Simple Summary
Tumor cells receive signals from the surrounding extracellular matrix that affect their growth and survival. An important component of the extracellular matrix is the large polysaccharide hyaluronan, which binds and activates certain receptors at the cell surface, including CD44. Activation of CD44 initiates several signaling pathways; one of them involves the cleavage of CD44 by proteases, leading to the release of the intracellular domain of CD44, which after translocation to the nucleus affects the transcription of certain genes. In the present report, we elucidate the mechanism by which CD44 is cleaved, and show that this occurs at an increased rate in stem-like tumor cells grown in spheres. We also show that CD44 cleavage promotes the migration of tumor cells. Since the mechanism we have elucidated promotes tumorigenesis, it is possible that inhibition of this pathway may be beneficial in the treatment of tumor patients.
Abstract
The hyaluronan receptor CD44 can undergo proteolytic cleavage in two steps, leading to the release of its intracellular domain; this domain is translocated to the nucleus, where it affects the transcription of target genes. We report that CD44 cleavage in A549 lung cancer cells and other cells is promoted by transforming growth factor-beta (TGFβ) in a manner that is dependent on ubiquitin ligase tumor necrosis factor receptor-associated factor 4 or 6 (TRAF4 or TRAF6, respectively). Stem-like A549 cells grown in spheres displayed increased TRAF4-dependent expression of CD44 variant isoforms, CD44 cleavage, and hyaluronan synthesis. Mechanistically, TRAF4 activated the small GTPase RAC1. CD44-dependent migration of A549 cells was inhibited by siRNA-mediated knockdown of TRAF4, which was rescued by the transfection of a constitutively active RAC1 mutant. Our findings support the notion that TRAF4/6 mediates pro-tumorigenic effects of CD44, and suggests that inhibitors of CD44 signaling via TRAF4/6 and RAC1 may be beneficial in the treatment of tumor patients.
1. Introduction
Hyaluronan, a prominent constituent of the extracellular matrix, is synthesized by three hyaluronan synthase isoforms, i.e., HAS1, HAS2, and HAS3, and is catabolized by hyaluronidases, including HYAL1, HYAL2, and TMEM2. In addition to its structural role in the extracellular matrix, hyaluronan exerts signaling effects via binding to specific cell surface receptors, such as CD44 [1,2,3], which is a transmembrane receptor that mediates signals affecting cell proliferation and migration [4,5]. The human CD44 gene is composed of 19 exons, i.e., 10 “constant exons” encoding the standard CD44 molecule (CD44s) and nine “variable exons”, which in different combinations can be inserted in the membrane-proximal stem region through alternative splicing to generate CD44v isoforms [6]. Certain CD44v isoforms bind chondroitin sulfate and heparan sulfate in addition to hyaluronan. Importantly, expression of both CD44s and CD44v isoforms has been linked to tumor progression [7,8,9].
CD44 isoforms interact with certain adaptor proteins and receptors [3,10,11], and consist of different functional domains, including the N-terminal extracellular domain that binds various ligands; the stem domain, which possesses proteolytic cleavage sites for matrix metalloproteases (MMPs) and ADAM-like proteases [12,13]; a transmembrane domain; and an intracellular domain. The MMP-mediated cleavage of the CD44 ectodomain allows the release of cancer cells bound to hyaluronan, promoting their migration through hyaluronan-containing extracellular matrix (ECM) and facilitating cancer cell dissemination [12,13]. The resulting transmembrane and intracellular domain (CD44 TM-ICD) is subject to a second cleavage in the transmembrane domain by presenilin 1, the catalytic subunit of γ-secretase, releasing the CD44 cytoplasmic region (CD44 ICD) [14]. CD44′s intracellular domain (ICD) possesses signaling functions through interactions with key regulatory molecules, such as Ezrin, radixin and moesin (ERM) proteins and small Rho GTPases [5,15,16,17]. Furthermore, the CD44 ICD regulates the transcription of target genes by forming complexes with cAMP response element-binding protein (CREB)-binding protein (CBP)/p300 or STAT3 [9,18,19,20,21].
Transforming growth factor β (TGFβ) exerts its cellular effects by forming a complex of TGFβ type I (TβRI) and type II (TβRII) receptors [22]. Within the complex, TβRII phosphorylates and activates TβRI, promoting the phosphorylation of Smad molecules that are translocated to the nucleus, where they regulate the transcription of certain genes. In addition, TGFβ activates several non-Smad signaling pathways, including MAP kinase, phosphatidyl-inositol-3′-kinase (PI3-kinase), the tyrosine kinase Src, and similar to CD44, release the intracellular domain of TβRI (TβRI-ICD) [23], which acts as a transcription factor in the nucleus [24]. Some of the non-Smad pathways, including p38 and JNK MAP kinases [25,26], PI3-kinase [27], and release of TβRI-ICD [23], are activated through mechanisms involving ubiquitin ligase tumor necrosis factor receptor associated factor (TRAF) 6.
The TRAF family consists of seven members (TRAF1–TRAF7), which play important roles in signaling events and are implicated in many diseases, including inflammation and cancer [28,29]. Their main characteristics are a RING domain that has E3 ligase activity, followed by a zinc-finger motif and two TRAF domains that function as scaffolds in signaling via Toll-like receptors and receptors for TNF, TGFβ, and cytokines [30,31].
We present here a TRAF4/6-dependent mechanism for how CD44 is cleaved, and demonstrate that the cleavage occurs at increased rate in stem-like A549 lung cancer cells grown in spheres. Moreover, we show that migration of A549 cells is dependent on TRAF4 and its downstream effector RAC1.
2. Results
2.1. Cleavage of CD44 in Response to External Stimuli
TGFβ promoted the cleavage of CD44 and formation of the CD44 TM-ICD domain in a time-dependent manner in the lung adenocarcinoma A549 cell line (Figure 1a), as did several other growth factors (Figure S1a). We also investigated the effect of proteasomal (MG132) and lysosomal (chloroquine (CQ)) inhibitors on the accumulation of CD44 cleavage products in untreated and TGFβ-treated cells. In TGFβ stimulated cultures, the CD44 TM-ICD and CD44 ICD products increased, indicating that they are unstable and subject to degradation in lysosomes and proteasomes (Figure 1a,b). The CD44 TM-ICD and CD44 ICD products were absent in CD44-depleted cells (Figure 1b), and TGFβ promoted CD44 cleavage in a concentration-dependent manner (Figure 1c). Following CD44 cleavage, translocation of CD44 ICD to the nucleus was detected in untreated cells consistent with its role in transcription regulation; TGFβ and TPA stimulation further enhanced its translocation (Figure 1d). The constitutive, TGFβ- and TPA-mediated generation of CD44 ICD was abolished after incubation with the γ-secretase inhibitor DAPT. Certain smaller CD44 ICD-like bands were detected; it is possible that these are generated due to less specific proteolytic cleavage performed by presenilin 1/γ-secretase activity and/or because CD44 ICD is highly susceptible to degradation (Figure 1c,d,e). Similarly, in TGFβ-treated glioblastoma U251MG cell cultures, the CD44 TM-ICD and CD44 ICD products were clearly detected in TGFβ-stimulated cultures after the inhibition of lysosomal or proteasomal degradation (Figure 1e). In U251MG cells treated with the γ-secretase inhibitor N-[N-3,5-Difluorophenacetyl)-L-alanyl]-S-phenylglycine t-butyl ester (DAPT), the induction of CD44 TM-ICD product was increased—whereas, as expected, no CD44 ICD was detected (Figure 1e). Moreover, as in A549 cells, the proteolytic cleavage products of CD44 in U251MG cells were sensitive to lysosomal and proteasomal degradation. TPA (12-O-tetradecanoylphorbol-13-acetate), a protein kinase C activator that has been demonstrated to promote the proteolytic cleavage of several cell surface receptors [32], was used as a positive control. Since both A549 and U251MG cells synthesize hyaluronan, we investigated the effect of exogenous hyaluronan on CD44 cleavage; both fragmented and high-molecular-mass hyaluronan promoted CD44 cleavage in both cell lines (Figure S1b). TGFβ also induced proteolytic cleavage of CD44 in another non-small-cell lung cancer cell line, H1299, in a time course- and concentration-dependent manner (Figure S1c). The proteolytic cleavage of CD44 was high in U251MG, A549, and H1299 cell lines, compared to other cancer cell lines tested (Figure S1d); these cell lines were therefore selected for further studies.
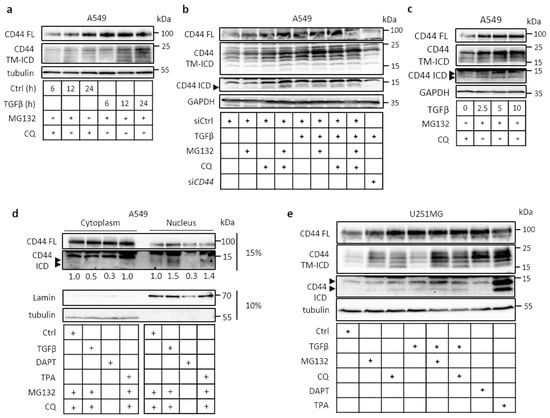
Figure 1.
Transforming growth factor β (TGFβ) induces ectodomain and intramembranous cleavage of CD44 in a dose- and time-dependent manners. (a,b) Immunoblot analysis of full-length (FL) and cleavage products (transmembrane and intracellular domain (TM-ICD) and ICD) of CD44 in A549 cells after treatment or no treatment with TGFβ (5 ng/mL) for the indicated time periods; knockdown of CD44 by siRNA was performed to confirm the specificity of observed bands (b). (c) Immunoblotting of FL CD44, as well as the TM-ICD and ICD of CD44 after 24 h of treatment with different concentrations of TGFβ. (d), A549 cells treated or not with TGFβ (5 ng/ml), DAPT (40 μM), MG132 and CQ for 24 h, or TPA (80 nM) for 1 h, were subjected to nuclear/cytoplasmic fractionation and subjected to SDS gel electrophoresis in 15% or 10% polyacrylamide gels, followed by immunoblotting for FL, ICD of CD44, lamin, and tubulin. Quantification of CD44-ICD bands was performed via ImageJ after normalizing to CD44 FL (e), Immunoblot analysis of FL CD44, as well as the TM-ICD and ICD of CD44 in U251MG cells treated or not treated with TGFβ for 24 h, or 12-O-tetradecanoylphorbol-13-acetate (TPA) (80 nM) for 1 h. The cultures were treated with the γ-secretase inhibitor DAPT (40 μM)), the proteasomal inhibitor MG132, and the lysosomal inhibitor chloroquine (CQ) for 24 h; tubulin or glyceraldehyde 3-phosphate dehydrogenase (GAPDH) were used as loading controls in the same immunoblots. Arrowheads indicate CD44 ICD-like products.
2.2. Stem-Like Cells Show Enhanced Expression of CD44v and HAS2 Genes and Cleavage of CD44
To investigate a possible correlation between CD44 cleavage and stemness in A549 cells, we compared cells cultured in anchorage-dependent monolayer conditions with cells cultured in ultra-low attachment plates, in which tumor cells form spheres enriched in stem-like cancer cells (Figure 2a,b). A549-derived spheres exhibited greater expression of mRNA for the stem cell markers NANOG and OCT4, compared to cells grown in a monolayer, whereas mRNA for another stem cell marker, SOX2, was not increased in the spheres compared to cells grown under adhesive conditions (Figure 2b). The mRNA for the CD44 variant isoforms increased in A549 cell spheres (Figure 2c); however, the transcript for the CD44 standard isoform did not (Figure 2d). Moreover, the expression of CD44v isoforms and the formation of CD44 TM-ICD and CD44 ICD increased in non-adherent A549 cell spheres compared to cells grown in a monolayer (Figure 2e). These findings are consistent with previous reports, which have shown that the cleavage of CD44s and CD44v isoforms are linked to cancer stem-like characteristics [13,33,34,35]. As in A549 cells, the stem cell markers NANOG and OCT4 were also increased in H1299 cells grown under non-adhesive conditions, but the expression levels were about 10-fold lower compared to those in A549 cultures (Figure S2a). Importantly to mention that the expression levels of the two CD44v transcripts (v3 and v6) were lower compared to A549 cells, whereas the mRNA levels for CD44s were similar under adherent and non-adherent conditions (Figure S2b,c); this highlights the correlation of CD44v status with stem cell potential. Quantification of mRNA for HAS isoforms in A549 cell spheroids revealed a 30-fold increase in HAS2 mRNA; the mRNAs for HAS1 and HAS3 also increased, but to lesser degrees (Figure 2f). Analysis of the mRNA for hyaluronidase family members revealed that the most abundant one, TMEM2, did not increase in the spheroids; however, HYAL1 and HYAL2 increased about three-fold (Figure 2f). An about two-fold increase in hyaluronan synthesis in the spheroids, compared to monolayer cell cultures, was observed (Figure 2g). Thus, in stem-like A549 cells grown as spheres, both HAS2-synthesized hyaluronan and CD44v isoforms increased.
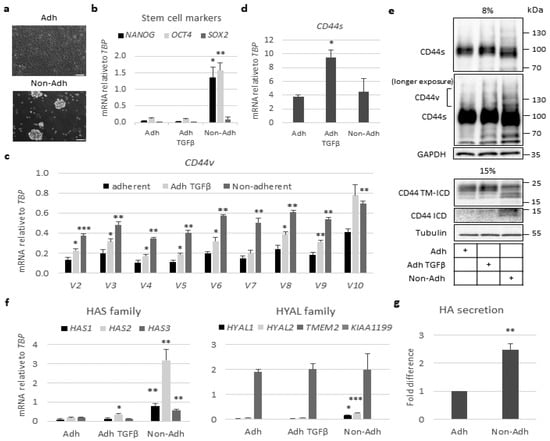
Figure 2.
CD44 cleavage increases in stem-like A549 cells grown in spheres. (a) Phase contrast microscopy of A549 grown in adherent (Adh) or low-attachment conditions (Non-adh) for 96 h. Zeiss Axiovision was employed to take the micrographs (bars = 100 μm). (b–d) A549 cells were cultured under adherent conditions or as spheres. Adherent cells were stimulated or not with TGFβ for 24 h. The mRNA levels of transcription factors NANOG, OCT4, and SOX2 (b), as well as CD44v isoforms (c) and CD44s (d) were determined by real-time qPCR and normalized to TBP mRNA levels. (e) Lysates from adherent A549 cells, stimulated or not with TGFβ for 24 h, and non-adherent A549 cells were subjected to SDS-PAGE using 8% and 15% polyacrylamide gels, followed by immunoblotting for CD44s and CD44v, CD44 TM-ICD, and CD44 ICD; GAPDH and tubulin were used as loading controls. (f) The mRNA levels of hyaluronan synthase (HAS1, HAS2, HAS3) and hyaluronidase (HYAL1, HYAL2, TMEM2, KIAA1199) family members were determined by real-time qPCR and normalized to TBP. (g), Hyaluronan levels in the culture medium of adherent and non-adherent A549 cells were evaluated by an enzyme-linked immunosorbent assay (ELISA)-like assay and normalized against protein amounts; values are expressed as fold difference. Asterisks illustrate significant differences between the different conditions compared to the respective adherent ones: * p < 0.05, ** p < 0.01, *** p < 0.001.
Stimulation with TGFβ, which potently induces HAS2-mediated hyaluronan production in several cell types [36,37], enhanced the expressions of mRNAs for CD44s and CD44v, as well as HAS2 in A549 cells grown in a monolayer (Figure 2c,d,f).
2.3. Involvement of TRAF Family Members in CD44 Cleavage
The cleavage of TβRI by ADAM17 and presenilin 1 occurs in a TRAF6-dependent manner [23]. This observation prompted us to investigate whether the cleavage of CD44 is also dependent on TRAF family members. In mouse embryo fibroblasts (MEFs) depleted of Traf6 (MEF Traf6-/-), the formation of CD44 TM-ICD was significantly reduced compared to wild-type MEFs (Figure S3a). The ectopic expression of wild-type Traf6 rescued the cleavage products of Cd44 TM-ICD, but the expression of the enzymatically inactive mutant Traf6 C70A did not (Figure S3b,c), suggesting that the cleavage of CD44 is dependent on Traf6-induced ubiquitination. We next investigated the expression levels of mRNA for TRAF family members in A549 and U251MG cells. TRAF2, TRAF4, TRAF6, and TRAF7 were abundantly expressed in A549 cells, whereas TRAF4 dominated in U251MG cells (Figure S4a,b); TGFβ stimulation did not affect the expression levels of TRAF family members in either of the cell lines. Silencing of TRAF6 in A549 cells suppressed the formation of CD44 TM-ICD; however, the silencing of TRAF4 resulted in an even more prominent reduction of the formation of CD44 TM-ICD (Figure 3a). A decrease in CD44 cleavage was also detected in H1299 cells depleted of TRAF4 (Figure S4c).
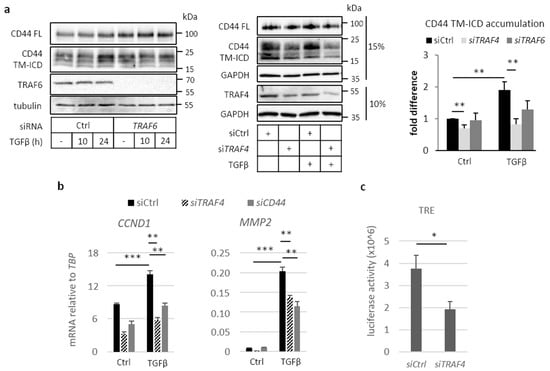
Figure 3.
Tumor necrosis factor receptor associated factor (TRAF) family members mediate TGFβ-induced CD44 cleavage. (a) A549 cells were transfected with non-targeting siRNA (siCtrl) or siRNAs against TRAF6 or TRAF4, and stimulated or not with TGFβ for up to 24 h, followed by immunoblotting for CD44 FL, TM-ICD, TRAF6, and TRAF4; tubulin and GAPDH served as loading controls. CD44 TM-ICD bands were quantified using the ImageJ software and normalized to either tubulin or GAPDH; values are depicted as fold difference; lysates were run at either 10% or 15% polyacrylamide gels, as depicted. (b), A549 cells were transfected with control siRNA or siRNAs targeting TRAF4 or CD44, and subsequently treated or not with TGFβ (5 ng/mL) for 24 h. Relative mRNA levels of CCND1 and MMP2 were quantified by real-time qPCR and normalized to TBP. (c), A549 cells were co-transfected with scrambled control siRNA or siRNAs against TRAF4, a β-gal reporter, and a TPA-responsive element (TRE) luciferase reporter, after which luciferase activity was measured and normalized to β-gal activity. All graph bars are shown as the average ± standard error of the mean (SEM), based on at least three independent experiments, unless mentioned otherwise. Asterisks illustrate significant differences between the conditions indicated with lines; * p < 0.05, ** p < 0.01, *** p < 0.001.
The expression of cyclin D1 (CCND1) and MMP2 have been shown to be dependent on CD44 [21,38,39]. We found that knockdown of TRAF4 in A549 cells suppressed the TGFβ-induced expression of CCND1, as well as MMP2, as did knockdown of CD44 (Figure 3b). TRAF4 and CD44 silencing efficiency is depicted in Figure S4d. The translocation of CD44 ICD to the nucleus has been shown to promote activation of the TPA-responsive element (TRE) luciferase reporter [19]; knockdown of TRAF4 reduced the activation of the TRE reporter by 50% (Figure 3c). The results support the notion that TRAF4 is important for CD44 signaling in A549 cells.
2.4. TRAF4 Plays an Important Role for CD44 Expression in A549 Cell Spheres
We analyzed the importance of TRAF4 and TRAF6 for the expression of CD44s and CD44v, and mRNAs for CD44, HAS2, and the hyaluronidase TMEM2 in A549 cell spheres. We found that silencing of TRAF4 by siRNA, but not the silencing of TRAF6, led to about a 40% reduction in the expression of the CD44s isoform (Figure 4a). Furthermore, the expression of mRNAs for CD44, HAS2, and TMEM2 was suppressed after silencing of TRAF4 (Figure 4b,c), resulting in a 30% decrease in hyaluronan production (Figure 4d). In contrast, knockdown of TRAF6 did not significantly affect the expression of CD44, HAS2, or TMEM2, or the level of secreted hyaluronan (Figure 4b,d, respectively).
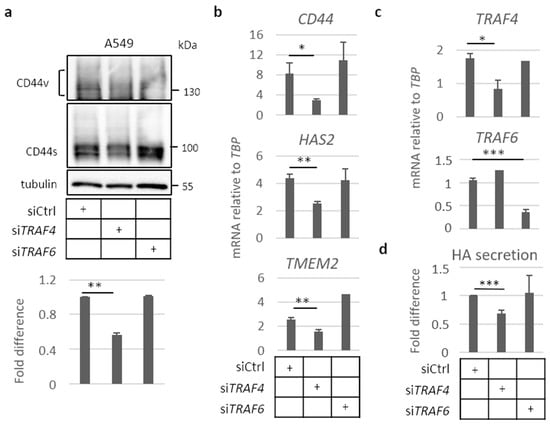
Figure 4.
TRAF4, but not TRAF6, is necessary for expression of CD44 and HAS2 in A549 cells grown in spheres. A549 cells were transfected with control siRNA or siRNAs targeting TRAF4 or TRAF6, and were thereafter seeded in non-adherent conditions for 96 h. (a), Cells were collected, and samples were resolved in 8% SDS-PAGE, followed by immunoblotting for CD44s and v isoforms. CD44 bands were quantified by the software ImageJ and normalized to tubulin levels and expressed as fold difference. (b–c) mRNA expression of CD44, HAS2, and TMEM2 (b), as well as TRAF4 and TRAF6 (c) were determined by real-time qPCR and normalized to TBP mRNA. (d) Hyaluronan levels in the culture medium of cells in low-attachment conditions was quantified by an ELISA-like assay; the results are given a fold difference after normalization to total protein levels of the collected cells. All graph bars are shown as the average ± SEM based from at least three independent experiments. Asterisks illustrate significant differences compared to the siCtrl condition: * p < 0.05, ** p < 0.01, *** p < 0.001.
2.5. TRAF4 Is Required for TGFβ-Induced RAC1 Activation
To further address the mechanisms regulating signaling via CD44 and TRAF4, we investigated the involvement of the small GTPase RAC1, which has been demonstrated to promote the cleavage of CD44 in TPA- [12] or EGF-stimulated [40] U251MG cells. TGFβ stimulation of A549 cells activated RAC1, first observed after 30 min and sustained over several hours, as demonstrated by the pull-down of GTP-RAC1 by glutathione S-transfarase-tagged p21-activated kinase (GST-PAK1) (Figure 5a). Silencing of TRAF4 by siRNA inhibited the activation of RAC1 (Figure 5b). Transfection of a dominant-negative RAC1 mutant (MYC-RAC1 T17N) suppressed the TGFβ-induced formation of CD44 TM-ICD (Figure 5c), as did knockdown of TRAF4 (Figure 5d). In contrast, transfection of a constitutively active RAC1 mutant (MYC-RAC1 Q61L) rescued the formation of CD44 TM-ICD, in the absence or presence of TGFβ (Figure 5d). A similar dependence on TRAF4 for RAC1 activation was also observed in H1299 cells (Figure S5). Moreover, TGFβ-mediated cleavage of CD44 was suppressed by the dominant-negative mutant MYC-RAC1 N17 (Figure S5c).
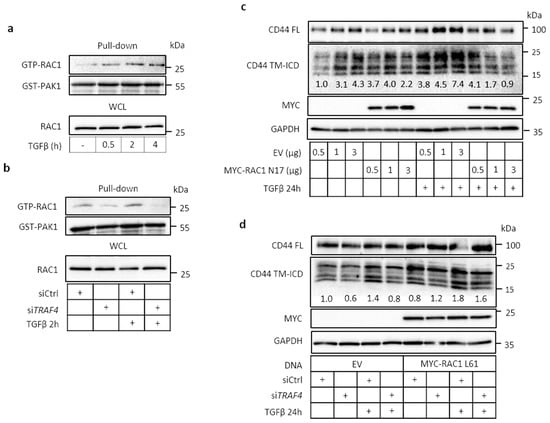
Figure 5.
TRAF4 is required for TGFβ-induced RAC1 activity and for CD44 cleavage. (a) A549 cells were treated with TGFβ (5 ng/mL) for the indicated time periods, and RAC1 activity was assessed after pull-down with GST-PAK1, followed by immunoblotting for RAC1 and GST-PAK1; in addition, whole cell lysate was immunoblotted for RAC1. (b), A549 cells were transfected with control siRNA or TRAF4 siRNA, and treated with TGFβ (5 ng/mL) for 2 h. RAC1 activity was determined as in panel a. (c,d), A549 cells were transfected with either empty vector, different amounts of RAC1 dominant-negative mutant (MYC-RAC1 N17) (c), or MYC-tagged RAC1 constitutively active mutant (MYC-RAC1 L61) (d), followed by treatment with or without TGFβ (5 ng/mL) for 24 h. Cell lysates were subjected to immunoblotting for MYC, FL CD44 and CD44 TM-ICD. GAPDH was used as loading control. Cells transfected with non-targeting siRNA or siRNAs against TRAF4 were also analyzed (d). CD44 TM-ICD bands were quantified using the ImageJ software and normalized to GAPDH; values are shown as fold-difference.
2.6. Migration of A549 Cells Is Dependent on CD44, TRAF4, and RAC1
In order to determine if CD44-mediated migration of A549 cells requires TRAF4 and RAC1, we employed a cell culture scratch assay. The silencing of either CD44 or TRAF4 decreased the migration of A549 cells significantly; ectopic expression of the constitutively active RAC1 L61 rescued the migration of TRAF4-depleted cells and enhanced the migration of control cells (Figure 6). This observation suggests that the TRAF4–RAC1 axis might be important for CD44-dependent A549 cell motility. The migration of H1299 cells was significantly reduced by knockdown of CD44; a reduced migration, albeit not significant, was also observed in TRAF4-depleted cells, probably due to insufficient silencing efficiency (Figure S6).
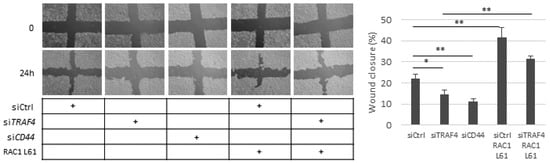
Figure 6.
Knockdown of TRAF4 diminishes cell motility, which is rescued by constitutively active RAC1. A549 cells were transfected with control siRNA or siRNAs targeting TRAF4 or CD44 and co-transfected with either empty vector or the constitutively active RAC1 L61 mutant. Cell migration was evaluated using a cell culture wound healing assay. Pictures of the cell cultures were taken at 0 h and at 24 h. The results are presented as a percentage of wound closure and illustrate the average values ± SEM out of three independent experiments. Asterisks illustrate significant differences between the conditions indicated with lines: * p < 0.05, ** p < 0.01.
3. Discussion
We show in the present communication that TRAF4 is needed for the activation of RAC1 and cleavage of CD44, resulting in the formation of CD44 TM-ICD and CD44 ICD and promoting the migration of A549 cells. It is possible that TRAF4 activation and other signaling pathways lead to RAC1-mediated activation of MMPs, enhancing CD44 cleavage (Figure 7) [14]. TRAF4 is also required for the increased expression of CD44 variant isoforms and HAS2 in stem-like cells, grown as spheres, leading to increased synthesis of hyaluronan.
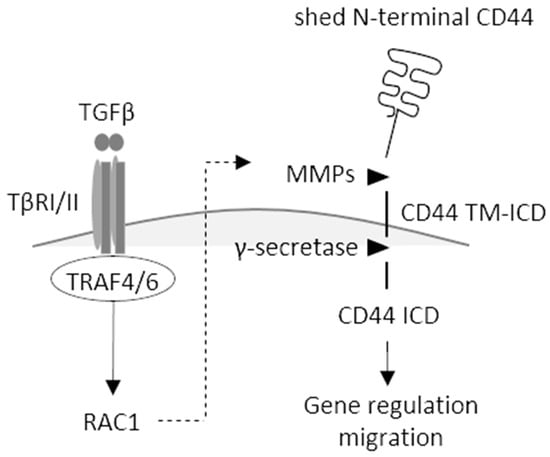
Figure 7.
Schematic illustration of TGFβ-induced activation of TRAF4/6 and RAC1, leading to CD44 cleavage, enhanced migration, and altered gene regulation. Binding of TGFβ to its type II receptor (TβII) recruits and activates the type I receptor (TβRI), activating TRAF4/6 and RAC1, leading to the activation of MMPs, resulting in the formation of CD44 TM-ICD; after further processing by γ-secretase, CD44 ICD is translocated to the nucleus, where it regulates transcription and promotes cell migration.
TRAF6 has been shown to be essential for TGFβ-stimulated, two-step cleavage of TβRI, resulting in the release of TβRI-ICD [23]. We found that TRAF6 is important also for the cleavage of CD44 and release of its intracellular domain in MEFs, as well as in A549 cells; however, in A549 cells, TRAF4 was found to be more important (Figure 3). The expression of full-length (FL) CD44 in MEF Traf6-/- was lower than in MEF wild-type (Wt) (Figure S3). According to previous reports, upon ICD translocation to the nucleus, CD44 is among the positively regulated genes. Due to stable depletion of Traf6, CD44 cleavage is constantly impaired, therefore disrupting the positive feedback loop replenishing Cd44 FL. Thus, it is possible that this would lead to a significant downregulation of Cd44 in Traf6-/- cells. Since TRAF4 and TRAF6 are structurally related to each other, it is possible that they have partly redundant functions, and that their relative expression levels determine which of the two isoforms is the most important for the cleavage of CD44 in different cell types. TRAF4 has been found to promote TGFβ signaling via Smad and non-Smad pathways [28], and to promote the migration of hepatocellular carcinoma [41], as well as endothelial and epithelial cells [42,43]. Moreover, TRAF4 has been found to be amplified during breast cancer progression [44]. Thus, TRAF4 has pro-tumorigenic effects in advanced cancer.
Cleavage of CD44 has been observed in several types of tumors, and increases in response to stimulation with fragmented hyaluronan, activation of protein kinase C (PKC), and activation of RAS [45,46,47], and CD44 ICD has been shown to promote epithelial–mesenchymal transition and the stemness of tumor cells [34]. Furthermore, CD44 ICD drives the proliferation of thyroid cancer by increasing cyclin D1 expression and the activity of CREB [38]. Mechanistically, CD44 ICD has been shown to bind to a consensus sequence, CCTGCG, found for example in the promoter regions of the MMP9 and HIF1a genes, and to interact with the co-activators CBP/p300 [19,38,48]. It has also been reported that CD44 ICD confers hypoxia-inducible factor-2α (HIF2α) stabilization, leading to an enhanced hypoxia-related gene signature [49]. Moreover, a recent study highlighted the importance of CD44 and its cleavage products in driving PD-L1 expression in A549 cells [50]. Taken together, these observations support the notion that CD44 ICD has a tumor promoting role. We observed that both TRAF4 and CD44 are needed for TGFβ stimulated induction of CCND1 (Figure 3). Our findings support the notion that TRAF4/6 promote pro-tumorigenic effects of CD44.
The expression of CD44v isoforms is linked to tumor growth, metastasis, and stemness [35], and importantly, CD44v isoforms exhibit increased proteolytic cleavage compared with CD44s in tumors [47]. CD44v6 is a biomarker associated with colon cancer stemness and tumor progression [51,52], and hyaluronan-engaged CD44v3 induces the expression of stem cell markers, NANOG, SOX2, and OCT4, promoting tumor progression and chemoresistance of head and neck squamous cell carcinomas [18,53]. Consistent with these observations, we found that CD44v isoforms are overexpressed in A549 cells grown as spheres and enriched for stemness markers, such as NANOG and OCT4 (Figure 2). We also found that A549 cells grown in spheres overexpress HAS2 and secrete more hyaluronan than cells grown under adhesive conditions (Figure 2f,g). Interestingly, hyaluronan synthesis was found to be increased in spheroids compared to adherent cultures of malignant mesothelioma [54]. These observations are consistent with the possibility that hyaluronan contributes to the stem cell niche [55].
The findings reported in this communication support the notion that TRAF4 mediates CD44 cleavage through the activation of RAC1, which is required for the migration of A549 cells. We also demonstrate a TRAF4-dependent association between tumor cell stemness, CD44v expression, and increased synthesis of hyaluronan. Our results suggest that the inhibition of CD44 signaling via TRAF4 and RAC1 may be beneficial in tumor treatment.
4. Materials and Methods
4.1. Cell Culture, Transfections, Plasmids, and Reagents
Human lung adenocarcinoma A549 cells (kindly provided by A. Moustakas, Uppsala University, purchased from American Type Culture Collection (ATCC)), non-small-cell lung carcinoma H1299 cells [56], glioblastoma U251MG cells [57], and prostate cancer PC3U cells [23] were grown in RPMI-1640 (Sigma-Aldrich Sweden AB, Stockholm, Sweden), whereas wild-type (Wt) or Traf6-deficient (Traf6-/-) mouse embryonic fibroblasts [23], human breast cancer Hs578 cells [56], and a bone-metastasizing clone of human breast cancer MDA MB 231 cells [58] were cultured in Dulbecco’s Modified Eagle Medium (DMEM) (Sigma-Aldrich Sweden AB, Stockholm, Sweden), all in the presence of 10% fetal bovine serum (FBS) (Biowest, Biotech-IgG AB, Lund, Sweden), penicillin and streptomycin (100 X/mL), and 5 mM L-glutamine (Sigma-Aldrich Sweden AB, Stockholm, Sweden). Human colon cancer HCT-116 and CACO-2 cells were grown in McCoy′s 5a medium (Sigma-Aldrich Sweden AB, Stockholm, Sweden) and in Minimum Essential Medium Eagle (Sigma-Aldrich Sweden AB, Stockholm, Sweden), respectively (both cell lines were kindly provided by Dr I. Ferby, Uppsala University, being purchased from ATCC). Ras-transformed premalignant MCF10AneoT (MII) cells [59] were cultured in DMEM/F12 (Gibco, Life Technologies Europe BV, Stockholm, Sweden). Cells were maintained at 5% CO2 in a humidified atmosphere at 37 °C.
For culturing in non-adherent conditions, A549 or H1299 cells were grown for 96 h in ultra-low-attachment six-well plates (Corning, Sigma-Aldrich Sweden AB, Stockholm, Sweden) in DMEM/F12 medium, supplemented with B27-containing vitamin A (Invitrogen, Life Technologies Europe BV, Stockholm, Sweden), also including basic fibroblast growth factor (20 ng/mL), EGF (20 ng/mL), insulin (4 μg/mL), and TGFβ (10 ng/mL) (PeproTech EC Ltd. Nordic, Stockholm, Sweden). Treatments with tumor necrosis factor alpha, hepatocyte growth factor, stem cell factor, and platelet-derived growth factor-BB (all from PeproTech EC Ltd. Nordic, Stockholm, Sweden; or Biosource Inc., Dacula, GA, United States) were at 10 ng/mL. The siRNAs (Non-targeting control, TRAF4, TRAF6, and CD44 (all ON-TARGET plus) were purchased from Dharmacon, Thermo Fischer Scientific (Gothenburg, Sweden), and transiently transfected on two consecutive days, 30 nM each time. Silentfect (Biorad Laboratories AB, Sundbyberg, Sweden) was used as a liposome reagent, according to the manufacturer’s instructions. For cDNA transfections, Lipofectamine 3000 reagent (Thermo Fischer Scientific, Gothenburg, Sweden) was employed, following the manufacturer’s protocol. FLAG-tagged TRAF6 Wt and TRAF6 C70A plasmids were kind gifts from Dr M. Landström, Umeå University. MYC-tagged RAC1 T17N, which is a nucleotide binding-defective mutant, hence acting in a dominant-negative (DN) manner, and RAC1 Q61L, which is a defective GTPase and thus acts in a constitutively active (CA) manner [60,61], were kind gifts from Dr P. Aspenström, Uppsala University. The proteasomal inhibitor MG132 was purchased from Calbiochem, Merck, Germany, and the lysosomal inhibitor chloroquine, the γ-secretase inhibitor DAPT, and 12-O-tetradecanoylphorbol-13-acetate (TPA) from Sigma-Aldrich Sweden AB, Stockholm, Sweden. High-molecular-weight hyaluronan (HMW HA; 1000 kDa) was kindly provided by Dr. Ove Wik (Q-Med, Uppsala, Sweden), as was 200 kDA of fragmented hyaluronan by Dr A. Passi (Insurbia University, Varese, Italy).
4.2. Immunoblotting
Cells were lysed in buffer containing 50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% NP-40, 0.1% sodium dodecyl sulfate (SDS), and 0.5% sodium deoxycholate, supplemented with HALT protease and a phosphatase inhibitor cocktail (Thermo Fischer Scientific, Gothenburg, Sweden); protein concentration was measured by utilizing a Bicinchoninic acid (BCA) assay (Thermo Fischer Scientific, Gothenburg, Sweden). Samples with equal protein content were subjected to SDS–polyacrylamide gel electrophoresis (SDS-PAGE), followed by wet transfer to nitrocellulose membrane (Amersham, GE Healthcare, Uppsala, Sweden) and blocking in 5% non-fat milk in Tris buffered saline (TBS), supplemented with 1% Tween 20. Subsequently, the membranes were incubated at 4 °C overnight with antibodies against CD44 (ab157107), lamin (ab16048) (Abcam, Cambridge, United Kingdom), α-tubulin (TU-02), MYC (SC-40), RAC1 (C-11), GST (B-14) (Santa Cruz Biotechnology Inc., Santa Cruz, CA, United States), GAPDH (D16H11) and phospho-SMAD3 (9520) (Cell Signaling Technology, Leiden, The Netherlands), TRAF4 (BD Biosciences, Stockholm, Sweden), or TRAF6 (Invitrogen, Life Technologies Europe BV, Stockholm, Sweden)), followed by incubation with horseradish peroxidase-conjugated secondary antibodies (1:10,000; Invitrogen, Life Technologies Europe BV, Stockholm, Sweden) for 1 h at room temperature, and development by chemiluminescence (Millipore, MA, United States). Band intensity quantification was performed by using the ImageJ software.
4.3. Nuclear–Cytoplasmic Fractionation
For the isolation of cytoplasmic and nuclear fractions from A549 cell cultures, NuCLEAR Extraction Kit (Sigma-Aldrich Sweden AB, Stockholm, Sweden) was employed following the manufacturer’s instructions. Briefly, cells from one 10-cm petri dish per condition were centrifuged, and cell pellets were incubated with 300 μL hypotonic lysis buffer (cytoplasmic fraction). Nuclei were resuspended in 100 μL of nuclear extraction buffer. For immunoblotting, 30 μL of the cytoplasmic fraction and 10 μL of the nuclear were subjected to electroforesis in either 10% or 15% polyacrylamide gels. Lamin B1 and tubulin were used as markers for nuclear and cytoplasmic compartments, respectively.
4.4. RAC1 Activity Assay
After the indicated treatments, A549 cells were washed once in ice-cold PBS and lysed in 50 mM Tris-HCl, pH 7.5, 1% Triton X-100, 0.5% deoxycholate, 500 mM NaCl, and 10 mM MgCl2 supplemented with protease and phosphatase inhibitors. Cell lysates were centrifuged at 13,000 rpm for 5 min; thereafter, supernatants were incubated with GST-fused PAK-1 (kindly provided by Dr P. Aspenström, Uppsala University) isolated from bacteria after pulling down with glutathione-fused agarose beads for 10 min at 4 °C, end over end. Afterwards, the beads were washed three times with wash buffer (50 mM Tris-HCl, pH 7.5, 1% Triton X-100, 150 mM NaCl, 10 mM MgCl2) and eluents were retrieved in 2x Laemmli sample buffer. Pulled down GTP-bound RAC1 was detected by immunoblotting analysis.
4.5. RNA Extraction and Real-Time qPCR
RNA was extracted from cells by using the RNeasy kit (QIAGEN AB, Sollentuna, Sweden) according to the manufacturer’s instructions. An iScript DNA synthesis kit (Biorad, Biorad Laboratories AB, Sundbyberg, Sweden) was used to reverse-transcribe 1 μg of total RNA to cDNA. KAPA Sybr Fast (PCR biosystems, Techtum Lab AB, Umeå, Sweden) was employed to perform real-time qPCR in triplicate (95 °C, 2 min; 40 × (95 °C, 10 s; 60 °C, 30 s)). Primers used for gene detection are summarized in Table S1. Gene expression was normalized to the housekeeping gene TATA-Box Binding Protein (TBP).
4.6. Luciferase Assay
A549 cells were co-transfected with siRNA targeting TRAF4 or control siRNA, together with the 5xTRE reporter plasmid and pCMV-β-gal (a β-galactosidase expression plasmid), kind gifts from Drs H. van Dam (Leiden University, Holland) and A. Moustakas (Uppsala University, Sweden), respectively. The β-galactosidase assay was performed by mixing the cell lysate with 100 mM sodium phosphate pH 7.3, 1 mM MgCl2, 50 mM β-mercaptoethanol, and 0.67 mg/mL of 2-nitrophenyl β-D-galactopyranoside, followed by monitoring of the absorbance at 420 nm. Luciferase reporter assays were performed with the firefly luciferase assay kit from Biotium (Fremont, CA, United States (BTIU30003-2)), according to the protocol of the manufacturer.
4.7. Phase Contrast Microscopy
Cells were subjected to phase contrast microscopy by employing a Zeiss Axiovision 40 microscope (Carl Zeiss) at the indicated time points and conditions. Bars at the respective micrographs represent 100 μm.
4.8. In Vitro Wound Healing Assay
A549 or H1299 cells were treated with siRNAs as indicated, and then grown to confluency. One vertical and three parallel horizontal wounds were then introduced by using a 200 μL pipette tip. Fresh medium containing 5% FBS was added and wound closure was monitored with a Zeiss Axiovert 40 phase-contrast microscope after 24 h, and quantified by the ImageJ software.
4.9. Hyaluronan Assay
Cell culture media from A549 and U251MG cultures, conditioned for 24 h, were collected after the indicated treatments, and hyaluronan concentration was measured and normalized to 1 μg of total protein extracted from the cells, as previously described [37].
4.10. Statistical Analysis
Graphs showing the mean ± SEM and are based on at least three independent experiments, unless stated otherwise. Two-paired Student’s t-test was used to calculate significance; three significance levels are indicated, i.e., * p < 0.05, ** p < 0.01, and *** p < 0.001.
5. Conclusions
The cleavage of the hyaluronan receptor CD44 is mediated by TRAF4 via the activation of RAC1, promoting migration of A549 lung cancer cells. The expression of CD44 variant isoforms, the cleavage of CD44, and the production of hyaluronan increases in stem-like tumor cells grown in spheres.
Supplementary Materials
The following are available online at https://www.mdpi.com/2072-6694/13/5/1021/s1, Figure S1: Screening for CD44 cleavage in different cell lines and in response to various external stimuli; Figure S2: NANOG, OCT4, and CD44v3 and v6 increase in stem-like H1299 cells grown in low-attachment conditions; Figure S3: The catalytic activity of TRAF6 is indispensable for TGFβ-induced CD44 cleavage in mouse embryo fibroblasts (MEFs); Figure S4: Importance of TRAF4 for the cleavage of CD44 in H1299 cells and expression of TRAF family members in lung cancer and glioma cells; Figure S5: TRAF4 is required for TGFβ-induced RAC1 activity and subsequent CD44 cleavage in H1299; Figure S6: Knockdown of CD44 decreases cell motility of H1299, Table S1: Primers used in this study.
Author Contributions
C.K., C.-H.H., and P.H. designed the study. C.K. and A.C. performed the experiments. P.H. and C.-H.H. supervised the study. C.K., A.C., S.S.S., C.-H.H., and P.H. interpretated data and wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work as supported by grants from the Swedish Cancer Society (180657 and 190066Pj01H).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
No data more than what is shown in this study were created or analyzed. Data sharing is not applicable to this article.
Acknowledgments
We thank P. Aspenström, I. Ferby, M. Landström, H. Van Dam, A. Passi, O. Wik, and A. Moustakas for cell lines and reagents.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Goodison, S.; Urquidi, V.; Tarin, D. CD44 cell adhesion molecules. Mol. Pathol. 1999, 52, 189–196. [Google Scholar] [CrossRef]
- Jalkanen, S.T.; Bargatze, R.F.; Herron, L.R.; Butcher, E.C. A lymphoid cell surface glycoprotein involved in endothelial cell recognition and lymphocyte homing in man. Eur. J. Immun. 1986, 16, 1195–1202. [Google Scholar] [CrossRef]
- Orian-Rousseau, V.; Ponta, H. Adhesion proteins meet receptors: A common theme? Adv. Cancer Res. 2008, 101, 63–92. [Google Scholar]
- Orian-Rousseau, V. CD44 acts as a signaling platform controlling tumor progression and metastasis. Front. Immunol. 2015, 6, 154. [Google Scholar] [CrossRef] [PubMed]
- Ponta, H.; Sherman, L.; Herrlich, P.A. CD44: From adhesion molecules to signalling regulators. Nat. Rev. Mol. Cell Biol. 2003, 4, 33–45. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, L.A.; Zhou, D.F.; Picker, L.J.; Minty, C.N.; Bargatze, R.F.; Ding, J.F.; Butcher, E.C. A human lymphocyte homing receptor, the hermes antigen, is related to cartilage proteoglycan core and link proteins. Cell 1989, 56, 1063–1072. [Google Scholar] [CrossRef]
- Matsumura, Y.; Tarin, D. Significance of Cd44 Gene-Products for Cancer-Diagnosis and Disease Evaluation. Lancet 1992, 340, 1053–1058. [Google Scholar] [CrossRef]
- Orian-Rousseau, V. CD44, a therapeutic target for metastasising tumours. Eur. J. Cancer 2010, 46, 1271–1277. [Google Scholar] [CrossRef]
- Su, Y.J.; Lai, H.M.; Chang, Y.W.; Chen, G.Y.; Lee, J.L. Direct reprogramming of stem cell properties in colon cancer cells by CD44. EMBO J. 2011, 30, 3186–3199. [Google Scholar] [CrossRef]
- Porsch, H.; Mehic, M.; Olofsson, B.; Heldin, P.; Heldin, C.H. Platelet-derived Growth Factor beta-Receptor, Transforming Growth Factor beta Type I Receptor, and CD44 Protein Modulate Each Other’s Signaling and Stability. J. Biol. Chem. 2014, 289, 19747–19757. [Google Scholar] [CrossRef] [PubMed]
- Bourguignon, L.Y.; Singleton, P.A.; Zhu, H.; Zhou, B. Hyaluronan promotes signaling interaction between CD44 and the transforming growth factor beta receptor I in metastatic breast tumor cells. J. Biol. Chem. 2002, 277, 39703–39712. [Google Scholar] [CrossRef]
- Okamoto, I.; Kawano, Y.; Tsuiki, H.; Sasaki, J.; Nakao, M.; Matsumoto, M.; Suga, M.; Ando, M.; Nakajima, M.; Saya, H. CD44 cleavage induced by a membrane-associated metalloprotease plays a critical role in tumor cell migration. Oncogene 1999, 18, 1435–1446. [Google Scholar] [CrossRef]
- Nagano, O.; Murakami, D.; Hartmann, D.; de Strooper, B.; Saftig, P.; Iwatsubo, T.; Nakajima, M.; Shinohara, M.; Saya, H. Cell-matrix interaction via CD44 is independently regulated by different metal loproteinases activated in response to extracellular Ca2+ influx and PKC activation. J. Cell Biol. 2004, 165, 893–902. [Google Scholar] [CrossRef] [PubMed]
- Nagano, O.; Saya, H. Mechanism and biological significance of CD44 cleavage. Cancer Sci. 2004, 95, 930–935. [Google Scholar] [CrossRef]
- Bourguignon, L.Y.; Gilad, E.; Rothman, K.; Peyrollier, K. Hyaluronan-CD44 interaction with IQGAP1 promotes Cdc42 and ERK signaling, leading to actin binding, Elk-1/estrogen receptor transcriptional activation, and ovarian cancer progression. J. Biol. Chem. 2005, 280, 11961–11972. [Google Scholar] [CrossRef] [PubMed]
- Bourguignon, L.Y. Hyaluronan-mediated CD44 activation of RhoGTPase signaling and cytoskeleton function promotes tumor progression. Semin. Cancer Biol. 2008, 18, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Skandalis, S.S.; Kozlova, I.; Engstrom, U.; Hellman, U.; Heldin, P. Proteomic identification of CD44 interacting proteins. IUBMB Life 2010, 62, 833–840. [Google Scholar] [CrossRef] [PubMed]
- Bourguignon, L.Y.; Earle, C.; Wong, G.; Spevak, C.C.; Krueger, K. Stem cell marker (Nanog) and Stat-3 signaling promote MicroRNA-21 expression and chemoresistance in hyaluronan/CD44-activated head and neck squamous cell carcinoma cells. Oncogene 2012, 31, 149–160. [Google Scholar] [CrossRef]
- Okamoto, I.; Kawano, Y.; Murakami, D.; Sasayama, T.; Araki, N.; Miki, T.; Wong, A.J.; Saya, H. Proteolytic release of CD44 intracellular domain and its role in the CD44 signaling pathway. J. Cell Biol. 2001, 155, 755–762. [Google Scholar] [CrossRef]
- Pietras, A.; Katz, A.M.; Ekstrom, E.J.; Wee, B.; Halliday, J.J.; Pitter, K.L.; Werbeck, J.L.; Amankulor, N.M.; Huse, J.T.; Holland, E.C. Osteopontin-CD44 signaling in the glioma perivascular niche enhances cancer stem cell phenotypes and promotes aggressive tumor growth. Cell Stem Cell 2014, 14, 357–369. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.L.; Wang, M.J.; Chen, J.Y. Acetylation and activation of STAT3 mediated by nuclear translocation of CD44. J. Cell Biol. 2009, 185, 949–957. [Google Scholar] [CrossRef]
- Heldin, C.H.; Moustakas, A. Signaling Receptors for TGF-beta Family Members. Cold Spring Harb. Perspect. Biol. 2016, 8, a022053. [Google Scholar] [CrossRef] [PubMed]
- Mu, Y.; Sundar, R.; Thakur, N.; Ekman, M.; Gudey, S.K.; Yakymovych, M.; Hermansson, A.; Dimitriou, H.; Bengoechea-Alonso, M.T.; Ericsson, J.; et al. TRAF6 ubiquitinates TGFbeta type I receptor to promote its cleavage and nuclear translocation in cancer. Nat. Commun. 2011, 2, 330. [Google Scholar] [CrossRef] [PubMed]
- Derynck, R.; Budi, E.H. Specificity, versatility, and control of TGF-beta family signaling. Sci. Signal. 2019, 12, eaav5183. [Google Scholar] [CrossRef] [PubMed]
- Sorrentino, A.; Thakur, N.; Grimsby, S.; Marcusson, A.; von Bulow, V.; Schuster, N.; Zhang, S.; Heldin, C.H.; Landstrom, M. The type I TGF-beta receptor engages TRAF6 to activate TAK1 in a receptor kinase-independent manner. Nat. Cell Biol. 2008, 10, 1199–1207. [Google Scholar] [CrossRef]
- Yamashita, M.; Fatyol, K.; Jin, C.; Wang, X.; Liu, Z.; Zhang, Y.E. TRAF6 mediates Smad-independent activation of JNK and p38 by TGF-beta. Mol. Cell 2008, 31, 918–924. [Google Scholar] [CrossRef]
- Hamidi, A.; Song, J.; Thakur, N.; Itoh, S.; Marcusson, A.; Bergh, A.; Heldin, C.H.; Landstrom, M. TGF-beta promotes PI3K-AKT signaling and prostate cancer cell migration through the TRAF6-mediated ubiquitylation of p85 alpha. Sci. Signal 2017, 10, eaal4186. [Google Scholar] [CrossRef]
- Zhang, L.; Zhou, F.F.; de Vinuesa, A.G.; de Kruijf, E.M.; Mesker, W.E.; Hui, L.; Drabsch, Y.; Li, Y.H.; Bauer, A.; Rousseau, A.; et al. TRAF4 Promotes TGF-beta Receptor Signaling and Drives Breast Cancer Metastasis. Mol. Cell 2013, 51, 559–572. [Google Scholar] [CrossRef] [PubMed]
- Zapata, J.M.; Lefebvre, S.; Reed, J.C. Targeting TRAFs for therapeutic intervention. Adv. Exp. Med. Biol. 2007, 597, 188–201. [Google Scholar]
- Chung, J.Y.; Park, Y.C.; Ye, H.; Wu, H. All TRAFs are not created equal: Common and distinct molecular mechanisms of TRAF-mediated signal transduction. J. Cell Sci. 2002, 115, 679–688. [Google Scholar]
- Bradley, J.R.; Pober, J.S. Tumor necrosis factor receptor-associated factors (TRAFs). Oncogene 2001, 20, 6482–6491. [Google Scholar] [CrossRef]
- Werb, Z. ECM and cell surface proteolysis: Regulating cellular ecology. Cell 1997, 91, 439–442. [Google Scholar] [CrossRef]
- Takamune, Y.; Ikebe, T.; Nagano, O.; Shinohara, M. Involvement of NF-kappaB-mediated maturation of ADAM-17 in the invasion of oral squamous cell carcinoma. Biochem. Biophys. Res. Commun. 2008, 365, 393–398. [Google Scholar] [CrossRef]
- Cho, Y.; Lee, H.W.; Kang, H.G.; Kim, H.Y.; Kim, S.J.; Chun, K.H. Cleaved CD44 intracellular domain supports activation of stemness factors and promotes tumorigenesis of breast cancer. Oncotarget 2015, 6, 8709–8721. [Google Scholar] [CrossRef]
- Nishino, M.; Ozaki, M.; Hegab, A.E.; Hamamoto, J.; Kagawa, S.; Arai, D.; Yasuda, H.; Naoki, K.; Soejima, K.; Saya, H.; et al. Variant CD44 expression is enriching for a cell population with cancer stem cell-like characteristics in human lung adenocarcinoma. J. Cancer 2017, 8, 1774–1785. [Google Scholar] [CrossRef]
- Porsch, H.; Bernert, B.; Mehic, M.; Theocharis, A.D.; Heldin, C.H.; Heldin, P. Efficient TGFbeta-induced epithelial-mesenchymal transition depends on hyaluronan synthase HAS2. Oncogene 2013, 32, 4355–4365. [Google Scholar] [CrossRef] [PubMed]
- Kolliopoulos, C.; Lin, C.Y.; Heldin, C.H.; Moustakas, A.; Heldin, P. Has2 natural antisense RNA and Hmga2 promote Has2 expression during TGFbeta-induced EMT in breast cancer. Matrix Biol. 2019, 80, 29–45. [Google Scholar] [CrossRef]
- De Falco, V.; Tamburrino, A.; Ventre, S.; Castellone, M.D.; Malek, M.; Manie, S.N.; Santoro, M. CD44 Proteolysis Increases CREB Phosphorylation and Sustains Proliferation of Thyroid Cancer Cells. Cancer Res. 2012, 72, 1449–1458. [Google Scholar] [CrossRef]
- Takahashi, K.; Eto, H.; Tanabe, K.K. Involvement of CD44 in matrix metalloproteinase-2 regulation in human melanoma cells. Int. J. Cancer 1999, 80, 387–395. [Google Scholar] [CrossRef]
- Murai, T.; Miyauchi, T.; Yanagida, T.; Sako, Y. Epidermal growth factor-regulated activation of Rac GTPase enhances CD44 cleavage by metalloproteinase disintegrin ADAM10. Biochem. J. 2006, 395, 65–71. [Google Scholar] [CrossRef]
- Liu, K.R.; Wu, X.L.; Zang, X.; Huang, Z.J.; Lin, Z.Y.; Tan, W.L.; Wu, X.; Hu, W.R.; Li, B.Q.; Zhang, L. TRAF4 Regulates Migration, Invasion, and Epithelial-Mesenchymal Transition via PI3K/AKT Signaling in Hepatocellular Carcinoma. Oncol. Res. 2017, 25, 1329–1340. [Google Scholar] [CrossRef]
- Wang, X.; Jin, C.; Tang, Y.; Tang, L.Y.; Zhang, Y.E. Ubiquitination of tumor necrosis factor receptor-associated factor 4 (TRAF4) by Smad ubiquitination regulatory factor 1 (Smurf1) regulates motility of breast epithelial and cancer cells. J. Biol. Chem. 2013, 288, 21784–21792. [Google Scholar] [CrossRef] [PubMed]
- Rousseau, A.; McEwen, A.G.; Poussin-Courmontagne, P.; Rognan, D.; Nomine, Y.; Rio, M.C.; Tomasetto, C.; Alpy, F. TRAF4 Is a Novel Phosphoinositide-Binding Protein Modulating Tight Junctions and Favoring Cell Migration. PLoS Biol. 2013, 11, e1001726. [Google Scholar] [CrossRef]
- Tomasetto, C.; Regnier, C.; Moog-Lutz, C.; Mattei, M.G.; Chenard, M.P.; Lidereau, R.; Basset, P.; Rio, M.C. Identification of four novel human genes amplified and overexpressed in breast carcinoma and localized to the q11-q21.3 region of chromosome 17. Genomics 1995, 28, 367–376. [Google Scholar] [CrossRef]
- Kawano, Y.; Okamoto, I.; Murakami, D.; Itoh, H.; Yoshida, M.; Ueda, S.; Saya, H. Ras oncoprotein induces CD44 cleavage through phosphoinositide 3-OH kinase and the rho family of small G proteins. J. Biol. Chem. 2000, 275, 29628–29635. [Google Scholar] [CrossRef]
- Sugahara, K.N.; Murai, T.; Nishinakamura, H.; Kawashima, H.; Saya, H.; Miyasaka, M. Hyaluronan oligosaccharides induce CD44 cleavage and promote cell migration in CD44-expressing tumor cells. J. Biol. Chem. 2003, 278, 32259–32265. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, I.; Tsuiki, H.; Kenyon, L.C.; Godwin, A.K.; Emlet, D.R.; Holgado-Madruga, M.; Lanham, I.S.; Joynes, C.J.; Vo, K.T.; Guha, A.; et al. Proteolytic cleavage of the CD44 adhesion molecule in multiple human tumors. Am. J. Pathol. 2002, 160, 441–447. [Google Scholar] [CrossRef]
- Miletti-Gonzalez, K.E.; Murphy, K.; Kumaran, M.N.; Ravindranath, A.K.; Wernyj, R.P.; Kaur, S.; Miles, G.D.; Lim, E.; Chan, R.; Chekmareva, M.; et al. Identification of function for CD44 intracytoplasmic domain (CD44-ICD): Modulation of matrix metalloproteinase 9 (MMP-9) transcription via novel promoter response element. J. Biol. Chem. 2012, 287, 18995–19007. [Google Scholar] [CrossRef]
- Johansson, E.; Grassi, E.S.; Pantazopoulou, V.; Tong, B.; Lindgren, D.; Berg, T.J.; Pietras, E.J.; Axelson, H.; Pietras, A. CD44 Interacts with HIF-2 alpha to Modulate the Hypoxic Phenotype of Perinecrotic and Perivascular Glioma Cells. Cell Rep. 2017, 20, 1641–1653. [Google Scholar] [CrossRef]
- Kong, T.; Ahn, R.; Yang, K.; Zhu, X.; Fu, Z.; Morin, G.; Bramley, R.; Cliffe, N.C.; Xue, Y.; Kuasne, H.; et al. CD44 Promotes PD-L1 Expression and Its Tumor-Intrinsic Function in Breast and Lung Cancers. Cancer Res. 2020, 80, 444–457. [Google Scholar] [CrossRef]
- Todaro, M.; Gaggianesi, M.; Catalano, V.; Benfante, A.; Iovino, F.; Biffoni, M.; Apuzzo, T.; Sperduti, I.; Volpe, S.; Cocorullo, G.; et al. CD44v6 Is a Marker of Constitutive and Reprogrammed Cancer Stem Cells Driving Colon Cancer Metastasis. Cell Stem Cell 2014, 14, 342–356. [Google Scholar] [CrossRef]
- Wang, Z.; Zhao, K.; Hackert, T.; Zoller, M. CD44/CD44v6 a Reliable Companion in Cancer-Initiating Cell Maintenance and Tumor Progression. Front. Cell Dev. Biol. 2018, 6, 97. [Google Scholar] [CrossRef]
- Bourguignon, L.Y.W.; Earle, C.; Shiina, M. Activation of Matrix Hyaluronan-Mediated CD44 Signaling, Epigenetic Regulation and Chemoresistance in Head and Neck Cancer Stem Cells. Int. J. Mol. Sci. 2017, 18, 1849. [Google Scholar] [CrossRef]
- Ohno, Y.; Shingyoku, S.; Miyake, S.; Tanaka, A.; Fudesaka, S.; Shimizu, Y.; Yoshifuji, A.; Yamawaki, Y.; Yoshida, S.; Tanaka, S.; et al. Differential regulation of the sphere formation and maintenance of cancer-initiating cells of malignant mesothelioma via CD44 and ALK4 signaling pathways. Oncogene 2018, 37, 6357–6367. [Google Scholar] [CrossRef] [PubMed]
- Skandalis, S.S.; Karalis, T.T.; Chatzopoulos, A.; Karamanos, N.K. Hyaluronan-CD44 axis orchestrates cancer stem cell functions. Cell. Signal. 2019, 63, 109377. [Google Scholar] [CrossRef]
- Mehic, M.; de Sa, V.K.; Hebestreit, S.; Heldin, C.H.; Heldin, P. The deubiquitinating enzymes USP4 and USP17 target hyaluronan synthase 2 and differentially affect its function. Oncogenesis 2017, 6, e348. [Google Scholar] [CrossRef]
- Eger, G.; Papadopoulos, N.; Lennartsson, J.; Heldin, C.H. NR4A1 Promotes PDGF-BB-Induced Cell Colony Formation in Soft Agar. PLoS ONE 2014, 9, e109047. [Google Scholar] [CrossRef]
- Bernert, B.; Porsch, H.; Heldin, P. Hyaluronan synthase 2 (HAS2) promotes breast cancer cell invasion by suppression of tissue metalloproteinase inhibitor 1 (TIMP-1). J. Biol. Chem. 2011, 286, 42349–42359. [Google Scholar] [CrossRef] [PubMed]
- Sundqvist, A.; Vasilaki, E.; Voytyuk, O.; Bai, Y.; Morikawa, M.; Moustakas, A.; Miyazono, K.; Heldin, C.H.; Ten Dijke, P.; van Dam, H. TGFbeta and EGF signaling orchestrates the AP-1- and p63 transcriptional regulation of breast cancer invasiveness. Oncogene 2020, 39, 4436–4449. [Google Scholar] [CrossRef] [PubMed]
- Aspenstrom, P. The Intrinsic GDP/GTP Exchange Activities of Cdc42 and Rac1 Are A Critical Determinants for Their Specific Effects on Mobilization of the Actin Filament System. Cells 2019, 8, 759. [Google Scholar] [CrossRef]
- Aspenstrom, P.; Fransson, A.; Saras, J. Rho GTPases have diverse effects on the organization of the actin filament system. Biochem. J. 2004, 377, 327–337. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).