Abstract
Faba bean is gaining attention in Australia as a rotation grain legume where most of the country’s produce is exported. Rust, caused by Uromyces viciae-fabae, is a major constraint to Faba bean production in eastern Australia and its chemical control results in increased cost of production. The deployment of diverse sources of resistance in new cultivars underpins economic and eco-friendly control of rust diseases of crops. A selection from cultivar Doza (Doza#12034) and a European accession Ac1655 exhibited seedling rust resistance against U. viciae-fabae pathotype 24–40. Doza#12034 and Ac1655 were crossed with a susceptible genotype Fiord and recombinant inbred line (RIL) F6 populations were generated. Rust tests on Fiord/Doza#12034 and Fiord/Ac1655 F4 and F6 populations demonstrated monogenic inheritance of resistance in both crosses and the underlying resistance loci were named Uvf-2 and Uvf-3, respectively. Genetic mapping of both RIL populations located Uvf-2 and Uvf-3 in chromosomes III and V, respectively. The SNPs that showed linkage with Uvf-2 and Uvf-3 were converted into kompititive allele specific PCR (KASP) assays. Markers KASP_Vf_0703 and KASP_C250539 flanked Uvf-2 at 4.9 cM and 2.9 cM distances, whereas Uvf-3 was flanked by KASP_Ac×F165 (2.5 cM) and KASP_vf_1090 (10.1 cM). Markers KASP_Vf_0703 and KASP_Ac×F165 can be used for marker-assisted selection of Uvf-2 and Uvf-3, respectively, after confirming parental polymorphisms.
1. Introduction
Faba bean (Vicia faba L.) is an early domesticated crop species and a source of protein for humans and animals. It is known to have originated from three primary centers in the Fertile Crescent; Near East, Iraq and Iran with unknown wild progenitor(s) and all hybridization efforts with other Vicia species have proved futile [1,2,3]. In the absence of wild relatives of modern faba bean, genetic diversity is restricted to the available primary gene pool.
China, Ethiopia and Australia contribute more than 70% to the global faba bean production [4]. It was introduced in Australia in the late 20th century as a rotational crop after wheat and since been gradually established as an important legume crop because of its increasing export potential [5]. Australia is the world’s leading exporter of faba bean with an average of 218 thousand tons (69% of total produce) to Egypt and other Middle Eastern countries [6]. It plays a key role in soil fertility restoration through nitrogen fixation by its successful association with Rhizobium sp. (root nodulation bacterium), therefore making a significant contribution to agricultural sustainability [7].
The key objectives in faba bean improvement programs include resistance to biotic and abiotic factors, matching phenology to targeted cropping regions and incorporation of better agronomic traits for higher yield [8,9]. In Australia, faba bean production is threatened by fungal and viral diseases. Faba bean rust, caused by Uromyces viciae-fabae, is the most commonly occurring disease in major cultivation sites of northern New South Wales (NSW) and southern Queensland (QLD) [10]. Rust can appear throughout the crop season, but early onset of the disease can destroy 70–80% of the crop [11,12]. Unlike cereals, none of the faba bean genotypes is immune to rust [5,13]. However, a late necrosis observed in some cases [13,14] can reduce disease severity.
Sustained release of rust resistant cultivars of faba bean relies on the availability of genetically diverse sources of resistance. No wild species is sufficiently close to allow inter-specific gene transfer, which restricts genetic diversity to the primary gene pool [15]. Pioneering work on exploring inheritance of rust resistance dates back to the 1980s when Canadian researchers identified and named three to seven resistance genes in various studies [16,17,18]. A similar set of Canadian genotypes showed resistance when tested under field and greenhouse conditions in Spain [9,19] and Australia [5,13]. Two independently inherited dominant rust resistance genes with possible additivity, and a complimentary gene pair were reported against Australian faba bean rust isolates [13,20]. Although various sources of rust resistance were reported, none of these were precisely characterised and genetically mapped. Only Uvf-1 was mapped using Randomly Amplified Polymorphic DNA (RAPD) markers [21] and information on the use of this RAPD marker in faba bean breeding is lacking.
Genetic mapping leads to relative ordering of various loci on chromosomes of an organism determined through recombination [22]. It is a powerful tool that facilitates the development of closely linked DNA markers with a particular phenotype/locus [23]. The application of trait-linked markers to improve selection efficiency in breeding programs is highly desirable to avoid expensive and time-consuming phenotypic tests, particularly in the case of quantitatively inherited traits. Simple sequence repeat (SSR) markers have been and are still being used for mapping of economic traits and their marker-assisted selection (MAS) in many crop plants. Genotyping-by-sequencing based on transcriptome (GBS-t) has maximized the capacity of marker identification in plants species with large genome sizes in the absence of whole genome sequences [24]. Therefore, it has become the method of choice for marker development in legume crops such as field pea [25], yellow lupin (Lupinus luteus L.) [26], lentil [27], chickpea (Cicer arientinum L.) [28], and faba bean [29,30]. GBS-t enables the discovery of trait-linked SNP markers which are co-dominant in nature and amenable to high throughput systems for selection in crop improvement programs [31].
The moderately rust resistant cultivar Doza was released in 2008 for cultivation in northern NSW and southern Queensland regions [6]. Doza was heterogenous for rust response at the time of release. Adhikari et al. [13] reported three single rust resistant plants (#12034, #12035 and #14916), selected by Mr. J. van Leur, under field conditions. Doza#12034 expressed strong chlorosis leading to the death of infected leaves on the first and the second nodes within 2–3 days post-sporulation in the seedling tests [20]. The faba bean accession Ac1655 was reported to exhibit IT 2+ in greenhouse against rust isolates collected from Cordoba, Spain [14].
The objectives of this study were to: (1) characterise seedling rust resistance in Fiord/Doza#12034 and Fiord/Ac1655 recombinant inbred line (RIL) populations, (2) determine genomic locations of resistance in Doza#12034 and Ac1655 through molecular mapping, (3) develop KASP markers linked with the underlying resistance loci and (4) validate resistance linked markers on a set of local and exotic cultivars/genotypes to demonstrate their utility in marker-assisted breeding.
2. Materials and Methods
2.1. Plant Material and Development of RIL Populations
Two rust resistant faba bean genotypes, a single plant selection #12034 from an Australian faba bean cultivar Doza and the European line Ac1655 (V-300) from Spain (originally from central Europe), were selected for the present study. The RIL populations were developed by crossing resistant parents Doza#12034 and Ac1655 with the susceptible cultivar Fiord at the University of Sydney Plant Breeding Institute, Narrabri under controlled conditions to avoid cross pollination. One hundred-forty and 145 F2 seeds were harvested from Doza#12034/Fiord and Ac1655/Fiord crosses, respectively. A single pod from each F2 plant was harvested and used for generation advancement. This process was repeated for four generations and finally F6 population of 104 Fiord/Doza#12034 and 120 Fiord/Ac1655 individuals were harvested. Both resistant parents, Doza#12034 and Ac1655, were also crossed with each other to study allelic relationship of the underlying genes and to understand their interaction. From this cross, 92 F2 seeds were harvested and no further population advancement was carried out.
2.2. Pathogen Material and Inheritance Studies
The rust isolate-8 collected from Breeza, New South Wales, named pathotype 24–40 [32], was used to study inheritance of rust resistance in Fiord/Doza#12034 and Fiord/Ac1655 F4 and F6 RILs and Doza#12034/Ac1655 F2 population under controlled environment conditions at the University of Sydney Plant Breeding Institute, Cobbitty. Parents Doza#12034, Ac1655 and Fiord were planted as controls with each experiment. Ten seeds of each RIL and five seeds of each parent were sown under greenhouse conditions and inoculated according to the procedure described by Adhikari et al. [13]. The wheat rust infection type (0–4) scale was used, where 0 = no visible uredinia, ; = hypersensitive pin head flecks, 1 = small uredinia with necrosis, 2 = small to medium uredinia with by green island and necrosis/chlorosis, 3 = medium sized uredinia with or without chlorosis and 4 = large uredinia without chlorosis. The F4 populations were classified as homozygous resistant (HR), segregating (SEG) and homozygous susceptible (HS), whereas F6 RIL populations were categorized as HR and HS. Chi-squared (χ2) analyses were performed to determine the number of genes involved in conditioning rust resistance in Doza#12034 and Ac1655.
2.3. DNA and RNA Extraction
DNA was extracted from seedlings of Fiord/Doza#12034 and Fiord/Ac1655 RILs, parents (Doza#12034, Ac1655 and Fiord) and a set of 40 genotypes (Supplementary Table S1) using modified Cetyltrimethylammonium bromide (CTAB) method [33] with an addition of 2% Polyvinylpyrrolidone (PVP) in extraction buffer. The quality of DNA was analysed on 1% agarose gel, quantification was performed using Nanodrop spectrophotometer (Nanodrop Technologies) and working dilutions of 30 ng/µL concentration were prepared for marker genotyping. All DNA samples were stored at −20 °C.
Fiord/Doza#12034 (F6) and Fiord/Ac1655 (F4) populations were sown in a tray (54 cm × 28 cm) and leaf tissue from each line (two replicates) was harvested and immediately transferred to liquid nitrogen before storage at −80 °C. Total RNA was extracted from all samples using QIAGEN RNeasy 96 kit following manufacturer’s instructions.
2.4. Genotyping-by-Sequencing and SNP Discovery
Fiord/Doza#12034 and Fiord/Ac1655 F4 populations were genotyped with Illumina Infinium 1536 -SNP array. In addition, RNAseq-based genotyping-by-sequencing (GBS-t) was performed on Fiord/Ac1655 F4 and Fiord/Doza#12034 F6 RIL populations.
Following mRNA enrichment in both populations, the sequencing library preparation was performed using the SureSelect stranded RNA library preparation kit (Agilent Technologies, Santa Clara, CA, USA) following the manufacturer’s instructions. Libraries were evaluated using the TapeStation 2200 platform (Agilent Technologies, Santa Clara, United States), pooled, and quantified using the KAPA library quantification kit (KAPA Biosystems, Wilmington, NC, USA). Sequencing data were generated using Illumina HiSeq 3000 (Illumina Inc., San Diego, CA, USA). The reference transcriptome of Doza published by Braich et al. [34] was used for SNP discovery, whereas a less comprehensive sequence was used in the case of Ac1655.
2.5. Construction of Linkage Map
The SNP alleles were named as ‘A’: Doza or Ac1655 (in respective population), ‘B’: Fiord, ‘H’: heterozygote and ‘-’ for missing values. Genotypic data were tested for segregation distortion of markers using χ2 test and markers with distorted segregation were removed from further analysis. Monomorphic markers, redundant markers and markers with more than 10% missing data were also excluded. Mapping was performed using Kosambi [35] mapping function with Map Manager version QTXb20 [36] for the Fiord/Doza#12034 RIL population. The linkage groups (LGs) were generated using the R/ASMap package [37] and linkage maps and the joint linkage map diagrams were drawn using the MapChart software [38].
2.6. KASP Genotyping
Rust resistance-linked SNPs from both RIL populations were converted into Kompetitive Allele Specific PCR (KASP) assays. The primers were designed using online tool BatchPrimer3 [39]. The primers with GC content ~50% were picked to construct allele specific-1(A-1), allele specific-2 (A-2), and common (C) primers (Supplementary Table S2). The KASP primers were assayed on the entire respective RIL population and parents using KASP technology (LGC genomics, UK) described in Nsabiyera et al. [40]. The closely linked KASP markers were also assayed on a set of 40 genotypes to test polymorphism (Supplementary Table S1).
3. Results
3.1. Genetic Analysis
The resistant parents Doza#12034 and Ac1655 produced infection types (ITs) 1C and 12C, respectively, and the susceptible parent Fiord expressed IT 33+, when tested against rust pathotype 24-40 (Figure 1). Screening of F4 generations of both crosses showed monogenic segregation for rust resistance. Forty-four Fiord/Doza#12034 F4 lines were scored HR, 24 SEG and 42 HS (χ2 3:2:3 = 0.64, non-significant at p = 0.05 and 2 d.f.). Similarly, the Fiord/Ac1655 F4 population included 34 HR and 17 SEG and 32 HS lines (χ2 3:2:3 = 0.97, non- significant at p = 0.05 and 2 d.f.). Monogenic segregation for rust resistance was confirmed through tests on F6 generations of both crosses (Table 1).

Figure 1.
Infection types produced by parental genotypes when inoculated with the U. viciae-fabae pathotype 24–40.

Table 1.
Seedling rust response variation among Fiord/Doza#12034, Fiord/Ac1655 RIL, and Doza#12034/Ac1655 populations when tested with Uromyces viciae-fabae pathotype 20–40 under greenhouse conditions.
The F1 seedlings of Doza#12034/Ac1655 cross produced IT ;1=. The F2 population showed digenic segregation (15 resistant: 1 susceptible) to demonstrate that the two sources of resistance carry genetically independent genes (Table 1). The ITs among resistant plants varied from ;1= to 12C. These results confirmed the independent and dominant inheritance of resistance in both parents. The rust resistance loci from Doza#12034 and Ac1655 were named Uvf-2 and Uvf-3, respectively.
3.2. Molecular Mapping of Rust Resistance
3.2.1. Construction of Linkage Maps
A total of 409 and 557 markers were used to generate linkage groups for Fiord/Doza#12034 and Fiord/Ac1655 F4 populations using JoinMap 4 software. The Fiord/Doza#12034 linkage map covered 969 cM and the Fiord/Ac1655 spanned 1434 cM. QTL detection was performed using simple interval mapping (SIM) and composite interval mapping (CIM).
3.2.2. Mapping of Uvf-2
Single genomic region on chromosome III of Fiord/Doza#12034 showed association with rust resistance (Figure 2a) and it was named q_rust_Doza. It was flanked by SNP markers Vf_0685 and Vf_0035. Six SNPs were converted into KASP assays. Four KASP markers (KASP_Vf_0703, KASP_Vf_0493, KASP_Vf_0198 and KASP_Vf_0032) that showed polymorphisms between parents Doza#12034 and Fiord were mapped on 104 lines of the Fiord/Doza#12034 F6 RIL population and covered a 37.1 cM interval. Rust resistance gene Uvf-2 mapped 4.9 cM away from the marker KASP_Vf_0703 (Figure 2b).

Figure 2.
Linkage maps of chromosome III of the Fiord/Doza#12034 RIL population: (a) location of QTL for rust resistance in F4 and (b) association of KASP markers converted from SNP in the QTL region with rust resistance gene Uvf-2.
A total of 15,672 high-confidence SNPs were identified by alignment of GBS-t reads against the reference transcriptome data of Doza. Of those, 20% (3138) showed distorted segregation and 7988 had more than 10% missing data. These markers were excluded from the analysis. Redundant markers were filtered out from the remaining 4546 polymorphic SNPs resulting in 3924 that were used for constructing the genetic linkage map. In total, 2784 (out of 3924) SNP markers were distributed across 12 linkage groups (LGs) covering 1026.59 cM with an average marker density of 0.369 cM (Supplementary Table S3 and Supplementary Figure S1) and 1140 SNPs remained unlinked.
In GBS-t data, 14 SNPs showed strong association with rust response of 88 Fiord/Doza#12034 RILs. Of these 14 SNPs, seven (C250539, C241629, C150949, C244859, C208897, C246203 and C229467) co-segregated and mapped at 1.4 cM away from Uvf-2 and the remaining seven SNPs (C198227, C233649, C225873, scaffold3124, C241205, C174045 and C239085) mapped at 0.7 cM from this cluster (Figure 3a). The KASP markers designed from these 14 SNPs (Supplementary Table S2) were genotyped on 104 Fiord/Doza#12034 RILs. The marker KASP_C250539 mapped closest to Uvf-2 at a distance of 2.9 cM (Figure 3b). The joint linkage map was also constructed by combining flanking KASPs developed from GBS-t (Figure 3b) and Illumina array (Figure 2b) to refine the map location of rust resistance gene Uvf-2 (Figure 3c). Markers KASP_Vf_0703 and KASP_C250539 flanked Uvf-2.
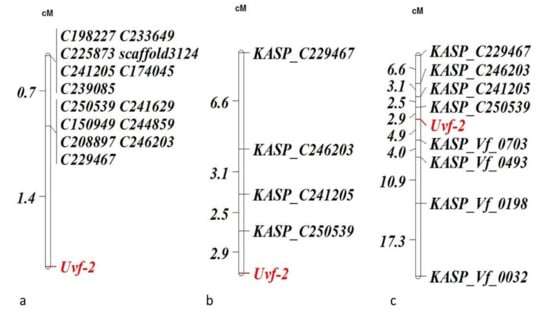
Figure 3.
Linkage maps of chromosome III of Fiord/Doza#12034 population showing association of markers linked with rust resistance gene Uvf-2: (a) constructed from SNPs identified through GBS-t (88 RILs), (b) based on KASP markers developed for SNPs detected from GBS-t and (c) joint linkage map constructed with KASPs derived from 1536 Illumina and GBS-t.
3.2.3. Mapping of Uvf-3
QTL analysis of rust resistance showed association of markers from chromosome V with rust resistance in the Fiord/Ac1655 F4 population and it was designated q_rust_Ac1655 (Figure 4a). It was delimited by markers AC×F138 and Vf_1318. Twenty SNP markers (Vf_0096, Vf_0110, Vf_0565, AC×F213, AC×F40, AC×F260, AC×F275, AC×F165, Vf_0592, Vf_1319, AC×F140, AC×135, VF_1090, AC×F138, VF_1318, Vf_0257, Vf_023, AC×F157, AC×F327 and Vf_0457) from the QTL interval were converted into KASP assays (Supplementary Table S2). Three KASP markers (KASP_AC×F165, KASP_Vf_1090 and KASP_Vf_0203) showed polymorphisms between parents and were assayed on the entire Fiord/Ac1655 F6 RIL population. These markers resulted in a genetic map with resistance gene Uvf-3 spanning across 20 cM (Figure 4b). The KASP markers KASP_AC×F165 and KASP_Vf_1090 flanked Uvf-3 at 2.5 cM 10.1 cM, respectively (Figure 4b).

Figure 4.
Linkage maps of chromosome V of the Fiord/Ac1655 RIL population: (a) location of QTL for rust resistance in 1536 Illumina and GBS-t based map and (b) association of KASP markers converted from SNP in the QTL region with Uvf-3.
3.3. Validation of KASP Markers
3.3.1. Uvf-2
The flanking markers KASP_Vf_0703 and KASP_C250539, which showed close linkage with Uvf-2 were evaluated on a set of 40 genotypes (Table 2). The KASP markers KASP_Vf_0703 and KASP_C250539 amplified the Uvf-2 linked alleles G:G and A:A, respectively, in resistant parent (Doza#12034). KASP_Vf_0703 amplified the Fiord allele (A:A) in all the tested genotypes, except Doza#14916, which is another selection from cultivar Doza. Similarly, KASP_C250539 also produced resistance linked allele in Doza#14916, but it amplified resistance linked allele in genotypes known not to carry Uvf-2 (PBA Warda, IX585c/1-11 and IX524Rb-2-1), indicating loose genetic association with resistance. These results indicated that KASP_Vf_0703 predicted the presence of Uvf-2 accurately among the tested genotypes.

Table 2.
Validation of Uvf-2 and Uvf-3-linked KASP markers on a set of local and exotic cultivars/genotypes.
3.3.2. Uvf-3
The closely linked marker KASP_AC×F165 (2.5 cM) produced ‘C:C’ allele in resistant parent Ac1655 and ‘T:T’ allele in susceptible parent Fiord. Four cultivars, 10 breeding lines, four field selections, and nine germplasm accessions produced the ‘T:T’ allele similar to the susceptible parent Fiord (Table 2).
This marker amplified the Uvf-3-linked allele ‘C:C’ in genotypes (IX474/4-3, IX561f-4-2, IX552Rb-2-4, IX553Rc-2-4, AF10089 Ac1231#14905, Acc740, Ac1866#15013) known not to possess this resistance locus. In addition, three cultivars Doza, PBA Warda and PBA Nasma were mixed (‘T:C’ allele). KASP_AC×F165 marker was polymorphic in 32 out of 40 genotypes (80%) tested in this study. We recommend the use of KASP_AC×F165 for marker-assisted selection of Uvf-3 after testing the parental polymorphism.
4. Discussion
The dawn of next generation sequencing (NGS) technologies has enabled plant scientists to generate transcriptome sequence assemblies to study crops where whole genome sequences are unavailable and faba bean represents this category [34,41,42,43]. Although QTL for several desired economic traits of faba bean have been reported [30,44], markers linked with rust resistance did not get attention. Faba bean rust is an important disease and is commercially managed by spraying fungicides. Breeding rust resistant cultivars is the best disease management strategy where success relies on the identification of new resistance sources and their deployment into high yielding and well adapted agronomic backgrounds [20]. It is difficult to ascertain the presence of more than one gene using conventional selection technologies and the development of molecular markers closely linked with disease resistance genes can overcome this hurdle [45].
Monogenic inheritance of rust resistance in Doza#12034 and Ac1655 was demonstrated. Adhikari et al. [13] also reported the presence of single resistance genes in another Doza selection (#12035) and Ac1655. These inheritance studies further confirmed the previous findings by Adhikari et al. [13] and permanent names Uvf-2 and Uvf-3 were assigned to resistance loci present in Doza#12034 and Ac1655, respectively. It was also concluded that Doza derived lines #12034 and #12035 possessed Uvf-2.
The mapping of Uvf-2 and Uvf-3 at higher genetic distances in Fiord/Doza#12034 and Fiord/Ac1655 F4 populations, respectively from closely associated markers KASP_Vf_0703 and KASP_AC×F165, could be attributed to the robust phenotyping and genotyping data generated on F6 RILs. The marker-trait association was reported in a range of crops during the past decade; the validation of such associations is often missing [45]. This is the first time markers linked with rust resistance were validated across a set of diverse genotypes of faba bean. This validation test demonstrated the value of KASP markers for MAS.
Two modern technologies, GBS-t and Illumina SNP array, were employed to identify SNPs linked with rust resistance gene Uvf-2 and Uvf-3. Our assumption was that GBS-t may result in the identification of more robust markers; however, this was not the case. The SNP array is rather more straightforward and one-step ahead in comparison to GBS-t and detected better trait-marker association. The higher cost of genotyping is a bottleneck for adopting GBS-t technology in faba bean. Genotyping with 90K SNP Array [46] and DArTseq array is routinely used to map economic traits in wheat. Chhetri et al. [47] mapped stripe rust resistance gene Yr58 in chromosome 3B using the DArTseq array and Qureshi et al. [48] developed markers linked with Yr34 using a combination of different technologies. Both DArTseq and SNP genotyping technologies have also been used to determine marker-trait associations in durum wheat [49]. Wheat, being a major crop, attracts more research funds; hence, large sample numbers can be genotyped. However, developing large SNP arrays is not feasible in faba bean because genotyping a small sample number is economically unattractive, as compared to wheat, making GBS-t a close alternative.
Deployment of single resistance genes through conventional breeding is easy, but durability of any resistant variety carrying single resistance gene is always doubtful, especially where pathogen can evolve to acquire virulence [50]. Ijaz et al. [20] reported that the longevity of rust resistance in faba bean can be achieved when multiple resistance genes are deployed together. Combination of rust resistance genes Uvf-2 and Uvf-3 produced lower IT ;1= compared to IT 1C and IT 12C displayed individually by these genes. These results confirmed additivity of resistance conferred by Uvf-2 and Uvf-3 in the seedling stage. This finding implied the practical significance of gene pyramiding for enhanced resistance. In addition, both resistance genes Uvf-2 and Uvf-3 expressed an incomplete non-hypersensitive resistance as cell death remained undetected in histopathological studies that showed race-specificity in multi-pathotype tests [32]. The expression of incomplete seedling resistance is not new to faba bean because immunity to rust has never been described [13,14].
As of now, three rust resistance genes—Uvf-1 [22], Uvf-2, and Uvf-3 (present study) have been identified in faba bean. The Uvf-1 conferring hypersensitivity against single isolate 96-Cord-2 (characterized as race-1) of U. viciae-fabae, was reported from Spain [22] in accession 2N52 with unknown chromosome location. The nomenclature of naming rust resistance genes Uvf-2 and Uvf-3 in this study was adopted from Avila et al. [22]. The faba bean genotype 2N52 is recorded in Australia under accession number Ac1653 [21]. AC1653 needs to be tested with Uvf-2 and Uvf-3 linked KASP markers designed in this study to recognise the uniqueness of Uvf-1.
The development of KASP markers for rust resistance genes Uvf-2 and Uvf-3 in this study has successfully filled the gap in the integration of marker technologies for faba bean breeding since the report by Avila et al. [21]. Significant achievements have been made in marker development for rust resistance in other legumes such as Rpp1, Rpp2, Rpp3, Rpp4, Rpp5 and Rpp6 [51,52] in soybean [Glycine max (L.) Merr.] against Phakospora pachyrhizi Syd. & P. Syd., and Ur-3, Ur-4, Ur-5 and Ur-9 in common bean (Phaseolus vulgaris L.) against Uromyces appendiculatus Pers. [45]. Like cereals, the examples of marker development in other legumes [45] and the present study will allow a shift of focus for rust resistance breeding in legumes from field/glasshouse based phenotypic selection to marker-assisted selection.
Rust resistance is of high importance for Australian faba bean breeders. Although, Adhikari et al. [13] and van Leur et al. [53] identified various rust resistant faba bean genotypes in Australia, precise characterisation and molecular mapping was not conducted. The present study reported molecular markers linked with rust resistance genes Uvf-2 and Uvf-3 for their MAS in breeding programs. We preferred the KASP assay technology because of its user-friendly and cost-effective implementation in breeding programs. In Australia, the genes Uvf-2 and Uvf-3 provide resistance against a range of U. viciae-fabae pathotypes 0–10, 0–46, 40–31, 40–55, and 24–40 [32]. Therefore, considering the benefits of additivity and effectiveness of rust resistance against a range of pathogen isolates, both genes Uvf-2 and Uvf-3 should be pyramided in future cultivars through MAS to provide durable protection against faba bean rust.
Supplementary Materials
The following supplementary information is available online at https://www.mdpi.com/article/10.3390/agronomy11071370/s1, Table S1. Details of a set of 40 faba bean genotypes collected from six countries and used in this study; Table S2. Primer sequences of allele specific markers used for mapping of Uvf-2 and Uvf-; Table S3. Linkage group (LG) statistics for Fiord/Doza#12034 RIL population forSNPs detected from RNA-Seq; Figure S1. GBS-t based linkage groups (LG) of Fiord/Doza#12034 RIL population.
Author Contributions
K.A. developed early generation populations; U.I. and A.S. developed recombinant inbred line populations; U.I., U.B. and H.B. conducted rust screening of mapping populations; U.I. and S.S. conducted molecular mapping; U.B. and S.K. planned and supervised molecular aspects of project; U.I. wrote manuscript; H.B., U.B., S.K. and K.A. edited the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
The first author was awarded the University of Sydney International Postgraduate Research Scholarship and the research was also supported by the Australian Grains Research and Development Corporation grant UA00163.
Data Availability Statement
Data are available from the first and the last authors.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Cubero, J.I. Evolutionary trends in Vicia faba L. Theor. Appl. Genet. 1973, 43, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Cubero, J.I. On the evolution of Vicia faba L. Theor. Appl. Genet. 1974, 45, 47–51. [Google Scholar] [CrossRef] [PubMed]
- Maxted, N.; Callimassia, M.; Bennett, M. Cytotaxonomic studies of Eastern mediterranean Vicia species (Leguminosae). Plant Syst. Evol. 1991, 177, 221–234. [Google Scholar] [CrossRef]
- FAOSTAT. World Statistics on Faba Bean; FAO: Rome, Italy, 2017; Available online: http://faostat.fao.org/ (accessed on 25 March 2021).
- Ijaz, U.; Adhikari, K.N. Understanding genetics of seedling rust resistance in faba bean. In Proceedings of the Australian Pulse Conference, Tamworth, NSW, Australia, 12–14 September 2016; pp. 21–22. [Google Scholar]
- Pulse-Australia. The Australian Industry Faba Bean (Pulse.com.au); PBA-Australia: Barton, ACT, Australia, 2016. [Google Scholar]
- Chen, Y.X.; Zou., L.; Penttinen, P.; Chen, Q.; Li, Q.Q.; Wang, C.Q.; Xu, K.W. Faba Bean (Vicia faba L.) nodulating Rhizobia in Panxi, China, are diverse at species, plant growth promoting ability, and symbiosis related gene levels. Front. Microbiol. 2016, 9. [Google Scholar] [CrossRef]
- Gnanasambandam, A.; Paull, J.; Torres, A.; Kaur, S.; Leonforte, T.; Li, H.; Zong, X.; Yang, T.; Materne, M. Impact of molecular technologies on faba bean (Vicia faba L.) breeding strategies. Agronomy 2012, 2, 132–166. [Google Scholar] [CrossRef] [Green Version]
- Sillero, J.C.; Villegas-Fernández, A.M.; Thomas, J.; Rojas-Molina, M.M.; Emeran, A.A.; Fernández-Aparicio, M.; Rubiales, D. Faba bean breeding for disease resistance. Field Crops Res. 2010, 115, 297–307. [Google Scholar] [CrossRef] [Green Version]
- Ijaz, U.; Adhikari, K.N.; Bansal, U.; Bariana, H.; Trethowan, R. Virulence status of Uromyces viciae-fabae pathotypes in Australia. In Proceedings of the Australian Pulse Conference, Tamworth, NSW, Australia, 12–14 September 2016; p. 66. [Google Scholar]
- Liang, X. Faba bean diseases in China. FABIS Newsl. Faba Bean Inf. Serv. ICARDA 1986, 15, 49–51. [Google Scholar]
- Rashid, K.Y.; Bernier, C.C. The effect of rust on yield of faba bean cultivars and slow-rusting populations. Can. J. Plant Sci. 1991, 71, 967–972. [Google Scholar] [CrossRef] [Green Version]
- Adhikari, K.N.; Zhang, P.; Sadeque, A.; Hoxha, S.; Trethowan, R. Single independent genes confer resistance to faba bean rust (Uromyces viciae-fabae) in the current Australian cultivar Doza and a central European line Ac1655. Crop Pasture Sci. 2016, 67, 649–654. [Google Scholar] [CrossRef]
- Sillero, J.; Moreno, M.; Rubiales, D. Characterization of new sources of resistance to Uromyces viciae-fabae in a germplasm collection of Vicia faba. Plant Pathol. 2000, 49, 389–395. [Google Scholar] [CrossRef]
- Cubero, J.I. The faba beans: A historic perspective. Grain Legumes 2011, 56, 5–7. [Google Scholar]
- Conner, R.; Bernier, C. Inheritance of rust resistance in inbred lines of Vicia faba. Phytopathology 1982, 72, 1555–1557. [Google Scholar] [CrossRef]
- Conner, R.; Bernier, C. Race identification in Uromyces viciae-fabae. Can. J. Plant Pathol. 1982, 4, 157–160. [Google Scholar] [CrossRef]
- Rashid, K.; Bernier, C. The genetics of resistance in Vicia faba to two races of Uromyces viciae-fabae from Manitoba. Can. J. Plant Pathol. 1986, 8, 317–322. [Google Scholar] [CrossRef]
- Sillero, J.C.; Rojas-Molina, M.M.; Emeran, A.A.; Rubiales, D. Rust resistance in faba beans. Grain Legumes Mag. 2011, 56, 27–28. [Google Scholar]
- Ijaz, U.; Adhikari, K.N.; Stoddard, F.L.; Trethowan, R.M. Rust resistance in faba bean (Vicia faba L.): Status and strategies for improvement. Australas. Plant Pathol. 2018, 47, 71–81. [Google Scholar] [CrossRef] [Green Version]
- Avila, C.; Sillero, J.; Rubiales, D.; Moreno, M.; Torres, A. Identification of RAPD markers linked to the Uvf-1 gene conferring hypersensitive resistance against rust (Uromyces viciae-fabae) in Vicia faba L. Theor. Appl. Genet. 2003, 107, 353–358. [Google Scholar] [CrossRef]
- Singh, B.; Singh, A. Linkage mapping of molecular markers and oligogenes. Marker-Assisted Plant Breeding: Principles and Practices; Springer: Berlin/Heidelberg, Germany, 2015; pp. 151–183. [Google Scholar]
- Collard, B.C.; Mackill, D.J. Marker-assisted selection: An approach for precision plant breeding in the twenty-first century. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2008, 363, 557–572. [Google Scholar] [CrossRef] [Green Version]
- Malmberg, M.M.; Pembleton, L.W.; Baillie, R.C.; Drayton, M.C.; Sudheesh, S.; Kaur, S.; Shinozuka, H.; Verma, P.; Spangenberg, G.C.; Daetwyler, H.D. Genotyping-by-sequencing through transcriptomics: Implementation in a range of crop species with varying reproductive habits and ploidy levels. Plant Biotechnol. J. 2018, 16, 877–889. [Google Scholar] [CrossRef]
- Franssen, S.U.; Shrestha, R.P.; Bräutigam, A.; Bornberg-Bauer, E.; Weber, A.P. Comprehensive transcriptome analysis of the highly complex Pisum sativum genome using next generation sequencing. BMC Genomics. 2011, 12, 227. [Google Scholar] [CrossRef] [Green Version]
- Parra-González, L.B.; Aravena-Abarzúa, G.A.; Navarro-Navarro, C.S.; Udall, J.; Maughan, J.; Peterson, L.M.; Salvo-Garrido, H.E.; Maureira-Butler, I.J. Yellow lupin (Lupinus luteus L.) transcriptome sequencing: Molecular marker development and comparative studies. BMC Genom. 2012, 13, 425. [Google Scholar] [CrossRef] [Green Version]
- Verma, P.; Shah, N.; Bhatia, S. Development of an expressed gene catalogue and molecular markers from the de novo assembly of short sequence reads of the lentil (Lens culinaris Medik.) transcriptome. Plant Biotechnol. J. 2013, 11, 894–905. [Google Scholar] [CrossRef]
- Garg, R.; Patel, R.K.; Tyagi, A.K.; Jain, M. De novo assembly of chickpea transcriptome using short reads for gene discovery and marker identification. DNA Res. 2011, 18, 53–63. [Google Scholar] [CrossRef] [Green Version]
- Kaur, S.; Kimber, R.B.; Cogan, N.O.; Materne, M.; Forster, J.W.; Paull, J.G. SNP discovery and high-density genetic mapping in faba bean (Vicia faba L.) permits identification of QTLs for ascochyta blight resistance. Plant Sci. 2014, 217, 47–55. [Google Scholar] [CrossRef]
- Ocaña, S.; Seoane, P.; Bautista, R.; Palomino, C.; Claros, G.M.; Torres, A.M.; Madrid, E. Large-scale transcriptome analysis in faba bean (Vicia faba L.) under Ascochyta fabae infection. PLoS ONE 2015, 10, e0135143. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef]
- Ijaz, U. Molecular Mapping and Microscopic Analysis of Faba Bean-Uromyces viciae-fabae Host-Pathogen Interaction. Ph.D. Thesis, The University of Sydney, Sydney, Australia, 2018. [Google Scholar]
- Bansal, U.K.; Kazi, A.G.; Singh, B.; Hare, R.A.; Bariana, H.S. Mapping of durable stripe rust resistance in a durum wheat cultivar Wollaroi. Mol. Breed. 2014, 33, 51–59. [Google Scholar] [CrossRef]
- Braich, S.; Sudheesh, S.; Forster, J.W.; Kaur, S. Characterisation of Faba Bean (Vicia faba L.) Transcriptome Using RNA-Seq: Sequencing, De Novo assembly, annotation, and expression analysis. Agronomy 2017, 7, 53. [Google Scholar] [CrossRef] [Green Version]
- Kosambi, D.D. The estimation of map distances from recombination values. Ann. Eugen. 1943, 12, 172–175. [Google Scholar] [CrossRef]
- Manly, K.F.; Cudmore, R.H., Jr.; Meer, J.M. Map Manager QTX, cross-platform software for genetic mapping. Mamm. Genome. 2001, 12, 930–932. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J. Efficient Linkage Map Construction Using R/ASMap. 2015. Available online: https://cran.r-project.org/web/packages/ASMap/vignettes/asmapvignette.pdf (accessed on 10 November 2017).
- Voorrips, R. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- You, F.M.; Huo, N.; Gu, Y.Q.; Luo, M.-C.; Ma, Y.; Hane, D.; Lazo, G.R.; Dvorak, J.; Anderson, O.D. BatchPrimer3: A high throughput web application for PCR and sequencing primer design. BMC Bioinform. 2008, 9, 253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nsabiyera, V.; Qureshi, N.; Bariana, H.S.; Wong, D.; Forrest, K.L.; Hayden, M.J.; Bansal, U.K. Molecular markers for adult plant leaf rust resistance gene Lr48 in wheat. Mol. Breed. 2016, 36, 65. [Google Scholar] [CrossRef]
- Cooper, J.W.; Wilson, M.H.; Derks, M.F.; Smit, S.; Kunert, K.J.; Cullis, C.; Foyer, C.H. Enhancing faba bean (Vicia faba L.) genome resources. J. Exp. Bot. 2017, 68, 1941–1953. [Google Scholar] [CrossRef] [Green Version]
- O’Sullivan, D.M.; Angra, D. Advances in faba bean genetics and genomics. Front. Genet. 2016, 7, 150. [Google Scholar] [CrossRef] [Green Version]
- Kaur, S.; Cogan, N.O.; Forster, J.W.; Paull, J.G. Assessment of genetic diversity in faba bean based on single nucleotide polymorphism. Diversity 2014, 6, 88–101. [Google Scholar] [CrossRef] [Green Version]
- Atienza, S.; Palomino, C.; Gutiérrez, N.; Alfaro, C.; Rubiales, D.; Torres, A.; Ávila, C. QTLs for ascochyta blight resistance in faba bean (Vicia faba L.): Validation in field and controlled conditions. Crop Pasture Sci. 2016, 67, 216–224. [Google Scholar] [CrossRef]
- Bariana, H.S.; Bansal, U.K. Breeding for disease resistance. In Encyclopedia of Applied Plant Sciences, 2nd ed.; Murray, B.G., Murphy, D.J., Eds.; Academic Press: Oxford, UK, 2017; pp. 69–76. [Google Scholar]
- Wang, S.; Wong, D.; Forrest, K.; Allen, A.; Chao, S.; Huang, B.E.; Maccaferri, M.; Salvi, S.; Milner, S.G.; Cattivelli, L. Characterization of polyploid wheat genomic diversity using a high-density 90,000 single nucleotide polymorphism array. Plant Biotechnol. J. 2014, 12, 787–796. [Google Scholar] [CrossRef] [Green Version]
- Chhetri, M.; Bariana, H.; Kandiah, P.; Bansal, U. Yr58: A new stripe rust resistance gene and its interaction with Yr46 for enhanced resistance. Phytopathology 2016, 106, 1530–1534. [Google Scholar] [CrossRef] [Green Version]
- Qureshi, N.; Bariana, H.; Zhang, P.; McIntosh, R.; Bansal, U.; Wong, D.; Hayden, M.; Dubcovsky, J.; Shankar, M. Genetic relationship of stripe rust resistance genes Yr34 and Yr48 in wheat and identification of linked KASP markers. Plant Dis. 2018, 102, 413–420. [Google Scholar] [CrossRef] [Green Version]
- Qureshi, N.; Bariana, H.; Kolmer, J.A.; Miah, H.; Bansal, U. Genetic and molecular characterization of leaf rust resistance in two durum wheat landraces. Phytopathology 2017, 107, 1381–1387. [Google Scholar] [CrossRef] [Green Version]
- Pink, D.A. Strategies using genes for non-durable disease resistance. Euphytica 2002, 124, 227–236. [Google Scholar] [CrossRef]
- Garcia, A.; Calvo, É.S.; de Souza Kiihl, R.A.; Harada, A.; Hiromoto, D.M.; Vieira, L.G.E. Molecular mapping of soybean rust (Phakopsora pachyrhizi) resistance genes: Discovery of a novel locus and alleles. Theor. Appl. Genet. 2008, 117, 545–553. [Google Scholar] [CrossRef]
- Yu, N.; Kim, M.; King, Z.R.; Harris, D.K.; Buck, J.W.; Li, Z.; Diers, B.W. Fine mapping of the Asian soybean rust resistance gene Rpp2 from soybean PI 230970. Theor. Appl. Genet. 2015, 128, 387–396. [Google Scholar] [CrossRef]
- van Leur, J.; Paull, J.; Kimber, R.; Robertson, L. Disease resistance and susceptibility in Ecuadorian faba bean germplasm. In Proceedings of the Australian Pulse Conference, Tamworth, NSW, Australia, 12–14 September 2016; p. 110. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).