Abstract
Gossypium hirsutum L. is a worldwide economical crop; however, premature leaf senescence reduces its production and quality which is regulated by stresses, hormones, and genes. DNA binding with the one zinc finger (Dof) transcription factors (TFs) participate widely in plant development and responses to biotic and abiotic stresses, but there have been few reports of these TFs in cotton. Here, we perform a genome-wide study of G. Hirsutum L. Dof (GhDof) genes and analyze their phylogeny, duplication, and expression. In total, 114 GhDof genes have been identified and classified into nine subgroups (A, B1, B2.2, B2.1, C1, C2.1, C2.2, D1, and D2) based on phylogenetic analysis. An MCScanX analysis showed that the GhDof genes expanded due to segmental duplications. Quantitative real-time polymerase chain reaction (qRT-PCR) analysis showed that GhDofD9.6 was not only differentially expressed between CCRI10 (with premature senescence) and Liao4086 (without premature senescence) but also responded to salinity stress; GhDofA5.7, GhDofA7.4, GhDofA8.2, GhDof11.1, GhDofD7.2, and GhDofD11.3 signfificantly responded to cold (4 °C) stress. This work lays the foundation for further analysis of the function of GhDof genes in G. hirsutum, which will be helpful for improving the production and quality of cotton.
1. Introduction
In plants, transcriptional regulation plays an important role in the regulation of many biological processes, such as morphogenesis, signal transduction, and responses to abiotic and biotic stresses. Transcription factors (TFs) bind cis-acting elements of gene promoters to activate or repress gene expression. The DNA binding with the one zinc finger (Dof) family includes plant-specific TFs that contain an N-terminal Dof domain and a C-terminal transcriptional regulation domain [1]. The conserved Dof domain consists of 52 amino acids (aa) with a CX2CX21CX2C motif in the N-terminal domains, of which four Cys residues and Zn2+ form a zinc finger domain that functions as a Cys2/Cys2 domain [2]. The C-terminal domain is less well conserved. The Dof domain and some specific aa of the chain domain in the C-terminal domain also bind to the DNA sequence to regulate gene expression, which confers the diverse functions of Dof proteins [3].
The first Dof proteinwas identified in maize and named ZmDof1, whichspecifically interacts with AAGG motifs and is involved in a wide range of biological processes [4]. Dof genes have been subsequently identified, and their functions have been characterized in many species.
Previous studies have reported that Dof proteins are involved in photoperiodic flowering, such as OsDof4, which affects the flowering of rice under long- and short-day photoperiods [5], and CYCLING DOF FACTOR 2 (CDF2), which acts redundantly to reduce the expression of CONSTANS and affects photoperiodic flowering in Arabidopsis thaliana [6]. Dof proteins participate in secondary metabolism. FaDOF2 plays a subsidiary regulatory role with FaEOBII in the expression of genes encoding enzymes that control eugenol production in strawberries [7]. ZmDof3 functions as a positive regulator of starch accumulation and aleurone development [8].
Nitrogen is one of the most important mineral elements for plant growth. In rice, OsDof18 functions in ammonium uptake and nitrogen distribution by repressing ammonium transporter genes [9]. The overexpression of a pine Dof TF (PpDof5) in poplars was associated with higher growth and enhanced nitrogen uptake compared to those of untransformed controls [10].
Dof proteins participate in plant growth. The Dof-type transcription regulator OBF binding protein 4 (OBP4) represses cell growth by repressing the transcription of ROOT HAIR DEFECTIVE 6-LIKE 2 (RSL2), which contributes to abscisic acid (ABA)-dependent inhibition in A. thaliana [11]. The Dof TF STOMATAL CARPENTER 1 (SCAP1) is involved in stomata patterning by activating a series of guard cell-specific genes [12]. The overexpression lines of AtDof4.7 are associated with lower expression levels of the abscission-related ARABIDOPSIS DEHISCENCE ZONE POLYGALACTURONASE 2 (ADPG2) gene and the deficiency of floral organ abscission [13]. The AtDof5.4/OBP4 protein negatively regulates cell cycle progression and cell expansion [14]. AtDof5.8 is expressed early in provascular cells under the control of an auxin response factor (ARF5), which inhibits the formation of higher-order veins in cotyledons and leaves [15]. MaDof23 is a repressor of ripening-related genes in bananas [16].
Dof proteins participate in the tolerance of stress. CDF3, an A. thaliana Dof gene, functions in abiotic stress responses and the flowering process by controlling the CBF/DREB2A-CRT/DRE and ZAT10/12 modules [17]. The overexpression of GhDof1 in G. hirsutum L. could increase tolerance to salt and cold stresses [18]. Previously, it was shown that SiDof22 transgenic tomato lines have a higher tolerance to salt stress, which was achieved by accumulating ascorbate [19].
Overall, Dof TFs participate widely in plant photoperiodic regulation, responses to and tolerance of environmental stress, secondary metabolism, and plant growth regulation, making them some of the most important TFs in plants. G. hirsutum L. is one of the most important economic crops in the world, and its genome has been sequenced [20,21]. In recent years, premature senescence has been occurring with increasing frequency around the world, which has reduced the cotton lint yield and is associated with poor fiber properties [22]. Short-season cotton cultivars and transgenic Bacillus thuringiensis (Bt) cotton cultivars usually accompany premature senescence, and adverse environments induce premature senescence [23]. Dof, as an important TF, has rarely been studied in G. hirsutum. In this work, we identified 114 GhDof proteins and analyzed their gene numbers, physical locations, phylogeny, and duplication. Furthermore, expression patterns of GhDof TFs in different tissues and abiotic stresses were studied. This research provides an important foundation for future studies on GhDof gene cloning and functional characterization of Dofs in G. hirsutum.
2. Materials and Methods
2.1. Identification and Annotation of Dof Gene Family Members in G. hirsutum
A. thaliana Dof gene family members were downloaded from The A. Information Resource (TAIR). The genome sequences of G. hirsutum L. were downloaded from the CottonGene database [20]. We used AtDof protein sequences as query sequences to search against the G. hirsutum L. protein sequences using a local BLAST search, considering those with an E-value of less than e-10 as GhDof protein sequences [24]. We used a profile Hidden Markov Model (HMM) implemented with default parameters in HMMER v3.2.1 for Windows to search for GhDof proteins, such as G. hirsutum L. proteins with the Dof domain (PF02701) of zf-Dof.hmm in the Pfam database. Each of the retrieved sequences was manually checked based on the features of the Dof domain structure [25,26]. To analyze the number of gene introns, we used coding sequences (CDS) of all the candidate GhDof genes to BLAST against the G. hirsutum L. genome sequences. We conducted blastp of the GhDof proteins against nonredundant protein sequences (nr) on the National Center for Biotechnology Information (NCBI) to find homologs (listed in Supplementary Material 1).
All of the candidate GhDof genes were named based on their physical location, and their distribution on the 26 G. hirsutum L. chromosomes was visualized by using MapChart 2.2.
The open reading frame (ORF), molecular weight (MW), isoelectric point (pI), instability index, aliphatic index, and grand average of hydropathicity (GRAVY) were predicted using the ProtParam tool through default parameters. Protein subcellular localization was predicted by the online software WoLF PSORT.
2.2. GhDof Protein Alignment and Phylogenetic Analysis
The complete GhDof and AtDof protein sequences were aligned by ClustalX (University College Dublin, Dublin, Ireland) with default settings. Subsequently, MEGA 5.2 software (Arizona State Universit, Arizona, AZ, USA) was employed to construct an unrooted phylogenetic tree based on alignments using the neighbor-joining (NJ) method [27].
2.3. Duplication and Synteny Analysis of GhDof Genes
We used MCScanX software (University of Georgia, Athens, GA, USA) with the default parameters to analyze the synteny relationship and duplication events of GhDof genes in the G. hirsutum L. genome [28] and the results were visualized with Circos software (Canada’s Michael Smith Genome Sciences Center, Vancouver, BC, Canada) [29].
2.4. Plant Materials, Treatments, and Collections
To assess the tissue-specific expression of the GhDof genes, we sampled the roots, stems, leaves, and shoot apexes when the plants had two fully expanded true leaves and the buds were at a length of 6 mm [30]. To measure the gene expression patterns during the leaf senescence process, the short-season cultivars CCRI10 (premature senescence) and Liao4086 (without premature senescence) [31] were planted in a field at the same depth to ensure that the seeds germinated at the same time. Once the cotyledons flattened, samples were collected every two weeks until the cotyledons senesced, for a total of four collections [24]. The tissues and cotyledon samples were preserved in a −80 °C freezer.
For the abiotic stress treatment, Liao4086 seeds were germinated on 1/2 Murashige and Skoog (MS) medium, and the seedlings were then transferred onto each stress medium. For the ABA stress, seedlings were kept on 1/2 MS medium with 50 μmol L−1 ABA and a pH of 5.8 at 25 °C for ten days. For the pH stress, seedlings were kept on 1/2 MS medium with a pH of 11.0 at 25 °C for ten days. For the salinity stress, seedlings were kept on 1/2 MS medium with 200 mmol/L salt and a pH of 5.8 at 25 °C for ten days. For the cold stress, seedlings were kept on 1/2 MS medium with a pH of 5.8 at 4 °C for ten days. Seedlings kept on 1/2 MS medium with a pH of 5.8 at 25 °C for ten days were treated as the control (CK) [32]. The CK and stressed experiments were done with three technical repeats. Cotyledons of the CK and stressed plants were used for RNA extraction and subsequent qRT-PCR experiments.
2.5. RNA Isolation and qRT-PCR Experiments
Total RNA was isolated by an RNA-prep Pure Plant Kit (Tiangen, Beijing, China), which is specifically for polysaccharide- and polyphenol-rich plants, to ensure that all of the OD260/280 values ranged from 1.8 to 2.1 and the OD260/230 value was more than 1.8. cDNA synthesis was performed with a PrimeScriptTM RT reagent Kit with a gDNA Eraser (Perfect Real Time, Code No. RR047A) (TaKaRa, Shiga, Japan) which can digest the residual DNA in the RNA extractions to enhance the accuracy of qRT-PCR. qRT-PCR with the SYBR Premix Ex TaqTM Kit (Code No. RR420A, TaKaRa, Shiga, Japan) was used to measure the gene expression patterns. The threshold cycle (Ct) values of all of the genes were normalized to the internal reference gene—a house-keeping gene (GhHis) [33]. The relative gene expression fold-changes were calculated by the 2−ΔΔCt method [34]. The primers were designed by the Oligo primer analysis software version 7 (MBI, California, CA, USA) and are listed in Supplementary Material 2. The qRT-PCR experiments were carried out with three independent total RNA samples, and each sample had three technical repeats.
All the data were analyzed by one-way ANOVA in SPSS 17.0; p < 0.05 was considered to represent a statistically significant difference, and p < 0.01 was considered to represent an extremely significant difference.
3. Results
3.1. Identification of GhDof Transcription Factor Gene Family Members in G. hirsutum
In the G. hirsutum L. genome, 114 genes have been identified as possible members of the GhDof gene family. Of these, 102 Dof genes could be mapped onto the 26 G. hirsutum L. chromosomes and were named based on their order on the chromosomes as GhDofA1.1 to GhDofD13.1 (Supplementary Material 1, Figure 1). Twelve GhDof genes could be mapped on scaffolds and were named GhDofs1 to GhDofs12.
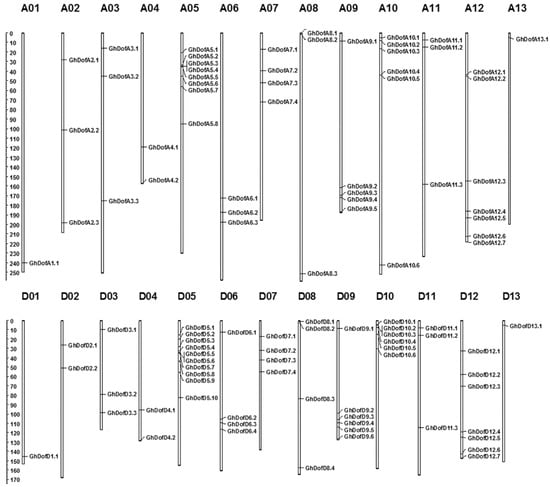
Figure 1.
Chromosomal locations of GhDof genes on 26 chromosomes. A1–A13 and D1–D13 indicate the A-subgenome and D-subgenome, respectively.
Furthermore, we analyzed the physicochemical properties of GhDof proteins, as shown in Supplementary material 1, where the ORFs of the 114 annotated GhDof genes ranged from 167 to 510 aa, and the molecular weights (MWs) of the proteins encoded by these genes varied from 18,453.03 to 55,388.97 Da. Based on the pI analysis, there were 79 GhDof TFs with a pI greater than 7.0 that were basic in nature, and 35 GhDof TFs with a pI lower than 7.0 that were acidic in nature. The instability index varied from 34.4 to 69.19. The predicted subcellular localizations showed that 107 GhDof proteins were located in the nucleus, five GhDof proteins (GhDofs2, GhDofA6.3, GhDofA10.2, GhDofD10.4, and GhDofA10.1) were located in the chloroplast, and two GhDof proteins (GhDofA12.1 and GhDofD12.1) were located in the mitochondria.
3.2. Phylogenetic Analysis of GhDof TFs
Based on the complete alignment of the sequences of Dof TFs from G. hirsutum L. and A. thaliana, a total of 117 GhDof TFs were classified into nine subgroups, including the classes A, B1, B2.2, B2.1, C1, C2.1, C2.2, D1, and D2 [35,36] The number of GhDof TFs in each subgroup is shown in Supplementary material 1. The phylogenetic tree (Figure 2) shows the phylogenetic relationships of Dof TFs between G. hirsutum L. and A. thaliana.
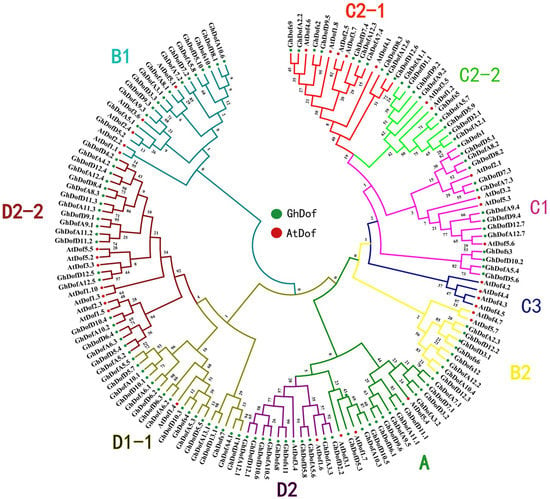
Figure 2.
Phylogenetic tree of Dof proteins in G. hirsutum L. and A. thaliana. The full-length amino acid sequences of 114 GhDofs and 36 AtDofs were aligned by ClustalX, and the phylogenetic tree was constructed using MEGA 5.2 by the neighbor-joining method with 1000 bootstrap replicates. Each Dof subgroup is indicated by a specific color.
3.3. Gene Duplication and Synteny Analysis of GhDofs
As a result of the whole genome analysis of G. hirsutum, 5492 (7.79%) genes were identified as tandem duplications, and 51,329 (72.83%) were identified as segmental duplications (Supplementary Materials 3 and 4). Among the Dof gene family members in G. hirsutum, none were tandem repeats, while 103 (90.35%) GhDof genes (Figure 3) were segmental repeats, which indicates that the GhDof genes in G. hirsutum L. expand only due to the segmental duplication of chromosomes.
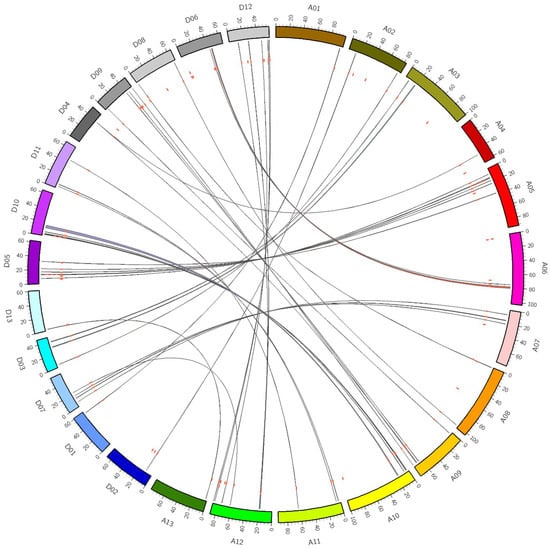
Figure 3.
Synteny analysis of GhDof genes in G. Hirsutum L. The chromosomes of G. hirsutum L. are described in different colors in circle form. The red dots represent the distribution of each GhDof gene and the colored curves indicate the syntenic regions around the GhDof genes in G. hirsutum L. genomes.
3.4. Gene Expression Analysis
To evaluate the potential functions of GhDof genes, we searched 114 GhDof genes in the transcriptomes of collections made during cotton leaf senescence and the transcriptome profiles of four tissues including the root, stem, leaf, and embryo in cotton [23,37]. According to the expression abundance of GhDofs in the transcriptomes, we selected 14 GhDof genes and used qRT-PCR to check theirrelative expression patterns in tissues (root, stem, bud, shoot apex, and leaf) and under abiotic stresses (ABA, cold, pH, and salinity).
3.4.1. Tissue-Specific Expression
As shown in Figure 4, GhDofA5.7 and GhDofA2.2 were significantly highly expressed in the stem and bud. GhDofA2.2 and GhDofA7.4 had significantly lower expression levels in the shoot apex and leaf than in the root. GhDofA8.2 and GhDofA11.1 had significantly higher expression values in the stem than in the other tissues. GhDofD5.5 and GhDofD9.6 had significantly higher expression values in the leaf than in the root.
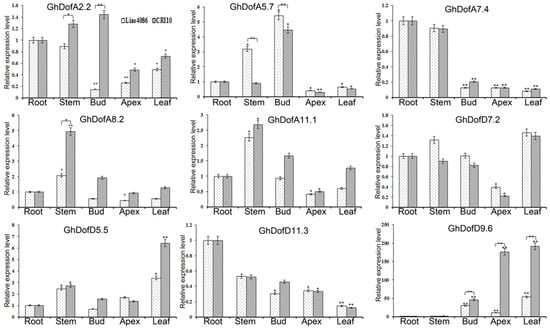
Figure 4.
GhDof relative expression levels in the root, stem, bud, shoot apex, and leaf. The X-axis indicates tissues, and the Y-axis indicates relative expression values. The Y-axis indicates the relative expression level. The threshold cycle (Ct) values of all of the genes were normalized to the internal reference gene—a house-keeping gene (GhHis); the relative expression level was calculated by the 2−ΔΔCt method. The bars indicate the standard deviation (SD). The brackets indicate the comparison between CCRI10 and Liao4086. * indicates a significant difference at the p < 0.05 level. ** indicates an extremely significant difference at the p < 0.01 level.
As GhDofD5.5 and GhDofD9.6 had high expression values in the leaves, we measured their expression patterns during leaf senescence (Figure 5). We compared their expression trends between CCRI10 (premature senescence) and Liao4086 (without premature senescence) during the leaf senescence process. The leaf senescence process is a continuous process: stage 1 to stage 2 is the young to mature stage, stages 2–3 represent the leaves beginning to yellow, and stage 4 is when the leaf is almost at its final stage. Accompanied by leaf development from the young to senescence stages, GhDofD5.5 was upregulated, and then downregulated at the last stage; between CCRI10 and Liao4086, no significant difference existed. GhDofD9.6 was downregulated during cotyledon development, with significant higher relative expression levels in Liao4086 than CCRI10 at the young and mature stages. Based on their expression patterns, we inferred that these two genes may play a function in leaf senescence.
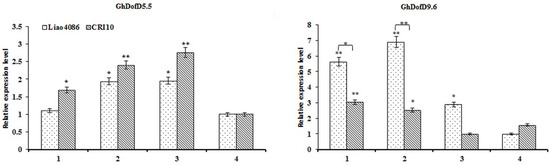
Figure 5.
Gene expression patterns during cotyledon development in G. Hirsutum L. The X-axis indicates the cotyledon development stages. The numbers 1, 2, 3, and 4 indicate the samples of flattened cotyledons taken in the first week, the third week, the fifth week, and the seventh week, respectively. The Y-axis indicates relative expression level. The threshold cycle (Ct) values of all of the genes were normalized to the internal reference gene—a house-keeping gene (GhHis); the relative expression level was calculated by the 2−ΔΔCt method. The bars indicate SDs. The brackets indicate the comparison between CCRI10 and Liao4086. * indicates significant differences at the p < 0.05 level. ** indicates extreme significant differences at the p < 0.01 level.
3.4.2. Analysis of Gene Expression under Abiotic Stresses
The expression patterns of the 14 GhDof genes in response to ABA, cold temperature (4 °C), high alkalinity (pH = 11.0), and salinity stresses were analyzed. There were six GhDof genes (GhDofA5.7, GhDofA7.4, GhDofA8.2, GhDofA11.1, GhDofD7.2, and GhDofD11.3) that responded only to cold stress, and under such stress, they had extremely significantly high expression (Figure 6).
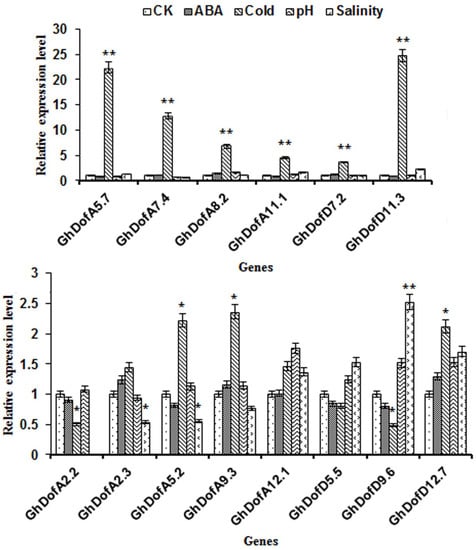
Figure 6.
GhDof gene expression under ABA, cold temperature, pH, and salinity stresses. The X-axis indicates the GhDof genes.The Y-axis indicates relative expression level. The threshold cycle (Ct) values of all of the genes were normalized to the internal reference gene—a house-keeping gene (GhHis), the relative expression level was calculated by the 2−ΔΔCt method. Abscisic acid (ABA), cold, pH, salinity, and control (CK) seedlings were kept on 1/2 MS medium with 50 μmol L−1 ABA, a temperature of 4 °C, a pH of 11.0, 200 mmol/L NaCl, or no stress for ten days. The bars indicate SDs. * indicates significant differences at the p < 0.05 level. ** indicates extremely significant differences at the p < 0.01 level.
Compared with that in the CK treatment, GhDofA2.2 expression under cold stress was significantly lower, GhDofA2.3 expression under salinity stress was significantly lower, GhDofA5.2 expression under cold stress was significantly higher but under salinity stress was significantly lower, GhDofA9.3 and GhDofD12.7 expression under cold stress was significantly higher, and GhDofD9.6 expression under cold stress was significantly lower and under salinity stress was much higher (Figure 6).
4. Discussion
Premature cotton senescence has occurred with increasing frequency in recent years, which has reduced the lint yield and quality [23]. To elucidate the mechanism of premature senescence and improve the cotton quality, we studied the GhDof gene family in cotton. Dof TFs are one of the most important regulators of gene expression. In G. hirsutum, 114 GhDof genes and their physicochemical properties were analyzed. Based on the instability index analysis, most of the proteins had an instability index value of more than 40.0, except for 12 GhDof proteins, including GhDofD10.4, GhDofA5.2, GhDofs5, GhDofD13.1, GhDofA5.7, GhDofD5.9, GhDofA6.3, GhDofD5.4, GhDofA12.1, GhDofD12.1, GhDofD10.6, and GhDofA10.5, each of which had an instability index value of less than 40.0. A protein whose instability index was smaller than 40 with an in vivo half-life more than 16 h was predicated as stable, a protein whose instability index was bigger than 40 with an in vivo half-life was predicated as unstable less than 5 h [24]. Subcellular localization analysis indicated that most of the GhDof proteins can be transported from the cytoplasm to nuclear via nuclear transportation signal domains. Some of them may play functions through modifications such as phosphorylation, acetylation, methylation, or other specific reactions that cause them to move from the cytoplasm to the nucleus to carry out their functions by binding to the promoter regions of their targets for transcriptional activation or regulation [25].
4.1. GhDof Genes Are Amplified by Segmental Duplication
The 114 GhDof proteins were classified into A, B1, B2.2, B2.1, C1, C2.1, C2.2, D1, and D2subgroups based on their phylogenetic relationships with the Dof proteins in A. thaliana. As shown in Supplementary material 1, which illustrates the intron/exon gene structure, the positions and phases of most Dof gene introns are conserved between sorghum and cotton [38]. There are 78 GmDof genes in soybean (genome size 1100 Mb) [39], 34 SlDof genes in tomato (genome size 900 Mb) [40], 76 BraDof genes in Chinese cabbage (genome size 485 Mb) [41], 35 StDof genes in potato (genome size 844 Mb) [42], 42 MtDof genes in Medicago truncatula (genome size 500 Mb) [43], and 46 DcDof genes in carrot (genome size 421.5 Mb) [44], while the number of known GhDof genes in G. hirsutum L. has increased to 114. G. hirsutum L. (AD1, 2n = 4x = 52, genome size 2.5 Gb) is an allotetraploid plant species with a D-subgenome and A-subgenome. The genome size is bigger than the others and shows many repeat sequences in the G. hirsutum L. genome, accounting for 67.2% of the AD genome [21]. The repeat sequences consist of tandem repeats and segmental duplications. Based on our MCScanX results, in G. hirsutum, tandem duplications account for 7.79% of the repeat sequences, and segmental duplications accountfor 72.83%. Thus, segmental duplication was the main cause of gene duplication in G. hirsutum. While no tandem duplication exists in GhDof genes, segmental duplication accounts for 90.35% of the repeats in the GhDof genes, which indicates that the GhDof gene members increase by segmental duplication.
4.2. GhDof Genes Might Play Functions in G. hirsutum L. Leaf Senescence
As Dof genes widely participate in the regulation of photoperiodic flowering [5], nitrogen metabolism [9], and plant growth [11], we searched the GhDof homologs, and their functions are listed in Supplementary material 1. Cotton premature senescence has occurred with increasing frequency in recent years and is regulated by stresses, hormones, and genes [25]. In cotton breeding, short season cotton cultivars are usually accompanied by premature senescence, so comparing gene expression patterns between cultivars with premature senescence and without premature senescence symptoms is a way to determine which candidate genes are involved in premature senescence; therefore, we compared the gene expression differences between CCRI10 (premature senescence) and Liao4086 (without premature senescence). According to expression analysis in the tissues, we conclude that GhDofD9.6 may function in the leaf development process.
Furthermore, in cotton production, abiotic stresses, such as alkalinity (pH = 11.0), cold, drought, and high salinity are environmental limiting factors for crop growth and yield [35]; therefore, we checked GhDof gene expression patterns under ABA, cold, pH, and salinity stresses. GhDofA5.7, GhDofA7.4, GhDofA8.2, GhDof11.1, GhDofD7.2, and GhDofD11.3 may play important functions in responding to cold, and Zheng et al. (2017) reported that cold responsive proteins were involved in leaf senescence in cotton [22]. According to tissue-specific and leaf development analysis, GhDofD9.6 not only was expressed with differences between CCRI10 and Liao4086 but also responded to salinity stress which indicates that GhDofD9.6 may participate in the crosstalk of leaf senescence and salinity stress.
5. Conclusions
In this study, we identified a total of 114 GhDof proteins that were classified into A, B1, B2.2, B2.1, C1, C2.1, C2.2, D1, and D2 subgroups based on their phylogenetic relationships with AtDof proteins. The numbers of GhDof genes is high and increased by segmental duplication during the G. hirsutum L. evolution process. Gene expression analysis showed that GhDofD9.6 may function in the leaf senescent process, with GhDofA5.7, GhDofA7.4, GhDofA8.2, GhDof11.1, GhDofD7.2, and GhDofD11.3 significantly responding to cold stress, and GhDofD9.6 responding to salinity stress.
Supplementary Materials
The following are available online at http://www.mdpi.com/2073-4395/8/9/186/s1, Supplementary Material 1: The GhDof transcription factor family in G. hirsutum. Notes: MW, molecular weight; GRAVY, grand average of hydropathicity; nucl, nucleus; chlo, chloroplast. mito, mitochondria. Supplementary Material 2: Primer sequences for qRT-PCR. Supplementary Material 3: List of segmentally duplicated gene pairs in the G. hirsutum L. genome, along with their E-value identified from MCScanX software. Supplementary Material 4: List of tandemly and segmentally duplicated genes in the G. hirsutum L. genome identified from MCScanX software.
Author Contributions
Formal analysis, R.X.; Investigation, Q.Z.; Resources, W.L.; Software, P.W. and X.P.; Visualization, Y.L. and Z.R.; Writing—original draft, L.D.; Writing—review & editing, H.L.
Funding
This research was sponsored by State Key Laboratory of Cotton Biology Open Fund, grant number CB2018A07.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Yanagisawa, S. The Dof Family of Plant Transcription Factors. Trends Plant Sci. 2002, 7, 555–560. [Google Scholar] [CrossRef]
- Umemura, Y.; Ishiduka, T.; Yamamoto, R.; Esaka, M. The Dof domain, a zinc finger DNA-binding domain conserved only in higher plants, truly functions as a Cys2/Cys2 Zn finger domain. Plant J. 2004, 37, 741–749. [Google Scholar] [CrossRef] [PubMed]
- Yanagisawa, S.; Schmidt, R.J. Diversity and similarity among recognition sequences of Dof transcription factors. Plant J. 1999, 17, 209–214. [Google Scholar] [CrossRef] [PubMed]
- Yanagisawa, S. A Novel DNA-binding domain that may form a single zinc finger motif. Nucleic Acids Res. 1995, 23, 3403–3410. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Liu, X.; Yin, D.; Yuan, H.; Xie, Q.; Zhao, X.; Li, X.; Zhu, L.; Li, S.; Li, D. Constitutive expression of OsDof4, encoding a C2-C2 zinc finger transcription factor, confesses its distinct flowering effects under long- and short-day photoperiods in rice (Oryza Sativa L.). BMC Plant Biol. 2017, 17, 166. [Google Scholar] [CrossRef] [PubMed]
- Fornara, F.; Panigrahi, K.C.; Gissot, L.; Sauerbrunn, N.; Ruhl, M.; Jarillo, J.A.; Coupland, G. Arabidopsis Dof transcription factors act redundantly to reduce constans expression and are essential for a photoperiodic flowering response. Dev. Cell 2009, 17, 75–86. [Google Scholar] [CrossRef] [PubMed]
- Molina-Hidalgo, F.J.; Medina-Puche, L.; Canete-Gomez, C.; Franco-Zorrilla, J.M.; Lopez-Vidriero, I.; Solano, R.; Caballero, J.L.; Rodriguez-Franco, A.; Blanco-Portales, R.; Munoz-Blanco, J.; et al. The fruit-specific transcription factor FadoF2 regulates the production of eugenol in ripe fruit receptacles. J. Exp. Bot. 2017, 68, 4529–4543. [Google Scholar] [CrossRef] [PubMed]
- Qi, X.; Li, S.; Zhu, Y.; Zhao, Q.; Zhu, D.; Yu, J. ZmDof3, a maize endosperm-specific Dof protein gene, regulates starch accumulation and aleurone development in maize endosperm. Plant Mol. Biol. 2017, 93, 7–20. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Yang, W.; Wei, J.; Yoon, H.; An, G. Transcription factor OsDOF18 controls ammonium uptake by inducing ammonium transporters in rice roots. Mol. Cells 2017, 40, 178–185. [Google Scholar] [PubMed]
- Rueda-Lopez, M.; Pascual, M.B.; Pallero, M.; Henao, L.M.; Lasa, B.; Jauregui, I.; Aparicio-Tejo, P.M.; Canovas, F.M.; Avila, C. Overexpression of a pine Dof transcription factor in hybrid poplars: A comparative study in trees growing under controlled and natural conditions. PLoS ONE 2017, 12, e0174748. [Google Scholar] [CrossRef] [PubMed]
- Rymen, B.; Kawamura, A.; Schafer, S.; Breuer, C.; Iwase, A.; Shibata, M.; Ikeda, M.; Mitsuda, N.; Koncz, C.; Ohme-Takagi, M.; et al. ABA suppresses root hair growth via OBP4 transcriptional-regulator of the RSL2 promoter. Plant Physiol. 2017, 173, 1750–1762. [Google Scholar] [CrossRef] [PubMed]
- Castorina, G.; Fox, S.; Tonelli, C.; Galbiati, M.; Conti, L. A novel role for stomatal carpenter 1 in stomata patterning. BMC Plant Biol. 2016, 16, 172. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.-Q.; Wei, P.-C.; Tan, F.; Yu, M.; Zhang, X.-Y.; Chen, Q.-J.; Wang, X.-C. The transcription Factor AtDOF4.7 is involved in ethylene- and IDA-mediated organ abscission in Arabidopsis. Front. Plant Sci. 2016, 7, 863. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Chen, H.; Ying, L.; Cai, W. AtDOF5.4/OBP4, a DOF transcription factor gene that negatively regulates cell cycle progression and cell expansion in Arabidopsis thaliana. Sci. Rep. 2016, 6, 27705. [Google Scholar] [CrossRef] [PubMed]
- Konishi, M.; Yanagisawa, S. Transcriptional repression caused by Dof5.8 is involved in proper vein network formation in Arabidopsis thaliana Leaves. J. Plant Res. 2015, 128, 643–652. [Google Scholar] [CrossRef] [PubMed]
- Feng, B.H.; Han, Y.C.; Xiao, Y.Y.; Kuang, J.F.; Fan, Z.Q.; Chen, J.Y.; Lu, W.J. The banana fruit Dof transcription factor Madof23 acts as a repressor and interacts with Maerf9 in regulating ripening-related genes. J. Exp. Bot. 2016, 67, 2263–2275. [Google Scholar] [CrossRef] [PubMed]
- Corrales, A.R.; Carrillo, L.; Lasierra, P.; Nebauer, S.G.; Dominguez-Figueroa, J.; Renau-Morata, B.; Pollmann, S.; Granell, A.; Molina, R.V.; Vicente-Carbajosa, J.; et al. Multifaceted role of cycling Dof factor 3 (CDF3) in the regulation of flowering time and abiotic stress responses in Arabidopsis. Plant Cell Environ. 2017, 40, 748–764. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Liang, W.; Liu, Z.; Wang, Y.; Zhao, Y.; Ijaz, B.; Hua, J. Overexpression of GhDOF1 improved salt and cold tolerance and seed oil content in Gossypium Hirsutum. J. Plant Physiol. 2017, 218, 222–234. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Zhang, C.; Shu, W.; Ye, Z.; Li, H.; Zhang, Y. The transcription factor Sldof22 involved in ascorbate accumulation and salinity stress in tomato. Biochem. Biophys. Res. Commun. 2016, 474, 736–741. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Hu, Y.; Jiang, W.; Fang, L.; Guan, X.; Chen, J.; Zhang, J.; Saski, C.A.; Scheffler, B.E.; Stelly, D.M.; et al. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 2015, 33, 531–537. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Fan, G.; Lu, C.; Xiao, G.; Zou, C.; Kohel, R.J.; Ma, Z.; Shang, H.; Ma, X.; Wu, J.; et al. Genome sequence of cultivated upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat. Biotechnol. 2015, 33, 524–530. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Fan, S.; Wei, H.; Tao, C.; Ma, Q.; Zhang, S.; Li, H.; Pang, C.; Yu, S. Itraq-based quantitative proteomic analysis reveals cold responsive proteins involved in leaf senescence in upland cotton (Gossypium hirsutum L.). Int. J. Mol. Sci. 2017, 18, 1984. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Wei, H.; Song, M.; Pang, C.; Liu, J.; Wang, L.; Zhang, J.; Fan, S.; Yu, S. Transcriptome profiling analysis reveals that flavonoid and ascorbate-glutathione cycle are important during another development in upland cotton. PLoS ONE 2012, 7, e49244. [Google Scholar] [CrossRef] [PubMed]
- Dou, L.; Zhang, X.; Pang, C.; Song, M.; Wei, H.; Fan, S.; Yu, S. Genome-wide analysis of the Wrky gene family in cotton. Mol. Genet. Genom. 2014, 289, 1103–1121. [Google Scholar] [CrossRef] [PubMed]
- Hir, R.L.; Bellini, C. The plant-specific Dof transcription factors family: New players involved in vascular system development and functioning in Arabidopsis. Front. Plant Sci. 2013, 4, 164. [Google Scholar] [PubMed]
- Shigyo, M.; Tabei, N.; Yoneyama, T.; Yanagisawa, S. Evolutionary processes during the formation of the plant-specific Dof transcription factor family. Plant Cell Physiol. 2007, 48, 179–185. [Google Scholar] [CrossRef] [PubMed]
- Wen, C.L.; Cheng, Q.; Zhao, L.; Mao, A.; Yang, J.; Yu, S.; Weng, Y.; Xu, Y. Identification and characterisation of Dof transcription factors in the cucumber genome. Sci. Rep. 2016, 6, 23072. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, C.; Pang, C.; Wei, H.; Wang, H.; Song, M.; Fan, S.; Yu, S. Characterization and functional analysis of PEBP family genes in upland cotton (Gossypium hirsutum L.). PLoS ONE 2016, 11, e0161080. [Google Scholar] [CrossRef] [PubMed]
- Song, M.; Fan, S.; Pang, C.; Wei, H.; Yu, S. Genetic analysis of the antioxidant enzymes, methane dicarboxylic aldehyde (MDA) and chlorophyll content in leaves of the short season cotton (Gossypium hirsutum L.). Euphytica 2014, 198, 153–162. [Google Scholar] [CrossRef]
- Zhu, Y.N.; Shi, D.Q.; Ruan, M.B.; Zhang, L.L.; Meng, Z.H.; Liu, J.; Yang, W.C. Transcriptome analysis reveals crosstalk of responsive genes to multiple abiotic stresses in cotton (Gossypium hirsutum L.). PLoS ONE 2013, 8, e80218. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Han, X.; Lv, J.; Zhao, L.; Xu, X.; Zhang, T.; Guo, W. Structure, expression differentiation and evolution of duplicated fiber developmental genes in Gossypium barbadense and G. Hirsutum. BMC Plant Biol. 2011, 11, 40. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.; Schmittgen, T. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta c(t)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Cheng, J.; Cui, J.; Xu, X.; Liang, G.; Luo, X.; Chen, X.; Tang, X.; Hu, K.; Qin, C. Genome-wide identification and expression profile of Dof transcription factor gene family in pepper (Capsicum annuum L.). Front. Plant Sci. 2016, 7, 574. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Risueno, M.A.; Martinez, M.; Vicente-Carbajosa, J.; Carbonero, P. The family of DOF transcription factors: From green unicellular algae to vascular plants. Mol. Genet. Genom. 2007, 277, 379–390. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.; Pang, C.; Fan, S.; Song, M.; Wei, H.; Yu, S. Global analysis of the Gossypium Hirsutum L. Transcriptome during leaf senescence by RNA-Seq. BMC Plant Biol. 2015, 15, 43. [Google Scholar] [CrossRef] [PubMed]
- Kushwaha, H.; Gupta, S.; Singh, V.; Rastogi, S.; Yadav, D. Genome wide identification of dof transcription factor gene family in sorghum and its comparative phylogenetic analysis with rice and Arabidopsis. Mol. Biol. Rep. 2011, 38, 5037–5053. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Qiu, L.-J. Retraction: Genome-wide analysis of the Dof transcription factor gene family reveals soybean-specific duplicable and functional characteristics. PLoS ONE 2016, 11, e0167019. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Zhang, Y.; Zhang, C.; Zhang, T.; Hu, T.; Ye, J.; Zhang, J.; Wang, T.; Li, H.; Ye, Z. Genome-wide analysis of plant-specific Dof transcription factor family in tomato. J. Plant Biol. 2013, 55, 552–566. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Li, M.Y.; Wang, F.; Tang, J.; Xiong, A.S. Genome-wide analysis of Dof family transcription factors and their responses to abiotic stresses in Chinese cabbage. BMC Genom. 2015, 16, 33. [Google Scholar] [CrossRef] [PubMed]
- Venkatesh, J.; Park, S.; Se, W. Genome-wide analysis and expression profiling of DNA-binding with one zinc finger (Dof) transcription factor family in potato. Plant Physiol. Biochem. 2015, 94, 73–86. [Google Scholar] [CrossRef] [PubMed]
- Shu, Y.; Song, L.L.; Zhang, J.; Liu, Y.; Guo, C.H. Genome-wide identification and characterization of the Dof gene family in Medicago truncatula. Genet. Mol. Res. 2015, 14, 10645–10657. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Huang, Y.; Li, M.Y.; Wang, F.; Xu, Z.S.; Xiong, A.S. Dof transcription factors in carrot: Genome-wide analysis and their response to abiotic stress. Biotechnol. Lett. 2016, 38, 145–155. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
