Abstract
Leishmaniasis is a neglected parasitic disease caused by various Leishmania species. The discovery of new protozoa drugs makes it easier to treat the disease; but, conventional clinical issues like drug resistance, cumulative toxicity, and target selectivity are also getting attention. So, there is always a need for new therapeutics to treat Leishmaniasis. Here, we have reported 2,3-dihydroquinazolin-4(1H)-one derivative as a new class of anti-leishmanial agents. Two derivatives, 3a (6,8-dinitro-2,2-disubstituted-2,3-dihydroquinazolin-4(1H)-ones) and 3b (2-(4-chloro-3-nitro-phenyl)-2-methyl-6,8-dinitro-2,3-dihydro-1H-quinazolin-4-one) were prepared that show promising in silico anti-leishmanial activities. Molecular docking was performed against the Leishmanial key proteins including Pyridoxal Kinase and Trypanothione Reductase. The stability of the ligand-protein complexes was further studied by 100 ns MD simulations and MM/PBSA calculations for both compounds. 3b has been shown to be a better anti-leishmanial candidate. In vitro studies also agree with the in-silico results where IC50 for 3a and 3b was 1.61 and 0.05 µg/mL, respectively.
1. Introduction
Leishmaniasis is caused by a protozoan parasite of the Leishmania genus that has become a serious public health concern regarding diagnosis, prevention, and cure [1,2]. Biting over 90 species of female sandfly vectors of the genera Lutzomyia and Phlebotomus are known to transfer these parasites from one host by infecting the host while taking a blood meal. Leishmania disease has been classified as a neglected tropical disease associated with population displacement, malnutrition, weak immune system, and poor housing. It is estimated that around 1 million new cases of the contagious disease occur every year [3,4]. The number increases due to the association and co-infection with the human immunodeficiency virus (HIV) and Leishmania. The situation aggravates due to the non-availability of effective therapy or treatment for such patients [5,6].
Leishmaniasis exists in many forms that depend on species of parasite infecting the host. Clinically, the disease exists in three different forms, namely mucocutaneous (skin, nose, and mouth), cutaneous (skin ulcers), and visceral leishmaniasis (starting from skin ulcers causing fever, enlargement of liver/spleen, and low red blood cells) [7]. The later form of Leishmaniasis is chronic, mainly caused by L. donovani, L. infantum, and L. chagasi, resulting in debilitating infection of the reticuloendothelial system and is fatal if left untreated [8,9]. Currently, there are no approved vaccines or prophylactic drugs against any form of leishmaniasis [10]. Thus, conventional chemotherapy is used to control and manage disease by using pentavalent antimonials like organic complexes with SbV including meglumine antimoniate (Glucantime®) and sodium stibogluconate (Pentostam®) through multiple parenteral administrations. Still, there are reports of considerable toxicity and reduced activity due to induced resistance [11,12].
Other second-line therapeutic agents include amphotericin B and its lipid formulations, Miltefosine, pentamidine, and a phosphocholine analog, but these drugs have also developed resistance, are too toxic, or too expensive for common patients in the developing countries [13,14]. Apropos, the discovery of economical and efficient chemotherapy for leishmaniasis is still awaited, so the development of new efficient, safe and cheap therapeutic alternatives is urgently needed. Several researchers have reported the anti-leishmanial activity of versatile synthetic compounds [15,16,17,18,19,20,21] and the potential of alternative entities and new strategies [22,23].
Since the last couple of decades, medicinal scientists have switched their attention to using different enzyme inhibitors to control and treat various psychological disorders and health implications resulting from malfunction or over-activity of enzymes. Various enzyme inhibitors have been designed and reported to control different diseases and disorders [24,25,26,27,28,29,30].
Quinazolines show promising antileishmanial, antimalarial, antibacterial, antidiabetic, antifungal, anthelmintic, cardiotonic, anticonvulsant, anti-inflammatory, antiviral, analgesic, antidiuretic, cytotoxicity, antitumor, and many other biological activities [31,32,33,34,35,36,37,38,39,40,41,42]. Several researchers have synthesized and documented the potential of quinazoline derivatives as anti-leishmanial agents [16,43,44,45,46,47,48,49,50,51]. There are more than 150 naturally occurring alkaloids with the Quinazoline scaffold, including echinozolinone, glucosamine, rutaecarpine, and deoxyvascinone have been reported from animals, microorganisms, and plants kingdoms are structural subunits of this scaffold [52].
Quinazoline scaffolds also display immense importance for designing and developing some clinically proven and commercially available drugs such as Gefitinib (used against lung cell cancer), Proquazone (a non-steroidal anti-inflammatory drug), Doxazosin (an antihypertensive agent), Prazosin (used to treat high blood pressure), Erlotinib (useful for pancreatic cancer, small cell lung cancer and several other types of cancer), Quinethazone (used to treat hypertension), Febrifugine (used as antimalarial), Afloqualone (used as sedative and muscle relaxant), Fenquizone (used as antidiuretic), and many other pharmacological active drug candidates (Figure 1) [53,54].
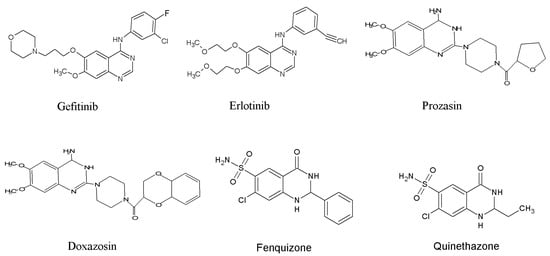
Figure 1.
Quinazoline-based drugs.
In a previous study, we have reported the synthesis and cholinesterase inhibitory activity of several 2,3-dihydroquinazolin-4(1H)-ones derivatives [26]. Keeping in view the versatile nature of quinazoline moiety and pursuing the strategy of developing potent anti-leishmanial agents, we have designed our research project to evaluate the biological potential of a relatively new class of di-substituted 2,3-dihydroquinazolin-4(1H)-one moiety through molecular docking and in vitro assay study.
2. Materials and Methods
Analytical-grade chemicals, reagents, and solvents used for quinazoline derivatives preparation were of analytical grade as received from suppliers. Anthranilamide, ethyl methyl ketone, diethyl ketone, methyl-(4-chloro phenyl) ketone, triethyl amine (Et3N), hydrochloric acid (HCl), nitric acid (HNO3), and DCM was purchased from Merck Millipore and Sigma-Aldrich, UK. Melting points were measured by using an SMP30 Stuart Scientific melting point apparatus. The 1H and 13C NMR spectra were recorded using Bruker DRX 400 MHz NMR spectrometers using CDCl3 as solvent. All chemical shifts are reported with reference to SiMe4 and corresponding residual 1H and 13C solvent peaks. Bruker Smart APEX II diffractometer was used for collecting X-ray diffraction data of the crystals. Data reduction was carried out using the SAINT program, and structure solution and refinements were performed with the SHELXL-2013 program package.
2.1. General Procedure for the Synthesis of 6,8-dinitro-2,2-disubstituted-2,3-dihydroquinazolin-4(1H)-ones (3a and 3b)
The ligands 3a and 3b were synthesized and characterized as stated previously [55] wherein a solvent-free condensation of anthranilamide1 and ketone 2 was carried out by adding a few drops of conc. HNO3 and 2,3-dihydroquinazolin-4(1H)-one derivatives were prepared by heating the reaction mixture under reflux conditions for 30 min. The intermediate compound was concentrated on a rotary evaporator and cooled down. Further, this compound (100 mM) was dissolved in conc. H2SO4 (10 mL) and conc. HNO3 (0.5 mL) was added dropwise while stirring in an ice bath. Stirring was continued for 6 h, and the reaction mixture was poured on crushed ice. Precipitates so formed were filtered, washed with water, and dried in an oven. Recrystallization with ethanol afforded yellowish crystals of 6,8-dinitro-2,2-disubstituted-2,3-dihydroquinazolin-4(1H)-one which was characterized by X-ray crystallographic and NMR techniques.
2.2. Synthesis of 6,8-dinitro-2,2-diethyl-2,3-dihydroquinazolin-4(1H)-one (3a)
As stated in [55], synthesis of compound 3a was achieved from reaction of 1 (13.6 g, 100 mmol) and diethyl ketone (21.5 mL, 200 mmol) by adopting general procedure as yellow crystalline solid. (98.5% yield); m.p. 200–202 °C; CCDC No. 1568517; 1H NMR (400 MHz, CDCl3, 25 °C): δ = 9.21 (d, 3J = 8 Hz, 1H, ArH), 9.02 (d, 3J = 8 Hz, 1H, ArH), 8.52 (s, 1H, NH), 6.67 (s, 1H, NH), 1.95–1.88 (m, 4H, 2 × CH2), 1.05 (t, 3J = 7 Hz, 6H, 2 × CH3) ppm. 13C NMR (100 MHz, CDCl3, 25 °C), δ = 165.4, 144.2, 138.6, 134.5, 129.2, 123.1, 118.9, 66.3, 34.9, 6.4.
2.3. Synthesis of 2-(4-chloro-3-nitro-phenyl)-2-methyl-6,8-dinitro-2,3-dihydro-1H-quinazolin-4-one (3b)
As stated in [55], 3b was prepared from a mixture of 1 (6.8 g, 50 mmol) and 4-chloro acetophenone (13.0 mL, 100 mmol) by utilizing general synthesis procedure as yellow crystalline solid. (95.8% yield); m.p. 200–202 °C; CCDC 1508840; 1H NMR (400 MHz, CDCl3, 25 °C): δ = 9.21 (d, 3J = 8 Hz, 1H, ArH), 9.07 (d, 3J = 8 Hz, 1H, ArH), 8.36 (s, 1H, NH), 8.14 (s, 1H, ArH), 7.63 (d, 3J = 8 Hz, 1H, ArH), 7.57 (d, 3J = 8 Hz, 1H, ArH), 6.45 (s, 1H, NH), 2.12 (s, 3H, CH3) ppm. 13C NMR (100 MHz, CDCl3, 25 °C), δ = 168.2, 149.1, 143.5, 141.6, 138.4, 134.5, 133.6, 129.8, 129.2, 127.1, 124.6, 123.0, 119.6, 66.8, 30.4.
2.4. X-rays Crystallographic Study
Suitable crystals of the synthesized compounds were obtained by slowly evaporating a solution of individual compounds in ethyl acetate/DCM/methanol (2:1:1). Bruker KAPPA Apex II diffractometer having graphite-monochromatized Mo Ka radiation, λMo = 0.710 73 Å at 100 K, was used to analyze crystals. Data reduction and structure refinement were achieved using SAINT and SHELXL-2013 program packages. Material for publication was prepared by using PLATON software [56,57]. Finalized crystal structure data was deposited with Cambridge Crystallographic Data Centre (CCDC).
2.5. Anti-Leishmanial Activity
The 3a and 3b compounds were screened for anti-leishmanial activity in an in vitro assay, where Amphotericin B deoxycholate and Miltefosine were used as the reference drugs [58]. In vitro inhibition of Leishmania growth was assessed using a 96-well growth plate assay by modifying s previous method [58]. The promastigotes of L. amazonensis (1 × 106 cells) were cultivated with various 3a and 3b concentrations for 48 h at 24 °C. Cell viability was observed by MTT assay [58]. The IC50 was determined using sigmoidal regression of the concentration-response curves, and results were obtained in triplicates.
2.6. Molecular Docking and Molecular Dynamic Simulations
Molecular docking was performed to map the ligand interactions with Leishmanial key protein involved in parasite viability. Protein structures for Pyridoxal Kinase and Trypanothione Reductase from Leishmania sp. were obtained from Protein Data Bank (PDB) as PDB IDs 6K92 and 6T98, respectively. The 3a and 3b ligands were optimized using the MM4 and screened against the proteins using a modified AutoDock-LGA module while applying AMBER03-FF in YASARA Structure version 20.7.4 (available at Dr. Fayyaz’s Computational Biology Lab) [59] as described earlier [60,61]. Binding energies were tabulated along with dissociation constants, and LigPlus [62] was used to find ligand-protein interactions. Molecular Dynamics (MD) were performed using AMBER14 as a force field in a 20 Å water-filled space around the protein-ligand complexes obtained from molecular docking. The same condition was used as described earlier [61,63,64] and 100 ns MD simulations were obtained. Prism GraphPad ver 7.0 [65] was used to analyze the data. MMPBSA calculations were employed by the binding energy module in YASARA using the following equation.
Binding Energy = EpotRecept + EsolvRecept + EpotLigand + EsolvLigand − EpotComplex − EsolvComplex
2.7. Determination of In Silico Pharmacokinetic Properties
The 3a and 3b ligands were screened for their pharmacokinetic potential and drug-likeness using the SwissADME server (http://www.swissadme.ch/, accessed on 20 September 2021).
3. Results and Discussions
Condensation of anthranilamide with ketone was achieved under Lewis acid catalysis to produce intermediate quinazoline moiety, which was further treated with a nitrating mixture. Resultantly, compounds (3a and 3b) were synthesized by reacting the unsubstituted phenyl ring of 2,3-dihydroquinazolin-4(1H)-one core with conc. HNO3 in the presence of H2SO4 (Figure 2).
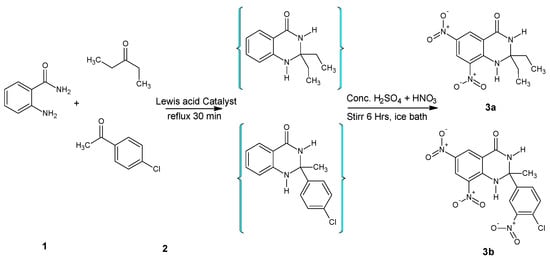
Figure 2.
Synthesis of 6,8-dinitro-2,3-dihydroquinazolin-4(1H)-ones.
3.1. NMR and XRD Study
1H NMR spectra of 6,8-dinitro-2,2-disubstituted-2,3-dihydroquinazolin-4(1H)-ones were elucidated with respect to corresponding chemical shift values, proton signals/peaks in aromatic or aliphatic regions, and their coupling constants. In 1H NMR spectra, two highly deshielded proton signals appeared in the range of 9.21–9.02 ppm (d, J = 8 Hz) due to aromatic protons of the pyrimidine phenyl ring. Two doublets (Jortho = 8 Hz) were assigned to protons of phenyl ring attached to C-2 of compound 3b. Two broad singlets ranging from 6.67–6.45 ppm and 8.52–8.36 ppm were interpreted for the –NH protons at position-1 and 3 of the quinazoline ring. 1H-NMR signals due to other protons of methyl and ethyl groups were assigned at different chemical shift values [26].
Structures of the synthesized compounds were confirmed by performing single-crystal X-ray diffraction (XRD) analysis by using suitable crystals of individual compounds grown in ethyl acetate. Ortep diagrams of compounds 3a and 3b are shown in Figure 3 and Figure 4, whereas crystal data is presented in Table 1 and Table 2, respectively.
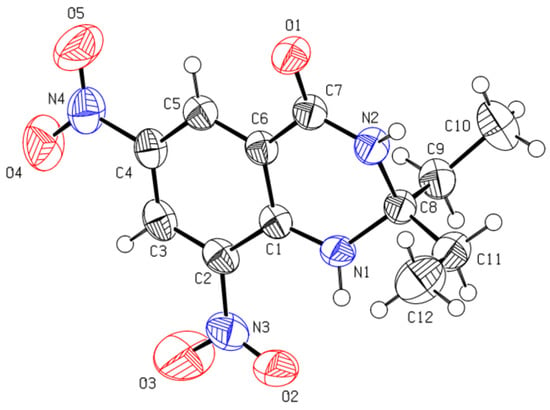
Figure 3.
Ortep diagram of compound 3a (CCDC No. 1568517).
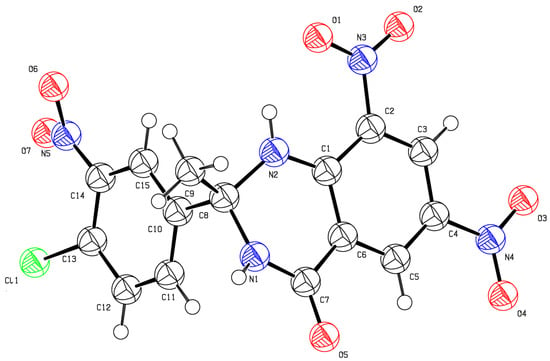
Figure 4.
Ortep diagram of compound 3b (CCDC No. 1508840).

Table 1.
Crystal structure data of the compound 6,8-dinitro-2,2-diethyl-2,3-dihydroquinazolin-4(1H)-one (3a).

Table 2.
Crystal structure data of the compound (2-(4-chloro-3-nitro-phenyl)-2-methyl-6,8-dinitro-2,3-dihydro-1H-quinazolin-4-one (3b).
H-bonding presents in the 3a, and 3b has been shown by the crystal (Figure 5). In the case of 3b, the intramolecular hydrogen bonding was present between oxygen in the phenyl ring (position 8) and H-N1. The intermolecular H-bonding is also observed between the oxygen of –C=O and H-N3 (Figure 5).
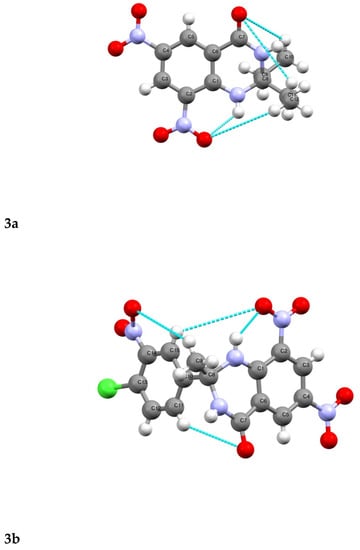
Figure 5.
Hydrogen bonding in 3a and 3b (colored lines show H-bonding).
3.2. Docking Studies, MMPBSA Calculations and MD Simaultions
Both ligands 3a and 3b were found to interact with Pyridoxal Kinase (PDK) and Trypanothione Reductase (TPR) from Leishmania sp. PDK is involved in phosphorylation of B6 vitamers into pyridoxal-5-phosphate (PLP) [66]. PDK is an interesting target for anti-leishmanial activity as it is involved in drug resistance especially Miltefosine [67].
3a interacts with Pyridoxal Kinase with a binding energy of −9.84 kcal/mol and a dissociation constant of 60.90 nM, while 1b interacts with Pyridoxal Kinase with a binding energy of −8.07 kcal/mol and dissociation constant 1.21 µM (Table 3).

Table 3.
Docking of the 3a and 3b against novel targets of Leishmania proteins.
The best binding site for the 3a ligand includes Val19, Asp119, Val121, Asp124, Tyr129, Asn151, Tyr152, Glu154, Lys187, Ser188, Leu198, Val219, Tyr226, Thr227, Gly228, Thr229, Gly230, Asp231, Phe233, Met254, Leu257, and Gln258. Here, the binding site residues contain 228GTGD231 motif and Gln258, which are conserved key residues in PDK [68]. For 3b, the binding site overlapped the 3a active site and consisted of amino acids including, Asn151, Lys187, Ser188, Leu198, Val219, Pro220, Tyr221, His222, Tyr226, Thr229, Gly230, Phe233, Met254, Leu257, Gln258, and Ile261 (Figure 6). The 3b ligand contains 228GTGD231 motif residues and conserved Gln258 and Ile261 residues. Ser188, another conserved residue present in the binding site of both ligands, is involved in the α or β phosphate binding in ATP [68]. MM/PBSA energy calculations using YASARA Structure show a free energy of −94 and −113 KJ/mol for 3a and 3b complex with PDK (Table 3).
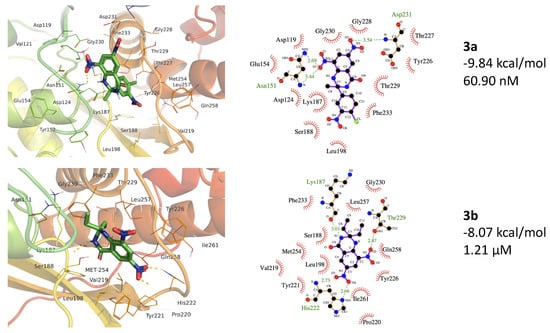
Figure 6.
The 3a and 3b binding to the protein structure of Pyridoxal Kinase (PDB ID 6K92). The top panels show 3a forms H-bonds with Asn151 (2.69 and 3.44 Å) and Asp231 (3.44 Å). The 3b forms H-bonds with Lys187 (3.01 Å), His222 (2.66 and 2.73 Å), and Thr22 (2.87 Å). (Hydrogen bonding is shown by light green color dotted lines).
In a previous study, a total of 5587 ligands were screened against PDK, and more than 1000 compounds were found to have binding properties to the enzyme [69]. A ligand, DNDI1103666, was ranked highest in enzyme binding. This ligand shares the enzyme binding site with 3a and 3b ligands in this study. The binding site overlapping residues include the Val19, Asp124, Asn151, Tyr152, Lys187, Ser188, His222, and Tyr226 [69].
The MD simulations in the case of 3a (active site binding) show that equilibrium was achieved after 60 ns (Figure 7). Compared with the native receptor, the 3a protein complex shows a lot of fluctuations while calculating RMSD-Cα. In the case of 3a, fluctuations in the RMSD-Cα are observed throughout the protein while these were higher in the region of amino acids 9–54 (Figure 7), while four different regions show fluctuations in the RMSD and RMSF values. These fluctuations also confirm ligand interactions with the protein receptor. These regions include the amino acids 14–34, 44–54, 95–115, 225–232, and 255–282 (Figure 8). This confirms the ligand interactions with the receptor.
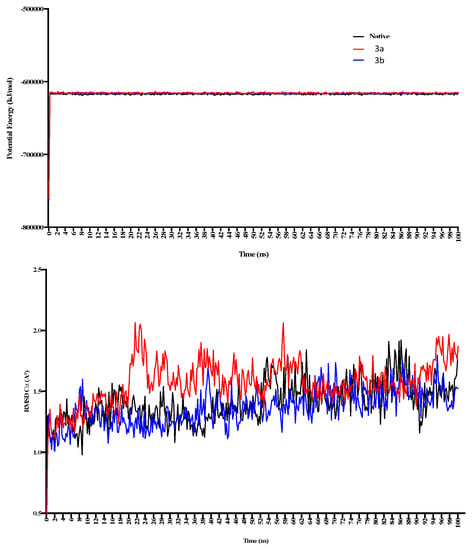
Figure 7.
The 100 ns MD simulations for 3a and 3b show potential energy in the top panel, while RMSD-Cα is shown in the bottom panel. 3a shows comparatively more significant fluctuations in the complex form during 20–50 ns of simulation.
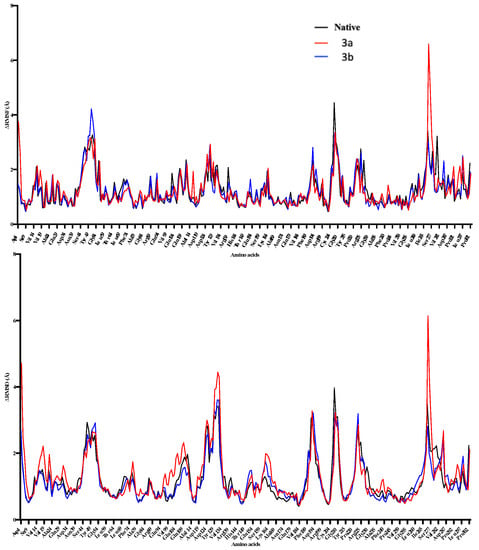
Figure 8.
The 100 ns MD simulations for 3a and 3b, where potential energy has been shown in the top panel, while RMSD-Cα is shown in the bottom panel. 3a shows comparatively more significant fluctuations in the complex form during 20–50 ns of simulation.
In the case of 3b, the calculated potential energy is comparatively equal to the native receptor and 3a. RMSD-Cα was not found to fluctuate as in the case of the 3a-receptor complex (Figure 6). RMSD and RMSF were found changing in the regions of 50–59, 149–169, 225–245, and 254–260 (Figure 8). The region consisting of 225–231 consists of 228GTGD231 motif, and fluctuation in this part of receptor show ligand interactions with these particular amino acids.
Trypanothione Reductase (TPR) maintains the redox balance and converts trypanothione disulfide to the reduced trypanothione dithiol. Anti-leishmanial agents like artonin B have shown strong enzyme inhibition in the in silico studies [70]. The 3a has shown very strong interactions with TPR showing binding energy of −11.35 kcal/mol and dissociation constant of 4.78 nM (Table 2). The binding site includes the amino acids, Leu10, Gly11, Val34, Asp35, Val36, Gly125, Phe126, Gly127, Glu141, Thr160, Trp163, Pro289, Arg290, Ser291, Gln292, Ala293, and Leu294 (Figure 8).
Ligand 3b also shows TPR binding with comparatively less affinity than 3a. The binding energy for 3b was −8.75 kcal/mol, and the dissociation constant was calculated as 383.72 nM (Table 3). The 3b ligand binding site occupies a different binding site than 3a consisting of Gly11, Gly13, Ser14, Gly15, Gly16, Ala46, Gly49, Gly50, Thr51, Cys52, Val53, Ala159, Thr160, Gly161, Ser162, Arg287, Arg290, Gly326, Asp327, Leu334, Thr335, and Ala338 (Figure 9). Here, Cys52 is a catalytic residue, while the FAD-binding domain consists of 1–160 residues, the NADPH-binding domain compasses 161 to 288 amino acids [71]. Aryl-substituted imidazole has been found to bind to the same binding site in TPR as 3a and 3b, where one ligand showed binding energy of −11.30 kcal/mol and dissociation of 5.19 nM [72]. Chromene-2-thione analogs are also found potent agents while inhibiting TPR with binding affinities of −9.20 to −6.82 kcal/mol [73].
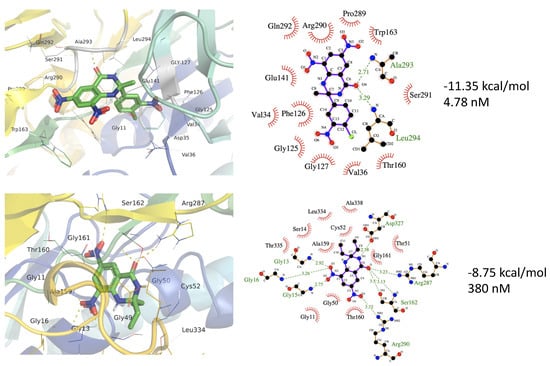
Figure 9.
The 3a and 3b binding to the protein structure of Trypanothione Reductase (PDB ID 6T98). The top panels show 3a forms H-bonds with Ala293 (2.71 Å) and Leu294 (3.29 Å). The 3b forms H-bonds with Gly15 (2.75 Å), Gly16 (3.26 Å), Ser162 (3.5 and 3.13 Å), Arg287 (2.23 Å), and Arg290 (3.52 Å). (Hydrogen bonding is shown by light green color dotted lines).
In a previous study, 600,000 ZINC compounds were screened against leishmanial trypanothione reductase 20 potential ligands with binding energies of −10.27 to −5.29 kcal/mol were reported [71]. Fucosterol was found to bind trypanothione reductase with a binding energy of −8.0 kcal/mol. MM/PBSA energy calculation using YASARA Structure shows a free energy of −32 and −61 KJ/mol for 3a and 3b complex with PDK (Table 3).
The ligand-protein complexes for 3a, 3b, and native proteins show a comparatively same potential energy, whereas the 3a complex shows the least fluctuations for RMSD-Cα (Figure 10). 3b protein complex shows higher RMSD-Cα changes throughout the 100 ns simulations. Both complexes show RMSD and RMSF changes in the 72–94 amino acids region. However, the active sites of both ligands don’t overlap, and the primary binding site residues don’t include these amino acids. This shows that ligand binding might be affecting this particular region in the enzyme. 3b shows higher fluctuations in RMSF in the 324–335 amino acid region showing ligand interactions with the part of the enzyme binding site (Figure 11). This also indicates that ligands 3a and 3b binding induce some conformational changes in the enzyme.
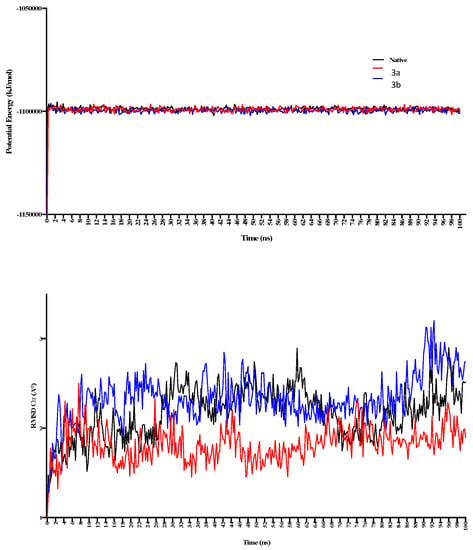
Figure 10.
The 100 ns MD simulations for 3a and 3b, where potential energy has been shown in the top panel, while RMSD-Cα is shown in the bottom panel. 3a shows comparatively larger fluctuations in the complex during 20–50 ns of simulations.
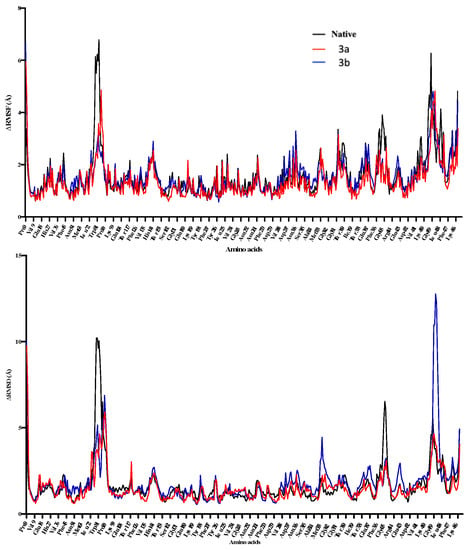
Figure 11.
The 3a and 3b binding to the protein structure of Pyridoxal Kinase (PDB ID 6K92). The top panels show 3a forms H-bonds with Asn151 (2.69 and 3.44 Å) and Asp231 (3.44 Å). The 3b forms H-bonds with Lys187 (3.01 Å), His222 (2.66 and 2.73 Å), and Thr22 (2.87 Å). (Hydrogen bonding is shown by light green color dotted lines).
3.3. Anti-Leishmanial Activity
Depending upon the IC50 values, anti-leishmanial activities of 3a and 3b were calculated by MTT assay using the L. amazonensis promastigotes. The intracellular macrophage susceptibility assay shows 3a and 3b having the IC50 values of 1.164 and 0.085 µg/mL, respectively (Table 4). The standard drugs show the IC50 values of 0.046 and 3.191 µg/mL.

Table 4.
Anti-leishmanial activity of the 3a and 3b ligands.
The ADMET properties analyses show that 3a and 3b possess good physicochemical characteristics and can be considered as potential drug candidates (Table 5).

Table 5.
Pharmacokinetics properties of the 3a and 3b.
4. Conclusions
The ligands 3a and 3b, derivatives of 2,3-dihydroquinazolin-4(1H)-one, have been found to show promising in silico and in vitro anti-leishmanial activities. Both ligands have shown a strong comparative binding to Pyridoxal Kinase and Trypanothione Reductase from Leishmania sp. by in silico studies. Some in vivo studies are further required to explore the potential of the ligands 3a and 3b as non-toxic potent candidates for their use as leishmanial agents.
Author Contributions
Conceptualization, M.S., M.I.T. and S.X.; methodology, M.S., N.S., F.K. and S.A.; formal analysis, M.S., S.A., M.F.u.R. and M.J.; investigation, M.S., M.F.u.R., H.E., C.W. and F.K.; resources, M.F.u.R., M.I.T. and S.X.; writing—original draft preparation, M.S. and M.F.u.R.; writing—review and editing, F.K., S.X., M.J., C.W. and M.F.u.R.; supervision, M.I.T., N.S. and M.F.u.R.; funding acquisition, S.X. and F.K.; computational methods including docking and MD simulations, M.F.u.R. and S.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Renslo, A.; McKerrow, J.H. Drug discovery and development for neglected parasitic diseases. Nat. Chem. Biol. 2006, 2, 701–710. [Google Scholar] [CrossRef] [PubMed]
- Akopyants, N.S.K.N.; Secundino, N.; Patrick, R.; Peters, N.; Lawyer, P.; Dobson, D.E.; Beverley, S.M.; Sacks, D.L. Demonstration of Genetic Exchange During Cyclical Development of Leishmania in the Sand Fly Vector. Science 2009, 324, 265–268. [Google Scholar] [CrossRef] [PubMed]
- Desjeux, P. Leishmaniasis: Public health aspects and control. Clin. Dermatol. 1996, 14, 417–423. [Google Scholar] [CrossRef]
- WHO World Health Organization Zoonotic Disease: Emerging Public Health Threats in the Region. Available online: http://www.emro.who.int/about-who/rc61/zoonotic-diseases.html (accessed on 18 November 2021).
- Nascimento, M.M.; Queiroz, J.W.; Barroso, A.W.; Araujo, A.F.; Rego, E.F.; Wilson, M.E.; Pearson, R.D.; Jeronimo, S.M. The emergence of concurrent HIV-1/AIDS and visceral leishmaniasis in Northeast Brazil. Trans. R. Soc. Trop. Med. Hyg. 2011, 105, 298–300. [Google Scholar] [CrossRef]
- Aronson, N.; Herwaldt, B.L.; Libman, M.; Pearson, R.; Lopez-Velez, R.; Weina, P.; Carvalho, E.; Ephros, M.; Jeronimo, S.; Magill, A. Diagnosis and Treatment of Leishmaniasis: Clinical Practice Guidelines by the Infectious Diseases Society of America (IDSA) and the American Society of Tropical Medicine and Hygiene (ASTMH). Am. J. Trop. Med. Hyg. 2017, 96, 24–45. [Google Scholar] [CrossRef]
- Leon, L.L.; Carvalho-Paes, L.E.; Grimaldi, J.G. Antigenic differences of Leishmania amazonensis isolate causing diffuse cutaneous leishmaniasis. Trans. R. Soc. Trop. Med. Hyg. 1990, 84, 678–680. [Google Scholar] [CrossRef]
- Chappuis, F.S.; Hailu, A.; Ghalib, H.; Rijal, S.; Peeling, R.W.; Alvar, J.; Boelaert, M. Visceral leishmaniasis: What are the needs for diagnosis, treatment and control? Nat. Rev. Microbiol. 2007, 5, S7–S16. [Google Scholar] [CrossRef]
- Pearson, R.D.S. Clinical spectrum of Leishmaniasis. Clin. Infect. Dis. 1996, 22, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Modabber, F. Leishmaniasis vaccines: Past, present and future. Int. J. Antimicrob. Agents 2010, 36, S58–S61. [Google Scholar] [CrossRef]
- Wise, E.S.; Armstrong, M.; Watson, J.; Lockwood, D.N.J. Monitoring Toxicity Associated with Parenteral Sodium Stibogluconate in the Day-Case Management of Returned Travellers with New World Cutaneous Leishmaniasi. PLoS Negl. Trop. Dis. 2012, 6, e1688. [Google Scholar] [CrossRef]
- Sundar, S.; More, D.K.; Singh, M.K.; Singh, V.P.; Sharma, S.; Makharia, A.; Kumar, P.C.K.; Murray, H.W. Failure of Pentavalent Antimony in Visceral Leishmaniasis in India: Report from the Center of the Indian Epidemic. Clin. Infect. Dis. 2000, 31, 1104–1107. [Google Scholar] [CrossRef]
- Singh, N.; Kumar, M.; Singh, R.K. Leishmaniasis: Current status of available drugs and new potential drug targets. Asian Pac. J. Trop. Med. 2012, 5, 485–497. [Google Scholar] [CrossRef]
- Bray, P.G.; Barrett, M.; Ward, S.; de Koning, H.P. Pentamidine uptake and resistance in pathogenic protozoa: Past, present and future. Trends Parasitol. 2003, 19, 232–239. [Google Scholar] [CrossRef]
- Torres-Guerrero, E.; Ruiz-Esmenjaud, J.; Arenas, R. Leishmaniasis: A review. F1000Res 2017, 6, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Birhan, Y.S.; Bekhit, A.A.; Hymete, A. Synthesis and antileishmanial evaluation of some 2,3-disubstituted-4(3H)-quinazolinone derivatives. Org. Med. Chem. Lett. 2014, 4, 10. [Google Scholar] [CrossRef][Green Version]
- Bonano, V.I.Y.-Y.; Miguel, D.C.; Jones, S.A.; Dodge, J.A.; Uliana, S.R.B. Discovery of Synthetic Leishmania Inhibitors by Screening of a 2-Arylbenzothiophene Library. Chem. Biol. Drug Des. 2014, 81, 658–665. [Google Scholar]
- Coimbra, E.S.A.; Silva, A.D.; Bispo, M.L.F.; Kaiser, C.R.; De Souza, M.V.N. 7-Chloro-4-quinolinyl hydrazones: A promising and potent class of antileishmanial compounds. Chem. Biol. Drug. Des. 2013, 81, 658–665. [Google Scholar] [CrossRef]
- Gómez-Pérez, V.; Manzano, J.I.; García-Hernández, R.; Castanys, S.; Rosa, J.M.C.; Gamarro, F. 4-Amino Bis-Pyridinium Derivatives as Novel Antileishmanial Agents. Antimicrob. Agents Chemother. 2014, 58, 4103–4112. [Google Scholar] [CrossRef]
- Gellis, A.D.; Lanzada, G.; Hutter, S.; Ollivier, E.; Vanelle, P.; Azas, N. Preparation and antiprotozoal evaluation of promising β-carboline alkaloids. Biomed. Pharmacother. 2012, 66, 339–347. [Google Scholar] [CrossRef]
- Machado, P.A.H.; Carvalho, L.O.; Silveira, M.L.T.; Alves, R.B.; Freitas, R.P.; Coimbra, E.S. Effect of 3-Alkylpyridine marine alkaloid analogues in Leishmania species related to American cutaneous lLeishmaniasis. Chem. Biol. Drug Des. 2012, 80, 745–751. [Google Scholar] [CrossRef]
- Roatt, B.M.; De Brito, R.C.F.; Coura-Vital, W.; de Oliveira Aguiar-Soares, R.D.; Reis, A.B. Recent advances and new strategies on leishmaniasis treatment. Appl. Microbiol. Biotechnol. 2020, 104, 8965–8977. [Google Scholar] [CrossRef] [PubMed]
- Tiuman, T.S.; Santos, A.O.; Ueda-Nakamura, T.; Filho, B.P.D.; Nakamura, C.V. Recent advances in leishmaniasis treatment. Int. J. Infect. Dis. 2011, 15, e525–e532. [Google Scholar] [CrossRef] [PubMed]
- Ramsay, R.R.; Tipton, K.F. Assessment of Enzyme Inhibition: A Review with Examples from the Development of Monoamine Oxidase and Cholinesterase Inhibitory Drugs. Molecules 2017, 22, 1192. [Google Scholar] [CrossRef]
- Balbaa, M.; El Ashry, E.S.H. Enzyme Inhibitors as Therapeutic Tools. Biochem. Physiol. Open Access 2012, 1, 13. [Google Scholar] [CrossRef]
- Akram, M.S.; Pery, N.; Butler, L.; Shafiq, M.I.; Batool, N.; Rehman, M.F.U.; Grahame-Dunn, L.G.; Yetisen, A.K. Challenges for biosimilars: Focus on rheumatoid arthritis. Crit. Rev. Biotechnol. 2021, 41, 121–153. [Google Scholar] [CrossRef]
- Sarfraz, M.; Tariq, M.I. Synthesis, in silico study and cholinesterases inhibition activity of 2-substituted 2,3-dihydroquinazolin-4(1H)-one derivatives. Rev. Roum. Chim. 2018, 63, 227–234. [Google Scholar]
- Tyagi, R.; Elfawal, M.A.; Wildman, S.A.; Helander, J.; Bulman, C.A.; Sakanari, J.; Rosa, B.A.; Brindley, P.J.; Janetka, J.W.; Aroian, R.V.; et al. Identification of small molecule enzyme inhibitors as broad-spectrum anthelmintics. Sci. Rep. 2019, 9, 9085. [Google Scholar] [CrossRef]
- Sultana, N.; Sarfraz, M.; Tanoli, S.T.; Akram, M.S.; Sadiq, A.; Rashid, U.; Tariq, M.I. Synthesis, crystal structure determination, biological screening and docking studies of N1-substituted derivatives of 2,3-dihydroquinazolin-4(1H)-one as inhibitors of cholinesterase. Bioorg. Chem. 2017, 72, 256–267. [Google Scholar] [CrossRef]
- Sarfraz, M.; Rashid, U.; Sultana, N.; Tariq, M.I. Synthesis, X-Rays Analysis, Docking Study and Cholinesterase Inhibition Activity of 2, 3-dihydroquinazolin-4 (1H)-one Derivatives. Iran. J. Chem. Chem. Eng. 2019, 38, 213–227. [Google Scholar]
- El-Azab, A.S.E.; Eltahir, K.E.H. Design and synthesis of novel 7-aminoquinazoline derivatives: Antitumor and anticonvulsant activities. Bioorg. Med. Chem. Lett. 2012, 22, 1879–1885. [Google Scholar] [CrossRef]
- Saravanan, G.A.; Prakash, C.R. Design, synthesis and anticonvulsant activities of novel 1-(substituted/unsubstituted benzylidene)-4-(4-(6,8-dibromo-2-(methyl/ phenyl)-4-oxoquinazolin-3(4H)-yl)phenyl) semicarbazide derivatives. Bioorg. Med. Chem. Lett. 2012, 22, 3072–3078. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.W.; Wang, M.; Yao, X.; Li, Y.; Tan, J.; Wang, L.; Qiao, W.; Geng, Y.; Liu, Y.; Wang, Q. Design, synthesis and antiviral activity of novel quinazolinones. Eur. J. Med. Chem. 2012, 53, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Abbas, S.E.; Awadallah, F.; Ibrahin, N.A.; Said, E.; Kamel, G. New quinazolinone–pyrimidine hybrids: Synthesis, anti-inflammatory, and ulcerogenicity studies. Eur. J. Med. Chem. 2012, 53, 141–149. [Google Scholar] [CrossRef]
- Al-Omary, F.A.; Abou-Zeid, L.A.; Nagi, M.N.; Habib, E.-S.E.; Abdel-Aziz, A.A.-M.; El-Azab, A.S.; Abdel-Hamide, S.G.; Al-Omar, M.A.; Al-Obaid, A.M.; El-Subbagh, H.I. Non-classical antifolates. Part 2: Synthesis, biological evaluation, and molecular modeling study of some new 2,6-substituted-quinazolin-4-ones. Bioorg. Med. Chem. 2010, 18, 2849–2863. [Google Scholar] [CrossRef] [PubMed]
- Amin, K.; Kamel, M.; Anwar, M.; Khedr, M.; Syam, Y. Synthesis, biological evaluation and molecular docking of novel series of spiro [(2H,3H) quinazoline-2,1′- cyclohexan]-4(1H)- one derivatives as anti-inflammatory and analgesic agents. Eur. J. Med. Chem. 2010, 45, 2117–2131. [Google Scholar] [CrossRef]
- Mohameda, M.S.K.M.; Kassem, E.M.M.; Abotaleb, N.; AbdEl-moez, S.I.; Ahmeda, M.F. Novel 6,8-dibromo-4(3H)-quinazolinone derivatives of anti-bacterial and anti-fungal activities. Eur. J. Med. Chem. 2010, 45, 3311–3319. [Google Scholar] [CrossRef]
- Gemma, S.C.; Brindisi, M.; Brogi, S.; Kukreja, G.; Kunjir, S.; Gabellieri, E.; Lucantoni, L.; Habluetzel, A.; Taramelli, D.; Basilico, N.; et al. Mimicking the Intramolecular Hydrogen Bond: Synthesis, Biological Evaluation, and Molecular Modeling of Benzoxazines and Quinazolines as Potential Antimalarial Agents. J. Med. Chem. 2012, 55, 10387–10404. [Google Scholar] [CrossRef]
- Rudolph, J.; O’connor, S.; Coish, P.D.; Wickens, P.L.; Brands, M.; Bierer, D.E.; Bloomquist, B.T.; Bondar, G.; Chen, L.; Chuang, C.Y.; et al. Quinazolinone derivatives as orally available ghrelin receptor antagonists for the treatment of diabetes and obesity. J. Med. Chem. 2007, 50, 5202–5216. [Google Scholar] [CrossRef]
- Rhee, H.-K.; Yoo, J.H.; Lee, E.; Kwon, Y.J.; Seo, H.-R.; Lee, Y.-S.; Choo, H.-Y.P. Synthesis and cytotoxicity of 2-phenylquinazolin-4(3H)-one derivatives. Eur. J. Med. Chem. 2011, 46, 3900–3908. [Google Scholar] [CrossRef]
- Radwan, A.A.; Alanazi, F.K. Biological Activity of Quinazolinones, Quinazolinone and Quinazoline Derivatives; IntechOpen: London, UK, 2020. [Google Scholar]
- Faisal, M.; Saeed, A. Chemical Insights into the Synthetic Chemistry of Quinazolines: Recent Advances. Front. Chem. 2021, 8, 594717. [Google Scholar] [CrossRef]
- Agarwal, K.; Sharma, V.; Shakya, N.; Gupta, S. Design and synthesis of novel substituted quinazoline derivatives as antileishmanial agents. Bioorg. Med. Chem. Lett. 2009, 19, 5474–5477. [Google Scholar] [CrossRef] [PubMed]
- Fleita, D.H.; Mohareb, R.M.; Sakka, O.K. Antitumor and antileishmanial evaluation of novel heterocycles derived from quinazoline scaffold: A molecular modeling approach. Med. Chem. Res. 2013, 22, 2207–2221. [Google Scholar] [CrossRef]
- Tesfahunegn, R.G. Synthesis and Anti-leishmanial Activity Evaluation of Some 2, 3-Disubstituted Quinazoline-4(3H)-Ones Bearing Quinoline and Pyrazole Moieties. EC Pharma. Sci. 2015, 1, 153–164. [Google Scholar]
- Shaterian, H.R.; Ali Reza, O.; Moones, H. Synthesis of 2,3-Dihydroquinazoline-4(1H)-ones. Synth. Commun. 2010, 40, 1231–1242. [Google Scholar] [CrossRef]
- Khattab, S.N.; Haiba, N.S.; Asal, A.M.; Bekhit, A.A.; Guemei, A.A.; Amer, A.; El-Faham, A. Study of antileishmanial activity of 2-aminobenzoyl amino acid hydrazides and their quinazoline derivatives. Bioorg. Med. Chem. Lett. 2017, 27, 918–921. [Google Scholar] [CrossRef]
- Asif, M. Chemical Characteristics, Synthetic Methods, and Biological Potential of Quinazoline and Quinazolinone Derivatives. Int. J. Med. Chem. 2014, 2014, 1–27. [Google Scholar] [CrossRef]
- Badolato, M.; Aiello, F.; Neamati, N. 2,3-Dihydroquinazolin-4(1H)-one as a privileged scaffold in drug design. RSC Adv. 2018, 8, 20894–20921. [Google Scholar] [CrossRef]
- Van Horn, K.S.; Zhu, X.; Pandharkar, T.; Yang, S.; Vesely, B.; Vanaerschot, M.; Dujardin, J.-C.; Rijal, S.; Kyle, D.E.; Wang, M.Z.; et al. Antileishmanial Activity of a Series of N2,N4-Disubstituted Quinazoline-2,4-diamines. J. Med. Chem. 2014, 57, 5141–5156. [Google Scholar] [CrossRef]
- Prinsloo, I.F.; Zuma, N.H.; Aucamp, J.; N’Da, D.D. Synthesis and in vitro antileishmanial efficacy of novel quinazolinone derivatives. Chem. Biol. Drug Des. 2021, 97, 383–398. [Google Scholar] [CrossRef]
- Kshirsagar, U.A. Recent developments in the chemistry of quinazolinone alkaloids. Org. Biomol. Chem. 2015, 13, 9336–9352. [Google Scholar] [CrossRef]
- Prashant, S.A.; Gatish, T.P.G. Recent advances in the pharmacological diversification of quinazoline/quinazolinone hybrids. RSC Adv. 2020, 10, 41353–41392. [Google Scholar]
- Nayyar, P.; Arpana, R.; Mohd, I. An updated review: Newer quinazoline derivatives under clinical trial. Int. J. Pharm. Biol. Arch. 2011, 2, 1651–1657. [Google Scholar]
- Sarfraz, M.; Sultana, N.; Rashid, U.; Akram, M.S.; Sadiq, A.; Tariq, M.I. Synthesis, biological evaluation and docking studies of 2,3-dihydroquinazolin-4(1H)-one derivatives as inhibitors of cholinesterases. Bioorg. Chem. 2017, 70, 237–244. [Google Scholar] [CrossRef] [PubMed]
- Jamil, M.; Sultana, N.; Ashraf, R.; Bashir, M.; Rehman, M.; Kanwal, F.; Ellahi, H.; Lu, C.; Zhang, W.; Tariq, M. Bis (Diamines) Cu and Zn Complexes of Flurbiprofen as Potential Cholinesterase Inhibitors: In Vitro Studies and Docking Simulations. Crystals 2021, 11, 208. [Google Scholar] [CrossRef]
- Tariq, M.I.; Noreen, T.; Tahir, M.N.; Ahmad, S.; Fayyaz-ur-Rehman, M. 2-(2,3-Dimethylphenyl)-1H-isoindole-1,3 (2H)-dione. Acta Cryst. Sect. E Struct. Rep. Online 2010, 66, 2440. [Google Scholar] [CrossRef]
- Lavorato, S.N.; Duarte, M.C.; De Andrade, P.H.R.; Coelho, E.A.F.; Alves, R.J. Synthesis, antileishmanial activity and QSAR studies of 2-chloro- N -arylacetamides. Braz. J. Pharm. Sci. 2017, 53, 16067. [Google Scholar] [CrossRef]
- Krieger, E.; Vriend, G. YASARA View—Molecular graphics for all devices—From smartphones to workstations. Bioinformatics 2014, 30, 2981–2982. [Google Scholar] [CrossRef]
- Bilal, S.; Hassan, M.M.; Rehman, M.F.; Nasir, M.; Sami, A.J.; Hayat, A. An insect acetylcholinesterase biosensor utilizing WO3/g-C3N4 nanocomposite modified pencil graphite electrode for phosmet detection in stored grains. Food Chem. 2021, 346, 128894. [Google Scholar] [CrossRef]
- Rehman, M.F.U.; Akhter, S.; Batool, A.I.; Selamoglu, Z.; Sevindik, M.; Eman, R.; Mustaqeem, M.; Akram, M.S.; Kanwal, F.; Lu, C.; et al. Effectiveness of Natural Antioxidants against SARS-CoV-2? Insights from the In-Silico World. Antibiotics 2021, 10, 1011. [Google Scholar] [CrossRef]
- Laskowski, R.A.; Swindells, M.B. LigPlot+: Multiple ligand-protein interaction diagrams for drug discovery. Chem. Inf. Model. 2011, 51, 2778–2786. [Google Scholar] [CrossRef]
- Fernandez-Poza, S.; Padros, A.; Thompson, R.; Butler, L.; Islam, M.; Mosely, J.; Scrivens, J.H.; Rehman, M.F.; Akram, M.S. Tailor-made recombinant prokaryotic lectins for characterisation of glycoproteins. Anal. Chim. Acta 2021, 1155, 338352. [Google Scholar] [CrossRef]
- Bilal, S.; Sami, A.J.; Hayat, A.; Rehman, M.F.U. Assessment of pesticide induced inhibition of Apis mellifera (honeybee) acetylcholinesterase by means of N-doped carbon dots/BSA nanocomposite modified electrochemical biosensor. Bioelectrochemistry 2021, 144, 107999. [Google Scholar] [CrossRef] [PubMed]
- Motulsky, H. Prism 4 Statistics Guide—Statistical Analyses for Laboratory and Clinical Researchers; GraphPad Software Inc.: San Diego, CA, USA, 2003; pp. 122–126. [Google Scholar]
- Fitzpatrick, T.B.; Amrhein, N.; Kappes, B.; Macheroux, P.; Tews, I.; Raschle, T. Two independent routes of de novo vitamin B6 biosynthesis: Not that different after all. Biochem. J. 2007, 407, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Coelho, A.C.; Boisvert, S.; Mukherjee, A.; Leprohon, P.; Corbeil, J.; Ouellette, M. Multiple Mutations in Heterogeneous Miltefosine-Resistant Leishmania major Population as Determined by Whole Genome Sequencing. PLoS Negl. Trop. Dis. 2012, 6, e1512. [Google Scholar] [CrossRef] [PubMed]
- Are, S.; Gatreddi, S.; Jakkula, P.; Qureshi, I.A. Structural attributes and substrate specificity of pyridoxal kinase from Leishmania donovani. Int. J. Biol. Macromol. 2020, 152, 812–827. [Google Scholar] [CrossRef]
- Alfadhel, S. Virtual screening of Leishmanial pyridoxal kinase enzyme inhibitors by repurposed anti-Trypanosomal libraries reveals two core scaffolds. Latin Am. J. Pharm. 2021, 40, 69–75. [Google Scholar]
- Battista, T.; Colotti, G.; Ilari, A.; Fiorillo, A. Targeting Trypanothione Reductase, a Key Enzyme in the Redox Trypanosomatid Metabolism, to Develop New Drugs against Leishmaniasis and Trypanosomiases. Molecules 2020, 25, 1924. [Google Scholar] [CrossRef]
- Matadamas-Martínez, F.; Hernández-Campos, A.; Téllez-Valencia, A.; Vázquez-Raygoza, A.; Comparán-Alarcón, S.; Yépez-Mulia, L.; Castillo, R. Leishmania mexicana Trypanothione Reductase Inhibitors: Computational and Biological Studies. Molecules 2019, 24, 3216. [Google Scholar] [CrossRef]
- Vargas, J.; López, A.; Vargas, A.; Fidalgo, L.; Froeyen, M. In Vitro Evaluation and Molecular Docking Studies of Aryl-Substituted Imidazoles against Leishmania Amazonensis. Int. J. Trop. Dis. 2021, 4, 50. [Google Scholar]
- Verma, R.K.; Prajapati, V.K.; Verma, G.K.; Chakraborty, D.; Sundar, S.; Rai, M.; Dubey, V.K.; Singh, M.S. Molecular Docking and In Vitro Antileishmanial Evaluation of Chromene-2-thione Analogues. ACS Med. Chem. Lett. 2012, 3, 243–247. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).