Simple Summary
We present an exhaustive review of the literature that has evaluated the role of ctDNA analysis in upper gastrointestinal tumors, including gastroesophageal adenocarcinoma (GEC), biliary tract cancer (BTC) and pancreatic ductal adenocarcinoma (PADC). We describe the implications of ctDNA from early diagnosis to molecular characterization and follow-up of tumor genomic evolution, from a current point of view and debating strengths and weaknesses.
Abstract
Circulating tumor DNA (ctDNA) has emerged as a promising non-invasive source to characterize genetic alterations related to the tumor. Upper gastrointestinal cancers, including gastroesophageal adenocarcinoma (GEC), biliary tract cancer (BTC) and pancreatic ductal adenocarcinoma (PADC) are poor prognostic malignancies, usually diagnosed at advanced stages when no longer amenable to surgical resection and show a poor prognosis even for resected patients. In this sense, ctDNA has emerged as a promising non-invasive tool with different applications, from early diagnosis to molecular characterization and follow-up of tumor genomic evolution. In this manuscript, novel advances in the field of ctDNA analysis in upper gastrointestinal tumors are presented and discussed. Overall, ctDNA analyses can help in early diagnosis, outperforming current diagnostic approaches. Detection of ctDNA prior to surgery or active treatment is also a prognostic marker that associates with worse survival, while ctDNA detection after surgery is indicative of minimal residual disease, anticipating in some cases the imaging-based detection of progression. In the advanced setting, ctDNA analyses characterize the genetic landscape of the tumor and identify patients for targeted-therapy approaches, and studies show variable concordance levels with tissue-based genetic testing. In this line, several studies also show that ctDNA serves to follow responses to active therapy, especially in targeted approaches, where it can detect multiple resistance mechanisms. Unfortunately, current studies are still limited and observational. Future prospective multi-center and interventional studies, carefully designed to assess the value of ctDNA to help clinical decision-making, will shed light on the real applicability of ctDNA in upper gastrointestinal tumor management. This manuscript presents a review of the evidence available in this field up to date.
1. Introduction
Currently, histotype diagnosis on tumor tissue is the gold standard for tumor characterization and first therapeutic approach guidance. Moreover, tissue analyses are needed to assess the local tumor microenvironment, for example to know the programmed death ligand-1 (PD-L1) status or the tumor-infiltrating lymphocyte (TIL) characterization. For a further tumor genomic characterization, tissue genomic testing in tissue is the gold standard to identify genomic alterations, assess their prognostic value, select patients for matched targeted therapy and monitor their responses. However, tissue-based genomic testing presents important limitations; mainly, it requires an invasive procedure, tissue may not be available depending on tumor location, and it may not recover full tumor heterogeneity [1,2]. In this scenario, genomic testing in ctDNA arises as a promising strategy to overcome these pitfalls. Importantly, as aforementioned, ctDNA analysis overcomes the limitations of tissue testing in terms of polyclonal heterogeneity and tissue availability; moreover, due to its minimally invasive nature, it allows for repetitive testing, providing real-time information on the tumor biology [3].
Liquid biopsy is the concept of analyzing biologic material derived from tumor cells into bodily fluids. Among different biologic materials found in liquid biopsies, cell free DNA (cfDNA) is fragmented DNA that can originate from, potentially, any cell of the organism. Circulating tumor DNA (ctDNA), is the fraction of cfDNA released from tumor cells and may reflect the genomic as well as epigenomic landscape of the tumor [4].
2. Technical Aspects of ctDNA Analysis
Detailed technical aspects of ctDNA analysis are beyond the scope of this manuscript, and only some important considerations will be highlighted; for further information, readers are referred to recent publications in the field [5,6]. Two main approaches are used for the detection of genetic alterations in ctDNA. Advanced polymerase chain reaction (PCR)-based techniques are allele-specific approaches that interrogate previously known mutations with high sensitivity [5]. Alternatively, next-generation sequencing (NGS)-based techniques analyze several alterations in a unique experiment, although with lower sensitivity, and they can detect de novo alterations including copy number variations and rearrangements. NGS analyses can focus on a panel of selected genes or perform whole-exome or genome sequencing [7,8]. Each approach can be complementary and useful depending on the context and purpose of the assay. PCR-based techniques may be more suitable for the detection of a common, previously identified mutation, while NGS techniques cover the entire molecular characterization and over-time tumor clonality of patients receiving targeted therapy [5,6]. Finally, low-coverage whole-exome or genome sequencing have been employed for the detection of copy number variations (CNVs) [9].
An important technical limitation of ctDNA analysis relates to the low amounts of DNA present in the sample in some settings, preventing detection of present alterations with current methods. In line with this, detection of CNVs and genomic rearrangements in ctDNA with current technologies is also considered suboptimal [5,10]. Moreover, some tumors, such as primary brain tumors, renal, prostate and thyroid cancers, are regarded as non-shedders and may not be amenable to ctDNA analysis [11]. Finally, blood cells accumulate somatic mutations with age in a process named clonal hematopoiesis of indeterminate potential, which represents a source of false positives in ctDNA analysis [12].
Figure 1 presents the characteristics of genomic testing in tissue and ctDNA of liquid biopsy. In line with this, a recent study that included 1021 patients with different solid tumors revealed that ctDNA sequencing detected a considerable amount of targetable alterations not present in tissue (9% ESCAT-tier I/II, 14% ESCAT-tier III, and 6% ESCAT-tier IV), while in 7% of the patients the alterations were only found in tissue. This study suggests that ctDNA genetic testing can complement and even replace tissue testing [2]. Indeed, the ESMO has recently published their recommendations for genetic testing in ctDNA in several solid tumors [10], essentially when rapid results are needed and tissue is unavailable.
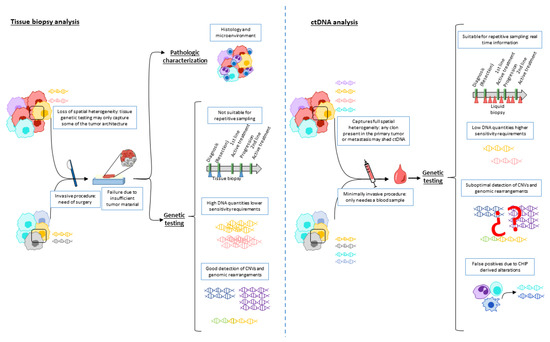
Figure 1.
Characteristics of genomic testing in tissue and ctDNA of liquid biopsy. Tissue analysis allows for histotyping and tumor microenvironment characterization. Genetic testing in tissue biopsy (left panel) needs an invasive procedure, thus preventing repetitive testing. It may also show high failure rates due to insufficient tumor material and may only represent part of the entire tissue architecture. On the other hand, high DNA quantities are usually extracted, which eases detection of genetic alteration including CNVs and genomic rearrangements. Genetic testing ctDNA (right panel) is minimally invasive, allowing repetitive testing, and can cover full tumor heterogeneity. Nevertheless, low DNA quantities require highly sensitive technologies, CNV and genomic rearrangement detection is still unsatisfactory, and CHIP may lead to false positives. Abbreviations: CNV: copy number variation; ctDNA: circulating tumoral DNA; CHIP: clonal hematopoiesis of indeterminate potential.
3. Clinical Applications of ctDNA Analysis in GEC, BTC and PDAC
The analysis of ctDNA and its clinical applicability is most advanced in lung, breast and colon cancer. Indeed, accumulating evidence supports the clinical usefulness of this approach at different time points along disease progression, including early diagnosis, detection of minimal residual disease (MRD) after surgery, molecular characterization for selection of targeted therapy, patient prognosis, and monitoring of treatment response and emergence of resistance to targeted therapy in the advanced setting [2,3,6,13,14]. In the present review, recent advances in the applications and usefulness of ctDNA in upper gastrointestinal cancers, namely, gastroesophageal adenocarcinoma (GEC), biliary tract cancer (BTC) and pancreatic ductal adenocarcinoma (PDAC) will be presented and discussed. Of note, these malignancies represent hard-to-treat cancers with poor prognosis, which would potentially benefit from the implementation of ctDNA analysis in different clinical contexts [6,15,16].
Importantly, GEC, BTC and PDAC are usually diagnosed at advanced stages, when no longer amenable to surgical resection. In this context, it is critical to characterize the genomic landscape of the tumor, with the aim of identifying targetable alterations and selecting patients who would potentially benefit from matched targeted therapies [16]. Overall, targeted therapy approaches improve patient survival compared to conventional chemotherapy [17,18,19,20,21]. Nevertheless, distinguishing the real clinical value of a given genomic alteration remains challenging [22,23]. Along this line, important scientific societies have proposed guidelines in order to help in the interpretation of the potential clinical utility of genomic alterations in solid tumors [24,25,26]. The European Society of Medical Oncology (ESMO) proposed a classification system for genomic alterations based on their clinical evidence of actionability [27]. The ESMO Scale of Actionability of molecular Targets (ESCAT) establishes six progressive levels according to the existence of matched drugs for a given genomic alteration and their reported clinical benefit: from tier I, in which alteration-drug match is associated with improved outcome in clinical trials, to tier IV, where only preclinical evidence of actionability is reported. Tier IV and X mean a lack of clinical benefit or evidence of actionability, respectively [27]. Interestingly, in a study assessing the clinical applicability of this scale that included 552 patients with different solid tumors in the advanced setting, 67% of the patients showed at least one actionable genetic alteration, and 27% of those were treated with a matched therapy. Importantly, patients harboring alterations of a stronger clinical evidence (tiers I and II) and treated with a matched drug showed longer progression-free survival (PFS) compared to those harboring alterations with a weaker clinical meaning (tiers III and IV) [22]. In this scenario, the ESMO has recently reviewed the genetic landscape and recommendations for genetic testing in several tumors in the advanced setting [28]. Focusing on gastrointestinal malignancies, the ESMO recommends genetic testing of specific alterations in GEC (ERBB2 amplifications, MSI-H, and NTRK 1,2,3 fusions), BTC (IDH1 mutations, FGFR2 fusions, MSI-H, and NTRK 1,2,3 fusions) and PDAC (MSI-H and NTRK 1,2,3 fusions) [28]. Similarly, the American Society of Clinical Oncology (ASCO) has recently proposed their Provisional Clinical Opinion (PCO), a panel of recommendations addressing specific clinical questions related to genomic testing in solid tumors. Overall, genomic testing is recommended for tumors with biomarker-based therapeutic indications and multigene testing when more than one targeted therapeutic indication is available [29].
The evidence of the clinical usefulness of ctDNA in GEC, BTC and PDAC is still limited, and prospective multi-center studies are needed in order to shed light on its real clinical applicability in the next future. In the following lines, pan-cancer studies including GEC, BTC and PDAC patients as well as small studies focused on each of the malignancies will be discussed.
Analyzing methylation profiles in ctDNA has been proposed as a valuable tool for early cancer diagnosis, as epigenetic changes, mainly methylation, could precede genetic changes. Pan-cancer studies including thousands of patients with different malignancies have proposed cfDNA concentrations or methylation panels with high diagnostic capacities and the ability to distinguish primary tumor sites [11,30,31,32,33].
In the advanced setting, the SCRUM-Japan GOZILA study assessed ctDNA-based genetic testing utility for targeted-therapy trial enrolment in patients with advanced gastrointestinal cancers (including colorectal carcinoma, GEC, squamous cell carcinoma, BTC and PDAC) and compared its performance to tissue genetic testing. Importantly, ctDNA-based genetic testing shortened enrolment time and improved enrolment rate without influencing response rates [34]. ctDNA could also be used to predict treatment response and monitor relapses. In two proof-of-concept studies assessing ctDNA levels in pan-cancer patients treated with immunotherapy, ctDNA analysis had a prognostic value and could predict responses [35,36]. Finally, ctDNA analysis has the capability to monitor polyclonal resistance mechanisms arising in targeted therapy-treated patients. A study including 42 patients with advanced gastrointestinal tumors that developed resistance upon targeted therapy identified novel resistance alterations not found in tissue biopsies for 78% of the cases. Importantly, among the 23 patients analyzed at tissue and ctDNA level, tissue biopsy identified multiple resistance mechanisms in 9% of patients, while by ctDNA this percentage rose to 40% [37]. Table 1 summarizes key findings and study designs of selected studies assessing the role of ctDNA in different types of cancer.

Table 1.
Key findings and study design of main studies assessing the role of ctDNA in different types of cancer.
4. Gastroesophageal Cancer (GEC)
Gastric cancer represents a global healthcare challenge. With an estimated 1,089,103 new cases and 768,793 new deaths in 2020, this tumor type ranks fifth in incidence and fourth in mortality. Further, the gastroesophagogastric junction adenocarcinoma incidence is increasing [38]. GEC, referring to both tumors together, has been historically referred as one unique entity, although it comprises four different molecular subtypes [39,40,41] that recognize the well-described interpatient heterogeneity, a major cause of failure of phase II-III clinical trials with targeted therapies [20,42]. Additionally, variations within the same tumor (intrapatient-intratumoral-heterogeneity) have also been described, from a spatial and temporal perception [43,44,45]. In this regard, especially in GEC, ctDNA appears as a novel approach to overcome this obstacle.
4.1. Screening and Diagnosis
Except in high-risk East Asian countries (China, Japan and South Korea), there are no population GEC-screening strategies, and most GEC patients are diagnosed with an advanced disease [46]. The first approach to tumor diagnosis and characterization is based on the tissue histotype, and the performance of genetic testing adds value for prognostic and treatment purposes. Multiple (5–8) biopsies should be carried out to ensure enough material for a first and necessary histological and molecular interpretation [47,48]. Sequencing the tumor tissue also requires an invasive procedure in order to obtain enough tumor cells. In this regard, ctDNA analyses could cover both limitations, i.e., facilitate the invasive procedures and overcome the spatial heterogeneity. Some studies have shown that ctDNA can be detected in patients with GEC more often than in patients with benign pathologies of the stomach or in healthy controls [33,49], thus proposing it as a potential non-invasive tool for massive screening. Furthermore, and as mentioned before, the use of DNA methylation patterns for screening purposes has also been also investigated in GEC as in other tumors, although the sensitivity reported is low for early tumor stages [32]. Prospective studies evaluating the potential use of ctDNA for early diagnosis of GEC are currently ongoing in South Korea (NCT04665687), US (NCT04241796) and UK [50].
4.2. MRD and Recurrence Monitoring
For patients with potentially resectable GEC, standard treatment includes peri-operative combination chemotherapy and surgery in Western countries [51]. Adjuvant chemotherapy is indicated regardless of the tumor response to the neoadjuvant treatment, and there is a lack of reliable programs to monitor recurrence [46]. The use of ctDNA to identify GEC patients at risk of recurrence has been limited to small cohorts, varying assays and time points. Different studies have confirmed the association of pre-operative ctDNA levels with different tumor stages [11,32], and how the detection of ctDNA in the immediate post-operative period correlates with eventual recurrences [33,52,53,54,55,56,57,58]. Notably, a subanalysis of 50 patients included in the phase III CRITICS study, which randomized patients to receive pre-operative chemotherapy and surgery plus post-operative chemotherapy vs. post-operative chemoradiotherapy, demonstrated that the presence of ctDNA within nine weeks of surgery predicted recurrence [59]. Additionally, a retrospective analysis of real-world data including 295 patients showed that ctDNA detection at any time point after surgery or during the surveillance period was associated with shorter recurrence-free survival [60]. Currently, there are different ongoing studies prospectively assessing the value of the MRD-ctDNA detection as a key tool for deciding on the adjuvant treatment indication, in patients with GEC (NTC04510285, NCT02674373).
4.3. Metastatic Disease Monitoring
Advanced GEC patients show a very poor prognosis with a 5-year relative survival rate of 6%. Treatment with chemotherapy has been shown to improve overall survival (OS) and quality of life, compared to best supportive care alone [61]. For the time being, only two tissue biomarker subpopulations have been identified for targeted treatment: the human epidermal growth factor (HER2)-positive and PD-L1-positive subpopulations, with potential benefit from the addition of trastuzumab and anti-PD1 agents to the first-chemotherapy line of treatment, respectively [62,63,64]. As in other tumors, ctDNA detection and sequencing in GEC patients could provide valuable genetic information and allow to follow up on the tumor genomic evolution without the need for serial tissue biopsies. Again, one should recognize the intrinsic limitations related to the analyses of circulating genomics but not local peri-tumor protein expression. One of the largest retrospective studies of GEC patients demonstrated how ctDNA can potentially recover the temporal and spatial molecular heterogeneity, and how temporal changes in the tumor somatic variant allelic frequency (VAF) correlate with prognosis in patients receiving chemotherapy and immunotherapy [52]. These findings have been corroborated by other smaller studies [65], including the association of basal ctDNA levels with prognosis [66], and how ctDNA changes over time correlate with response to different treatments [66,67,68].
When considering the HER2-positive subpopulation of GEC, the detection of HER2 amplification in ctDNA has been proposed as an optimal tool to overcome the challenge of tumor temporal and spatial heterogeneity, thus being able to predict and monitor responses to anti-HER2 therapies, and to inform about possible mechanisms of resistance [45,52,69,70,71,72]. Ongoing observational and interventional studies would probably validate these findings (NTC04520295, NTC03409848).
Blood samples from Epstein Barr Virus (EBV)-positive GEC tumors have been analyzed. Although the plasma EBV-DNA load has been identified only in half of EBV-(tissue)-positive cases, it could correlate with response to treatment [73]. Furthermore, detection of FGFR2b amplification in ctDNA has been partially associated with responses to anti-FGFR2b targeted therapies [74,75], but not EGFR amplifications [76].
Concerning historical approaches to individual molecular alterations focusing on single targeted therapies, sequencing techniques could lead to a multiplex approach to define the best personalized treatment algorithm, and ctDNA analysis arises as the best approach to follow up on clonal tumor evolution. The VIKTORY trial [77] was an umbrella trial conducted in South Korea, which assigned patients with metastatic GEC to one of 10 phase II molecularly-driven clinical trials for a second-line treatment, depending on eight different tumor biomarkers on tissue-based sequencing, although ctDNA was analyzed at baseline and longitudinally. Patients receiving the biomarker-selected therapy presented prolonged PFS and OS compared with patients receiving conventional treatment, and reduction of ctDNA levels correlated with response to treatment. The PANGEA trial [45] considered the first three lines of treatment, with optimally sequenced chemotherapy plus different monoclonal antibodies, depending on the molecular findings in tissue and ctDNA. Genomic discordance was observed in tissue between primary and metastatic tumors in 35% of patients, with better concordance when comparing results of metastatic tissue and ctDNA. Additionally, tumor changes after a first- and second-targeted line of treatment was identified in 50% of the treated patients. Meeting its primary endpoint of OS, this study again confirms the spatial and temporal heterogeneity of GEC and demonstrates the feasibility and efficacy of molecular approaches. Finally, and from a country approximation, the already mentioned GOZILA initiative [78] performed comprehensive ctDNA sequencing to rapidly screen cancer patients for trial eligibility, including GEC patients; the authors demonstrated that massive ctDNA genotyping unveils the presence of rare molecular targetable alterations in these patients, including tumors harboring neurotropic receptor tyrosine kinase 1 (NTRK1) fusions. The results of these trials highlight the potential clinical utility of ctDNA analysis in selecting patients for personalized treatment. The incorporation of ctDNA analysis as a complement in the comprehensive tumor characterization in the majority of clinical trials in patients with GEC will confirm its real value. Table 2 summarizes key findings and study designs of selected studies assessing the role of ctDNA in GEC.

Table 2.
Key findings and study design of main studies assessing the role of ctDNA in GEC.
5. Biliary Tract Cancer (BTC)
BTC is a group of heterogeneous malignancies including mainly intra- and extrahepatic cholangiocarcinomas (CCA), as well as gallbladder cancer (GBC). The incidence of these tumors is relatively low in western countries, but significantly higher in certain geographic areas such as China and Thailand for CCA, and Chile for GBC. Moreover, the incidence of intrahepatic CCA is increasing worldwide [38,79]. When possible, surgery is the unique option of cure, although tumor recurrences are frequent. Even though the majority of patients are diagnosed in advanced stages and prognosis remains dismal, identification of distinct patient subgroups harboring unique molecular alterations with corresponding targeted therapies is improving the treatment paradigm of these patients [79,80].
5.1. Screening and Early Diagnosis
Early diagnosis of BTC is challenging. BTC usually present with unspecific symptoms and can be confused with benign biliary disorders that may also cause biliary stenosis {Valle, 2021, Biliary tract cancer}. In this regard, cfDNA concentration, CNV scores and/or methylation scores analyzed on ctDNA are increased in patients with CCA and/or GBC and serve to distinguish them from healthy controls and patients with benign biliary lesions [81,82,83,84]. Interestingly, in the context of BTC and PDAC, bile arises as a novel source for ctDNA analysis. Several studies have assessed ctDNA in bile collected after endoscopic biliary drainage for early diagnosis and tumor molecular characterization. Overall, ctDNA found in bile is characterized by larger fragments and shows a better correlation with tumor tissue than plasma [85,86,87,88,89]. Interestingly, bile ctDNA shows promising diagnostic capacities that outperform current diagnostic strategies [86,90]. However, the procedure to obtain bile is invasive, which prevents its use for repetitive testing. A meta-analysis comparing the diagnostic efficacy of cfDNA analysis in plasma and bile found that ctDNA detection in blood was more sensible than in bile [91]. Nevertheless, the studies included in this meta-analysis measured different cfDNA or ctDNA-related characteristics and were not homogenous in terms of the characteristics of the patients included.
5.2. Metastatic Disease Monitoring
As previously mentioned, median OS of patients with advanced disease is poor, and treatment with up to two lines of chemotherapy has shown modest efficacy [79,92]. Integration of NGS techniques identifying distinct genomic alterations that underlie disease progression [93] has accelerated the treatment paradigm of BTC. Patients with intrahepatic CCA bear genetic alterations in FGFR2 and IDH1 mutations and benefit from matched targeted therapy [94], while in patients with extrahepatic CCA and GBC ERBB2 amplifications are more commonly identified [95]. Overall, low numbers of patients with BTC have shown benefit from immunotherapy and NTRK-targeted therapy in clinical trials {Valle, 2021, Biliary tract cancer}. Indeed, the ESMO recommends multigene testing in patients with advanced CCA, in order to detect the full picture of targetable alterations [28]. In this line, a study including 327 patients with BTC revealed that patients receiving genetic alteration-matched targeted therapy in the second line showed better survival than patients without targetable alterations. Particularly, survival was better in patients with ESCAT I-II than the ESCAT III-IV alteration [24].
Different studies have assessed the feasibility of ctDNA genetic testing in BTC and compared its performance to tissue genetic testing. Overall, a similar genetic landscape is identified with both approaches, albeit with different concordance levels depending on experiment settings and testing technologies [96,97,98,99,100,101,102,103,104]. The largest study in this regard included 1671 patients and detected ctDNA in 84% of them, of whom 44% harbored targetable alterations. This study highlights a good concordance level between ctDNA and tissue NGS, and the advantage of ctDNA in terms of repeated sampling to follow tumor clonality and arising resistance-mechanisms [99]. Another study analyzed genetic alterations in 121 patients by tissue or ctDNA NGS, showing better survival in patients treated with matched targeted therapies than in patients with un-matched treatment [98]. Moreover, this work reported a better concordance of genetic alterations found in ctDNA with metastatic lesions than with the primary tumor, suggesting that novel alterations may arise in metastasis and that ctDNA analysis may be more informative in the selection of patients for matched targeted therapy. In this regard, a recent study reported a lower failure rate with blood genetic testing than with tissue (15.4% of no detectable alterations in blood vs. 26.8% in tissue), insufficient tumor tissue being the most common cause for tissue testing failure. Importantly, ctDNA-based genetic testing could be an alternative in this situation [102]. Of note, another study focused on early-onset BTC revealed a distinct genetic landscape in patients with early-onset BTC compared to older patients [104].
5.3. Prognosis and Disease Monitoring
Additionally, ctDNA analysis holds a prognostic value [105,106,107]. Overall, higher VAF of the dominant genetic alteration prior to treatment associates with worse clinical outcomes than PFS or a higher tumor burden [99,103,105,107]. Intriguingly, some other studies report no association between VAF and clinical outcomes [102,106]. A recent study proposes a CNV score based on plasma ctDNA analysis that is able to predict response to immunotherapy in patients with hepatobiliary malignancies. Among patients treated with immune checkpoint inhibitors, those with lower CNV risk scores had longer OS and PFS than those with high CNV risk scores [107].
Finally, the analysis of ctDNA to detect genetic alterations conferring acquired resistance to targeted therapy has also been assessed in BTC. Two studies have monitored resistance mechanisms in FGFR2-positive-patients treated with FGFR2 inhibitors by serial ctDNA analysis. Importantly, ctDNA was able to identify multiple resistance mechanisms not detected by tissue biopsy, even before detection of progression by radiologic imaging [108,109]. Table 3 summarizes key findings and designs of selected studies assessing the role of ctDNA in BTC.

Table 3.
Key findings and study design of main studies assessing the role of ctDNA in BTC.
6. Pancreatic Ductal Adenocarcinoma (PDAC)
PDAC is an aggressive and fatal malignancy. It ranks seventh among cancer deaths worldwide, with an estimated 495,773 new cases and 466,003 deaths in 2020 [38]. Nevertheless, it is much more common in highly developed countries, in which it will probably surpass breast cancer as the third leading cause of cancer death by 2025 [110].
Around 5–10% of PDAC comprise germline alterations, BRCA1/BRCA2 being the most commonly mutated genes [111,112], thus identifying a subset of patients with a potential benefit of PARP inhibitors [112,113]. Finally, the most common driver mutated genes in PDAC are non-druggable, and chemotherapy constitutes the unique therapeutic approach in the metastatic setting [111,114]. Considering KRAS as the most frequently mutated gene [115,116,117,118,119,120,121,122], multigene NGS evaluation could be of interest in those non-mutated cases for targeted treatment considerations [10]. Pan-cancer studies have reported promising results with targeted therapy for PDAC patients with MSI-H and NTRK fusions [123,124].
6.1. Screening and Diagnosis
Screening for PDAC is not recommended except for those individuals with a family history or presenting several associated conditions [111]. In this sense, due to the unspecific symptoms, PDAC is normally detected in advance stages and only 20% of cases are surgically treatable [114]. The definitive diagnosis needs an invasive procedure ordinarily performed with a fine needle aspiration cytology of the primary tumor, which relies on sufficient tumor tissue for an accurate molecular analysis [111]. The protein cancer antigen (CA) 19.9, which is measured in clinical practice, is commonly increased not only in an advanced stage of disease but also parallel to bilirubin levels, resulting in an unspecific marker with frequent false-positive results and not useful for initial diagnosis [111]. This highlights the importance of finding a reliable marker to detect the disease at early stages. In patients with resectable tumors, the presence of preoperative ctDNA was associated with larger tumor size, lymph node positivity and the presence of microscopic lymphovascular invasion [125]. In this sense, some studies have shown that levels of ctDNA are significantly greater in those patients with PDAC compared with healthy controls [118,126] but not with chronic pancreatitis [119,127]. Moreover, it has been observed that both ctDNA detection and the number of somatic alterations are higher in those patients with metastatic disease in comparison with patients with resectable locally advanced or early-stage tumors [116,121,128].
6.2. Prognosis
The prognostic value of ctDNA has been assessed in both early and advanced PDAC. Several studies report that the detection of KRAS mutations in ctDNA prior to active treatment or surgery associates with worse survival rates, as well as with certain clinico-pathological features. Particularly, KRAS mutations in ctDNA associates with tumor location in tail and neck, advanced stages, diagnosis of liver metastasis and high numbers of circulating regulatory T-cells [119,120,122,125,126,127,128,129,130,131,132,133,134,135,136,137,138,139,140]. Interestingly, in some studies, detection of KRAS mutations in tissue does not show prognostic significance, while ctDNA-based testing does [120]. Genetic alterations in other genes or ctDNA-related factors may also hold a prognostic value. In this regard, alterations in chromatin-regulating genes are linked to better outcomes [115] while higher ctDNA concentration, ERBB2 mutations or methylation sites in ctDNA associate with worse outcomes [141,142,143]. Studies that assess the genetic concordance between ctDNA and tissue biopsy obtained varying results [115,119,120,128,129,139,141].
6.3. Disease Monitoring
Besides its diagnostic properties, ctDNA also serves to monitor patient response in different settings. In resectable PDAC, it has been observed that both neoadjuvant therapy and surgery reduce the levels of ctDNA [125,139], and its detection after surgery could be predictive of an early relapse and worse outcome [115,116,125,134,136,139,144,145,146]. Indeed, some studies, including small subsets of patients, suggest that detection of MRD by ctDNA analysis can anticipate imaging-based diagnosis of relapse [115,116,125], and it also outperforms the predictive capacities of protein markers such as CA 19.9 [125,131,134,139,140,145,146]. Similarly, in metastatic PDAC, ctDNA analysis during chemotherapy serves as a marker to monitor response, as it associates with worse survival [117,118,130,145,147]. In this regard, a recent clinical trial assessed the role of ctDNA to monitor responses to second-line treatment based on enzyme administration, showing that ctDNA evolution after treatment correlated with outcomes [117]. Table 4 summarizes key findings and study design of selected studies assessing the role of ctDNA in PDAC.

Table 4.
Key findings and study design of main studies assessing the role of ctDNA in PDAC.
7. Conclusions and Future Directions
Analyses of ctDNA have emerged as a revolutionary tool for the management of patients with cancer over the past decade. Although the gold standard for tumor histotype characterization and first-line therapeutic approaches relies on the analysis of the tumor tissue, ctDNA constitutes an alternative source of tumor-derived DNA when tumor tissue sampling is challenging. In non-small-cell lung cancer, for instance, ctDNA analysis has become part of the armamentarium for treatment decision-making. Of note, ctDNA is unable to define either the tumor histology or the local microenvironment, but it does challenge the genomic testing. Considering upper gastrointestinal malignancies, several studies have shown the utility of ctDNA analysis, although further prospective validation is needed. Advantages over genetic testing in traditional tissue biopsy include the minimally invasive nature of the test and its ability to reveal a more holistic overview of the tumor genomic landscape, thus covering the temporal and spatial heterogeneity of tumor biology.
Studies in GEC, BTC and PDAC have shown how liquid biopsy can potentially impact early cancer detection and prognostication in all tumor stages and may identify the progression earlier than imaging techniques. Additionally, in the advanced setting, ctDNA analysis serves as a non-invasive tool for genetic testing and targeted therapy selection as well as for monitoring therapy response, being able to detect multiple resistance mechanisms. With such an amount of evidence, data generated adds convincement to researchers to rely on the potential role of liquid biopsy in these lethal tumors. In the era of personalized medicine, ctDNA analysis in liquid biopsy emerges as a highly valuable paradigm. Nevertheless, most of the studies are still observational and include small cohorts of patients. Future, prospective multi-center and interventional studies, carefully designed to base clinical decision-making on ctDNA results, will unveil the real clinical role of ctDNA analysis for upper gastrointestinal tumors and allow its integration into the therapeutic armamentarium.
Author Contributions
Conceptualization and design: I.L., A.E.H. and M.A.; Literature review: I.L., A.E.H. and M.A.; writing and revision: I.L., A.EH., V.A., I.H.-G., E.M., D.G., H.A., R.V. and M.A. All authors have read and agreed to the published version of the manuscript.
Funding
L.I. financed by National Agency of Research (AEI) in the “Juan de la Cierva-Postdoctoral formación” (8FJC2021-046521-I); A.H. is supported by the Clínico Junior 2019 scholarship from the Spanish Association Against Cancer (AECC) (CLJUN19010ARAS); A.M. is partially financed by the Government of Navarra in the La Caixa Program.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data supporting reported results are the references described above.
Conflicts of Interest
L.I., H.A.E. and G.D. declare no conflict of interest. A.V. has been involved as a consultant for advisory roles and received speaker honoraria from MSD, Bristol, Lilly, Astra-Zeneca and Pierre-Fabre. H-G.I. has received speaker honoraria from Astra Zeneca. M.E. has been involved as a consultant for advisory roles with Servier, Roche and Merck Sharp. A.H. has been involved as a consultant for advisory roles from Astra Zeneca and for trial coordination from Ferrer Farma. V.R has been involved as a consultant for advisory roles with Servier, Roche and Merck Sharp and has received speaker honoraria from Roche, Amgen, Merck Sharp and Dohme, Astra Zeneca. A.M. has been involved as a consultant for advisory roles with Amgen, BMS, MSD, Lilly and Servier.
Abbreviations
| ASCO | American Society of Clinical Oncology |
| BTC | Biliary tract cancer |
| CCA | Cholangiocarcinoma |
| cfDNA | Circulating free DNA |
| ctDNA | Circulating tumor DNA |
| ESCAT | ESMO Scale of Actionability of Molecular Targets |
| ESMO | European Society of Medical Oncology |
| ERBB2 | Epidermal growth factor receptor 2 |
| FGFR2 | Fibroblast growth factor receptor 2 |
| GBC | Gallbladder cancer |
| GEC | Gastroesophageal adenocarcinoma |
| MRD | Minimal residual disease |
| MSI-H | Microsatellite instability—high |
| NGS | Next-generation sequencing |
| NTRK | Neurotrophic tyrosine receptor kinase |
| OS | Overall survival |
| PCR | Polymerase chain reaction |
| PDAC | Pancreatic ductal adenocarcinoma |
| PFS | Progression-free survival |
| VAF | Tumor somatic variant allelic frequency |
References
- Gouda, M.A.; Huang, H.J.; Piha-Paul, S.A.; Call, S.G.; Karp, D.D.; Fu, S.; Naing, A.; Subbiah, V.; Pant, S.; Dustin, D.J.; et al. Longitudinal Monitoring of Circulating Tumor DNA to Predict Treatment Outcomes in Advanced Cancers. JCO Precis. Oncol. 2022, 6, e2100512. [Google Scholar] [CrossRef]
- Bayle, A.; Peyraud, F.; Belcaid, L.; Brunet, M.; Aldea, M.; Clodion, R.; Dubos, P.; Vasseur, D.; Nicotra, C.; Geraud, A.; et al. Liquid versus tissue biopsy for detecting actionable alterations according to ESCAT in patients with advanced cancer: A study from the French National Center for Precision Medicine (PRISM). Ann. Oncol. 2022, 33, 1328–1331. [Google Scholar] [CrossRef]
- Corcoran, R.B.; Chabner, B.A. Application of Cell-free DNA Analysis to Cancer Treatment. N. Engl. J. Med. 2018, 379, 1754–1765. [Google Scholar] [CrossRef]
- Diehl, F.; Schmidt, K.; Choti, M.A.; Romans, K.; Goodman, S.; Li, M.; Thornton, K.; Agrawal, N.; Sokoll, L.; Szabo, S.A.; et al. Circulating mutant DNA to assess tumor dynamics. Nat. Med. 2008, 14, 985–990. [Google Scholar] [CrossRef]
- Wen, X.; Pu, H.; Liu, Q.; Guo, Z.; Luo, D. Circulating Tumor DNA—A Novel Biomarker of Tumor Progression and Its Favorable Detection Techniques. Cancers 2022, 14, 6025. [Google Scholar] [CrossRef]
- Alese, O.B.; Cook, N.; Ortega-Franco, A.; Ulanja, M.B.; Tan, L.; Tie, J. Circulating Tumor DNA: An Emerging Tool in Gastrointestinal Cancers. Am. Soc. Clin. Oncol. Educ. Book 2022, 42, 279–298. [Google Scholar] [CrossRef]
- Wong, S.Q.; Fellowes, A.; Doig, K.; Ellul, J.; Bosma, T.J.; Irwin, D.; Vedururu, R.; Tan, A.Y.-C.; Weiss, J.; Chan, K.S.; et al. Assessing the clinical value of targeted massively parallel sequencing in a longitudinal, prospective population-based study of cancer patients. Br. J. Cancer 2015, 112, 1411–1420. [Google Scholar] [CrossRef]
- Wong, K.H.; Jin, Y.; Moqtaderi, Z. Multiplex Illumina Sequencing Using DNA Barcoding. Curr. Protoc. Mol. Biol. 2013, 101, 7.11.1–7.11.11. [Google Scholar]
- Xi, R.; Hadjipanayis, A.G.; Luquette, L.J.; Kim, T.-M.; Lee, E.; Zhang, J.; Johnson, M.D.; Muzny, D.M.; Wheeler, D.A.; Gibbs, R.A.; et al. Copy number variation detection in whole-genome sequencing data using the Bayesian information criterion. Proc. Natl. Acad. Sci. USA 2011, 108, E1128–E1136. [Google Scholar] [CrossRef]
- Pascual, J.; Attard, G.; Bidard, F.-C.; Curigliano, G.; De Mattos-Arruda, L.; Diehn, M.; Italiano, A.; Lindberg, J.; Merker, J.; Montagut, C.; et al. ESMO recommendations on the use of circulating tumour DNA assays for patients with cancer: A report from the ESMO Precision Medicine Working Group. Ann. Oncol. 2022, 33, 750–768. [Google Scholar] [CrossRef]
- Bettegowda, C.; Sausen, M.; Leary, R.J.; Kinde, I.; Wang, Y.; Agrawal, N.; Bartlett, B.R.; Wang, H.; Luber, B.; Alani, R.M.; et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 2014, 6, 224. [Google Scholar] [CrossRef]
- Razavi, P.; Li, B.T.; Brown, D.N.; Jung, B.; Hubbell, E.; Shen, R.; Abida, W.; Juluru, K.; De Bruijn, I.; Hou, C.; et al. High-intensity sequencing reveals the sources of plasma circulating cell-free DNA variants. Nat. Med. 2019, 25, 1928–1937. [Google Scholar] [CrossRef]
- Reichert, Z.; Morgan, T.; Li, G.; Castellanos, E.; Snow, T.; Dall’Olio, F.; Madison, R.; Fine, A.; Oxnard, G.; Graf, R.; et al. Prognostic value of plasma circulating tumor DNA fraction across four common cancer types: A real-world outcomes study. Ann. Oncol. 2022, 34, 111–120. [Google Scholar] [CrossRef]
- Rothwell, D.G.; Ayub, M.; Cook, N.; Thistlethwaite, F.; Carter, L.; Dean, E.; Smith, N.; Villa, S.; Dransfield, J.; Clipson, A.; et al. Utility of ctDNA to support patient selection for early phase clinical trials: The TARGET study. Nat. Med. 2019, 25, 738–743. [Google Scholar] [CrossRef]
- Wu, C.; Zhang, J.; Li, H.; Xu, W.; Zhang, X. The potential of liquid biopsies in gastrointestinal cancer. Clin. Biochem. 2020, 84, 1–12. [Google Scholar] [CrossRef]
- Siravegna, G.; Mussolin, B.; Venesio, T.; Marsoni, S.; Seoane, J.; Dive, C.; Papadopoulos, N.; Kopetz, S.; Corcoran, R.; Siu, L.; et al. How liquid biopsies can change clinical practice in oncology. Ann. Oncol. 2019, 30, 1580–1590. [Google Scholar] [CrossRef]
- Baselga, J.; Swain, S.M. Novel anticancer targets: Revisiting ERBB2 and discovering ERBB3. Nat. Rev. Cancer 2009, 9, 463–475. [Google Scholar] [CrossRef]
- Holderfield, M.; Deuker, M.M.; McCormick, F.; McMahon, M. Targeting RAF kinases for cancer therapy: BRAF-mutated melanoma and beyond. Nat. Rev. Cancer 2014, 14, 455–467. [Google Scholar] [CrossRef]
- Nigro, O.; Chini, C.; Proserpio, I. Molecularly targeted therapy for advanced gastrointestinal noncolorectal cancer treatment: How to choose? Past, present, future. Anti-Cancer Drugs 2021, 32, 593–601. [Google Scholar] [CrossRef]
- Nakamura, Y.; Kawazoe, A.; Lordick, F.; Janjigian, Y.Y.; Shitara, K. Biomarker-targeted therapies for advanced-stage gastric and gastro-oesophageal junction cancers: An emerging paradigm. Nat. Rev. Clin. Oncol. 2021, 18, 473–487. [Google Scholar] [CrossRef]
- Massard, C.; Michiels, S.; Ferté, C.; Le Deley, M.-C.; Lacroix, L.; Hollebecque, A.; Verlingue, L.; Ileana, E.; Rosellini, S.; Ammari, S.; et al. High-Throughput Genomics and Clinical Outcome in Hard-to-Treat Advanced Cancers: Results of the MOSCATO 01 Trial. Cancer Discov. 2017, 7, 586–595. [Google Scholar] [CrossRef]
- Martin-Romano, P.; Mezquita, L.; Hollebecque, A.; Lacroix, L.; Rouleau, E.; Gazzah, A.; Bahleda, R.; Planchard, D.; Varga, A.; Baldini, C.; et al. Implementing the European Society for Medical Oncology Scale for Clinical Actionability of Molecular Targets in a Comprehensive Profiling Program: Impact on Precision Medicine Oncology. JCO Precis. Oncol. 2022, 6, e2100484. [Google Scholar] [CrossRef]
- Moscow, J.A.; Fojo, T.; Schilsky, R.L. The evidence framework for precision cancer medicine. Nat. Rev. Clin. Oncol. 2017, 15, 183–192. [Google Scholar] [CrossRef]
- Verdaguer, H.; Saurí, T.; Acosta, D.A.; Guardiola, M.; Sierra, A.; Hernando, J.; Nuciforo, P.; Miquel, J.M.; Molero, C.; Peiró, S.; et al. ESMO Scale for Clinical Actionability of Molecular Targets Driving Targeted Treatment in Patients with Cholangiocarcinoma. Clin. Cancer Res. 2022, 28, 1662–1671. [Google Scholar] [CrossRef]
- Wagner, A.H.; Walsh, B.; Mayfield, G.; Tamborero, D.; Sonkin, D.; Krysiak, K.; Deu-Pons, J.; Duren, R.P.; Gao, J.; McMurry, J.; et al. A harmonized meta-knowledgebase of clinical interpretations of somatic genomic variants in cancer. Nat. Genet. 2020, 52, 448–457. [Google Scholar] [CrossRef]
- Li, M.M.; Datto, M.; Duncavage, E.J.; Kulkarni, S.; Lindeman, N.I.; Roy, S.; Tsimberidou, A.M.; Vnencak-Jones, C.L.; Wolff, D.J.; Younes, A.; et al. Standards and Guidelines for the Interpretation and Reporting of Sequence Variants in Cancer: A Joint Consensus Recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J. Mol. Diagn. 2017, 19, 4–23. [Google Scholar] [CrossRef]
- Mateo, J.; Chakravarty, D.; Dienstmann, R.; Jezdic, S.; Gonzalez-Perez, A.; Lopez-Bigas, N.; Ng, C.; Bedard, P.; Tortora, G.; Douillard, J.-Y.; et al. A framework to rank genomic alterations as targets for cancer precision medicine: The ESMO Scale for Clinical Actionability of molecular Targets (ESCAT). Ann. Oncol. 2018, 29, 1895–1902. [Google Scholar] [CrossRef]
- Mosele, F.; Remon, J.; Mateo, J.; Westphalen, C.; Barlesi, F.; Lolkema, M.; Normanno, N.; Scarpa, A.; Robson, M.; Meric-Bernstam, F.; et al. Recommendations for the use of next-generation sequencing (NGS) for patients with metastatic cancers: A report from the ESMO Precision Medicine Working Group. Ann. Oncol. 2020, 31, 1491–1505. [Google Scholar] [CrossRef]
- Chakravarty, D.; Johnson, A.; Sklar, J.; Lindeman, N.I.; Moore, K.; Ganesan, S.; Lovly, C.M.; Perlmutter, J.; Gray, S.W.; Hwang, J.; et al. Somatic Genomic Testing in Patients With Metastatic or Advanced Cancer: ASCO Provisional Clinical Opinion. J. Clin. Oncol. 2022, 40, 1231–1258. [Google Scholar] [CrossRef]
- Chen, X.; Gole, J.; Gore, A.; He, Q.; Lu, M.; Min, J.; Yuan, Z.; Yang, X.; Jiang, Y.; Zhang, T.; et al. Non-invasive early detection of cancer four years before conventional diagnosis using a blood test. Nat. Commun. 2020, 11, 3475. [Google Scholar] [CrossRef]
- Liu, M.C.; Oxnard, G.R.; Klein, E.A.; Swanton, C.; Seiden, M.V.; CCGA Consortium. Sensitive and specific multi-cancer detection and localization using methylation signatures in cell-free DNA. Ann. Oncol. 2020, 31, 745–759. [Google Scholar] [CrossRef]
- Klein, E.; Richards, D.; Cohn, A.; Tummala, M.; Lapham, R.; Cosgrove, D.; Chung, G.; Clement, J.; Gao, J.; Hunkapiller, N.; et al. Clinical validation of a targeted methylation-based multi-cancer early detection test using an independent validation set. Ann. Oncol. 2021, 32, 1167–1177. [Google Scholar] [CrossRef]
- Lan, Y.-T.; Chen, M.-H.; Fang, W.-L.; Hsieh, C.-C.; Lin, C.-H.; Jhang, F.-Y.; Yang, S.-H.; Lin, J.-K.; Chen, W.-S.; Jiang, J.-K.; et al. Clinical relevance of cell-free DNA in gastrointestinal tract malignancy. Oncotarget 2016, 8, 3009–3017. [Google Scholar] [CrossRef]
- Nakamura, Y.; Taniguchi, H.; Ikeda, M.; Bando, H.; Kato, K.; Morizane, C.; Esaki, T.; Komatsu, Y.; Kawamoto, Y.; Takahashi, N.; et al. Clinical utility of circulating tumor DNA sequencing in advanced gastrointestinal cancer: SCRUM-Japan GI-SCREEN and GOZILA studies. Nat. Med. 2020, 26, 1859–1864. [Google Scholar] [CrossRef]
- Zhang, Q.; Luo, J.; Wu, S.; Si, H.; Gao, C.; Xu, W.; Abdullah, S.E.; Higgs, B.W.; Dennis, P.A.; van der Heijden, M.S.; et al. Prognostic and Predictive Impact of Circulating Tumor DNA in Patients with Advanced Cancers Treated with Immune Checkpoint Blockade. Cancer Discov. 2020, 10, 1842–1853. [Google Scholar] [CrossRef]
- Bratman, S.V.; Yang, S.Y.C.; Iafolla, M.A.J.; Liu, Z.; Hansen, A.R.; Bedard, P.L.; Lheureux, S.; Spreafico, A.; Razak, A.A.; Shchegrova, S.; et al. Personalized circulating tumor DNA analysis as a predictive biomarker in solid tumor patients treated with pembrolizumab. Nat. Cancer 2020, 1, 873–881. [Google Scholar] [CrossRef]
- Parikh, A.R.; Leshchiner, I.; Elagina, L.; Goyal, L.; Levovitz, C.; Siravegna, G.; Livitz, D.; Rhrissorrakrai, K.; Martin, E.E.; Van Seventer, E.E.; et al. Liquid versus tissue biopsy for detecting acquired resistance and tumor heterogeneity in gastrointestinal cancers. Nat. Med. 2019, 25, 1415–1421. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014, 513, 202–209. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network. Integrated genomic characterization of oesophageal carcinoma. Nature 2017, 541, 169–175. [Google Scholar] [CrossRef]
- Cristescu, R.; Lee, J.; Nebozhyn, M.; Kim, K.-M.; Ting, J.C.; Wong, S.S.; Liu, J.; Yue, Y.G.; Wang, J.; Yu, K.; et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat. Med. 2015, 21, 449–456. [Google Scholar] [CrossRef]
- Alsina, M.; Arrazubi, V.; Diez, M.; Tabernero, J. Current developments in gastric cancer: From molecular profiling to treatment strategy. Nat. Rev. Gastroenterol. Hepatol. 2022. Epub ahead of print. [Google Scholar] [CrossRef]
- Alsina, M.; Gullo, I.; Carneiro, F. Intratumoral heterogeneity in gastric cancer: A new challenge to face. Ann. Oncol. 2017, 28, 912–913. [Google Scholar] [CrossRef]
- Pectasides, E.; Stachler, M.D.; Derks, S.; Liu, Y.; Maron, S.; Islam, M.; Alpert, L.; Kwak, H.; Kindler, H.; Polite, B.; et al. Genomic Heterogeneity as a Barrier to Precision Medicine in Gastroesophageal Adenocarcinoma. Cancer Discov. 2018, 8, 37–48. [Google Scholar] [CrossRef]
- Catenacci, D.V.T.; Moya, S.; Lomnicki, S.; Chase, L.M.; Peterson, B.F.; Reizine, N.; Alpert, L.; Setia, N.; Xiao, S.-Y.; Hart, J.; et al. Personalized Antibodies for Gastroesophageal Adenocarcinoma (PANGEA): A Phase II Study Evaluating an Individualized Treatment Strategy for Metastatic Disease. Cancer Discov. 2021, 11, 308–325. [Google Scholar] [CrossRef]
- Lordick, F.; Carneiro, F.; Cascinu, S.; Fleitas, T.; Haustermans, K.; Piessen, G.; Vogel, A.; Smyth, E. Gastric cancer: ESMO Clinical Practice Guideline for diagnosis, treatment and follow-up. Ann. Oncol. 2022, 33, 1005–1020. [Google Scholar] [CrossRef]
- Gullo, I.; Grillo, F.; Molinaro, L.; Fassan, M.; De Silvestri, A.; Tinelli, C.; Rugge, M.; Fiocca, R.; Mastracci, L. Minimum biopsy set for HER2 evaluation in gastric and gastro-esophageal junction cancer. Endosc. Int. Open 2015, 3, E165–E170. [Google Scholar] [CrossRef]
- Tominaga, N.; Gotoda, T.; Hara, M.; Hale, M.; Tsuchiya, T.; Matsubayashi, J.; Kono, S.; Kusano, C.; Itoi, T.; Fujimoto, K.; et al. Five biopsy specimens from the proximal part of the tumor reliably determine HER2 protein expression status in gastric cancer. Gastric Cancer 2015, 19, 553–560. [Google Scholar] [CrossRef]
- Qian, C.; Ju, S.; Qi, J.; Zhao, J.; Shen, X.; Jing, R.; Yu, J.; Li, L.; Shi, Y.; Zhang, L.; et al. Alu-based cell-free DNA: A novel biomarker for screening of gastric cancer. Oncotarget 2016, 8, 54037–54045. [Google Scholar] [CrossRef]
- Swanton, C.; Neal, R.D.; Johnson, P.W.M.; Dur, C.C.; Hamilton, S.A.; Zhang, N.; Kumar, H.; Sasieni, P. NHS-Galleri Trial Design: Equitable study recruitment tactics for targeted population-level screening with a multi-cancer early detection (MCED) test. J. Clin. Oncol. 2022, 40, TPS6606. [Google Scholar] [CrossRef]
- Al-Batran, S.-E.; Homann, N.; Pauligk, C.; Goetze, T.O.; Meiler, J.; Kasper, S.; Kopp, H.-G.; Mayer, F.; Haag, G.M.; Luley, K.; et al. Perioperative chemotherapy with fluorouracil plus leucovorin, oxaliplatin, and docetaxel versus fluorouracil or capecitabine plus cisplatin and epirubicin for locally advanced, resectable gastric or gastro-oesophageal junction adenocarcinoma (FLOT4): A randomised, phase 2/3 trial. Lancet 2019, 393, 1948–1957. [Google Scholar]
- Maron, S.B.; Chase, L.M.; Lomnicki, S.; Kochanny, S.; Moore, K.L.; Joshi, S.S.; Landron, S.; Johnson, J.; Kiedrowski, L.A.; Nagy, R.J.; et al. Circulating Tumor DNA Sequencing Analysis of Gastroesophageal Adenocarcinoma. Clin. Cancer Res. 2019, 25, 7098–7112. [Google Scholar] [CrossRef]
- Ococks, E.; Frankell, A.; Soler, N.M.; Grehan, N.; Northrop, A.; Coles, H.; Redmond, A.; Devonshire, G.; Weaver, J.; Hughes, C.; et al. Longitudinal tracking of 97 esophageal adenocarcinomas using liquid biopsy sampling. Ann. Oncol. 2020, 32, 522–532. [Google Scholar] [CrossRef]
- Yang, J.; Gong, Y.; Lam, V.K.; Shi, Y.; Guan, Y.; Zhang, Y.; Ji, L.; Chen, Y.; Zhao, Y.; Qian, F.; et al. Deep sequencing of circulating tumor DNA detects molecular residual disease and predicts recurrence in gastric cancer. Cell Death Dis. 2020, 11, 346. [Google Scholar] [CrossRef]
- Openshaw, M.R.; Mohamed, A.A.; Ottolini, B.; Fernandez-Garcia, D.; Richards, C.J.; Page, K.; Guttery, D.S.; Thomas, A.L.; Shaw, J.A. Longitudinal monitoring of circulating tumour DNA improves prognostication and relapse detection in gastroesophageal adenocarcinoma. Br. J. Cancer 2020, 123, 1271–1279. [Google Scholar] [CrossRef]
- Kim, Y.W.; Kim, Y.H.; Song, Y.; Kim, H.S.; Sim, H.W.; Poojan, S.; Eom, B.W.; Kook, M.C.; Joo, J.; Hong, K.M. Monitoring circulating tumor DNA by analyzing personalized cancer-specific rearrangements to detect recurrence in gastric cancer. Exp. Mol. Med. 2019, 51, 1–10. [Google Scholar] [CrossRef]
- Wo, J.Y.; Clark, J.W.; Eyler, C.E.; Mino-Kenudson, M.; Klempner, S.J.; Allen, J.N.; Keane, F.K.; Parikh, A.R.; Roeland, E.; Drapek, L.C.; et al. Results and Molecular Correlates from a Pilot Study of Neoadjuvant Induction FOLFIRINOX Followed by Chemoradiation and Surgery for Gastroesophageal Adenocarcinomas. Clin. Cancer Res. 2021, 27, 6343–6353. [Google Scholar] [CrossRef]
- Fedyanin, M.; Ignatova, E.; Boyarskikh, U.; Polyanskaya, E.; Kechin, A.; Osccorobin, I.; Shamovskaya, D.; Popova, A.; Trigolosov, A.; Nikulin, M.; et al. 137P Clinical utility of circulating tumour DNA (ctDNA) in resectable gastric cancer (GC). Ann. Oncol. 2020, 31, S1295. [Google Scholar] [CrossRef]
- Leal, A.; van Grieken, N.C.T.; Palsgrove, D.N.; Phallen, J.; Medina, J.E.; Hruban, C.; Broeckaert, M.A.M.; Anagnostou, V.; Adleff, V.; Bruhm, D.C.; et al. White blood cell and cell-free DNA analyses for detection of residual disease in gastric cancer. Nat. Commun. 2020, 11, 525. [Google Scholar] [CrossRef]
- Huffman, B.M.; Aushev, V.N.; Budde, G.L.; Chao, J.; Dayyani, F.; Hanna, D.; Botta, G.P.; Catenacci, D.V.; Maron, S.B.; Krinshpun, S.; et al. Analysis of Circulating Tumor DNA to Predict Risk of Recurrence in Patients with Esophageal and Gastric Cancers. JCO Precis. Oncol. 2022, 6, e2200420. [Google Scholar] [CrossRef]
- Wagner, A.D.; Syn, N.L.; Moehler, M.; Grothe, W.; Yong, W.P.; Tai, B.C.; Ho, J.; Unverzagt, S. Chemotherapy for advanced gastric cancer. Cochrane Database Syst. Rev. 2017, 8, CD004064. [Google Scholar] [CrossRef]
- Bang, Y.-J.; Van Cutsem, E.; Feyereislova, A.; Chung, H.C.; Shen, L.; Sawaki, A.; Lordick, F.; Ohtsu, A.; Omuro, Y.; Satoh, T.; et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): A phase 3, open-label, randomised controlled trial. Lancet 2010, 376, 687–697. [Google Scholar] [CrossRef]
- Janjigian, Y.Y.; Shitara, K.; Moehler, M.; Garrido, M.; Salman, P.; Shen, L.; Wyrwicz, L.; Yamaguchi, K.; Skoczylas, T.; Bragagnoli, A.C.; et al. First-line nivolumab plus chemotherapy versus chemotherapy alone for advanced gastric, gastro-oesophageal junction, and oesophageal adenocarcinoma (CheckMate 649): A randomised, open-label, phase 3 trial. Lancet 2021, 3, 27–40. [Google Scholar] [CrossRef]
- Sun, J.-M.; Shen, L.; Shah, M.A.; Enzinger, P.; Adenis, A.; Doi, T.; Kojima, T.; Metges, J.-P.; Li, Z.; Kim, S.-B.; et al. Pembrolizumab plus chemotherapy versus chemotherapy alone for first-line treatment of advanced oesophageal cancer (KEYNOTE-590): A randomised, placebo-controlled, phase 3 study. Lancet 2021, 398, 759–771. [Google Scholar] [CrossRef]
- Jin, Y.; Chen, D.-L.; Wang, F.-H.; Yang, C.-P.; Chen, X.-X.; You, J.-Q.; Huang, J.-S.; Shao, Y.; Zhu, D.-Q.; Ouyang, Y.-M.; et al. The predicting role of circulating tumor DNA landscape in gastric cancer patients treated with immune checkpoint inhibitors. Mol. Cancer 2020, 19, 154. [Google Scholar] [CrossRef]
- Davidson, M.; Barber, L.J.; Woolston, A.; Cafferkey, C.; Mansukhani, S.; Griffiths, B.; Moorcraft, S.-Y.; Rana, I.; Begum, R.; Assiotis, I.; et al. Detecting and Tracking Circulating Tumour DNA Copy Number Profiles during First Line Chemotherapy in Oesophagogastric Adenocarcinoma. Cancers 2019, 11, 736. [Google Scholar] [CrossRef]
- Kim, S.T.; Cristescu, R.; Bass, A.J.; Kim, K.-M.; Odegaard, J.I.; Kim, K.; Liu, X.Q.; Sher, X.; Jung, H.; Lee, M.; et al. Comprehensive molecular characterization of clinical responses to PD-1 inhibition in metastatic gastric cancer. Nat. Med. 2018, 24, 1449–1458. [Google Scholar] [CrossRef]
- Chen, Z.; Zhang, C.; Zhang, M.; Li, B.; Niu, Y.; Chen, L.; Yang, J.; Lu, S.; Gao, J.; Shen, L. Chromosomal instability of circulating tumor DNA reflect therapeutic responses in advanced gastric cancer. Cell Death Dis. 2019, 10, 697. [Google Scholar] [CrossRef]
- Wang, H.; Li, B.; Liu, Z.; Gong, J.; Shao, L.; Ren, J.; Niu, Y.; Bo, S.; Li, Z.; Lai, Y.; et al. HER2 copy number of circulating tumour DNA functions as a biomarker to predict and monitor trastuzumab efficacy in advanced gastric cancer. Eur. J. Cancer 2018, 88, 92–100. [Google Scholar] [CrossRef]
- Wang, D.-S.; Liu, Z.-X.; Lu, Y.-X.; Bao, H.; Wu, X.; Zeng, Z.-L.; Liu, Z.; Zhao, Q.; He, C.-Y.; Lu, J.-H.; et al. Liquid biopsies to track trastuzumab resistance in metastatic HER2-positive gastric cancer. Gut 2019, 68, 1152–1161. [Google Scholar] [CrossRef]
- Shoda, K.; Ichikawa, D.; Fujita, Y.; Masuda, K.; Hiramoto, H.; Hamada, J.; Arita, T.; Konishi, H.; Komatsu, S.; Shiozaki, A.; et al. Monitoring the HER2 copy number status in circulating tumor DNA by droplet digital PCR in patients with gastric cancer. Gastric Cancer 2016, 20, 126–135. [Google Scholar] [CrossRef]
- Kim, S.T.; Banks, K.C.; Pectasides, E.; Kim, K.; Lanman, R.; Talasaz, A.; An, J.; Choi, M.; Lee, J.; Sohn, T.; et al. Impact of genomic alterations on lapatinib treatment outcome and cell-free genomic landscape during HER2 therapy in HER2+ gastric cancer patients. Ann. Oncol. 2018, 29, 1037–1048. [Google Scholar] [CrossRef]
- Qiu, M.; Miaozhen, Q.; Lu, S.; Guan, W.; Wang, F.; Wang, X.; Jin, Y.; Wang, F.; Li, Y.; Shao, J.; et al. Prospective observation: Clinical utility of plasma Epstein–Barr virus DNA load in EBV-associated gastric carcinoma patients. Int. J. Cancer 2019, 146, 272–280. [Google Scholar] [CrossRef]
- Bang, Y.-J.; Van Cutsem, E.; Mansoor, W.; Petty, R.D.; Chao, Y.; Cunningham, D.; Ferry, D.; Landers, D.; Stockman, P.; Smith, N.R.; et al. A randomized, open-label phase II study of AZD4547 (AZD) versus Paclitaxel (P) in previously treated patients with advanced gastric cancer (AGC) with Fibroblast Growth Factor Receptor 2 (FGFR2) polysomy or gene amplification (amp): SHINE study. J. Clin. Oncol. 2015, 33, 4014. [Google Scholar] [CrossRef]
- Wainberg, Z.A.; Enzinger, P.C.; Kang, Y.-K.; Qin, S.; Yamaguchi, K.; Kim, I.-H.; Saeed, A.; Oh, S.C.; Li, J.; Turk, H.M.; et al. Bemarituzumab in patients with FGFR2b-selected gastric or gastro-oesophageal junction adenocarcinoma (FIGHT): A randomised, double-blind, placebo-controlled, phase 2 study. Lancet Oncol. 2022, 23, 1430–1440. [Google Scholar] [CrossRef]
- Smyth, E.C.; Vlachogiannis, G.; Hedayat, S.; Harbery, A.; Hulkki-Wilson, S.; Salati, M.; Kouvelakis, K.; Fernandez-Mateos, J.; Cresswell, G.D.; Fontana, E.; et al. EGFR amplification and outcome in a randomised phase III trial of chemotherapy alone or chemotherapy plus panitumumab for advanced gastro-oesophageal cancers. Gut 2020, 70, 1632–1641. [Google Scholar] [CrossRef]
- Lee, J.; Kim, S.T.; Kim, K.; Lee, H.; Kozarewa, I.; Mortimer, P.G.; Odegaard, J.I.; Harrington, E.A.; Lee, J.; Lee, T.; et al. Tumor Genomic Profiling Guides Patients with Metastatic Gastric Cancer to Targeted Treatment: The VIKTORY Umbrella Trial. Cancer Discov. 2019, 9, 1388–1405. [Google Scholar] [CrossRef]
- Nakamura, Y.; Fujisawa, T.; Taniguchi, H.; Bando, H.; Okamoto, W.; Tsuchihara, K.; Yoshino, T.; Ohtsu, A. SCRUM-Japan GI-SCREEN and MONSTAR-SCREEN: Path to the realization of biomarker-guided precision oncology in advanced solid tumors. Cancer Sci. 2021, 112, 4425–4432. [Google Scholar] [CrossRef]
- Lamarca, A.; Palmer, D.H.; Wasan, H.S.; Ross, P.J.; Ma, Y.T.; Arora, A.; Falk, S.; Gillmore, R.; Wadsley, J.; Patel, K.; et al. Second-line FOLFOX chemotherapy versus active symptom control for advanced biliary tract cancer (ABC-06): A phase 3, open-label, randomised, controlled trial. Lancet Oncol. 2021, 22, 690–701. [Google Scholar] [CrossRef]
- Izquierdo-Sanchez, L.; Lamarca, A.; La Casta, A.; Buettner, S.; Utpatel, K.; Klümpen, H.-J.; Adeva, J.; Vogel, A.; Lleo, A.; Fabris, L.; et al. Cholangiocarcinoma landscape in Europe: Diagnostic, prognostic and therapeutic insights from the ENSCCA Registry. J. Hepatol. 2022, 76, 1109–1121. [Google Scholar] [CrossRef]
- Wang, X.; Fu, X.-H.; Qian, Z.-L.; Zhao, T.; Duan, A.-Q.; Ruan, X.; Zhu, B.; Yin, L.; Zhang, Y.-J.; Yu, W.-L. Non-invasive detection of biliary tract cancer by low-coverage whole genome sequencing from plasma cell-free DNA: A prospective cohort study. Transl. Oncol. 2020, 14, 100908. [Google Scholar] [CrossRef]
- Wasenang, W.; Chaiyarit, P.; Proungvitaya, S.; Limpaiboon, T. Serum cell-free DNA methylation of OPCML and HOXD9 as a biomarker that may aid in differential diagnosis between cholangiocarcinoma and other biliary diseases. Clin. Epigenetics 2019, 11, 39. [Google Scholar] [CrossRef]
- Kumari, S.; Tewari, S.; Husain, N.; Agarwal, A.; Pandey, A.; Singhal, A.; Lohani, M. Quantification of Circulating Free DNA as a Diagnostic Marker in Gall Bladder Cancer. Pathol. Oncol. Res. 2016, 23, 91–97. [Google Scholar] [CrossRef]
- Kumari, S.; Husain, N.; Agarwal, A.; Neyaz, A.; Gupta, S.; Chaturvedi, A.; Lohani, M.; Sonkar, A.A. Diagnostic Value of Circulating Free DNA Integrity and Global Methylation Status in Gall Bladder Carcinoma. Pathol. Oncol. Res. 2018, 25, 925–936. [Google Scholar] [CrossRef]
- Han, J.-Y.; Ahn, K.S.; Kim, T.-S.; Kim, Y.H.; Cho, K.B.; Shin, D.W.; Baek, W.-K.; Suh, S.-I.; Jang, B.-C.; Kang, K.J. Liquid Biopsy from Bile-Circulating Tumor DNA in Patients with Biliary Tract Cancer. Cancers 2021, 13, 4581. [Google Scholar] [CrossRef]
- Arechederra, M.; Rullán, M.; Amat, I.; Oyon, D.; Zabalza, L.; Elizalde, M.; Latasa, M.U.; Mercado, M.R.; Ruiz-Clavijo, D.; Saldaña, C.; et al. Next-generation sequencing of bile cell-free DNA for the early detection of patients with malignant biliary strictures. Gut 2021, 71, 1141–1151. [Google Scholar] [CrossRef]
- Shen, N.; Zhang, D.; Yin, L.; Qiu, Y.; Liu, J.; Yu, W.; Fu, X.; Zhu, B.; Xu, X.; Duan, A.; et al. Bile cell-free DNA as a novel and powerful liquid biopsy for detecting somatic variants in biliary tract cancer. Oncol. Rep. 2019, 42, 549–560. [Google Scholar] [CrossRef]
- Kinugasa, H.; Nouso, K.; Ako, S.; Dohi, C.; Matsushita, H.; Matsumoto, K.; Kato, H.; Okada, H. Liquid biopsy of bile for the molecular diagnosis of gallbladder cancer. Cancer Biol. Ther. 2018, 19, 934–938. [Google Scholar] [CrossRef]
- Gou, Q.; Zhang, C.; Sun, Z.; Wu, L.; Chen, Y.; Mo, Z.; Mai, Q.; He, J.; Zhou, Z.; Shi, F.; et al. Cell-free DNA from bile outperformed plasma as a potential alternative to tissue biopsy in biliary tract cancer. ESMO Open 2021, 6, 100275. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, Z.; Tang, W.; Wang, X.; Liu, R.; Bao, H.; Chen, X.; Wei, Y.; Wu, S.; Bao, H.; et al. Ultrasensitive and affordable assay for early detection of primary liver cancer using plasma cell-free DNA fragmentomics. Hepatology 2021, 76, 317–329. [Google Scholar] [CrossRef]
- Singh, A.; Dwivedi, A. Circulating miRNA and cell-free DNA as a potential diagnostic tool in early detection of biliary tract cancer: A meta-analysis. Biomarkers 2022, 27, 399–406. [Google Scholar] [CrossRef]
- Valle, J.; Wasan, H.; Palmer, D.H.; Cunningham, D.; Anthoney, A.; Maraveyas, A.; Madhusudan, S.; Iveson, T.; Hughes, S.; Pereira, S.P.; et al. Cisplatin plus Gemcitabine versus Gemcitabine for Biliary Tract Cancer. N. Engl. J. Med. 2010, 362, 1273–1281. [Google Scholar] [CrossRef]
- Nakamura, H.; Arai, Y.; Totoki, Y.; Shirota, T.; ElZawahry, A.; Kato, M.; Hama, N.; Hosoda, F.; Urushidate, T.; Ohashi, S.; et al. Genomic spectra of biliary tract cancer. Nat. Genet. 2015, 47, 1003–1010. [Google Scholar] [CrossRef]
- Banales, J.M.; Marin, J.J.G.; Lamarca, A.; Rodrigues, P.M.; Khan, S.A.; Roberts, L.R.; Cardinale, V.; Carpino, G.; Andersen, J.B.; Braconi, C.; et al. Cholangiocarcinoma 2020: The next horizon in mechanisms and management. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 557–588. [Google Scholar] [CrossRef]
- Valle, J.W.; Lamarca, A.; Goyal, L.; Barriuso, J.; Zhu, A.X. New Horizons for Precision Medicine in Biliary Tract Cancers. Cancer Discov. 2017, 7, 943–962. [Google Scholar] [CrossRef]
- Zill, O.A.; Greene, C.; Sebisanovic, D.; Siew, L.M.; Leng, J.; Vu, M.; Hendifar, A.E.; Wang, Z.; Atreya, C.E.; Kelley, R.K.; et al. Cell-Free DNA Next-Generation Sequencing in Pancreatobiliary Carcinomas. Cancer Discov. 2015, 5, 1040–1048. [Google Scholar] [CrossRef]
- Kim, S.T.; Lira, M.; Deng, S.; Lee, S.; Park, Y.S.; Lim, H.Y.; Kang, W.K.; Mao, M.; Heo, J.S.; Kwon, W.; et al. PIK3CA mutation detection in metastatic biliary cancer using cell-free DNA. Oncotarget 2015, 6, 40026–40035. [Google Scholar] [CrossRef]
- Okamura, R.; Kurzrock, R.; Mallory, R.J.; Fanta, P.T.; Burgoyne, A.M.; Clary, B.M.; Kato, S.; Sicklick, J.K. Comprehensive genomic landscape and precision therapeutic approach in biliary tract cancers. Int. J. Cancer 2020, 148, 702–712. [Google Scholar] [CrossRef]
- Berchuck, J.; Facchinetti, F.; DiToro, D.; Baiev, I.; Majeed, U.; Reyes, S.; Chen, C.; Zhang, K.; Sharman, R.; Junior, P.U.; et al. The clinical landscape of cell-free DNA alterations in 1671 patients with advanced biliary tract cancer. Ann. Oncol. 2022, 33, 1269–1283. [Google Scholar] [CrossRef]
- Chen, C.; Wang, T.; Yang, M.; Song, J.; Huang, M.; Bai, Y.; Su, H. Genomic Profiling of Blood-Derived Circulating Tumor DNA from Patients with Advanced Biliary Tract Cancer. Pathol. Oncol. Res. 2021, 27, 1609879. [Google Scholar] [CrossRef]
- Csoma, S.L.; Bedekovics, J.; Veres, G.; Árokszállási, A.; András, C.; Méhes, G.; Mokánszki, A. Circulating Cell-Free DNA-Based Comprehensive Molecular Analysis of Biliary Tract Cancers Using Next-Generation Sequencing. Cancers 2022, 14, 233. [Google Scholar] [CrossRef]
- Lamarca, A.; Kapacee, Z.; Breeze, M.; Bell, C.; Belcher, D.; Staiger, H.; Taylor, C.; McNamara, M.G.; Hubner, R.A.; Valle, J.W. Molecular Profiling in Daily Clinical Practice: Practicalities in Advanced Cholangiocarcinoma and Other Biliary Tract Cancers. J. Clin. Med. 2020, 9, 2854. [Google Scholar] [CrossRef]
- Ettrich, T.J.; Schwerdel, D.; Dolnik, A.; Beuter, F.; Blätte, T.J.; Schmidt, S.A.; Stanescu-Siegmund, N.; Steinacker, J.; Marienfeld, R.; Kleger, A.; et al. Genotyping of circulating tumor DNA in cholangiocarcinoma reveals diagnostic and prognostic information. Sci. Rep. 2019, 9, 13261. [Google Scholar] [CrossRef]
- Mody, K.; Kasi, P.M.; Yang, J.; Surapaneni, P.K.; Bekaii-Saab, T.; Ahn, D.H.; Mahipal, A.; Sonbol, M.B.; Starr, J.S.; Roberts, A.; et al. Circulating Tumor DNA Profiling of Advanced Biliary Tract Cancers. JCO Precis. Oncol. 2019, 3, 1–9. [Google Scholar] [CrossRef]
- Junior, P.L.S.U.; Majeed, U.; Yin, J.; Botrus, G.; Sonbol, M.B.; Ahn, D.H.; Starr, J.S.; Jones, J.C.; Babiker, H.; Inabinett, S.R.; et al. Cell-Free Tumor DNA Dominant Clone Allele Frequency Is Associated With Poor Outcomes in Advanced Biliary Cancers Treated With Platinum-Based Chemotherapy. JCO Precis. Oncol. 2022, 6, e2100274. [Google Scholar] [CrossRef]
- Lapin, M.; Huang, H.J.; Chagani, S.; Javle, M.; Shroff, R.T.; Pant, S.; Gouda, M.A.; Raina, A.; Madwani, K.; Holley, V.R.; et al. Monitoring of Dynamic Changes and Clonal Evolution in Circulating Tumor DNA From Patients With IDH-Mutated Cholangiocarcinoma Treated With Isocitrate Dehydrogenase Inhibitors. JCO Precis. Oncol. 2022, 6, e2100197. [Google Scholar] [CrossRef]
- Yang, X.; Hu, Y.; Yang, K.; Wang, D.; Lin, J.; Long, J.; Xie, F.; Mao, J.; Bian, J.; Guan, M.; et al. Cell-free DNA copy number variations predict efficacy of immune checkpoint inhibitor-based therapy in hepatobiliary cancers. J. Immunother. Cancer 2021, 9, e001942. [Google Scholar] [CrossRef]
- Goyal, L.; Saha, S.K.; Liu, L.Y.; Siravegna, G.; Leshchiner, I.; Ahronian, L.G.; Lennerz, J.K.; Vu, P.; Deshpande, V.; Kambadakone, A.; et al. Polyclonal Secondary FGFR2 Mutations Drive Acquired Resistance to FGFR Inhibition in Patients with FGFR2 Fusion–Positive Cholangiocarcinoma. Cancer Discov. 2017, 7, 252–263. [Google Scholar] [CrossRef]
- Varghese, A.M.; Patel, J.; Janjigian, Y.Y.; Meng, F.; Selcuklu, S.D.; Iyer, G.; Houck-Loomis, B.; Harding, J.J.; O’Reilly, E.M.; Abou-Alfa, G.K.; et al. Noninvasive Detection of Polyclonal Acquired Resistance to FGFR Inhibition in Patients With Cholangiocarcinoma Harboring FGFR2 Alterations. JCO Precis. Oncol. 2021, 5, 44–50. [Google Scholar] [CrossRef]
- Ferlay, J.; Partensky, C.; Bray, F. More deaths from pancreatic cancer than breast cancer in the EU by 2017. Acta Oncol. Stockh. Swed. 2016, 55, 1158–1160. [Google Scholar] [CrossRef]
- Ducreux, M.; Cuhna, A.S.; Caramella, C.; Hollebecque, A.; Burtin, P.; Goéré, D.; Seufferlein, T.; Haustermans, K.; Van Laethem, J.L.; Conroy, T.; et al. Cancer of the pancreas: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2015, 26 (Suppl. S5), v56–v68. [Google Scholar] [CrossRef]
- Kindler, H.L.; Hammel, P.; Reni, M.; Van Cutsem, E.; Macarulla, T.; Hall, M.J.; Park, J.O.; Hochhauser, D.; Arnold, D.; Oh, D.-Y.; et al. Overall Survival Results From the POLO Trial: A Phase III Study of Active Maintenance Olaparib Versus Placebo for Germline BRCA-Mutated Metastatic Pancreatic Cancer. J. Clin. Oncol. 2022, 40, 3929–3939. [Google Scholar] [CrossRef]
- Golan, T.; Hammel, P.; Reni, M.; Van Cutsem, E.; Macarulla, T.; Hall, M.J.; Park, J.-O.; Hochhauser, D.; Arnold, D.; Oh, D.-Y.; et al. Maintenance Olaparib for Germline BRCA-Mutated Metastatic Pancreatic Cancer. N. Engl. J. Med. 2019, 381, 317–327. [Google Scholar] [CrossRef]
- Gómez-España, M.A.; Montes, A.F.; Garcia-Carbonero, R.; Mercadé, T.M.; Maurel, J.; Martín, A.M.; Pazo-Cid, R.; Vera, R.; Carrato, A.; Feliu, J. SEOM clinical guidelines for pancreatic and biliary tract cancer (2020). Clin. Transl. Oncol. 2021, 23, 988–1000. [Google Scholar] [CrossRef]
- Sausen, M.; Phallen, J.; Adleff, V.; Jones, S.; Leary, R.J.; Barrett, M.T.; Anagnostou, V.; Parpart-Li, S.; Murphy, D.; Kay Li, Q.; et al. Clinical implications of genomic alterations in the tumour and circulation of pancreatic cancer patients. Nat. Commun. 2015, 6, 7686. [Google Scholar] [CrossRef]
- Pietrasz, D.; Pécuchet, N.; Garlan, F.; Didelot, A.; Dubreuil, O.; Doat, S.; Imbert-Bismut, F.; Karoui, M.; Vaillant, J.-C.; Taly, V.; et al. Plasma Circulating Tumor DNA in Pancreatic Cancer Patients Is a Prognostic Marker. Clin. Cancer Res. 2017, 23, 116–123. [Google Scholar] [CrossRef]
- Bachet, J.-B.; Blons, H.F.; Hammel, P.; El Hariry, I.; Portales, F.; Mineur, L.; Metges, J.-P.; Mulot, C.; Bourreau, C.; Cain, J.; et al. Circulating Tumor DNA Is Prognostic and Potentially Predictive of Eryaspase Efficacy in Second-Line in Patients with Advanced Pancreatic Adenocarcinoma. Clin. Cancer Res. 2020, 26, 5208–5216. [Google Scholar] [CrossRef]
- Wei, T.; Zhang, Q.; Li, X.; Su, W.; Li, G.; Ma, T.; Gao, S.; Lou, J.; Que, R.; Zheng, L.; et al. Monitoring Tumor Burden in Response to FOLFIRINOX Chemotherapy Via Profiling Circulating Cell-Free DNA in Pancreatic Cancer. Mol. Cancer Ther. 2019, 18, 196–203. [Google Scholar] [CrossRef]
- Adamo, P.; Cowley, C.M.; Neal, C.P.; Mistry, V.; Page, K.; Dennison, A.R.; Isherwood, J.; Hastings, R.; Luo, J.; Moore, D.A.; et al. Profiling tumour heterogeneity through circulating tumour DNA in patients with pancreatic cancer. Oncotarget 2017, 8, 87221–87233. [Google Scholar] [CrossRef]
- Kinugasa, H.; Nouso, K.; Miyahara, K.; Morimoto, Y.; Dohi, C.; Tsutsumi, K.; Kato, H.; Okada, H.; Yamamoto, K. 761 Detection of K-RAS Gene Mutation by Liquid Biopsy in Patients With Pancreatic Cancer. Gastroenterology 2015, 121, 2271–2280. [Google Scholar] [CrossRef]
- Mohan, S.; Ayub, M.; Rothwell, D.G.; Gulati, S.; Kilerci, B.; Hollebecque, A.; Leong, H.S.; Smith, N.K.; Sahoo, S.; Descamps, T.; et al. Analysis of circulating cell-free DNA identifies KRAS copy number gain and mutation as a novel prognostic marker in Pancreatic cancer. Sci. Rep. 2019, 9, 11610. [Google Scholar] [CrossRef]
- Strijker, M.; Soer, E.C.; Pastena, M.; Creemers, A.; Balduzzi, A.; Beagan, J.J.; Busch, O.R.; Delden, O.M.; Halfwerk, H.; Hooft, J.E.; et al. Circulating tumor DNA quantity is related to tumor volume and both predict survival in metastatic pancreatic ductal adenocarcinoma. Int. J. Cancer 2019, 146, 1445–1456. [Google Scholar] [CrossRef]
- Doebele, R.C.; Drilon, A.; Paz-Ares, L.; Siena, S.; Shaw, A.T.; Farago, A.F.; Blakely, C.M.; Seto, T.; Cho, B.C.; Tosi, D.; et al. Entrectinib in patients with advanced or metastatic NTRK fusion-positive solid tumours: Integrated analysis of three phase 1–2 trials. Lancet Oncol. 2020, 21, 271–282. [Google Scholar] [CrossRef]
- Le, D.T.; Durham, J.N.; Smith, K.N.; Wang, H.; Bartlett, B.R.; Aulakh, L.K.; Lu, S.; Kemberling, H.; Wilt, C.; Luber, B.S.; et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 2017, 357, 409–413. [Google Scholar] [CrossRef]
- Groot, V.P.; Mosier, S.; Javed, A.A.; Teinor, J.A.; Gemenetzis, G.; Ding, D.; Haley, L.M.; Yu, J.; Burkhart, R.A.; Hasanain, A.; et al. Circulating Tumor DNA as a Clinical Test in Resected Pancreatic Cancer. Clin. Cancer Res. 2019, 25, 4973–4984. [Google Scholar] [CrossRef]
- Lapin, M.; Oltedal, S.; Tjensvoll, K.; Buhl, T.; Smaaland, R.; Garresori, H.; Javle, M.; Glenjen, N.I.; Abelseth, B.K.; Gilje, B.; et al. Fragment size and level of cell-free DNA provide prognostic information in patients with advanced pancreatic cancer. J. Transl. Med. 2018, 16, 300. [Google Scholar] [CrossRef]
- Singh, N.; Gupta, S.; Pandey, R.M.; Chauhan, S.S.; Saraya, A. High Levels of Cell-Free Circulating Nucleic Acids in Pancreatic Cancer are Associated With Vascular Encasement, Metastasis and Poor Survival. Cancer Investig. 2015, 33, 78–85. [Google Scholar] [CrossRef]
- Kim, M.K.; Woo, S.M.; Park, B.; Yoon, K.-A.; Kim, Y.-H.; Joo, J.; Lee, W.J.; Han, S.-S.; Park, S.-J.; Kong, S.-Y. Prognostic Implications of Multiplex Detection of KRAS Mutations in Cell-Free DNA from Patients with Pancreatic Ductal Adenocarcinoma. Clin. Chem. 2018, 64, 726–734. [Google Scholar] [CrossRef]
- Lin, M.; Alnaggar, M.; Liang, S.; Chen, J.; Xu, K.; Dong, S.; Du, D.; Niu, L. Circulating Tumor DNA as a Sensitive Marker in Patients Undergoing Irreversible Electroporation for Pancreatic Cancer. Cell. Physiol. Biochem. 2018, 47, 1556–1564. [Google Scholar] [CrossRef]
- Tjensvoll, K.; Lapin, M.; Buhl, T.; Oltedal, S.; Berry, K.S.-O.; Gilje, B.; Søreide, J.A.; Javle, M.; Nordgård, O.; Smaaland, R. Clinical relevance of circulating KRAS mutated DNA in plasma from patients with advanced pancreatic cancer. Mol. Oncol. 2015, 10, 635–643. [Google Scholar] [CrossRef]
- Hadano, N.; Murakami, Y.; Uemura, K.; Hashimoto, Y.; Kondo, N.; Nakagawa, N.; Sueda, T.; Hiyama, E. Prognostic value of circulating tumour DNA in patients undergoing curative resection for pancreatic cancer. Br. J. Cancer 2016, 115, 59–65. [Google Scholar] [CrossRef]
- Earl, J.; Garcia-Nieto, S.; Martinez-Avila, J.C.; Montans, J.; Sanjuanbenito, A.; Rodríguez-Garrote, M.; Lisa, E.; Mendía, E.; Lobo, E.; Malats, N.; et al. Circulating tumor cells (CTC) and KRAS mutant circulating free DNA (cfDNA) detection in peripheral blood as biomarkers in patients diagnosed with exocrine pancreatic cancer. BMC Cancer 2015, 15, 797. [Google Scholar] [CrossRef]
- Chen, H.; Tu, H.; Meng, Z.; Chen, Z.; Wang, P.; Liu, L. K-ras mutational status predicts poor prognosis in unresectable pancreatic cancer. Eur. J. Surg. Oncol. (EJSO) 2010, 36, 657–662. [Google Scholar] [CrossRef]
- Hussung, S.; Akhoundova, D.; Hipp, J.; Follo, M.; Klar, R.F.U.; Philipp, U.; Scherer, F.; von Bubnoff, N.; Duyster, J.; Boerries, M.; et al. Longitudinal analysis of cell-free mutated KRAS and CA 19–9 predicts survival following curative resection of pancreatic cancer. BMC Cancer 2021, 21, 49. [Google Scholar] [CrossRef]
- Bernard, V.; Kim, D.U.; Lucas, F.A.S.; Castillo, J.; Allenson, K.; Mulu, F.C.; Stephens, B.M.; Huang, J.; Semaan, A.; Guerrero, P.A.; et al. Circulating Nucleic Acids Are Associated With Outcomes of Patients With Pancreatic Cancer. Gastroenterology 2019, 156, 108–118.e4. [Google Scholar] [CrossRef]
- Lee, B.; Lipton, L.; Cohen, J.; Tie, J.; Javed, A.; Li, L.; Goldstein, D.; Burge, M.; Cooray, P.; Nagrial, A.; et al. Circulating tumor DNA as a potential marker of adjuvant chemotherapy benefit following surgery for localized pancreatic cancer. Ann. Oncol. 2019, 30, 1472–1478. [Google Scholar] [CrossRef]
- Guo, S.; Shi, X.; Shen, J.; Gao, S.; Wang, H.; Shen, S.; Pan, Y.; Li, B.; Xu, X.; Shao, Z.; et al. Preoperative detection of KRAS G12D mutation in ctDNA is a powerful predictor for early recurrence of resectable PDAC patients. Br. J. Cancer 2020, 122, 857–867. [Google Scholar] [CrossRef]
- Cheng, H.; Luo, G.; Jin, K.; Fan, Z.; Huang, Q.; Gong, Y.; Xu, J.; Yu, X.; Liu, C. Kras mutation correlating with circulating regulatory T cells predicts the prognosis of advanced pancreatic cancer patients. Cancer Med. 2020, 9, 2153–2159. [Google Scholar] [CrossRef]
- Jiang, J.; Ye, S.; Xu, Y.; Chang, L.; Hu, X.; Ru, G.; Guo, Y.; Yi, X.; Yang, L.; Huang, D. Circulating Tumor DNA as a Potential Marker to Detect Minimal Residual Disease and Predict Recurrence in Pancreatic Cancer. Front. Oncol. 2020, 10, 1220. [Google Scholar] [CrossRef]
- Toledano-Fonseca, M.; Cano, M.T.; Inga, E.; Rodríguez-Alonso, R.; Gómez-España, M.A.; Guil-Luna, S.; Mena-Osuna, R.; De La Haba-Rodríguez, J.R.; Rodríguez-Ariza, A.; Aranda, E. Circulating Cell-Free DNA-Based Liquid Biopsy Markers for the Non-Invasive Prognosis and Monitoring of Metastatic Pancreatic Cancer. Cancers 2020, 12, 1754. [Google Scholar] [CrossRef]
- Patel, H.; Okamura, R.; Fanta, P.; Patel, C.; Lanman, R.B.; Raymond, V.M.; Kato, S.; Kurzrock, R. Clinical correlates of blood-derived circulating tumor DNA in pancreatic cancer. J. Hematol. Oncol. 2019, 12, 130. [Google Scholar] [CrossRef]
- Pietrasz, D.; Wang-Renault, S.; Taieb, J.; Dahan, L.; Postel, M.; Durand-Labrunie, J.; Le Malicot, K.; Mulot, C.; Rinaldi, Y.; Phelip, J.-M.; et al. Prognostic value of circulating tumour DNA in metastatic pancreatic cancer patients: Post-hoc analyses of two clinical trials. Br. J. Cancer 2021, 126, 440–448. [Google Scholar] [CrossRef]
- Cheng, H.; Liu, C.; Jiang, J.; Luo, G.; Lu, Y.; Jin, K.; Guo, M.; Zhang, Z.; Xu, J.; Liu, L.; et al. Analysis of ctDNA to predict prognosis and monitor treatment responses in metastatic pancreatic cancer patients. Int. J. Cancer 2017, 140, 2344–2350. [Google Scholar] [CrossRef]
- Nakano, Y.; Kitago, M.; Matsuda, S.; Nakamura, Y.; Fujita, Y.; Imai, S.; Shinoda, M.; Yagi, H.; Abe, Y.; Hibi, T.; et al. KRAS mutations in cell-free DNA from preoperative and postoperative sera as a pancreatic cancer marker: A retrospective study. Br. J. Cancer 2018, 118, 662–669. [Google Scholar] [CrossRef]
- Watanabe, F.; Suzuki, K.; Tamaki, S.; Abe, I.; Endo, Y.; Takayama, Y.; Ishikawa, H.; Kakizawa, N.; Saito, M.; Futsuhara, K.; et al. Longitudinal monitoring of KRAS-mutated circulating tumor DNA enables the prediction of prognosis and therapeutic responses in patients with pancreatic cancer. PLoS ONE 2019, 14, e0227366. [Google Scholar] [CrossRef]
- Kruger, S.; Heinemann, V.; Ross, C.; Diehl, F.; Nagel, D.; Ormanns, S.; Liebmann, S.; Prinz-Bravin, I.; Westphalen, C.; Haas, M.; et al. Repeated mutKRAS ctDNA measurements represent a novel and promising tool for early response prediction and therapy monitoring in advanced pancreatic cancer. Ann. Oncol. 2018, 29, 2348–2355. [Google Scholar] [CrossRef]
- Del Re, M.; Vivaldi, C.; Rofi, E.; Vasile, E.; Miccoli, M.; Caparello, C.; D’Arienzo, P.D.; Fornaro, L.; Falcone, A.; Danesi, R. Early changes in plasma DNA levels of mutant KRAS as a sensitive marker of response to chemotherapy in pancreatic cancer. Sci. Rep. 2017, 7, 7931. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).