Gelsolin Contributes to the Motility of A375 Melanoma Cells and This Activity Is Mediated by the Fibrous Extracellular Matrix Protein Profile
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells and Culture Conditions
2.2. Coating of the Plates and Dishes with ECM Proteins
2.3. GSN Knockout with CRISPR/Cas9(D10A) Technique
2.4. gDNA Analysis
2.5. Western Blot Analysis (WB)
2.6. Cell Proliferation Assay
2.7. Cell Confluence Analysis
2.8. Immunocytochemistry and Fluorescence Microscopy
2.9. Structured Illuminated Microscopy (SIM)
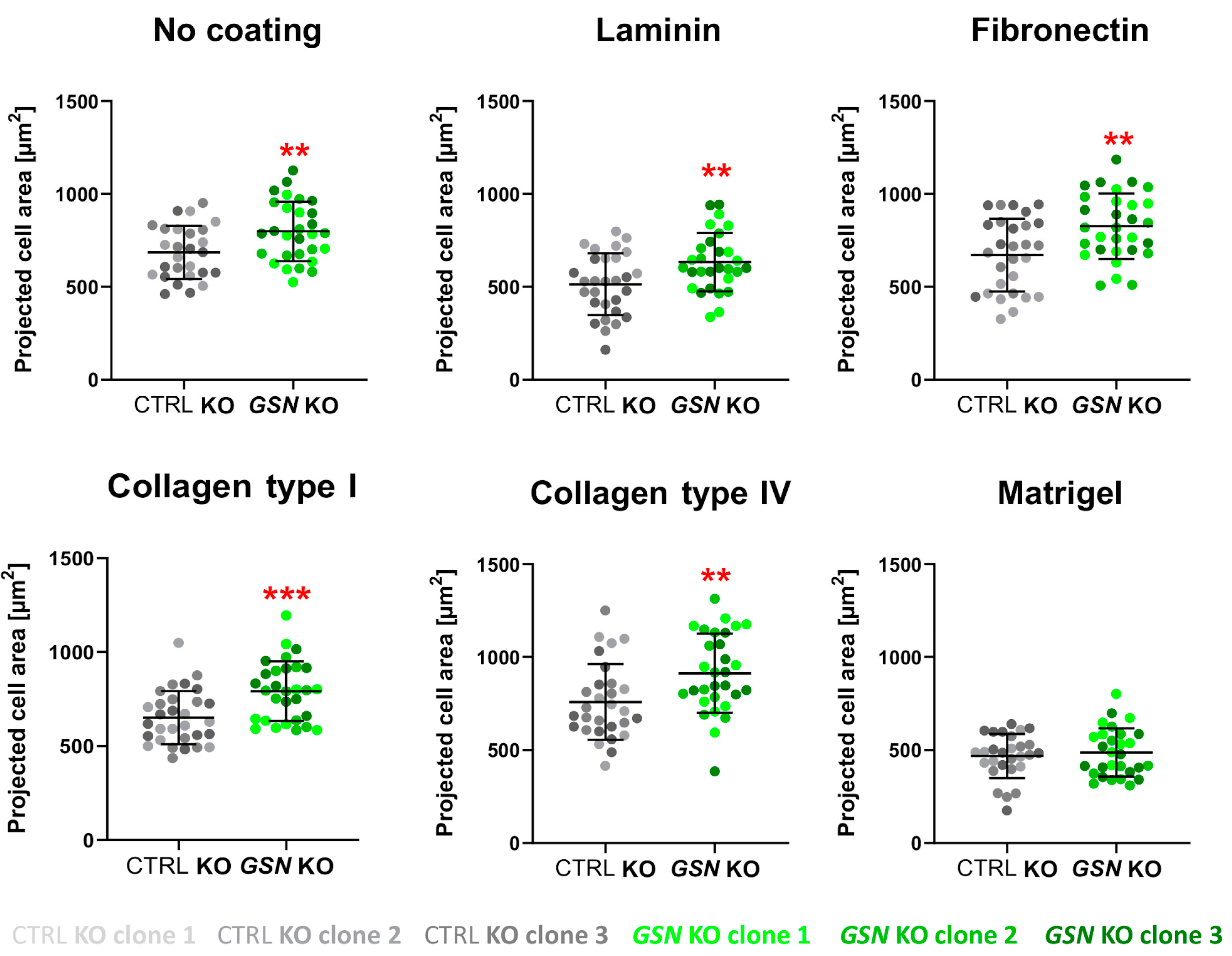
2.10. Projected Cell’s Area Assessment
2.11. Stress Fiber Number Evaluation
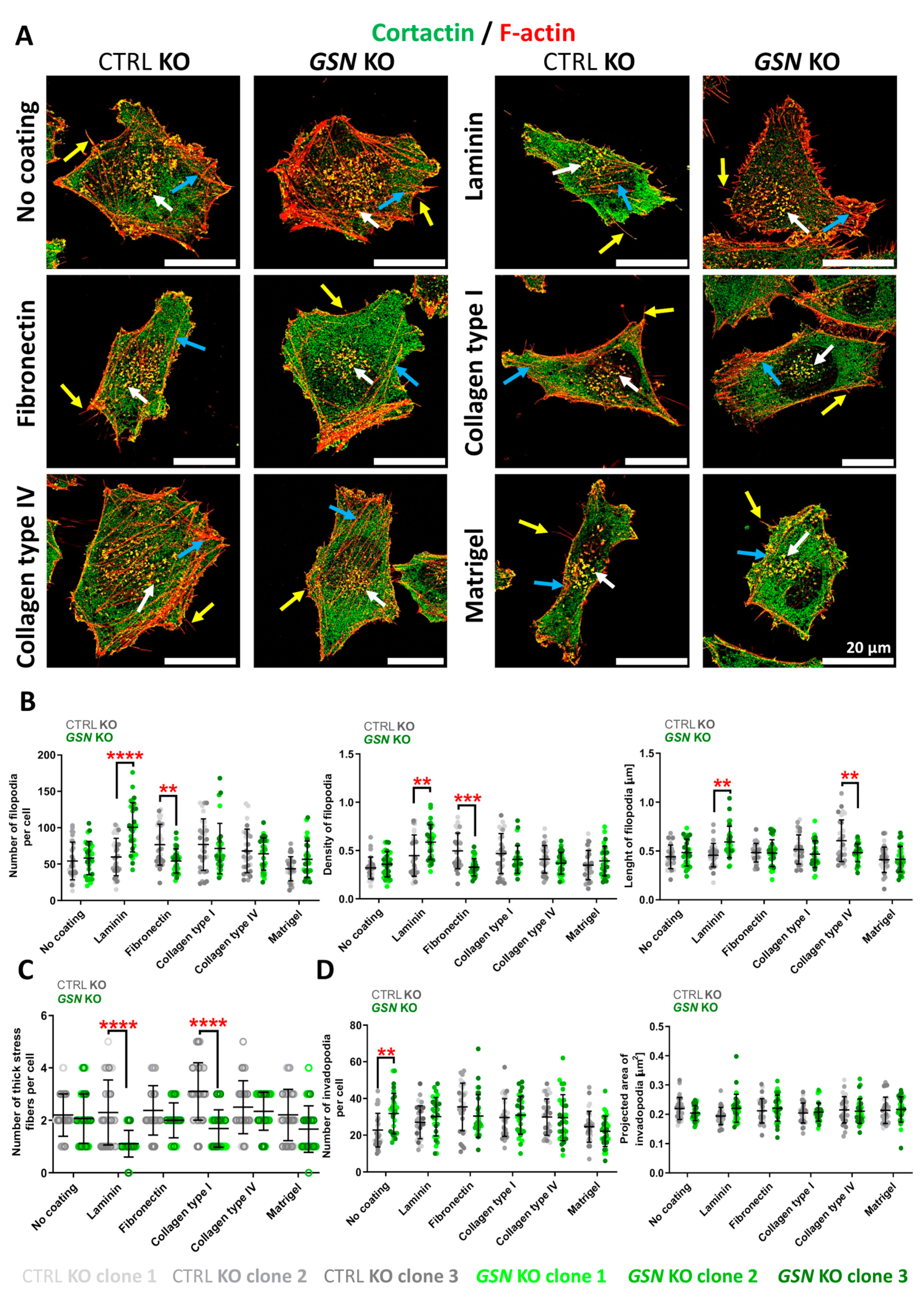
2.12. Filopodia Number, Length, and Density Evaluation
2.13. Invadopodia Number and Size Calculation
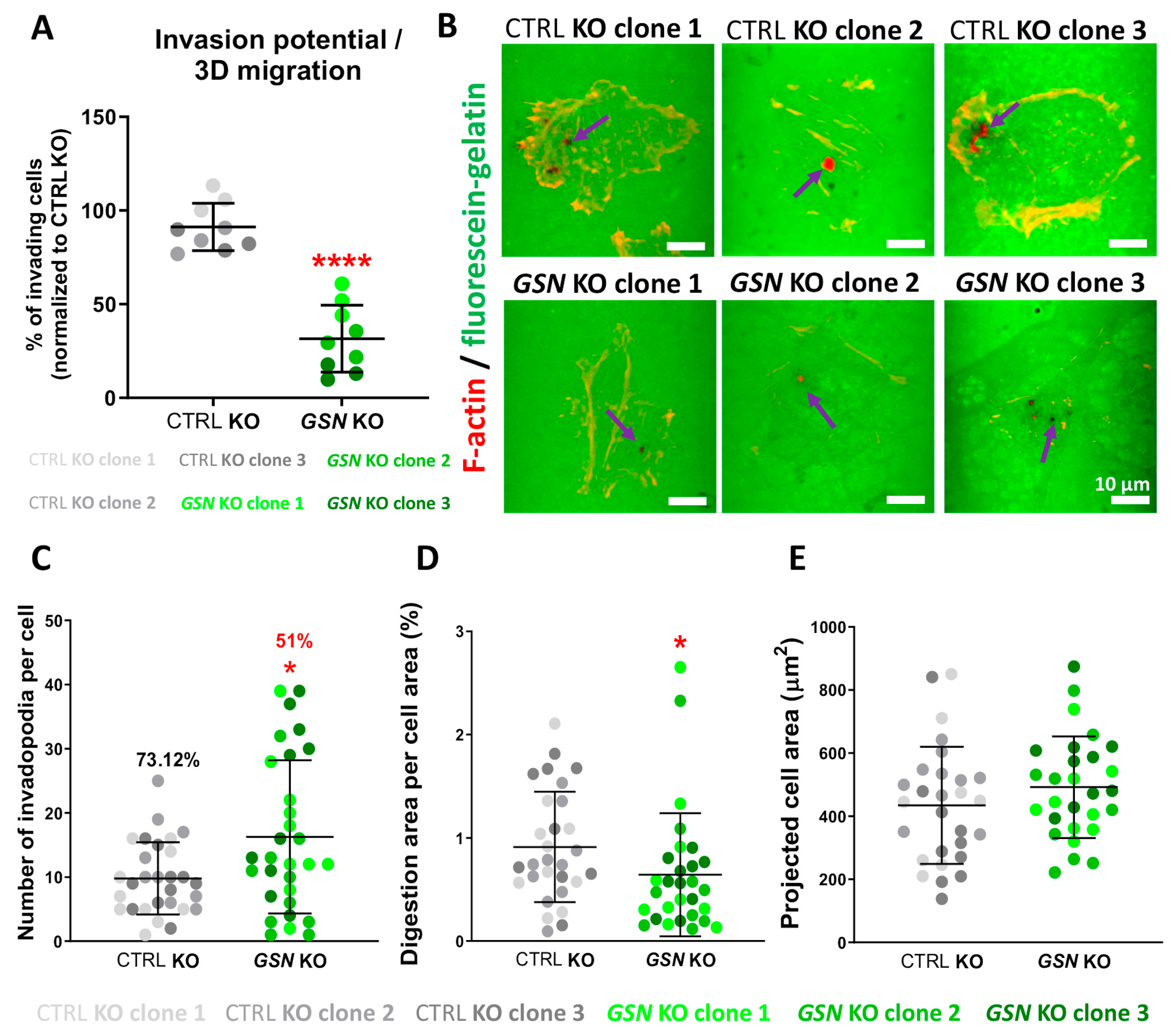
2.14. Invasion Assay
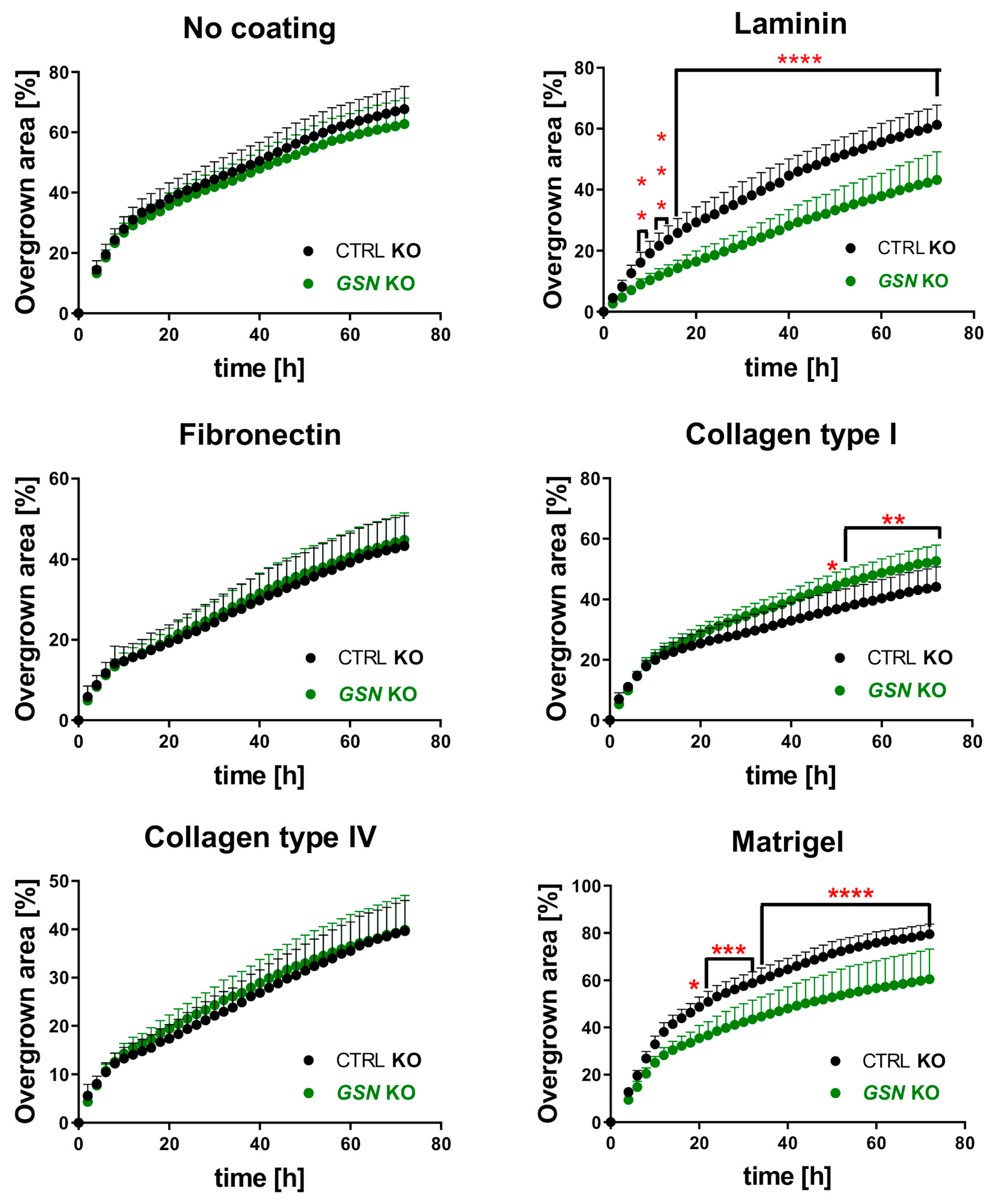
2.15. Cell Migration Assays
2.16. Gelatin Digestion Assay
2.17. Statistical Analysis
3. Results
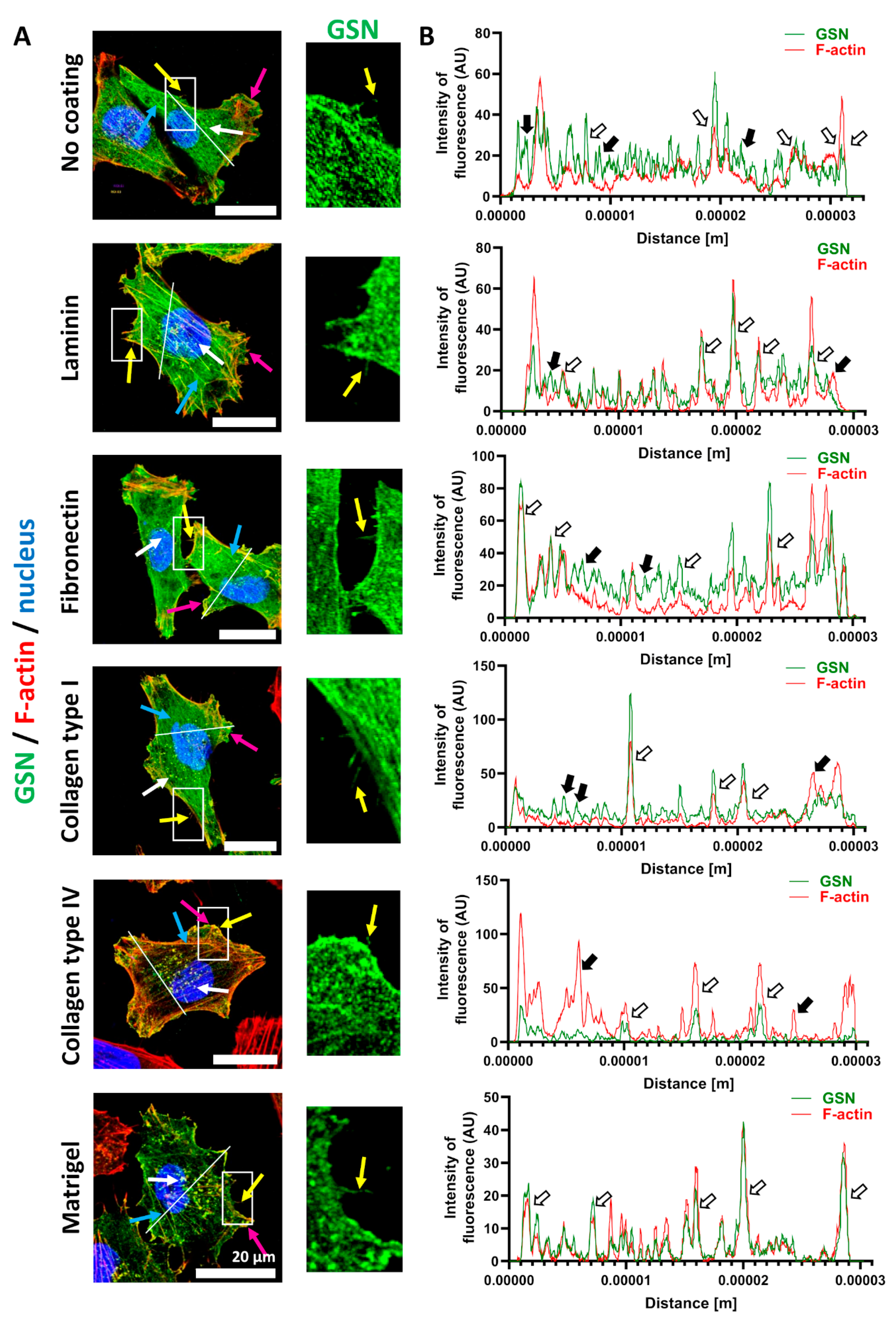
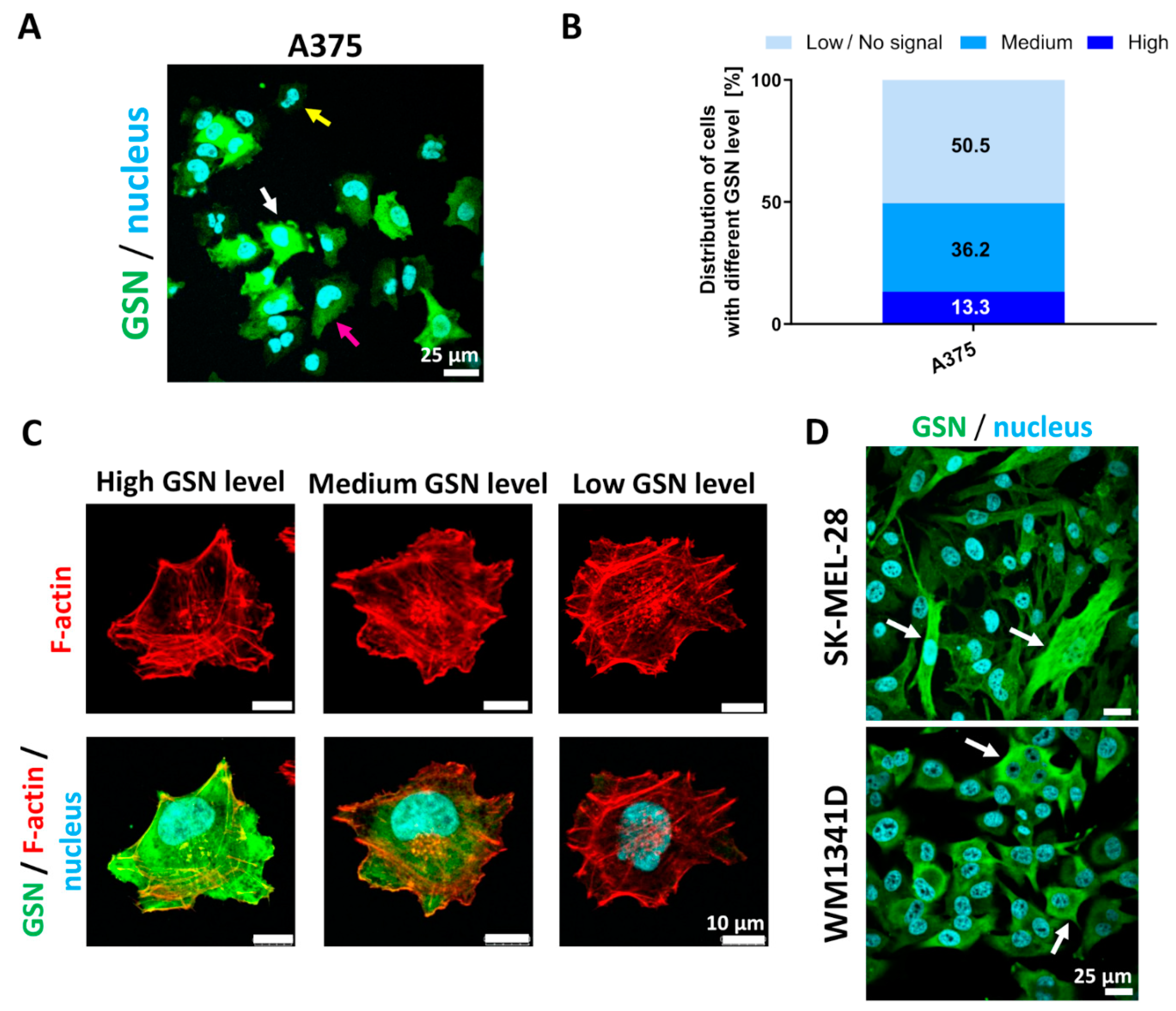
3.1. GSN Is Found in Motile Structures of A375 Cells When Cultured on ECM Proteins, and GSN Expression Is Not Homogenous in Melanoma Cell Lines
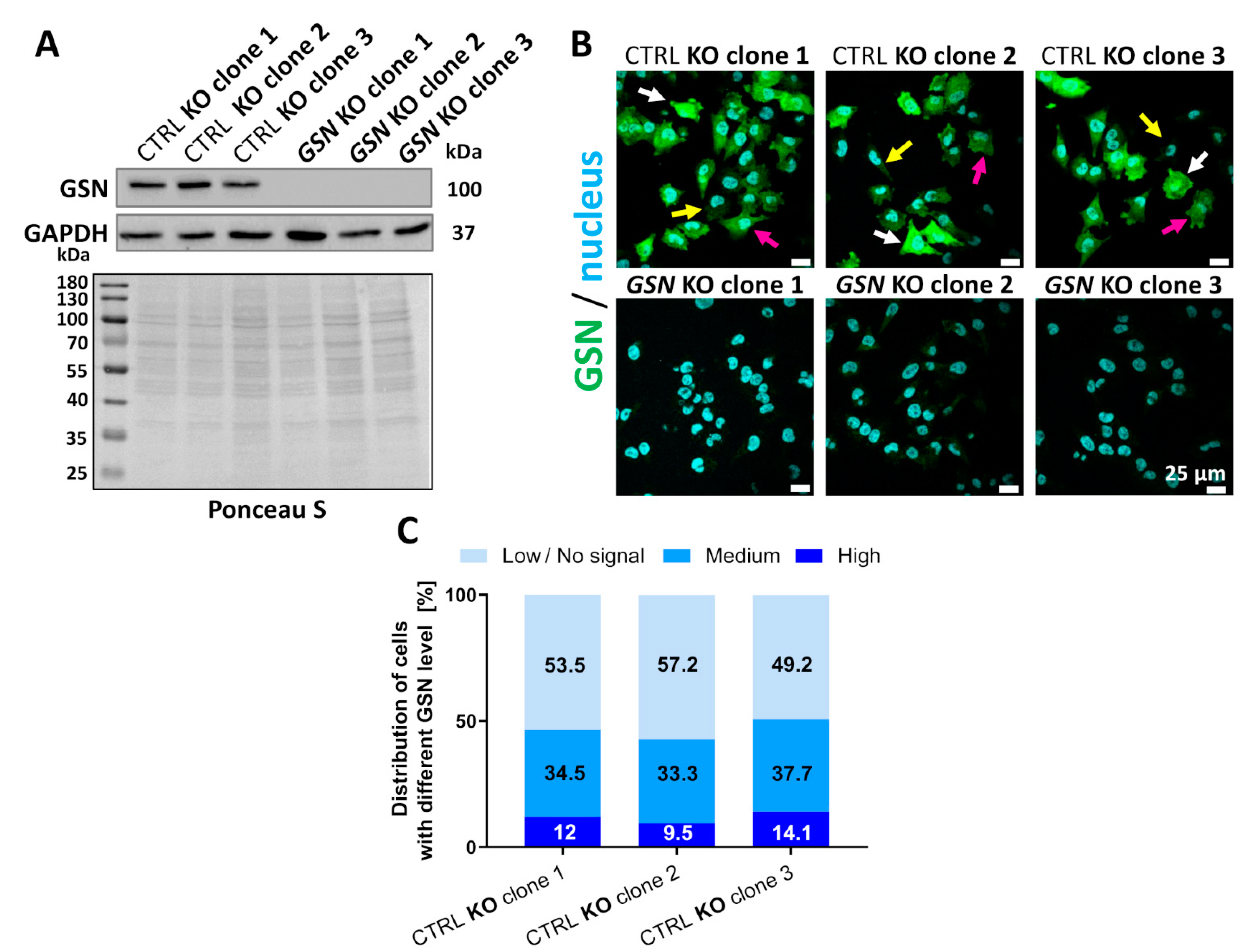
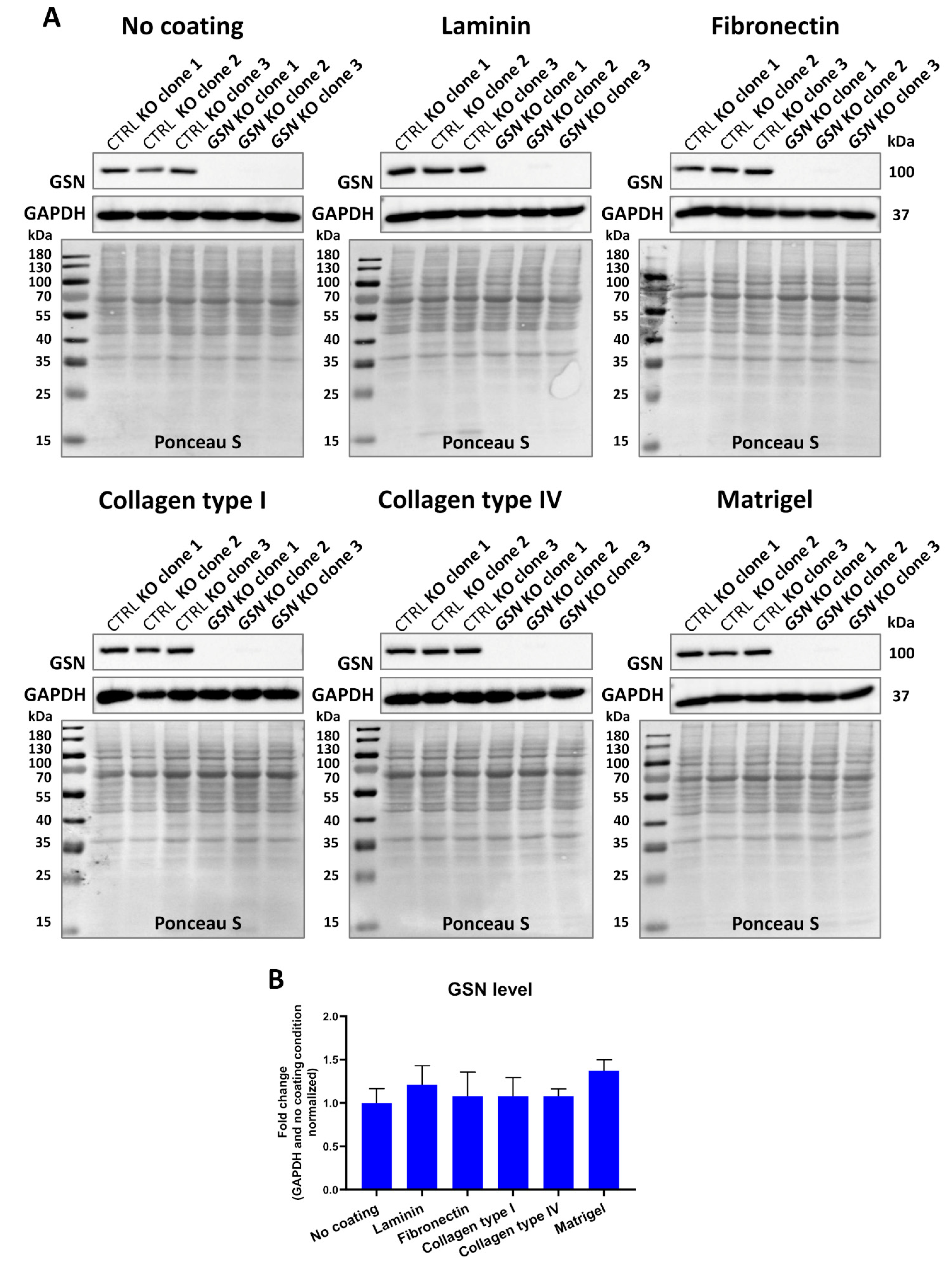
3.2. Knockout of GSN in Melanoma A375 Cells
3.3. GSN Knockout Does Not Change the Proliferation Rate of A375 Cells
3.4. Control and GSN KO A375 Cells Have Changed Morphology and Formation of F-Actin-Rich Protrusive Structures When Growing on Some ECM Proteins
3.5. GSN-Depletion Results in Impaired 2-D Migration of A375 Cells on Selected ECM Proteins
3.6. GSN-Depletion Heavily Impairs Invasion Abilities of A375 Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BM | basement membrane |
| CTRL KO | control cells |
| GSN KO | cells with knocked out GSN |
| CRISPR | Clustered Regularly Interspaced Short Palindromic Repeats |
| ECM | extracellular matrix |
| F-actin | filamentous actin |
| FBS | fetal bovine serum |
| gDNA | genomic DNA |
| GSN | gene coding for gelsolin |
| GSN | gelsolin |
| MMPs | metalloproteases |
| SIM | structured illuminated microscopy |
References
- Nag, S.; Larsson, M.; Robinson, R.C.; Burtnick, L.D. Gelsolin: The tail of a molecular gymnast. Cytoskeleton 2013, 70, 360–384. [Google Scholar] [CrossRef] [PubMed]
- Li, G.H.; Arora, P.D.; Chen, Y.; McCulloch, C.A.; Liu, P. Multifunctional roles of gelsolin in health and diseases. Med. Res. Rev. 2012, 32, 999–1025. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.L.; Stossel, T.P. Control of cytoplasmic actin gel–sol transformation by gelsolin, a calcium-dependent regulatory protein. Nature 1979, 281, 583–586. [Google Scholar] [CrossRef] [PubMed]
- Piktel, E.; Levental, I.; Durnaś, B.; Janmey, P.; Bucki, R. Plasma Gelsolin: Indicator of Inflammation and Its Potential as a Diagnostic Tool and Therapeutic Target. Int. J. Mol. Sci. 2018, 19, 2516. [Google Scholar] [CrossRef] [PubMed]
- Schneider, U.W.; Garreis, F.; Welss, J.; Feldt, J.; Paulsen, F.; Schicht, M. Structure, regulation and related diseases of the actin-binding protein gelsolin. Expert Rev. Mol. Med. 2019, 20, e7. [Google Scholar]
- Litwin, M.; Nowak, D.; Mazur, A.J.; Baczyńska, D.; Mannherz, H.G.; Malicka-Błaszkiewicz, M. Gelsolin affects the migratory ability of human colon adenocarcinoma and melanoma cells. Life Sci. 2012, 90, 851–861. [Google Scholar] [CrossRef] [PubMed]
- Mazur, A.J.; Radaszkiewicz, T.; Makowiecka, A.; Malicka-Błaszkiewicz, M.; Mannherz, H.G.; Nowak, D. Gelsolin interacts with LamR, hnRNP U, nestin, Arp3 and β-tubulin in human melanoma cells as revealed by immunoprecipitation and mass spectrometry. Eur. J. Cell Biol. 2016, 95, 26–41. [Google Scholar] [CrossRef]
- Furukawa, H.; Fujita, H.; Kokubu, I.; Yamamoto, Y.; Sasaki, S.; Chodon, T.; Okubo, Y.; Sugihara, T.; Kuzumaki, N. Identification of a novel gelsolin truncate in the vertical and metastatic phase malignant melanomas. Melanoma Res. 2002, 12, 523–528. [Google Scholar] [CrossRef]
- Fujita, H.; Okada, F.; Hamada, J.; Hosokawa, M.; Moriuchi, T.; Koya, R.C.; Kuzumaki, N. Gelsolin functions as a metastasis suppressor in B16-BL6 mouse melanoma cells and requirement of the carboxyl-terminus for its effect. Int. J. Cancer 2001, 93, 773–780. [Google Scholar] [CrossRef]
- Khalil, D.N.; Carvajal, R.D. Treatments for noncutaneous melanoma. Hematol. Oncol. Clin. N. Am. 2014, 28, 507–521. [Google Scholar] [CrossRef]
- Rabbie, R.; Ferguson, P.; Molina-Aguilar, C.; Adams, D.J.; Robles-Espinoza, C.D. Melanoma subtypes: Genomic profiles, prognostic molecular markers and therapeutic possibilities. J. Pathol. 2019, 247, 539–551. [Google Scholar] [CrossRef] [PubMed]
- Bellew, S.; Del Rosso, J.Q. Should all patients with melanoma between 1 and 2mm breslow thickness undergo sentinel lymph node biopsy? Yearb. Dermatol. Dermatol. Surg. 2011, 2011, 405–407. [Google Scholar] [CrossRef]
- Clark, W.H.; Elder, D.E.; Guerry, D.; Epstein, M.N.; Greene, M.H.; Van Horn, M. A study of tumor progression: The precursor lesions of superficial spreading and nodular melanoma. Hum. Pathol. 1984, 15, 1147–1165. [Google Scholar] [CrossRef]
- Breitkreutz, D.; Koxholt, I.; Thiemann, K.; Nischt, R. Skin Basement Membrane: The foundation of epidermal integrity-bm functions and diverse roles of bridging molecules nidogen and perlecan. BioMed Res. Int. 2013, 2013, 179784. [Google Scholar] [CrossRef] [PubMed]
- Sibony-Benyamini, H.; Gil-Henn, H. Invadopodia: The leading force. Eur. J. Cell Biol. 2012, 91, 896–901. [Google Scholar] [CrossRef] [PubMed]
- Quintero-Fabián, S.; Arreola, R.; Becerril-Villanueva, E.; Torres-Romero, J.C.; Arana-Argáez, V.; Lara-Riegos, J.; Ramírez-Camacho, M.A.; Alvarez-Sánchez, M.E. Role of matrix metalloproteinases in angiogenesis and cancer. Front. Oncol. 2019, 9, 1370. [Google Scholar] [CrossRef] [PubMed]
- Rottner, K.; Schaks, M. Assembling actin filaments for protrusion. Curr. Opin. Cell Biol. 2019, 56, 53–63. [Google Scholar] [CrossRef]
- Blanchoin, L.; Boujemaa-Paterski, R.; Sykes, C.; Plastino, J. Actin dynamics, architecture, and mechanics in cell motility. Physiol. Rev. 2014, 94, 235–263. [Google Scholar] [CrossRef] [PubMed]
- Bergert, M.; Chandradoss, S.D.; Desai, R.A.; Paluch, E. Cell mechanics control rapid transitions between blebs and lamellipodia during migration. Proc. Natl. Acad. Sci. USA 2012, 109, 14434–14439. [Google Scholar] [CrossRef]
- Kular, J.K.; Basu, S.; Sharma, R.I. The extracellular matrix: Structure, composition, age-related differences, tools for analysis and applications for tissue engineering. J. Tissue Eng. 2014, 5, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Fyrand, O. Studies on fibronectin in the skin. Arch. Dermatol. Res. 1979, 266, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Kleinman, H.K.; McGarvey, M.L.; Hassell, J.R.; Star, V.L.; Cannon, F.B.; Laurie, G.W.; Martin, G.R. Basement membrane complexes with biological activity. Biochemistry 1986, 25, 312–318. [Google Scholar] [CrossRef] [PubMed]
- Kleinman, H.K.; Martin, G.R. Matrigel: Basement membrane matrix with biological activity. Semin. Cancer Biol. 2005, 15, 378–386. [Google Scholar] [CrossRef] [PubMed]
- De Wever, O.; Hendrix, A.; De Boeck, A.; Westbroek, W.; Braems, G.; Emami, S.; Sabbah, M.; Gespach, C.; Bracke, M. Modeling and quantification of cancer cell invasion through collagen type I matrices. Int. J. Dev. Biol. 2010, 54, 887–896. [Google Scholar] [CrossRef] [PubMed]
- Malek, N.; Mrówczyńska, E.; Michrowska, A.; Mazurkiewicz, E.; Pavlyk, I.; Mazur, A.J. Knockout of ACTB and ACTG1 with CRISPR/Cas9(D10A) technique shows that non-muscle β and γ actin are not equal in relation to human melanoma cells’ motility and focal adhesion formation. Int. J. Mol. Sci. 2020, 21, 2746. [Google Scholar] [CrossRef] [PubMed]
- Jacquemet, G.; Paatero, I.; Carisey, A.F.; Padzik, A.; Orange, J.S.; Hamidi, H.; Ivaska, J. FiloQuant reveals increased filopodia density during breast cancer progression. J. Cell Biol. 2017, 216, 3387–3403. [Google Scholar] [CrossRef]
- Kyykallio, H.; Oikari, S.; Álvez, M.B.; Dodd, C.J.G.; Capra, J.; Rilla, K. The Density and length of filopodia associate with the activity of hyaluronan synthesis in tumor cells. Cancers 2020, 12, 1908. [Google Scholar] [CrossRef] [PubMed]
- Makowiecka, A.; Simiczyjew, A.; Nowak, D.; Mazur, A.J. Varying effects of EGF, HGF and TGFγ on formation of invadopodia and invasiveness of melanoma cell lines of different origin. Eur. J. Histochem. 2016, 60, 230–238. [Google Scholar] [CrossRef] [PubMed]
- Mazurkiewicz, E.; Mrówczyńska, E.; Simiczyjew, A.; Nowak, D.; Mazur, A.J. A Fluorescent Gelatin Degradation Assay to Study Melanoma Breakdown of Extracellular Matrix; Humana: New York, NY, USA, 2021; pp. 47–63. [Google Scholar]
- Corning® Matrigel® Matrix. Available online: https://www.corning.com/catalog/cls/documents/faqs/CLS-DL-CC-026.pdf (accessed on 1 June 2021).
- Malek, N.; Michrowska, A.; Mazurkiewicz, E.; Mrówczyńska, E.; Mackiewicz, P.; Mazur, A.J. The origin of the expressed retrotransposed gene ACTBL2 and its influence on human melanoma cells’ motility and focal adhesion formation. Sci. Rep. 2021, 11, 3329. [Google Scholar] [CrossRef]
- Lehtimäki, J.; Hakala, M.; Lappalainen, P. Actin Filament Structures in Migrating Cells. In The Actin Cytoskeleton; Springer: Cham, Switzerland, 2016; pp. 123–152. [Google Scholar]
- Bergman, A.; Condeelis, J.S.; Gligorijevic, B. Invadopodia in context. Cell Adh. Migr. 2014, 8, 1–7. [Google Scholar] [CrossRef]
- Makowiecka, A.; Mazurkiewicz, E.; Mrówczyńska, E.; Malek, N.; Battistella, A.; Lazzarino, M.; Nowak, D.; Mazur, A.J. Changes in biomechanical properties of A375 cells due to the silencing of TMSB4X expression are not directly correlated with alterations in their stemness features. Cells 2021, 10, 769. [Google Scholar] [CrossRef] [PubMed]
- Mishra, H.; Mishra, P.K.; Ekielski, A.; Jaggi, M.; Iqbal, Z.; Talegaonkar, S. Melanoma treatment: From conventional to nanotechnology. J. Cancer Res. Clin. Oncol. 2018, 144, 2283–2302. [Google Scholar] [CrossRef] [PubMed]
- Simiczyjew, A.; Dratkiewicz, E.; Mazurkiewicz, J.; Ziętek, M.; Matkowski, R.; Nowak, D. The influence of tumor microenvironment on immune escape of melanoma. Int. J. Mol. Sci. 2020, 21, 8359. [Google Scholar] [CrossRef] [PubMed]
- Mazurkiewicz, J.; Simiczyjew, A.; Dratkiewicz, E.; Ziętek, M.; Matkowski, R.; Nowak, D. Stromal cells present in the melanoma niche affect tumor invasiveness and its resistance to therapy. Int. J. Mol. Sci. 2021, 22, 529. [Google Scholar] [CrossRef] [PubMed]
- Dratkiewicz, E.; Simiczyjew, A.; Mazurkiewicz, J.; Ziętek, M.; Matkowski, R.; Nowak, D. Hypoxia and extracellular acidification as drivers of melanoma progression and drug resistance. Cells 2021, 10, 862. [Google Scholar] [CrossRef] [PubMed]
- Falcone, I.; Conciatori, F.; Bazzichetto, C.; Ferretti, G.; Cognetti, F.; Ciuffreda, L.; Milella, M. Tumor microenvironment: Implications in melanoma resistance to targeted therapy and immunotherapy. Cancers 2020, 12, 2870. [Google Scholar] [CrossRef]
- Hirata, E.; Girotti, M.R.; Viros, A.; Hooper, S.; Spencer-Dene, B.; Matsuda, M.; Larkin, J.; Marais, R.; Sahai, E. Intravital imaging reveals how BRAF inhibition generates drug-tolerant microenvironments with high integrin β1/FAK Signaling. Cancer Cell 2015, 27, 574–588. [Google Scholar] [CrossRef]
- Muncie, J.M.; Weaver, V.M. The physical and biochemical properties of the extracellular matrix regulate cell fate. Curr. Top. Dev. Biol. 2018, 130, 1–37. [Google Scholar]
- Christofori, G. Changing neighbours, changing behaviour: Cell adhesion molecule-mediated signalling during tumour progression. EMBO J. 2003, 22, 2318–2323. [Google Scholar] [CrossRef] [PubMed]
- Saladi, S.; Keenen, B.; Marathe, H.G.; Qi, H.; Chin, K.-V.; de la Serna, I.L. Modulation of extracellular matrix/adhesion molecule expression by BRG1 is associated with increased melanoma invasiveness. Mol. Cancer 2010, 9, 280. [Google Scholar] [CrossRef]
- Schmoeckel, C.; Stolz, W.; Sakai, L.Y.; Burgeson, R.E.; Timpl, R.; Krieg, T. Structure of basement membranes in malignant melanoma and nevocytic nevi. J. Investig. Dermatol. 1989, 92, 663–668. [Google Scholar] [CrossRef]
- Schaumburg-Lever, G.; Lever, I.; Fehrenbacher, B.; Möller, H.; Bischof, B.; Kaiserling, E.; Garbe, C.; Rassner, G. Melanocytes in nevi and melanomas synthesize basement membrane and basement membrane-like material. An immunohistochemical and electron microscopic study including immunoelectron microscopy. J. Cutan. Pathol. 2000, 27, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Kramer, R.H.; Vogel, K.G. Selective degradation of basement membrane macromolecules by metastatic melanoma cells. J. Natl. Cancer Inst. 1984, 72, 889–899. [Google Scholar] [PubMed]
- Pasco, S.; Ramont, L.; Maquart, F.X.; Monboisse, J.C. Control of melanoma progression by various matrikines from basement membrane macromolecules. Crit. Rev. Oncol. Hematol. 2004, 49, 221–223. [Google Scholar] [CrossRef] [PubMed]
- Boussadia, Z.; Lamberti, J.; Mattei, F.; Pizzi, E.; Puglisi, R.; Zanetti, C.; Pasquini, L.; Fratini, F.; Fantozzi, L.; Felicetti, F.; et al. Acidic microenvironment plays a key role in human melanoma progression through a sustained exosome mediated transfer of clinically relevant metastatic molecules. J. Exp. Clin. Cancer Res. 2018, 37, 1–15. [Google Scholar] [CrossRef]
- Haga, K.; Fujita, H.; Nomoto, M.; Sazawa, A.; Nakagawa, K.; Harabayashi, T.; Shinohara, N.; Takimoto, M.; Nonomura, K.; Kuzumaki, N. Gelsolin gene silencing involving unusual hypersensitivities to dimethylsulfate and KMnO4in vivo footprinting on its promoter region. Int. J. Cancer 2004, 111, 873–880. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gallaher, J.A.; Brown, J.S.; Anderson, A.R.A. The impact of proliferation-migration tradeoffs on phenotypic evolution in cancer. Sci. Rep. 2019, 9, 2425. [Google Scholar] [CrossRef]
- Deng, L.; Liu, H. MicroRNA-506 suppresses growth and metastasis of oral squamous cell carcinoma via targeting GATA6. Int J. Clin. Exp. Med. 2015, 8, 1862–1870. [Google Scholar] [PubMed]
- Miskolczi, Z.; Smith, M.P.; Rowling, E.J.; Ferguson, J.; Barriuso, J.; Wellbrock, C. Collagen abundance controls melanoma phenotypes through lineage-specific microenvironment sensing. Oncogene 2018, 37, 3166–3182. [Google Scholar] [CrossRef]
- Lugassy, C.; Shahsafaei, A.; Bonitz, P.; Busam, K.J.; Barnhill, R.L. Tumor microvessels in melanoma express the beta-2 chain of laminin. Implications for melanoma metastasis. J. Cutan. Pathol. 1999, 26, 222–226. [Google Scholar] [CrossRef]
- Oikawa, Y.; Hansson, J.; Sasaki, T.; Rousselle, P.; Domogatskaya, A.; Rodin, S.; Tryggvason, K.; Patarroyo, M. Melanoma cells produce multiple laminin isoforms and strongly migrate on α5 laminin(s) via several integrin receptors. Exp. Cell Res. 2011, 317, 1119–1133. [Google Scholar] [CrossRef]
- Givant-horwitz, V.; Davidson, B.; Reich, R. Laminin-induced signaling in tumor cells: The role of the Mr 67,000 laminin receptor. Cancer Res. Baltim. 2004, 64, 3572–3579. [Google Scholar] [CrossRef]
- Chung, H.; Suh, E.K.; Han, I.O.; Oh, E.S. Keratinocyte-derived laminin-332 promotes adhesion and migration in melanocytes and melanoma. J. Biol. Chem. 2011, 286, 13438–13447. [Google Scholar] [CrossRef] [PubMed]
- Berno, V.; Porrini, D.; Castiglioni, F.; Campiglio, M.; Casalini, P.; Pupa, S.M.; Balsari, A.; Ménard, S.; Tagliabue, E. The 67 kDa laminin receptor increases tumor aggressiveness by remodeling laminin-1. Endocr. Relat. Cancer 2005, 12, 393–406. [Google Scholar] [CrossRef]
- Welf, E.S.; Driscoll, M.K.; Sapoznik, E.; Murali, V.S.; Weems, A.; Roh-Johnson, M.; Dean, K.M.; Fiolka, R.; Danuser, G. Worrying drives cell migration in mechanically unrestrained environments. bioRxiv 2020. [Google Scholar] [CrossRef]
- Van Kempen, L.C.L.T.; Rijntjes, J.; Mamor-Cornelissen, I.; Vincent-Naulleau, S.; Gerritsen, M.J.P.; Ruiter, D.J.; Van Dijk, M.C.R.F.; Geffrotin, C.; Van Muijen, G.N.P. Type I collagen expression contributes to angiogenesis and the development of deeply invasive cutaneous melanoma. Int. J. Cancer 2008, 122, 1019–1029. [Google Scholar] [CrossRef] [PubMed]
- Friedl, P.; Maaser, K.; Klein, C.E.; Niggemann, B.; Krohne, G.; Zänker, K.S. Migration of highly aggressive MV3 melanoma cells in 3-dimensional collagen lattices results in local matrix reorganization and shedding of α2 and β1 integrins and CD44. Cancer Res. 1997, 57, 2061–2070. [Google Scholar]
- Chelberg, M.K.; Tsilibary, E.C.; Hauser, A.R.; Mccarthy, J.B. Type IV collagen-mediated melanoma cell adhesion and migration: Involvement of multiple, distinct domains of the collagen molecule1. Cancer Res. 1989, 49, 4796–4802. [Google Scholar]
- Eble, J.A.; Golbik, R.; Mann, K.; Kühn, K. The alpha 1 beta 1 integrin recognition site of the basement membrane collagen molecule [alpha 1(IV)]2 alpha 2(IV). EMBO J. 1993, 12, 4795–4802. [Google Scholar] [CrossRef]
- Lauer-Fields, J.L.; Malkar, N.B.; Richet, G.; Drauz, K.; Fields, G.B. Melanoma cell CD44 interaction with the α1(IV)1263-1277 region from basement membrane collagen is modulated by ligand glycosylation. J. Biol. Chem. 2003, 278, 14321–14330. [Google Scholar] [CrossRef]
- Lauer, J.L.; Gendron, C.M.; Fields, G.B. Effect of ligand conformation on melanoma cell α3β1 integrin- mediated signal transduction events: Implications for a collagen structural modulation mechanism of tumor cell invasion. Biochemistry 1998, 37, 5279–5287. [Google Scholar] [CrossRef]
- Hodgson, L.; Henderson, A.J.; Dong, C. Melanoma cell migration to type IV collagen requires activation of NF-κB. Oncogene 2003, 22, 98–108. [Google Scholar] [CrossRef] [PubMed]
- Langereis, J.D.; Koenderman, L.; Huttenlocher, A.; Ulfman, L.H. Gelsolin expression increases β1 -integrin affinity and L1210 cell adhesion. Cytoskeleton 2013, 70, 385–393. [Google Scholar] [CrossRef]
- Qian, F.; Zhang, Z.C.; Wu, X.F.; Li, Y.P.; Xu, Q. Interaction between integrin α5 and fibronectin is required for metastasis of B16F10 melanoma cells. Biochem. Biophys. Res. Commun. 2005, 333, 1269–1275. [Google Scholar] [CrossRef]
- Li, B.; Shen, W.; Peng, H.; Li, Y.; Chen, F.; Zheng, L.; Xu, J.; Jia, L. Fibronectin 1 promotes melanoma proliferation and metastasis by inhibiting apoptosis and regulating EMT. Onco. Targets. Ther. 2019, 12, 3207–3221. [Google Scholar] [CrossRef] [PubMed]
- Jacquemet, G.; Hamidi, H.; Ivaska, J. Filopodia in cell adhesion, 3D migration and cancer cell invasion. Curr. Opin. Cell Biol. 2015, 36, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Nobes, C.D.; Hall, A. Rho, Rac, and Cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell 1995, 81, 53–62. [Google Scholar] [CrossRef]
- Xue, F.; Janzen, D.M.; Knecht, D.A. Contribution of filopodia to cell migration: A mechanical link between protrusion and contraction. Int. J. Cell Biol. 2010, 2010, 507821. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.; Witke, W.; Kwiatkowski, D.J.; Kosik, K.S. Delayed retraction of filopodia in gelsolin null mice. J. Cell Biol. 1997, 138, 1279–1287. [Google Scholar] [CrossRef]
- Huang, G.W.; Liao, L.D.; Li, E.M.; Xu, L.Y. SiRNA induces gelsolin gene transcription activation in human esophageal cancer cell. Sci. Rep. 2015, 5, 7901. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Sancho, A.; Chung, W.-L.; Vinik, Y.; Groll, J.; Zick, Y.; Medalia, O.; Bershadsky, A.D.; Geiger, B. Differential cellular responses to adhesive interactions with galectin-8- and fibronectin-coated substrates. J. Cell Sci. 2021, 134, jcs252221. [Google Scholar] [CrossRef] [PubMed]
- Gupton, S.L.; Waterman-Storer, C.M. Spatiotemporal Feedback between Actomyosin and Focal-Adhesion Systems Optimizes Rapid Cell Migration. Cell 2006, 125, 1361–1374. [Google Scholar] [CrossRef] [PubMed]
- Langereis, J.D.; Prinsen, B.H.C.M.T.; de Sain-van der Velden, M.G.M.; Coppens, C.J.C.; Koenderman, L.; Ulfman, L.H. A 2D-DIGE approach to identify proteins involved in inside-out control of integrins. J. Proteome Res. 2009, 8, 3824–3833. [Google Scholar] [CrossRef]
- Yamaguchi, H.; Lorenz, M.; Kempiak, S.; Sarmiento, C.; Coniglio, S.; Symons, M.; Segall, J.; Eddy, R.; Miki, H.; Takenawa, T.; et al. Molecular mechanisms of invadopodium formation: The role of the N-WASP-Arp2/3 complex pathway and cofilin. J. Cell Biol. 2005, 168, 441–452. [Google Scholar] [CrossRef] [PubMed]
- Stock, K.; Borrink, R.; Mikesch, J.H.; Hansmeier, A.; Rehkämper, J.; Trautmann, M.; Wardelmann, E.; Hartmann, W.; Sperveslage, J.; Steinestel, K. Overexpression and Tyr421-phosphorylation of cortactin is induced by three-dimensional spheroid culturing and contributes to migration and invasion of pancreatic ductal adenocarcinoma (PDAC) cells. Cancer Cell Int. 2019, 19, 77. [Google Scholar] [CrossRef] [PubMed]
- Zeugolis, D.I.; Khew, S.T.; Yew, E.S.Y.; Ekaputra, A.K.; Tong, Y.W.; Yung, L.Y.L.; Hutmacher, D.W.; Sheppard, C.; Raghunath, M. Electro-spinning of pure collagen nano-fibres—Just an expensive way to make gelatin? Biomaterials 2008, 29, 2293–2305. [Google Scholar] [CrossRef] [PubMed]
- Cooper, J.; Giancotti, F.G. Integrin signaling in cancer: Mechanotransduction, stemness, epithelial plasticity, and therapeutic resistance. Cancer Cell 2019, 35, 347–367. [Google Scholar] [CrossRef]
- Giancotti, F.G.; Ruoslahti, E. Integrin signaling. Science 1999, 285, 1028–1032. [Google Scholar] [CrossRef] [PubMed]
- Multhaupt, H.A.B.; Leitinger, B.; Gullberg, D.; Couchman, J.R. Extracellular matrix component signaling in cancer. Adv. Drug Deliv. Rev. 2016, 97, 28–40. [Google Scholar] [CrossRef] [PubMed]


| Coating | No Coating | Laminin | Fibronectin | Collagen Type I | Collagen Type IV | Matrigel | |
|---|---|---|---|---|---|---|---|
| Feature | |||||||
| Proliferation |  |  |  |  |  |  | |
| Projected cell area |  |  |  |  |  |  | |
| Number of filopodia |  |  |  |  |  |  | |
| Number of filopodia |  |  |  |  |  |  | |
| Length of filopodia |  |  |  |  |  |  | |
| Spontaneous migration |  |  |  |  |  |  | |
| Collective migration |  |  |  |  |  |  | |
| Spontaneous migration |  |  |  |  |  |  | |
| Collective migration |  |  |  |  |  |  | |
| Mixed Coatings | |||||||
| Active invadopodia |  | ||||||
| Invasion potential |  | ||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mazurkiewicz, E.; Makowiecka, A.; Mrówczyńska, E.; Kopernyk, I.; Nowak, D.; Mazur, A.J. Gelsolin Contributes to the Motility of A375 Melanoma Cells and This Activity Is Mediated by the Fibrous Extracellular Matrix Protein Profile. Cells 2021, 10, 1848. https://doi.org/10.3390/cells10081848
Mazurkiewicz E, Makowiecka A, Mrówczyńska E, Kopernyk I, Nowak D, Mazur AJ. Gelsolin Contributes to the Motility of A375 Melanoma Cells and This Activity Is Mediated by the Fibrous Extracellular Matrix Protein Profile. Cells. 2021; 10(8):1848. https://doi.org/10.3390/cells10081848
Chicago/Turabian StyleMazurkiewicz, Ewa, Aleksandra Makowiecka, Ewa Mrówczyńska, Iryna Kopernyk, Dorota Nowak, and Antonina Joanna Mazur. 2021. "Gelsolin Contributes to the Motility of A375 Melanoma Cells and This Activity Is Mediated by the Fibrous Extracellular Matrix Protein Profile" Cells 10, no. 8: 1848. https://doi.org/10.3390/cells10081848
APA StyleMazurkiewicz, E., Makowiecka, A., Mrówczyńska, E., Kopernyk, I., Nowak, D., & Mazur, A. J. (2021). Gelsolin Contributes to the Motility of A375 Melanoma Cells and This Activity Is Mediated by the Fibrous Extracellular Matrix Protein Profile. Cells, 10(8), 1848. https://doi.org/10.3390/cells10081848






