Simple Summary
The gut microbiota is a commensal microbiota living in the human intestine. Its status and composition have a profound impact on human antitumor immunity. Gut microbiota and its metabolites can influence tumor immune escape through immune cells and inflammatory factors, changing the patient’s response to immunotherapy. Protecting normal gut microbiota or optimizing its composition can improve the effects of tumor immunotherapy and bring new hope for cancer treatment.
Abstract
The gut microbiota is a large symbiotic community of anaerobic and facultative aerobic bacteria inhabiting the human intestinal tract, and its activities significantly affect human health. Increasing evidence has suggested that the gut microbiome plays an important role in tumor-related immune regulation. In the tumor microenvironment (TME), the gut microbiome and its metabolites affect the differentiation and function of immune cells regulating the immune evasion of tumors. The gut microbiome can indirectly influence individual responses to various classical tumor immunotherapies, including immune checkpoint inhibitor therapy and adoptive immunotherapy. Microbial regulation through antibiotics, prebiotics, and fecal microbiota transplantation (FMT) optimize the composition of the gut microbiome, improving the efficacy of immunotherapy and bringing a new perspective and hope for tumor treatment.
1. Introduction
The gut microbiota comprises numerous anaerobic and facultative aerobic bacteria living in the human gastrointestinal tract [1]. A stable gut microbiota is indispensable for maintaining the normal physiological function of the human body, directly or indirectly involved in the body’s digestion, metabolism, immunity, and inflammation, etc. [2]. Abnormal changes in gut microbiota have been shown to induce or promote a variety of diseases and pathological conditions, including cancer [3,4,5,6]. With the in-depth study of the microbiota, researchers have proposed several ways through which gut microbiota affects distant organs, such as the Microbiota–Gut–Brain Axis [7], the Microbiota–Gut–Bone Axis [8], and the Microbiota–Gut–Liver Axis [9]. These findings have enriched our understanding of the mechanisms by which gut microbiota affects disease states. The important role of gut microbiota in the initiation and progression of cancer has also been gradually discovered. The gut microbiota and its metabolites were found to promote the occurrence and development of colon cancer and hepatocellular carcinoma (HCC) [10,11], and the gut microbiota may increase the risk of a poor prognosis for breast cancer patients by affecting estrogen metabolism [12].
The imbalance of gut microbiota is mainly the decrease of good bacteria and the increase of bad bacteria. Therefore, to change this dysregulation, increase the proportion of anti-tumor beneficial bacteria and restore the intestinal beneficial bacteria ecology, such as prebiotics and FMT [13,14], can fight against the occurrence and development of tumors. Diet therapy is also a highly concerned treatment. High cholesterol diet can induce dysbiosis of the microbiota, leading to the progression of hepatocellular carcinoma (HCC). Reducing dietary cholesterol intake in HCC patients may be beneficial to the treatment of HCC [15]. High dietary fiber diet is also beneficial to the gut microbiota ecology of cancer patients. Dietary fiber supplementation can improve the dysregulation of intestinal flora and improve the effect of tumor immunotherapy [16].
With the development of immune surveillance theory, tumor immune escape has been recognized as an indispensable mechanism of tumorigenesis [17,18]. Tumor immune escape refers to the process in which tumor cells evade the surveillance of the immune system, avoiding the clearance. Tumor immune escape is achieved through the interaction between three factors: the tumor microenvironment (TME), the tumor cells, and the immune status of the body [19,20,21]. Gut microbiota is closely related to the human immune system. It can affect tumor immune escape by its own structure and metabolites, which is an important reason for tumor progression and poor prognosis [22,23].
The goal of tumor immunotherapy is to restore antitumor immunity to control and eliminate tumors. Tumor immunotherapy includes cytokine therapy, adoptive immune cell therapy, and immune checkpoint inhibitor therapy, etc. [24]. Since gut microbiota affects the efficacy of tumor immunotherapy by influencing the function of immune cells and the release of cytokines and chemokines, microbiota regulatory strategies, such as fecal microbiota transplantation (FMT), antibiotic cocktails, and prebiotics, are potential adjuvants to tumor immunotherapy [25,26,27].
2. Gut Microbiota Influences Tumor Immune Escape through Tumor Microenvironment
The tumor microenvironment (TME) contains components that inhibit/promote the proliferation and invasion of tumor cells [28]. Treg cells, TAMs (tumor-associated macrophages), MDSCs (myeloid-derived suppressor cells), immunosuppressive molecules, and immunoregulatory enzymes promote tumor growth, proliferation, and metastasis, while immune effector cells and immune effector molecules inhibit them. In the TME, the immunosuppressive network composed of immunosuppressive cells and their secreted immunosuppressive molecules is the most important reason for tumor immune escape and resistance to antitumor therapy [29]. Gut microbiota can influence the status of key components in the TME, affecting tumor immune escape.
Inflammation plays an important role in tumor immune evasion triggered by gut microbiota. As early as 1863, Virchow et al. observed that malignancy tends to occur at sites of chronic inflammation [30], suggesting a correlation between inflammation and tumorigenesis. Subsequently, epidemiological data strongly suggested that chronic inflammation is associated with an increased risk of cancer [31,32]. At the same time, there is inflammation in TME, where infiltrating cells such as macrophages, neutrophils, monocytes, and mast cells together with the secretion of cytokines and the interaction with the tumor constitute the complex inflammation of the TME and are the foundation of tumor immune escape. Recent studies have reported that gut microbiota can invade colonic epithelial cells and activate the intracellular signaling system to trigger the host inflammatory response [33,34]. These inflammatory reactions are closely related to the occurrence of gastrointestinal tumors [35,36].
Particular attention should be paid to the fact that angiogenesis is a common feature of the inflammatory response and tumor progression. The inflammatory microenvironment promotes tumor development by accelerating angiogenesis and disrupting adaptive immune response [37,38]. It has long been noted that colonizing gut microbiota can induce intestinal angiogenesis in germ-free mice [39]. Later studies demonstrated that this angiogenesis is mediated through the tissue factor (TF) [40], a membrane receptor that initiates an exogenous coagulation pathway and can promote tumor angiogenesis [41,42]. Gut microbiota closely links inflammation to tumor progression by promoting angiogenesis. Metabolites of gut microbiota promote specific angiogenesis in a NOD-like receptor-dependent manner and are associated with chronic intestinal inflammation [43]. Mucosal Escherichia coli expressing the AFA-1 operon upregulates the expression of HIF-1α in the intestinal epithelial cells, promotes angiogenesis, and induces inflammation, leading to tumor progression [44,45,46].
Because of the temporal and spatial proximity, colorectal cancer (CRC) is currently considered to be the cancer most affected by gut microbiota that is closely related to biological dysregulation, extensive changes in gut microbiota, and enrichment of certain microbial strains [47,48]. Gut microbiota plays a role in maintaining the homeostasis of the normal intestinal environment. However, once the intestinal environment is changed under the induction of internal or external factors or the composition of gut microbiota is unbalanced, gut microbiota may aggravate the inflammatory reaction and release inflammatory factors. These inflammatory factors influence the TME, promoting the occurrence and development of CRC [49]. In addition, the microbiota located in the gut can also affect tumors outside the gut through the maturation and migration of the body’s own immune system, such as in leukemia [50,51,52] and lymphoma [53,54,55].
2.1. Gut Microbiota, Th17 Cells, and IL-17 Families
The accumulation of T helper 17 (Th17) cells in a variety of tumors promotes the occurrence and development of tumors [56,57]. The interleukin-(IL-)17 cytokine family consists of six members named IL-17A, IL-17B, IL-17C, IL-17D, IL-17E, and IL-17F. Th17 cells primarily produce IL-17A (also known as IL-17), IL-17F, and IL-22, cytokines that play an important role in inflammation [58]. It is estimated that almost two-thirds of primary sporadic CRCs have increased IL17 expression [59]. IL-17A levels are elevated in the stroma and intestinal epithelium of CRC in patients with colorectal adenoma throughout all stages of cancer [60]. Chronic IL-17 mucosal production may be the result of specific microorganisms or communities’ stimulation [61]. Segmented filamentous bacteria (SFB), a microbe of mice, has been found to adhere to the intestinal surface of mice, contributing to the differentiation of Th17 cells in the lamina propria of the mouse small intestine and inducing IL17 expression [62,63]. Bile acid metabolites have a similar effect, which may promote the malignant behavior of tumors. Bile acids undergo gut microbiota-mediated transformation in the intestine to produce many bioactive molecules, such as lithocholic acid (LCA) derivatives, which can reduce Th17 in the intestinal lamina propria and increase Treg cell differentiation [64].
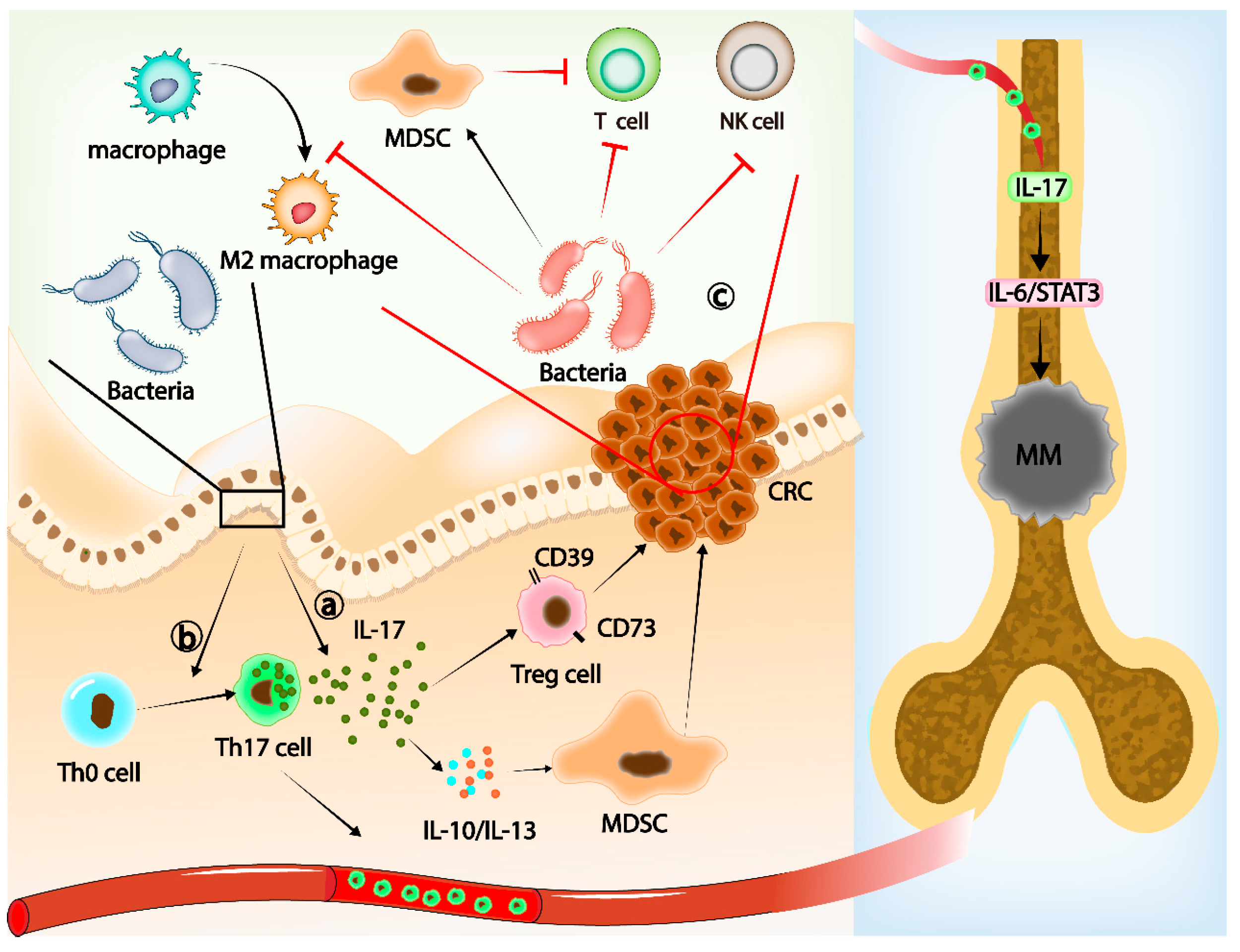
IL-17 can induce and change the signaling pathway in colonic epithelial cells (CECs). Furthermore, it promotes the activation of T cells and stimulates epithelial cells, endothelial cells, and fibroblasts to produce a variety of cytokines, such as IL-6, IL-8, granulocyte-macrophage colony-stimulating factor (GM-CSF), and chemical activator and cellular adhesion molecule-1 (CAM-1). This leads to inflammation [65,66]. The present study confirmed a causal link between gut microbiota, IL-17-mediated chronic inflammation, and cancer [67,68,69]. Yang Z et al. reported that IL-17 upregulated the expression of CD39 and CD73 in Treg cells in the TME, enhancing the immunosuppressive function of Treg cells, and promoted the immunosuppressive ability of myeloid-derived suppressor cells (MDSCs) by upregulating IL-10 and IL-13, which promoted the immune escape of tumors [70] (Figure 1a). These observations support the conclusion that gut microbiota may regulate IL-17 production by participating in the differentiation of Th17 cells, promote the immunosuppressive function of MDSC and Treg cells, and, ultimately, lead to the immune evasion of CRC.
Figure 1.
Gut microbiota, Th17 cells, and IL-17 families. (a) The increase in IL-17 induced by gut microbiota can upregulate the expression of CD39 and CD73 in Treg cells and enhance the immunosuppressive ability of Treg cells. Meanwhile, IL-17 promotes the immunosuppressive ability of MDCS by upregulating IL-10 and IL-13, thus promoting tumor immune evasion. (b) Gut microbiota induces Th0 cells to differentiate into Th17 cells and migrate into the bone marrow. Il-17 released from Th17 cells in the bone marrow promotes MM progression through the IL-6–STAT3 axis. (c) In CRC, gut microbiota promotes the function of MDSCs, induces T-cell apoptosis, inhibits NK cell function, and promotes M2 polarization of macrophages.
Th17 cells are also important mediators of gut microbiota affecting the TME of extramucosal tumors. Arianna et al. found that Prevotella heparinolytica, a commensal bacterium that promotes local and distant Th17 differentiation [71], may have a significant effect on the invasiveness of extramucosal tumors by affecting the differentiation of Th17 cells in the intestinal epithelium, independent of intestinal inflammation [72]. Prevotella heparinolytica promotes the differentiation of Th17 cells in the gut and their migration into the bone marrow (BM) of mice, where Th17 releases IL-17, contributing to the progression of multiple myeloma (MM). Multiple myeloma (MM) is a clonal malignancy of plasma cells characterized by the abnormal proliferation of clonal plasma cells in the bone marrow and overproduction of monoclonal antibodies [73]. IL-17 produced after Prevotella heparinolytica stimulation affects Th17 differentiation and migration, which induces STAT3 phosphorylation in the plasma cells of MM patients and promotes MM progression [72] (Figure 1b). Related to this, IL-17 was found to promote tumor growth through the IL-6–STAT3 signaling pathway [74]. A recent study has also reported that IL-17A can increase MM cell viability by upregulating Syk expression and activating the NF-κB signaling pathway [75].
In addition to IL-17A, other cytokines in the IL-17 family may also be involved in the gut microbiota-induced tumor progression. IL-17C can regulate the innate immune function of epithelial cells [76], and its upregulation promotes the progression of human CRC. Song et al. reported that changes in the gut microbiota caused the upregulation of IL-17C in intestinal epithelial cells and induced the expression of Bcl-2 and Bcl-xL, which ultimately supported the development of CRC [77]. IL-17RA belongs to the IL-17 receptor subfamily. IL-17A, IL-17B, IL-17C, and IL-17E (also known as IL-25) all activate IL-17 signaling through IL-17RA [78]. Therefore, further understanding of IL-17RA and other IL-17 receptor subfamily members can help us better understand the role of different cytokines in the gut microbiota–immune–cancer axis [79]. Targeting IL-17 and Th17 cells has yielded some results in the treatment of chronic inflammation [80]. We look forward to its success in tumor immunotherapy in the future.
2.2. Gut Microbiota and TAMs
The tumor-associated macrophage (TAM) is a main type of infiltrating immune cell in the TME [81]. TAMs are not a single cell population in phenotype and biological activity. In the TME, hypoxia, metabolic reprogramming, fibrosis, and various inflammatory factors can significantly alter the phenotype of TAMs [82,83,84,85,86]. Because of the heterogeneity and plasticity of TAMs, they may vary among different cancer types and even among different individuals of the same cancer [87]. The broad consensus is that macrophages can be divided into two types according to their functions: classically activated M1 or alternatively activated M2 phenotypes [88]. M1 macrophages target neoplastic cells and mediate antibody-dependent cellular cytotoxicity (ADCC) [89]. At the same time, M1 macrophages initiate an inflammatory response and promote antitumor immunity [90,91]. M2 macrophages can inhibit inflammation, induce angiogenesis, and promote tumor invasion and migration [92,93]. Unfortunately, during tumorigenesis, TAMs are more likely to be polarized into a protumor phenotype [94]. M2-polarized TAMs can inhibit cytotoxic T lymphocytes (CTL) [95], recruit Tregs [96], and release PGE2 and TGF-β [97] to change the TME to an immunosuppressive microenvironment [98], allowing tumor cells to escape immune surveillance (Figure 2b).
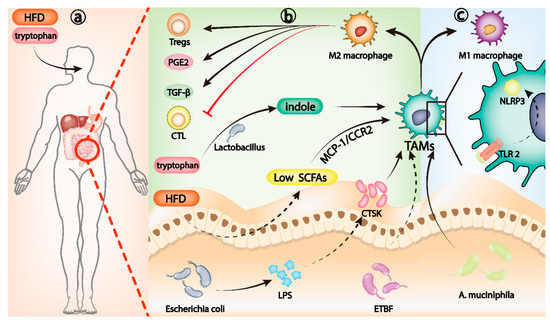
Figure 2.
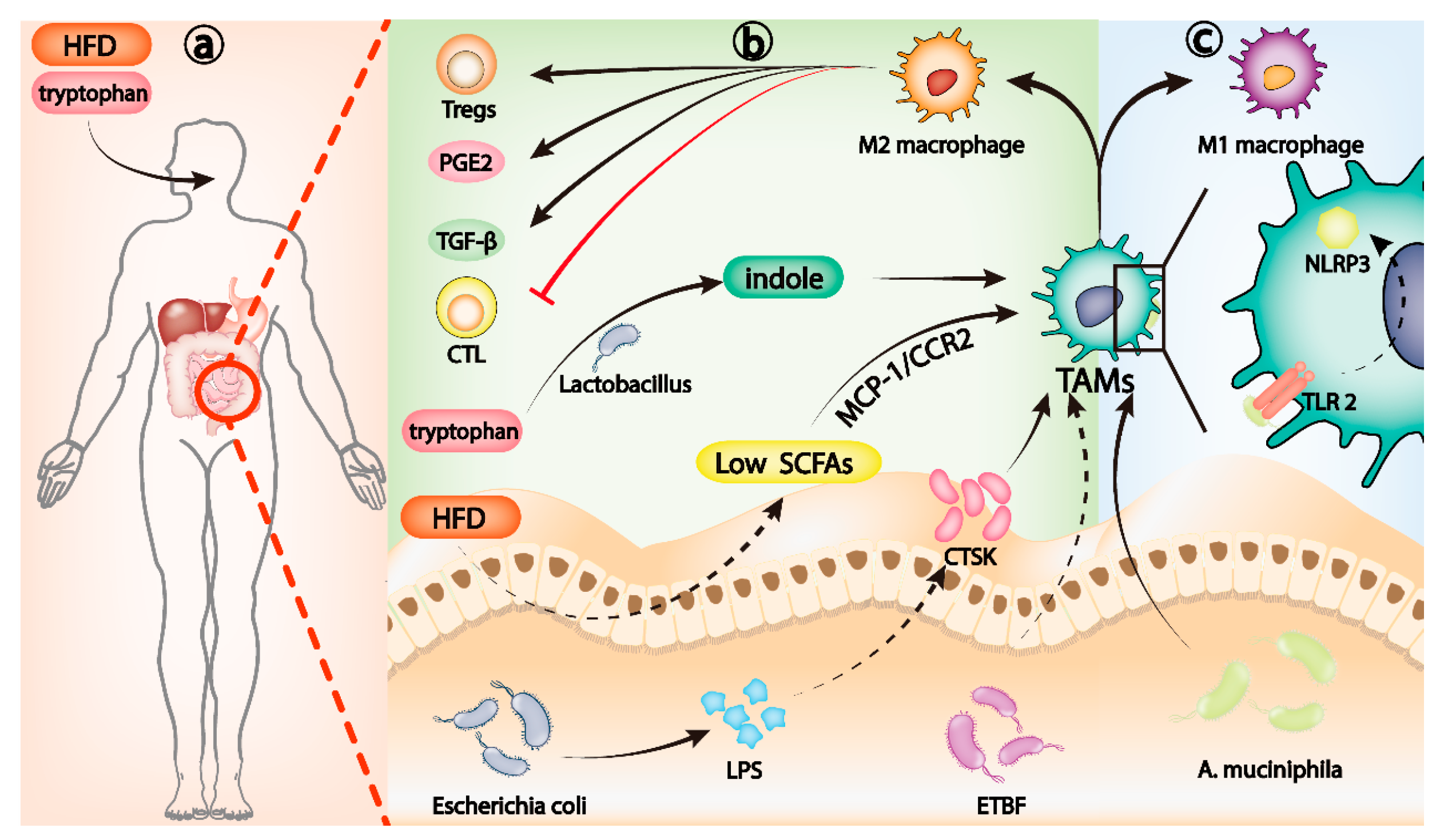
Gut microbiota, Th17 cells, and IL-17 families. (a) HFD can induce the dysregulation of gut microbiota and reduce the level of SCFAs. Low levels of SCFAs activate the MCP-1/CCR2 axis, which promotes the recruitment and polarization of M2 TAMs. Lactobacillus metabolizes dietary tryptophan into indole, which can activate aryl hydrocarbon receptor (AhR) to transform TAMs to immunosuppressive phenotype. (b) LPS released by Escherichia coli can upregulate CTSK secretion and induce M2 polarization of TAMs. Dysregulation of ETBF can also induce M2 polarization of TAMs. M2-polarized TAMs can inhibit cytotoxic T lymphocytes (CTL), recruit Tregs, and release PGE2 and TGF-β to change the TME to an immunosuppressive microenvironment. (c) A. muciniphila induces TLR2/NLRP3-mediated recruitment and polarization of M1 TAMs.
Gut microbiota can induce the polarization of macrophages to M1 or M2, affecting the process of many inflammatory diseases [99,100,101]. In the tumor microenvironment, the recruitment and differentiation of TAMs influenced by intestinal microbiota play a crucial role in evading the immune surveillance of tumor cells. High expression of cathepsin K (CTSK), a lysosomal cysteine protease, is associated with poor clinical prognosis in a variety of tumors [102,103,104]. Rui Li et al. reported that imbalance of gut microbiota, mainly the excessive proliferation of Escherichia coli, can promote the late progression and poor prognosis of CRC. Further studies revealed that as Escherichia coli increased in dominance, its release of lipopolysaccharide (LPS), a bacterial antigen, could upregulate CTSK secretion by CRC cells. In CRC mice, CTSK upregulation of CRC caused by intestinal microbiota imbalance can induce M2 polarization of TAMs, leading to rapid CRC progression [105]. Enterotoxigenic Bacteroides fragilis (ETBF) dysregulation can also induce M2 polarization of TAMs and promote immune escape of mouse CRC cells. In germ-free mice, ETBF colonization promotes the formation of a chronic inflammatory and immunosuppressive microenvironment by stimulating p-STAT3-mediated polarization of M2 macrophages, which in turn accelerates the progression of CRC [106]. Dysregulation of gut microbiota is closely related to M2 polarization of TAMs that contribute to tumor immune escape, and it is well-known that diet is a key determinant of gut microbiota homeostasis [107]. A high-fat diet (HFD) is an established risk factor for microbiota dysregulation and tumor progression [108]. In Apcmin/+ mice, a HFD can induce the dysregulation of gut microbiota and reduce the level of short chain fatty acids (SCFAs), which mainly include acetate, propionate, and butyrate and are metabolites of gut microbiota [109]. Low levels of SCFAs activate the MCP-1/CCR2 axis, which promotes the recruitment and polarization of M2 TAMs and ultimately CRC progression [110] (Figure 2a). In another particular example, Lactobacillus metabolizes dietary tryptophan into indole. Activation of aryl hydrocarbon receptor (AhR) can promote TAMs from pancreatic ductal adenocarcinoma (PDAC) to immunosuppressive phenotype and inhibit the accumulation of CD8+ T cells in the tumor [111] (Figure 2a).
The role of TAMs in the development of tumor immune escape is so critical that researchers have increasingly focused on them as a target for cancer therapy [90,112]. Selective depletion of TAMs can increase CD8+ T-cell infiltration in the TME, stimulate antitumor immune response, and improve the efficacy of chemotherapy in PDAC and breast cancer mice in vivo [113,114]. A variety of drugs and therapies targeting TAMs have been developed, and remarkable results have been achieved in animal models [115,116,117,118]. Gut microbiota also has potential as a therapeutic approach for targeting TAMs. Supplementation of ApcMin/+ mice with Akkermansia muciniphila (A. muciniphila) inhibited the progression of CRC. This is caused by induction of TLR2/ NLRP3-mediated recruitment and polarization of M1 TAMs by A. muciniphila [119] (Figure 2c). Perhaps in the future, we can target TAMs as a powerful adjunct to antitumor therapy by regulating the gut microbiota through dietary therapy, FMT, antibiotic cocktails, or prebiotics.
2.3. Gut Microbiota and Other Immune Cells in the TME
Fusobacterium nucleatum (F. nucleatum) is a component of the human oral and intestinal microbiota. Metagenomic analysis showed that F. nucleatum was abundant in CRC tissues and in metastatic lymph nodes [120]. F. nucleatum has been shown to promote CRC progression through multiple mechanisms, and a high abundance of F. nucleatum in human CRC tissues is associated with a low density of CD4+ T cells [120,121,122]. F. nucleatum was found to expand MDSCs, inhibit T-cell proliferation, and induce T-cell apoptosis in CRC [123]. Many studies have shown that F. nucleatum has immunosuppressive activity by inhibiting the response of human T cells to mitogens and antigens [124,125,126]. Tumors can use the Fap2 protein of F. nucleatum to inhibit the function of NK T cells through the T-cell immunoreceptor with immunoglobulin and ITIM domain (TIGIT), so that tumors can evade immune surveillance [127]. In addition, F. nucleatum also promotes M2 polarization of macrophages in F. nucleatum-related CRCs, which has an immunosuppressive effect [59] (Figure 1c).
In fact, the development of the mucosal immune system depends in part on innate immunity triggered by commensal microbes in the gut [128]. The colonization of Bacteroides fragilis (B. fragilis) and the production of polysaccharide A (PSA) in the mouse intestine promoted the development and differentiation of T cells [129]. At the same time, PSA recognized by toll-like receptors 2 (TLR2) on CD4+ T cells induces the development of Tregs [130]. Tregs can inhibit the function of tumor-specific T cells by secreting the inhibitory factors IL-10 and TGF-β [131] and activating the CTLA-4 pathway [132] to promote tumor immune escape. In CRC mice, the gut microbiota was found to downregulate CD8+ T-cell infiltration in the TME through activation of myeloid calcineurin, leading to tumor immune escape [133]. In addition, differences in T-cell differentiation caused by changes in gut microbiota can influence the effect of tumor chemoradiotherapy and regulate the response of patients to immunotherapy [134,135]. The role of gut microbiota in T-cell-centric immunotherapies such as CAR-T therapies and immune checkpoint inhibitors cannot be ignored. Elucidating the complex association and interaction between gut microbiota and immune cells, especially T cells, may be the key to resolving the nonresponse and side effects of immunotherapy in the future.
2.4. Gut Microbiota and PGE2
In the existing research, gut microbiota has been found to directly affect the immune evasion of intestinal tumors (mainly CRC) through inflammation and the tumor microenvironment. However, many studies have demonstrated that gut microbiota can affect gastrointestinal malignancies, but its driving mechanism in non-gastrointestinal malignancies remains to be elucidated. In addition to the gut microbiota mentioned above, which affects the differentiation of Th17 cells and leads to immune evasion of distant MM, gut microbiota can also affect the occurrence of nondigestive tract tumors through its metabolites.
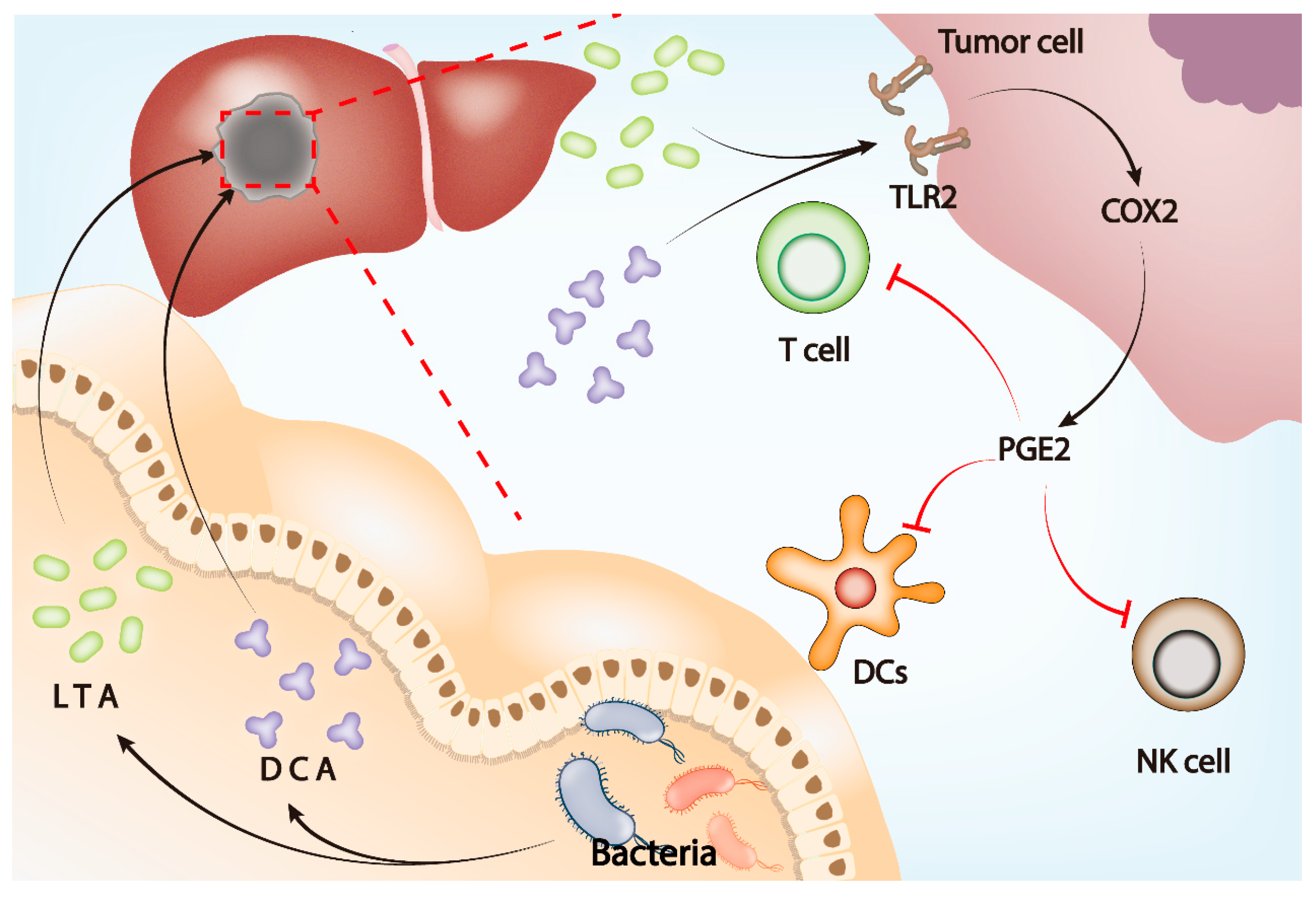
Prostaglandin E2 (PGE2) is an important inflammatory factor in the TME, affecting the occurrence of tumors through a wide range of mechanisms and exerting immunosuppressive effects [136,137,138]. In HCC, gut microbiota can promote tumor development by increasing PGE2 in the TME. Metabolites of gut microbiota including lipoteichoic acid (LTA) and deoxycholic acid upregulate cyclooxygenase-2 (COX2) expression through toll-like receptor 2 (TLR2) on tumor cell membranes. At the same time, the expression of PGE2 increases and inhibits antitumor immunity under COX2 [139]. The activity of the COX2–PGE2 signaling pathway can inhibit dendritic cells (DCs), natural killer cells (NKs), and T cells, thereby promoting tumor immune escape [140]. Previous studies confirmed that COX2 inhibitors can inhibit tumor immune escape [138]. Therefore, gut microbiota may affect key immune cells in the TME through the TLR2–COX2–PGE2 signaling pathway and promote immune escape of HCC (Figure 3).
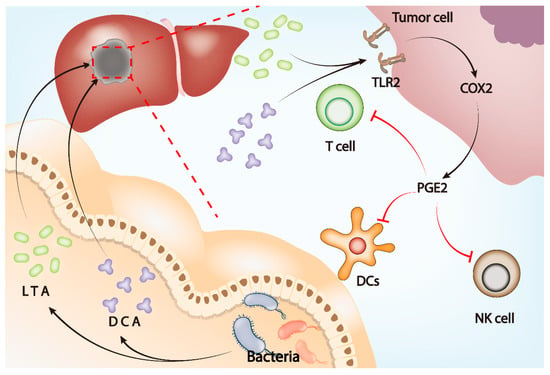
Figure 3.
Gut microbiota influences tumor microenvironment through the gut–liver axis. LTA and DCA, metabolites of gut microbiota, upregulate the expression of COX2 by activating TLR2 on the tumor cell membrane, leading to the upregulation of PGE2 in the tumor microenvironment. Elevated PGE2 inhibits the function of DCs, T cells, and NK cells and ultimately induces an immunosuppressive microenvironment to assist tumor immune escape.
In addition to changes in the microbiota colonizing the gut affecting tumor immune escape through contact and noncontact modes, the microbiota colonizing other organs can also affect tumor progression. The sources of these organ microbiomes include gut microbiota migration and bacterial colonization from in vitro, which together with gut microbiota, play important roles in the process of tumor evasion from immune surveillance. In recent years, a variety of previously considered sterile tumors were found to have their own unique microbial composition, and tumor-resident intracellular microbiota has entered scientific view [141,142]. In pancreatic cancer, researchers have used antibiotic cocktail therapy and fecal microbiota transplantation (FMT) to demonstrate that the pancreatic cancer microbiome promotes tumor immune evasion by inducing innate and adaptive immunity [143]. The lung cancer microbiome promotes immunosuppression of lung cancer by inducing IL-17A production by γδT cells [144]. The tumor microbiome is increasingly recognized as an indispensable part of the TME, which may serve as a bridge between gut microbiota and tumor immune evasion and is expected to become a new target for cancer treatment. These findings suggest a complex relationship between gut microbiota, inflammation, and the TME. Their interactions constitute an immunosuppressive network that links gut microbiota with tumor immune escape. Understanding the relationship between microbes, immune cells, and inflammatory factors in the TME will provide a new perspective and method for tumor immunotherapy.
3. Gut Microbiota and Tumor Immunotherapy
The proliferation of tumor cells is the result of the tumor evading the surveillance of the host immune system through a variety of mechanisms, such as the induction of immunosuppression in the TME and the downregulation of the expression of target antigens [145]. In recent years, the enhanced understanding of the tumor immune escape mechanism has led to the rapid development of tumor immunotherapy [146]. Tumor immunotherapy can help the body rebuild or strengthen antitumor immunity through various ways to cure or delay the tumor process [147,148]. Tumor immunotherapy includes chimeric antigen receptor (CAR)-T-cell therapy, immune checkpoint inhibitors (ICIs), CpG oligonucleotides, etc. Gut microbiota has been found to influence the response to immunotherapy and the occurrence of complications by modulating the infiltration degree of immune cells and the secretion of inflammatory factors in the TME [149,150,151]. We believe that further elucidation of the involvement of gut microbiota in the mechanism of tumor immune evasion will guide the development of tumor immunotherapy.
3.1. Gut Microbiota Affects the Efficacy of Immune Checkpoint Inhibitors
ICIs target the regulatory pathways of T cells and re-expose tumor cells that have escaped immune surveillance to enhance antitumor immune responses [152]. Antibodies that block cytotoxic T lymphocyte-associated protein 4 (CTLA-4) or programmed cell death 1 (PD-1) pathways are the most commonly used ICIs [153,154]. ICIs have been shown to have good efficacy in melanoma, nonsmall cell lung cancer, and other tumors, but the interpatient variability of the treatment effect is still a concern [155].
In an ICI treatment targeting the PD-1/PD-L1 axis in melanoma patients, Response Evaluation Criteria in Solid Tumors (RECIST) were used to distinguish melanoma patients with or without a clinical response to ICI treatment, and they were labeled as responders or nonresponders. There was a significant difference in fecal microbial composition between responders and nonresponders. When fecal microbiota from responders and nonresponders were transplanted into melanoma mice that received PD-1 blockade treatment, the response was significantly enhanced in mice that underwent responder feces, while the response was not observed in mice that underwent nonresponders [156]. This suggests that the gut microbiota includes both favorable and unfavorable microbiota, and the composition of gut microbiota plays a role in regulating the therapeutic effect. Analysis of the TME revealed that this effect was due to enhanced activation of tumor antigen-specific CD8+ T cells [157,158]. Higher concentrations of CD8+ T cells were clustered in the TME of mice receiving fecal microbial transplantation from “responders”, and the infiltration degree of CD8+ T bacteria was positively correlated with Faecalibacterium [159]. Bifidobacterium can directly induce DC maturation and cytokine synthesis and increase the number of CD8+ T cells in peripheral areas and in the TME [160]. In addition to CD8+ T cells, favorable flora can also increase the number and function of CD4+ T cells in the body. An abundance of Faecalibacterium cells can improve the number of circulating CD4+ T cells and promote cytokine response [159]. In view of the effects of gut microbiota on T cells, artificial regulation of microbiota composition is an effective strategy to assist ICI. Oral administration of Akkermansia muciniphila could induce the secretion of IL-12 from DCs and increase the accumulation of CCR9+CXCR3+CD4+ T lymphocytes in mouse tumors, thus recovering the efficacy of the PD-1 blockade [161]. The mechanism of gut microbiota influencing the CTLA-4 blockade effect is similar to that of the PD-1 blockade. Bacteroides species play an important role in it. Fecal microbial transplantation from humans to mice confirmed that B. fragilis and B. thetaiotaomicron promote tumor CTLA-4 blockade reactivity by promoting the IL-12-dependent TH1 immune response [162].
The mechanism by which favorable and unfavorable microbiota can regulate the efficacy of ICI in the intestinal tract has not been clearly reported, which may be related to the metabolic function of the microbiota. Favorable microbiota can synthesize a variety of metabolites that promote host immune function, while the catabolic function of unfavorable microbiota has the opposite effect [163]. In CRC model mice, inosine produced by the metabolism of B. pseudolongum was confirmed to enhance antitumor immunity induced by anti-CTLA-4 treatment, mediated by T cells [164]. In another study in mice, short chain fatty acids (SCFAs) produced by gut microbiota inhibited CD80/CD86 upregulation on dendritic cells induced by anti-CTLA-4 treatment, limiting the efficacy of anti-CTLA-4 in mice with metastatic melanoma [165]. The roles of favorable and unfavorable microbiota are not static. For example, in MSS-type CRC tumor-bearing mice, abundant Bacteroides would lead to increased Tregs and MDSCs in circulation and decreased CD8+ T-cell infiltration in the tumor bed, which would decrease the cytokine response and the therapeutic effect of the PD-1 blockade [166]. Bacteroides has the role of promoting the therapeutic effect in the CTLA-4 blockade mentioned above [162], and the reason for the difference between the two remains to be discussed.
3.2. Gut Microbiota and Antibiotic Use in ICI Therapy
In cancer patients, antibiotics are commonly used to prevent and treat a range of potentially life-threatening bacterial infections, which complicates cancer treatment and worsens the prognosis of patients with tumors [167,168,169]. It is a clinically obvious phenomenon that the long-term application of broad-spectrum antibiotics can lead to the irreversible destruction of gut microbiota ecology, which might result in diarrhea, double infection, and even the selection of drug-resistant strains [170,171,172]. In addition to increasing the complexity and difficulty of tumor treatment, dysregulated gut microbiota can also cause resistance to chemotherapy and immunotherapy in cancer patients [173,174]. A meta-analysis of 23 eligible studies showed that antibiotic use before or during ICI treatment resulted in a reduction in median OS for more than six months [175]. A retrospective study suggested that early use of antibiotics may be more harmful, while use of antibiotics during ICI treatment may be safer [176].
At present, it seems reasonable to avoid long-term and broad-spectrum antibiotics before starting ICI treatment, if possible. However, for some patients with malignant tumors, the use of antibiotics to prevent and treat infection is an irreplaceable choice because of long-term consumption and the decline of immunity [177,178]. For patients who do require antibiotics prior to ICI treatment, favorable strategies may be receiving FMT [157,179] or probiotics combined with dietary fiber therapy [180] in an attempt to eliminate the possible harmful effects of broad-spectrum antibiotics in ICI treatment. Another valuable direction would be to reduce the concentration of antibiotics in the gut without affecting their plasma pharmacokinetics. For example, nonspecific adsorbents can be applied to the patient’s intestine [181,182,183]. Oral β-lactamases target β-lactam antibiotics [184,185,186], significantly reducing the damaging effects of antibiotics on gut microbiota.
3.3. Gut Microbiota Affects the Efficacy of CAR T-Cell Therapy
Chimeric antigen receptor T (CAR-T) cell therapy refers to the modification of immunoreactive T cells derived from tumor patients with the chimeric antigen receptor (CAR), in vitro expansion, and then transfusion back into patients. The aim is to establish a long-term specific antitumor immune effect [187]. Therapeutic efficacy is usually limited by treatment-related toxicity, T-cell dysfunction, the TME, tumor antigen loss, and other reasons [188]. Antibiotic use is closely related to CAR-T efficacy, which is mediated by gut microbiota. In mice treated with broad-spectrum antibiotics, the tumor-suppressive effect of CAR-T was decreased [158]. Vancomycin-treated mice were more responsive than untreated mice to CAR-T treatment because vancomycin-screened gut microbiota induced an increase in systemic CD8α + DC and maintained adoptively transferred antitumor T cells in an IL-12-dependent manner [134].
The efficacy of CAR-T is affected by lymphocyte infiltration in the TME and the function of effector molecules. Vancomycin treatment increased the levels of granzyme B, perforin 1, IL-12, and IFN-γ in the TME of mice, which are all antitumor effector molecules [134]. The expression of antitumor effector molecules is affected by the metabolites of gut microbiota. Butyrate affects the gene expression of effector molecules by inhibiting histone deacetylase (HDAC) in CD8+ T cells. Higher concentrations of acetate can promote IFN-γ production by CD8+ T cells by regulating cell metabolism and mTOR activity, both of which affect the efficacy of CAR-T in mice [189]. In addition to effector molecules, gut microbiota can also modulate tumor-infiltrating lymphocytes (TILs). In human CRC, the gut microbiota stimulates tumor cells to produce chemokines to recruit antitumor T cells into the tumor tissue. Among them, the abundance of Rikenellaceae, Ruminococcace, and Lachnospiracee is significantly correlated with the expression levels of chemokines CCL5, CCL20, and CXCL11 [190].
A better understanding of the mechanisms by which the gut microbiome regulates the effects of CAR-T and ICI therapies will help maximize the effectiveness of immunotherapies in the future and reduce immunotherapy-related side effects [191]. A serious side effect of CTLA-4 blockade is autoimmunity, inducing severe inflammation. An experiment on mice showed that Bifidobacterium can relieve the toxicity of CTLA-4 blockade by affecting the metabolic function of Treg cells without affecting antitumor immunity [192]. Many immunotherapy strategies have been practiced and validated in experiments or clinical practice. More attention has been paid to the relationship between gut microbiota and antitumor immunotherapy. However, the role of gut microbiota in other immunotherapies, such as cytokine therapy and immune vaccines, remains poorly investigated. We believe that the regulation of gut microbiota will become an effective and necessary means of adjuvant antitumor immunotherapy with the further elucidation of the influence of gut microbiota on tumor immune escape.
4. Conclusions and Future Directions
Tumor immune escape is the process of tumor cells escaping from monitoring and killing by the immune system, while antitumor immunotherapy is the process of assisting the immune system to monitor and eliminate tumor cells. There is a complex crosstalk between gut microbiota, inflammation, and the TME. The colonization and metabolites of gut microbiota affect tumor escape from immune surveillance through complex mechanisms and the effect and complications of antitumor immunotherapy. The mechanism by which gut microbiota helps tumor cells escape immune surveillance has been identified, but many questions remain unanswered. Tumor cells themselves can escape the attack of tumor killer cells through a variety of ways, such as immune editing, low expression of MHC class I molecules, abnormal costimulatory signals (decreased expression of CD86 and upregulated expression of PD-L1), and expression or secretion of immunosuppressive factors TGF-β and IL-10 [193]. Studies have confirmed that gut microbiota can promote or inhibit tumor progression by activating intracellular signaling pathways through metabolites [22,194,195]. This suggests that gut microbiota may have a mechanism that directly enables tumor cells to evade immune surveillance. However, current studies on the regulation of antitumor immunity by gut microbiota have mainly focused on the regulation of immune cells and inflammatory factors outside tumor cells. There is no report of gut microbiota causing changes in tumor cells to escape immune surveillance. We look forward to further research revealing the status and role of gut microbiota in the network of tumor immune evasion.
The metabolites of gut microbiota are important mediators in the regulation of antitumor immunity. In the process of exploring the relationship between gut microbiota and tumor immunity, researchers have found a variety of metabolites that can affect tumor progression and antitumor immunity. These metabolites can still play an antitumor immune role or assist in enhancing the effect of immunotherapy in the absence of gut microbiota [164,196]. Elucidating the mechanism by which these metabolites act as antitumor drugs and developing targeted drugs or metabolite analogues are the two foreseeable industrial transformation directions regarding gut microbiota. At the same time, the development of inhibitors or antagonists [197] targeting tumor-promoting gut microbial metabolites may also be a promising prospect for adjuvant tumor immunotherapy.
The composition of gut microbiota plays an important role in tumor progression. Therefore, manipulating gut microbiota may be a new approach to improve tumor immunotherapy. At present, some microbiota interventions, such as FMT, prebiotics, probiotics, and antibiotic cocktail therapy, have been developed. Manipulation can be used to study the gut microbiota or as an adjunct to the experimental treatment of tumors. These strategies have shown promise as modulators of the gut microbiome. In general, gut microbiota can affect the process of tumor immune escape and the efficacy of tumor immunotherapy. Targeting gut microbiota and its metabolites to detect, block, adjust, or transplant is important for tumor immunotherapy intervention and prognosis assessment.
Author Contributions
Y.H. and J.H. (Jinliang Huang) wrote the manuscript; H.L., J.H. (Jiao Hu), H.D. and Q.L. created the figures and wrote the abstract; W.X., C.Z., X.Z. and Z.L. gave constructive feedback and guidance; J.C., Z.Y., J.X., Z.X. and C.Q. made critical revisions and proofread the manuscript. All the authors read and approved the final manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (81873626, 81902592, 82070785), the Hunan Natural Science Foundation (2020JJ5884), and the Hunan Province Young Talents Program (2021RC3027).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
TME, tumor microenvironment; CAR-T, chimeric antigen receptor T; ICI, immune checkpoint inhibitor; IRES, internal ribosome entry sites; FMT, fecal microbiota transplantation; Th17, T helper 17; SFB, segmented filamentous bacteria; CECs, colonic epithelial cells; GM-CSF, granulocyte-macrophage colony-stimulating factor; CAM-1, cellular adhesion molecule-1; MDSC, myeloid-derived suppressor cells; CRC, colorectal cancer; BM, bone marrow; MM, multiple myeloma; F. nucleatum, Fusobacterium nucleatum; TIGIT, T cell immunoreceptor with immunoglobulin and ITIM domain; PSA, polysaccharide A; B. fragilis, Bacteroides fragilis; PGE2, prostaglandin E2; LTA, lipoteichoic acid; COX2, cyclooxygenase-2; DCs, dendritic cells; CTLA-4, cytotoxic T lymphocyte-associated protein 4; PD-1, programmed cell death 1; ICI, immune checkpoint inhibitors; HDAC, histone deacetylase; TILT, tumor-infiltrating lymphocyte; LPS, lipopolysaccharide; CTSK, cathepsin K; ADCC, antibody-dependent cellular cytotoxicity; TAMs, tumor-associated macrophages; ETBF, enterotoxigenic Bacteroides fragilis; SCFAs, short chain fatty acids; AhR, aryl hydrocarbon receptor; PDAC, pancreatic ductal adenocarcinoma.
References
- Clemente, J.C.; Ursell, L.K.; Parfrey, L.W.; Knight, R. The impact of the gut microbiota on human health: An integrative view. Cell 2012, 148, 1258–1270. [Google Scholar] [CrossRef] [PubMed]
- Chow, J.; Lee, S.M.; Shen, Y.; Khosravi, A.; Mazmanian, S.K. Host-bacterial symbiosis in health and disease. Adv. Immunol. 2010, 107, 243–274. [Google Scholar] [CrossRef] [PubMed]
- Sen, P.; Molinero-Perez, A.; O’Riordan, K.J.; McCafferty, C.P.; O’Halloran, K.D.; Cryan, J.F. Microbiota and sleep: Awakening the gut feeling. Trends Mol. Med. 2021, 27, 935–945. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Du, J.; Wu, X.; Abdelrehem, A.; Ren, Y.; Liu, C.; Zhou, X.; Wang, S. Crosstalk between autophagy and microbiota in cancer progression. Mol. Cancer 2021, 20, 163. [Google Scholar] [CrossRef]
- Cullin, N.; Azevedo Antunes, C.; Straussman, R.; Stein-Thoeringer, C.K.; Elinav, E. Microbiome and cancer. Cancer Cell 2021, 39, 1317–1341. [Google Scholar] [CrossRef]
- Schwabe, R.F.; Jobin, C. The microbiome and cancer. Nat. Rev. Cancer 2013, 13, 800–812. [Google Scholar] [CrossRef]
- Agirman, G.; Yu, K.B.; Hsiao, E.Y. Signaling inflammation across the gut-brain axis. Science 2021, 374, 1087–1092. [Google Scholar] [CrossRef]
- Villa, C.R.; Ward, W.E.; Comelli, E.M. Gut microbiota-bone axis. Crit. Rev. Food Sci. Nutr. 2017, 57, 1664–1672. [Google Scholar] [CrossRef]
- Beyaz Coşkun, A.; Sağdiçoğlu Celep, A.G. Therapeutic modulation methods of gut microbiota and gut-liver axis. Crit. Rev. Food Sci. Nutr. 2022, 62, 6505–6515. [Google Scholar] [CrossRef]
- Yang, J.; Wei, H.; Zhou, Y.; Szeto, C.H.; Li, C.; Lin, Y.; Coker, O.O.; Lau, H.C.H.; Chan, A.W.H.; Sung, J.J.Y.; et al. High-Fat Diet Promotes Colorectal Tumorigenesis Through Modulating Gut Microbiota and Metabolites. Gastroenterology 2022, 162, 135–149.e132. [Google Scholar] [CrossRef]
- Ohtani, N.; Hara, E. Gut-liver axis-mediated mechanism of liver cancer: A special focus on the role of gut microbiota. Cancer Sci. 2021, 112, 4433–4443. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Xia, Y.; Sun, J. Breast and gut microbiome in health and cancer. Genes Dis. 2021, 8, 581–589. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Wu, J.; Jin, D.; Wang, B.; Cao, H. Fecal microbiota transplantation in cancer management: Current status and perspectives. Int. J. Cancer 2019, 145, 2021–2031. [Google Scholar] [CrossRef]
- Eslami, M.; Yousefi, B.; Kokhaei, P.; Hemati, M.; Nejad, Z.R.; Arabkari, V.; Namdar, A. Importance of probiotics in the prevention and treatment of colorectal cancer. J. Cell. Physiol. 2019, 234, 17127–17143. [Google Scholar] [CrossRef]
- Zhang, X.; Coker, O.O.; Chu, E.S.; Fu, K.; Lau, H.C.H.; Wang, Y.X.; Chan, A.W.H.; Wei, H.; Yang, X.; Sung, J.J.Y.; et al. Dietary cholesterol drives fatty liver-associated liver cancer by modulating gut microbiota and metabolites. Gut 2021, 70, 761–774. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.L.; Mao, Y.Q.; Zhang, Z.Y.; Li, Z.M.; Kong, C.Y.; Chen, H.L.; Cai, P.R.; Han, B.; Ye, T.; Wang, L.S. Pectin supplement significantly enhanced the anti-PD-1 efficacy in tumor-bearing mice humanized with gut microbiota from patients with colorectal cancer. Theranostics 2021, 11, 4155–4170. [Google Scholar] [CrossRef]
- Chen, D.S.; Mellman, I. Elements of cancer immunity and the cancer-immune set point. Nature 2017, 541, 321–330. [Google Scholar] [CrossRef]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef]
- Gajewski, T.F.; Schreiber, H.; Fu, Y.X. Innate and adaptive immune cells in the tumor microenvironment. Nat. Immunol. 2013, 14, 1014–1022. [Google Scholar] [CrossRef]
- Jhunjhunwala, S.; Hammer, C.; Delamarre, L. Antigen presentation in cancer: Insights into tumour immunogenicity and immune evasion. Nat. Rev. Cancer 2021, 21, 298–312. [Google Scholar] [CrossRef]
- Zhang, J.; Huang, D.; Saw, P.E.; Song, E. Turning cold tumors hot: From molecular mechanisms to clinical applications. Trends Immunol. 2022, 43, 523–545. [Google Scholar] [CrossRef] [PubMed]
- Kadosh, E.; Snir-Alkalay, I.; Venkatachalam, A.; May, S.; Lasry, A.; Elyada, E.; Zinger, A.; Shaham, M.; Vaalani, G.; Mernberger, M.; et al. The gut microbiome switches mutant p53 from tumour-suppressive to oncogenic. Nature 2020, 586, 133–138. [Google Scholar] [CrossRef] [PubMed]
- Matson, V.; Chervin, C.S.; Gajewski, T.F. Cancer and the Microbiome-Influence of the Commensal Microbiota on Cancer, Immune Responses, and Immunotherapy. Gastroenterology 2021, 160, 600–613. [Google Scholar] [CrossRef] [PubMed]
- Houot, R.; Schultz, L.M.; Marabelle, A.; Kohrt, H. T-cell-based Immunotherapy: Adoptive Cell Transfer and Checkpoint Inhibition. Cancer Immunol. Res. 2015, 3, 1115–1122. [Google Scholar] [CrossRef] [PubMed]
- El-Salhy, M.; Hatlebakk, J.G.; Gilja, O.H.; Bråthen Kristoffersen, A.; Hausken, T. Efficacy of faecal microbiota transplantation for patients with irritable bowel syndrome in a randomised, double-blind, placebo-controlled study. Gut 2020, 69, 859–867. [Google Scholar] [CrossRef]
- Arias-Borrego, A.; Selma-Royo, M.; Collado, M.C.; Abril, N.; García-Barrera, T. Impact of “chemical cocktails” exposure in shaping mice gut microbiota and the role of selenium supplementation combining metallomics, metabolomics, and metataxonomics. J. Hazard. Mater. 2022, 438, 129444. [Google Scholar] [CrossRef] [PubMed]
- Sanders, M.E.; Merenstein, D.J.; Reid, G.; Gibson, G.R.; Rastall, R.A. Probiotics and prebiotics in intestinal health and disease: From biology to the clinic. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 605–616. [Google Scholar] [CrossRef] [PubMed]
- Hinshaw, D.C.; Shevde, L.A. The Tumor Microenvironment Innately Modulates Cancer Progression. Cancer Res. 2019, 79, 4557–4566. [Google Scholar] [CrossRef]
- Hegde, P.S.; Chen, D.S. Top 10 Challenges in Cancer Immunotherapy. Immunity 2020, 52, 17–35. [Google Scholar] [CrossRef]
- Balkwill, F.; Mantovani, A. Inflammation and cancer: Back to Virchow? Lancet 2001, 357, 539–545. [Google Scholar] [CrossRef]
- Wang, L.; Jiang, Y.; Zhang, Y.; Wang, Y.; Huang, S.; Wang, Z.; Tian, B.; Yang, Y.; Jiang, W.; Pang, D. Association analysis of IL-17A and IL-17F polymorphisms in Chinese Han women with breast cancer. PLoS ONE 2012, 7, e34400. [Google Scholar] [CrossRef] [PubMed]
- Grote, V.A.; Kaaks, R.; Nieters, A.; Tjønneland, A.; Halkjær, J.; Overvad, K.; Skjelbo Nielsen, M.R.; Boutron-Ruault, M.C.; Clavel-Chapelon, F.; Racine, A.; et al. Inflammation marker and risk of pancreatic cancer: A nested case-control study within the EPIC cohort. Br. J. Cancer 2012, 106, 1866–1874. [Google Scholar] [CrossRef] [PubMed]
- Kipanyula, M.J.; Seke Etet, P.F.; Vecchio, L.; Farahna, M.; Nukenine, E.N.; Nwabo Kamdje, A.H. Signaling pathways bridging microbial-triggered inflammation and cancer. Cell Signal 2013, 25, 403–416. [Google Scholar] [CrossRef] [PubMed]
- Ohkusa, T.; Yoshida, T.; Sato, N.; Watanabe, S.; Tajiri, H.; Okayasu, I. Commensal bacteria can enter colonic epithelial cells and induce proinflammatory cytokine secretion: A possible pathogenic mechanism of ulcerative colitis. J. Med. Microbiol 2009, 58, 535–545. [Google Scholar] [CrossRef] [PubMed]
- Jia, W.; Xie, G.; Jia, W. Bile acid-microbiota crosstalk in gastrointestinal inflammation and carcinogenesis. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 111–128. [Google Scholar] [CrossRef]
- Man, S.M. Inflammasomes in the gastrointestinal tract: Infection, cancer and gut microbiota homeostasis. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 721–737. [Google Scholar] [CrossRef]
- Mantovani, A.; Allavena, P.; Sica, A.; Balkwill, F. Cancer-related inflammation. Nature 2008, 454, 436–444. [Google Scholar] [CrossRef]
- Karki, R.; Kanneganti, T.D. Diverging inflammasome signals in tumorigenesis and potential targeting. Nat. Rev. Cancer 2019, 19, 197–214. [Google Scholar] [CrossRef]
- Stappenbeck, T.S.; Hooper, L.V.; Gordon, J.I. Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proc. Natl. Acad. Sci. USA 2002, 99, 15451–15455. [Google Scholar] [CrossRef]
- Reinhardt, C.; Bergentall, M.; Greiner, T.U.; Schaffner, F.; Ostergren-Lundén, G.; Petersen, L.C.; Ruf, W.; Bäckhed, F. Tissue factor and PAR1 promote microbiota-induced intestinal vascular remodelling. Nature 2012, 483, 627–631. [Google Scholar] [CrossRef]
- Eisenreich, A.; Zakrzewicz, A.; Huber, K.; Thierbach, H.; Pepke, W.; Goldin-Lang, P.; Schultheiss, H.P.; Pries, A.; Rauch, U. Regulation of pro-angiogenic tissue factor expression in hypoxia-induced human lung cancer cells. Oncol. Rep. 2013, 30, 462–470. [Google Scholar] [CrossRef] [PubMed]
- Van den Berg, Y.W.; van den Hengel, L.G.; Myers, H.R.; Ayachi, O.; Jordanova, E.; Ruf, W.; Spek, C.A.; Reitsma, P.H.; Bogdanov, V.Y.; Versteeg, H.H. Alternatively spliced tissue factor induces angiogenesis through integrin ligation. Proc. Natl. Acad. Sci. USA 2009, 106, 19497–19502. [Google Scholar] [CrossRef] [PubMed]
- Schirbel, A.; Kessler, S.; Rieder, F.; West, G.; Rebert, N.; Asosingh, K.; McDonald, C.; Fiocchi, C. Pro-angiogenic activity of TLRs and NLRs: A novel link between gut microbiota and intestinal angiogenesis. Gastroenterology 2013, 144, 613–623.e619. [Google Scholar] [CrossRef] [PubMed]
- Prorok-Hamon, M.; Friswell, M.K.; Alswied, A.; Roberts, C.L.; Song, F.; Flanagan, P.K.; Knight, P.; Codling, C.; Marchesi, J.R.; Winstanley, C.; et al. Colonic mucosa-associated diffusely adherent afaC+ Escherichia coli expressing lpfA and pks are increased in inflammatory bowel disease and colon cancer. Gut 2014, 63, 761–770. [Google Scholar] [CrossRef] [PubMed]
- Cane, G.; Ginouvès, A.; Marchetti, S.; Buscà, R.; Pouysségur, J.; Berra, E.; Hofman, P.; Vouret-Craviari, V. HIF-1alpha mediates the induction of IL-8 and VEGF expression on infection with Afa/Dr diffusely adhering E. coli and promotes EMT-like behaviour. Cell. Microbiol. 2010, 12, 640–653. [Google Scholar] [CrossRef]
- Waldner, M.J.; Wirtz, S.; Jefremow, A.; Warntjen, M.; Neufert, C.; Atreya, R.; Becker, C.; Weigmann, B.; Vieth, M.; Rose-John, S.; et al. VEGF receptor signaling links inflammation and tumorigenesis in colitis-associated cancer. J. Exp. Med. 2010, 207, 2855–2868. [Google Scholar] [CrossRef]
- Lin, S.-C.; Lo, Y.-C.; Wu, H. Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signalling. Nature 2010, 465, 885–890. [Google Scholar] [CrossRef]
- Valkov, E.; Stamp, A.; Dimaio, F.; Baker, D.; Verstak, B.; Roversi, P.; Kellie, S.; Sweet, M.J.; Mansell, A.; Gay, N.J.; et al. Crystal structure of Toll-like receptor adaptor MAL/TIRAP reveals the molecular basis for signal transduction and disease protection. Proc. Natl. Acad. Sci. USA 2011, 108, 14879–14884. [Google Scholar] [CrossRef]
- Del Prete, A.; Allavena, P.; Santoro, G.; Fumarulo, R.; Corsi, M.M.; Mantovani, A. Molecular pathways in cancer-related inflammation. Biochem. Med. 2011, 21, 264–275. [Google Scholar] [CrossRef]
- Ozdemir, H.; Ciftçi, E.; Karbuz, A.; Oktay, G.; Aysev, D.; Yavuz, G.; Ince, E. Successful treatment of multidrug-resistant Escherichia coli bacteremia with tigecycline in an acute myeloid leukemia child. Turk. J. Pediatr. 2012, 54, 59–60. [Google Scholar]
- Kurucu, N.; Kul, S.; Tosun, I.; Erduran, E.; Köksal, I. Fungemia and renal fungus ball formation with Candida norvegensis in a child with acute lymphoblastic leukemia. Turk. J. Pediatr. 2011, 53, 448–451. [Google Scholar] [PubMed]
- De Carvalho Parahym, A.M.R.; da Silva, C.M.; Leão, M.P.C.; Macario, M.C.; Filho, G.A.d.T.M.H.; de Oliveira, N.T.; Neves, R.P. Invasive infection in an acute myeloblastic leukemia patient due to triazole-resistant Candida tropicalis. Diagn. Microbiol. Infect. Dis. 2011, 71, 291–293. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gogia, A.; Bakhshi, S. Plasmablastic lymphoma of oral cavity in a HIV-negative child. Pediatr. Blood Cancer 2010, 55, 390–391. [Google Scholar] [CrossRef] [PubMed]
- Marteau, P.; Chaput, U. Bacteria as trigger for chronic gastrointestinal disorders. Dig. Dis. 2011, 29, 166–171. [Google Scholar] [CrossRef]
- Khan, M.A.; Jakate, S.; Komanduri, S. Rare AIDS-associated plasmablastic lymphoma as the initial presentation of AIDS. Clin. Adv. Hematol. Oncol. 2010, 8, 55–57. [Google Scholar]
- McAllister, F.; Bailey, J.M.; Alsina, J.; Nirschl, C.J.; Sharma, R.; Fan, H.; Rattigan, Y.; Roeser, J.C.; Lankapalli, R.H.; Zhang, H.; et al. Oncogenic Kras activates a hematopoietic-to-epithelial IL-17 signaling axis in preinvasive pancreatic neoplasia. Cancer Cell 2014, 25, 621–637. [Google Scholar] [CrossRef]
- Tartour, E.; Fossiez, F.; Joyeux, I.; Galinha, A.; Gey, A.; Claret, E.; Sastre-Garau, X.; Couturier, J.; Mosseri, V.; Vives, V.; et al. Interleukin 17, a T-cell-derived cytokine, promotes tumorigenicity of human cervical tumors in nude mice. Cancer Res. 1999, 59, 3698–3704. [Google Scholar]
- Gaffen, S.L.; Jain, R.; Garg, A.V.; Cua, D.J. The IL-23-IL-17 immune axis: From mechanisms to therapeutic testing. Nat. Rev. Immunol. 2014, 14, 585–600. [Google Scholar] [CrossRef]
- Tosolini, M.; Kirilovsky, A.; Mlecnik, B.; Fredriksen, T.; Mauger, S.; Bindea, G.; Berger, A.; Bruneval, P.; Fridman, W.-H.; Pagès, F.; et al. Clinical impact of different classes of infiltrating T cytotoxic and helper cells (Th1, th2, treg, th17) in patients with colorectal cancer. Cancer Res. 2011, 71, 1263–1271. [Google Scholar] [CrossRef]
- Cui, G.; Yuan, A.; Goll, R.; Florholmen, J. IL-17A in the tumor microenvironment of the human colorectal adenoma-carcinoma sequence. Scand. J. Gastroenterol. 2012, 47, 1304–1312. [Google Scholar] [CrossRef]
- Hurtado, C.G.; Wan, F.; Housseau, F.; Sears, C.L. Roles for Interleukin 17 and Adaptive Immunity in Pathogenesis of Colorectal Cancer. Gastroenterology 2018, 155, 1706–1715. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, I.I.; Atarashi, K.; Manel, N.; Brodie, E.L.; Shima, T.; Karaoz, U.; Wei, D.; Goldfarb, K.C.; Santee, C.A.; Lynch, S.V.; et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 2009, 139, 485–498. [Google Scholar] [CrossRef] [PubMed]
- Gaboriau-Routhiau, V.; Rakotobe, S.; Lécuyer, E.; Mulder, I.; Lan, A.; Bridonneau, C.; Rochet, V.; Pisi, A.; De Paepe, M.; Brandi, G.; et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity 2009, 31, 677–689. [Google Scholar] [CrossRef] [PubMed]
- Hang, S.; Paik, D.; Yao, L.; Kim, E.; Trinath, J.; Lu, J.; Ha, S.; Nelson, B.N.; Kelly, S.P.; Wu, L.; et al. Bile acid metabolites control T(H)17 and T(reg) cell differentiation. Nature 2019, 576, 143–148. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Jiang, X.; Xiao, Y.; Zhou, T.; Guo, Y.; Wang, R.; Zhao, Z.; Xiao, H.; Hou, C.; Ma, L.; et al. IL-17A signaling in colonic epithelial cells inhibits pro-inflammatory cytokine production by enhancing the activity of ERK and PI3K. PLoS ONE 2014, 9, e89714. [Google Scholar] [CrossRef]
- Chung, L.; Thiele Orberg, E.; Geis, A.L.; Chan, J.L.; Fu, K.; DeStefano Shields, C.E.; Dejea, C.M.; Fathi, P.; Chen, J.; Finard, B.B.; et al. Bacteroides fragilis Toxin Coordinates a Pro-carcinogenic Inflammatory Cascade via Targeting of Colonic Epithelial Cells. Cell Host Microbe 2018, 23, 203–214.e205. [Google Scholar] [CrossRef]
- Rutkowski, M.R.; Stephen, T.L.; Svoronos, N.; Allegrezza, M.J.; Tesone, A.J.; Perales-Puchalt, A.; Brencicova, E.; Escovar-Fadul, X.; Nguyen, J.M.; Cadungog, M.G.; et al. Microbially driven TLR5-dependent signaling governs distal malignant progression through tumor-promoting inflammation. Cancer Cell 2015, 27, 27–40. [Google Scholar] [CrossRef]
- Grivennikov, S.I.; Wang, K.; Mucida, D.; Stewart, C.A.; Schnabl, B.; Jauch, D.; Taniguchi, K.; Yu, G.-Y.; Osterreicher, C.H.; Hung, K.E.; et al. Adenoma-linked barrier defects and microbial products drive IL-23/IL-17-mediated tumour growth. Nature 2012, 491, 254–258. [Google Scholar] [CrossRef]
- Wu, S.; Rhee, K.-J.; Albesiano, E.; Rabizadeh, S.; Wu, X.; Yen, H.-R.; Huso, D.L.; Brancati, F.L.; Wick, E.; McAllister, F.; et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat. Med. 2009, 15, 1016–1022. [Google Scholar] [CrossRef]
- Yang, Z.; Zhang, B.; Li, D.; Lv, M.; Huang, C.; Shen, G.X.; Huang, B. Mast cells mobilize myeloid-derived suppressor cells and Treg cells in tumor microenvironment via IL-17 pathway in murine hepatocarcinoma model. PLoS ONE 2010, 5, e8922. [Google Scholar] [CrossRef]
- De Aquino, S.G.; Abdollahi-Roodsaz, S.; Koenders, M.I.; van de Loo, F.A.J.; Pruijn, G.J.M.; Marijnissen, R.J.; Walgreen, B.; Helsen, M.M.; van den Bersselaar, L.A.; de Molon, R.S.; et al. Periodontal pathogens directly promote autoimmune experimental arthritis by inducing a TLR2- and IL-1-driven Th17 response. J. Immunol. 2014, 192, 4103–4111. [Google Scholar] [CrossRef] [PubMed]
- Calcinotto, A.; Brevi, A.; Chesi, M.; Ferrarese, R.; Garcia Perez, L.; Grioni, M.; Kumar, S.; Garbitt, V.M.; Sharik, M.E.; Henderson, K.J.; et al. Microbiota-driven interleukin-17-producing cells and eosinophils synergize to accelerate multiple myeloma progression. Nat. Commun. 2018, 9, 4832. [Google Scholar] [CrossRef] [PubMed]
- Hideshima, T.; Mitsiades, C.; Tonon, G.; Richardson, P.G.; Anderson, K.C. Understanding multiple myeloma pathogenesis in the bone marrow to identify new therapeutic targets. Nat. Rev. Cancer 2007, 7, 585–598. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Yi, T.; Kortylewski, M.; Pardoll, D.M.; Zeng, D.; Yu, H. IL-17 can promote tumor growth through an IL-6-Stat3 signaling pathway. J. Exp. Med. 2009, 206, 1457–1464. [Google Scholar] [CrossRef]
- Wang, S.; Ma, Y.; Wang, X.; Jiang, J.; Zhang, C.; Wang, X.; Jiang, Y.; Huang, H.; Hong, L. IL-17A Increases Multiple Myeloma Cell Viability by Positively Regulating Syk Expression. Transl. Oncol. 2019, 12, 1086–1091. [Google Scholar] [CrossRef]
- Ramirez-Carrozzi, V.; Sambandam, A.; Luis, E.; Lin, Z.; Jeet, S.; Lesch, J.; Hackney, J.; Kim, J.; Zhou, M.; Lai, J.; et al. IL-17C regulates the innate immune function of epithelial cells in an autocrine manner. Nat. Immunol. 2011, 12, 1159–1166. [Google Scholar] [CrossRef]
- Song, X.; Gao, H.; Lin, Y.; Yao, Y.; Zhu, S.; Wang, J.; Liu, Y.; Yao, X.; Meng, G.; Shen, N.; et al. Alterations in the microbiota drive interleukin-17C production from intestinal epithelial cells to promote tumorigenesis. Immunity 2014, 40, 140–152. [Google Scholar] [CrossRef]
- McGeachy, M.J.; Cua, D.J.; Gaffen, S.L. The IL-17 Family of Cytokines in Health and Disease. Immunity 2019, 50, 892–906. [Google Scholar] [CrossRef]
- Brevi, A.; Cogrossi, L.L.; Grazia, G.; Masciovecchio, D.; Impellizzieri, D.; Lacanfora, L.; Grioni, M.; Bellone, M. Much More Than IL-17A: Cytokines of the IL-17 Family Between Microbiota and Cancer. Front. Immunol. 2020, 11, 565470. [Google Scholar] [CrossRef]
- Miossec, P.; Kolls, J.K. Targeting IL-17 and TH17 cells in chronic inflammation. Nat. Rev. Drug Discov. 2012, 11, 763–776. [Google Scholar] [CrossRef]
- Chanmee, T.; Ontong, P.; Konno, K.; Itano, N. Tumor-associated macrophages as major players in the tumor microenvironment. Cancers 2014, 6, 1670–1690. [Google Scholar] [CrossRef] [PubMed]
- Dong, F.; Qin, X.; Wang, B.; Li, Q.; Hu, J.; Cheng, X.; Guo, D.; Cheng, F.; Fang, C.; Tan, Y.; et al. ALKBH5 Facilitates Hypoxia-Induced Paraspeckle Assembly and IL8 Secretion to Generate an Immunosuppressive Tumor Microenvironment. Cancer Res. 2021, 81, 5876–5888. [Google Scholar] [CrossRef] [PubMed]
- Wenes, M.; Shang, M.; Di Matteo, M.; Goveia, J.; Martín-Pérez, R.; Serneels, J.; Prenen, H.; Ghesquière, B.; Carmeliet, P.; Mazzone, M. Macrophage Metabolism Controls Tumor Blood Vessel Morphogenesis and Metastasis. Cell Metab. 2016, 24, 701–715. [Google Scholar] [CrossRef] [PubMed]
- Ahirwar, D.K.; Charan, M.; Mishra, S.; Verma, A.K.; Shilo, K.; Ramaswamy, B.; Ganju, R.K. Slit2 Inhibits Breast Cancer Metastasis by Activating M1-Like Phagocytic and Antifibrotic Macrophages. Cancer Res. 2021, 81, 5255–5267. [Google Scholar] [CrossRef]
- Espindola, M.S.; Habiel, D.M.; Narayanan, R.; Jones, I.; Coelho, A.L.; Murray, L.A.; Jiang, D.; Noble, P.W.; Hogaboam, C.M. Targeting of TAM Receptors Ameliorates Fibrotic Mechanisms in Idiopathic Pulmonary Fibrosis. Am. J. Respir. Crit. Care Med. 2018, 197, 1443–1456. [Google Scholar] [CrossRef]
- Torres, S.; Bartolomé, R.A.; Mendes, M.; Barderas, R.; Fernandez-Aceñero, M.J.; Peláez-García, A.; Peña, C.; Lopez-Lucendo, M.; Villar-Vázquez, R.; de Herreros, A.G.; et al. Proteome profiling of cancer-associated fibroblasts identifies novel proinflammatory signatures and prognostic markers for colorectal cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2013, 19, 6006–6019. [Google Scholar] [CrossRef]
- Ruffell, B.; Coussens, L.M. Macrophages and therapeutic resistance in cancer. Cancer Cell 2015, 27, 462–472. [Google Scholar] [CrossRef]
- Italiani, P.; Boraschi, D. From Monocytes to M1/M2 Macrophages: Phenotypical vs. Functional Differentiation. Front. Immunol. 2014, 5, 514. [Google Scholar] [CrossRef]
- Bruns, H.; Büttner, M.; Fabri, M.; Mougiakakos, D.; Bittenbring, J.T.; Hoffmann, M.H.; Beier, F.; Pasemann, S.; Jitschin, R.; Hofmann, A.D.; et al. Vitamin D-dependent induction of cathelicidin in human macrophages results in cytotoxicity against high-grade B cell lymphoma. Sci. Transl. Med. 2015, 7, 282ra247. [Google Scholar] [CrossRef]
- Noy, R.; Pollard, J.W. Tumor-associated macrophages: From mechanisms to therapy. Immunity 2014, 41, 49–61. [Google Scholar] [CrossRef]
- Pan, Y.; Yu, Y.; Wang, X.; Zhang, T. Tumor-Associated Macrophages in Tumor Immunity. Front. Immunol. 2020, 11, 583084. [Google Scholar] [CrossRef] [PubMed]
- Martinez, F.O.; Helming, L.; Gordon, S. Alternative activation of macrophages: An immunologic functional perspective. Annu. Rev. Immunol. 2009, 27, 451–483. [Google Scholar] [CrossRef] [PubMed]
- Van Dyken, S.J.; Locksley, R.M. Interleukin-4- and interleukin-13-mediated alternatively activated macrophages: Roles in homeostasis and disease. Annu. Rev. Immunol. 2013, 31, 317–343. [Google Scholar] [CrossRef] [PubMed]
- Redente, E.F.; Dwyer-Nield, L.D.; Merrick, D.T.; Raina, K.; Agarwal, R.; Pao, W.; Rice, P.L.; Shroyer, K.R.; Malkinson, A.M. Tumor progression stage and anatomical site regulate tumor-associated macrophage and bone marrow-derived monocyte polarization. Am. J. Pathol. 2010, 176, 2972–2985. [Google Scholar] [CrossRef]
- Kuang, D.M.; Zhao, Q.; Peng, C.; Xu, J.; Zhang, J.P.; Wu, C.; Zheng, L. Activated monocytes in peritumoral stroma of hepatocellular carcinoma foster immune privilege and disease progression through PD-L1. J. Exp. Med. 2009, 206, 1327–1337. [Google Scholar] [CrossRef]
- Curiel, T.J.; Coukos, G.; Zou, L.; Alvarez, X.; Cheng, P.; Mottram, P.; Evdemon-Hogan, M.; Conejo-Garcia, J.R.; Zhang, L.; Burow, M.; et al. Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival. Nat. Med. 2004, 10, 942–949. [Google Scholar] [CrossRef]
- Torroella-Kouri, M.; Silvera, R.; Rodriguez, D.; Caso, R.; Shatry, A.; Opiela, S.; Ilkovitch, D.; Schwendener, R.A.; Iragavarapu-Charyulu, V.; Cardentey, Y.; et al. Identification of a subpopulation of macrophages in mammary tumor-bearing mice that are neither M1 nor M2 and are less differentiated. Cancer Res. 2009, 69, 4800–4809. [Google Scholar] [CrossRef]
- Sami, E.; Paul, B.T.; Koziol, J.A.; ElShamy, W.M. The Immunosuppressive Microenvironment in BRCA1-IRIS-Overexpressing TNBC Tumors Is Induced by Bidirectional Interaction with Tumor-Associated Macrophages. Cancer Res. 2020, 80, 1102–1117. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, X.; Zhu, L.; Yang, X.; He, F.; Wang, T.; Bao, T.; Lu, H.; Wang, H.; Yang, S. Inulin alleviates inflammation of alcoholic liver disease via SCFAs-inducing suppression of M1 and facilitation of M2 macrophages in mice. Int. Immunopharmacol. 2020, 78, 106062. [Google Scholar] [CrossRef]
- Wang, L.; Gong, Z.; Zhang, X.; Zhu, F.; Liu, Y.; Jin, C.; Du, X.; Xu, C.; Chen, Y.; Cai, W.; et al. Gut microbial bile acid metabolite skews macrophage polarization and contributes to high-fat diet-induced colonic inflammation. Gut Microbes 2020, 12, 1819155. [Google Scholar] [CrossRef]
- Long, J.; Liu, X.K.; Kang, Z.P.; Wang, M.X.; Zhao, H.M.; Huang, J.Q.; Xiao, Q.P.; Liu, D.Y.; Zhong, Y.B. Ginsenoside Rg1 ameliorated experimental colitis by regulating the balance of M1/M2 macrophage polarization and the homeostasis of intestinal flora. Eur. J. Pharmacol. 2022, 917, 174742. [Google Scholar] [CrossRef] [PubMed]
- Kleer, C.G.; Bloushtain-Qimron, N.; Chen, Y.H.; Carrasco, D.; Hu, M.; Yao, J.; Kraeft, S.K.; Collins, L.C.; Sabel, M.S.; Argani, P.; et al. Epithelial and stromal cathepsin K and CXCL14 expression in breast tumor progression. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2008, 14, 5357–5367. [Google Scholar] [CrossRef] [PubMed]
- Novinec, M.; Lenarčič, B. Cathepsin K: A unique collagenolytic cysteine peptidase. Biol. Chem. 2013, 394, 1163–1179. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Ma, Y.; Zhang, Y.; Pan, Z.; Lu, Y.; Liu, P.; Lu, B. Identification of Cathepsin K in the Peritoneal Metastasis of Ovarian Carcinoma Using In-silico, Gene Expression Analysis. J. Cancer 2016, 7, 722–729. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Zhou, R.; Wang, H.; Li, W.; Pan, M.; Yao, X.; Zhan, W.; Yang, S.; Xu, L.; Ding, Y.; et al. Gut microbiota-stimulated cathepsin K secretion mediates TLR4-dependent M2 macrophage polarization and promotes tumor metastasis in colorectal cancer. Cell Death Differ. 2019, 26, 2447–2463. [Google Scholar] [CrossRef]
- Chai, N.; Xiong, Y.; Zhang, Y.; Cheng, Y.; Shi, W.; Yao, Y.; Sui, H.; Zhu, H. YYFZBJS inhibits colorectal tumorigenesis by remodeling gut microbiota and influence on M2 macrophage polarization in vivo and in vitro. Am. J. Cancer Res. 2021, 11, 5338–5357. [Google Scholar] [CrossRef]
- Zmora, N.; Suez, J.; Elinav, E. You are what you eat: Diet, health and the gut microbiota. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 35–56. [Google Scholar] [CrossRef]
- O’Keefe, S.J.; Li, J.V.; Lahti, L.; Ou, J.; Carbonero, F.; Mohammed, K.; Posma, J.M.; Kinross, J.; Wahl, E.; Ruder, E.; et al. Fat, fibre and cancer risk in African Americans and rural Africans. Nat. Commun. 2015, 6, 6342. [Google Scholar] [CrossRef]
- Flint, H.J.; Duncan, S.H.; Scott, K.P.; Louis, P. Links between diet, gut microbiota composition and gut metabolism. Proc. Nutr. Soc. 2015, 74, 13–22. [Google Scholar] [CrossRef]
- Liu, T.; Guo, Z.; Song, X.; Liu, L.; Dong, W.; Wang, S.; Xu, M.; Yang, C.; Wang, B.; Cao, H. High-fat diet-induced dysbiosis mediates MCP-1/CCR2 axis-dependent M2 macrophage polarization and promotes intestinal adenoma-adenocarcinoma sequence. J. Cell. Mol. Med. 2020, 24, 2648–2662. [Google Scholar] [CrossRef]
- Hezaveh, K.; Shinde, R.S.; Klötgen, A.; Halaby, M.J.; Lamorte, S.; Ciudad, M.T.; Quevedo, R.; Neufeld, L.; Liu, Z.Q.; Jin, R.; et al. Tryptophan-derived microbial metabolites activate the aryl hydrocarbon receptor in tumor-associated macrophages to suppress anti-tumor immunity. Immunity 2022, 55, 324–340.e328. [Google Scholar] [CrossRef] [PubMed]
- Coussens, L.M.; Zitvogel, L.; Palucka, A.K. Neutralizing tumor-promoting chronic inflammation: A magic bullet? Science 2013, 339, 286–291. [Google Scholar] [CrossRef] [PubMed]
- Mitchem, J.B.; Brennan, D.J.; Knolhoff, B.L.; Belt, B.A.; Zhu, Y.; Sanford, D.E.; Belaygorod, L.; Carpenter, D.; Collins, L.; Piwnica-Worms, D.; et al. Targeting tumor-infiltrating macrophages decreases tumor-initiating cells, relieves immunosuppression, and improves chemotherapeutic responses. Cancer Res. 2013, 73, 1128–1141. [Google Scholar] [CrossRef] [PubMed]
- DeNardo, D.G.; Brennan, D.J.; Rexhepaj, E.; Ruffell, B.; Shiao, S.L.; Madden, S.F.; Gallagher, W.M.; Wadhwani, N.; Keil, S.D.; Junaid, S.A.; et al. Leukocyte complexity predicts breast cancer survival and functionally regulates response to chemotherapy. Cancer Discov. 2011, 1, 54–67. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Niu, M.; O’Mary, H.; Cui, Z. Targeting of tumor-associated macrophages made possible by PEG-sheddable, mannose-modified nanoparticles. Mol. Pharm. 2013, 10, 3525–3530. [Google Scholar] [CrossRef] [PubMed]
- Nagai, T.; Tanaka, M.; Tsuneyoshi, Y.; Xu, B.; Michie, S.A.; Hasui, K.; Hirano, H.; Arita, K.; Matsuyama, T. Targeting tumor-associated macrophages in an experimental glioma model with a recombinant immunotoxin to folate receptor beta. Cancer Immunol. Immunother. CII 2009, 58, 1577–1586. [Google Scholar] [CrossRef]
- Lin, Y.; Wei, C.; Liu, Y.; Qiu, Y.; Liu, C.; Guo, F. Selective ablation of tumor-associated macrophages suppresses metastasis and angiogenesis. Cancer Sci. 2013, 104, 1217–1225. [Google Scholar] [CrossRef]
- Cieslewicz, M.; Tang, J.; Yu, J.L.; Cao, H.; Zavaljevski, M.; Motoyama, K.; Lieber, A.; Raines, E.W.; Pun, S.H. Targeted delivery of proapoptotic peptides to tumor-associated macrophages improves survival. Proc. Natl. Acad. Sci. USA 2013, 110, 15919–15924. [Google Scholar] [CrossRef]
- Fan, L.; Xu, C.; Ge, Q.; Lin, Y.; Wong, C.C.; Qi, Y.; Ye, B.; Lian, Q.; Zhuo, W.; Si, J.; et al. A. Muciniphila Suppresses Colorectal Tumorigenesis by Inducing TLR2/NLRP3-Mediated M1-Like TAMs. Cancer Immunol. Res. 2021, 9, 1111–1124. [Google Scholar] [CrossRef]
- Yang, Y.; Weng, W.; Peng, J.; Hong, L.; Yang, L.; Toiyama, Y.; Gao, R.; Liu, M.; Yin, M.; Pan, C.; et al. Fusobacterium nucleatum Increases Proliferation of Colorectal Cancer Cells and Tumor Development in Mice by Activating Toll-Like Receptor 4 Signaling to Nuclear Factor-κB, and Up-regulating Expression of MicroRNA-21. Gastroenterology 2017, 152, 851–866.e824. [Google Scholar] [CrossRef]
- Hong, J.; Guo, F.; Lu, S.Y.; Shen, C.; Ma, D.; Zhang, X.; Xie, Y.; Yan, T.; Yu, T.; Sun, T.; et al. F. nucleatum targets lncRNA ENO1-IT1 to promote glycolysis and oncogenesis in colorectal cancer. Gut 2021, 70, 2123–2137. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Fan, L.; Lin, Y.; Shen, W.; Qi, Y.; Zhang, Y.; Chen, Z.; Wang, L.; Long, Y.; Hou, T.; et al. Fusobacterium nucleatum promotes colorectal cancer metastasis through miR-1322/CCL20 axis and M2 polarization. Gut Microbes 2021, 13, 1980347. [Google Scholar] [CrossRef]
- Mima, K.; Sukawa, Y.; Nishihara, R.; Qian, Z.R.; Yamauchi, M.; Inamura, K.; Kim, S.A.; Masuda, A.; Nowak, J.A.; Nosho, K.; et al. Fusobacterium nucleatum and T Cells in Colorectal Carcinoma. JAMA Oncol. 2015, 1, 653–661. [Google Scholar] [CrossRef] [PubMed]
- Nosho, K.; Sukawa, Y.; Adachi, Y.; Ito, M.; Mitsuhashi, K.; Kurihara, H.; Kanno, S.; Yamamoto, I.; Ishigami, K.; Igarashi, H.; et al. Association of Fusobacterium nucleatum with immunity and molecular alterations in colorectal cancer. World J. Gastroenterol. 2016, 22, 557–566. [Google Scholar] [CrossRef] [PubMed]
- Borowsky, J.; Haruki, K.; Lau, M.C.; Dias Costa, A.; Väyrynen, J.P.; Ugai, T.; Arima, K.; da Silva, A.; Felt, K.D.; Zhao, M.; et al. Association of Fusobacterium nucleatum with Specific T-cell Subsets in the Colorectal Carcinoma Microenvironment. Clin. Cancer Res. 2021, 27, 2816–2826. [Google Scholar] [CrossRef] [PubMed]
- Parhi, L.; Alon-Maimon, T.; Sol, A.; Nejman, D.; Shhadeh, A.; Fainsod-Levi, T.; Yajuk, O.; Isaacson, B.; Abed, J.; Maalouf, N.; et al. Breast cancer colonization by Fusobacterium nucleatum accelerates tumor growth and metastatic progression. Nat. Commun. 2020, 11, 3259. [Google Scholar] [CrossRef] [PubMed]
- Gur, C.; Ibrahim, Y.; Isaacson, B.; Yamin, R.; Abed, J.; Gamliel, M.; Enk, J.; Bar-On, Y.; Stanietsky-Kaynan, N.; Coppenhagen-Glazer, S.; et al. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity 2015, 42, 344–355. [Google Scholar] [CrossRef]
- Chu, H.; Mazmanian, S.K. Innate immune recognition of the microbiota promotes host-microbial symbiosis. Nat. Immunol. 2013, 14, 668–675. [Google Scholar] [CrossRef]
- Round, J.L.; Lee, S.M.; Li, J.; Tran, G.; Jabri, B.; Chatila, T.A.; Mazmanian, S.K. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science 2011, 332, 974–977. [Google Scholar] [CrossRef]
- Mazmanian, S.K.; Liu, C.H.; Tzianabos, A.O.; Kasper, D.L. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 2005, 122, 107–118. [Google Scholar] [CrossRef]
- Jarnicki, A.G.; Lysaght, J.; Todryk, S.; Mills, K.H.G. Suppression of antitumor immunity by IL-10 and TGF-beta-producing T cells infiltrating the growing tumor: Influence of tumor environment on the induction of CD4+ and CD8+ regulatory T cells. J. Immunol. 2006, 177, 896–904. [Google Scholar] [CrossRef] [PubMed]
- Wing, K.; Onishi, Y.; Prieto-Martin, P.; Yamaguchi, T.; Miyara, M.; Fehervari, Z.; Nomura, T.; Sakaguchi, S. CTLA-4 control over Foxp3+ regulatory T cell function. Science 2008, 322, 271–275. [Google Scholar] [CrossRef] [PubMed]
- Peuker, K.; Strigli, A.; Tauriello, D.V.F.; Hendricks, A.; von Schönfels, W.; Burmeister, G.; Brosch, M.; Herrmann, A.; Krüger, S.; Nitsche, J.; et al. Microbiota-dependent activation of the myeloid calcineurin-NFAT pathway inhibits B7H3- and B7H4-dependent anti-tumor immunity in colorectal cancer. Immunity 2022, 55, 701–717.e707. [Google Scholar] [CrossRef] [PubMed]
- Uribe-Herranz, M.; Bittinger, K.; Rafail, S.; Guedan, S.; Pierini, S.; Tanes, C.; Ganetsky, A.; Morgan, M.A.; Gill, S.; Tanyi, J.L.; et al. Gut microbiota modulates adoptive cell therapy via CD8α dendritic cells and IL-12. JCI Insight 2018, 3, e94952. [Google Scholar] [CrossRef] [PubMed]
- Uribe-Herranz, M.; Rafail, S.; Beghi, S.; Gil-de-Gómez, L.; Verginadis, I.; Bittinger, K.; Pustylnikov, S.; Pierini, S.; Perales-Linares, R.; Blair, I.A.; et al. Gut microbiota modulate dendritic cell antigen presentation and radiotherapy-induced antitumor immune response. J. Clin. Investig. 2020, 130, 466–479. [Google Scholar] [CrossRef]
- Wang, D.; Dubois, R.N. Eicosanoids and cancer. Nat. Rev. Cancer 2010, 10, 181–193. [Google Scholar] [CrossRef]
- Mao, Y.; Poschke, I.; Wennerberg, E.; Pico de Coaña, Y.; Egyhazi Brage, S.; Schultz, I.; Hansson, J.; Masucci, G.; Lundqvist, A.; Kiessling, R. Melanoma-educated CD14+ cells acquire a myeloid-derived suppressor cell phenotype through COX-2-dependent mechanisms. Cancer Res. 2013, 73, 3877–3887. [Google Scholar] [CrossRef]
- O’Callaghan, G.; Ryan, A.; Neary, P.; O’Mahony, C.; Shanahan, F.; Houston, A. Targeting the EP1 receptor reduces Fas ligand expression and increases the antitumor immune response in an in vivo model of colon cancer. Int. J. Cancer 2013, 133, 825–834. [Google Scholar] [CrossRef]
- Loo, T.M.; Kamachi, F.; Watanabe, Y.; Yoshimoto, S.; Kanda, H.; Arai, Y.; Nakajima-Takagi, Y.; Iwama, A.; Koga, T.; Sugimoto, Y.; et al. Gut Microbiota Promotes Obesity-Associated Liver Cancer through PGE-Mediated Suppression of Antitumor Immunity. Cancer Discov. 2017, 7, 522–538. [Google Scholar] [CrossRef]
- Mizuno, R.; Kawada, K.; Sakai, Y. Prostaglandin E2/EP Signaling in the Tumor Microenvironment of Colorectal Cancer. Int. J. Mol. Sci. 2019, 20, 6254. [Google Scholar] [CrossRef]
- Fu, A.; Yao, B.; Dong, T.; Chen, Y.; Yao, J.; Liu, Y.; Li, H.; Bai, H.; Liu, X.; Zhang, Y.; et al. Tumor-resident intracellular microbiota promotes metastatic colonization in breast cancer. Cell 2022, 185, 1356–1372.e1326. [Google Scholar] [CrossRef] [PubMed]
- Nejman, D.; Livyatan, I.; Fuks, G.; Gavert, N.; Zwang, Y.; Geller, L.T.; Rotter-Maskowitz, A.; Weiser, R.; Mallel, G.; Gigi, E.; et al. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science 2020, 368, 973–980. [Google Scholar] [CrossRef] [PubMed]
- Pushalkar, S.; Hundeyin, M.; Daley, D.; Zambirinis, C.P.; Kurz, E.; Mishra, A.; Mohan, N.; Aykut, B.; Usyk, M.; Torres, L.E.; et al. The Pancreatic Cancer Microbiome Promotes Oncogenesis by Induction of Innate and Adaptive Immune Suppression. Cancer Discov. 2018, 8, 403–416. [Google Scholar] [CrossRef] [PubMed]
- Jin, C.; Lagoudas, G.K.; Zhao, C.; Bullman, S.; Bhutkar, A.; Hu, B.; Ameh, S.; Sandel, D.; Liang, X.S.; Mazzilli, S.; et al. Commensal Microbiota Promote Lung Cancer Development via γδ T Cells. Cell 2019, 176, 998–1013.e1016. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Cao, X. Immunosuppressive cells in tumor immune escape and metastasis. J. Mol. Med. 2016, 94, 509–522. [Google Scholar] [CrossRef]
- Minton, K. Immunotherapy: Cytokine boost for CAR T cells. Nat. Rev. Immunol. 2018, 18, 150–151. [Google Scholar] [CrossRef]
- Pulendran, B.; Davis, M.M. The science and medicine of human immunology. Science 2020, 369. [Google Scholar] [CrossRef]
- Demaria, O.; Cornen, S.; Daëron, M.; Morel, Y.; Medzhitov, R.; Vivier, E. Harnessing innate immunity in cancer therapy. Nature 2019, 574, 45–56. [Google Scholar] [CrossRef]
- Gopalakrishnan, V.; Helmink, B.A.; Spencer, C.N.; Reuben, A.; Wargo, J.A. The Influence of the Gut Microbiome on Cancer, Immunity, and Cancer Immunotherapy. Cancer Cell 2018, 33, 570–580. [Google Scholar] [CrossRef]
- Iida, N.; Dzutsev, A.; Stewart, C.A.; Smith, L.; Bouladoux, N.; Weingarten, R.A.; Molina, D.A.; Salcedo, R.; Back, T.; Cramer, S.; et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science 2013, 342, 967–970. [Google Scholar] [CrossRef]
- Di Modica, M.; Gargari, G.; Regondi, V.; Bonizzi, A.; Arioli, S.; Belmonte, B.; De Cecco, L.; Fasano, E.; Bianchi, F.; Bertolotti, A.; et al. Gut Microbiota Condition the Therapeutic Efficacy of Trastuzumab in HER2-Positive Breast Cancer. Cancer Res. 2021, 81, 2195–2206. [Google Scholar] [CrossRef]
- Sharma, P.; Allison, J.P. The future of immune checkpoint therapy. Science 2015, 348, 56–61. [Google Scholar] [CrossRef] [PubMed]
- Ribas, A.; Wolchok, J.D. Cancer immunotherapy using checkpoint blockade. Science 2018, 359, 1350–1355. [Google Scholar] [CrossRef]
- Carlino, M.S.; Larkin, J.; Long, G.V. Immune checkpoint inhibitors in melanoma. Lancet 2021, 398, 1002–1014. [Google Scholar] [CrossRef]
- Pan, C.; Liu, H.; Robins, E.; Song, W.; Liu, D.; Li, Z.; Zheng, L. Next-generation immuno-oncology agents: Current momentum shifts in cancer immunotherapy. J. Hematol. Oncol. 2020, 13, 29. [Google Scholar] [CrossRef]
- Matson, V.; Fessler, J.; Bao, R.; Chongsuwat, T.; Zha, Y.; Alegre, M.L.; Luke, J.J.; Gajewski, T.F. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science 2018, 359, 104–108. [Google Scholar] [CrossRef]
- Davar, D.; Dzutsev, A.K.; McCulloch, J.A.; Rodrigues, R.R.; Chauvin, J.M.; Morrison, R.M.; Deblasio, R.N.; Menna, C.; Ding, Q.; Pagliano, O.; et al. Fecal microbiota transplant overcomes resistance to anti-PD-1 therapy in melanoma patients. Science 2021, 371, 595–602. [Google Scholar] [CrossRef] [PubMed]
- Paulos, C.M.; Wrzesinski, C.; Kaiser, A.; Hinrichs, C.S.; Chieppa, M.; Cassard, L.; Palmer, D.C.; Boni, A.; Muranski, P.; Yu, Z.; et al. Microbial translocation augments the function of adoptively transferred self/tumor-specific CD8+ T cells via TLR4 signaling. J. Clin. Investig. 2007, 117, 2197–2204. [Google Scholar] [CrossRef]
- Gopalakrishnan, V.; Spencer, C.N.; Nezi, L.; Reuben, A.; Andrews, M.C.; Karpinets, T.V.; Prieto, P.A.; Vicente, D.; Hoffman, K.; Wei, S.C.; et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 2018, 359, 97–103. [Google Scholar] [CrossRef]
- Lee, S.H.; Cho, S.Y.; Yoon, Y.; Park, C.; Sohn, J.; Jeong, J.J.; Jeon, B.N.; Jang, M.; An, C.; Lee, S.; et al. Bifidobacterium bifidum strains synergize with immune checkpoint inhibitors to reduce tumour burden in mice. Nat. Microbiol. 2021, 6, 277–288. [Google Scholar] [CrossRef]
- Routy, B.; Le Chatelier, E.; Derosa, L.; Duong, C.P.M.; Alou, M.T.; Daillère, R.; Fluckiger, A.; Messaoudene, M.; Rauber, C.; Roberti, M.P.; et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 2018, 359, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Vétizou, M.; Pitt, J.M.; Daillère, R.; Lepage, P.; Waldschmitt, N.; Flament, C.; Rusakiewicz, S.; Routy, B.; Roberti, M.P.; Duong, C.P.; et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 2015, 350, 1079–1084. [Google Scholar] [CrossRef] [PubMed]
- Clavel, T.; Gomes-Neto, J.C.; Lagkouvardos, I.; Ramer-Tait, A.E. Deciphering interactions between the gut microbiota and the immune system via microbial cultivation and minimal microbiomes. Immunol. Rev. 2017, 279, 8–22. [Google Scholar] [CrossRef]
- Mager, L.F.; Burkhard, R.; Pett, N.; Cooke, N.C.A.; Brown, K.; Ramay, H.; Paik, S.; Stagg, J.; Groves, R.A.; Gallo, M.; et al. Microbiome-derived inosine modulates response to checkpoint inhibitor immunotherapy. Science 2020, 369, 1481–1489. [Google Scholar] [CrossRef]
- Coutzac, C.; Jouniaux, J.M.; Paci, A.; Schmidt, J.; Mallardo, D.; Seck, A.; Asvatourian, V.; Cassard, L.; Saulnier, P.; Lacroix, L.; et al. Systemic short chain fatty acids limit antitumor effect of CTLA-4 blockade in hosts with cancer. Nat. Commun. 2020, 11, 2168. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Lv, J.; Guo, F.; Li, J.; Jia, Y.; Jiang, D.; Wang, N.; Zhang, C.; Kong, L.; Liu, Y.; et al. Gut Microbiome Influences the Efficacy of PD-1 Antibody Immunotherapy on MSS-Type Colorectal Cancer via Metabolic Pathway. Front. Microbiol. 2020, 11, 814. [Google Scholar] [CrossRef]
- Holland, T.; Fowler, V.G., Jr.; Shelburne, S.A., 3rd. Invasive gram-positive bacterial infection in cancer patients. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2014, 59 (Suppl. 5), S331–S334. [Google Scholar] [CrossRef]
- Bucaneve, G.; Micozzi, A.; Menichetti, F.; Martino, P.; Dionisi, M.S.; Martinelli, G.; Allione, B.; D’Antonio, D.; Buelli, M.; Nosari, A.M.; et al. Levofloxacin to prevent bacterial infection in patients with cancer and neutropenia. N. Engl. J. Med. 2005, 353, 977–987. [Google Scholar] [CrossRef]
- Attiê, R.; Chinen, L.T.; Yoshioka, E.M.; Silva, M.C.; de Lima, V.C. Acute bacterial infection negatively impacts cancer specific survival of colorectal cancer patients. World J. Gastroenterol. 2014, 20, 13930–13935. [Google Scholar] [CrossRef]
- Johanesen, P.A.; Mackin, K.E.; Hutton, M.L.; Awad, M.M.; Larcombe, S.; Amy, J.M.; Lyras, D. Disruption of the Gut Microbiome: Clostridium difficile Infection and the Threat of Antibiotic Resistance. Genes 2015, 6, 1347–1360. [Google Scholar] [CrossRef]
- Leffler, D.A.; Lamont, J.T. Clostridium difficile infection. N. Engl. J. Med. 2015, 372, 1539–1548. [Google Scholar] [CrossRef] [PubMed]
- Lastours, V.D.; Fantin, B. Resistance to fluoroquinolones in 2013: What are the consequences in internal medicine? Rev. Med. Interne 2014, 35, 601–608. [Google Scholar] [PubMed]
- Galloway-Peña, J.; Brumlow, C.; Shelburne, S. Impact of the Microbiota on Bacterial Infections during Cancer Treatment. Trends Microbiol. 2017, 25, 992–1004. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.B.; Zhou, Y.L.; Fang, J.Y. Gut Microbiota in Cancer Immune Response and Immunotherapy. Trends Cancer 2021, 7, 647–660. [Google Scholar] [CrossRef] [PubMed]
- Lurienne, L.; Cervesi, J.; Duhalde, L.; de Gunzburg, J.; Andremont, A.; Zalcman, G.; Buffet, R.; Bandinelli, P.A. NSCLC Immunotherapy Efficacy and Antibiotic Use: A Systematic Review and Meta-Analysis. J. Thorac. Oncol. Off. Publ. Int. Assoc. Study Lung Cancer 2020, 15, 1147–1159. [Google Scholar] [CrossRef]
- Pinato, D.J.; Howlett, S.; Ottaviani, D.; Urus, H.; Patel, A.; Mineo, T.; Brock, C.; Power, D.; Hatcher, O.; Falconer, A.; et al. Association of Prior Antibiotic Treatment With Survival and Response to Immune Checkpoint Inhibitor Therapy in Patients With Cancer. JAMA Oncol. 2019, 5, 1774–1778. [Google Scholar] [CrossRef]
- Lehrnbecher, T.; Fisher, B.T.; Phillips, B.; Alexander, S.; Ammann, R.A.; Beauchemin, M.; Carlesse, F.; Castagnola, E.; Davis, B.L.; Dupuis, L.L.; et al. Guideline for Antibacterial Prophylaxis Administration in Pediatric Cancer and Hematopoietic Stem Cell Transplantation. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2020, 71, 226–236. [Google Scholar] [CrossRef]
- Goldman, J.D.; Gallaher, A.; Jain, R.; Stednick, Z.; Menon, M.; Boeckh, M.J.; Pottinger, P.S.; Schwartz, S.M.; Casper, C. Infusion-Compatible Antibiotic Formulations for Rapid Administration to Improve Outcomes in Cancer Outpatients With Severe Sepsis and Septic Shock: The Sepsis STAT Pack. J. Natl. Compr. Cancer Netw. JNCCN 2017, 15, 457–464. [Google Scholar] [CrossRef]
- Baruch, E.N.; Youngster, I.; Ben-Betzalel, G.; Ortenberg, R.; Lahat, A.; Katz, L.; Adler, K.; Dick-Necula, D.; Raskin, S.; Bloch, N.; et al. Fecal microbiota transplant promotes response in immunotherapy-refractory melanoma patients. Science 2021, 371, 602–609. [Google Scholar] [CrossRef]
- Spencer, C.N.; McQuade, J.L.; Gopalakrishnan, V.; McCulloch, J.A.; Vetizou, M.; Cogdill, A.P.; Khan, M.A.W.; Zhang, X.; White, M.G.; Peterson, C.B.; et al. Dietary fiber and probiotics influence the gut microbiome and melanoma immunotherapy response. Science 2021, 374, 1632–1640. [Google Scholar] [CrossRef]
- de Gunzburg, J.; Ghozlane, A.; Ducher, A.; Le Chatelier, E.; Duval, X.; Ruppé, E.; Armand-Lefevre, L.; Sablier-Gallis, F.; Burdet, C.; Alavoine, L.; et al. Protection of the Human Gut Microbiome From Antibiotics. J. Infect. Dis. 2018, 217, 628–636. [Google Scholar] [CrossRef] [PubMed]
- Khoder, M.; Tsapis, N.; Domergue-Dupont, V.; Gueutin, C.; Fattal, E. Removal of residual colonic ciprofloxacin in the rat by activated charcoal entrapped within zinc-pectinate beads. Eur. J. Pharm. Sci. Off. J. Eur. Fed. Pharm. Sci. 2010, 41, 281–288. [Google Scholar] [CrossRef]
- de Gunzburg, J.; Ducher, A.; Modess, C.; Wegner, D.; Oswald, S.; Dressman, J.; Augustin, V.; Feger, C.; Andremont, A.; Weitschies, W.; et al. Targeted adsorption of molecules in the colon with the novel adsorbent-based medicinal product, DAV132: A proof of concept study in healthy subjects. J. Clin. Pharmacol. 2015, 55, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Léonard, F.; Andremont, A.; Leclerq, B.; Labia, R.; Tancrède, C. Use of beta-lactamase-producing anaerobes to prevent ceftriaxone from degrading intestinal resistance to colonization. J. Infect. Dis. 1989, 160, 274–280. [Google Scholar] [CrossRef] [PubMed]
- Stiefel, U.; Pultz, N.J.; Harmoinen, J.; Koski, P.; Lindevall, K.; Helfand, M.S.; Donskey, C.J. Oral administration of beta-lactamase preserves colonization resistance of piperacillin-treated mice. J. Infect. Dis. 2003, 188, 1605–1609. [Google Scholar] [CrossRef][Green Version]
- Stiefel, U.; Nerandzic, M.M.; Koski, P.; Donskey, C.J. Orally administered beta-lactamase enzymes represent a novel strategy to prevent colonization by Clostridium difficile. J. Antimicrob. Chemother. 2008, 62, 1105–1108. [Google Scholar] [CrossRef]
- Rosenberg, S.A.; Restifo, N.P. Adoptive cell transfer as personalized immunotherapy for human cancer. Science 2015, 348, 62–68. [Google Scholar] [CrossRef]
- Verdegaal, E.M. Adoptive cell therapy: A highly successful individualized therapy for melanoma with great potential for other malignancies. Curr. Opin. Immunol. 2016, 39, 90–95. [Google Scholar] [CrossRef]
- Luu, M.; Weigand, K.; Wedi, F.; Breidenbend, C.; Leister, H.; Pautz, S.; Adhikary, T.; Visekruna, A. Regulation of the effector function of CD8(+) T cells by gut microbiota-derived metabolite butyrate. Sci. Rep. 2018, 8, 14430. [Google Scholar] [CrossRef]
- Cremonesi, E.; Governa, V.; Garzon, J.F.G.; Mele, V.; Amicarella, F.; Muraro, M.G.; Trella, E.; Galati-Fournier, V.; Oertli, D.; Däster, S.R.; et al. Gut microbiota modulate T cell trafficking into human colorectal cancer. Gut 2018, 67, 1984–1994. [Google Scholar] [CrossRef]
- Snyder, A.; Pamer, E.; Wolchok, J. IMMUNOTHERAPY. Could microbial therapy boost cancer immunotherapy? Science 2015, 350, 1031–1032. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Yin, Q.; Chen, L.; Davis, M.M. Bifidobacterium can mitigate intestinal immunopathology in the context of CTLA-4 blockade. Proc. Natl. Acad. Sci. USA 2018, 115, 157–161. [Google Scholar] [CrossRef] [PubMed]
- Vinay, D.S.; Ryan, E.P.; Pawelec, G.; Talib, W.H.; Stagg, J.; Elkord, E.; Lichtor, T.; Decker, W.K.; Whelan, R.L.; Kumara, H.; et al. Immune evasion in cancer: Mechanistic basis and therapeutic strategies. Semin. Cancer Biol. 2015, 35, S185–S198. [Google Scholar] [CrossRef]
- Han, B.; Sivaramakrishnan, P.; Lin, C.J.; Neve, I.A.A.; He, J.; Tay, L.W.R.; Sowa, J.N.; Sizovs, A.; Du, G.; Wang, J.; et al. Microbial Genetic Composition Tunes Host Longevity. Cell 2017, 169, 1249–1262.e1213. [Google Scholar] [CrossRef]
- Giménez-Bastida, J.A.; Ávila-Gálvez, M.; Espín, J.C.; González-Sarrías, A. The gut microbiota metabolite urolithin A, but not other relevant urolithins, induces p53-dependent cellular senescence in human colon cancer cells. Food Chem. Toxicol. Int. J. Publ. Br. Ind. Biol. Res. Assoc. 2020, 139, 111260. [Google Scholar] [CrossRef] [PubMed]
- Sugimura, N.; Li, Q.; Chu, E.S.H.; Lau, H.C.H.; Fong, W.; Liu, W.; Liang, C.; Nakatsu, G.; Su, A.C.Y.; Coker, O.O.; et al. Lactobacillus gallinarum modulates the gut microbiota and produces anti-cancer metabolites to protect against colorectal tumourigenesis. Gut 2021, 71, 2011–2021. [Google Scholar] [CrossRef]
- Muir, R.M.; Ibáñez, A.M.; Uratsu, S.L.; Ingham, E.S.; Leslie, C.A.; McGranahan, G.H.; Batra, N.; Goyal, S.; Joseph, J.; Jemmis, E.D.; et al. Mechanism of gallic acid biosynthesis in bacteria (Escherichia coli) and walnut (Juglans regia). Plant Mol. Biol. 2011, 75, 555–565. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).