Simple Summary
Why does gastric cancer frequently recur despite aggressive treatment? The answer may lie in a small but stubborn population of cells hidden deep within the tumor, the cancer stem cells (CSCs). These cells possess self-renewal capacity, resist conventional therapies and drive relapse and metastasis, making them critical determinants of patient outcomes. Their persistence highlights the urgent need to understand the molecular mechanisms sustaining their survival. One emerging explanation comes from the field of epitranscriptomics, which investigates chemical modifications of RNA that regulate gene expression post-transcriptionally. Among these modifications, N6-methyladenosine (m6A) has attracted particular attention. It serves as a dynamic regulator of RNA metabolism, controlling stability, translation and splicing, and has been shown to promote stemness and aggressiveness in gastric cancer stem cells (GCSCs). Dissecting the m6A-dependent regulatory networks in CSCs paves the way for novel diagnostic biomarkers and therapeutic strategies designed to eliminate the root of gastric cancer recurrence and progression.
Abstract
Cancer stem cells (CSCs) represent a small but critical subpopulation of tumor cells that drive therapy resistance, relapse and metastasis. Gastric cancer stem cells (GCSCs) have been identified through surface markers and transcriptional signatures, revealing their central role in tumor progression. Recently, N6-methyladenosine (m6A) RNA modification has emerged as a crucial epitranscriptomic regulator of CSC biology. The m6A machinery, including “writers” (METTL3, METTL14, WTAP, VIRMA), “erasers” (FTO, ALKBH5) and “readers” (YTHDFs/ YTHDCs, IGF2BPs, hnRNPA2B1), orchestrates RNA stability, splicing, translation and decay, thereby influencing self-renewal and oncogenic signaling. In GCSCs, m6A controls pluripotency factors, oncogenic transcripts and non-coding RNAs, collectively reinforcing stemness and malignant potential. Mounting evidence implicates dysregulated m6A effectors as not only key drivers of GCSC biology but also as promising biomarkers for patient stratification and therapeutic targets capable of selectively eliminating CSCs. Harnessing this knowledge could enable earlier diagnosis, more accurate patient stratification and more precise treatments. However, challenges remain regarding the resolution of m6A profiling, therapeutic selectivity to avoid unwanted toxicity and biomarker validation for clinical use. This review summarizes the discovery and features of CSCs, highlights the functional role of m6A in GCSCs, and explores diagnostic and therapeutic opportunities while outlining key difficulties for clinical translation.
1. Introduction
Gastric cancer remains one of the most serious health challenges, ranking among the top five most common and deadly cancers in the world [1]. Despite advances, both in early detection tools and therapeutic strategies, patient prognosis remains poor [2]. Additionally, no two gastric cancers are the same, reflecting the disease’s heterogeneity across distinct molecular subtypes and variations within the tumor microenvironment, which complicates diagnosis and treatment [3]. Within this complexity, cancer stem cells (CSCs) have emerged as key drivers of tumor initiation, progression, metastasis and relapse even after a seemingly successful treatment [4]. Gastric cancer stem cells (GCSCs) have shown to be resistant to conventional therapies and capable of renewing tumors, making them a critical therapeutic target. Several surface markers, including CD44, ALDH1, LGR5 and CD133, along with transcription factors such as SOX2 and HMGA1, are commonly used to identify and isolate GCSCs [5,6,7,8]. Understanding the molecular vulnerabilities of these cells, as well as the regulatory networks that sustain their stem-like properties, is essential for the development of precise diagnostic tools and personalized biomarker-driven therapies that can improve patient outcomes [5,6,9]. In recent years, a new layer of cancer biology has captured attention—RNA modifications—adding a new dimension to our understanding of cancer biology [10,11]. Among these, N6-methyladenosine (m6A) has emerged as the most prevalent and functionally significant internal mRNA modification in eukaryotic cells [11]. The m6A landscape is dynamically regulated by three classes of proteins: “writers” (e.g., METTL3, METTL14, WTAP and VIRMA), “erasers” (e.g., FTO and ALKBH5) and “readers” (e.g., YTHDF, YTHDC, IGF2BP family members and hnRNPA2B1) [12,13]. These factors collectively control key aspects of RNA metabolism, including splicing, export, stability and translation, influencing cell differentiation, development and disease progression [14]. Dysregulation of the m6A machinery has been increasingly linked to tumor growth, therapy resistance and the maintenance of stem-like traits in various cancers [15,16]. While m6A biology has been explored extensively in other tumor types, its role in GCSCs is only beginning to be unraveled. Aberrant expression of m6A-related enzymes has already been associated with aggressive tumor behavior, poor prognosis and treatment resistance in gastric cancer [17]. However, the precise molecular mechanisms by which m6A influences GCSC biology remain incompletely understood. This review aims to bring together the current knowledge on the intersection between m6A RNA methylation and GCSCs, highlighting how this emerging epitranscriptomic layer shapes stemness and tumor behavior. We also explore the therapeutic potential of targeting m6A regulators as a novel strategy to eradicate GCSCs and ultimately improve treatment outcomes for gastric cancer patients.
2. Gastric Cancer Stem Cells Overview
In the past three decades, CSCs have attracted significant attention due to their role in tumor biology. In 1964, Kleinsmith and Pierce demonstrated that a single mouse embryonal carcinoma cell, derived from a teratocarcinoma, was capable of regenerating the complete histopathological features of the original tumor [18]. Notably, embryonal carcinoma cells represent a small stem cell–like subpopulation within teratocarcinomas. These defining properties—tumor regeneration from a single cell and the presence of a self-renewing subpopulation—constitute the fundamental hallmarks of CSCs. The first evidence for the existence of human CSCs came in 1994, when Lapidot and colleagues identified a rare CD34+/CD38− cell subpopulation in acute myeloid leukemia capable of initiating disease in immunodeficient mice [19]. A few years later, Bonnet and Dick confirmed that these cells alone could reproduce the disease, establishing the concept that only a fraction of cancer cells possess the potential to initiate the tumorigenic process [20]. This discovery was soon extended to solid tumors, where it was validated that only specific subsets of cells were able to initiate tumors in vivo [21,22]. Since then, CSCs have been described in a wide range of cancers and are now recognized as key contributors to tumor development, therapy resistance, metastasis and relapse [23]. Biologically, CSCs are defined by their ability to self-renew, to differentiate into diverse tumor cell lineages (asymmetric division) and to adapt under stressful conditions such as chemotherapy, radiotherapy or hypoxia [24]. They are usually identified by a combination of functional assays and cell-surface markers, which vary according to tumor type. For example, in breast cancer, CD44+/CD24+ enriched cells displayed cancer stem-like features while CD133 is frequently used in colon and brain tumors and CD90 in liver tumors [25,26,27,28]. Importantly, the transplantation of only a few hundred CSCs into immunodeficient mice can be sufficient to regenerate an entire tumor, illustrating their remarkable tumorigenic potential [24].
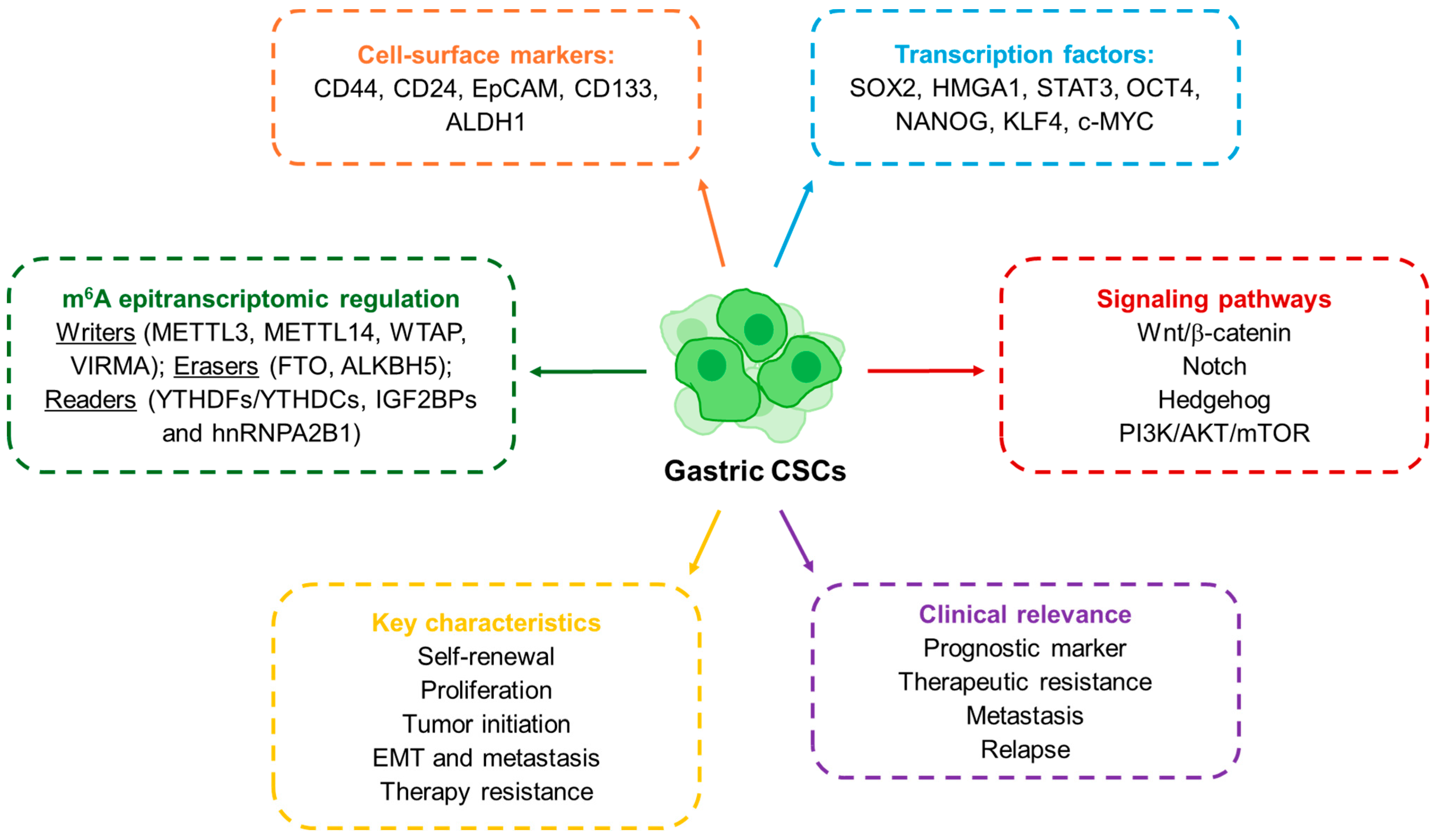
GCSCs share these core CSC properties but also exhibit features unique to gastric cancer biology. They were first characterized in 2007, when Takaishi and colleagues reported that CD44+ cells exhibited spheroid-forming ability and could initiate tumors in vivo, fulfilling the criteria of cancer stem-like cells [29]. Since then, several markers have been proposed to define gastric GCSCs (Figure 1). CD44 remains the most widely studied, but other molecules such as CD24, EpCAM, CD54, CD90, CD133, CXCR4, Lgr5 and ALDH1 have also been associated with GCSC populations [6,9,30]. In parallel, transcription factors central to pluripotency such as SOX2, OCT4 and NANOG have been shown to regulate GCSCs characteristics, while signaling pathways like WNT, NOTCH, Hedgehog, NF-κB and TGF-β/SMAD provide the molecular framework that sustains their activity [6,31,32]. Thus, these molecules are also key to identifying and studying GCSCs. For instance, using the SORE6-GFP reporter system, engineered to detect transcriptional activity of SOX2 and OCT4, we were able to identify and isolate GCSCs (SORE6+ cells). These cells exhibited enhanced stemness characteristics, including increased self-renewal capacity, elevated tumor-initiating potential and resistance to conventional chemotherapeutic agents such as 5-fluorouracil (5-FU) [7]. Clinically, the expression of CSC-associated markers is linked to more aggressive disease and worse outcomes: for instance, CD44 has been validated as an independent prognostic factor, while high levels of OCT4 or SOX2 correlate with metastasis and poor survival [33,34,35,36]. A more recent layer of complexity has been added by the discovery of the role of RNA modifications in CSC regulation. Among them, N6-methyladenosine (m6A) has emerged as the most abundant internal modification in eukaryotic mRNA, regulating RNA metabolism, including stability, splicing and translation [14]. The deposition and interpretation of m6A marks depend on a set of enzymes commonly referred to as “writers” (METTL3, METTL14, WTAP), “erasers” (FTO, ALKBH5), and “readers” (YTHDF, IGF2BP and hnRNPA2B1 proteins) [37]. Studies in several cancers show that m6A modifications are central for the maintenance of stem-like features and therapy resistance in CSCs [38,39]. In gastric cancer, the relevance of m6A regulation is becoming increasingly clear. Elevated expression of METTL3 or WTAP, as well as decreased levels of METTL14, have been associated with poor patient prognosis [40,41]. More strikingly, recent work has shown that m6A modification of long non-coding RNAs in GCSCs can promote stemness and tumorigenicity by stabilizing transcripts and enhancing proliferative pathways [41]. Taken together, the evidence underscores how the CSC concept has evolved: from its first description in leukemia, to its establishment in solid tumors, and more recently to its epitranscriptomic regulation. In gastric cancer, the identification of CSCs through surface markers and transcriptional networks has already provided important insights into disease aggressiveness and therapeutic resistance. The emerging role of m6A adds a powerful new dimension, suggesting that RNA modifications could serve not only as biomarkers for GCSCs but also as actionable targets in the development of future therapies.
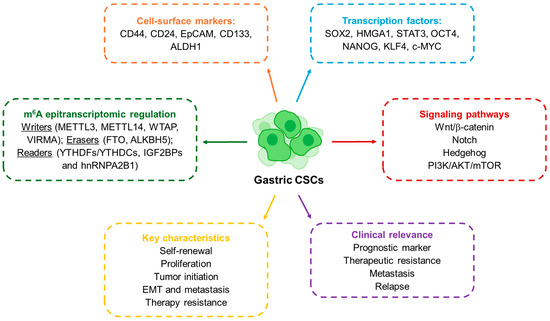
Figure 1.
Schematic representation of the molecular and functional features of GCSCs.
3. The m6A Modification Machinery in the Gastric Cancer Stem Cell Phenotype
Among the regulatory layers that shape CSC biology, RNA modifications, prominently m6A, have recently come into the spotlight. In gastric cancer, increasing evidence indicates that m6A modifications not only correlate with tumor progression but also play a role in maintaining the malignant phenotype of GCSCs [42]. The enzymes that establish, remove, and interpret this mark, often referred to as the m6A machinery, form a dynamic system that can profoundly influence cell fate decisions. An overview of their roles is provided below and in Table 1.

Table 1.
The core components of the m6A modification machinery, categorized as “writers,” “erasers,” and “readers”, implicated in GCSC maintenance and therapy resistance, highlighting their potential as diagnostic biomarkers and therapeutic targets. Their canonical functions in RNA metabolism are outlined alongside their roles in GCSCs features.
3.1. “Writers”
The “writers” of m6A marks are methyltransferase complexes that deposit the modification on specific adenosine residues. Central to this activity are METTL3 and METTL14 methyltransferases, stabilized by the cofactor WTAP, along with accessory proteins such as RBM15, KIAA1429 (VIRMA) and ZC3H13, which confer substrate specificity [96,97,98]. These enzymes are responsible for the global landscape of m6A in cells, and their dysregulation has been linked to stemness, tumor initiation, metastasis and resistance to therapy in multiple cancers. Overall, a consistent pattern emerges, METTL3 generally functions as an oncogenic driver that enhances stem-like properties through m6A-dependent stabilization of key transcripts. In gastric cancer, METTL3-driven m6A methylation at specific adenosines on the long non-coding RNAs (lncRNAs) PSMA3-AS1 (A1225) and MIR22HG (A2041) is enriched in GCSCs compared with non-stem cells [41]. Silencing METTL3 reduced GCSC viability, induced G0/G1 arrest, and promoted apoptosis, highlighting the role of m6A in stemness maintenance. Remarkably, site-specific methylation rescue restored proliferative and tumorigenic potential by stabilizing EEF1A1 and LRPPRC, thereby suppressing apoptosis and reestablishing self-renewal in vivo [41,43]. These findings identify site-specific m6A methylation of lncRNAs as a molecular switch sustaining GCSC survival and malignant progression [41]. Moreover, studies with oxaliplatin sensitive and resistant gastric cancer organoids revealed that CD133+ CSCs acquire resistance through METTL3-mediated m6A modification of PARP1 mRNA. This modification stabilizes PARP1, enhances DNA repair, and promotes oxaliplatin resistance, advancing our understanding of drug resistance mechanisms in gastric cancer [44]. Evidence from other tumors supports the notion that METTL3 often acts as an oncogenic driver of CSC phenotypes. For example, in colorectal CSCs, METTL3 promotes WNT/β-catenin signaling, sustaining stemness [99]. In glioblastoma CSCs, METTL3-mediated m6A methylation enhances SOX2 expression, reinforcing self-renewal and tumorigenic capacity [39]. In leukemia, overexpression of METTL3 has been shown to increase MYC pathway activity, supporting leukemic CSCs survival [100]. These studies highlight METTL3 as a master regulator that reinforces CSC phenotypes through transcript stabilization and pathway activation. In contrast, METTL14 exhibits context-dependent and often opposing roles. In gastric cancer, METTL14 suppresses stemness, and low METTL14 expression correlates with poor survival. Mechanistically, it promotes m6A-dependent degradation of ATF5 mRNA and restrain the WDR74/β-catenin axis [48]. Also, loss of METTL14 suppresses differentiation and promotes stem-like traits in acute myeloid leukemia [101]. This points to a nuanced role of “writer” components depending on tumor type and cellular context. Finally, in what concerns WTAP, it stabilizes the METTL3/METTL14 complex, and its overexpression in gastric cancer predicts poor prognosis and therapy resistance [50,51].
Taken together, these studies reveal that the interplay between METTL3, METTL14 and WTAP fine-tunes m6A methylation activity, influencing the balance between CSC maintenance and differentiation across tumor types.
3.2. “Erasers”
Counterbalancing the “writers” are the “erasers”, enzymes that remove m6A and thereby restore transcripts to an unmethylated state. The first to be identified, fat mass-and obesity-associated protein (FTO), and later AlkB homolog 5 (ALKBH5), provide CSCs with plasticity, allowing dynamic remodeling of the transcriptome in response to stress, therapy or microenvironmental cues [102,103]. Recent studies showed that FTO promotes stem-like properties and lymph node metastasis in gastric cancer, while its knockdown suppresses proliferation, migration, and invasion [55,56]. Mechanistically, FOXA2 suppresses FTO transcription, while FTO promotes stemness by stabilizing MYC mRNA and acting through SOX2 [55,57]. ALKBH5 plays a context-dependent role in gastric cancer, reported as either upregulated or downregulated across studies [61,64,65]. Functionally, it promotes cisplatin resistance via the HIF-1α pathway and enhances N-Methyl-N’-nitro-N-nitrosoguanidine (MNNG)-induced stemness and invasion through activation of the ZKSCAN3–VEGFA axis [61]. In addition, lncNRON facilitates ALKBH5 recognition of m6A-modified RNAs, stabilizing NANOG expression [62]. Collectively, modulation of FTO, ALKBH5, or their downstream targets restores drug sensitivity and limits tumor relapse in preclinical models. FTO has also been implicated in leukemic CSCs survival and resistance to differentiation therapies [104]. By demethylating transcripts such as ASB2 and RARA, FTO promotes survival and resistance to differentiation therapies in acute myeloid leukemia [104]. ALKBH5 supports the maintenance of glioblastoma cancer stem-like cells by stabilizing FOXM1 transcripts [103]. In breast cancer, ALKBH5 has been shown to enhance NANOG expression, contributing to hypoxia-induced CSC enrichment [105]. All these findings illustrate how “erasers” allow CSCs to adapt to tumor microenvironmental cues, such as low oxygen levels or chemotherapeutic stress and the therapeutic potential of targeting epitranscriptomic modifications.
3.3. “Readers”
Finally, the functional impact of m6A is determined by “readers”, proteins that recognize methylated transcripts and dictate their fate. The YTH (YT521-B homology) domain family proteins (YTHDF1/2/3 and YTHDC1/2) are the best characterized, with roles in translation promotion, mRNA decay and nuclear processing, shaping the balance between differentiation and self-renewal [106]. Another important reader family, the Insulin-like growth factor 2 mRNA-binding proteins (IGF2BP1–3), stabilizes methylated mRNAs and enhances their translation, frequently promoting oncogenic programs [80]. Among these m6A “readers”, YTHDF1/2 and IGF2BP2/3 regulate mRNA stabilization and translation of stemness-related genes in gastric cancer. YTHDF2 destabilizes ONECUT2 mRNA, indirectly activating TFPI and promoting stemness and oxaliplatin resistance [70]. IGF2BP2 stabilizes colony stimulating factor 2 (CSF2) mRNA, driving MSC reprogramming and enhancing stemness, proliferation, and invasion via Notch suppression [86]. HNRNPA2B1 (Heterogeneous nuclear ribonucleoproteins A2/B1) has emerged as another critical reader, promoting metabolic rewiring and therapy resistance [93]; its knockdown reduces tumorsphere formation, stemness marker expression, and increases response to cisplatin [94]. Mechanistically, hnRNPA2B1 regulates BIRC5 splicing, stabilizes NEAT1, and activates Wnt/β-catenin signaling, thereby sustaining CSC traits and chemoresistance [95]. Collectively, these m6A “readers” act as post-transcriptional regulators that reinforce oncogenic programs driving CSC identity. In hepatocellular carcinoma, IGF2BP1 stabilizes c-MYC mRNA in an m6A-dependent manner, sustaining cancer stem-like features [80]. IGF2BPs also contribute to chemoresistance and metastasis in multiple cancers, underscoring their role as oncogenic amplifiers [107]. Through all these “readers”, CSCs exploit m6A marks to amplify pathways such as WNT, NOTCH, or MYC signaling, sustaining self-renewal and therapy resistance.
3.4. Clinical Relevance of the m6A Molecular Pathway
Evidence across tumor types highlights the versatility of the m6A pathway in CSC biology and shows that CSCs rely on the flexibility of its components to maintain their identity and adapt under selective pressures. The clinical relevance of this pathway is increasingly evident, as aberrant expression of m6A regulators such as METTL3, FTO, ALKBH5, or IGF2BPs correlates with poor prognosis, advanced disease stage, therapy resistance and immune evasion, in multiple cancer types [108,109,110,111]. In gastric cancer, global dysregulation of the m6A machinery has also been documented, with METTL3, FTO, YTHDFs, IGF2BPs and hnRNPA2B1 frequently upregulated in patient tissues and associated with poor prognosis, advanced stages and metastasis [42]. Conversely, downregulation of METTL14 expression correlates with unfavorable outcomes [42].
Beyond prognostic implications, the m6A machinery presents promising therapeutic opportunities. Strategies aimed at selectively targeting “writer”’s activity, disrupting “readers” or interfering with their RNA interactions are under active investigation [112,113,114,115,116]. By inhibiting METTL3, m6A marks on oncogenic mRNAs are reduced, destabilizing them and suppressing gastric cancer growth [117]. The METTL3 small molecule inhibitor STM2457 was firstly studied as a therapeutic approach in myeloid leukemia [118]. Importantly, STM2457 shows antitumor activity in preclinical models of gastric cancer, when combined with anti–PD-1 therapy [119]. Small-molecule inhibitors of FTO have also shown promise in gastric cancer, allowing the increase in m6A levels and regulating Wnt/PI3K-Akt signaling [120]. One of these inhibitors, meclofenamic acid, is already being used in a clinical trial enrolling patients with recurrent or progressive brain metastasis from solid primary tumors (NCT02429570) and in preclinical leukemia models [121].
The m6A RNA modifications and their regulators—including METTL3, ALKBH5, FTO, YTHDFs, hnRNPA2B1, and IGF2BP2—have particularly emerged as key drivers of chemotherapy resistance in gastric cancer [86,94,122]. For instance, METTL3 “writer” is upregulated in oxaliplatin-resistant gastric cancer cells and promotes resistance via the DNA repair pathway [44,47]. The “eraser” ALKBH5 has been associated with chemotherapy resistance and stemness maintenance, suggesting that its expression levels might predict therapeutic response [94]. Equally important are the m6A “readers”, including YTHDF1-3, IGF2BP1-3 and hnRNPA2B1 proteins, which bind methylated transcripts to regulate their stability or translation to confer chemoresistance [94,95]. Elevated IGF2BP expression has been generally observed in gastric cancer and shown to stabilize oncogenic mRNAs, reinforcing stem-like traits [86]. These insights suggest that combining m6A modulators with chemotherapy may enhance therapeutic efficacy.
Besides driving CSC maintenance and therapeutic resistance, imbalanced m6A regulators promote immune evasion by modulating cytokine signaling, antigen presentation, and immune checkpoint expression, fostering an immunosuppressive tumor microenvironment. High expression of METTL3, METTL14, FTO, YTDHF1-2 and IGF2BP1, in gastric cancer, facilitates tumor immune evasion by maintaining the stability and expression of PD-L1 transcripts [123,124,125,126,127]. Increased activity of the IGF2BP1 “reader”, for instance, amplifies tumor proliferation and dampens CD8+ T-cell–mediated cytotoxicity, correlating with unfavorable patient outcomes. In contrast, its downregulation disrupts PD-L1–dependent immune suppression, thereby restoring antitumor immunity [127]. Loss of other m6A readers, such as YTHDF1, further influence immune modulation by enhancing dendritic cell recruitment and antigen presentation [128,129]. Demethylases such as ALKBH5 and FTO also reshape cytokine networks in gastric cancer [130]. ALKBH5 has been shown to modulate immune evasion by regulating CD8+ T-cell infiltration, CD58 expression, dendritic cell recruitment, and broader immune cell interactions [131,132]. FTO instead regulates TGF-β expression, but also correlates with poor immune infiltration [133]. Evidence shows dynamic interactions between CSCs and immune cells in the tumor microenvironment, with certain immune cells promoting CSC expansion while enabling immune evasion [134].
Collectively, these findings underscore the m6A-CSC axis as a pivotal regulator linking stemness, therapeutic resistance and immune escape (Figure 2). The m6A modification machinery provides CSCs with a powerful mechanism to control RNA fate and thereby sustain stemness, plasticity, and survival. While different cancers exploit different nodes of this machinery, the principle remains consistent: the balance between “writers”, “erasers”, and “readers” dictates CSC function and tumor behavior. As our understanding advances, targeting the m6A RNA methylation pathway, through pharmacological inhibition or silencing of its machinery, has emerged as a potential strategy to eliminate CSCs, overcome therapy resistance and enhance responses to chemotherapy and immunotherapy, offering new avenues for precision treatment also in gastric cancer [110,123,125].
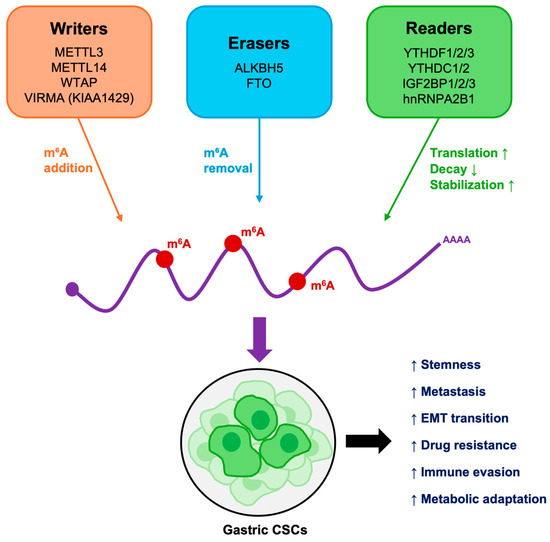
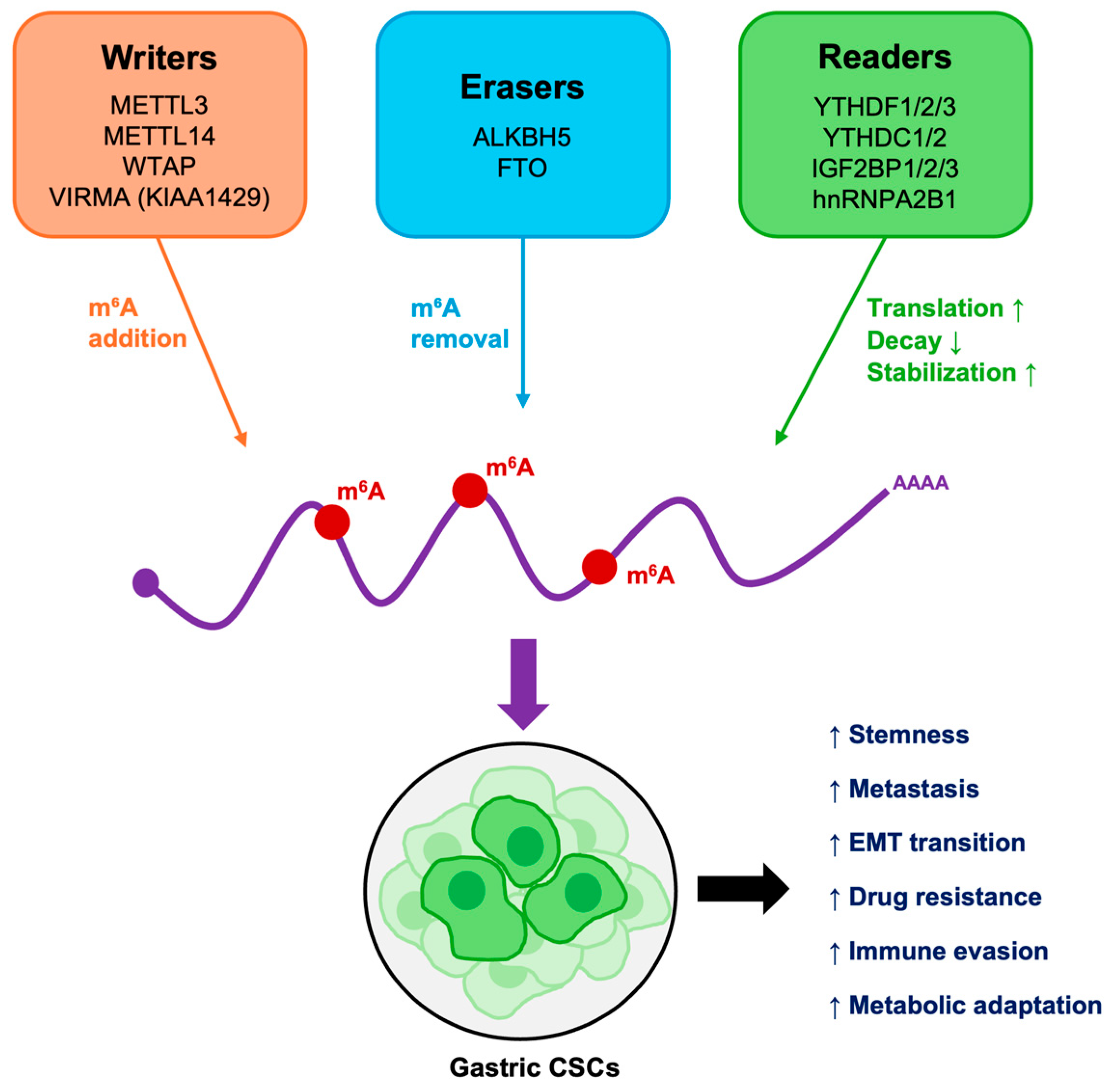
Figure 2.
Functional overview of m6A-mediated regulation in GCSCs. m6A writers, erasers, and readers cooperatively regulate mRNA stability, translation, and degradation. Their dysregulation sustains stemness, promotes epithelial–mesenchymal transition (EMT), alters metabolism and immune responses, and drives therapy resistance and metastasis.
4. Functional Role of m6A in Gastric Cancer Stem Cells
Functionally, METTL3, one of the best characterized m6A “writers”, appears to contribute to gastric cancer progression by stabilizing oncogenic transcripts and modulating key signaling pathways. Reported mechanisms include: (i) stabilization of HDGF mRNA via IGF2BP3, which enhances glycolysis and angiogenesis [45]; (ii) methylation of ZMYM1, which contributes to the inactivation of the RAS/ERK/c-FOS pathway and reduces E-cadherin expression, thereby facilitating EMT and metastasis [135]; (iii) promotion of ADAMTS9 degradation via YTHDF2, resulting in PI3K/AKT pathway activation and enhanced tumor progression [136]. Collectively, these processes may reinforce of a GCSC phenotype, characterized by increased therapy resistance, invasiveness and metastatic potential. Overall, current evidence suggests that m6A modifications in GCSCs function across multiple regulatory layers, from the fine-tuned editing of non-coding RNAs to the broader remodeling of oncogenic signaling networks [137]. These findings underscore that GCSCs are particularly reliant on m6A-driven transcriptional and post-transcriptional programs, which sustain their stem-like properties and underlie relapse and metastasis [17,41]. This dependency makes m6A regulators potential vulnerabilities of CSCs. Yet, because m6A also plays essential roles in normal physiology, therapeutic strategies must carefully exploit CSC-specific epitranscriptomic dependencies to achieve selectivity [138,139].
Additionally, several studies have also highlighted the diagnostic potential of m6A regulators. Unlike CSCs conventional markers such as CD44, CD133 and EpCAM, which often lack specificity and can be expressed in normal progenitor cells, m6A-associated signatures may offer a more dynamic and mechanistic readout of tumor aggressiveness [140,141]. For instance, the role of METTL3 in stabilizing oncogenic transcripts such as HDGF and long non-coding RNAs (e.g., PSMA3-AS1, MIR22HG) is strongly linked to GCSC self-renewal and tumor initiation capacity [41,45]. Measuring METTL3 expression or its downstream m6A-dependent RNA modifications could thus serve as a biomarker for the presence of GCSC populations. Their diagnostic relevance lies not only in tissue expression patterns but also in their potential detection in liquid biopsies (e.g., circulating tumor cells, exosomal RNA), making them attractive candidates for non-invasive cancer monitoring [142,143]. What makes m6A particularly compelling in diagnostics is its contextual plasticity. Unlike static genetic mutations, RNA modifications are reversible and dynamically reflect cellular states, including stress, hypoxia, and therapy exposure. This means that an m6A-based diagnostic approach could capture the evolving nature of GCSCs during disease progression and treatment, enabling more precise patient stratification [144]. Recent advances in m6A profiling technologies, such as MeRIP-seq, m6A-CLIP, and nanopore direct RNA sequencing, are accelerating the identification of GCSC-specific methylation signatures (Table 2) [145]. These methods have enabled the identification, quantification and functional characterization of m6A marks across the transcriptome with increasing accuracy and depth.

Table 2.
Summary of the main techniques used for m6A detection and their translational relevance.
Early detection methods, such as MeRIP-seq and m6A-seq, relied on antibody-based RNA immunoprecipitation, followed by next-generation sequencing, providing the first transcriptome-wide m6A maps in mammalian cells [145,149]. In gastric cancer, MeRIP-seq has been instrumental in revealing that METTL3- and METTL14-mediated m6A methylation regulates oncogenic lncRNA, such as PSMA3-AS1 and MIR22HG, whose site-specific m6A modifications enhance transcript stability and promote proliferation and stemness while suppressing apoptosis in GCSCs but were limited in resolution (~100–200 nt) [41,158]. Subsequent advances, including miCLIP and m6A-CLIP, achieved single-nucleotide precision, enabling a more accurate characterization of m6A topology in cancer-related transcripts [146,159]. More recently, nanopore-based direct RNA sequencing and targeted assays such as SELECT and SCARLET have improved the sensitivity and clinical applicability of m6A detection. Notably, these techniques have validated lncRNA methylation sites that are associated with poor prognosis and chemoresistance in gastric cancer patients [41]. Integrated with computational modeling, single-cell, and spatial transcriptomic analyses, these approaches are now enlightening how m6A dynamics influence intratumoral heterogeneity, cancer stem cell maintenance, and therapy response, marking a significant step towards translational and precision applications in gastric cancer [159,160,161]. These approaches may eventually enable clinicians to better distinguish aggressive, stem-like gastric cancers from less malignant subtypes, potentially complementing or even enhancing current molecular diagnostic panels [138,139,146]. Thereby, the same m6A signatures that maintain GCSC plasticity might also serve as dynamic biomarkers, capturing the evolving states of stemness, drug resistance, and tumor aggressiveness [80]. By integrating epitranscriptomic readouts into diagnostic platforms, whether through tissue analysis or liquid biopsy approaches, it may become possible to identify high-risk patients earlier, monitor therapeutic responses in real time, and stratify patients more precisely than is achievable with conventional static markers [140,141,162]. In this way, the functional role of m6A in GCSCs extends beyond fundamental tumor biology, suggesting that epitranscriptomic markers may represent a promising bridge between mechanistic insight and clinical application.
Interestingly, similar m6A-dependent mechanisms have been reported in esophageal cancer, an anatomically and molecularly related malignancy [163]. METTL3 is frequently upregulated and promotes tumor initiation, proliferation, and metastasis through the AKT and EGR1/Snail pathways, while also enhancing glutamine metabolism [163,164,165,166]. Other “writers” (METTL16, WTAP, KIAA1429, RBM15) are likewise overexpressed, suggesting a broader activation of the m6A writing machinery, whereas METTL14 loss correlates with poor differentiation and aggressive behavior suggesting a tumor-suppressive role similar to that seen in gastric cancer [164,165]. Among “readers,” YTHDF1, YTHDF3, and hnRNPA2B1 facilitate metastasis, proliferation, and lipid metabolism, and “erasers” (ALKBH5, FTO) influence cell cycle control and therapy resistance [163,167,168]. This conserved epitranscriptomic machinery in gastric and esophageal cancer appears to support cancer stem-like properties and therapy resistance and may hold potential as a basis for developing improved diagnostic and therapeutic strategies in upper gastrointestinal malignancies.
5. Challenges and Future Directions
Although significant advances have been made in understanding the role of m6A modifications in GCSCs, several challenges remain before these findings can be effectively translated into clinical practice. One of the major limitations arises from the technical constraints of current profiling methods. Standard approaches such as MeRIP-seq and m6A-CLIP provide only population-level and low-resolution data, obscuring the heterogeneity of m6A signatures across individual CSCs [147,166]. Recently, single-cell m6A sequencing and direct RNA nanopore approaches have begun to address this issue, offering unprecedented resolution of isoform-specific and cell-specific methylation patterns; however, these technologies remain costly, technically demanding and are not yet standardized for clinical application [169,170,171]. Tools such as CRISPR/dCas13-based targeted m6A editing now make it possible to experimentally manipulate RNA methylation at single sites, offering unprecedented precision to dissect how these modifications influence CSC biology. Another important challenge concerns the pleiotropic nature of m6A regulators. Core enzymes such as METTL3, ALKBH5 and IGF2BP proteins play essential roles not only in cancer but also in normal tissue homeostasis, raising concerns about toxicity and selectivity when targeting these proteins therapeutically [10,167]. Preclinical studies, such as the development of the first-in-class METTL3 inhibitor STM2457 in leukemia, have demonstrated that pharmacological disruption of m6A machinery can impair cancer stemness [172]. Nevertheless, extending these approaches to solid tumors, including gastric cancer, will require strategies that exploit CSC-specific or context-dependent vulnerabilities, thereby minimizing adverse effects [118,139,173]. The dynamic and reversible nature of m6A modifications presents both opportunities and difficulties for their use as biomarkers. On one hand, their plasticity reflects the evolving states of cancer stemness, drug resistance and adaptation to microenvironmental stressors such as hypoxia. On the other hand, this variability complicates the establishment of stable diagnostic markers. Liquid biopsy approaches, including circulating RNA and exosomal RNA analysis, hold promise for tracking such changes in real time, but robust validation in large patient cohorts remains lacking [80,168]. Looking forward, the field is moving toward integrative and translational approaches. Multi-omics frameworks that combine epitranscriptomic signatures with genomic, epigenetic, proteomic, and metabolomic data may provide more robust predictive models of GCSC biology and disease progression [174]. Advances in artificial intelligence and machine learning are expected to accelerate this process by enabling the discovery of complex m6A-based patterns that predict relapse or therapeutic response [175,176,177]. At the same time, the translational gap must be bridged through prospective studies, standardized detection protocols and early-phase clinical trials designed to test m6A modulators in combination with established therapies. In summary, while challenges remain in terms of resolution, therapeutic selectivity, and biomarker validation, the study of m6A modifications in GCSCs represents a rapidly evolving frontier. With continued technological innovation and clinical validation, the epitranscriptomic regulation of GCSCs is poised to transition from a mechanistic insight into a clinically actionable paradigm for the diagnosis and treatment of gastric cancer.
6. Conclusions
Over time, CSCs have shifted from a theoretical concept to a clinically relevant driver of relapse and therapy resistance. In gastric cancer, they represent a crucial challenge, yet also an opportunity for targeted intervention. The discovery of m6A RNA modification as a master regulator of GCSC biology has opened new lines of research linking epitranscriptomics to cancer progression, diagnostics and therapy. Current evidence indicates that GCSCs are particularly dependent on m6A-driven programs controlling stemness, signaling and adaptation to stress, making m6A regulators compelling candidates for selective therapeutic targeting. At the same time, the widespread physiological roles of m6A highlight the need for precision strategies that exploit GCSC-specific dependencies while minimizing toxicity. Advances in single-cell sequencing, liquid biopsy technologies and selective small-molecule inhibitors are expected to accelerate the translation of these insights into the clinic. Taken together, integrating epitranscriptomic knowledge into CSCs biology offers a promising path towards more accurate diagnostics and effective therapies for gastric cancer.
Funding
This work was supported by Portuguese funds through the FCT—Fundação para a Ciência e a Tecnologia/Ministério da Ciência, Tecnologia e Inovação in the framework of the project “Institute for Research and Innovation in Health Sciences” (POCI-01-0145-FEDER-007274).
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| ALKBH5 | AlkB homolog 5 |
| CSC | Cancer stem cell |
| EMT | Epithelial–mesenchymal transition |
| GCSC | Gastric cancer stem cell |
| FTO | Fat mass-and obesity-associated protein |
| hnRNPA2B1 | Heterogeneous nuclear ribonucleoproteins A2/B1 |
| IGF2BP1/2 | Insulin-like growth factor 2 mRNA-binding proteins |
| lncRNA | Long non-coding RNAs |
| m6A | N6-methyladenosine |
| METTL3 | Methyltransferase 3 |
| METTL14 | Methyltransferase 14 |
| WTAP | Wilms tumor 1-associated protein |
| VIRMA | vir like m6A methyltransferase associated |
| YTHD | YT521-B homology domain |
References
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef]
- Park, J.Y.; Georges, D.; Alberts, C.J.; Bray, F.; Clifford, G.; Baussano, I. Global lifetime estimates of expected and preventable gastric cancers across 185 countries. Nat. Med. 2025, 31, 3020–3027. [Google Scholar] [CrossRef]
- Lin, X.; Yang, P.; Wang, M.; Huang, X.; Wang, B.; Chen, C.; Xu, A.; Cai, J.; Khan, M.; Liu, S.; et al. Dissecting gastric cancer heterogeneity and exploring therapeutic strategies using bulk and single-cell transcriptomic analysis and experimental validation of tumor microenvironment and metabolic interplay. Front. Pharmacol. 2024, 15, 1355269. [Google Scholar] [CrossRef]
- El-Tanani, M.; Rabbani, S.A.; Satyam, S.M.; Rangraze, I.R.; Wali, A.F.; El-Tanani, Y.; Aljabali, A.A.A. Deciphering the Role of Cancer Stem Cells: Drivers of Tumor Evolution, Therapeutic Resistance, and Precision Medicine Strategies. Cancers 2025, 17, 382. [Google Scholar] [CrossRef]
- Rao, X.; Zhang, C.; Luo, H.; Zhang, J.; Zhuang, Z.; Liang, Z.; Wu, X. Targeting Gastric Cancer Stem Cells to Enhance Treatment Response. Cells 2022, 11, 2828. [Google Scholar] [CrossRef]
- Pádua, D.; Figueira, P.; Ribeiro, I.; Almeida, R.; Mesquita, P. The Relevance of Transcription Factors in Gastric and Colorectal Cancer Stem Cells Identification and Eradication. Front. Cell Dev. Biol. 2020, 8, 442. [Google Scholar] [CrossRef]
- Pádua, D.; Barros, R.; Amaral, A.L.; Mesquita, P.; Freire, A.F.; Sousa, M.; Maia, A.F.; Caiado, I.; Fernandes, H.; Pombinho, A.; et al. A SOX2 Reporter System Identifies Gastric Cancer Stem-Like Cells Sensitive to Monensin. Cancers 2020, 12, 495. [Google Scholar] [CrossRef] [PubMed]
- Pádua, D.; Figueira, P.; Pombinho, A.; Monteiro, I.; Pereira, C.F.; Almeida, R.; Mesquita, P. HMGA1 stimulates cancer stem-like features and sensitivity to monensin in gastric cancer. Exp. Cell Res. 2024, 442, 114257. [Google Scholar] [CrossRef]
- Fu, L.; Bu, L.; Yasuda, T.; Koiwa, M.; Akiyama, T.; Uchihara, T.; Baba, H.; Ishimoto, T. Gastric Cancer Stem Cells: Current Insights into the Immune Microenvironment and Therapeutic Targets. Biomedicines 2020, 8, 7. [Google Scholar] [CrossRef] [PubMed]
- Qiu, L.; Jing, Q.; Li, Y.; Han, J. RNA modification: Mechanisms and therapeutic targets. Mol. Biomed. 2023, 4, 25. [Google Scholar] [CrossRef] [PubMed]
- Haruehanroengra, P.; Zheng, Y.Y.; Zhou, Y.; Huang, Y.; Sheng, J. RNA modifications and cancer. RNA Biol. 2020, 17, 1560–1575. [Google Scholar] [CrossRef] [PubMed]
- Petri, B.J.; Klinge, C.M. m6A readers, writers, erasers, and the m6A epitranscriptome in breast cancer. J. Mol. Endocrinol. 2023, 70, e220110. [Google Scholar] [CrossRef]
- Jiang, X.; Liu, B.; Nie, Z.; Duan, L.; Xiong, Q.; Jin, Z.; Yang, C.; Chen, Y. The role of m6A modification in the biological functions and diseases. Signal Transduct. Target. Ther. 2021, 6, 74. [Google Scholar] [CrossRef]
- Liu, W.W.; Zhang, Z.Y.; Wang, F.; Wang, H. Emerging roles of m6A RNA modification in cancer therapeutic resistance. Exp. Hematol. Oncol. 2023, 12, 21. [Google Scholar] [CrossRef]
- Shen, L.T.; Che, L.R.; He, Z.; Lu, Q.; Chen, D.F.; Qin, Z.Y.; Wang, B. Aberrant RNA m(6)A modification in gastrointestinal malignancies: Versatile regulators of cancer hallmarks and novel therapeutic opportunities. Cell Death Dis. 2023, 14, 236. [Google Scholar] [CrossRef]
- Pinello, N.; Sun, S.; Wong, J.J. Aberrant expression of enzymes regulating m(6)A mRNA methylation: Implication in cancer. Cancer Biol. Med. 2018, 15, 323–334. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Li, C.; Zhou, H.; Yang, Y.; Liang, N.; Fu, Y.; Luo, Q.; Zhan, Y. The regulatory mechanism of m6A modification in gastric cancer. Discov. Oncol. 2024, 15, 283. [Google Scholar] [CrossRef] [PubMed]
- Kleinsmith, L.J.; Pierce, G.B., Jr. Multipotentiality of single embryonal carcinoma cells. Cancer Res. 1964, 24, 1544–1551. [Google Scholar]
- Lapidot, T.; Sirard, C.; Vormoor, J.; Murdoch, B.; Hoang, T.; Caceres-Cortes, J.; Minden, M.; Paterson, B.; Caligiuri, M.A.; Dick, J.E. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature 1994, 367, 645–648. [Google Scholar] [CrossRef]
- Bonnet, D.; Dick, J.E. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat. Med. 1997, 3, 730–737. [Google Scholar] [CrossRef]
- Dirks, P.B. Brain tumor stem cells: The cancer stem cell hypothesis writ large. Mol. Oncol. 2010, 4, 420–430. [Google Scholar] [CrossRef]
- Singh, S.K.; Hawkins, C.; Clarke, I.D.; Squire, J.A.; Bayani, J.; Hide, T.; Henkelman, R.M.; Cusimano, M.D.; Dirks, P.B. Identification of human brain tumour initiating cells. Nature 2004, 432, 396–401. [Google Scholar] [CrossRef]
- Visvader, J.E.; Lindeman, G.J. Cancer stem cells in solid tumours: Accumulating evidence and unresolved questions. Nat. Rev. Cancer 2008, 8, 755–768. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Kim, B.; Park, J.; Park, S.; Yoo, G.; Yum, S.; Kang, W.; Lee, J.-M.; Youn, H.; Youn, B. Cancer stem cells: Landscape, challenges and emerging therapeutic innovations. Signal Transduct. Target. Ther. 2025, 10, 248. [Google Scholar] [CrossRef] [PubMed]
- Al-Hajj, M.; Wicha, M.S.; Benito-Hernandez, A.; Morrison, S.J.; Clarke, M.F. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. USA 2003, 100, 3983–3988. [Google Scholar] [CrossRef]
- Horst, D.; Kriegl, L.; Engel, J.; Kirchner, T.; Jung, A. CD133 expression is an independent prognostic marker for low survival in colorectal cancer. Br. J. Cancer 2008, 99, 1285–1289. [Google Scholar] [CrossRef]
- Li, B.; McCrudden, C.M.; Yuen, H.F.; Xi, X.; Lyu, P.; Chan, K.W.; Zhang, S.D.; Kwok, H.F. CD133 in brain tumor: The prognostic factor. Oncotarget 2017, 8, 11144–11159. [Google Scholar] [CrossRef]
- Yamashita, T.; Honda, M.; Nakamoto, Y.; Baba, M.; Nio, K.; Hara, Y.; Zeng, S.S.; Hayashi, T.; Kondo, M.; Takatori, H.; et al. Discrete nature of EpCAM+ and CD90+ cancer stem cells in human hepatocellular carcinoma. Hepatology 2013, 57, 1484–1497. [Google Scholar] [CrossRef] [PubMed]
- Takaishi, S.; Okumura, T.; Tu, S.; Wang, S.S.; Shibata, W.; Vigneshwaran, R.; Gordon, S.A.; Shimada, Y.; Wang, T.C. Identification of gastric cancer stem cells using the cell surface marker CD44. Stem Cells 2009, 27, 1006–1020. [Google Scholar] [CrossRef]
- Hsieh, H.L.; Yu, M.C.; Cheng, L.C.; Yeh, T.S.; Tsai, M.M. Molecular mechanism of therapeutic approaches for human gastric cancer stem cells. World J. Stem Cells 2022, 14, 76–91. [Google Scholar] [CrossRef]
- Wuputra, K.; Ku, C.-C.; Pan, J.-B.; Liu, C.-J.; Liu, Y.-C.; Saito, S.; Kato, K.; Lin, Y.-C.; Kuo, K.-K.; Chan, T.-F.; et al. Stem Cell Biomarkers and Tumorigenesis in Gastric Cancer. J. Pers. Med. 2022, 12, 929. [Google Scholar] [CrossRef]
- Chen, Z.; Gao, Y.; Zhang, P.; Liu, Y.; Wei, B.; Chen, L.; Xi, H. Identification of gastric cancer stem cells with CD44 and Lgr5 double labelling and their initial roles on gastric cancer malignancy and chemotherapy resistance. Cell Biol. Toxicol. 2024, 41, 12. [Google Scholar] [CrossRef]
- Chen, Y.; Fu, Z.; Xu, S.; Xu, Y.; Xu, P. The prognostic value of CD44 expression in gastric cancer: A meta-analysis. Biomed. Pharmacother. 2014, 68, 693–697. [Google Scholar] [CrossRef] [PubMed]
- Tang, K.; Liu, J.; Liu, B.; Meng, C.; Liao, J. SOX2 contributes to invasion and poor prognosis of gastric cancer: A meta-analysis. Medicine 2022, 101, e30559. [Google Scholar] [CrossRef] [PubMed]
- Basati, G.; Mohammadpour, H.; Emami Razavi, A. Association of High Expression Levels of SOX2, NANOG, and OCT4 in Gastric Cancer Tumor Tissues with Progression and Poor Prognosis. J. Gastrointest. Cancer 2020, 51, 41–47. [Google Scholar] [CrossRef]
- El-Guindy, D.M.; Wasfy, R.E.; Abdel Ghafar, M.T.; Ali, D.A.; Elkady, A.M. Oct4 expression in gastric carcinoma: Association with tumor proliferation, angiogenesis and survival. J. Egypt. Natl. Cancer Inst. 2019, 31, 3. [Google Scholar] [CrossRef]
- Malvi, P.; Ball, P.; Gupta, R.; Wajapeyee, N. m(6)A-driven transcriptomic rewiring in tumor immune surveillance. J. Immunother. Cancer 2025, 13, e012744. [Google Scholar] [CrossRef] [PubMed]
- Lan, Q.; Liu, P.Y.; Haase, J.; Bell, J.L.; Hüttelmaier, S.; Liu, T. The Critical Role of RNA m6A Methylation in Cancer. Cancer Res. 2019, 79, 1285–1292. [Google Scholar] [CrossRef]
- Visvanathan, A.; Patil, V.; Arora, A.; Hegde, A.S.; Arivazhagan, A.; Santosh, V.; Somasundaram, K. Essential role of METTL3-mediated m6A modification in glioma stem-like cells maintenance and radioresistance. Oncogene 2018, 37, 522–533. [Google Scholar] [CrossRef]
- Okugawa, Y.; Toiyama, Y.; Yin, C.; Ruiya, M.; Goel, A.; Ichikawa, T.; Imaoka, H.; Kitajima, T.; Shimura, T.; Kawamura, M.; et al. Prognostic potential of METTL3 expression in patients with gastric cancer. Oncol. Lett. 2023, 25, 64. [Google Scholar] [CrossRef]
- Ci, Y.; Zhang, Y.; Zhang, X. Methylated lncRNAs suppress apoptosis of gastric cancer stem cells via the lncRNA-miRNA/protein axis. Cell Mol. Biol. Lett. 2024, 29, 51. [Google Scholar] [CrossRef]
- Li, P.; Fang, X.; Huang, D. Exploring m6A modifications in gastric cancer: From molecular mechanisms to clinical applications. Eur. J. Med. Res. 2025, 30, 98. [Google Scholar] [CrossRef] [PubMed]
- Ji, G.; Wang, X.; Xi, H. METTL3-mediated m(6)A modification of lncRNA SNHG3 accelerates gastric cancer progression by modulating miR-186-5p/cyclinD2 axis. Int. J. Immunopathol. Pharmacol. 2023, 37, 3946320231204694. [Google Scholar] [CrossRef]
- Li, H.; Wang, C.; Lan, L.; Yan, L.; Li, W.; Evans, I.; Ruiz, E.J.; Su, Q.; Zhao, G.; Wu, W.; et al. METTL3 promotes oxaliplatin resistance of gastric cancer CD133+ stem cells by promoting PARP1 mRNA stability. Cell Mol. Life Sci. 2022, 79, 135. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Chen, C.; Ding, Q.; Zhao, Y.; Wang, Z.; Chen, J.; Jiang, Z.; Zhang, Y.; Xu, G.; Zhang, J.; et al. METTL3-mediated m(6)A modification of HDGF mRNA promotes gastric cancer progression and has prognostic significance. Gut 2020, 69, 1193–1205. [Google Scholar] [CrossRef]
- Yang, D.D.; Chen, Z.H.; Yu, K.; Lu, J.H.; Wu, Q.N.; Wang, Y.; Ju, H.Q.; Xu, R.H.; Liu, Z.X.; Zeng, Z.L. METTL3 Promotes the Progression of Gastric Cancer via Targeting the MYC Pathway. Front. Oncol. 2020, 10, 115. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Hong, Z.; Song, J.; Zhong, P.; Lin, L. METTL3 promotes drug resistance to oxaliplatin in gastric cancer cells through DNA repair pathway. Front. Pharmacol. 2023, 14, 1257410. [Google Scholar] [CrossRef]
- Zhang, P.; Xiang, H.; Peng, Q.; Ma, L.; Weng, C.; Liu, G.; Lu, L. METTL14 attenuates cancer stemness by suppressing ATF5/WDR74/β-catenin axis in gastric cancer. Cancer Sci. 2025, 116, 112–127. [Google Scholar] [CrossRef]
- Liu, X.; Xiao, M.; Zhang, L.; Li, L.; Zhu, G.; Shen, E.; Lv, M.; Lu, X.; Sun, Z. The m6A methyltransferase METTL14 inhibits the proliferation, migration, and invasion of gastric cancer by regulating the PI3K/AKT/mTOR signaling pathway. J. Clin. Lab. Anal. 2021, 35, e23655. [Google Scholar] [CrossRef]
- Cao, D.; Wang, M.; Wang, Y.; Hong, X.; Liu, X.; Si, W. WTAP-mediated m6A regulation in digestive system cancers: From molecular mechanisms to therapeutic strategies. Am. J. Cancer Res. 2025, 15, 3661–3677. [Google Scholar] [CrossRef]
- Liu, Y.; Da, M. Wilms tumor 1 associated protein promotes epithelial mesenchymal transition of gastric cancer cells by accelerating TGF-β and enhances chemoradiotherapy resistance. J. Cancer Res. Clin. Oncol. 2023, 149, 3977–3988. [Google Scholar] [CrossRef]
- Yu, H.; Zhao, K.; Zeng, H.; Li, Z.; Chen, K.; Zhang, Z.; Li, E.; Wu, Z. N(6)-methyladenosine (m(6)A) methyltransferase WTAP accelerates the Warburg effect of gastric cancer through regulating HK2 stability. Biomed. Pharmacother. 2021, 133, 111075. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Su, Q.; Li, B.; Lan, L.; Wang, C.; Li, W.; Wang, G.; Chen, W.; He, Y.; Zhang, C. High expression of WTAP leads to poor prognosis of gastric cancer by influencing tumour-associated T lymphocyte infiltration. J. Cell Mol. Med. 2020, 24, 4452–4465. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Zhang, C.; Zhang, L. WTAP-Involved the m6A Modification of lncRNA FAM83H-AS1 Accelerates the Development of Gastric Cancer. Mol. Biotechnol. 2024, 66, 1883–1893. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Jiang, X.; Zhang, Z.; Zhao, Z.; Xing, W.; Liu, Y.; Jiang, X.; Zhao, H. HDAC3-dependent transcriptional repression of FOXA2 regulates FTO/m6A/MYC signaling to contribute to the development of gastric cancer. Cancer Gene Ther. 2021, 28, 141–155. [Google Scholar] [CrossRef]
- Xu, D.; Shao, W.; Jiang, Y.; Wang, X.; Liu, Y.; Liu, X. FTO expression is associated with the occurrence of gastric cancer and prognosis. Oncol. Rep. 2017, 38, 2285–2292. [Google Scholar] [CrossRef]
- Li, M.; Wu, X.; Li, G.; Lv, G.; Wang, S. FTO Promotes the Stemness of Gastric Cancer Cells. DNA Cell Biol. 2023, 42, 411–420. [Google Scholar] [CrossRef]
- Liu, N.; Liu, C.; Wang, Z.; Wang, L.; Wang, J.; Kong, J. FTO demethylates m6A modifications in CDKAL1 mRNA and promotes gastric cancer chemoresistance by altering mitochondrial dynamics. Clin. Exp. Pharmacol. Physiol. 2023, 50, 307–315. [Google Scholar] [CrossRef]
- Zeng, X.; Lu, Y.; Zeng, T.; Liu, W.; Huang, W.; Yu, T.; Tang, X.; Huang, P.; Li, B.; Wei, H. RNA demethylase FTO participates in malignant progression of gastric cancer by regulating SP1-AURKB-ATM pathway. Commun. Biol. 2024, 7, 800. [Google Scholar] [CrossRef]
- Fang, M.; Ye, L.; Zhu, Y.; Huang, L.; Xu, S. M6A Demethylase ALKBH5 in Human Diseases: From Structure to Mechanisms. Biomolecules 2025, 15, 157. [Google Scholar] [CrossRef]
- Wang, Q.; Huang, Y.; Jiang, M.; Tang, Y.; Wang, Q.; Bai, L.; Yu, C.; Yang, X.; Ding, K.; Wang, W.; et al. The demethylase ALKBH5 mediates ZKSCAN3 expression through the m(6)A modification to activate VEGFA transcription and thus participates in MNNG-induced gastric cancer progression. J. Hazard. Mater. 2024, 473, 134690. [Google Scholar] [CrossRef]
- Wang, S.; Wang, Y.; Zhang, Z.; Zhu, C.; Wang, C.; Yu, F.; Zhao, E. Long Non-Coding RNA NRON promotes Tumor Proliferation by regulating ALKBH5 and Nanog in Gastric Cancer. J. Cancer 2021, 12, 6861–6872. [Google Scholar] [CrossRef]
- Chen, C.; Zhai, E.; Liu, Y.; Qian, Y.; Zhao, R.; Ma, Y.; Liu, J.; Huang, Z.; Chen, J.; Cai, S. ALKBH5-mediated CHAC1 depletion promotes malignant progression and decreases cisplatin-induced oxidative stress in gastric cancer. Cancer Cell Int. 2023, 23, 293. [Google Scholar] [CrossRef]
- Hu, Y.; Gong, C.; Li, Z.; Liu, J.; Chen, Y.; Huang, Y.; Luo, Q.; Wang, S.; Hou, Y.; Yang, S.; et al. Demethylase ALKBH5 suppresses invasion of gastric cancer via PKMYT1 m6A modification. Mol. Cancer 2022, 21, 34. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Lin, F.; Zhao, B.; Chen, G.; Wei, C.; Chen, X.; Nie, R.; Zhang, R.; Zhao, Z.; Zhou, Z.; et al. ALKBH5 suppresses gastric cancer tumorigenesis and metastasis by inhibiting the translation of uncapped WRAP53 RNA isoforms in an m6A-dependent manner. Mol. Cancer 2025, 24, 19. [Google Scholar] [CrossRef]
- Chen, D.; Cheung, H.; Lau, H.C.-H.; Yu, J.; Wong, C.C. N6-Methyladenosine RNA-Binding Protein YTHDF1 in Gastrointestinal Cancers: Function, Molecular Mechanism and Clinical Implication. Cancers 2022, 14, 3489. [Google Scholar] [CrossRef] [PubMed]
- Jang, D.; Hwa, C.; Kim, S.; Oh, J.; Shin, S.; Lee, S.J.; Kim, J.; Lee, S.E.; Yang, Y.; Kim, D.; et al. RNA N(6)-Methyladenosine-Binding Protein YTHDFs Redundantly Attenuate Cancer Immunity by Downregulating IFN-γ Signaling in Gastric Cancer. Adv. Sci. 2025, 12, e2410806. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.Y.; Liang, R.; Yi, Y.C.; Fan, H.N.; Chen, M.; Zhang, J.; Zhu, J.S. The m(6)A Reader YTHDF1 Facilitates the Tumorigenesis and Metastasis of Gastric Cancer via USP14 Translation in an m(6)A-Dependent Manner. Front. Cell Dev. Biol. 2021, 9, 647702. [Google Scholar] [CrossRef]
- Pi, J.; Wang, W.; Ji, M.; Wang, X.; Wei, X.; Jin, J.; Liu, T.; Qiang, J.; Qi, Z.; Li, F.; et al. YTHDF1 Promotes Gastric Carcinogenesis by Controlling Translation of FZD7. Cancer Res. 2021, 81, 2651–2665. [Google Scholar] [CrossRef]
- Fan, X.; Han, F.; Wang, H.; Shu, Z.; Qiu, B.; Zeng, F.; Chen, H.; Wu, Z.; Lin, Y.; Lan, Z.; et al. YTHDF2-mediated m(6)A modification of ONECUT2 promotes stemness and oxaliplatin resistance in gastric cancer through transcriptionally activating TFPI. Drug Resist. Updat. 2025, 79, 101200. [Google Scholar] [CrossRef]
- Fang, Y.; Wu, X.; Gu, Y.; Shi, R.; Yu, T.; Pan, Y.; Zhang, J.; Jing, X.; Ma, P.; Shu, Y. LINC00659 cooperated with ALKBH5 to accelerate gastric cancer progression by stabilising JAK1 mRNA in an m(6) A-YTHDF2-dependent manner. Clin. Transl. Med. 2023, 13, e1205. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Chen, Y.; He, Y.; Da, M. YTHDF2 promotes gastric cancer progression and enhances chemoradiotherapy resistance. Drug Dev. Res. 2024, 85, e22179. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Han, S.; Liu, K.; Si, Y.; Xuan, M.; Zhang, R.; Jiao, J.; Liu, T.; Yu, W.; Wang, J. YTHDF3 promotes the progression of gastric cancer by activating Wnt/β-catenin signaling pathway via targeting NEK7. bioRxiv 2025. [Google Scholar] [CrossRef]
- Yu, Y.; Meng, L.L.; Chen, X.Y.; Fan, H.N.; Chen, M.; Zhang, J.; Zhu, J.S. m(6)A reader YTHDF3 is associated with clinical prognosis, related RNA signatures and immunosuppression in gastric cancer. Cell Signal 2023, 108, 110699. [Google Scholar] [CrossRef]
- Yuan, W.; Chen, S.; Li, B.; Han, X.; Meng, B.; Zou, Y.; Chang, S. The N6-methyladenosine reader protein YTHDC2 promotes gastric cancer progression via enhancing YAP mRNA translation. Transl. Oncol. 2022, 16, 101308. [Google Scholar] [CrossRef] [PubMed]
- Niu, L.; Li, Y.; Huang, G.; Huang, W.; Fu, J.; Feng, L. FAM120A deficiency improves resistance to cisplatin in gastric cancer by promoting ferroptosis. Commun. Biol. 2024, 7, 399. [Google Scholar] [CrossRef]
- Zhou, H.; Sun, Q.; Feng, M.; Gao, Z.; Jia, S.; Cao, L.; Yu, X.; Gao, S.; Wu, H.; Li, K. Regulatory mechanisms and therapeutic implications of insulin-like growth factor 2 mRNA-binding proteins, the emerging crucial m(6)A regulators of tumors. Theranostics 2023, 13, 4247–4265. [Google Scholar] [CrossRef]
- Ramesh-Kumar, D.; Guil, S. The IGF2BP family of RNA binding proteins links epitranscriptomics to cancer. Semin. Cancer Biol. 2022, 86, 18–31. [Google Scholar] [CrossRef]
- Wang, Q.; Chen, C.; Xu, X.; Shu, C.; Cao, C.; Wang, Z.; Fu, Y.; Xu, L.; Xu, K.; Xu, J.; et al. APAF1-Binding Long Noncoding RNA Promotes Tumor Growth and Multidrug Resistance in Gastric Cancer by Blocking Apoptosome Assembly. Adv. Sci. 2022, 9, e2201889. [Google Scholar] [CrossRef]
- Huang, H.; Weng, H.; Sun, W.; Qin, X.; Shi, H.; Wu, H.; Zhao, B.S.; Mesquita, A.; Liu, C.; Yuan, C.L.; et al. Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 2018, 20, 285–295. [Google Scholar] [CrossRef]
- Xiao, J.; Lin, L.; Luo, D.; Shi, L.; Chen, W.; Fan, H.; Li, Z.; Ma, X.; Ni, P.; Yang, L.; et al. Long noncoding RNA TRPM2-AS acts as a microRNA sponge of miR-612 to promote gastric cancer progression and radioresistance. Oncogenesis 2020, 9, 29. [Google Scholar] [CrossRef]
- Myint, K.; Chuang, L.S.H.; Teh, Y.X.; Mawan, N.A.; Shi, E.J.; Mok, M.M.H.; Nuttonmanit, N.; Matsuo, J.; Li, Y.; Yang, H.; et al. Oncofetal protein IGF2BP1 regulates IQGAP3 expression to maintain stem cell potential in cancer. iScience 2022, 25, 105194. [Google Scholar] [CrossRef]
- Zhao, T.; You, J.; Wang, C.; Li, B.; Liu, Y.; Shao, M.; Zhao, W.; Zhou, C. Cell-based immunotherapies for solid tumors: Advances, challenges, and future directions. Front. Oncol. 2025, 15, 1551583. [Google Scholar] [CrossRef]
- Yang, F.; Xue, X.; Zheng, L.; Bi, J.; Zhou, Y.; Zhi, K.; Gu, Y.; Fang, G. Long non-coding RNA GHET1 promotes gastric carcinoma cell proliferation by increasing c-Myc mRNA stability. FEBS J. 2014, 281, 802–813. [Google Scholar] [CrossRef] [PubMed]
- Ding, N.; Cao, G.; Wang, Z.; Xu, S.; Chen, W. Tumor suppressive function of IGF2BP1 in gastric cancer through decreasing MYC. Cancer Sci. 2024, 115, 427–438. [Google Scholar] [CrossRef]
- Ji, R.; Wu, C.; Yao, J.; Xu, J.; Lin, J.; Gu, H.; Fu, M.; Zhang, X.; Li, Y.; Zhang, X. IGF2BP2-meidated m6A modification of CSF2 reprograms MSC to promote gastric cancer progression. Cell Death Dis. 2023, 14, 693. [Google Scholar] [CrossRef]
- Shen, H.; Zhu, H.; Chen, Y.; Shen, Z.; Qiu, W.; Qian, C.; Zhang, J. ZEB1-induced LINC01559 expedites cell proliferation, migration and EMT process in gastric cancer through recruiting IGF2BP2 to stabilize ZEB1 expression. Cell Death Dis. 2021, 12, 349. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, J.; Li, J.; Li, D.; Jiang, J.; Hao, T.; Xia, Y.; Lu, X.; Zhang, C.; He, Y. IGF2BP2 Promotes Epithelial to Mesenchymal Transition and Metastasis through Stabilizing HMGA1 mRNA in Gastric Cancer. Cancers 2022, 14, 5381. [Google Scholar] [CrossRef]
- Zeng, X.; Zhang, Y.; Shapaer, T.; Abudoukelimu, A.; Zhao, Z.; Ma, B. IGF2BP3 prefers to regulate alternative splicing of genes associated with the progression of gastric cancer in AGS cells. Discov. Oncol. 2025, 16, 235. [Google Scholar] [CrossRef]
- Zhou, Y.; Huang, T.; Siu, H.L.; Wong, C.C.; Dong, Y.; Wu, F.; Zhang, B.; Wu, W.K.; Cheng, A.S.; Yu, J.; et al. IGF2BP3 functions as a potential oncogene and is a crucial target of miR-34a in gastric carcinogenesis. Mol. Cancer 2017, 16, 77. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.; Ran, L.; Zhao, H.; Yin, P.; Li, W.; Lin, J.; Mao, H.; Cai, D.; Ma, Q.; Pan, X.; et al. Circular RNA circ-TNPO3 suppresses metastasis of GC by acting as a protein decoy for IGF2BP3 to regulate the expression of MYC and SNAIL. Mol. Ther. Nucleic Acids 2021, 26, 649–664. [Google Scholar] [CrossRef]
- Jiang, L.; Li, Y.; He, Y.; Wei, D.; Yan, L.; Wen, H. Knockdown of m6A Reader IGF2BP3 Inhibited Hypoxia-Induced Cell Migration and Angiogenesis by Regulating Hypoxia Inducible Factor-1α in Stomach Cancer. Front. Oncol. 2021, 11, 711207. [Google Scholar] [CrossRef]
- Yu, M.; Fei, B.; Chu, S. Targeting HNRNPA2B1 to overcome chemotherapy resistance in gastric cancer stem cells: Mechanisms and therapeutic potential. J. Biol. Chem. 2025, 301, 108234. [Google Scholar] [CrossRef]
- Peng, W.-z.; Zhao, J.; Liu, X.; Li, C.-f.; Si, S.; Ma, R. hnRNPA2B1 regulates the alternative splicing of BIRC5 to promote gastric cancer progression. Cancer Cell Int. 2021, 21, 281. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, J.; Liu, H.; Meng, L.; Gao, X.; Zhao, Y.; Wang, C.; Gao, X.; Fan, A.; Cao, T.; et al. N6-methyladenosine reader hnRNPA2B1 recognizes and stabilizes NEAT1 to confer chemoresistance in gastric cancer. Cancer Commun. 2024, 44, 469–490. [Google Scholar] [CrossRef] [PubMed]
- Mathoux, J.; Henshall, D.C.; Brennan, G.P. Regulatory Mechanisms of the RNA Modification m(6)A and Significance in Brain Function in Health and Disease. Front. Cell Neurosci. 2021, 15, 671932. [Google Scholar] [CrossRef] [PubMed]
- Yue, Y.; Liu, J.; Cui, X.; Cao, J.; Luo, G.; Zhang, Z.; Cheng, T.; Gao, M.; Shu, X.; Ma, H.; et al. VIRMA mediates preferential m6A mRNA methylation in 3′UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 2018, 4, 10. [Google Scholar] [CrossRef]
- Knuckles, P.; Lence, T.; Haussmann, I.U.; Jacob, D.; Kreim, N.; Carl, S.H.; Masiello, I.; Hares, T.; Villaseñor, R.; Hess, D.; et al. Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the m(6)A machinery component Wtap/Fl(2)d. Genes Dev. 2018, 32, 415–429. [Google Scholar] [CrossRef]
- Xu, J.; Chen, Q.; Tian, K.; Liang, R.; Chen, T.; Gong, A.; Mathy, N.W.; Yu, T.; Chen, X. m6A methyltransferase METTL3 maintains colon cancer tumorigenicity by suppressing SOCS2 to promote cell proliferation. Oncol. Rep. 2020, 44, 973–986. [Google Scholar] [CrossRef] [PubMed]
- Barbieri, I.; Tzelepis, K.; Pandolfini, L.; Shi, J.; Millán-Zambrano, G.; Robson, S.C.; Aspris, D.; Migliori, V.; Bannister, A.J.; Han, N.; et al. Promoter-bound METTL3 maintains myeloid leukaemia by m6A-dependent translation control. Nature 2017, 552, 126–131. [Google Scholar] [CrossRef]
- Weng, H.; Huang, H.; Wu, H.; Qin, X.; Zhao, B.S.; Dong, L.; Shi, H.; Skibbe, J.; Shen, C.; Hu, C.; et al. METTL14 Inhibits Hematopoietic Stem/Progenitor Differentiation and Promotes Leukemogenesis via mRNA m(6)A Modification. Cell Stem Cell 2018, 22, 191–205.e199. [Google Scholar] [CrossRef]
- Jia, G.; Fu, Y.; Zhao, X.; Dai, Q.; Zheng, G.; Yang, Y.; Yi, C.; Lindahl, T.; Pan, T.; Yang, Y.-G.; et al. N6-Methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011, 7, 885–887. [Google Scholar] [CrossRef]
- Zhang, S.; Zhao, B.S.; Zhou, A.; Lin, K.; Zheng, S.; Lu, Z.; Chen, Y.; Sulman, E.P.; Xie, K.; Bögler, O.; et al. m6A Demethylase ALKBH5 Maintains Tumorigenicity of Glioblastoma Stem-like Cells by Sustaining FOXM1 Expression and Cell Proliferation Program. Cancer Cell 2017, 31, 591–606.e596. [Google Scholar] [CrossRef]
- Li, Z.; Weng, H.; Su, R.; Weng, X.; Zuo, Z.; Li, C.; Huang, H.; Nachtergaele, S.; Dong, L.; Hu, C.; et al. FTO Plays an Oncogenic Role in Acute Myeloid Leukemia as a N(6)-Methyladenosine RNA Demethylase. Cancer Cell 2017, 31, 127–141. [Google Scholar] [CrossRef]
- Zhang, C.; Samanta, D.; Lu, H.; Bullen, J.W.; Zhang, H.; Chen, I.; He, X.; Semenza, G.L. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m6A-demethylation of NANOG mRNA. Proc. Natl. Acad. Sci. USA 2016, 113, E2047–E2056. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Lu, Z.; Gomez, A.; Hon, G.C.; Yue, Y.; Han, D.; Fu, Y.; Parisien, M.; Dai, Q.; Jia, G.; et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 2014, 505, 117–120. [Google Scholar] [CrossRef] [PubMed]
- Zeng, F.; Chen, L.; Li, J.; Yu, W.; Sa, N.; Zhang, K.; Qu, C.; Wen, D. A pan-cancer analysis reveals the oncogenic and immunological role of insulin-like growth factor 2 mRNA-binding protein family members. Discov. Oncol. 2025, 16, 323. [Google Scholar] [CrossRef]
- Lan, Q.; Liu, P.Y.; Bell, J.L.; Wang, J.Y.; Hüttelmaier, S.; Zhang, X.D.; Zhang, L.; Liu, T. The Emerging Roles of RNA m(6)A Methylation and Demethylation as Critical Regulators of Tumorigenesis, Drug Sensitivity, and Resistance. Cancer Res. 2021, 81, 3431–3440. [Google Scholar] [CrossRef]
- Deng, X.; Qing, Y.; Horne, D.; Huang, H.; Chen, J. The roles and implications of RNA m(6)A modification in cancer. Nat. Rev. Clin. Oncol. 2023, 20, 507–526. [Google Scholar] [CrossRef] [PubMed]
- Shi, T.; Zhang, H.; Chen, Y. The m6A revolution: Transforming tumor immunity and enhancing immunotherapy outcomes. Cell Biosci. 2025, 15, 27. [Google Scholar] [CrossRef]
- Tang, H.; Zhang, R.; Zhang, A. Small-Molecule Inhibitors Targeting RNA m(6)A Modifiers for Cancer Therapeutics: Latest Advances and Future Perspectives. J. Med. Chem. 2025, 68, 18114–18142. [Google Scholar] [CrossRef]
- Zeng, C.; Huang, W.; Li, Y.; Weng, H. Roles of METTL3 in cancer: Mechanisms and therapeutic targeting. J. Hematol. Oncol. 2020, 13, 117. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Xia, W.; Dong, Z.; Yang, C.-G. Chemical Inhibitors Targeting the Oncogenic m6A Modifying Proteins. Acc. Chem. Res. 2023, 56, 3010–3022. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Ge, R. Roles and drug development of METTL3 (methyltransferase-like 3) in anti-tumor therapy. Eur. J. Med. Chem. 2022, 230, 114118. [Google Scholar] [CrossRef]
- Shimura, T.; Kandimalla, R.; Okugawa, Y.; Ohi, M.; Toiyama, Y.; He, C.; Goel, A. Novel evidence for m(6)A methylation regulators as prognostic biomarkers and FTO as a potential therapeutic target in gastric cancer. Br. J. Cancer 2022, 126, 228–237. [Google Scholar] [CrossRef]
- Deng, L.-J.; Deng, W.-Q.; Fan, S.-R.; Chen, M.-F.; Qi, M.; Lyu, W.-Y.; Qi, Q.; Tiwari, A.K.; Chen, J.-X.; Zhang, D.-M.; et al. m6A modification: Recent advances, anticancer targeted drug discovery and beyond. Mol. Cancer 2022, 21, 52. [Google Scholar] [CrossRef]
- He, H.; Wu, W.; Sun, Z.; Chai, L. MiR-4429 prevented gastric cancer progression through targeting METTL3 to inhibit m(6)A-caused stabilization of SEC62. Biochem. Biophys. Res. Commun. 2019, 517, 581–587. [Google Scholar] [CrossRef]
- Yankova, E.; Blackaby, W.; Albertella, M.; Rak, J.; De Braekeleer, E.; Tsagkogeorga, G.; Pilka, E.S.; Aspris, D.; Leggate, D.; Hendrick, A.G.; et al. Small-molecule inhibition of METTL3 as a strategy against myeloid leukaemia. Nature 2021, 593, 597–601. [Google Scholar] [CrossRef]
- Fang, M.; Li, Y.; Wang, P.; Wang, Y.; Wang, X.; Wa, X.; Zhang, Y.; He, Z.; Li, J.; Li, L.; et al. METTL3 Inhibition Restores PD-L1 Expression and CD8+ T-cell Cytotoxic Function in Immunotherapy-Treated Gastric Cancer. Cancer Immunol. Res. 2025, 13, 1037–1052. [Google Scholar] [CrossRef]
- Huff, S.; Kummetha, I.R.; Zhang, L.; Wang, L.; Bray, W.; Yin, J.; Kelley, V.; Wang, Y.; Rana, T.M. Rational Design and Optimization of m6A-RNA Demethylase FTO Inhibitors as Anticancer Agents. J. Med. Chem. 2022, 65, 10920–10937. [Google Scholar] [CrossRef] [PubMed]
- Elcheva, I.A.; Wood, T.; Chiarolanzio, K.; Chim, B.; Wong, M.; Singh, V.; Gowda, C.P.; Lu, Q.; Hafner, M.; Dovat, S.; et al. RNA-binding protein IGF2BP1 maintains leukemia stem cell properties by regulating HOXB4, MYB, and ALDH1A1. Leukemia 2020, 34, 1354–1363. [Google Scholar] [CrossRef]
- Xu, Z.; Sun, B.; Wang, W.; Fan, Y.; Su, J.; Sun, J.; Gu, X. Research progress on m6A and drug resistance in gastrointestinal tumors. Front. Pharmacol. 2025, 16, 1565738. [Google Scholar] [CrossRef] [PubMed]
- Kumari, S.; Kumar, S.; Muthuswamy, S. RNA N6-methyladenosine modification in regulating cancer stem cells and tumor immune microenvironment and its implication for cancer therapy. J. Cancer Res. Clin. Oncol. 2023, 149, 1621–1633. [Google Scholar] [CrossRef]
- Xu, Z.; Chen, Q.; Shu, L.; Zhang, C.; Liu, W.; Wang, P. Expression profiles of m6A RNA methylation regulators, PD-L1 and immune infiltrates in gastric cancer. Front. Oncol. 2022, 12, 970367. [Google Scholar] [CrossRef]
- Peng, C.; Xiong, F.; Pu, X.; Hu, Z.; Yang, Y.; Qiao, X.; Jiang, Y.; Han, M.; Wang, D.; Li, X. m(6)A methylation modification and immune cell infiltration: Implications for targeting the catalytic subunit m(6)A-METTL complex in gastrointestinal cancer immunotherapy. Front. Immunol. 2023, 14, 1326031. [Google Scholar] [CrossRef]
- Chen, S.; Su, X.; Wang, J.; Zheng, N.; Tang, Y.; Peng, G.; Dong, R.; Lu, F.; Liu, M.; Zhao, Y.; et al. Identification and Validation of METTL3-Related Molecules for Predicting Prognosis and Efficacy of Immunotherapy in Gastric Cancer Based on m6A Methylome and Transcriptome Sequencing Analysis. Front. Oncol. 2022, 12, 935239. [Google Scholar] [CrossRef]
- Jiang, T.; Xia, Y.; Li, Y.; Lu, C.; Lin, J.; Shen, Y.; Lv, J.; Xie, L.; Gu, C.; Xu, Z.; et al. TRIM29 promotes antitumor immunity through enhancing IGF2BP1 ubiquitination and subsequent PD-L1 downregulation in gastric cancer. Cancer Lett. 2024, 581, 216510. [Google Scholar] [CrossRef]
- Bai, X.; Wong, C.C.; Pan, Y.; Chen, H.; Liu, W.; Zhai, J.; Kang, W.; Shi, Y.; Yamamoto, M.; Tsukamoto, T.; et al. Loss of YTHDF1 in gastric tumors restores sensitivity to antitumor immunity by recruiting mature dendritic cells. J. Immunother. Cancer 2022, 10, e003663. [Google Scholar] [CrossRef] [PubMed]
- Liao, Q.; Xiong, J. YTHDF1 regulates immune cell infiltration in gastric cancer via interaction with p53. Exp. Ther. Med. 2024, 27, 255. [Google Scholar] [CrossRef]
- Xu, X.; Zhou, E.; Zheng, J.; Zhang, C.; Zou, Y.; Lin, J.; Yu, J. Prognostic and Predictive Value of m6A “Eraser” Related Gene Signature in Gastric Cancer. Front. Oncol. 2021, 11, 631803. [Google Scholar] [CrossRef] [PubMed]
- Suo, D.; Gao, X.; Chen, Q.; Zeng, T.; Zhan, J.; Li, G.; Zheng, Y.; Zhu, S.; Yun, J.; Guan, X.Y.; et al. HSPA4 upregulation induces immune evasion via ALKBH5/CD58 axis in gastric cancer. J. Exp. Clin. Cancer Res. 2024, 43, 106. [Google Scholar] [CrossRef]
- Ji, T.; Gao, X.; Li, D.; Huai, S.; Chi, Y.; An, X.; Ji, W.; Yang, S.; Li, J. Identification and validation of signature for prognosis and immune microenvironment in gastric cancer based on m6A demethylase ALKBH5. Front. Oncol. 2022, 12, 1079402. [Google Scholar] [CrossRef]
- Lai, H.; Hu, N.; Zhang, M.; Jiang, W.; Han, Y.; Mao, C.; Zhou, K.; Zhang, J.; Hong, Y.; Wu, F. FTO is Associated with Patient Prognosis and Immune Infiltrates in Gastric Cancer and Regulates TGF-β Expression. Comb. Chem. High Throughput Screen. 2024, 28, 1911–1926. [Google Scholar] [CrossRef]
- Lei, M.M.L.; Lee, T.K.W. Cancer Stem Cells: Emerging Key Players in Immune Evasion of Cancers. Front. Cell Dev. Biol. 2021, 9, 692940. [Google Scholar] [CrossRef]
- Yue, B.; Song, C.; Yang, L.; Cui, R.; Cheng, X.; Zhang, Z.; Zhao, G. METTL3-mediated N6-methyladenosine modification is critical for epithelial-mesenchymal transition and metastasis of gastric cancer. Mol. Cancer 2019, 18, 142. [Google Scholar] [CrossRef]
- Wang, N.; Huo, X.; Zhang, B.; Chen, X.; Zhao, S.; Shi, X.; Xu, H.; Wei, X. METTL3-Mediated ADAMTS9 Suppression Facilitates Angiogenesis and Carcinogenesis in Gastric Cancer. Front. Oncol. 2022, 12, 861807. [Google Scholar] [CrossRef]
- Liu, Y.; Ao, X.; Wang, Y.; Li, X.; Wang, J. Long Non-Coding RNA in Gastric Cancer: Mechanisms and Clinical Implications for Drug Resistance. Front. Oncol. 2022, 12, 841411. [Google Scholar] [CrossRef]
- Zaccara, S.; Ries, R.J.; Jaffrey, S.R. Publisher Correction: Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell Biol. 2023, 24, 770. [Google Scholar] [CrossRef] [PubMed]
- Barbieri, I.; Kouzarides, T. Role of RNA modifications in cancer. Nat. Rev. Cancer 2020, 20, 303–322. [Google Scholar] [CrossRef] [PubMed]
- Batlle, E.; Clevers, H. Cancer stem cells revisited. Nat. Med. 2017, 23, 1124–1134. [Google Scholar] [CrossRef]
- Shibue, T.; Weinberg, R.A. EMT, CSCs, and drug resistance: The mechanistic link and clinical implications. Nat. Rev. Clin. Oncol. 2017, 14, 611–629. [Google Scholar] [CrossRef]
- Jiang, N.; Pan, J.; Fang, S.; Zhou, C.; Han, Y.; Chen, J.; Meng, X.; Jin, X.; Gong, Z. Liquid biopsy: Circulating exosomal long noncoding RNAs in cancer. Clin. Chim. Acta 2019, 495, 331–337. [Google Scholar] [CrossRef]
- Allen, T.A. The Role of Circulating Tumor Cells as a Liquid Biopsy for Cancer: Advances, Biology, Technical Challenges, and Clinical Relevance. Cancers 2024, 16, 1377. [Google Scholar] [CrossRef] [PubMed]
- Han, M.; Zhang, M.; Qi, M.; Zhou, Y.; Li, F.; Fang, S. Regulatory mechanism and promising clinical application of exosomal circular RNA in gastric cancer. Front. Oncol. 2023, 13, 1236679. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Liang, H.; Wan, A.H.; Xiao, M.; Sun, L.; Yu, Y.; Yan, S.; Deng, Y.; Liu, R.; Fang, J.; et al. Decoding the m6A epitranscriptomic landscape for biotechnological applications using a direct RNA sequencing approach. Nat. Commun. 2025, 16, 798. [Google Scholar] [CrossRef]
- Yang, Y.; Lu, Y.; Wang, Y.; Wen, X.; Qi, C.; Piao, W.; Jin, H. Current progress in strategies to profile transcriptomic m(6)A modifications. Front. Cell Dev. Biol. 2024, 12, 1392159. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Xia, W.; Li, J.; Zhang, L.; Qian, C.; Li, H.; Huang, B. Comparative analysis of improved m6A sequencing based on antibody optimization for low-input samples. Sci. Rep. 2025, 15, 1058. [Google Scholar] [CrossRef]
- Xia, W.; Guo, L.; Su, H.; Li, J.; Lu, J.; Li, H.; Huang, B. A low-cost, low-input method establishment for m6A MeRIP-seq. Biosci. Rep. 2024, 44, 1430. [Google Scholar] [CrossRef]
- Zhong, Z.-D.; Xie, Y.-Y.; Chen, H.-X.; Lan, Y.-L.; Liu, X.-H.; Ji, J.-Y.; Wu, F.; Jin, L.; Chen, J.; Mak, D.W.; et al. Systematic comparison of tools used for m6A mapping from nanopore direct RNA sequencing. Nat. Commun. 2023, 14, 1906. [Google Scholar] [CrossRef]
- Teng, H.; Stoiber, M.; Bar-Joseph, Z.; Kingsford, C. Detecting m6A RNA modification from nanopore sequencing using a semisupervised learning framework. Genome Res. 2024, 34, 1987–1999. [Google Scholar] [CrossRef] [PubMed]
- Katopodi, X.-L.; Begik, O.; Novoa Eva, M. Toward the use of nanopore RNA sequencing technologies in the clinic: Challenges and opportunities. Nucleic Acids Res. 2025, 53, gkaf128. [Google Scholar] [CrossRef]
- Murakami, S.; Jaffrey, S.R. Hidden codes in mRNA: Control of gene expression by m6A. Mol. Cell 2022, 82, 2236–2251. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, Z.; Sepich-Poore, C.; Zhang, L.; Xiao, Y.; He, C. LEAD-m(6) A-seq for Locus-Specific Detection of N(6)-Methyladenosine and Quantification of Differential Methylation. Angew. Chem. Int. Ed. Engl. 2021, 60, 873–880. [Google Scholar] [CrossRef]
- Tegowski, M.; Meyer, K.D. Studying m(6)A in the brain: A perspective on current methods, challenges, and future directions. Front. Mol. Neurosci. 2024, 17, 1393973. [Google Scholar] [CrossRef]
- Ren, Z.; He, J.; Huang, X.; Gao, Y.; Wei, C.; Wu, Z.; Guo, W.; Wang, F.; Zhao, Q.; Sun, X.; et al. Isoform characterization of m6A in single cells identifies its role in RNA surveillance. Nat. Commun. 2025, 16, 5828. [Google Scholar] [CrossRef]
- Zou, Y.; Ahsan, M.U.; Chan, J.; Meng, W.; Gao, S.J.; Huang, Y.; Wang, K. A Comparative Evaluation of Computational Models for RNA modification detection using Nanopore sequencing with RNA004 Chemistry. bioRxiv 2025. [Google Scholar] [CrossRef]
- Duan, D.; Tang, W.; Wang, R.; Guo, Z.; Feng, H. Evaluation of epitranscriptome-wide N6-methyladenosine differential analysis methods. Brief. Bioinform. 2023, 24, bbad139. [Google Scholar] [CrossRef]
- Clyde, D. New tools for transcriptome-wide mapping of m6A. Nat. Rev. Genet. 2020, 21, 387. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Lou, Y.; Li, W.; Guo, H.; Truong Nguyen, L.X.; Chen, Z. RNA m6A modification: A key regulator in normal and malignant processes. Cell Investig. 2025, 1, 100023. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Xu, X.; Huang, Y.; Shang, S.; Ma, L. RNA methylation: A new promising biomaker in cancer liquid biopsy. Biochim. Biophys. Acta (BBA) Rev. Cancer 2025, 1880, 189337. [Google Scholar] [CrossRef] [PubMed]
- Jin, R.; Zou, Q.; Luo, X. From Detection to Prediction: Advances in m6A Methylation Analysis Through Machine Learning and Deep Learning with Implications in Cancer. Int. J. Mol. Sci. 2025, 26, 6701. [Google Scholar] [CrossRef]
- Gu, C.; Shi, X.; Dai, C.; Shen, F.; Rocco, G.; Chen, J.; Huang, Z.; Chen, C.; He, C.; Huang, T.; et al. RNA m(6)A Modification in Cancers: Molecular Mechanisms and Potential Clinical Applications. Innovation 2020, 1, 100066. [Google Scholar] [CrossRef]
- Wen, L.; Fu, J.; Wang, Z.; Xie, R.; Tang, S.; Yu, L.; Zhou, H. Regulatory mechanisms of m6A RNA methylation in esophageal cancer: A comprehensive review. Front. Genet. 2025, 16, 1561799. [Google Scholar] [CrossRef]
- Meng, W.; Han, Y.; Li, B.; Li, H. The diverse role of RNA methylation in esophageal cancer. Acta Biochim. Biophys. Sin. 2023, 55, 988–1000. [Google Scholar] [CrossRef]
- Guan, Q.; Lin, H.; Miao, L.; Guo, H.; Chen, Y.; Zhuo, Z.; He, J. Functions, mechanisms, and therapeutic implications of METTL14 in human cancer. J. Hematol. Oncol. 2022, 15, 13. [Google Scholar] [CrossRef] [PubMed]
- McIntyre, A.B.R.; Gokhale, N.S.; Cerchietti, L.; Jaffrey, S.R.; Horner, S.M.; Mason, C.E. Limits in the detection of m(6)A changes using MeRIP/m(6)A-seq. Sci. Rep. 2020, 10, 6590. [Google Scholar] [CrossRef]
- Dai, W.; Qiao, X.; Fang, Y.; Guo, R.; Bai, P.; Liu, S.; Li, T.; Jiang, Y.; Wei, S.; Na, Z.; et al. Epigenetics-targeted drugs: Current paradigms and future challenges. Signal Transduct. Target. Ther. 2024, 9, 332. [Google Scholar] [CrossRef]
- Xu, Z.; Peng, B.; Cai, Y.; Wu, G.; Huang, J.; Gao, M.; Guo, G.; Zeng, S.; Gong, Z.; Yan, Y. N6-methyladenosine RNA modification in cancer therapeutic resistance: Current status and perspectives. Biochem. Pharmacol. 2020, 182, 114258. [Google Scholar] [CrossRef]
- Xie, Y.-Y.; Zhong, Z.-D.; Chen, H.-X.; Ren, Z.-H.; Qiu, Y.-T.; Lan, Y.-L.; Wu, F.; Kong, J.-W.; Luo, R.-J.; Zhang, D.; et al. Single-molecule direct RNA sequencing reveals the shaping of epitranscriptome across multiple species. Nat. Commun. 2025, 16, 5119. [Google Scholar] [CrossRef]
- Hendra, C.; Pratanwanich, P.N.; Wan, Y.K.; Goh, W.S.S.; Thiery, A.; Göke, J. Detection of m6A from direct RNA sequencing using a multiple instance learning framework. Nat. Methods 2022, 19, 1590–1598. [Google Scholar] [CrossRef]
- Xu, X.; Huang, J.; Ocansey, D.K.W.; Xia, Y.; Zhao, Z.; Xu, Z.; Yan, Y.; Zhang, X.; Mao, F. The Emerging Clinical Application of m6A RNA Modification in Inflammatory Bowel Disease and Its Associated Colorectal Cancer. J. Inflamm. Res. 2021, 14, 3289–3306. [Google Scholar] [CrossRef] [PubMed]
- Sturgess, K.; Yankova, E.; Vijayabaskar, M.S.; Isobe, T.; Rak, J.; Kucinski, I.; Barile, M.; Webster, N.A.; Eleftheriou, M.; Hannah, R.; et al. Pharmacological inhibition of METTL3 impacts specific haematopoietic lineages. Leukemia 2023, 37, 2133–2137. [Google Scholar] [CrossRef]
- Deng, X.; Su, R.; Weng, H.; Huang, H.; Li, Z.; Chen, J. RNA N(6)-methyladenosine modification in cancers: Current status and perspectives. Cell Res. 2018, 28, 507–517. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Jia, W. Multi-omics profiling: The way towards precision medicine in metabolic diseases. J. Mol. Cell Biol. 2021, 13, 576–593. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zhang, Y.; Li, Q.; Zhang, A.; Xu, J.; Li, Y.; Li, W.; Tang, L.; Yang, F.; Meng, J. N6-methyladenosine (m6A) in cancer therapeutic resistance: Potential mechanisms and clinical implications. Biomed. Pharmacother. 2023, 167, 115477. [Google Scholar] [CrossRef]
- Song, N.; Cui, K.; Zhang, K.; Yang, J.; Liu, J.; Miao, Z.; Zhao, F.; Meng, H.; Chen, L.; Chen, C.; et al. The Role of m6A RNA Methylation in Cancer: Implication for Nature Products Anti-Cancer Research. Front. Pharmacol. 2022, 13, 933332. [Google Scholar] [CrossRef] [PubMed]
- Blanco-González, A.; Cabezón, A.; Seco-González, A.; Conde-Torres, D.; Antelo-Riveiro, P.; Piñeiro, Á.; Garcia-Fandino, R. The Role of AI in Drug Discovery: Challenges, Opportunities, and Strategies. Pharmaceuticals 2023, 16, 891. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).