ctDNA Is Useful to Detect Mutations at Codon 641 of Exon 16 of EZH2, a Biomarker for Relapse in Patients with Diffuse Large B-Cell Lymphoma
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Selection
2.2. Nucleic Acid Extraction
2.3. PCR Amplification
2.4. Sanger Sequencing
2.5. Droplet Digital PCR (ddPCR)
2.6. Statistical Analysis
3. Results
3.1. Patients
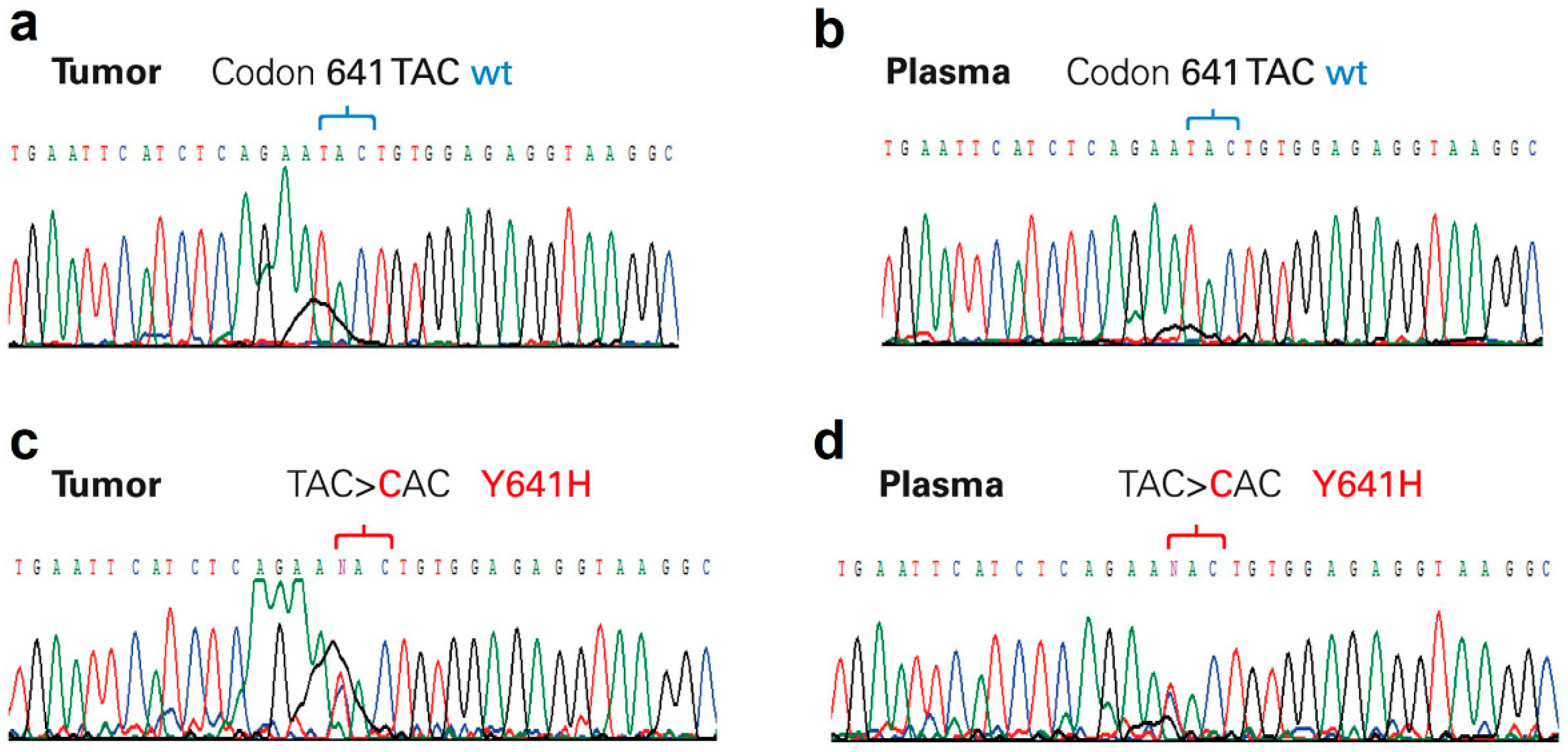
3.2. Analysis of EZH2 Mutations
3.3. Clinical Response
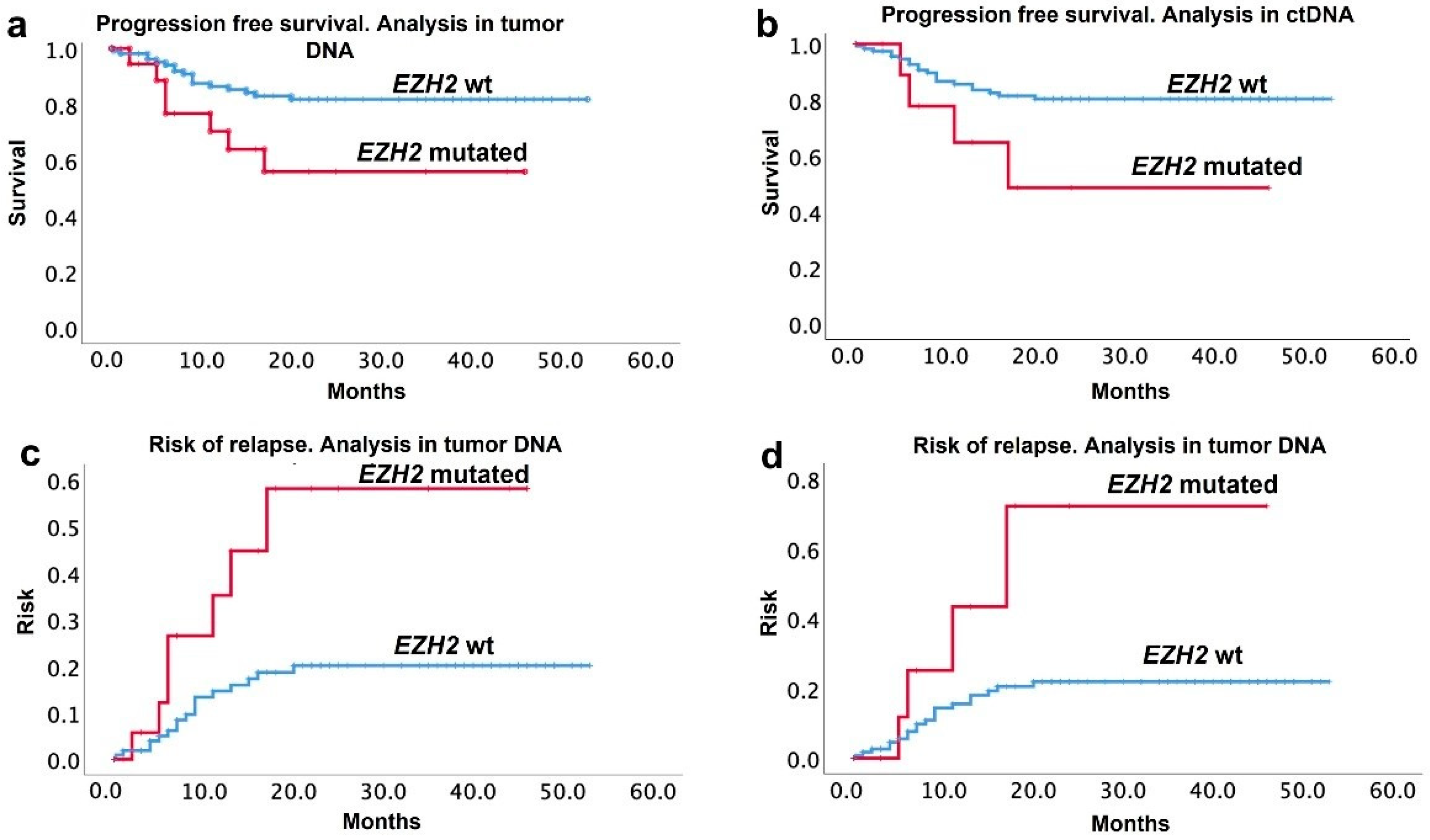
3.4. Relapse and Survival
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zelenetz, A.D.; Abramson, J.S.; Advani, R.H.; Andreadis, C.B.; Bartlett, N.; Bellam, N.; Byrd, J.C.; Czuczman, M.S.; Fayad, L.E.; Glenn, M.J.; et al. Non-Hodgkin’s Lymphomas. J. Natl. Compr. Cancer Netw. 2011, 9, 484–560. [Google Scholar] [CrossRef]
- Nguyen, L.; Papenhausen, P.; Shao, H. The Role of C-MYC in B-Cell Lymphomas: Diagnostic and Molecular Aspects. Genes 2017, 8, 116. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.-T.; Lu, L.; Xu, W.; Li, J.-Y. Circulating Tumor DNA: Clinical Roles in Diffuse Large B Cell Lymphoma. Ann. Hematol. 2019, 98, 255–269. [Google Scholar] [CrossRef] [PubMed]
- Singh, K.; Gollapudi, S.; Mittal, S.; Small, C.; Kumar, J.; Ohgami, R.S. Point Mutation Specific Antibodies in B-Cell and T-Cell Lymphomas and Leukemias: Targeting IDH2, KRAS, BRAF and Other Biomarkers RHOA, IRF8, MYD88, ID3, NRAS, SF3B1 and EZH2. Diagnostics 2021, 11, 600. [Google Scholar] [CrossRef] [PubMed]
- Chapuy, B.; Stewart, C.; Dunford, A.J.; Kim, J.; Kamburov, A.; Redd, R.A.; Lawrence, M.S.; Roemer, M.G.M.; Li, A.J.; Ziepert, M.; et al. Molecular Subtypes of Diffuse Large B Cell Lymphoma Are Associated with Distinct Pathogenic Mechanisms and Outcomes. Nat. Med. 2018, 24, 679–690. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, R.; Wright, G.W.; Huang, D.W.; Johnson, C.A.; Phelan, J.D.; Wang, J.Q.; Roulland, S.; Kasbekar, M.; Young, R.M.; Shaffer, A.L.; et al. Genetics and Pathogenesis of Diffuse Large B-Cell Lymphoma. N. Engl. J. Med. 2018, 378, 1396–1407. [Google Scholar] [CrossRef]
- Müller, J.; Hart, C.M.; Francis, N.J.; Vargas, M.L.; Sengupta, A.; Wild, B.; Miller, E.L.; O’Connor, M.B.; Kingston, R.E.; Simon, J.A. Histone Methyltransferase Activity of a Drosophila Polycomb Group Repressor Complex. Cell 2002, 111, 197–208. [Google Scholar] [CrossRef]
- Czermin, B.; Melfi, R.; McCabe, D.; Seitz, V.; Imhof, A.; Pirrotta, V. Drosophila Enhancer of Zeste/ESC Complexes Have a Histone H3 Methyltransferase Activity That Marks Chromosomal Polycomb Sites. Cell 2002, 111, 185–196. [Google Scholar] [CrossRef]
- Martín-Subero, J.I.; Kreuz, M.; Bibikova, M.; Bentink, S.; Ammerpohl, O.; Wickham-Garcia, E.; Rosolowski, M.; Richter, J.; Lopez-Serra, L.; Ballestar, E.; et al. New Insights into the Biology and Origin of Mature Aggressive B-Cell Lymphomas by Combined Epigenomic, Genomic, and Transcriptional Profiling. Blood 2009, 113, 2488–2497. [Google Scholar] [CrossRef] [PubMed]
- Bödör, C.; O’Riain, C.; Wrench, D.; Matthews, J.; Iyengar, S.; Tayyib, H.; Calaminici, M.; Clear, A.; Iqbal, S.; Quentmeier, H.; et al. EZH2 Y641 Mutations in Follicular Lymphoma. Leukemia 2011, 25, 726–729. [Google Scholar] [CrossRef] [PubMed]
- Oñate-Ocaña, L.F.; Ponce-Martínez, M.; Taja-Chayeb, L.; Gutiérrez-Hernández, O.; Avilés-Salas, A.; Cantú-de-León, D.; Dueñas-González, A.; Candelaria-Hernández, M. A Cohort Study of the Prognostic Impact of Exon-16 EZH2 Mutations in a Mexican-Mestizo Population of Patients with Diffuse Large B-Cell Lymphoma. Rev. Investig. Clin. 2021, 73, 362–370. [Google Scholar] [CrossRef] [PubMed]
- Yap, D.B.; Chu, J.; Berg, T.; Schapira, M.; Cheng, S.-W.G.; Moradian, A.; Morin, R.D.; Mungall, A.J.; Meissner, B.; Boyle, M.; et al. Somatic Mutations at EZH2 Y641 Act Dominantly through a Mechanism of Selectively Altered PRC2 Catalytic Activity, to Increase H3K27 Trimethylation. Blood 2011, 117, 2451–2459. [Google Scholar] [CrossRef]
- Wigle, T.J.; Knutson, S.K.; Jin, L.; Kuntz, K.W.; Pollock, R.M.; Richon, V.M.; Copeland, R.A.; Scott, M.P. The Y641C Mutation of EZH2 Alters Substrate Specificity for Histone H3 Lysine 27 Methylation States. FEBS Lett. 2011, 585, 3011–3014. [Google Scholar] [CrossRef] [PubMed]
- Sneeringer, C.J.; Scott, M.P.; Kuntz, K.W.; Knutson, S.K.; Pollock, R.M.; Richon, V.M.; Copeland, R.A. Coordinated Activities of Wild-Type Plus Mutant EZH2 Drive Tumor-Associated Hypertrimethylation of Lysine 27 on Histone H3 (H3K27) in Human B-Cell Lymphomas. Proc. Natl. Acad. Sci. USA 2010, 107, 20980–20985. [Google Scholar] [CrossRef] [PubMed]
- Lue, J.K.; Amengual, J.E. Emerging EZH2 Inhibitors and Their Application in Lymphoma. Curr. Hematol. Malig. Rep. 2018, 13, 369–382. [Google Scholar] [CrossRef] [PubMed]
- Melani, C.; Wilson, W.H.; Roschewski, M. Monitoring Clinical Outcomes in Aggressive B-Cell Lymphoma: From Imaging Studies to Circulating Tumor DNA. Best Pract. Res. Clin. Haematol. 2018, 31, 285–292. [Google Scholar] [CrossRef] [PubMed]
- Arzuaga-Mendez, J.; Prieto-Fernández, E.; Lopez-Lopez, E.; Martin-Guerrero, I.; García-Ruiz, J.C.; García-Orad, A. Cell-Free DNA as a Biomarker in Diffuse Large B-Cell Lymphoma: A Systematic Review. Crit. Rev. Oncol. Hematol. 2019, 139, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Kallam, A.; Adusumalli, J.; Fu, K.; Armitage, J.O. Circulating Tumor DNA in Lymphomas: Era of Precision Medicine. Adv. Cell Gene Ther. 2020, 3, e81. [Google Scholar] [CrossRef]
- Cheson, B.D.; Fisher, R.I.; Barrington, S.F.; Cavalli, F.; Schwartz, L.H.; Zucca, E.; Lister, T.A.; Alliance, Australasian Leukaemia and Lymphoma Group; Eastern Cooperative Oncology Group; European Mantle Cell Lymphoma Consortium; et al. Recommendations for Initial Evaluation, Staging, and Response Assessment of Hodgkin and Non-Hodgkin Lymphoma: The Lugano Classification. J. Clin. Oncol. 2014, 32, 3059–3068. [Google Scholar] [CrossRef] [PubMed]
- Oken, M.M.; Creech, R.H.; Tormey, D.C.; Horton, J.; Davis, T.E.; McFadden, E.T.; Carbone, P.P. Toxicity and Response Criteria of the Eastern Cooperative Oncology Group. Am. J. Clin. Oncol. 1982, 5, 649–655. [Google Scholar] [CrossRef]
- Hans, C.P.; Weisenburger, D.D.; Greiner, T.C.; Gascoyne, R.D.; Delabie, J.; Ott, G.; Müller-Hermelink, H.K.; Campo, E.; Braziel, R.M.; Jaffe, E.S.; et al. Confirmation of the Molecular Classification of Diffuse Large B-Cell Lymphoma by Immunohistochemistry Using a Tissue Microarray. Blood 2004, 103, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Solís-Armenta, R.; Cacho-Díaz, B.; Gutiérrez-Hernández, O.; Candelaria-Hernández, M. Central Nervous System International Prognostic Index Impacts Overall Survival in Diffuse Large B-Cell Lymphoma Treated with R-Chop in a Third Level Cancer Center from Mexico: A Survey of 642 Patients. Rev. Investig. Clin. 2021, 73, 231–237. [Google Scholar] [CrossRef] [PubMed]
- Alcaide, M.; Yu, S.; Bushell, K.; Fornika, D.; Nielsen, J.S.; Nelson, B.H.; Mann, K.K.; Assouline, S.; Johnson, N.A.; Morin, R.D. Multiplex Droplet Digital PCR Quantification of Recurrent Somatic Mutations in Diffuse Large B-Cell and Follicular Lymphoma. Clin. Chem. 2016, 62, 1238–1247. [Google Scholar] [CrossRef]
- Rossi, D.; Spina, V.; Bruscaggin, A.; Gaidano, G. Liquid Biopsy in Lymphoma. Haematologica 2019, 104, 648–652. [Google Scholar] [CrossRef] [PubMed]
- Roschewski, M.; Dunleavy, K.; Pittaluga, S.; Moorhead, M.; Pepin, F.; Kong, K.; Shovlin, M.; Jaffe, E.S.; Staudt, L.M.; Lai, C.; et al. Circulating Tumour DNA and CT Monitoring in Patients with Untreated Diffuse Large B-Cell Lymphoma: A Correlative Biomarker Study. Lancet Oncol. 2015, 16, 541–549. [Google Scholar] [CrossRef]
- Condoluci, A.; Rossi, D. The Future of Cell-Free DNA Testing to Guide Therapeutic Decisions in B-Cell Lymphomas. Curr. Opin. Hematol. 2019, 26, 281–287. [Google Scholar] [CrossRef] [PubMed]
- Hohaus, S.; Giachelia, M.; Massini, G.; Mansueto, G.; Vannata, B.; Bozzoli, V.; Criscuolo, M.; D’Alò, F.; Martini, M.; Larocca, L.M.; et al. Cell-Free Circulating DNA in Hodgkin’s and Non-Hodgkin’s Lymphomas. Ann. Oncol. 2009, 20, 1408–1413. [Google Scholar] [CrossRef] [PubMed]
- Camus, V.; Stamatoullas, A.; Mareschal, S.; Viailly, P.-J.; Sarafan-Vasseur, N.; Bohers, E.; Dubois, S.; Picquenot, J.M.; Ruminy, P.; Maingonnat, C.; et al. Detection and Prognostic Value of Recurrent Exportin 1 Mutations in Tumor and Cell-Free Circulating DNA of Patients with Classical Hodgkin Lymphoma. Haematologica 2016, 101, 1094–1101. [Google Scholar] [CrossRef]
- Schwarzenbach, H.; Hoon, D.S.B.; Pantel, K. Cell-Free Nucleic Acids as Biomarkers in Cancer Patients. Nat. Rev. Cancer 2011, 11, 426–437. [Google Scholar] [CrossRef]
- Bohers, E.; Viailly, P.-J.; Becker, S.; Marchand, V.; Ruminy, P.; Maingonnat, C.; Bertrand, P.; Etancelin, P.; Picquenot, J.-M.; Camus, V.; et al. Non-Invasive Monitoring of Diffuse Large B-Cell Lymphoma by Cell-Free DNA High-Throughput Targeted Sequencing: Analysis of a Prospective Cohort. Blood Cancer J. 2018, 8, 74. [Google Scholar] [CrossRef] [PubMed]
- Kurtz, D.M.; Scherer, F.; Jin, M.C.; Soo, J.; Craig, A.F.M.; Esfahani, M.S.; Chabon, J.J.; Stehr, H.; Liu, C.L.; Tibshirani, R.; et al. Circulating Tumor DNA Measurements as Early Outcome Predictors in Diffuse Large B-Cell Lymphoma. J. Clin. Oncol. 2018, 36, 2845–2853. [Google Scholar] [CrossRef] [PubMed]
- Scherer, F.; Kurtz, D.M.; Newman, A.M.; Stehr, H.; Craig, A.F.M.; Esfahani, M.S.; Lovejoy, A.F.; Chabon, J.J.; Klass, D.M.; Liu, C.L.; et al. Distinct Biological Subtypes and Patterns of Genome Evolution in Lymphoma Revealed by Circulating Tumor DNA. Sci. Transl. Med. 2016, 8, 364ra155. [Google Scholar] [CrossRef] [PubMed]
- Rossi, D.; Diop, F.; Spaccarotella, E.; Monti, S.; Zanni, M.; Rasi, S.; Deambrogi, C.; Spina, V.; Bruscaggin, A.; Favini, C.; et al. Diffuse Large B-Cell Lymphoma Genotyping on the Liquid Biopsy. Blood 2017, 129, 1947–1957. [Google Scholar] [CrossRef]
- Assouline, S.E.; Nielsen, T.H.; Yu, S.; Alcaide, M.; Chong, L.; MacDonald, D.; Tosikyan, A.; Kukreti, V.; Kezouh, A.; Petrogiannis-Haliotis, T.; et al. Phase 2 Study of Panobinostat with or without Rituximab in Relapsed Diffuse Large B-Cell Lymphoma. Blood 2016, 128, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Kurtz, D.M. Prognostication with Circulating Tumor DNA: Is It Ready for Prime Time? Hematol. Am. Soc. Hematol. Educ. Program 2019, 2019, 47–52. [Google Scholar] [CrossRef]
- Coccaro, N.; Anelli, L.; Zagaria, A.; Perrone, T.; Specchia, G.; Albano, F. Molecular Complexity of Diffuse Large B-Cell Lymphoma: Can It Be a Roadmap for Precision Medicine? Cancers 2020, 12, 185. [Google Scholar] [CrossRef]
- Rossi, D.; Kurtz, D.M.; Roschewski, M.; Cavalli, F.; Zucca, E.; Wilson, W.H. The Development of Liquid Biopsy for Research and Clinical Practice in Lymphomas: Report of the 15-ICML Workshop on CtDNA. Hematol. Oncol. 2020, 38, 34–37. [Google Scholar] [CrossRef]
- Camus, V.; Sarafan-Vasseur, N.; Bohers, E.; Dubois, S.; Mareschal, S.; Bertrand, P.; Viailly, P.-J.; Ruminy, P.; Maingonnat, C.; Lemasle, E.; et al. Digital PCR for Quantification of Recurrent and Potentially Actionable Somatic Mutations in Circulating Free DNA from Patients with Diffuse Large B-Cell Lymphoma. Leuk. Lymphoma 2016, 57, 2171–2179. [Google Scholar] [CrossRef]
- Dubois, S.; Mareschal, S.; Picquenot, J.-M.; Viailly, P.-J.; Bohers, E.; Cornic, M.; Bertrand, P.; Veresezan, E.L.; Ruminy, P.; Maingonnat, C.; et al. Immunohistochemical and Genomic Profiles of Diffuse Large B-Cell Lymphomas: Implications for Targeted EZH2 Inhibitor Therapy? Oncotarget 2015, 6, 16712–16724. [Google Scholar] [CrossRef][Green Version]
- Morin, R.D.; Johnson, N.A.; Severson, T.M.; Mungall, A.J.; An, J.; Goya, R.; Paul, J.E.; Boyle, M.; Woolcock, B.W.; Kuchenbauer, F.; et al. Somatic Mutation of EZH2 (Y641) in Follicular and Diffuse Large B-Cell Lymphomas of Germinal Center Origin. Nat. Genet. 2010, 42, 181–185. [Google Scholar] [CrossRef]
- Caganova, M.; Carrisi, C.; Varano, G.; Mainoldi, F.; Zanardi, F.; Germain, P.-L.; George, L.; Alberghini, F.; Ferrarini, L.; Talukder, A.K.; et al. Germinal Center Dysregulation by Histone Methyltransferase EZH2 Promotes Lymphomagenesis. J. Clin. Investig. 2013, 123, 5009–5022. [Google Scholar] [CrossRef] [PubMed]
- Nagy, Á.; Bátai, B.; Balogh, A.; Illés, S.; Mikala, G.; Nagy, N.; Kiss, L.; Kotmayer, L.; Matolcsy, A.; Alpár, D.; et al. Quantitative Analysis and Monitoring of EZH2 Mutations Using Liquid Biopsy in Follicular Lymphoma. Genes 2020, 11, 785. [Google Scholar] [CrossRef] [PubMed]
- Duan, R.; Du, W.; Guo, W. EZH2: A Novel Target for Cancer Treatment. J. Hematol. Oncol. 2020, 13, 104. [Google Scholar] [CrossRef] [PubMed]
- Tuaeva, N.O.; Falzone, L.; Porozov, Y.B.; Nosyrev, A.E.; Trukhan, V.M.; Kovatsi, L.; Spandidos, D.A.; Drakoulis, N.; Kalogeraki, A.; Mamoulakis, C.; et al. Translational Application of Circulating DNA in Oncology: Review of the Last Decades Achievements. Cells 2019, 8, 1251. [Google Scholar] [CrossRef] [PubMed]
| n (%) wt | n (%) Mutated | p | |
|---|---|---|---|
| Total | 118 (85.5) | 20 (14.5) | - |
| Gender Female Male | 52 (37.7) 66 (47.8) | 12 (8.7) 8 (5.8) | 0.18 |
Mean age (25–75 IQR) | 60.46 (50–71) | 58.55 (52.5–66) | 0.68 |
| ECOG * 0–1 >2 | 86 (62.3) 32 (23.2) | 12 (8.7) 8 (5.8) | 0.097 |
| Presence of B symptoms Yes No | 51 (37) 67 (48.5) | 12 (8.7) 8 (5.8) | 0.164 |
| Presence of Bulky disease Yes No | 43 (31.2) 76 (55.0) | 11 (8.0) 8 (5.8) | 0.076 |
| Clinical stage I–II III–IV | 40 78 | 2 18 | 0.01 |
| IPI score+ Low Intermediate-low Intermediate-high High | 38 (27.5) 14 (10.1) 17 (12.3) 49 (35.5) | 3 (2.2) 2 (1.5) 3 (2.2) 12 (8.7) | 0.38 |
| Cell of origin GC ** Non-GC *** | 78 (56.5) 40 (29.0) | 20 (14.5) 0 | 0.008 |
| Double hit | 12 (8.7) | 3 (2.2) | 0.52 |
| Extranodal sites 0–1 ≥2 | 97 21 | 11 9 | 0.33 |
| ß2 microglobulin Normal Increased | 39 (28.3) 79 (57.2) | 6 (4.4) 14(10.1) | 0.51 |
| Tumor | ctDNA | ctDNA | |
|---|---|---|---|
| Mutation | Sanger sequencing | Sanger sequencing | ddPCR * |
| N = 20 n (%) | N = 11 n (%) | N = 9 n (%) | |
| Y641F | 7 (5.1) | 4 (2.9) | 3 (2.1) |
| Y641N | 4 (2.9) | 2 (1.4) | 2 (1.4) |
| Y641H | 3 (2.2) | 2 (1.4) | 1 (0.7) |
| Y641S | 3 (2.2) | 1 (0.7) | 2 (1.4) |
| I638T | 1 (0.7) | ----- | n.d. |
| Y641N + F637L | 1 (0.7) | 1 (0.7) | n.d |
| Y641S + Y641F | 1(0.7) | ----- | 1 (0.7) |
| Y641C | ----- | 1 (0.7) | n.d. |
| Factor | % Response | p * | |
|---|---|---|---|
| Bulky | Yes | 48.1 | 0.03 |
| No | 66.2 | ||
| Lugano clinical stage | I–II | 71.2 | 0.024 |
| III–IV | 54.1 | ||
| Molecular type | GC ** | 68.1 | 0.24 |
| Non-GC *** | 66.0 | ||
| IPI score | I–II | 75.4 | 0.03 |
| III–IV | 48 |
| Factor | Risk | 95 % Confidence Interval | p * |
|---|---|---|---|
| ECOG (>2) | 1.95 | 0.572–1.33 | 0.42 |
| B symptoms | 1.26 | 0.324–1.740 | 0.44 |
| Bulky disease | 1.27 | 0.526–1.560 | 0.95 |
| Lugano Clinical stage (III–IV) | 1.24 | 1.21–1.266 | 0.022 |
| Cell of origin (Non-GC) ** | 1.38 | 0.96–1.54 | 0.58 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Díaz-Chávez, J.; Gutiérrez-Hernández, O.; Taja-Chayeb, L.; Gutiérrez-Chavarría, S.; Avilés-Salas, A.; Candelaria, M. ctDNA Is Useful to Detect Mutations at Codon 641 of Exon 16 of EZH2, a Biomarker for Relapse in Patients with Diffuse Large B-Cell Lymphoma. Cancers 2022, 14, 4650. https://doi.org/10.3390/cancers14194650
Díaz-Chávez J, Gutiérrez-Hernández O, Taja-Chayeb L, Gutiérrez-Chavarría S, Avilés-Salas A, Candelaria M. ctDNA Is Useful to Detect Mutations at Codon 641 of Exon 16 of EZH2, a Biomarker for Relapse in Patients with Diffuse Large B-Cell Lymphoma. Cancers. 2022; 14(19):4650. https://doi.org/10.3390/cancers14194650
Chicago/Turabian StyleDíaz-Chávez, José, Olga Gutiérrez-Hernández, Lucia Taja-Chayeb, Sindy Gutiérrez-Chavarría, Alejandro Avilés-Salas, and Myrna Candelaria. 2022. "ctDNA Is Useful to Detect Mutations at Codon 641 of Exon 16 of EZH2, a Biomarker for Relapse in Patients with Diffuse Large B-Cell Lymphoma" Cancers 14, no. 19: 4650. https://doi.org/10.3390/cancers14194650
APA StyleDíaz-Chávez, J., Gutiérrez-Hernández, O., Taja-Chayeb, L., Gutiérrez-Chavarría, S., Avilés-Salas, A., & Candelaria, M. (2022). ctDNA Is Useful to Detect Mutations at Codon 641 of Exon 16 of EZH2, a Biomarker for Relapse in Patients with Diffuse Large B-Cell Lymphoma. Cancers, 14(19), 4650. https://doi.org/10.3390/cancers14194650






