Abstract
Melon is an important horticultural crop worldwide. The high diversity of melon makes it a model plant for various different properties. Some quantitative trait loci or candidates have been discovered, but few were verified as limiting genetic transformation and genome editing systems. Identifying new genetic resources with resistance and special fruit quality traits is imperative to develop effective and useful breeding technologies in melon. This review describes the advances in genetics, genomics, and the breeding of melon and puts forward some recommendations in these areas.
1. Introduction
Melon (Cucumis melo L.), an important crop in the Cucurbitaceae family, is cultivated worldwide, with more than 28 million tons produced in 2020 (United Nations Food and Agriculture Organization (FAO) statistics). It is highly diverse in fruit type and other properties, such as fruit size, peel and flesh color, and flavor []. Based on ovary pubescence, melon has been classified into two subspecies, C. melo subsp. melo (hereafter as melo) and C. melo subsp. agrestis (hereafter as agrestis). The two subspecies can be further divided into 16 groups or varieties, five in agrestis and eleven in melo []. Recently, the melon was described as 19 groups of wild, feral, and domesticated melons, and some of them of sub-groups []. The diversity in phenotype and genetics of melon make it feasible as a model cucurbit for studying sex expression [,] and flower and fruit development [,,].
The previous researchers have identified a few accessions with resistance or special characters, and several elite varieties developed using these accessions. In addition, some quantitative trait loci or candidates for important characters in melon have been discovered. Furthermore, next-generation sequencing technologies in the past years have allowed unprecedented access to draft genome sequences for the main crops and plants. Since the first melon genome sequence was released [], more and more data from divergent genotypes by de novo sequencing and re-sequencing have become available. The advances in genetics and genomics in melon have accelerated the development of melon breeding. This paper will review the advances in genetics, genomics, and breeding in melon.
2. The Genetic Maps and Genetics of Related Characters in Melon
Dissecting the genetic base of characters is the foundation for utilizing germplasm innovation and breeding. In previous studies, several genetic and physical maps were conducted, and considerable genetic research on agronomic characteristics of melon was performed, including sex determination, fruit quality, fruit development, and resistance. The main QTLs and genes for melon reported in previous research are discussed in detail below (Table 1).
2.1. Genetic Map
Considerable melon genetic maps have been reported since the first melon genetic linkage map constructed in 1991 [] based on relatively few markers as amplified fragment length polymorphisms (AFLPs) [], random amplified polymorphic DNA (RAPD) [] and simple sequence repeats (SSRs) []. Then, the first consensus linkage map for melon was integrated under the International Cucurbit Genomics Initiative (ICuGI) framework, including the position of QTLs related to important agronomic traits []. However, due to the position of ICuGI markers in the genome remaining unknown and different marker sets used, it is essential to develop an integrated genetic map with the physical position of different makers after the melon genome was released in 2012 []. Diaz et al. [] integrated a total of 1850 previously reported markers in the genome sequence, which has been used as a reference genetic map for research. In recent years, with the development of next-generation sequencing (NGS), high-resolution genetic maps have been constructed using single nucleotide polymorphisms (SNPs) from genotyping-by-sequencing (GBS) and re-sequencing data for melon [,,,,,].
2.2. Genetics of Related Characters in Melon
2.2.1. Sex Expression
Melon is considered the model plant for studying sex determination [,]. The flowers of melon can be divided into male flowers, female flowers, and bisexual flowers. Based on the distribution and ratio of different flowers on the same plant, gynoecy, androecy, hermaphrodite, monoecy, and andromonoecy were classified as melon []. In melon, sex determination is governed by two major genes, andromonoecious (a) and gynoecious (g), which correspond to CmACS-7 and CmWIP1 [,], respectively. The dominant G allele governs the production of a separate male flower by suppressing carpel development [,]. In contrast, a gene encoding an ACC synthase inhibits the development of the male organs in female flowers []. The transition between monoecy and andromonoecy is conferred by a single substitution in CmACS-7, which leads to an inactive form of this key enzyme (1-aminocyclopropane-1-carboxylic acid synthase) in the ethylene biosynthesis []. Monoecious (A-G-) and andromonoecious (aaG-) individuals bear male flowers on the main stem and female or hermaphrodite flowers on axillary branches, respectively, whereas gynoecious (AAgg) and hermaphrodite (aagg) individuals only bear female and hermaphrodite flowers, respectively []. Additionally, a third locus having the recessive m allele (CmACS11) is essential for the production of stable gynoecious phenotypes []. However, sex expression in melon is quite plastic and collectively determined through mutual interaction of the hormonal, environmental, and development aspects [].
This plasticity requires signal perception/integration, changing gene expression in response to those signals, and then the maintenance of that response until conditions change again []. Epigenetic mechanisms provide a molecular memory that underpins the maintenance phase of these responses. The perception of the sex-determinative signals and their translation at the product level in flowers or plants might have been regulated by some epigenetic mechanisms [] and have been described previously []. In the gynoecious genotype, the transition from male to female flowers results from epigenetic changes in the CmWIP1 promoter caused by the insertion of a transposon, Gyno-hAT [,]. The female-promoting gene, CmACS11, represses the expression of the male-promoting gene CmWIP1 via the deposition of H3K27me3 []. The genes of ACS7, ACS11, and WIP1, in an epistatic or hypostatic manner, along with the recruitment of H3K9ac and H3K27me3, determine sex expression epigenetically [].
2.2.2. Sugar Content
The sensory quality of fruit is largely determined by its sugar and organic acid levels, in addition to the volatile aromatic components. Sugar content is not only the major determinant of both fruit quality and consumer acceptance. Still, it is also a primary target for crop improvement in melon [], mainly comprised of sucrose, glucose, and fructose. Significantly, the increase in total sugar content in mature melon fruit is particularly due to sucrose accumulation during the final stages of fruit development. As sucrose is the dominant factor in sugar content, understanding the molecular mechanisms involved in the process of sucrose accumulation is essential for possible genetic improvements.
The metabolic pathway of sucrose accumulation was involved in nearly 20 enzymes, especially invertases (INV), sucrose synthase (SS), and sucrose phosphate synthase (SPS), and the balance between sucrose breakdown and sucrose synthesis activities was considered the crucial factor in final sucrose accumulation. Forty-two genes encoding the enzymatic reactions of the sugar metabolism pathway in melon were reported []. There has been some progress in studying the genetic control of sugar accumulation. Burger et al. [] considered that a single major gene, suc, controls sucrose accumulation. However, Harel-Beja et al. [] reported six significant QTLs on chromosomes 2, 3, 5, and 8 for sugar content. Diaz et al. [] further developed a consensus linkage map of melon combining the previously reported QTLs from 18 mapping experiments, which consisted of more than 10 QTLs for Brix, sugars, and sucrose. Though many QTLs have been reported, few candidate genes have been identified. Recently, three important clusters related to sugar content have been identified, of which a major QTL, SUCQSC5.1, reducing soluble solids content (SSC) and sucrose content was detected, and MELO3C014519 was considered as the candidate gene []. Additionally, some new different-expression genes responsible for the sucrose content were detected for melon, such as CmINH3, CmTPP1, and CmTPS5, CmTPS7, CmTPS9 []. Sugars are synthesized in mesophyll cells and translated into the other parts of the plant, particularly sink organs in plants. Therefore, sugar accumulation in melon fruit depends not only on sugar synthesis but also on sugar transport. Three tonoplast sugar transporters (TSTs), CmTST1, CmTST2, and CmTST3, were isolated from melon, but only CmTST2 plays an important role in sugar accumulation in melon fruit []. The considerable QTLs for sugar content detected in melon fruit is probably due to the diverse backgrounds in these research or the complex metabolic pathways in sugar. It is a challenge to discover and functionally validate the major QTLs or crucial genes for sugar accumulation and transportation.
2.2.3. Acidity
Acidity is a major determinant of the taste and quality of most fruits, in combination with sugars and flavor volatiles. Melon is fairly unique among fleshy fruit in that they have very low acidity besides a few varieties []. Fruit acidity is due to the presence of organic acids, and malic and citric acids are the main acids found in most ripe fruits, measured by titratable acidity and PH. In melon, citric acid is the predominant organic acid present throughout fruit development and is positively correlated with titratable acidity [].
A single dominant locus for the sourness or acidity of melon flesh was identified based on different populations and termed So or PH [,,,], which encodes a transmembrane transporter []. Surprisingly, the nucleosides adenosine and the major organic acids citrate and malate in melon fruits were detected. The PH protein harboring in the endoplasmic reticulum may indirectly play a role via modification of a proton gradient or nucleoside and acid metabolism []. A 12-bp insertion was detected occurring in non-acidic melon accessions of the melo group but not consistent in the agrestis group. Intriguingly, MELO3C011482, which encodes ATP-citrate synthase, was identified on chromosome 3 in the agrestis group []. The results also support the hypothesis that the melo and agrestis groups were domesticated independently.
2.2.4. Peel Color
Peel color is an important fruit quality trait that influences the choice of the consumer and the acceptability of the melon []. The primary peel colors of commercial melons are green, white, or yellow, which are conferred by distinct pigments accumulation, mainly by switching from green rind containing chlorophyll to various peel colors possessing kinds of combinations of chlorophylls, carotenoids, and flavonoids [,].
Green peel is a dominant epistatic to non-green (white and yellow) peel by analyzing an F2 segregating population from a cross between green peel and yellow peel lines []. The candidate gene MELO3C003375 on chromosome 4 was detected associating with green peel formation of melon and fruit pigment accumulation in watermelon []. In fact, besides the primary color, the secondary color of the mature fruit is also present in some special melon accessions, such as speckles, stripes, and spots. Recently, two dominant epistatic genes (CmMt1 and CmMt2) for mottled peel and one gene (st3) for striped peel were reported. CmMt1 and CmMt2 could control the content of chlorophyll and the different ratios in chlorophyll accumulation conjointly. Interestingly, CmMt1 has been confirmed as MELO3C003375 [], and the position of st3 harboring on the chromosome is close to MELO3C003375 [], which was suggested that MELO3C003375 plays a central role on regulating the accumulation of pigment. We also detected a gene (MELO3C003097) encoding the protein SLOW GREEN 1, which is required for chloroplast development on chromosome 8 and is associated with the green peel trait and maybe a minor gene involved in the formation of rind color []. Naringenin chalcone, a kind of yellow flavonoid pigment, was found to be one of the major pigments infecting the melon rind color in ‘canary yellow’ type melons [], which is independent of carotenoids and chlorophyll pigments []. Additionally, CmKFB, a kelch domain-containing F-box protein-coding gene located on chromosome 10, has been identified to be a negative regulator for naringenin chalcone accumulation [].
Many genes controlling the rind color have been identified, and the genetic mechanism is complex. The functional verification of these genes and their application in breeding remains to be developed.
2.2.5. Flesh Color
Flesh color is one of the most important traits of melon. The different flesh colors not only affect the preference of consumers but also mean different nutrients. The primary flesh colors in commercial varieties contain orange, white and green, largely governed by two major genes, white flesh (wf) and green flesh (gf). Orange flesh is determined by Gf and is dominant to green flesh (gf). Fruit with the genotype of gfgf has either green (Wf-) or white flesh (wfwf) []. Recently, the two major genes for flesh color in melon were identified. The candidate gene CmOr associated with carotenoid accumulation in melon fruit flesh was discovered, which is identical to the previously described gf locus []. Combining the QTL with GWAS results, a 96-kb overlapping interval containing 11 protein-coding genes was detected []. However, a previously reported candidate gene MELO3C003069 for Wf is 202 kb away from the 96-kb interval, and thus MELO3C003097 was considered a strong candidate for the Wf locus [,].
2.2.6. Resistance to Abiotic and Biotic Stresses in Melon
Plants suffer from abiotic and biotic stresses frequently during their development. The occurrence of powdery mildew, downy mildew, fusarium wilt, gummy stem blight, virus, and aphis gossypii reduce melon yield and quality worldwide []. Much research has been conducted on the genetics of resistance to abiotic and biotic stresses in melon.
Powdery Mildew
Powdery mildew (PM) caused by Podosphaera xanthii and Golovinomyces cichoracearum is an important foliar disease in melon, which includes seven races of Podosphaera xanthii and two of Golovinomyces cichoracearum [,,,,,]. The diverse climatic conditions result in differentiation in the dominant physiological race of powdery mildew around the world. Several QTLs associating resistance to different races of powdery mildew have been identified from the different resistant accessions in previous research, including Pm-1 from PMR 45 [], Pm-2 from PMR 5 and PMR 6 [], Pm-3 and Pm-6 from PI 124111F [], Pm-4 and Pm-5 from PI 124112 [,], Pm-w from WMR 29 [], Pm-x from PI 414723 and Pm-y from VA 435 [], Pm-R from TGR-1551 [], PmV.1 and PmXII.1 from PI 124112 [], BPm12.1 from MR-1 [], PmEdisto47–1 from Edisto47 [], Pm-2F from K7–1 []. The different results obtained in inheritance patterns and QTL mapping may be due to the diverse physiological races of powdery mildew and resistant accessions used. Unfortunately, there are not so many functional genes reported so far.
Gummy Stem Blight
Gummy stem blight (GSB) is one of the most serious diseases causing enormous losses to melon production worldwide []. In previous studies, genetic analyses have explored several independent GSB resistance loci from diverse cultigens of melon. The first resistant gene for GSB, Gsb-1, was reported to be a monogenic dominant locus in PI 140471 []. The other four resistant genes, Gsb-2, Gsb-3, Gsb-4, and Gsb-5, were considered to govern the resistance of PI 157082, PI 511890, PI 482398, PI 482399 to GSB, respectively []. Globally, the previous studies suggested GBS is governed by a single gene in each of the resistant melon accessions. Gsb-1, Gsb-2, Gsb-3, Gsb-4 and Gsb-6 are dominant loci, except for Gsb-5 [,,]. In addition to MELO03C012987, few candidates were identified through several loci reported [].
Fusarium Wilt
Fusarium wilt (FW) caused by Fusarium oxysporum (Fom) is one of the destructive soil-borne diseases resulting in economic damage in a large number of melon-producing countries []. Four races of Fom have been described in melon: 0, 1, 2, and 1, 2 based on resistance genes. Genetic studies on the inheritance of resistance to the different races of Fom have been described in previous studies [,]. Fom-2 was identified and considered as the locus conferring resistance to Fom races 0 and 1 []. Fom-1 locus was mapped and conferred as resistance to race 2 []. Fom-4 conferred resistance to races 0 and 2 and was found to be a recessive gene closely linked to Fom-1 []. Interestingly, Fom-3 was found to control resistance to races 0 and 2, and it is possible allelism with Fom-1 []. Recently, another single dominant gene for FW resistance and defined as Fom-5 []. Fom-1, Fom-2, Fom-3 and Fom-5 are four single dominant genes associated with FW resistance besides Fom-4.
Virus Disease
Cucumber mosaic virus (CMV), Zucchini yellow mosaic virus (ZYMV), Cucumber green mottle virus (CGMV), and Melon necrotic spot virus (MNSV) is the main virus diseases in melon [,,]. To date, around 200 viral-resistant genes have been found, of which more than half are recessive genes, part of the recessive genes encoding eukaryotic translation initiation factors (eIF) that control the viral resistance in melon [,,]. The Cm-eIF4E knockdown melon plants possess resistance to MNSV, ZYMV, Cucumber vein yellowing virus (CVYV), and Moroccan watermelon mosaic virus (MWMV) []. The resistance to CMV in melon accession PI 161375 is governed by one gene and at least two quantitative trait loci []. The major gene CmVPS41 was reported as a general gatekeeper for resistance to CMV phloem entry in melon []. ZYMV resistance in melon PI 414723 is controlled by a dominant allele at the Zym locus []. Nucleotide-binding leucine-rich-repeat (NB–LRR) genes are one vital resistance (R) gene in melon and have been found to resist many diseases and insect pests []. Vat belonging to an NBS-LRR gene is considered the aphid resistance gene []. Most NB-LRR genes are specialized in resistance. However, plant disease resistance is easily lost with the mutation of pathogenic bacteria []. Therefore, exploring and cloning disease resistance genes with a broad spectrum will be an important part of the research of NBS disease resistance genes.
Abiotic Stress
Salinity, drought, and temperature extremes are the main abiotic in melon. Abiotic stress tolerance is complex because the sensitivity of many crops to a particular abiotic stress varies depending on their developmental stage. Therefore, the mechanisms of resistance or tolerance to abiotic stress are poorly understood. Compared to biotic stresses, the genetics of abiotic stress tolerance have been paid little attention in melon. The genetics of abiotic stresses in melon potassium is a major factor in resistance to salinity. Shaker-like K+ outward rectifying channel (SKOR) is participated in the long-distance distribution of K+ from roots to the upper parts of the plant []. The previous study indicated that CmSKOR might play a role in distributing K+ to the shoot in melon and improving saline tolerance in Arabidopsis []. CmLOX10 is crucial in regulating tolerance to drought in melon seedlings by promoting JA accumulation and stomatal closure []. Both CmLOX08 and CmCADs were considered the key players in abiotic stress responses, including drought and salt [,]. CmNCED3 plays a crucial role in low-temperature stress response, besides drought and salt stresses [].

Table 1.
The main QTLs and genes for melon were reported in previous research.
Table 1.
The main QTLs and genes for melon were reported in previous research.
| Traits | QTLs or Gene | Chromosome | Function of QTLs and Genes | Reference |
|---|---|---|---|---|
| Sex expression | CmACS-7 (a) | 2 | Inhibiting stamen development of female flowers | [] |
| CmWIP1 (g) | 10 | Suppressing female | [] | |
| CmACS11 | 3 | Negatively regulating expression of CmWIP1 | [] | |
| Sugar content | SUCQSC5.1 | 5 | Sucrose metabolism | [] |
| suc | Sucrose metabolism | [] | ||
| Acidity | PH | 8 | Flesh acidity | [] |
| Peel color | MELO3C003375 | 4 | Green peel | [] |
| CmKFB | 10 | Yellow peel color | [] | |
| Flesh color | CmOr | 9 | Orange flesh color | [] |
| MELO3C003097 | 8 | White and green flesh color | [] | |
| Powdery mildew | Pm-1 | 9 | Resistance to P. xanthii race 1 | [] |
| Pm-2 | - | Conferring resistance to PMR 5 and PMR 6 | [] | |
| Pm-3 | - | Conferring resistance to PI 124111F | [] | |
| Pm-4 | 5 | Conferring resistance to PI 124112 | [] | |
| Pm-5 | 5 | Conferring resistance to PI 124112 | [] | |
| Pm-w | 5 | Resistant to races 1 and 2 | [] | |
| Pm-x | - | Resistant to race 2F | [] | |
| Pm-y | 12 | Resistant to P. xanthii race 2 | [] | |
| Pm-R | 5 | Resistant to races 1, 2, and 5 | [] | |
| PmV.1 and PmXII.1 | 12 | Conferring resistance to PI 124112 | [] | |
| BPm12.1 | 12 | Resistant to P. xanthii race 1 | [] | |
| PmEdisto47–1 | 2 | resistance to P. xanthii race 1 | [] | |
| Pm-2F | 2 | Resistant to P. xanthii race 2F | [] | |
| Gummy stem blight | Gsb-1 | 1 | Conferring resistance to PI 1401471 | [] |
| Gsb-2 | - | Conferring resistance to PI 157082 | [] | |
| Gsb-3 | - | Conferring resistance to PI 511890 | [] | |
| Gsb-4 | - | Conferring resistance to PI 482398 | [] | |
| Gsb-5 | - | Conferring resistance to PI 482399 | [] | |
| Fusarium wilt | Fom-1 | 9 | Resistance to race 2 | [] |
| Fom-2 | 11 | Resistance to race 0 and 1 | [] | |
| Fom-3 | - | Resistance to races 0 and 2 | [] | |
| Fom-4 | - | Resistance to race 0 and 2 | [] | |
| Virus disease | Cm-eIF4E | 12 | Resistance to virus | [] |
| CmVPS41 | 12 | Resistance to CMV | [] | |
| Aphis gossypii | Vat | 5 | Resistance to aphid | [] |
| Abiotic stress | CmSKOR | 9 | Tolerance to salt | [] |
| CmLOX10 | 5 | Tolerance to drought | [] | |
| CmLOX08 | 10 | Tolerance to drought and salt | [] | |
| CmCADs | - | Tolerance to drought and salt | [] | |
| CmNCED3 | 7 | Tolerance to drought, salt, and low temperature | [] |
3. Genomics of Melon
The reference genome containing the whole genome sequence is the premise for genomics research and the utilization of plants. The first reference genome (Version 3.5) of melon with 375 Mb total length and 27,427 protein-coding genes was released in 2012, which was derived from a double-haploid line by crossing two phylogenetically distant melon cultivars from melo and agrestis []. However, the ratio of oriented scaffold assembly of just 80.8% in the first released melon genome limits its application. Subsequently, the quantity of anchored and oriented melon scaffold genome assembly was significantly improved by targeted SNP selection and defined as version 3.5.1 []. In order to update the previous annotation version 3.5.1, an improved assembly (Version 3.6.1) of the melon genome and new genome annotation (Version 4.0) were reported, which corrected the order and the orientation of 21 previous scaffolds and identified 8000 new genes []. Further, Castanera et al. [] improved the melon genome assembly (version 4.0) with the PacBio single-molecule real-time (SMRT) sequencing technology, reduced the unassigned sequences substantially, made a great effort to distinguish new gene or transposon variants related to important phenotypes. Recently, several melon genome assemblies by de novo sequencing were published based on genetically diverse individuals, which provide insights into genome structures, genome evolution, diversification, and identified candidate genes for several agronomic traits of melon [,,]. Structural variation (SV), including copy number variation (CNV) and presence/absence variation (PAV), has been shown to be frequent in plant species []. Transposons may be at the origin of an important fraction of the variability in melon besides SV []. Melon research has entered into post-genomic generation since the genome sequences were released. The analysis of genome variability using re-sequencing data has been used to shed light on the domestication history. Based on the re-sequencing of the melon genome, a comprehensive variation map of melon was constructed, and the domestication history and loci influencing agronomic traits were identified [,,,,].
4. Breeding of Melon
Melon breeding has been around for hundred years. The breeding objectives in melons have developed from enhanced yield, shelf life, disease resistance, and resistance to abiotic stress to improve fruit quality.
Crop plants encounter various biotic and abiotic stresses that hinder life throughout their growth and development. Therefore, resistance is a major objective in crop breeding. Several varieties with resistance to powdery mildew in the USA have developed by using exotic accessions from India. From the previous reports related to resistance evaluation, most accessions with resistance to powdery mildew and downy mildew derive from the momordica group and acidulus group. Additionally, there are some Turkish melon accessions for resistance to ZYMV and WMV []. PI 161375 from the conomon group was identified as a resistant accession to CMV and aphids []. Most accessions with resistance in melon come from the primary and secondary diversity centers and could be considered important germplasm reservoirs for melon breeders. Therefore, exploiting the accessions for resistance improvement in melon is imperative. In the future, there will be more and more challenges from diseases, pests, and potentially extreme weather for us. Developing cultivars with high resistance to biotic and abiotic stresses is necessary.
With the improvement of people’s living standards, fruit quality has become one of the major objectives in breeding programs as it influences fruit marketability. Fruit quality consists of many attributes, including internal quality, such as sugar and acid contents, flesh texture, and flavor, and external features, such as size, shape, and rind color. The candidates and molecular markers have been identified for the traits of rind color, flesh color, and acid content, which will benefit early selection in breeding. However, it is complex for the genetic basis of most traits related to fruit quality, especially flavor. It is an efficient strategy to dissect the genetic basis and discover the candidates of fruit flavor traits by using the comprehensive analysis of genetics, transcriptomics, and metabolomics, which has been reported in tomatoes in recent years [,].
Heterosis results in the phenotypic superiority of a hybrid over its parents with respect to traits such as growth rate, reproductive success, and yield []. The accessions of different horticultural groups in melo had high nucleotide diversity. Therefore, it is an alternative strategy to cross the melon accessions from the divergent horticultural group for germplasm innovation. Conversely, though agrestis accessions are morphological variables in fruit, it is observed that they had quite a low nucleotide diversity []. This is consistent with the fact that there was no obvious heterosis in hybrid by crossing two cultivated agrestis accessions. However, there is an obvious differentiation in the two melon subspecies, melo, and agrestis, not only for morphological characteristics, but also for ecological adaptation. It indicates that we can acquire high heterosis and diversity by inter-subspecies crossing in melon breeding, especially for the agrestis population.
Traditional breeding based on crossing and selection remains important for crop improvement. Although the efficiency of crossing and selection has been improved using marker-assisted selection, it faces limitations in crops with complex genetics. With the increase of re-sequencing data in melon, more and more polymorphic SNPs have been identified. It is feasible to construct the platform based on whole genome selection. Genome editing is expected to be a powerful tool to create desirable variation using molecular scissors and artificially engineered nucleases. The utilization of CRISPR/Cas editing can accelerate melon improvement through the introduction of genetic variation in a targeted manner []. The application of genome editing is based on an effective and stable genetic transformation system. Nevertheless, though some research related to melon genetic transformation has been reported, the efficiency and universality of distinct genotypes is the limiting factor. Fortunately, the breakthrough of genetic transformation assisted by genes encoding developmental regulators in watermelons could provide a good reference for melons [].
5. Concluding Remarks
Melon is an important horticultural crop worldwide with high diversity. In the future, resistance to biotic and abiotic stresses and fruit quality will be the most important traits for melon breeding. Identifying new genetic resources with horizontal resistance or special quality traits of melon fruit is needed. However, the genetics of the traits associated with resistance and fruit quality are always complex and are controlled by multiple loci. Though several QTLs have been identified, few candidate genes were reported. It is becoming a challenge for us to discover the causative genes and pivotal variations for these complex traits. This might be facilitated by a comprehensive analysis of genomics, transcriptomics, metabonomics, and bioinformatics.
Most modern elite varieties were developed by conventional breeding, which may not be able to meet current demands. The new strategies, such as whole genome selection, genetic transformation, and genome editing, need to be actively utilized in current breeding programs, which will provide powerful opportunities for genetic improvement of fruit quality and accelerate the process of future breeding of melon (Figure 1).
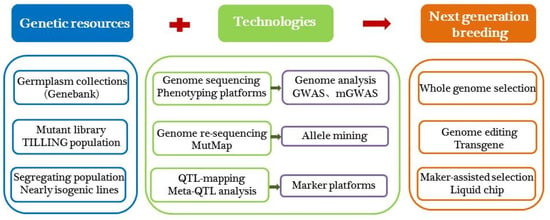
Figure 1.
The key elements of ‘Next generation breeding’.
Author Contributions
L.X. and Y.H.: writing the original draft; L.T. and Y.X.: revising the manuscript; G.Z.: writing, reviewing, revising, and finally approving the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (32172581), the Key Research and Development Program of Hainan (ZDYF2021XDNY164), the Agricultural Science and Technology Innovation Program (CAAS-ASTIP-2016-ZFRI-06), the China Agriculture Research System (CARS-25), and the Major Science and Technology Projects of Henan Province (221100110400).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Nunez-Palenius, H.G.; Gomez-Lim, M.; Ochoa-Alejo, N.; Grumet, R.; Lester, G.; Cantliffe, D.J. Melon fruits: Genetic diversity, physiology, and biotechnology features. Crit. Rev. Biotechnol. 2008, 28, 13–55. [Google Scholar] [CrossRef] [PubMed]
- Pitrat, M.; Hanelt, P.; Hammer, K. Some comments on infraspecific classification of cultivars ofmelon. Acta Hortic. 2000, 510, 29–36. [Google Scholar] [CrossRef]
- Pitrat, M. Melon genetic resources: Phenotypic diversity and horticultural taxonomy. In Genetics and Genomics of Cucurbitaceae; Springer: Berlin/Heidelberg, Germany, 2016; pp. 25–60. [Google Scholar]
- Latrasse, D.; Rodriguez-Granados, N.Y.; Veluchamy, A.; Mariappan, K.G.; Bevilacqua, C.; Crapart, N.; Camps, C.; Sommard, V.; Raynaud, C.; Dogimont, C.; et al. The quest for epigenetic regulation underlying unisexual flower development in Cucumis melo. Epigenetics Chromatin. 2017, 10, 22. [Google Scholar] [CrossRef]
- Ezura, H.; Owino, W.O. Melon, an alternative model plant for elucidating fruit ripening. Plant Sci. 2008, 175, 121–129. [Google Scholar] [CrossRef]
- Gao, P.; Sheng, Y.Y.; Luan, F.S.; Ma, H.Y.; Liu, S. RNA-Seq transcriptome profiling reveals differentially expressed genes involved in sex expression in melon. Crop Sci. 2015, 55, 1686–1695. [Google Scholar] [CrossRef]
- Garcia-Mas, J.; Benjak, A.; Sanseverino, W.; Bourgeois, M.; Mir, G.; Gonzalez, V.M.; Henaff, E.; Camara, F.; Cozzuto, L.; Lowy, E. The genome of melon (Cucumis melo L.). Proc. Natl. Acad. Sci. USA 2012, 109, 11872–11877. [Google Scholar] [CrossRef] [PubMed]
- Pitrat, M. Linkage groups in Cucumis melo L. J. Hered. 1991, 82, 406–411. [Google Scholar] [CrossRef]
- Wang, Y.H.; Thomas, C.E.; Dean, R.A. A genetic map of melon (Cucumis melo L.) based on amplified fragment length polymorphism (AFLP) markers. Theor. Appl. Genet. 1997, 95, 791–798. [Google Scholar] [CrossRef]
- Baudracco-Arnas, S.; Pitrat, M. A genetic map of melon (Cucumis melo L.) with RFLP, RAPD, isozyme, disease resistance and morphological markers. Theor. Appl. Genet. 1996, 93, 57–64. [Google Scholar] [CrossRef]
- Gonzalo, M.J.; Oliver, M.; Garcia-Mas, J.; Monfort, A.; Dolcet-Sanjuan, R.; Katzir, N.; Arus, P.; Monforte, A.J. Simple-sequence repeat markers used in merging linkage maps of melon (Cucumis melo L.). Theor. Appl. Genet. 2005, 110, 802–811. [Google Scholar] [CrossRef]
- Diaz, A.; Fergany, M.; Formisano, G.; Ziarsolo, P.; Blanca, J.; Fei, Z.; Staub, J.E.; Zalapa, J.E.; Cuevas, H.E.; Dace, G. A consensus linkage map for molecular markers and quantitative trait loci associated with economically important traits in melon (Cucumis melo L.). BMC Plant Biol. 2011, 11, 111. [Google Scholar] [CrossRef] [PubMed]
- Diaz, A.; Forment, J.; Argyris, J.M.; Fukino, N.; Tzuri, G.; Harel-Beja, R.; Katzir, N.; Garcia-Mas, J.; Monforte, A.J. Anchoring the consensus ICuGI genetic map to the melon (Cucumis melo L.) genome. Mol. Breed. 2015, 35, 188. [Google Scholar] [CrossRef]
- Deleu, W.; Esteras, C.; Roig, C.; Gonzalez-To, M.; Fernandez-Silva, I.; Gonzalez-Ibeas, D.; Blanca, J.; Aranda, M.A.; Arus, P.; Nuez, F. A set of EST-SNPs for map saturation and cultivar identification in melon. BMC Plant Biol. 2009, 9, 90. [Google Scholar] [CrossRef] [PubMed]
- Amanullah, S.; Liu, S.; Gao, P.; Zhu, Z.C.; Zhu, Q.L.; Fan, C.; Luan, F.S. QTL mapping for melon (Cucumis melo L.) fruit traits by assembling and utilization of novel SNPs based CAPS markers. Sci. Hortic. 2018, 236, 18–29. [Google Scholar] [CrossRef]
- Pereira, L.; Ruggieri, V.; Perez, S.; Alexiou, K.G.; Fernandez, M.; Jahrmann, T.; Pujol, M.; Garcia-Mas, J. QTL mapping of melon fruit quality traits using a high-density GBS-based genetic map. BMC Plant Biol. 2018, 18, 324. [Google Scholar] [CrossRef]
- Wang, P.Y.; Xu, X.J.; Zhao, G.W.; He, Y.H.; Hou, C.; Kong, W.H.; Zhang, J.; Liu, S.M.; Xu, Y.Y.; Xu, Z.H. Genetic mapping and candidate gene analysis for melon resistance to Phytophthora capsici. Sci. Rep. 2020, 10, 20456. [Google Scholar] [CrossRef]
- Lian, Q.; Fu, Q.S.; Xu, Y.Y.; Hu, Z.C.; Zheng, J.; Zhang, A.A.; He, Y.H.; Wang, C.S.; Xu, C.Q.; Chen, B.X.; et al. QTLs and candidate genes analyses for fruit size under domestication and differentiation in melon (Cucumis melo L.) based on high resolution maps. BMC Plant Biol. 2021, 21, 126. [Google Scholar] [CrossRef]
- Aamir, M.; Karmakar, P.; Singh, V.K.; Kashyap, S.P.; Pandey, S.; Singh, B.K.; Singh, P.M.; Singh, J. A novel insight into transcriptional and epigenetic regulation underlying sex expression and flower development in melon (Cucumis melo L.). Physiol. Plant. 2021, 173, 1729–1764. [Google Scholar] [CrossRef]
- Boualem, A.; Fergany, M.; Fernandez, R.; Troadec, C.; Martin, A.; Morin, H.; Sari, M.A.; Collin, F.; Flowers, J.M.; Pitrat, M.; et al. A conserved mutation in an ethylene biosynthesis enzyme leads to andromonoecy in melons. Science 2008, 321, 836–838. [Google Scholar] [CrossRef]
- Martin, A.; Troadec, C.; Boualem, A.; Rajab, M.; Fernandez, R.; Morin, H.; Pitrat, M.; Dogimont, C.; Bendahmane, A. A transposon-induced epigenetic change leads to sex determination in melon. Nature 2009, 461, 1135–1138. [Google Scholar] [CrossRef]
- Boualem, A.; Troadec, C.; Camps, C.; Lemhemdi, A.; Morin, H.; Sari, M.A.; Fraenkel-Zagouri, R.; Kovalski, I.; Dogimont, C.; Perl-Treves, R.; et al. A cucurbit androecy gene reveals how unisexual flowers develop and dioecy emerges. Science 2015, 350, 688–691. [Google Scholar] [CrossRef] [PubMed]
- Baulcombe, D.C.; Dean, C. Epigenetic regulation in plant responses to the environment. Cold Spring Harb. Perspect. Biol. 2014, 6, a019471. [Google Scholar] [CrossRef] [PubMed]
- Piferrer, F. Epigenetics of sex determination and gonadogenesis. Dev. Dyn. 2013, 242, 360–370. [Google Scholar] [CrossRef] [PubMed]
- Argyris, J.M.; Diaz, A.; Ruggieri, V.; Fernandez, M.; Jahrmann, T.; Gibon, Y.; Pico, B.; Martin-Hernandez, A.M.; Monforte, A.J.; Garcia-Mas, J. QTL analyses in multiple populations employed for the fine mapping and identification of candidate genes at a locus affecting sugar accumulation in melon (Cucumis melo L.). Front. Plant Sci. 2017, 8, 1679. [Google Scholar] [CrossRef] [PubMed]
- Dai, N.; Cohen, S.; Portnoy, V.; Tzuri, G.; Harel-Beja, R.; Pompan-Lotan, M.; Schaffer, A.A. Metabolism of soluble sugars in developing melon fruit- a global transcriptional view of the metabolic transition to sucrose accumulation. Plant Mol. Biol. 2011, 76, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Burger, Y.; Saar, U.; Katzir, N.; Paris, H.S.; Yeselson, Y.; Levin, I.; Schaffer, A.A. A single recessive gene for sucrose accumulation in Cucumis melo fruit. J. Am. Soc. Hortic. Sci. 2002, 127, 938–943. [Google Scholar] [CrossRef]
- Harel-Beja, R.; Tzuri, G.; Portnoy, V.; Lotan-Pompan, M.; Lev, S.; Cohen, S.; Dai, N.; Yeselson, L.; Meir, A.; Libhaber, S. A genetic map of melon highly enriched with fruit quality QTLs and EST markers, including sugar and carotenoid metabolism genes. Theor. Appl. Genet. 2010, 121, 511–533. [Google Scholar] [CrossRef]
- Schemberger, M.O.; Stroka, M.A.; Reis, L.; Los, K.K.D.S.; de Araujo, G.A.T.; Sfeir, M.Z.T.; Galvao, C.W.; Etto, R.M.; Baptistao, A.R.G.; Ayub, R.A. Transcriptome profiling of non-climacteric ‘yellow’ melon during ripening: Insights on sugar metabolism. BMC Genom. 2020, 21, 262. [Google Scholar] [CrossRef]
- Cheng, J.; Wen, S.; Xiao, S.; Lu, B.; Ma, M.; Bie, Z. Overexpression of the tonoplast sugar transporter CmTST2 in melon fruit increases sugar accumulation. J. Exp. Bot. 2018, 69, 511–523. [Google Scholar] [CrossRef]
- Burger, Y.; Sa’ar, U.; Distelfeld, A.; Katzir, N.; Yeselson, Y.; Shen, S.; Schaffer, A.A. Development of sweet melon (Cucumis melo) genotypes combining high sucrose and organic acid content. J. Am. Soc. Hortic. Sci. 2003, 128, 537–540. [Google Scholar] [CrossRef]
- Tang, M.; Bie, Z.L.; Wu, M.Z.; Yi, H.P.; Feng, J.X. Changes in organic acids and acid metabolism enzymes in melon fruit during development. Sci. Hortic. 2010, 123, 360–365. [Google Scholar] [CrossRef]
- Kubicki, B. Inheritance of some characters in muskmelons (Cucumis melo). Genet. Pol. 1962, 3, 265. [Google Scholar]
- Cohen, S.; Tzuri, G.; Harel-Beja, R.; Itkin, M.; Portnoy, V.; Sa’Ar, U.; Lev, S.; Yeselson, L.; Petrikov, M.; Rogachev, I. Co-mapping studies of QTLs for fruit acidity and candidate genes of organic acid metabolism and proton transport in sweet melon (Cucumis melo L.). Theor. Appl. Genet. 2012, 125, 343–353. [Google Scholar] [CrossRef] [PubMed]
- Cohen, S.; Itkin, M.; Yeselson, Y.; Tzuri, G.; Portnoy, V.; Harel-Baja, R.; Lev, S.; Sa’ar, U.; Davidovitz-Rikanati, R.; Baranes, N.; et al. The PH gene determines fruit acidity and contributes to the evolution of sweet melons. Nat. Commun. 2014, 5, 4026. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.; Lian, Q.; Zhang, Z.; Fu, Q.; He, Y.; Ma, S.; Ruggieri, V.; Monforte, A.J.; Wang, P.; Julca, I.; et al. A comprehensive genome variation map of melon identifies multiple domestication events and loci influencing agronomic traits. Nat. Genet. 2019, 51, 1607–1615. [Google Scholar] [CrossRef] [PubMed]
- Tadmor, Y.; Burger, J.; Yaakov, I.; Feder, A.; Libhaber, S.E.; Portnoy, V.; Meir, A.; Tzuri, G.; Sa’ar, U.; Rogachev, I.; et al. Genetics of flavonoid, carotenoid, and chlorophyll pigments in melon fruit rinds. J. Agric. Food Chem. 2010, 58, 10722–10728. [Google Scholar] [CrossRef]
- Feder, A.; Burger, J.; Gao, S.; Lewinsohn, E.; Katzir, N.; Schaffer, A.A.; Meir, A.; Davidovich-Rikanati, R.; Portnoy, V.; Gal-On, A.; et al. A Kelch domain-containing F-Box coding gene negatively regulates flavonoid accumulation in muskmelon. Plant Physiol. 2015, 169, 1714–1726. [Google Scholar]
- Burger, Y.; Sa’ar, U.; Paris, H.S.; Lewinsohn, E.; Katzir, N.; Tadmor, Y.; Schaffer, A. Genetic variability for valuable fruit quality traits in Cucumis melo. Isr. J. Plant Sci. 2006, 54, 233–242. [Google Scholar] [CrossRef]
- Oren, E.; Tzuri, G.; Vexler, L.; Dafna, A.; Meir, A.; Faigenboim, A.; Kenigswald, M.; Portnoy, V.; Schaffer, A.A.; Levi, A.; et al. The multi-allelic APRR2 gene is associated with fruit pigment accumulation in melon and watermelon. J. Exp. Bot. 2019, 70, 3781–3794. [Google Scholar] [CrossRef]
- Shen, J.; Xu, X.; Zhang, Y.; Niu, X.; Shou, W. Genetic Mapping and identification of the candidate genes for mottled rind in Cucumis melo L. Front. Plant Sci. 2021, 12, 2563. [Google Scholar] [CrossRef]
- Liu, L.; Sun, T.; Liu, X.; Guo, Y.; Huang, X.; Gao, P.; Wang, X. Genetic analysis and mapping of a striped rind gene (st3) in melon (Cucumis melo L.). Euphytica 2019, 215, 1–12. [Google Scholar] [CrossRef]
- Clayberg, C.D. Interaction and linkage test of flesh colour genes in Cucumis melo L. Cucurbit Genet. Coop. 1992, 15, 53. [Google Scholar]
- Tzuri, G.; Zhou, X.; Chayut, N.; Yuan, H.; Portnoy, V.; Meir, A.; Sa’ar, U.; Baumkoler, F.; Mazourek, M.; Lewinsohn, E.; et al. A ‘golden’ SNP in CmOr governs the fruit flesh color of melon (Cucumis melo). Plant J. 2015, 82, 267–279. [Google Scholar] [CrossRef] [PubMed]
- Galpaz, N.; Gonda, I.; Shem-Tov, D.; Barad, O.; Tzuri, G.; Lev, S.; Fei, Z.; Xu, Y.; Mao, L.; Jiao, C.; et al. Deciphering genetic factors that determine melon fruit-quality traits using RNA-Seq-based high-resolution QTL and eQTL mapping. Plant J. 2018, 94, 169–191. [Google Scholar] [CrossRef] [PubMed]
- McGrath, M.T. Powdery Mildew of Cucurbits; New York State IPM Program: Geneva, NY, USA, 1997. [Google Scholar]
- Perchepied, L.; Bardin, M.; Dogimont, C.; Pitrat, M. Relationship between loci conferring downy mildew and powdery mildew resistance in melon assessed by quantitative trait loci mapping. Phytopathology 2005, 95, 556–565. [Google Scholar] [CrossRef]
- Sowell, G., Jr.; Corley, W.L. Severity of race 2 of Sphaerotheca fuliginea (Schlecht.) Poll. on muskmelon introductions reported resistant to powdery mildew. HortScience 1974, 9, 398–399. [Google Scholar] [CrossRef]
- McCreight, J.D.; Coffey, M.D.; Turini, T.A.; Matheron, M.E. Field evidence for a new race of powdery mildew on melon. Hortscience 2005, 40, 888. [Google Scholar] [CrossRef]
- Křístková, E.; Lebeda, A.; Sedláková, B. Temporal and spatial dynamics of powdery mildew species on cucurbits in the Czech Republic. Acta Hortic. 2007, 731, 337–343. [Google Scholar] [CrossRef]
- Kenigsbuch, D.; Cohen, Y. Independent inheritance of resistance to race 1 and race 2 of Sphaerotheca fuliginea in muskmelon. Plant Dis. 1989, 73, 206–208. [Google Scholar] [CrossRef]
- Ning, X.; Wang, X.; Gao, X.; Zhang, Z.; Zhang, L.; Yan, W.; Li, G. Inheritances and location of powdery mildew resistance gene in melon Edisto47. Euphytica 2014, 195, 345–353. [Google Scholar] [CrossRef]
- Jagger, I.C.; Whitaker, T.W.; Porter, D.R. Inheritance in Cucumis melo of resistance to powdery mildew (Erysiphe cichoracerarum). Phytopathology 1938, 28, 671. [Google Scholar]
- Cohen, Y.; Eyal, H. Reaction of Muskmelon Genotypes to Races 1 and 2 of Sphaerotheca fuliginea in Israel. Cucurbit Genet. Coop. 1988, 11, 4–49. [Google Scholar]
- Thomas, C.; Charleston, S.C. Inheritance and allelism of genes for resistance to races 1 and 2 of Sphaerotheca fuliginea in muskmelon. Plant Dis. 1992, 76, 626–629. [Google Scholar]
- Harwood, R.; Markarian, D. The inheritance of resistance to powdery mildew in the cantaloupe variety Seminole. J. Hered. 1968, 59, 126–130. [Google Scholar] [CrossRef]
- Yuste-Lisbona, F.J.; Capel, C.; Gómez-Guillamón, M.L.; Capel, J.; López-Sesé, A.I.; Lozano, R. Codominant PCR-based markers and candidate genes for powdery mildew resistance in melon (Cucumis melo L.). Theor. Appl. Genet. 2011, 122, 747–758. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Zhao, Y.; Zhu, Q.; Zhang, Z.; Fan, C.; Amanullah, S.; Gao, P.; Luan, F. Mapping of powdery mildew resistance genes in melon (Cucumis melo L.) by bulked segregant analysis. Sci. Hortic. 2017, 220, 160–167. [Google Scholar] [CrossRef]
- Zhang, C.; Ren, Y.; Guo, S.; Zhang, H.; Gong, G.; Du, Y.; Xu, Y. Application of comparative genomics in developing markers tightly linked to the Pm-2F gene for powdery mildew resistance in melon (Cucumis melo L.). Euphytica 2013, 190, 157–168. [Google Scholar] [CrossRef]
- Keinath, A.; Farnham, M.; Zitter, T. Morphological, pathological, and genetic differentiation of Didymella bryoniae and Phoma spp. isolated from cucurbits. Phytopathology 1995, 85, 364–369. [Google Scholar] [CrossRef]
- Prasad, K.; Norton, J.D. Inheritance of resistance to Mycosphaerella citrullina in muskmelon. J. Am. Soc. Hort. Sci. 1967, 91, 396–400. [Google Scholar]
- Frantz, J.D.; Jahn, M.M. Five independent loci each control monogenic resistance to gummy stem blight in melon (Cucumis melo L.). Theor. Appl. Genet. 2004, 108, 1033–1038. [Google Scholar] [CrossRef]
- Santos, L.S.; Candido, S.; Rabelo, H.; Marin, M.V.; Gaion, L.A.; Gomes, R.F.; Camargo, M.; Braz, L.T. Reaction of melon genotypes to Didymella bryoniae (Fuckel) Rehm. Chil. J. Agric. Res. 2017, 77, 71–77. [Google Scholar] [CrossRef][Green Version]
- Virtuoso, M.C.S.; Valente, T.S.; Silva, E.H.C.; Braz, L.T.; Cassia-Panizzi, R.; Vargas, P.F. Implications of the inoculation method and environment in the selection of melon genotypes resistant to Didymella bryoniae. Sci. Hortic. 2022, 300, 111066. [Google Scholar] [CrossRef]
- Hassan, M.Z.; Rahim, M.A.; Natarajan, S.; Robin, A.H.K.; Kim, H.T.; Park, J.I.; Nou, I.S. Gummy stem blight resistance in melon: Inheritance pattern and development of molecular markers. Int. J. Mol. Sci. 2018, 19, 2914. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Deng, G.; Mou, H.; Xu, Y.; Chen, L.; Yang, J.; Zhang, M. A re-sequencing-based ultra-dense genetic map reveals a gummy stem blight resistance-associated gene in Cucumis melo. DNA Res. 2018, 25, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Oumouloud, A.; Álvarez, J.M. Breeding and genetics of resistance to Fusarium wilt in melon. In Advances in Plant Breeding Strategies: Agronomic, Abiotic and Biotic Stress Traits; Springer: Berlin/Heidelberg, Germany, 2016; pp. 601–626. [Google Scholar]
- Risser, G.; Davis, D.W. A proposed nomenclature of Fusarium oxysporum f. sp. melonis races and resistance genes in Cucumis melo. Phytopathology 1976, 66, 1105–1106. [Google Scholar] [CrossRef]
- Zink, F.; Thomas, C. Genetics of resistance to Fusarium oxysporum f. sp. melonis races 0, 1, and 2 in muskmelon line MR-1. Phytopathology 1990, 80, 1230–1232. [Google Scholar] [CrossRef]
- Joobeur, T.; King, J.J.; Nolin, S.J.; Thomas, C.E.; Dean, R.A. The Fusarium wilt resistance locus Fom-2 of melon contains a single resistance gene with complex features. Plant J. 2004, 39, 283–297. [Google Scholar] [CrossRef]
- Tezuka, T.; Waki, K.; Yashiro, K.; Kuzuya, M.; Ishikawa, T.; Takatsu, Y.; Miyagi, M. Construction of a linkage map and identification of DNA markers linked to Fom-1, a gene conferring resistance to Fusarium oxysporum f. sp. melonis race 2 in melon. Euphytica 2009, 168, 177–188. [Google Scholar] [CrossRef]
- Oumouloud, A.; Arnedo-Andrés, M.; González-Torres, R.; Alvarez, J.M. Inheritance of resistance to Fusarium oxysporum f. sp. melonis races 0 and 2 in melon accession Tortuga. Euphytica 2010, 176, 183–189. [Google Scholar] [CrossRef]
- Zink, F.; Gubler, W.D. Inheritance of resistance in muskmelon to Fusarium wilt. J. Am. Soc. Hortic. Sci. 1985, 110, 600–604. [Google Scholar] [CrossRef]
- Deol, J.K.; Sharma, S.P.; Rani, R.; Kalia, A.; Chhuneja, P.; Sarao, N.K. Inheritance analysis and identification of SSR markers associated with fusarium wilt resistance in melon. J. Hortic. Sci. Biotechnol. 2022, 97, 66–74. [Google Scholar] [CrossRef]
- Thakur, H.; Sharma, S.; Thakur, M. Biotechnology: Recent trends in muskmelon (Cucumis melo L.) research: An overview. J. Hortic. Sci. Biotechnol. 2019, 94, 533–547. [Google Scholar] [CrossRef]
- Argyris, J.M.; Pujol, M.; Martín-Hernández, A.M.; Garcia-Mas, J. Combined use of genetic and genomics resources to understand virus resistance and fruit quality traits in melon. Physiol. Plant. 2015, 155, 4–11. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Hernandez, A.M.; Gosalvez, B.; Sempere, R.N.; Burgos, L.; Aranda, M.A.; Truniger, V. Melon RNA interference (RNAi) lines silenced for Cm-eIF4E show broad virus resistance. Mol. Plant Pathol. 2012, 13, 755–763. [Google Scholar] [CrossRef] [PubMed]
- Truniger, V.; Aranda, M.A. Recessive resistance to plant viruses. Adv. Virus Res. 2009, 75, 119–231. [Google Scholar] [PubMed]
- Diaz-Pendon, J.A.; Truniger, V.; Nieto, C.; Garcia-Mas, J.; Bendahmane, A.; Aranda, M.A. Advances in understanding recessive resistance to plant viruses. Mol. Plant Pathol. 2004, 5, 223–233. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Johansen, E.; Eyers, S.; Thomas, C.L.; Noel Ellis, T.; Maule, A.J. The potyvirus recessive resistance gene, sbm1, identifies a novel role for translation initiation factor eIF4E in cell-to-cell trafficking. Plant J. 2004, 40, 376–385. [Google Scholar] [CrossRef]
- Guiu-Aragonés, C.; Monforte, A.J.; Saladié, M.; Corrêa, R.X.; Garcia-Mas, J.; Martín-Hernández, A.M. The complex resistance to cucumber mosaic cucumovirus (CMV) in the melon accession PI161375 is governed by one gene and at least two quantitative trait loci. Mol. Breed. 2014, 34, 351–362. [Google Scholar] [CrossRef]
- Pascual, L.; Yan, J.; Pujol, M.; Monforte, A.J.; Picó, B.; Martín-Hernández, A.M. CmVPS41 is a general gatekeeper for resistance to Cucumber mosaic virus phloem entry in melon. Front. Plant Sci. 2019, 10, 1219. [Google Scholar] [CrossRef]
- Adler-Berke, N.; Goldenberg, Y.; Brotman, Y.; Kovalski, I.; Gal-On, A.; Doniger, T.; Harel-Beja, R.; Troadec, C.; Bendahmane, A.; Pitrat, M.; et al. The melon zym locus conferring resistance to ZYMV: High resolution mapping and candidate gene identification. Agronomy 2021, 11, 2427. [Google Scholar] [CrossRef]
- Marone, D.; Russo, M.A.; Laidò, G.; De Leonardis, A.M.; Mastrangelo, A.M. Plant nucleotide binding site-leucine-rich repeat (NBS-LRR) genes: Active guardians in host defense responses. Int. J. Mol. Sci. 2013, 14, 7302–7326. [Google Scholar] [CrossRef] [PubMed]
- Tian, D.; Traw, M.; Chen, J.; Kreitman, M.; Bergelson, J. Fitness costs of R-gene-mediated resistance in Arabidopsis thaliana. Nature 2003, 423, 74–77. [Google Scholar] [CrossRef] [PubMed]
- Johansson, I.; Wulfetange, K.; Porée, F.; Michard, E.; Gajdanowicz, P.; Lacombe, B.; Sentenac, H.; Thibaud, J.B.; Mueller-Roeber, B.; Blatt, M.R.; et al. External K+ modulates the activity of the Arabidopsis potassium channel SKOR via an unusual mechanism. Plant J. 2006, 46, 269–281. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.T.; Zhao, L.N.; Gao, L.W.; Very, A.A.; Sentenac, H.; Zhang, Y.D. Constitutive expression of CmSKOR, an outward K+ channel gene from melon, in Arabidopsis thaliana involved in saline tolerance. Plant Sci. 2018, 274, 492–502. [Google Scholar]
- Xing, Q.; Liao, J.; Cao, S.; Li, M.; Lv, T.; Qi, H. CmLOX10 positively regulates drought tolerance through jasmonic acid-mediated stomatal closure in oriental melon (Cucumis melo var. makuwa makino). Sci. Rep. 2020, 10, 17452. [Google Scholar] [CrossRef]
- Wang, C.; Gao, G.; Cao, S.; Xie, Q.; Qi, H. Isolation and functional validation of the CmLOX08 promoter associated with signaling molecule and abiotic stress responses in oriental melon (Cucumis melo var. makuwa Makino). BMC Plant Biol. 2019, 19, 75. [Google Scholar]
- Liu, W.; Jin, Y.; Li, M.; Dong, L.; Guo, D.; Lu, C.; Qi, H. Analysis of CmCADs and three lignifying enzymes in oriental melon (CaiHong7) seedlings in response to three abiotic stresses. Sci. Hortic. 2018, 237, 257–268. [Google Scholar] [CrossRef]
- Chevilly, S.; Dolz-Edo, L.; Martínez-Sánchez, G.; Morcillo, L.; Vilagrosa, A.; López-Nicolás, J.M.; Blanca, J.; Yenush, L.; Mulet, J.M. Distinctive traits for drought and salt stress tolerance in melon (Cucumis melo L.). Front Plant Sci. 2021, 12, 777060. [Google Scholar] [CrossRef]
- Argyris, J.M.; Ruiz-Herrera, A.; Madriz-Masis, P.; Sanseverino, W.; Morata, J.; Pujol, M.; Ramos-Onsins, S.E.; Garcia-Mas, J. Use of targeted SNP selection for an improved anchoring of the melon (Cucumis melo L.) scaffold genome assembly. BMC Genom. 2015, 16, 4. [Google Scholar] [CrossRef]
- Ruggieri, V.; Alexiou, K.G.; Morata, J.; Argyris, J.; Pujol, M.; Yano, R.; Nonaka, S.; Ezura, H.; Latrasse, D.; Boualem, A. An improved assembly and annotation of the melon (Cucumis melo L.) reference genome. Sci. Rep. 2018, 8, 8088. [Google Scholar] [CrossRef]
- Castanera, R.; Ruggieri, V.; Pujol, M.; Garcia-Mas, J.; Casacuberta, J.M. An improved melon reference genome with single-molecule sequencing uncovers a recent burst of transposable elements with potential impact on genes. Front. Plant Sci. 2020, 10, 1815. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Deng, G.; Lian, J.; Garraway, J.; Niu, Y.; Hu, Z.; Yu, J.; Zhang, M. The chromosome-scale genome of melon dissects genetic architecture of important agronomic Traits. iScience 2020, 23, 101422. [Google Scholar] [CrossRef] [PubMed]
- Shin, A.Y.; Koo, N.; Kim, S.; Sim, Y.M.; Choi, D.; Kim, Y.M.; Kwon, S.Y. Draft genome sequences of two oriental melons, Cucumis melo L. var. makuwa. Sci. Data 2019, 6, 220. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Li, X.; Yu, H.; Zhang, Y.; Li, M.; Wang, H.; Wang, D.; Wang, H.; Fu, Q.; Liu, M.; et al. A high-quality melon genome assembly provides insights into genetic basis of fruit trait improvement. iScience 2019, 22, 16–27. [Google Scholar] [CrossRef]
- Saxena, R.K.; Edwards, D.; Varshney, R.K. Structural variations in plant genomes. Brief. Funct. Genom. 2014, 13, 296–307. [Google Scholar] [CrossRef] [PubMed]
- Sanseverino, W.; Hénaff, E.; Vives, C.; Pinosio, S.; Burgos-Paz, W.; Morgante, M.; Ramos-Onsins, S.E.; Garcia-Mas, J.; Gasacuberta, J.M. Transposon insertions, structural variations, and SNPs contribute to the evolution of the melon genome. Mol. Biol. Evol. 2015, 32, 2760–2774. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Gao, P.; Zhu, Q.; Zhu, Z.; Liu, H.; Wang, X.; Weng, Y.; Gao, M.; Luan, F. Resequencing of 297 melon accessions reveals the genomic history of improvement and loci related to fruit traits in melon. Plant Biotechnol. J. 2020, 18, 2545–2558. [Google Scholar] [CrossRef] [PubMed]
- Nimmakayala, P.; Tomason, Y.R.; Abburi, V.L.; Alvarado, A.; Saminathan, T.; Vajja, V.G.; Salazar, G.; Panicker, G.K.; Levi, A.; Wechteret, W.P.; et al. Genome-wide differentiation of various melon horticultural groups for use in GWAS for fruit firmness and construction of a high resolution genetic map. Front. Plant Sci. 2016, 7, 1437. [Google Scholar] [CrossRef]
- Natarajan, S.; Kim, H.T.; Thamilarasan, S.K.; Veerappan, K.; Park, J.I.; Nou, I.S. Whole genome re-sequencing and characterization of powdery mildew disease-associated allelic variation in melon. PLoS ONE 2016, 11, e0157524. [Google Scholar] [CrossRef] [PubMed]
- Kishor, D.S.; Noh, Y.; Song, W.H.; Lee, G.P.; Park, Y.; Jung, J.K.; Shim, E.J.; Sim, S.C.; Chung, S.M. SNP marker assay and candidate gene identification for sex expression via genotyping-by-sequencing-based genome-wide associations (GWAS) analyses in Oriental melon (Cucumis melo L. var. makuwa). Sci. Hortic. 2021, 276, 109711. [Google Scholar] [CrossRef]
- Ekbic, E.; Fidan, H.; Yildiz, M.; Kazim, A. Screening of Turkish melon accessions for resistance to ZYMV, WMV and CMV. Not. Sci. Biol. 2010, 2, 55–57. [Google Scholar] [CrossRef]
- Moing, A.; Allwood, J.W.; Aharoni, A.; Baker, J.; Beale, M.H.; Ben-Dor, S.; Biais, B.; Brigante, F.; Burger, Y.; Deborde, C.; et al. Comparative metabolomics and molecular phylogenetics of melon (Cucumis melo, Cucurbitaceae) biodiversity. Metabolites 2020, 10, 121. [Google Scholar] [CrossRef] [PubMed]
- Tieman, D.; Zhu, G.; Resende, M.F., Jr.; Lin, T.; Nguyen, C.; Bies, D.; Rambla, J.L.; Beltran, K.S.O.; Taylor, M.; Zhang, B.; et al. A chemical genetic roadmap to improved tomato flavor. Science 2017, 355, 391–394. [Google Scholar] [CrossRef] [PubMed]
- Lippman, Z.B.; Zamir, D. Heterosis: Revisiting the magic. Trends Genet. 2007, 23, 60–66. [Google Scholar] [CrossRef]
- Pan, W.; Cheng, Z.; Han, Z.; Yang, H.; Zhang, W.; Zhang, H. Efficient genetic transformation and CRISPR/Cas9-mediated genome editing of watermelon assisted by genes encoding developmental regulators. J. Zhejiang Univ. Sci. B 2022, 23, 339–344. [Google Scholar] [CrossRef]
- Feng, Q.; Xiao, L.; He, Y.; Liu, M.; Wang, J.; Tian, S.; Zhang, X.; Yuan, L. Highly efficient, genotype-independent transformation and gene editing in watermelon (Citrullus lanatus) using a chimeric ClGRF4-GIF1 gene. J. Integr. Plant Biol. 2021, 63, 2038–2042. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).