Abstract
A gene encoding LgEstI was cloned from a bacterial fish pathogen, Lactococcus garvieae. Sequence and bioinformatic analysis revealed that LgEstI is close to the acetyl esterase family and had maximum similarity to a hydrolase (UniProt: Q5UQ83) from Acanthamoeba polyphaga mimivirus (APMV). Here, we present the results of LgEstI overexpression and purification, and its preliminary X-ray crystallographic analysis. The wild-type LgEstI protein was overexpressed in Escherichia coli, and its enzymatic activity was tested using p-nitrophenyl of varying lengths. LgEstI protein exhibited higher esterase activity toward p-nitrophenyl acetate. To better understand the mechanism underlying LgEstI activity and subject it to protein engineering, we determined the high-resolution crystal structure of LgEstI. First, the wild-type LgEstI protein was crystallized in 0.1 M Tris-HCl buffer (pH 7.1), 0.2 M calcium acetate hydrate, and 19% (w/v) PEG 3000, and the native X-ray diffraction dataset was collected up to 2.0 Å resolution. The crystal structure was successfully determined using a molecular replacement method, and structure refinement and model building are underway. The upcoming complete structural information of LgEstI may elucidate the substrate-binding mechanism and provide novel strategies for subjecting LgEstI to protein engineering.
1. Introduction
Esterases (E.C. 3.1.1.X) catalyze the hydrolysis of various substrates containing ester groups. Esterases are serine hydrolases that contain a conserved Ser-Asp/Glu-His catalytic triad with an α/β hydrolase fold. Although esterases harbor the same α/β hydrolase fold and have high sequence homology, they have different substrate specificities and perform varying biological functions. Recently, esterases of microbial origin have gained significant interest in scientific research because of their potential applications in the biotechnology industry [1,2]. Therefore, extensive efforts are being made to identify unique esterases with higher activity, improved stability, and broad substrate specificity from newly found microorganism genomes as well as metagenomes. Such esterases can be further subjected to protein engineering to generate esterases with a precise structure and desirable functions [3].
To date, many microbial esterases have been discovered and characterized [4,5]. In particular, pathogenic bacteria have been well-known for producing various extracellular esterases. These secreted esterases may be important for the virulence and pathogenesis of some bacteria. In addition, some pathogenic bacterial esterases are involved in the drug resistance mechanism by enzymatic cleavage of antibiotics [6,7]. However, detailed enzymatic activity differences and the structural information of pathogenic bacterial esterases are still unclear. Recently, the complete genome sequence information of a major fish pathogen bacteria, Lactococcus garvieae, has been published. Lactococcus garvieae is a Gram-positive bacterium with a sphere-shape (cocci) [8].
In this study, several putative esterase encoding genes of Lactococcus garvieae were identified as targets and sequentially tested. In this study, we cloned the LgEstI of L. garvieae into a plasmid vector to overexpress the LgEstI protein in Escherichia coli. To obtain structural information, we crystallized LgEstI to perform initial X-ray crystallographic experiments and successfully determined its structure. Further structural refinements and model building are underway. We believe that structural analysis of LgEstI in the near future will add further value to our biochemical analysis and facilitate a better understanding of the potential application of LgEstI, as well as provide new insights for further engineering of this protein.
2. Materials and Methods
2.1. Protein Clustering
ProtBLAST/PSI-BLAST was used to detect the remote homologs [9]. Initially, the LgEstI sequence was blasted against the Protein Data Bank (PDB) database, and the result (E-value cutoff for reporting = 1 × 10−10) was reloaded for performing a second blast against the Uniprot_sport database. Finally, the data with full-length sequences were forwarded to CLANS [10] for clustering by sequence similarity. The data were visualized with connecting lines, and shorter lines indicated higher sequence similarity.
2.2. Esterase Activity
p-Nitrophenyl esters were purchased from Sigma-Aldrich (Sigma, St. Louis, MO, USA) and used as substrates to assay LgEstI activity. LgEstI activity with different lengths of acyl carbon chains was determined by monitoring the production of p-nitrophenol (PNP) spectrophotometrically as previously described, with minor modifications [11].
2.3. Gene Cloning, Expression, and Purification of Recombinant LgEstI Protein
Genomic DNA of L. garvieae was isolated using a DNA extraction kit according to the manufacturer’s instructions (Bioneer, Daejeon, Korea). The CDS of LgEstI (NCBI accession number: WP_042219410.1) was amplified by performing PCR using appropriate primers. The PCR product was cloned into the pET21a vector between the NdeI and XhoI restriction sites. For protein expression, cells of the E. coli BL21 (DE3) strain were transformed with the plasmid harboring LgEstI (Table 1). Fresh transformants were grown at 37 °C in 4 L of Luria–Bertani medium containing 50 μg/mL ampicillin. When the O.D.600 reached 0.5, protein overexpression was induced by adding 1.0 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) to the cell culture, which was then continued for 20 h at 20 °C. Next, cells were pelleted by 20 min of centrifugation at 4000 rpm, resuspended in a lysis buffer (NPI-20, 50 mM NaH2PO4, 300 mM NaCl, and 20 mM imidazole), and disrupted by ultrasonication at 30% amplitude. An ice bath was used to maintain the temperature below 40 °C. The cell debris was removed by 1 h of centrifugation at 16,000 rpm and 4 °C, and the supernatant was used for the purification step.

Table 1.
Recombinant LgEstI protein production information.
The recombinant LgEstI was purified using gravity-flow purification as the initial step. To eliminate the remaining free nickel, a pre-packed column with His-tagged resin was washed with 5 mL of NPI-20. The supernatant containing recombinant LgEstI was slowly loaded onto the column and then washed by applying 10 CV with NPI-20. Next, the purified recombinant LgEstI was eluted with NPI-300 (50 mM NaH2PO4, 300 mM NaCl, and 300 mM imidazole). The elution was concentrated to 5 mL and incubated with thrombin for 2 days in a rotating incubator at 4 °C to cleave the His tag. For the second purification, the tag-removed sample was loaded onto a Superdex prep grade column (HiLoad® 16/600 Superdex® 200 pg) equilibrated with 20 mM Tris-HCl (pH 8.0), 200 mM NaCl, and 1 mM TCEP (Tris(2-carboxyethyl)phosphine hydrochloride) for SDS-PAGE. The Bradford protein assay was performed to confirm the purity and concentration of the recombinant LgEstI.
2.4. LgEstI Crystallization, Data Collection, and Phasing
More than 1000 different crystallization screening solutions were screened for the initial crystallization trial using recombinant LgEstI (25 mg/mL) by the sitting-drop vapor diffusion method. The crystallization conditions established from the initial screening were further optimized to generate diffraction-quality crystals (Table 2). Diffraction data were collected at the 5C beamline of the PLS, Korea. A single square pillar crystal was mounted on a goniometer equipped with a nitrogen stream with perfluoropolyether cryo-oil (Hampton Research, Laguna, CA, USA) as a cryoprotectant, with an oscillation range of 1°. Native data diffracting to 2.0 Å were collected, and processing and reduction were performed using XDS [12]. The LgEstI phase was obtained by molecular replacement using the I-TASSER model [13]. Next, the coordinates of LgEstI were built by iterative model building using a combination of Coot and Autobuild. Refinement of LgEstI was performed using phenix.refine from Phenix. The details of selected X-ray data collection statistics are listed in Table 3.

Table 2.
Crystallization details.

Table 3.
X-ray diffraction data collection statistics.
3. Results
3.1. Clustering Analysis of LgEstI
Clustering analysis of LgEstI was performed against the PDB and UniProt databases for primary and secondary blasting, respectively, using the MPI Bioinformatics Toolkit. Available online: https://toolkit.tuebingen.mpg.de (accessed on 6 December 2021) [9]. In total, 170 proteins were detected and aligned to visualize these relationships. Four main clusters were identified from the clustering analysis. The acetyl esterase family is tightly clustered and shows a high degree of sequence similarity. The second cluster included a combination of carboxylesterase, acetylcholinesterase, and para-nitrobenzyl esterase. Arylacetamide deacetylases and tuliposide A-converting enzymes were grouped into separate clusters. Interestingly, LgEstI could not be assigned to any of these clusters and showed maximum similarity to a hydrolase (UniProt: Q5UQ83) of Acanthamoeba polyphaga mimivirus (APMV) within the UniProt database. Based on the length of the connection and the color indicating sequence similarity, LgEstI appeared close to the acetyl esterase group but was not part of it. The clustering analysis confirmed that LgEstI has a unique sequence and may have a distinctive function from the other enzyme clusters (Figure 1).
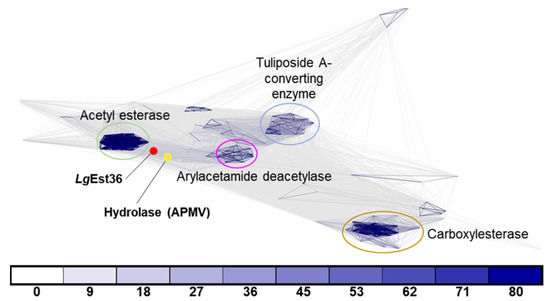
Figure 1.
Sequences of orthologs from different species were used for clustering by CLANS clustering. ProtBLAST/PSI-BLAST was used to detect distantly related proteins using the Protein Data Bank (PDB) and Uniprot_sport databases. LgEstI and the hydrolase (UniProt: Q5UQ83) from Acanthamoeba polyphaga mimivirus (APMV) within the UniProt database are indicated by red and yellow dots, respectively. Connecting lines of darker intensity and shorter length indicate higher sequence similarity. Connections with a p-value better than 1 × 10−x are drawn in the corresponding color (x = number below).
3.2. Purification and Biochemical Characterization of LgEstI
To conduct a preliminary study for understanding the detailed functional mechanisms of LgEstI, we cloned the LgEstI of L. garvieae into a plasmid vector and overexpressed the LgEstI protein in E. coli. Purified LgEstI protein was used to perform preliminary X-ray crystallography. Two conventional purification steps, namely tag-affinity purification and size-exclusion chromatography, were applied to purify the recombinant LgEstI protein (>95%), and the final protein purity was checked on SDS-PAGE. The molecular weight of LgEstI was approximately 37 kDa, which is similar to the calculated molecular weight based on the amino acid sequence (Figure 2A). The enzyme activity assay using purified LgEstI protein with various p-nitrophenyl esters showed that LgEstI had a narrow substrate specificity, as it was active only against p-nitrophenyl acetate (Figure 2B). Thus, the clustering and enzyme activity in combination indicated that LgEstI exhibits specific activity against analogs of acetate.
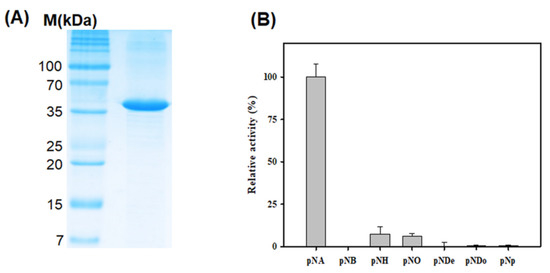
Figure 2.
Purification and activity analysis of LgEstI using various p-nitrophenyl esters. (A) SDS-PAGE analysis of LgEstI purified by tag-affinity purification and size-exclusion chromatography. (B) The relative activities of LgEstI against p-nitrophenyl esters, expressed as percentages. This activity assay was carried out using various substrates (pNA: p-nitrophenyl acetate; pNB: p-nitrophenyl butyrate; pNH: p-nitrophenyl hexanoate; pNO: p-nitrophenyl octanoate; pNDe: p-nitrophenyl decanoate; pNDo: p-nitrophenyl dodecanoate; pNP: p-nitrophenyl phosphate). The activity of LgEstI with p-nitrophenyl acetate was defined as 100%.
3.3. X-ray Crystallographic Study of LgEstI
To obtain the factual structural information of LgEstI, we performed crystallization and initial X-ray crystallographic experiments. The crystallization conditions for LgEstI protein were screened using more than 1000 different crystallization buffers. After optimizing the crystallization conditions by changing the precipitant concentration and pH in the reservoir solution and drops, rhombus-shaped crystals (approximately 100 × 200 μm) were obtained using 0.1 M Tris-HCl (pH 7.1), 0.2 M calcium acetate hydrate, and 19% (w/v) PEG 3000 (Figure 3A). However, X-ray diffraction of LgEstI crystals resulted in a poor diffraction pattern with a resolution of approximately 4 Å. Instead of searching for new crystallization conditions where LgEstI could be better diffracted, we aimed to optimize the cryoprotectant conditions. LgEstI crystals soaked in solutions containing perfluoropolyether cryo-oil (Hampton Research, Laguna, CA, USA) for 10 s exhibited a dramatic improvement in diffraction quality (~2 Å), whereas the crystals cryoprotected with general cryoprotectants (e.g., glycerol and PEG) showed low-resolution diffraction patterns (Figure 3B). Briefly, we tested more than 50 LgEstI crystals to find the optimal cryoprotectant solution. LgEstI crystals were very unstable in glycerol or PEG-containing cryoprotectant solutions, resulting in the LgEstI crystals melting and cracking. However, LgEstI crystals were more stable for a long time (more than 5 min) without melting or cracking in oil-based cryoprotectant solutions (Paratone-N oil and perfluoropolyether cryo-oil). We used the same conditions and similar sized crystals grown under the same crystallization conditions for this experiment. Finally, we obtained the best LgEstI crystals for X-ray diffraction data collection under the conditions with perfluoropolyether cryo-oil. During X-ray diffraction data collection, flash cooling the crystal in a nitrogen gas stream at 100 K prevented crystal cracking. We inferred that an oil-based solution could be a suitable cryoprotectant for LgEstI. All the diffraction data (completeness: 98.7%) were collected at a resolution of 2.0 Å. The data processing program XDS was used to index, integrate, and scale the acquired diffraction data [12].
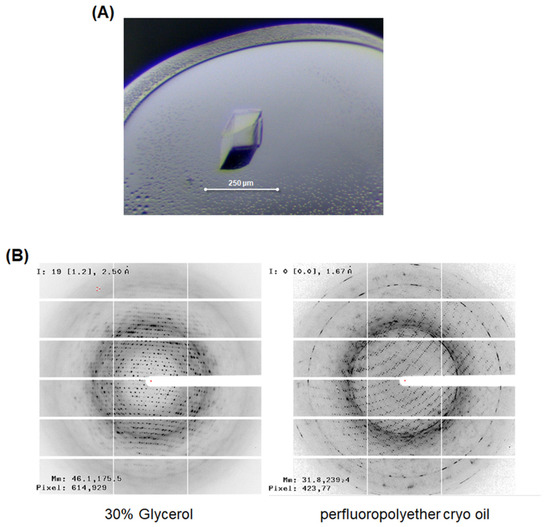
Figure 3.
X-ray crystallographic study of LgEstI. (A) Crystal of LgEstI protein grown in 0.1 M Tris-HCl (pH 7.1), 0.2 M calcium acetate hydrate, and 19% (w/v) PEG 3000. (B) Cryoprotectant optimization for the X-ray diffraction study. LgEstI crystals soaked in solutions containing perfluoropolyether cryo-oil for 10 s and flash-frozen under a nitrogen stream are shown (right).
The initial structure of LgEstI was successfully determined. For phasing LgEstI, we first generated the LgEstI model based on the homologous structures using the protein structure prediction server I-TASSER [13]. The quality of the model was confirmed to have a C-score of 1.40 and a TM-score of 0.91 ± 0.06, respectively. Next, the LgEstI model was used to overcome the phase problem using the molecular replacement method. Further structural refinements and model building are underway in our laboratory. We believe that a structural analysis of LgEstI in the near future will facilitate a better understanding of our biochemical analysis results, as well as providing valuable insights for further engineering of LgEstI protein.
Author Contributions
Conceptualization, J.H.L. and T.D.K.; methodology, H.D., Y.W. and C.W.L.; validation, W.Y., S.J., J.H., M.J.L. and H.-W.K.; resources, M.J.L. and H.-W.K.; writing—original draft preparation, H.D., Y.W. and K.K.K.; writing—review and editing, J.H.L. and T.D.K.; funding acquisition, H.-W.K., J.H.L. and T.D.K. All authors have read and agreed to the published version of the manuscript.
Funding
This work was financially supported by an academic grant (NRF-2021R1F1A1048135) from the National Research Foundation of Korea (T.D.K). This research was also supported by a National Research Foundation of Korea Grant from the Korean Government (MSIT; the Ministry of Science and ICT), NRF-2021M1A5A1075524) (KOPRI-PN21014) (H.-W.K). This research was a part of the project titled “Development of potential antibiotic compounds using polar organism resources (15250103, KOPRI Grant PM21030)”, funded by the Ministry of Oceans and Fisheries, Korea (J.H.L).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Fazary, A.E.; Ju, Y.H. Feruloyl Esterases as Biotechnological Tools: Current and Future Perspectives. Acta Biochim. Biophys. Sin. 2007, 39, 811–828. [Google Scholar] [CrossRef] [PubMed]
- Celligoi, M.A.P.C.; Baldo, C.; De Melo, M.R.; Gasparin, F.G.M.; Marques, T.A.; De Barros, M. Lipase Properties, Functions and Food Applications, Microb. In Microbial Enzyme Technology in Food Applications; CRC Press: Boca Raton, FL, USA, 2017; pp. 214–240. [Google Scholar] [CrossRef]
- Pleiss, J.; Fischer, M.; Peiker, M.; Thiele, C.; Schmid, R.D. Lipase Engineering Database. J. Mol. Catal. B Enzym. 2000, 10, 491–508. [Google Scholar] [CrossRef]
- Bornscheuer, U.T. Microbial carboxyl esterases: Classification, properties and application in biocatalysis. FEMS Microbiol. Rev. 2002, 26, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Barzkar, N.; Sohail, M.; Tamadoni Jahromi, S.; Gozari, M.; Poormozaffar, S.; Nahavandi, R.; Hafezieh, M. Marine Bacterial Esterases: Emerging Biocatalysts for Industrial Applications. Appl. Biochem. Biotechnol. 2021, 193, 1187–1214. [Google Scholar] [CrossRef] [PubMed]
- Larsen, E.M.; Johnson, R.J. Microbial esterases and ester prodrugs: An unlikely marriage for combating antibiotic resistance. Drug Dev. Res. 2019, 80, 33–47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwon, S.; Yoo, W.; Kim, Y.O.; Kim, K.K.; Kim, T.D. Molecular Characterization of a Novel Family VIII Esterase with β-Lactamase Activity (PsEstA) from Paenibacillus sp. Biomolecules 2019, 9, 786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morita, H.; Toh, H.; Oshima, K.; Yoshizaki, M.; Kawanishi, M.; Nakaya, K.; Suzuki, T.; Miyauchi, E.; Ishii, Y.; Tanabe, S.; et al. Complete genome sequence and comparative analysis of the fish pathogen Lactococcus garvieae. PLoS ONE 2011, 6, e23184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gabler, F.; Nam, S.Z.; Till, S.; Mirdita, M.; Steinegger, M.; Söding, J.; Lupas, A.N.; Alva, V. Protein Sequence Analysis Using the MPI Bioinformatics Toolkit. Curr. Protoc. Bioinform. 2020, 72, e108. [Google Scholar] [CrossRef] [PubMed]
- Frickey, T.; Lupas, A. CLANS: A Java Application for Visualizing Protein Families Based on Pairwise Similarity. Bioinformatics 2004, 20, 3702–3704. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.W.; Kwon, S.; Park, S.H.; Kim, B.Y.; Yoo, W.; Ryu, B.H.; Kim, H.W.; Shin, S.C.; Kim, S.; Park, H.; et al. Crystal Structure and Functional Characterization of an Esterase (EaEST) from Exiguobacterium antarcticum. PLoS ONE 2017, 12, e0169540. [Google Scholar] [CrossRef] [PubMed]
- Kabsch, W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 125–132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.; Yan, R.; Roy, A.; Xu, D.; Poisson, J.; Zhang, Y. The I-TASSER Suite: Protein Structure and Function Prediction. Nat. Methods 2015, 12, 7–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karplus, P.A.; Diederichs, K. Linking Crystallographic Model and Data Quality. Science 2012, 336, 1030–1033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).