Abstract
In the last decade, an increasing number of studies have demonstrated that non-coding RNA (ncRNAs) cooperate in the gene regulatory networks with other biomolecules, including coding RNAs, DNAs and proteins. Among them, microRNAs (miRNAs), long non-coding RNAs (lncRNAs) and circular RNAs (circRNAs) are involved in transcriptional and translation regulation at different levels. Intriguingly, ncRNAs can be packed in vesicles, released in the extracellular space, and finally internalized by receiving cells, thus affecting gene expression also at distance. This review focuses on the mechanisms through which the ncRNAs can be selectively packaged into extracellular vesicles (EVs).
1. Introduction
1.1. Extracellular Vesicles (EVs)
EVs are a family of membrane-coated vesicles with different proteomic and lipidomic profile, as well as different size.
In their study, Pan and Johnstone firstly described extracellular vesicles containing the transferrin receptor during reticulocytes maturation [1]. In the beginning, the scientific community thought that extracellular vesicles were containers of waste products of cellular metabolism that the cell wanted to remove. Nowadays the role of EVs is emerging as an important new area of biomedical research [2]. It is well known that EVs are actively secreted by cells and that their content can affect activities and functions of receiving cells, in physiological as well as in pathological conditions [3,4,5].
Due to their biogenesis, we can identify two different types of EVs: exosomes and microvesicles (MV) [6]. The last ones originate by direct budding of the plasma membrane with a rearrangement of the cytoskeleton [7]. On the contrary, the biogenesis and release of exosomes is a multi-step process. In brief, it begins with the inward budding of the plasma membrane to form the early endosome that, passing through the late endosome, maturates to multivesicular bodies (MVBs) [8]. These incorporate, through the endosomal sorting complexes required for transport (ESCRT), the intraluminal vesicles (ILs), which contain different cytosolic components, including proteins and nucleic acids. After the fusion of the MVBs with cell membrane, the ILs are released outside the cells as exosomes [9,10].
The different biogenesis of the EVs also affects the composition of the vesicles’ membrane. The invagination process of ILs determines a specific lipidic and protein composition of the exosome’s membranes, which gives them a distinctive molecular profile. Exosomes’ membranes contain ESCRT components and associated proteins, as TSG101 and Alix. Moreover, the participation of tetraspanins in exosomes formation determines an abundance of these proteins in the exo-membranes [11].
MV as shed directly from the plasma membrane, hold the annexin A1 [12] and are enriched in specific phospholipids such as phosphatidylserine (PS), which promotes uptake by recipient cells, or sphingolipids as well as ceramide and cholesterol, that allow MV formation [13].
Finally, EVs are characterized by their size; generally, microvesicles range from 200 to 2000 nm while exosomes from 30 to 150/200 nm. Recently the ISEV society established that it is better to prefer other methods of classification because many vesicles are quite similar in size range [14,15].
Many studies have focused on the biological effects of EVs in cell-cell communication both between neighboring cells and between cells in distant body districts. Biological fluids carry the EVs released in the extracellular space until they find a landing place where dump their content through the fusion of plasma membranes or endocytosis [16]. Recently, a great number of data support the transport of ncRNAs among cells, demonstrating that they can exert special functional roles [17]. This review aims to shed light and collect the latest published data about the mechanisms of RNA-loading in extracellular vesicles. To this end, we searched for papers indexed on the PubMed database by combining the following keywords: non-coding RNAs, EVs, and RNA-loading. The time limit was set from 2015 to date, however, we listed a few older documents since noteworthy as pioneering works. Among the emerged manuscripts, we summarized here those providing molecular and functional data on the RNA loading.
1.2. RNA Families in Extracellular Vesicles
Much of the recent interest in EVs was triggered by the discovery of their involvement in horizontal transfer of secreted extracellular RNA (exRNA). From the first evidence of functional mRNA in EVs, a huge number of studies revealed a significant assortment of ncRNAs in EVs. The deep sequencing RNA technique allows demonstrating a selective enrichment of small ncRNAs in human-derived extracellular vesicles, isolated from different cytotypes [18,19]. Among them, small RNA families are the most abundant, these include small nuclear RNAs, small nucleolar RNAs, ribosomal RNAs, transfer RNAs, miRNAs. However, also larger RNAs groups, including mitochondrial RNAs, Y RNA, vault RNA, piwi RNA and long non-coding RNA have been found in EVs, for a detailed review of the different RNAs families in EV see [20]. A recent work of Mosbach et al. demonstrated that RNA sorting depends on its size but also on its origin, the authors in fact revealed that RNA polymerase III transcripts are preferentially associated with EVs [21].
As pointed out by [20] although several types of RNAs have been identified in EVs, only some of these have been demonstrated functioning in the recipient cell, i.e., miRNAs and lncRNAs. For other RNA molecules, such as tRNAs, can be assumed a possible activity in the cytoplasm of the recipient cells while the role of other RNA molecules such as mRNA fragments or ribosomal RNAs remains unclear (Figure 1).
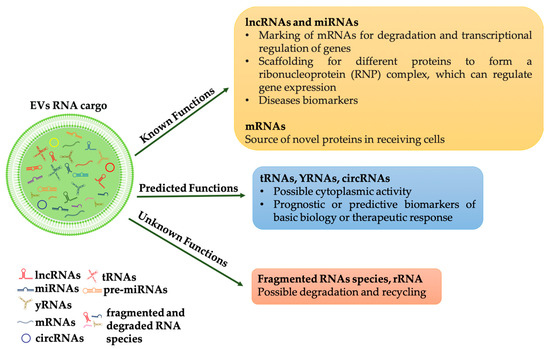
Figure 1.
RNAs species into EVs and their functions. RNAs in extracellular vesicles can be classified into three types: (1) RNAs that have known function when internalized into target cells, such as mRNAs, miRNAs and lncRNAs; (2) RNAs that are predicted to be functional (for example, tRNAs, YRNAs, circRNAs); (3) RNAs with unknown functions (for example, fragments RNAs and rRNAs), some of which may be functional, but others may be non-functional degradation products.
Still controversial is the presence of miRNA precursors: their identification, together with Dicer and Argonaute 2, let scientists to suppose that complete silencing machinery could be transferred by EVs [22,23]. However, the hypothesis was not largely confirmed by other investigators.
Other conflicting observations in studying exRNAs are due to the different isolation strategies. For example, ultracentrifugation, the gold standard method for EVs purification, biases RNAs data. It in fact does not allow to separate vesicles from free ribonucleoproteins or lipoproteins both associated with RNAs [24].
To overcome this limitation, Lässer group developed a method combining size exclusion chromatography with a density cushion; this allows to isolate and characterize EVs from blood with minimal contamination by plasma proteins and lipoprotein particles [25].
Meanwhile, Jeppesen et al. used high-resolution density gradient fractionation to separate EVs from non-vesicular material, then, with a direct immunoaffinity capture (DIC) targeting classical exosomal tetraspanins, exosomes were specifically isolated from other types of EVs [12]. Nucleic acid analysis revealed that exRNAs are differentially expressed between EVs and non-vesicle compartments. Interestingly the authors demonstrated that miRNAs were mainly associated with extracellular non-vesicular fractions while the EVs were enriched in transfer RNA (tRNA) fragments. In addition, YRNA and vault RNA were particularly enriched in non-vesicular fractions [12].
New isolation techniques have allowed more accurate identification of the RNA families transported through the EVs, further confirming the role of vesicles in the horizontal transfer of this nucleic acid.
2. The Effects of the Horizontal Transfer of EVs Derived RNAs
Many studies demonstrated the functional role of the RNA-cargo that, selectively packaged inside EVs, are transferred to target cells. Firstly, the transfer of exosomal mRNAs and miRNAs was reported by Valadi et al. in 2007, which revealed a “novel mechanism of genetic exchange between cells” [26]. Since that study, numerous papers have demonstrated that the horizontal transfer of exosomes cargo can modulate target cells behaviors.
Studies on tumor cells revealed as exosome-mediated RNA transfer may control tumor growth, affect its microenvironment, and promote metastases.
It is well known that a central event in tumor progression is the induction of angiogenesis. Focusing on this pathway it has been demonstrated that glioblastoma, chronic myelogenous leukemia, and breast cancer derived EVs can reprogram endothelial cells through horizontal transfer of their miRNAs cargo [27,28].
The involvement of cancer derived EVs in facilitating brain infiltration is worth mentioning. Lu and collaborators have recently demonstrated that exosomes derived by highly brain metastatic breast cancer cells are able to destroy the blood–brain barrier through its lncRNA GS1-600G8.5 [29,30].
Exosomal miRNAs have a role also in tumor drug resistance. Mao et al. support this hypothesis demonstrating that Adriamycin-resistant breast cancer cells deliver specific miRNAs through exosomes thus promoting drug resistance in neighboring cells [31]. Quin and collaborators demonstrated that exosomes derived from cisplatin-resistant lung cancer cell line A549 induce drug resistance in receiving cells. MiRNA profile identified the miR100-5p as the mediator of this process [32].
In addition, exosome-transported lncRNAs may participate in drug resistance induction. A recent paper by Wang et al. proved that the exosome-mediated transfer of lncRNA H19 induces doxorubicin resistance in breast cancer [33]. The authors showed that drug-resistant cells release exosomes enriched in lncH19; these exosomes increase the chemoresistance of doxorubicin once internalized by sensitive cells. Moreover, downregulation of H19 in sensitive cells ablated this effect thus confirming the direct role of the long non-coding RNA [33]. EVs released into the tumor microenvironment strongly affect metastatic niche. For example, the prometastatic miRNA miR-9 and miR-155, carried respectively by breast cancer derived exosomes and pancreatic cancer derived microvesicles, are able to reprogram fibroblast to cancer associated fibroblast (CAF) phenotype thus promoting tumor progression [34,35]. While the EV-mediated delivery of the miR-105 and -miR-122 reprogram CAFs metabolism to sustain tumor growth [36,37]. Moreover, cancer derived EVs can reprogram immune cells thus preventing immunosurveillance and promoting immunotolerance in cancer microenvironment, as revised by Graner [38]. In addition, EVs derived RNAs profiling can be useful as a prognostic indicator to therapeutic response. For example, miR196a-5p and miR-501-3p were significantly downregulated in exosomes isolated from the urine of prostate cancer patients [39], while let7-b and miR-18a, isolated from plasma of multiple myeloma patients, were associated with overall survival [40]. As expected, the effects of the EV-transported RNA are not limited to the tumor context.
Barile and colleagues revealed the cardioprotective role of the different miRNAs transported by EVs derived from cardiac progenitor cells (CPC). They demonstrated that miR-210 inhibits cardiomyocyte apoptosis by targeting Ephrin A3 (cell surface GPI-bound ligand for Eph receptors) and PTP1b (protein-tyrosine phosphatase 1b). Moreover, EVs derived miR132 stimulates angiogenesis acting on RasGAP-p120, Ras GTPase activating protein p120 [41]. Furthermore, CPC derived exosomal miR21 exert similar effects preventing cell apoptosis by targeting PDCD4 (Programmed Cell Death 4) [42]. Similarly, Gray and collaborators, through microarray analysis of exosomes derived from hypoxic CPC, identified 11 miRNAs that improve cardiac function stimulating tube formation of endothelial cells and reducing fibrosis [43].
Regarding differentiation, it has been demonstrated that exosomes released from Neural stem/progenitor cells (NPCs) have an important role in neurogenesis; Ma and collaborators demonstrated that mouse cortical NPCs, isolated from fetal brain, promote neuronal differentiation through exosomal miR-21a [44].
Adipose tissue is an excellent resource for circulating exosomal miRNAs that have a role in regulating liver gene expression, as well as affecting obesity or diabetes. Adipose-derived exosomal miR-99b has been demonstrated to control in vivo fibroblast growth factor 21 (FGF21) production [45]. While exosomes secreted by adipose tissue macrophages transfer miRNAs modulating, in vivo and in vitro, insulin sensitivity and glucose homeostasis [46].
All these experiments suggest that exosomes may be a vehicle of therapeutic non-coding RNAs in physiological and pathological conditions as well as in the field of regenerative therapy (Figure 2) (Table 1). To this end, a comprehensive analysis of the RNA loading mechanisms is appropriate.
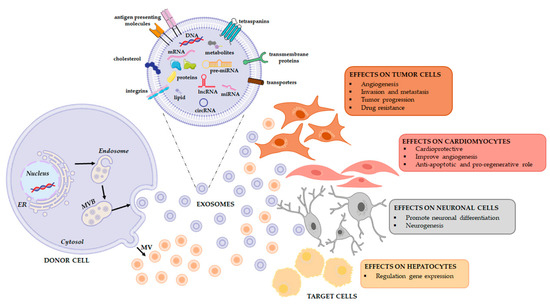
Figure 2.
Extracellular vesicles release and their functional effects on target cells. EVs are a heterogeneous population, both in form and content since their cargo is strictly dependent on the pathophysiological conditions of the cell at the exact moment in which it produces the vesicle. When studying the complexity of the EV-mediated cell-cell communication, it is necessary to evaluate that the same vesicle, i.e., the same message, can be interpreted differently depending on the cytotype that receives it. This will depend in good part on the gene expression profile of the recipient cell.

Table 1.
Functional effects of ncRNAs shuttled by extracellular vesicles.
3. Loading of EVs and Cargo Sorting
3.1. RNA Binding Protein-Mediated Loading
Recent evidence highlighted the main role of RNA-binding proteins in RNA sorting and loading in EVs. Santangelo et al. have identified the RNA binding protein SYNCRIP (synaptotagmin-binding cytoplasmic RNA-interacting protein; also known as hnRNPQ or NSAP1) as a component of the hepatocyte exosomal miRNA sorting machinery. They showed that SYNCRIP knockdown impairs the internalization of miRNAs in exosomes [47]. Subsequently, Hobor et al., identified that SYNCRIP contains a sequence called NURR (N-terminal unit for RNA recognition) that recognizes and bind the motif GGCU/A in miRNAs. This interaction guides miRNA loading into exosomes [48].
Recently, Temoche-Diaz and collaborators showed that the metastatic breast cancer cell line MDA-MB-231 releases two sub-populations of EVs, called vesicular low density (vLD) and vesicular high density (vHD) EVs. Mass spectrometry analysis revealed that CD63, known as an exosomal marker, was enriched in the vHD subpopulation, as well as other endosome-associated proteins. MiRNA analysis identified five miRNAs that were enriched in vHD vesicles and not in vLD ones, thus suggesting that there is a specific mechanism of miRNA sorting for the vHD subpopulation [49]. Interestingly, miR-122, a well-known prognostic biomarker for metastasis in breast cancer patients [50,51] was identified. The authors demonstrated that the RNA binding protein Lupus La drives the selective miR-122 vHD enrichment in breast cancer cell lines [49].
RNA binding proteins not only promote RNA loading but also contribute to RNA function into target cells. Chen et al. studying bladder cancer metastatization identified an exosomal lncRNA, termed lymph node metastasis-associated transcript 2 (LNMAT2) that promotes lymphangiogenesis. Interestingly, they found that LNMAT2 was loaded into exosomes by the heterogeneous nuclear ribonucleoprotein A2/B1 (HNRNPA2B1). The complex LNMAT2/hnRNPA2B1, once in receiving cells, can migrate into the nucleus and interact with PROX1 promoter. Here HNRNPA2B1 mediates H3 lysine 4 trimethylation (H3K4me3) and subsequent activation of PROX1 which, in turn, regulates endothelial cell differentiation and metastatic dissemination [52].
Often post-translational modifications of RNA binding proteins, such as SUMOylation, phosphorylation or glycosylation, contribute to regulating exosome loading and release [53,54].
Regarding non-coding RNAs loading, the sumoylation of heterogeneous nuclear ribonucleoproteins (HNRNPs) is one of the proposed mechanisms. HNRNPA1, after sumoylation, recognizes the long non-coding RNA ELNAT1 thus mediating its packaging into EVs [55]. While HNRNPA2B1, after sumoylation, binds miRNAs recognizing the EXOmotif GGAG (conserved motifs that allow the specific miRNAs sorting into exosomes) and this interaction is responsible for miRNAs loading into exosomes [56].
The ribonucleoprotein HNRNPA2B1, again, through GlcNAcylation, can regulate the internalization of specific miRNAs into EVs. Recently, Lee and collaborators demonstrated that Caveolin1 (CAV1), after its tyrosine phosphorylation (Y14), interacts with HNRNPA2B1, promoting HNRNPA2B1 O-GlcNAcylation on two serine domains. This modified complex directly controls the packaging of specific miRNAs into EVs [57]. Another study by McKenzie and colleagues highlighted that Ago2 phosphorylation could control loading of miRNAs in exosomes [58].
In a recent work, Robinson and coworkers have identified HNRNPK as a CAV1-regulated microRNA binding protein. They observed that CAV1 drives HNRPNK localization to MVB, which brings the miRNA containing the AsUGnA motif. Moreover, they found that membrane-rafts take part in this transport [59]. Export of miRNAs into exosomes and their subsequent release, in fact, can occur also through a lipid rafts dependent mechanism, as discussed later.
Again, HNRNPK participates in EVs loading and secretion of non-coding RNAs, through an alternative pathway mediated by LC3-conjugation machinery and called LDELS, LC3 dependent EV loading and secretion [60].
Leidal et al. demonstrated that lipidated LC3-II is involved in the loading of specific proteins into intraluminal vesicles (ILVs) for their subsequent release as EVs. Interestingly the process is distinct from classical macroautophagy/autophagy because it requires components of the LC3 conjugation machinery, but not other ATGs involved in autophagosome formation. By proteomic analyses of LC3-conjugated protein obtained from secretome, the authors demonstrated that 33% of identified proteins have been previously associated with EVs. Moreover, after sucrose density gradient purification, they found that endogenous LC3-II co-fractionated with well-defined EV markers. This result further confirms the presence of LC3-II residues inside the lumen of EVs. Through different knock out models, the authors identified several RNA binding proteins that require LC3 to be internalized in EVs, among these the HNRNPK and scaffold-attachment Factor B (SAFB). Interestingly the deficiency in LC3-conjugation machinery affects the amounts of RNA loaded in EV. In particular, snoRNAs and miRNAs were reduced with an increase in tRNAs while no effects were found in large RNA levels [60].
New evidence from Arabidopsis proteomic analysis revealed that also plant- derived EVs contain several RNA-binding proteins, as Argonaute 1 (AGO1) and RNA helicases (RHs). These proteins selectively associate with EV-enriched small RNAs, thus suggesting their involvement in specific loading of sRNAs into EVs also in plants [61].
In conclusion, several RNA binding proteins, alone or in combination with other molecular interactors may control RNA sorting inside EV (Figure 3).
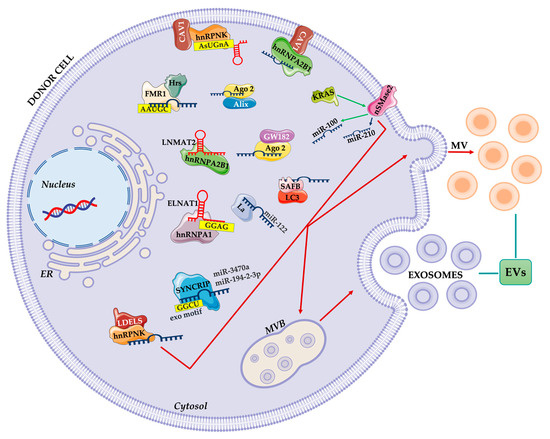
Figure 3.
Summary of proteins involved in the ncRNA packaging into EVs. RNA binding proteins alone or in cooperation with other proteins bind specific ncRNAs and selectively transport them into EVs. Membrane proteins are also involved in the ncRNA loading EVs mechanism.
3.2. Other Carriers for RNA Loading
It is well known that the ESCRT pathway is responsible for protein sorting into EVs [62,63].
Alix is an adaptor protein involved in EVs biogenesis and cargo sorting through an ESCRT dependent pathway [64]. However, new evidence indicated that Alix is involved in miRNAs loading into EVs. Co-immunoprecipitation experiments revealed a direct interaction between Alix and the RNA binding protein Ago2, commonly involved in miRNA transport and processing. This complex drives Alix with Ago2-associated miRNAs into EVs [65,66]. A further connection between ESCRT complex and selective RNA loading was confirmed by Wozniak et al. [67]. In particular, the authors demonstrated that the RBP fragile X mental retardation 1 (FMR1) interacts with the hepatocyte growth factor-regulated tyrosine kinase substrate (Hrs), a component of the ESCRT complex. The RNA binding protein FMR1 acts as a chaperone that recognizes a specific sequence in miRNA (AAUGC) while Hrs allows complex internalization. Interestingly, inflammosome activation mediates this interaction, through the cleavage of the trafficking adaptor protein RILP (Rab-interacting lysosomal protein) that works as ride [67].
Cargo sorting could also be driven via ESCRT-independent pathways, e.g., through the neutral sphingomyelinase, phospholipase or other lipids and associated protein such as tetraspanin [9,10,60,68].
Janas et al. have proposed that the RNA loading into exosomes could be mediated by the direct interaction between RNAs and the lipid raft of MVB membrane [69]. Moreover, in their recent studies, through bioinformatic analysis, they identified four raft RNA motifs that are frequent in the exosomal pro-tumoral miRNAs transferred from cancer cells to immune cells [70].
Other recent evidence suggests that neutral sphingomyelinase 2 (nSMase 2), via ceramide production, regulates EVs cargo loading. Kosaka and coworkers demonstrated that nSMase2 promotes the miR-210 packaging in EVs that finally affect the initiation of metastatic process through the induction of angiogenesis [71]. Cha et al. compared small RNAs from isogenic cell lines, that differ only in KRAS status, to demonstrate a KRAS dependent sorting of miRNAs into EVs. In mutant KRAS-derived EVs authors showed enrichment of tumor-suppressive miR-100 while the oncomiR miR-10b was preferentially secreted in EVs derived from wild type KRAS cell line; these data demonstrate a key role for KRAS in orchestrating RNA trafficking. Interestingly nSMase2 is the mediator of this mechanism since its inhibition caused the miR-100 confinement in KRAS mutated cells [72].
Sphingomyelinase takes part also in the transport of viral RNA mediated by EVs. Zhou et al. demonstrated that ZIKA viruses, which infection induces severe neurological manifestations, use exosomes as mediators of viral transmission between neurons. The authors demonstrated that cortical neuronal cell-derived exosomes contain ZIKA virus RNAs and proteins and that the treatment with GW4869, nSMase-2/SMPD3 specific inhibitor, significantly reduced the loading of viral molecules. Interestingly, viral RNA positively participates in transmission promoting in neuronal cells the expression and activity of the nSMase-2 [73].
Finally, it has to consider the possibility that the RNAs are not passively loaded into the exosomes but may themselves participate in vesicle formation. This is the case of lncRNA plasmacytoma variant translocation 1 (PVT1) that, highly expressed in several cancers including pancreatic cancer, promotes exosome secretion [74]. Sun et al. demonstrated that PVT1 promotes the docking of MVBs by altering RAB7 expression and localization. Moreover, PVT1 promotes the palmitoylation of the v-SNARE homolog YKT6 and its colocalization with vesicle-associated membrane protein 3 (VAMP3) thus determining the fusion of MVBs with the plasma membrane. The involvement of lncRNA in exosomes formation or release let also suppose a direct role of these molecules in transporting other smaller RNAs.
In conclusion, cargo sorting could be mediated by carriers commonly involved in EV biogenesis or in miRNA transport and processing. Cargo sorting could also be driven via ESCRT dependent or via ESCRT-independent pathways, e.g., through the neutral sphingomyelinase, phospholipase or other lipids and associated proteins (Figure 2).
3.3. Engeneering Vesicles for RNA Loading
Several characteristics of EVs make them an interesting candidate for RNA delivery. EVs in fact can cross biological barriers, avoid toxicity and immunogenicity, and have an endogenous targeting ability. However, not all EVs show the same properties.
A recent paper by Murphy et al. formally demonstrated that EVs possess higher RNA delivery efficiency than synthetic RNA delivery systems, used for clinical delivery of siRNAs [75]. To evaluate the efficiency of transported RNAs, the authors developed the CRISPR Operated Stoplight System for Functional Intercellular RNA Exchange (CROSS-FIRE) reporter system that can be activated only by the functional transfer of a specific single-guide RNA (sgRNA). Interestingly, comparing the sgRNA-delivery among EVs and synthetic systems the authors confirmed the highest efficiency of EVs. Moreover, comparative analyses among EVs from different cell lines highlighted the divergence in the efficiency of encapsulating RNA by up to 30 times [75].
Several groups are developing engineering strategies to facilitate loading to develop systemic delivery of siRNA or miRNA by EVs. The ultra-thermostable pRNA-3WJ core has been used in many applications including gene therapy, target specific delivery, controlled drug release, and image-guided diagnostics [76]. Recently Pi et al. demonstrated that the arrow-shaped pRNA-3WJ offers the opportunity to control either partial loading of RNA into EVs or decoration of ligands on the surface of EVs. With cholesterol placed on the arrow-tail of the 3WJ, in fact, the RNA is mainly located on the surface of the EVs, while, placing the cholesterol at the arrowhead resulted in partial loading of RNA nanoparticles into the extracellular vesicles [77].
Specific loading of miRNA or siRNA in EVs has been performed through overexpression of the interested RNA in EV producing cells; however, recently the manuscript of Sork et al. demonstrated this strategy as a “wasteful way of loading miRNA to EVs”. The authors transiently overexpressed two different pri-miRNAs in HEK293-T cells and quantified the respective mature miRNA levels in the EV and non-EV portion separated by size-exclusion chromatography. The analysis revealed that most overexpressed mature transcripts were secreted in the non-EV fraction, whereas the EV fraction contained <2% of the respective miRNAs [78].
The new generation of loading techniques uses RNA binding proteins to guide the internalization of specific RNA sequences. Kojima et al. [79] among EXOtic devices able to boost exosome production and application, developed a specific RNA packaging system using the archaeal ribosomal protein L7Ae which binds to the C/D box RNA structure. They conjugated L7Ae to the C-terminus of CD63, thus allowing its localization on the exosomal membrane, and inserted a C/D box into the 3′-untranslated region (3′-UTR) of the reporter gene [79].
Wang et al. took advantage of the use of the transactivator of transcription (Tat) protein, which binds specifically to the stem-loop-containing trans-activating response (TAR) element RNA. They added TAR directly to the 5′ end of a cargo mRNA and fused Tat peptide directly to the C-terminus of ARRDC1, the Arrestin domain containing protein 1 [80].
Despite numerous attempts to engineer vesicles with specific RNAs, the major limitation is still the partial knowledge of the sorting and loading mechanisms in nature.
4. Conclusions
Recent investigations have shown the central role of ncRNAs in gene regulation at different levels, including the ‘remote’ regulation mediated by EVs.
Here we summarized the newest studies describing the RNA binding proteins and other interactors controlling the RNA sorting inside EVs. However, many aspects of RNA trafficking need to be further investigated. Moreover, it remains unclear why cells selectively load specific RNAs into subclasses of extracellular vesicles unless we still want to convince ourselves that it is only a strategy for discarding excess materials.
Detailed knowledge of the loading mechanisms may point towards the wider use of EVs as diagnostic and prognostic tools. Moreover, it can provide essential indications for enhancing the use of EVs as drug delivery systems.
Author Contributions
Conceptualization, C.C., M.M.B. and A.C., writing—original draft preparation, C.C., M.M.B., C.Z., review and editing, R.A. and A.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by AIRC under MFAG 2017—ID. 19982 project—P.I. Alice Conigliaro‘.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the writing of the manuscript, or in the decision to publish it.
References
- Pan, B.T.; Johnstone, R.M. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: Selective externalization of the receptor. Cell 1983, 33, 967–978. [Google Scholar] [CrossRef]
- Boriachek, K.; Islam, M.N.; Moller, A.; Salomon, C.; Nguyen, N.T.; Hossain, M.S.A.; Yamauchi, Y.; Shiddiky, M.J.A. Biological Functions and Current Advances in Isolation and Detection Strategies for Exosome Nanovesicles. Small 2018, 14, 1702153. [Google Scholar] [CrossRef]
- Martellucci, S.; Orefice, N.S.; Angelucci, A.; Luce, A.; Caraglia, M.; Zappavigna, S. Extracellular Vesicles: New Endogenous Shuttles for miRNAs in Cancer Diagnosis and Therapy? Int. J. Mol. Sci. 2020, 21, 6486. [Google Scholar] [CrossRef]
- Hafiane, A.; Daskalopoulou, S.S. Extracellular vesicles characteristics and emerging roles in atherosclerotic cardiovascular disease. Metabolism 2018, 85, 213–222. [Google Scholar] [CrossRef] [PubMed]
- Hosseinkhani, B.; Kuypers, S.; van den Akker, N.M.S.; Molin, D.G.M.; Michiels, L. Extracellular Vesicles Work as a Functional Inflammatory Mediator Between Vascular Endothelial Cells and Immune Cells. Front. Immunol. 2018, 9, 1789. [Google Scholar] [CrossRef] [Green Version]
- Van Niel, G.; D’Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef]
- Li, B.; Antonyak, M.A.; Zhang, J.; Cerione, R.A. RhoA triggers a specific signaling pathway that generates transforming microvesicles in cancer cells. Oncogene 2012, 31, 4740–4749. [Google Scholar] [CrossRef] [Green Version]
- Hessvik, N.P.; Llorente, A. Current knowledge on exosome biogenesis and release. Cell Mol. Life Sci. 2018, 75, 193–208. [Google Scholar] [CrossRef] [Green Version]
- Pegtel, D.M.; Gould, S.J. Exosomes. Annu. Rev. Biochem. 2019, 88, 487–514. [Google Scholar] [CrossRef] [PubMed]
- Russell, A.E.; Sneider, A.; Witwer, K.W.; Bergese, P.; Bhattacharyya, S.N.; Cocks, A.; Cocucci, E.; Erdbrugger, U.; Falcon-Perez, J.M.; Freeman, D.W.; et al. Biological membranes in EV biogenesis, stability, uptake, and cargo transfer: An ISEV position paper arising from the ISEV membranes and EVs workshop. J. Extracell. Vesicles 2019, 8, 1684862. [Google Scholar] [CrossRef] [Green Version]
- Deng, F.; Miller, J. A review on protein markers of exosome from different bio-resources and the antibodies used for characterization. J. Histotechnol. 2019, 42, 226–239. [Google Scholar] [CrossRef]
- Jeppesen, D.K.; Fenix, A.M.; Franklin, J.L.; Higginbotham, J.N.; Zhang, Q.; Zimmerman, L.J.; Liebler, D.C.; Ping, J.; Liu, Q.; Evans, R.; et al. Reassessment of Exosome Composition. Cell 2019, 177, 428–445. [Google Scholar] [CrossRef] [Green Version]
- Sedgwick, A.E.; D’Souza-Schorey, C. The biology of extracellular microvesicles. Traffic 2018, 19, 319–327. [Google Scholar] [CrossRef]
- Shao, H.; Im, H.; Castro, C.M.; Breakefield, X.; Weissleder, R.; Lee, H. New Technologies for Analysis of Extracellular Vesicles. Chem. Rev. 2018, 118, 1917–1950. [Google Scholar] [CrossRef] [PubMed]
- Thery, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef]
- Xie, Y.; Dang, W.; Zhang, S.; Yue, W.; Yang, L.; Zhai, X.; Yan, Q.; Lu, J. The role of exosomal noncoding RNAs in cancer. Mol. Cancer 2019, 18, 37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lunavat, T.R.; Cheng, L.; Kim, D.K.; Bhadury, J.; Jang, S.C.; Lasser, C.; Sharples, R.A.; Lopez, M.D.; Nilsson, J.; Gho, Y.S.; et al. Small RNA deep sequencing discriminates subsets of extracellular vesicles released by melanoma cells—Evidence of unique microRNA cargos. RNA Biol. 2015, 12, 810–823. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Q.; Yang, J.; Zheng, J.; Hsueh, C.; Guo, Y.; Zhou, L. Characterization of selective exosomal microRNA expression profile derived from laryngeal squamous cell carcinoma detected by next generation sequencing. Oncol. Rep. 2018, 40, 2584–2594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Brien, K.; Breyne, K.; Ughetto, S.; Laurent, L.C.; Breakefield, X.O. RNA delivery by extracellular vesicles in mammalian cells and its applications. Nat. Rev. Mol. Cell Biol. 2020, 21, 585–606. [Google Scholar] [CrossRef] [PubMed]
- Mosbach, M.-L.; Pfafenrot, C.; von Strandmann, E.P.; Bindereif, A.; Preußer, C. Molecular Determinants for RNA Release into Extracellular Vesicles. Cells 2021, 10, 2674. [Google Scholar] [CrossRef]
- Clancy, J.W.; Zhang, Y.; Sheehan, C.; D’Souza-Schorey, C. An ARF6-Exportin-5 axis delivers pre-miRNA cargo to tumour microvesicles. Nat. Cell Biol. 2019, 21, 856–866. [Google Scholar] [CrossRef] [PubMed]
- Melo, S.A.; Sugimoto, H.; O’Connell, J.T.; Kato, N.; Villanueva, A.; Vidal, A.; Qiu, L.; Vitkin, E.; Perelman, L.T.; Melo, C.A.; et al. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell 2014, 26, 707–721. [Google Scholar] [CrossRef] [Green Version]
- Van Deun, J.; Mestdagh, P.; Sormunen, R.; Cocquyt, V.; Vermaelen, K.; Vandesompele, J.; Bracke, M.; de Wever, O.; Hendrix, A. The impact of disparate isolation methods for extracellular vesicles on downstream RNA profiling. J. Extracell. Vesicles 2014, 3, 24858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karimi, N.; Cvjetkovic, A.; Jang, S.C.; Crescitelli, R.; Hosseinpour Feizi, M.A.; Nieuwland, R.; Lotvall, J.; Lasser, C. Detailed analysis of the plasma extracellular vesicle proteome after separation from lipoproteins. Cell Mol. Life Sci. 2018, 75, 2873–2886. [Google Scholar] [CrossRef] [Green Version]
- Valadi, H.; Ekstrom, K.; Bossios, A.; Sjostrand, M.; Lee, J.J.; Lotvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [Green Version]
- Lucero, R.; Zappulli, V.; Sammarco, A.; Murillo, O.D.; Cheah, P.S.; Srinivasan, S.; Tai, E.; Ting, D.T.; Wei, Z.; Roth, M.E.; et al. Glioma-Derived miRNA-Containing Extracellular Vesicles Induce Angiogenesis by Reprogramming Brain Endothelial Cells. Cell Rep. 2020, 30, 2065–2074. [Google Scholar] [CrossRef] [Green Version]
- Liu, Q.; Peng, F.; Chen, J. The Role of Exosomal MicroRNAs in the Tumor Microenvironment of Breast Cancer. Int. J. Mol. Sci. 2019, 20, 3884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, Y.; Chen, L.; Li, L.; Cao, Y. Exosomes Derived from Brain Metastatic Breast Cancer Cells Destroy the Blood-Brain Barrier by Carrying lncRNA GS1-600G8.5. Biomed. Res. Int. 2020, 2020, 7461727. [Google Scholar] [CrossRef]
- Yi, Y.; Wu, M.; Zeng, H.; Hu, W.; Zhao, C.; Xiong, M.; Lv, W.; Deng, P.; Zhang, Q.; Wu, Y. Tumor-Derived Exosomal Non-Coding RNAs: The Emerging Mechanisms and Potential Clinical Applications in Breast Cancer. Front. Oncol. 2021, 11, 738945. [Google Scholar] [CrossRef]
- Mao, L.; Li, J.; Chen, W.X.; Cai, Y.Q.; Yu, D.D.; Zhong, S.L.; Zhao, J.H.; Zhou, J.W.; Tang, J.H. Exosomes decrease sensitivity of breast cancer cells to adriamycin by delivering microRNAs. Tumour Biol. 2016, 37, 5247–5256. [Google Scholar] [CrossRef]
- Qin, X.; Yu, S.; Zhou, L.; Shi, M.; Hu, Y.; Xu, X.; Shen, B.; Liu, S.; Yan, D.; Feng, J. Cisplatin-resistant lung cancer cell-derived exosomes increase cisplatin resistance of recipient cells in exosomal miR-100-5p-dependent manner. Int. J. Nanomed. 2017, 12, 3721–3733. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Pei, X.; Guo, G.; Qian, X.; Dou, D.; Zhang, Z.; Xu, X.; Duan, X. Exosome-mediated transfer of long noncoding RNA H19 induces doxorubicin resistance in breast cancer. J. Cell Physiol. 2020, 235, 6896–6904. [Google Scholar] [CrossRef]
- Baroni, S.; Romero-Cordoba, S.; Plantamura, I.; Dugo, M.; D’Ippolito, E.; Cataldo, A.; Cosentino, G.; Angeloni, V.; Rossini, A.; Daidone, M.G.; et al. Exosome-mediated delivery of miR-9 induces cancer-associated fibroblast-like properties in human breast fibroblasts. Cell Death Dis. 2016, 7, e2312. [Google Scholar] [CrossRef] [PubMed]
- Pang, W.; Su, J.; Wang, Y.; Feng, H.; Dai, X.; Yuan, Y.; Chen, X.; Yao, W. Pancreatic cancer-secreted miR-155 implicates in the conversion from normal fibroblasts to cancer-associated fibroblasts. Cancer Sci. 2015, 106, 1362–1369. [Google Scholar] [CrossRef]
- Yan, W.; Wu, X.; Zhou, W.; Fong, M.Y.; Cao, M.; Liu, J.; Liu, X.; Chen, C.H.; Fadare, O.; Pizzo, D.P.; et al. Cancer-cell-secreted exosomal miR-105 promotes tumour growth through the MYC-dependent metabolic reprogramming of stromal cells. Nat. Cell Biol. 2018, 20, 597–609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fong, M.Y.; Zhou, W.; Liu, L.; Alontaga, A.Y.; Chandra, M.; Ashby, J.; Chow, A.; O’Connor, S.T.; Li, S.; Chin, A.R.; et al. Breast-cancer-secreted miR-122 reprograms glucose metabolism in premetastatic niche to promote metastasis. Nat. Cell Biol. 2015, 17, 183–194. [Google Scholar] [CrossRef] [Green Version]
- Graner, M.W.; Schnell, S.; Olin, M.R. Tumor-derived exosomes, microRNAs, and cancer immune suppression. Semin. Immunopathol. 2018, 40, 505–515. [Google Scholar] [CrossRef]
- Rodriguez, M.; Bajo-Santos, C.; Hessvik, N.P.; Lorenz, S.; Fromm, B.; Berge, V.; Sandvig, K.; Line, A.; Llorente, A. Identification of non-invasive miRNAs biomarkers for prostate cancer by deep sequencing analysis of urinary exosomes. Mol. Cancer 2017, 16, 156. [Google Scholar] [CrossRef]
- Manier, S.; Liu, C.J.; Avet-Loiseau, H.; Park, J.; Shi, J.; Campigotto, F.; Salem, K.Z.; Huynh, D.; Glavey, S.V.; Rivotto, B.; et al. Prognostic role of circulating exosomal miRNAs in multiple myeloma. Blood 2017, 129, 2429–2436. [Google Scholar] [CrossRef]
- Barile, L.; Lionetti, V.; Cervio, E.; Matteucci, M.; Gherghiceanu, M.; Popescu, L.M.; Torre, T.; Siclari, F.; Moccetti, T.; Vassalli, G. Extracellular vesicles from human cardiac progenitor cells inhibit cardiomyocyte apoptosis and improve cardiac function after myocardial infarction. Cardiovasc. Res. 2014, 103, 530–541. [Google Scholar] [CrossRef]
- Xiao, J.; Pan, Y.; Li, X.H.; Yang, X.Y.; Feng, Y.L.; Tan, H.H.; Jiang, L.; Feng, J.; Yu, X.Y. Cardiac progenitor cell-derived exosomes prevent cardiomyocytes apoptosis through exosomal miR-21 by targeting PDCD4. Cell Death Dis. 2016, 7, e2277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gray, W.D.; French, K.M.; Ghosh-Choudhary, S.; Maxwell, J.T.; Brown, M.E.; Platt, M.O.; Searles, C.D.; Davis, M.E. Identification of therapeutic covariant microRNA clusters in hypoxia-treated cardiac progenitor cell exosomes using systems biology. Circ. Res. 2015, 116, 255–263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, Y.; Li, C.; Huang, Y.; Wang, Y.; Xia, X.; Zheng, J.C. Exosomes released from neural progenitor cells and induced neural progenitor cells regulate neurogenesis through miR-21a. Cell Commun. Signal. 2019, 17, 96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomou, T.; Mori, M.A.; Dreyfuss, J.M.; Konishi, M.; Sakaguchi, M.; Wolfrum, C.; Rao, T.N.; Winnay, J.N.; Garcia-Martin, R.; Grinspoon, S.K.; et al. Adipose-derived circulating miRNAs regulate gene expression in other tissues. Nature 2017, 542, 450–455. [Google Scholar] [CrossRef] [PubMed]
- Ying, W.; Riopel, M.; Bandyopadhyay, G.; Dong, Y.; Birmingham, A.; Seo, J.B.; Ofrecio, J.M.; Wollam, J.; Hernandez-Carretero, A.; Fu, W.; et al. Adipose Tissue Macrophage-Derived Exosomal miRNAs Can Modulate In Vivo and In Vitro Insulin Sensitivity. Cell 2017, 171, 372–384. [Google Scholar] [CrossRef] [Green Version]
- Santangelo, L.; Giurato, G.; Cicchini, C.; Montaldo, C.; Mancone, C.; Tarallo, R.; Battistelli, C.; Alonzi, T.; Weisz, A.; Tripodi, M. The RNA-Binding Protein SYNCRIP Is a Component of the Hepatocyte Exosomal Machinery Controlling MicroRNA Sorting. Cell Rep. 2016, 17, 799–808. [Google Scholar] [CrossRef] [Green Version]
- Hobor, F.; Dallmann, A.; Ball, N.J.; Cicchini, C.; Battistelli, C.; Ogrodowicz, R.W.; Christodoulou, E.; Martin, S.R.; Castello, A.; Tripodi, M.; et al. A cryptic RNA-binding domain mediates Syncrip recognition and exosomal partitioning of miRNA targets. Nat. Commun. 2018, 9, 831. [Google Scholar] [CrossRef]
- Temoche-Diaz, M.M.; Shurtleff, M.J.; Nottingham, R.M.; Yao, J.; Fadadu, R.P.; Lambowitz, A.M.; Schekman, R. Distinct mechanisms of microRNA sorting into cancer cell-derived extracellular vesicle subtypes. Elife 2019, 8, e47544. [Google Scholar] [CrossRef]
- Saleh, A.A.; Soliman, S.E.; Habib, M.S.E.; Gohar, S.F.; Abo-Zeid, G.S. Potential value of circulatory microRNA122 gene expression as a prognostic and metastatic prediction marker for breast cancer. Mol. Biol. Rep. 2019, 46, 2809–2818. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, X. miR-122-5p promotes aggression and epithelial-mesenchymal transition in triple-negative breast cancer by suppressing charged multivesicular body protein 3 through mitogen-activated protein kinase signaling. J. Cell Physiol. 2020, 235, 2825–2835. [Google Scholar] [CrossRef]
- Chen, C.; Luo, Y.; He, W.; Zhao, Y.; Kong, Y.; Liu, H.; Zhong, G.; Li, Y.; Li, J.; Huang, J.; et al. Exosomal long noncoding RNA LNMAT2 promotes lymphatic metastasis in bladder cancer. J. Clin. Investig. 2020, 130, 404–421. [Google Scholar] [CrossRef] [PubMed]
- Kunadt, M.; Eckermann, K.; Stuendl, A.; Gong, J.; Russo, B.; Strauss, K.; Rai, S.; Kugler, S.; Falomir Lockhart, L.; Schwalbe, M.; et al. Extracellular vesicle sorting of alpha-Synuclein is regulated by sumoylation. Acta Neuropathol. 2015, 129, 695–713. [Google Scholar] [CrossRef] [PubMed]
- Atukorala, I.; Mathivanan, S. The Role of Post-Translational Modifications in Targeting Protein Cargo to Extracellular Vesicles. Subcell. Biochem. 2021, 97, 45–60. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Zheng, H.; Luo, Y.; Kong, Y.; An, M.; Li, Y.; He, W.; Gao, B.; Zhao, Y.; Huang, H.; et al. SUMOylation promotes extracellular vesicle-mediated transmission of lncRNA ELNAT1 and lymph node metastasis in bladder cancer. J. Clin. Investig. 2021, 131, e146431. [Google Scholar] [CrossRef]
- Villarroya-Beltri, C.; Gutierrez-Vazquez, C.; Sanchez-Cabo, F.; Perez-Hernandez, D.; Vazquez, J.; Martin-Cofreces, N.; Martinez-Herrera, D.J.; Pascual-Montano, A.; Mittelbrunn, M.; Sanchez-Madrid, F. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat. Commun. 2013, 4, 2980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, H.; Li, C.; Zhang, Y.; Zhang, D.; Otterbein, L.E.; Jin, Y. Caveolin-1 selectively regulates microRNA sorting into microvesicles after noxious stimuli. J. Exp. Med. 2019, 216, 2202–2220. [Google Scholar] [CrossRef]
- McKenzie, A.J.; Hoshino, D.; Hong, N.H.; Cha, D.J.; Franklin, J.L.; Coffey, R.J.; Patton, J.G.; Weaver, A.M. KRAS-MEK Signaling Controls Ago2 Sorting into Exosomes. Cell Rep. 2016, 15, 978–987. [Google Scholar] [CrossRef] [Green Version]
- Robinson, H.; Ruelcke, J.E.; Lewis, A.; Bond, C.S.; Fox, A.H.; Bharti, V.; Wani, S.; Cloonan, N.; Lai, A.; Margolin, D.; et al. Caveolin-1-driven membrane remodelling regulates hnRNPK-mediated exosomal microRNA sorting in cancer. Clin. Transl. Med. 2021, 11, e381. [Google Scholar] [CrossRef]
- Leidal, A.M.; Huang, H.H.; Marsh, T.; Solvik, T.; Zhang, D.; Ye, J.; Kai, F.; Goldsmith, J.; Liu, J.Y.; Huang, Y.H.; et al. The LC3-conjugation machinery specifies the loading of RNA-binding proteins into extracellular vesicles. Nat. Cell Biol. 2020, 22, 187–199. [Google Scholar] [CrossRef]
- He, B.; Cai, Q.; Qiao, L.; Huang, C.Y.; Wang, S.; Miao, W.; Ha, T.; Wang, Y.; Jin, H. RNA-binding proteins contribute to small RNA loading in plant extracellular vesicles. Nat. Plants 2021, 7, 342–352. [Google Scholar] [CrossRef]
- Katzmann, D.J.; Babst, M.; Emr, S.D. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 2001, 106, 145–155. [Google Scholar] [CrossRef] [Green Version]
- Migliano, S.M.; Teis, D. ESCRT and Membrane Protein Ubiquitination. Prog. Mol. Subcell. Biol. 2018, 57, 107–135. [Google Scholar] [CrossRef]
- Larios, J.; Mercier, V.; Roux, A.; Gruenberg, J. ALIX- and ESCRT-III-dependent sorting of tetraspanins to exosomes. J. Cell Biol. 2020, 219, 1–22. [Google Scholar] [CrossRef] [Green Version]
- Iavello, A.; Frech, V.S.; Gai, C.; Deregibus, M.C.; Quesenberry, P.J.; Camussi, G. Role of Alix in miRNA packaging during extracellular vesicle biogenesis. Int. J. Mol. Med. 2016, 37, 958–966. [Google Scholar] [CrossRef] [Green Version]
- Ye, Z.L.; Huang, Y.; Li, L.F.; Zhu, H.L.; Gao, H.X.; Liu, H.; Lv, S.Q.; Xu, Z.H.; Zheng, L.N.; Liu, T.; et al. Argonaute 2 promotes angiogenesis via the PTEN/VEGF signaling pathway in human hepatocellular carcinoma. Acta Pharmacol. Sin. 2015, 36, 1237–1245. [Google Scholar] [CrossRef]
- Wozniak, A.L.; Adams, A.; King, K.E.; Dunn, W.; Christenson, L.K.; Hung, W.T.; Weinman, S.A. The RNA binding protein FMR1 controls selective exosomal miRNA cargo loading during inflammation. J. Cell Biol. 2020, 219, e201912074. [Google Scholar] [CrossRef]
- Babst, M. MVB vesicle formation: ESCRT-dependent, ESCRT-independent and everything in between. Curr. Opin. Cell Biol. 2011, 23, 452–457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janas, T.; Janas, M.M.; Sapon, K.; Janas, T. Mechanisms of RNA loading into exosomes. FEBS Lett. 2015, 589, 1391–1398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janas, T.; Janas, P.; Sapon, K.; Janas, T. Binding of RNA Aptamers to Membrane Lipid Rafts: Implications for Exosomal miRNAs Transfer from Cancer to Immune Cells. Int. J. Mol. Sci. 2020, 21, 8503. [Google Scholar] [CrossRef] [PubMed]
- Kosaka, N.; Iguchi, H.; Hagiwara, K.; Yoshioka, Y.; Takeshita, F.; Ochiya, T. Neutral sphingomyelinase 2 (nSMase2)-dependent exosomal transfer of angiogenic microRNAs regulate cancer cell metastasis. J. Biol. Chem. 2013, 288, 10849–10859. [Google Scholar] [CrossRef] [Green Version]
- Cha, D.J.; Franklin, J.L.; Dou, Y.; Liu, Q.; Higginbotham, J.N.; Demory Beckler, M.; Weaver, A.M.; Vickers, K.; Prasad, N.; Levy, S.; et al. KRAS-dependent sorting of miRNA to exosomes. Elife 2015, 4, e07197. [Google Scholar] [CrossRef]
- Zhou, W.; Woodson, M.; Sherman, M.B.; Neelakanta, G.; Sultana, H. Exosomes mediate Zika virus transmission through SMPD3 neutral Sphingomyelinase in cortical neurons. Emerg. Microbes Infect. 2019, 8, 307–326. [Google Scholar] [CrossRef] [PubMed]
- Schubert, G. The position of the advice concerning regulations for setting free genetically-engineered microorganisms and viruses in West Germany in light of environmental genetic regulations. Schriftenr. Ver. Wasser Boden Lufthyg. 1988, 78, 295–304. [Google Scholar] [PubMed]
- Hoy, S.M. Patisiran: First Global Approval. Drugs 2018, 78, 1625–1631. [Google Scholar] [CrossRef] [PubMed]
- Shu, D.; Shu, Y.; Haque, F.; Abdelmawla, S.; Guo, P. Thermodynamically stable RNA three-way junction for constructing multifunctional nanoparticles for delivery of therapeutics. Nat. Nanotechnol. 2011, 6, 658–667. [Google Scholar] [CrossRef] [Green Version]
- Pi, F.; Binzel, D.W.; Lee, T.J.; Li, Z.; Sun, M.; Rychahou, P.; Li, H.; Haque, F.; Wang, S.; Croce, C.M.; et al. Nanoparticle orientation to control RNA loading and ligand display on extracellular vesicles for cancer regression. Nat. Nanotechnol. 2018, 13, 82–89. [Google Scholar] [CrossRef]
- Sork, H.; Conceicao, M.; Corso, G.; Nordin, J.; Lee, Y.X.F.; Krjutskov, K.; Orzechowski Westholm, J.; Vader, P.; Pauwels, M.; Vandenbroucke, R.E.; et al. Profiling of Extracellular Small RNAs Highlights a Strong Bias towards Non-Vesicular Secretion. Cells 2021, 10, 1543. [Google Scholar] [CrossRef]
- Kojima, R.; Bojar, D.; Rizzi, G.; Hamri, G.C.; El-Baba, M.D.; Saxena, P.; Auslander, S.; Tan, K.R.; Fussenegger, M. Designer exosomes produced by implanted cells intracerebrally deliver therapeutic cargo for Parkinson‘s disease treatment. Nat. Commun. 2018, 9, 1305. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Yu, J.; Kadungure, T.; Beyene, J.; Zhang, H.; Lu, Q. ARMMs as a versatile platform for intracellular delivery of macromolecules. Nat. Commun. 2018, 9, 960. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).