Abstract
Enzymes are powerful biological catalysts for natural substrates but they have low catalytic efficiency for non-natural substrates. Protein engineering can be used to optimize enzymes for catalysis and stability. 3α-Hydroxysteroid dehydrogenase/carbonyl reductase (3α-HSD/CR) catalyzes the oxidoreduction reaction of NAD+ with androsterone. Based on the structure and catalytic mechanism, we mutated the residues of T11, I13, D41, A70, and I112 and they interacted with different portions of NAD+ to switch cofactor specificity to biomimetic cofactor nicotinamide mononucleotide (NMN+). Compared to wild-type 3α-HSD/CR, the catalytic efficiency of these mutants for NAD+ decreased significantly except for the T11 mutants but changed slightly for NMN+ except for the A70K mutant. The A70K mutant increased the catalytic efficiency for NMN+ by 8.7-fold, concomitant with a significant decrease in NAD+ by 1.4 × 104-fold, resulting in 9.6 × 104-fold cofactor specificity switch toward NMN+ over NAD+. Meanwhile, the I112K variant increased the thermal stability and changed to a three-state transition from a two-state transition of thermal unfolding of wild-type 3α-HSD/CR by differential scanning fluorimetry. Molecular docking analysis indicated that mutations on these residues affect the position and conformation of the docked NAD+ and NMN+, thereby affecting their activity. A70K variant sterically blocks the binding with NAD+, restores the H-bonding interactions of catalytic residues of Y155 and K159 with NMN+, and enhances the catalytic efficiency for NMN+.
1. Introduction
Enzyme engineering is important for practical applications by increasing the thermal stability in the presence or absence of an organic solvent, enhancing the catalytic properties, and modifying the catalytic specificity for their cofactor or substrate, producing new products using alternate substrates [1,2]. Dehydrogenases utilizing NAD(P)+/NAD(P)H cofactor for oxidoreductive reactions with high efficiency, specificity, and enantioselectivity are used to perform various reactions in biotechnology [3]. To reduce the stoichiometric amount of NAD(P)H and its cost in the oxidative reaction, the cofactor is regenerated by a coupling reaction [4]. This constant supply of regenerated cofactor, in addition to the removal of the product, helps to push the reaction to completion. Meanwhile, the reversal of specificity from NADP+ to NAD+ is used for non-cell-based industrial applications due to the lower cost and better stability of NAD+ [5,6]. A computer program, CSR-SALAD (Coenzyme Specificity Reversal–Structural Analysis and Library Design), based on a cofactor-bound structure was designed to generate a mutant library to reverse coenzyme specificity from NADP+ toward NAD+ [7]. Furthermore, the replacement of natural cofactor with biomimetic nicotinamide cofactor, such as nicotinamide mononucleotide (NMN+), has been used to develop bioorthogonal redox systems in vivo without interfering with native biochemical processes [8]. The structure of NMN+ has a nicotinamide ring to participate in an oxidoreduction reaction but lacks the AMP portion of NAD+ (Figure 1). However, when alcohol dehydrogenase from Pyrococcus furiosus is engineered to act on NMN+, the enzyme activity remains low [9]. Other mimetics of NAD+ have been developed for the engineered enzymes to switch the cofactor specificity [10]. Enzymatic conversions performed with such biomimetic cofactors are far less efficient than those performed with native cofactors due to the significant differences in structures and sizes between natural and biomimetic cofactors. Recently, engineered glucose dehydrogenase through molecular modeling for target residues showed a 107-fold cofactor specificity switch toward NMN+ over NADP+ [11].
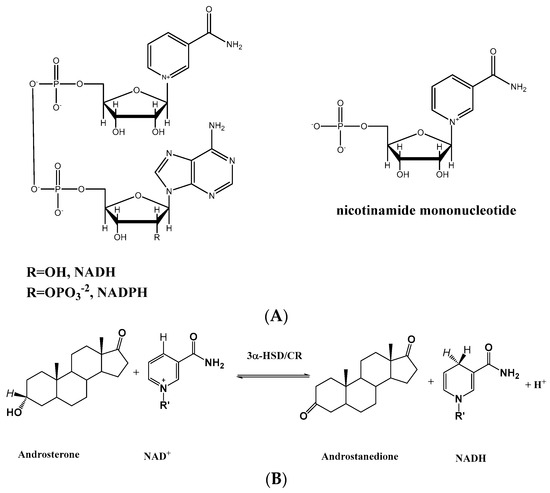
Figure 1.
(A) The chemical structures of NAD(P)+ and NMN+, and (B) the oxidoreductive reaction of androsterone with NAD+ catalyzed by 3α-HSD/CR to form androstandione and NADH.
Enzyme engineering can be performed using directed evolution, semi-rational design, and rational design to optimize enzymes for practical applications [12,13]. Directed evolution generates a mutant library by an error-prone PCR or gene shuffling for whole-gene randomization. The mutant library is then screened with increased stringency for improved properties via iterative rounds of mutagenesis in laboratory evolution [14,15]. Semi-rational design creates a mutant library by site-saturation mutagenesis at specific residues of the targeted enzymes identified by structure or computer modeling. These residues are replaced with any of the other 19 natural amino acids for screening and functional assays. The disadvantages of directed evolution and semi-rational design are that they are time-consuming and costly processes that require many rounds of mutagenesis and functional screening and selection to reach the target level of enzyme performance [16]. For example, engineering a bacterial phosphotriesterase (PTE) to an aryl carboxyesterase involves 18 rounds of mutagenesis and screening to improve 105-fold with a non-cognate aryl-ester substrate [17]. While rational design is based on the structures of enzyme–substrate complexes and related catalytic mechanisms, specific residues of the targeted enzymes are replaced by site-directed mutagenesis to obtain mutants with the desired properties.
Understanding how enzymes work is important for the rational design of a biocatalyst. 3α-HSD/CR from Comamonas testosteroni belonging to the short-chain dehydrogenase/reductase (SDR) superfamily catalyzes the stereospecific oxidoreductive reaction of androsterone with NAD+ to form androstanedione and NADH. Most SDR enzymes require NAD(H) or NADP(H) as a cofactor and are involved in the metabolism of steroid hormones, prostaglandins, retinoids, lipids, and xenobiotics [18,19]. The chemical mechanism of hydride transfer and the role of catalytic tetrad of Y155-K159-S114-N86 have been elucidated and characterized [20,21]. The catalytic efficiency of 3α-HSD/CR for androsterone is close to the diffusion rate, and the rate-limiting step is the release of NADH in the enzyme-catalyzed reaction [21,22,23]. Therefore, the evolution of 3α-HSD/CR may result in a reduction in activation energy and may promote the catalytic efficiency of 3α-HSD/CR for androsterone, close to a perfect enzyme. However, 3α-HSD/CR activity significantly decreased when NAD+ was replaced by a truncated biomimetic cofactor NMN+. The structure of 3α-HSD/CR is homodimer with a typical Rossmann fold consisting of βαβ units in binding with a nucleotide cofactor [24]. The NAD+ cofactor is bound at the C-terminus of the central parallel β-strands in the 3α-HSD/CR. Structural information on the NAD+ binding site provides a basis for the rational design of enzyme engineering for switching cofactor specificity. In this report, we present the rational design of an engineered 3α-HSD/CR to switch cofactor specificity toward NMN+ from NAD+. We plan to replace the residues that interact with NAD+ in the active site of 3α-HSD/CR to destabilize the AMP portion of NAD+ and stabilize the interactions with the pyrophosphate and ribose parts of NAD+. Single mutations of the T11A, T11K, T11R, I13A, I13K, I13R, D41I, D41Q, A70I, A70Q, A70K, I112A, and I112K 3α-HSD/CR variants were prepared in homogeneity and characterized kinetically. Among these mutants, the A70K mutant showed a 9.6 × 104-fold cofactor specificity switch toward NMN+ over NAD+. Meanwhile, the substitution of I112 with lysine significantly enhanced the thermal stability of 3α-HSD/CR as determined by differential scanning fluorimetry (DSF).
2. Results
2.1. Residue Selection of 3α-HSD/CR Engineering by Rational Design
We explored the structure of the E-NAD+ complex of 3α-HSD/CR (pdb:1fk8) for enzyme engineering to switch cofactor preference from NAD+ to the biomimetic cofactor of NMN+. The crystal structure of the binary complex of 3α-HSD/CR with NAD+, the residues within 4 Å of NAD+, and the residues of 3α-HSD/CR that interacted with NAD+ using the LigPlot program are shown in Figure 2. The residues closed to the various portions of NAD+ were the following: nicotinamide portion of NAD+: I112 and P185GAT188; the ribose of nicotinamide ribose: C69, S113, Y155, and K159; pyrophosphate: A10, G12, I13, and G14; ribose of adenosine: G8, A10, T11, D32 and G71; and adenine: I33, A40, D41, L42, A70, L72, and V85. Multiple sequence alignment of the short-chain dehydrogenase/reductase superfamily showed that the most conserved residues were G8, G12, G14, D41, A70, G71, N86, S114, Y155, K159, P185, P212, and P219 (Figure 3). The catalytic tetrad of Y155, K159, S114, and N86 plays an important role in catalysis [20,21,25], while the P185 and P212 in 3α-HSD/CR are situated at the hinge region of the loop and involved in the conformational flexibility of the substrate binding loop which appears as an unresolved region of T188-K208 for binding with androsterone [22]. These key residues are retained for their functions in androsterone binding and catalysis. Meanwhile, the conserved residues GXXXGXG (G8CATG12IG14 in 3α-HSD/CR) are the nicotinamide-coenzyme binding motif in the SDR family [26].
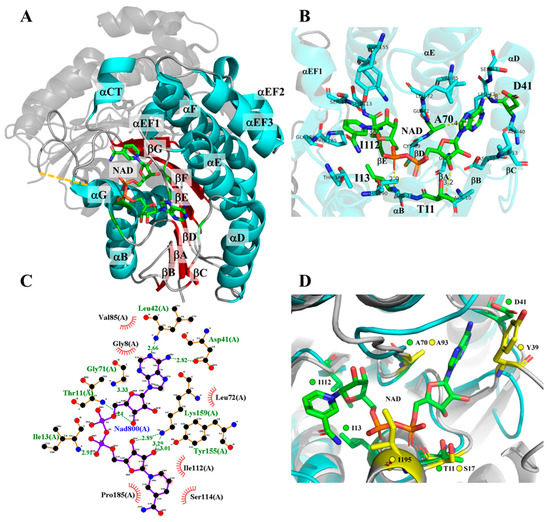
Figure 2.
Structure of 3α-HSD/CR and interactions of the selected residues with NAD+. (A) 3α-HSD/CR (pdb:1fk8 [24]) is a dimeric enzyme with an unresolved loop (dashed line). NAD+ is bound with the C-terminal end of the central parallel β-sheets in the Rossmann fold. (B) Close-up of the active site of 3α-HSD/CR with the residues within 4 Å of NAD+, where the selected residues of T11, I13, D41, A70, and I112 for mutagenesis are labeled. Residues D41, A70, and I112 are located near the C-terminal end of βC, βD, βE, respectively. Residues T11 and I13 are located at the glycine-rich of nucleotide binding motif (GXXXGXG) situated in the tight loop of βA and αB. (C) H-bonding and hydrophobic interactions of the residues with NAD+ were plotted using LigPlot. (D) The binding pocket of NAD+ in 3α-HSD/CR was structurally aligned with glucose dehydrogenase. The structure of glucose dehydrogenase (grey; residues in yellow) was obtained using the program AlphaFold2 and aligned with the structure of 3α-HSD/CR (blue; residues in green). A glucose dehydrogenase variant with I195R/A93K/Y39Q/S17E mutations has been shown to switch cofactor specificity from NAD+ to NMN+ [11].
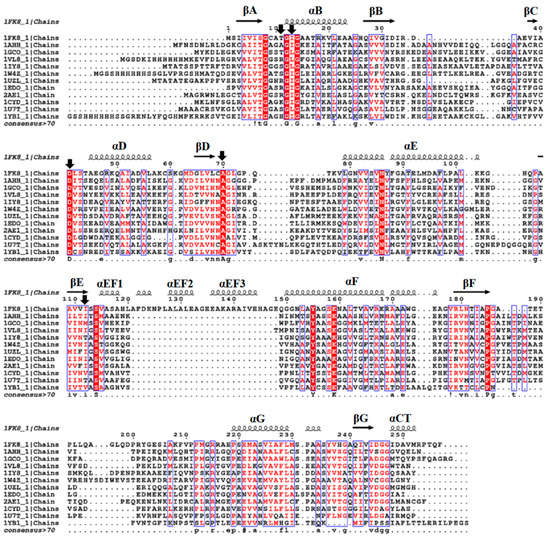
Figure 3.
Sequence alignment of the SDR superfamily. Multiple sequence alignment was performed using T-COFFEE and the ESPript 3.0. program to obtain aligned sequences with similarities and secondary structure elements [27,28]. The secondary structural elements of 1fk8 are indicated by arrows for β-strands and as squiggles for α-helices. Mutated residues are indicated by black arrows. Abbreviations and Protein Data Bank accession numbers are as follows:1FK8, 3α-hydroxysteroid dehydrogenase/carbonyl reductase from Comamonas testosteroni; 1AHH, 7α-hydroxysteroid dehydrogenase from Escherichia coli; 1GCO, glucose dehydrogenase from Bacillus megaterium; 1VL8, Gluconate 5-dehydrogenase from Thermotoga maritima; 1IY8, levodione reductase from Leifsonia aquatica; 1W4Z, ketoacyl reductase from Streptomyces coelicolor; 1UZL, 3-oxoacyl-[acyl-carrier protein] reductase from Mycobacterium tuberculosis; 1EDO, β-keto acyl carrier protein reductase from Brassica napus; 2AE1, tropinone reductase-II from Datura stramonium; 1CYD, carbonyl reductase from Mus musculus; 1U7T, 3-hydroxyacyl-CoA dehydrogenase type II from Homo sapiens; 1YB1, 17β-hydroxysteroid dehydrogenase type XI from Homo sapiens.
We selected the candidate residues T11, I13, D41, A70, and I112 for mutagenesis to switch the cofactor specificity by rational design. Structurally, the T11 and I13 residues are located in the glycine-rich nicotinamide-coenzyme binding motif. The D41 and A70 residues are conserved in the SDR superfamily, where D41 forms H-bonding with the adenine portion of NAD+, and the NNAG motif (CLA70G in 3α-HSD/CR) maintains the structure of the central β-sheets [26]. The I112 residue is located at C-terminus of βE and interacted with the nicotinamide portion of the NAD+. Black et al. demonstrated that engineered glucose dehydrogenase with a mutation at I195R/A93K/Y39Q/S17E switches cofactor specificity by 107-fold toward NMN+ over NAD+ [11]. To understand how these residues interacted with NAD+, we ran the program AlphaFold2 in Google Colaboratory (https://bit.ly/alphafoldcolab, accessed on 9 December 2021) to obtain the predicted protein structure of glucose dehydrogenase and to align it with the structure of 3α-HSD/CR (Figure 2D). Residues T11 and A70 of 3α-HSD/CR were structurally aligned with S17 and A93 of glucose dehydrogenase, respectively. The residues I13 and D41 of 3α-HSD/CR were near to I195 and Y39 of glucose dehydrogenase, respectively.
Therefore, we selected these residues that interacted with different parts of NAD+ for mutation through site-directed mutagenesis (Figure 2B). Our strategy was to replace the residues to destabilize the AMP portion of NAD+ and stabilize the interactions with the pyrophosphate and ribose parts of NAD+ to switch cofactor specificity toward NMN+ from NAD+. To improve the chances of the successful switching of cofactor specificity from NAD+ to NMN+, we adopted the following strategy (Table 1): 1. Substitution of the residues T11, D41, I13, and I112 with alanine increases the size of active site. These mutants of T11A, I13A, and I112A were expected to relax cofactor specificity by allowing binding with cofactor mimetics. 2. Substitution of the residues T11, D41, I13, and I112 with a larger size to decrease the size of active site. These D41I, D41Q, A70I, and A70Q mutants were expected to sterically interfere with NAD+ binding and decrease cofactor specificity for NAD+. 3. Polar interactions for NMN+ were created by adding a charge or polar group to the residue. Hence, mutants of T11K, T11R, I13K, I13R, A70K, and I112K 3α-HSD/CR were expected to form salt bridges with the phosphate moiety of NMN+. Therefore, we prepared mutants of T11A, T11K, T11R, I13A, I13K, I13R, D41I, D41Q, A70I, A70K, A70Q, I112A, and I112K and kinetically characterized their cofactor specificity.

Table 1.
Rationale for selecting the residues for mutagenesis.
2.2. Protein Preparation and Characterization
2.2.1. Site-Directed Mutagenesis
Mutagenetic replacements were performed using Quickchange Site-Directed Mutagenesis kit (Agilent Technologies, Santa Clara, CA, USA) using Pfu polymerase and pet15b-3alphaHSD/CR plasmid as a template. Mutants of T11A, T11K, T11R, I13A, I13K, I13R, D41I, D41Q, A70I, A70K, A70Q, I112A, and I112K were cloned and transformed into BL21 E. coli. DNA sequencing was performed and confirmed the required mutants.
2.2.2. Overexpression and Purification of Wild-Type and Mutant 3α-HSD/CRs
Wild-type and mutants of T11A, T11K, T11R, I13A, I13K, I13R, D41I, D41Q, A70I, A70Q, A70K, I112A, and I112K 3α-HSD/CRs were overexpressed in E. coli BL21 and purified by metal-chelate chromatography. The yields of purification are shown in Table S1. SDS-PAGE analysis of the expression and purification of a typical mutant is shown in Figure S1. Each expressed enzyme showed homogeneity according to the SDS-PAGE results (Figure 4). A significant amount of inclusion bodies was observed for the I13K, I13R, D41I, D41Q, and A70I mutants during expression and purification. In a previous study, we demonstrated that the yield of soluble 17β-HSD increased through induction by IPTG at lower temperatures overnight [29]. Hence, we changed the condition for inducing the expression of I13K, I13R, D41Q, and A70I mutants by IPTG at 14 °C for 14 h., leading to a significant decrease in the formation of inclusion bodies and the yields for the purified I13K, I13R, D41Q, and A70I mutants increased by 6-, 15-, 6-, and 30-fold, respectively.
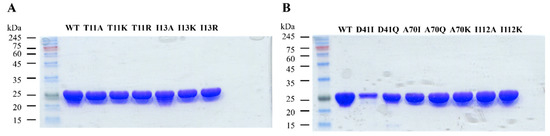
Figure 4.
SDS-PAGE of wild-type and mutant 3α-HSD/CRs. (A) SDS-PAGE analysis of wild-type (WT), T11A, T11K, T11R, I13A, I13K, and I13R mutants, and (B) WT, D41I, D41Q, A70I, A70Q, A70K, I112A, and I112K mutants. Purity was analyzed after metal-chelate chromatography. A total of 10 μg of the sample was loaded on lanes with Coomassie staining. The molecular weights (kDa) of the standard proteins are labeled.
2.2.3. Protein Intrinsic Fluorescence
The intrinsic fluorescence spectra for the wild-type and mutant 3α-HSD/CRs were measured at pH 7.5 at 25 °C with the excitation at 295 nm. 3α-HSD/CR has a single tryptophan residue (W173) contributing to the observed intrinsic tryptophan fluorescence signal. The fluorescence spectra for the wild-type and mutant 3α-HSD/CRs are shown in Figure 5. Tryptophan residue is sensitive to its environment. Mutants of T11A, T11K, I13A, I13K, I13R, A70Q, A70K, I112A, and I112K seemed to have no effect on the local environment of W173, since the same maximum wavelength at 329 nm in the intrinsic protein fluorescence emission of W173 for the wild-type and mutant enzymes was observed. Mutants of T11R, D41I, D41Q, and A70I displayed a red shift in the maximum wavelength, in which the D41I mutant shifted to a maximum wavelength at 336 nm (Table S2). The red shift of the emission spectrum of the mutated enzyme indicated that the local structure was perturbed to a more hydrophilic environment on the W173. The fluorescence intensity decreased in the following order: I112K~I112A > WT > A70Q~I13R~T11A~T11K > T11R > A70K~A70I > I13K > I13A > D41Q > D41I. Meanwhile, the mutation on D41 increased the quenching of the W173 fluorescence.
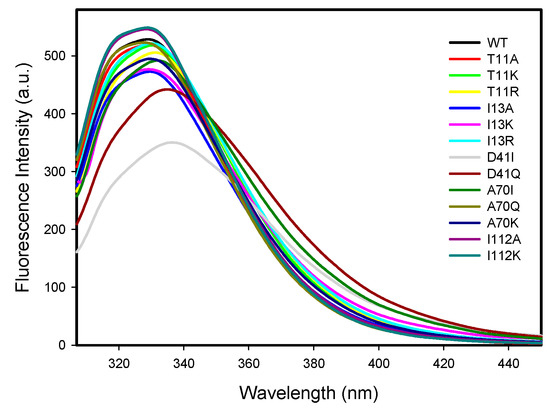
Figure 5.
Fluorescence emission spectra of wild-type and mutant 3α-HSD/CRs. The mutants of T11A, T11K, I13A, I13K, I13R, A70Q, A70K, I112A, and I112K 3α-HSD/CRs had a similar maximum wavelength of 329 nm as the wild-type, whereas the T11R, D41I, D41Q, and A70I variants displayed a red shift in the maximum wavelength. Intrinsic tryptophan fluorescence of the D41I mutant was quenched and shifted to a maximum wavelength of 336 nm. The protein fluorescence spectra were measured at 4 μM enzyme in 0.1 M Hepes at pH 7.5, 25 °C. The excitation wavelength was 295 nm, and the emission range was recorded from 300 to 500 nm.
2.3. Steady-State Kinetics of Wild-Type and Mutant 3α-HSD/CRs
2.3.1. Determine the Cofactor Specificity of Wild-Type and Mutant 3α-HSD/CRs for NAD+
The oxidation of androsterone with NAD+ catalyzed by wild-type and mutant 3α-HSD/CRs was studied by varying the concentration of NAD+ in the presence of 30 μM androsterone at pH 9.0, following the formation of NADH spectrophotometrically at 340 nm. The initial rates as a function of the concentration of NAD+ showed a saturation curve for wild-type and mutants of T11A, T11K, T11R, I13A, I13K, I13R, D41Q, I112A, and I112K 3α-HSD/CRs (Figure 6A). Data were fitted to Equation (1) to obtain kinetic parameters of kcat, KNAD, and kcat/KNAD. While the D41I, A70I, A70Q, and A70K mutants could not be saturated with the concentration of NAD+ tested, the kinetic parameter of kcat/KNAD was determined from the linear relationship between the initial rate and the concentration of NAD+ (Figure 6B). Data were fitted to Equation (2). The obtained kinetic parameters of kcat, KNAD, and kcat/KNAD of wild-type and mutant 3α-HSD/CRs are shown in Table 2. Most mutants caused an increase in KNAD and a decrease in kcat/KNAD, especially for mutants of I13R, A70K, and I112K, which showed a significant decrease in kcat/KNAD by over 105-fold compared to that of the wild-type enzyme. The kcat/KNAD was similar for the wild-type with mutants of T11A and T11K 3α-HSD/CRs. Meanwhile, mutants of T11A, T11K, D41Q, and I112A increased the catalytic constants kcat.
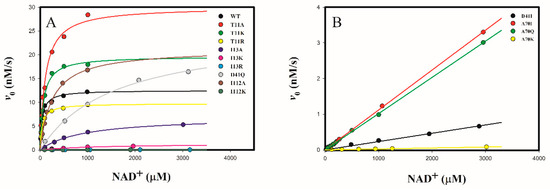
Figure 6.
Initial rates for wild-type and mutant 3α-HSD/CRs as a function of NAD+ concentration. (A) Wild-type and mutants of T11A, T11K, T11R, I13A, I13K, I13R, D41Q, I112A, and I112K 3α-HSD/CRs showed a hyperbolic saturation curve with respect to NAD+. The activity of the I13R and I112K mutants decreased significantly by over 105-fold in kcat/KNAD compared to that of the wild-type enzyme. The lines represent a fit of data to Equation (1). (B) D41I, A70I, A70Q, and A70K mutants were not saturated with the concentration of NAD+ tested. The reaction rate was linearly proportional to the NAD+ concentration. The lines represent a fit of data to Equation (2). The reactions catalyzed by wild-type and mutant 3α-HSD/CRs were measured by varying the concentrations of NAD+ at 30 μM androsterone, 0.1 M Ches at pH 9.0. Error bars represent the standard derivation from at least two independent measurements.

Table 2.
Kinetic parameters for wild-type and mutant 3α-HSD/CRs that catalyzed the oxidation of androsterone with NAD+ and NMN+ a,b,c,d.
2.3.2. Determine the Cofactor Specificity of Wild-Type and Mutant 3α-HSD/CRs for NMN+
Wild-type 3α-HSD/CR showed a low activity toward the oxidation of androsterone with NMN+ by following the formation of NMNH at 336 nm in the presence of 30 μM androsterone at varied concentrations of NMN+ at pH 9.0 (Figure 7). The data were fitted to Equation (1), giving kcat, kcat/KNMN, and KNMN values of 0.025 ± 0.003 s−1, 1.46 ± 0.06 M−1s−1, and 17.1 ± 2.6 mM for the wild-type enzyme, respectively. Comparing the values of kcat/KNMN with kcat/KNAD for wild-type enzyme, the catalytic specificity ratio (CSR) was 4.6 × 10−7. These results indicated that NMN+ is a poor cofactor for 3α-HSD/CR. No activity was observed for mutants of I13A, I13K, I13R, and D41I 3α-HSD/CRs acting on NMN+. Mutants of T11A, T11K, T11R, D41I, D41Q, A70I, A70Q, and I112A 3α-HSD/CRs could not be saturated at the concentration of 12 mM NMN+ tested. Therefore, the initial rate proportional to the concentration of NMN+ was measured (Figure 7B). Data were fitted into Equation (2) to obtain kinetic parameter kcat/KNMN (Table 2). Significantly, the A70K mutant showed a saturation curve with the concentration of NMN+ (Figure 7A). Data were fitted to Equation (1), giving the kinetic parameters of kcat, kcat/KNMN and KNMN of 0.19 ± 0.01 s−1, 12.7 ± 0.7 M−1s−1, and 14 ± 2 mM, respectively. In comparison with wild-type 3α-HSD/CR, kcat/KNMN was increased in the A70K mutant, but decreased in the T11K, T11R, D41I, D41Q A70I, A70Q, and I112A mutants. The preference for NMN+ over NAD+ for wild-type 3α-HSD/CR and mutant variants (CSR) with their relative cofactor specificity (RCS) are compared in Table 2. The catalytic efficiency of the A70K mutant for NMN+ was 0.044-fold lower than that for NAD+ (CSR in Table 2). However, compared to wild-type, the A70K mutant increased kcat and kcat/KNMN by 7.6- and 8.7-fold, respectively, and decreased kcat/KNAD by 1.1 × 104-fold, giving a 9.6 × 104-fold cofactor specificity switch toward NMN+ over NAD+ (RCS in Table 2).
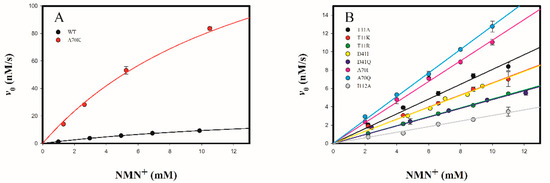
Figure 7.
Initial rates for wild-type and mutant 3α-HSD/CRs as a function of NMN+ concentration. (A) Saturation curve of the initial rate as a function of the concentration of NMN+ for wild-type and A70K mutant 3α-HSD/CRs. The line represents a fit of data to Equation (1). (B) Wild-type and mutants of T11A, T11K, T11R, D41I, D41Q, A70I, A70Q, and I112A 3α-HSD/CRs showed that the initial rate was linearly related to the concentration of NMN+. The lines represent a fit of data to Equation (2). Wild-type and mutant 3α-HSD/CRs-catalyzed reactions were measured by varying the concentrations of NMN+ at 30 μM androsterone, 0.1 M Ches at pH 9.0. Error bars represent the standard derivations from at least two independent measurements.
2.4. Protein Thermal Stability
The protein thermal stability for 3α-HSD/CR variants was studied by differential scanning fluorimetry (DSF) at pH 7.0. The DSF monitors the thermal unfolding of proteins in the presence of a fluorescent dye. We observed a sigmoidal curve for thermal unfolding of wild-type, I13R, D41I, D41Q, A70Q, A70K, and I112A 3α-HSD/CR. The ratio of unfolding ((yt − yn)/(yu − yn)) as a function of temperature is plotted in Figure 8. The results indicate the cooperative unfolding status of the protein and suggest a two-state transition from the native state (N) to the denatured state (D). The midpoint of the transition curve is the melting temperature (Tm), at which 50% of protein is in a denatured state [30,31]. Data were fitted to Equation (6) to obtain Tm. The Tm values for wild-type, I112A, A70Q, D41I, D41Q, A70K, and I13R were 51.9, 51.4, 50.8, 48.3, 46.8, 46.6, and 35.4, respectively. The T11A, T11K, T11R, I13A, I13K, A70I, and I112K mutant enzymes showed a three-state transition with a small change in fluorescence intensity in the first transition, following the major transition in the second phase. This result suggests that an intermediate formed during the thermal unfolding of the mutants. Data were fitted to Equation (7) to obtain Tm1 and Tm2 for the transition from a native state to an intermediate state and an intermediate state to an unfolded state, respectively (Table S2). Compared to the Tm value and the Tm2 value in the second transition, the order of thermostability for wild-type and mutant 3α-HSD/CRs at pH 7.0 was I13R< I13K~A70I < I13A <D41Q~A70K < D41I~T11K < T11R < T11A~A70Q < I112A~wild-type < I112K, where mutant I112K was the most stable and I13R was the least stable in the thermal unfolding.
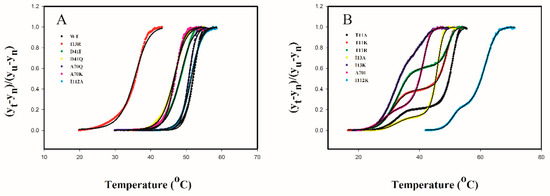
Figure 8.
Differential scanning fluorimetry curves for the thermal unfolding of wild-type and mutant 3α-HSD/CRs. (A) The thermal stability of the wild-type and mutants of I112A, A70Q, D41I, D41Q, A70K, and I13R 3α-HSD/CRs showed a two-state transition, and (B) mutants of T11A, T11K, T11R, I13A, I13K, A70I, and I112K 3α-HSD/CR showed a three-state transition at pH 7.0. The relative order of thermostability for the wild-type (WT) and mutant 3α-HSD/CRs at pH 7.0 was I13R < I13K~A70I < I13A < D41Q~A70K < D41I~T11K < T11R < T11A~A70Q < I112A~WT < I112K. The curve lines represent a fit of data to Equations (6) and (7) for two-state transition and three-state transition, respectively. The Tm values are listed in Table S2. yt, yn, and yu are the fluorescence intensities of the observed, native, and unfolded states of the protein during the thermal unfolding by DSF.
2.5. Molecular Docking Analysis of NAD+ and NMN+ for Wild-Type and Mutant 3α-HSD/CRs
To assess the molecular basis of the activity of wild-type and mutant 3α-HSD/CRs with NAD+ and NMN+, molecular modeling of the structures of mutant 3α-HSD/CRs and the docking of NAD+ and NMN+ into the active site of enzymes were carried out in BIOVIA Discovery Studio. The highest scoring pose of the cofactor (NAD+ or NMN+) at the active site of wild-type and mutant 3α-HSD/CRs was selected. The molecular modeling structures showed a slight change in the overall structures of mutants of T11A, T11K, T11R, I13A, I13K, I13R, D41I, D41Q, A70I, A70Q, A70K, I112A, and I112K 3α-HSD/CRs. However, compared to the structure of the binary complex of NAD+-bound wild-type 3α-HSD/CR, the docking of NAD+ in the active site of mutant variants showed a change in both the conformation of NAD+ and its binding interactions with residues. Docking of NMN+ in the wild-type and mutant variants also displayed different positions and conformations of NMN+ in the binding pocket of the enzyme. The docked NAD+ or NMN+ in the active site of the binary complex and the binding interactions with enzyme residues are shown in Figure S2.
To further explore the molecular mechanisms of the enhanced kcat/KNMN for A70K mutant, the structures of the binary complexes of wild-type with NAD+ and NMN+, wild-type and A70K with NAD+, and wild-type and A70K mutant with NMN+ were aligned to compare the binding interactions, respectively, with their LigPlot interaction maps shown in Figure 9. Compared to the interactions of NAD+ with the residues of wild-type 3α-HSD/CR, NMN+ formed additional H-bonding of G8 and G14 with phosphate moiety and T188 with amide group of NMN+ but lost the AMP portions of NAD+ interacted with enzyme and H-bonding with catalytic residues of K159 and Y155. The pose of the docked NAD+ in the binding pocket of the A70K mutant was different from that in the wild-type 3α-HSD/CR and appeared as a compact and folded conformation, which may have caused difficulty in hydride transfer and led to a significant decrease in activity. The docked NMN+ in the active site of the A70K mutant showed restored interactions between the catalytic residues of K159 and Y155.
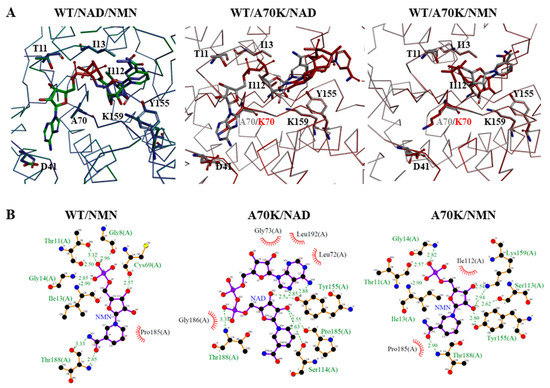
Figure 9.
Molecular docking of wild-type and A70K mutant 3α-HSD/CRs interacted with cofactors of NAD+ and NMN+. (A) Overlapping structures of the binary complexes of wild-type 3α-HSD/CR with NAD+ and NMN+, the NAD+-bound wild-type and A70K mutant enzymes, and the NMN+-bound wild-type and A70K mutant enzymes. Docked NAD+ and NMN+ in the binding pocket of the A70K mutant are shown in red. The enzyme is shown as a Cα backbone, and the ligand is shown in stick mode. The NAD+-bound binary complex of wild-type enzymes is from the crystal structure (pdb:1fk8) [24]. Molecular docking was performed using BIOVIA Discovery Studio. (B) LigPlot showing the H-bonding and hydrophobic interactions of the residues of wild-type enzyme with NMN+, A70K mutant with NAD+ and NMN+, respectively. The interacting residues are labeled. Binding interactions are shown as dashed lines indicating salt bridges and hydrogen bonding interactions with labeled distances.
3. Discussion
Dehydrogenases that catalyze the oxidoreductive reactions with NAD(P)+/NAD(P)H have been engineered to perform various reactions for practical applications [3,8]. The reversal of NADP+ toward NAD+ specificity has been applied to non-cell-based industrial applications because of the lower cost and better stability of NAD+. Bioorthogonal redox systems with biomimetic nicotinamide cofactors of NMN+ by the replacement of natural cofactors in vivo have been developed to prevent interfering with native biochemical processes [11]. However, shifting the cofactor preference of enzymes toward biomimetic redox cofactors remains a challenging task. By changing the cofactor from NAD+ to NMN+, the activity of 3α-HSD/CR that catalyzed the oxidation of androsterone significantly decreased. The kcat and kcat/Km of 3α-HSD/CR acting on NMN+ decreased by 5.4 × 103- and 2.2 × 106-fold at pH 9.0, respectively, and the value of Km for NAD+ was 41 μM and increased to 18 mM for NMN+. We externally added AMP into the reaction mixtures of androsterone with NMN+ catalyzed by 3α-HSD/CR to rescue the lost activity of 3α-HSD/CR without success. Compared to the interactions of NAD+ with the residues of wild-type 3α-HSD/CR, the docked NMN+ lost the interactions of the AMP portions of NAD+ with the enzyme and H-bonding with catalytic residues of K159 and Y155. The binding interactions between the residues in the active site and ligands can be utilized to correctly position the reactive portion of the substrate relative to the active site functional groups to facilitate the reaction. Hence, the loss of activity and the increased Km for wild-type 3α-HSD/CR acting on NMN+ can be attributed to the loss of the remote binding interactions with the residues in the active site of 3α-HSD/CR and the interactions with the catalytic groups of K159 and Y155.
Enzyme engineering through either directed evolution or semi-rational design for the designed properties is usually a time-consuming and costly process. Therefore, we adopted a rational design for the cofactor specificity engineering of 3α-HSD/CR to accelerate the feasibility of switching the cofactor specificity from NAD+ to NMN+ when 3α-HSD/CR catalyzed the oxidation of androsterone. We focused on the residues in the NAD+-binding pocket of 3α-HSD/CR for site-directed mutagenesis, resulting a smaller sequence space of mutations to improve the enzyme. Furthermore, the remote binding energy of the residues with the non-reactive portions of the substrate can assist in its binding and induce a conformational change to make synergistic group contributions to catalysis [23,32,33,34,35,36,37,38,39]. Hence, residues that interact with remote portions of NAD+ were targeted for enzyme engineering using biomimetic NMN+. The crystal structure of the NAD+-bound binary complex of 3α-HSD/CR showed that the residues T11 and I13 were located at the coenzyme binding motif of G8LAT11GI13G14 at the N-terminal region, D41 and A70 interacted with adenosine portion of NAD+, and I112 interacted with the nicotinamide portion of NAD+. In addition, the residues I13, D41, A70, and I112 were conserved residues based on the multiple sequence alignment of SDR enzymes. These residues were replaced with other amino acids of different sizes and charges. The rationale for selecting the residues for mutagenesis is described in Table 1. Substitution of T11, I13, and I112 with alanine can increase the space of the active site to allow for binding with the biomimetic cofactor to relax cofactor specificity. Replacement of D41 and A70 by Ile and Gln increased the size of residue, resulting in a size reduction in the cofactor binding pocket. Meanwhile, the replacement of T11, I13, A70, and I112 by Lys and Arg can induce polar interactions for NMN+ by incorporating a charge group. We prepared the mutants of T11A, T11K, T11R, I13A, I13K, I13R, D41I, D41Q, A70I, A70Q, A70K, I112A, and I112K 3α-HSD/CRs to switch cofactor specificity toward NMN+ from NAD+. As expected, these mutants caused a significant decrease in catalytic efficiency for NAD+ except for T11 mutants, but an unexpected slight decrease for NMN+, except for the A70K mutant, in the steady-state kinetic study. The A70K mutant improved the kcat/KNMN by 8.7-fold. The catalytic specificity preference for NMN+ over NAD+ for wild-type 3α-HSD/CR was poor with 4.6 × 10−7, but increased to 1.0 × 10−4, 2.7 × 10−6, 9.1 × 10−5, 1.1 × 10−4, and 4.4 × 10−2 for mutants of D41I, D41Q, A70I, A70Q, and A70K, respectively. Among these mutants, A70K mutant was the best variant showing an overall specificity switch of 9.6 × 104-fold from NAD+ toward NMN+, and the I112K variant was the most stable in the thermal unfolding by DSF.
3.1. Roles of T11 and I13 in 3α-HSD/CR Catalysis and Cofactor Specificity
T11 and I13 residues are located in the coenzyme-binding motif of G8LAT11GI13G14. The residue of T11 is located in the connecting loop between the first β-strand and the α-helix and I13 is located at the N-terminus of αB. Mutation of I13 caused a more deleterious effect on 3α-HSD/CR catalysis than the mutation of T11. Mutation of T11 had a slight effect on catalytic efficiency for NAD+, either by replacing a smaller size with alanine or increasing the size and charge of lysine and arginine. The catalytic efficiency of T11 mutants for NMN+ was lower than that of the wild-type enzyme. The catalytic specificity of NMN+ for T11A, T11K, and T11R was lower than that of the wild-type enzyme by 1.8-, 2.3-, and 3.1-fold, respectively. Hence, the mutation of T11 by changing the size and charge cannot improve the cofactor specificity toward NMN+. The T11A and T11K mutants increased the KNAD and kcat. The rate-limiting step for the wild-type enzyme that catalyzed the reaction of androsterone with NAD+ was the release of NADH. An increase in kcat was observed for the T188A mutant, in which the replacement of T188 by alanine disrupted H-bonding with nicotinamide of NADH, leading to an increase in the dissociation constant of NADH and kcat [22]. Substitution of alanine or lysine for T11 may increase NADH release and kcat. The kcat/KNAD decreased by 40-, 290-, and 2.9 × 104-fold for I13A, I13K, and I13R mutants, respectively. No activity was observed with NMN+ as an alternate cofactor. Substitution of I13 with alanine may decrease the hydrophobic interactions with NAD+, while the positive charge of K13 and R13 of mutants at N-terminus of αB may destabilize the helix structure due to the dipolar property of α-helix, and interfere with the cofactor binding motif, leading to a decrease in activity and thermal stability. Mutations in T11 and I13 decreased thermal stability and formed intermediate during thermal unfolding. Hence, mutations on T11 and I13 located at glycine rich nucleotide binding motif are not suitable for switching the cofactor specificity.
3.2. Roles of D41 in 3α-HSD/CR Catalysis and Cofactor Specificity
D41 is a conserved aspartic acid residue in the SDR enzyme, located in the loop between βC and αD, and is required for the stabilization of the adenine-binding pocket [13,24]. The carboxylate is hydrogen-bonded with the adenine of NAD+. Substitution of D41 with isoleucine disrupted the H-bonding, leading to a significant decrease in the catalytic efficiency for NAD+ by 500-fold, but a slightly decrease for NMN+ by 2.3-fold, giving the increase in the relative cofactor specificity of D41I over wild-type enzymes toward NMN+ by 217-fold. The D41I mutant could not be saturated by either the NAD+ or NMN+ tested. The substitution of D41 with Gln restored H-bonding with NAD+ but not with NMN+, leading to a decrease in the catalytic efficiency for NAD+ and NMN+ by 19-, and 3.2-fold, respectively. The relative cofactor specificity of the D41Q and wild-type enzymes toward NMN+ was only slightly increased by 5.9-fold. Hence, the formation of hydrogen bonds between the D41 residue and the adenine of NAD+ is important for binding the NAD+ cofactor at the canonical positions to facilitate the hydride transfer reaction. Mutation of D41 significantly affects the catalytic efficiency of NAD+ but cannot improve the catalytic efficiency of NMN+.
3.3. Roles of A70 in 3α-HSD/CR Catalysis and Cofactor Specificity
The conserved NNAG motif of the SDR superfamily plays a role in stabilizing the central β-sheets [26]. The A70 residue is located at the C-terminus of βD and interacts with the adenine moiety of NAD+. Mutation of A70 in 3α-HSD/CR caused a decrease in kcat/KNAD. Compared to the wild-type enzyme, the kcat/KNAD decreased by 267-, 291-, and 1.1 × 104-fold for A70I, A70Q, and A70K mutants, respectively, and slightly decreased in kcat/KNMN by 1.4- and 1.2-fold for A70I and A70Q mutants, respectively. The kcat/KNMN of the A70K mutant increased by 8.7-fold compared to that for wild-type enzyme. The relative cofactor specificity of the A70I, A70Q, and A70K enzymes toward NMN+ increased by 200-, 240-, and 9.6 × 104-fold, respectively. Molecular docking was performed to elucidate the underlying mechanisms of wild-type and mutant enzyme activities. Instead of the extended conformation of NAD+ in the active site of the wild-type 3α-HSD/CR, the pose of the docked NAD+ in the binding pocket of the A70K mutant appeared to have a compact and folded conformation. It seems a bulk and positive charge of K70 blocked the AMP portion of NAD+ from binding at the canonical positions, resulting in the bending of the pyrophosphate group to interact with nicotinamide moieties and turning AMP portion into the opposite site interacting with Y155. The roles of K159 in the 3α-HSD/CR-catalyzed reaction involved the proton relay system and the lowering of the pKa of Y155 as a general base through electrostatic interactions [20,21]. The folded compact conformations and different orientation of NAD+ in A70K mutant led to a significant decrease in activity. Meanwhile, the docked NMN+ in the active site of A70K mutant showed the restored interactions of catalytic residues of K159 and Y155 in comparison to the structure of the binary complex of NMN+-bound wild-type 3α-HSD/CR, thereby increasing the catalytic efficiency of NMN+ for mutant A70K.
3.4. Roles of I112 in 3α-HSD/CR Catalysis and Cofactor Specificity
I112 is located near the C-terminus of βE and has hydrophobic interactions with the nicotinamide ring of NAD+. Thermal unfolding of wild-type and I112A enzymes was a two-state transition with Tm of 51.9 °C and 51.6 °C, respectively, but it changed to a three-state transition for the I112K mutant with the midpoint temperature of 51.6 °C and 60.4 °C for the first transition and the second transition, respectively. We used the MODELER program to model the structures of the I112A and I112K mutants, which displayed a structure similar to that of the wild-type enzyme. Therefore, the enhanced thermal stability of I112K mutant may have been due to the positive charge of K112, which formed the salt bridges with the residues to form a stable intermediate during the process of thermal unfolding. However, substitution of I112 with Lys significantly decreased the activity. The pose of the docked NAD+ in the binding pocket of I112K showed similar interactions of the adenosine portion of NAD+ with that for the wild-type enzyme but shifted the position and orientation of the nicotinamide portion of NAD+ to form a new H-bonding with Q194, which may have caused difficulty in the hydride transfer and loss of the activity.
4. Materials and Methods
4.1. Site-Directed Mutagenesis, and the Overexpression and Purification of 3α-HSD/CR Variants
We performed the mutagenetic replacements for mutant enzymes of T11A, T11K, T11R, I13A, I13K, I13R, D41I, D41Q, A70I, A70Q, A70K, I112A, and I112K by using Pfu polymerase and pet15b-3αHSD/CR plasmid as templates through Quickchange Site-Directed Mutagenesis kit (Agilent Technologies, Santa Clara, CA, USA). The kits utilized PfuUltra HF DNA polymerase to extend the oligonucleotide primers during the temperature cycle. The primers used to create the cDNA encoding the 3α-HSD/CR mutants are shown in Table S3. The parental DNA template was digested by Dpn1 endonuclease, which is specific for methylated and hemimethylated DNA. The resulting nicked vector DNA incorporating the desired mutations was then transformed into DH5α supercompetent cells. The selected colonies were picked, and the plasmid from DH5α cell was purified. The mutant vectors were used to transform competent Escherichia coli BL21(DE3) cells. The entire mutants of 3α-HSD/CR cDNA were sequenced to ensure their fidelity.
4.2. Overexpression and Purification of Wild-Type and Mutant 3α-HSD/CRs
Wild-type and mutants of T11A, T11K, T11R, I13A, I13K, I13R, D41I, D41Q, A70I, A70Q, A70K, I112A, and I112K 3α-HSD/CRs were overexpressed in BL21(DE3) cells grown in 1 L LB medium by the addition of 0.5 mM IPTG when the culture had reached an optical density at 600 nm of 0.6–1. After 3.5 h, the cells were harvested. For the formation of inclusion bodies, BL21(DE3) cells were induced by 0.5 mM IPTG at 14 °C for overnight. The cells were lysed by sonication for 5 min with 10 s on and 10 s off cycles. The cell suspension was kept on ice during sonication. The resulting supernatant after ultracentrifuge at 8000× g was loaded to the His-bind Ni+-NTA resin column (Cytiva). After a washing step, the protein was eluted with a stepwise gradient of 50–500 mM imidazole. Each variant showed a homogeneous protein confirmed by separation on 15% SDS/PAGE. The protein concentration was determined by measuring the absorbance at 280 nm with an extinction coefficient ε280 of 22,920 calculated from the ExPASy ProtParam tool (https://web.expasy.org/protparam/, accessed on 16 April 2022).
4.3. Steady-State Kinetics
The oxidation of androsterone with NAD+ catalyzed by wild-type and mutant 3α-HSD/CRs was monitored by the formation of NADH spectrophotometrically at 340 nm, using ε340 = 6220 M−1cm−1, while the oxidation of androsterone with NMN+ catalyzed by wild-type and mutant 3α-HSD/CRs was monitored spectrophotometrically at 336 nm, using ε336 = 6800 M−1cm−1 [40]. Typically, initial rates were measured by adding wild-type or mutant 3α-HSD/CRs in the presence of 30 μM androsterone at varied concentrations of NAD+ or NMN+ in 0.1 mg/mL BSA, 0.1 M Ches at pH 9.0 at 25 °C.
4.4. Protein Intrinsic Tryptophan Fluorescence
Fluorescence measurements for wild-type and mutant enzymes were performed using an LS55 Luminescence spectrometer (Perkin Elmer). Enzymes were measured at a final concentration of 4 μM in 0.1 M Hepes at pH 7.5. The excitation wavelength was set at 295 nm and the emission range was recorded from 300 to 500 nm at 25 °C with the slit width of 7.0 nm for both emission and excitation. The spectra were averaged with three consecutive measurements and corrected for background by subtracting the buffer spectra.
4.5. Thermal Denaturation Assay by Differential Scanning Fluorimetry
Wild-type and mutant 3α-HSD/CRs are diluted to a final concentration of 2 μM with 0.1 M Hepes pH 7.0 in the presence of 5 × SYPRO Orange (stock 5000× from Invitrogen, Waltham, MA, USA). A StepOnePlus system RT-PCR (Applied BiosystemsTM, Waltham, MA, USA) using the ROX channel as the detector was used to collect the DSF data. Protein samples were dispensed into a 96-well PCR plate and overlaid with a clear thermal-seal film to prevent evaporation. The temperature was increased in 0.5 °C/min increments from 4 °C to 85 °C. The data files obtained for each 96 well plate from the RT-PCR runs were imported into SigmaPlot 11 for the analysis.
4.6. Molecular Docking Analysis
The modeled structure of 3α-HSD/CR was generated from the crystal structure of NAD+ bound binary complex of 3α-HSD/CR (pdb: 1fk8) using a homology models protocol (BIOVIA Discovery Studio 2019) to build a missing substrate binding loop. Modeling of the mutant structure was performed by using the MODELER program to mutate residues to the designed amino acids. The conformation of the mutated residues and any surrounding residues at the cut-off distance of 4.5 Å were optimum using the default value of the parameters with 5 modelers created in which the lowest Discrete Optimized Protein Energy (DOPE) score was selected. Minimization was performed to refine the conformations by using a Smart Minimizer Algorithm with a CHARMM force field. To perform molecular docking of wild-type and mutant 3α-HSD/CRs with the ligand, the structure of NAD+ was obtained from the binary complex of NAD+-bound 3α-HSD/CR, whereas the NMN+ ligand was obtained from ″PubChem″ (http://pubchem.ncbi.nlm.nih.gov/, accessed on 25 July 2022). Protein docking of wild-type and mutant 3α-HSD/CRs was conducted using the LibDock program. Minimization was performed to refine the docked ligand poses using a CHARMM force field.
4.7. Data Analysis
Data for a substrate saturation curve at a fixed concentration of the second substrate were fitted using Equation (1). In Equation (1), v and V represent the initial and maximum velocities and KA is the Michaelis constant for substrate A. V is equal to kcat [E]t, where kcat and [E]t are the catalytic constant and the total concentration of enzyme in the reaction solution, respectively. In case that the concentration of A is far less than KA ([A]<<KA), Equation (1) is deduced to Equation (2), and data were fitted to Equation (2) to obtain V/KA. The degree of success for switching the cofactor specificity from NAD+ to NMN+ was evaluated by comparing the cofactor specificity ratio (CSR, Equation (3)), reflecting the degree of preference of the target cofactor in the mutated enzyme, and the relative cofactor specificity (RCS, Equation (4)), which compares the cofactor specificity between the mutated and wild type enzymes. For a two-state unfolding model (N ⇌ U), the equilibrium constant K is equal to exp(−ΔG/RT), where R, T, and ΔG are gas constant (1.987 cal mol−1 K−1), temperature in Kelvin, and the free energy difference between unfolding and native state, respectively. Fluorescence intensity data obtained from the DSF were fitted to Equation (6) using the Gibbs–Helmholtz equation (Equation (5)) to determine the melting temperature Tm and the enthalpy change (ΔHm) of the protein. In case of a three-state unfolding model (N ⇌ I ⇌ U), the observed fluorescence intensity data (yt) were fitted to Equation (7) to determine the melting temperature Tm1 and ΔHm1 of protein for the first transition (N ⇌ I) with equilibrium constant K1, and Tm2 and ΔHm2 for the second transition (I ⇌ U) with equilibrium constant K2. yn, yi, and yu are the fluorescence intensities of native (N), intermediate (I), and unfolded (U) states of protein during the thermal unfolding by DSF, respectively.
v = VA/(KA + A)
v = (V/KA) A
CSR = (kcat/KM)NMN/(kcat/KM)NAD
RCS = ((kcat/KM)NMN/(kcat/KM)NAD)mut/((kcat/KM)NMN/(kcat/KM)NAD)WT
ΔG(T) = ΔHm (1 − T/Tm) + ΔCp (T − Tm − T ln(T/Tm))
yt = ynfn + yufu = (yn + yuK)/(1 + K)
yt = ynfn + yifi + yufu = (yn + yIK1 + yu K1K2)/(1 + K1 + K1K2)
5. Conclusions
3α-HSD/CR catalyzed the reaction of androsterone with NAD+ with a rate-limiting step in the release of NADH. 3α-HSD/CR was engineered to switch the cofactor specificity from NAD+ to biomimetic NMN+. The conserved residues of T11, I13, D41, A70, and I112 interacted with different portions of NAD+ and were mutated to change the size of the active site for binding with the truncated NMN+ but blocked the NAD+ from binding and increased the polar and hydrophobic interactions with NMN+. Engineering 3α-HSD/CR by increasing the rate of dissociation of NADH can increase the catalytic constant as observed for the mutants of T11A, T11K, D41Q, and I112A. However, mutation on the conserved residues that interact with NAD+ by changing a size or charge affected the binding interactions with NAD+, resulting in a change in the position, conformation, and orientation of NAD+, thereby significantly decreasing the catalytic efficiency for NAD+. By switching to the truncated NMN+ analog as a cofactor, the remote binding interactions of the AMP moiety and H-bonding with the catalytic groups of Y155 and K159 were lost; thus, the activity decreased. Mutations at the T11, I13, D41, A70, and I112 residues could not restore these interactions, except for the A70K mutant, which maintained the interactions with catalytic residues of Y155 and K159. By destabilizing the interactions with NAD+ and enhancing the interactions with NMN+, the cofactor specificity preference toward NMN+ over NAD+ for A70K mutant compared with wild-type 3α-HSD/CR increased by 9.6 × 104-fold.
These studies shed light on a strategy for coenzyme engineering by enhancing the binding interactions with biomimetic NMN+ and maintaining the functions of catalytic residues. Engineering 3α-HSD/CR for developing a biomimetic redox cofactor system as a green and sustainable biocatalyst can support diverse redox chemistries. The results presented here suggest an approach to improve activity for biomimetic cofactor and stability for 3α-HSD/CR that may be applicable to other SDR enzymes.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/catal12101094/s1, Figure S1: the SDS-PAGE analysis of mutant 3α-HSD/CR expressed in E. coli BL21(DE3); Figure S2: Molecular docking analysis of NAD+ and NMN+ for wild-type and mutant 3α HSD/CRs; Table S1: Purification of wild-type and mutant 3α HSD/CRs; Table S2: The Tm values and protein tryptophan intrinsic fluorescence of wild-type and mutant 3α HSD/CRs; Table S3: Oligonucleotide primers used for site directed.
Author Contributions
Conceptualization, Y.-L.C., Y.-H.C., S.-J.C. and C.-C.H.; Methodology, Y.-L.C., Y.-H.C. and C.-C.H.; Software, Y.-L.C. and Y.-H.C.; Validation, Y.-L.C., Y.-H.C. and C.-C.H.; Formal Analysis, Y.-L.C., Y.-H.C. and C.-C.H.; Investigation, Y.-L.C. and Y.-H.C.; Resources, C.-C.H.; Data Curation, Y.-L.C., Y.-H.C. and C.-L.H.; Writing—Original Draft Preparation, C.-C.H.; Writing—Review and Editing, S.-J.C., T.-P.W. and C.-C.H. Visualization, Y.-L.C., Y.-H.C., C.-L.H. and S.-J.C.; Supervision, C.-C.H.; Funding Acquisition, C.-C.H. All authors have read and agreed to the published version of the manuscript.
Funding
The study was supported by grants from KMU to C.-C.H. (Grant no: KMU-M109013; KMU-M110020) and the Ministry of Science and Technology (MOST) to C.-C.H. (Grant no: MOST 110-2311-B-037-001-).
Data Availability Statement
The data presented in this study are available upon request from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
3α-HSD/CR, 3α-hydroxysteroid dehydrogenase/carbonyl reductase; SDR, short-chain dehydrogenase/reductase; NMN+, nicotinamide mononucleotide; Tm, midpoint temperature; DSF, differential scanning fluorimetry.
References
- Sharma, A.; Gupta, G.; Ahmad, T.; Mansoor, S.; Kaur, B. Enzyme engineering: Current trends and future perspectives. Food Rev. Int. 2021, 37, 121–154. [Google Scholar] [CrossRef]
- Sheldon, R.A.; Pereira, P.C. Biocatalysis engineering: The big picture. Chem. Soc. Rev. 2017, 46, 2678–2691. [Google Scholar] [CrossRef]
- Vidal, L.S.; Kelly, C.L.; Mordaka, P.M.; Heap, J.T. Review of NAD(P)H-dependent oxidoreductases: Properties, engineering and application. BBA-Proteins Proteom. 2018, 1866, 327–347. [Google Scholar] [CrossRef] [PubMed]
- Mordhorst, S.; Andexer, J.N. Round, round we go—Strategies for enzymatic cofactor regeneration. Nat. Prod. Rep. 2020, 37, 1316–1333. [Google Scholar] [CrossRef] [PubMed]
- Graef, M.R.D.; Alexeeva, S.; Snoep, J.L.; Mattos, M.J.T.d. The steady-state internal redox state (NADH/NAD) reflects the external redox state and is correlated with catabolic adaptation in Escherichia coli. J. Bacteriol. 1999, 181, 2351–2357. [Google Scholar] [CrossRef]
- Wang, M.; Chen, B.; Fang, Y.; Tan, T. Cofactor engineering for more efficient production of chemicals and biofuels. Biotechnol. Adv. 2017, 35, 1032–1039. [Google Scholar] [CrossRef]
- Cahn, J.K.B.; Werlang, C.A.; Baumschlager, A.; Brinkrnann-Chen, S.; Mayo, S.L.; Arnold, F.H. A general tool for engineering the NAD/NADP cofactor preference of oxidoreductases. Acs Synth. Biol. 2017, 6, 326–333. [Google Scholar] [CrossRef]
- You, C.; Huang, R.; Wei, X.L.; Zhu, Z.G.; Zhang, Y.-H.P. Protein engineering of oxidoreductases utilizing nicotinamide-based coenzymes, with applications in synthetic biology. Synth. Syst. Biotechnol. 2017, 2, 208–218. [Google Scholar] [CrossRef]
- Campbell, E.; Meredith, M.; Minteer, S.D.; Banta, S. Enzymatic biofuel cells utilizing a biomimetic cofactor. Chem. Commun. 2012, 48, 1898–1900. [Google Scholar] [CrossRef]
- King, E.; Maxel, S.; Li, H. Engineering natural and noncanonical nicotinamide cofactor-dependent enzymes: Design principles and technology development. Curr. Opin. Biotechnol. 2020, 66, 217–226. [Google Scholar] [CrossRef]
- Black, W.B.; Zhang, L.; Mak, W.S.; Maxel, S.; Cui, Y.; King, E.; Fong, B.; Martinez, A.S.; Siegel, J.B.; Li, H. Engineering a nicotinamide mononucleotide redox cofactor system for biocatalysis. Nat. Chem. Biol. 2020, 16, 87–94. [Google Scholar] [CrossRef] [PubMed]
- Bloom, J.D.; Meyer, M.M.; Meinhold, P.; Otey, C.R.; MacMillan, D.; Arnold, F.H. Evolving strategies for enzyme engineering. Curr. Opin. Struct. Biol. 2005, 15, 447–452. [Google Scholar] [CrossRef] [PubMed]
- Zeymer, C.; Hilvert, D. Directed evolution of protein catalysts. Annu. Rev. Biochem. 2018, 87, 131–157. [Google Scholar] [CrossRef] [PubMed]
- Kuchner, O.; Arnold, F.H. Directed evolution of enzyme catalysts. Trends Biotechnol. 1997, 15, 523–530. [Google Scholar] [CrossRef]
- Chen, K.; Arnold, F.H. Tuning the activity of an enzyme for unusual environments: Sequential random mutagenesis of subtilisin E for catalysis in dimethylformamide. Proc. Natl. Acad. Sci. USA 1993, 90, 5618–5622. [Google Scholar] [CrossRef]
- Goldsmith, M.; Tawfik, D.S. Enzyme engineering: Reaching the maximal catalytic efficiency peak. Curr. Opin. Struct. Biol. 2017, 47, 140–150. [Google Scholar] [CrossRef]
- Tokuriki, N.; Jackson, C.J.; Afriat-Jurnou, L.; Wyganowski, K.T.; Tang, R.M.; Tawfik, D.S. Diminishing returns and tradeoffs constrain the laboratory optimization of an enzyme. Nat. Commun. 2012, 3, 1257. [Google Scholar] [CrossRef]
- Persson, B.; Kallberg, Y.; Bray, J.E.; Bruford, E.; Dellaporta, S.L.; Favia, A.D.; Duarte, R.G.; Jornvall, H.; Kavanagh, K.L.; Kedishvili, N.; et al. The SDR (short-chain dehydrogenase/reductase and related enzymes) nomenclature initiative. Chem. Biol. Interact. 2009, 178, 94–98. [Google Scholar] [CrossRef]
- Kavanagh, K.L.; Jornvall, H.; Persson, B.; Oppermann, U. Medium- and short-chain dehydrogenase/reductase gene and protein families: The SDR superfamily: Functional and structural diversity within a family of metabolic and regulatory enzymes. Cell. Mol. Life Sci. 2008, 65, 3895–3906. [Google Scholar] [CrossRef]
- Hwang, C.C.; Chang, Y.H.; Hsu, C.N.; Hsu, H.H.; Li, C.W.; Pon, H.I. Mechanistic roles of Ser-114, Tyr-155, and Lys-159 in 3alpha-hydroxysteroid dehydrogenase/carbonyl reductase from Comamonas testosteroni. J. Biol. Chem. 2005, 280, 3522–3528. [Google Scholar] [CrossRef]
- Chang, Y.H.; Chuang, L.Y.; Hwang, C.C. Mechanism of proton transfer in the 3alpha-hydroxysteroid dehydrogenase/carbonyl reductase from Comamonas testosteroni. J. Biol. Chem. 2007, 282, 34306–34314. [Google Scholar] [CrossRef] [PubMed]
- Hwang, C.C.; Chang, Y.H.; Lee, H.J.; Wang, T.P.; Su, Y.M.; Chen, H.W.; Liang, P.H. The catalytic roles of P185 and T188 and substrate-binding loop flexibility in 3 alpha-hydroxysteroid dehydrogenase/carbonyl reductase from Comamonas testosteroni. PLoS ONE 2013, 8, e63594. [Google Scholar]
- Hwang, C.C.; Chang, P.R.; Wang, T.P. Contribution of remote substrate binding energy to the enzymatic rate acceleration for 3 alpha-hydroxysteroid dehydrogenase/carbonyl reductase. Chem.-Biol. Interact. 2017, 276, 133–140. [Google Scholar] [CrossRef] [PubMed]
- Grimm, C.; Maser, E.; Mobus, E.; Klebe, G.; Reuter, K.; Ficner, R. The crystal structure of 3alpha-hydroxysteroid dehydrogenase/carbonyl reductase from Comamonas testosteroni shows a novel oligomerization pattern within the short chain dehydrogenase/reductase family. J. Biol. Chem. 2000, 275, 41333–41339. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.H.; Huang, T.J.; Chuang, L.Y.; Hwang, C.C. Role of S114 in the NADH-induced conformational change and catalysis of 3alpha-hydroxysteroid dehydrogenase/carbonyl reductase from Comamonas testosteroni. Biochim. Biophys. Acta-Proteins Proteom. 2009, 1794, 1459–1466. [Google Scholar] [CrossRef]
- Filling, C.; Berndt, K.D.; Benach, J.; Knapp, S.; Prozorovski, T.; Nordling, E.; Ladenstein, R.; Jornvall, H.; Oppermann, U. Critical residues for structure and catalysis in short-chain dehydrogenases/reductases. J. Biol. Chem. 2002, 277, 25677–25684. [Google Scholar] [CrossRef]
- Notredame, C.; Higgins, D.G.; Heringa, J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000, 302, 205–217. [Google Scholar] [CrossRef]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef]
- Chang, Y.H.; Wang, Y.L.; Lin, J.Y.; Chuang, L.Y.; Hwang, C.C. Expression, purification, and characterization of a human recombinant 17 beta-hydroxysteroid dehydrogenase type 1 in Escherichia coli. Mol. Biotechnol. 2010, 44, 133–139. [Google Scholar] [CrossRef]
- Gao, K.; Oerlemans, R.; Groves, M.R. Theory and applications of differential scanning fluorimetry in early-stage drug discovery. Biophys. Rev. 2020, 12, 85–104. [Google Scholar] [CrossRef]
- Matulis, D.; Kranz, J.K.; Salemme, F.R.; Todd, M.J. Thermodynamic stability of carbonic anhydrase: Measurements of binding affinity and stoichiometry using ThermoFluor. Biochemistry 2005, 44, 5258–5266. [Google Scholar] [CrossRef] [PubMed]
- Wolfenden, R.; Snider, M.J. The depth of chemical time and the power of enzymes as catalysts. Acc. Chem. Res. 2001, 34, 938–945. [Google Scholar] [CrossRef] [PubMed]
- Amyes, T.L.; Richard, J.P.; Tait, J.J. Activation of orotidine 5′-monophosphate decarboxylase by phosphite dianion: The whole substrate is the sum of two parts. J. Am. Chem. Soc. 2005, 127, 15708–15709. [Google Scholar] [CrossRef] [PubMed]
- Amyes, T.L.; Richard, J.P. Specificity in transition state binding: The Pauling model revisited. Biochemistry 2013, 52, 2021–2035. [Google Scholar] [CrossRef] [PubMed]
- Jencks, W.P. Binding energy, specificity, and enzymic catalysis: The circe effect. Adv. Enzymol. Relat. Areas Mol. Biol. 1975, 43, 219–410. [Google Scholar]
- Yao, X.; Bleile, D.W.; Yuan, Y.; Chao, J.; Sarathy, K.P.; Sanders, D.A.; Pinto, B.M.; O’Neill, M.A. Substrate directs enzyme dynamics by bridging distal sites: UDP-galactopyranose mutase. Proteins 2009, 74, 972–979. [Google Scholar] [CrossRef]
- Chen, C.H.; Namanja, A.T.; Chen, Y. Conformational flexibility and changes underlying activation of the SUMO-specific protease SENP1 by remote substrate binding. Nat. Commun. 2014, 5, 4968. [Google Scholar] [CrossRef]
- Johnson, T.A.; McLeod, M.J.; Holyoak, T. Utilization of substrate intrinsic binding energy for conformational change and catalytic function in phosphoenolpyruvate carboxykinase. Biochemistry 2016, 55, 575–587. [Google Scholar] [CrossRef]
- Hwang, C.-C.; Chang, P.-R.; Hsieh, C.-L.; Chou, Y.-H.; Wang, T.-P. Thermodynamic analysis of remote substrate binding energy in 3alpha-hydroxysteroid dehydrogenase/carbonyl reductase catalysis. Chem.-Biol. Interact. 2019, 302, 183–189. [Google Scholar] [CrossRef]
- Sicsic, S.; Durand, P.; Langrene, S.; Legoffic, F. A new approach for using cofactor dependent enzymes—Example of alcohol-dehydrogenase. FEBS Lett. 1984, 176, 321–324. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).