Preclinical Testing Techniques: Paving the Way for New Oncology Screening Approaches
Abstract
Simple Summary
Abstract
1. Introduction
2. Spheroids
2.1. Limitations of 2D Cell Culture
2.2. Spheroids for Preclinical Drug Development
2.3. Spheroids for Drug Efficacy
2.4. Spheroids for Drug Toxicity
3. Organoids
Organoids for Preclinical Drug Development
4. Organ-on-a-Chip
4.1. Single-Organ and Multi-Organ Chips
4.2. OoC Architecture
4.3. OoC for Preclinical Drug Development
4.4. Vascularization Capabilities of OoCs
5. Three-Dimensional Bioprinting
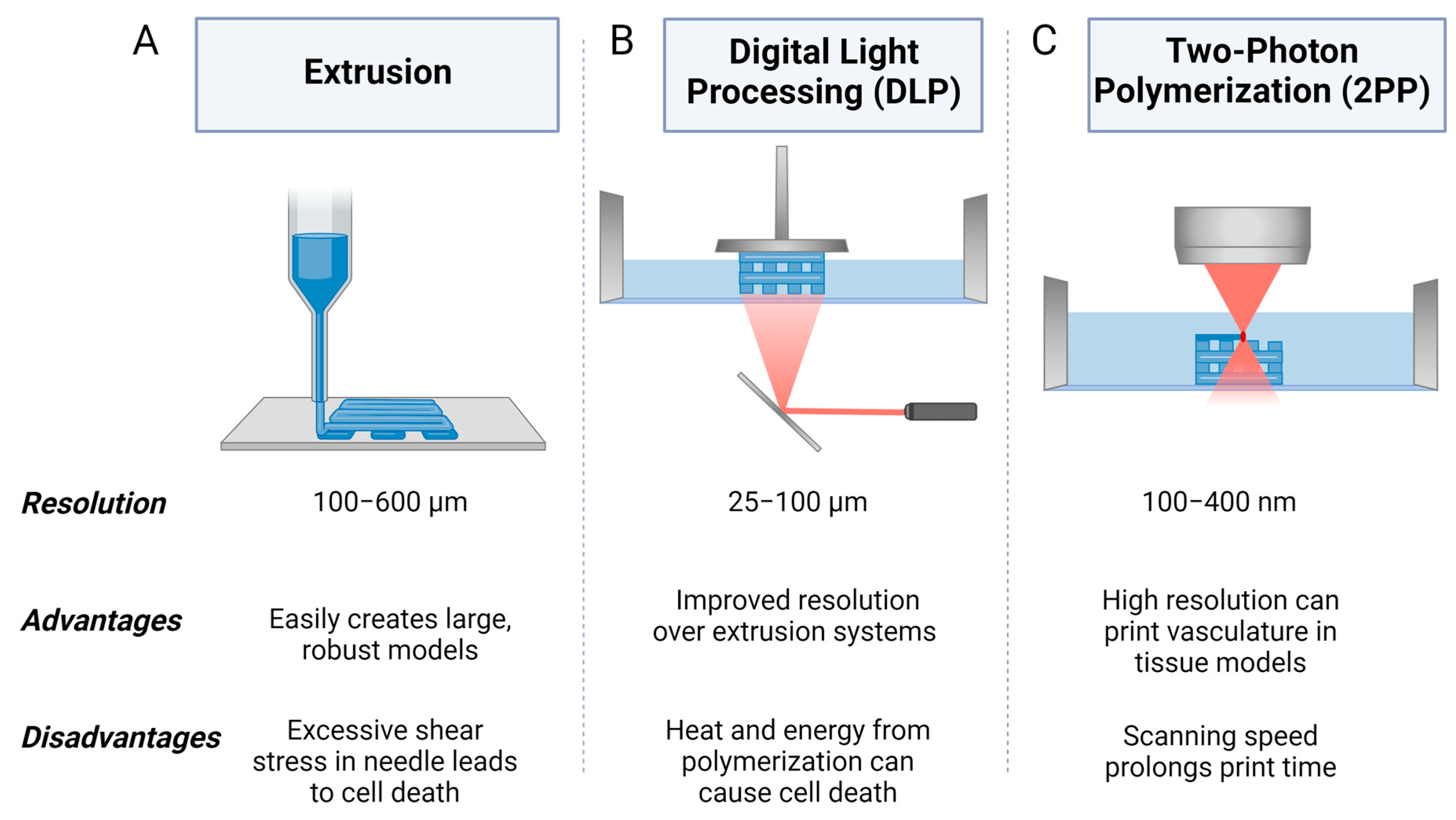
5.1. Extrusion 3D Bioprinting
5.2. Digital Light Processing (DLP) 3D Bioprinting
5.3. Two-Photon Polymerization (2PP) 3D Bioprinting
5.4. Three-Dimensional Bioprinting Biomaterials
5.5. Three-Dimensional Bioprinting for Preclinical Drug Development
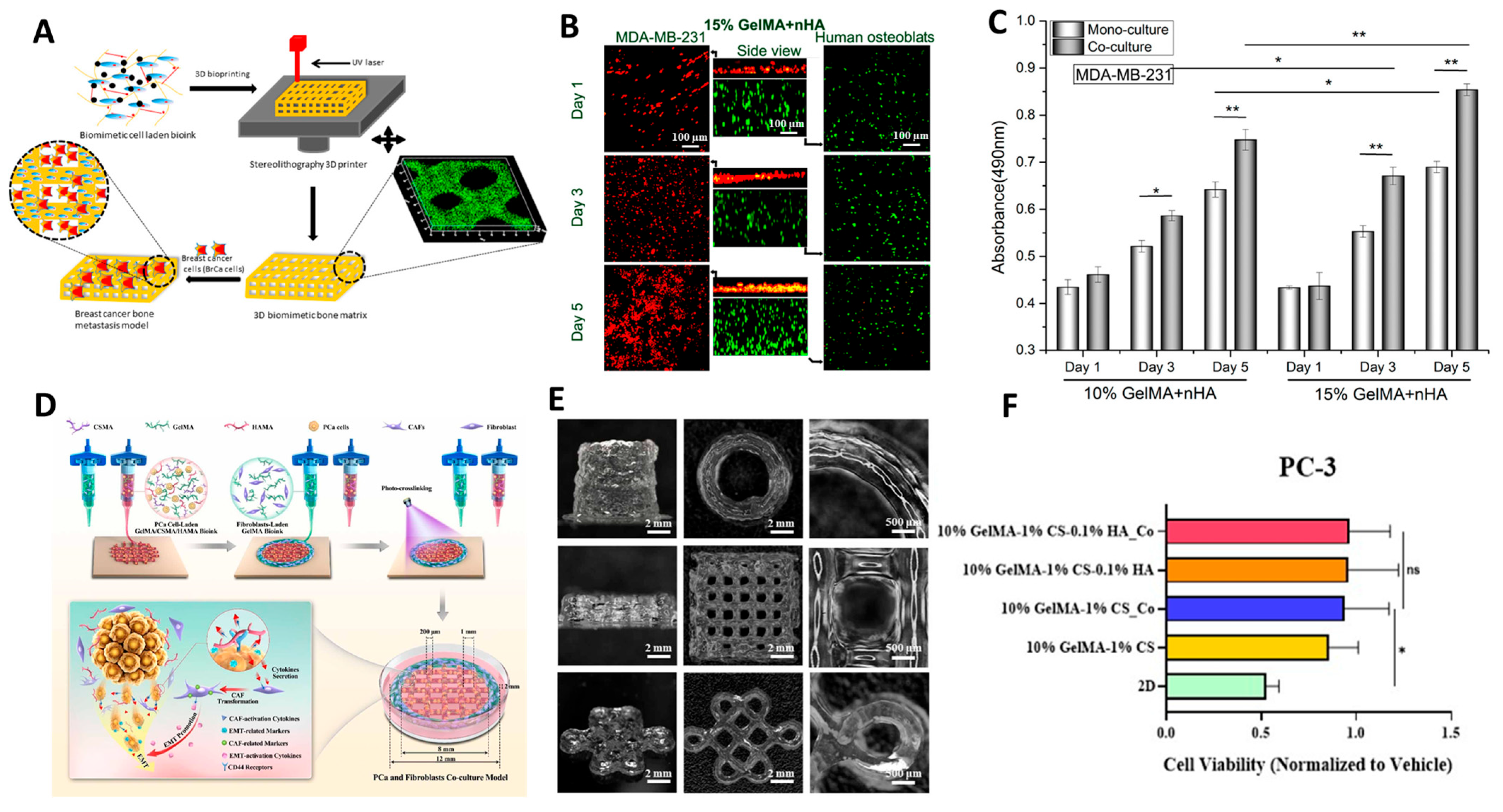
5.6. Three-Dimensional Bioprinting for Modelling Metastasis
5.7. Vascularization Capabilities of 3D Bioprinting
6. Conclusions and Future Outlook
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bailey, A.M.; Mendicino, M.; Au, P. An FDA Perspective on Preclinical Development of Cell-Based Regenerative Medicine Products. Nat. Biotechnol. 2014, 32, 721–723. [Google Scholar] [CrossRef]
- Yang, W.; Meng, L.; Chen, K.; Tian, C.; Peng, B.; Zhong, L.; Zhang, C.; Yang, X.; Zou, J.; Yang, S.; et al. Preclinical Pharmacodynamic Evaluation of a New Src/FOSL 1 Inhibitor, LY-1816, in Pancreatic Ductal Adenocarcinoma. Cancer Sci. 2019, 110, 1408–1419. [Google Scholar] [CrossRef] [PubMed]
- Lovitt, C.; Shelper, T.; Avery, V. Advanced Cell Culture Techniques for Cancer Drug Discovery. Biology 2014, 3, 345–367. [Google Scholar] [CrossRef]
- Ikeda, K.; Iwasaki, Y. Edaravone, a Free Radical Scavenger, Delayed Symptomatic and Pathological Progression of Motor Neuron Disease in the Wobbler Mouse. PLoS ONE 2015, 10, e0140316. [Google Scholar] [CrossRef]
- Sun, D.; Gao, W.; Hu, H.; Zhou, S. Why 90% of Clinical Drug Development Fails and How to Improve It? Acta Pharm. Sin. B 2022, 12, 3049–3062. [Google Scholar] [CrossRef] [PubMed]
- Valente, K.P.; Khetani, S.; Kolahchi, A.R.; Sanati-Nezhad, A.; Suleman, A.; Akbari, M. Microfluidic Technologies for Anticancer Drug Studies. Drug Discov. Today 2017, 22, 1654–1670. [Google Scholar] [CrossRef] [PubMed]
- Unger, C.; Kramer, N.; Walzl, A.; Scherzer, M.; Hengstschläger, M.; Dolznig, H. Modeling Human Carcinomas: Physiologically Relevant 3D Models to Improve Anti-Cancer Drug Development. Adv. Drug Deliv. Rev. 2014, 79, 50–67. [Google Scholar] [CrossRef]
- Darrow, J.J.; Avorn, J.; Kesselheim, A.S. FDA Approval and Regulation of Pharmaceuticals, 1983–2018. JAMA 2020, 323, 164. [Google Scholar] [CrossRef] [PubMed]
- van Norman, G.A. Drugs, Devices, and the FDA: Part 1: An Overview of Approval Processes for Drugs. JACC Basic Transl. Sci. 2016, 1, 170–179. [Google Scholar] [CrossRef]
- S.5002—117th Congress (2021–2022): FDA Modernization Act 2.0. Available online: https://www.congress.gov/bill/117th-congress/senate-bill/5002/text (accessed on 30 August 2023).
- Andersen, T.; Auk-Emblem, P.; Dornish, M. 3D Cell Culture in Alginate Hydrogels. Microarrays 2015, 4, 133–161. [Google Scholar] [CrossRef]
- Gong, X.; Lin, C.; Cheng, J.; Su, J.; Zhao, H.; Liu, T.; Wen, X.; Zhao, P. Generation of Multicellular Tumor Spheroids with Microwell-Based Agarose Scaffolds for Drug Testing. PLoS ONE 2015, 10, e0130348. [Google Scholar] [CrossRef] [PubMed]
- Tuveson, D.; Clevers, H. Cancer Modeling Meets Human Organoid Technology. Science 2019, 364, 952–955. [Google Scholar] [CrossRef] [PubMed]
- Drasdo, D.; Höhme, S. A Single-Cell-Based Model of Tumor Growth In Vitro: Monolayers and Spheroids. Phys. Biol. 2005, 2, 133–147. [Google Scholar] [CrossRef]
- Barisam, M.; Saidi, M.; Kashaninejad, N.; Nguyen, N.-T. Prediction of Necrotic Core and Hypoxic Zone of Multicellular Spheroids in a Microbioreactor with a U-Shaped Barrier. Micromachines 2018, 9, 94. [Google Scholar] [CrossRef]
- Glicklis, R.; Merchuk, J.C.; Cohen, S. Modeling Mass Transfer in Hepatocyte Spheroids via Cell Viability, Spheroid Size, and Hepatocellular Functions. Biotechnol. Bioeng. 2004, 86, 672–680. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Luo, Z.; Lee, J.; Kim, H.-J.; Lee, K.; Tebon, P.; Feng, Y.; Dokmeci, M.R.; Sengupta, S.; Khademhosseini, A. Organ-on-a-Chip for Cancer and Immune Organs Modeling. Adv. Healthc. Mater. 2019, 8, 1801363. [Google Scholar] [CrossRef]
- Ma, C.; Peng, Y.; Li, H.; Chen, W. Organ-on-a-Chip: A New Paradigm for Drug Development. Trends Pharmacol. Sci. 2021, 42, 119–133. [Google Scholar] [CrossRef]
- Thompson, C.L.; Fu, S.; Heywood, H.K.; Knight, M.M.; Thorpe, S.D. Mechanical Stimulation: A Crucial Element of Organ-on-Chip Models. Front. Bioeng. Biotechnol. 2020, 8, 602646. [Google Scholar] [CrossRef]
- Kaarj, K.; Yoon, J.Y. Methods of Delivering Mechanical Stimuli to Organ-on-a-Chip. Micromachines 2019, 10, 700. [Google Scholar] [CrossRef]
- Satoh, T.; Sugiura, S.; Shin, K.; Onuki-Nagasaki, R.; Ishida, S.; Kikuchi, K.; Kakiki, M.; Kanamori, T. A Multi-Throughput Multi-Organ-on-a-Chip System on a Plate Formatted Pneumatic Pressure-Driven Medium Circulation Platform. Lab Chip 2018, 18, 115–125. [Google Scholar] [CrossRef]
- Danku, A.E.; Dulf, E.-H.; Braicu, C.; Jurj, A.; Berindan-Neagoe, I. Organ-on-a-Chip: A Survey of Technical Results and Problems. Front. Bioeng. Biotechnol. 2022, 10, 840674. [Google Scholar] [CrossRef]
- Datta, P.; Dey, M.; Ataie, Z.; Unutmaz, D.; Ozbolat, I.T. 3D Bioprinting for Reconstituting the Cancer Microenvironment. NPJ Precis. Oncol. 2020, 4, 18. [Google Scholar] [CrossRef]
- Unagolla, J.M.; Jayasuriya, A.C. Hydrogel-Based 3D Bioprinting: A Comprehensive Review on Cell-Laden Hydrogels, Bioink Formulations, and Future Perspectives. Appl. Mater. Today 2020, 18, 100479. [Google Scholar] [CrossRef] [PubMed]
- Jia, L.; Zhang, Y.; Yao, L.; Zhang, P.; Ci, Z.; Zhang, W.; Miao, C.; Liang, X.; He, A.; Liu, Y.; et al. Regeneration of Human-Ear-Shaped Cartilage with Acellular Cartilage Matrix-Based Biomimetic Scaffolds. Appl. Mater. Today 2020, 20, 100639. [Google Scholar] [CrossRef]
- Elomaa, L.; Keshi, E.; Sauer, I.M.; Weinhart, M. Development of GelMA/PCL and DECM/PCL Resins for 3D Printing of Acellular in Vitro Tissue Scaffolds by Stereolithography. Mater. Sci. Eng. C 2020, 112, 110958. [Google Scholar] [CrossRef] [PubMed]
- Lewicki, J.; Bergman, J.; Kerins, C.; Hermanson, O. Optimization of 3D Bioprinting of Human Neuroblastoma Cells Using Sodium Alginate Hydrogel. Bioprinting 2019, 16, e00053. [Google Scholar] [CrossRef]
- Kingsley, D.M.; Roberge, C.L.; Rudkouskaya, A.; Faulkner, D.E.; Barroso, M.; Intes, X.; Corr, D.T. Laser-Based 3D Bioprinting for Spatial and Size Control of Tumor Spheroids and Embryoid Bodies. Acta Biomater. 2019, 95, 357–370. [Google Scholar] [CrossRef]
- Hauptmann, N.; Lian, Q.; Ludolph, J.; Rothe, H.; Hildebrand, G.; Liefeith, K. Biomimetic Designer Scaffolds Made of D,L-Lactide-ɛ-Caprolactone Polymers by 2-Photon Polymerization. Tissue Eng. Part B Rev. 2019, 25, 167–186. [Google Scholar] [CrossRef]
- Potter, R.F.; Groom, A.C. Capillary Diameter and Geometry in Cardiac and Skeletal Muscle Studied by Means of Corrosion Casts. Microvasc. Res. 1983, 25, 68–84. [Google Scholar] [CrossRef]
- Schwab, A.; Levato, R.; D’Este, M.; Piluso, S.; Eglin, D.; Malda, J. Printability and Shape Fidelity of Bioinks in 3D Bioprinting. Chem. Rev. 2020, 120, 11028–11055. [Google Scholar] [CrossRef]
- Hinton, T.J.; Lee, A.; Feinberg, A.W. 3D Bioprinting from the Micrometer to Millimeter Length Scales: Size Does Matter. Curr. Opin. Biomed. Eng. 2017, 1, 31–37. [Google Scholar] [CrossRef]
- Pawlicki, M.; Collins, H.A.; Denning, R.G.; Anderson, H.L. Two-Photon Absorption and the Design of Two-Photon Dyes. Angew. Chem. Int. Ed. 2009, 48, 3244–3266. [Google Scholar] [CrossRef] [PubMed]
- Limberg, D.K.; Kang, J.-H.; Hayward, R.C. Triplet–Triplet Annihilation Photopolymerization for High-Resolution 3D Printing. J. Am. Chem. Soc. 2022, 144, 5226–5232. [Google Scholar] [CrossRef] [PubMed]
- Valente, F.; Hepburn, M.S.; Chen, J.; Aldana, A.A.; Allardyce, B.J.; Shafei, S.; Doyle, B.J.; Kennedy, B.F.; Dilley, R.J. Bioprinting Silk Fibroin Using Two-Photon Lithography Enables Control over the Physico-Chemical Material Properties and Cellular Response. Bioprinting 2022, 25, e00183. [Google Scholar] [CrossRef]
- Richter, M.; Piwocka, O.; Musielak, M.; Piotrowski, I.; Suchorska, W.M.; Trzeciak, T. From Donor to the Lab: A Fascinating Journey of Primary Cell Lines. Front. Cell Dev. Biol. 2021, 9, 711381. [Google Scholar] [CrossRef]
- Saji Joseph, J.; Tebogo Malindisa, S.; Ntwasa, M. Two-Dimensional (2D) and Three-Dimensional (3D) Cell Culturing in Drug Discovery. In Cell Culture; IntechOpen: London, UK, 2019. [Google Scholar]
- Anton, D.; Burckel, H.; Josset, E.; Noel, G. Three-Dimensional Cell Culture: A Breakthrough In Vivo. Int. J. Mol. Sci. 2015, 16, 5517–5527. [Google Scholar] [CrossRef]
- Yue, B. Biology of the Extracellular Matrix: An Overview. J. Glaucoma 2014, 23, S20–S23. [Google Scholar] [CrossRef]
- Kim, B.-S.; Nikolovski, J.; Bonadio, J.; Smiley, E.; Mooney, D.J. Engineered Smooth Muscle Tissues: Regulating Cell Phenotype with the Scaffold. Exp. Cell Res. 1999, 251, 318–328. [Google Scholar] [CrossRef]
- Yin, Z.; Dong, C.; Jiang, K.; Xu, Z.; Li, R.; Guo, K.; Shao, S.; Wang, L. Heterogeneity of Cancer-Associated Fibroblasts and Roles in the Progression, Prognosis, and Therapy of Hepatocellular Carcinoma. J. Hematol. Oncol. 2019, 12, 711381. [Google Scholar] [CrossRef]
- Prager, B.C.; Xie, Q.; Bao, S.; Rich, J.N. Cancer Stem Cells: The Architects of the Tumor Ecosystem. Cell Stem Cell 2019, 24, 41–53. [Google Scholar] [CrossRef]
- Jong, B.K. Three-Dimensional Tissue Culture Models in Cancer Biology. Semin. Cancer Biol. 2005, 15, 365–377. [Google Scholar]
- Białkowska, K.; Komorowski, P.; Bryszewska, M.; Miłowska, K. Spheroids as a Type of Three-Dimensional Cell Cultures—Examples of Methods of Preparation and the Most Important Application. Int. J. Mol. Sci. 2020, 21, 6225. [Google Scholar] [CrossRef] [PubMed]
- Tancioni, I.; Miller, N.L.G.; Uryu, S.; Lawson, C.; Jean, C.; Chen, X.L.; Kleinschmidt, E.G.; Schlaepfer, D.D. FAK Activity Protects Nucleostemin in Facilitating Breast Cancer Spheroid and Tumor Growth. Breast Cancer Res. 2015, 17, 47. [Google Scholar] [CrossRef] [PubMed]
- Del Duca, D.; Werbowetski, T.; Del Maestro, R.F. Spheroid Preparation from Hanging Drops: Characterization of a Model of Brain Tumor Invasion. J. Neurooncol. 2004, 67, 295–303. [Google Scholar] [CrossRef] [PubMed]
- Foty, R. A Simple Hanging Drop Cell Culture Protocol for Generation of 3D Spheroids. J. Vis. Exp. 2011, 51, e2720. [Google Scholar] [CrossRef]
- Mirab, F.; Kang, Y.J.; Majd, S. Preparation and Characterization of Size-Controlled Glioma Spheroids Using Agarose Hydrogel Microwells. PLoS ONE 2019, 14, e0211078. [Google Scholar] [CrossRef]
- Tang, Y.; Liu, J.; Chen, Y. Agarose Multi-Wells for Tumour Spheroid Formation and Anti-Cancer Drug Test. Microelectron. Eng. 2016, 158, 41–45. [Google Scholar] [CrossRef]
- Kang, S.; Kim, D.; Lee, J.; Takayama, S.; Park, J.Y. Engineered Microsystems for Spheroid and Organoid Studies. Adv. Healthc. Mater. 2021, 10, 2001284. [Google Scholar] [CrossRef]
- Lin, R.-Z.; Chang, H.-Y. Recent Advances in Three-Dimensional Multicellular Spheroid Culture for Biomedical Research. Biotechnol. J. 2008, 3, 1172–1184. [Google Scholar] [CrossRef]
- Kim, S.; Kim, E.M.; Yamamoto, M.; Park, H.; Shin, H. Engineering Multi-Cellular Spheroids for Tissue Engineering and Regenerative Medicine. Adv. Healthc. Mater. 2020, 9, 2000608. [Google Scholar] [CrossRef]
- Ryu, J.H.; Kim, M.S.; Lee, G.M.; Choi, C.Y.; Kim, B.-S. The Enhancement of Recombinant Protein Production by Polymer Nanospheres in Cell Suspension Culture. Biomaterials 2005, 26, 2173–2181. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.-Y.; Park, J.; Jang, H.-K.; Lee, T.-J.; La, W.-G.; Bhang, S.H.; Kwon, I.K.; Kwon, O.H.; Kim, B.-S. Efficient Formation of Cell Spheroids Using Polymer Nanofibers. Biotechnol. Lett. 2012, 34, 795–803. [Google Scholar] [CrossRef] [PubMed]
- Anil-Inevi, M.; Yaman, S.; Yildiz, A.A.; Mese, G.; Yalcin-Ozuysal, O.; Tekin, H.C.; Ozcivici, E. Biofabrication of in Situ Self Assembled 3D Cell Cultures in a Weightlessness Environment Generated Using Magnetic Levitation. Sci. Rep. 2018, 8, 7239. [Google Scholar] [CrossRef]
- Souza, G.R.; Molina, J.R.; Raphael, R.M.; Ozawa, M.G.; Stark, D.J.; Levin, C.S.; Bronk, L.F.; Ananta, J.S.; Mandelin, J.; Georgescu, M.-M.; et al. Three-Dimensional Tissue Culture Based on Magnetic Cell Levitation. Nat. Nanotechnol. 2010, 5, 291–296. [Google Scholar] [CrossRef]
- Türker, E.; Demirçak, N.; Arslan-Yildiz, A. Scaffold-Free Three-Dimensional Cell Culturing Using Magnetic Levitation. Biomater. Sci. 2018, 6, 1745–1753. [Google Scholar] [CrossRef] [PubMed]
- Baek, N.; Seo, O.W.; Lee, J.; Hulme, J.; An, S.S.A. Real-Time Monitoring of Cisplatin Cytotoxicity on Three-Dimensional Spheroid Tumor Cells. Drug Des. Dev. Ther. 2016, 10, 2155–2165. [Google Scholar] [CrossRef]
- Wong, C.W.; Han, H.W.; Tien, Y.W.; Hsu, S.H. Biomaterial Substrate-Derived Compact Cellular Spheroids Mimicking the Behavior of Pancreatic Cancer and Microenvironment. Biomaterials 2019, 213, 119202. [Google Scholar] [CrossRef]
- Antunes, J.; Gaspar, V.M.; Ferreira, L.; Monteiro, M.; Henrique, R.; Jerónimo, C.; Mano, J.F. In-Air Production of 3D Co-Culture Tumor Spheroid Hydrogels for Expedited Drug Screening. Acta Biomater. 2019, 94, 392–409. [Google Scholar] [CrossRef]
- Culp, M.B.B.; Soerjomataram, I.; Efstathiou, J.A.; Bray, F.; Jemal, A. Recent Global Patterns in Prostate Cancer Incidence and Mortality Rates. Eur. Urol. 2020, 77, 38–52. [Google Scholar] [CrossRef]
- Weydert, Z.; Lal-Nag, M.; Mathews-Greiner, L.; Thiel, C.; Cordes, H.; Küpfer, L.; Guye, P.; Kelm, J.M.; Ferrer, M. A 3D Heterotypic Multicellular Tumor Spheroid Assay Platform to Discriminate Drug Effects on Stroma versus Cancer Cells. SLAS Discov. 2020, 25, 265–276. [Google Scholar] [CrossRef]
- Courau, T.; Bonnereau, J.; Chicoteau, J.; Bottois, H.; Remark, R.; Assante Miranda, L.; Toubert, A.; Blery, M.; Aparicio, T.; Allez, M.; et al. Cocultures of Human Colorectal Tumor Spheroids with Immune Cells Reveal the Therapeutic Potential of MICA/B and NKG2A Targeting for Cancer Treatment. J. Immunother. Cancer 2019, 7, 74. [Google Scholar] [CrossRef] [PubMed]
- Drost, J.; Clevers, H. Organoids in Cancer Research. Nat. Rev. Cancer 2018, 18, 407–418. [Google Scholar] [CrossRef] [PubMed]
- Lancaster, M.A.; Knoblich, J.A. Organogenesis in a Dish: Modeling Development and Disease Using Organoid Technologies. Science 2014, 345, 1247125. [Google Scholar] [CrossRef] [PubMed]
- Quadrato, G.; Brown, J.; Arlotta, P. The Promises and Challenges of Human Brain Organoids as Models of Neuropsychiatric Disease. Nat. Med. 2016, 22, 1220–1228. [Google Scholar] [CrossRef] [PubMed]
- Aisenbrey, E.A.; Murphy, W.L. Synthetic Alternatives to Matrigel. Nat. Rev. Mater. 2020, 5, 539–551. [Google Scholar] [CrossRef]
- Jiang, S.; Zhao, H.; Zhang, W.; Wang, J.; Liu, Y.; Cao, Y.; Zheng, H.; Hu, Z.; Wang, S.; Zhu, Y.; et al. An Automated Organoid Platform with Inter-Organoid Homogeneity and Inter-Patient Heterogeneity. Cell Rep. Med. 2020, 1, 100161. [Google Scholar] [CrossRef]
- Maenhoudt, N.; Defraye, C.; Boretto, M.; Jan, Z.; Heremans, R.; Boeckx, B.; Hermans, F.; Arijs, I.; Cox, B.; Van Nieuwenhuysen, E.; et al. Developing Organoids from Ovarian Cancer as Experimental and Preclinical Models. Stem Cell Rep. 2020, 14, 717–729. [Google Scholar] [CrossRef]
- Steele, N.G.; Chakrabarti, J.; Wang, J.; Biesiada, J.; Holokai, L.; Chang, J.; Nowacki, L.M.; Hawkins, J.; Mahe, M.; Sundaram, N.; et al. An Organoid-Based Preclinical Model of Human Gastric Cancer. Cell Mol. Gastroenterol. Hepatol. 2019, 7, 161–184. [Google Scholar] [CrossRef]
- Schmitz, C.H.J.; Rowat, A.C.; Köster, S.; Weitz, D.A. Dropspots: A Picoliter Array in a Microfluidic Device. Lab Chip 2009, 9, 44–49. [Google Scholar] [CrossRef]
- Whitesides, G.M. The Origins and the Future of Microfluidics. Nature 2006, 442, 368–373. [Google Scholar] [CrossRef]
- Ronaldson-Bouchard, K.; Teles, D.; Yeager, K.; Tavakol, D.N.; Zhao, Y.; Chramiec, A.; Tagore, S.; Summers, M.; Stylianos, S.; Tamargo, M.; et al. A Multi-Organ Chip with Matured Tissue Niches Linked by Vascular Flow. Nat. Biomed. Eng. 2022, 6, 351–371. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.-D.; Wang, Y.-T.; Wang, J.-R.; Wu, J.-L.; Meng, X.-S.; Hu, P.; Mu, X.; Liang, Q.-L.; Luo, G.-A. Design and Fabrication of a Liver-on-a-Chip Platform for Convenient, Highly Efficient, and Safe in Situ Perfusion Culture of 3D Hepatic Spheroids. Lab Chip 2018, 18, 2547–2562. [Google Scholar] [CrossRef] [PubMed]
- Leung, C.M.; de Haan, P.; Ronaldson-Bouchard, K.; Kim, G.A.; Ko, J.; Rho, H.S.; Chen, Z.; Habibovic, P.; Jeon, N.L.; Takayama, S.; et al. A Guide to the Organ-on-a-Chip. Nat. Rev. Methods Primers 2022, 2, 33. [Google Scholar] [CrossRef]
- Xu, M.; Shaw, G.; Murphy, M.; Barry, F. Induced Pluripotent Stem Cell-Derived Mesenchymal Stromal Cells Are Functionally and Genetically Different from Bone Marrow-Derived Mesenchymal Stromal Cells. Stem Cells 2019, 37, 754–765. [Google Scholar] [CrossRef]
- Diederichs, S.; Tuan, R.S. Functional Comparison of Human-Induced Pluripotent Stem Cell-Derived Mesenchymal Cells and Bone Marrow-Derived Mesenchymal Stromal Cells from the Same Donor. Stem Cells Dev. 2014, 23, 1594–1610. [Google Scholar] [CrossRef] [PubMed]
- van den Berg, A.; Mummery, C.L.; Passier, R.; van der Meer, A.D. Personalised Organs-on-Chips: Functional Testing for Precision Medicine. Lab Chip 2019, 19, 198–205. [Google Scholar] [CrossRef]
- Ronaldson-Bouchard, K.; Vunjak-Novakovic, G. Organs-on-a-Chip: A Fast Track for Engineered Human Tissues in Drug Development. Cell Stem Cell 2018, 22, 310–324. [Google Scholar] [CrossRef]
- Maoz, B.M.; Herland, A.; FitzGerald, E.A.; Grevesse, T.; Vidoudez, C.; Pacheco, A.R.; Sheehy, S.P.; Park, T.-E.; Dauth, S.; Mannix, R.; et al. A Linked Organ-on-Chip Model of the Human Neurovascular Unit Reveals the Metabolic Coupling of Endothelial and Neuronal Cells. Nat. Biotechnol. 2018, 36, 865–874. [Google Scholar] [CrossRef]
- da Ponte, R.M.; Gaio, N.; van Zeijl, H.; Vollebregt, S.; Dijkstra, P.; Dekker, R.; Serdijn, W.A.; Giagka, V. Monolithic Integration of a Smart Temperature Sensor on a Modular Silicon-Based Organ-on-a-Chip Device. Sens. Actuators A Phys. 2021, 317, 112439. [Google Scholar] [CrossRef]
- Quirós-Solano, W.F.; Gaio, N.; Stassen, O.M.J.A.; Arik, Y.B.; Silvestri, C.; Van Engeland, N.C.A.; Van der Meer, A.; Passier, R.; Sahlgren, C.M.; Bouten, C.V.C.; et al. Microfabricated Tuneable and Transferable Porous PDMS Membranes for Organs-on-Chips. Sci. Rep. 2018, 8, 13524. [Google Scholar] [CrossRef]
- Campbell, S.B.; Wu, Q.; Yazbeck, J.; Liu, C.; Okhovatian, S.; Radisic, M. Beyond Polydimethylsiloxane: Alternative Materials for Fabrication of Organ-on-a-Chip Devices and Microphysiological Systems. ACS Biomater. Sci. Eng. 2021, 7, 2880–2899. [Google Scholar] [CrossRef] [PubMed]
- Hirama, H.; Satoh, T.; Sugiura, S.; Shin, K.; Onuki-Nagasaki, R.; Kanamori, T.; Inoue, T. Glass-Based Organ-on-a-Chip Device for Restricting Small Molecular Absorption. J. Biosci. Bioeng. 2019, 127, 641–646. [Google Scholar] [CrossRef]
- Fritschen, A.; Bell, A.K.; Königstein, I.; Stühn, L.; Stark, R.W.; Blaeser, A. Investigation and Comparison of Resin Materials in Transparent DLP-Printing for Application in Cell Culture and Organs-on-a-Chip. Biomater. Sci. 2022, 10, 1981–1994. [Google Scholar] [CrossRef]
- Warr, C.; Valdoz, J.C.; Bickham, B.P.; Knight, C.J.; Franks, N.A.; Chartrand, N.; Van Ry, P.M.; Christensen, K.A.; Nordin, G.P.; Cook, A.D. Biocompatible PEGDA Resin for 3D Printing. ACS Appl. Bio Mater. 2020, 3, 2239–2244. [Google Scholar] [CrossRef] [PubMed]
- Alsharhan, A.T.; Acevedo, R.; Warren, R.; Sochol, R.D. 3D Microfluidics via Cyclic Olefin Polymer-Based in Situ Direct Laser Writing. Lab Chip 2019, 19, 2799–2810. [Google Scholar] [CrossRef] [PubMed]
- Busek, M.; Nøvik, S.; Aizenshtadt, A.; Amirola-Martinez, M.; Combriat, T.; Grünzner, S.; Krauss, S. Thermoplastic Elastomer (TPE)–Poly(Methyl Methacrylate) (PMMA) Hybrid Devices for Active Pumping PDMS-Free Organ-on-a-Chip Systems. Biosensors 2021, 11, 162. [Google Scholar] [CrossRef]
- Nguyen, T.; Jung, S.H.; Lee, M.S.; Park, T.-E.; Ahn, S.; Kang, J.H. Robust Chemical Bonding of PMMA Microfluidic Devices to Porous PETE Membranes for Reliable Cytotoxicity Testing of Drugs. Lab Chip 2019, 19, 3706–3713. [Google Scholar] [CrossRef]
- Regehr, K.J.; Domenech, M.; Koepsel, J.T.; Carver, K.C.; Ellison-Zelski, S.J.; Murphy, W.L.; Schuler, L.A.; Alarid, E.T.; Beebe, D.J. Biological Implications of Polydimethylsiloxane-Based Microfluidic Cell Culture. Lab Chip 2009, 9, 2132. [Google Scholar] [CrossRef]
- Lee, H.-M.; Yu, M.-S.; Kazmi, S.R.; Oh, S.Y.; Rhee, K.-H.; Bae, M.-A.; Lee, B.H.; Shin, D.-S.; Oh, K.-S.; Ceong, H.; et al. Computational Determination of HERG-Related Cardiotoxicity of Drug Candidates. BMC Bioinform. 2019, 20, 250. [Google Scholar] [CrossRef]
- Chramiec, A.; Teles, D.; Yeager, K.; Marturano-Kruik, A.; Pak, J.; Chen, T.; Hao, L.; Wang, M.; Lock, R.; Tavakol, D.N.; et al. Integrated Human Organ-on-a-Chip Model for Predictive Studies of Anti-Tumor Drug Efficacy and Cardiac Safety. Lab Chip 2020, 20, 4357–4372. [Google Scholar] [CrossRef]
- Liu, Y.; Sakolish, C.; Chen, Z.; Phan, D.T.T.; Bender, R.H.F.; Hughes, C.C.W.; Rusyn, I. Human in Vitro Vascularized Micro-Organ and Micro-Tumor Models Are Reproducible Organ-on-a-Chip Platforms for Studies of Anticancer Drugs. Toxicology 2020, 445, 152601. [Google Scholar] [CrossRef] [PubMed]
- Hachey, S.J.; Movsesyan, S.; Nguyen, Q.H.; Burton-Sojo, G.; Tankazyan, A.; Wu, J.; Hoang, T.; Zhao, D.; Wang, S.; Hatch, M.M.; et al. An: In Vitro Vascularized Micro-Tumor Model of Human Colorectal Cancer Recapitulates In Vivo Responses to Standard-of-Care Therapy. Lab Chip 2021, 21, 1333–1351. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Lee, S.; Song, J.; Lee, S.R.; Kim, S.; Choi, H.; Kang, H.; Hwang, Y.; Hong, Y.K.; Jeon, N.L. Perfusable Micro-Vascularized 3D Tissue Array for High-Throughput Vascular Phenotypic Screening. Nano Converg. 2022, 9, 16. [Google Scholar] [CrossRef] [PubMed]
- Shahrubudin, N.; Lee, T.C.; Ramlan, R. An Overview on 3D Printing Technology: Technological, Materials, and Applications. Procedia Manuf. 2019, 35, 1286–1296. [Google Scholar] [CrossRef]
- Hwang, S.; Reyes, E.I.; Moon, K.; Rumpf, R.C.; Kim, N.S. Thermo-Mechanical Characterization of Metal/Polymer Composite Filaments and Printing Parameter Study for Fused Deposition Modeling in the 3D Printing Process. J. Electron. Mater. 2015, 44, 771–777. [Google Scholar] [CrossRef]
- Valino, A.D.; Dizon, J.R.C.; Espera, A.H.; Chen, Q.; Messman, J.; Advincula, R.C. Advances in 3D Printing of Thermoplastic Polymer Composites and Nanocomposites. Prog. Polym. Sci. 2019, 98, 101162. [Google Scholar] [CrossRef]
- Gopinathan, J.; Noh, I. Recent Trends in Bioinks for 3D Printing. Biomater. Res. 2018, 22, 11. [Google Scholar] [CrossRef]
- Zhu, Y.; Joralmon, D.; Shan, W.; Chen, Y.; Rong, J.; Zhao, H.; Xiao, S.; Li, X. 3D Printing Biomimetic Materials and Structures for Biomedical Applications. Bio-Des. Manuf. 2021, 4, 405–428. [Google Scholar] [CrossRef]
- Zhang, B.; Pei, X.; Zhou, C.; Fan, Y.; Jiang, Q.; Ronca, A.; D’Amora, U.; Chen, Y.; Li, H.; Sun, Y.; et al. The Biomimetic Design and 3D Printing of Customized Mechanical Properties Porous Ti6Al4V Scaffold for Load-Bearing Bone Reconstruction. Mater. Des. 2018, 152, 30–39. [Google Scholar] [CrossRef]
- Zennifer, A.; Manivannan, S.; Sethuraman, S.; Kumbar, S.G.; Sundaramurthi, D. 3D Bioprinting and Photocrosslinking: Emerging Strategies & Future Perspectives. Biomater. Adv. 2022, 134, 112576. [Google Scholar] [CrossRef]
- Ouyang, L.; Highley, C.B.; Sun, W.; Burdick, J.A. A Generalizable Strategy for the 3D Bioprinting of Hydrogels from Nonviscous Photo-crosslinkable Inks. Adv. Mater. 2017, 29, 1604983. [Google Scholar] [CrossRef]
- Wilson, S.A.; Cross, L.M.; Peak, C.W.; Gaharwar, A.K. Shear-Thinning and Thermo-Reversible Nanoengineered Inks for 3D Bioprinting. ACS Appl. Mater. Interfaces 2017, 9, 43449–43458. [Google Scholar] [CrossRef]
- Highley, C.B.; Rodell, C.B.; Burdick, J.A. Direct 3D Printing of Shear-Thinning Hydrogels into Self-Healing Hydrogels. Adv. Mater. 2015, 27, 5075–5079. [Google Scholar] [CrossRef] [PubMed]
- Kuzucu, M.; Vera, G.; Beaumont, M.; Fischer, S.; Wei, P.; Shastri, V.P.; Forget, A. Extrusion-Based 3D Bioprinting of Gradients of Stiffness, Cell Density, and Immobilized Peptide Using Thermogelling Hydrogels. ACS Biomater. Sci. Eng. 2021, 7, 2192–2197. [Google Scholar] [CrossRef]
- Li, W.; Wang, M.; Ma, H.; Chapa-Villarreal, F.A.; Lobo, A.O.; Zhang, Y.S. Stereolithography Apparatus and Digital Light Processing-Based 3D Bioprinting for Tissue Fabrication. iScience 2023, 26, 106039. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Ma, X.; Zhu, W.; Wang, P.; Miller, K.L.; Stupin, J.; Koroleva-Maharajh, A.; Hairabedian, A.; Chen, S. Scanningless and Continuous 3D Bioprinting of Human Tissues with Decellularized Extracellular Matrix. Biomaterials 2019, 194, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Godar, D.E.; Gurunathan, C.; Ilev, I. 3D Bioprinting with UVA1 Radiation and Photoinitiator Irgacure 2959: Can the ASTM Standard L929 Cells Predict Human Stem Cell Cytotoxicity? Photochem. Photobiol. 2019, 95, 581–586. [Google Scholar] [CrossRef]
- Gittard, S.D. Two-Photon Polymerization Microstructuring in Regenerative Medicine. Front. Biosci. 2013, E5, 602–609. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tromayer, M.; Gruber, P.; Markovic, M.; Rosspeintner, A.; Vauthey, E.; Redl, H.; Ovsianikov, A.; Liska, R. A Biocompatible Macromolecular Two-Photon Initiator Based on Hyaluronan. Polym. Chem. 2017, 8, 451–460. [Google Scholar] [CrossRef]
- Li, Z.; Torgersen, J.; Ajami, A.; Mühleder, S.; Qin, X.; Husinsky, W.; Holnthoner, W.; Ovsianikov, A.; Stampfl, J.; Liska, R. Initiation Efficiency and Cytotoxicity of Novel Water-Soluble Two-Photon Photoinitiators for Direct 3D Microfabrication of Hydrogels. RSC Adv. 2013, 3, 15939–15946. [Google Scholar] [CrossRef]
- Masuma, R.; Kashima, S.; Kurasaki, M.; Okuno, T. Effects of UV Wavelength on Cell Damages Caused by UV Irradiation in PC12 Cells. J. Photochem. Photobiol. B 2013, 125, 202–208. [Google Scholar] [CrossRef] [PubMed]
- Moon, S.H.; Choi, H.N.; Yang, Y.J. Natural/Synthetic Polymer Materials for Bioink Development. Biotechnol. Bioprocess. Eng. 2022, 27, 482–493. [Google Scholar] [CrossRef]
- Muthusamy, S.; Kannan, S.; Lee, M.; Sanjairaj, V.; Lu, W.F.; Fuh, J.Y.H.; Sriram, G.; Cao, T. 3D Bioprinting and Microscale Organization of Vascularized Tissue Constructs Using Collagen-based Bioink. Biotechnol. Bioeng. 2021, 118, 3150–3163. [Google Scholar] [CrossRef] [PubMed]
- Shoulders, M.D.; Raines, R.T. Collagen Structure and Stability. Annu. Rev. Biochem. 2009, 78, 929–958. [Google Scholar] [CrossRef] [PubMed]
- Kadler, K.E.; Baldock, C.; Bella, J.; Boot-Handford, R.P. Collagens at a Glance. J. Cell Sci. 2007, 120, 1955–1958. [Google Scholar] [CrossRef]
- Sun, M.; Sun, X.; Wang, Z.; Guo, S.; Yu, G.; Yang, H. Synthesis and Properties of Gelatin Methacryloyl (GelMA) Hydrogels and Their Recent Applications in Load-Bearing Tissue. Polymers 2018, 10, 1290. [Google Scholar] [CrossRef]
- Wang, X.; Ao, Q.; Tian, X.; Fan, J.; Tong, H.; Hou, W.; Bai, S. Gelatin-Based Hydrogels for Organ 3D Bioprinting. Polymers 2017, 9, 401. [Google Scholar] [CrossRef]
- Foox, M.; Zilberman, M. Drug Delivery from Gelatin-Based Systems. Expert Opin. Drug Deliv. 2015, 12, 1547–1563. [Google Scholar] [CrossRef]
- Lee, B.H.; Shirahama, H.; Cho, N.-J.; Tan, L.P. Efficient and Controllable Synthesis of Highly Substituted Gelatin Methacrylamide for Mechanically Stiff Hydrogels. RSC Adv. 2015, 5, 106094–106097. [Google Scholar] [CrossRef]
- Sharifi, S.; Sharifi, H.; Akbari, A.; Chodosh, J. Systematic Optimization of Visible Light-Induced Crosslinking Conditions of Gelatin Methacryloyl (GelMA). Sci. Rep. 2021, 11, 23276. [Google Scholar] [CrossRef]
- Zhu, M.; Wang, Y.; Ferracci, G.; Zheng, J.; Cho, N.-J.; Lee, B.H. Gelatin Methacryloyl and Its Hydrogels with an Exceptional Degree of Controllability and Batch-to-Batch Consistency. Sci. Rep. 2019, 9, 6863. [Google Scholar] [CrossRef] [PubMed]
- Kneser, U.; Voogd, A.; Ohnolz, J.; Buettner, O.; Stangenberg, L.; Zhang, Y.H.; Stark, G.B.; Schaefer, D.J. Fibrin Gel-Immobilized Primary Osteoblasts in Calcium Phosphate Bone Cement: In Vivo Evaluation with Regard to Application as Injectable Biological Bone Substitute. Cells Tissues Organs 2005, 179, 158–169. [Google Scholar] [CrossRef] [PubMed]
- Abelseth, E.; Abelseth, L.; De la Vega, L.; Beyer, S.T.; Wadsworth, S.J.; Willerth, S.M. 3D Printing of Neural Tissues Derived from Human Induced Pluripotent Stem Cells Using a Fibrin-Based Bioink. ACS Biomater. Sci. Eng. 2019, 5, 234–243. [Google Scholar] [CrossRef] [PubMed]
- Piard, C.; Baker, H.; Kamalitdinov, T.; Fisher, J. Bioprinted Osteon-like Scaffolds Enhance in Vivo Neovascularization. Biofabrication 2019, 11, 025013. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.-W.; Zhang, X.-W.; Mi, C.-H.; Qi, X.-Y.; Zhou, J.; Wei, D.-X. Recent Advances in Hyaluronic Acid-Based Hydrogels for 3D Bioprinting in Tissue Engineering Applications. Smart Mater. Med. 2023, 4, 59–68. [Google Scholar] [CrossRef]
- Collins, M.N.; Birkinshaw, C. Hyaluronic Acid Based Scaffolds for Tissue Engineering—A Review. Carbohydr. Polym. 2013, 92, 1262–1279. [Google Scholar] [CrossRef] [PubMed]
- Perng, C.-K.; Wang, Y.-J.; Tsi, C.-H.; Ma, H. In Vivo Angiogenesis Effect of Porous Collagen Scaffold with Hyaluronic Acid Oligosaccharides. J. Surg. Res. 2011, 168, 9–15. [Google Scholar] [CrossRef]
- Wang, X.; He, J.; Wang, Y.; Cui, F.-Z. Hyaluronic Acid-Based Scaffold for Central Neural Tissue Engineering. Interface Focus 2012, 2, 278–291. [Google Scholar] [CrossRef]
- Mazzocchi, A.; Devarasetty, M.; Huntwork, R.; Soker, S.; Skardal, A. Optimization of Collagen Type I-Hyaluronan Hybrid Bioink for 3D Bioprinted Liver Microenvironments. Biofabrication 2018, 11, 015003. [Google Scholar] [CrossRef]
- Khunmanee, S.; Jeong, Y.; Park, H. Crosslinking Method of Hyaluronic-Based Hydrogel for Biomedical Applications. J. Tissue Eng. 2017, 8, 204173141772646. [Google Scholar] [CrossRef]
- Wu, Q.; Therriault, D.; Heuzey, M.-C. Processing and Properties of Chitosan Inks for 3D Printing of Hydrogel Microstructures. ACS Biomater. Sci. Eng. 2018, 4, 2643–2652. [Google Scholar] [CrossRef] [PubMed]
- Taghizadeh, M.; Taghizadeh, A.; Yazdi, M.K.; Zarrintaj, P.; Stadler, F.J.; Ramsey, J.D.; Habibzadeh, S.; Hosseini Rad, S.; Naderi, G.; Saeb, M.R.; et al. Chitosan-Based Inks for 3D Printing and Bioprinting. Green Chem. 2022, 24, 62–101. [Google Scholar] [CrossRef]
- He, Y.; Derakhshanfar, S.; Zhong, W.; Li, B.; Lu, F.; Xing, M.; Li, X. Characterization and Application of Carboxymethyl Chitosan-Based Bioink in Cartilage Tissue Engineering. J. Nanomater. 2020, 2020, 2057097. [Google Scholar] [CrossRef]
- Ahmad Raus, R.; Wan Nawawi, W.M.F.; Nasaruddin, R.R. Alginate and Alginate Composites for Biomedical Applications. Asian J. Pharm. Sci. 2021, 16, 280–306. [Google Scholar] [CrossRef]
- Emami, Z.; Ehsani, M.; Zandi, M.; Foudazi, R. Controlling Alginate Oxidation Conditions for Making Alginate-Gelatin Hydrogels. Carbohydr. Polym. 2018, 198, 509–517. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, K.; Hu, J.; Zhang, Y.; Dai, Y.; Xia, F. Role of a High Calcium Ion Content in Extending the Properties of Alginate Dual-Crosslinked Hydrogels. J. Mater. Chem. A Mater. 2020, 8, 25390–25401. [Google Scholar] [CrossRef]
- Lee, K.Y.; Mooney, D.J. Alginate: Properties and Biomedical Applications. Prog. Polym. Sci. 2012, 37, 106–126. [Google Scholar] [CrossRef]
- Keane, T.J.; Swinehart, I.T.; Badylak, S.F. Methods of Tissue Decellularization Used for Preparation of Biologic Scaffolds and in Vivo Relevance. Methods 2015, 84, 25–34. [Google Scholar] [CrossRef]
- Hussey, G.S.; Dziki, J.L.; Badylak, S.F. Extracellular Matrix-Based Materials for Regenerative Medicine. Nat. Rev. Mater. 2018, 3, 159–173. [Google Scholar] [CrossRef]
- Kim, B.S.; Das, S.; Jang, J.; Cho, D.-W. Decellularized Extracellular Matrix-Based Bioinks for Engineering Tissue- and Organ-Specific Microenvironments. Chem. Rev. 2020, 120, 10608–10661. [Google Scholar] [CrossRef]
- Ma, B.; Wang, X.; Wu, C.; Chang, J. Crosslinking Strategies for Preparation of Extracellular Matrix-Derived Cardiovascular Scaffolds. Regen. Biomater. 2014, 1, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Capuana, E.; Lopresti, F.; Ceraulo, M.; La Carrubba, V. Poly-l-Lactic Acid (PLLA)-Based Biomaterials for Regenerative Medicine: A Review on Processing and Applications. Polymers 2022, 14, 1153. [Google Scholar] [CrossRef] [PubMed]
- Donate, R.; Monzón, M.; Alemán-Domínguez, M.E. Additive Manufacturing of PLA-Based Scaffolds Intended for Bone Regeneration and Strategies to Improve Their Biological Properties. e-Polymers 2020, 20, 571–599. [Google Scholar] [CrossRef]
- Muthe, L.P.; Pickering, K.; Gauss, C. A Review of 3D/4D Printing of Poly-Lactic Acid Composites with Bio-Derived Reinforcements. Compos. Part C Open Access 2022, 8, 100271. [Google Scholar] [CrossRef]
- Tan, Y.J.; Tan, X.; Yeong, W.Y.; Tor, S.B. Hybrid Microscaffold-Based 3D Bioprinting of Multi-Cellular Constructs with High Compressive Strength: A New Biofabrication Strategy. Sci. Rep. 2016, 6, 39140. [Google Scholar] [CrossRef]
- Choe, G.; Lee, M.; Oh, S.; Seok, J.M.; Kim, J.; Im, S.; Park, S.A.; Lee, J.Y. Three-Dimensional Bioprinting of Mesenchymal Stem Cells Using an Osteoinductive Bioink Containing Alginate and BMP-2-Loaded PLGA Nanoparticles for Bone Tissue Engineering. Biomater. Adv. 2022, 136, 212789. [Google Scholar] [CrossRef]
- Gao, G.; Lee, J.H.; Jang, J.; Lee, D.H.; Kong, J.-S.; Kim, B.S.; Choi, Y.-J.; Jang, W.B.; Hong, Y.J.; Kwon, S.-M.; et al. Tissue Engineered Bio-Blood-Vessels Constructed Using a Tissue-Specific Bioink and 3D Coaxial Cell Printing Technique: A Novel Therapy for Ischemic Disease. Adv. Funct. Mater. 2017, 27, 1700798. [Google Scholar] [CrossRef]
- Naseri, E.; Butler, H.; MacNevin, W.; Ahmed, M.; Ahmadi, A. Low-Temperature Solvent-Based 3D Printing of PLGA: A Parametric Printability Study. Drug Dev. Ind. Pharm. 2020, 46, 173–178. [Google Scholar] [CrossRef]
- Arcaute, K.; Mann, B.; Wicker, R. Stereolithography of Spatially Controlled Multi-Material Bioactive Poly(Ethylene Glycol) Scaffolds. Acta Biomater. 2010, 6, 1047–1054. [Google Scholar] [CrossRef]
- Tu, X.; Wang, L.; Wei, J.; Wang, B.; Tang, Y.; Shi, J.; Zhang, Z.; Chen, Y. 3D Printed PEGDA Microstructures for Gelatin Scaffold Integration and Neuron Differentiation. Microelectron. Eng. 2016, 158, 30–34. [Google Scholar] [CrossRef]
- Bandyopadhyay, A.; Mandal, B.B.; Bhardwaj, N. 3D Bioprinting of Photo-crosslinkable Silk Methacrylate (SilMA)-polyethylene Glycol Diacrylate (PEGDA) Bioink for Cartilage Tissue Engineering. J. Biomed. Mater. Res. A 2022, 110, 884–898. [Google Scholar] [CrossRef]
- Fathi, A.; Kermani, F.; Behnamghader, A.; Banijamali, S.; Mozafari, M.; Baino, F.; Kargozar, S. Three-Dimensionally Printed Polycaprolactone/Multicomponent Bioactive Glass Scaffolds for Potential Application in Bone Tissue Engineering. Biomed. Glas. 2020, 6, 57–69. [Google Scholar] [CrossRef]
- Elomaa, L.; Kokkari, A.; Närhi, T.; Seppälä, J.V. Porous 3D Modeled Scaffolds of Bioactive Glass and Photocrosslinkable Poly(ε-Caprolactone) by Stereolithography. Compos. Sci. Technol. 2013, 74, 99–106. [Google Scholar] [CrossRef]
- Yang, X.; Wang, Y.; Zhou, Y.; Chen, J.; Wan, Q. The Application of Polycaprolactone in Three-Dimensional Printing Scaffolds for Bone Tissue Engineering. Polymers 2021, 13, 2754. [Google Scholar] [CrossRef]
- Cai, Z.; Wan, Y.; Becker, M.L.; Long, Y.-Z.; Dean, D. Poly(Propylene Fumarate)-Based Materials: Synthesis, Functionalization, Properties, Device Fabrication and Biomedical Applications. Biomaterials 2019, 208, 45–71. [Google Scholar] [CrossRef] [PubMed]
- Walker, J.M.; Bodamer, E.; Krebs, O.; Luo, Y.; Kleinfehn, A.; Becker, M.L.; Dean, D. Effect of Chemical and Physical Properties on the In Vitro Degradation of 3D Printed High Resolution Poly(Propylene Fumarate) Scaffolds. Biomacromolecules 2017, 18, 1419–1425. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Lu, L.; Yaszemski, M.J. Bone-Tissue-Engineering Material Poly(Propylene Fumarate): Correlation between Molecular Weight, Chain Dimensions, and Physical Properties. Biomacromolecules 2006, 7, 1976–1982. [Google Scholar] [CrossRef]
- Bordonaro, M.; Lazarova, D.L.; Augenlicht, L.H.; Sartorelli, A.C. Estimates of the World-Wide Prevalence of Cancer for 25 Sites in the Adult Population. Int. J. Cancer 2002, 97, 72–81. [Google Scholar] [CrossRef]
- Sztankovics, D.; Moldvai, D.; Petővári, G.; Gelencsér, R.; Krencz, I.; Raffay, R.; Dankó, T.; Sebestyén, A. 3D Bioprinting and the Revolution in Experimental Cancer Model Systems—A Review of Developing New Models and Experiences with in Vitro 3D Bioprinted Breast Cancer Tissue-Mimetic Structures. Pathol. Oncol. Res. 2023, 29, 1610996. [Google Scholar] [CrossRef]
- Zhou, X.; Zhu, W.; Nowicki, M.; Miao, S.; Cui, H.; Holmes, B.; Glazer, R.I.; Zhang, L.G. 3D Bioprinting a Cell-Laden Bone Matrix for Breast Cancer Metastasis Study. ACS Appl. Mater. Interfaces 2016, 8, 30017–30026. [Google Scholar] [CrossRef]
- Xu, K.; Huang, Y.; Wu, M.; Yin, J.; Wei, P. 3D Bioprinting of Multi-Cellular Tumor Microenvironment for Prostate Cancer Metastasis. Biofabrication 2023, 15, 035020. [Google Scholar] [CrossRef] [PubMed]
- Pulido, C.; Vendrell, I.; Ferreira, A.R.; Casimiro, S.; Mansinho, A.; Alho, I.; Costa, L. Bone Metastasis Risk Factors in Breast Cancer. Ecancermedicalscience 2017, 11, 715. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.Q.; Qu, X.; Liu, J.; Chen, S. 3D Printing of Biomimetic Microstructures for Cancer Cell Migration. Biomed. Microdevices 2014, 16, 127–132. [Google Scholar] [CrossRef] [PubMed]
- Ning, L.; Shim, J.; Tomov, M.L.; Liu, R.; Mehta, R.; Mingee, A.; Hwang, B.; Jin, L.; Mantalaris, A.; Xu, C.; et al. A 3D Bioprinted in Vitro Model of Neuroblastoma Recapitulates Dynamic Tumor-Endothelial Cell Interactions Contributing to Solid Tumor Aggressive Behavior. Adv. Sci. 2022, 9, e2200244. [Google Scholar] [CrossRef]

| Spheroid Fabrication Method | Overview | Advantages | Drawbacks | References |
|---|---|---|---|---|
| Hanging drop | A drop of cell suspension is placed onto the inside of a cell culture plate lid, which is then inverted without disturbing the droplets held by surface tension. Over time, cells are concentrated and cluster into a spheroid at the bottom of the hanging droplet. |
|
| [46,47] |
| Liquid overlay | Cell suspension is seeded onto a non-adherent surface with recesses that promote cell aggregation. |
|
| [48,49] |
| Rotary cell culture | Cells are cultured in a container with an agitator that disrupts the cells’ ability to adhere to the substrate, forcing them to self-assemble into spheroids. |
|
| [44,50,51] |
| Nanofiber cell suspension | Adding polymer nanofibers to the cell suspension increases spheroid production due to cells interacting with the nanofibers. |
|
| [52,53,54] |
| Magnetic levitation | Magnetic particles are combined with cells, and a magnetic force is introduced. Negative magnetophoresis induces a weightless environment where cell aggregation is promoted. |
|
| [55,56,57] |
| Material | Fabrication Method | Advantages | Drawbacks | References |
|---|---|---|---|---|
| Polydimethylsiloxane (PDMS) | Soft Lithography |
|
| [82,83,90] |
| Polymethylmethacrylate (PMMA) | Injection Molding |
|
| [88,89] |
| Cyclic olefin copolymer (COC) | Injection Molding |
|
| [87] |
| Silicon | Photolithography |
|
| [81] |
| Glass | Etching |
|
| [84] |
| Resins | 3D Printing |
|
| [85,86] |
| Material | Overview | Properties for Bioinks | Crosslinking Mechanisms | References |
|---|---|---|---|---|
| Collagen | Triple helical protein for tissue scaffolding and tensile strength in tendon, cartilage, bone, and skin |
| Covalent bonding of fibrils | [115,116,117] |
| Gelatin | Hydrogel from the hydrolysis of collagen, solid when cooled, and can be used to synthesize gelatin methacryloyl (GelMA) |
| Gelation under cold temperatures | [118,119,120,121] |
| Gelatin Methacryloyl (GelMA) | Gelatin derivative with methacrylated functional groups; mechanically stable after photocrosslinking |
| Photocrosslinked under UV light exposure | [122,123] |
| Fibrin | High viscosity; insoluble biopolymer that allows for paracrine signalling due to non-linear elasticity |
| Cleaved by thrombin which induces polymerization | [124,125,126] |
| Hyaluronic Acid | Bioresorbable material found in mammalian ECM; maintains a hydrated environment |
| Enzyme-crosslinking, Schiff base reaction, thiol-modified HA crosslinking, Diels–Alder click crosslinking, ionic crosslinking, and photo-crosslinking | [127,128,129,130,131,132] |
| Chitosan | Polysaccharide derived from chitin deacetylation with solubility at low pH levels |
| Chemical crosslinking with glutaraldehyde (amine groups) or citric acid (covalent) | [133,134,135] |
| Alginate | Polymer derived from brown algae, can form hydrogels that mimic the ECM, and can be crosslinked through its aldehyde groups |
| Ionically crosslinked with divalent cations | [136,137,138,139] |
| Decellularized ECM | Produced by removing cellular components from tissues via chemical or physical processes |
| Glutaraldehyde; thermal gelation | [140,141,142,143] |
| Material | Overview | Properties for Bioinks | References |
|---|---|---|---|
| Polylactic acid (PLA) | Semi-crystalline structure with high molecular weight, used in extrusion-based bioprinting |
| [144,145,146] |
| Poly(lactic-co-glycolic acid) (PLGA) | Synthesized through co-polymerization of both glycolic acid and lactic acid |
| [147,148,149,150] |
| Poly(ethylene glycol) diacrylate (PEGDA) | Long-chain photo-crosslinkable monomer that forms hydrogels |
| [151,152,153] |
| Poly e-caprolactone (PCL) | Semi-crystalline thermoplastic with high thermal stability and long degradation rate |
| [154,155,156] |
| Poly(propylene fumarate) (PPF) | Linear unsaturated polyester with fumaric acid backbone chains |
| [157,158,159] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
van Rijt, A.; Stefanek, E.; Valente, K. Preclinical Testing Techniques: Paving the Way for New Oncology Screening Approaches. Cancers 2023, 15, 4466. https://doi.org/10.3390/cancers15184466
van Rijt A, Stefanek E, Valente K. Preclinical Testing Techniques: Paving the Way for New Oncology Screening Approaches. Cancers. 2023; 15(18):4466. https://doi.org/10.3390/cancers15184466
Chicago/Turabian Stylevan Rijt, Antonia, Evan Stefanek, and Karolina Valente. 2023. "Preclinical Testing Techniques: Paving the Way for New Oncology Screening Approaches" Cancers 15, no. 18: 4466. https://doi.org/10.3390/cancers15184466
APA Stylevan Rijt, A., Stefanek, E., & Valente, K. (2023). Preclinical Testing Techniques: Paving the Way for New Oncology Screening Approaches. Cancers, 15(18), 4466. https://doi.org/10.3390/cancers15184466




