Abstract
Efficient utilization of genetic variation in plant germplasm collections is impeded by large collection size, uneven characterization of traits, and unpredictable apportionment of allelic diversity among heterogeneous accessions. Distributing compact subsets of the complete collection that contain maximum allelic diversity at functional loci of interest could streamline conventional and precision breeding. Using heterogeneous population samples from Arabidopsis, Populus and sorghum, we show that genomewide single nucleotide polymorphism (SNP) data permits the capture of 3–78 fold more haplotypic diversity in subsets than geographic or environmental data, which are commonly used surrogate predictors of genetic diversity. Using a large genomewide SNP data set from landrace sorghum, we demonstrate three bioinformatic approaches to extract functional genetic diversity. First, in a “candidate gene” approach, we assembled subsets that maximized haplotypic diversity at 135 putative lignin biosynthetic loci, relevant to biomass breeding programs. Secondly, we applied a keyword search against the Gene Ontology to identify 1040 regulatory loci and assembled subsets capturing genomewide regulatory gene diversity, a general source of phenotypic variation. Third, we developed a machine-learning approach to rank semantic similarity between Gene Ontology term definitions and the textual content of scientific publications on crop adaptation to climate, a complex breeding objective. We identified 505 sorghum loci whose defined function is semantically-related to climate adaptation concepts. The assembled subsets could be used to address climatic pressures on sorghum production. To face impending agricultural challenges and foster rapid extraction and use of novel genetic diversity resident in heterogeneous germplasm collections, whole genome resequencing efforts should be prioritized.
1. Introduction
Plant germplasm collections safeguarded in gene banks conserve the raw materials necessary to confront agricultural challenges. Agricultural challenges come in many forms, from the immediate—disease outbreaks [1,2]; to the persistent—greater yield from fewer inputs (e.g., in maize, [3,4,5]); to future unknowns—the adaptation of cropping systems to no-analog climates and pest assemblages [6,7]. Gene banks conserve DNA sequence variation packaged into reproductive propagules. This DNA sequence variation forms the material basis of the potential phenotypic variation available in a collection that can be used to address challenges.
In plant populations, whether natural or artificial (as in the case of gene bank accessions), segregating DNA sequence variation consists of haplotype blocks—contiguous spans of sequence that are inherited as a unit. Extraction of improved or novel traits from a collection depends on the mobilization of haplotype blocks covering a desired set of genes into breeding lines, and eventually, to elite cultivars. This process has been aided by use of “core collections”, subsets of the broader collection designed to contain maximum genetic variation in a compact number of accessions [8]. These subsets decrease the number of crosses, and hence the scale of experiments, necessary to explore the potential phenotypic variation held in a collection.
Germplasm subsets have often been assembled to maximize variation in traits, using phenotypic measurements from field evaluations [9]. The degree to which phenotypic trait variation can be used to capture, and ultimately distribute, underlying genetic diversity is not known although it is unlikely to be a very effective proxy. Much functional genetic diversity is cryptic with respect to phenotype, observable only when recombined into a different genetic background [10,11,12,13]. Molecular marker data is a popular alternative for guiding subset assembly [14,15,16]. While shown to capture more allelic diversity than random sampling [17], typical marker data sets are sparsely distributed across the genome thus haplotype capture at particular loci of agronomic importance may fail [18]. In cases where trait evaluation data and/or molecular marker data are incomplete or unavailable, geographic variation (samples come from widely dispersed sites) and environmental variation (samples come from different environments) have been treated as surrogates for genetic variation and likewise maximized [19,20,21,22]. Reeves and Richards [23,24] showed that the use of geographic and environmental surrogates produces subsets with little added genomewide haplotypic diversity compared to subsets selected at random and, moreover, that the use of these surrogates results in biased capture of important functional genetic variation.
In order to deliver breeding subsets enriched in haplotypic diversity, we support the notion that germplasm collections should be curated and accessed with a central focus on DNA sequence variation [25]. In this study, using three exemplar data sets, we quantify the improvement in genomewide haplotype capture that can be gained by using dense single nucleotide polymorphism (SNP) data sets instead of geographic or environmental surrogate predictors of genetic diversity. Using landrace sorghum we demonstrate three bioinformatic procedures that might be used to assemble subsets enriched in haplotypic diversity at targeted sets of functional loci. This includes a novel machine-learning approach which, by leveraging the information encoded in written language, transduces scientific publications related to complex breeding objectives into pertinent functional genes, at which haplotypic diversity can be efficiently captured in subsets.
2. Material and Methods
Genomewide SNP data was acquired from published studies on wild European Arabidopsis thaliana (L.) Heynh., a model species, wild North American Populus trichocarpa Torr. & Gray, a source of wood fiber, and landrace African Sorghum bicolor (L.) Moench., a grain/feedstock commonly cultivated in arid areas [26,27,28]. Many published data sets of genomewide SNP variation sample a single individual per accession. Such data are not suitable for representing the genetic diversity present in heterogeneous germplasm collections of wild and landrace material. Therefore, populations/accessions used here were subsampled from the original studies. All contained > 4 individuals, equivalent to accepting a worst-case minor allele frequency (MAF) of 0.125. The Arabidopsis data set contained 40 populations with 441 individuals and 214,051 SNPs on 5 chromosomes; Populus, 56 populations, 251 individuals, 32,860 SNPs on 19 chromosomes; sorghum, 113 individuals, 22 accessions, 404,614 SNPs on 10 chromosomes. For Populus and sorghum, haplotype phase and missing genotypes were imputed using fastPHASE [29]. Provenance details and maps displaying the geographic distribution of the populations/accessions used here are contained in prior publications [23,24,26,27,28].
Geographic coordinates (latitude/longitude) at site of origin of each population/accession were used to extract 19 environmental variables expressing precipitation and temperature regimes at the sampling site from the BIOCLIM set of data layers (http:/www.worldclim.org). Altitude was also included as an environmental variable. The continuous geographic and environmental variables were quantized into categorical states using the hierarchical method described in Reeves and Richards [24] in order to assemble subsets.
To simulate varying levels of linkage disequilibrium (LD) across the genome, SNP data were recoded into haplotype blocks using the software HAPLOTYPISTA (https://github.com/NCGRP/haplotypista), which calls alleles according to the genotype induced by concatenating adjacent SNPs. By varying the number of concatenated SNPs from 2 to 50, we explored average physical block lengths from 0.555 ± 1.46 Kbp (kilobase pairs) (± 1 sd) to 27.2 ± 17.5 Kbp for Arabidopsis, 10.4 ± 53.1 Kbp to 550 ± 489 Kbp for Populus, and 1.63 ± 14.5 Kbp to 79.8 ± 140 Kbp for sorghum. The number of haplotype blocks likewise varied: 107,024 to 4280, Arabidopsis; 16,425 to 647, Populus; 202,304 to 8088, sorghum.
Loci of interest were identified in heterogeneous landrace sorghum using three scenarios applicable to breeding programs. Scenario one, a candidate gene approach, used an expert-curated list of 135 genes belonging to 10 gene families involved in lignin biosynthesis, derived from Xu et al. [30]. Genomic intervals containing these genes were identified using the Sorghum v1.4 annotation and were found on all 10 sorghum chromosomes. Eighteen intervals contained no SNPs in our 22 accession data set so were not used, leaving 117. We call the set of 117 loci of interest for scenario one “lignin biosynthetic genes”.
In the second scenario, we used a keyword search strategy against the Gene Ontology to identify genes with regulatory properties. A body of text (“corpus”) containing the GO term ID, name, namespace, and definition lines from the human-readable ontology description file ‘go-basic.obo’ (release 2016-12-20) was created for sorghum. In this corpus, a single GO term ID/name/namespace/defline combination is treated as a “document”. The corpus contained 2697 documents, each corresponding to a GO term occurring in Sorghum v1.4, as determined from a custom gene association file (which excluded annotation qualifiers ‘not’, ‘contributes_to’, and ‘colocalizes_with’) that contained 58,610 GO annotations of 16,262 named sorghum genes. A keyword search for the root word ‘regulat’ returned 161 terms (151 biological process, 8 molecular function, 2 cellular component) meant to encompass broad-sense regulatory gene functional diversity in sorghum. The set of loci of interest for scenario two, called “regulatory genes”, was made up of 1040 genomic intervals containing genes annotated to 161 GO terms.
In the third scenario we used natural language processing to transduce the written language of scientific publications into relevant genomic intervals. For proof of concept we chose the topic “crop adaptation to climate in the United States”, a concept for which breeding objectives are complex. We used two highly-cited journal articles on the subject: “Nonlinear temperature effects indicate severe damages to U.S. crop yields under climate change” by Schlenker and Roberts [31], and “Greater sensitivity to drought accompanies maize yield increase in the U.S. Midwest” by Lobell et al. [32], plus a primer published as United States Department of Agriculture Technical Bulletin 1935 entitled “Climate change and agriculture in the United States: effects and adaptation” [33]. Documents were rendered as plain text and cleaned of low-relevance and unwanted information (e.g., author affiliations, citations). Latent semantic analysis [34,35], a natural language processing procedure, was performed using the machine-learning Python toolkit Scikit-learn [36]. Latent semantic analysis processes a text corpus into a system of vectors using singular value decomposition (SVD). A query, in this case the combined text of the scientific publications, is then fit to the vector system so that a distance between the query and each document in the corpus can be calculated based on similarity in word occurrence frequency.
Cellular component GO terms were removed from the corpus prior to vectorization using TfidfVectorizer(), configured with preprocessing directives to eliminate common English stop words (“it”, “and”, “the”, etc.) and normalize the character set by stripping accents, punctuation, and other non-standard characters. Single word and adjacent two-word pairs were permitted as “tokens”. Tokens were “tf-idf” weighted (“term frequency-inverse document frequency”) which scales the importance of a token (here, a word or two word phrase) in relation to its frequency of occurrence in a document (here, a single GO term) and to the inverse of its frequency in the corpus (all GO terms). A matrix containing the weighted frequency of words and word pairs for each GO term was then subject to singular value decomposition for reduction into a vector system, allowing up to 2500 component axes such that the distance between each individual GO term and query could be computed without the progressive reduction of corpus size usually necessary to achieve stable document-query ranking. TruncatedSVD() was used with ten randomized iterations, method fit_transform() created the vector system for the corpus, and method transform() mapped the query text onto that system. To express similarity in concepts, cosine distances between the query and each GO term were calculated using sklearn.metrics.pairwise_distances() then ranked. The top 50 GO terms most semantically related to the query text were identified (Table 1), resulting in a set containing 505 genes, here called “climate adaptation genes”.

Table 1.
Top 50 Gene Ontology terms ranked by semantic similarity to scientific publications on crop adaptation to climate in the United States. Rank 1–25, left; 26–50, right.
The software M+ (https://github.com/NCGRP/Mplus) [18,23] was used to assemble optimized subsets of populations/accessions containing maximum diversity in reference data sets. M+ is a parallelized implementation of the M strategy, a commonly used subset optimization algorithm that outperforms stratified (population structure-based) sampling procedures for retaining allelic diversity [17]. The M strategy focuses only on retaining the maximum number of distinct alleles in a minimized subset relative to the complete collection. It does not attempt to maintain representative allele frequencies across a stratified sample as they occur in the complete collection. Both subset optimization philosophies are useful, but the former quantity, raw allelic diversity or high allele count, is the quantity of interest here. Reference data sets included geographic, environmental, and whole genome haplotypic diversity, as well as, for sorghum, the lignin biosynthetic gene, regulatory gene, and the climate adaptation gene sets. M+ returns a normalized metric of diversity captured at user-defined variables (here, loci or quantized geographic/environmental data) in optimized subsets (mopt) as well as in subsets that are randomly assembled (mran). To assign a value indicating the capacity of a reference set of information to predict diversity at each SNP position in the genome, we calculated the deviation mopt − mran for each genomic interval defined by a haplotype block, for all possible subset sizes. We have previously termed this value the “diversity retention index” (DRI, details [24]). DRI > 0 indicates that allelic diversity at target loci can be predicted (and therefore captured) better than random by using reference data. DRI ≤ 0 indicates that the reference data contain no useful information to predict allelic diversity at the target loci. Using this analytical construct we can estimate the relative enrichment of allelic diversity in subsets targeting the whole genome or specific functional gene sets of interest. By varying haplotype block length, we can also evaluate the effect of the extent of LD on those estimates. Annotated bioinformatic pipelines and other resources relevant to performing the analyses have been archived at https://github.com/NCGRP/agr1suppl.
3. Results
Genomewide SNP data broadly outperformed geographic and environmental data in capturing haplotypes across the genome (Figure 1). Using genomewide SNP data allowed more haplotypic diversity to be captured across 99% of the Arabidopsis genome (98%, Populus; 89%, sorghum). In all three taxa, genomewide SNP data allowed capture of significantly more haplotypic diversity than geographic and environmental data (t-test, p < 2E−16; Figure 2). In Arabidopsis, maximizing geographic and environmental diversity resulted in the capture of less haplotypic diversity than would be expected in random subsets. In Populus, genomewide SNP data led to the capture of 3–9 fold more haplotypic diversity than geographic and environmental data. In sorghum, genomewide SNP data captured 19–78 fold more haplotypic diversity while geographic and environmental data types captured little more than random.
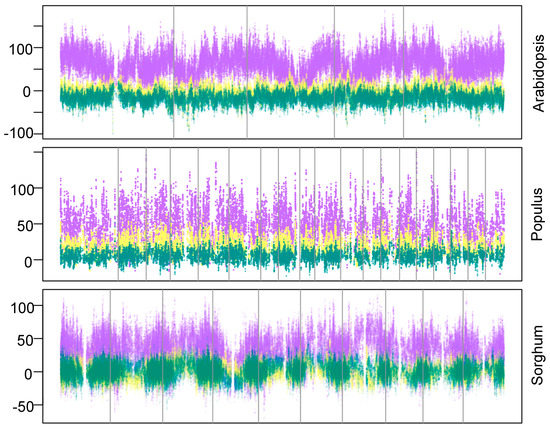
Figure 1.
Relative improvement in genomewide haplotype capture achieved by using genomewide single nucleotide polymorphism (SNP) data instead of geographic or environmental data. Y-axis measures relative enrichment, where zero is the haplotypic diversity recovered in random subsets. Vertical lines mark chromosome boundaries. Points show haplotypic diversity captured at each SNP position by maximizing genomewide diversity (purple), geographic diversity (yellow), or environmental diversity (teal) in subsets. Subsets that maximize geographic or environmental diversity capture less haplotypic diversity than randomly assembled subsets across much of the genome.
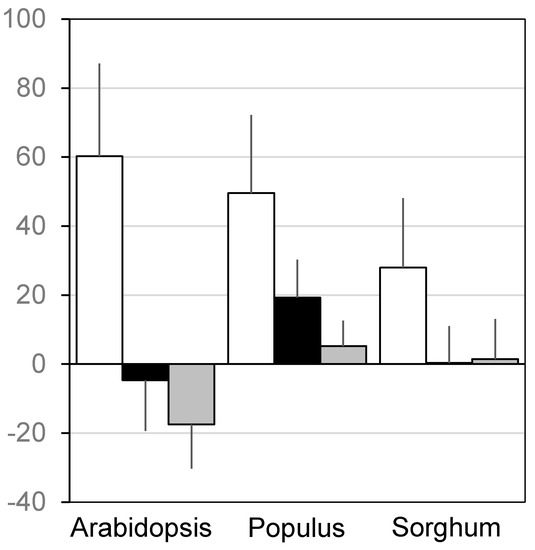
Figure 2.
Average haplotype capture in subsets assembled using genomewide single nucleotide polymorphism (SNP), geographic, and environmental data. Y-axis measures relative enrichment, zero is haplotypic diversity of random subsets. Error bar shows one standard deviation from the mean. White bars, genomewide SNP data; black, geographic data; grey, environmental data. All pairwise comparisons within taxa differ significantly (t-test, p < 2E−16).
In sorghum, we evaluated the ability of reference data sets comprised of genomewide SNP data, geographic data, or environmental data to capture diversity at loci in the lignin biosynthetic gene, regulatory gene, and climate adaptation gene sets. Genomewide SNP data showed superior ability to capture haplotypes at all target gene sets relative to geographic and environmental data (Figure 3). This was true whether the haplotypic diversity used for subset optimization was derived from the whole genome or solely from the haplotype blocks covering the target genes. Geographic and environmental data performed little better than random, regardless of the LD extent simulated. When the lignin biosynthetic gene set was used as reference there was a modest advantage to using short block lengths (<~10 Kbp), consistent with estimates of the rate of LD decay in sorghum [37] (Figure 3a). A related pattern was observed for the regulatory genes, where, for long LD, subsets that maximized whole genome haplotypic diversity captured more regulatory gene haplotypes than subsets that maximized haplotypic diversity in the regulatory genes themselves. For short LD, regulatory gene haplotype capture was equivalent. Regardless of LD extent, climate adaptation gene haplotypes were better captured by maximizing diversity in the target gene set itself rather than the whole genome.
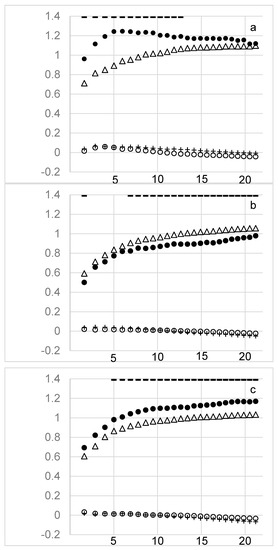
Figure 3.
Haplotype capture at three sets of target genes using genomewide single nucleotide polymorphism (SNP), geographic, and environmental data to assemble subsets in sorghum. Target gene sets are (a) lignin biosynthetic genes, (b) regulatory genes, (c) climate adaptation genes. Plotted points are coded by reference data source: triangles, whole genome; closed circle, target genes; plus sign, geography; open circle, environment. X-axis, average physical length of haplotype blocks (Kbp). Y-axis, rescaled relative enrichment of haplotypic diversity at target gene sets, zero is random expectation. Dashes at top indicate significant difference between whole genome (triangle) vs. target gene (closed circle) reference (Holm-Bonferroni corrected p-value < 0.05).
We also evaluated the ability of the target gene sets, used as reference, to capture haplotypes genomewide, again varying the maximum block length to simulate different levels of LD around the loci. This is a measure of genomewide haplotypic diversity that might be “lost” if subset optimization only considered the target gene sets. Target gene sets captured significantly fewer haplotypes genomewide than genomewide SNP data, but significantly more than geographic and environmental data (t-test, Holm-Bonferroni corrected p < 2E−33) (Figure 4). Geographic and environmental data captured slightly more haplotypic diversity genomewide than random subsets when LD was short, and similar levels to random when LD was long.
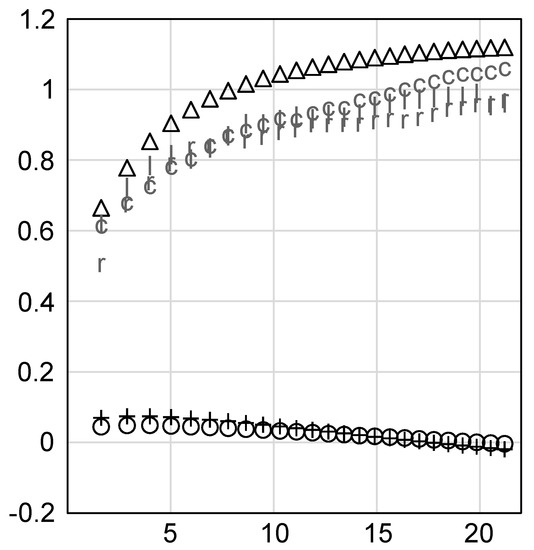
Figure 4.
Genomewide haplotype capture in sorghum using genomewide SNPs (single nucleotide polymorphisms), three sets of target genes, geographic data, and environmental data to assemble subsets. X-axis, average physical length of haplotype blocks (Kbp). Y-axis, rescaled relative enrichment of haplotypic diversity genomewide, zero is random expectation. Points are coded by reference data source: triangle, whole genome; ‘l’, lignin biosynthetic genes; ‘r’, regulatory genes; ‘c’, climate adaptation genes; plus sign, geography; circle, environment.
4. Discussion
Germplasm collections are complex assemblages of genetic material and associated information. “Accessions”, the basic organizational unit in most collections, may derive from elite cultivars, breeding lines, unimproved landraces, wild populations, or single individuals. Collections are sometimes large: 21,393 accessions of Hordeum vulgare at the Leibniz-Institut für Pflanzengenetik und Kulturpflanzenforschung (IPK), 53,777 accessions of Sorghum bicolor in the US National Plant Germplasm System (NPGS), 124,378 accessions of Oryza sativa at the International Rice Research Institute (IRRI). Within a species, genetic diversity is unevenly partitioned across accessions, with ample evidence of redundancy [38,39]. Phenotypic characterization may also be uneven (Figure 5). A deluge of genomewide DNA sequence data is underway, greatly increasing the informational complexity of collections (e.g., barley, 20,458 accessions, 306,049 SNPs, [40]; sorghum, 10,323 accessions, 459,304 SNPs, [41]; rice, 3010 accessions, 160,267 SNPs, [42]).
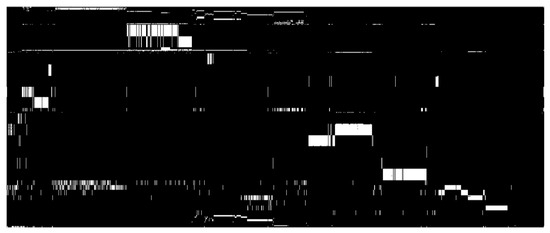
Figure 5.
Uneven phenotyping in United States sorghum collection. Columns contain 2378 accessions with genomewide SNP data. Rows contain one trait measured in one study (787 rows). The same trait may occur multiple times in the matrix, measured in independent studies. White indicates a measurement, black, missing data. Horizontal white traces indicate traits that have been measured for many accessions in a single study. Vertical traces mark accessions that have been measured for many traits.
Breeder access to these large, complex collections is dependent on the generation and distribution of manageable subsets that contain genetic diversity relevant to a breeding objective. Subsets drawn from germplasm collections for breeding purposes are often ad hoc assemblages based on experience with the collection, intuition, imperfect phenotypic data, and some expression of diversity in provenance such as geographic (source locality) or environmental (cultivation conditions) variation, intended as a surrogate for molecular genetic diversity. Given limited information, these are all valuable factors. Increasingly, breeders may possess a set of candidate genes that, effectively, defines the breeding objective, e.g., in cases like metabolic pathway engineering [43] or NBS-LRR gene-mediated disease resistance breeding [44]. Alternatively, they may have a general idea of categories of genes relevant to the breeding objective, e.g., drought resistance genes [45], secondary cell wall deposition genes [46], photosynthetic genes [47]. Or, they may face complex, multidimensional challenges where many genes are likely to be involved, e.g., yield increase [48,49], generalized stress resistance [50,51], or adaptation to new climates [52].
The purpose of this study is to demonstrate the superiority of genomewide DNA sequence data for navigating complex germplasm collections to produce subsets containing genetic diversity appropriate to a breeding objective. Using genomewide SNP data sets from three heterogeneous species samples, we visualized haplotype capture at each SNP position across the genome (Figure 1). We found geographic and environmental surrogate predictors of genetic diversity to be profoundly limited in their ability to capture haplotypic diversity genomewide. Subsets optimized using genomewide SNP data should be expected to contain an order of magnitude or more genetic diversity than those produced using geographic or environmental data alone (Figure 2). We have made this observation in a general sense before [23,24] but here illustrate it with previously unobtained granularity, down to the level of the nucleotide position. Geography and environment remain useful for assembling subsets for ecological conservation (landscape restoration, reintroductions, etc.) [53], where the object is to mimic natural distributions in allele frequency, because geography and environment are good predictors of allele frequency differences across species ranges. However, they do not appear useful for producing subsets with maximum haplotypic diversity, as would be desirable for crop breeding purposes.
Some level of phenotypic characterization is available for many species held in gene banks thus phenotypic data is an important component of the informational complexity of a collection. In this study we have not quantified the utility of phenotypic data. Phenotypic data are often incomplete, inapplicable to a breeding objective, or imperfectly sampled. In the US sorghum collection, for example, 94 traits have been measured in 101 studies over 26 years at 12 locations (9 US, 2 Mexico, 1 Brazil). In much of the collection (45396 accessions) at least one trait has been measured, but few traits have been measured for broad swaths of the collection and few accessions have been phenotyped for multiple traits (Figure 5). In order to be useful for assembling subsets from the entire collection, many traits would need to be phenotyped across most accessions at the same site in the same growing season. This is a practical impossibility for large collections. Moreover, in order to be especially valuable for breeding, accessions would have to be phenotyped at multiple sites over multiple growing seasons so that genotype by environment interactions affecting trait expression could be mitigated. Phenotypic data is laborious to obtain and, ultimately, is not a reliable predictor of trait values in different genetic backgrounds [10,13]. Assuming comprehensive genomewide sequence data is held by the gene bank, phenotyping is a responsibility that could logically rest with the user. Within a breeding program, sample sizes are manageable, and scoring of traits and site selection can be carefully tailored to the breeding objective.
We considered three scenarios for accessing germplasm collections bioinformatically instead of using passport (e.g., geographic or environmental variation) and evaluation data. In the first, where the user begins with a modest-sized list of candidate genes (here, putative lignin biosynthetic genes), we showed that much more haplotypic diversity can be captured in a subset by using SNPs in the candidate loci for the optimization, rather than surrogate predictors or even genomewide SNPs (Figure 3a). Thus breeders with sets of candidate loci can easily mine gene banks for haplotypic diversity of direct relevance to them, provided the collection has been characterized with genomewide DNA sequence data.
The second and third scenarios relied on the Gene Ontology to relate breeding objectives with genomic locations. The Gene Ontology is a controlled vocabulary designed to specify what genes “do” (molecular function and biological process terms) and where they act (cellular component terms). By associating each gene with a set of GO terms, genome annotations assign functional concepts to genomic intervals. GO term definitions contain human-readable, descriptive text passages organized into a formalized set of machine-readable fields so can be leveraged, computationally, to bridge the gap between breeding objective, expressed as phenotype, and genomic locations, which hold haplotypic diversity. The theory behind expressing phenotypes as GO terms was developed in the biomedical literature [54,55].
In scenario two, we imagined a subset containing the maximum potential to alter phenotypes in a general sense (i.e., no particular trait or set of traits is targeted). Regulatory genes, by definition, alter gene expression and are well-established generators of morphological trait variation [56,57,58], thus these regions should be included in such a subset. Most sequence variation is neutral with respect to phenotype, residing in non-functional, non-genic regions, thus these regions should be excluded. By identifying Gene Ontology terms with a regulatory connotation then extracting the associated genomic regions in sorghum, we were able to assemble subsets that specifically maximized regulatory gene haplotypic diversity while excluding non-regulatory and non-genic regions from the optimization. Subsets enriched in regulatory gene diversity could contribute greater general phenotypic variation to a breeding program than those that maximize haplotypic diversity genomewide since the latter include much non-functional variation.
In the third scenario we posited a situation where the breeding objective is sufficiently complex that an intuition-based query of the Gene Ontology would be ill-advised. For this we used a natural language processing procedure, latent semantic analysis, to transduce concepts presented in scientific publications into GO terms, which map to specific regions of the genome that can be targeted, as proposed by Reeves and Richards [24]. Scientific knowledge exists in two places: the mind (as thoughts) and publications (as written words). In cases where problems contain too many variables for human thought processes to represent them adequately, machine learning procedures can help. Computers excel at multidimensional problems and, due to rapid advances in artificial intelligence, can now readily parse written words into basic “meaning” [59]. As proof of concept, we chose the subject “crop adaptation to climate in the United States”, which represents a long-standing breeding objective (crop variety development for new growing regions) coupled with the contemporary challenge of mitigating climate change effects on agriculture (crop variety development for new growing conditions in the same regions). We processed three well-regarded scientific publications on the subject [31,32,33] into GO terms and the associated intervals in the sorghum genome. The 50 GO terms most semantically-related to the textual content of the publications are shown in Table 1. While we have no objective means at present to test their validity for producing subsets that maximize phenotypic variation in traits related to climate adaptation, the GO terms found by latent semantic analysis are intuitively reasonable, representing responses to changing moisture and temperature regimes as well as other plant stressors.
The three scenarios demonstrate that, when armed with genomewide SNP data, one can produce efficient breeding subsets from heterogeneous collections that specifically target genes of interest. More importantly, the procedure to produce such subsets can be adapted to any breeding objective and even automated, to the point of requiring very little prior scientific knowledge on the part of the collection curator or gene bank user (e.g., scenario three). Accordingly, we support the proposal that germplasm collections be curated and accessed with a central focus on DNA sequence diversity [25], rather than phenotypic variation or associated passport data. DNA sequence diversity forms the underlying physical basis of the potential phenotypic variation held in collections and, now that large scale resequencing is practical (e.g., [40,41]), should be used as the primary feature by which collections are interrogated. Genomewide sequence data sets provide the added opportunity to catalog allelic variants at loci of interest and enter them directly into crop improvement programs utilizing transgenics or genome editing, thereby circumventing traditional breeding and associated linkage drag lag time [60,61].
Our proposed methods for accessing functional genetic diversity in plant germplasm collections come with caveats, the most important of which is that the correlation between haplotypic diversity at genes identified by GO term, and phenotypic diversity at traits implied by GO term, is untested. It could be tested with large phenotype-genotype data sets from model organisms (e.g., [62]), but ultimately should be worked out in the field using the crop of interest because genetic background and environmental conditions are essential components of phenotypic expression. A technical caveat concerns the capture of low frequency variants. Genomewide resequencing projects typically define a MAF cutoff (often 5%) in order to exclude singletons and sequencing errors from the final data set. Low frequency SNP variants may not be directly observable in such data sets and thus rare haplotypes could be excluded from subset optimizations. Decreasing the MAF cutoff necessitates increasing sequence read depth, which increases costs. Additionally, the causal genetic variation underlying phenotypic variation may, for some traits, be attributable to copy number variation or indels (CNV) rather than nucleotide substitutions. By simulating haplotype blocks prior to subset assembly this concern is likely mitigated by virtue of linkage between SNPs and adjacent CNVs. Advances in pangenome analysis [63,64,65,66] may further ameliorate this concern because CNVs can be used in subset optimization the same as SNPs. The same caveat holds for non-coding regulatory sequence variation, such as that in promoter and enhancer sequences. This variation would be excluded, by definition, from subsets assembled to maximize variation in gene sets discovered by interrogating the Gene Ontology. Operationally, the possibility of “missing” any of this esssential variation would be partly mitigated by linkage with adjacent, included, genic regions, and could be explicitly tackled by extending the genomic interval contained in the gene definition to include putative cis-regulatory sequence when maximizing haplotypic diversity. Sequence variation in distant enhancer elements could still be overlooked.
5. Conclusions
The bioinformatic procedures described here support the proposal to use DNA sequence as the primary means to organize plant germplasm collections, and bioinformatics as the primary means of access [25]. Implicit in that support is an advocacy for systematic whole genome resequencing projects and software development (e.g., DivSeek, GOBII, InterMine initiatives) directed at these carefully curated and maintained public resources. Increased focus on the fundamental unit of conservation, the haplotype block, and increased utilization of artificial intelligence approaches to translate complex genomic information into manageable, relevant, and distributable genetic diversity will be necessary to address future agricultural challenges.
Author Contributions
Conceptualization, P.A.R. and C.M.R.; Software, P.A.R.; Formal analysis, P.A.R. and H.M.T.; Investigation, P.A.R.; Resources, H.M.T.; Supervision, C.M.R.; Writing—original draft preparation, P.A.R.; Writing—review and editing, H.M.T., C.M.R. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the United States Department of Agriculture, Agricultural Research Service, project number 3012-21000-015-00-D.
Acknowledgments
We thank María Muñoz-Amatriaín for helpful comments on the manuscript. This research used resources provided by the SCINet project of the USDA Agricultural Research Service, ARS project number 0500-00093-001-00-D. USDA is an equal opportunity provider and employer.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Vasudevan, K.; Vera Cruz, C.M.; Gruissem, W.; Bhullar, N.K. Large scale germplasm screening for identification of novel rice blast resistance sources. Front. Plant Sci. 2014, 5, 505. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.; Sun, J.; Matthews, A.; Armas-Egas, L.; Chen, N.; Hamill, S.; Mintoff, S.; Tran-Nguyen, L.T.T.; Batley, J.; Aitken, E.A.B. Assessing variations in host resistance to Fusarium oxysporum f sp. cubense race 4 in Musa species, with a focus on the subtropical race 4. Front. Microbiol. 2019, 10, A1062. [Google Scholar] [CrossRef]
- Mansfield, B.D.; Mumm, R.H. Survey of plant density tolerance in U.S. maize germplasm. Crop. Sci. 2014, 54, 157–173. [Google Scholar] [CrossRef]
- Kurtz, B.; Gardner, C.A.C.; Millard, M.J.; Nickson, T.; Smith, J.S.C. Global access to maize germplasm provided by the US National Plant Germplasm System and by US plant breeders. Crop. Sci. 2016, 56, 931–941. [Google Scholar] [CrossRef]
- Mastrodomenico, A.T.; Hendrix, C.C.; Below, F.E. Nitrogen use efficiency and the genetic variation of maize expired Plant Variety Protection germplasm. Agriculture 2018, 8, 3. [Google Scholar] [CrossRef]
- Williams, J.W.; Jackson, S.T. Novel climates, no analog communities, and ecological surprises. Front. Ecol. Environ. 2007, 5, 475–482. [Google Scholar] [CrossRef]
- Murdock, T.Q.; Taylor, S.W.; Flower, A.; Mehlenbacher, A.; Montenegro, A.; Zwiers, F.W.; Alfaro, R.; Spittlehouse, D.L. Pest outbreak distribution and forest management impacts in a changing climate in British Columbia. Environ. Sci. Policy 2012, 26, 75–89. [Google Scholar] [CrossRef]
- Brown, A.H.D. Core collections: A practical approach to genetic resources management. Genome 1989, 31, 818–824. [Google Scholar] [CrossRef]
- Upadhyaya, H.D.; Pundir, R.P.S.; Dwivedi, S.L.; Gowda, C.L.L.; Reddy, V.G.; Singh, S. Developing a mini core collection of sorghum for diversified utilization of germplasm. Crop. Sci. 2009, 49, 1769–1780. [Google Scholar] [CrossRef]
- Tanksley, S.D.; McCouch, S.R. Seed banks and molecular maps: Unlocking genetic potential from the wild. Science 1997, 22, 1063–1066. [Google Scholar] [CrossRef]
- Lauter, N.; Doebley, J. Genetic variation for phenotypically invariant traits detected in teosinte: Implications for the evolution of novel forms. Genetics 2002, 160, 333–342. [Google Scholar] [PubMed]
- Le Rouzic, A.; Carlborg, Ö. Evolutionary potential of hidden genetic variation. Trends Ecol. Evolut. 2008, 23, 33–37. [Google Scholar] [CrossRef]
- Paaby, A.B.; Rockman, M.V. Cryptic genetic variation: Evolution’s hidden substrate. Nat. Rev. Genet. 2014, 15, 247–258. [Google Scholar] [CrossRef]
- McKhann, H.I.; Camilleri, C.; Bérard, A.; Bataillon, T.; David, J.L.; Reboud, X.; Le Corre, V.; Caloustian, C.; Gut, I.G.; Brunel, D. Nested core collections maximizing genetic diversity in Arabidopsis thaliana. Plant J. 2004, 38, 193–202. [Google Scholar] [CrossRef] [PubMed]
- Belaj, A.; Dominguez-Garcia, M.C.; Atienza, S.G.; Urdíroz, N.M.; De la Rosa, R.; Satovic, Z.; Martín, A.; Kilian, A.; Trujillo, I.; Valpuesta, V.; et al. Developing a core collection of olive (Olea europaea L.) based on molecular markers (DArTs, SSRs, SNPs) and agronomic traits. Tree Genet. Genomes 2012, 8, 365–378. [Google Scholar] [CrossRef]
- Gross, B.L.; Volk, G.M.; Richards, C.M.; Reeves, P.A.; Henk, A.D.; Forsline, P.L.; Szewc-McFadden, A.; Fazio, G.; Chao, C.T. Diversity captured in the USDA-ARS national plant germplasm system apple core collection. J. Amer. Soc. Hort. Sci. 2013, 138, 375–381. [Google Scholar] [CrossRef]
- Bataillon, T.M.; David, J.L.; Schoen, D.J. Neutral genetic markers and conservation genetics: Simulated germplasm collections. Genetics 1996, 144, 409–417. [Google Scholar]
- Reeves, P.A.; Panella, L.W.; Richards, C.M. Retention of agronomically important variation in germplasm core collections: Implications for allele mining. Theor. Appl. Genet. 2012, 124, 1155–1171. [Google Scholar] [CrossRef]
- Upadhyaya, H.D.; Bramel, P.J.; Singh, S. Development of a chickpea core subset using geographic distribution and quantitative traits. Crop. Sci. 2001, 41, 206–210. [Google Scholar] [CrossRef]
- Upadhyaya, H.D.; Gowda, C.L.L.; Pundir, R.P.S.; Reddy, V.G.; Singh, S. Development of core subset of finger millet germplasm using geographical origin and data on 14 quantitative traits. Genet. Resour. Crop. Evol. 2006, 53, 679–685. [Google Scholar] [CrossRef]
- Yan, W.; Rutger, J.N.; Bryant, R.J.; Bockelman, H.E.; Fjellstrom, R.G.; Chen, M.H.; Tai, T.H.; McClung, A.M. Development and evaluation of core subset of the USDA rice germplasm collection. Crop. Sci. 2007, 47, 869–878. [Google Scholar] [CrossRef]
- Parra-Quijano, M.; Iriondo, J.M.; Torres, E. Improving representativeness of genebank collections through species distribution models, gap analysis and ecogeographical maps. Biodivers Conserv. 2012, 21, 79–96. [Google Scholar] [CrossRef]
- Reeves, P.A.; Richards, C.M. Capturing haplotypes in germplasm core collections using bioinformatics. Genet. Resour. Crop. Evol. 2017, 64, 1821–1828. [Google Scholar] [CrossRef]
- Reeves, P.A.; Richards, C.M. Biases induced by using geography and environment to guide ex situ conservation. Conserv. Genet. 2018, 19, 1281–1293. [Google Scholar] [CrossRef]
- Mascher, M.; Schreiber, M.; Scholz, U.; Graner, A.; Reif, J.C.; Stein, N. Genebank genomics bridges the gap between the conservation of crop diversity and plant breeding. Nat. Genet. 2019, 51, 1076–1081. [Google Scholar] [CrossRef]
- Lasky, J.R.; Des Marais, D.L.; McKay, J.K.; Richards, J.H.; Juenger, T.E.; Keitt, T.H. Characterizing genomic variation of Arabidopsis thaliana: The roles of geography and climate. Mol. Ecol. 2012, 21, 5512–5529. [Google Scholar] [CrossRef]
- Geraldes, A.; Farzaneh, N.; Grassa, C.J.; McKown, A.D.; Guy, R.D.; Mansfield, S.D.; Douglas, C.J.; Cronk, Q.C.B. Landscape genomics of Populus trichocarpa: The role of hybridization, limited gene flow, and natural selection in shaping patterns of population structure. Evolution 2014, 68, 3260–3280. [Google Scholar] [CrossRef]
- Lasky, J.R.; Upadhyaya, H.D.; Ramu, P.; Deshpande, S.; Hash, C.T.; Bonnette, J.; Juenger, T.E.; Hyma, K.; Acharya, C.; Mitchell, S.E.; et al. Genome environment associations in sorghum landraces predict adaptive traits. Sci. Adv. 2015, 1, e1400218. [Google Scholar] [CrossRef]
- Scheet, P.; Stephens, M. A fast and flexible statistical model for large scale population genotype data: Applications to inferring missing genotypes and haplotypic phase. Am. J. Hum. Genet. 2006, 78, 629–644. [Google Scholar] [CrossRef]
- Xu, Z.; Zhang, D.; Hu, J.; Zhou, X.; Ye, X.; Reichel, K.L.; Stewart, N.R.; Syrenne, R.D.; Yang, X.; Gao, P.; et al. Comparative genome analysis of lignin biosynthesis gene families across the plant kingdom. BMC Bioinformatics 2009, 10 (Suppl. S11), S3. [Google Scholar] [CrossRef]
- Schlenker, W.; Roberts, M.J. Nonlinear temperature effects indicate severe damages to U.S. crop yields under climate change. Proc. Nat. Acad. Sci. USA 2009, 106, 15594–15598. [Google Scholar] [CrossRef] [PubMed]
- Lobell, D.B.; Roberts, M.J.; Schlenker, W.; Braun, N.; Little, B.B.; Rejesus, R.M.; Hammer, G.L. Greater sensitivity to drought accompanies maize yield increase in the U.S. Midwest. Sci. 2014, 344, 516–519. [Google Scholar] [CrossRef] [PubMed]
- Walthall, C.L.; Hatfield, J.; Backlund, P.; Lengnick, L.; Marshall, E.; Walsh, M.; Adkins, S.; Aillery, M.; Ainsworth, E.A.; Ammann, C.; et al. Climate Change and Agriculture in the United States: Effects and Adaptation. USDA Technical Bulletin 1935; USDA: Washington, DC, USA, 2012.
- Deerwester, S.; Dumais, S.T.; Furnas, G.W.; Landauer, T.K.; Harshman, R. Indexing by latent semantic analysis. J. Am. Soc. Inf. Sci. 1990, 41, 391–407. [Google Scholar] [CrossRef]
- Landauer, T.K.; Foltz, P.W.; Laham, D. Introduction to latent semantic analysis. Discourse Process. 1998, 25, 259–284. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine learning in Python. J. Mach. Learn Res. 2011, 12, 2825–2830. [Google Scholar]
- Hamblin, M.T.; Salas Fernandez, M.G.; Casa, A.M.; Mitchell, S.E.; Paterson, A.H.; Kresovich, S. Equilibrium processes cannot explain high levels of short- and medium-range linkage disequilibrium in the domesticated grass Sorghum bicolor. Genetics 2005, 171, 1247–1256. [Google Scholar] [CrossRef]
- Van Treuren, R.; van Hintum, T.J.L. Marker assisted reduction of redundancy in germplasm collections: Genetic and economic aspects. Acta. Hort. 2003, 623, 139–149. [Google Scholar] [CrossRef]
- Singh, N.; Wu, S.; Raupp, W.J.; Sehgal, S.; Arora, S.; Tiwari, V.; Vikram, P.; Singh, S.; Chhuneja, P.; Gill, B.S.; et al. Efficient curation of genebanks using next generation sequencing reveals substantial duplication of germplasm accessions. Sci. Rep. 2019, 9, 650. [Google Scholar] [CrossRef]
- Milner, S.G.; Jost, M.; Taketa, S.; Mazón, E.R.; Himmelbach, A.; Oppermann, M.; Weise, S.; Knüpffer, H.; Basterrechea, M.; König, P.; et al. Genebank genomics highlights the diversity of a global barley collection. Nat. Genet. 2019, 51, 319–326. [Google Scholar] [CrossRef]
- Hu, Z.; Olatoye, M.O.; Marla, S.; Morris, G.P. An integrated genotyping by sequencing polymorphism map for over 10,000 sorghum genotypes. Plant Genome 2019, 12, 180044. [Google Scholar] [CrossRef]
- Wang, W.; Mauleon, R.; Hu, Z.; Chebotarov, D.; Tai, S.; Wu, Z.; Li, M.; Zheng, T.; Fuentes, R.R.; Zhang, F.; et al. Genomic variation in 3,010 diverse accessions of Asian cultivated rice. Nature 2018, 557, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Oksman-Caldentey, K.M.; Saito, K. Integrating genomics and metabolomics for engineering plant metabolic pathways. Curr. Opin. Biotechnol. 2005, 16, 174–179. [Google Scholar] [CrossRef]
- Jupe, F.; Witek, K.; Verweij, W.; Sliwka, J.; Pritchard, L.; Etherington, G.J.; Maclean, D.; Cock, P.J.; Leggett, R.M.; Bryan, G.J.; et al. Resistance gene enrichment sequencing (RenSeq) enables reannotation of the NB-LRR gene family from sequenced plant genomes and rapid mapping of resistance loci in segregating populations. Plant J. 2013, 76, 530–544. [Google Scholar] [CrossRef] [PubMed]
- Valliyodan, B.; Nguyen, H.T. Understanding regulatory networks and engineering for enhanced drought tolerance in plants. Curr. Opin. Plant Biol. 2006, 9, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Loqué, D.; Scheller, H.V.; Pauly, M. Engineering of plant cell walls for enhanced biofuel production. Curr. Opin. Plant Biol. 2015, 25, 151–161. [Google Scholar] [CrossRef] [PubMed]
- Ort, D.R.; Merchant, S.S.; Alric, J.; Barkan, A.; Blankenship, R.E.; Bock, R.; Croce, R.; Hanson, M.R.; Hibberd, J.M.; Long, S.P.; et al. Redesigning photosynthesis to sustainably meet global food and bioenergy demand. Proc. Natl. Acad. Sci. USA 2015, 28, 8529–8536. [Google Scholar] [CrossRef] [PubMed]
- Wallace, D.H.; Ozbun, J.L.; Munger, H.M. Physiological genetics of crop yield. Adv. Agron. 1972, 24, 97–146. [Google Scholar]
- Gur, A.; Zamir, D. Unused natural variation can lift yield barriers in plant breeding. PLoS Biol. 2004, 2, e245. [Google Scholar] [CrossRef]
- Ashraf, M.; Harris, P.J.C. Abiotic Stresses. Plant Resistance Through Breeding and Molecular Approaches; Haworth Press: New York, NY, USA, 2005. [Google Scholar]
- Das, G.; Rao, G.J.N. Molecular marker assisted gene stacking for biotic and abiotic stress resistance genes in an elite rice cultivar. Front. Plant Sci. 2015, 6, 698. [Google Scholar] [CrossRef]
- Ceccarelli, S.; Grando, S.; Maatougui, M.; Michael, M.; Slash, M.; Haghparast, R.; Rahmanian, M.; Taheri, A.; Al-Yassin, A.; Benbelkacem, A.; et al. Plant breeding and climate changes. J. Agric. Sci. 2010, 148, 627–637. [Google Scholar] [CrossRef]
- Hanson, J.O.; Rhodes, J.R.; Riginos, C.; Fuller, R.A. Environmental and geographic variables are effective surrogates for genetic variation in conservation planning. Proc. Nat. Acad. Sci. USA 2017, 114, 12755–12760. [Google Scholar] [CrossRef] [PubMed]
- Friedman, C.; Borlawsky, T.; Shagina, L.; Xing, H.R.; Lussier, Y.A. Bio Ontology and text: Bridging the modeling gap. Bioinformatics 2006, 22, 2421–2429. [Google Scholar] [CrossRef]
- Sam, L.T.; Mendonça, E.A.; Li, J.; Blake, J.; Friedman, C.; Lussier, Y.A. PhenoGO: An integrated resource for the multiscale mining of clinical and biological data. BMC Bioinform. 2009, 10, S8. [Google Scholar] [CrossRef] [PubMed]
- Doebley, J.; Lukens, L. Transcriptional regulators and the evolution of plant form. Plant Cell 1998, 10, 1075–1082. [Google Scholar] [CrossRef] [PubMed]
- Wray, G.A. The evolutionary significance of cis-regulatory mutations. Nat. Rev. Genet. 2007, 8, 206–216. [Google Scholar] [CrossRef] [PubMed]
- Carroll, S.B. Evo-devo and an expanding evolutionary synthesis: A genetic theory of morphological evolution. Cell 2008, 134, 25–36. [Google Scholar] [CrossRef]
- Cambria, E.; White, B. Jumping NLP curves: A review of natural language processing research. IEEE Comput. Intell. Mag. 2014, 9, 48–57. [Google Scholar] [CrossRef]
- Rodríguez-Leal, D.; Lemmon, Z.H.; Man, J.; Bartlett, M.E.; Lippman, Z.B. Engineering quantitative trait variation for crop improvement by genome editing. Cell 2017, 171, 470–480. [Google Scholar] [CrossRef]
- Lemmon, Z.H.; Reem, N.T.; Dalrymple, J.; Soyk, S.; Swartwood, K.E.; Rodriguez-Leal, D.; Van Eck, J.; Lippman, Z.B. Rapid improvement of domestication traits in an orphan crop by genome editing. Nat. Plants 2018, 4, 766–770. [Google Scholar] [CrossRef] [PubMed]
- Atwell, S.; Huang, Y.S.; Vilhjálmsson, B.J.; Willems, G.; Horton, M.; Li, Y.; Meng, D.; Platt, A.; Tarone, A.M.; Hu, T.T.; et al. Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature 2010, 465, 627–631. [Google Scholar] [CrossRef]
- Golicz, A.A.; Batley, J.; Edwards, D. Towards plant pangenomics. Plant Biotechnol. J. 2016, 14, 1099–1105. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Gonda, I.; Sun, H.; Ma, Q.; Bao, K.; Tieman, D.M.; Burzynski-Chang, E.A.; Fish, T.L.; Stromberg, K.A.; Sacks, G.L.; et al. The tomato pan-genome uncovers new genes and a rare allele regulating fruit flavor. Nat. Genet. 2019, 51, 1044–1051. [Google Scholar] [CrossRef]
- Hübner, S.; Bercovich, N.; Todesco, M.; Mandel, J.R.; Odenheimer, J.; Ziegler, E.; Lee, J.S.; Baute, G.J.; Owens, G.L.; Grassa, C.J.; et al. Sunflower pan-genome analysis shows that hybridization altered gene content and disease resistance. Nat. Plants 2019, 5, 54–62. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.; Zhao, X.; Mace, E.; Henry, R.; Jordan, D. Exploring and exploiting pan-genomics for crop improvement. Mol. Plant 2019, 12, 156–169. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).