Utility of Same-Modality, Cross-Domain Transfer Learning for Malignant Bone Tumor Detection on Radiographs: A Multi-Faceted Performance Comparison with a Scratch-Trained Model
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population and Dataset
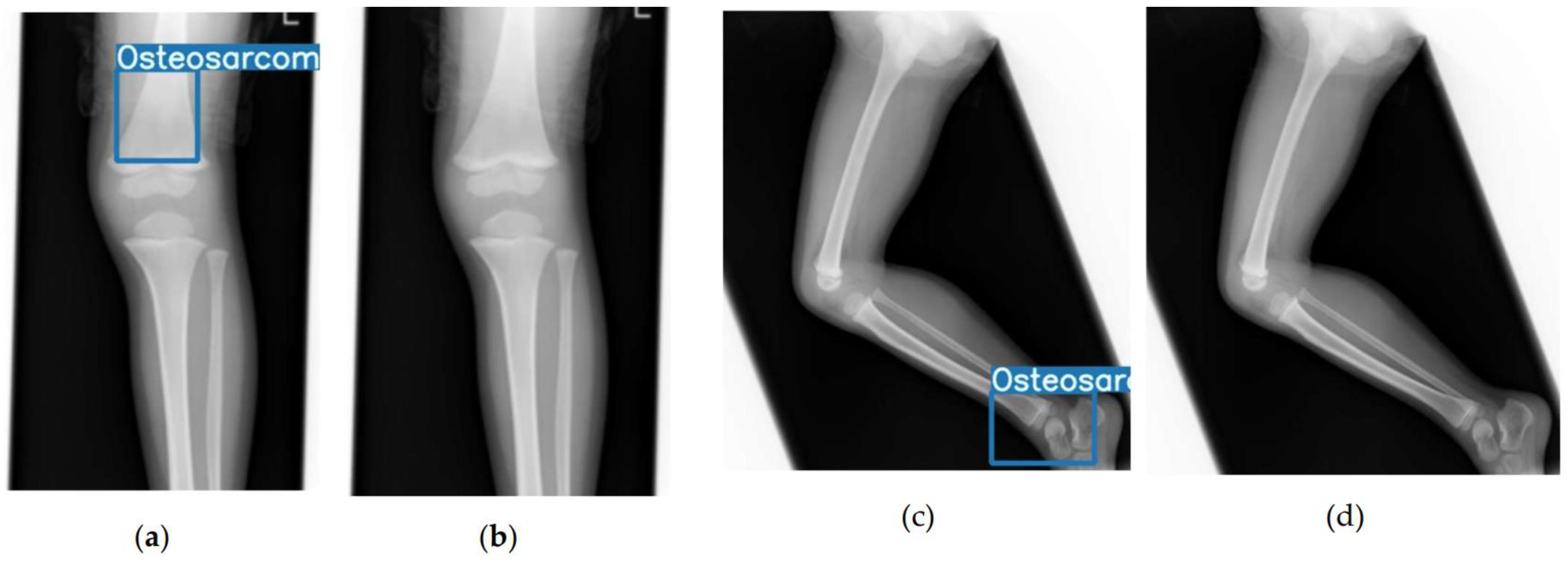
2.2. Model Development and Annotation
2.3. Statistical Analysis
2.3.1. Diagnostic Performance Metrics
2.3.2. Definition of Clinical Operating Points
- Youden-optimal point: The threshold that maximizes Youden’s index (Sensitivity + Specificity − 1), representing the optimal balance between sensitivity and specificity.
- High-sensitivity point: The threshold that maximizes specificity while maintaining a sensitivity of at least 0.90, designed for screening scenarios where minimizing missed cases is the priority.
- High-specificity point: The threshold that maximizes sensitivity while maintaining a specificity of at least 0.90, designed for scenarios where minimizing false-positive results and subsequent unnecessary workups is crucial.
2.3.3. Statistical Comparison
2.3.4. Supplementary Performance and Utility Analyses
- PR Analysis: We generated PR curves and calculated the Average Precision (AP) to evaluate model performance, taking into account the dataset’s class imbalance (prevalence).
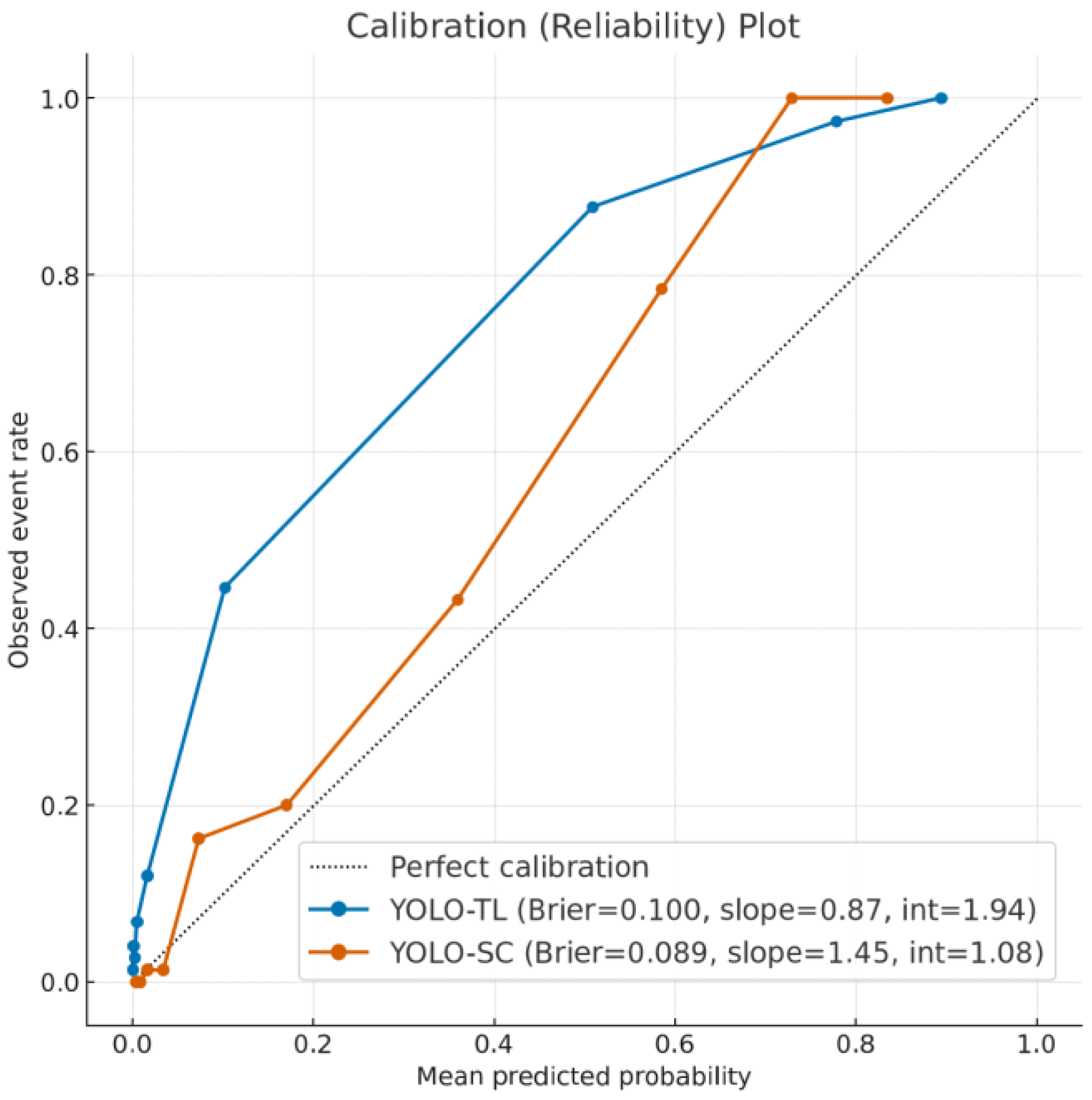
- Calibration Assessment: We assessed the reliability of the models’ predictive scores using calibration plots, generated by plotting observed event rates against predicted probabilities within 10 decile bins. The Brier score was calculated to quantify the overall prediction error.
- Decision Curve Analysis (DCA): We evaluated the clinical utility of the models by calculating the net benefit across a range of threshold probabilities. DCA visualizes the clinical value of a model by weighing the benefits of true positives against the harms of false positives, allowing for a comparison against default strategies of treating all or no patients.
2.4. Ethical Considerations
3. Results
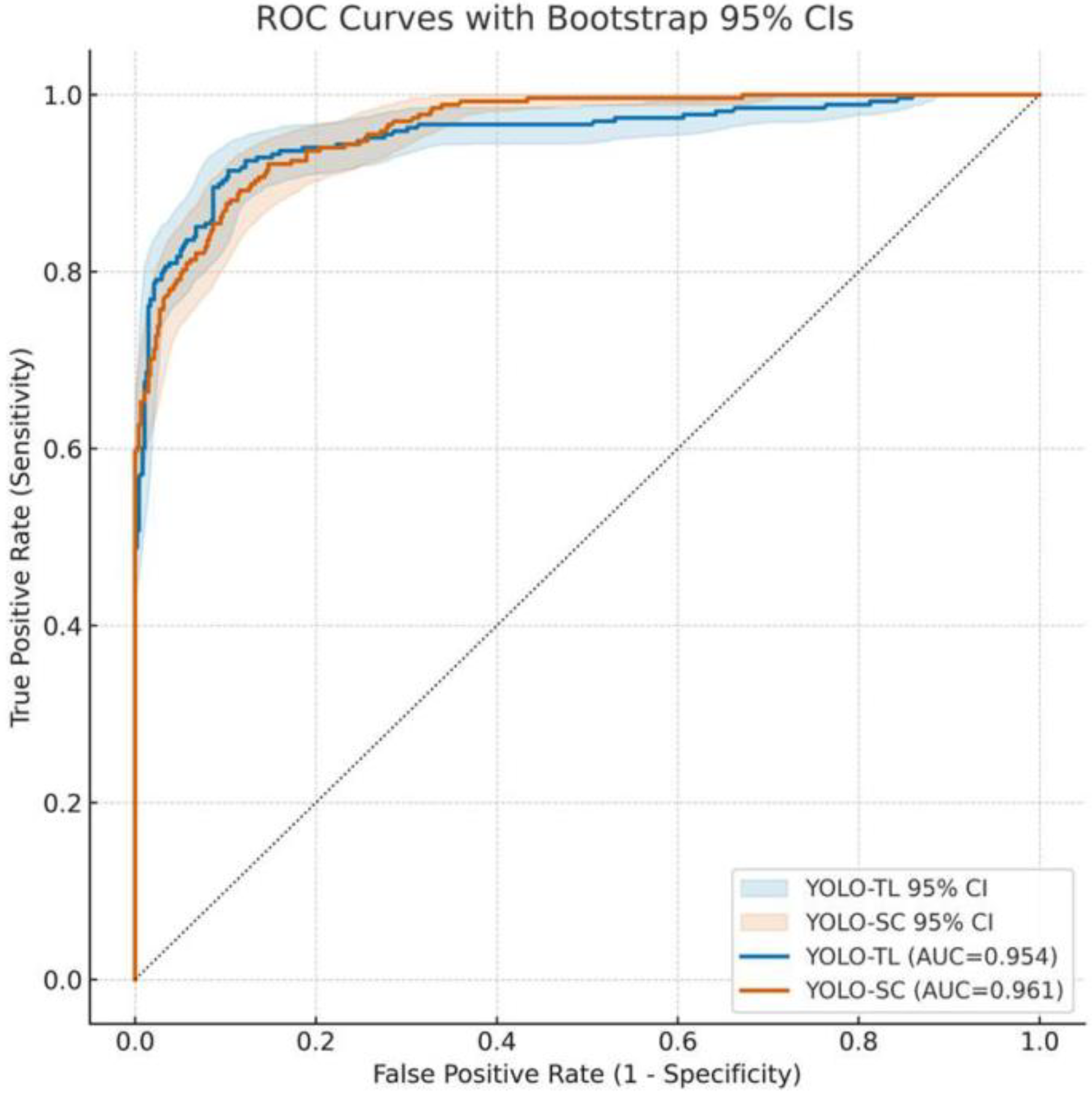
3.1. Overall Discriminative Performance of Models: ROC Curve and AUC
3.2. Diagnostic Performance for Clinical Operation
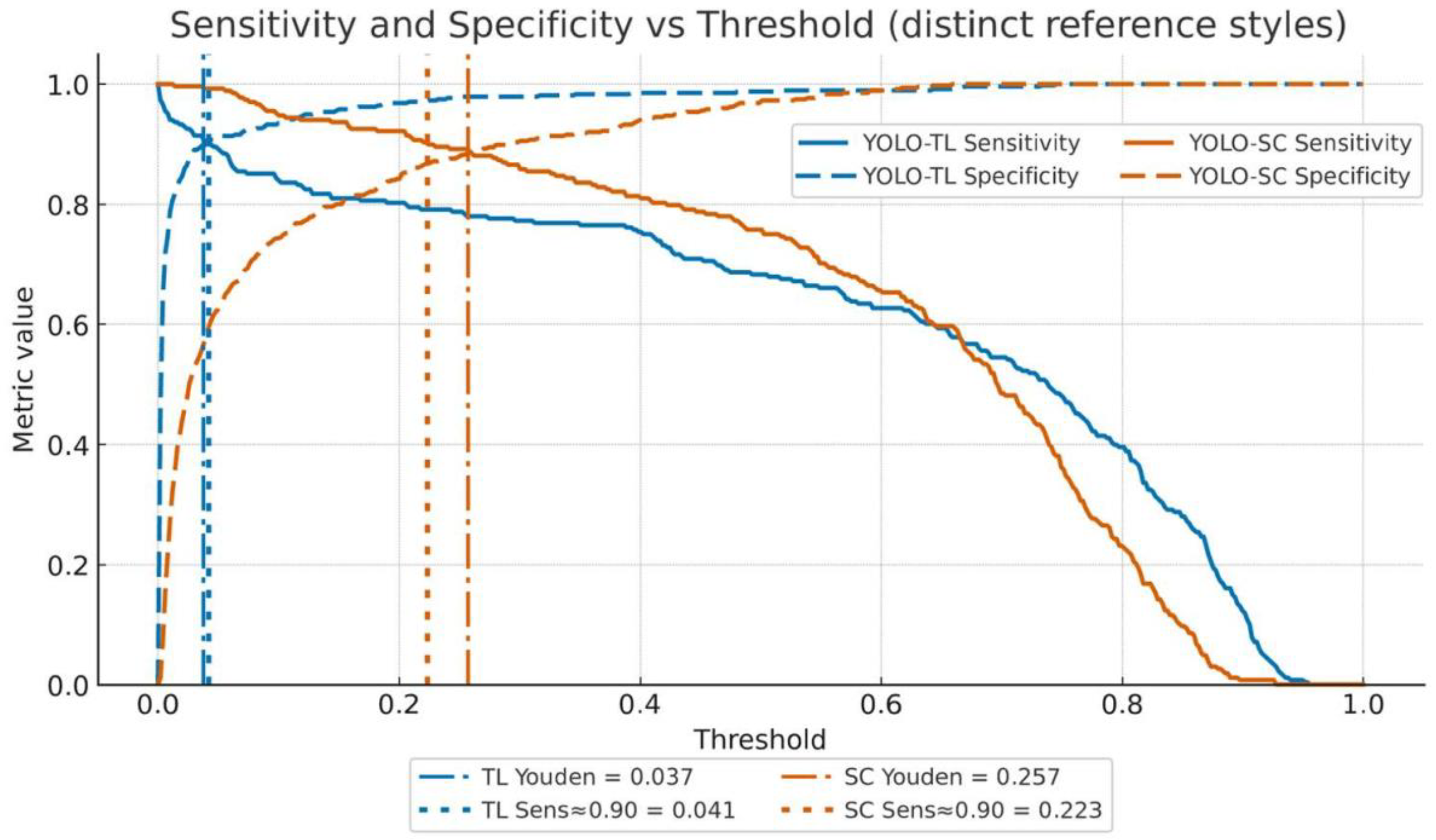
3.2.1. Sensitivity and Specificity
3.2.2. Youden’s Index Maximization Point (Balanced)
3.2.3. High-Sensitivity Operating Point (Missed-Case-Suppression Focused)
3.2.4. High-Specificity Operating Point (False-Positive-Suppression Focused)
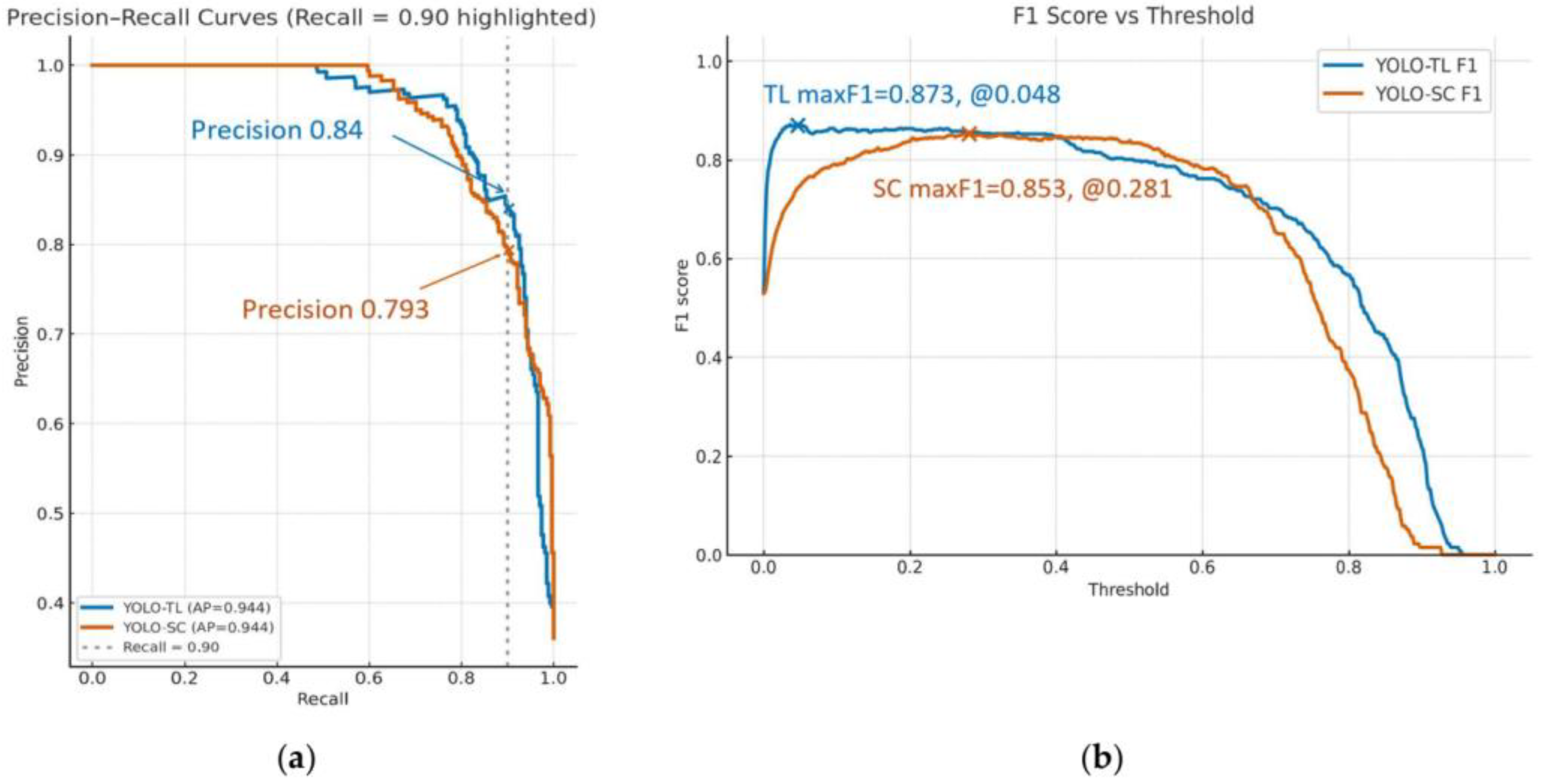
3.3. Detection Performance and Balance for Positive Cases
3.4. Reliability of Prediction Scores: Calibration Analysis
3.5. Evaluation of Clinical Utility: DCA
4. Discussion
4.1. The Performance Difference Induced by Transfer Learning and Its Mechanism
4.2. Clinical Significance and Operational Implications
4.3. Relation to Prior Work and Originality of This Study
4.4. Limitations and Future Perspectives
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Choi, J.H.; Ro, J.Y. The 2020 WHO Classification of Tumors of Bone: An Updated Review. Adv. Anat. Pathol. 2021, 28, 119–138. [Google Scholar] [CrossRef]
- Beird, H.C.; Bielack, S.S.; Flanagan, A.M.; Gill, J.; Heymann, D.; Janeway, K.A.; Livingston, J.A.; Roberts, R.D.; Strauss, S.J.; Gorlick, R. Osteosarcoma. Nat. Rev. Dis. Primers 2022, 8, 77. [Google Scholar] [CrossRef] [PubMed]
- Cole, S.; Gianferante, D.M.; Zhu, B.; Mirabello, L. Osteosarcoma: A Surveillance, Epidemiology, and End Results Program-Based Analysis from 1975 to 2017. Cancer 2022, 128, 2107–2118. [Google Scholar] [CrossRef] [PubMed]
- Mirabello, L.; Troisi, R.J.; Savage, S.A. International Osteosarcoma Incidence Patterns in Children and Adolescents, Middle Ages and Elderly Persons. Int. J. Cancer 2009, 125, 229–234. [Google Scholar] [CrossRef]
- Longhi, A.; Errani, C.; De Paolis, M.; Mercuri, M.; Bacci, G. Primary Bone Osteosarcoma in the Pediatric Age: State of the Art. Cancer Treat. Rev. 2006, 32, 423–436. [Google Scholar] [CrossRef]
- Goedhart, L.M.; Gerbers, J.G.; Ploegmakers, J.J.W.; Jutte, P.C. Delay in Diagnosis and Its Effect on Clinical Outcome in High-Grade Sarcoma of Bone: A Referral Oncological Centre Study. Orthop. Surg. 2016, 8, 122–128. [Google Scholar] [CrossRef]
- Bielack, S.S.; Kempf-Bielack, B.; Delling, G.; Exner, G.U.; Flege, S.; Helmke, K.; Kotz, R.; Salzer-Kuntschik, M.; Werner, M.; Winkelmann, W.; et al. Prognostic Factors in High-Grade Osteosarcoma of the Extremities or Trunk: An Analysis of 1,702 Patients Treated on Neoadjuvant Cooperative Osteosarcoma Study Group Protocols. J. Clin. Oncol. 2002, 20, 776–790. [Google Scholar] [CrossRef]
- Yoshida, S.; Celaire, J.; Pace, C.; Taylor, C.; Kaneuchi, Y.; Evans, S.; Abudu, A. Delay in Diagnosis of Primary Osteosarcoma of Bone in Children: Have We Improved in the Last 15 Years and What Is the Impact of Delay on Diagnosis? J. Bone Oncol. 2021, 28, 100359. [Google Scholar] [CrossRef]
- Anderson, P.G.; Tarder-Stoll, H.; Alpaslan, M.; Keathley, N.; Levin, D.L.; Venkatesh, S.; Bartel, E.; Sicular, S.; Howell, S.; Lindsey, R.V.; et al. Deep Learning Improves Physician Accuracy in the Comprehensive Detection of Abnormalities on Chest X-Rays. Sci. Rep. 2024, 14, 25151. [Google Scholar] [CrossRef]
- Wang, C.; Ma, J.; Zhang, S.; Shao, J.; Wang, Y.; Zhou, H.-Y.; Song, L.; Zheng, J.; Yu, Y.; Li, W. Development and Validation of an Abnormality-Derived Deep-Learning Diagnostic System for Major Respiratory Diseases. npj Digit. Med. 2022, 5, 124. [Google Scholar] [CrossRef]
- Nabulsi, Z.; Sellergren, A.; Jamshy, S.; Lau, C.; Santos, E.; Kiraly, A.P.; Ye, W.; Yang, J.; Pilgrim, R.; Kazemzadeh, S.; et al. Deep Learning for Distinguishing Normal versus Abnormal Chest Radiographs and Generalization to Two Unseen Diseases Tuberculosis and COVID-19. Sci. Rep. 2021, 11, 15523. [Google Scholar] [CrossRef] [PubMed]
- Najjar, R. Redefining Radiology: A Review of Artificial Intelligence Integration in Medical Imaging. Diagnostics 2023, 13, 2760. [Google Scholar] [CrossRef] [PubMed]
- Nagaraj, M.; Rodriguez, A.; Wahl, T. Regional Modeling of Storm Surges Using Localized Features and Transfer Learning. J. Geophys. Res. Mach. Learn. Comput. 2025, 2, e2025JH000650. [Google Scholar] [CrossRef]
- Morid, M.A.; Borjali, A.; Del Fiol, G. A Scoping Review of Transfer Learning Research on Medical Image Analysis Using ImageNet. Comput. Biol. Med. 2021, 128, 104115. [Google Scholar] [CrossRef]
- Niu, Z.; Ouyang, S.; Xie, S.; Chen, Y.-W.; Lin, L. A Survey on Domain Generalization for Medical Image Analysis. arXiv 2024, arXiv:2402.05035. [Google Scholar] [CrossRef]
- Yoon, J.S.; Oh, K.; Shin, Y.; Mazurowski, M.A.; Suk, H.-I. Domain Generalization for Medical Image Analysis: A Review. Proc. IEEE 2024, 112, 1583–1609. [Google Scholar] [CrossRef]
- Kim, H.E.; Cosa-Linan, A.; Santhanam, N.; Jannesari, M.; Maros, M.E.; Ganslandt, T. Transfer Learning for Medical Image Classification: A Literature Review. BMC Med. Imaging 2022, 22, 69. [Google Scholar] [CrossRef]
- Hosseinzadeh Taher, M.R.; Haghighi, F.; Gotway, M.B.; Liang, J. Large-Scale Benchmarking and Boosting Transfer Learning for Medical Image Analysis. Med. Image Anal. 2025, 102, 103487. [Google Scholar] [CrossRef]
- Yu, X.; Wang, J.; Hong, Q.-Q.; Teku, R.; Wang, S.-H.; Zhang, Y.-D. Transfer Learning for Medical Images Analyses: A Survey. Neurocomputing 2022, 489, 230–254. [Google Scholar] [CrossRef]
- Napravnik, M.; Hržić, F.; Urschler, M.; Miletić, D.; Štajduhar, I. Lessons Learned from RadiologyNET Foundation Models for Transfer Learning in Medical Radiology. Sci. Rep. 2025, 15, 21622. [Google Scholar] [CrossRef]
- Hasei, J.; Nakahara, R.; Otsuka, Y.; Nakamura, Y.; Hironari, T.; Kahara, N.; Miwa, S.; Ohshika, S.; Nishimura, S.; Ikuta, K.; et al. High-Quality Expert Annotations Enhance Artificial Intelligence Model Accuracy for Osteosarcoma X-Ray Diagnosis. Cancer Sci. 2024, 115, 3695–3704. [Google Scholar] [CrossRef]
- Hasei, J.; Nakahara, R.; Otsuka, Y.; Nakamura, Y.; Ikuta, K.; Osaki, S.; Hironari, T.; Miwa, S.; Ohshika, S.; Nishimura, S.; et al. The Three-Class Annotation Method Improves the AI Detection of Early-Stage Osteosarcoma on Plain Radiographs: A Novel Approach for Rare Cancer Diagnosis. Cancers 2024, 17, 29. [Google Scholar] [CrossRef]
- Martinović, I.; Mao, S.; Dousty, M.; Li, W.; Đukanović, M.; Colak, E.; Sejdić, E. X-Ray Modalities in the Era of Artificial Intelligence: Overview of Self-Supervised Learning Approach. FACETS 2025, 10, 1–17. [Google Scholar] [CrossRef]
- Liu, H.; Sun, F.; Gu, J.; Deng, L. SF-YOLOv5: A Lightweight Small Object Detection Algorithm Based on Improved Feature Fusion Mode. Sensors 2022, 22, 5817. [Google Scholar] [CrossRef] [PubMed]
- Loshchilov, I.; Hutter, F. SGDR: Stochastic Gradient Descent with Warm Restarts. arXiv 2016, arXiv:1608.03983. [Google Scholar]
- Smith, L.N. Cyclical Learning Rates for Training Neural Networks. In Proceedings of the 2017 IEEE Winter Conference on Applications of Computer Vision (WACV), Santa Rosa, CA, USA, 24–31 March 2017. [Google Scholar]
- Stokel-Walker, C. AI in Medicine: UK Is Pursuing “Middle Path” in Adoption and Regulation. BMJ 2025, 388, r240. [Google Scholar] [CrossRef]
- McKinney, S.M.; Sieniek, M.; Godbole, V.; Godwin, J.; Antropova, N.; Ashrafian, H.; Back, T.; Chesus, M.; Corrado, G.S.; Darzi, A.; et al. Addendum: International Evaluation of an AI System for Breast Cancer Screening. Nature 2020, 586, E19. [Google Scholar] [CrossRef]
- Shen, Y.; Shamout, F.E.; Oliver, J.R.; Witowski, J.; Kannan, K.; Park, J.; Wu, N.; Huddleston, C.; Wolfson, S.; Millet, A.; et al. Artificial Intelligence System Reduces False-Positive Findings in the Interpretation of Breast Ultrasound Exams. Nat. Commun. 2021, 12, 5645. [Google Scholar] [CrossRef]
- Achour, N.; Zapata, T.; Saleh, Y.; Pierscionek, B.; Azzopardi-Muscat, N.; Novillo-Ortiz, D.; Morgan, C.; Chaouali, M. The Role of AI in Mitigating the Impact of Radiologist Shortages: A Systematised Review. Health Technol. 2025, 15, 489–501. [Google Scholar] [CrossRef]
- Jing, A.B.; Garg, N.; Zhang, J.; Brown, J.J. AI Solutions to the Radiology Workforce Shortage. npj Health Syst. 2025, 2, 20. [Google Scholar] [CrossRef]
- Zha, D.; Lai, K.-H.; Yang, F.; Zou, N.; Gao, H.; Hu, X. Data-Centric AI: Techniques and Future Perspectives. In Proceedings of the 29th ACM SIGKDD Conference on Knowledge Discovery and Data Mining, Long Beach, CA, USA, 6–10 August 2023; Association for Computing Machinery: New York, NY, USA, 2023; pp. 5839–5840. [Google Scholar]
- Singh, P. Systematic Review of Data-Centric Approaches in Artificial Intelligence and Machine Learning. Data Sci. Manag. 2023, 6, 144–157. [Google Scholar] [CrossRef]
- Hamid, O.H. From Model-Centric to Data-Centric AI: A Paradigm Shift or Rather a Complementary Approach? In Proceedings of the 2022 8th International Conference on Information Technology Trends (ITT), Dubai, United Arab Emirates, 25–26 May 2022; pp. 196–199. [Google Scholar]
- Zha, D.; Bhat, Z.P.; Lai, K.-H.; Yang, F.; Jiang, Z.; Zhong, S.; Hu, X. Data-Centric Artificial Intelligence: A Survey. ACM Comput. Surv. 2025, 57, 129. [Google Scholar] [CrossRef]
- Vidhya, G.; Nirmala, D.; Manju, T. Quality Challenges in Deep Learning Data Collection in Perspective of Artificial Intelligence. J. Inf. Technol. Comput. 2023, 4, 46–58. [Google Scholar] [CrossRef]

| Operating Point | Model | Threshold | Sensitivity | Specificity | PPV | NPV | False Positives | False Negatives |
|---|---|---|---|---|---|---|---|---|
| Youden-optimal | YOLO-TL | 0.0371 | 0.914 (0.879–0.944) | 0.897 (0.865–0.925) | 0.833 | 0.949 | 49 | 23 |
| YOLO-SC | 0.2568 | 0.892 (0.855–0.925) | 0.884 (0.850–0.913) | 0.813 | 0.936 | 55 | 29 | |
| High Sensitivity (≥0.90) | YOLO-TL | 0.0413 | 0.903 (0.868–0.935) | 0.903 (0.873–0.929) | 0.840 (0.805–0.872) | 0.943 (0.918–0.963) | 46 | 26 |
| YOLO-SC | 0.2230 | 0.903 (0.868–0.935) | 0.867 (0.834–0.896) | 0.793 (0.755–0.829) | 0.941 (0.916–0.961) | 63 | 26 | |
| High Specificity (≥0.90) | YOLO-TL | 0.0413 | 0.798 (0.752–0.840) | 0.903 (0.873–0.929) | — | — | 46 | 54 |
| YOLO-SC | 0.2903 | 0.764 (0.716–0.808) | 0.902 (0.872–0.928) | — | — | 47 | 63 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hasei, J.; Nakahara, R.; Otsuka, Y.; Takeuchi, K.; Nakamura, Y.; Ikuta, K.; Osaki, S.; Tamiya, H.; Miwa, S.; Ohshika, S.; et al. Utility of Same-Modality, Cross-Domain Transfer Learning for Malignant Bone Tumor Detection on Radiographs: A Multi-Faceted Performance Comparison with a Scratch-Trained Model. Cancers 2025, 17, 3144. https://doi.org/10.3390/cancers17193144
Hasei J, Nakahara R, Otsuka Y, Takeuchi K, Nakamura Y, Ikuta K, Osaki S, Tamiya H, Miwa S, Ohshika S, et al. Utility of Same-Modality, Cross-Domain Transfer Learning for Malignant Bone Tumor Detection on Radiographs: A Multi-Faceted Performance Comparison with a Scratch-Trained Model. Cancers. 2025; 17(19):3144. https://doi.org/10.3390/cancers17193144
Chicago/Turabian StyleHasei, Joe, Ryuichi Nakahara, Yujiro Otsuka, Koichi Takeuchi, Yusuke Nakamura, Kunihiro Ikuta, Shuhei Osaki, Hironari Tamiya, Shinji Miwa, Shusa Ohshika, and et al. 2025. "Utility of Same-Modality, Cross-Domain Transfer Learning for Malignant Bone Tumor Detection on Radiographs: A Multi-Faceted Performance Comparison with a Scratch-Trained Model" Cancers 17, no. 19: 3144. https://doi.org/10.3390/cancers17193144
APA StyleHasei, J., Nakahara, R., Otsuka, Y., Takeuchi, K., Nakamura, Y., Ikuta, K., Osaki, S., Tamiya, H., Miwa, S., Ohshika, S., Nishimura, S., Kahara, N., Yoshida, A., Kondo, H., Fujiwara, T., Kunisada, T., & Ozaki, T. (2025). Utility of Same-Modality, Cross-Domain Transfer Learning for Malignant Bone Tumor Detection on Radiographs: A Multi-Faceted Performance Comparison with a Scratch-Trained Model. Cancers, 17(19), 3144. https://doi.org/10.3390/cancers17193144







