Abstract
Ginseng (Panax ginseng C.A. Meyer), commonly known as Korean or Asian ginseng, is a perennial herb native to Korea and China. There has been limited research effort to analyze the genetic diversity and population structure of ginseng germplasm because of its growth habits. In the present study, genetic diversity and population structure of ginseng germplasm conserved in the National Agrobiodiversity Center (NAC) of South Korea were analyzed to provide basic data for future preservation and breeding of ginseng genetic resources. Seventeen simple sequence repeat (SSR) markers were used to assess the genetic diversity and population structure of 1109 ginseng accessions. Among 1109 ginseng accessions, 1042 (94.0%) accessions were landraces and 66 (6.0%) accessions were breeding lines (61 accessions, 5.5%) or cultivars (5 accessions, 0.5%). SSR markers revealed 56 different alleles with an average of 3.29 alleles per locus. The average gene diversity was 0.49. Analysis of molecular variance showed that 91% of allelic diversity was attributed to individual accessions within clusters while only 9% was distributed among clusters. Using discriminant analysis of principal components, 12 clusters were detected in 1109 ginseng accessions. The results of this study provide molecular evidence for the narrow genetic base of ginseng germplasm in NAC. For the broad understanding and efficient use of ginseng germplasm, it is necessary to analyze functional factors and to evaluate morphological traits.
1. Introduction
Ginseng (Panax ginseng Meyer, 2n = 4x = 48), a medicinal herb, has long been used in the Far East, particularly in Korea and China as a respected herbal medicine for maintaining physical vitality [1]. The main physiologically active components of ginseng are saponins, phenols, polyacetylenes, alkaloids, and polysaccharides [2]. These major components are known to have beneficial properties for the central nervous system, cardiovascular system, endocrine system, and immune systems [3,4].
Ginseng has been one of the most important trade goods for health care and treatment of diseases in Asia including Korea and China and is currently distributed to 35 countries around the world [5]. Among ginseng production countries, China has the most production, 44,749 tons in 2008, followed by South Korea with 27,480 tons [5]. As of 2017, the annual production of ginseng is 23,310 tons and domestic distribution size is approximately $1584 million in terms of the amount based on ginseng root and processed products in South Korea [6]. With regard to ginseng production, cultivation is focused on harvesting four to six-year-old roots in South Korea and China [7].
The hybridization breeding of ginseng is difficult because of slow growth (~four years/generation) and few seeds (~40 seeds/plant) [8]. Ginseng is originally a self-pollination plant and starts to bloom at its third-year growth stage. In general, its seeds are obtained from a fourth-year growth plant, and flower buds are removed for seeds and root growth. Seeds are harvested from the red fruit having 2 pale yellow seeds in it. At harvesting time the seeds are in immature condition, which does not form a clear embryo. Ginseng seeds need a dormant period for maturity that enables germination [9]. In addition, ginseng is sensitive to environmental factors such as light and soil moisture, so maintaining individual plants requires considerable effort [8].
Analysis of genetic diversity and population structure of the existing population is required for the purpose of conservation and reintroduction of rare and endangered species [10]. A species with little genetic variability may suffer from reduced fitness in its current environment. It may not have the evolutionary potential necessary for a changing environment. Genetic variability is critical for a species to adapt to environmental changes and survive in the long term [11]. Knowledge of genetic diversity within a population and among populations is important for conservation management, especially for identifying genetically unique structural units within a species and determining populations that need protection [12].
Genetic diversity of germplasm can be investigated using various techniques, including analysis of morphological and agricultural traits [13], isozymes [14], biochemical characteristics [15], and molecular markers [14,15,16]. In recent years, the development of various chemical and molecular techniques for differentiation among P. ginseng samples has received attention [10,17,18,19,20]. Gepts has reported that DNA markers are the most reliable ones for distinguishing variations of alleles because these markers are not influenced by environmental or physiological factors [21].
Crawford has reported that efficient population genetics studies could be performed using a high level of polymorphic markers [22]. Previous studies have performed genetic analysis of wild ginseng using random amplified polymorphic DNA and allozyme, respectively, and detected a low level of genetic polymorphisms in their samples, meaning that effective conservation strategies would be difficult to design using these markers [23,24]. Using amplified fragment length polymorphism markers, genetic diversity and genetic structure of P. ginseng have been analyzed [25,26,27]. Among various molecular markers, simple sequence repeat (SSR) markers have been successfully used to described population genetic diversity because they showed codominance and high polymorphism [7,10,20,28,29].
Since the 21st century, the number of publications on P.ginseng has exponentially increased; more than 3400 articles, including 242 reviews, have focused on P. ginseng. However, the main leading research subjects and keywords were pharmacology (39%) and functional materials such as ginsenoside and saponin, with little research on the genetic diversity of P. ginseng [30]. Understanding the genetic diversity on plant germplasm is important as plant germplasm provides an opportunity for plant breeders to develop new and improved cultivars with desirable characteristics, which include both the farmer and breeder’s preferred traits [31]. The objective of the present study was to analyze genetic diversity and population structure of 1109 ginseng accessions conserved in the National Agrobiodiversity Center (NAC) in South Korea to provide basic data for future preservation and breeding of ginseng genetic resources.
2. Materials and Methods
2.1. Plant Materials
A total of 1109 ginseng accessions were obtained from the National Agrobiodiversity Center (NAC) at the Rural Development Administration in South Korea (Table S1). All ginseng accessions have been separately conserved at three institutes (109 accessions Chungcheongbuk-do national University (CB); 160 accessions, Chungcheongnam-do Agricultural Research & Extension Service (CN); and 840 accessions, National Institute of Horticultural and Herbal Science (NIHHS)) designed by NAC.
2.2. DNA Extraction
Genomic DNA was extracted from 100 mg of freeze-dried ginseng leaves in each ginseng accession (4~5 plants per accession) using a DNeasy plant mini kit (Qiagen, Hilden, Germany). DNA quality and quantity were measured using 1% (w/v) agarose gel and a spectrophotometer (Epoch, BioTek, Winooski, VT, USA). Extracted DNA was diluted to 30ng/ul and stored at −20 °C until further PCR amplification.
2.3. SSR Genotyping
For SSR analysis, a total of 17 SSRs were selected from the previously reported study [28] and labeled fluorescently (6-FAM, HEX, and NED) (Table S2). PCR reactions were carried out using 25 ul reaction mixture, containing 30 ng template DNA, 10x PCR buffer, 1.5 mM MgCl2, 0.2 mM of each dNTPs, 0.5 uM of each primer, and 1 U Taq polymerase (Inclone, Korea). The amplification was performed with the following cycling conditions: initial denaturation at 94 °C for 5 min, followed by 35 cycles of denaturation at 95 °C for 30 s, annealing at 55 °C for 30 s, extension at 72 °C for 1 min, and a final extension step at 72 °C for 10 min. Each amplicon was resolved on the ABI prism 3500 DNA sequence (ABI3500, Thermo Fisher Scientific Inc., Wilmington, DE, USA). Individual tetraploid genotypes were scored from the electropherograms following the microsatellite DNA allele counting-peak ratios (MAC-PR) method of Esselink et al. [32] using Gene Mapper Software (Version 4.0, Thermo Fisher Scientific Inc., Wilmington, DE, USA).
2.4. Population Structure and Genetic Diversity
The number of alleles (Na), Shannon index (I), Nei’s unbiased gene diversity (GD), and Evenness were calculated using poppr package for R software [33]. The analysis of molecular variance (AMOVA) and calculation of the coefficient of genetic differentiation among populations (PhiPT) were done using GenAlEx software (v. 6.5) with 999 permutations [34].
Population structure was analyzed with a DAPC using adegenet package for R software [35,36]. The find.clusters function was used to detect the number of clusters in the population. It used K-means clustering which decomposed the total variance of a variable into between-group and within-group components. The best number of subpopulations had the lowest associated Bayesian Information Criterion (BIC). A cross-validation function (Xval. dapc) was used to confirm the correct number of principal components (PC) to be retained. In this analysis, data were divided into two sets: a training set (90% of the data) and a validation set (10% of the data). Members of each group were selected by stratified random sampling, ensuring that at least one member of each group or population in the original data was represented in both training and validation sets. DAPC was carried out on the training set by retaining variable numbers of PCs at a degree to which the analysis was able to accurately predict the group membership of excluded individuals (those in the validation set) was used to identify the optimal number of PCs to be retained. At each level of PC retention, the sampling and DAPC procedures were repeated many times [37]. The best number of PCs that should be retained is associated with the lowest root mean square error. Resultant clusters were plotted in a scatter plot of the first and second linear discriminants of DAPC.
Bayesian-based clustering was performed using STRUCTURE v.2.3.4 [38], testing three independent runs with K from 1 to 15, each run with a burn-in period of 50,000 iterations and 500,000 Monte Carlo Markov iterations, assuming an admixture model. The most likely K value was processed with STRUCTURE HARVESTER v.0.9.94 [39] and was detected using the Evanno transformation method [40]. To assign samples to clusters, a membership coefficient q > 0.8 was used, while coefficients ≤0.8 were considered “genetically admixed.”
3. Results
Among 1109 ginseng accessions, 1,042 (94.0%) accessions were landraces and 66 (6.0%) accessions were breeding lines (61 accessions, 5.5%) or cultivars (5 accessions, 0.5%). Of 1109 ginseng accessions, 899 (81.2%) were from Korea and 202 (18.0%) were from China (Figure S1A).
CN collected 73 ginseng accessions from Chungnam province from 1997 to 2000, 56 accessions from China, and 21 accessions from Korea from 2013 to 2016 (Table S1 and Figure S1B). NIHHS obtained approximately 300 ginseng accessions from Korea Tobacco and Ginseng Corporation (KT&G), 40 accessions from China, and 435 from Korea from 2003 to 2012. CB collected 90 accessions from China and 10 from Korea from 2007 to 2010.
SSR locus diversity data are summarized in Table 1. A total of 56 alleles were detected among 1109 ginseng accessions by using 17 polymorphic SSR markers. On average, 3.29 alleles varying from 2 to 6 were amplified by each marker. Shannon index (I) varied from 0.58 to 0.82 among 17 SSR markers. The average gene diversity (GD) was estimated to be 0.49 (range, 0.39 to 0.50). The average evenness (E) was 0.96 (range, 0.81 to 1.00). A total of 22 rare alleles (Ra) ranging from 1 to 4 with each allele had a frequency of less than 5%. Six SSR loci (PES0024, PES0021, PES0018, PES0019, PES0005, and PES0007) did not contain rare alleles. Common alleles (Ca) ranged from 1 to 2 with a total of 24, showing a frequency of 5%–50%.

Table 1.
Statistics of genetic diversity across 17 simple sequence repeat (SSR) loci in 1109 ginseng accessions.
Five from the 56 alleles were identified to private alleles which are detected in only one cluster (Table S3). Specifically, KT&G and GG showed two private alleles in locus PES0092 and PES0012 and PES0032 and PES0040, respectively, while GW possesses one private allele in locus PES0039.
A total of 942 multilocus genotypes (MLGs) were detected among 1109 ginseng accessions (Table 2). CN, CB, and NIHHS clusters had 148, 104, and 707 MLGs, respectively. The numbers of average alleles (Na) of CN, CB, and NIHHS were 1.77, 2.08, and 2.62, respectively. The Shannon–Wiener index (H) of NIHHS was the highest (6.45), while that of CN was similar to that of CB (4.97 and 4.63, respectively). Nei’s genetic diversity (GD) was similar to each other for the three clusters.

Table 2.
Summary of cluster diversity indices averaged over 17 loci according to conservation institutes.
Analysis of molecular variance (AMOVA) on genetic differentiation among and within clusters of ginseng accessions was conducted. The results are shown in Table 3. Findings from AMOVA revealed that 91% of total genetic variations were contributed by differences within clusters, which was notably and significantly higher than that among clusters (only 9% of total genetic variation was due to differences among clusters). PhiPT and gene flow (Nm) for 1109 ginseng accessions were 0.094 (p < 0.001) and 4.822, respectively. Pairwise cluster PhiPT values for the three institutes ranged from 0.081 (CN–NIHHS) to 0.113 (CB–NIHHS) (Table 4). Pairwise cluster estimates of gene flow (Nm) for the three institutes ranged from 3.940 to 5.656 migrants per cluster.

Table 3.
Analysis of molecular variance (AMOVA) within/among ginseng clusters based on 17 SSR data according to conservation institutes.

Table 4.
Pairwise cluster PhiPT values and Nm values based on 999 permutations from AMOVA according to conservation institutes (all PhiPT values were significantly greater than 0, p < 0.0001).
To understand the pattern of the genetic structure, a Bayesian clustering analysis in STRUCTURE and complementary coordination analysis by Discriminant Analysis of Principal Components (DAPC) were performed. STRUCTURE results suggested the best grouping number (K = 2) based on delta K (Figure S2). Clusters 1 and 2 consisted of 359 and 460 accessions, respectively. Two hundred and ninety accessions were identified in the admixed population (Figure 1A). All (100%) ginseng accessions of cluster 1 and admix were all in NIHHS while 53.8% and 30.7% of population 1 and admix cluster were from KT&G, respectively. Cluster 2 contained 191 accessions from NIHHS and all accessions of CN and CB and 80.3% of cluster 2 were from China.
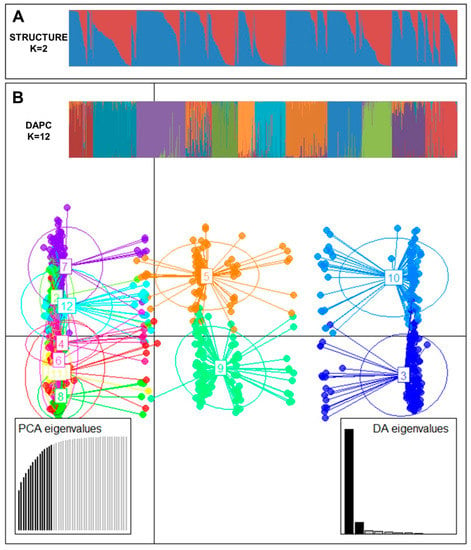
Figure 1.
(A) Population structure analysis of 1109 ginseng accessions inferred using STRUCTURE software based on 17 SSR markers for delta K = 2. (B) Discriminant analysis of principal components (DAPC) for 1109 ginseng accessions. Axes represent the first two linear discriminants (LD). Each circle represents a cluster and each dot represents an individual. Numbers represent different clusters identified by DAPC analysis.
The number of detected clusters was 12 in concordance with the lowest BIC value obtained using the find.clusters function. DAPC analysis was carried out using the detected number of clusters (Figure 1B). Fifteen first PCs (90.9% of variance conserved) of PCA and three discriminant eigenvalues were retained. These values were confirmed by cross-validation analysis.
Sources of genetic differentiation were revealed among 12 clusters based on DAPC analysis using the AMOVA method. Results indicated that 12% and 88% of variations could be attributed to differentiation among clusters and within inferred clusters, respectively (Table 5). PhiPT and gene flow (Nm) were 0.120 (p < 0.001) and 3.678, respectively. Pairwise cluster PhiPT values for 12 clusters ranged from 0.015 (C6–C7) to 0.224 (C4–C12) (Table 6). Pairwise cluster estimates of gene flow (Nm) for 12 clusters ranged from 1.734 (C4–C12) to 33.651 (C6–C7) migrants per clusters.

Table 5.
Analysis of molecular variance (AMOVA) within/among twelve clusters based on DAPC analysis.

Table 6.
Pairwise cluster PhiPT values (above diagonal) and Nm values based on 999 permutations (below diagonal) from AMOVA (all PhiPT values were significantly greater than 0, p < 0.0001).
4. Discussion
Among collected and preserved 1109 ginseng accessions in this study, 94.0% were landraces, with only 6% being breeding lines. This might be due to the collection strategy of ginseng germplasm. Three institutes collected ginseng accessions from all parts of South Korea since 1997 to develop new ginseng cultivars. In general, ginseng cultivars have been bred by selecting individual plants from arable fields of mixed local landrace population rather than by crossbreeding [41]. It is known that breeding of ginseng is difficult because of its long generation period and small number of seeds, although it is originally a self-pollination plant [9]. For these reasons, outstanding individual plants were selected and registered as cultivars during the cultivation of a mixed population of local landrace in Korea.
Ginseng is cultivated from 36° to 38° N and can still be found sometimes in mountainous areas in Korea [42]. In general, Ginseng farmers have traditionally kept various kinds of seeds by conserving and sowing the seeds that have been harvested from 3-year-old ginseng. In the case of large-scale ginseng farming (about 1 ha or more), farmers purchase seed because home seed production alone lacks it. However, most of the small farms in South Korea cultivate ginseng using their own seed from home seed production. Ginseng farmers choose and preserve seeds very carefully due to long-term (4~6 years) characteristics of ginseng cultivation. For this reason, various Korean ginseng genotypes are being conserved by the ginseng farmers in South Korea [43]. In this study, we used ginseng landrace based on the information of the genebank management system in NAC. Landrace defined by the FAO is “a crop that has developed its unique characteristics through repeated in situ grower selection and that has never been subjected to formal plant breeding” [44]. In terms of the definition, ginseng accessions collected from various local farms in Korea are likely to be landrace although their information except the geographical data is lack. Nevertheless, the question of whether the accessions are landraces still remains. Therefore, additional research on their genetic information will be needed to efficiently manage the ginseng accessions.
Among 1109 ginseng accessions, 167 (15.1%) (12 accessions in CN, five accessions in CB, and 133 accessions in NIHHS) might be duplicated (Table 2). It is important to identify these duplicated accessions, due to the limitation of funding and resources in genebanks [45]. Although not all duplicate accessions are worthless, identifying unique accessions within and across the genebanks will facilitate the better use of plant germplasms [45,46]. In this study, 167 ginseng accessions identified will also require the management of duplicate accessions using various additional methods such as evaluation of morphological and/or biochemical traits.
Results of the present study showed a similar number of alleles (56) and average number of alleles (3.29) to results of Choi et al. [28] (number of alleles = 51; average number of alleles = 3.0), while genetic diversity (0.490) in the present study was higher than (0.398) in the previous study. Reunova et al. [12] have reported similar results (average number of alleles: 2.49; expected heterozygosity (He): 0.453) in 139 individual ginseng plants using 11 SSR primers. Otherwise, Bang et al. [7] have reported a higher average number of alleles (4.3) and He (0.627) in 17 ginseng accessions using 8 SSR markers. Gene diversity of a locus, also known as expected heterozygosity, is a fundamental measure of genetic variation in a population. It describes the proportion of heterozygosis expected under the Hardy–Weinberg (HW) equilibrium [47]. Previous studies have confirmed the excess heterozygosity in their ginseng populations [7,12,28,48]. Indeed, the present study also showed excess heterozygosity in these ginseng accessions (Table 1). Causes of the excess of heterozygosity in natural plant populations are not well known yet, although reasons causing a deficit of heterozygotes have been well explained [12]. Previous studies have guessed that the excess of heterozygosity might result from a small reproductive population size [49,50,51]. When only a few breeders contribute to the next generation, allelic frequencies can differ between male and female parents by chance alone [49,50]. This effect can be reinforced for loci with many rare alleles. Moreover, in small sexual or self-incompatible populations, the fact that individuals cannot reproduce with themselves decreases the probability of creating homozygote offspring [52,53]. Meanwhile, Reunova et al. [12] have mentioned that high levels of observed heterozygosity might be associated with population structure and reflect the history of its formation under the influence of global climatic and local human-induced impacts.
In this study, 171 ginseng accessions showed rare alleles (Table S1). Rare alleles may be lost due to natural and management-originated bottleneck effects [54]. Although they are not likely to have major importance in the conservation of endangered species, rare alleles may be important for plant breeding [55]. They may be present or even fixed in certain populations because of genetic drift or their relationship with fitness in specific environments. Such uniqueness makes them prone to be absent in whole collections and their subsets, albeit their potential importance as a source of important traits for plant breeding. Novel strategies in germplasm management are needed to preserve those alleles, prone to be lost, which can be potentially useful for plant breeding, especially in a scenario of rapid climate change that represents serious threats to worldwide food production [56].
In addition, five private alleles were detected after screening the collection areas or institutes (Table S3). Private alleles are important in plant breeding and conservation as they are present only in a single population among a broader collection of populations [57]. However, all five private alleles observed in this study were present at very low frequencies (<0.9%), suggested that they could not be suitable for the ginseng breeding program. On the other hand, three ginseng accessions, IT272054, IT272375, and IT288747, with unique alleles might be used as a valuable source for selecting genetic variants related to useful chemical compounds such as saponins (Table S1).
To analyze population structure, various methods such as STRUCTURE and principal coordinates analysis have been used for ginseng germplasm [19,20,58]. In this study, STRUCTURE and DAPC used to analyze the population structure of 1109 ginseng accessions appeared to provide complementary information. Results of STRUCTURE and DAPC divided 1109 ginseng accessions into two and twelve clusters, respectively. The DAPC method is being used instead of STRUCTURE software as it does not require that populations should be in HW equilibrium. In addition, it can handle large sets of data without using parallel processing software [59]. Deperi et al. [60] have mentioned that DAPC analysis can divide the population into well-defined clusters associated with provenance, ploidy, taxonomy, and breeding program of genotypes related to their genetic structure. Rosyara et al. [61] have reported that DAPC is slightly better than the STRUCTURE leading to better separation among potato germplasms. Campoy et al. [59] have also reported that DAPC analysis provides more detailed clustering among cherry populations compared to STRUCTURE analysis. More detailed clustering was also obtained from DAPC analysis. Although 99.2% of 1109 ginseng accessions were collected from Korea (899 accessions) and China (202 accessions) (Table S1), 12 clusters of DAPC analysis showed that ginseng accessions from two countries were separated into all clusters. Also, low PhiPT (0.036) and high Nm (13.280) were detected between Korean and Chinese accessions. This means that there are high genetic similarities in ginseng accessions collected from these two countries. According to NIHHS [62], the genetic similarity of cultivated ginsengs between Korea and China was more than 70%, due to the import of ginseng seeds from South Korea to China and the cultivation and production of them in the ginseng main production area of China such as Gillim Province.
The reason why 1109 ginseng accessions were divided into twelve clusters on DAPC analysis was not clear. It might be due to different methods of cultivation, seed gathering, and seed distribution for ginseng in Korea. Since new ginseng cultivars, “Chunpoong” and “Yunpoong,” were developed in 2002, a total of 21 ginseng cultivars have been developed. However, farmers are still cultivating, self-seed-gathering, and seed-selling ginseng landraces not ginseng cultivars [63,64]. With the exception of some contract cultivation, ginseng landrace seeds would have been traded in the Korean ginseng market. As a result, ginseng cultivated in farms is expected to be genetically mixed regardless of their cultivation region.
5. Conclusions
In this study, genetic diversity and population structure of 1109 ginseng accessions collected and conserved in NAC were analyzed using 17 SSR markers. The results of this study provided molecular evidence for the narrow genetic base of 1109 ginseng germplasm in NAC. It was not enough to understand the genetic diversity of ginseng germplasm because there was a lack of sufficient information about these collected ginseng accessions, although the aim of the present study was to analyze genetic diversity in larger ginseng accessions than those from previous studies [7,20,52], In Korea, the majority of new ginseng cultivars are developed by selecting individual plants from ginseng landraces, although some research institutes have performed development and selection of breeding lines. To have a broad understanding and efficient use of ginseng germplasm, it is necessary to analyze functional factors and evaluate their morphological traits.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4395/10/1/68/s1, Figure S1: Geographic distribution of 1109 ginseng accessions. Pie charts on the map represent the collected province of the accessions and the colors in each chart represent the number of ginseng accessions indicated by conserved three institutes. (A) Original countries of ginseng accessions. (B) Original province of ginseng accessions in South Korea, Figure S2: Relationship between delta K and K as revealed by STRUCTURE harvester, Table S1: List of 1109 ginseng accessions used in this study, Table S2: List of 17 SSR primers used in this study, Table S3: Allele frequency based on collection area in ginseng accessions.
Author Contributions
Conceptualization, K.J.L.; data curation, K.J.L.; formal analysis, K.J.L. and R.S.; Investigation, J.-R.L. and G.-T.C.; project administration, D.Y.H.; resources, J.-R.L. and G.-T.C.; writing—original draft, K.J.L.; writing—review and editing, D.Y.H. All authors have read and agreed to the published version of the manuscript.
Funding
This work was carried out with the support of the “Research Program for Agricultural Science & Technology Development (Project No. PJ01355701),” National Institute of Agricultural Sciences, Rural Development Administration, Republic of Korea.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Yun, T.K. Brief Introduction of Panax ginseng C.A. Meyer. J. Korea Med. Sci. 2001, 16, S3–S5. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.J.; Kim, E.H.; Rhee, D.K. Effect of Panax ginseng on stress. J. Ginseng Res. 2008, 32, 8–14. [Google Scholar]
- Attele, A.S.; Wu, J.A.; Yuan, C.-S. Ginseng pharmacology: Multiple constituents and multiple actions. Biochem. Pharm. 1999, 58, 1685–1693. [Google Scholar] [CrossRef]
- Nam, M.H.; Kim, S.I.; Liu, J.R.; Yang, D.C.; Lim, Y.P.; Kwon, K.-H.; Yoo, J.S.; Park, Y.M. Proteomic analysis of Korean ginseng (Panax ginseng C.A. Meyer). J. Chromatogr. B 2005, 815, 147–155. [Google Scholar] [CrossRef] [PubMed]
- Baeg, I.-H.; So, S.-H. The world ginseng market and the ginseng (Korea). J. Ginseng Res. 2013, 37, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Ministry of Agriculture Food and Rural Affairs. Ginseng Annual Report; Ministry of Agriculture Food and Rural Affair: Sejong, South Korea, 2018.
- Bang, K.-H.; Jo, I.-H.; Chung, J.-W.; Kim, Y.-C.; Lee, J.-W.; Seo, A.Y.; Park, J.-H.; Kim, O.; Hyun, D.-Y.; Kim, D.-H.; et al. Analysis of Genetic Polymorphism of Korean Ginseng Cultivars and Foreign Accessions using SSR Markers. Korean J. Med. Crop. Sci. 2011, 19, 347–353. [Google Scholar] [CrossRef][Green Version]
- Jang, W.; Kim, N.-H.; Lee, J.; Waminal, N.; Lee, S.-C.; Jayakodi, M.; Choi, H.-I.; Park, J.Y.; Lee, J.-E.; Yang, T.-J. A Glimpse of Panax ginseng Genome Structure Revealed from Ten BAC Clone Sequences Obtained by SMRT Sequencing Platform. Plant Breed. Biotechnol. 2017, 5, 25–35. [Google Scholar] [CrossRef]
- Choi, K.T. Botanical characteristics, pharmacological effects and medicinal components of Korean Panax ginseng C A Meyer. Acta Phamacol. Sin. 2008, 29, 1109–1118. [Google Scholar] [CrossRef]
- Zhuravlev, Y.N.; Reunova, G.D.; Kats, I.L.; Muzarok, T.I.; Bondar, A.A. Genetic variability and population structure of endangered Panax ginseng in the Russian Primorye. Chin. Med. 2010, 5, 21. [Google Scholar] [CrossRef]
- Reed, D.H.; Frankham, R. Correlation between Fitness and Genetic Diversity. Conserv. Biol. 2003, 17, 230–237. [Google Scholar] [CrossRef]
- Reunova, G.; Koren, O.; Muzarok, T.I.; Zhuravlev, Y. Microsatellite Analysis of Panax ginseng Natural Populations in Russia. Chin. Med. 2014, 5, 231–243. [Google Scholar] [CrossRef][Green Version]
- Sinkovi, L.; Pipan, B.; Sinkovi, E.; Megli, V. Morphological Seed Characterization of Common (Phaseolus vulgaris L.) and Runner (Phaseolus coccineus L.) Bean Germplasm: A Slovenian Gene Bank Example. Biomed Res. Int. 2019, 2019, 13. [Google Scholar]
- Jain, A.; Roy, A.K.; Kaushal, P.; Malaviya, D.R.; Zadoo, S.N. Isozyme Banding Pattern and Estimation of Genetic Diversity among Guinea Grass Germplasm. Genet. Resour. Crop. Evol. 2006, 53, 339–347. [Google Scholar] [CrossRef]
- Kottawa-Arachchi, J.; Gunasekare, M.; Ranatunga, M.; Punyasiri, N.; Jayasinghe, L.; Karunagoda, R.P. Biochemical Characteristics of Tea (Camellia L. spp.) Germplasm Accessions in Sri Lanka: Correlation between Black Tea Quality Parameters and Organoleptic Evaluation. Int. J. Tea Sci. 2014, 10, 3–13. [Google Scholar]
- Halász, J.; Kodad, O.; Galiba, G.M.; Skola, I.; Ercisli, S.; Ledbetter, C.A.; Hegedűs, A. Genetic variability is preserved among strongly differentiated and geographically diverse almond germplasm: An assessment by simple sequence repeat markers. Tree Genet. Genome 2019, 15, 12. [Google Scholar] [CrossRef]
- Cho, C.-W.; Kim, Y.-C.; Rhee, Y.K.; Lee, Y.-C.; Kim, K.-T.; Hong, H.-D. Chemical composition characteristics of Korean straight ginseng products. J. Ethn. Foods 2014, 1, 24–28. [Google Scholar] [CrossRef]
- Shin, B.-K.; Kwon, S.W.; Park, J.H. Chemical diversity of ginseng saponins from Panax ginseng. J. Ginseng Res. 2015, 39, 287–298. [Google Scholar] [CrossRef]
- Li, M.-R.; Shi, F.-X.; Li, Y.-L.; Jiang, P.; Jiao, L.; Liu, B.; Li, L.-F. Genome-Wide Variation Patterns Uncover the Origin and Selection in Cultivated Ginseng (Panax ginseng Meyer). Genome Biol. Evol. 2017, 9, 2159–2169. [Google Scholar] [CrossRef]
- An, H.; Park, J.-H.; Hong, C.E.; Sebastin, R.; Lee, Y.; Jo, I.-H.; Chung, J.-W. Genetic diversity and population structure of Chinese ginseng accessions using SSR markers. J. Plant Biotechnol. 2017, 44, 312–319. [Google Scholar] [CrossRef]
- Gepts, P. The Use of Molecular and Biochemical Markers in Crop Evolution Studies. In Evolutionary Biology; Hecht, M.K., MacIntyre, R.J., Clegg, M.T., Eds.; Springer: Boston, MA, USA, 1993; Volume 27, pp. 51–94. [Google Scholar]
- Crawford, D.J. Molecular markers for the study of genetic variation within and between populations of rare plants. Opera Bot. 1997, 132, 149–157. [Google Scholar]
- Zhuravlev, Y.; Koren, O.; Reunova, G.D.; Artyukova, E.; Kozyrenko, M.; Muzarok, T.I.; Kats, I.L. Ginseng Conservation Program in Russian Primorye: Genetic Structure of Wild and Cultivated Populations. J. Ginseng Res. 2004, 28, 60–66. [Google Scholar]
- Koren, O.; Potenko, V.V.; Zhuravlev, Y. Inheritance and Variation of Allozymes in Panax ginseng C.A. Meyer (Araliaceae). Int. J. Plant Sci. 2003, 164, 189–195. [Google Scholar] [CrossRef]
- Reunova, G.D.; Kats, I.L.; Muzarok, T.I.; Nguen, C.T.; Dang, T.T.; Zhuravlev, I.N. Population Genetic Structure of Wild-Growing Ginseng (Planax ginseng CA Meyer) Assessed Using AFLP Markers. Genetika 2012, 48, 340–351. [Google Scholar] [PubMed]
- Zhuravlev, Y.; Koren, O.; Reunova, G.D.; Muzarok, T.I.; Gorpenchenko, T.; Kats, I.L.; Khrolenko, Y. Panax ginseng natural populations: Their past, current state and perspectives. Acta Pharmacol. Sin. 2008, 29, 1127–1136. [Google Scholar] [CrossRef]
- Zhuravlev, Y.; Reunova, G.D.; Kats, I.L.; Muzarok, T.I.; Bondar, A.A. Molecular variation of wild Panax ginseng C.A. Meyer (Araliaceae) by AFLP markers. Chin. Med. 2010, 5, 21. [Google Scholar] [CrossRef]
- Choi, H.-I.; Kim, N.H.; Kim, J.H.; Choi, B.S.; Ahn, I.-O.; Lee, J.-S.; Yang, T.-J. Development of Reproducible EST-derived SSR Markers and Assessment of Genetic Diversity in Panax ginseng Cultivars and Related Species. J. Ginseng Res. 2011, 35, 399–412. [Google Scholar] [CrossRef]
- Kalia, R.; Rai, M.; Kalia, S.; Singh, R.; Dhawan, A. Microsatellite markers: An overview of the recent progress in plants. Euphytica 2011, 177, 309–334. [Google Scholar] [CrossRef]
- Xu, W.; Choi, H.-K.; Huang, L. State of Panax ginseng Research: A Global Analysis. Molecules 2017, 22, 1518. [Google Scholar] [CrossRef]
- Govindaraj, M.; Vetriventhan, M.; Srinivasan, M. Importance of Genetic Diversity Assessment in Crop Plants and Its Recent Advances: An Overview of Its Analytical Perspectives. Genet. Res. Int. 2015, 2015, 431487. [Google Scholar] [CrossRef]
- Esselink, G.D.; Nybom, H.; Vosman, B. Assignment of allelic configuration in polyploids using the MAC-PR (microsatellite DNA allele counting—peak ratios) method. Appl. Genet. 2004, 109, 402–408. [Google Scholar] [CrossRef]
- Kamvar, Z.N.; Tabima, J.F.; Grünwald, N.J. Poppr: An R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2014, 2, e281. [Google Scholar] [CrossRef] [PubMed]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research±an update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [PubMed]
- Ivandic, V.; Hackett, C.A.; Nevo, E.; Keith, R.; Thomas, W.T.B.; Forster, B.P. Analysis of simple sequence repeats (SSRs) in wild barley from the Fertile Crescent: Associations with ecology, geography and flowering time. Plant Mol. Biol. 2002, 48, 511–527. [Google Scholar] [CrossRef] [PubMed]
- Jombart, T. adegenet: A R package for the multivariate analysis of genetic markers. Bioinformatics 2008, 24, 1403–1405. [Google Scholar] [CrossRef]
- Jombart, T.; Collins, C. A Tutorial for Discriminant Analysis of Principal Components (DAPC) Using adegenet 2.0.0. Available online: http://adegenet.r-forge.r-project.org/files/tutorial-dapc.pdf (accessed on 9 November 2019).
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar]
- Earl, D.A.; Vonholdt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Lee, S.-S.; Lee, J.-H.; Ahn, I.-O. Characteristics of new cultivars in Panax ginseng CA Meyer. In Proceedings of the 9th International Symposium on Ginseng, Chungnam, South Korea, 25 September 2006. [Google Scholar]
- Proctor, J.T.A.; Lee, J.C.; Lee, S.-S. Ginseng Production in Korea. HortScience 1990, 25, 746–750. [Google Scholar] [CrossRef]
- FAO. Korean Ginseng Agriculture System; FAO: Rome, Italy, 2018. [Google Scholar]
- Maxted, N.; Magos Brehm, J.; Kell, S. Resource Book for Preparation of National Conservation Plans for Crop Wild Relatives and Landraces; Food and Agriculture Organization of the United Nations: Rome, Italy, 2013. [Google Scholar]
- Singh, N.; Wu, S.; Raupp, W.J.; Sehgal, S.; Arora, S.; Tiwari, V.; Vikram, P.; Singh, S.; Chhuneja, P.; Gill, B.S.; et al. Efficient curation of genebanks using next generation sequencing reveals substantial duplication of germplasm accessions. Sci. Rep. 2019, 9, 650. [Google Scholar] [CrossRef]
- Van Hintum, T.J.L.; Knüpffer, H. Duplication within and between germplasm collections. Genet. Res. Crop Evol. 1995, 42, 127–133. [Google Scholar] [CrossRef]
- Harris, A.M.; DeGiorgio, M. An Unbiased Estimator of Gene Diversity with Improved Variance for Samples Containing Related and Inbred Individuals of any Ploidy. G3 Genes Genomes Genet. 2017, 7, 671–691. [Google Scholar] [CrossRef] [PubMed]
- Van Dan, N.; Ramchiary, N.; Choi, S.R.; Uhm, T.S.; Yang, T.-J.; Ahn, I.-O.; Lim, Y.P. Development and characterization of new microsatellite markers in Panax ginseng (C.A. Meyer) from BAC end sequences. Conserv. Genet. 2010, 11, 1223–1225. [Google Scholar] [CrossRef]
- Pudovkin, A.I.; Zaykin, D.V.; Hedgecock, D. On the potential for estimating the effective number of breeders from heterozygote-excess in progeny. Genetics 1996, 144, 383–387. [Google Scholar] [PubMed]
- Rasmussen, D.I. Sibling Clusters and Genotypic Frequencies. Am. Nat. 1979, 113, 948–951. [Google Scholar] [CrossRef]
- Stoeckel, S.; Ggange, J.; Fernandez-manjarres, J.F.; Bilger, I.; Frascaria-lacoste, N.; Mariette, S. Heterozygote excess in a self-incompatible and partially clonal forest tree species—Prunus avium L. Mol. Ecol. 2006, 15, 2109–2118. [Google Scholar] [CrossRef]
- Balloux, F. Heterozygote excess in small populations and the heterozygote-excess effective population size. Evolution 2004, 58, 1891–1900. [Google Scholar] [CrossRef]
- Mark Welch, D.B.; Meselson, M. Evidence for the Evolution of Bdelloid Rotifers without Sexual Reproduction or Genetic Exchange. Science 2000, 288, 1211–1215. [Google Scholar] [CrossRef]
- Hartl, D.L.; Clark, A.G.; Clark, A.G. Principles of Population Genetics; Sinauer Associates: Sunderland, MA, USA, 1997; Volume 116. [Google Scholar]
- Brown, A.; Briggs, J. Sampling strategies for genetic variation in ex situ collections of endangered plant species. In Genetics and Conservation of Rare Plants; Falk, D., Holsinger, K., Eds.; Oxford University Press: Oxford, UK, 1991; pp. 99–119. [Google Scholar]
- Reyes-Valdés, M.H.; Burgueño, J.; Singh, S.; Martínez, O.; Sansaloni, C.P. An informational view of accession rarity and allele specificity in germplasm banks for management and conservation. PLoS ONE 2018, 13, e0193346. [Google Scholar] [CrossRef]
- Kalinowski, S.T. Counting alleles with rarefaction: Private alleles and hierarchical sampling designs. Conserv. Genet. 2004, 5, 539–543. [Google Scholar] [CrossRef]
- Li, S.; Li, J.; Yang, X.-L.; Cheng, Z.; Zhang, W.-J. Genetic diversity and differentiation of cultivated ginseng (Panax ginseng C. A. Meyer) populations in North-east China revealed by inter-simple sequence repeat (ISSR) markers. Genet. Res. Crop Evol. 2011, 58, 815–824. [Google Scholar] [CrossRef]
- Campoy, J.A.; Lerigoleur-Balsemin, E.; Christmann, H.; Beauvieux, R.; Girollet, N.; Quero-García, J.; Dirlewanger, E.; Barreneche, T. Genetic diversity, linkage disequilibrium, population structure and construction of a core collection of Prunus avium L. landraces and bred cultivars. BMC Plant Biol. 2016, 16, 49. [Google Scholar] [CrossRef] [PubMed]
- Deperi, S.I.; Tagliotti, M.E.; Bedogni, M.C.; Manrique-Carpintero, N.C.; Coombs, J.; Zhang, R.; Douches, D.; Huarte, M.A. Discriminant analysis of principal components and pedigree assessment of genetic diversity and population structure in a tetraploid potato panel using SNPs. PLoS ONE 2018, 13, e0194398. [Google Scholar] [CrossRef] [PubMed]
- Rosyara, U.R.; De Jong, W.S.; Douches, D.S.; Endelman, J.B. Software for Genome-Wide Association Studies in Autopolyploids and Its Application to Potato. Plant Genome 2016, 9. [Google Scholar] [CrossRef] [PubMed]
- NIHHS. Distribution Status of Panax ginseng in China and Comparative Analysis on the Characteristics of Korean and China ginseng; RDA: Jeonju, South Korea, 2016. [Google Scholar]
- Horticulture Business Division. 2017 Ginseng Data Sheet; Ministory of Agriculture Food and Rural Affairs: Sejong, South Korea, 2018.
- Yun, Y.M. A Study on the Production and Distribution of Domestic Korea Ginseng Seeds—A Case Study of Korea Ginseng Seeds in Chungnam Province. Master’s Thesis, Kongju National University, Yesan, South Korea, 2015. [Google Scholar]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).