Abstract
Cultivated peanut (Arachis hypogaea L.) is one of the most important oilseed crops worldwide. Pod-related traits, including pod length (PL), pod width (PW), ratio of PL to PW (PL/W) and 100-pod weight (100-PW), are crucial factors for pod yield and are key target traits for selection in peanut breeding. However, the studies on the natural variation and genetic mechanism of pod-related traits are not clear in peanut. In this study, we phenotyped 136 peanut accessions for four pod-related traits in two consecutive years and genotyped the population using a re-sequencing technique. Based on 884,737 high-quality single nucleotide polymorphisms (SNPs), genome-wide association studies (GWAS) were conducted for four pod-related traits using a fixed and random model uniform cyclic probability (FarmCPU) model. The results showed that a total of 36 SNPs were identified by GWAS, among which twenty-one, fourteen and one SNPs were significantly associated with PL, PL/W and 100-PW, respectively. The candidate regions where the four peak SNPs (10_76084075, 11_138356586, 16_64420451, and 18_126782541) were located were used for searching genes, and nineteen candidate genes for pod-related traits were preliminarily predicted based on functional annotations. In addition, we also compared the expression patterns of these nineteen candidate genes in different tissues of peanut, and we found that eight genes were specifically highly expressed in tender fruit, immature pericarp, or seed, so we considered these genes to be the potential candidate genes for pod-related traits. These results enriched the understanding of the genetic basis of pod-related traits and provided an important theoretical basis for subsequent gene cloning and marker-assisted selection (MAS) breeding in peanut.
1. Introduction
Peanut (Arachis hypogaea L.) is one of the most important oil crops in the world, providing rich edible oil and protein for human beings. Cultivated peanut is an allotetraploid (AABB, 2n = 40) crop formed by natural interspecific hybridization between A. duranensis (AA genome) and A. ipaensis (BB genome) [1,2]. It is widely cultivated in more than 100 countries throughout the world and has been used for processing food, extracting vegetable oil and feed, etc., and has significant economic value [3,4,5]. With the increasing demand for peanut products and edible oil, the global peanut harvested area and annual production have increased rapidly from 2010 (26.14 Mha and 43.45 Mt) to 2020 (31.57 Mha and 53.64 Mt) (http://faostat.fao.org/, accessed on 20 March 2023). Nevertheless, peanut production is still struggling to meet the growing consumption demand [6]. Increased yield has always been the primary goal for peanut breeding. Genetic improvement of yield-related traits is the key to increase the yield potential of new peanut varieties and contribute to achieving the goal of sustainable agriculture [7,8].
Yield is a complex quantitative trait, and for peanut, pod- and kernel-related traits are the direct factors affecting yield. The pod is the harvesting organ of peanut, and pod-related traits-pod length (PL), pod width (PW), ratio of PL to PW (PL/W) and 100-pod weight (100-PW)-directly determine the yield and commerciality of peanut [9,10,11]. It is generally believed that small pods or low 100-PW is the main factor of low average yield per unit area of peanut, and breeding high–yielding varieties with large pods is one of the main goals of peanut genetic improvement [3,11,12]. However, according to market consumption demand, different pod types have different uses. For example, when peanut is used for extracting vegetable oil or as fresh food, large-pod varieties tend to be more economical, whereas in the roasted shelled peanut processing industry, the market prefers peanut varieties with smaller pods. Pod size and weight are important for peanut breeding and production, so a better understanding of the genetic mechanisms for pod-related traits would benefit the genetic improvement of these traits.
Pod-related traits are complex quantitative traits controlled by multiple quantitative trait loci (QTLs) / genes and are influenced by environmental factors. QTL mapping based on bi-parental population is a traditional method to analyze the genetic basis of target traits [6]. In recent years, with the development of molecular marker detection techniques and the improvement of QTL detection methods, many important advances have been made in the research on the QTL mapping of pod-related traits in peanut [3,11,12,13,14,15]. Although QTLs for pod-related traits have been extensively studied in previous studies, due to the few molecular markers and low polymorphism, the mapping accuracy of QTLs is not high enough [6]. Therefore, it is difficult to apply these QTLs to subsequent gene cloning and molecular breeding.
With the development of whole genome re-sequencing technology, the acquisition of high–throughput SNP markers has become more and more viable, which provides strong support for the study of the genetic basis of quantitative genetic traits at the molecular level [16]. A genome-wide association study (GWAS) is an effective method developed in recent years for studying complicated traits [17]. Traditional QTL mapping usually requires the construction of bi-parental populations, which takes a long time and has low positioning accuracy [18]. However, GWAS based on natural populations can be directly used for the mapping of target traits, which saves time, simultaneously detects multiple alleles and has the advantages of high positioning accuracy [19,20,21]. In addition, natural populations have a wide range of genetic variation, and GWAS makes full use of a large number of historical recombination events in natural populations, with a higher detection accuracy than linkage mapping by detecting genome-wide high–density molecular markers associated with phenotypes [22]. At present, GWAS has been successfully applied in order to research agronomic traits in wheat [23], rice [24,25], maize [26,27], and soybean [28]. In recent years, as the genomes of different peanut materials have been reported, the research on the GWAS of peanut has gradually increased, especially for yield-related traits [6,8,17,29,30,31]. Zhao et al. (2017) utilized 554 SSR markers to perform an association analysis on four seed-related traits in 104 peanut accessions, and identified 30 significant associated markers, which accounted for phenotypic variation ranging from 11.22% to 32.30% [6]. Wang et al. (2019) conducted a GWAS on seven yield-related traits in 195 peanut accessions and identified 93 candidate regions and 36 candidate genes related to hundred-seed weight (100-SW), hundred-pod weight (100-PW), yield per plant (YP), and pod branch number per plant (PBP) [32]. Gangurde et al. (2020) utilized the NAM method to perform GWAS analysis on two yield-related traits and revealed candidate regions and candidate genes that could potentially contribute to pod and seed weight [33]. Recently, Patel et al. (2022) performed an association analysis using the Axiom_Arachis array on the U.S. peanut mini-core collection and identified various genomic regions associated with mature nucleus, seed weight, molting rate, seed germination, and dormancy [34]. Although many studies have been carried out for pod-related traits, their genetic mechanism remains unclear.
In order to elucidate the genetic mechanism of pod-related traits in peanut, 136 peanut accessions were genotyped by whole–genome re-sequencing in this study; GWASs were performed on four pod-related traits in two environments to identify genomic regions that were significantly associated with pod-related traits. In addition, combined with the functional annotation and expression profile of genes in the PeanutBase database, candidate genes of important associated SNP loci were discovered. This study provides a theoretical basis for the genetic improvement and marker-assisted selection (MAS) breeding of pod-related traits in peanut.
2. Materials and Methods
2.1. Plant Materials
The experimental material used for association mapping was a natural population consisted of 136 peanut accessions collected from 16 provinces in China. The natural population included 82 landraces and 54 cultivars. Most of these cultivars were excellent accessions from different provinces in northern China, some of which were used for large-scale production and generally belong to the large pod type. The landraces were mostly from different provinces in southern China, were planted in special areas, and generally belong to the small pod type. This population consisted of six A. hypogaea var. hypogaea, 99 var. vulgaris, eight var. hirsuta, 19 var. fastigiata and four irregular types. These irregular types are different from the above four types and may be intermediate types produced by hybridization. The detailed information of each accession was also listed in Table S1.
2.2. Field Trials and Phenotypic Statistics
All materials were planted in Hangzhou, Zhejiang Province (120.10° E, 30.16° N) in 2021, and in Lingshui, Hainan Province (110.04° E, 18.51° N) in 2022. E1 and E2 represent the environments of Hangzhou in 2021 and Lingshui in 2022, respectively. A randomized block design with three replicates was adopted for the experiment. Two rows were planted with a length of 2.0 m and a spacing of 30.0 cm. After maturation, the pods were dried and phenotypic identification of four pod-related traits was performed according to the investigation criteria [35].
The MEANS procedure of SAS was used for statistical analysis of phenotypic data in the two environments [36]. Analysis of variance (ANOVA) and correlation analysis of four pod-related traits were performed based on SAS PROC generalized linear model (GLM) and PROC CORR programs, respectively. The function lmer from the package lme4 in R was used to estimate the best linear unbiased predictive value (BLUP) for the four traits [37]. The BLUP values were used for association mapping and hunting the genetic loci as phenotypic data. Broad-sense heritability (h2) was calculated in R. The formula for h2 was as follows:
where σ2g is the genotype variance, σ2ge is the interaction of genotype and environment variance, σ2e is the residual error, n is the number of environments and r is the number of replicates within the environment.
2.3. Whole-Genome Re-Sequencing and SNP Genotyping
Genomic DNA of 136 peanut accessions was extracted by cetyltrimethyl ammonium bromide method. The DNA sample from each sample was used to construct a library with an insert size of approximately 350 bp. Paired-end sequencing libraries were constructed using Truseq Nano DNA HT Sample preparation Kit (Illumina, San Diego, California, USA) following manufacturer’s recommendations. The 136 peanut accessions were re-sequenced based on the Illumina Novaseq 6000 platform with 5 × coverage. To avoid reads with artificial bias, adaptors and low–quality reads were removed using strictly followed filtering criteria: (1) reads with ≥10% unidentified nucleotides; (2) reads with >10 nucleotides aligned to the adapter, allowing ≤10% mismatches; (3) reads with >50% bases had a Phred score < 5; and (4) there were putative PCR duplicates generated by PCR amplification in the library construction process. High-quality reads were compared with the Tifrunner reference genome (arahy.Tifrunner.gnm2) using burrows wheeler aligner (BWA) software (v.0.7.8) [38]. The results were converted into BAM files and indexed using SAMtools software (v.1.3) [39]. In order to obtain high–quality SNPs, the screening parameters were set as follows: coverage depth ≥ 2, minor allele frequency (MAF) > 0.01, and missing ratio ≤ 20%. All high–quality SNPs were annotated using ANNOVAR [40].
2.4. Phylogenetic Tree and Population Structure Analysis
In order to clarify the phylogenetic relationships among the materials in the population, the neighbor-joining (NJ) tree was constructed using TreeBeST software (v.1.9.2) [41]. The population structure of 136 individuals was analyzed based on Admixture (ver. 1.3.0) [42]. Principal component analysis (PCA) of peanut population was performed by GCTA software (ver. 1.92.0) [43].
2.5. Linkage-Disequilibrium Analysis
We used the Haploview software (v.4.2) to calculate the square correlation coefficient (r2) between pairwise SNPs to assess linkage disequilibrium (LD) of this population. The program takes ‘-n-dprime -min MAF 0.01’ as a parameter. Average r2 values for pairwise markers were calculated in a 500 kb window and averaged across the genome.
2.6. Genome-Wide Association Analysis
GWAS was carried out with a fixed random model cyclic probability unification (FarmCPU) model by GAPIT in the R package [44]. The principal components and the kinship matrix were used to control the influence of population structure on association analysis. Based on 884,737 high-quality SNPs, the multi-locus linear mixed model FarmCPU was used to identify significant SNPs for the four pod-related traits. The threshold for significant associations was set to 1/n, where n is the number of markers and the p value is <1/n, or –log10 (p) ≥ 5.95 [45]. The visualization of Manhattan plot and Q-Q plot was realized by using the R package qqman.
2.7. Candidate Genes Prediction
To reduce false positives, we selected peak SNPs that strongly associated with target traits and searched for genes within relevant candidate regions based on LD decay distance (±160.3 kb). Furthermore, we predicted candidate genes of target traits based on functional annotations of genes in the PeanutBase database (https://www.peanutbase.org/, accessed on 6 July 2023). In addition, the expression patterns of candidate genes in different peanut tissues in the PeanutBase database were downloaded to further identify possible candidate genes. The peanut reference genome was arahy.Tifrunner.gnm2.
3. Results
3.1. Analysis of Phenotypic Variations
In this study, the phenotypic identification of four pod-related traits was performed in 136 peanut accessions for two consecutive years to evaluate the phenotypic variation of pod-related traits. The results showed that the four pod-related traits showed large phenotypic variation, and the variation values of each trait were different in different environments (Table 1). For example, the phenotypic variation for PL in the E1 and E2 environments ranged from 20.70 to 56.47 mm and 24.19 to 49.63 mm, respectively. The four pod-related traits of the 136 peanut accessions showed continuous variation, showing approximately normal distribution, which was consistent with the genetic characteristics of quantitative traits (Figure 1).

Table 1.
Phenotypic variation for four pod-related traits of 136 peanut accessions under two environments.
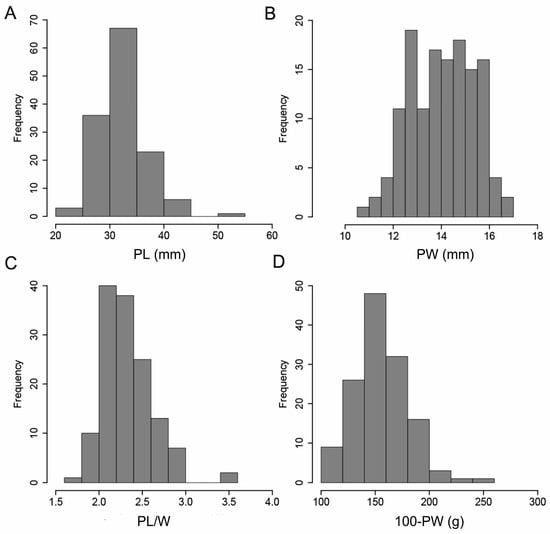
Figure 1.
Frequency distribution of phenotypic variation of four pod-related traits in 136 peanut accessions. (A–D) represent the frequency distributions of PL, PW, PL/W, and 100-PW, respectively. The averaged phenotype value of two environments for each trait was used.
The results of the ANOVA showed that PL, PL/W and 100-PW had significant differences in genotype, environment and genotype–environment interaction, while PW only had significant differences in genotype and genotype–environment interaction but no significant differences in environment. These results indicate that PL, PL/W and 100-PW are more affected by environmental factors. The broad-sense heritability of the four pod-related traits ranged from 68.22% to 90.38%, indicating that genetic effect was the main factor affecting phenotypic variation. The results of the correlation analysis showed that, except for the extremely significant negative correlation between PW and PL/W, there was a significant positive correlation between any two pairs of other pod-related traits (Table 2).

Table 2.
Correlation analysis of four pod-related traits under two environments.
3.2. Genomic Variation
By re-sequencing the whole genome of 136 peanut accessions, a total of 2082.07 Gb of high–quality sequences were obtained, in which the effective rate range was 99.27–99.73%, and the average sequencing depth reached 4.81× . The Q30 percentages for each accession ranged from 87.86% to 92.43%, and the GC content ranged from 35.74% to 39.31%. Compared with the peanut reference genome (arahy.Tifrunner.gnm2), the effective mapping rate ranged from 95.16% to 99.85%, and the 4× genome coverage ranged from 7.29 to 70.64%. The re-sequencing quality analysis of 136 peanut accessions was shown in Table S2. The high-quality sequencing data in this study ensured the accuracy and reliability of subsequent analyses.
After using the following filtering parameters: depth ≥ 2, MAF > 0.01, and missing rate ≤ 20%, a total of 904,183 high-quality SNPs were obtained (Table 3). These SNPs were mapped to 20 chromosomes of peanut, and 884,737 SNPs were used for subsequent GWAS analysis. The 884,737 SNPs were unevenly distributed on 20 chromosomes of peanut, and the number of SNP markers on each chromosome ranged from 14,421 to 76,254, with an average of 44,236 (Figure 2A,B and Table S3). In the whole genome of peanut, the average marker density was 384.28 SNPs/Mb, the highest marker density was 478.5 SNPs/Mb on chromosome 19, and the lowest marker density was 279.86 SNPs/Mb on chromosome 8. The average MAF of all markers was 0.13, and 26.96% SNPs exhibited an MAF higher than 0.2 (Figure 3A). The polymorphism information content (PIC) ranged from 0.02 to 0.38, with an average of 0.15 (Figure 3B and Table S3).

Table 3.
Summary of categorized SNPs in peanut population.
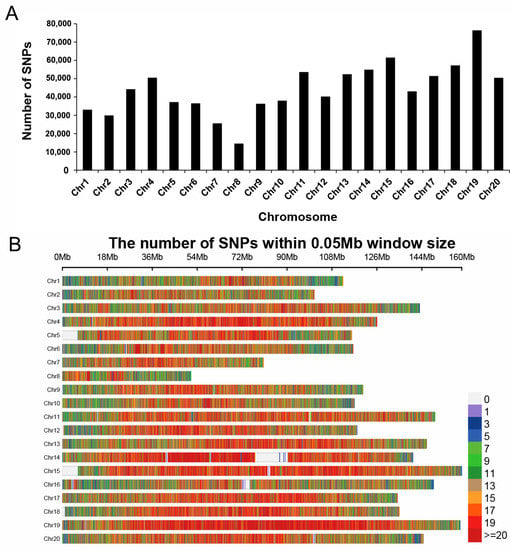
Figure 2.
SNP markers density and distribution across the genome of peanut. (A) The distribution of SNP markers on 20 chromosomes of peanut. (B) SNP markers density on each chromosome. The horizontal axis shows chromosome length (Mb), and the vertical axis shows the 20 chromosomes.
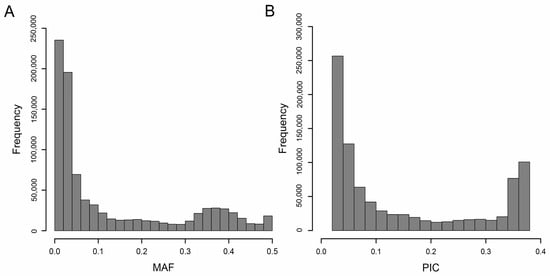
Figure 3.
Minor allelic frequency (MAF) and polymorphism information content (PIC) analysis based on 884,737 SNPs in 136 peanut accessions. (A) Distribution of minor allele frequency (MAF). (B) Polymorphism information content (PIC).
3.3. Population Structure and Linkage Disequilibrium Analysis
To elucidate the genetic and evolutionary relationships among 136 peanut accessions, population structure analysis, phylogenetic analysis, principal component analysis and linkage disequilibrium attenuation analysis were conducted based on 884,737 SNPs. The Admixture software (ver. 1.3.0) was used to evaluate the population structure of 136 peanut accessions with K values ranging from 2 to 8. When K = 5, the cross validation (CV) error value was at the inflection point and showed the lowest value, indicating that the population could be divided into five subpopulations (Figure 4A,B and Figure S1). The five subpopulations were named A, B, C, D and E, among which the A subpopulation contained 28 peanut accessions, and 25 belonged to A. hypogaea var. vulgaris. Among the 23 peanut accessions contained in B subpopulation, 22 belonged to var. fastigiata. Among the eleven peanut accessions contained in C subpopulation, seven belonged to var. hirsuta. D is the mixed subpopulation, which consists of eighteen var. fastigiata, seventeen var. vulgaris and seven var. hirsuta. Among the 27 peanut accessions contained in subpopulation E, 23 belonged to var. vulgaris. It can be seen that there is a certain correlation between population structure and peanut subspecies classification. In addition, phylogenetic tree and principal component analysis showed that the results were basically consistent with the population structure (Figure 4C,D). Therefore, in order to improve operational efficiency, the principal component analysis matrix will be used to replace the Q matrix at K = 5 for subsequent GWAS.
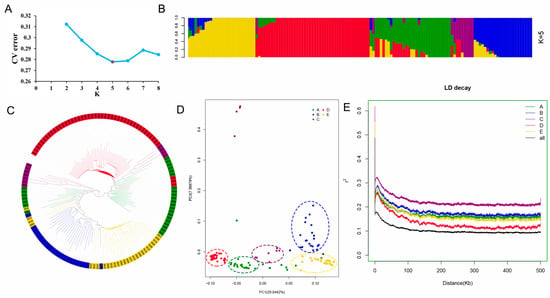
Figure 4.
Population structure and LD analysis of 136 peanut accessions. (A) Calculation of the true K value in the association population. (B) Population structure analysis with K = 5, each accession is represented by a single vertical line and colors represent ancestries. (C) A maximum likelihood neighbor-joining tree of the 136 peanut accessions. (D) Principal component analysis (PCA) for all peanut accessions. Each dot represents one accession, and the colors of the virtual circle match the structure grouping. (E) LD decay plots for different subgroups (A–E) and all peanut accessions.
In addition, PopLDdecay software was used to calculate the r2 values of all pairs of SNPs, and the LD levels of peanut accessions population were estimated according to the r2 values. When r2 reduced by half, the average LD decay distance was 160.3 kb (Figure 4E). Further LD analysis of the five subpopulations showed that the LD decay of subpopulation D was faster than that of the other four subpopulations (Figure 4E).
3.4. Genome-Wide Association Analysis of Pod-Related Traits
In total, 884,737 high-quality SNPs were used to perform a GWAS for four pod-related traits using the BLUP values across two environments using FarmCPU model by GAPIT in the R package. Using −log10 (p) ≥ 5.94 as the significance threshold, 36 SNPs passed the threshold and were significantly associated with three pod-related traits, including twenty-one, fourteen, and one SNPs for PL, PL/W, and 100-PW, respectively (Figure 5 and Table 4). The 21 SNPs significantly associated with PL were mainly distributed on 12 chromosomes of peanut, including chromosomes 7, 10, 12, 14, 18, etc. For the PW trait, we did not find any SNPs that passed the threshold and were significantly associated (Figure S2). A total of 14 SNPs located on chromosomes 4, 10, and 16, respectively, were indicated to be associated with the trait PL/W. For the 100-PW trait, the SNP 11_138356586 was present on chromosome 11. Among these SNPs, 18_126782541 for PL, 10_76084075 and 16_64420451 for PL/W, and 11_138356586 for 100-PW were peak SNPs that strongly associated with these traits in the Manhattan plot (Figure 5). Therefore, these four SNP loci have the potential to be used for regulating peanut pod-related traits for further analysis.
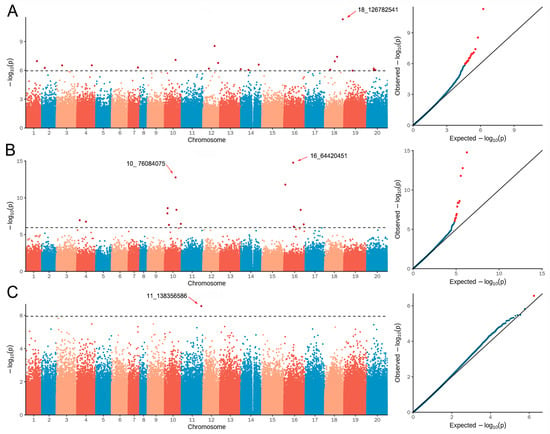
Figure 5.
Presentation of Manhattan and Q-Q plots for the three pod-related traits in peanut. (A–C) represent association mapping of PL, PL/W, and 100-PW based on the BLUP values, respectively. For each trait, the Q-Q plot from FarmCPU model is also shown in the right. The horizontal dashed line indicates the genome-wide significance threshold (−log10 (p) ≥ 5.95), and the four peak SNPs are shown through the red arrows.

Table 4.
GWAS significant SNPs associated with pod-related traits in peanut.
3.5. Prediction of Candidate Genes
According to the average LD decay distance (LD ± 160.3 kb), genes were searched in the candidate regions where the four significant association peak SNPs (10_76084075, 11_138356586, 16_64420451, and 18_126782541) were located. According to the functional annotation of the genes, a total of nineteen candidate genes were found for pod-related traits, including six, five, and eight candidate genes for PL, PL/W, and 100-PW, respectively. These candidate genes mainly encode the F-box/kelch-repeat protein, argonaute family protein, zinc knuckle family protein, pentatricopeptide repeat (PPR) superfamily protein, calmodulin-binding family protein, glutathione S-transferase family protein, and cell wall protein. Detailed functional annotation information for these candidate genes is shown in Table 5.

Table 5.
Functional annotation of 19 candidate genes for pod-related traits in peanut.
The expression patterns of these candidate genes in different peanut tissues were downloaded from the peanut database, and comparative analysis showed that eight candidate genes were highly expressed in tender fruit, immature pericarp or seed (Figure 6). Among these candidate genes, three genes (arahy.0DV40D, arahy.N83A6V and arahy.AAI9J8) were specifically highly expressed in tender fruit and immature pericarp, and two genes (arahy.693TR4 and arahy.UCE8LX) were specifically highly expressed in immature seed. The candidate gene arahy.EJ6VAJ was specifically highly expressed in immature pericarp and the root tissues of peanut, and the candidate gene arahy.WI6ZIF was specifically highly expressed in leaves and the immature pericarp tissues of peanut. In addition, the candidate gene arahy.6HP7HZ was specifically highly expressed in tender fruit.
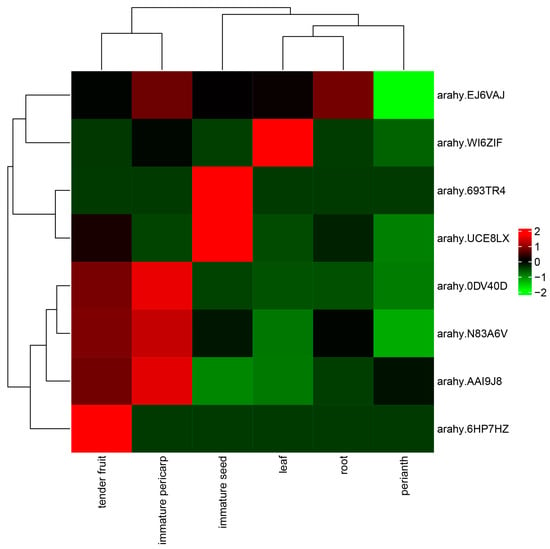
Figure 6.
Heatmap profiles of the candidate genes in different tissues of peanut. The color scale represents increased (red) and decreased (green) fold change in the levels of candidate gene transcripts. The horizontal axis represents different peanut tissues (immature seed, root, tender fruit, immature pericarp, leaf and perianth), and the vertical axis stands for different candidate genes.
4. Discussion
Germplasm resources are the basic materials of peanut genetic breeding research. Fully exploiting and utilizing the excellent traits and genes in peanut germplasm resources is the basis and key to breeding new peanut varieties. With the development of high-throughput sequencing technology, the acquisition cost of molecular markers has been continuously reduced, and the analysis of the genetic basis of quantitative traits in peanut has been accelerated [47]. Zhang et al. (2017) genotyped 158 peanut materials by the SLAF-seq method, obtaining a total of 17,338 high-quality SNPs, and analyzed the genetic basis of 11 domestication-related traits by GWAS [48]. Genotyping-by-sequencing (GBS) technology was used to genotype 195 peanut materials and perform GWAS analysis on seven yield-related traits [32]. Zou et al. (2020) genotyped 384 peanut germplasm resources using 58 K SNP array to assess the evolutionary relationship between different materials and to detect the genetic loci related to seed aspect ratio [49]. Recently, Liu et al. (2022) resequenced 203 peanut materials and analyzed the molecular basis related to seed traits through GWAS and transgenic experiments. The sequencing depth of each material in the re-sequencing reached 14.16×, which is the highest sequencing depth of peanut so far [4]. In this study, 136 peanut accessions were genotyped using whole–genome re-sequencing technology, with an average sequencing depth of 4.81×, which has a high sequencing depth with regard to the current peanut genome research. After that, the obtained genotypic data were filtered through coverage depth ≥ 2, MAF > 0.01 and missing rate ≤ 20%, resulting in 884,737 high-quality SNPs. With the re-sequencing, assembly and detailed annotation of reference genomes of different cultivated peanuts, the efficiency of GWAS research has been greatly improved [2,50,51]. Therefore, the genetic information obtained from the 136 peanut accessions based on whole–genome re-sequencing has laid a certain foundation for peanut genomics and genetic research.
Previous studies have confirmed that pod-related traits are important components of peanut yield, which are multi-gene controlled quantitative traits and are susceptible to environmental factors [9,10]. There were extensive phenotypic variations in the four pod-related traits in the 136 peanut accessions, and the generalized heritability ranged from 68.22 to 90.38%. The generalized heritability of 100-PW was the smallest, and that of PL was the largest. Compared with previous studies, there was no significant difference in the heritability of those traits, except for 100-PW, which may be related to the greater influence of 100-PW by environmental factors [11,46]. Correlation analysis showed that in addition to the extremely significant negative correlation between PW and PL/W, a high positive correlation was observed in other pairs of the four pod-related traits (Table 2). This indicates that increasing PL and PW can improve 100-PW to a certain extent in peanut. Through the analysis of phenotypic variation, we believed that this population was suitable for the genetic analysis of the four pod-related traits.
According to previous reports, QTLs for pod-related traits were mostly obtained through linkage mapping, and were mainly distributed on chromosomes A02, A05, A06, A07, A07, B04, B05, B06, and B07 of peanut [11,12,16,33,46]. However, linkage mapping usually involves only two parents and can detect less genetic variation, making it difficult to detect QTL and genetic variation comprehensively [17]. In contrast, a GWAS uses a large number of recombination events in natural populations to scan the markers associated with the phenotype of target traits in the whole genome, and the detection accuracy is higher [22]. In this research, the GWAS revealed 36 SNPs significantly associated with three pod-related traits in peanut using the FarmCPU model. However, we indeed failed to detect the SNP loci that were significantly associated with the PW trait (Figure S2). We speculate that the genetic complexity of the PW trait and the lack of a major QTL are the main reasons for the absence of significantly associated SNPs and may also be related to the strict threshold applied in this study [28]. Among the SNPs significantly associated with PL, two SNPs (7_66473592 and 7_66473593) located on chromosome 7 are within three reported QTLs of other pod-related traits (qPLA07, qPAA07_B07.3, and qHPWA07.1) [11,12,46]. The SNP 10_76084075 on chromosome 10 was colocalized with the pod width QTL (qPWA10.2) reported by Chen et al. (2021). The SNP 14_124629353 on chromosome 14 was located within two QTLs that have been reported to be associated with pod weight [46]. For PL/W, The SNP 4_19502508 on chromosome 4 was located within the QTL (qPA04.2) that has been reported to be associated with pod area, and two SNPs (16_116682980 and 16_137494047) located on chromosome 16 were located within four reported QTLs for pod- and kernel-related traits [16,33]. In summary, we detected thirty-six SNPs that were significantly associated with the three pod-related traits in this study, only seven were located within QTLs of reported pod-related traits, and twenty-nine were new loci identified in this study, which may be related to the population and environment used in association mapping (Table 4).
The candidate interval was determined by the LD decay distance, which is related to species reproduction mode, population size, artificial selection and other factors [52]. The average LD decay distance of the population used in this study was 160.3 kb, which was lower than the LD decay distance of the reported peanut populations, and the positioning accuracy was good [6,17,53]. The 136 peanut accessions were geographically widely sourced, which may be the reason for the relatively low LD in this study. In addition, population size and molecular marker density are also two important factors affect the accuracy of a GWAS [27,54]. Theoretically, the larger population size and higher accuracy are the result of the GWAS, but in practice, the population can not be infinite expanded. Although the population used for association mapping in this study was not large, the application of high–density SNPs to a certain extent compensated the shortcomings of the study. According to the association mapping results in this study, four peak SNPs (10_76084075, 11_138356586, 16_64420451, and 18_126782541) for pod-related traits were used to search genes in the candidate region (LD ± 160.3 kb), and 19 candidate genes were preliminarily identified based on functional annotation.
Further, we compared the expression patterns of these candidate genes in different tissues of peanut and found that eight genes were specifically highly expressed in tender fruit, immature pericarp or seed; so, we believe that these eight genes have great potential as candidate genes for pod-related traits. The gene arahy.EJ6VAJ encodes mRNA-decapping enzyme-like protein, which has been reported to regulate wheat root development by participating in ABA signal transduction [55,56] and may affect peanut PL through ABA regulation and root development. Argonaute (AGO) proteins interact with miRNAs to form silencing complexes that regulate gene expression [57]. In rice, OsAGO17 might be a critical protein in the sRNA pathway and could positively improve grain size and weight and promote stem development [58]. The gene arahy.WI6ZIF encodes AGO protein, which may regulate peanut PL development through the sRNA pathway. The F-box gene family is one of the largest superfamilies of regulatory proteins that notably regulate various biological processes related to plant organ growth development, hormone signaling pathways and stress responses [59,60]. The gene arahy.AAI9J8 encodes F-box/kelch-repeat protein, which could regulate the development of floral organs and pods through hormones, and it may eventually lead to changes in PL.
The genes arahy.UCE8LX and arahy.693TR4 are PL/W candidate genes that encode the kinase interacting (KIP1-like) family protein and MYB/SANT-like DNA-binding domain protein, respectively. In tomato, the SD1 gene encodes a kinase-interacting protein, which positively regulates stem development, and there is a significant correlation between stem diameter and fruit size [61]. The Lefsm1 gene contains the MYB/SANT domain, whose ectopic expression leads to developmental alterations in tomato and Arabidopsis. It may also be involved in the regulation of plant-specific developmental programs [62]. The genes arahy.UCE8LX and arahy.693TR4 may control PL/W through stem development and the early stages of seed development.
For 100-PW, there are three potential candidate genes, arahy.6HP7HZ, arahy.N83A6V and arahy.0DV40D, which encode pentatricopeptide repeat (PPR) superfamily protein, calmodulin-binding family protein and peroxidase superfamily protein, respectively. In maize, the Dek39 gene encodes an E sub-class PPR protein that is involved in the RNA editing of multiple sites and is necessary for seed development and reduced kernel size [63]. IQM4 is a novel calmodulin-binding protein that participates in seed dormancy and germination by controlling ABA biosynthesis genes and seed maturation regulators in Arabidopsis [64]. In Arabidopsis, APx-R is a plastidial ascorbate-independent peroxidase regulated by photomorphogenesis, which is involved in seed germination by controlling the rate of seed germination [65]. Therefore, these three candidate genes may control the development of the pod through RNA editing and the regulation of hormone levels, thus increasing 100-PW.
5. Conclusions
In this study, we used a natural population of 136 peanut accessions as experimental materials, we used the FarmCPU model to perform association mapping for pod-related traits, and we identified associated loci of three pod-related traits in the whole genome. We selected four peak SNPs and searched for genes in their candidate regions, and we identified a total of 19 candidate genes for pod-related traits, combined with gene functional annotation. A further comparison of the expression patterns of these candidate genes in different tissues of peanut revealed that eight genes were specifically highly expressed in tender fruit, immature pericarp or seed. These genes were considered to be the potential candidate genes, but further functional studies are needed to confirm this hypothesis. The significant associated SNPs and candidate genes identified in this study will help to analyze the genetic mechanism of pod-related traits, and they lay a theoretical foundation for the genetic improvement and marker-assisted selection breeding of pod-related traits in peanut.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/agronomy13071863/s1, Table S1: Information of 136 peanut accessions in the present study. Table S2: Re-sequencing quality of the 136 peanut accessions. Table S3: Summary of the polymorphic markers used for association mapping on the 20 chromosomes of peanut. Figure S1: Structure clustering results obtained at K = 2 to K = 8 of the 136 peanut accessions. Figure S2: Presentation of Manhattan and Q-Q plots for PW in peanut. The horizontal dashed line indicates the genome-wide significance threshold (−log10 (p) ≥ 5.95).
Author Contributions
Conceptualization, X.Z., L.Z. and M.R.; investigation, C.X. and D.S.; resources, X.T. and Y.X.; formal analysis, X.Z.and L.Z.; data curation, L.Z. and M.R.; methodology, X.Z.; software, X.Z. and L.Z.; funding acquisition, X.Z. and F.L.; writing—original draft, X.Z.; writing—review and editing, X.Z. and F.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (32201797), International Cooperation Project of Zhejiang Academy of Agricultural Sciences (peanut).
Data Availability Statement
The datasets supporting the results of this article are included within the main text and the Supplementary Materials.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Fávero, A.P.; Simpson, C.E.; Valls, M.; Velo, N.A. Study of evolution of cultivated peanut through cross ability studies among Arachis ipaensis, A. duranensis and A. hypogaea. Crop Sci. 2016, 46, 1546–1552. [Google Scholar] [CrossRef]
- Zhuang, W.; Chen, H.; Yang, M.; Wang, J.; Pandey, M.K.; Zhang, C.; Chang, W.; Zhang, L.; Zhang, X.; Tang, R.; et al. The Arachis hypogaea genome elucidates legume karyotypes, polyploid evolution and crop domestication. Nat. Genet. 2019, 51, 865–876. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; Ren, X.; Li, Z.; Xu, Z.; Li, X.; Huang, L.; Zhou, X.; Chen, Y.; Chen, W.; Lei, Y.; et al. Co-localization of major quantitative trait loci for pod size and weight to a 3.7 cM interval on chromosome A05 in cultivated peanut (Arachis hypogaea L.). BMC Genom. 2017, 18, 58. [Google Scholar] [CrossRef]
- Liu, Y.; Shao, L.; Zhou, J.; Li, R.; Pandey, M.K.; Han, Y.; Cui, F.; Zhang, J.; Gao, F.; Chen, J.; et al. Genomic insights into the genetic signatures of selection and seed trait loci in cultivated peanut. J. Adv. Res. 2022, 42, 237–248. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Luo, H.; Yu, B.; Huang, L.; Liu, N.; Chen, W.; Liao, B.; Lei, Y.; Huai, D.; Guo, P.; et al. Genetic dissection of fatty acid components in the Chinese peanut (Arachis hypogaea L.) mini-core collection under multi-environments. PLoS ONE 2022, 17, e0279650. [Google Scholar] [CrossRef]
- Zhao, J.; Huang, L.; Ren, X.; Manish, K.P.; Wu, B.; Chen, Y.; Zhou, X.; Chen, W.; Xia, Y.; Li, Z.; et al. Genetic variation and association mapping of seed-related traits in cultivated peanut (Arachis hypogaea L.) using single locus simple sequence repeat markers. Front. Plant Sci. 2017, 8, 2105. [Google Scholar] [CrossRef]
- Varshney, R.K.; Mohan, S.M.; Gaur, P.M.; Gangarao, N.V.; Pandey, M.K.; Bohra, A.; Sawargaonkar, S.L.; Chitikineni, A.; Kimurto, P.K.; Janila, P.; et al. Achievements and prospects of genomics-assisted breeding in three legume crops of the semi-arid tropics. Biotechnol. Adv. 2013, 31, 1120–1134. [Google Scholar] [CrossRef]
- Zhang, S.; Hu, X.; Miao, H.; Chu, Y.; Cui, F.; Yang, W.; Wang, C.; Shen, Y.; Xu, T.; Zhao, L.; et al. QTL identification for seed weight and size based on a high-density SLAF-seq genetic map in peanut (Arachis hypogaea L.). BMC Plant Biol. 2019, 19, 537. [Google Scholar] [CrossRef]
- Lucia, R.; Gomes, F.; Ângela Celis Lopes, A. Correlations and path analysis in peanut. Crop Breed. Appl. Biotechnol. 2005, 5, 105–110. [Google Scholar] [CrossRef]
- Selvaraj, M.G.; Narayana, M.; Schubert, A.M.; Ayers, J.L.; Baring, M.R.; Burow, M.D. Identification of QTLs for pod and kernel traits in cultivated peanut by bulked segregant analysis. Electron. J. Biotechnol. 2009, 12, 1–10. [Google Scholar] [CrossRef]
- Chen, W.; Jiao, Y.; Cheng, L.; Huang, L.; Liao, B.; Tang, M.; Ren, X.; Zhou, X.; Chen, Y.; Jiang, H. Quantitative trait locus analysis for pod- and kernel-related traits in the cultivated peanut (Arachis hypogaea L.). BMC Genet. 2016, 17, 25. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; Guo, J.; Ren, X.; Chen, W.; Huang, L.; Zhou, X.; Chen, Y.; Liu, N.; Xiong, F.; Lei, Y.; et al. Chromosomes A07 and A05 associated with stable and major QTLs for pod weight and size in cultivated peanut (Arachis hypogaea L.). Theor. Appl. Genet. 2018, 131, 267–282. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; He, H.; Chen, W.; Ren, X.; Chen, Y.; Zhou, X.; Xia, Y.; Wang, X.; Jiang, X.; Liao, B.; et al. Quantitative trait locus analysis of agronomic and quality-related traits in cultivated peanut (Arachis hypogaea L.). Theor. Appl. Genet. 2015, 128, 1103–1115. [Google Scholar] [CrossRef] [PubMed]
- Alyr, M.H.; Pallu, J.; Sambou, A.; Nguepjop, J.R.; Seye, M.; Tossim, H.A.; Sane, D.; Rami, J.F.; Fonceka, D. Fine-mapping of a wild genomic region involved in pod and seed size reduction on chromosome A07 in peanut (Arachis hypogaea L.). Genes 2020, 11, 1402. [Google Scholar] [CrossRef]
- Chu, Y.; Chee, P.; Isleib, T.G.; Holbrook, C.C.; Ozias-Akins, P. Major seed size QTL on chromosome A05 of peanut (Arachis hypogaea) is conserved in the US mini core germplasm collection. Mol. Breed. 2020, 40, 6. [Google Scholar] [CrossRef]
- Zhou, X.; Guo, J.; Pandey, M.K.; Varshney, R.K.; Huang, L.; Luo, H.; Liu, N.; Chen, W.; Lei, Y.; Liao, B.; et al. Dissection of the genetic basis of yield-related traits in the Chinese peanut mini-core collection through genome-wide association studies. Front. Plant Sci. 2021, 12, 637284. [Google Scholar] [CrossRef]
- Flint-Garcia, S.A.; Thornsberry, J.M.; Buckler, E.S. Structure of linkage disequilibrium in plants. Annu. Rev. Plant Biol. 2003, 54, 357–374. [Google Scholar] [CrossRef]
- Rafalski, J.A. Association genetics in crop improvement. Curr. Opin. Plant Biol. 2010, 13, 174–180. [Google Scholar] [CrossRef]
- Gupta, P.K.; Rustgi, S.; Kulwal, P.L. Linkage disequilibrium and association studies in higher plants: Present status and future prospects. Plant Mol. Biol. 2005, 57, 461–485. [Google Scholar] [CrossRef]
- Mackay, I.; Powell, W. Methods for linkage disequilibrium mapping in crops. Trends Plant Sci. 2007, 12, 57–63. [Google Scholar] [CrossRef]
- Li, H.; Ren, X.; Zhang, X.; Chen, Y.; Jiang, H. Association analysis of agronomic traits and resistance to Aspergillus flavus in the ICRISAT peanut mini-core collection. Acta. Agron. Sin. 2012, 38, 935–946. [Google Scholar]
- Visscher, P.M.; Wray, N.R.; Zhang, Q.; Sklar, P.; McCarthy, M.I.; Brown, M.A.; Yang, J. 10 years of GWAS discovery: Biology, function, and translation. Am. J. Hum. Genet. 2017, 101, 5–22. [Google Scholar] [CrossRef] [PubMed]
- Schilling, S.; Kennedy, A.; Pan, S.; Jermiin, L.S.; Melzer, R. Genome-wide analysis of MIKC-type MADS-box genes in wheat: Pervasive duplications, functional conservation and putative neofunctionalization. New Phytol. 2020, 225, 511–529. [Google Scholar] [CrossRef]
- Huang, X.; Wei, X.; Sang, T.; Zhao, Q.; Feng, Q.; Zhao, Y.; Li, C.; Zhu, C.; Lu, T.; Zhang, Z.; et al. Genome-wide association studies of 14 agronomic traits in rice landraces. Nat. Genet. 2010, 42, 961–967. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Gao, Y.; Xie, W.; Gong, L.; Lu, K.; Wang, W.; Li, Y.; Liu, X.; Zhang, H.; Dong, H.; et al. Genome-wide association analyses provide genetic and biochemical insights into natural variation in rice metabolism. Nat. Genet. 2014, 46, 714–721. [Google Scholar] [CrossRef] [PubMed]
- Buckler, E.S.; Holland, J.B.; Bradbury, P.J.; Acharya, C.B.; Brown, P.J.; Browne, C.; Ersoz, E.; Flint-Garcia, S.; Garcia, A.; Glaubitz, J.C.; et al. The genetic architecture of maize flowering time. Science 2009, 325, 714–718. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Peng, Z.; Yang, X.; Wang, W.; Fu, J.; Wang, J.; Han, Y.; Chai, Y.; Guo, T.; Yang, N.; et al. Genome-wide association study dissects the genetic architecture of oil biosynthesis in maize kernels. Nat. Genet. 2013, 45, 43–50. [Google Scholar] [CrossRef]
- Fang, C.; Ma, Y.; Wu, S.; Liu, Z.; Wang, Z.; Yang, R.; Hu, G.; Zhou, Z.; Yu, H.; Zhang, M.; et al. Genome-wide association studies dissect the genetic networks underlying agronomical traits in soybean. Genome Biol. 2017, 18, 161. [Google Scholar] [CrossRef]
- Pandey, M.K.; Upadhyaya, H.D.; Rathore, A.; Vadez, V.; Sheshshayee, M.S.; Sriswathi, M.; Govil, M.; Kumar, A.; Gowda, M.V.; Sharma, S.; et al. Genome wide association studies for 50 agronomic traits in peanut using the ‘reference set’ comprising 300 genotypes from 48 countries of the semi-arid tropics of the world. PLoS ONE 2014, 9, 105228. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, M.; Dang, P.; Jiang, T.; Zhao, S.; Lamb, M.; Chen, C. Identification of potential QTLs and genes associated with seed composition traits in peanut (Arachis hypogaea L.) using GWAS and RNA-Seq analysis. Gene 2021, 769, 145215. [Google Scholar] [CrossRef]
- Li, L.; Cui, S.; Dang, P.; Yang, X.; Wei, X.; Chen, K.; Chen, C. GWAS and bulked segregant analysis reveal the loci controlling growth habit-related traits in cultivated peanut (Arachis hypogaea L.). BMC Genom. 2022, 23, 403. [Google Scholar] [CrossRef]
- Wang, J.; Yan, C.; Li, Y.; Li, C.; Zhao, X.; Yuan, C.; Sun, Q.; Shan, S. GWAS discovery of candidate genes for yield-related traits in peanut and support from earlier QTL mapping studies. Genes 2019, 10, 803. [Google Scholar] [CrossRef] [PubMed]
- Gangurde, S.S.; Wang, H.; Yaduru, S.; Pandey, M.K.; Fountain, J.C.; Chu, Y.; Isleib, T.; Holbrook, C.C.; Xavier, A.; Culbreath, A.K.; et al. Nested-association mapping (NAM)-based genetic dissection uncovers candidate genes for seed and pod weights in peanut (Arachis hypogaea). Plant Biotechnol. J. 2020, 18, 1457–1471. [Google Scholar] [CrossRef] [PubMed]
- Patel, J.D.; Wang, M.L.; Dang, P.; Butts, C.; Lamb, M.; Chen, C.Y. Insights into the genomic architecture of seed and pod quality traits in the U.S. peanut mini-core diversity panel. Plants 2022, 11, 837. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Duan, N.; Ren, X. Descriptors and Data Standard for Peanut (Arachis spp.); China Agriculture Press: Beijing, China, 2006; pp. 26–27. [Google Scholar]
- Schlotzhauer, S.D.; Littell, R.C. SAS System for Elementary Statistical Analysis; SAS Institute Inc.: Cary, NC, USA, 1997. [Google Scholar]
- Bates, D.; Maechler, M.; Bolker, B.; Walker, S. LME4: Linear Mixed-Effects Models Using Eigen and S4; R Package Version 1; R Foundation for Statistical Computing: Vienna, Austria, 2014; pp. 1–4. [Google Scholar]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef]
- Vilella, A.J.; Severin, J.; Ureta-Vidal, A.; Heng, L.; Durbin, R.; Birney, E. EnsemblCompara GeneTrees: Complete, duplication-aware phylogenetic trees in vertebrates. Genome Res. 2009, 19, 327–335. [Google Scholar] [CrossRef]
- Alexander, D.H.; Novembre, J.; Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009, 19, 1655–1664. [Google Scholar] [CrossRef]
- Yang, J.; Lee, S.H.; Goddard, M.E.; Visscher, P.M. GCTA: A tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2021, 88, 76–82. [Google Scholar] [CrossRef]
- Liu, X.; Huang, M.; Fan, B.; Buckler, E.S.; Zhang, Z. Iterative Usage of Fixed and Random Effect Models for Powerful and Efficient Genome-Wide Association Studies. PLoS Genet. 2016, 12, 427–433. [Google Scholar] [CrossRef] [PubMed]
- Yang, N.; Lu, Y.; Yang, X.; Huang, J.; Zhou, Y.; Ali, F.; Wen, W.; Liu, J.; Li, J.; Yan, J. Genome wide association studies using a new nonparametric model reveal the genetic architecture of 17 agronomic traits in an enlarged maize association panel. PLoS Genet. 2014, 10, e1004573. [Google Scholar] [CrossRef] [PubMed]
- Chavarro, C.; Chu, Y.; Holbrook, C.; Isleib, T.; Bertioli, D.; Hovav, R.; Hovav, R.; Butts, C.; Lamb, M.; Sorensen, R.; et al. Pod and seed trait QTL identification to assist breeding for peanut market preferences. G3 Genes Genomes Genet. 2020, 10, 2297–2315. [Google Scholar] [CrossRef]
- Brown, N.; Branch, W.D.; Johnson, M.; Wallace, J. Genetic diversity assessment of Georgia peanut cultivars developed during ninety years of breeding. Plant Genome 2021, 14, e20141. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, J.; He, X.; Wang, Y.; Ma, X.; Yin, D. Genome-wide association study of major agronomic traits related to domestication in peanut. Front. Plant Sci. 2017, 8, 1611. [Google Scholar] [CrossRef] [PubMed]
- Zou, K.; Kim, K.S.; Kim, K.; Kang, D.; Park, Y.H.; Sun, H.; Ha, B.K.; Ha, J.; Jun, T.H. Genetic diversity and genome-wide association study of seed aspect ratio using a high-density SNP array in peanut (Arachis hypogaea L.). Genes 2020, 12, 2. [Google Scholar] [CrossRef]
- Bertioli, D.J.; Jenkins, J.; Clevenger, J.; Gao, D.; Dudchenko, O.; Seijo, G.; Leal-Bertioli, S.C.; Ren, L.; Farmer, A.D.; Pandey, M.K.; et al. The genome sequence of peanut (Arachis hypogaea), a segmental allotetraploid. Nat. Genet. 2019, 51, 877–884. [Google Scholar] [CrossRef]
- Chen, X.; Lu, Q.; Liu, H.; Zhang, J.; Hong, Y.; Lan, H.; Li, H.; Wang, J.; Liu, H.; Li, S.; et al. Sequencing of cultivated peanut, Arachis hypogaea, yields insights into genome evolution and oil improvement. Mol. Plant 2019, 12, 920–934. [Google Scholar] [CrossRef]
- Li, Y.; Reif, J.C.; Hong, H.; Li, H.; Liu, Z.; Ma, Y.; Li, J.; Tian, Y.; Li, Y.; Li, W.; et al. Genome-wide association mapping of QTL underlying seed oil and protein contents of a diverse panel of soybean accessions. Plant Sci. 2018, 266, 95–101. [Google Scholar] [CrossRef]
- Liu, N.; Huang, L.; Chen, W.; Wu, B.; Pandey, M.K.; Luo, H.; Zhou, X.; Guo, J.; Chen, H.; Huai, D.; et al. Dissection of the genetic basis of oil content in Chinese peanut cultivars through association mapping. BMC Genet. 2020, 21, 60. [Google Scholar] [CrossRef]
- Atwell, S.; Huang, Y.S.; Vilhjálmsson, B.J.; Willems, G.; Horton, M.; Li, Y.; Meng, D.; Platt, A.; Tarone, A.M.; Hu, T.T.; et al. Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature 2010, 465, 627–631. [Google Scholar] [CrossRef] [PubMed]
- Munoz-Bertomeu, J.; Cascales-Minana, B.; Mulet, J.M.; Baroja-Fernandez, E.; Pozueta-Romero, J.; Kuhn, J.M.; Segura, J.; Ros, R. Plastidial glyceraldehyde-3-phosphate dehydrogenase deficiency leads to altered root development and affects the sugar and amino acid balance in Arabidopsis. Plant Physiol. 2009, 151, 541–558. [Google Scholar] [CrossRef] [PubMed]
- Akram, S.; Ghaffar, M.; Wadood, A.; Shokat, S.; Hameed, A.; Waheed, M.Q.; Arif, M.A.R. A GBS-based genome-wide association study reveals the genetic basis of salinity tolerance at the seedling stage in bread wheat (Triticum aestivum L.). Front. Genet. 2022, 13, 997901. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Zhang, Q.Q.; Zhou, H.Y.; Ni, F.R.; Wu, X.Y.; Qi, Y.J. Rice microRNA effector complexes and targets. Plant Cell 2009, 21, 3421–3435. [Google Scholar] [CrossRef]
- Zhong, J.; He, W.J.; Peng, Z.; Zhang, H.; Li, F.; Yao, J.L. A putative AGO protein, OsAGO17, positively regulates grain size and grain weight through OsmiR397b in rice. Plant Biotechnol. J. 2020, 18, 916–928. [Google Scholar] [CrossRef]
- Abd-Hamid, N.A.; Ahmad-Fauzi, M.I.; Zainal, Z.; Ismail, I. Diverse and dynamic roles of F-box proteins in plant biology. Planta 2020, 251, 68. [Google Scholar] [CrossRef]
- Hong, M.J.; Kim, J.B.; Seo, Y.W.; Kim, D.Y. F-Box genes in the wheat genome and expression profiling in wheat at different developmental stages. Genes 2020, 11, 1154. [Google Scholar] [CrossRef]
- Ye, J.; Tian, R.W.; Meng, X.F.; Tao, P.W.; Li, C.X.; Liu, G.Z.; Chen, W.F.; Wang, Y.; Li, H.X.; Ye, Z.B.; et al. Tomato SD1, encoding a kinase-interacting protein, is a major locus controlling stem development. J. Exp. Bot. 2020, 71, 3575–3587. [Google Scholar] [CrossRef]
- Barg, R.; Sobolev, I.; Eilon, T.; Shabtai, S.; Gur, A.; Velichko, O.; Grotewold, E.; Salts, Y. The tomato early fruit specific gene Lefsm1 defines a novel class of plant-specific SANT/MYB domain proteins. Planta 2005, 221, 197–211. [Google Scholar] [CrossRef]
- Li, X.J.; Gu, W.; Sun, S.L.; Chen, Z.L.; Chen, J.; Song, W.B.; Zhao, H.M.; Lai, J.S. Defective Kernel 39 encodes a PPR protein required for seed development in maize. J. Integr. Plant Biol. 2018, 60, 45–64. [Google Scholar] [CrossRef]
- Zhou, Y.P.; Wu, J.H.; Xiao, W.H.; Chen, W.; Chen, Q.H.; Fan, T.; Xie, C.P.; Tian, C.E. Arabidopsis IQM4, a novel calmodulin-binding protein, is involved with seed dormancy and germination in Arabidopsis. Front. Plant Sci. 2018, 9, 721. [Google Scholar] [CrossRef] [PubMed]
- Lazzarotto, F.; Wahni, K.; Piovesana, M.; Maraschin, F.; Messens, J.; Margis-Pinheiro, M. Arabidopsis APx-R is a plastidial ascorbate-independent peroxidase regulated by photomorphogenesis. Antioxidants 2021, 10, 65. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).