Simple Summary
Ewing sarcoma (EwS) is a highly aggressive and metastatic cancer in children and adolescents. Canonical therapy mainly comprises the combination of intensive chemotherapy, radiation, and local surgery, which give rise to acute and chronic adverse effects. Drugs targeting EwS without side effects are in urgent demand. Genetically, EwS is characterized by chromosomal translocations with a low mutation burden. As a result, the chimeric protein EWS-ETS, mainly EWS-FLI1(85%), is critical for the malignancy of EwS. EWS-FLI1 directly binds to GGAA microsatellites in enhancers and promotors of the target genes and recruits multiple transcription factors or epigenetic regulators to reprogramme the epigenome. Direct targeting EWS-FLI1 is difficult due to the disordered structure, we mainly review the current knowledge of EWS-FLI1 property, the EWS-FLI1 protein complex, and the downstream pathways, we also summarize the targeted therapy of EwS by taking advantage of the EWS-FLI1 protein complex and the immunotherapy of the genes activated by EWS-FLI1.
Abstract
Ewing sarcoma (EwS) is a rare and predominantly pediatric malignancy of bone and soft tissue in children and adolescents. Although international collaborations have greatly improved the prognosis of most EwS, the occurrence of macrometastases or relapse remains challenging. The prototypic oncogene EWS-FLI1 acts as an aberrant transcription factor that drives the cellular transformation of EwS. In addition to its involvement in RNA splicing and the DNA damage response, this chimeric protein directly binds to GGAA repeats, thereby modifying the transcriptional profile of EwS. Direct pharmacological targeting of EWS-FLI1 is difficult because of its intrinsically disordered structure. However, targeting the EWS-FLI1 protein complex or downstream pathways provides additional therapeutic options. This review describes the EWS-FLI1 protein partners and downstream pathways, as well as the related target therapies for the treatment of EwS.
1. Introduction
Ewing sarcoma (EwS) is a poorly differentiated malignancy that mainly arises in bone and soft tissue and is more prevalent among those in the second decade of life [1]. Extraosseous EwS is more common in adults [2]. The cellular origin of EwS remains controversial, although it is speculated that it arises from neuroectodermal cells or primitive mesenchymal stem cells (MSC) [3,4,5]. Despite considerable improvements in overall survival achieved using a multimodal approach, including intensive chemotherapy for localized disease [6], the prognosis of patients who develop metastatic disease or relapse remains dismal [7,8]. Approximately 20–25% of patients are diagnosed with early micro-dissemination [6,9].
Dissecting the pioneer molecular events provides a precision approach to the treatment of cancers including EwS. EwS is genetically characterized by chromosomal translocation, which generates the EWS-ETS (85% is EWS-FLI1) chimeric protein accompanied by a quiet genomic background [10,11]. EWS-ETS governs the transcriptome to drive malignant transformation and serves as the most reliable diagnostic marker of EwS, as well as providing a therapeutic target. Directly targeting EWS-ETS is difficult because of its intrinsically disordered structure [12,13]; therefore, understanding the EWS-ETS protein complex and the downstream molecular events involved in EwS is essential for the design of therapeutic strategies.
EwS is genetically distinct because of a balanced chromosomal translocation that joins the N-terminus of EWS (on chromosome 22) with a gene from the C-terminus of the E-twenty-six (ETS) family of transcription factors (FLI1, ERG, ETV1, ETV5, and FEV), predominantly FLI1 (85%, on chromosome 11) and ERG (10%, on chromosome 21) [14]. This has been well reviewed in the literature [1,15,16,17].
The EWS protein consists of an N-terminal transcription-activation domain (Exons 1 to 7) and C-terminal RNA-binding domains (Exons 8 to 17) [18]. The N-terminus contains a serine–tyrosine–glycine–glutamine (SYGQ) domain that has low-complexity and is intrinsically disordered. The C-terminus contains three RNA-binding arginine–glycine–glycine- (RGG-) motifs, the three RGG- motifs are separated by an RNA-recognition motif (RRM) and a zinc finger motif (ZF) [19]. EWS functions in transcription, RNA processing, transportation, alternative splicing, DNA repair, and homologous recombination [20,21,22,23,24]. Because of these multiple functions, EWS is essential for the central nervous system [25], meiosis, B-lymphocyte development, and hematopoietic stem-cell self-renewal [22]. The ETS domain of the ETS protein binds to DNA sequences with a central GGAA/T core motif [26], and the protein is thus involved in cell-cycle regulation and differentiation [27,28].
Chromosomal translocation results in the formation of a chimeric protein because the RNA-binding domain of EWS is replaced by a DNA-binding domain [14]. The formation of this chimeric protein is thought to be the initial event in the tumorigenesis and malignancy of EwS.
2. Involvement of EWS-FLI1 in Transcription, Epigenetic Reprogramming, and Alternative Splicing in EwS
2.1. EWS-FLI1 in Transcription and Epigenetic Reprogramming
EWS-FLI1 is an aberrant transcription factor that drives cellular transformation by rewiring the epigenome to induce a large number of oncogenes. The N-terminus of EWSR1 contains a prion-like domain, characterized by an intrinsically disordered structure and low complexity. This domain has phase transition properties and manipulates multiple proteins involved in epigenome reprogramming and epigenetic alterations [29,30,31,32,33,34,35,36]. In addition to the canonical ETS-binding sites, EWS-FLI1 binds to DNA sequences at the GGAA/T core motif [37,38,39] via a conserved ETS domain. It regulates multiple proteins through its prion-like domain to tumor-specific enhancers and promotors, recruiting acetyltransferases and establishing de novo enhancers by generating H3K27ac, thus opening the chromosomal architecture, which contributes to the activation of target genes [29,30,37,39]. The EWS-FLI1 protein complex includes RNA polymerase II [23,40], the core subunit hsRBP7 (human RNA polymerase II) [41,42], E2F3 [43,44], EWSR1 [45], CBP/p300 [46], WDR5, ASH2, MLL [30], and the BAF complex (mammalian SWI/SNF complex) [29,47]. The threshold of GGAA motifs optimal for maximal expression is 20–26 [48], which differs from that in wild-type FLI1. Super-enhancer-associated MEIS1 and RING1B also contribute to the chromatin reprogramming through co-localization with EWS-FLI1 at the active enhancers to drive the malignancy of EwS [49,50]. This specific coupling results in the activation of many genes (Figure 1), such as NKX2.2 [51], NROB1 [52,53,54], IGF1R [55], BCL11B [56], EZH2 [36], VRK1 [30], GLI1 [57], PTPL1 [58], PPPR1A [59], ERG2 [60], GSTM4 [61], PAX7 [62], CHM1 [63], REST [64], PHF19 [32], STEAP1 [65,66], SLFN11(Schlafen 11) [67], HDAC3 [68], TNC [69], APCDD1 [49], IL1RAP [70,71], MYC [72], and PRC1 (protein regulator of cytokinesis 1) [73].
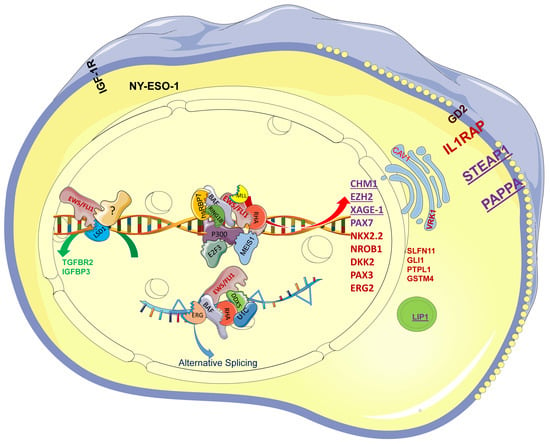
Figure 1.
The EWS-FLI1 protein complex drives the specific transcription profile of EwS. EWS-FLI1 recruits E2F3, hsRBP7, BAF, RING1B, RHA, P300, and MEIS1, among others, to GGAA repeats and further activates CHM1, EZH2, PAX7, NKX2.2, NROB1, and STEAP1, among others. EWS-FLI1 functions as a protein complex with ERG, BAF, RHA, DDX5, and U1C to drive alternative splicing. Among the genes, the purple ones, such as CHM1, could serve as TCR-based immunotherapy targets; the red ones, such as NKX2-2, serve as diagnostic markers in clinic diagnosis. EWS/FLI1 recruits LSD1 and unknown transcription factors (?) to repress TGFBR1 and IGFBP3, which still needs further research.
Among the direct targets of EWS-FLI1, NKX2-2 mediates oncogenic transformation via transcriptional repression and is necessary and sufficient for the oncogenic phenotype of EwS [74]. Further work demonstrates that NKX2-2, KLF15, and TCF4 occupy similar super-enhancers and promoters, forming an inter-connected auto-regulatory loop and occupying 77.2% of promoters and 55.6% of enhancers shared with EWS-FLI1 [75], such as STEAP1 [76]; this kind of coordinate regulation drives the proliferation of EwS. NROB1 directly interacts with EWS-FLI1 to modulate multiple gene expressions and mediate the oncogenic phenotype of EwS [77]. SLFN11 is a putative DNA/RNA helicase that recruits to the stressed replication fork and irreversibly triggers replication block and cell death. Overexpression of SLFN11 is associated with resistance to topoisomerase I inhibitors and poly (ADP-ribose) polymerase (PARP) inhibitor combinations [67,78]. STEAP1 and IL1RAP are vital for the redox homeostasis of EwS [65,70]. APCDD1, PHF19, GSTM4, and PTPL1 are genes that are involved in the proliferation of EwS.
EWS-FLI1 is also involved in transcriptional repression of tumor suppressors such as IGFBP3 [79] and PHLDA1 [53] to drive oncogenic transformation [31,80]. The nucleosome remodeling and deacetylase (NuRD) complex is a typical ATP-dependent chromatin remodeling complex [81] that plays a critical role in transcription and determines differentiation and development [82]. EWS-FLI1 recruits the NuRD-LSD1 complex to repress LOX and TGFBR2 [80,83]. It also affects the transcriptional activation of AP-1 [33] and MRTFB [84] and binds to the promotor of FOXO1 to repress its expression, thereby increasing tumor growth [85]. EWS-FLI1 promotes the phosphorylation of cyclin-dependent kinase-2 and AKT to inhibit the activity of FOXO1, thus rewiring transcriptional repression [85]. EWS-FLI1 is also involved in the regulation of microRNAs (miRNAs) [86]. It downregulates miRNA-145 to initiate mesenchymal stem-cell reprogramming toward EwS stem cells [87] and represses miR-708, which induces the overexpression of EYA3 and contributes to the chemoresistance to etoposide and doxorubicin [88].
The histone methyltransferase EZH2 exhibits silencing activity via methylation of H3K27 [89]. In EwS, EWS-FLI1 upregulates EZH2 expression by interacting with the EZH2 promoter, thereby promoting tumor growth/metastasis and blocking endothelial/neuro-ectodermal differentiation [36].
MiR-34a inhibits the proliferation and increases the sensitivity of EwS to doxorubicin and vincristine and is a strong predictor of a favorable prognosis in EwS [90]. However, the exact mechanism underlying its downregulation remains elusive. Exportin 5 (XPO5), which mediates the nuclear export of pre-miRNAs and short hairpin RNAs [91,92,93], interacts with EWS-FLI1 based on mass spectrometry [94]. XPO5 is highly expressed in various cancers including EwS (Supplementary Figure S1A). Furthermore, the phosphorylation of XPO5 alters the nucleus and cytoplasm shift [95]. Investigating XPO5 and its relationship with EWS-FLI1 may offer new insights into the therapy of EwS. Post-translational modifications of EWS-FLI1 modulate its transcriptional activity. Phosphorylation and O-GlcNAcylation of the N-terminus of EWSR1 [96,97,98], as well as acetylation of the C-terminal FLI1 domain by PCAF (KAT2B, lysine acetyltransferase 2B), enhance the transcriptional activity of EWS-FLI1 [99]. However, PCAF expression is lower in EwS tissues, which is a common feature of cancer (Supplementary Figure S1B).
2.2. EWS-FLI1 in Alternative Splicing
Pre-mRNA splicing is critical for gene expression, and most protein-encoding transcripts are alternatively spliced to provide diverse functions [100,101]. The N-terminus of EWSR1 interacts with the hyperphosphorylated RNA polymerase II and recruits serine-arginine (SR) through its C-terminus. After chromosome translocation, the C-terminus of wild-type EWSR1 is replaced by FLI1, which hinders the recruitment of SR-splicing factors and interferes with mRNA splicing [23], thus demonstrating the negative property of this chimeric protein [102]. This leads to comprehensive alternative splicing of numerous genes. Meanwhile, EWS-FLI1 interacts with the splicing components (snRNP) U1C and SF1 to modulate pre-mRNA splicing [103]. It also recruits the BAF complex to drive the alternative splicing of ARID1A and the preferential splicing of ARID1A-L, which is necessary for tumor growth [104]. Work by Selvanathan [94] demonstrates that EWS-FLI1 is involved in the alternative splicing of CLK1, CASP3, PPFIBP1, and TERT, which potentially regulate the oncogenesis of EwS.
3. The Regulation of EWS-FLI1
Transcription and post-transcriptional modifications are involved in the regulation of expression and activity of EWS-FLI1. Although the transcriptional regulation of EWS-FLI1 remains elusive, the BRD4 inhibitor JQ1 suppresses this activity [32,34,105]. HDAC6 deacetylates specificity protein 1 (SP1), thereby inhibiting the recruitment of the SP1/P300 complex to the promoters of EWSR1 and EWS-FLI1 and downregulating EWS-FLI1 [106]. MiR-145 and let-7 repress EWS-FLI1 by targeting its mRNA [87,107,108,109] and inhibit the proliferation of EwS. The RNA-binding protein LIN28B interacts with EWS-FLI1 transcripts to maintain the stability and ensure the expression of EWS-FLI1 to enhance the tumorigenicity of the self-renewal of EwS [108]. At the post-transcriptional level, EWS-FLI1 degradation is proteasome dependent, and the protein has a half-life of 2–4 h [110]. This process can be protected by the action of ubiquitin-specific protease 19 (USP19) at the N-terminus [111] and accelerated by tripartite-motif-containing 8 (TRIM8) at K334 [112]; however, USP19 is expressed at low levels in EwS (Supplementary Figure S2). Casein kinase 1 (CK1)-mediated phosphorylation of the VTSSS degron in the FLI1 domain activates speckle-type POZ protein (SPOP) activity, which degrades EWS-FLI1. In contrast, OTU-domain-containing protein 7A (OTUD7A) participates in the deubiquitination of the C-terminus and stabilizes EWS-FLI1 [113]. The inhibitor of chromosomal maintenance 1 (CRM1 and XPO1), KPT-330 [114], and IFN-γ [115] suppress expression of EWS-FLI1 at the protein level. FOXM1, a downstream factor of EWS-FLI1, upregulates its expression [116]. Cytosine arabinoside (ARA-C) downregulates EWS-FLI1 at the protein level and inhibits tumor growth [117]; however, it shows hematologic toxicity and minimal activity in patients [118].
STAG2 (stromal antigen 2) is a core subunit of the cohesion complex and is frequently mutated in multiple cancers [119] including EwS [10,120]. Mutation of STAG2 in EwS is associated with poor outcomes by improving metastasis [10]. Mechanically, in addition to the disruption of PRC2-mediated regulation of gene expression in EwS [121], the inactivation of STAG2 strongly altered CTCF-anchored loop extrusion and decreases promotor-enhancer interactions. As a result, the cis-mediated EWS-FLI1 activity at GGAA microsatellite neo-enhancers is downregulated and the cells are enhanced in their migration and invasion properties [121,122].
Unlike STAG2, there was no evidence showing mutations of the ETS transcription factor ETV6 in EwS [123]. ETV6 does not change the expression of EWS-FLI1 but co-occupy loci genome wide at the short consecutive GGAA repeats and constrains the transcriptional activity of EWS-FLI1 [124,125]. Upon inactivating ETV6, EWS-FLI1 overtakes and activates these cis-elements to promote mesenchymal differentiation by upregulating the expression of SOX11 [125].
4. EWS-FLI1 in the Metastasis and DNA Damage Repair of EwS
4.1. EWS-FLI1 in the Metastasis of EwS
Tumor metastasis is a major cause of death in cancer patients, and the survival of EwS patients with early metastasis is poor [126,127], especially those with bone metastasis alone or with multiple metastatic sites [9,128]. In addition to the tumor microenvironment, the differences in EWS-FLI1 expression levels determine the balance between proliferation and metastasis in EwS [129,130]. EWS-FLI1high cells promote proliferation, whereas EWS-FLI1low cells tend to shift to a cell-matrix adhesion phenotype, which gives rise to invasion/metastasis by altering the structure of the cytoskeleton [84,129,131,132] and eventually drives metastatic-related death. Many genes repressed by EWS-FLI1 are enriched in focal adhesion, including zyxin and α5-integrin [133]. EWS-FLI1 also suppresses the MRTFB/YAP/TAZ/TEAD regulatory pathway by inhibiting the Rho-F-actin signal, resulting in the inhibition of the invasive properties of EwS [84,131]. On the other hand, the canonical targets of EWS-FLI1 such as DKK2 [134], CAV1 [135,136], EZH2 [36], CHM1 [63], GPR64 [137], PPP1R1A [59], and TNC [69] are involved in the invasive properties of EwS.
Hypoxia is a common feature of cancers [138], and hypoxia-inducible factor-1a (HIF-1a) is highly expressed in EwS and contributes to the metastasis of EwS [139]. HIF-1a also activates the transcription of EWS-FLI1. However, cells induced by hypoxia are mainly enriched in an HIF-1a signature in EwS, indicating that the treatment of EwS should not only focus on EWS-FLI1, but also on the microenvironmental conditions such as hypoxia [139].
AMER2 is a direct target of EWS-FLI1 [108] (Figure 2A), and knockdown of EWS-FLI1 inhibits the expression of AMER2 based on GSE176190 (Figure 2B). AMER2 is highly expressed in EwS compared to other cancers based on the CCLE database (Supplementary Figure S3) and normal tissues based on GSE17679 (Figure 2C). Activation of Wnt/β-catenin signaling is a common feature in multiple metastatic cancers [140,141], whereas AMER2 works as a negative regulator of Wnt/β-catenin signaling [142]. The knockdown of EWS-FLI1 might activate Wnt/β-catenin and drive the metastasis of EwS (Figure 2D), but the specific mechanisms are still to be explored.
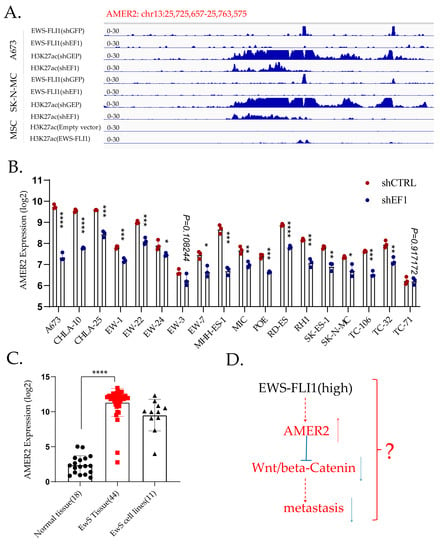
Figure 2.
AMER2 is a direct target of EWS-FLI1. (A). CHIP-sequencing data demonstrate that AMER2 is a direct target of EWS-FLI1. In A673 and SK-N-MC cells, EWS-FLI1 binds to the promotor of AMER2 directly, and the H3K27ac level is lower after EWS-FLI1 knockdown. In MSC, transduction of EWS-FLI1 enhances the H3K27ac level in the promotor region of AMER2. (B). Knockdown of EWS-FLI1 downregulates AMER2 in EwS cell lines apart from the TC-71 cell line. (C). AMER2 expression is higher in EwS tissues than in normal tissues. (D). Potential mechanism of AMER2 in the metastasis of EwS. *, p < 0.01; **, p < 0.05; ***, p < 0.001; ****, p < 0.0001.
4.2. EWS-FLI1 in DNA Damage Repair
In addition to its involvement in regulating and driving EwS, EWS-FLI1 causes DNA damage [24], leading to growth arrest and apoptosis [112,143,144,145]. EWS-FLI1 binds to the promotor of caspase-3; conditional expression of EWS-FLI1 in embryos increases caspase-3 expression and triggers early onset of apoptosis and acute lethality in mouse kidney [144]. The hypersensitivity to radiation [146] and DNA-damaging agents such as etoposide [24,147,148] provides further evidence that tailored therapies based on DNA-damaging agents [6] are needed, despite the fact that few mutations have been identified that participate in DNA damage repair [149].
The role of PARP in the DNA damage response has been studied extensively, and its inhibition is widely used in breast and ovarian cancer with BRCA1/BRCA2 mutations [150,151,152]; PARP inhibitors mainly target defective homologous recombination [153]. In EwS, EWS-FLI1 maintains the expression of PAPR1 and also forms a protein complex with PARP1, which promotes oncogene-dependent sensitivity to PARP1 inhibition [148]. EWS-FLI1 also inhibits EWSR1 in a dominant–negative manner [154,155]. EWSR1 interacts with PARylated PARP1 through RGG domains at DNA damage sites and suppresses DNA damage by preventing excessive PARP1 accumulation. However, in EwS, the presence of EWS-FLI1 leads to the hyper-PARylation and accumulation of PARP1 [156]. On the other hand, EWS-FLI1 promotes the aberrant transcription by reversing EWSR1-dependent inhibition of CDK9 activity, resulting in the accumulation of R-loops and blocking the release of BRCA1 from transcription complexes, finally leading to transcription stress. This mimics BRCA1 deficiency and impairs homologous recombination repair [24]. Paronetto et al. demonstrated that EWSR1 depletion results in the alternative splicing of genes involved in DNA repair and genotoxic stress signaling, including ABL1, CHEK2, and MAP4K2, which decreases cell viability and proliferation upon UV irradiation [157]. After chromosomal rearrangement, wild-type EWSR1 is replaced by EWS-FLI1, which might mimic the depletion of EWSR1. EWS-FLI1 dysregulates the expression and the function of PARP1 and is involved in the hypersensitivity of EwS to DNA-damaging agents.
EWSR1 binds to negatively charged poly-ADP ribose molecules via positively charged RGG repeats [158,159] and then recruits to the DNA double strand; however, after the formation of EWS-FLI1, the RGG repeats in the C terminus are replaced. Menon et al.’s work shows that EwS are distinct from BRCA1/2 mutant tumors and do not display the genomic scars of homologous recombination [160]. The aberrant recruitment of EWS-FLI1 to sites of DNA damage can disrupt physiologic DNA double-strand break repair. They also found that EWS-FLI1 suppressed the activation of DNA damage sensor ATM; meanwhile, a collateral dependence on the ATR signaling axis was induced by EWS-FLI1, and EWS-FLI1 increased sensitivity to inhibitors of both ATR and its key downstream target, CHK1 [160].
In brief, EWS-FLI1 enhances genotoxic stress signaling by disrupting physiologic DNA repair networks.
5. Killing the Driver or Taking Advantage of the Driver
5.1. Targeting the EWS-FLI1-Related Protein Complex
The treatment of EwS typically consists of radiotherapy, surgical resection, and intensive induction chemotherapy [1,15], which has significantly improved overall survival. However, acute and chronic adverse effects can negatively affect the quality of life of survivors [1].
Knockdown or overexpression of EWS-FLI1 leads to mitotic defects and apoptosis [79,102,112], suggesting that a dynamic fluctuation in EWS-FLI1 expression is essential for EwS cell survival. Therefore, EWS-FLI1 downregulation or overexpression are two potential strategies for EwS.
Targeting EWS-FLI1 directly remains challenging; however, pharmacological targeting of the EWS-FLI1 protein complex, for example using the YK-4-279 and its analog TK-216, is a potentially promising strategy. YK-4-279 blocks the interaction of EWS-FLI1 with helicase A [161,162] or DDX5 [94] and reverses the transcriptional activation of EWS-FLI1. YK-4-279 also leads to an isoform shift from ARID1A-L to ARID1A-S and prevents the interaction between EWS-FLI1 and ARID1A-L [104]. This was originally thought to mimic the inhibition of EWS-FLI1 and showed a synergistic effect with vincristine [163]. However, further work demonstrated that YK-4-279 affects alternative splicing rather than directly inhibiting the transcriptional activity of EWS-FLI1, which mimics the reduction in EWS-FLI1 [94]. Despite the occurrence of drug resistance, as detected in preclinical experiments [164,165], the combination of TK-216 and vincristine showed anti-tumor activity in specific population in a Phase I clinical trial [166] and is well tolerated. A further Phase II clinical trial (NCT02657005) is ongoing. In addition to EWS-FLI1-positive cells, TK216 also exhibits toxicity to other cancer cell lines, such as thyroid cancer [167] and melanoma [168]. Based on the property of cytotoxicity to multiple cancers, Povedano et al. revealed that TK216 binds to distinct sites compared with vincristine and acts as a microtubule-destabilizing agent, such as by providing an additional mechanistic explanation for the clinical activity of this combination [169].
Mithramycin suppresses the transcription of EWS-FLI1 [71]; however, its hepatotoxicity and the narrow therapeutic window results in the discontinuation of a clinical trial (clinical trial: NCT01610570) [170]. Semi-synthetic analogues of mithramycin, such as MTMSA-Trp and MTMSA-Phe, are more selective for EWS-FLI1-positive cell lines [171]. Although mithramycin A does not increase the level of DNA double-strand breaks, it inhibits DNA double-strand break repair and further radiosensitizes EwS [172].
Pretreatment with mithramycin A for 24 h before irradiation significantly reduced clonogenic survival in vitro and delayed tumor regrowth in vivo, prolonging survival of EWS-FLI-positive tumor-bearing mice. An alternative strategy takes advantage of the chimeric protein, leading to R-loops and the positive feedback loop with PARP1 and increasing the sensitivity to PARP inhibitors [24,148]. Mechanistically, EWS-FLI1 blocks the BRCA1, thus mimicking BRCA1 deficiency [24]. However, clinical trials did not meet expectations (clinical trial: NCT01583543) [173]. A clinical trial (NCT01858168) combining olaparib and temozolomide in adults with recurrent/metastatic EwS is ongoing. Chemical genomics screening performed by Iniguez et al. [174] showed that the CDK12/13 inhibitor THZ531 impairs DNA damage repair in a EWS-FLI1-dependent manner, leading to synthetic lethality with PARP inhibitors.
5.2. Targeting EWS-FLI1-Related Transcription
In addition to the EWS-FLI1 protein partners, the pharmaceutical targeting of EWS-FLI1-based transcriptome is worth further exploration [33,175]. For example, chemotherapeutic agents targeting LSD1, such as SP-2509, could modify the transcriptional signature of EWS-FLI1 and regulate the proliferation of EwS [31,176]. These agents also show synergic effects with HDAC inhibitors [177,178]. Seclidemstat (SP-2577), a reversible oral LSD1 inhibitor, showed manageable safety and positive preliminary activity in a Phase I trial (NCT03600649) [179], and patients are being recruited for a Phase I/II clinical trial (NCT05266196). EwS is highly sensitive to HDAC inhibition [68,175,180,181,182]. Clinical trials (NCT04308330; NCT00020579) of the VIT (vincristine, irinotecan, and temozolomide) combination have been conducted. However, pan-HDAC inhibitors are highly toxic owing to unspecific effects [183], underscoring the need to develop specific HDAC inhibitors. HDAC1 is critical for the malignancy of EWS-FLI1-mediated transcription and posttranscriptional modifications of EwS [175,184]. Agents targeting Class I/II HDACs show synergistic effects with doxorubicin [106,175], although further clinical research is necessary to avoid potential side effects.
Ecteinascidin 743 (ET-743, trabectedin) interferes with the activity of EWS-FLI1 [185]. However, single-agent trabectedin showed negative results in a clinical trial (NCT00070109) [185,186]. A synergistic effect with SN38 was demonstrated [187], and the combination of trabectedin with irinotecan for the treatment of EwS is being tested in clinical trials (NCT04067115 and NCT02509234). Additionally, a multicohort trial of trabectedin combined with low-dose radiation therapy in advanced/metastatic sarcomas including EwS is also recruiting new patients (NCT05131386).
GSK-J4, an H3K27 demethylase inhibitor, can reverse the H3K27ac driven by EWS-FLI1 at enhancers and shows a synergistic cytotoxic effect when combined with the CDK7 inhibitor THZ1 [188]. BET proteins, particularly BRD4, are also essential for the EWS-FLI1 transcription signature and may be involved in the sensitivity of EwS to JQ1 [32]. The combination of JQ1 with a CDK9 inhibitor, CDKI-73, showed a synergistic effect [34]. The RUVBL1 inhibitor CB6644 also exhibits a synergistic cytotoxic effect with JQ1 in EwS [189].
5.3. EWS-FLI1-Dependent Immunotherapy of Ewing Sarcoma
EwS is a ‘cold tumor’ with a low mutation burden, and it is characterized by an immunosuppressive microenvironment [190]. This environment inhibits the infiltration of effective cytotoxic T-cells, which limits the response to clinical treatments. Immunotherapies for EwS mainly consist of immune checkpoint inhibitors and adoptive T-cell transfer [191,192,193,194].
5.4. TCR-Based T-Cell Therapy
Peptides derived from the EWS-FLI1 chimeric protein provide unique targets for the TCR-based T-cell therapy, although there is no conclusive evidence supporting their clinical significance [195]. Alternative strategies based on targeting peptides derived from the downstream proteins of EWS-FLI1, such as the CHM1-derived peptide VIMPCSWWV [196,197], have shown clinical regression in EwS patients [193]. The T-cell-targeting LIPI-derived peptides LDYTDAKFV and NLLKHGASL [198], STEAP1-derived YLPGVIAAI [192,194] and MIAVFLPIV [199], PAPPA-derived IILPMNVTV [200], EZH2-derived YMCSFLFNL [197], PAX3-derived QLMAFNHLI, and the modified version, QLMAFNHLV [201], showed effective killing of HLA-A*02:01+ ES cell lines. XAGE-1A is a direct downstream target of EWS-FLI1 (Supplementary Figure S4A) and serves as a potential target in TCR-based T-cell therapy [202,203], although no killing of targeting EwS has been identified yet [204].
6. CAR-T Therapy
Unlike TCR-based T-cell therapy, which is limited to specific HLA restriction and deficient HLA expression in EwS [205] because of the presence of myeloid-derived suppressor cells, F2 fibrocytes, and M2-like macrophages in the microenvironment [190], CAR-engineered T-cell therapy can target specific cell-surface antigens in tumors, independent of HLA. VEGFR2 is a potential target for CAR-T-cell therapy in EwS [206]. In addition to TCR-T-cells, CAR-T-cells [207] can target STEAP1, which is involved in the malignant phenotype of EwS [65].
CAR-T targeting GPR64, ROR1, and IGF1R, which are highly expressed in EwS [137,208], leads to a selective killing of EwS in vivo [209]. LINGO1, which is highly expressed in EwS [210], is a direct target of EWS-FLI1 (Supplementary Figure S4B). EZH2 inhibition by GSK-126 induces GD2 surface expression in EwS [211], and the combination of CAR-T therapy targeting GD2 and EZH2 inhibitors have synthetic cytotoxic in the treatment of EwS; this kind of combination provides new options for the clinical application. IL1RAP, a direct target of EWS-FLI1, is highly expressed in EwS, but minimally expressed in normal tissues, which makes it a promising surface target for EwS [70] and a potential candidate for advanced CAR-T therapy. ICAM-1 can promote tumor cell/T-cell interaction and T-cell activation, and the knockdown of EWS-FLI1 upregulates ICAM-1 expression and leads to the upregulation of PD-L1 and PD-L2, both proteins that inhibit the activity of T-cells [115]. Blocking PD-1 with a checkpoint inhibitor could increase the T-cell-mediated killing of EwS cells with low expression of EWS-FLI1.
7. Perspectives
Strategies targeting EWS-FLI1 for the treatment of EwS need to be established. Direct targeting of EWS-FLI1 or therapies related to EWS-FLI1 protein are both viable options. In the ‘EWS-FLI1 toxicity’ strategy, targeting EWS-FLI1 offers a wide therapeutic window because of its unique expression pattern. However, further work is necessary to develop specific inhibitors. Additionally, a better understanding of the cytotoxicity and chemoresistance of YK-4-279, such as its destabilization in microtubules, is necessary to optimize its clinical application. EWS-FLI1 contributes to the sensitivity of EwS to radiotherapy and PARP inhibitors and is involved in DNA damage. Therefore, further research on the EWS-FLI1 protein complex is necessary to elucidate its role in chemoresistance, particularly the dynamic balance of EWS-FLI1 with wild-type EWSR1 and their related phase transitions.
Targeting the downstream factors of EWS-FLI1 with TCR- or CAR-T-based immunotherapy is based on the role of EWS-FLI1 in modulating CHM1, IL1RAP, and other molecules (Figure 3A). This type of immunotherapy has revolutionized cancer treatment, whereas canonical methods of retro- or lentivirally induced TCR may disrupt TCR dynamics and lead to atypical T-cell function.
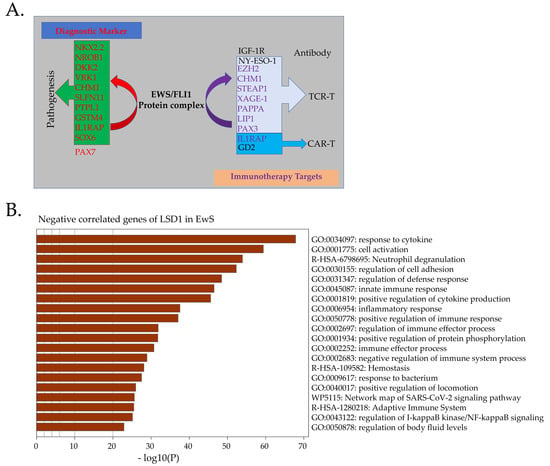
Figure 3.
Immuno-targeting therapy for EwS. (A). Direct downstream targets of EWS-FLI1, such as CHM1, STEAP1, and PAPPA, could serve as TCR-based immunotherapy. IL1RAP and STEPA1 are potential targets for CAR-T therapy. NY-ESO-1 is not a direct target of EWS-FLI1, but it is also a good target for TCR-T therapy. NKX2-2, NROB1, and PAX7 are reliable diagnostic markers of EwS. (B). Negatively correlated genes of LSD1 (p < 0.05) in EwS samples are mainly enriched in immune responses.
Recent work from Schober et al. demonstrated that orthotopic TCR replacement using nonviral methods results in near-physiological T-cell function [212], thereby preventing potential mispairing of endogenous and extraneous TCR. Our work also provides evidence that CRISPR/Cas9-engineered TCR replacement by TCR targeting CHM1 in T-cells could enhance the longevity of the cytotoxic effect of targeting EwS compared with the transgenic T-cells via retrovirus [213]. Metabolic or epigenetic reprogramming to enhance the function of T-cells provides another therapeutic option [214]. Apart from leading apoptosis, the PARP inhibitor modulates the tumor microenvironment by activating the cGAS-STING pathway and, thus, enables low-dose CAR-T-cells with higher efficacy [215,216]. Olaparib also suppresses myeloid-derived suppressor cell migration via the SDF1α/CXCR4 axis and promotes the survival of CD8+ T-cells in tumor tissue [217]. The combination of PARP inhibitor and CAR-T/TCR-T therapy is worthy of further research in EwS.
Through multiple pathways, the purpose of T-cell therapy is to achieve the apoptosis of cancer cells [216]. LSD1 inhibition promotes tumor immunogenicity and T-cell infiltration [218]. Genes negatively correlated with LSD1 in EwS are mainly associated with immune responses (Figure 3B). LSD1 expression in EwS is the highest among all cancers, and LSD1 inhibition leads to robust apoptosis through the endoplasmic reticulum (ER) stress pathway in EwS [176]. Exploring the combination of LSD1 inhibition with immunotherapy targeting the downstream of EWS-FLI1 could lead to important discoveries.
8. Conclusions
EWS-FLI1 was identified three decades ago (1992), 70 years after James Ewing first defined Ewing sarcoma (1921). Since then, this chimeric protein has been widely accepted as the oncogenic driver and master regulator of EwS because of its ability to modify the transcriptome. However, pharmacologically targeting EWS-FLI1 remains challenging because of its disordered structure. The clinical treatment of EwS mainly consists of radiotherapy, surgical resection, and intensive induction cytotoxic chemotherapy, which can lead to acute and chronic adverse effects.
Two strategies have been proposed based on the concept of precision medicine: targeting EWS-FLI1 directly or developing methods that indirectly exploit this dominant oncogene. This can involve targeting EWS-FLI1 in DNA damage or immunotherapy targeting the downstream transcripts such as CHM1. In this review, we summarized the current knowledge of EWS-FLI1 and the advances in related therapies, thereby providing the basis for further research.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cancers15164035/s1, Figure S1: High expression of XPO5 and low expression of PCAF in EwS. Figure S2: Low expression of USP19 in EwS. Figure S3: High expression of AMER2 in EwS. Figure S4: EWS-FLI1 binds to the promotor of XAGE-1A and LINGO1.
Author Contributions
Writing—original draft preparation, H.G., B.X. and Y.L.; writing—review and editing, H.G., B.X., J.R., G.P. and Y.L. All authors have read and agreed to the published version of the manuscript.
Funding
The China Scholarship Council (No. 201908210290) supported this work.
Acknowledgments
ChIP-sequencing data (GSE61944: GSM15175472, GSM1517573, GSM1517546, GSM1517547, GSM1517555, GSM1517556, GSM1517569, GSM1517570, GSM1517577, and GSM1517581) were downloaded from the GEO database, processed and displayed in the IGV browser [219]. Expression data were either from GSE17679, GSE63157, GSE34620, and GSE176190 or from Sangerbox [220]. For the informatics analysis of LSD1 negatively correlated genes, correlations based on GSE34620 with a p value < 0.05 were considered differentially expressed and Enrichr [221] was used for GO analysis. Part of the job is included in the B.X.’s doctoral thesis.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Grunewald, T.G.P.; Cidre-Aranaz, F.; Surdez, D.; Tomazou, E.M.; de Alava, E.; Kovar, H.; Sorensen, P.H.; Delattre, O.; Dirksen, U. Ewing sarcoma. Nat. Rev. Dis. Primers 2018, 4, 5. [Google Scholar] [CrossRef]
- Jahanseir, K.; Folpe, A.L.; Graham, R.P.; Giannini, C.; Robinson, S.I.; Sukov, W.; Fritchie, K. Ewing Sarcoma in Older Adults: A Clinicopathologic Study of 50 Cases Occurring in Patients Aged >/=40 Years, With Emphasis on Histologic Mimics. Int. J. Surg. Pathol. 2020, 28, 352–360. [Google Scholar] [CrossRef] [PubMed]
- Riggi, N.; Suva, M.L.; Stamenkovic, I. Ewing’s sarcoma origin: From duel to duality. Expert Rev. Anticancer Ther. 2009, 9, 1025–1030. [Google Scholar] [CrossRef] [PubMed]
- Tirode, F.; Laud-Duval, K.; Prieur, A.; Delorme, B.; Charbord, P.; Delattre, O. Mesenchymal stem cell features of Ewing tumors. Cancer Cell 2007, 11, 421–429. [Google Scholar] [CrossRef] [PubMed]
- Kovar, H. Blocking the road, stopping the engine or killing the driver? Advances in targeting EWS/FLI-1 fusion in Ewing sarcoma as novel therapy. Expert Opin. Ther. Targets 2014, 18, 1315–1328. [Google Scholar] [CrossRef] [PubMed]
- Gaspar, N.; Hawkins, D.S.; Dirksen, U.; Lewis, I.J.; Ferrari, S.; Le Deley, M.C.; Kovar, H.; Grimer, R.; Whelan, J.; Claude, L.; et al. Ewing Sarcoma: Current Management and Future Approaches Through Collaboration. J. Clin. Oncol. 2015, 33, 3036–3046. [Google Scholar] [CrossRef]
- Haeusler, J.; Ranft, A.; Boelling, T.; Gosheger, G.; Braun-Munzinger, G.; Vieth, V.; Burdach, S.; van den Berg, H.; Juergens, H.; Dirksen, U. The value of local treatment in patients with primary, disseminated, multifocal Ewing sarcoma (PDMES). Cancer 2010, 116, 443–450. [Google Scholar] [CrossRef]
- Zollner, S.K.; Amatruda, J.F.; Bauer, S.; Collaud, S.; de Alava, E.; DuBois, S.G.; Hardes, J.; Hartmann, W.; Kovar, H.; Metzler, M.; et al. Ewing Sarcoma-Diagnosis, Treatment, Clinical Challenges and Future Perspectives. J. Clin. Med. 2021, 10, 1685. [Google Scholar] [CrossRef]
- Ladenstein, R.; Potschger, U.; Le Deley, M.C.; Whelan, J.; Paulussen, M.; Oberlin, O.; van den Berg, H.; Dirksen, U.; Hjorth, L.; Michon, J.; et al. Primary disseminated multifocal Ewing sarcoma: Results of the Euro-EWING 99 trial. J. Clin. Oncol. 2010, 28, 3284–3291. [Google Scholar] [CrossRef]
- Tirode, F.; Surdez, D.; Ma, X.; Parker, M.; Le Deley, M.C.; Bahrami, A.; Zhang, Z.; Lapouble, E.; Grossetete-Lalami, S.; Rusch, M.; et al. Genomic landscape of Ewing sarcoma defines an aggressive subtype with co-association of STAG2 and TP53 mutations. Cancer Discov. 2014, 4, 1342–1353. [Google Scholar] [CrossRef]
- Lawrence, M.S.; Stojanov, P.; Mermel, C.H.; Robinson, J.T.; Garraway, L.A.; Golub, T.R.; Meyerson, M.; Gabriel, S.B.; Lander, E.S.; Getz, G. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 2014, 505, 495–501. [Google Scholar] [CrossRef] [PubMed]
- Uren, A.; Tcherkasskaya, O.; Toretsky, J.A. Recombinant EWS-FLI1 oncoprotein activates transcription. Biochemistry 2004, 43, 13579–13589. [Google Scholar] [CrossRef] [PubMed]
- Erkizan, H.V.; Uversky, V.N.; Toretsky, J.A. Oncogenic partnerships: EWS-FLI1 protein interactions initiate key pathways of Ewing’s sarcoma. Clin. Cancer Res. 2010, 16, 4077–4083. [Google Scholar] [CrossRef] [PubMed]
- Delattre, O.; Zucman, J.; Plougastel, B.; Desmaze, C.; Melot, T.; Peter, M.; Kovar, H.; Joubert, I.; de Jong, P.; Rouleau, G.; et al. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature 1992, 359, 162–165. [Google Scholar] [CrossRef] [PubMed]
- Riggi, N.; Suva, M.L.; Stamenkovic, I. Ewing‘s Sarcoma. N. Engl. J. Med. 2021, 384, 154–164. [Google Scholar] [CrossRef] [PubMed]
- Janknecht, R. EWS-ETS oncoproteins: The linchpins of Ewing tumors. Gene 2005, 363, 1–14. [Google Scholar] [CrossRef]
- Li, M.; Chen, C.W. Epigenetic and Transcriptional Signaling in Ewing Sarcoma-Disease Etiology and Therapeutic Opportunities. Biomedicines 2022, 10, 1325. [Google Scholar] [CrossRef]
- Plougastel, B.; Zucman, J.; Peter, M.; Thomas, G.; Delattre, O. Genomic structure of the EWS gene and its relationship to EWSR1, a site of tumor-associated chromosome translocation. Genomics 1993, 18, 609–615. [Google Scholar] [CrossRef]
- Morohoshi, F.; Ootsuka, Y.; Arai, K.; Ichikawa, H.; Mitani, S.; Munakata, N.; Ohki, M. Genomic structure of the human RBP56/hTAFII68 and FUS/TLS genes. Gene 1998, 221, 191–198. [Google Scholar] [CrossRef]
- Kovar, H. Dr. Jekyll and Mr. Hyde: The Two Faces of the FUS/EWS/TAF15 Protein Family. Sarcoma 2011, 2011, 837474. [Google Scholar] [CrossRef]
- Ohno, T.; Ouchida, M.; Lee, L.; Gatalica, Z.; Rao, V.N.; Reddy, E.S. The EWS gene, involved in Ewing family of tumors, malignant melanoma of soft parts and desmoplastic small round cell tumors, codes for an RNA binding protein with novel regulatory domains. Oncogene 1994, 9, 3087–3097. [Google Scholar] [PubMed]
- Li, H.; Watford, W.; Li, C.; Parmelee, A.; Bryant, M.A.; Deng, C.; O’Shea, J.; Lee, S.B. Ewing sarcoma gene EWS is essential for meiosis and B lymphocyte development. J. Clin. Investig. 2007, 117, 1314–1323. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Chansky, H.A.; Hickstein, D.D. EWS.Fli-1 fusion protein interacts with hyperphosphorylated RNA polymerase II and interferes with serine-arginine protein-mediated RNA splicing. J. Biol. Chem. 2000, 275, 37612–37618. [Google Scholar] [CrossRef] [PubMed]
- Gorthi, A.; Romero, J.C.; Loranc, E.; Cao, L.; Lawrence, L.A.; Goodale, E.; Iniguez, A.B.; Bernard, X.; Masamsetti, V.P.; Roston, S.; et al. EWS-FLI1 increases transcription to cause R-loops and block BRCA1 repair in Ewing sarcoma. Nature 2018, 555, 387–391. [Google Scholar] [CrossRef]
- Yoon, Y.; Park, H.; Kim, S.; Nguyen, P.T.; Hyeon, S.J.; Chung, S.; Im, H.; Lee, J.; Lee, S.B.; Ryu, H. Genetic Ablation of EWS RNA Binding Protein 1 (EWSR1) Leads to Neuroanatomical Changes and Motor Dysfunction in Mice. Exp. Neurobiol. 2018, 27, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Hollenhorst, P.C.; McIntosh, L.P.; Graves, B.J. Genomic and biochemical insights into the specificity of ETS transcription factors. Annu. Rev. Biochem. 2011, 80, 437–471. [Google Scholar] [CrossRef] [PubMed]
- Tomlins, S.A.; Rhodes, D.R.; Perner, S.; Dhanasekaran, S.M.; Mehra, R.; Sun, X.W.; Varambally, S.; Cao, X.; Tchinda, J.; Kuefer, R.; et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science 2005, 310, 644–648. [Google Scholar] [CrossRef]
- Chen, Z.; Arai, E.; Khan, O.; Zhang, Z.; Ngiow, S.F.; He, Y.; Huang, H.; Manne, S.; Cao, Z.; Baxter, A.E.; et al. In vivo CD8(+) T cell CRISPR screening reveals control by Fli1 in infection and cancer. Cell 2021, 184, 1262–1280.e1222. [Google Scholar] [CrossRef]
- Boulay, G.; Sandoval, G.J.; Riggi, N.; Iyer, S.; Buisson, R.; Naigles, B.; Awad, M.E.; Rengarajan, S.; Volorio, A.; McBride, M.J.; et al. Cancer-Specific Retargeting of BAF Complexes by a Prion-like Domain. Cell 2017, 171, 163–178.e119. [Google Scholar] [CrossRef]
- Riggi, N.; Knoechel, B.; Gillespie, S.M.; Rheinbay, E.; Boulay, G.; Suva, M.L.; Rossetti, N.E.; Boonseng, W.E.; Oksuz, O.; Cook, E.B.; et al. EWS-FLI1 utilizes divergent chromatin remodeling mechanisms to directly activate or repress enhancer elements in Ewing sarcoma. Cancer Cell 2014, 26, 668–681. [Google Scholar] [CrossRef]
- Sankar, S.; Theisen, E.R.; Bearss, J.; Mulvihill, T.; Hoffman, L.M.; Sorna, V.; Beckerle, M.C.; Sharma, S.; Lessnick, S.L. Reversible LSD1 inhibition interferes with global EWS/ETS transcriptional activity and impedes Ewing sarcoma tumor growth. Clin. Cancer Res. 2014, 20, 4584–4597. [Google Scholar] [CrossRef] [PubMed]
- Gollavilli, P.N.; Pawar, A.; Wilder-Romans, K.; Natesan, R.; Engelke, C.G.; Dommeti, V.L.; Krishnamurthy, P.M.; Nallasivam, A.; Apel, I.J.; Xu, T.; et al. EWS/ETS-Driven Ewing Sarcoma Requires BET Bromodomain Proteins. Cancer Res. 2018, 78, 4760–4773. [Google Scholar] [CrossRef] [PubMed]
- Tomazou, E.M.; Sheffield, N.C.; Schmidl, C.; Schuster, M.; Schonegger, A.; Datlinger, P.; Kubicek, S.; Bock, C.; Kovar, H. Epigenome mapping reveals distinct modes of gene regulation and widespread enhancer reprogramming by the oncogenic fusion protein EWS-FLI1. Cell Rep. 2015, 10, 1082–1095. [Google Scholar] [CrossRef] [PubMed]
- Richter, G.H.S.; Hensel, T.; Schmidt, O.; Saratov, V.; von Heyking, K.; Becker-Dettling, F.; Prexler, C.; Yen, H.Y.; Steiger, K.; Fulda, S.; et al. Combined Inhibition of Epigenetic Readers and Transcription Initiation Targets the EWS-ETS Transcriptional Program in Ewing Sarcoma. Cancers 2020, 12, 304. [Google Scholar] [CrossRef]
- Sheffield, N.C.; Pierron, G.; Klughammer, J.; Datlinger, P.; Schonegger, A.; Schuster, M.; Hadler, J.; Surdez, D.; Guillemot, D.; Lapouble, E.; et al. DNA methylation heterogeneity defines a disease spectrum in Ewing sarcoma. Nat. Med. 2017, 23, 386–395. [Google Scholar] [CrossRef]
- Richter, G.H.; Plehm, S.; Fasan, A.; Rossler, S.; Unland, R.; Bennani-Baiti, I.M.; Hotfilder, M.; Lowel, D.; von Luettichau, I.; Mossbrugger, I.; et al. EZH2 is a mediator of EWS/FLI1 driven tumor growth and metastasis blocking endothelial and neuro-ectodermal differentiation. Proc. Natl. Acad. Sci. USA 2009, 106, 5324–5329. [Google Scholar] [CrossRef]
- Gangwal, K.; Sankar, S.; Hollenhorst, P.C.; Kinsey, M.; Haroldsen, S.C.; Shah, A.A.; Boucher, K.M.; Watkins, W.S.; Jorde, L.B.; Graves, B.J.; et al. Microsatellites as EWS/FLI response elements in Ewing’s sarcoma. Proc. Natl. Acad. Sci. USA 2008, 105, 10149–10154. [Google Scholar] [CrossRef]
- Gangwal, K.; Lessnick, S.L. Microsatellites are EWS/FLI response elements: Genomic “junk” is EWS/FLI’s treasure. Cell Cycle 2008, 7, 3127–3132. [Google Scholar] [CrossRef]
- Guillon, N.; Tirode, F.; Boeva, V.; Zynovyev, A.; Barillot, E.; Delattre, O. The oncogenic EWS-FLI1 protein binds in vivo GGAA microsatellite sequences with potential transcriptional activation function. PLoS ONE 2009, 4, e4932. [Google Scholar] [CrossRef]
- Ahmed, N.S.; Harrell, L.M.; Wieland, D.R.; Lay, M.A.; Thompson, V.F.; Schwartz, J.C. Fusion protein EWS-FLI1 is incorporated into a protein granule in cells. RNA 2021, 27, 920–932. [Google Scholar] [CrossRef]
- Petermann, R.; Mossier, B.M.; Aryee, D.N.; Khazak, V.; Golemis, E.A.; Kovar, H. Oncogenic EWS-Fli1 interacts with hsRPB7, a subunit of human RNA polymerase II. Oncogene 1998, 17, 603–610. [Google Scholar] [CrossRef]
- Zhou, H.; Lee, K.A. An hsRPB4/7-dependent yeast assay for trans-activation by the EWS oncogene. Oncogene 2001, 20, 1519–1524. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bilke, S.; Schwentner, R.; Yang, F.; Kauer, M.; Jug, G.; Walker, R.L.; Davis, S.; Zhu, Y.J.; Pineda, M.; Meltzer, P.S.; et al. Oncogenic ETS fusions deregulate E2F3 target genes in Ewing sarcoma and prostate cancer. Genome Res. 2013, 23, 1797–1809. [Google Scholar] [CrossRef] [PubMed]
- Schwentner, R.; Papamarkou, T.; Kauer, M.O.; Stathopoulos, V.; Yang, F.; Bilke, S.; Meltzer, P.S.; Girolami, M.; Kovar, H. EWS-FLI1 employs an E2F switch to drive target gene expression. Nucleic Acids Res. 2015, 43, 2780–2789. [Google Scholar] [CrossRef]
- Mertens, F.; Antonescu, C.R.; Mitelman, F. Gene fusions in soft tissue tumors: Recurrent and overlapping pathogenetic themes. Genes Chromosomes Cancer 2016, 55, 291–310. [Google Scholar] [CrossRef]
- Ramakrishnan, R.; Fujimura, Y.; Zou, J.P.; Liu, F.; Lee, L.; Rao, V.N.; Reddy, E.S. Role of protein-protein interactions in the antiapoptotic function of EWS-Fli-1. Oncogene 2004, 23, 7087–7094. [Google Scholar] [CrossRef] [PubMed]
- Harlow, M.L.; Chasse, M.H.; Boguslawski, E.A.; Sorensen, K.M.; Gedminas, J.M.; Kitchen-Goosen, S.M.; Rothbart, S.B.; Taslim, C.; Lessnick, S.L.; Peck, A.S.; et al. Trabectedin Inhibits EWS-FLI1 and Evicts SWI/SNF from Chromatin in a Schedule-dependent Manner. Clin. Cancer Res. 2019, 25, 3417–3429. [Google Scholar] [CrossRef]
- Monument, M.J.; Johnson, K.M.; McIlvaine, E.; Abegglen, L.; Watkins, W.S.; Jorde, L.B.; Womer, R.B.; Beeler, N.; Monovich, L.; Lawlor, E.R.; et al. Clinical and biochemical function of polymorphic NR0B1 GGAA-microsatellites in Ewing sarcoma: A report from the Children’s Oncology Group. PLoS ONE 2014, 9, e104378. [Google Scholar] [CrossRef]
- Lin, L.; Huang, M.; Shi, X.; Mayakonda, A.; Hu, K.; Jiang, Y.Y.; Guo, X.; Chen, L.; Pang, B.; Doan, N.; et al. Super-enhancer-associated MEIS1 promotes transcriptional dysregulation in Ewing sarcoma in co-operation with EWS-FLI1. Nucleic Acids Res. 2019, 47, 1255–1267. [Google Scholar] [CrossRef]
- Sanchez-Molina, S.; Figuerola-Bou, E.; Blanco, E.; Sanchez-Jimenez, M.; Taboas, P.; Gomez, S.; Ballare, C.; Garcia-Dominguez, D.J.; Prada, E.; Hontecillas-Prieto, L.; et al. RING1B recruits EWSR1-FLI1 and cooperates in the remodeling of chromatin necessary for Ewing sarcoma tumorigenesis. Sci. Adv. 2020, 6. [Google Scholar] [CrossRef]
- Smith, R.; Owen, L.A.; Trem, D.J.; Wong, J.S.; Whangbo, J.S.; Golub, T.R.; Lessnick, S.L. Expression profiling of EWS/FLI identifies NKX2.2 as a critical target gene in Ewing’s sarcoma. Cancer Cell 2006, 9, 405–416. [Google Scholar] [CrossRef]
- Kinsey, M.; Smith, R.; Lessnick, S.L. NR0B1 is required for the oncogenic phenotype mediated by EWS/FLI in Ewing’s sarcoma. Mol. Cancer Res. 2006, 4, 851–859. [Google Scholar] [CrossRef] [PubMed]
- Boro, A.; Pretre, K.; Rechfeld, F.; Thalhammer, V.; Oesch, S.; Wachtel, M.; Schafer, B.W.; Niggli, F.K. Small-molecule screen identifies modulators of EWS/FLI1 target gene expression and cell survival in Ewing’s sarcoma. Int. J. Cancer 2012, 131, 2153–2164. [Google Scholar] [CrossRef]
- Garcia-Aragoncillo, E.; Carrillo, J.; Lalli, E.; Agra, N.; Gomez-Lopez, G.; Pestana, A.; Alonso, J. DAX1, a direct target of EWS/FLI1 oncoprotein, is a principal regulator of cell-cycle progression in Ewing’s tumor cells. Oncogene 2008, 27, 6034–6043. [Google Scholar] [CrossRef]
- Cironi, L.; Riggi, N.; Provero, P.; Wolf, N.; Suva, M.L.; Suva, D.; Kindler, V.; Stamenkovic, I. IGF1 is a common target gene of Ewing’s sarcoma fusion proteins in mesenchymal progenitor cells. PLoS ONE 2008, 3, e2634. [Google Scholar] [CrossRef] [PubMed]
- Wiles, E.T.; Lui-Sargent, B.; Bell, R.; Lessnick, S.L. BCL11B is up-regulated by EWS/FLI and contributes to the transformed phenotype in Ewing sarcoma. PLoS ONE 2013, 8, e59369. [Google Scholar] [CrossRef]
- Beauchamp, E.; Bulut, G.; Abaan, O.; Chen, K.; Merchant, A.; Matsui, W.; Endo, Y.; Rubin, J.S.; Toretsky, J.; Uren, A. GLI1 is a direct transcriptional target of EWS-FLI1 oncoprotein. J. Biol. Chem. 2009, 284, 9074–9082. [Google Scholar] [CrossRef] [PubMed]
- Abaan, O.D.; Levenson, A.; Khan, O.; Furth, P.A.; Uren, A.; Toretsky, J.A. PTPL1 is a direct transcriptional target of EWS-FLI1 and modulates Ewing’s Sarcoma tumorigenesis. Oncogene 2005, 24, 2715–2722. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Xu, C.; Ayello, J.; Dela Cruz, F.; Rosenblum, J.M.; Lessnick, S.L.; Cairo, M.S. Protein phosphatase 1 regulatory subunit 1A in ewing sarcoma tumorigenesis and metastasis. Oncogene 2018, 37, 798–809. [Google Scholar] [CrossRef]
- Grunewald, T.G.; Bernard, V.; Gilardi-Hebenstreit, P.; Raynal, V.; Surdez, D.; Aynaud, M.M.; Mirabeau, O.; Cidre-Aranaz, F.; Tirode, F.; Zaidi, S.; et al. Chimeric EWSR1-FLI1 regulates the Ewing sarcoma susceptibility gene EGR2 via a GGAA microsatellite. Nat. Genet. 2015, 47, 1073–1078. [Google Scholar] [CrossRef]
- Luo, W.; Gangwal, K.; Sankar, S.; Boucher, K.M.; Thomas, D.; Lessnick, S.L. GSTM4 is a microsatellite-containing EWS/FLI target involved in Ewing’s sarcoma oncogenesis and therapeutic resistance. Oncogene 2009, 28, 4126–4132. [Google Scholar] [CrossRef]
- Charville, G.W.; Wang, W.L.; Ingram, D.R.; Roy, A.; Thomas, D.; Patel, R.M.; Hornick, J.L.; van de Rijn, M.; Lazar, A.J. EWSR1 fusion proteins mediate PAX7 expression in Ewing sarcoma. Mod. Pathol. 2017, 30, 1312–1320. [Google Scholar] [CrossRef]
- Von Heyking, K.; Calzada-Wack, J.; Gollner, S.; Neff, F.; Schmidt, O.; Hensel, T.; Schirmer, D.; Fasan, A.; Esposito, I.; Muller-Tidow, C.; et al. The endochondral bone protein CHM1 sustains an undifferentiated, invasive phenotype, promoting lung metastasis in Ewing sarcoma. Mol. Oncol. 2017, 11, 1288–1301. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Yu, L.; Kleinerman, E.S. EWS-FLI-1 regulates the neuronal repressor gene REST, which controls Ewing sarcoma growth and vascular morphology. Cancer 2014, 120, 579–588. [Google Scholar] [CrossRef]
- Grunewald, T.G.; Diebold, I.; Esposito, I.; Plehm, S.; Hauer, K.; Thiel, U.; da Silva-Buttkus, P.; Neff, F.; Unland, R.; Muller-Tidow, C.; et al. STEAP1 is associated with the invasive and oxidative stress phenotype of Ewing tumors. Mol. Cancer Res. 2012, 10, 52–65. [Google Scholar] [CrossRef]
- Grunewald, T.G.P.; Ranft, A.; Esposito, I.; da Silva-Buttkus, P.; Aichler, M.; Baumhoer, D.; Schaefer, K.L.; Ottaviano, L.; Poremba, C.; Jundt, G.; et al. High STEAP1 expression is associated with improved outcome of Ewing’s sarcoma patients. Ann. Oncol. 2012, 23, 2185–2190. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.W.; Bilke, S.; Cao, L.; Murai, J.; Sousa, F.G.; Yamade, M.; Rajapakse, V.; Varma, S.; Helman, L.J.; Khan, J.; et al. SLFN11 Is a Transcriptional Target of EWS-FLI1 and a Determinant of Drug Response in Ewing Sarcoma. Clin. Cancer Res. 2015, 21, 4184–4193. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Baltezor, M.; Rajewski, L.; Crow, J.; Samuel, G.; Staggs, V.S.; Chastain, K.M.; Toretsky, J.A.; Weir, S.J.; Godwin, A.K. Targeted inhibition of histone deacetylase leads to suppression of Ewing sarcoma tumor growth through an unappreciated EWS-FLI1/HDAC3/HSP90 signaling axis. J. Mol. Med. 2019, 97, 957–972. [Google Scholar] [CrossRef]
- He, S.; Huang, Q.; Hu, J.; Li, L.; Xiao, Y.; Yu, H.; Han, Z.; Wang, T.; Zhou, W.; Wei, H.; et al. EWS-FLI1-mediated tenascin-C expression promotes tumour progression by targeting MALAT1 through integrin alpha5beta1-mediated YAP activation in Ewing sarcoma. Br. J. Cancer 2019, 121, 922–933. [Google Scholar] [CrossRef]
- Zhang, H.F.; Hughes, C.S.; Li, W.; He, J.Z.; Surdez, D.; El-Naggar, A.M.; Cheng, H.; Prudova, A.; Delaidelli, A.; Negri, G.L.; et al. Proteomic screens for suppressors of anoikis identify IL1RAP as a promising surface target in Ewing sarcoma. Cancer Discov. 2021, 11, 2884–2903. [Google Scholar] [CrossRef]
- Grohar, P.J.; Woldemichael, G.M.; Griffin, L.B.; Mendoza, A.; Chen, Q.R.; Yeung, C.; Currier, D.G.; Davis, S.; Khanna, C.; Khan, J.; et al. Identification of an inhibitor of the EWS-FLI1 oncogenic transcription factor by high-throughput screening. J. Natl. Cancer Inst. 2011, 103, 962–978. [Google Scholar] [CrossRef] [PubMed]
- Bailly, R.A.; Bosselut, R.; Zucman, J.; Cormier, F.; Delattre, O.; Roussel, M.; Thomas, G.; Ghysdael, J. DNA-binding and transcriptional activation properties of the EWS-FLI-1 fusion protein resulting from the t(11;22) translocation in Ewing sarcoma. Mol. Cell. Biol. 1994, 14, 3230–3241. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Ohmura, S.; Marchetto, A.; Orth, M.F.; Imle, R.; Dallmayer, M.; Musa, J.; Knott, M.M.L.; Holting, T.L.B.; Stein, S.; et al. Therapeutic targeting of the PLK1-PRC1-axis triggers cell death in genomically silent childhood cancer. Nat. Commun. 2021, 12, 5356. [Google Scholar] [CrossRef]
- Owen, L.A.; Kowalewski, A.A.; Lessnick, S.L. EWS/FLI mediates transcriptional repression via NKX2.2 during oncogenic transformation in Ewing’s sarcoma. PLoS ONE 2008, 3, e1965. [Google Scholar] [CrossRef]
- Shi, X.; Zheng, Y.; Jiang, L.; Zhou, B.; Yang, W.; Li, L.; Ding, L.; Huang, M.; Gery, S.; Lin, D.C.; et al. EWS-FLI1 regulates and cooperates with core regulatory circuitry in Ewing sarcoma. Nucleic Acids Res. 2020, 48, 11434–11451. [Google Scholar] [CrossRef]
- Markey, F.B.; Romero, B.; Parashar, V.; Batish, M. Identification of a New Transcriptional Co-Regulator of STEAP1 in Ewing’s Sarcoma. Cells 2021, 10, 1300. [Google Scholar] [CrossRef] [PubMed]
- Kinsey, M.; Smith, R.; Iyer, A.K.; McCabe, E.R.; Lessnick, S.L. EWS/FLI and its downstream target NR0B1 interact directly to modulate transcription and oncogenesis in Ewing’s sarcoma. Cancer Res. 2009, 69, 9047–9055. [Google Scholar] [CrossRef]
- Tang, S.W.; Thomas, A.; Murai, J.; Trepel, J.B.; Bates, S.E.; Rajapakse, V.N.; Pommier, Y. Overcoming Resistance to DNA-Targeted Agents by Epigenetic Activation of Schlafen 11 (SLFN11) Expression with Class I Histone Deacetylase Inhibitors. Clin. Cancer Res. 2018, 24, 1944–1953. [Google Scholar] [CrossRef]
- Prieur, A.; Tirode, F.; Cohen, P.; Delattre, O. EWS/FLI-1 silencing and gene profiling of Ewing cells reveal downstream oncogenic pathways and a crucial role for repression of insulin-like growth factor binding protein 3. Mol. Cell. Biol. 2004, 24, 7275–7283. [Google Scholar] [CrossRef]
- Sankar, S.; Bell, R.; Stephens, B.; Zhuo, R.; Sharma, S.; Bearss, D.J.; Lessnick, S.L. Mechanism and relevance of EWS/FLI-mediated transcriptional repression in Ewing sarcoma. Oncogene 2013, 32, 5089–5100. [Google Scholar] [CrossRef]
- Clapier, C.R.; Cairns, B.R. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 2009, 78, 273–304. [Google Scholar] [CrossRef] [PubMed]
- Lai, A.Y.; Wade, P.A. Cancer biology and NuRD: A multifaceted chromatin remodelling complex. Nat. Rev. Cancer 2011, 11, 588–596. [Google Scholar] [CrossRef] [PubMed]
- Agra, N.; Cidre, F.; Garcia-Garcia, L.; de la Parra, J.; Alonso, J. Lysyl oxidase is downregulated by the EWS/FLI1 oncoprotein and its propeptide domain displays tumor supressor activities in Ewing sarcoma cells. PLoS ONE 2013, 8, e66281. [Google Scholar] [CrossRef]
- Katschnig, A.M.; Kauer, M.O.; Schwentner, R.; Tomazou, E.M.; Mutz, C.N.; Linder, M.; Sibilia, M.; Alonso, J.; Aryee, D.N.T.; Kovar, H. EWS-FLI1 perturbs MRTFB/YAP-1/TEAD target gene regulation inhibiting cytoskeletal autoregulatory feedback in Ewing sarcoma. Oncogene 2017, 36, 5995–6005. [Google Scholar] [CrossRef]
- Niedan, S.; Kauer, M.; Aryee, D.N.; Kofler, R.; Schwentner, R.; Meier, A.; Potschger, U.; Kontny, U.; Kovar, H. Suppression of FOXO1 is responsible for a growth regulatory repressive transcriptional sub-signature of EWS-FLI1 in Ewing sarcoma. Oncogene 2014, 33, 3927–3938. [Google Scholar] [CrossRef]
- Dylla, L.; Moore, C.; Jedlicka, P. MicroRNAs in Ewing Sarcoma. Front. Oncol. 2013, 3, 65. [Google Scholar] [CrossRef] [PubMed]
- Riggi, N.; Suva, M.L.; De Vito, C.; Provero, P.; Stehle, J.C.; Baumer, K.; Cironi, L.; Janiszewska, M.; Petricevic, T.; Suva, D.; et al. EWS-FLI-1 modulates miRNA145 and SOX2 expression to initiate mesenchymal stem cell reprogramming toward Ewing sarcoma cancer stem cells. Genes Dev. 2010, 24, 916–932. [Google Scholar] [CrossRef]
- Robin, T.P.; Smith, A.; McKinsey, E.; Reaves, L.; Jedlicka, P.; Ford, H.L. EWS/FLI1 regulates EYA3 in Ewing sarcoma via modulation of miRNA-708, resulting in increased cell survival and chemoresistance. Mol. Cancer Res. 2012, 10, 1098–1108. [Google Scholar] [CrossRef]
- Sparmann, A.; van Lohuizen, M. Polycomb silencers control cell fate, development and cancer. Nat. Rev. Cancer 2006, 6, 846–856. [Google Scholar] [CrossRef]
- Nakatani, F.; Ferracin, M.; Manara, M.C.; Ventura, S.; Del Monaco, V.; Ferrari, S.; Alberghini, M.; Grilli, A.; Knuutila, S.; Schaefer, K.L.; et al. miR-34a predicts survival of Ewing’s sarcoma patients and directly influences cell chemo-sensitivity and malignancy. J. Pathol. 2012, 226, 796–805. [Google Scholar] [CrossRef]
- Bohnsack, M.T.; Czaplinski, K.; Gorlich, D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA 2004, 10, 185–191. [Google Scholar] [CrossRef] [PubMed]
- Yi, R.; Qin, Y.; Macara, I.G.; Cullen, B.R. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003, 17, 3011–3016. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Lee, J.E.; Riemondy, K.; Yu, Y.; Marquez, S.M.; Lai, E.C.; Yi, R. XPO5 promotes primary miRNA processing independently of RanGTP. Nat. Commun. 2020, 11, 1845. [Google Scholar] [CrossRef] [PubMed]
- Selvanathan, S.P.; Graham, G.T.; Erkizan, H.V.; Dirksen, U.; Natarajan, T.G.; Dakic, A.; Yu, S.; Liu, X.; Paulsen, M.T.; Ljungman, M.E.; et al. Oncogenic fusion protein EWS-FLI1 is a network hub that regulates alternative splicing. Proc. Natl. Acad. Sci. USA 2015, 112, E1307–E1316. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.L.; Cui, R.; Zhou, J.; Teng, K.Y.; Hsiao, Y.H.; Nakanishi, K.; Fassan, M.; Luo, Z.; Shi, G.; Tili, E.; et al. ERK Activation Globally Downregulates miRNAs through Phosphorylating Exportin-5. Cancer Cell 2016, 30, 723–736. [Google Scholar] [CrossRef] [PubMed]
- Klevernic, I.V.; Morton, S.; Davis, R.J.; Cohen, P. Phosphorylation of Ewing’s sarcoma protein (EWS) and EWS-Fli1 in response to DNA damage. Biochem. J. 2009, 418, 625–634. [Google Scholar] [CrossRef]
- Bachmaier, R.; Aryee, D.N.; Jug, G.; Kauer, M.; Kreppel, M.; Lee, K.A.; Kovar, H. O-GlcNAcylation is involved in the transcriptional activity of EWS-FLI1 in Ewing’s sarcoma. Oncogene 2009, 28, 1280–1284. [Google Scholar] [CrossRef][Green Version]
- Olsen, R.J.; Hinrichs, S.H. Phosphorylation of the EWS IQ domain regulates transcriptional activity of the EWS/ATF1 and EWS/FLI1 fusion proteins. Oncogene 2001, 20, 1756–1764. [Google Scholar] [CrossRef]
- Schlottmann, S.; Erkizan, H.V.; Barber-Rotenberg, J.S.; Knights, C.; Cheema, A.; Uren, A.; Avantaggiati, M.L.; Toretsky, J.A. Acetylation Increases EWS-FLI1 DNA Binding and Transcriptional Activity. Front. Oncol. 2012, 2, 107. [Google Scholar] [CrossRef]
- Maniatis, T.; Tasic, B. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature 2002, 418, 236–243. [Google Scholar] [CrossRef]
- Eymin, B. Targeting the spliceosome machinery: A new therapeutic axis in cancer? Biochem. Pharmacol. 2021, 189, 114039. [Google Scholar] [CrossRef] [PubMed]
- Embree, L.J.; Azuma, M.; Hickstein, D.D. Ewing sarcoma fusion protein EWSR1/FLI1 interacts with EWSR1 leading to mitotic defects in zebrafish embryos and human cell lines. Cancer Res. 2009, 69, 4363–4371. [Google Scholar] [CrossRef] [PubMed]
- Knoop, L.L.; Baker, S.J. The splicing factor U1C represses EWS/FLI-mediated transactivation. J. Biol. Chem. 2000, 275, 24865–24871. [Google Scholar] [CrossRef]
- Selvanathan, S.P.; Graham, G.T.; Grego, A.R.; Baker, T.M.; Hogg, J.R.; Simpson, M.; Batish, M.; Crompton, B.; Stegmaier, K.; Tomazou, E.M.; et al. EWS-FLI1 modulated alternative splicing of ARID1A reveals novel oncogenic function through the BAF complex. Nucleic Acids Res. 2019, 47, 9619–9636. [Google Scholar] [CrossRef] [PubMed]
- Hensel, T.; Giorgi, C.; Schmidt, O.; Calzada-Wack, J.; Neff, F.; Buch, T.; Niggli, F.K.; Schafer, B.W.; Burdach, S.; Richter, G.H. Targeting the EWS-ETS transcriptional program by BET bromodomain inhibition in Ewing sarcoma. Oncotarget 2016, 7, 1451–1463. [Google Scholar] [CrossRef]
- García-Domínguez, D.J.; Hajji, N.; Sánchez-Molina, S.; Figuerola-Bou, E.; de Pablos, R.M.; Espinosa-Oliva, A.M.; Andrés-León, E.; Terrón-Camero, L.C.; Flores-Campos, R.; Pascual-Pasto, G.; et al. HDAC6 regulates expression of the oncogenic driver EWSR1-FLI1 through the <em>EWSR1</em> promoter in Ewing sarcoma. bioRxiv 2021. [Google Scholar] [CrossRef]
- Ban, J.; Jug, G.; Mestdagh, P.; Schwentner, R.; Kauer, M.; Aryee, D.N.; Schaefer, K.L.; Nakatani, F.; Scotlandi, K.; Reiter, M.; et al. Hsa-mir-145 is the top EWS-FLI1-repressed microRNA involved in a positive feedback loop in Ewing’s sarcoma. Oncogene 2011, 30, 2173–2180. [Google Scholar] [CrossRef]
- Keskin, T.; Bakaric, A.; Waszyk, P.; Boulay, G.; Torsello, M.; Cornaz-Buros, S.; Chevalier, N.; Geiser, T.; Martin, P.; Volorio, A.; et al. LIN28B Underlies the Pathogenesis of a Subclass of Ewing Sarcoma LIN28B Control of EWS-FLI1 Stability. Cell Rep. 2020, 30, 4567–4583.e5. [Google Scholar] [CrossRef]
- De Vito, C.; Riggi, N.; Suva, M.L.; Janiszewska, M.; Horlbeck, J.; Baumer, K.; Provero, P.; Stamenkovic, I. Let-7a is a direct EWS-FLI-1 target implicated in Ewing’s sarcoma development. PLoS ONE 2011, 6, e23592. [Google Scholar] [CrossRef]
- Gierisch, M.E.; Pfistner, F.; Lopez-Garcia, L.A.; Harder, L.; Schafer, B.W.; Niggli, F.K. Proteasomal Degradation of the EWS-FLI1 Fusion Protein Is Regulated by a Single Lysine Residue. J. Biol. Chem. 2016, 291, 26922–26933. [Google Scholar] [CrossRef]
- Gierisch, M.E.; Pedot, G.; Walser, F.; Lopez-Garcia, L.A.; Jaaks, P.; Niggli, F.K.; Schafer, B.W. USP19 deubiquitinates EWS-FLI1 to regulate Ewing sarcoma growth. Sci. Rep. 2019, 9, 951. [Google Scholar] [CrossRef]
- Seong, B.K.A.; Dharia, N.V.; Lin, S.; Donovan, K.A.; Chong, S.; Robichaud, A.; Conway, A.; Hamze, A.; Ross, L.; Alexe, G.; et al. TRIM8 modulates the EWS/FLI oncoprotein to promote survival in Ewing sarcoma. Cancer Cell 2021, 39, 1262–1278.e1267. [Google Scholar] [CrossRef] [PubMed]
- Su, S.; Chen, J.; Jiang, Y.; Wang, Y.; Vital, T.; Zhang, J.; Laggner, C.; Nguyen, K.T.; Zhu, Z.; Prevatte, A.W.; et al. SPOP and OTUD7A Control EWS-FLI1 Protein Stability to Govern Ewing Sarcoma Growth. Adv. Sci. 2021, 8, e2004846. [Google Scholar] [CrossRef]
- Sun, H.; Lin, D.C.; Cao, Q.; Guo, X.; Marijon, H.; Zhao, Z.; Gery, S.; Xu, L.; Yang, H.; Pang, B.; et al. CRM1 Inhibition Promotes Cytotoxicity in Ewing Sarcoma Cells by Repressing EWS-FLI1-Dependent IGF-1 Signaling. Cancer Res. 2016, 76, 2687–2697. [Google Scholar] [CrossRef] [PubMed]
- Bailey, K.M.; Julian, C.M.; Klinghoffer, A.N.; Bernard, H.; Lucas, P.C.; McAllister-Lucas, L.M. EWS-FLI1 low Ewing sarcoma cells demonstrate decreased susceptibility to T-cell-mediated tumor cell apoptosis. Oncotarget 2019, 10, 3385–3399. [Google Scholar] [CrossRef] [PubMed]
- Sengupta, A.; Rahman, M.; Mateo-Lozano, S.; Tirado, O.M.; Notario, V. The dual inhibitory effect of thiostrepton on FoxM1 and EWS/FLI1 provides a novel therapeutic option for Ewing’s sarcoma. Int. J. Oncol. 2013, 43, 803–812. [Google Scholar] [CrossRef]
- Stegmaier, K.; Wong, J.S.; Ross, K.N.; Chow, K.T.; Peck, D.; Wright, R.D.; Lessnick, S.L.; Kung, A.L.; Golub, T.R. Signature-based small molecule screening identifies cytosine arabinoside as an EWS/FLI modulator in Ewing sarcoma. PLoS Med. 2007, 4, e122. [Google Scholar] [CrossRef]
- DuBois, S.G.; Krailo, M.D.; Lessnick, S.L.; Smith, R.; Chen, Z.; Marina, N.; Grier, H.E.; Stegmaier, K.; Children’s Oncology, G. Phase II study of intermediate-dose cytarabine in patients with relapsed or refractory Ewing sarcoma: A report from the Children’s Oncology Group. Pediatr. Blood Cancer 2009, 52, 324–327. [Google Scholar] [CrossRef]
- Chu, Z.; Gu, L.; Hu, Y.; Zhang, X.; Li, M.; Chen, J.; Teng, D.; Huang, M.; Shen, C.H.; Cai, L.; et al. STAG2 regulates interferon signaling in melanoma via enhancer loop reprogramming. Nat. Commun. 2022, 13, 1859. [Google Scholar] [CrossRef]
- Bailey, M.H.; Tokheim, C.; Porta-Pardo, E.; Sengupta, S.; Bertrand, D.; Weerasinghe, A.; Colaprico, A.; Wendl, M.C.; Kim, J.; Reardon, B.; et al. Comprehensive Characterization of Cancer Driver Genes and Mutations. Cell 2018, 174, 1034–1035. [Google Scholar] [CrossRef]
- Adane, B.; Alexe, G.; Seong, B.K.A.; Lu, D.; Hwang, E.E.; Hnisz, D.; Lareau, C.A.; Ross, L.; Lin, S.; Dela Cruz, F.S.; et al. STAG2 loss rewires oncogenic and developmental programs to promote metastasis in Ewing sarcoma. Cancer Cell 2021, 39, 827–844.e810. [Google Scholar] [CrossRef] [PubMed]
- Surdez, D.; Zaidi, S.; Grossetete, S.; Laud-Duval, K.; Ferre, A.S.; Mous, L.; Vourc’h, T.; Tirode, F.; Pierron, G.; Raynal, V.; et al. STAG2 mutations alter CTCF-anchored loop extrusion, reduce cis-regulatory interactions and EWSR1-FLI1 activity in Ewing sarcoma. Cancer Cell 2021, 39, 810–826.e819. [Google Scholar] [CrossRef] [PubMed]
- Crompton, B.D.; Stewart, C.; Taylor-Weiner, A.; Alexe, G.; Kurek, K.C.; Calicchio, M.L.; Kiezun, A.; Carter, S.L.; Shukla, S.A.; Mehta, S.S.; et al. The genomic landscape of pediatric Ewing sarcoma. Cancer Discov. 2014, 4, 1326–1341. [Google Scholar] [CrossRef]
- Lu, D.Y.; Ellegast, J.M.; Ross, K.N.; Malone, C.F.; Lin, S.; Mabe, N.W.; Dharia, N.V.; Meyer, A.; Conway, A.; Su, A.H.; et al. The ETS transcription factor ETV6 constrains the transcriptional activity of EWS-FLI to promote Ewing sarcoma. Nat. Cell Biol. 2023, 25, 285–297. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; He, X.Y.; Wu, X.S.; Huang, Y.H.; Toneyan, S.; Ha, T.; Ipsaro, J.J.; Koo, P.K.; Joshua-Tor, L.; Bailey, K.M.; et al. ETV6 dependency in Ewing sarcoma by antagonism of EWS-FLI1-mediated enhancer activation. Nat. Cell Biol. 2023, 25, 298–308. [Google Scholar] [CrossRef] [PubMed]
- Miser, J.S.; Goldsby, R.E.; Chen, Z.; Krailo, M.D.; Tarbell, N.J.; Link, M.P.; Fryer, C.J.; Pritchard, D.J.; Gebhardt, M.C.; Dickman, P.S.; et al. Treatment of metastatic Ewing sarcoma/primitive neuroectodermal tumor of bone: Evaluation of increasing the dose intensity of chemotherapy—A report from the Children’s Oncology Group. Pediatr. Blood Cancer 2007, 49, 894–900. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Yang, J.; Ma, X.; Wang, X. Risk factors for metastasis and poor prognosis of Ewing sarcoma: A population based study. J. Orthop. Surg. Res. 2020, 15, 88. [Google Scholar] [CrossRef]
- Ye, C.; Dai, M.; Zhang, B. Risk Factors for Metastasis at Initial Diagnosis With Ewing Sarcoma. Front. Oncol. 2019, 9, 1043. [Google Scholar] [CrossRef] [PubMed]
- Franzetti, G.A.; Laud-Duval, K.; van der Ent, W.; Brisac, A.; Irondelle, M.; Aubert, S.; Dirksen, U.; Bouvier, C.; de Pinieux, G.; Snaar-Jagalska, E.; et al. Cell-to-cell heterogeneity of EWSR1-FLI1 activity determines proliferation/migration choices in Ewing sarcoma cells. Oncogene 2017, 36, 3505–3514. [Google Scholar] [CrossRef]
- Li, M.; Chen, C. Regulation of Metastasis in Ewing Sarcoma. Cancers 2022, 14, 4902. [Google Scholar] [CrossRef]
- Bierbaumer, L.; Katschnig, A.M.; Radic-Sarikas, B.; Kauer, M.O.; Petro, J.A.; Hogler, S.; Gurnhofer, E.; Pedot, G.; Schafer, B.W.; Schwentner, R.; et al. YAP/TAZ inhibition reduces metastatic potential of Ewing sarcoma cells. Oncogenesis 2021, 10, 2. [Google Scholar] [CrossRef]
- Gonzalez, I.; Vicent, S.; de Alava, E.; Lecanda, F. EWS/FLI-1 oncoprotein subtypes impose different requirements for transformation and metastatic activity in a murine model. J. Mol. Med. 2007, 85, 1015–1029. [Google Scholar] [CrossRef] [PubMed]
- Chaturvedi, A.; Hoffman, L.M.; Jensen, C.C.; Lin, Y.C.; Grossmann, A.H.; Randall, R.L.; Lessnick, S.L.; Welm, A.L.; Beckerle, M.C. Molecular dissection of the mechanism by which EWS/FLI expression compromises actin cytoskeletal integrity and cell adhesion in Ewing sarcoma. Mol. Biol. Cell 2014, 25, 2695–2709. [Google Scholar] [CrossRef] [PubMed]
- Hauer, K.; Calzada-Wack, J.; Steiger, K.; Grunewald, T.G.; Baumhoer, D.; Plehm, S.; Buch, T.; Prazeres da Costa, O.; Esposito, I.; Burdach, S.; et al. DKK2 mediates osteolysis, invasiveness, and metastatic spread in Ewing sarcoma. Cancer Res. 2013, 73, 967–977. [Google Scholar] [CrossRef]
- Sainz-Jaspeado, M.; Lagares-Tena, L.; Lasheras, J.; Navid, F.; Rodriguez-Galindo, C.; Mateo-Lozano, S.; Notario, V.; Sanjuan, X.; Garcia Del Muro, X.; Fabra, A.; et al. Caveolin-1 modulates the ability of Ewing’s sarcoma to metastasize. Mol. Cancer Res. 2010, 8, 1489–1500. [Google Scholar] [CrossRef]
- Lagares-Tena, L.; Garcia-Monclus, S.; Lopez-Alemany, R.; Almacellas-Rabaiget, O.; Huertas-Martinez, J.; Sainz-Jaspeado, M.; Mateo-Lozano, S.; Rodriguez-Galindo, C.; Rello-Varona, S.; Herrero-Martin, D.; et al. Caveolin-1 promotes Ewing sarcoma metastasis regulating MMP-9 expression through MAPK/ERK pathway. Oncotarget 2016, 7, 56889–56903. [Google Scholar] [CrossRef] [PubMed]
- Richter, G.H.; Fasan, A.; Hauer, K.; Grunewald, T.G.; Berns, C.; Rossler, S.; Naumann, I.; Staege, M.S.; Fulda, S.; Esposito, I.; et al. G-Protein coupled receptor 64 promotes invasiveness and metastasis in Ewing sarcomas through PGF and MMP1. J. Pathol. 2013, 230, 70–81. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Aryee, D.N.; Niedan, S.; Kauer, M.; Schwentner, R.; Bennani-Baiti, I.M.; Ban, J.; Muehlbacher, K.; Kreppel, M.; Walker, R.L.; Meltzer, P.; et al. Hypoxia modulates EWS-FLI1 transcriptional signature and enhances the malignant properties of Ewing’s sarcoma cells in vitro. Cancer Res. 2010, 70, 4015–4023. [Google Scholar] [CrossRef]
- DiMeo, T.A.; Anderson, K.; Phadke, P.; Fan, C.; Perou, C.M.; Naber, S.; Kuperwasser, C. A novel lung metastasis signature links Wnt signaling with cancer cell self-renewal and epithelial-mesenchymal transition in basal-like breast cancer. Cancer Res. 2009, 69, 5364–5373. [Google Scholar] [CrossRef]
- Yu, F.; Yu, C.; Li, F.; Zuo, Y.; Wang, Y.; Yao, L.; Wu, C.; Wang, C.; Ye, L. Wnt/beta-catenin signaling in cancers and targeted therapies. Signal Transduct. Target. Ther. 2021, 6, 307. [Google Scholar] [CrossRef] [PubMed]
- Pfister, A.S.; Tanneberger, K.; Schambony, A.; Behrens, J. Amer2 protein is a novel negative regulator of Wnt/beta-catenin signaling involved in neuroectodermal patterning. J. Biol. Chem. 2012, 287, 1734–1741. [Google Scholar] [CrossRef] [PubMed]
- Deneen, B.; Denny, C.T. Loss of p16 pathways stabilizes EWS/FLI1 expression and complements EWS/FLI1 mediated transformation. Oncogene 2001, 20, 6731–6741. [Google Scholar] [CrossRef] [PubMed]
- Sohn, E.J.; Li, H.; Reidy, K.; Beers, L.F.; Christensen, B.L.; Lee, S.B. EWS/FLI1 oncogene activates caspase 3 transcription and triggers apoptosis in vivo. Cancer Res. 2010, 70, 1154–1163. [Google Scholar] [CrossRef]
- Su, X.A.; Ma, D.; Parsons, J.V.; Replogle, J.M.; Amatruda, J.F.; Whittaker, C.A.; Stegmaier, K.; Amon, A. RAD21 is a driver of chromosome 8 gain in Ewing sarcoma to mitigate replication stress. Genes Dev. 2021, 35, 556–572. [Google Scholar] [CrossRef]
- Ewing, J. Classics in oncology. Diffuse endothelioma of bone. James Ewing. Proceedings of the New York Pathological Society, 1921. CA Cancer J. Clin. 1972, 22, 95–98. [Google Scholar] [CrossRef]
- Garnett, M.J.; Edelman, E.J.; Heidorn, S.J.; Greenman, C.D.; Dastur, A.; Lau, K.W.; Greninger, P.; Thompson, I.R.; Luo, X.; Soares, J.; et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature 2012, 483, 570–575. [Google Scholar] [CrossRef]
- Brenner, J.C.; Feng, F.Y.; Han, S.; Patel, S.; Goyal, S.V.; Bou-Maroun, L.M.; Liu, M.; Lonigro, R.; Prensner, J.R.; Tomlins, S.A.; et al. PARP-1 inhibition as a targeted strategy to treat Ewing’s sarcoma. Cancer Res. 2012, 72, 1608–1613. [Google Scholar] [CrossRef]
- Brohl, A.S.; Patidar, R.; Turner, C.E.; Wen, X.; Song, Y.K.; Wei, J.S.; Calzone, K.A.; Khan, J. Frequent inactivating germline mutations in DNA repair genes in patients with Ewing sarcoma. Genet Med. 2017, 19, 955–958. [Google Scholar] [CrossRef]
- Gonzalez-Martin, A.; Pothuri, B.; Vergote, I.; DePont Christensen, R.; Graybill, W.; Mirza, M.R.; McCormick, C.; Lorusso, D.; Hoskins, P.; Freyer, G.; et al. Niraparib in Patients with Newly Diagnosed Advanced Ovarian Cancer. N. Engl. J. Med. 2019, 381, 2391–2402. [Google Scholar] [CrossRef]
- Banerjee, S.; Gonzalez-Martin, A.; Harter, P.; Lorusso, D.; Moore, K.N.; Oaknin, A.; Ray-Coquard, I. First-line PARP inhibitors in ovarian cancer: Summary of an ESMO Open—Cancer Horizons round-table discussion. ESMO Open 2020, 5, e001110. [Google Scholar] [CrossRef]
- O’Shaughnessy, J.; Osborne, C.; Pippen, J.E.; Yoffe, M.; Patt, D.; Rocha, C.; Koo, I.C.; Sherman, B.M.; Bradley, C. Iniparib plus chemotherapy in metastatic triple-negative breast cancer. N. Engl. J. Med. 2011, 364, 205–214. [Google Scholar] [CrossRef] [PubMed]
- Bryant, H.E.; Schultz, N.; Thomas, H.D.; Parker, K.M.; Flower, D.; Lopez, E.; Kyle, S.; Meuth, M.; Curtin, N.J.; Helleday, T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 2005, 434, 913–917. [Google Scholar] [CrossRef]
- Spahn, L.; Siligan, C.; Bachmaier, R.; Schmid, J.A.; Aryee, D.N.; Kovar, H. Homotypic and heterotypic interactions of EWS, FLI1 and their oncogenic fusion protein. Oncogene 2003, 22, 6819–6829. [Google Scholar] [CrossRef]
- Gorthi, A.; Bishop, A.J.R. Ewing sarcoma fusion oncogene: At the crossroads of transcription and DNA damage response. Mol. Cell. Oncol. 2018, 5, e1465014. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.G.; Kim, N.; Kim, S.M.; Park, I.B.; Kim, H.; Kim, S.; Kim, B.G.; Hwang, J.M.; Baek, I.J.; Gartner, A.; et al. Ewing sarcoma protein promotes dissociation of poly(ADP-ribose) polymerase 1 from chromatin. EMBO Rep. 2020, 21, e48676. [Google Scholar] [CrossRef] [PubMed]
- Paronetto, M.P.; Minana, B.; Valcarcel, J. The Ewing sarcoma protein regulates DNA damage-induced alternative splicing. Mol. Cell 2011, 43, 353–368. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, J.C.; Cech, T.R.; Parker, R.R. Biochemical Properties and Biological Functions of FET Proteins. Annu. Rev. Biochem. 2015, 84, 355–379. [Google Scholar] [CrossRef]
- Altmeyer, M.; Neelsen, K.J.; Teloni, F.; Pozdnyakova, I.; Pellegrino, S.; Grofte, M.; Rask, M.D.; Streicher, W.; Jungmichel, S.; Nielsen, M.L.; et al. Liquid demixing of intrinsically disordered proteins is seeded by poly(ADP-ribose). Nat. Commun. 2015, 6, 8088. [Google Scholar] [CrossRef]
- Menon, S.; Breese, M.R.; Lin, Y.P.; Allegakoen, H.; Perati, S.; Heslin, A.; Horlbeck, M.A.; Weissman, J.; Sweet-Cordero, E.A.; Bivona, T.G.; et al. FET fusion oncoproteins disrupt physiologic DNA repair networks in cancer. bioRxiv 2023. [Google Scholar] [CrossRef]
- Erkizan, H.V.; Kong, Y.; Merchant, M.; Schlottmann, S.; Barber-Rotenberg, J.S.; Yuan, L.; Abaan, O.D.; Chou, T.H.; Dakshanamurthy, S.; Brown, M.L.; et al. A small molecule blocking oncogenic protein EWS-FLI1 interaction with RNA helicase A inhibits growth of Ewing’s sarcoma. Nat. Med. 2009, 15, 750–756. [Google Scholar] [CrossRef]
- Barber-Rotenberg, J.S.; Selvanathan, S.P.; Kong, Y.; Erkizan, H.V.; Snyder, T.M.; Hong, S.P.; Kobs, C.L.; South, N.L.; Summer, S.; Monroe, P.J.; et al. Single enantiomer of YK-4-279 demonstrates specificity in targeting the oncogene EWS-FLI1. Oncotarget 2012, 3, 172–182. [Google Scholar] [CrossRef] [PubMed]
- Zollner, S.K.; Selvanathan, S.P.; Graham, G.T.; Commins, R.M.T.; Hong, S.H.; Moseley, E.; Parks, S.; Haladyna, J.N.; Erkizan, H.V.; Dirksen, U.; et al. Inhibition of the oncogenic fusion protein EWS-FLI1 causes G2-M cell cycle arrest and enhanced vincristine sensitivity in Ewing’s sarcoma. Sci. Signal. 2017, 10, eaam8429. [Google Scholar] [CrossRef] [PubMed]
- Lamhamedi-Cherradi, S.E.; Menegaz, B.A.; Ramamoorthy, V.; Aiyer, R.A.; Maywald, R.L.; Buford, A.S.; Doolittle, D.K.; Culotta, K.S.; O’Dorisio, J.E.; Ludwig, J.A. An Oral Formulation of YK-4-279: Preclinical Efficacy and Acquired Resistance Patterns in Ewing Sarcoma. Mol. Cancer Ther. 2015, 14, 1591–1604. [Google Scholar] [CrossRef]
- Conn, E.; Hour, S.; Allegakoen, D.; Graham, G.; Petro, J.; Kouassi-Brou, M.; Hong, S.H.; Selvanathan, S.; Celik, H.; Toretsky, J.; et al. Development of an Ewing sarcoma cell line with resistance to EWSFLI1 inhibitor YK4279. Mol. Med. Rep. 2020, 21, 1667–1675. [Google Scholar] [CrossRef]
- Ludwig, J.; Federman, N.; Anderson, P.; Macy, M.; Davis, L.; Riedel, R.; Muscal, J.; Daw, N.; Ratan, R.; Toretsky, J. 1620O Phase I study of TK216, a novel anti-ETS agent for Ewing sarcoma. Ann. Oncol. 2020, 31, S972. [Google Scholar] [CrossRef]
- Xue, J.; Li, S.; Shi, P.; Chen, M.; Yu, S.; Hong, S.; Li, Y.; Liu, R.; Xiao, H. The ETS Inhibitor YK-4-279 Suppresses Thyroid Cancer Progression Independent of TERT Promoter Mutations. Front. Oncol. 2021, 11, 649323. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Zhai, Y.; La, J.; Lui, J.W.; Moore, S.P.G.; Little, E.C.; Xiao, S.; Haresi, A.J.; Brem, C.; Bhawan, J.; et al. Targeting Pan-ETS Factors Inhibits Melanoma Progression. Cancer Res. 2021, 81, 2071–2085. [Google Scholar] [CrossRef]
- Povedano, J.M.; Li, V.; Lake, K.E.; Bai, X.; Rallabandi, R.; Kim, J.; Xie, Y.; De Brabander, J.K.; McFadden, D.G. TK216 targets microtubules in Ewing sarcoma cells. Cell Chem. Biol. 2022, 29, 1325–1332.e1324. [Google Scholar] [CrossRef]
- Grohar, P.J.; Glod, J.; Peer, C.J.; Sissung, T.M.; Arnaldez, F.I.; Long, L.; Figg, W.D.; Whitcomb, P.; Helman, L.J.; Widemann, B.C. A phase I/II trial and pharmacokinetic study of mithramycin in children and adults with refractory Ewing sarcoma and EWS-FLI1 fusion transcript. Cancer Chemother. Pharmacol. 2017, 80, 645–652. [Google Scholar] [CrossRef]
- Mitra, P.; Eckenrode, J.M.; Mandal, A.; Jha, A.K.; Salem, S.M.; Leggas, M.; Rohr, J. Development of Mithramycin Analogues with Increased Selectivity toward ETS Transcription Factor Expressing Cancers. J. Med. Chem. 2018, 61, 8001–8016. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.Y.; Damron, T.A.; Oest, M.E.; Horton, J.A. Mithramycin A Radiosensitizes EWS:Fli1(+) Ewing Sarcoma Cells by Inhibiting Double Strand Break Repair. Int. J. Radiat. Oncol. Biol. Phys. 2021, 109, 1454–1471. [Google Scholar] [CrossRef] [PubMed]
- Choy, E.; Butrynski, J.E.; Harmon, D.C.; Morgan, J.A.; George, S.; Wagner, A.J.; D’Adamo, D.; Cote, G.M.; Flamand, Y.; Benes, C.H.; et al. Phase II study of olaparib in patients with refractory Ewing sarcoma following failure of standard chemotherapy. BMC Cancer 2014, 14, 813. [Google Scholar] [CrossRef]
- Iniguez, A.B.; Stolte, B.; Wang, E.J.; Conway, A.S.; Alexe, G.; Dharia, N.V.; Kwiatkowski, N.; Zhang, T.; Abraham, B.J.; Mora, J.; et al. EWS/FLI Confers Tumor Cell Synthetic Lethality to CDK12 Inhibition in Ewing Sarcoma. Cancer Cell 2018, 33, 202–216.e206. [Google Scholar] [CrossRef]
- Schmidt, O.; Nehls, N.; Prexler, C.; von Heyking, K.; Groll, T.; Pardon, K.; Garcia, H.D.; Hensel, T.; Gurgen, D.; Henssen, A.G.; et al. Class I histone deacetylases (HDAC) critically contribute to Ewing sarcoma pathogenesis. J. Exp. Clin. Cancer Res. 2021, 40, 322. [Google Scholar] [CrossRef] [PubMed]
- Pishas, K.I.; Drenberg, C.D.; Taslim, C.; Theisen, E.R.; Johnson, K.M.; Saund, R.S.; Pop, I.L.; Crompton, B.D.; Lawlor, E.R.; Tirode, F.; et al. Therapeutic Targeting of KDM1A/LSD1 in Ewing Sarcoma with SP-2509 Engages the Endoplasmic Reticulum Stress Response. Mol. Cancer Ther. 2018, 17, 1902–1916. [Google Scholar] [CrossRef]
- Garcia-Dominguez, D.J.; Hontecillas-Prieto, L.; Rodriguez-Nunez, P.; Pascual-Pasto, G.; Vila-Ubach, M.; Garcia-Mejias, R.; Robles, M.J.; Tirado, O.M.; Mora, J.; Carcaboso, A.M.; et al. The combination of epigenetic drugs SAHA and HCI-2509 synergistically inhibits EWS-FLI1 and tumor growth in Ewing sarcoma. Oncotarget 2018, 9, 31397–31410. [Google Scholar] [CrossRef]
- Welch, D.; Kahen, E.; Fridley, B.; Brohl, A.S.; Cubitt, C.L.; Reed, D.R. Small molecule inhibition of lysine-specific demethylase 1 (LSD1) and histone deacetylase (HDAC) alone and in combination in Ewing sarcoma cell lines. PLoS ONE 2019, 14, e0222228. [Google Scholar] [CrossRef]
- Reed, D.R.; Chawla, S.P.; Setty, B.; Mascarenhas, L.; Meyers, P.A.; Metts, J.; Harrison, D.J.; Loeb, D.; Crompton, B.D.; Wages, D.S.; et al. Phase 1 trial of seclidemstat (SP-2577) in patients with relapsed/refractory Ewing sarcoma. J. Clin. Oncol. 2021, 39, 11514. [Google Scholar] [CrossRef]
- Jaboin, J.; Wild, J.; Hamidi, H.; Khanna, C.; Kim, C.J.; Robey, R.; Bates, S.E.; Thiele, C.J. MS-27-275, an inhibitor of histone deacetylase, has marked in vitro and in vivo antitumor activity against pediatric solid tumors. Cancer Res. 2002, 62, 6108–6115. [Google Scholar]
- Souza, B.K.; da Costa Lopez, P.L.; Menegotto, P.R.; Vieira, I.A.; Kersting, N.; Abujamra, A.L.; Brunetto, A.T.; Brunetto, A.L.; Gregianin, L.; de Farias, C.B.; et al. Targeting Histone Deacetylase Activity to Arrest Cell Growth and Promote Neural Differentiation in Ewing Sarcoma. Mol. Neurobiol. 2018, 55, 7242–7258. [Google Scholar] [CrossRef] [PubMed]
- Pattenden, S.G.; Simon, J.M.; Wali, A.; Jayakody, C.N.; Troutman, J.; McFadden, A.W.; Wooten, J.; Wood, C.C.; Frye, S.V.; Janzen, W.P.; et al. High-throughput small molecule screen identifies inhibitors of aberrant chromatin accessibility. Proc. Natl. Acad. Sci. USA 2016, 113, 3018–3023. [Google Scholar] [CrossRef] [PubMed]
- Hontecillas-Prieto, L.; Flores-Campos, R.; Silver, A.; de Alava, E.; Hajji, N.; Garcia-Dominguez, D.J. Synergistic Enhancement of Cancer Therapy Using HDAC Inhibitors: Opportunity for Clinical Trials. Front. Genet. 2020, 11, 578011. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, X.; Fan, G.; Fukushi, J.; Matsumoto, Y.; Iwamoto, Y.; Zhu, Y. Impairment of p53 acetylation by EWS-Fli1 chimeric protein in Ewing family tumors. Cancer Lett. 2012, 320, 14–22. [Google Scholar] [CrossRef]
- Grohar, P.J.; Griffin, L.B.; Yeung, C.; Chen, Q.R.; Pommier, Y.; Khanna, C.; Khan, J.; Helman, L.J. Ecteinascidin 743 interferes with the activity of EWS-FLI1 in Ewing sarcoma cells. Neoplasia 2011, 13, 145–153. [Google Scholar] [CrossRef]
- Baruchel, S.; Pappo, A.; Krailo, M.; Baker, K.S.; Wu, B.; Villaluna, D.; Lee-Scott, M.; Adamson, P.C.; Blaney, S.M. A phase 2 trial of trabectedin in children with recurrent rhabdomyosarcoma, Ewing sarcoma and non-rhabdomyosarcoma soft tissue sarcomas: A report from the Children’s Oncology Group. Eur. J. Cancer 2012, 48, 579–585. [Google Scholar] [CrossRef]
- Grohar, P.J.; Segars, L.E.; Yeung, C.; Pommier, Y.; D’Incalci, M.; Mendoza, A.; Helman, L.J. Dual targeting of EWS-FLI1 activity and the associated DNA damage response with trabectedin and SN38 synergistically inhibits Ewing sarcoma cell growth. Clin. Cancer Res. 2014, 20, 1190–1203. [Google Scholar] [CrossRef]
- Heisey, D.A.R.; Jacob, S.; Lochmann, T.L.; Kurupi, R.; Ghotra, M.S.; Calbert, M.L.; Shende, M.; Maves, Y.K.; Koblinski, J.E.; Dozmorov, M.G.; et al. Pharmaceutical Interference of the EWS-FLI1-driven Transcriptome By Cotargeting H3K27ac and RNA Polymerase Activity in Ewing Sarcoma. Mol. Cancer Ther. 2021, 20, 1868–1879. [Google Scholar] [CrossRef]
- Li, M.; Yang, L.; Chan, A.K.N.; Pokharel, S.P.; Liu, Q.; Mattson, N.; Xu, X.; Chang, W.H.; Miyashita, K.; Singh, P.; et al. Epigenetic Control of Translation Checkpoint and Tumor Progression via RUVBL1-EEF1A1 Axis. Adv. Sci. 2023, 10, e2206584. [Google Scholar] [CrossRef]
- Morales, E.; Olson, M.; Iglesias, F.; Dahiya, S.; Luetkens, T.; Atanackovic, D. Role of immunotherapy in Ewing sarcoma. J. Immunother. Cancer 2020, 8, e000653. [Google Scholar] [CrossRef]
- Thanindratarn, P.; Dean, D.C.; Nelson, S.D.; Hornicek, F.J.; Duan, Z. Advances in immune checkpoint inhibitors for bone sarcoma therapy. J. Bone Oncol. 2019, 15, 100221. [Google Scholar] [CrossRef]
- Schirmer, D.; Grunewald, T.G.; Klar, R.; Schmidt, O.; Wohlleber, D.; Rubio, R.A.; Uckert, W.; Thiel, U.; Bohne, F.; Busch, D.H.; et al. Transgenic antigen-specific, HLA-A*02:01-allo-restricted cytotoxic T cells recognize tumor-associated target antigen STEAP1 with high specificity. Oncoimmunology 2016, 5, e1175795. [Google Scholar] [CrossRef] [PubMed]
- Thiel, U.; Schober, S.J.; Einspieler, I.; Kirschner, A.; Thiede, M.; Schirmer, D.; Gall, K.; Blaeschke, F.; Schmidt, O.; Jabar, S.; et al. Ewing sarcoma partial regression without GvHD by chondromodulin-I/HLA-A*02:01-specific allorestricted T cell receptor transgenic T cells. Oncoimmunology 2017, 6, e1312239. [Google Scholar] [CrossRef] [PubMed]
- Schober, S.J.; Thiede, M.; Gassmann, H.; Prexler, C.; Xue, B.; Schirmer, D.; Wohlleber, D.; Stein, S.; Grunewald, T.G.P.; Busch, D.H.; et al. MHC Class I-Restricted TCR-Transgenic CD4(+) T Cells Against STEAP1 Mediate Local Tumor Control of Ewing Sarcoma In Vivo. Cells 2020, 9, 1581. [Google Scholar] [CrossRef] [PubMed]
- Evans, C.H.; Liu, F.; Porter, R.M.; O’Sullivan, R.P.; Merghoub, T.; Lunsford, E.P.; Robichaud, K.; Van Valen, F.; Lessnick, S.L.; Gebhardt, M.C.; et al. EWS-FLI-1-targeted cytotoxic T-cell killing of multiple tumor types belonging to the Ewing sarcoma family of tumors. Clin. Cancer Res. 2012, 18, 5341–5351. [Google Scholar] [CrossRef]
- Blaeschke, F.; Thiel, U.; Kirschner, A.; Thiede, M.; Rubio, R.A.; Schirmer, D.; Kirchner, T.; Richter, G.H.S.; Mall, S.; Klar, R.; et al. Human HLA-A*02:01/CHM1+ allo-restricted T cell receptor transgenic CD8+ T cells specifically inhibit Ewing sarcoma growth in vitro and in vivo. Oncotarget 2016, 7, 43267–43280. [Google Scholar] [CrossRef]
- Thiel, U.; Pirson, S.; Muller-Spahn, C.; Conrad, H.; Busch, D.H.; Bernhard, H.; Burdach, S.; Richter, G.H. Specific recognition and inhibition of Ewing tumour growth by antigen-specific allo-restricted cytotoxic T cells. Br. J. Cancer 2011, 104, 948–956. [Google Scholar] [CrossRef][Green Version]
- Mahlendorf, D.E.; Staege, M.S. Characterization of Ewing sarcoma associated cancer/testis antigens. Cancer Biol. Ther. 2013, 14, 254–261. [Google Scholar] [CrossRef]
- Rodeberg, D.A.; Nuss, R.A.; Elsawa, S.F.; Celis, E. Recognition of six-transmembrane epithelial antigen of the prostate-expressing tumor cells by peptide antigen-induced cytotoxic T lymphocytes. Clin. Cancer Res. 2005, 11, 4545–4552. [Google Scholar] [CrossRef]
- Kirschner, A.; Thiede, M.; Grunewald, T.G.; Alba Rubio, R.; Richter, G.H.; Kirchner, T.; Busch, D.H.; Burdach, S.; Thiel, U. Pappalysin-1 T cell receptor transgenic allo-restricted T cells kill Ewing sarcoma in vitro and in vivo. Oncoimmunology 2017, 6, e1273301. [Google Scholar] [CrossRef]
- Rodeberg, D.A.; Nuss, R.A.; Elsawa, S.F.; Erskine, C.L.; Celis, E. Generation of tumoricidal PAX3 peptide antigen specific cytotoxic T lymphocytes. Int. J. Cancer 2006, 119, 126–132. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.F.; Helman, L.J.; Yeung, C.; Bera, T.K.; Lee, B.; Pastan, I. XAGE-1, a new gene that is frequently expressed in Ewing’s sarcoma. Cancer Res. 2000, 60, 4752–4755. [Google Scholar] [PubMed]
- Zendman, A.J.; Van Kraats, A.A.; Weidle, U.H.; Ruiter, D.J.; Van Muijen, G.N. The XAGE family of cancer/testis-associated genes: Alignment and expression profile in normal tissues, melanoma lesions and Ewing’s sarcoma. Int. J. Cancer 2002, 99, 361–369. [Google Scholar] [CrossRef] [PubMed]
- Altvater, B.; Kailayangiri, S.; Theimann, N.; Ahlmann, M.; Farwick, N.; Chen, C.; Pscherer, S.; Neumann, I.; Mrachatz, G.; Hansmeier, A.; et al. Common Ewing sarcoma-associated antigens fail to induce natural T cell responses in both patients and healthy individuals. Cancer Immunol. Immunother. 2014, 63, 1047–1060. [Google Scholar] [CrossRef] [PubMed]
- Berghuis, D.; de Hooge, A.S.; Santos, S.J.; Horst, D.; Wiertz, E.J.; van Eggermond, M.C.; van den Elsen, P.J.; Taminiau, A.H.; Ottaviano, L.; Schaefer, K.L.; et al. Reduced human leukocyte antigen expression in advanced-stage Ewing sarcoma: Implications for immune recognition. J. Pathol. 2009, 218, 222–231. [Google Scholar] [CrossRef]
- Englisch, A.; Altvater, B.; Kailayangiri, S.; Hartmann, W.; Rossig, C. VEGFR2 as a target for CAR T cell therapy of Ewing sarcoma. Pediatr. Blood Cancer 2020, 67, e28313. [Google Scholar] [CrossRef]
- Challita-Eid, P.M.; Morrison, K.; Etessami, S.; An, Z.; Morrison, K.J.; Perez-Villar, J.J.; Raitano, A.B.; Jia, X.C.; Gudas, J.M.; Kanner, S.B.; et al. Monoclonal antibodies to six-transmembrane epithelial antigen of the prostate-1 inhibit intercellular communication in vitro and growth of human tumor xenografts in vivo. Cancer Res. 2007, 67, 5798–5805. [Google Scholar] [CrossRef]
- Huang, X.; Park, H.; Greene, J.; Pao, J.; Mulvey, E.; Zhou, S.X.; Albert, C.M.; Moy, F.; Sachdev, D.; Yee, D.; et al. IGF1R- and ROR1-Specific CAR T Cells as a Potential Therapy for High Risk Sarcomas. PLoS ONE 2015, 10, e0133152. [Google Scholar] [CrossRef]
- Schirmer, D.; Storz, I.; Wisskirchen, K.; Feederle, R.; Schmidt, O.; Abken, H.; Protzer, U.; Burdach, S.; Richter, G.H.S. GPR64-specific CAR-transgenic T cells selectively kill Ewing sarcoma in vivo. Oncol. Res. Treat. 2018, 41, 130–131. [Google Scholar] [CrossRef]
- Town, J.; Pais, H.; Harrison, S.; Stead, L.F.; Bataille, C.; Bunjobpol, W.; Zhang, J.; Rabbitts, T.H. Exploring the surfaceome of Ewing sarcoma identifies a new and unique therapeutic target. Proc. Natl. Acad. Sci. USA 2016, 113, 3603–3608. [Google Scholar] [CrossRef]
- Kailayangiri, S.; Altvater, B.; Lesch, S.; Balbach, S.; Gottlich, C.; Kuhnemundt, J.; Mikesch, J.H.; Schelhaas, S.; Jamitzky, S.; Meltzer, J.; et al. EZH2 Inhibition in Ewing Sarcoma Upregulates G(D2) Expression for Targeting with Gene-Modified T Cells. Mol. Ther. 2019, 27, 933–946. [Google Scholar] [CrossRef] [PubMed]
- Schober, K.; Muller, T.R.; Gokmen, F.; Grassmann, S.; Effenberger, M.; Poltorak, M.; Stemberger, C.; Schumann, K.; Roth, T.L.; Marson, A.; et al. Orthotopic replacement of T-cell receptor alpha- and beta-chains with preservation of near-physiological T-cell function. Nat. Biomed. Eng. 2019, 3, 974–984. [Google Scholar] [CrossRef] [PubMed]
- Xue, B.; von Heyking, K.; Gassmann, H.; Poorebrahim, M.; Thiede, M.; Schober, K.; Mautner, J.; Hauer, J.; Ruland, J.; Busch, D.H.; et al. T Cells Directed against the Metastatic Driver Chondromodulin-1 in Ewing Sarcoma: Comparative Engineering with CRISPR/Cas9 vs. Retroviral Gene Transfer for Adoptive Transfer. Cancers 2022, 14, 5485. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; Su, J.; Sun, R.; Sun, Y.; Wang, Y.; Dong, Y.; Shi, B.; Jiang, H.; Li, Z. Coexpression of IL7 and CCL21 Increases Efficacy of CAR-T Cells in Solid Tumors without Requiring Preconditioned Lymphodepletion. Clin. Cancer Res. 2020, 26, 5494–5505. [Google Scholar] [CrossRef]
- Ji, F.; Zhang, F.; Zhang, M.; Long, K.; Xia, M.; Lu, F.; Li, E.; Chen, J.; Li, J.; Chen, Z.; et al. Targeting the DNA damage response enhances CD70 CAR-T cell therapy for renal carcinoma by activating the cGAS-STING pathway. J. Hematol. Oncol. 2021, 14, 152. [Google Scholar] [CrossRef]
- Lee, Y.G.; Yang, N.; Chun, I.; Porazzi, P.; Carturan, A.; Paruzzo, L.; Sauter, C.T.; Guruprasad, P.; Pajarillo, R.; Ruella, M. Apoptosis: A Janus bifrons in T-cell immunotherapy. J. Immunother. Cancer 2023, 11, e005967. [Google Scholar] [CrossRef]
- Sun, R.; Luo, H.; Su, J.; Di, S.; Zhou, M.; Shi, B.; Sun, Y.; Du, G.; Zhang, H.; Jiang, H.; et al. Olaparib Suppresses MDSC Recruitment via SDF1alpha/CXCR4 Axis to Improve the Anti-tumor Efficacy of CAR-T Cells on Breast Cancer in Mice. Mol. Ther. 2021, 29, 60–74. [Google Scholar] [CrossRef]
- Sheng, W.; LaFleur, M.W.; Nguyen, T.H.; Chen, S.; Chakravarthy, A.; Conway, J.R.; Li, Y.; Chen, H.; Yang, H.; Hsu, P.H.; et al. LSD1 Ablation Stimulates Anti-tumor Immunity and Enables Checkpoint Blockade. Cell 2018, 174, 549–563.e519. [Google Scholar] [CrossRef]
- Robinson, J.T.; Thorvaldsdottir, H.; Turner, D.; Mesirov, J.P. igv.js: An embeddable JavaScript implementation of the Integrative Genomics Viewer (IGV). Bioinformatics 2023, 39, btac830. [Google Scholar] [CrossRef]
- Shen, W.; Song, Z.; Zhong, X.; Huang, M.; Shen, D.; Gao, P.; Qian, X.; Wang, M.; He, X.; Wang, T.; et al. Sangerbox: A comprehensive, interaction-friendly clinical bioinformatics analysis platform. Imeta 2022, 1, e36. [Google Scholar] [CrossRef]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).