Abstract
Red raspberry (Rubus idaeus L.) is an expanding high-value berry crop worldwide. The presence of prickles, outgrowths of epidermal tissues lacking vasculature, on the canes, petioles, and undersides of leaves complicates both field management and harvest. The utilization of cultivars with fewer prickles or prickle-free canes simplifies production. A previously generated population segregating for prickles utilizing the s locus between the prickle-free cultivar Joan J (ss) and the prickled cultivar Caroline (Ss) was analyzed to identify the genomic region associated with prickle development in red raspberry. Genotype by sequencing (GBS) was combined with a genome-wide association study (GWAS) using fixed and random model circulating probability unification (FarmCPU) to analyze 8474 single nucleotide polymorphisms (SNPs) and identify significant markers associated with the prickle-free trait. A total of four SNPs were identified on chromosome 4 that were associated with the phenotype and were located near or in annotated genes. This study demonstrates how association genetics can be used to decipher the genetic control of important horticultural traits in Rubus, and provides valuable information about the genomic region and potential genes underlying the prickle-free trait.
1. Introduction
Red raspberry belongs to the genus Rubus, which is a member of the family Rosaceae and subfamily Rosoideae. Rubus is one of the most diverse genera comprising 12 subgenera with the overall number of species in Rubus estimated to be between 600 and 800 [1]. The most commonly cultivated types worldwide are red raspberry (R. ideaus L.), black raspberry (R. occidentalis L.), and various blackberry types (R. hybrid) [2,3]. Red raspberry is a globally commercialized specialty crop and is the most economically important species in the genus. It is diploid (2n = 2x = 14) with a basic set of seven chromosomes [1] and a nuclear haploid genome of approximately 280 MB in size [4]. R. idaeus is highly heterozygous in its wild state due to the presence of dominant self-incompatibility alleles that limit pollen tube formation in the style [5]. Although germplasm used in breeding programs has been selected for self-compatibility, inbreeding depression can be very severe, which constrains population development for both breeding and molecular studies [6] thus complicating many genetic studies. Recurrent mass selection is used as the breeding approach with only limited self-pollination and backcrossing for specific traits, thus inbred lines for developing study populations are unavailable.
A plant’s epidermis plays several important roles, from protection against pathogens and predators to controlling vital exchange of gas, water and nutrients with the environment. Epidermal structures like the cuticle, hairs, waxy exudates, trichomes, spines, thorns and prickles help in the protection of plants in several ways [7]. Trichomes, simple hair-like structures, in addition to providing protection against biotic and abiotic stresses [8,9,10], also moderate surface temperatures and help reduce the transpiration rate by creating a barrier between the epidermis and the environment [11]. Thorns (modified branches containing vasculature), spines (modified leaves containing vasculature) and prickles (epidermal outgrowths lacking vasculature), while botanically different organs, all provide additional mechanical protection from herbivory and mechanical damage [12,13,14,15,16].
Additionally, the presence of glandular hairs (pubescence) has been found to be associated with resistance to some fungal diseases [17]. Raspberry cultivars with fine hairs on the canes (pubescent canes) are known to be more resistant to spur blight (Didymella applanata (Niessl) Sacc.) and cane botrytis (Botrytis cinerea) [18,19,20] but are more susceptible to powdery mildew (Sphaerotheca macularis), yellow rust (Phragmidium rubi-idaei) and cane spot (Elsinoe veneta) [5,21,22,23,24]. Pubescence in raspberry is determined by gene H manifested as glandular hairs, which are distinct from prickles with the recessive allele resulting in glabrous canes in the homozygous state (genotype hh) [17]. Gene H has been mapped to linkage group (LG) 2 [25]. Moreover, besides cane pubescence, gene H is known to have other linked pleiotropic effects including a slight increase in prickle density (the term spine is used in the literature) and a decrease in prickle size [22,26]. Since both hairs and prickles are glandular epidermal outgrowths, their early development is likely inter-related [27], suggesting the gene H may be regulated similarly to the early development of Rubus prickles and affect several cell characteristics. Similarly, among several genes known to confer prickles in various Rubus species, the s locus in red raspberry is a simple dominant/recessive locus for the presence of prickles, and has yet to be genetically characterized [28,29]. Other major loci conferring the prickle-free trait in various Rubus species are STE [30], Sf [31], and SfL [32], which all confer prickle-free in the dominant state and originate in blackberry germplasm not generally utilized in breeding due to the unpredictable nature of the loci and ploidy incompatibility.
In rose, the prickle-free trait is also recessive and has been mapped to a major locus on linkage group 3 (LG3) in an interspecific population with three quantitative trait loci (QTLs) on LG3, LG4 and LG1 associated with prickle density [33]. However, segregation for prickles in this population was highly skewed with the prickle-free trait deviating significantly from simple Mendelian inheritance suggesting genetic anomalies or incompatibilities in the interspecific population studied.
Despite the fact that prickles help in protection against mechanical damage, natural predators, and fungal pathogens, it is an undesirable trait for commercial production as a complicating factor in pruning, trellising, and harvest. Hence, the prickle-free trait in Rubus cultivars is one of the most sought after traits for breeders and commercial industries. Several programs have made efforts in incorporating the prickle-free phenotype in their programs, resulting in cultivars such as ‘Joan J’ and ‘Glen Ample’ red raspberry [34], and ‘Natchez’, ‘Chester’ [35], ‘Apache’, and ‘Triple Crown’ blackberry, among others. However, it is time consuming and expensive to integrate the prickle-free trait with other important traits through conventional breeding approaches. For instance, the prickle-free trait has been transferred into black raspberry (R. occidentalis L.) germplasm from related red raspberry germplasm through a multigenerational process spanning more than a decade [36] but has yet to result in a commercial quality cultivar. Additionally, cultivated blackberries are bred at the tetraploid or higher ploidy level, which makes the production of prickle-free homozygous recessive types for evaluation extremely difficult when utilizing the majority of the prickled germplasm available to breeders [14]. The development of molecular tools will expedite both the breeding process and the development of prickle-free cultivars through alternative methods.
In this study, an F1 population segregating for prickles [37] controlled by the s locus conferring the prickle-free trait in the homozygous recessive state [28,29] and segregating in a standard 1:1 Mendelian ratio was examined. As such, the trait is simple and unambiguous to score starting at the cotyledon stage and throughout the life cycle of the raspberry plant. Although a molecular marker for the gene is not necessary for selecting prickle-free canes, it can simplify breeding by eliminating the need for test crosses with prickled parental selections. Additionally, precise genomic data is needed if gene editing techniques are to be utilized to modify important existing cultivars to a prickle-free version.
Currently, the information on the underlying genomics of prickle development in R. idaeus is limited. Genome-wide association studies (GWAS) are powerful tools for mapping complex traits down to the sequence level, aiding in the identification of genes associated with important horticultural traits and biological processes [38,39]. GWAS identifies the association between loci and a particular trait by examining single nucleotide polymorphisms (SNPs) throughout an entire genome. As selected loci and SNPs are associated with a target trait, GWAS can be used for the development of useful molecular markers. GWAS identifies associated SNPs of the target trait using both genotypic and phenotypic data and various statistical methods, which can then be linked to genomic sequence data to identify gene candidates within a specific genomic region. The present study utilizes genome-wide association analysis to elucidate the genetic control of prickle development in red raspberry by identifying significant SNPs associated with the trait, linking those SNPs to a specific chromosomal region, and identifying putative candidate genes underlying the trait.
2. Materials and Methods
2.1. Plant Materials
A previously described population from a controlled hybridization between prickle-free ‘Joan J’ (ss) and prickled ‘Caroline’ (Ss) was phenotyped for cane prickles [37]. ‘Caroline’ is derived from the hybridization between GE01 × ‘Heritage’, in which GE01 is a cross between ‘Autumn Bliss’ × ‘Glen Moy’. ‘Heritage’ and ‘Autumn Bliss’ are homozygous for prickles and ‘Glen Moy’ is prickle-free. Similarly, the source of the prickle-free alleles in ‘Joan J’ is also ‘Glen Moy’ through a complex set of hybridizations [40]. Plants were grown in a growth chamber prior transfer to a greenhouse for phenotyping and tissue collection. Leaf tissue was collected from 90 F1 progeny along with both the parents (92 total) for DNA extraction.
2.2. DNA Extraction
Genomic DNA was isolated from parents and progeny using a Qiagen DNeasy 96 Plant Extraction Kit (Qiagen Inc., Valencia, CA, USA) following the manufacturer’s protocol. DNA concentration and purity were assessed using a NanoDrop ND -1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). DNA contamination and degradation were assessed on 1.0% agarose gels.
2.3. Genotype by Sequencing (GBS)
Genotype by sequencing (GBS) (Novogene, Beijing, China) was used to develop SNP markers on a population segregating for prickles. Briefly, DNA samples from the 92 samples were digested with MseI and EcoRI. Then, a 95-plex GBS sequencing library was prepared by ligating the digested DNA to unique barcode adapters. PCR amplification was performed according to a standard PCR protocol [41], and then all the samples were pooled and size-selected for the required fragments to complete the library construction. Sequencing was performed using an Illumina HiSeq 2500 platform (Illumina, San Diego, CA, USA).
2.4. Preprocessing
After sequencing, the raw reads were demultiplexed according to the barcode sequences and trimmed using the Illumina pipeline CASAVA v1.8.2 software. The sequences and corresponding quality information were stored in 92 separate FASTQ files. It is common for the quality of bases to decrease at the ends of Illumina reads, so the ends were trimmed at the point where the Phred quality score dropped below Q = 20 (or 0.05 probability of error). Additionally, all 5′ and 3′ stretches of ambiguous N nucleotides were trimmed. Poor quality sequence reads and reads shorter than 25 bases were discarded.
2.5. Sequence Alignment, SNP Calling, and SNP Imputation
The sequencing reads were processed with the GBS Discovery Pipeline for species with a reference genome implemented in TASSEL Version 3.0 [42] and following the pipeline documentation [43]. The Burrows–Wheeler Aligner (BWA) program [44] (parameters: mem — t 4 — k 32 — M) was used to align the clean reads to an unpublished version of the red raspberry (R. idaeus) genome as the reference genome. The TASSEL 3.0 Discovery SNP Caller was implemented to align the multiple sequence tags from the same physical locations across the genome, to call SNPs at these locations across the individual samples. Quality control checks were performed by eliminating markers with minor allele frequency (MAF) < 5%, and markers with a missing rate higher than 10%. Marker data imputation was then applied using the FILLIN algorithm [45] implemented in TASSEL Version 5.0 [42]. The final GBS SNPs were plotted against their respective physical positions on each chromosome to examine their density and distribution with ggplot2 [46]. Identification of significant SNPs from the remaining 8474 SNPs was completed after filtration and imputation.
2.6. Genome‑Wide Association Analysis
To discover associations between the genome-wide GBS SNPs and the prickle phenotype in our population, a genome-wide association study (GWAS) was conducted. The general linear (GLM) and mixed linear models (MLM) are the most commonly used models for association analysis. GLM accounts for population structure only and a MLM accounts for the population structure and the family relatedness [47,48]. Compared to GLM, the incorporation of population structure and family relatedness in MLM in the association tests helps control false positives; however, true positives can be compromised by these adjustments [49]. Therefore both GLM and MLM show weaknesses in the proclivity to induce false negatives due to overfitting of the model so that important associations can be missed. Fixed and random model circulating probability unification (FarmCPU) works to correct false positives while identifying true positives [49]. In this study, GLM, MLM, and FarmCPU models were compared using phenotypic data and results were evaluated based on quantile–quantile (Q–Q) plots. Subsequently, the FarmCPU model proved to be the most accurate model for association analysis of the data from the raspberry population studied here.
In FarmCPU, the multiple loci linear mixed model (MLMM) is divided into two parts: a fixed effect model (FEM) and a random effect model (REM), which are used iteratively [49]. To avoid model overfitting, REM estimates the multiple associated markers that are used to obtain kinship. The FEM tests markers, one at a time, and kinship from REM as covariates to control false positives and false negatives. At each iteration, p-values of testing markers and multiple associated markers are unified. GWAS was conducted using the genome association and prediction integrated tool (GAPIT) R package [50] in R v.3.0.2. The analysis was composed of 8474 SNPs, each with a minor allele frequency greater than or equal to 0.05. A threshold value -Log p-value ≥ 5.98 was used to declare a significant association of SNPs with the prickle phenotype.
2.7. Candidate Gene Identification
SNPs at level of −Log10 p ≥ 5.98 were considered highly significant and were characterized in silico for their genomic position and functional effect. Candidate genes surrounding significantly associated SNPs were annotated using the Blast2GO tool with BLASTp search against the non-redundant protein database [51]. Candidate genes with a possible connection to prickle, trichome, or epidermal cell growth regulation were taken into consideration.
3. Results
3.1. Phenotype Descriptions
The gross morphology of prickled versus prickle-free raspberry has been previously described [37] and is demonstrated in Figure 1. There are clear morphological differences between the two types, prickled plants having a mixture of the prickle development phases from recently initiated to mature lignified prickles on their stem, petioles and leaves, and prickle-free plants having smooth epidermal surfaces with only simple trichomes. The ‘Joan J’ × ‘Caroline’ population segregated almost perfectly in the predicted Mendelian ratio (1:1) (44 prickled:46 prickle-free) for the presence/absence of prickles (Figure 2A).

Figure 1.
Gross morphology (A,B) and micrographs (C,D) showing prickles and trichomes on the surface of prickled and prickle-free Rubus idaeus L.: (A) prickle-free; (B) prickled; (C) micrograph of trichomes on prickle-free stem; and (D) micrograph of prickles and trichomes on the prickled stem.
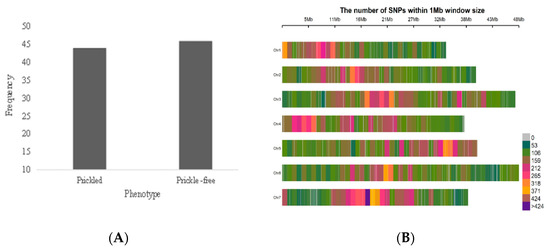
Figure 2.
(A) Distribution of prickles in F1 progeny generated from hybridization between prickle-free ‘Joan J’ (ss) and prickled ‘Caroline’ (Ss) (46:44) and (B) distribution of single nucleotide polymorphisms (SNPs) in 1 Mb window size across the raspberry chromosomes. The x-axis represents the distance in base pairs.
3.2. Genome‑Wide Association Analysis
A total of 8474 SNPs were used for association analysis to identify significant SNPs. The final GBS SNPs plotted against their respective physical positions on each chromosome to examine their density and distribution with ggplot2 is shown in Figure 2B [46]. A comparison of FarmCPU, GLM, and MLM models was completed using the prickle data across all parents and progenies. The quantile–quantile (Q–Q) plot assesses how well the GWAS model accounts for population structure and kinship (familial relatedness). The negative logarithms of the p-values from the models fitted in GWAS are plotted against their expected value under the null hypothesis of no association with the trait. The majority of the dotted lines or the points should lie on the diagonal line, since most of the SNPs tested are not associated with the trait. It is expected to see the deviation from the diagonal line at the right tail area, which suggests the association of those SNPs with the trait. The Q–Q plot of the FarmCPU model resulted in a sharp deviation from the expected negative base 10 logarithm of the p-value distribution in the tail area, indicating the association of the SNPs with the trait, and that false positives and negatives were adequately controlled. However, Q–Q plots from MLM and GLM models did not show a sharp deviation (Figure 3). With MLM and GLM most of the SNPs did not lie on the diagonal line indicating that FarmCPU was a better choice than MLM and GLM model for association testing with this data set.
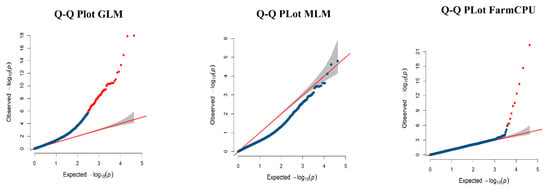
Figure 3.
Quantile–quantile (Q–Q) plot of the general linear model (GLM), mixed linear model (MLM) and fixed and random model circulating probability unification (FarmCPU) model using prickle data. A Q–Q plot represents the extent to which the observed p-value distribution of the test statistic follows the null hypothesis i.e., expected p-value distribution (indicated by the red line). All points are on or near the middle red line if the observed values correspond to the expected values. Points deviating from the expected values under null hypothesis in the tail area indicate significant association. The Q–Q plots of the GLM and MLM models did not show sharp deviation from the expected p-value distribution in the tail area while the Q–Q plot of FarmCPU model showed a sharp deviation indicating adequate control of false positives and negatives. The Q–Q plot indicated that the population structure and kinship were well controlled for GWAS in the FarmCPU model. Shaded gray area represents 95% confidence interval.
GWAS using the FarmCPU model was conducted on 8474 SNPs (MAF ≥ 0.05) that segregated within this population. Four SNPs significantly associated with the prickle phenotype were identified with association analysis at the level of −Log10 p ≥ 5.98 (Figure 4) that delimit a 1604 kb physical region (33,543,882–35,148,226) on the distal portion of chromosome 4. On chromosome 4 the allelic effect for these significant SNPs ranged from −0.38 to 0.20 as seen in Table 1. A significant SNP 4_34738035 (5.08 × 10−23) contributed negatively to the trait indicating a major allele (i.e., more common) that favored the prickle-free phenotype, followed by two other significant SNPs, 4_33543882 (2.80 × 10−11) and 4_34134523 (2.22 × 10−18), which also contributed negatively to the trait (i.e., favored prickle-free). SNP 4_35148226 (3.61 × 10−13) contributed positively to the trait indicating a minor allele (i.e., less common) favored the prickle-free trait.
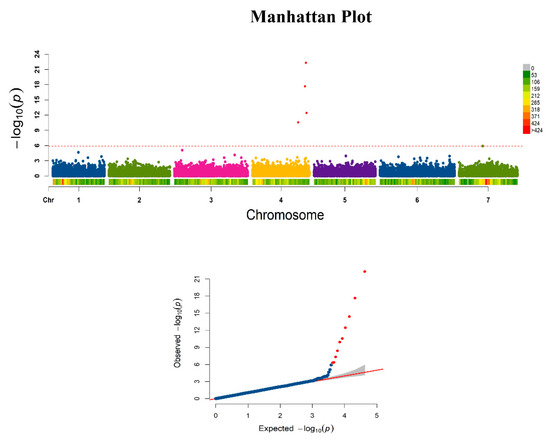
Figure 4.
Manhattan plots of −Log10 (P) vs. chromosomal position of significant SNP associations and respective quantile–quantile (Q–Q) plots on prickle data using FarmCPU model. Each chromosome is colored differently. The red dotted line in the Manhattan plot represents the association threshold (−Log10 p ≥ 5.98). SNPs reaching the genome-wide significance threshold are represented by red dots. A total of four significant SNPs were identified on the distal portion of chromosome 4 that were associated with prickle phenotype in the association analysis. The Q–Q plot of the FarmCPU model using prickle data indicated adequate control of false positives and negatives.

Table 1.
List of significant SNPs associated with the prickle phenotype using the FarmCPU model with a threshold of −Log10 p ≥ 5.98.
3.3. Candidate Gene Identification
Candidate genes flanking the four SNPs significantly associated with the trait based on the annotation of the draft red raspberry genome were identified (Table 2). All together 98 transcripts were identified flanking 100 kb (up and downstream) of all the associated SNPs. Among the protein-coding genes identified was the transcription factor (TF) MYB16-like (MIXTA-like R2R3-MYB family member), which regulates conical cell outgrowth and trichome initiation in diverse plant species [52]. R2R3-MYB TFs have been determined to function in trichome development via incorporation into the TTG1-bHLH-MYB regulatory complexes [53].

Table 2.
Top SNPs associated with prickle phenotype using the FarmCPU model and associated transcripts and putative candidate genes annotated and identified using the Blast2GO tool with BLASTp search against the non-redundant protein database.
Other transcripts identified in the nearby region of the SNPs included Rosa chinensis axial regulator YABBY 4, Rosa chinensis MLP-like protein 31, Rosa chinensis agamous-like MADS- box protein AGL30, Rosa chinensis ubiquitin-like-specific protease 1D, and Rosa chinensis protein trichome birefringence-like 2. The accession numbers for the genes flanking each marker are listed in Table 2.
4. Discussion
This research evaluated prickle development in a segregating F1 population and the parental genotypes in red raspberry. GBS and GWAS was adapted and applied utilizing a draft red raspberry reference genome to identify candidate genes associated with prickle formation. This genetic information may be useful for future modification of important prickled Rubus cultivars into prickle-free versions utilizing gene editing techniques. GWAS has the advantage over traditional QTL mapping in that variations can be mapped down to the nucleotide level. In this study, four highly significant SNPs were identified on the distal portion of chromosome 4 that delimited a 1604 kb region spanning the physical position between 33,543,882 and 35,148,226.
The significant SNP 4_35148226 was identified to contribute positively to the trait while all the remaining significant SNPs contributed negatively to the trait. Positive and negative contributions do not refer to the percentage of the genetic variation caused by the SNPs. It identifies the common and uncommon alleles associated with the phenotype we are looking for, i.e., prickle-free in this case. A positive value indicates that the minor allele (i.e., less common allele) was the allele associated with the prickle-free phenotype (favorable or sought after phenotype) while a negative value indicates that the major allele (i.e., more common allele) was the allele associated with the prickle-free phenotype. Alleles from either the major class (the common allele/variation) or minor class (less common allele/variation) were considered as favorable if they were associated with the prickle-free phenotype (i.e., allelic effect favored the sought after phenotype, prickle-free in this case).
Understanding of the genetic and physiological control of prickle development in Rubus at the molecular level is limited. A genetic linkage map constructed using a population derived from the prickle-free “Glen Moy” and the homozygous prickled “Latham”, (the terms spine-free and spiny were used in the study) measured varying prickle density among the progeny [54]. This trait mapped to LG2. A QTL for prickle density was previously mapped to LG6 and three QTL for fungal resistance that overlapped this same region [25].
Another study constructed a genetic linkage map and performed QTL mapping of prickle density and other traits using a (R. parvifolius × “Tulameen”) × “Qualicum” population [55]. Both parents exhibited prickled canes, however prickle density ranged from highly prickled canes to prickle-free canes. The study identified two QTL linked to prickle density, one associated with linkage group 4 (LG4), present at the distal end, accounting for approximately 84% of the variation with high LOD score, and second QTL associated with LG6 accounting for less than 10% of the variation. The segregation pattern followed a single gene model with 3:1 segregation, indicating both parents were heterozygous at the prickle locus. The same QTL on LG4 and LG6 were detected when scores for prickle-free individuals were removed and only prickle density was examined, accounting for 46.3% and 16.7% of the variation, respectively. When the trait was scored as qualitative (0 as prickle-free and 1 as prickled on canes), the trait mapped to the QTL region on the distal end of LG4 and was designated gene s.
In the present study, a significant locus mapped to the distal end of LG4 (chromosome 4) associated with the trait corroborates the previous study [55]. A total of 98 transcripts flanking 100 kb upstream and downstream of the four associated SNPs were identified (Table 2). Among these transcripts, there were a few that are known to be directly or indirectly associated with trichome initiation/early stage development. There were 16 genes flanking 100 kb upstream and downstream of Marker 4_33543882, notably, Rosa chinensis transcription factor MYB16-like (Accession: XP_024196462.1) and Rosa chinensis agamous-like MADS-box protein AGL30 (Accession: XP_011463061.1) (Table 2), which are potential candidate genes associated with prickle development. The MYB16-like (MIXTA-like R2R3-MYB) transcription factor is of particular interest as transcriptome analysis showed it to be significantly down regulated in prickle-free plants [56].
As further evidence supporting this candidate gene, a transcription factor MYB16-like belonging to the MIXTA-like R2R3-MYB TF family regulates trichome development in other plant species [52]. The contribution to trichome development of MIXTA-like R2R3-MYB family members has been identified in snapdragon (Antirrhinum majus L.) [57], and its homologs in Arabidopsis thaliana (L.) Heynh., cucumber (Cucumis sativus L.), cotton (Gossypium hirsutum L.), and many other species [58,59,60].
In cucumber, the MIXTA-like homolog CsMYB6 has been determined to regulate epidermal cell differentiation, cuticle wax biosynthesis, and trichome morphogenesis [61,62,63]. Moreover, a study conducted to identify and characterize the genome-wide R2R3-MYB family in three species in the Rosaceae family: Malus domestica Borkh., Prunus persica (L.) Batsch, and Fragaria vesca L., identified 44 functional subgroups with seven unique to the Rosaceae family [64]. In the study, functional analysis of the TFs were performed based on the clustering of R2R3-MYB genes of Arabidopsis, which identified two R2R3-MYB genes; Arabidopsis transcription factor AtMYB5 in subgroup 16 and AtMYB106/NOK and AtMYB16/MIXTA in subgroup 5. AtMYB5 is known to regulate trichome morphogenesis and mucilage synthesis [65]. AtMYB106/NOK and AtMYB16/MIXTA are known to participate in trichome development [58]. Therefore, a MIXTA-like R2R3-MYB family member could be one of the key regulators of prickle development.
Similarly, MADS-box genes are the key members of regulatory networks behind multiple developmental pathways. These genes regulate the networks involved in plant responses to stress and the plant developmental plasticity response to seasonal fluctuations [66]. Transcriptome study on Solanum viarum Dunal prickles concluded that the development related transcription factor MADS-box play a role in prickle development in addition to R2R3-MYB, REM, and DRL1 [67].
A total of 19 genes flanking 100 kb upstream and downstream of Marker 4_34134523 were identified, notably Rosa chinensis Jacq. MLP-like protein 423 (Accession: XP_024192632.1), Rosa chinensis WAT1-related protein At1g70260 (Accession: XP_004297654.1), and Rosa chinensis kinesin-like protein KIN-4C (Accession: XP_024191881.1) (Table 2). For Marker 4_34738035, a total of 32 genes flanking 100 kb upstream and downstream were identified, including Rosa chinensis homeobox protein knotted-1-like 1 (Accession: XP_024192540.1), Rosa chinensis probable UDP-3-O-acyl-N-acetylglucosamine deacetylase 2, mitochondrial (Accession: XP_024169277.1), and Rosa chinensis microtubule-associated protein 70-1-like (Accession: XP_024194091.1) (Table 2).
Finally, a total of 31 genes flanking 100 kb upstream and downstream of Marker 4_35148226 were identified. Among these 31 genes was Rosa chinensis protein trichome birefringence-like 2 (Accession: XP_024197787.1) (Table 2), which is a putative candidate gene for prickle development in Rubus. Cellulose is one of the major components of the plant cell wall and has diverse functions [68]. The cell shape and development patterns are mainly determined by the cellulose microfibrils [69]. A secondary cell wall, which is the major component in wood and plant fibers, is deposited once the plant cells stop expanding [70]. This secondary cell wall is mainly composed of crystalline cellulose, microfibrils, lignin, and non-cellulosic polysaccharides [69,71,72]. The plant cellulose synthase (CESA) genes function as cellulose synthases and mutants in primary CESA genes exhibit reduced cellulose levels and dwarf growth phenotypes [73,74,75]. The gene that controls a trait referred to as TRICHOME BIREFRINGENCE (TBR) is also an important component required for secondary cell wall cellulose synthesis [76]. Wild type Arabidopsis trichomes display strong birefringence under polarized light whereas the tbr mutant has severely reduced crystalline cellulose content in trichomes and lacks such birefringence [76]. TBR belongs to the TRICHOME BIREFRINGENCE-Like (TBL) gene family. Members of the TBL protein family influence the resistance to pathogens, tolerance to freezing, and synthesis of cellulose on the secondary cell wall [77]. In trichome differentiation, the gene TBR has a fundamental function in the cellulose content, but additionally regulates the density of trichomes on the epidermal surface [76,77]. Therefore, Rosa chinensis protein trichome birefringence-like 2 could play a potential role in prickle development in Rubus.
5. Conclusions
Prickle-free cultivars facilitate fruit harvesting, cultivation, and management and, as such, are highly desirable. However, development of new prickle-free cultivars through conventional techniques is time consuming and difficult due to the limited prickle-free germplasm and the complex nature of other desirable traits, especially fruit quality traits. The development of prickle-free versions of economically important cultivars through gene editing techniques will provide both desirable new planting material and expand the available germplasm for future breeding efforts. An understanding of the genetic and molecular processes behind prickle development is necessary to facilitate this process. Genotype by sequencing (GBS) combined with genome-wide association mapping study (GWAS) identified four SNPs on chromosome 4 associated with the phenotype. Transcripts flanking ±100 kb of the 4 associated SNPs were identified, which provided potential candidate genes for prickle production in red raspberry including MYB16-like (MIXTA-like R2R3-MYB), MADS-box protein AGL30, and trichome birefringence-like 2 protein. Specific sequence information from these targets could then use gene editing techniques to identify the gene responsible for the prickle development and further development of prickle-free cultivars. With the expanding market for fresh raspberries and blackberries, prickle-free cultivars can improve efficiency of production practices thus reducing labor costs required for plant management and harvest.
Author Contributions
A.K. conducted the research in this manuscript as partial requirement for her PhD degree program at Cornell University under the advisement of C.A.W. who contributed to the project design, data analysis and interpretation, and editorial guidance. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in writing of the manuscript, or the decision to publish the results.
References
- Thompson, M.M. Chromosome numbers of Rubus species at the national clonal germplasm repository. HortScience 1995, 30, 1447–1452. [Google Scholar] [CrossRef]
- Deighton, N.; Brennan, R.; Finn, C.; Davies, H.V. Antioxidant properties of domesticated and wild Rubus species. J. Sci. Food Agric. 2000, 80, 1307–1313. [Google Scholar] [CrossRef]
- Yousefi, G.; Yousefi, S.; Emam-Djomeh, Z. A comparative study on different concentration methods of extracts obtained from two raspberries (Rubus idaeus L.) cultivars: Evaluation of anthocyanins and phenolics contents and antioxidant activity. Int. J. Food Sci. Tech. 2013, 48, 1179–1186. [Google Scholar] [CrossRef]
- Arumuganathan, K.; Earle, E. Nuclear DNA content of some important plant species. Plant Mol. Biol. Rep. 1991, 9, 208–218. [Google Scholar] [CrossRef]
- Keep, E. Incompatibility in Rubus with special reference to R. idaeus L. Can. J. Genet. Cytol. 1968, 10, 253–262. [Google Scholar] [CrossRef]
- Keep, E. Dwarfing in the raspberry, Rubus idaeus L. Euphytica 1969, 18, 256–276. [Google Scholar]
- Doughari, J. An overview of plant immunity. J. Plant Pathol. Microbiol. 2015, 6. [Google Scholar] [CrossRef]
- Barton, K.E. Prickles, latex, and tolerance in the endemic Hawaiian prickly poppy (Argemone glauca): Variation between populations, across ontogeny, and in response to abiotic factors. Oecologia 2014, 174, 1273–1281. [Google Scholar] [CrossRef]
- Halpern, M.; Raats, D.; Lev-Yadun, S. The potential anti-herbivory role of microorganisms on plant thorns. Plant Sign. Behav. 2007, 2, 503–504. [Google Scholar] [CrossRef]
- Szymanski, D.B.; Lloyd, A.M.; Marks, M.D. Progress in the molecular genetic analysis of trichome initiation and morphogenesis in Arabidopsis. Trends Plant Sci. 2000, 5, 214–219. [Google Scholar] [CrossRef]
- Schuepp, P. Tansley review No. 59. Leaf boundary layers. New Phytol. 1993, 125, 477–507. [Google Scholar] [CrossRef]
- Bieniek, M.E.; Millington, W. Differentiation of lateral shoots as thorns in Ulex europaeus. Amer. J. Bot. 1967, 54, 61–70. [Google Scholar] [CrossRef]
- Blaser, H.W. Morphology of the determinate thorn-shoots of Gleditsia. Am. J. Bot. 1956, 43, 22–28. [Google Scholar] [CrossRef]
- Coyner, M.; Skirvin, R.M.; Norton, M.; Otterbacher, A. Thornlessness in blackberries: A review. Small Fruits Rev. 2005, 4, 83–106. [Google Scholar] [CrossRef]
- Esau, K. Anatomy of Seed Plants; John Wiley & Sons, Inc.: New York, NY, USA, 1977; pp. 455–500. [Google Scholar]
- Posluszny, U.; Fisher, J.B. Thorn and hook ontogeny in Artabotrys hexapetalus (Annonaceae). Am. J. Bot. 2000, 87, 1561–1570. [Google Scholar] [CrossRef]
- Jennings, D. Balanced lethals and polymorphism in Rubus idaeus. Heredity 1967, 22, 465–479. [Google Scholar] [CrossRef]
- Jennings, D. Resistance to Didymella applanata in red raspberry and some related species. Ann. Appl. Biol. 1982, 101, 331–337. [Google Scholar] [CrossRef]
- Jennings, D.; Brydon, E. Further studies on resistance to Leptosphaeria coniothyrium in the red raspberry and related species. Ann. Appl. Biol. 1989, 115, 499–506. [Google Scholar] [CrossRef]
- Knight, R.; Keep, E. Developments in soft fruit breeding at East Malling. In Report of East Malling Research Station for 1957; 1958; pp. 62–67. [Google Scholar]
- Anthony, V.M.; Williamson, B.; Jennings, D.; Shattock, R. Inheritance of resistance to yellow rust (Phragmidium rubi-idaei) in red raspberry. Ann. Appl. Biol. 1986, 109, 365–374. [Google Scholar] [CrossRef]
- Jennings, D. Some evidence on the influence of the morphology of raspberry canes upon their liability to be attacked by certain fungi. Hort. Res. 1962, 1, 100–111. [Google Scholar]
- Jennings, D.; McGregor, G. Resistance to cane spot (Elsinoe veneta) in the red raspberry and its relationship to resistance to yellow rust (Phragmidium rubi-idaei). Euphytica 1988, 37, 173–180. [Google Scholar] [CrossRef]
- Keep, E. Progress in Rubus breeding at East Malling. Acta Hortic. 1976, 60, 123–128. [Google Scholar] [CrossRef]
- Graham, J.; Smith, K.; Tierney, I.; MacKenzie, K.; Hackett, C. Mapping gene H controlling cane pubescence in raspberry and its association with resistance to cane botrytis and spur blight, rust and cane spot. Theor. Appl. Genet. 2006, 112, 818–831. [Google Scholar] [CrossRef] [PubMed]
- Keep, E.; Knight, V.H.; Parker, J.H. Rubus coreanus as donor of resistance to cane diseases and mildew in red raspberry breeding. Euphytica 1977, 26, 505–510. [Google Scholar] [CrossRef]
- Peitersen, A.K. Blackberries of New England-genetic status of the plants. Vermont Agric. Expt. Stn. Bull. 1921, 218, 1–34. [Google Scholar]
- Jennings, D.; Ingram, R. Hybrids of Rubus parviflorus (Nutt.) with raspberry and blackberry and the inheritance of spinelessness derived from this species. Crop Res. 1983, 23, 95–101. [Google Scholar]
- Jennings, D.L. Raspberries and Blackberries: Their Breeding, Diseases and Growth; Academic Press: London, UK, 1988; 230p. [Google Scholar]
- Hall, H.; Quazi, M.; Skirvin, R. Isolation of a pure thornless loganberry by meristem tip culture. Euphytica 1986, 35, 1039–1044. [Google Scholar] [CrossRef]
- Rosati, P.; Gaggioli, D.; Giunchi, L. Genetic stability of micropropagated loganberry plants. J. Hortic. Sci. 1986, 61, 33–41. [Google Scholar] [CrossRef]
- Rosati, P.; Hall, H.; Jennings, D.; Gaggioli, D. A dominant gene for thornlessness obtained from the chimeral thornless loganberry. HortScience 1988, 23, 899–902. [Google Scholar]
- Zhou, N.; Tang, K.; Jeauffre, J.; Thouroude, T.; Arias, D.L.; Foucher, F.; Hibrand-Saint Oyant, L. Genetic determinism of prickles in rose. Theor. Appl. Genet. 2020, 133, 3017–3035. [Google Scholar] [CrossRef]
- Finn, C.; Moore, P.; Kempler, C. Raspberry Cultivars: What’s New? What’s Succeeding? Where are the Breeding Programs Headed? Acta Hortic. 2008, 777, 33–40. [Google Scholar] [CrossRef]
- Clark, J.R.; Moore, J.N. ‘Natchez’thornless blackberry. HortScience 2008, 43, 1897–1899. [Google Scholar] [CrossRef]
- Weber, C. In Introgression of spine-free and primocane fruiting from red raspberry (Rubus idaeus L.) to black raspberry (R. occidentalis L.). Acta Hortic. 2019, 1277, 17–24. [Google Scholar]
- Khadgi, A.; Weber, C.A. Morphological characterization of prickled and prickle-free Rubus using scanning electron microscopy. HortScience 2020, 55, 676–683. [Google Scholar] [CrossRef]
- Nordborg, M.; Borevitz, J.O.; Bergelson, J.; Berry, C.C.; Chory, J.; Hagenblad, J.; Kreitman, M.; Maloof, J.N.; Noyes, T.; Oefner, P.J. The extent of linkage disequilibrium in Arabidopsis thaliana. Nat. Genet. 2002, 30, 190–193. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Gore, M.; Buckler, E.S.; Yu, J. Status and prospects of association mapping in plants. Plant Genom. 2008, 1, 5–20. [Google Scholar] [CrossRef]
- Jennings, D.L. Raspberry plant named ‘Joan J’. 2008. US Patent and Trademark Office. Available online: www.uspto.gov (accessed on 23 December 2020).
- Elshire, R.J.; Glaubitz, J.C.; Sun, Q.; Poland, J.A.; Kawamoto, K.; Buckler, E.S.; Mitchell, S.E. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 2011, 6, e19379. [Google Scholar] [CrossRef] [PubMed]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef] [PubMed]
- Glaubitz, J.C.; Casstevens, T.M.; Lu, F.; Harriman, J.; Elshire, R.J.; Sun, Q.; Buckler, E.S. TASSEL-GBS: A high capacity genotyping by sequencing analysis pipeline. PLoS ONE 2014, 9, e90346. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Swarts, K.; Li, H.; Romero Navarro, J.A.; An, D.; Romay, M.C.; Hearne, S.; Acharya, C.; Glaubitz, J.C.; Mitchell, S.; Elshire, R.J. Novel methods to optimize genotypic imputation for low-coverage, next-generation sequence data in crop plants. Plant Genome 2014, 7, 1–12. [Google Scholar] [CrossRef]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis: Use R; Springer: New York, NY, USA, 2009. [Google Scholar]
- Yu, J.; Pressoir, G.; Briggs, W.H.; Bi, I.V.; Yamasaki, M.; Doebley, J.F.; McMullen, M.D.; Gaut, B.S.; Nielsen, D.M.; Holland, J.B. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 2006, 38, 203–208. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Ersoz, E.; Lai, C.Q.; Todhunter, R.J.; Tiwari, H.K.; Gore, M.A.; Bradbury, P.J.; Yu, J.; Arnett, D.K.; Ordovas, J.M. Mixed linear model approach adapted for genome-wide association studies. Nat. Genet. 2010, 42, 355–360. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Huang, M.; Fan, B.; Buckler, E.S.; Zhang, Z. Iterative usage of fixed and random effect models for powerful and efficient genome-wide association studies. PLoS Genet. 2016, 12, e1005767. [Google Scholar] [CrossRef]
- Lipka, A.E.; Tian, F.; Wang, Q.; Peiffer, J.; Li, M.; Bradbury, P.J.; Gore, M.A.; Buckler, E.S.; Zhang, Z. GAPIT: Genome association and prediction integrated tool. Bioinformatics 2012, 28, 2397–2399. [Google Scholar] [CrossRef]
- Götz, S.; García-Gómez, J.M.; Terol, J.; Williams, T.D.; Nagaraj, S.H.; Nueda, M.J.; Robles, M.; Talón, M.; Dopazo, J.; Conesa, A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucl. Acids Res. 2008, 36, 3420–3435. [Google Scholar] [CrossRef]
- Oshima, Y.; Mitsuda, N. The MIXTA-like Transcription factor MYB16 is a major regulator of cuticle formation in vegetative organs. Plant Sign. Behav. 2013, 8, e26826. [Google Scholar] [CrossRef]
- Liang, G.; He, H.; Li, Y.; Ai, Q.; Yu, D. MYB82 functions in regulation of trichome development in Arabidopsis. J. Exp. Bot. 2014, 65, 3215–3223. [Google Scholar] [CrossRef]
- Graham, J.; Smith, K.; MacKenzie, K.; Jorgenson, L.; Hackett, C.; Powell, W. The construction of a genetic linkage map of red raspberry (Rubus idaeus subsp. idaeus) based on AFLPs, genomic-SSR and EST-SSR markers. Theor. Appl. Genet. 2004, 109, 740–749. [Google Scholar] [CrossRef]
- Molina-Bravo, R.; Fernandez, G.E.; Sosinski, B.R. Quantitative trait locus analysis of tolerance to temperature fluctuations in winter, fruit characteristics, flower color, and prickle-free canes in raspberry. Mol. Breed. 2014, 33, 267–280. [Google Scholar] [CrossRef]
- Khadgi, A.; Weber, C.A. RNA-Seq analysis of prickled and prickle-free epidermis provides insight into the genetics of prickle development in red raspberry (Rubus ideaus L.). Agronomy 2020, 10, 1904. [Google Scholar] [CrossRef]
- Noda, K.I.; Glover, B.J.; Linstead, P.; Martin, C. Flower colour intensity depends on specialized cell shape controlled by a Myb-related transcription factor. Nature 1994, 369, 661–664. [Google Scholar] [CrossRef]
- Baumann, K.; Perez-Rodriguez, M.; Bradley, D.; Venail, J.; Bailey, P.; Jin, H.; Koes, R.; Roberts, K.; Martin, C. Control of cell and petal morphogenesis by R2R3 MYB transcription factors. Development 2007, 134, 1691–1701. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Yang, S.S.; Liang, Z.; Feng, B.R.; Liu, L.; Huang, Y.B.; Tang, Y.X. Genome-wide analysis of the MYB transcription factor superfamily in soybean. BMC Plant Biol. 2012, 12, 106. [Google Scholar] [CrossRef]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Zhang, C.; Li, J.; Wang, L.; Ren, Z. Genome-wide identification and characterization of R2R3MYB family in Cucumis sativus. PLoS ONE 2012, 7, e47576. [Google Scholar] [CrossRef]
- Matus, J.T.; Aquea, F.; Acre-Johnson, P. Analysis of the grape MYB R2R3 subfamily reveals expanded wine quality-related clades and conserved gene structure organization across Vitis and Arabidopsis genomes. BMC Plant Biol. 2008, 8, 83. [Google Scholar] [CrossRef]
- Wilkins, O.; Nahal, H.; Foong, J.; Provart, N.J.; Campbell, M.M. Expansion and diversification of the Populus R2R3-MYB family of transcription factors. Plant Phys. 2009, 149, 981–993. [Google Scholar] [CrossRef]
- González, M.; Carrasco, B.; Salazar, E. Genome-wide identification and characterization of R2R3MYB family in Rosaceae. Genome. Data 2016, 9, 50–57. [Google Scholar] [CrossRef]
- Sun, Y.; Zhou, Q.; Zhang, W.; Fu, Y.; Huang, H. Asymmetric Leaves1, an Arabidopsis gene that is involved in the control of cell differentiation in leaves. Planta 2002, 214, 694–702. [Google Scholar] [CrossRef]
- Castelán-Muñoz, N.; Herrera, J.; Cajero-Sánchez, W.; Arrizubieta, M.; Trejo, C.; Garcia-Ponce, B.; Sánchez, M.D.L.P.; Álvarez-Buylla, E.R.; Garay-Arroyo, A. MADS-box genes are key components of genetic regulatory networks involved in abiotic stress and plastic developmental responses in plants. Front. Plant Sci. 2019, 10, 853. [Google Scholar]
- Pandey, S.; Goel, R.; Bhardwaj, A.; Asif, M.H.; Sawant, S.V.; Misra, P. Transcriptome analysis provides insight into prickle development and its link to defense and secondary metabolism in Solanum viarum Dunal. Sci. Rep. 2018, 8, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Shedletzky, E.; Shmuel, M.; Trainin, T.; Kalman, S.; Delmer, D. Cell wall structure in cells adapted to growth on the cellulose-synthesis inhibitor 2, 6-dichlorobenzonitrile: A comparison between two dicotyledonous plants and a graminaceous monocot. Plant Phys. 1992, 100, 120–130. [Google Scholar] [CrossRef] [PubMed]
- Carpita, N.; Tierney, M.; Campbell, M. Molecular biology of the plant cell wall: Searching for the genes that define structure, architecture and dynamics. In Plant Cell Walls; Carpita, N., Tierney, M., Campbell, M., Eds.; Springer: Dordrecht, The Netherlands, 2001; pp. 1–5. [Google Scholar]
- Taylor, N.G.; Laurie, S.; Turner, S.R. Multiple cellulose synthase catalytic subunits are required for cellulose synthesis in Arabidopsis. Plant Cell 2000, 12, 2529–2539. [Google Scholar] [CrossRef] [PubMed]
- Brown, D.M.; Zeef, L.A.; Ellis, J.; Goodacre, R.; Turner, S.R. Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell 2005, 17, 2281–2295. [Google Scholar] [CrossRef]
- Ebringerová, A.; Heinze, T. Xylan and xylan derivatives–biopolymers with valuable properties, 1. Naturally occurring xylans structures, isolation procedures and properties. Macromol. Rap. Comm. 2000, 21, 542–556. [Google Scholar] [CrossRef]
- Arioli, T.; Peng, L.; Betzner, A.S.; Burn, J.; Wittke, W.; Herth, W.; Camilleri, C.; Höfte, H.; Plazinski, J.; Birch, R. Molecular analysis of cellulose biosynthesis in Arabidopsis. Science 1998, 279, 717–720. [Google Scholar] [CrossRef]
- Burn, J.E.; Hocart, C.H.; Birch, R.J.; Cork, A.C.; Williamson, R.E. Functional Analysis of the Cellulose Synthase GenesCesA1, CesA2, and CesA3 in Arabidopsis. Plant Phys. 2002, 129, 797–807. [Google Scholar] [CrossRef]
- Fagard, M.; Desnos, T.; Desprez, T.; Goubet, F.; Refregier, G.; Mouille, G.; McCann, M.; Rayon, C.; Vernhettes, S.; Höfte, H. PROCUSTE1 encodes a cellulose synthase required for normal cell elongation specifically in roots and dark-grown hypocotyls of Arabidopsis. Plant Cell 2000, 12, 2409–2423. [Google Scholar] [CrossRef]
- Potikha, T.; Delmer, D.P. A mutant of Arabidopsis thaliana displaying altered patterns of cellulose deposition. Plant J. 1995, 7, 453–460. [Google Scholar] [CrossRef]
- Bischoff, V.; Nita, S.; Neumetzler, L.; Schindelasch, D.; Urbain, A.; Eshed, R.; Persson, S.; Delmer, D.; Scheible, W.R. TRICHOME BIREFRINGENCE and its homolog AT5G01360 encode plant-specific DUF231 proteins required for cellulose biosynthesis in Arabidopsis. Plant Phys. 2010, 153, 590–602. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).