) TGF-β mRNA expression levels by RT-qPCR. Results are expressed in graph bars as mean ± SEM, representative of n = 4 animals per experimental group and expressed as fold change vs. 3-month-old cortical tissues from the WT mice. * p < 0.05 vs. respective WT, two-tailed Student’s t-test.
3.5. Inflamma-miRNAs Are Mainly Overexpressed in the Cortical Region of 3-Month-Old 3xTg-AD Mice, and Only miR-155 Remains Elevated at 9 Months
Inflamma-miRNAs, such as miR-124, miR-155 and miR-146a, are known to play a fundamental role in the regulation of microglial polarization by targeting specific molecules involved in key signaling pathways [32,48]. In conformity, it turned out to be interesting to explore the representation of such miRNAs in our samples, considering the downregulation of the unique inflammatory and microglia markers found in the early stages of AD progression in the 3xTg-AD mouse model.
Here, we observed a ~4-fold increase of miR-155 in the cortex of 3-month-old 3xTg-AD animals (p < 0.01) which was sustained at 9 months for both cortical and hippocampal areas (~2-fold, p < 0.05, Figure 7A). These results may concur with the decrease of its target SOCS1, above mentioned. Upregulation of miR-124 was also found in the cortical samples of 3xTg-AD mice at 3-month-old (~1.8-fold, p < 0.05, Figure 7B), but it was decreased at 9 months in both cortical and hippocampal regions (~30% and ~15%, respectively, p < 0.05). The upregulation of miR-124 in the cortex of 3xTg-AD at 3-month-old may justify the reduction we observed for its target C/EBP-α at this age. As for miR-146a, recently considered to oppose the pathological processes of AD [49], elevated expression was only found in the early 3 months stage in both the cortex and the hippocampus (~1.8- and 1.4-fold, respectively, p < 0.05, Figure 7C), significantly decreasing thereafter (~35% and ~25% for cortex and hippocampus, respectively, p < 0.05, Figure 7C), which may explain the increase of inflammatory molecules we noticed in 9-month-old 3xTg-AD animals. In summary, miR-155 can be sorted as an early and continuous biomarker involved in the emergence of AD pathological processes in both the cortex and the hippocampus to which the miR-146a switch from upregulated to downregulated expression levels may be accounted.
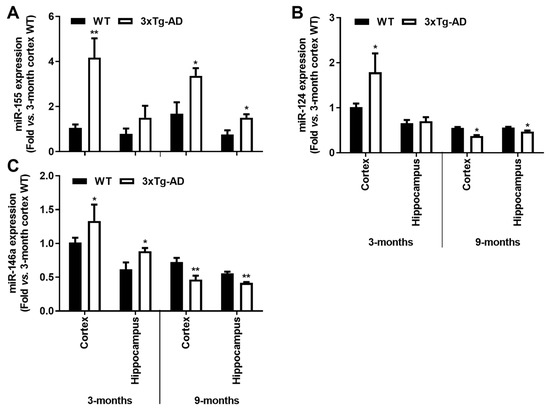
Figure 7.
Upregulation of inflammatory-associated miRNAs is a common feature in the brain cortex at the early-AD stage of the 3xTg-AD mice, and a single elevation of miR-155 characterizes both cortex and hippocampus at 9 months. Brain samples from wild-type (WT) and 3xTg-AD animals were collected at 3 and 9 months and analyzed for (A) miR-155, (B) miR-124 and (C) miR-146a expression levels by RT-qPCR. Results are expressed as mean ± SEM, representative of n = 4 animals per experimental group and expressed as fold change vs. 3-month-old cortical tissues from the WT mice. * p < 0.05 and ** p < 0.01 vs. respective WT, two-tailed Student’s t-test.
3.6. miR-155 Upregulation in the Cortical Region of 3xTg-AD Is Mainly Associated with a Downregulation of Genes Linked to Anti-Inflammatory and Neuroprotective Properties
As miR-155 was found to be a potential biomarker in the early stage of the AD disease, as shown by its cortical overexpression in the 3xTg-AD mice, we decided to explore the expression profile of its known target genes. We used cortical pooled samples from 3- and 9-month-old WT and 3xTg-AD mice and performed a PCR array analysis for the miR-155 targets. Looking at the expression of some already validated and predicted targets of miR-155, we found that, aside from all the individual variation, control animals clustered together, indicating expression profile similarities, followed by 3-month-old 3xTg-AD and then the 9-month-old transgenic animals. Data suggest that changes between the 3xTg-AD mice and the age-matched WT animals increase with age. Interestingly, results also showed that the elevation of miR-155 expression levels led to both up- and downregulation of several genes (Figure 8 and Figure 9).
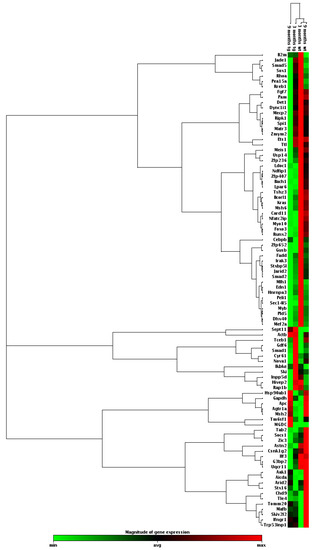
Figure 8.
Heat map of miR-155 targets by PCR array profiling for pooled samples from 3xTg-AD mice (tg) and age-matched wild-type (WT) mice. RNA was isolated from cortical brain samples of WT and 3xTg-AD animals at 3- and 9-month-old and analyzed for miR-155 target genes by RT-qPCR. Results are expressed as magnitude of gene expression from a pool of n = 4 animals per experimental group.
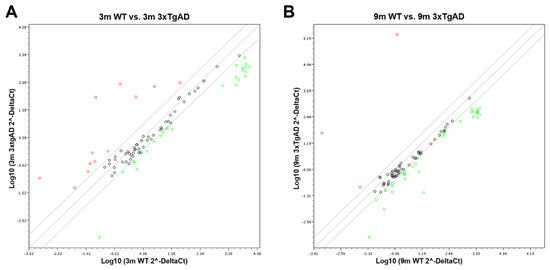
Figure 9.
Scatter plot representation of the expression of miR-155 targets in 3xTg-AD vs. wild-type (WT) mice. RNA was isolated from cortical brain samples of WT and 3xTg-AD animals at 3- (A) and 9-month-old (B) and analyzed for the expression of miR-155 target genes by RT-qPCR. Results are expressed as Log10 gene expression from a pool of n = 4 animals per experimental group.
The magnitude of the alterations obtained in cortical samples from the 3xTg-AD animals, as compared with age-matched WT mice, can be visualized in the heat map analysis depicted in Figure 8. Results obtained for the 9-month-old 3xTg-AD samples were those that mostly differed from the 3- and 9-month-old WT animals and included some upregulated genes but predominantly downregulated ones. However, increased, and decreased gene expression levels in the transgenic animals at 3-month-old, as compared with matched-age WT mice, were also clearly identified, and, again, a higher number of underexpressed than overexpressed genes were observed. To further explore differences between the samples collected from the cortex of 3xTg-AD mice at 3 months and 9 months with upregulated expression of miR-155, a more detailed scatter plot analysis of its targets was performed and compared with age-matched WT animals. At 3 months, the comparison of the 3xTg-AD vs. WT mice (Figure 9A) revealed the existence of 12 upregulated and 26 downregulated genes in the transgenic mice, which are listed in Table 3. Septin11 and Mafb were some of those that were increased, while Foxo3, Runx2 and C/ebpβ were found decreased.

Table 3.
Expression of miR-155 target genes in cortical samples from 3-month-old 3xTg-AD mice vs. respective wild-type (WT) animals.
Relative to the expression of miR-155 targets in the 9-month-old 3xTg-AD mice vs. WT animals (Figure 9B), the scatter plot analysis only identified three upregulated genes (e.g., Septin 11). In contrast, a large majority was found diminished. Runx2, Foxo3 and Socs1 were some examples among the 35 genes listed in Table 4 that we found to be downregulated. The Socs1 decrease corroborates our previous finding shown in Figure 6 for the 9-month-old cortical samples. Most of the downregulated genes are associated with deregulated neuro-immune interactions, microglia malfunction, synaptic connectivity deficits and amyloid pathology.

Table 4.
Expression of miR-155 target genes in cortical samples from 9-month-old 3xTg-AD mice vs. respective wild-type (WT) animals.
4. Discussion
Experiments in this study were established to identify initial biomarkers of neuro-immune deregulation and microglia malfunction using the cortex and the hippocampus of the 3xTg-AD mouse model of AD, which are two of the brain areas most affected by AD pathology [50]. Here, we observed that inflammatory mediators and microglia homeostatic/reactive markers are depressed early and can be detected at 3 months in our AD mouse model, suggesting that microglia and probably also astrocytes refrain their reactivity to hold the disease onset. In preceding studies, we found that microglia and astrocyte pro-inflammatory and reactive markers were also decreased at the presymptomatic stage in the SOD1G3A mice, a model commonly used to discriminate the pathological events of amyotrophic lateral sclerosis [45]. Actually, hypometabolism and bioenergetic deficits have been described as early emerging events in AD prodromal stage [51] and impaired glucose metabolism was found in the young 3xTg-AD mice [52]. This corroborates our present data showing a deficiency in CX3CR1, also obtained in AD mouse models in the past [53], and in MFG-E8, documented to be important in reversing microglia-induced astrocyte neurotoxicity [54]. Prior studies in our 3xTg-AD mouse model did not identify differences in the number or phenotype of microglia and astrocytes at 3-month-old, based on IBA-1 and GFAP immunostaining [16]. Validating the restricted neuro-immune balance in the 3-month-old 3xTg-AD mice it is also the low expression of HMGB1 protein levels, an alarmin indicated as mediating neuroinflammatory responses in brain injuries [55]. A decrease of HMGB1 was particularly noticed in the cortical region of the 3-month-old 3xTg-AD mice, where we also found an overexpression of IL-4 that was proposed as an endogenous defense mechanism in other studies [56]. Together with that, the IL-10 deficiency we also identified in the brain cortex at the early stage may additionally sustain the constrained neuro-immune balance once its decreased levels are associated with the preservation of synaptic integrity and cognitive function in the APP/PS1+ Il10−/− mouse model [57].
Once past reports demonstrated the existence of at least a sub-population of microglial cells with a restrictive phenotype [28], the gene assembly identified to be reduced at the early disease stage, together with miR-146a downregulation, validates the 3-month-old 3xTg-AD mouse as a good model to further explore the consequences of such homeostatic imbalance if immune-challenged. The cross-temporal upregulation of miR-155 can be sorted in this study as a most promising AD biomarker in the prodromal stage, requesting future studies to address this possibility in patients. miR-155 overexpression was also shown to trigger an increased number of downregulated genes, at least in the cortical region of the transgenic mice. It should be noted that such a neuro-immune depressed network may be considered a negative factor if reparative or insult-related responses are required. In such a case, the targeting of inhibitory checkpoint activation can have benefits against progressive neuropathological alterations in AD.
AD is a continuing and irreversible neurodegenerative disorder that gradually reduces memory and thinking skills, leading to the inability to perform simple daily tasks [23]. Inflammation in AD has been associated with circulating pro-inflammatory cytokines by stress and cellular senescence [58] and risk genes [59], mainly those expressed in microglia [60]. Microglial-derived cytokines are described to be neurotoxic, to support a chronic inflammatory milieu and to contribute to disease progression [26]. Reports also indicate that the secreted molecules can lead to divergent effects depending on the extent and type of microglial activation, thus proposing that a fine equilibrium should be preserved [61]. Thus, it is extremely important to understand the pathogenic processes in AD and to discover targets that may help in new drug discovery toward microglia homeostasis.
Animal models are important tools to identify the drivers behind the onset and progression of AD-related cognitive decline, as well as to assay potential pharmacological interventions. So, in our work, we used the 3xTg-AD mouse model of AD, which progressively develops both Aβ plaques and neurofibrillary tangles with a temporal and spatial distribution that closely mimics the human AD brain [14,15,38]. The sequence of neuropathological development in this model suggests that Aβ is the initiating trigger of cognitive decline [14]. The 3xTg-AD mice model develops AD pathology in an age-dependent and progressive manner, ahead of the associated behavioral changes [62], similar to human patients [63]. Intraneuronal Aβ accumulation was reported in the cortex of these animals at around 3-month-old, spreading to the hippocampus and amygdala by 6 months, together with diffuse amyloid plaques in the cerebral cortex [15,20]. However, more recently, Aβ accumulation was only reported in the hippocampus for 6-month-old animals and in the cortex for 12-month-old 3xTg-AD mice [38]. However, widespread senile plaques were only detected at 12 months [14,64], coupled with the emergence of conformational changes in tau [15]. Detection of APP in the 3xTg-AD animal at 3-, 6- and 9-month-old, although at levels below those reported [15], is in accordance with previous data in the same animal colony [16] and may correspond to the β-secretase-derived C-terminal fragment of APP found in the 3xTg-AD hippocampi [65]. In other studies, changes in APP levels were not found at 5–7 months, and not even observed between 12 and 18 months in the same model [66], representing either variation in the APP expression levels or in the antibody used to detect it among studies. Moreover, APP enrichment in exosomes was outlined as a sorting mechanism into the brain extracellular space [67].
An increased density of GFAP immunoreactive astrocytes and IBA1 immunoreactive microglia, as well as upregulation of inflammatory markers, were shown in 3xTg-AD mice compared with WT at 3 [68] and at 6/7 months [21,22,38], suggesting that it may be a response to Aβ accumulation. Curiously, we found a marked downregulation of the microglia homeostatic/reactive markers, CD11b, CX3CR1 and MFG-E8, at 3 months in the cortex and hippocampus of the 3xTg-AD mice, as compared with age-matched WT animals. Our findings corroborate the previous report by Janelsins and colleagues, showing decreased F4/80-positive microglia in the hippocampus of 3- and 6-month-old 3xTg-AD mice [69] and that of Rodriguez and colleagues mentioning an increased density of both resting/ramified and activated microglia in the hippocampus of 9-month-old 3xTg-AD animals prior to senile plaque spread [64]. This increased density of ramified microglia may be a preparation stage to refrain the Aβ-associated damage. Furthermore, a recent study showed that microglia reactivity is enhanced in the 3xTg-AD mice only following T. gondii infection at 5-, 6- or 11/12-month-old [70]. Finally, it should be noted that microglia have regional and age-dependent variations in microglial phenotypes and that the definition of a subtype requires the analysis of a set of genes [71]. Density of both resting and activated microglia was previously reported to significantly increase in 12- and 17-month-old 3xTg-AD mouse hippocampi but not at 9-month-old, only based on polyclonal affinity-purified rat antiserum raised against CD11b (MAC-1)-IR density with no further validation by gene signature [64]. Once increased, Cd11b microglia were observed near Aβ plaques by immunostaining [64,72]; such finding may justify the unchanged gene expression levels we obtained in our total homogenates. Moreover, it is documented that common microglial markers, such IBA1 and CD11b, stain both resting and activated cells and their determination was shown to be heterogeneous and to not demonstrate a consistent elevation in samples from the AD brain [73]. Moreover, CD11b was not found upregulated in the microglia of double APP/PS1 transgenic mice, while it was upregulated in the PS19 mouse model in the 3-month-old animals [72], corroborating the wide range of CD11b expression levels among AD models and its environment-dependent changes.
It has been described that microglia activation promotes the release of potentially cytotoxic molecules, which contribute to the onset and/or progression of AD [26]. Several cytokines, including IL-1β, IL-18 and TNF-α, were shown to be overexpressed in the AD brain [23,74]. In a previous study, we found that such pro-inflammatory cytokines are secreted by microglia upon the interaction with Aβ1-42 oligomers and fibrils [30]. However, expression levels in AD mouse models were usually found to be similar to WT animals [38,40], or even to be reduced [16,46]. In the present study, the diminished expression of CD11b, CX3CR1, MFG-E8, TNF-α and IL-1β gene levels in the brain cortices and hippocampi, with a more robust decrease for the last two genes, together with the decrease of HMGB1 protein and the pro-inflammatory genes IL-6 and IL-18 only in the cortex, validate the presence of an immunosuppressed microglia in the young 3xTg-AD animals and their failure in preventing AD plaque pathology [75]. The reduction of hippocampal expression levels of IL-6, TNF-α and IL-1β was also observed in the same 3xTg-AD at 3-month-old [16,69]. Moreover, while IL-10 was reduced in the cortex of 3-month-old 3xTg-AD animals, IL-4 was found enhanced. Interestingly, administration of IL-4 into an amyotrophic lateral sclerosis animal model was reported to induce microglia to adopt a slowly proliferating phenotype [76], which may corroborate a skewed microglia reactivity in these young animals. Curiously, IL-4 expression was deficient in the cortex of 9-month-old 3xTg-AD animals in opposition to upregulation of inflammatory markers, such as IL-1β and HMGB1. This inverse correlation was previously reported in the 3xTg-AD mouse model where hyperbaric oxygen therapy was shown to reduce gliosis in parallel with the decrease of pro-inflammatory cytokines (IL-1β and TNF-α) and the increase of Arginase 1 and anti-inflammatory cytokines (IL-4 and IL-10) [77].
In contrast, enhanced production of TNF-α and IL-1β was detected in the APP/PS1ΔE9 mouse model at 10-month-old [40] and upregulation of inflammatory genes at 4-6 months in the 5xFAD animals, namely in the hippocampus [39], which may, eventually, be linked to Aβ accumulation. Indeed, we found upregulated gene expression levels of TNF-α, IL-1β, IL-6 and HMGB1 in the hippocampus, with the last also increased in the cortex, in our 3xTg-AD mouse model. TNF-α and IL-1β have been described for a long time as mediators of AD pathogenesis [78]. HMGB1 was reported to accumulate extracellularly on Aβ plaques, being the protein level increased in AD brains [79]. More recently, HMGB1 was implicated in the pathogenesis of AD by causing a microglial Aβ phagocytosis dysfunction [80] and neurite degeneration [81]. Distinct microglia phenotypes were found in the frontal cortex samples of AD patients and associated with the presence of different Aβ species [82]. One with high expression levels of IL-1β, TNF-α and IL-12A typical of a pro-inflammatory phenotype related with oligomeric Aβ, the other located near senile plaques with upregulation of IL-1ra, Arginase 1 and FIZZ, is characteristic of a reparative phenotype.
MHC-II positive microglia participate in the adaptive immune response, though it is questionable whether it is a marker of amoeboid/activated or ramified/homeostatic microglia [83]. Unchanged levels of MHC-II gene expression were found in the cortex of the AD mouse model at 9 months. Decreased expression was present in the hippocampus at both 3 and 9 months and in the cortex only at 3 months. Other studies report no changes in the APP/PS1ΔE9 model [40]. C/EBP-α was described to regulate the transcription of the MHC-II gene in murine microglia [84]. Interestingly, it was found decreased in the cortex and hippocampus of 3-month-old 3xTg-AD animals but unaffected at 9 months. iNOS, the typical pro-inflammatory marker, was only upregulated at the hippocampus in the older animals, probably exerting cytotoxic effects since it uses arginine to produce nitric oxide [47]. Elevation of microglial iNOS was also observed in the APP/PS1 and APP/PS1ΔE9 models at 10-15 months [40].
It has been reported that SOCS1 is involved in the shift of macrophages from a pro-inflammatory to a reparative state [85], and SOCS1 was shown to attenuate Aβ-induced inflammation in a microglial murine cell line [86]. Decreased levels of SOCS1 were noticed in the cortex at both 3xTg-AD animal ages, though it was increased in the hippocampus at 9 months. In addition, Arginase 1 was shown to be upregulated in 9-months 3xTg-AD hippocampi and revealed to reduce Aβ plaque formation in conditions of IL-1β-dependent inflammation [87], as happened in our model with upregulated hippocampal IL-1β at this stage. TGF-β downregulation observed in the cortex of the young transgenic mice may facilitate Aβ-induced microglial chemotaxis, adding distinct microglial responses in the 3xTg-AD mice [88].
miRNAs regulate intracellular processes by targeting multiple mRNA molecules simultaneously, thus controlling immune cell phenotypes [16]. Deregulation of innate immune- and neuroinflammation-related miRNAs were identified in patient AD brains [89,90], together with disturbed microglia responses [91]. Here, we show that miR-146a is overexpressed as early as 3 months in the cortex and hippocampus of the 3xTg-AD mice, while similar findings for miR-155 and miR-124 only occurred in the cortical brain. Elevation of these miRNAs in AD patients and AD mouse models was previously documented [16,34,90,92,93,94,95]. Such early upregulation may represent compensatory mechanisms over the depressed neuro-immune axis in the transgenic mice at 3-month-old. Elevation of miR-155 can derive from upregulated HMGB1 [96] and may lead to decreased levels of SOCS1, its well-known target [97,98]. Indeed, it was recently shown that chronic treatment of 3xTg-AD mice with an anti-TNFSF10 monoclonal antibody inhibits retinal expression of miR-155, leading to SOCS1 upregulation [99]. A visible increase of miR-155 labelling was previously observed in the hippocampus region in 3-month-old 3xTg-AD mice, with a further strong enhancement at 12 months [16]. A decrease of miR-124 at 9 months may result from some neurodegenerative processes [32] and facilitate the pro-inflammatory status we found in the 3xTg-AD mice at such an age. Actually, Ponomarev and colleagues reported that the upregulation of miR-124 promoted microglia reparative processes by causing the downregulation of inflammatory-associated markers, such as TNF-α, iNOS and MHC-II [84]. Reduction of miR-146a contrasts with that of miR-155 upregulation and reinforces their opposite roles in immune response [100], pushing toward the pro-inflammatory signature and the increased expression of associated genes observed in the 3xTg-AD mice at 9-month-old.
The elevation of the miR-155 expression in the cortex at both 3 and 9 months was never explored in the 3xTg-AD mice for the alterations that it may induce in its multiple target genes. Looking at the expression of some miR-155-validated and predicted targets, we found that, aside from all the individual variation, control animals clustered together indicating expression profile similarities, followed by 3-month-old 3xTg-AD and then the 9-month-old transgenic animal. Data suggest that changes between the 3xTg-AD mice and the age-matched WT animals increase with age. As expected, most of the target genes were found decreased, i.e., 26 genes at 3 months and 35 genes at 9 months. However, some upregulation was also noticed, mainly in the younger transgenic animals, probably related with indirect effects to distal mRNAs. Septin11 was elevated at both ages and the Mafb gene only at 3 months in the 3xTg-AD mice. Genes from the Septin family are involved in vesicle trafficking and synaptic connectivity [101] and favor the β-amyloidogenic processing of APP through BACE1 accumulation [102], while the MafB gene is implicated in microglia inflammatory control [103]. MafB is a transcription factor involved in microglia differentiation [104] with an important role in the maintenance of homeostasis in adulthood if one considers that the knockout of microglia Mafb induces the expression of inflammation-related pathways [103]. So, MafB upregulation may be linked to the decrease of the inflammatory markers obtained in the 3-month-old transgenic mice. Relative to the downregulated genes, the C/EBP-β gene is, indeed, a direct target of miR-155 in this AD model [16], and its reduction is linked to a decrease in cell apoptosis [105] and delayed cell senescence [106]. On the other hand, the downregulation of Run2, at both 3 and 9 months in the transgenic animals may lead to a reduction in microglia phagocytic ability [107]. Relative to the Foxo3 gene, considered to be associated to longevity [108], its downregulation in 5xFAD mice also showed to exacerbate Aβ pathology and synapse loss by hampering the uptake of Aβ by astrocytes and microglia [109]. These findings open new avenues for further target evaluation concerning common and causal distinct stages of microglia and astrocyte dysfunctionalities in AD onset and progression. By showing divergent microglia phenotypes and inflammatory-associated features across AD state trajectory, the 3xTg-AD mice may be the appropriate model for fine-tuning the genetic signature of the brain’s immune cells at the different unbalanced homeostatic windows that characterize AD progression.
5. Conclusions
Overall, our results highlight a restricted microglia phenotype at both the cortex and hippocampus in the 3-month-old 3xTg-AD mice. This loss in microglia reactivity may be responsible for the reduced levels of tissue and microglia inflammatory mediators at such an age. Such homeostatic deregulation can be interpreted as a defensive mechanism to prevent Aβ accumulation, in accordance with the global downregulation of the expression of genes encoding proteins that are metastable to aggregation, as observed in another study [110]. However, the upregulation of miRNAs, mainly of miR-155, may counterbalance the defective neuro-immune response, though the downregulated miR-155 target genes seem to have dual intervention. On the one hand, they favor Aβ pathology by increased APP processing and decreased Aβ uptake. On the other hand, they support cell survival. Reduced expression levels of miR-146a and miR-124, considered to be anti-inflammatory [32], are believed to contribute to the upregulated immune system crosstalk we observed in 9-month-old 3xTg-AD mice. Upregulated inflammatory mediators predominate at the hippocampus at such an age. These age-dependent neuroinflammatory changes may be critical to the cognitive decline [111] and explain the controversies about the therapeutic efficacy of non-steroidal anti-inflammatory drugs (NSAIDs) in AD [112,113]. In summary, if the homeostatic microglia phenotype is lost early, the miR-155 overexpression and the upregulated APP processing mechanisms are among the first pathological features preceding AD onset. Future studies should explore which modulators for the identified targets are able to recover homeostasis and prevent the course of the disease.
Author Contributions
The study was conceived, designed and directed by D.B., who supervised C.C. (Cláudia Caldeira) and C.C. (Carolina Cunha). E.F. was responsible for animal breeding and sample collection. C.C. (Cláudia Caldeira), C.C. (Carolina Cunha), A.F. and A.R.V. planned and performed sample processing and data acquisition. Data analysis and manuscript writing were performed by A.F., C.C. (Cláudia Caldeira) and D.B. D.B. edited the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by FEDER/COMPETE/FCT—Fundação para a Ciência e a Tecnologia JPco-fuND/0003/2015, PTDC/MED-NEU/31395/2017 to DB, Programa Operacional Regional de Lisboa and the Programa Operacional Competitividade e Internacionalização (LISBOA- 01-0145-FEDER-031395) to DB, FCT- EXPL/NEU-NMC/1003/2013 to AF and by the prize Edgar Cruz e Silva 2012 from Grupo de Estudo do Envelhecimento Cerebral e Demência to DB, in part by UID/DTP/04138/2019 and UIDB/UIDP/04138/2020 to iMed.ULisboa, as well as by UID/NEU/04539/2019 and UIDB/04539/2020 to CNC and CEECIND/00322/2017 to EF.
Institutional Review Board Statement
Animal care followed the recommendations of the European Convention for the Protection of Vertebrate Animals Used for Experimental and Other Scientific Purposes (Council Directive 86/609/EEC) and National Law 1005/92 (rules for protection of experimental animals) under the approved license nº 520.000.000.2006 from the Portuguese animal welfare authorities.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding authors.
Acknowledgments
This work is part of an EU Joint Program—Neurodegenerative Disease Research (JPND) project, supported by FEDER/COMPETE/FCT—Fundação para a Ciência e a Tecnologia JPco-fuND/0003/2015 and PTDC/MED-NEU/31395/2017 to DB, FCT- EXPL/NEU-NMC/1003/2013 to AF, as well as by the prize Edgar Cruz e Silva 2012 from Grupo de Estudo do Envelhecimento Cerebral e Demência to DB. Also, by UID/DTP/04138/2019 and UIDB/UIDP/04138/2020 to iMed.ULisboa, as well as by UID/NEU/04539/2019 and UIDB/04539/2020 to CNC. EF is supported by an assistant researcher contract from FCT (CEECIND/00322/2017). We are grateful to Ana Cristina Rego and Ana Luísa Cardoso for providing additional samples from the 3xTg-AD colony that allowed the validation of some data.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study, in the collection, analyses or interpretation of data, in the writing of the manuscript or in the decision to publish the results.
References
- Vitek, M.P.; Araujo, J.A.; Fossel, M.; Greenberg, B.D.; Howell, G.R.; Rizzo, S.J.S.; Seyfried, N.T.; Tenner, A.J.; Territo, P.R.; Windisch, M.; et al. Translational animal models for Alzheimer’s disease: An Alzheimer’s Association Business Consortium Think Tank. Alzheimers Dement. 2020, 6, e12114. [Google Scholar] [CrossRef] [PubMed]
- Drummond, E.; Wisniewski, T. Alzheimer’s disease: Experimental models and reality. Acta Neuropathol. 2017, 133, 155–175. [Google Scholar] [CrossRef]
- Cummings, J.L.; Morstorf, T.; Zhong, K. Alzheimer’s disease drug-development pipeline: Few candidates, frequent failures. Alzheimer’s Res. Ther. 2014, 6, 37. [Google Scholar] [CrossRef]
- Zhang, L.; Chen, C.; Mak, M.S.; Lu, J.; Wu, Z.; Chen, Q.; Han, Y.; Li, Y.; Pi, R. Advance of sporadic Alzheimer’s disease animal models. Med. Res. Rev. 2020, 40, 431–458. [Google Scholar] [CrossRef] [PubMed]
- Duyckaerts, C.; Potier, M.C.; Delatour, B. Alzheimer disease models and human neuropathology: Similarities and differences. Acta Neuropathol. 2008, 115, 5–38. [Google Scholar] [CrossRef]
- Howlett, D.R.; Richardson, J.C. The pathology of APP transgenic mice: A model of Alzheimer’s disease or simply overexpression of APP? Histol. Histopathol. 2009, 24, 83–100. [Google Scholar]
- Morrissette, D.A.; Parachikova, A.; Green, K.N.; LaFerla, F.M. Relevance of transgenic mouse models to human Alzheimer disease. J. Biol. Chem. 2009, 284, 6033–6037. [Google Scholar] [CrossRef]
- Jankowsky, J.L.; Fadale, D.J.; Anderson, J.; Xu, G.M.; Gonzales, V.; Jenkins, N.A.; Copeland, N.G.; Lee, M.K.; Younkin, L.H.; Wagner, S.L.; et al. Mutant presenilins specifically elevate the levels of the 42 residue beta-amyloid peptide in vivo: Evidence for augmentation of a 42-specific gamma secretase. Hum. Mol. Genet. 2004, 13, 159–170. [Google Scholar] [CrossRef] [PubMed]
- Volianskis, A.; Kostner, R.; Molgaard, M.; Hass, S.; Jensen, M.S. Episodic memory deficits are not related to altered glutamatergic synaptic transmission and plasticity in the CA1 hippocampus of the APPswe/PS1deltaE9-deleted transgenic mice model of ss-amyloidosis. Neurobiol. Aging 2010, 31, 1173–1187. [Google Scholar] [CrossRef]
- Zou, C.; Shi, Y.; Ohli, J.; Schuller, U.; Dorostkar, M.M.; Herms, J. Neuroinflamm. impairs adaptive structural plasticity of dendritic spines in a preclinical model of Alzheimer’s disease. Acta Neuropathol. 2016, 131, 235–246. [Google Scholar] [CrossRef] [PubMed]
- Eimer, W.A.; Vassar, R. Neuron loss in the 5XFAD mouse model of Alzheimer’s disease correlates with intraneuronal Abeta42 accumulation and Caspase-3 activation. Mol. Neurodegener. 2013, 8, 2. [Google Scholar] [CrossRef]
- Oakley, H.; Cole, S.L.; Logan, S.; Maus, E.; Shao, P.; Craft, J.; Guillozet-Bongaarts, A.; Ohno, M.; Disterhoft, J.; Van Eldik, L.; et al. Intraneuronal beta-amyloid aggregates, neurodegeneration, and neuron loss in transgenic mice with five familial Alzheimer’s disease mutations: Potential factors in amyloid plaque formation. J. Neurosci. 2006, 26, 10129–10140. [Google Scholar] [CrossRef] [PubMed]
- Schwab, C.; Klegeris, A.; McGeer, P.L. Inflammation in transgenic mouse models of neurodegenerative disorders. Biochim. Biophys. Acta 2009, 1802, 889–902. [Google Scholar] [CrossRef] [PubMed]
- Oddo, S.; Caccamo, A.; Kitazawa, M.; Tseng, B.P.; LaFerla, F.M. Amyloid deposition precedes tangle formation in a triple transgenic model of Alzheimer’s disease. Neurobiol. Aging 2003, 24, 1063–1070. [Google Scholar] [CrossRef] [PubMed]
- Oddo, S.; Caccamo, A.; Shepherd, J.D.; Murphy, M.P.; Golde, T.E.; Kayed, R.; Metherate, R.; Mattson, M.P.; Akbari, Y.; LaFerla, F.M. Triple-transgenic model of Alzheimer’s disease with plaques and tangles: Intracellular Abeta and synaptic dysfunction. Neuron 2003, 39, 409–421. [Google Scholar] [CrossRef]
- Guedes, J.R.; Custodia, C.M.; Silva, R.J.; de Almeida, L.P.; Pedroso de Lima, M.C.; Cardoso, A.L. Early miR-155 upregulation contributes to neuroinflammation in Alzheimer’s disease triple transgenic mouse model. Hum. Mol. Genet. 2014, 23, 6286–6301. [Google Scholar] [CrossRef]
- Mota, S.I.; Ferreira, I.L.; Valero, J.; Ferreiro, E.; Carvalho, A.L.; Oliveira, C.R.; Rego, A.C. Impaired Src signaling and post-synaptic actin polymerization in Alzheimer’s disease mice hippocampus--linking NMDA receptors and the reelin pathway. Exp. Neurol. 2014, 261, 698–709. [Google Scholar] [CrossRef]
- Pardossi-Piquard, R.; Lauritzen, I.; Bauer, C.; Sacco, G.; Robert, P.; Checler, F. Influence of Genetic Background on Apathy-Like Behavior in Triple Transgenic AD Mice. Curr. Alzheimer Res. 2016, 13, 942–949. [Google Scholar] [CrossRef][Green Version]
- Gimenez-Llort, L.; Blazquez, G.; Canete, T.; Johansson, B.; Oddo, S.; Tobena, A.; LaFerla, F.M.; Fernandez-Teruel, A. Modeling behavioral and neuronal symptoms of Alzheimer’s disease in mice: A role for intraneuronal amyloid. Neurosci. Biobehav. Rev. 2007, 31, 125–147. [Google Scholar] [CrossRef]
- Billings, L.M.; Oddo, S.; Green, K.N.; McGaugh, J.L.; LaFerla, F.M. Intraneuronal Abeta causes the onset of early Alzheimer’s disease-related cognitive deficits in transgenic mice. Neuron 2005, 45, 675–688. [Google Scholar] [CrossRef]
- Janelsins, M.C.; Mastrangelo, M.A.; Park, K.M.; Sudol, K.L.; Narrow, W.C.; Oddo, S.; LaFerla, F.M.; Callahan, L.M.; Federoff, H.J.; Bowers, W.J. Chronic neuron-specific tumor necrosis factor-alpha expression enhances the local inflammatory environment ultimately leading to neuronal death in 3xTg-AD mice. Am. J. Pathol. 2008, 173, 1768–1782. [Google Scholar] [CrossRef] [PubMed]
- Caruso, D.; Barron, A.M.; Brown, M.A.; Abbiati, F.; Carrero, P.; Pike, C.J.; Garcia-Segura, L.M.; Melcangi, R.C. Age-related changes in neuroactive steroid levels in 3xTg-AD mice. Neurobiol. Aging 2013, 34, 1080–1089. [Google Scholar] [CrossRef]
- Rubio-Perez, J.M.; Morillas-Ruiz, J.M. A review: Inflammatory process in Alzheimer’s disease, role of cytokines. Sci. World J. 2012, 2012, 756357. [Google Scholar] [CrossRef] [PubMed]
- Krause, D.L.; Muller, N. Neuroinflammation, microglia and implications for anti-inflammatory treatment in Alzheimer’s disease. Int. J. Alzheimers Dis. 2010, 2010, 732806. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, M.; Butovsky, O.; Bruck, W.; Hanisch, U.K. Microglial phenotype: Is the commitment reversible? Trends Neurosci. 2006, 29, 68–74. [Google Scholar] [CrossRef] [PubMed]
- Lopategui Cabezas, I.; Herrera Batista, A.; Penton Rol, G. The role of glial cells in Alzheimer disease: Potential therapeutic implications. Neurologia 2014, 29, 305–309. [Google Scholar] [CrossRef]
- Mizuno, T. The biphasic role of microglia in Alzheimer’s disease. Int. J. Alzheimers Dis. 2012, 2012, 737846. [Google Scholar] [CrossRef]
- Keren-Shaul, H.; Spinrad, A.; Weiner, A.; Matcovitch-Natan, O.; Dvir-Szternfeld, R.; Ulland, T.K.; David, E.; Baruch, K.; Lara-Astaiso, D.; Toth, B.; et al. A Unique Microglia Type Associated with Restricting Development of Alzheimer’s Disease. Cell 2017, 169, 1276–1290. [Google Scholar] [CrossRef] [PubMed]
- Bachstetter, A.D.; Van Eldik, L.J.; Schmitt, F.A.; Neltner, J.H.; Ighodaro, E.T.; Webster, S.J.; Patel, E.; Abner, E.L.; Kryscio, R.J.; Nelson, P.T. Disease-related microglia heterogeneity in the hippocampus of Alzheimer’s disease, dementia with Lewy bodies, and hippocampal sclerosis of aging. Acta Neuropathol. Commun. 2015, 3, 32. [Google Scholar] [CrossRef]
- Caldeira, C.; Cunha, C.; Vaz, A.R.; Falcão, A.S.; Barateiro, A.; Seixas, E.; Fernandes, A.; Brites, D. Key Aging-Associated Alterations in Primary Microglia Response to Beta-Amyloid Stimulation. Front. Aging Neurosci. 2017, 9, 277. [Google Scholar] [CrossRef]
- Caldeira, C.; Oliveira, A.F.; Cunha, C.; Vaz, A.R.; Falcão, A.S.; Fernandes, A.; Brites, D. Microglia change from a reactive to an age-like phenotype with the time in culture. Front. Cell Neurosci. 2014, 8, 152. [Google Scholar] [CrossRef]
- Brites, D. Regulatory function of microRNAs in microglia. Glia 2020, 68, 1631–1642. [Google Scholar] [CrossRef]
- Richartz, E.; Stransky, E.; Batra, A.; Simon, P.; Lewczuk, P.; Buchkremer, G.; Bartels, M.; Schott, K. Decline of immune responsiveness: A pathogenetic factor in Alzheimer’s disease? J. Psychiatr. Res. 2005, 39, 535–543. [Google Scholar] [CrossRef]
- Fernandes, A.; Ribeiro, A.R.; Monteiro, M.; Garcia, G.; Vaz, A.R.; Brites, D. Secretome from SH-SY5Y APPSwe cells trigger time-dependent CHME3 microglia activation phenotypes, ultimately leading to miR-21 exosome shuttling. Biochimie 2018, 155, 67–82. [Google Scholar] [CrossRef]
- Pairojana, T.; Phasuk, S.; Suresh, P.; Huang, S.P.; Pakaprot, N.; Chompoopong, S.; Hsieh, T.C.; Liu, I.Y. Age and gender differences for the behavioral phenotypes of 3xTg alzheimer’s disease mice. Brain Res. 2021, 1762, 147437. [Google Scholar] [CrossRef] [PubMed]
- Simões, A.E.; Pereira, D.M.; Amaral, J.D.; Nunes, A.F.; Gomes, S.E.; Rodrigues, P.M.; Lo, A.C.; D’Hooge, R.; Steer, C.J.; Thibodeau, S.N.; et al. Efficient recovery of proteins from multiple source samples after TRIzol((R)) or TRIzol((R))LS RNA extraction and long-term storage. BMC Genom. 2013, 14, 181. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Belfiore, R.; Rodin, A.; Ferreira, E.; Velazquez, R.; Branca, C.; Caccamo, A.; Oddo, S. Temporal and regional progression of Alzheimer’s disease-like pathology in 3xTg-AD mice. Aging Cell 2019, 18, e12873. [Google Scholar] [CrossRef]
- Landel, V.; Baranger, K.; Virard, I.; Loriod, B.; Khrestchatisky, M.; Rivera, S.; Benech, P.; Feron, F. Temporal gene profiling of the 5XFAD transgenic mouse model highlights the importance of microglial activation in Alzheimer’s disease. Mol. Neurodegener. 2014, 9, 33. [Google Scholar] [CrossRef]
- Martin, E.; Boucher, C.; Fontaine, B.; Delarasse, C. Distinct inflammatory phenotypes of microglia and monocyte-derived macrophages in Alzheimer’s disease models: Effects of aging and amyloid pathology. Aging Cell 2017, 16, 27–38. [Google Scholar] [CrossRef]
- Dionisio-Santos, D.A.; Behrouzi, A.; Olschowka, J.A.; O’Banion, M.K. Evaluating the Effect of Interleukin-4 in the 3xTg Mouse Model of Alzheimer’s Disease. Front. Neurosci. 2020, 14, 441. [Google Scholar] [CrossRef]
- Roy, A.; Fung, Y.K.; Liu, X.; Pahan, K. Up-regulation of microglial CD11b expression by nitric oxide. J. Biol. Chem. 2006, 281, 14971–14980. [Google Scholar] [CrossRef]
- Limatola, C.; Ransohoff, R.M. Modulating neurotoxicity through CX3CL1/CX3CR1 signaling. Front. Cell Neurosci. 2014, 8, 229. [Google Scholar] [CrossRef]
- Neniskyte, U.; Brown, G.C. Lactadherin/MFG-E8 is essential for microglia-mediated neuronal loss and phagoptosis induced by amyloid beta. J. Neurochem. 2013, 126, 312–317. [Google Scholar] [CrossRef] [PubMed]
- Cunha, C.; Santos, C.; Gomes, C.; Fernandes, A.; Correia, A.M.; Sebastiao, A.M.; Vaz, A.R.; Brites, D. Downregulated Glia Interplay and Increased miRNA-155 as Promising Markers to Track ALS at an Early Stage. Mol. Neurobiol. 2018, 55, 4207–4224. [Google Scholar] [CrossRef]
- Iwahara, N.; Hisahara, S.; Kawamata, J.; Matsumura, A.; Yokokawa, K.; Saito, T.; Fujikura, M.; Manabe, T.; Suzuki, H.; Matsushita, T.; et al. Role of Suppressor of Cytokine Signaling 3 (SOCS3) in Altering Activated Microglia Phenotype in APPswe/PS1dE9 Mice. J. Alzheimer’s Dis. JAD 2017, 55, 1235–1247. [Google Scholar] [CrossRef]
- Colton, C.A. Heterogeneity of microglial activation in the innate immune response in the brain. J. Neuroimmune Pharm. 2009, 4, 399–418. [Google Scholar] [CrossRef] [PubMed]
- Ponomarev, E.D.; Veremeyko, T.; Weiner, H.L. MicroRNAs are universal regulators of differentiation, activation, and polarization of microglia and macrophages in normal and diseased CNS. Glia 2013, 61, 91–103. [Google Scholar] [CrossRef]
- Liang, C.; Zou, T.; Zhang, M.; Fan, W.; Zhang, T.; Jiang, Y.; Cai, Y.; Chen, F.; Chen, X.; Sun, Y.; et al. MicroRNA-146a switches microglial phenotypes to resist the pathological processes and cognitive degradation of Alzheimer’s disease. Theranostics 2021, 11, 4103–4121. [Google Scholar] [CrossRef] [PubMed]
- Braak, H.; Braak, E.; Bohl, J. Staging of Alzheimer-related cortical destruction. Eur. Neurol. 1993, 33, 403–408. [Google Scholar] [CrossRef]
- Caldwell, C.C.; Yao, J.; Brinton, R.D. Targeting the prodromal stage of Alzheimer’s disease: Bioenergetic and mitochondrial opportunities. Neurotherapeutics 2015, 12, 66–80. [Google Scholar] [CrossRef]
- Le Douce, J.; Maugard, M.; Veran, J.; Matos, M.; Jego, P.; Vigneron, P.A.; Faivre, E.; Toussay, X.; Vandenberghe, M.; Balbastre, Y.; et al. Impairment of Glycolysis-Derived l-Serine Production in Astrocytes Contributes to Cognitive Deficits in Alzheimer’s Disease. Cell Metab. 2020, 31, 503–517. [Google Scholar] [CrossRef]
- Lee, S.; Varvel, N.H.; Konerth, M.E.; Xu, G.; Cardona, A.E.; Ransohoff, R.M.; Lamb, B.T. CX3CR1 deficiency alters microglial activation and reduces beta-amyloid deposition in two Alzheimer’s disease mouse models. Am. J. Pathol. 2010, 177, 2549–2562. [Google Scholar] [CrossRef]
- Xu, X.; Zhang, A.; Zhu, Y.; He, W.; Di, W.; Fang, Y.; Shi, X. MFG-E8 reverses microglial-induced neurotoxic astrocyte (A1) via NF-kappaB and PI3K-Akt pathways. J. Cell. Physiol. 2018, 234, 904–914. [Google Scholar] [CrossRef] [PubMed]
- Paudel, Y.N.; Angelopoulou, E.; Piperi, C.; Othman, I.; Shaikh, M.F. HMGB1-Mediated Neuroinflammatory Responses in Brain Injuries: Potential Mechanisms and Therapeutic Opportunities. Int. J. Mol. Sci. 2020, 21, 4609. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Wang, H.; Sun, G.; Zhang, J.; Edwards, N.J.; Aronowski, J. Neuronal Interleukin-4 as a Modulator of Microglial Pathways and Ischemic Brain Damage. J. Neurosci. 2015, 35, 11281–11291. [Google Scholar] [CrossRef]
- Guillot-Sestier, M.V.; Doty, K.R.; Gate, D.; Rodriguez, J., Jr.; Leung, B.P.; Rezai-Zadeh, K.; Town, T. Il10 deficiency rebalances innate immunity to mitigate Alzheimer-like pathology. Neuron 2015, 85, 534–548. [Google Scholar] [CrossRef]
- Walker, K.A.; Ficek, B.N.; Westbrook, R. Understanding the Role of Systemic Inflammation in Alzheimer’s Disease. ACS Chem. Neurosci. 2019, 10, 3340–3342. [Google Scholar] [CrossRef] [PubMed]
- Bertram, L.; Tanzi, R.E. Alzheimer disease risk genes: 29 and counting. Nat. Rev. Neurol. 2019, 15, 191–192. [Google Scholar] [CrossRef]
- Hodges, A.K.; Piers, T.M.; Collier, D.; Cousins, O.; Pocock, J.M. Pathways linking Alzheimer’s disease risk genes expressed highly in microglia. Neuroimmunol. Neuroinflamm. 2021, 8. [Google Scholar] [CrossRef]
- Gray, S.C.; Kinghorn, K.J.; Woodling, N.S. Shifting equilibriums in Alzheimer’s disease: The complex roles of microglia in neuroinflammation, neuronal survival and neurogenesis. Neural Regen Res. 2020, 15, 1208–1219. [Google Scholar] [CrossRef] [PubMed]
- Sterniczuk, R.; Dyck, R.H.; Laferla, F.M.; Antle, M.C. Characterization of the 3xTg-AD mouse model of Alzheimer’s disease: Part 1. Circadian changes. Brain Res. 2010, 1348, 139–148. [Google Scholar] [CrossRef]
- Vandenberghe, R.; Tournoy, J. Cognitive aging and Alzheimer’s disease. Postgrad Med. J. 2005, 81, 343–352. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rodriguez, J.J.; Witton, J.; Olabarria, M.; Noristani, H.N.; Verkhratsky, A. Increase in the density of resting microglia precedes neuritic plaque formation and microglial activation in a transgenic model of Alzheimer’s disease. Cell Death Dis. 2010, 1, e1. [Google Scholar] [CrossRef]
- Lauritzen, I.; Pardossi-Piquard, R.; Bauer, C.; Brigham, E.; Abraham, J.D.; Ranaldi, S.; Fraser, P.; St-George-Hyslop, P.; Le Thuc, O.; Espin, V.; et al. The beta-secretase-derived C-terminal fragment of betaAPP, C99, but not Abeta, is a key contributor to early intraneuronal lesions in triple-transgenic mouse hippocampus. J. Neurosci. 2012, 32, 16243–1655a. [Google Scholar] [CrossRef]
- Oddo, S.; Caccamo, A.; Smith, I.F.; Green, K.N.; LaFerla, F.M. A dynamic relationship between intracellular and extracellular pools of Abeta. Am. J. Pathol. 2006, 168, 184–194. [Google Scholar] [CrossRef]
- Perez-Gonzalez, R.; Gauthier, S.A.; Kumar, A.; Levy, E. The exosome secretory pathway transports amyloid precursor protein carboxyl-terminal fragments from the cell into the brain extracellular space. J. Biol. Chem. 2012, 287, 43108–43115. [Google Scholar] [CrossRef]
- Di Benedetto, G.; Burgaletto, C.; Carta, A.R.; Saccone, S.; Lempereur, L.; Mulas, G.; Loreto, C.; Bernardini, R.; Cantarella, G. Beneficial effects of curtailing immune susceptibility in an Alzheimer’s disease model. J. Neuroinflamm. 2019, 16, 166. [Google Scholar] [CrossRef]
- Janelsins, M.C.; Mastrangelo, M.A.; Oddo, S.; LaFerla, F.M.; Federoff, H.J.; Bowers, W.J. Early correlation of microglial activation with enhanced tumor necrosis factor-alpha and monocyte chemoattractant protein-1 expression specifically within the entorhinal cortex of triple transgenic Alzheimer’s disease mice. J. Neuroinflamm. 2005, 2, 23. [Google Scholar] [CrossRef] [PubMed]
- Montacute, R.; Foley, K.; Forman, R.; Else, K.J.; Cruickshank, S.M.; Allan, S.M. Enhanced susceptibility of triple transgenic Alzheimer’s disease (3xTg-AD) mice to acute infection. J. Neuroinflamm. 2017, 14, 50. [Google Scholar] [CrossRef]
- Masuda, T.; Sankowski, R.; Staszewski, O.; Prinz, M. Microglia Heterogeneity in the Single-Cell Era. Cell Rep. 2020, 30, 1271–1281. [Google Scholar] [CrossRef] [PubMed]
- Edler, M.K.; Mhatre-Winters, I.; Richardson, J.R. Microglia in Aging and Alzheimer’s Disease: A Comparative Species Review. Cells 2021, 10, 1138. [Google Scholar] [CrossRef] [PubMed]
- Hopperton, K.E.; Mohammad, D.; Trepanier, M.O.; Giuliano, V.; Bazinet, R.P. Markers of microglia in post-mortem brain samples from patients with Alzheimer’s disease: A systematic review. Mol. Psychiatry 2018, 23, 177–198. [Google Scholar] [CrossRef] [PubMed]
- Ojala, J.; Alafuzoff, I.; Herukka, S.K.; van Groen, T.; Tanila, H.; Pirttila, T. Expression of interleukin-18 is increased in the brains of Alzheimer’s disease patients. Neurobiol. Aging 2009, 30, 198–209. [Google Scholar] [CrossRef]
- Sobue, A.; Komine, O.; Hara, Y.; Endo, F.; Mizoguchi, H.; Watanabe, S.; Murayama, S.; Saito, T.; Saido, T.C.; Sahara, N.; et al. Microglial gene signature reveals loss of homeostatic microglia associated with neurodegeneration of Alzheimer’s disease. Acta Neuropathol. Commun. 2021, 9, 1. [Google Scholar] [CrossRef]
- Rossi, C.; Cusimano, M.; Zambito, M.; Finardi, A.; Capotondo, A.; Garcia-Manteiga, J.M.; Comi, G.; Furlan, R.; Martino, G.; Muzio, L. Interleukin 4 modulates microglia homeostasis and attenuates the early slowly progressive phase of amyotrophic lateral sclerosis. Cell Death Dis. 2018, 9, 250. [Google Scholar] [CrossRef]
- Shapira, R.; Solomon, B.; Efrati, S.; Frenkel, D.; Ashery, U. Hyperbaric oxygen therapy ameliorates pathophysiology of 3xTg-AD mouse model by attenuating neuroinflammation. Neurobiol. Aging 2018, 62, 105–119. [Google Scholar] [CrossRef]
- Wang, W.Y.; Tan, M.S.; Yu, J.T.; Tan, L. Role of pro-inflammatory cytokines released from microglia in Alzheimer’s disease. Ann. Transl. Med. 2015, 3, 136. [Google Scholar] [CrossRef]
- Takata, K.; Kitamura, Y.; Kakimura, J.; Shibagaki, K.; Tsuchiya, D.; Taniguchi, T.; Smith, M.A.; Perry, G.; Shimohama, S. Role of high mobility group protein-1 (HMG1) in amyloid-beta homeostasis. Biochem. Biophys. Res. Commun. 2003, 301, 699–703. [Google Scholar] [CrossRef]
- Takata, K.; Takada, T.; Ito, A.; Asai, M.; Tawa, M.; Saito, Y.; Ashihara, E.; Tomimoto, H.; Kitamura, Y.; Shimohama, S. Microglial Amyloid-beta1-40 Phagocytosis Dysfunction Is Caused by High-Mobility Group Box Protein-1: Implications for the Pathological Progression of Alzheimer’s Disease. Int. J. Alzheimers Dis. 2012, 2012, 685739. [Google Scholar] [CrossRef] [PubMed]
- Fujita, K.; Motoki, K.; Tagawa, K.; Chen, X.; Hama, H.; Nakajima, K.; Homma, H.; Tamura, T.; Watanabe, H.; Katsuno, M.; et al. HMGB1, a pathogenic molecule that induces neurite degeneration via TLR4-MARCKS, is a potential therapeutic target for Alzheimer’s disease. Sci. Rep. 2016, 6, 31895. [Google Scholar] [CrossRef]
- Sudduth, T.L.; Schmitt, F.A.; Nelson, P.T.; Wilcock, D.M. Neuroinflammatory phenotype in early Alzheimer’s disease. Neurobiol. Aging 2013, 34, 1051–1059. [Google Scholar] [CrossRef] [PubMed]
- Walker, D.G.; Lue, L.F. Immune phenotypes of microglia in human neurodegenerative disease: Challenges to detecting microglial polarization in human brains. Alzheimer’s Res. Ther. 2015, 7, 56. [Google Scholar] [CrossRef] [PubMed]
- Ponomarev, E.D.; Veremeyko, T.; Barteneva, N.; Krichevsky, A.M.; Weiner, H.L. MicroRNA-124 promotes microglia quiescence and suppresses EAE by deactivating macrophages via the C/EBP-alpha-PU.1 pathway. Nat. Med. 2011, 17, 64–70. [Google Scholar] [CrossRef]
- Whyte, C.S.; Bishop, E.T.; Ruckerl, D.; Gaspar-Pereira, S.; Barker, R.N.; Allen, J.E.; Rees, A.J.; Wilson, H.M. Suppressor of cytokine signaling (SOCS)1 is a key determinant of differential macrophage activation and function. J. Leukoc. Biol. 2011, 90, 845–854. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Liang, Q.; Ge, G. Gypenoside Attenuates beta Amyloid-Induced Inflammation in N9 Microglial Cells via SOCS1 Signaling. Neural Plast. 2016, 2016, 6362707. [Google Scholar] [CrossRef]
- Cherry, J.D.; Olschowka, J.A.; O’Banion, M.K. Arginase 1+ microglia reduce Abeta plaque deposition during IL-1beta-dependent neuroinflammation. J. Neuroinflamm. 2015, 12, 203. [Google Scholar] [CrossRef]
- Huang, W.C.; Yen, F.C.; Shie, F.S.; Pan, C.M.; Shiao, Y.J.; Yang, C.N.; Huang, F.L.; Sung, Y.J.; Tsay, H.J. TGF-beta1 blockade of microglial chemotaxis toward Abeta aggregates involves SMAD signaling and down-regulation of CCL5. J. Neuroinflamm. 2010, 7, 28. [Google Scholar] [CrossRef]
- Li, Y.Y.; Cui, J.G.; Dua, P.; Pogue, A.I.; Bhattacharjee, S.; Lukiw, W.J. Differential expression of miRNA-146a-regulated inflammatory genes in human primary neural, astroglial and microglial cells. Neurosci. Lett. 2011, 499, 109–113. [Google Scholar] [CrossRef]
- Lukiw, W.J.; Zhao, Y.; Cui, J.G. An NF-kappaB-sensitive micro RNA-146a-mediated inflammatory circuit in Alzheimer disease and in stressed human brain cells. J. Biol. Chem. 2008, 283, 31315–31322. [Google Scholar] [CrossRef]
- Su, W.; Aloi, M.S.; Garden, G.A. MicroRNAs mediating CNS inflammation: Small regulators with powerful potential. Brain Behav. Immun. 2015, 52, 1–8. [Google Scholar] [CrossRef]
- Alexandrov, P.N.; Dua, P.; Hill, J.M.; Bhattacharjee, S.; Zhao, Y.; Lukiw, W.J. microRNA (miRNA) speciation in Alzheimer’s disease (AD) cerebrospinal fluid (CSF) and extracellular fluid (ECF). Int. J. Biochem. Mol. Biol. 2012, 3, 365–373. [Google Scholar] [PubMed]
- Culpan, D.; Kehoe, P.G.; Love, S. Tumour necrosis factor-alpha (TNF-alpha) and miRNA expression in frontal and temporal neocortex in Alzheimer’s disease and the effect of TNF-alpha on miRNA expression in vitro. Int. J. Mol. Epidemiol. Genet. 2011, 2, 156–162. [Google Scholar] [PubMed]
- Kou, X.; Chen, D.; Chen, N. The Regulation of microRNAs in Alzheimer’s Disease. Front. Neurol. 2020, 11, 288. [Google Scholar] [CrossRef]
- Garcia, G.; Pinto, S.; Cunha, M.; Fernandes, A.; Koistinaho, J.; Brites, D. Neuronal Dynamics and miRNA Signaling Differ between SH-SY5Y APPSwe and PSEN1 Mutant iPSC-Derived AD Models upon Modulation with miR-124 Mimic and Inhibitor. Cells 2021, 10, 2424. [Google Scholar] [CrossRef] [PubMed]
- Wen, Z.; Xu, L.; Chen, X.; Xu, W.; Yin, Z.; Gao, X.; Xiong, S. Autoantibody induction by DNA-containing immune complexes requires HMGB1 with the TLR2/microRNA-155 pathway. J. Immunol. 2013, 190, 5411–5422. [Google Scholar] [CrossRef]
- Cardoso, A.L.; Guedes, J.R.; Pereira de Almeida, L.; Pedroso de Lima, M.C. miR-155 modulates microglia-mediated immune response by down-regulating SOCS-1 and promoting cytokine and nitric oxide production. Immunology 2012, 135, 73–88. [Google Scholar] [CrossRef]
- Liu, G.; Abraham, E. MicroRNAs in immune response and macrophage polarization. Arter. Thromb Vasc. Biol. 2013, 33, 170–177. [Google Scholar] [CrossRef] [PubMed]
- Burgaletto, C.; Platania, C.B.M.; Di Benedetto, G.; Munafo, A.; Giurdanella, G.; Federico, C.; Caltabiano, R.; Saccone, S.; Conti, F.; Bernardini, R.; et al. Targeting the miRNA-155/TNFSF10 network restrains inflammatory response in the retina in a mouse model of Alzheimer’s disease. Cell Death Dis. 2021, 12, 905. [Google Scholar] [CrossRef]
- Testa, U.; Pelosi, E.; Castelli, G.; Labbaye, C. miR-146 and miR-155: Two Key Modulators of Immune Response and Tumor Development. Noncoding RNA 2017, 3, 22. [Google Scholar] [CrossRef]
- Roseler, S.; Sandrock, K.; Bartsch, I.; Busse, A.; Omran, H.; Loges, N.T.; Zieger, B. Lethal phenotype of mice carrying a Sept11 null mutation. Biol. Chem. 2011, 392, 779–781. [Google Scholar] [CrossRef] [PubMed]
- Kurkinen, K.M.; Marttinen, M.; Turner, L.; Natunen, T.; Makinen, P.; Haapalinna, F.; Sarajarvi, T.; Gabbouj, S.; Kurki, M.; Paananen, J.; et al. SEPT8 modulates beta-amyloidogenic processing of APP by affecting the sorting and accumulation of BACE1. J. Cell Sci. 2016, 129, 2224–2238. [Google Scholar] [CrossRef]
- Matcovitch-Natan, O.; Winter, D.R.; Giladi, A.; Vargas Aguilar, S.; Spinrad, A.; Sarrazin, S.; Ben-Yehuda, H.; David, E.; Zelada Gonzalez, F.; Perrin, P.; et al. Microglia development follows a stepwise program to regulate brain homeostasis. Science 2016, 353, aad8670. [Google Scholar] [CrossRef]
- Koshida, R.; Oishi, H.; Hamada, M.; Takahashi, S. MafB antagonizes phenotypic alteration induced by GM-CSF in microglia. Biochem. Biophys. Res. Commun. 2015, 463, 109–115. [Google Scholar] [CrossRef]
- Chen, X.; Lu, J.; Zhao, X.; Chen, C.; Qiao, D.; Wang, H.; Yue, X. Role of C/EBP-beta in Methamphetamine-Mediated Microglial Apoptosis. Front. Cell Neurosci. 2019, 13, 366. [Google Scholar] [CrossRef] [PubMed]
- Muller, C.; Zidek, L.M.; Ackermann, T.; de Jong, T.; Liu, P.; Kliche, V.; Zaini, M.A.; Kortman, G.; Harkema, L.; Verbeek, D.S.; et al. Reduced expression of C/EBPbeta-LIP extends health and lifespan in mice. Elife 2018, 7, e34985. [Google Scholar] [CrossRef]
- Nakazato, R.; Takarada, T.; Ikeno, S.; Nakamura, S.; Kutsukake, T.; Hinoi, E.; Yoneda, Y. Upregulation of Runt-Related Transcription Factor-2 Through CCAAT Enhancer Binding Protein-beta Signaling Pathway in Microglial BV-2 Cells Exposed to ATP. J. Cell. Physiol. 2015, 230, 2510–2521. [Google Scholar] [CrossRef]
- Morris, B.J.; Willcox, D.C.; Donlon, T.A.; Willcox, B.J. FOXO3: A Major Gene for Human Longevity--A Mini-Review. Gerontology 2015, 61, 515–525. [Google Scholar] [CrossRef]
- Du, S.; Jin, F.; Maneix, L.; Gedam, M.; Xu, Y.; Catic, A.; Wang, M.C.; Zheng, H. FoxO3 deficiency in cortical astrocytes leads to impaired lipid metabolism and aggravated amyloid pathology. Aging Cell 2021, 20, e13432. [Google Scholar] [CrossRef] [PubMed]
- Ciryam, P.; Kundra, R.; Freer, R.; Morimoto, R.I.; Dobson, C.M.; Vendruscolo, M. A transcriptional signature of Alzheimer’s disease is associated with a metastable subproteome at risk for aggregation. Proc. Natl. Acad. Sci. USA 2016, 113, 4753–4758. [Google Scholar] [CrossRef]
- Olah, M.; Biber, K.; Vinet, J.; Boddeke, H.W. Microglia phenotype diversity. CNS Neurol. Disord Drug Targets 2011, 10, 108–118. [Google Scholar] [CrossRef] [PubMed]
- Howard, R.; Zubko, O.; Bradley, R.; Harper, E.; Pank, L.; O’Brien, J.; Fox, C.; Tabet, N.; Livingston, G.; Bentham, P.; et al. Minocycline at 2 Different Dosages vs. Placebo for Patients with Mild Alzheimer Disease: A Randomized Clinical Trial. JAMA Neurol. 2020, 77, 164–174. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Wang, Y.; Wang, D.; Zhang, J.; Zhang, F. NSAID Exposure and Risk of Alzheimer’s Disease: An Updated Meta-Analysis from Cohort Studies. Front. Aging Neurosci. 2018, 10, 83. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).