Abstract
The yield of cowpea varieties is affected by environmental variability. Hence, candidate varieties must be tested for yield stability before release. This study assessed the impacts of genotypes, environments, and their interaction on the performance of elite cowpea lines for key adaptive, grain yield, and associated traits across different locations. A total of 42 elite genotypes were evaluated in five Nigerian environments, representing various savanna ecologies, during the 2021 growing season. The experimental design employed was an alpha lattice arrangement, with each genotype replicated three times. The results revealed significant differences among genotypes, environments, and genotype-by-environment interaction (G × E) for most traits, including days to maturity, 100-seed weight, and grain yield. The genotype and genotype-by-environment interaction (GGE) biplot showed G21 (IT14K-2111-2) and G25 (IT15K-2386-1) as the most stable genotypes across the five environments, G41 (IT11K-61-82) was best adapted to Ibadan and Shika, G5 (245-1) was best adapted to Bagauda and Gumel, and G30 (IT16K-2365-1) was best adapted to Bauchi. G21 (IT14K-2111-2) and G25 (IT15K-2386-1) could be recommended across the five test environments, whereas G41 (IT11K-61-82), G30 (IT16K-2365-1), and G5 (245-1) were specific to the adapted environments.
1. Introduction
Cowpea, Vigna unguiculata (L. Walp), is an important grain legume grown in the tropics, where it constitutes a valuable source of protein in the diets of millions of people [1]. The crop has become an essential nutritional component in the human diet due to its high protein content, carbohydrate composition that complements cereal grains, and relatively low fat content [2]. Improved cowpea varieties contain between 20 and 25 percent protein on a dry weight basis [3,4]. Smallholder farmers are the main cowpea producers in sub-Saharan Africa (SSA), where the crop is grown for various purposes, including tender leaves, green pods, grains, and fodder, which serve as both human food and livestock feed. Additionally, cowpea residues are utilized to replenish the soil, contributing to enhanced soil fertility in the region [5].
Cowpea also plays an important role in human nutrition, food security, and income generation for farmers and food vendors in SSA. Fresh leaves are used as pot herbs, especially in East Africa. In an earlier study, Duke (1990) [6] found that cowpea fodder could contain up to 18.6 g of protein per 100 g dry weight and that it plays a crucial role as a valuable and nourishing feed resource within crop–livestock systems. Additionally, it serves as a significant income source for various stakeholders in the value chain. Reports indicate that the price of cowpea fodder can range from 50% to 80% of the grain price [7], and in Nigeria, farmers who harvest and store cowpea fodder for sale at the peak of the dry season increased their annual income by 25% [8]. Depending on the region, seed coat color and texture could be very important to consumers. For example, in northern parts of Nigeria, where cowpea is generally produced because of favorable climatic conditions, white-colored grains are preferred by consumers, whereas in the southern parts of the country, brown-seeded types are preferred [1].
In Africa, particularly in West and Central Africa, Nigeria produces the highest quantity of cowpea grains annually at approximately 3.6 million metric tons; other major producers are Niger Republic and Burkina Faso, with an average of 2.6 and 0.660 million metric tons, respectively [9]. The crop is known to be relatively drought tolerant compared to other legumes and adapted to marginal soil due to its nitrogen-fixing ability, which makes it a useful staple crop for farmers in harsh environments under moisture stress and high temperatures [10,11]. It is widely cultivated and consumed, especially in the arid and semi-arid tropics and sub-Saharan Africa [12]. In recent times, the amount of rainfall received in the main producing areas has been declining and the distribution of the rains is irregular, especially during the early and late stages of the cropping seasons [1,13].
Although cowpea is a versatile grain legume, its productivity is hindered by various factors, both biotic and abiotic. Biotic factors such as weeds, insects, and diseases, as well as abiotic factors like soil type, altitude, and rainfall patterns, contribute to the low and unstable yields of cowpea across different environments and years [14,15,16]. In addition, the low yield by smallholder farmers has been partly attributed to the use of local varieties and poor agronomic practices, such as low plant density per hectare [17]. Likewise, the scarcity of widely adaptable and early-maturing varieties further exacerbates the problem. The productivity of the crop is highly influenced by the variability in environmental conditions, including location effects, seasonal fluctuations, and the interaction between these factors. These environmental variables play a crucial role in determining the actual yield potential of cowpea [18,19]. Studies on the crop have shown that genotype by environment (G × E) has a significant effect on trait performance [20,21,22,23]. It has been found that some cowpea genotypes are more stable in terms of agronomic trait performance across environments than others [20,24]. The variability in genotype performance is partially unpredictable, just as the response of genotypes to environmental change is not the same [24]. This agrees with [25,26] for cowpea.
The genotype-by-environment interaction (GEI) poses a significant challenge for plant breeders, as it complicates the process of recommending the best performing varieties. The inconsistency of genotypes that yield the highest results across different cropping environments and seasons adds to this challenge. Hence, the analysis of GEI is a fundamental requirement prior to recommending varieties for widespread cultivation since it serves as a crucial step in understanding the superiority and consistency of genotype performance across diverse geographic locations. This is essential because a genotype’s performance can be significantly influenced by its genetic worth, environmental conditions, or the interaction of both [20,21,23]. Often, the environment can mask the genetic potential of a genotype, leading to poor genetic gain from artificial selection, especially for quantitative traits such as grain yield [21]. Thus, GEI analysis is highly valuable during the final stages of selecting elite breeding materials, as it assists breeders in recommending candidate varieties as suitable for either location-specific adoption or wider geographic use [21,23]. Several techniques have been widely adapted to analyze and interpret G × E data for cowpea, including the main genotype effect plus genotype-by-environment interaction biplot (GGE biplot) analysis and the additive main effect and multiplicative interaction (AMMI) [21,22,27]. To mitigate the impact of GEI, researchers commonly repeat experiments in multiple sites within the same year or over multiple crop seasons in a single site, or sometimes both approaches are combined [28,29].
Therefore, evaluating the performance of improved cowpea in contrasting environments is imperative for the recommendation of the right genotype for a specific environment or wider use across different regions. In view of this, the objectives of the present study were to estimate the effects of genotype, environment, and genotype × environment interaction on key agronomic, grain yield, and yield-related traits of some elite cowpea lines and to assess the stability of improved lines for yield across different environments.
2. Materials and Methods
2.1. Genetic Materials
The genetic materials for this study were 40 test entries and 2 standard checks, which are presented in Table 1. The lines were recently developed by the cowpea breeding program at the International Institute of Tropical Agriculture (IITA), Ibadan, Kano, Kano State in Nigeria, and their response to various environments has not been documented in a published literature.

Table 1.
List and source of the genotypes.
2.2. Description of the Study Environments
The experiment was conducted in five locations in Nigeria in 2021 (Table 2). The locations were Bagauda in Kano state, situated on 499 m elevation and constituting a semi-arid/Sudan savanna with annual rainfall ranging from 552 to 1093 mm from June to November. The temperature ranges from 19 to 33 °C and the soil type is regosol. The second location was Ibadan in Oyo state, situated on 235 m elevation in derived savanna with an annual rainfall ranging from 939 to 1681mm from March to November. The temperature in this location ranges from 21 to 31 °C and the soil type is lixisol. Details of the environmental condition for the other locations, namely, Abubakar Tafawa Balewa University (ATBU), Gubi in Bauchi state, Gumel in Jigawa state, and Shika in Kaduna state, all in Nigeria, are presented in Table 2.

Table 2.
Description of the study environments.
The testing sites cut across three different agroecologies in Nigeria, which are also the key cowpea growing zones (Figure 1). These agroecological zones include derived Savanna, Northern Guinea savanna, and semi-arid/Sudan savanna.
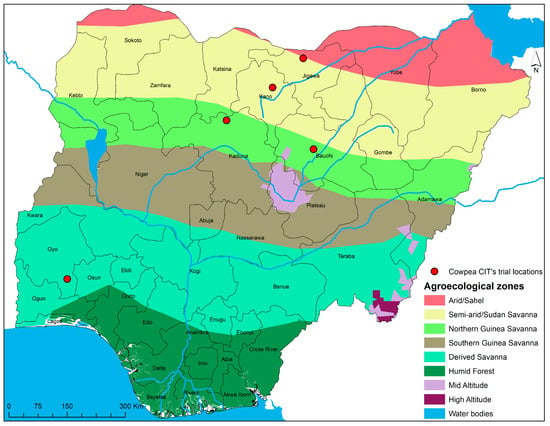
Figure 1.
Agroecological zone and site locations on a map of Nigeria. Source [30].
2.3. Experimental Layout and Data Collection
The experiment was laid out in a 7 × 6 alpha lattice design with three replications. Cowpea lines were established in four rows, each four meters long. Three to four seeds were planted per hill and later thinned down to two plants per hill. The intra- and inter-planting space was 20 cm and 75 cm, respectively. To control weeds, pre- and post-emergence herbicides—Lifeline [Cropserve (Pty) Ltd., Aston Manor, South Africa] with glufosinate ammonium (24.5%) as the active substance applied at a rate of 3 L/hectare and Raptor [BASF Agri-Production S.A.S, Gravelines, France] with an active substance of ammonium salt (40 g/L Imazamox), applied at a rate of 1.5 L per hectare, respectively—were applied. Fertilizer was applied at a rate of 100 kg NPK 15:15:15 (two bags), which supplied 15 kg each of nitrogen, phosphorus, and potassium, and 200 kg (4 bags) of single super phosphate, which supplied 30 kg P (P2O5) per hectare. All agronomic practices were carried out in accordance with recommendations for cowpea production [8].
The measured parameters included the number of plant hills, plant stands at harvest, days to first flowering, days to 50% flowering, days to first pod maturity, days to 95% pod maturity, bacteria blight score, grain yield (kg/ha), dried fodder yield (kg/ha) (weight of above-ground biomass taken from the two middle rows after removing all pods), and 100-seed weight (g). To minimize border effects, data were recorded from the net plot, consisting of the two middle rows.
3. Data Analysis
The analysis of variance was conducted using R software (version 4.2.3) [32]. Means were separated using the least significant difference (LSD), with significant differences found at a probability level of 5%. The following statistical measures were calculated in the analysis: phenotypic coefficient of variation (PCV), genotypic coefficient of variation (GCV), predicted genetic advance (GA), and genetic advance as percentage of mean (GAM), as per the formula listed below. Principal component analysis (PCA) was computed for yield and was completed using ViTSel [33] to further understand the genotypes’ stability across test locations. Additionally, stability parameters such as Wricke’s ecovalence [34], the cultivar superiority by Lin and Binns [35], the Shukla stability [36], and GGE biplot (displays the GGE of a genotype by two-way environmental data) were used. The GGE biplot methodology originates from graphical analysis of multi-environment variety trials, and Pearson correlations were computed among the traits measured using R [37].
The PCV, GCV, GA, and GAM were computed as follows;
Phenotypic coefficient of variation (PCV) was computed according to [38]:
The broad sense heritability (H2) was estimated as the ratio of genotypic variance to phenotypic variance and was expressed as a percentage [39,40,41].
where is the phenotypic variance and is the genotypic variance.
The genetic advance to be expected by selecting 5% of the superior progenies was calculated using the following formula, given by [42]:
i = standard selection differential for 5% selection intensity (=2.06), according to [43]. and H is the heritability.
Genetic advance as percentage of mean (GAM):
where GA is the genetic advance, and is the general mean. GAM was categorized as low (0–10%), moderate (10–20%), and high (>20%) following the recommendations in [44].
3.1. Stability Measures
- Wricke’s ecovalence (Wi) [34] is a quantitative assessment of the consistency of a specific genotype’s performance across test environments, where a low ecovalence value indicates greater stability, meaning that environmental variations influence the genotype’s performance less and consistently perform well under diverse conditions.where Wi = ecovalence of the i-th cultivar;
- Yij = the observed phenotypic value of the i-th cultivar in the j-th environment;
- Yi. = mean of i-th cultivar across the entire environment;
- Y.j = mean of j-th environment;
- Y.. = grand mean.
- The cultivar superiority (Pi) by Lin and Binns [35] is defined as the mean square distance between the cultivar’s response and the maximum response averaged over all locations.where Pi = superiority index of the i-th cultivar;
- Xij = yield of the i-th cultivar in the j-th environment;
- Mj = maximum response obtained among all the cultivars in the j-th environment;
- n = number of environments.
- Shukla’s stability (1972) [36] measure was calculated as the difference between the genotype’s observed performance and its expected performance across all environments divided by the overall mean performance across all genotypes and environments. The formula for Shukla’s stability measure is as follows:where S is the stability measure for genotype I;
- Yi. = the average performance of genotype i across all environments;
- = the overall mean performance across all genotypes and environments;
- Yij = the performance of genotype I in environment j.
3.2. Genotype Main Effect plus Genotype-by-Environment Interaction (GGE) Biplot Analysis
A GGE biplot generated from multivariate analysis was used to depict the associations between the genotypes and the specific testing environments [27].
4. Results
The results of the analysis of variance revealed significant differences for all the traits measured (Table 3). Significant differences were observed among the environments, genotypes, and genotype-by-environment interaction. The phenotypic coefficient of variation (PCV) consistently exhibited higher values compared to the genotypic coefficient of variation (GCV) for all the assessed traits. The heritability estimates ranged from 0.49 for grain yield to a maximum of 0.97 for 100-seed weight. Specifically, the GCV, which represents genotypic variability, ranged from 21.95 for days to maturity to 265.11 for dried-fodder weight. Conversely, the PCV, representing phenotypic variability, spanned from 26.82 for days to first flowering to 756.26 for dried fodder. Moreover, the expected genetic advance (GA) varied across traits, with the lowest value of 2.99 for days to first flowering and the highest value of 252.80 in dried-fodder weight. Additionally, the expected genetic advance as a percentage of the mean (GAM) was also found to differ across traits assessed, with values ranging from a minimum of 5.50 for days to 95% maturity to 43.16 for dried-fodder yield. These results provide valuable evidence of the diversity and heritability of the examined traits, which further indicates variation within the genotypes and environmental conditions.

Table 3.
Statistics summary, heritability estimates, and variation metrics for elite cowpea lines evaluated for key agronomic traits across various agroecologies.
Three stability measures were used to identify the most stable genotypes (Table 4). Genotypes G8 (IT07K-230-2-9), G16 (IT14K-1683-2), G5 (245-1), G26 (IT15K-2510-2), G25 (IT15K-2386-1), G20 (IT14K-2026), G19 (IT14K-1913), G12 (IT10K-863-11), G38 (IT17K-1589-2-1), and G36 (IT17K-1489-2-3) were the top 10 most stable based on Wricke’s ecovalence method. On the other hand, based on cultivar superiority, the top 10 most superior genotypes were G25 (IT15K-2386-1), G17 (IT14K-1813-1), G42 (StrigaMABC-42), G40 (IT17K-2515-3), G32 (IT16K-2597-2), G20 (IT14K-2026), G12 (IT10K-863-11), G14 (IT13K-1144-9), G19 (IT14K-1913), and G16 (IT14K-1683-2), whereas genotypes G27 (IT16K-1968-3), G28 (IT16K-1984-2), G23 (IT15K-2244-1), G37 (IT17K-1555-5-1), G21 (IT14K-2111-2), G5 (245-1), G19 (IT14K-1913), G6 (245-8), G41 (IT11K-61-82), and G18 (IT14K-1813-2) were detected via Shukla stability variance to be the most stable. Genotype G19 (IT14K-1913) was ranked in a top 10 position with all three stability measures, and G12 (IT10K-863-11), and G16 (IT14K-1683-2) were ranked among the top 10 genotypes with both Wricke’s and the cultivar superiority stability estimates.

Table 4.
Estimates of stability measures for cowpea genotypes using Wricke’s ecovalence, cultivar superiority, and Shukla stability variance.
4.1. Principal Components
Principal component 1 (PC1) and principal component 2 (PC2) together accounted for 79.5% of the total variation among the genotypes studied across the five environments (Figure 2).
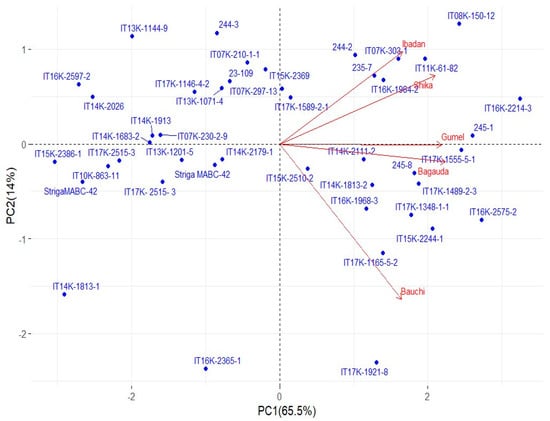
Figure 2.
PCA showing clustering of genotypes and relationships among the test locations for yield using ViTSel 2020. Locations are shown with red vector arrows emerging from the center and pointing towards each location. The size of the angle between any two location vectors determines the strength of correlation between the locations—that is, locations with narrow angles are closely related and vice versa. The blue dot represents each genotype tested; the proximity of a genotype to a specific location indicates its good performance in that location: Bagauda—Kano state; Bauchi—Abubakar Tafawa Balewa University (ATBU), Gubi in Bauchi state; Ibadan—Oyo state; Shika—Kaduna state; Gumel—Jigawa state.
PC1*PC2 indicated that the Ibadan and Shika environments were closely related to genotypes G41 (IT11K-61-82), G10 (IT07K-303-1), and G28 (IT16K-1984-2) (Table 1), suggesting that they are the most suitable for these environments. Gumel and Bagauda were also closely related, with genotypes G37 (IT17K-1555-5-1), G5 (245-1), and G6 (245-8) being the most adapted to these sites.
4.2. Main Genotype Effect plus Genotype by Environment Interaction (GGE) Biplot Analysis
The specific adaptation of the genotypes was assessed using the Which Won Where model of the GGE biplot (Figure 3). In this analysis, principal components 1 and 2 explained 79.69% of the total variation. Nine mega environments (sectors) were identified by the Which Won Where’ biplot, with Ibadan, Shika, Gumel, and Bagauda all falling within the same mega environment. Genotype G41 (IT11K-61-82), appeared to be the best for Ibadan and Shika, as it was located closer to these two environments on the biplot. On the other hand, genotype G5 (245-1) was the best for Gumel and Bagauda. Bauchi fell within another mega environment, and G30 (IT16K-2365-1) was the best genotype for that environment. These inferences are based on the proximity of each genotype to the respective environments in the biplot.
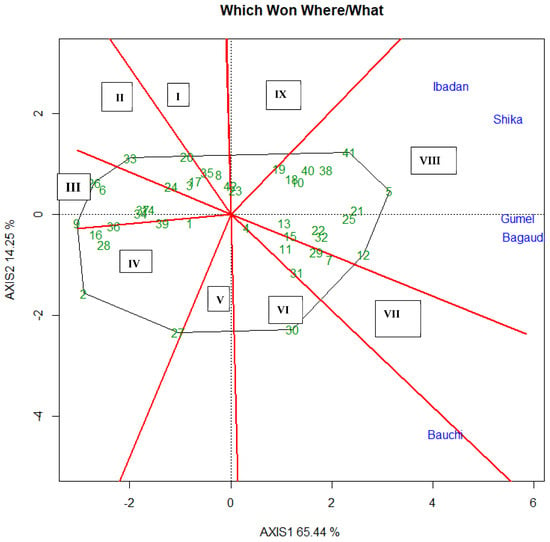
Figure 3.
GGE biplot—Which Won Where presenting the best genotypes in each environment. The green numbers are the codes for the genotypes tested; the blue text are the names of the locations: Bagauda—Kano state; Bauchi—Abubakar Tafawa Balewa University (ATBU), Gubi in Bauchi state; Ibadan—Oyo state; Shika—Kaduna state; Gumel—Jigawa state.
Figure 4 displays the results for the GGE biplot of mean versus stability. In this analysis, principal components 1 and 2 explained 79.69% of the total variation. The figure presents the mean yield and stability across the five test locations. G5 (245-1) had the highest mean yield, whereas G21 (IT14K-2111-2) and G25 (IT15K-2386-1) were the most stable, with the shortest perpendicular mark on the axis. G12 (IT10K-863-11) and G41 (IT11K-61-82) also had high mean yield values but were less stable, as indicated by the longer perpendicular marks on the axis.
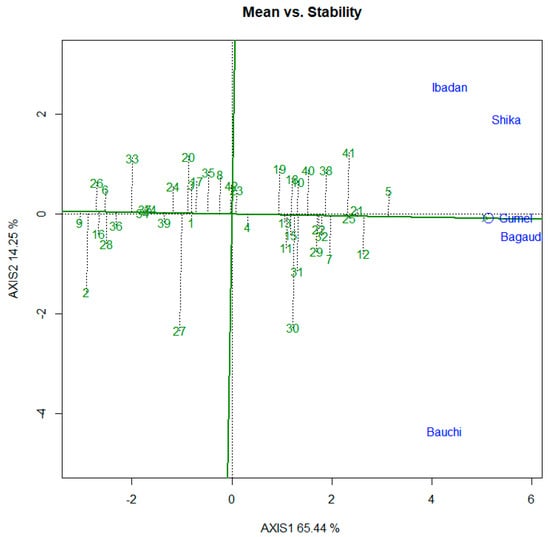
Figure 4.
GGE biplot presenting mean versus stability for grain yield data across five locations. The green numbers are the codes for the genotypes tested; the blue texts are the names of the locations: Bagauda—Kano state; Bauchi—Abubakar Tafawa Balewa University (ATBU), Gubi in Bauchi state; Ibadan—Oyo state; Shika—Kaduna state; Gumel—Jigawa state.
4.3. Correlations among Traits
There was a positive and significant correlation between the weight of dried fodder and several traits, including the number of plant hills, plant stands at harvest, days to 95% maturity, and grain yield in kg/ha (Figure 5). Of particular interest, significant positive correlations were detected among all maturity-related traits, days to first flower, days to 50% flowering and days to 95% pod maturity. In addition, days to 95% maturity was significantly correlated (r = 0.73) with 100-seed weight. However, grain yield had positive but weak correlation with maturity-related traits. These traits are expected to play a significant role in the selection process for dried-fodder weight, days to first flowering, days to 50% flowering, and 100-seed weight. Likewise, the 100-seed weight showed a weak negative relationship with bacterial blight, the number of plant hills, and the number of plant stands at harvest. Grain yield (kg/ha) also had a positive and significant correlation with dried-fodder weight, number of plants per hill, and number of plant stands at harvest.
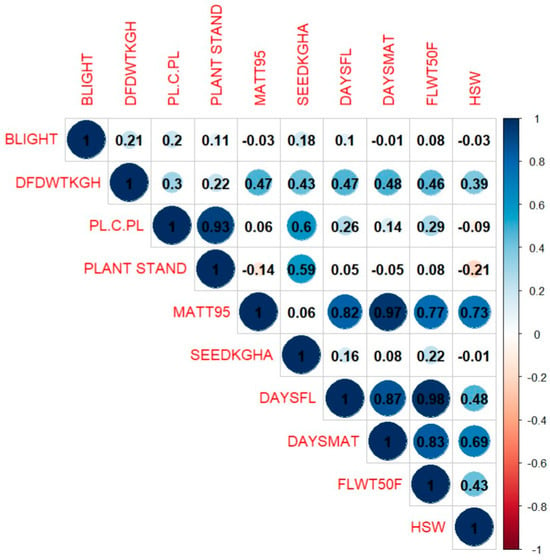
Figure 5.
Relationship between phenology, yield components, and bacteria blight incidence for elite cowpea lines evaluated across five locations in Nigeria. BLIGHT—bacteria blight, DFDWTKGH—dried-fodder weight kg/ha, PL.C.PI—number of plants per hill, PLANT STAND—number of plant stands at harvest, MATT95—days to 95% maturity, SEEDKGHA—grain yield kg/ha, DAYSFL—days to first flowering, DAYSMAT—days to first pod maturity, FLW50F—days to 50% flowering, HSW—100-seed weight. The scale on the right side of the figure indicates the strength and direction of the correlation; deep blue—highly positively correlated and deep red—highly negatively correlated. The size of the circles also reflects the strength of the correlation.
5. Discussion
The phenotypic coefficient of variation (PCV) was generally higher than the genotypic coefficient of variation (GCV) in all traits, suggesting that the environment played a major role in the performance of the genotypes across different locations. This means that the expression of the trait was more influenced by environmental conditions, such as temperature, humidity, nutrient availability, or other external factors, rather than solely by genetic differences, which agrees with the findings in [24]. High values of PCV and GCV observed in the present study suggest the reliability of effective selection for these traits. Furthermore, it also indicates the existence of substantial phenotypic variability among the tested cowpea genotypes. Similar reports of high PCV and GCV for agronomic traits, such as 100-seed weight and yield, were reported by [45,46,47,48,49].
Higher broad-sense heritability indicates that a large portion of variation may be heritable, especially if the additive component of the broad-sense heritability is very high. In this study, the highest broad-sense heritability was recorded in 100-seed weight, maturity related traits and dried-fodder weight.
The present study dissected the stability of advanced cowpea lines based on three stability measures. Wricke’s ecovalence stability measure [34] calculated the average deviation of a genotype’s performance from the average performance of all genotypes in each environment and then compared this deviation to a measure of the overall variation in performance across environments. A genotype is considered stable if its deviation is relatively small compared to the overall variation. In the present study, genotypes G8 (IT07K-230-2-9), G16 (IT14K-1683-2), G5 (245-1), G26 (IT15K-2510-2), and G25 (IT15K-2386-1) were the top five lines with very small mean yield deviation across the five environments and were considered to be stable across these environments. In agreement with this, [50,51] reported the highest yielding genotypes for cowpea and common bean, respectively, to have relatively low ecovalence values. In the present study, based on the ecovalence method, the most unstable genotypes were G30 (IT16K-2365-1), G38 (IT17K-1589-2-1), G27 (IT16K-1968-3), G28 (IT16K-1984-2), and G17 (IT14K-1813-1), which showed relatively higher ecovalence values for grain yield. According to [52], genotypes with a high ecovalence mean and large estimated values are suitable for high-input environments. Similarly, Abou-Khater et al., 2022 Ref. [53] also reported Wricke’s stability measures as an effective parameter in identifying stable and high-yielding genotypes in faba bean, which agrees with our findings.
Cultivar superiority (Pi) by Lin and Binns measures the most stable genotype as being the one with the least deviation from the maximum yield of each environment—that is, the one with the lowest (Pi) value. In the current investigation, the most stable genotype ranking first for Pi and for mean grain yield was G25 (IT15K-2386-1) followed by G17 (IT14K-1813-1), G42 (StrigaMABC-42), G40 (IT17K-2515-3), and G32 (IT16K-2597-2). These stable genotypes made the least contribution to the total variation due to the GEI. In contrast, G29 (IT16K-2214-3), G5 (245-1), G11 (IT08K-150-12), G31 (IT16K-2575-2), and G37 (IT17K-1555-5-1) were the most unstable genotypes, and they contributed a large portion of the total variation due to GEI; these results agree with the work in [51], which reported that the most stable cowpea genotypes had the lowest Pi value and highest mean grain yield.
Our study revealed significant G × E among cowpea genotypes for several agronomic traits, such as 100-seed weight, grain yield, days to flowering, and days to maturity, which was also reported by [45,54,55,56].
Overall, the GGE biplot provides a powerful visual representation of complex genotype-by-environment interactions and can help researchers to identify the genotypes that are most suited for specific environments, as well as the environments that are most similar to each other in terms of performance [57,58]. Hall et al. (1997) [59] reported that high heritability for the stay green trait and substantially low GEI for this trait in cowpea would enable the successful incorporation of this trait into improved varieties. The findings in [51] indicated that the yield performance of speckled bean genotypes was highly influenced by GE interaction effects; the magnitude of the environmental effect was about 2.88 times that of the genotypic effect. In agreement with our findings [60], in cowpea, it was also reported that the analysis of variance for each location and combined over five locations showed significant differences among the genotypes and environments for grain yield and most yield-related traits. The significant genotype × environment interaction effects indicates the inconsistent performance of genotypes across the tested environments and the differential discriminating ability of the tested environments. The significant effects of GEI on traits suggests the need to assess the stability of genotypes over different environments [50,61,62,63,64].
Grain (kg/ha) showed a positive significant correlation to days to 50% flowering, indicating that the longer it takes to attain 50% flowering, the higher the yield; this agreed with the findings in [65] for soybean that there was a positive significant correlation with grain yield and days to 50% flowering and days to maturity. However, our findings showed a negative, non-significant correlation between grain yield and 100-seed weight, which disagrees with the findings in [65]. The 100-seed weight showed a positive significant correlation with days to 50% flowering and days to 95% maturity. This was expected, as the longer grain-filling period resulted in larger seed size, which corroborates the findings in [66,67].
6. Conclusions
Our study reveals that the influence of genotypes, environments, and their interaction were highly significant for the grain yield of cowpea genotypes evaluated across five locations during the 2021 growing season in Nigeria. We employed various stability models to assess genotype stability, including Wricke’s ecovalence, cultivar superiority, Shukla stability variance, and a GGE biplot, and it was shown that the Wricke’s ecovalence and cultivar superiority methods were more dependable measures of stability than the Shukla method. This conclusion was drawn from the fact that the same five genotypes were consistently ranked as the most stable among the top 10 genotypes for both the Wricke’s ecovalence and cultivar superiority stability measures. Specifically, genotypes G16 (IT14K-1683-2), G25 (IT15K-2386-1), G20 (IT14K-2026), G12 (IT10K-863-11), and G19 (IT14K-1913) consistently ranked among the top 10 most stable genotypes based on both the Wricke’s ecovalence and the cultivar superiority measure. Furthermore, G19 (IT14K-1913) was the only genotype that appeared among the top 10 most stable genotypes in all three stability measures used in this study. The GGE biplot analysis revealed nine possible mega-environments, with four locations (Ibadan, Shika, Bagauda, and Gumel) falling within the same mega-environment (Sector VIII), whereas Bauchi constituted a distinct mega-environment. Based on the GGE biplot analysis, G21 (IT14K-2111-2) and G25 (IT15K-2386-1) emerged as the most stable genotypes across all five test locations, as indicated by the shortest perpendicular marks on the axis. As a result, we recommend these two genotypes for cultivation across the test locations.
Author Contributions
Conceptualization, B.O.P. and O.B.; methodology, B.O.P., P.O.O., O.B. and C.F.; software, B.O.P.; validation, B.O.P., P.O.O., A.T., S.B.M., O.B. and C.F.; formal analysis, B.O.P. and P.O.O.; investigation, B.O.P. and P.O.O.; resources, O.B. and C.F.; data capture and curation, B.O.P., P.O.O., A.T., D.J.I. and G.B.; writing—original draft preparation, B.O.P.; writing—review and editing, B.O.P., P.O.O., A.T., S.B.M., O.B. and C.F.; visualization, B.O.P. and P.O.O.; supervision, O.B. and C.F.; project administration, O.B.; funding acquisition, O.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Bill & Melinda Gates Foundation through the Accelerated Varietal Improvement and Seed Delivery of Legumes and Cereals in Africa (AVISA) project, grant #OPP1198373/INV-009649.
Data Availability Statement
All data reported in this study are provided in the text.
Acknowledgments
The authors appreciate the Bill & Melinda Gates Foundation (BMGF) through the Accelerated Varietal Improvement and Seed Delivery of Legumes and Cereals in Africa (AVISA) project and the Accelerated Breeding Initiative funds from the International Institute of Tropical Agriculture (IITA).
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Boukar, O.; Belko, N.; Chamarthi, S.; Togola, A.; Batieno, J.; Owusu, E.; Haruna, M.; Diallo, S.; Umar, M.L.; Olufajo, O.; et al. Cowpea (Vigna unguiculata): Genetics, genomics and breeding. Plant Breed. 2019, 138, 415–442. [Google Scholar] [CrossRef]
- Jayathilake, C.; Visvanathan, R.; Deen, A.; Bangamuwage, R.; Jayawardana, B.C.; Nammi, S.; Liyanage, R. Cowpea: An overview on its nutritional facts and health benefits. J. Sci. Food Agric. 2018, 98, 4793–4806. [Google Scholar] [CrossRef]
- Omoigui, L.O.; Kamara, A.Y.; Kamai, N.; Ekeleme, F.; Aliyu, K.T. Guide to Cowpea Production in Northern Nigeria; IITA: Ibadan, Nigeria, 2020; 48p. [Google Scholar]
- Samireddypalle, A.; Boukar, O.; Grings, E.; Fatokun, C.A.; Kodukula, P.; Devulapalli, R.; Okike, I.; Blummel, M. Cowpea and Groundnut Haulms Fodder Tradding and Its Lessons for Multidimensional Cowpea Improvement for Mixed crop Livestock Systems in West Africa. Front. Plant Sci. 2017, 8, 30. [Google Scholar] [CrossRef] [PubMed]
- Singh, B. Cowpea: The Food Legume of the 21st Century; Crop Science Society of America: Madison, WI, USA, 2014. [Google Scholar]
- Duke, J.A. Introduction to food legumes. In Insect Pests of Tropical Legumes; Singh, S.R., Ed.; John Wiley and Sons Ltd.: Chichester, UK, 1990; pp. 1–42. [Google Scholar]
- Singh, B.B.; Ajeigbe, H.A.; Tarawali, S.A.; Fernandez-Rivera, S.; Abubakar, M. Improving the production and utilization of cowpea as food and fodder. Field Crop. Res. 2003, 84, 169–177. [Google Scholar] [CrossRef]
- Dugje, I.Y.; Omoigui, L.O.; Ekeleme, F.; Kamara, A.Y.; Ajeigbe, H. Farmers’ Guide to Cowpea Production in West Africa; IITA: Ibadan, Nigeria, 2009; p. 20. [Google Scholar]
- FAOStat. FAOSTAT, Statistical Data Base; Food and Agricultural Organizations of the United Nations: Rome, Italy, 2021. [Google Scholar]
- Hill, S. Cowpea Adaptability to Southeastern Organic Farming Systems: Forage Productivity and Charcoal Rot Susceptibility. Master’s Thesis, University of Tennessee, Knoxville, TN, USA, 2015. [Google Scholar]
- Noubissietchiagm, J.B.; Bell, J.M.; Guissaibirwe, S.; Gonne, S.; Youmbi, E. Varietal response of cowpea (Vigna unguiculata L. Walp.) to striga gesnerioides (wild.) vatke race SG5 infestation. Horti Agrobot. Cluj-Napoca 2010, 38, 33–41. [Google Scholar]
- Baidoo, P.K.; Mochiah, M.B. Varietal susceptibility of improved cowpea Vigna unguiculata (L.) (walp.) cultivars to field and storage pests. Sustain. Agric. Res. 2014, 3, 69. [Google Scholar] [CrossRef][Green Version]
- Chamarthi, S.K.; Belko, N.; Togola, A.; Fatokun, C.A.; Boukar, O. Genomics-Assisted Breeding for Drought Tolerance in Cowpea. In Genomics Assisted Breeding of Crops for Abiotic Stress Tolerance, Vol. II; Sustainable Development and Biodiversity; Rajpal, V., Sehgal, D., Kumar, A., Raina, S., Eds.; Springer: Cham, Switzerland, 2019; pp. 187–209. [Google Scholar] [CrossRef]
- Horn, L.; Shimelis, H.; Laing, M. Participatory appraisal of production constraints, preferred traits and farming system of cowpea in the northern Namibia: Implications for breeding. Legume Res. 2015, 38, 691–700. [Google Scholar] [CrossRef]
- Maureen, F.A.K. Genetic Analysis of Traits Related to Biological Nitrogen Fixation in Cowpea [Vigna unguiculata L. WALP] under Low Soil Phosphorus. Ph.D. Thesis, University of Ghana, Legonm, Ghana, 2015. [Google Scholar]
- Mohammed, S.B.; Dzidzienyo, D.K.; Umar, M.L.; Ishiyaku, M.F.; Tongoona, P.B.; Gracen, V. Appraisal of cowpea cropping systems and farmers’ perceptions of production constraints and preferences in the dry savannah areas of Nigeria. CABI Agric. Biosci. 2021, 2, 25. [Google Scholar] [CrossRef]
- Olufajo, O.O.; Singh, B.B. Advances in cowpea cropping systems research. In Challenges and Opportunities for Enhancing Sustainable Cowpea Production; Fatokun, C.A., Tarawali, S.A., Singh, B.B., Kormawa, P.M., Tamo, M., Eds.; Institute of Tropic Agriculture, (IITA): Ibadan, Nigeria, 2002; Volume 4, pp. 267–277. [Google Scholar]
- Hall, A.E.; Cisse, N.; Thiaw, S.; Hassan, O.A.E.; Ehlers, J.D.; Ismail, A.M.; Fery, R.L.; Roberts, P.A.; Kitch, L.W.; Murdock, L.L.; et al. Development of cowpea cultivars and germplasm by the Bean/Cowpea CRSP. Field Crop. Res. 2003, 82, 103–134. [Google Scholar] [CrossRef]
- Adewale, B.D.; Adeigbe, O.; Aremu, C. Genetic distance and diversity among some cowpea (Vigna unguiculata L. Walp) genotypes. Int. J. Res. Pharm. Sci. 2011, 1, 9–14. [Google Scholar]
- Ali, Y.; Aslam, Z.; Hussain, F.; Shakur, A. Genotype and environmental interaction in cowpea (Vigna unguiculata-L) for yield and disease resistance. Int. J. Environ. Sci. Technol. 2004, 1, 119–123. [Google Scholar] [CrossRef][Green Version]
- Goa, Y.; Mohammed, H.; Worku, W.; Urage, E. Genotype by environment interaction and yield stability of cowpea (Vigna unguiculata (L.) Walp.) genotypes in moisture limited areas of Southern Ethiopia. Heliyon 2022, 8, e09013. [Google Scholar] [CrossRef] [PubMed]
- Gumede, M.T.; Gerrano, A.S.; Modi, A.T.; Thungo, Z. Influence of genotype and environment on grain yield among cowpea (Vigna unguiculata (L.) Walp) genotypes under dry land farming system. Acta Agric. Scand. Sect. B—Soil Plant Sci. 2022, 72, 709–719. [Google Scholar] [CrossRef]
- Horn, L.; Shimelis, H.; Sarsu, F.; Mwadzingeni, L.; Laing, M.D. Genotype-by-environment interaction for grain yield among novel cowpea (Vigna unguiculata L.) selections derived by gamma irradiation. Crop J. 2018, 6, 306–313. [Google Scholar] [CrossRef]
- Adewale, B.D.; Okonji, C.; Oyekanmi, A.A.; Akintobi DA, C.; Aremu, C.O. Genotypic variability and stability of some grain yield components of Cowpea. Afr. J. Agric. Res. 2010, 5, 874–880. [Google Scholar]
- Sarvamangala, C.; Uma, M.S.; Birada, S.; Salimath, P.M. Stability analysis for yield and yield components over seasons in cowpea (Vigna unguiculata L. Walp). Electron. J. Plant Breed. 2010, 1, 1392–1395. [Google Scholar]
- Nunes, H.F.; Filho, F.R.F.; Ribeiro, V.Q.; Gomes, R.L.F. Grain yield adaptability and stability of black-eyed cowpea genotypes under rainfed agriculture in Brazil. Afr. J. Agric. Res. 2014, 9, 255–261. [Google Scholar]
- Yan, W.; Kang, M.S.; Ma, B.; Woods, S.; Cornelius, P.L. GGE Biplot vs. AMMIAnalysis of Genotype-by-Environment Data. Crop Sci. 2007, 47, 643–653. [Google Scholar] [CrossRef]
- Gauch, H.; Zobel, W. AMMI analysis of yield trials. In Genotypes by Environment Interaction; Kang, S., Gauch, H., Eds.; CRC: New York, NY, USA, 1996. [Google Scholar]
- Bandeira, M.; Jackson, K.; de Moura, M.; Angelo, J.; Raphaelle, L. Genotype by environment interaction in cowpea lines using GGE biplot method. Rev. Caatinga 2018, 31, 64–71. [Google Scholar]
- International Institute of Tropical Agriculture (IITA). Geographic Information System (GIS); International Institute of Tropical Agriculture: Ibadan, Nigeria, 2023. [Google Scholar]
- FAO. Digital Soil Map of the World; FAO: Rome, Italy, 1991. [Google Scholar]
- R Core Team. A Language and Environment for Statistical Computing; R. Foundation for Statistical Computing: Vienna, Austria, 2023; Available online: http://www.R.Project.org (accessed on 23 November 2023).
- ViTSel, Version 1.0; Breeding and Germplasm Tool. Africa Rice Centre (AfricaRice), Centro Internacional de Mejoramiento de Maiz y Trigo (CIMMYT), and International Institute of Tropical Agriculture (IITA). 2020. Available online: https://www.iita.org/digital-tools/appdetails?app=ViTSel (accessed on 23 November 2023).
- Wricke, G. Übereine Methodezur Erfassung der ökologischen Streubreite in Feldversuchen. Z. Pflanzenzuecht. 1962, 47, 92–96. [Google Scholar]
- Lin, C.S.; Binns, M.R. A superiority measure of cultivar performance for cultivar x location data. Can. J. Plant Sci. 1988, 68, 193–198. [Google Scholar] [CrossRef]
- Shukla, G.K. Some statistical aspects of partitioning genotype environmental components of variability. Heredity 1972, 29, 237–245. [Google Scholar] [CrossRef] [PubMed]
- Dia, M.; Wehner, T.C.; Arellano, C. RGxE: An R Program for Genotype x Environment Interaction Analysis. Am. J. Plant Sci. 2017, 8, 1672–1698. [Google Scholar] [CrossRef]
- Singh, R.K.; Chaudhary, S.D. Biometrical Methods in Quantitative Genetic Analysis; Kalyan Publishers: New Delhi, India, 1985; pp. 205–214. [Google Scholar]
- Hanson, C.H.; Robinson, H.F.; Comstock, R.E. Biometrical studies of yield in segregating populations of Korean lespedeza 1. Agron. J. 1956, 48, 268–272. [Google Scholar] [CrossRef]
- Falconer, D.S.; Mackay, T.C.F. Introduction to Quantitative Genetics; Longman: London, UK, 1996. [Google Scholar]
- Lynch, M.; Walsh, B. Genetics and Analysis of Quantitative Traits; Sinauer Associates: Sunderland, MA, USA, 1998. [Google Scholar]
- Robinson, H.F.; Comstock, R.E.; Harvey, P.H. Estimates of heritability and the degree of dominance in corn 1. Agron. J. 1949, 41, 353–359. [Google Scholar] [CrossRef]
- Allard, R.W. Principles of Plant Breeding; John Wiley and Sons, Inc.: New York, NY, USA, 1960. [Google Scholar]
- Johnson, H.W.; Robinson, H.F.; Comstock, R.E. Estimates of genetic and environmental variability in soybeans 1. Agron. J. 1955, 47, 314–318. [Google Scholar] [CrossRef]
- Mofokeng, M.A.; Mashilo, J.; Rantso, P.; Shimelis, H. Genetic variation and genetic advance in cowpea based on yield and yield-related traits. Acta Agric. Scand. Sect. B Soil Plant Sci. 2020, 70, 381–391. [Google Scholar] [CrossRef]
- Manggoel, W.; Uguru, M.I.; Ndam, O.N.; Dasbak, M.A. Genetic Variability, Correlation and Path Coefficient Analysis of some Yield Components of Ten Cowpea [Vigna unguiculata (L.) Walp] accessions. J. Plant Breed. Crop Sci. 2002, 4, 80–86. [Google Scholar] [CrossRef]
- Kumawat, K.C.; Raje, R.S.; Kumhar, B.L. Genetic variation in yield and yield components in cowpea [Vigna unguiculata L. (Walp.)]. Ann. Agri-Bio Res. 2005, 10, 21–23. [Google Scholar]
- Narayanankutty, C.; Mili, R.; Jaikumaran, U. Variability and genetic divergence in vegetable cowpea. J Maharashtra Agric. Univ. 2003, 28, 26–29. [Google Scholar]
- Vidya, C.; Oommen, S.K.; Vijayaraghava, K. Genetic variability and heritability of yield and related characters in yard-longbean. J. Trop. Agric. 2002, 40, 11–13. [Google Scholar]
- Mohammed, I.; Victoria, M.Y.; Singh, B.B.; Olufajo, O.; Zaria, A.A. Phenotypic stability for selected traits of some cowpea lines in Nigerian agro-ecologies. Plant Biol. Biotechnol. 2017, 5, 67–77. [Google Scholar]
- Tadesse, T.; Sefera, G.; Asmare, B.; Teklaign, A. Application of AMMI for grain yield stability analysis in large speckled bean genotypes grown in midlands of bale zone. Chem. Biomol. Eng. 2018, 3, 17–21. [Google Scholar] [CrossRef]
- Asfaw, A.; Assefa, T.; Amsalu, B.; Negash k Alemayehu, F.; Gurum, F.; Rezene, Y.; Finenissa, C.; Atnafi, M.; Daba, C. Adaptation and yield stability of small red beans elite lines in Ethiopia. Int. J. Plant Breed. Genet. 2008, 2, 51–63. [Google Scholar] [CrossRef]
- Abou-Khater, L.; Maalouf, F.; Jighly, A.; Rubiales, D.; Kumar, S. Adaptability and Stability of Faba Bean (Vicia faba L.) Accessions under Diverse Environments and Herbicide Treatments. Plants 2022, 11, 251. [Google Scholar] [CrossRef] [PubMed]
- Gerrano, A.S.; van Rensburg, W.S.J.; Kutu, F.R. Agronomic evaluation and identification of potential cowpea (Vigna unguiculata L. Walp) genotypes in South Africa. Acta Agric. Scand. Sect. B Soil Plant Sci. 2019, 69, 295–303. [Google Scholar] [CrossRef]
- Martos-Fuentes, M.; Fernández, J.A.; Ochoa, J.; Carvalho, M.; Carnide, V.; Rosa, E.; Pereira, G.; Barcelos, C.; Bebeli, P.J.; Egea-Gilabert, C. Genotype by environment interactions in cowpea (Vigna unguiculata L. Walp.) grown in the Iberian Peninsula. Crop Past. Sci. 2017, 68, 924–931. [Google Scholar] [CrossRef]
- Ddamulira, G.; Santos, C.A.F.; Alanyo, M.; Ramathani, I.; Maphosa, M. Maturity, protein content and yield stability of cowpea in Uganda. SA J Plant Soil. 2017, 34, 255–261. [Google Scholar] [CrossRef]
- Yan, W.; Kang, M.S. GGE biplot analysis: A graphical tool for breeders. In Geneticists, and Agronomists; Kang, M.S., Ed.; CRC Press: Boca Raton, FL, USA, 2003; pp. 63–88. [Google Scholar]
- Yan, W.; Tinker, N.A. Biplot analysis of multi-environment trial data: Principles and applications. Can. J. Plant Sci. 2006, 86, 623–645. [Google Scholar] [CrossRef]
- Hall, A.E.; Thiaw, S.; Ismael, A.M.; Ehlers, J.D. Water-use efficiency and drought adaptation in cowpea. In Advances in Cowpea Research; Singh, B.B., Mohan Raj, D.R., Dashiell, K.E., Jackai, L.E.N., Eds.; Co-publication of International Institute of Tropical Agriculture (IITA) and Japan International Research Center for Agricultural Sciences (JIRCAS): Tsukuba, Japan; IITA: Ibadan, Nigeria, 1997; pp. 87–98. [Google Scholar]
- Kindie, Y.; Tesso, B.; Amsalu, B. Genotype X Environment Interaction and Yield Stability in Early-Maturing Cowpea (Vigna unguiculata (L.) Walp.) Landraces in Ethiopia. Adv. Agric. 2021, 2021, 3786945. [Google Scholar] [CrossRef]
- Santos, A.; Ceccn, G.; Rodrigues, E.V.; Teodoro, P.E.; Makimo, P.A.; Alves, V.B.; Silva, J.F.; Correa, A.M.; Alvares, R.C.F.; Torres, F.E. Adaptability and stability of cowpea genotypes to Brazilian Midwest. Afr. J. Agric. Res. 2015, 10, 3901–3908. [Google Scholar]
- Moges, A. Genotype by Environment Interaction on Yield and Micronutrient Concentration of Biofortified Common Bean (Phaseolus vulgaris L.) in Ethiopia. MSc Thesis, Haramaya University, Haramaya, Ethiopia, 2017. [Google Scholar]
- El-Shaieny, A.A.H.; Abdel-Ati, Y.A.A.; El-Damarany, M.E.D.; Rashwan, M.R. Stability analysis of components characters in cowpea (Vigna unguiculata (L.) Walp). J. Hortic. For. 2002, 7, 24–35. [Google Scholar] [CrossRef][Green Version]
- Firew, M. Simultaneous selection for high yield and stability in common bean (Phaseolus vulgaris) genotypes. J. Agric. Sci. 2002, 138, 249–253. [Google Scholar]
- Mesfin, H.M. Genotype x Environment Interaction and Stability Analysis in Soybean [Glycine max (L.) Merrill] for Grain Yield in Ethiopia. Master’s Thesis, Department of Horticulture and Plant Science, Jimma University College of Agriculture and Veterinary Medicine, Jimma University, Jimma, Ethiopia, 2017. [Google Scholar]
- Sharma, P.; Baranda, B.; Haritwal, S.; Sharma, M. Character association for seed yield and its components in cowpea [Vigna unguiculata (L.) Walp]. Int. J. Curr. Microbiol. Appl. Sci. 2017, 6, 967–975. [Google Scholar]
- Owusu, E.Y.; Karikari, B.; Kusi, F.; Haruna, M.; Richard AAmoah, R.A.; Attamah, P.; GAdazebra, G.; Sie, E.K.; Issahaku, M. Genetic variability, heritability and correlation analysis among maturity and yield traits in Cowpea (Vigna unguiculata (L) Walp) in Northern Ghana. Heliyon 2021, 7, e07890. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).