Identification and Validation of the Genomic Regions for Waterlogging Tolerance at Germination Stage in Wheat
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Evaluation of Waterlogging Tolerance
2.3. DNA Extraction and Genotyping
2.4. KASP Assay for SNPs
2.5. Statistical Analysis
3. Results
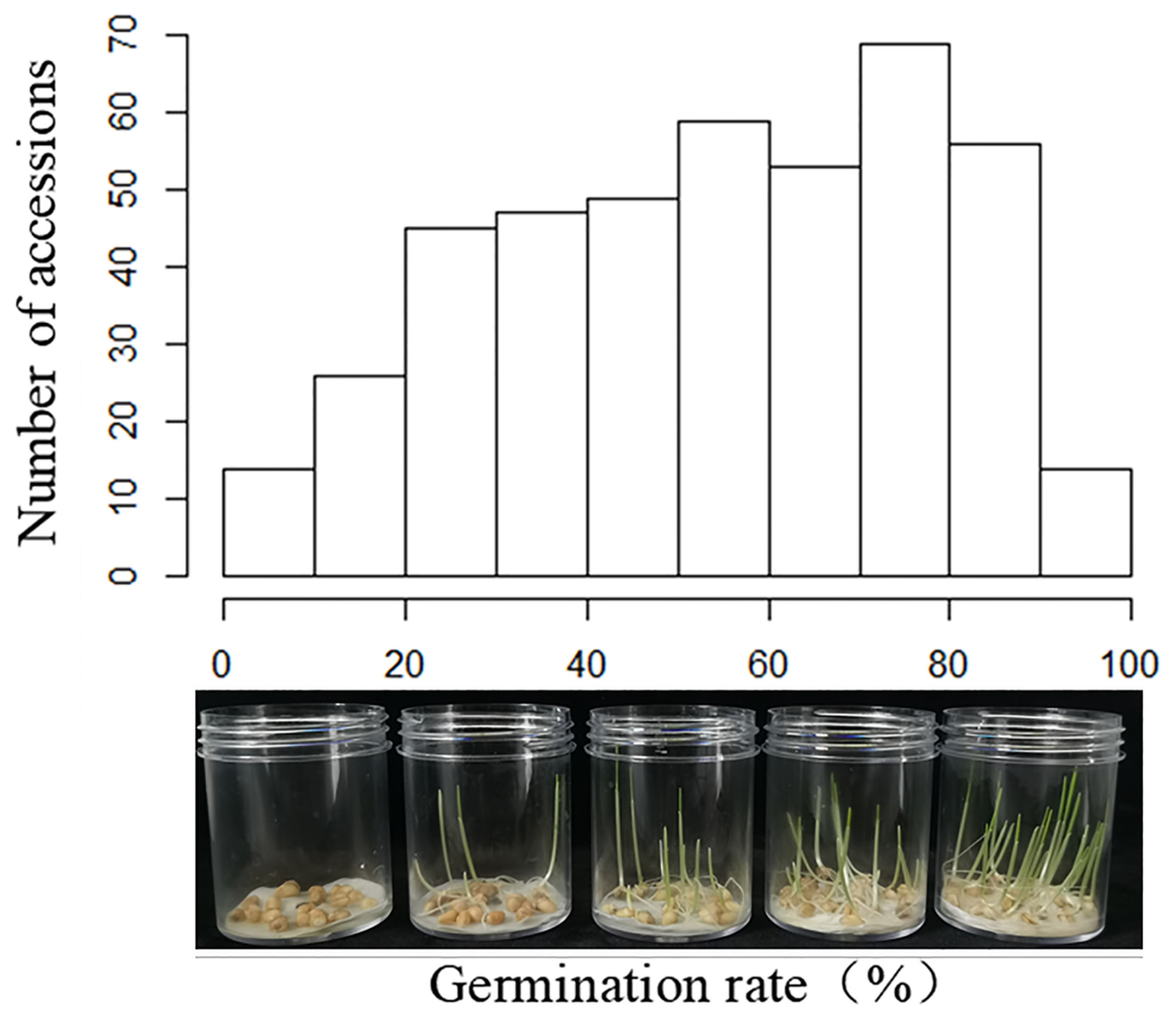
3.1. Variation of Waterlogging Tolerance among Wheat Varieties
3.2. Phenotyping of SN135/SY936 and HM18/CD1840 Populations and Extreme Bulks
3.3. Genotyping of Extreme Tolerant and Sensitive Bulks
3.4. Validation of QTL by KASP Assay
4. Discussion
4.1. SNP Array Combined with BSA Is Effective for QTL Identification
4.2. Comparison with Previous Reported Waterlogging Tolerance QTL
4.3. Candidate Genes Governing Waterlogging Tolerance
4.4. Germplasms for Wheat Waterlogging Tolerance Breeding
4.5. Cost-Effective Markers for Wheat Molecular Breeding
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Afzal, F.; Chaudhari, S.K.; Gul, A.; Farooq, A.; Ali, H.; Nisar, S.; Sarfraz, B.; Shehzadi, K.J.; Mujeeb-Kazi, A. Bread wheat (Triticum aestivum L.) under biotic and abiotic stresses: An overview. In Crop Production and Global Environmental Issues; Hakeem, K.R., Ed.; Springer International Publishing: Cham, Switzerland, 2015; pp. 293–317. [Google Scholar]
- Tong, C.; Hill, C.B.; Zhou, G.; Zhang, X.-Q.; Jia, Y.; Li, C. Opportunities for improving waterlogging tolerance in cereal crops—physiological traits and genetic mechanisms. Plants 2021, 10, 1560. [Google Scholar] [CrossRef] [PubMed]
- Shabala, S. Physiological and cellular aspects of phytotoxicity tolerance in plants: The role of membrane transporters and implications for crop breeding for waterlogging tolerance. New Phytol. 2011, 190, 289–298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaur, G.; Singh, G.; Motavalli, P.P.; Nelson, K.A.; Orlowski, J.M.; Golden, B.R. Impacts and management strategies for crop production in waterlogged or flooded soils: A review. Agron. J. 2020, 112, 1475–1501. [Google Scholar] [CrossRef] [Green Version]
- Ding, J.; Liang, P.; Wu, P.; Zhu, M.; Li, C.; Zhu, X.; Gao, D.; Chen, Y.; Guo, W. Effects of waterlogging on grain yield and associated traits of historic wheat cultivars in the middle and lower reaches of the Yangtze River, China. Field Crop. Res. 2020, 246, 107695. [Google Scholar] [CrossRef]
- Yang, H.; Zhai, S.; Li, Y.; Zhou, J.; He, R.; Liu, J.; Xue, Y.; Meng, Y. Waterlogging reduction and wheat yield increase through long-term ditch-buried straw return in a rice—wheat rotation system. Field Crop. Res. 2017, 209, 189–197. [Google Scholar] [CrossRef]
- Ghobadi, M.E.; Ghobadi, M.; Zebarjadi, A. Effect of waterlogging at different growth stages on some morphological traits of wheat varieties. Int. J. Biometeorol. 2017, 61, 635–645. [Google Scholar] [CrossRef]
- Mfarrej, M.F.B.; Wang, X.; Hamzah Saleem, M.; Hussain, I.; Rasheed, R.; Arslan Ashraf, M.; Iqbal, M.; Sohaib Chattha, M.; Nasser Alyemeni, M. Hydrogen sulphide and nitric oxide mitigate the negative impacts of waterlogging stress on wheat (Triticum aestivum L.). Plant Biol. 2022, 24, 670–683. [Google Scholar] [CrossRef]
- Arduini, I.; Orlandi, C.; Pampana, S.; Masoni, A. Waterlogging at tillering affects spike and spikelet formation in wheat. Crop Pasture Sci. 2016, 67, 703–711. [Google Scholar] [CrossRef]
- Ploschuk, R.A.; Miralles, D.J.; Striker, G.G. Early- and late-waterlogging differentially affect the yield of wheat, barley, oilseed rape and field pea through changes in leaf area index, radiation interception and radiation use efficiency. J. Agron. Crop Sci. 2021, 207, 504–520. [Google Scholar] [CrossRef]
- Ding, J.; Liang, P.; Wu, P.; Zhu, M.; Li, C.; Zhu, X.; Guo, W. Identifying the critical stage near anthesis for waterlogging on wheat yield and its components in the yangtze river basin, China. Agronomy 2020, 10, 130. [Google Scholar] [CrossRef] [Green Version]
- Xu, Z.; Shen, Q.; Zhang, G. The mechanisms for the difference in waterlogging tolerance among sea barley, wheat and barley. Plant Growth Regul. 2022, 96, 431–441. [Google Scholar] [CrossRef]
- Kulshreshtha, N.; Kumar, A.; Kumar, A.; Lata, C. Genetic Interventions to Improve Salt and Microelement Toxicity Tolerance in Wheat. In New Horizons in Wheat and Barley Research: Global Trends, Breeding and Quality Enhancement; Kashyap, P.L., Gupta, V., Prakash Gupta, O., Sendhil, R., Gopalareddy, K., Jasrotia, P., Singh, G.P., Eds.; Springer: Singapore, 2022; pp. 429–483. [Google Scholar]
- Herzog, M.; Striker, G.G.; Colmer, T.D.; Pedersen, O. Mechanisms of waterlogging tolerance in wheat—A review of root and shoot physiology. Plant Cell Environ. 2016, 39, 1068–1086. [Google Scholar] [CrossRef] [PubMed]
- Tian, L.X.; Zhang, Y.C.; Chen, P.L.; Zhang, F.F.; Li, J.; Yan, F.; Dong, Y.; Feng, B.L. How does the waterlogging regime affect crop yield? A global meta-analysis. Front. Plant Sci. 2021, 12, 634898. [Google Scholar] [CrossRef] [PubMed]
- Manik, S.M.N.; Pengilley, G.; Dean, G.; Field, B.; Shabala, S.; Zhou, M. Soil and crop management practices to minimize the impact of waterlogging on crop productivity. Front. Plant Sci. 2019, 10, 140. [Google Scholar] [CrossRef] [Green Version]
- Du, X.; He, W.; Wang, Z.; Xi, M.; Xu, Y.; Wu, W.; Gao, S.; Liu, D.; Lei, W.; Kong, L. Raised bed planting reduces waterlogging and increases yield in wheat following rice. Field Crop. Res. 2021, 265, 108119. [Google Scholar] [CrossRef]
- Ahmed, F.; Rafii, M.Y.; Ismail, M.R.; Juraimi, A.S.; Rahim, H.A.; Asfaliza, R.; Latif, M.A. Waterlogging tolerance of crops: Breeding, mechanism of tolerance, molecular approaches, and future prospects. Biomed Res. Int. 2013, 2013, 963525. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Mao, D.; Zhu, M.; Wang, X.; Li, C.; Zhu, X.; Guo, W.; Ding, J. Evaluating the waterlogging tolerance of wheat cultivars during the early growth stage using the comprehensive evaluation value and digital image analysis. Agriculture 2022, 12, 384. [Google Scholar] [CrossRef]
- Tanin, M.J.; Saini, D.K.; Sandhu, K.S.; Pal, N.; Gudi, S.; Chaudhary, J.; Sharma, A. Consensus genomic regions associated with multiple abiotic stress tolerance in wheat and implications for wheat breeding. bioRxiv 2022. [Google Scholar] [CrossRef]
- Burgos, M.S.; Messmer, M.M.; Stamp, P.; Schmid, J.E. Flooding tolerance of spelt (Triticum spelta L.) compared to wheat (Triticum aestivum L.)—A physiological and genetic approach. Euphytica 2001, 122, 287–295. [Google Scholar] [CrossRef]
- Yu, M.; Mao, S.L.; Chen, G.Y.; Liu, Y.X.; Li, W.; Wei, Y.M.; Liu, C.J.; Zheng, Y.L. QTLs for waterlogging tolerance at germination and seedling stages in population of recombinant inbred lines derived from a cross between synthetic and cultivated wheat genotypes. J. Integr. Agric. 2014, 13, 31–39. [Google Scholar] [CrossRef]
- Ballesteros, D.C.; Mason, R.E.; Addison, C.K.; Andrea Acuña, M.; Nelly Arguello, M.; Subramanian, N.; Miller, R.G.; Sater, H.; Gbur, E.E.; Miller, D.; et al. Tolerance of wheat to vegetative stage soil waterlogging is conditioned by both constitutive and adaptive QTL. Euphytica 2015, 201, 329–343. [Google Scholar] [CrossRef]
- Wei, M.; Li, X.; Yang, R.; Li, L.; Wang, Z.; Wang, X.; Sha, A. Novel insights into genetic responses for waterlogging stress in two local wheat cultivars in yangtze river basin. Front. Genet. 2021, 12, 681680. [Google Scholar] [CrossRef] [PubMed]
- Pang, Y.; Liu, C.; Wang, D.; St. Amand, P.; Bernardo, A.; Li, W.; He, F.; Li, L.; Wang, L.; Yuan, X.; et al. High-resolution genome-wide association study identifies genomic regions and candidate genes for important agronomic traits in wheat. Mol. Plant 2020, 13, 1311–1327. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, D.; Bajwa, G.S. Molecular marker techniques and recent advancements. In Genotyping by Sequencing for Crop Improvement; John Wiley & Sons: Hoboken, NJ, USA, 2022; pp. 1–21. [Google Scholar]
- Cavanagh, C.R.; Chao, S.; Wang, S.; Huang, B.E.; Stephen, S.; Kiani, S.; Forrest, K.; Saintenac, C.; Brown-Guedira, G.L.; Akhunova, A.; et al. Genome-wide comparative diversity uncovers multiple targets of selection for improvement in hexaploid wheat landraces and cultivars. Proc. Natl. Acad. Sci. USA 2013, 110, 8057–8062. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.; Kolmer, J.; Li, G.; Tan, C.; Carver, B.F.; Bian, R.; Bernardo, A.; Bai, G. Identification and characterization of the novel leaf rust resistance gene Lr81 in wheat. Theor. Appl. Genet. 2022, 135, 2725–2734. [Google Scholar] [CrossRef]
- Yu, M.; Chen, G.Y. Conditional QTL mapping for waterlogging tolerance in two RILs populations of wheat. SpringerPlus 2013, 2, 245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, R.; He, D.; Xu, L.; Zhou, M.; Li, C.; Wu, C.; Xu, Y.; Zhang, W. Proteomic analysis reveals response of differential wheat (Triticum aestivum L.) genotypes to oxygen deficiency stress. BMC Genom. 2019, 20, 60. [Google Scholar] [CrossRef] [Green Version]
- Wei, X.; Xu, H.; Rong, W.; Ye, X.; Zhang, Z. Constitutive expression of a stabilized transcription factor group VII ethylene response factor enhances waterlogging tolerance in wheat without penalizing grain yield. Plant Cell Environ. 2019, 42, 1471–1485. [Google Scholar] [CrossRef]
- Du, H.; Zhu, J.; Su, H.; Huang, M.; Wang, H.; Ding, S.; Zhang, B.; Luo, A.; Wei, S.; Tian, X.; et al. Bulked segregant RNA-seq reveals differential expression and SNPs of candidate genes associated with waterlogging tolerance in maize. Front. Plant Sci. 2017, 8, 1022. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Yang, B.; Xu, J.; Peng, L.; Sun, S.; Huang, Z.; Jiang, X.; He, Y.; Wang, Z. A genome-wide association study reveals that the 2-oxoglutarate/malate translocator mediates seed vigor in rice. Plant J. 2021, 108, 478–491. [Google Scholar] [CrossRef]
- Capo-chich, L.; Eldridge, S.; Elakhdar, A.; Kumamaru, T.; Anyia, A.O. Major QTLs for seedling traits in barley using a DArT-based linkage map. bioRxiv 2019, 12, 889899. [Google Scholar] [CrossRef]
- Hossain, M.A.; Uddin, S.N. Mechanisms of waterlogging tolerance in wheat: Morphological and metabolic adaptations under hypoxia or anoxia. Aust. J. Crop Sci. 2011, 5, 1094–1101. [Google Scholar]
- Pan, J.; Sharif, R.; Xu, X.; Chen, X. Mechanisms of waterlogging tolerance in plants: Research progress and prospects. Front. Plant Sci. 2021, 11, 627331. [Google Scholar] [CrossRef] [PubMed]
- Cai, B.; Tian, W.; Wang, X. Screening of wheat varieties tolerant to waterlogging at bud-seeding stage. J. Henan Agric. Sci. 2015, 44, 52–57. [Google Scholar] [CrossRef]
- Ma, B.; Li, Q.; Cai, J.; Zhou, Q.; Huang, M.; Dai, T.; Wang, X.; Jiang, D. Physiological mechanisms of pre-anthesis waterlogging priming on waterlogging stress tolerance under post-anthesis in wheat. Acta Agron. Sin. 2022, 48, 151–164. [Google Scholar] [CrossRef]
- Tong, H.; Liu, Y.; Zhu, Z.; Chen, L.; Zhang, Y.; Wu, B.; Gao, C. Morphological and metabolic adaptation of wheat root to waterlogging. Crops 2013, 5, 13–17. [Google Scholar] [CrossRef]
- He, C.; Holme, J.; Anthony, J. SNP genotyping: The KASP assay. In Crop Breeding: Methods and Protocols; Fleury, D., Whitford, R., Eds.; Springer: New York, NY, USA, 2014; pp. 75–86. [Google Scholar]



| Factors | Df | Sum Sq | Mean Sq | F Value | Pr (>F) | Heritability (h2) |
|---|---|---|---|---|---|---|
| Accessions | 431 | 30,012.3 | 69.6 | 3.8 | 1.7 × 10−41 | 0.79 |
| Replicates | 1 | 129.1 | 129.1 | 7.1 | 0.0080 | |
| Residues | 431 | 7845.9 | 18.2 |
| Population | QTL | Region | KASP a | Primer |
|---|---|---|---|---|
| SN135 × SY936 | qWlg5A | 659.6–709.1 | k5A6810 | F1: TTGCAGCGCTTCTTTTATCC |
| F2: TTGCAGCGCTTCTTTTATCG | ||||
| R: TTTACGATTTTGGTCGGGCC | ||||
| k5A6857 | F1: CAGGACACTACCGTCATGGA | |||
| F2: CAGGACACTACCGTCATGGG | ||||
| R: AGCACGGATAAGCAGGTGAC | ||||
| qWlg7B | 583.9–614.2 | k7B6029 | F1: TGGGAGCCGTAAATAAAGCA | |
| F2: TGGGAGCCGTAAATAAAGCG | ||||
| R: CTCCCTTCGAAGTTGGGCTT | ||||
| HM18 × CD1840 | qWlg2D | 570.4–619.0 | k2D5802 | F1:TCGAATGCCTACCGTGAATAAC |
| F2: TCGAATGCCTACCGTGAATAAT | ||||
| R: TGGGTGATATAGCGGCAAAGA | ||||
| k2D5904 | F1: GGACGTTGTCGGTACTAAAACT | |||
| F2: GGACGTTGTCGGTACTAAAACC | ||||
| R: TGATCCGAGATCCACAGCTC |
| Population | Factors | Df | Sum Sq | Mean Sq | F Value | Pr (>F) | R2 (%) |
|---|---|---|---|---|---|---|---|
| SN135 × SY936 | k5A6810 | 2 | 12,991.8 | 6495.9 | 10.65 | 4.3 × 10−5 | 10.6 |
| Residuals | 179 | 109,144.4 | 609.7 | ||||
| k5A6857. | 2 | 18,267.9 | 9134.0 | 15.82 | 4.9 × 10−7 | 15.3 | |
| Residuals | 175 | 101,070.3 | 577.5 | ||||
| k7B6029 | 2 | 24,980.7 | 12,490.3 | 23.01 | 1.3 × 10−9 | 20.5 | |
| Residuals | 179 | 97,155.6 | 542.8 | ||||
| HM18 × CD1840 | k2D5802 | 2 | 10,557.33 | 5278.7 | 8.66 | 0.0002 | 8.0 |
| Residuals | 200 | 121,909.4 | 609.5 | ||||
| k2D5904 | 2 | 6757.853 | 3378.9 | 5.38 | 0.0053 | 5.1 | |
| Residuals | 200 | 125,708.8 | 628.5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pang, Y.; Wang, X.; Zhao, M.; Lu, Y.; Yan, Q.; Sun, S.; Wang, Y.; Liu, S. Identification and Validation of the Genomic Regions for Waterlogging Tolerance at Germination Stage in Wheat. Agronomy 2022, 12, 1848. https://doi.org/10.3390/agronomy12081848
Pang Y, Wang X, Zhao M, Lu Y, Yan Q, Sun S, Wang Y, Liu S. Identification and Validation of the Genomic Regions for Waterlogging Tolerance at Germination Stage in Wheat. Agronomy. 2022; 12(8):1848. https://doi.org/10.3390/agronomy12081848
Chicago/Turabian StylePang, Yunlong, Xiaoqian Wang, Min Zhao, Yue Lu, Qiang Yan, Shanyi Sun, Yueping Wang, and Shubing Liu. 2022. "Identification and Validation of the Genomic Regions for Waterlogging Tolerance at Germination Stage in Wheat" Agronomy 12, no. 8: 1848. https://doi.org/10.3390/agronomy12081848
APA StylePang, Y., Wang, X., Zhao, M., Lu, Y., Yan, Q., Sun, S., Wang, Y., & Liu, S. (2022). Identification and Validation of the Genomic Regions for Waterlogging Tolerance at Germination Stage in Wheat. Agronomy, 12(8), 1848. https://doi.org/10.3390/agronomy12081848





