Abstract
Seeding rate for maximum grain yield can differ for diverse hard red spring wheat (HRSW) (Triticum aestivum L.) cultivars and is derived from a yield response curve to seeding rates. Six groups of HRSW cultivars with combinations of Rht-B, Rht-D, and Ppd-D genes were planted at five seeding rates in 21 environments during 2013–2015 throughout Minnesota and eastern North Dakota, USA. Seeding rates ranged from 1.59 to 5.55 million seeds ha−1 and planting timings were optimal and delayed dates. An analysis of covariance predictive model with 13 predetermined training environments was built for yield and tillering, and validated with eight predetermined environments. Optimal seeding rates from the yield model were not predictive for yield, with latitude of the environment negatively skewing the predictions from observed values. A second yield model fit to only the six lowest-yielding environments (<4.8 Mg ha−1) was more predictive (R2 = 0.44), and revealed yield response to seeding rate was influenced by cultivar traits for photoperiod response (Ppd-D gene) and plant stature (semi-dwarfing gene Rht-D). The tillering model was also predictive for the validation environments, with a R2 of 0.71. Using regression predictions for yield and tillering from training and validation datasets with HRSW genetic and geographic predictors shows promise to help recommend seeding rates for future environments.
1. Introduction
Plant density in cereals is important as grain yield is impacted by final plant stand [1,2,3,4,5]. A seeding rate by cultivar interaction is often found in cereal crop research [1,4,5,6,7,8]. Faris and DePauw [9] concluded that each new cultivar released from a breeding program should be tested at several seeding rates to fully understand the optimal seeding rate for maximum yield.
Cultivars of hard red spring wheat (HRSW) (Triticum aestivum L.) have genetic differences affecting plant height [10,11,12,13], tillering [6,14], photoperiod sensitivity [13,15], and yield [12], among many other genetically controlled traits. Spikes per plant in cereal grains is a quantitative trait affected by environment conditions, soil fertility, planting date, and other agronomic practices [16]. Li et al. [17] indicated that many quantitative trait loci (QTL) influence tillering. A region on the short arm of chromosome 6A has the greatest impact on stems per plant, with a region on the short arm of chromosome 1D and the region of Ppd-D1 also contributing.
Semi-dwarfing genes have been widely incorporated into HRSW cultivars, though yield improvements still remain dependent on environment [10,11]. Ciha [18] reinforced that the semi-dwarf growth habit has changed agronomic performance characteristics such as lodging and yield in HRSW. As the prevalence of standard height cultivars has declined, research has been conducted to find proper agronomic practices for semi-dwarf HRSW cultivars [1,3,5,6,9,18,19,20]. The genetic determinants of the semi-dwarf growth habit in HRSW are the Rht-8, Rht-B1, and Rht-D1 genes [21]. Rht-B1 and Rht-D1 occur on homeologous chromosomes 4B and 4D, respectively, while Rht-8 occurs on the short arm of chromosome 2D [22,23,24].
Photoperiod sensitivity in HRSW cultivars is imparted by the Ppd-A1, Ppd-B1, and Ppd-D1 genes [15]. Furthermore, these three genes have been ranked for their contribution to photoperiod sensitivity in the order Ppd-D1 > Ppd-B1 > Ppd-A1. These genes are located on the long arm of 2A for Ppd-A1, short arm of 2B for Ppd-B1, and long arm of 2D for Ppd-D1 [25]. For locus Ppd-D1, Ppd-D1a is the wild-type allele, while Ppd-D1b is the sensitive allele. Genes that govern response to changing daylight cycles are valuable tools with which to tailor varieties to environments [26]. This is especially of importance when considering flowering timing relative to environmental factors that greatly impact grain yield [15,27,28,29,30].
In cereal crops, a yield response curve is a useful statistical tool for analyzing the effect of seeding rates for a cultivar [31,32,33,34]. Holliday [32] found that with seeding rates, yield typically fits a quadratic response curve in cereal grains. As seeding rates increase, the yield response curve starts with a sharp incline to the optimal seeding rate, followed by a slow decline on the high side of the optimal seeding rate. Wiley and Heath [34] recognized that understanding the yield response curve to seeding rate was useful for making seeding rate decisions in different cropping systems and production settings.
The first objective of this research was to determine if traits such as plant stature, photoperiod sensitivity, and capacity to tiller, in addition to geographic indicators such as planting date, day length at planting, and latitude, can be used to predict the optimal seeding rate for maximum yield of a newly-released cultivar without the need for conducting classical seeding rate response experiments. This objective was investigated by building an analysis of covariance (ANCOVA) regression model for yield and tillering, using covariates as predictors with a training dataset. The second objective was to validate the regression model for yield and tillering with an out-of-sample validation dataset to gauge the predictive power of the model. The third objective was to use the entire dataset as a database to enter the known characteristics of a cultivar, expected planting date, and latitude of seeding location, to determine the optimal seeding rate for maximum yield of the cultivar.
2. Materials and Methods
2.1. Site Description
Field experiments were conducted from 2013 to 2015 in North Dakota (ND) and Minnesota (MN). In 2013, experiments were established in four locations: Prosper, ND, and Perley, Crookston, and Hallock, MN. Six locations were used in 2014 and 2015 at Prosper, ND, and Lamberton, Kimball, Perley, Crookston, and Hallock, MN. Table 1 lists the soil series, soil taxonomy, and slope at each location and year.

Table 1.
Soil series, taxonomy, and slope at 2013–2015 experiment locations.
2.2. Experimental Approach
The locations were set up in pairs, with Kimball, Perley, and Hallock being validation environments for the model training environments of Lamberton, Prosper, and Crookston, respectively. The experimental design at Lamberton, Prosper, and Crookston was a randomized complete block with a split-split plot arrangement and three replicates. The whole plot was planting date, the sub-plot was cultivar, and the sub-sub plot was seeding rate. In 2013, Prosper had no planting date factor, so the design was a split-plot. The trials at Kimball, Perley, and Hallock were a randomized complete block designwith a split-plot arrangement and three replicates. The whole plot was cultivar and the split-plot was seeding rate.
Cultivars were selected based on genetic characteristics for photoperiod sensitivity and plant stature (Table 2). Genotyping of the cultivars was done by the Wheat Genotyping Center at the USDA-ARS Cereal Crops Research utilizing polymerase chain reaction (PCR) methods [13].

Table 2.
Background genetic details of hard red spring wheat (HRSW) cultivars.
The germination percentage of each seed lot was determined using the ragdoll method [36]. Seed for planting was weighed out for individual experimental units of each cultivar using kernel weight, germination percentage, and plot size. The previous crop at all experiment sites was soybean [Glycine max (L.) Merr.]. Experiments were established and managed according to University of Minnesota and North Dakota State University Extension recommendations with regard to cultivation, fertilization, and pesticide application (Table 3) [37].

Table 3.
Site location and year characteristics for hard red spring wheat (HRSW) trials conducted in Minnesota and North Dakota, 2013–2015.
The plot size for the experimental unit at Prosper, ND was 1.24 m by 3.65 m, and had 7 rows of HRSW with 0.19 m spacing. The plot size at Kimball, Perley, Crookston, and Hallock was 1.52 m by 4.57 m and had 10 rows with 0.15 m row spacing. The plot size at Lamberton was 1.52 m by 2.43 m and had 8 rows with 0.19 m row spacing. Optimal seeding rates were determined using the first derivative of the regression equation for the quadratic yield response curves to seeding rate. For linear response curves, optimal seeding rate was determined by the seeding rate treatment for maximum grain yield.
2.3. Data Collection
Stand counts were recorded at approximately Feekes 1 to determine the established plant density [38]. Stem counts were evaluated at Feekes 11. These two measurements were used to calculate the number of stems per plant. Plant density and stem counts were obtained by counting plants and stems in two rows between two stakes that were placed 0.91 m apart in between two adjacent interior rows. Plot yield was collected from the entire plot with a small plot combine. Grain yield was adjusted to 13.5% moisture. Grain characteristics of moisture, test weight, and protein were collected at harvest or during post-harvest processing and adjusted to 13.5% and 12.5% grain moisture, respectively.
2.4. Statistical Analyses and Modeling
Statistical analyses were performed using SAS 9.4 software (SAS Institute, Cary, NC, USA). Seeding rate and cultivar were fixed effects, while replicate and environment were assigned as random effects in the initial analysis. Interactions of fixed effects were considered fixed, while any interaction with a random term was considered random. Environments were characterized by planting date, day length at planting, and latitude, to promote a robust model.
The models for yield (Mg ha−1) and tillers (stems per plant) as the dependent variables were built using unadjusted data from each plot. An ANCOVA model was built with data from 13 environments assigned as the a priori training dataset. The model was built with data from two random replicates, while the third replicate was used for validation. Models were fit in PROC MIXED, with Ppd-D, Rht-B, Rht-D, stems plant−1, geographic latitude of the field location, day length at planting (DL), and calendar days from January 1 (CD) as covariates. Method = type1 was the estimation method used to build the model, and parameter estimate solutions were found with the solutions htype = 1 statement. A model was built in the order of linear main effects, quadratic main effects, linear interactions, and quadratic interactions of all covariates. Effects that were non-significant at the 95% confidence level were removed from the end of the model to the beginning. Iterations of the model were run as effects were removed to ensure significance remained for effects below an eliminated term that may have partially explained the higher term.
Following the recommendations of Ngo [39], regression diagnostics from SAS output were used to assess model adequacy and assumptions as the training model was nearing the final iteration. The error mean square (EMS) was monitored to ensure it did not increase as the model was altered. Method = type3 was invoked to measure stability of the parameter estimates for the intercept and covariate terms as the last term in a model was added or removed. Residual plots were assessed to ensure stability in the model, and to identify any unusual data points that could potentially skew results. Residuals were monitored around absolute zero to screen for normality, skewness, and kurtosis. Tests were done for multicollinearity of the factors in the model using Pearson correlation coefficients from SAS (PROC CORR) as described by Ngo [39]. An R2 estimate was calculated using SAS (PROC MIXED) for the training data. Near completion, the model was further assessed for assumptions by running the validation dataset through the model and assessing the same regression diagnostics and assumptions as for the training model.
Model validation was performed for the training data ANCOVA model with an outpred = statement in PROC MIXED by assigning a missing data symbol ‘.’ as the dependent variable of the validation dataset. Predictions and residuals were calculated for each plot in the validation dataset to measure how predictive the model was of an out-of-sample dataset which was not used to build the model. The predictive nature of the model was assessed primarily using R2 of the observed versus predicted plot values. The residuals found by subtracting the observed minus predicted values were further assessed by running the validation data through the parameter estimates, as given by the solutions statement in SAS and plotting using PROC SGPLOT.
PROC REG in SAS was used for linear and quadratic regression analysis on the LSMEANS of the predicted outcomes of the yield response curve to seeding rate. The entire dataset (training and validation) was used for the analysis of the original model. The second model, built using only the six lowest-yielding environments (<4.8 Mg ha−1), was built and validated without a priori designation of training and validation environments. An alpha level of 0.05 was used for all hypothesis tests. Single degree of freedom linear and orthogonal contrasts were written to make comparisons for main effects and interaction terms for categorical covariates. Significance indicators from the contrast statement were used to declare differences between yield response curves for certain covariates.
3. Results
3.1. Original Yield Model
An ANCOVA regression model was built using the entire training dataset of 13 environments, and validated for the entire validation dataset of eight environments. The most stable model had 11 variables, including Ppd-D, Rht-B, Rht-D, seeding rate, CD, latitude of the environment, rate*rate, CD*CD, latitude*latitude, Ppd-D*Rht-B, and Ppd-D*Rht-D (Figure 1).

Figure 1.
Validation dataset yield predictions plotted against residual yields (observed minus predicted yield) for 2013–2015 locations near Kimball, Perley, and Hallock, Minnesota.
This model was not predictive of grain yield. The original regression model for the training environments had an R2 of 0.46 for observed versus predicted values. However, when this model was used to predict the yield of every plot from the eight validation environments, the R2 between observed and predicted values was only 0.01, and the residual values (difference between observed plot yield and predicted yield from the model) differed substantially from zero. The residuals were not centered on zero, and were upwards of 4 Mg ha−1 away from the observed plot yield, indicating an environmental effect that was reducing model stability (Figure 1).
Model Predictions
The yield model built from 13 environments and validated by eight environments was not predictive for out-of-sample data. However, some trends for the model covariates were found when all 21 environments were plotted. The four cultivars with Rht-Bb allele for semi-dwarf stature were higher-yielding at all seeding rates in comparison to the four semi-dwarf cultivars with Rht-Db allele or the four wild-type stature cultivars (Figure 2). The cultivars with the Rht-Db allele had a crossover response interaction with wild-type stature cultivars. These cultivar groups also differed in grain yield as Rht-Db cultivars peaked at 4.40 Mg ha−1, while the wild-type stature cultivars peaked at 3.41 Mg ha−1.
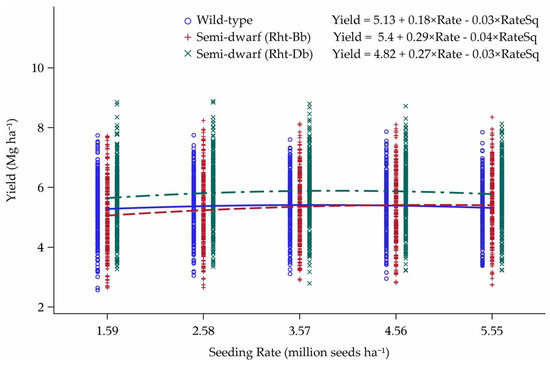
Figure 2.
Influence of Rht-Bb, Rht-Db, and Rht-a (wild-type stature) alleles on yield response to seeding rate over 21 environments in Minnesota and North Dakota, 2013–2015. Seeding rate for the peak of the curve: Wild-type = 3.41, Rht-Bb = 3.21, and Rht-Db = 4.40 million seeds ha−1.
The Ppd-Da allele for photoperiod insensitivity (PI) and Ppd-Db allele for photoperiod sensitivity (PS) imparted no yield advantage at any seeding rate when averaged over 21 environments for the six cultivars comprising each allele group (Table 4).

Table 4.
Regression summary of Ppd-D allele influence on yield response to seeding rate over 21 environments in Minnesota and North Dakota, 2013–2015.
There was an interaction of Ppd-D and Rht-B when combined over all 21 environments (Figure 3). Of the combinations of cultivars with different alleles for the two genes, the main effect of Rht-B is evident (Figure 2). The response curves were a converging interaction within the two subgroups of Rht-Ba and Rht-Bb. The wild-type stature cultivars yielded more at the lowest two seeding rates when the cultivar also had the PI allele Ppd-Da. The cultivars with semi-dwarfing gene Rht-Bb yielded more at the lowest seeding rates when the cultivar had the PS allele Ppd-Db. Within this group, the slope of the yield response curve was almost two times greater with PI, than with PS (Figure 3).
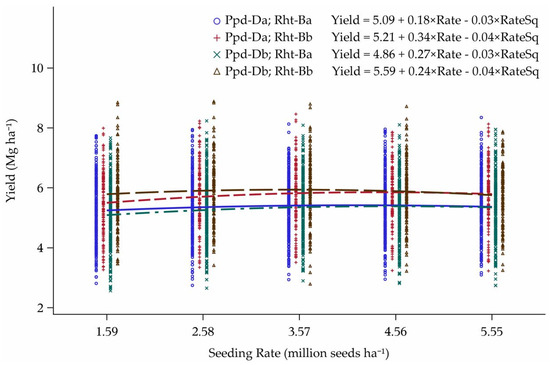
Figure 3.
Yield response to seeding rate for the interaction between Rht-B and Ppd-D alleles over 21 environments in Minnesota and North Dakota, 2013–2015. Seeding rate at peak of curve: Ppd-Da; Rht-Ba = 3.60, Ppd-Da; Rht-Bb = 3.88, Ppd-Db; Rht-Ba = 3.79, and Ppd-Db; Rht-Bb = 2.94 million seeds ha−1.
The PS cultivars (Ppd-Db) with semi-dwarfing allele Rht-Db did not have the same yield response to seeding rate as PS cultivars with semi-dwarfing allele Rht-Bb. These differences in response could be a consequence of a main effect from the Rht-Db allele, as cultivars with Rht-Db yielded lower than wild-type stature and Rht-Bb cultivars (data not shown). However, this is unlikely, as nearly identical yield response curves (similar slopes and peaks) were observed for cultivars with the Rht-Db allele in combination with the PS allele or PI allele.
Latitude had an impact on yield response to seeding rate when fit over all 21 environments (Figure 4). Although experiment sites differed geospatially from year to year, the difference in distance between environments at the same location was never more than about 8 km north to south (Hallock, Perley, and Kimball), and was typically only about 1 km difference (Prosper, Lamberton, and Crookston). The relative yield potential of the environments breaks out cleanly by location, in order from lowest to highest as: Prosper < Lamberton < Crookston < Kimball < Hallock < Perley. Environments with higher yield potential tended to require a lower seeding rate to reach maximum yield across the 12 diverse cultivars.
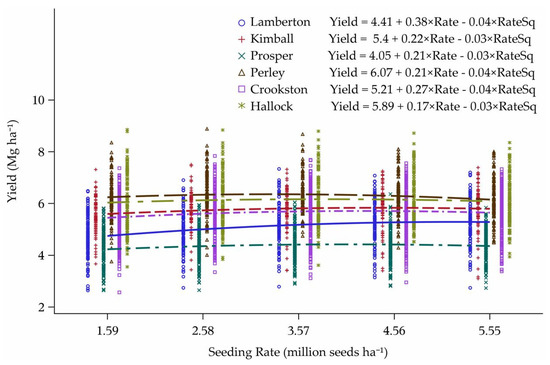
Figure 4.
Yield response to seeding rate for each location in Minnesota and North Dakota, 2013–2015. Seeding rate for the peak of the curve: Lamberton = 4.62, Kimball = 3.87, Prosper = 3.51, Perley = 2.68, Crookston = 3.73, and Hallock = 3.26 million seeds ha−1.
3.2. Tillering Model
With no model proving predictive for yield in the combined dataset, the predictive power of an ANCOVA model for stems per plant was built and assessed. The entire training dataset of 13 environments was used to build an ANCOVA regression model for tillering. This model was also validated using the entire dataset of eight validation environments. The most stable model (R2 = 0.63) for tillering (based on stems per plant) had 9 variables, including Ppd-D, Rht-B, seeding rate, CD, DL, latitude of environment, rate*rate, latitude*latitude, and rate*latitude (Figure 5).
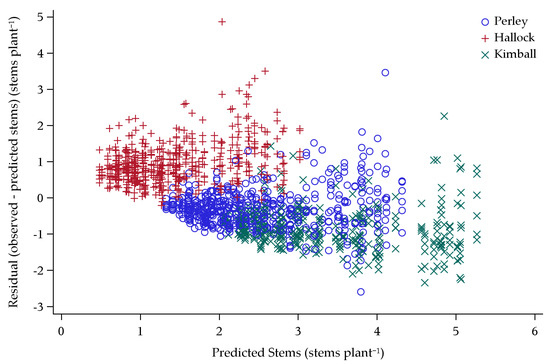
Figure 5.
Validation dataset stems per plant predictions plotted against residual stems per plant (observed minus predicted stems per plant) for 2013–2015 locations near Kimball, Perley, and Hallock, Minnesota.
When the validation dataset was fit by the tillering model, the resulting R2 was 0.71. The predicted and residual values were representative of expected output for a stable predictive model. However, the model was slightly biased due to autocorrelation and multicollinearity effects from latitude (Figure 5). Overall, the trends were indicative of a predictive model.
Model Predictions
The genes Rht-B, Rht-D, and Ppd-D had an influence on stems per plant across all 21 environments (Figure 6 and Figure 7). Rht-Bb cultivars had more stems per plant at the two lowest seeding rates compared to Rht-Db or wild-type stature cultivars, although all three categories plateaued towards the three highest seeding rates (Figure 6).
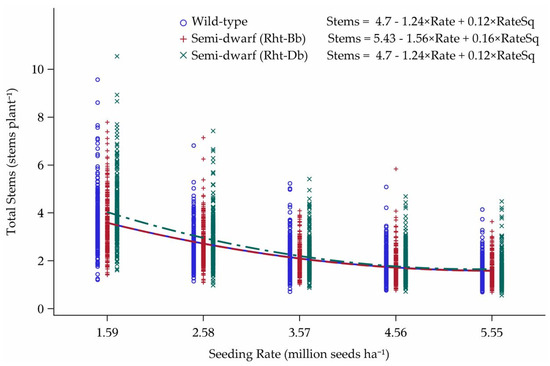
Figure 6.
Influence of Rht-Bb, Rht-Db, and Rht-a (wild-type stature) alleles on tillering response to seeding rate over 21 environments in Minnesota and North Dakota, 2013–2015.
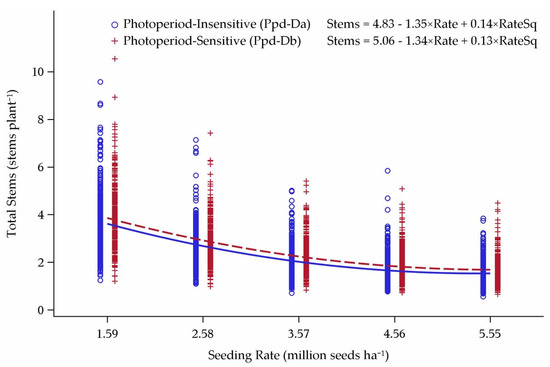
Figure 7.
Influence of Ppd-D alleles on tillering response to seeding rate over 21 environments in Minnesota and North Dakota, 2013–2015.
The PI cultivars consistently had fewer tillers than PS cultivars (Figure 7). The tillering response curves were mirrored between the cultivar groups, as slopes were very similar and differed only slightly in intercept. Interactions between Ppd-D and either semi-dwarfing gene provided only slight differences in stems per plant (data not shown). The result of PI cultivars having fewer stems per plant compared to PS cultivars was the main trend that came out of the interactions between photoperiod response and semi-dwarfing genes.
Latitude also had an impact on tillering response curves of all cultivars (Figure 8). In all years, Lamberton had the highest number of stems per plant for all cultivars, while Crookston had the lowest number of stems per plant. There was no noticeable interaction between the stems per plant response curves and latitude, revealing that cultivar tillering habit was consistent across locations. The intercepts and the slopes of each latitude gives an indication of how the locations separated out for tillering capacity.
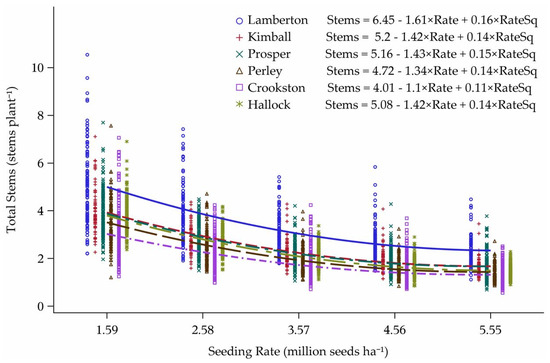
Figure 8.
Tillering response curves to seeding rate for each location in Minnesota and North Dakota, 2013–2015.
3.3. Reworked Yield Model
The original yield model was not predictive, and the tillering model was only moderately predictive. One problem that plagued the three years of seeding rate research was the lack of a yield response to seeding rate at many environments due to very productive HRSW growing seasons. The yield responses were minimal across the five seeding rates compared to what has previously been reported for this region [5]. The large difference in yield potential between the 21 environments is represented by mean grain yields in Table 3. The range in average yield of the 21 environments across all seeding rates and cultivars was from 3.62 Mg ha−1 to 7.27 Mg ha−1. When split into thirds based on yield, six environments comprised the bottom third (range: 3.62 to 4.83 Mg ha−1), ten environments comprised the middle third (range: 4.84 to 6.05 Mg ha−1), and five locations were in the top third (range: 6.06 to 7.27 Mg ha−1).
To account for this variability among environments, an ANCOVA model was built to test the predictive nature of the covariates for yield using only the six lowest-yielding environments where yield response curves to seeding rate were more pronounced. Thirteen covariates remained in the final model for the six lowest-yielding environments, including Ppd-D, Rht-B, Rht-D, seeding rate, DL, CD, latitude, rate*rate, CD*CD, latitude*latitude, Ppd-D*Rht-B, Rht-B*DL, and Rht-D*DL. The model for yield of the six lowest-yielding environments had a training R2 of 0.42. The model was validated three times, with one of the three replicates serving as the validation dataset, while two of the replicates were the training dataset. With replicate one, two, and three as the validation dataset the respective R2 values were very consistent at 0.43, 0.44, and 0.44 (Figure 9 and Figure 10). When the 16 lowest-yielding environments were run through the same scenario with a new model, the resulting R2 averaged 0.20. Therefore, the six lowest-yielding environments were the focus of this final model.
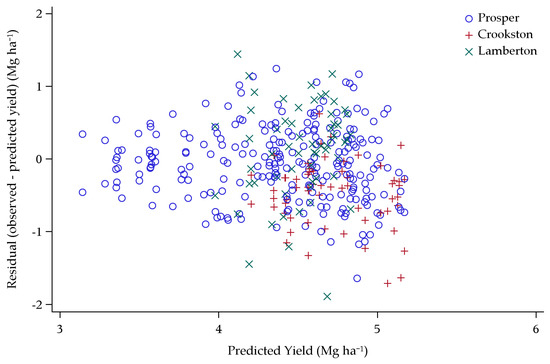
Figure 9.
Validation dataset yield predictions plotted against residual yields (observed minus predicted yield) over the six lowest-yielding environments in Minnesota and North Dakota, 2013–2015.
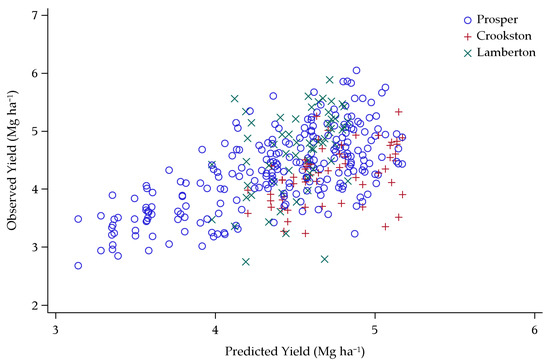
Figure 10.
Validation dataset yield predictions plotted against observed yields over the six lowest-yielding environments in Minnesota and North Dakota, 2013–2015.
Model Predictions
Semi-dwarf plant stature from the Rht-Bb allele was beneficial for yield over the 12 cultivars (Figure 11). The semi-dwarfing allele of Rht-B was represented by four cultivars in this study that yielded higher than the semi-dwarfing allele for Rht-D or wild-type stature cultivars. The slopes between the three classes of cultivars (based on genetic characteristics for semi-dwarfing genes) differed slightly. Seeding rate for maximum yield of Rht-Db cultivars required almost 1 million seeds ha−1 more than either Rht-Bb or wild-type stature cultivars.

Figure 11.
Influence of Rht-Bb, Rht-Db, and wild-type stature alleles on yield response to seeding rate over the six lowest-yielding environments in Minnesota and North Dakota, 2013–2015. Seeding rate for the peak of the curve: wild-type = 3.68, Rht-Bb = 3.75, and Rht-Db = 4.65 million seeds ha−1.
Cultivar response to photoperiod had an impact on the relative yield and yield response curve across the six lowest-yielding environments in this study (Figure 12). Photoperiod insensitive cultivars yielded consistently higher than PS cultivars across all five seeding rates. The seeding rate for maximum yield was higher for PI cultivars at 4.21 million seeds ha−1, compared to 3.71 million seeds ha−1 for PS cultivars. These results are a contrast from the original yield model, where PS and PI cultivars had nearly identical seeding rate response curve fits for yield (Table 4). The middle-third and top-third yielding environments had no apparent effect from the Ppd-D1 gene on grain yield (data not shown).
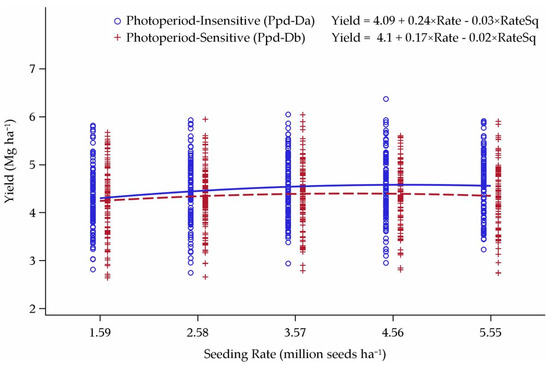
Figure 12.
Influence of Ppd-D alleles on yield response to seeding rate over the six lowest-yielding environments in Minnesota and North Dakota, 2013–2015. Seeding rate for the peak of the curve: Ppd-Da = 4.21 and Ppd-Db = 3.71 million seeds ha−1.
The yield advantage of Rht-Bb cultivars over Rht-Ba cultivars is evident in the interaction figure between Rht-B and Ppd-D (Figure 13). For wild-type stature cultivars, a yield advantage was found with PS cultivars compared to PI and there was no interaction between the two groups. However, when Rht-Bb cultivars were coupled with PS, the seeding rate for maximum yield was 1.59 million seeds ha−1 (lowest seeding rate treatment), compared to 4.26 million seeds ha−1 for Rht-Bb and PI combination cultivars. The crossover interaction occurred just above the second lowest seeding rate. The cultivars that possess both Rht-Bb and Ppd-Db (Albany and Faller) can both be planted at much lower seeding rates in the relatively low-yielding environments (<4.8 Mg ha−1) encountered in this study, though the exact reasons were not readily apparent.
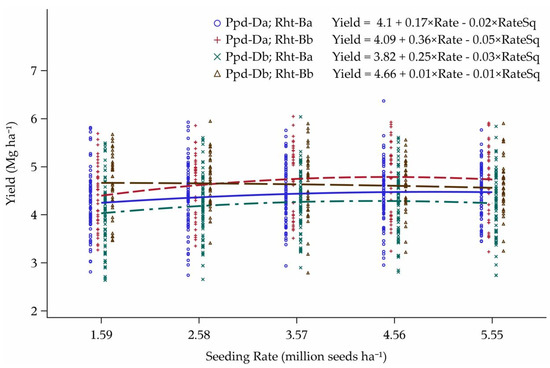
Figure 13.
Yield response curves to seeding rate for the interaction between Rht-B and Ppd-D alleles over the six lowest-yielding environments in Minnesota and North Dakota, 2013–2015. Seeding rate for the peak of the curve: Ppd-Da; Rht-Ba = 4.16, Ppd-Da; Rht-Bb = 4.26, Ppd-Db; Rht-Ba = 3.98, and Ppd-Db; Rht-Bb = 0.62 million seeds ha−1.
The interaction between Rht-D and Ppd-D breaks out first into main effects, with Rht-Db cultivars needing much higher seeding rates for maximum yield. Rht-Db cultivars also had lower relative yield across the six lowest-yielding environments in comparison to cultivars with wild-type stature (Figure 14). Photoperiod insensitive cultivars also had higher relative yield than PS cultivars, as was explained previously. Both interactions between the subgroups were diverging as the seeding rates increased.
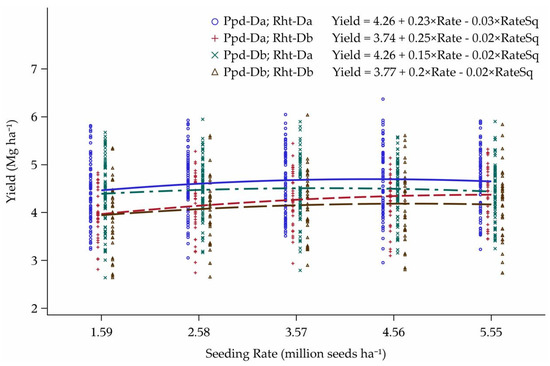
Figure 14.
Yield response curves to seeding rate for the interaction between Rht-D and Ppd-D alleles over the six lowest-yielding environments in Minnesota and North Dakota, 2013–2015. Seeding rate for the peak of the curve: Ppd-Da; Rht-Da = 3.83, Ppd-Da; Rht-Db = 5.16, Ppd-Db; Rht-Da = 3.36, and Ppd-Db; Rht-Db = 4.15 million seeds ha−1.
The latitude response for the six lowest-yielding environments was variable, as four environments comprised the Prosper yield response curve, but only one environment comprised both Lamberton and Crookston yield response curves (Table 5). Seeding rate for the peak of the curve at Lamberton, Prosper, and Crookston, was 4.76, 3.53, and 5.91 million seeds ha−1, respectively.

Table 5.
Regression summary of yield response to seeding rate over the six lowest-yielding environments in Minnesota and North Dakota, 2013–2015.
4. Discussion
4.1. Original Yield Model
Latitude greatly skewed predicted values in the original yield model that included all 21 environments. The validation environments at Kimball, MN are approximately 275 km away (1.77 degrees S) from the environments at Perley, MN, and Perley is approximately 179 km away (1.65 degrees S) from Hallock, MN. The yield trends from a limited dataset of 13 training environments did not accurately predict yields at eight out-of-sample validation environments that were relatively close in proximity to the training environments. One likely reason for the inaccurate predictions was that there were not enough environments to accurately represent differences in yield potential across the large geographic reference area in this study. Additionally, there were other potential in-season crop stressors (e.g., lodging, disease, water availability) that likely had an effect on yield, but were not included as covariates in the model. Regardless of the problems that latitude presented, the predictive power and R2 of the training model decreased greatly when latitude was not included as a covariate in the model and residuals were even more randomly distributed.
Twenty-one environments across three years provided a diversity of environments, where six early-maturing PI cultivars were balanced by six late-maturing PS cultivars (Table 2). This research found no yield advantage for either PI or PS cultivars. This is in contrast to Busch et al. [27], who found a 9% yield advantage from PI near isogenic lines (NILs) compared to PS NILs, and Marshall et al. [29] who found that yield for PI NILs was always equal to or greater than PS lines in similar geographies to this study. It is important to note that this contrasting result could be due to the limited sample size in this study.
Future research using this approach over a wide geographic reference area would likely benefit from including more environments spaced evenly and/or more closely together, or shrinking the overall size of the reference area. The conclusion for this first model for yield was that the training environments were not predictive of the validation environments.
4.2. Tillering Model
Predicting cultivar tillering can be useful as tillering gives HRSW adaptability to diverse geographies. Rht-Bb cultivars had more tillers than either Rht-Db or wild-type stature cultivars at the lowest two seeding rates in this study (Figure 6). This result is in-line with Sial et al. [40] who found the two cultivars with Rht-Bb to have significantly more stems per plant than Rht-Db cultivars. Contrary to both these results, Pinthus and Levy [41] found no difference in stems per plant between wild-type stature and semi-dwarf HRSW cultivars.
Latitude was a major factor in tillering when combined over all cultivars, and future analysis on the latitude by cultivar interaction for stems per plant would be important to explore with this data. There were no weather covariates taken for the environments, so the overall tillering capacity for each latitude cannot be fully explained. The trend of less tillers with increasing latitude in the geographic area could be useful to producers trying to predict the tillers they will have in their production region.
4.3. Yield Model Reworked
Although PS did not affect HRSW yield over 21 diverse geographies in eastern ND and MN, PI cultivars in environments with below average yields (<5.49 Mg ha−1) had increased yields compared to PS cultivars, likely a result of earlier heading and maturity. These results are similar to findings reported by Marshall et al. [29]. The increase in yield with PI cultivars was 0.05 Mg ha−1 at the lowest seeding rate, but was between 0.15 and 0.25 Mg ha−1 higher at the highest three seeding rate treatments. Photoperiod insensitive cultivars yielded 3.2% higher than PS cultivars when averaged over all five seeding rates. The sample of cultivars used in this research provided a lower increase for PI over PS cultivars, compared to the 9% increase from PI lines found by Busch et al. [27] in MN. Prior research on photoperiod response in HRSW in the Northern Plains region had used 10 [27] and 11 [29] pairs of NILs for Ppd-D1. This current research used six cultivars with each allele for Ppd-D1, instead of NILs, which is a slightly lower sample size and different methodology. However, the cultivars in this research were a random sampling of cultivars available to growers. Lower-yielding environments (<4.8 Mg ha−1) are subject to greater stresses, especially increased temperature that can be detrimental to a cool-season cereal crop. This current research found that the earlier maturing PI cultivars were advantageous in stressed environments and should be seeded at a rate that is 0.5 million seeds ha−1 higher than PS cultivars, to maximize yield in these environments.
With regards to the semi-dwarfing genes, it was clear that Rht-Db semi-dwarfs needed higher seeding rates to reach maximum yield, compared to wild-type stature or Rht-Bb semi-dwarfs (Figure 12, Figure 13 and Figure 14). Additionally, the relative yield of Rht-Bb cultivars in this study was higher than either Rht-Db or wild-type stature cultivars. However, this could have been a function of a small sample size and particularly high-yielding cultivars, Faller and Albany, making up two of the four Rht-Bb cultivars.
Predictive models have been attempted with varying success in a wide array of agricultural research, from QTL impacts, to water-use yield response curves and more. As far as we know, this is the first undertaking to try and use a rigorous experimental design and statistical approach to predict optimal seeding rates for maximum yield in HRSW using genetic traits. Although the predictive model was complicated by latitude and relative yield differences between environments, this was somewhat resolved by looking at the lowest-yielding environments.
5. Conclusions
Predicting the yield of plots in eight validation environments with a model built from 13 training environments proved difficult. Although the model was moderately predictive of yield for the training dataset (R2 = 0.46), the validation dataset was poorly fit, with a R2 of only 0.01. Relative yield levels and yield response curves were difficult to fit validation data due to geographic separation, not enough environments within a very large geographic reference area, stressors not included as covariates, and favorable growing conditions occurring in many environments.
A tillering model was developed and found to be predictive. Tillering is an important trait contributing to yield in wheat and it may be useful for a grower to know about this characteristic when planting a specific HRSW cultivar. The tillering model had an R2 of 0.63 for the training dataset, and 0.71 for the validation dataset. Photoperiod sensitive cultivars had more stems per plant than PI cultivars across all seeding rates. Latitude was predictive of stems per plant, with more southern environments having more stems per plant. Additionally, it was found that higher-yielding environments (>6.1 Mg ha−1) had less stems per plant.
The predictive power of a yield model based on the six lowest-yielding environments was tested and substantiated. An R2 of 0.42 for the training dataset and 0.44 for the validation dataset indicated the reworked model was more predictive than the original yield model. Another important finding from the reworked yield model was the influence that photoperiodism gene Ppd-D and semi-dwarfing gene Rht-D had on cultivar yield response to seeding rates. It was revealed that PI cultivars yielded higher than PS cultivars, and required a seeding rate of an additional 0.5 million seeds ha−1 to maximize yield. Additionally, cultivars with Rht-Db semi-dwarf stature required about 1.0 million more seeds ha−1 for maximum yield, compared to either wild-type stature or Rht-Bb semi-dwarfs.
The objective of this research to mesh genetic knowledge of a HRSW cultivar with agronomic practices in order to predict an optimal seeding rate for maximum yield. There proved to be predictive power by having knowledge of the genetic makeup of a cultivar coupled with known geographic and planting date information; this was more so for tillering than yield, but is beneficial information either way. The results are an important first step in developing predictive models for seeding rate decisions in HRSW with regards to genetic and location characteristics as covariates. For future research, more environments should be evaluated, and selected for geographic proximity. However, the unpredictable nature of weather and growing seasons can be expected to continue to provide a challenge.
Author Contributions
Conceptualization, G.H.M., J.J.W. and J.K.R.; methodology, G.H.M., J.J.W. and J.K.R.; validation, G.H.M.; formal analysis, G.H.M.; investigation, G.H.M.; resources, J.J.W. and J.K.R.; data curation, G.H.M.; writing—original draft preparation, G.H.M., J.J.W., J.D.S. and J.K.R.; writing—review and editing, G.H.M., J.D.S., J.J.W. and J.K.R.; visualization, G.H.M. and J.D.S.; supervision, J.J.W. and J.K.R.; project administration, G.H.M.; funding acquisition, G.H.M., J.J.W, J.D.S. and J.K.R. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Minnesota Wheat Research & Promotion Council.
Conflicts of Interest
The authors declare no conflict of interest. Author G.H.M. was employed by Bayer Crop Science after completion of the experiments. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Baker, R.J. Effect of seeding rate on grain yield, straw yield, and harvest index of eight spring wheat cultivars. Can. J. Plant Sci. 1982, 62, 285–291. [Google Scholar] [CrossRef]
- Guitard, A.A.; Newman, J.A.; Hoyt, P.B. The influence of seeding rate on the yield and the yield components of wheat, oats, and barley. Can. J. Plant Sci. 1961, 41, 750–758. [Google Scholar] [CrossRef]
- Hanson, B.K.; Lukach, J.R. Barley response to planting rate in northeastern North Dakota. North Dakota Farm Res. North Dakota Agri. Exper. Station 1992, 14–19. [Google Scholar]
- Pendleton, J.W.; Dungan, G.H. The effect of seeding rate of nitrogen application on winter wheat cultivars with different characteristics. Agron. J. 1960, 52, 310–312. [Google Scholar] [CrossRef]
- Wiersma, J.J. Determining an optimum seeding rate for spring wheat in Northwest Minnesota. Crop. Manage. 2002, 18, 1–7. [Google Scholar] [CrossRef]
- Anderson, W.K.; Barclay, J. Evidence for differences between three wheat cultivars in yield response to plant population. Aust. J. Agric. 1991, 42, 701–713. [Google Scholar] [CrossRef]
- Briggs, K.G.; Aytenfisu, A. The effects of seeding rate, seeding date and location on grain yield, maturity, protein percentage and protein yield of some spring wheats in central Alberta. Can. J. Plant Sci. 1979, 59, 1139–1145. [Google Scholar] [CrossRef]
- Faris, D.G.; De Pauw, R.M. Effect of seeding rate on growth and yield of three spring wheat cultivars. Field Crops Res. 1981, 3, 289–301. [Google Scholar] [CrossRef]
- Kirby, E.J.M. The effect of plant density upon growth and yield of barley. Cambridge J. of Agr. Sci. 1967, 68, 317–324. [Google Scholar] [CrossRef]
- Butler, J.D.; Byrne, P.F.; Mohammadi, V.; Chapman, P.L.; Haley, S.D. Agronomic performance of Rht alleles in a spring wheat population across a range of moisture levels. Crop. Sci. 2005, 45, 939–947. [Google Scholar] [CrossRef]
- Lanning, S.P.; Martin, J.M.; Stougaard, R.N.; Guillen-Portal, F.R.; Blake, N.K.; Sherman, J.D. Evaluation of near-isogenic lines for three height-reducing genes in hard red spring wheat. Crop. Sci. 2012, 52, 1145–1152. [Google Scholar] [CrossRef]
- North Dakota State University Extension. North Dakota Hard Red Spring Wheat Variety Trial Results and Selection Guide; North Dakota State University Extension: Fargo, ND, USA, 2015; A574-15. [Google Scholar]
- Wiersma, J.J. A pilot project for determining the optimum seeding rates for individual HRSW cultivars. Minn. Wheat Res. Promot. Council Res. 2012.
- Hucl, P.; Baker, R.J. An evaluation of common spring wheat germplasm for tillering. Can. J. Plant Sci. 1988, 68, 1119–1123. [Google Scholar] [CrossRef]
- Worland, A.J. The influence of flowering time genes on environmental adaptability in European wheats. Euphytica 1996, 89, 49–57. [Google Scholar] [CrossRef]
- Friend, D.J.C. Tillering and leaf production in wheat as affected by temperature and light intensity. Can. J. Bot. 1965, 43, 1063–1076. [Google Scholar] [CrossRef]
- Li, W.L.; Nelson, J.C.; Chu, C.Y.; Shi, L.H.; Huang, S.H.; Liu, D.J. Chromosomal locations and genetic relationships of tiller and spike characters in wheat. Euphytica 2002, 125, 357–366. [Google Scholar] [CrossRef]
- Ciha, A.J. Seeding rate and seeding date effects on spring seeded small grain cultivars. Agron. J. 1983, 75, 795–799. [Google Scholar] [CrossRef]
- Baker, R.J. Agronomic performance of semi-dwarf and normal height spring wheats seeded at different dates. Can. J. Plant Sci. 1990, 70, 295–298. [Google Scholar] [CrossRef]
- Miralles, D.J.; Slafer, G.A. Yield, biomass and yield components in dwarf, semi-dwarf, and tall isogenic lines of spring wheat under recommended and late sowing dates. Plant Breed. 1995, 114, 392–396. [Google Scholar] [CrossRef]
- Borojevic, K. The transfer and history of ‘reduced height genes’ (Rht) in wheat from Japan to Europe. J. Heredity. 2005, 96, 455–459. [Google Scholar] [CrossRef]
- Gale, M.D.; Law, C.N.; Worland, A.J. Chromosomal location of a major dwarfing gene from Norin 10 in new British semi-dwarf wheats. Heredity 1975, 35, 417–421. [Google Scholar] [CrossRef]
- Gale, M.D.; Marshall, G.A. Chromosomal location of Gai-1 and Rht-1, genes for gibberellin insensitivity and semi-dwarfism, in a derivative of Norin-10 wheat. Heredity 1976, 37, 283–289. [Google Scholar] [CrossRef]
- Gale, M.D.; Law, C.N.; Marshall, G.A.; Snape, J.W.; Worland, A.J. Analysis and evaluation of semi-dwarfing genes in wheat including a major height reducing gene in variety “Sava”. IAEA-Tecdoc: Semi-dwarf Cereal Mutants Use Cross Breed. 1982, 268, 7–23. [Google Scholar]
- Scarth, R.; Law, C.N. The control of the day-length response in wheat by the group 2 chromosomes. Z Pflanzenzuchtg 1984, 92, 140–150. [Google Scholar]
- Davidson, J.L.; Christian, K.R. Flowering in wheat. In Control of Crop Productivity; Pearson, C.J., Ed.; Academic Press: Sydney, Australia, 1984; pp. 112–126. [Google Scholar]
- Busch, R.H.; Elsayed, F.A.; Heiner, R.E. Effect of daylength insensitivity on agronomic traits and grain protein in hard red spring wheat. Crop. Sci. 1984, 24, 1106–1109. [Google Scholar] [CrossRef]
- Dyck, J.A.; Matus-Cádiz, M.A.; Hucl, P.; Talbert, L.; Hunt, T.; Dubec, J.P. Agronomic performance of hard red spring wheat isolines sensitive and insensitive to photoperiod. Crop. Sci. 2004, 44, 1979–1981. [Google Scholar] [CrossRef]
- Marshall, L.; Nusch, R.; Cholick, F.; Edwards, I.; Frohberg, R. Agronomic performance of spring wheat isolines differing for daylength response. Crop. Sci. 1989, 29, 752–757. [Google Scholar] [CrossRef]
- Worland, A.J.; Appendino, M.L.; Sayers, E.J. The distribution in European winter wheats, of genes that influence ecoclimatic adaptability whilst determining photoperiodic insensitivity and plant height. Euphytica 1994, 80, 219–228. [Google Scholar] [CrossRef]
- Donald, C.M. Competition among crop and pasture plants. Adv. Agron. 1963, 15, 1–118. [Google Scholar]
- Holliday, R. Plant population and crop yield: Part I. Field Crop. Abstr. 1960, 13, 159–167. [Google Scholar]
- Hudson, H.G. Population studies with wheat: III. Seed rates in nursery and field plots. J. Agric. Sci. 1941, 31, 138–144. [Google Scholar] [CrossRef]
- Willey, R.W.; Heath, S.B. The quantitative relationships between plant population and crop yield. Adv. Agron. 1969, 21, 281–321. [Google Scholar]
- United States Department of Agriculture, Natural Resources Conservation Service. Web Soil Survey. Available online: http://websoilsurvey.nrcs.usda.gov/app/WebSoilSurvey.aspx (accessed on 13 January 2020).
- Fabrizius, E. Home Germination Testing of Wheat Seed; Kansas State Univ.: Manhattan, KS, USA; Kansas State Univ. Ext.: Manhattan, KS, USA, 2007; e-Update 98. [Google Scholar]
- Wiersma, J.J.; Ransom, J.K. The Small Grains Field Guide; North Dakota State Univ.: Fargo, ND, USA; North Dakota State Univ. Ext.: Manhattan, KS, USA, 2012; p. A-290. [Google Scholar]
- Large, E.C. Growth Stages in Cereals, Illustration of the Feekes Scale. Plant Pathol. 1954, 3, 128–129. [Google Scholar] [CrossRef]
- Ngo, T.H.D. The steps to follow in a multiple regression analysis. In Proceedings of the SAS Global Forum 2012 Conference, Orlando, FL, USA, 22–25 April 2012; SAS Institute Inc.: Cary, NC, USA, 2012. Paper 333. [Google Scholar]
- Sial, M.A.; Arain, M.A.; Javed, M.A.; Jamali, K.D. Genetic impact of dwarfing genes (Rht1 and Rht2) for improving grain yield in wheat. Asian J. Plant Sci. 2002, 1, 254–256. [Google Scholar]
- Pinthus, M.J.; Levy, A.A. The relationship between the Rht1 and Rht2 dwarfing genes and grain weight in Triticum aestivum L. spring wheat. Theor. Appl. Genet. 1983, 66, 153–157. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).