Abstract
The small GTPase Rac1 has been implicated in a variety of dynamic cell biological processes, including cell proliferation, cell survival, cell-cell contacts, epithelial mesenchymal transition (EMT), cell motility, and invasiveness. These processes are orchestrated through the fine tuning of Rac1 activity by upstream cell surface receptors and effectors that regulate the cycling Rac1-GDP (off state)/Rac1-GTP (on state), but also through the tuning of Rac1 accumulation, activity, and subcellular localization by post translational modifications or recruitment into molecular scaffolds. Another level of regulation involves Rac1 transcripts stability and splicing. Downstream, Rac1 initiates a series of signaling networks, including regulatory complex of actin cytoskeleton remodeling, activation of protein kinases (PAKs, MAPKs) and transcription factors (NFkB, Wnt/β-catenin/TCF, STAT3, Snail), production of reactive oxygen species (NADPH oxidase holoenzymes, mitochondrial ROS). Thus, this GTPase, its regulators, and effector systems might be involved at different steps of the neoplastic progression from dysplasia to the metastatic cascade. After briefly placing Rac1 and its effector systems in the more general context of intestinal homeostasis and in wound healing after intestinal injury, the present review mainly focuses on the several levels of Rac1 signaling pathway dysregulation in colorectal carcinogenesis, their biological significance, and their clinical impact.
1. Introduction
Colorectal cancer (CRC) is a major cause of cancer morbidity and mortality in western countries. Nearly 140,000 and 450,000 individuals are diagnosed annually in United-States and in Europe, where this cancer is responsible of approximately 53,000 and 215,000 related deaths, respectively [1,2]. Colorectal cancers arise through the stepwise accumulation of genetic alterations, including the activation/overexpression of a series of (proto)-oncogenes, and the inactivation of tumor suppressor genes [3,4] and follow three molecular pathways to genome instability characterized by (i) chromosomal instability (CIN), (ii) high microsatellite instability (MSI-H), or (iii) CpG island methylator phenotype (CIMP). Mutations in APC, TP53, KRAS, PIK3CA, FBXW7, SMAD4, TCF7L2, and NRAS genes are the most frequently identified in CRC (cancer genome atlas network 2012). Activating mutations of KRAS, NRAS, and HRAS genes occurs in 33%, 3.7%, and 0.9% of CRC, respectively (URL http://cancer.sanger.ac.uk/, COSMIC v90, released 5 September 2019). Ras proteins belong to a superfamily of small GTPases composed of Ras, Rho, Ran, Rab, and Arf. These GTPases act as binary switches from active GTP (Guanosine triphosphate)-bound form, that interacts with effector molecules to initiate signaling, to the GDP (Guanosine diphosphate)-bound inactive form.
The Rho family of small GTPases are involved in the regulation of actin cytoskeleton remodeling, cell polarity, cell adhesion and migration, but also other processes including stem cell maintenance, cell proliferation and differentiation. Among the twenty members of this family, the best characterized GTPases are RhoA, Rac1, and Cdc42. Rac GTPases encompass 4 members: Rac1 which is ubiquitous, Rac2 mainly expressed in hematopoietic cells, Rac3 expressed in brain and testis, and RhoG present in fibroblasts, leukocytes, neuronal, and endothelial cells [5]. The GTP-bound state of Rac1 is accompanied by a conformational change in two regions, termed switch I and II (encompassing amino acids 25–40 and 60–76, respectively), which allows the selective interaction with diverse effectors that mediate downstream signaling cascade (Figure 1 and Figure 2). The activity of Rac1 is positively regulated by Guanine nucleotide Exchanges Factors (GEFs) favoring the GDP/GTP exchange, GTPase Activating proteins (GAPs) favoring the switch on/off (GTP/GDP), and Guanosine nucleotide Dissociation Inhibitor (GDI) which binds to the GDP-bound forms, preventing the GDP/GTP exchange (off-state) but also sequestering the small GTPase in the cytoplasm. More than 80 GEFs and 70 GAPs for the Rho GTPase family have been identified, highlighting the fine tuning of the level and the activity of the GTPases, and of the assembly and subcellular targeting of scaffolds involving these GTPases and their effector systems, to allow the selective activation of signaling cascades and to trigger appropriate cellular response [6]. Accordingly, many Rac1-GEFs are multi-domain proteins allowing the organization of signalosomes and driving their subcellular localization. For instance, the pleckstrin homology (PH) domain, that binds phosphatidylinositol 3,4,5 trisphosphate, in Tiam1, P-Rex1, and Vav1 allows the plasma membrane recruitment of these Rac1-selective GEFs following the activation of receptor tyrosine kinase and the downstream PI3 kinase activity. The selectivity of the downstream effectors driven by these GEFs is exemplified by the different interactomes involving Tiam1 and P-Rex1 that trigger two opposing Rac1 migratory responses. Tiam-1 proved to stabilize junctional complexes, whereas P-Rex1 stimulates cell motility [7,8].
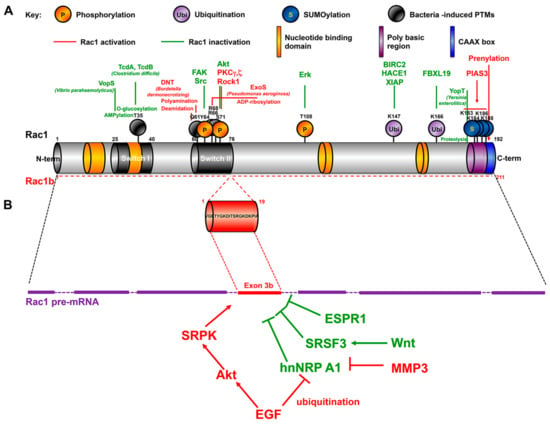
Figure 1.
Primary structure of Rac1 and Rac1b splice variant. (A) Posttranslational modifications (PTMs) of Rac1. In green are represented PTMs that inactivates Rac1, in Red those that stimulates the GTPase signaling. (B) Regulation of Rac1 splicing. For details see the text.
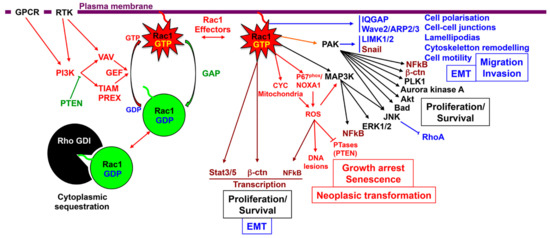
Figure 2.
Schematic representation of Rac1 signaling pathways and their biological significance. Rac1 upstream regulators: the Rac1 activating and inhibitory pathways are represented in red and in green respectively. Rac1 interacting partners and downstream effectors: transcription factors are represented in brown, effector molecules in black, pathways involved in cytoskeleton remodeling and cell migration in blue, and ROS pathway in Red.
The Rho family of GTPases is also regulated by posttranslational modifications, including prenylation of the C-terminal CAAX motif favoring membrane interaction, but also phosphorylation, SUMOylation, ubiquitination (Table 1, Figure 1 and see below).

Table 1.
Post translational modifications (PTMs) and functional consequences.
Rac1 initiates several signaling pathways, -including PAKs, NOX1 complex/reactive oxygen species (ROS) production, NFkB, members of MAPKs, Wnt/β-catenin/TCF, STAT3-, regulating membrane ruffling, cytoskeletal remodeling, cell adhesion, cell–cell contacts, cell polarity, cell motility, and transcriptional activity, leading to cell proliferation, epithelial mesenchymal transition (EMT), and invasiveness (Figure 2). Thus, this GTPase might be involved at different steps of the neoplastic progression from dysplasia to the metastatic cascade.
The hallmark of carcinogenesis relies on cancer cell invasiveness and dissemination. Rac1 activation and inactivation control plasticity of tumor cell movement [9]. Cancer cell invasiveness might involve individual (mesenchymal, amoeboid) or multi-cellular mode of migration. These modes of cell movement are interconvertible. Mesenchymal migration is driven by Rac1 via the WAVE regulatory complex that activates Arp2/3-dependent actin polymerization to generate membrane protrusions. Blocking Rac1 signaling suppresses mesenchymal movement and enhances amoeboid movement [9]. Amoeboid migration relies more on RhoA-ROCK signaling to promote actomyosin contractility and mechanical forces to move forward. This mode of moving takes advantage of the interstitial spaces, does not require extracellular matrix degradation or strong adhesion, and is independent from proteases and integrins. Collective migration is more representative of differentiated epithelial tumors, although some switch between these different modes of moving may timely operate [10].
2. Rac1 in Intestinal Physiology
2.1. Role of Rac1 in Intestinal Homeostasis
The implication of Rac1 in cell proliferation, cytoskeletal dynamic, junctional complexes activity, and cell motility make this small GTPase a critical effector in the maintenance of intestinal barrier integrity under physiological conditions and during tissue repair.
In human, the ontogeny of intestine occurs early during embryogenesis. In contrast, in mouse small intestine, the invagination of the intervillus epithelium as a specialized crypts compartment that contains stem cells, Paneth cells and progenitor cells occurs after birth (Figure 3A). A recent study uncovered the involvement of Rac1 signaling in this process. Conditional knockout of Rac1 in the intestine revealed that this GTPase is not required for initial crypt invagination, but later on, Rac1 depletion results in crypt expansion at the expense of villi, mimicking colon architecture [50]. The defect is not secondary to changes in cell proliferation or migration, but seems to be related to changes in cell shape at the crypt/villus boundaries due to increased hemidesmosomal α6/β4 integrin accumulation [50,51].
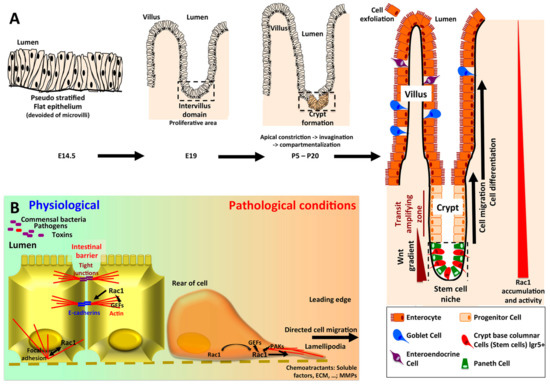
Figure 3.
(A) Ontogeny of the crypt-villus unit. The intestinal epithelium originates from endoderm, whereas the underlying mesenchymal and muscular layers derive from mesoderm. Intestinal ontogenesis follows an anteroposterior axis. In mouse, at embryonic day 14.5, the intestinal epithelium is pseudostratified and subjected to an active proliferation. By embryonic day 18, the villi have emerged in the duodenum and the proliferative area is restricted to the intervillus domain [64]. The crypts began to invaginate by day 5 post partum. First, myosin II-mediated apical constriction allows invagination of the crypt progenitor cells. Subsequently, a hinge region forms between crypts and villi. This process requires Rac1, which locally acts to suppress hemidesmosomal integrins, allowing cell shape changes and the formation of the hinge cells, leading to the compartmentalization of the crypt and villi [50]. In adult intestine, the stem cells -colocalized with Paneth cells in the bottom of the crypt- give rise to progenitor cells which will mainly differentiate into enterocytes, but also in goblet and enteroendocrine cells, and migrate along the villi to exfoliate at the top, whereas downward cell migration and differentiation allows to renew Paneth cells pool [65]. Wnt3 produced by Paneth cells, as well as EGF and Notch stimuli are required for the maintenance of stem cells. In the crypt villus unit, Rac1 expression and activity follow an increasing gradient from the top of the villi to the bottom of the crypts. As regards the colonic epithelium, the main characteristics rely on the absence of villi (flat mucosa), the absence of Paneth cell (substituted by Reg4+ deep crypt secretory cells), and the main type of differentiation being goblet cells [65]. (B) Rac1 in intestinal homeostasis and intestinal barrier repairs. By initiating the formation of E-cadherin complexes, Rac1 plays a critical role in the maintenance of tissue integrity. Upon wound formation, Rac1 localized to the leading edge of the cells, trigger lamellipodia formation through actin cystoskeleton remodeling, and directed cell migration.
In adult rat intestine, Rac1 protein levels as well as its GEF Tiam1 gradually increases from the villus tip to crypt compartment (Figure 3A) [52,53]. In this connection, a mouse Rac-FRET model further demonstrated that Rac1 activity is higher in cells at the base of the crypts than in distal cells [54]. Interestingly, the Leucine-rich repeat-containing G protein–coupled Receptor 5 (LGR5), a receptor for R-spondins strengthens cell–cell adhesion of stem cells through IQGAP/Rac1 interaction. LGR5 is a marker of adult stem cells, notably in the intestinal crypts and colon cancer cells that potentiates Wnt signaling via its interaction with the Wnt receptor frizzled. LGR5 favors Rac1-GTP in its competition with β-catenin to interact with IQGAP1, leading to enhanced linkage of E-cadherin junctional complexes to the cytoskeleton. This strong cell-cell adhesion has been proposed to retain stem cells within their niche at the crypt base [55]. Rac1 also enhances the nuclear formation of the β-catenin and LEF-1 complexes, providing the basis for the role of Rac1 in augmenting transactivation of Wnt target genes [56]. The Wnt/β-catenin pathway is a fundamental cornerstone for intestinal epithelial progenitor cell proliferation [57]. The dysregulation of this pathway occurs at high incidence and in the early steps of the neoplastic progression of intestinal epithelium. In mammalian as well as in drosophila, Rac1 proved to be necessary and sufficient to drive intestinal stem cells proliferation and regeneration in a ROS-dependent manner, suggesting the evolutionarily conserved role of Rac1 in intestinal homeostasis [58,59]. In these flies, the Rac1-JNK pathway controls the engulfment of dying intestinal stem cells by neighboring enterocytes [60].
Rac1 proved also to be involved in post-mitotic cells to control their terminal differentiation program. The expression of a constitutively active Rac1 mutant (Rac1Leu61) in mouse chimera induces precocious differentiation of members of the Paneth cells and enterocytic lineages, without affecting cell proliferation. It is worth noting that Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts [61,62]. On the other hand, the expression of a dominant negative form of Rac1 (Rac1Asn17) results in an inhibition of epithelial cell differentiation and a decrease in cell migration along the crypt–villus axis. Interestingly, none of these Rac1 mutations affect cell proliferation [63].
2.2. Intestinal Barrier Integrity/Mucosal Repair/Colitis
Intestinal epithelium does not only constitute a physical barrier against pathogen invasion but it also regulates nutrient uptake and innate immune function by avoiding the activation of mucosal immune responses. The barrier function in intestinal epithelia is carried out by specialized intercellular junctions, including tight junctions, zonula adherens, and desmosomes (Figure 3B). Tight junctions provide the paracellular permeability seal, through transmembrane proteins such as occludins and claudins, which are anchored to the actin cytoskeleton via scaffolding complexes of PDZ-containing proteins. Adherens junctions are composed by E-cadherin transmembrane proteins, anchored to the cytoskeleton through α-catenin, β-catenin, and p120ctn. Desmosomes containing desmocollin and desmoglein as transmembrane proteins, are essential to provide tissue integrity and to strength cell–cell junctions. These junctional complexes also participate in outside-in signaling, intestinal epithelial cell polarization, and differentiation [66].
The role of Rho/Rac1/CDC42 in intestinal barrier integrity is illustrated by the selective inhibition of these small GTPases by bacterial toxins leading to actin depolymerization, loss of cell-cell contacts, recruitment of immune cells and diarrhea (Figure 1A, Table 1) [67].
The activity of Rac1 is spatially and temporally orchestrated by scaffolding molecules, GEF and GAP regulatory proteins and effector proteins [68,69,70,71]. This concept is clearly demonstrated by the requirement of Rac1 for the initiation of the formation of E-cadherin-mediated cell-cell adherens junctions, which is linked to reduced cell motility and invasion.
Besides its critical role in the maintenance of cell-cell contacts and apical-basolateral cell polarization, a switch in Rac1 biological function from cell adherence to cell migration occurs following intestinal mucosal injury (Figure 3B). Rac1 triggers the formation of lamellipodia protrusions and epithelial cell migration at the leading-edge of a wound to promote wound closure. During this process, engagement of integrins with the extra cellular matrix leads to the recruitment and activation of Rac1, promoting the extension of membrane protrusion at the cell front. Downstream Rac1 effectors, such as WAVE complex, induce the polymerization of actin filaments in the vicinity of the leading-edge plasma membrane, creating the pushing forces required for membrane protrusion [72].
Rac1 activity appears to be critical for tissue repair after resolution of colitis. In this connection, ELMO1 -a component with Dock1 of a bipartite GEF for Rac1- is upregulated in inflamed intestinal tissue. In dextran sulphate sodium (DSS) experimental mouse model of colitis, systemic delivery of ELMO1- lentiviral vectors attenuated colonic inflammation and promoted recovery from colonic injury via Rac1 activation [73]. Lysophosphatidic acid (LPA) also promotes mucosal repair in DSS experimental mouse model of colitis, through the activation of Gαq/PLCβ1 -induced cell proliferation, and PLCβ2/Rac1 -induced cell migration [74]. The Rac1-PAK1-Akt signaling pathway alleviates DSS-induced colonic crypt damage and acute colitis via the preservation of survival and pluripotency of Lgr5+ colonic stem cells [75]. This protective effect is inhibited by the non-muscle-myosin-II heavy chain e9 that accumulates at epithelial injury sites [75]. The cellular inhibitor of apoptosis protein 2 (cIAP2/BIRC3) also upregulates and activates Rac1, and promotes cell migration and wound healing of human colon cancer Caco-2 cells in vitro [76]. In the same vein, the expression of cIAP2 increases in regenerating intestinal epithelial cells at the wound edge of mucosal biopsies in normal human colon in vivo, and returned to normal after reepithelialization.
On the other hand, the inflammatory response consecutive to intestinal barrier alteration might also contribute to wound repair through Rac1 mobilization. Accordingly, TNFα promotes EGFR/ Src pathways activation in intestinal epithelial cells, leading to the phosphorylation of Fak, further reinforced by the Rac1-induced ROS production and phosphatases inactivation [77].
Dietary factors might also modulate Rac1 activity, and consequently might strengthen intestinal barrier activity under physiological conditions or alleviate inflammatory severity and mucosal restitution following resolution of inflammation. In this context, Rutin-rich asparagus and black beans attenuate DSS-induced colitis by modulating the colonic microbiota, resulting in improved barrier integrity and upregulation of effectors of injury repair, including Rac1 [78,79].
Chronic intestinal inflammation, e.g., ulcerative colitis, is associated with an increased risk of colorectal cancer, due to excessive reactive oxygen and nitrogen species production, pro-inflammatory cytokines release, wound repair processes, and alteration in microbiota [80]. Thus, ROS production by the NOX1 complex in intestinal epithelial cells is tightly controlled at several levels including Rac1 ubiquination/degradation (see below) or impaired functional NOX1 complex formation through Rac1 interaction with apurinic/apyrimidinic endodeoxyribonuclease 1 (APE1) [81].
3. Rac1 in Colorectal Carcinogenesis
3.1. Rac1 Overexpression and Activation in CRC
Recurrent Rac1 P29S mutation in switch I domain, leads to gain-of-function, and is identified in 4.8% of skin tumors, and 9.2% of sun-exposed melanomas [82]. This markedly contrasts with the rare identification of Rac1 activating mutations in human colorectal cancers (0.89%). With regards to Rac1 copy number, an increase is identified in 2.1% of CRC (COSMIC v90). Nevertheless, Rac1 protein proved to be overexpressed in CRC and significantly associated with tumor stage [83]. Furthermore, Rac1 protein levels were higher in colon cancer liver metastases compared with primary colon tumor, and patients with high Rac1-expressing CRC showed shorter overall survival [83,84,85]. A recent meta-analysis of 14 studies including 1793 patients demonstrates that positive Rac1 expression correlated with tumor stage, blood vessel invasion, and lymph metastasis, but not with histological differentiation. This meta-analysis indicates that Rac1 could be used as a potential marker to predict CRC prognosis [86].
The increased Rac1 accumulation in CRC might account for an enhanced accumulation of Rac1 transcripts and/or transcription, or posttranslational modifications leading to Rac1 stabilization or decreased degradation. In this concern, Rac1 transcripts are direct target of miR-320a (a tumor suppressor miRNA), whose expression is inversely associated with CRC and cell line aggressiveness. In the human colon SW620 cancer cells, miR-320a efficiently inhibits cell migration/invasion, through Rac1 downregulation [87]. Interestingly, this miRNA might also indirectly target Rac1, through downregulation of PKCγ. PKCγ -induced Rac1 phosphorylation at Ser71 promotes the invasion of A431 cells [24]. MicroRNA-142-3p and miR-106b lead indirectly to upregulation of Rac1 in colonic cell lines, promoting EMT and cell invasiveness [83,88].
Besides accumulation, overactivation of Rac1 has been demonstrated in CRC [58,89]. High levels of Rac1-GTP were evidenced in colorectal cancerous tissues compared to control mucosa and these levels were significantly correlated with TNM stages, lymph node spread, and distant metastasis as well as patient survival [89].
The compartmentalization of Rac1 constitutes another important way in the regulation Rac1 signaling and biological processes [90]. Rac1 can be recruited in different subcellular plasma membrane subdomains, i.e., junctional complexes, leading-edge promoting antagonizing activity (see above). Rac1 is also recruited to the nucleus in cell cycle-dependent manner where it also regulates transcription factor activity, and in mitochondria where it interacts with Bcl2 and triggers ROS production. Thus, one might consider a role of Rac1 mislocalization in colorectal carcinogenesis. With this in mind, neutrophil gelatinase-associated lipocalin (NGAL)/lipocalin2 is overexpressed in CRC. In human colonic cell lines, lipocalin2 overexpression triggers relocalization of Rac1 from adherens junction to cell leading edge, and thus decreases E-cadherin-mediated cell–cell adhesion and increases cell motility and invasiveness [91].
3.2. Rac1 Post Translational Modifications (PTMs) in CRC
3.2.1. Rac1 SUMOylation
SUMOylation is an important PTM in the regulation of Rac1 signaling (Table 1). In response to hepatocyte growth factor treatment (HGF), the small ubiquitin-like modifier (SUMO) E3-ligase, PIAS3 interacts with Rac1 and triggers SUMO conjugation at Lys 188, 183, and 184 or 186 within the C-terminal polybasic region (PBR) of Rac1. This SUMOylation is required for increased Rac1 activation and for stimulating lamellipodia, cell migration and invasion [30]. In human colorectal cancers, Rac1 SUMOylation and activation correlates with mutant TP53 status. Accordingly, mutant TP53 competes with the SUMO-specific protease 1 (SENP1) for Rac1 binding, restraining the GTPase in a SUMOylated activated form [32]. In the human non-small cell lung carcinoma H1299 cell line, this competition promotes the metastatic phenotype. Thus, the frequent TP53 mutation identified in 43.5% of tumors (COSMIC v90) highlights the potential role Rac1 SUMOylation unbalance in CRC.
3.2.2. Rac1 Ubiquitination
Rac1 protein level is down-regulated by ligation of the protein modifiers ubiquitin and subsequent degradation by the proteasome (Table 1). Several E3 ligase, including HACE1, XIAP, c-IAP1 ubiquitinates Rac1 on Lysine residue 147, whereas FBXL19 ubiquitinates Lysine 166 [33,40,41]. It has been reported that ubiquinated Rac1 is also actively degraded in the nucleus by the proteasome [92].
In contrast to IAPs, which bind to Rac1 irrespective of its activation status, HACE1 preferentially interacts with the GTP-bound form. Notably, HACE1 targets Rac1 for degradation when Rac1 is localized to the NOX1 NADPH oxidase holoenzyme. This event blocks the ROS generation by NOX1 complex, and thereby confers cellular protection from ROS -induced DNA damage and cyclin D1-driven hyper-proliferation [34]. HACE1 activity is regulated by a negative feed-back loop involving the Ser/Thr kinase PAK1, a downstream effector of Rac1. HACE1 phosphorylation by PAK1 decreases its ability to interact with the plasma membrane and to ubiquitinate activated Rac1 [35]. HACE1 depletion is accompanied by increased total Rac1 level and Rac1 accumulation in membrane ruffles. Moreover, this depletion enhances cell migration independently of growth factor stimulation, which may have significance for malignant conversion [33]. Genetic inactivation of HACE1 in mice results in the development of spontaneous, late-onset cancer [38]. HACE1 is considered as a tumor suppressor and is frequently downregulated through hypermethylation of a CpG island in various human malignancies including colon, breast, liver, thyroid, kidney primary tumor and cell lines. In CRC, an aberrant methylation of HACE1 promoter is observed in 28% of tumors [39], whereas HACE1 mutation occurs in 4.4% of tumors (COSMIC v90).
3.2.3. Rac1 Phosphorylation
Carcinogenesis is associated with the activation of a series of Ser/Thr and Tyr protein kinases that might affect Rac1 subcellular localization and/or activity (Table 1). Phosphorylation of serine 71 by Akt inhibits GTP-binding activity without any significant change in GTPase activity [16]. This phosphorylation enhances Rac1 interaction and cytoplasmic sequestration by the 14-3-3 proteins [93]. Phosphorylation of Rac1 at serine 71 impairs its interaction with PAK1 and Sra-1, a component of the WAVE complex involved in actin cytoskeleton remodeling, JNK and SAPK activation, but did not affect NF-kappaB pathway [16,94]. This phosphorylation proved also to be essential for FBXL19-mediated Rac1 ubiquitination and depletion [40]. At functional level, phosphorylation of Ser71 by PKCγ or PKCζ stimulates cell migration and invasiveness in vitro [24,26]. Thus, this phosphorylation represents a reversible mechanism to shift specificity of Rac1/effector coupling, and to preferentially address selected downstream pathways.
Rac1 is also phosphorylated by Erk at threonine 108 in response to EGFR activation [19]. This phosphorylation decreases Rac1 activity, partially by inhibiting its interaction with phospholipase Cγ1, which might act as a GEF-, favors Rac1 nuclear translocation and impairs Rac1-induced cell motility.
Rac1 tyrosine residue 64 is a substrate for both Src and Fak that exerts a downward regulatory effect in Rac1 focal adhesions targeting and in cell spreading [22]. The phosphomimetic Rac1 mutant Y64D displays a lower affinity for PAK1, whereas the non-phosphorylable counterpart Rac1 mutant Y64F exhibits increased GTP-binding, increased association with βPIX, and reduced binding with RhoGDI as compared with wild type Rac1.
3.3. RAC1b Splice Variant
The incidence of KRAS activation in CRC led us to evaluate with Dr P. Jordan (Lisbon, Portugal) the status of other small GTPases and to identify a novel splice variant of Rac1 overexpressed in human colonic tumors that we designed Rac1b [13]. The ectopic expression of Rac1b has been later reported in various human malignancies, including breast, thyroid, ovarian, pancreatic, and lung cancers [29,95,96,97,98,99,100]. Rac1b is also expressed in human inflammatory colonic mucosa [101]. The ectopic expression of Rac1b in intestinal epithelial cells of transgenic mice increases cell proliferation and migration and contributes to intestinal wound-healing after acute inflammation [14].
Rac1b variant results from inclusion of exon 3b, which leads to a 19-amino acid in-frame insertion immediately C-terminal to the switch II domain (Figure 1B). The balance in Rac1/ Rac1b levels is regulated by splicing factors, that either induce skipping of the alternative exon 3b or favor its inclusion [102,103,104,105,106]. EGFR enhances Rac1b expression in two ways; (1) EGFR triggers a cascade of phosphorylation involving PI3K/Akt/SRPK leading the activation of SRSF1 splice factor that favors exon 3b maintenance; (2) EGFR induces the ubiquitination/inactivation of hnRNP A1, a splicing regulator that promotes exclusion of exon3b [103,104,106]. The Wnt signaling pathway favors exon 3b excision via the induction of SRSF3 expression [102].
Comparative to Rac1, Rac1b exhibits a number of distinctive features. This variant is preferentially in a GTP-bound active form, due to its reduced intrinsic GTPase activity and its impaired binding to Rho-GDI [107]. Unlike Rac1, Rac1b does not activate PAK1, Akt1, JNK, nor the transactivation activity of RelB-NF-kB2/p100 [15,108,109]. The inability of Rac1b to disrupt cell-cell contacts in keratinocytes seems to be related to Rac1b failure to stimulate PAK1 [110]. On the other hand, the extra 19-amino acid sequence enhances Rac1b binding to SmgGDS, RACK1, and p120 catenin, proteins involved in cell-cell adhesion, motility, and transcriptional regulation [111]. In this connection, the Rac1b-dependent motility and spreading of the mouse mammary epithelial SCp2 cells proved to require p120ctn. Rac1b has also been reported to interact with Dishevelled-3 and to form a tetramer with β-catenin/TCF that is recruited to the promoter of canonical Wnt target genes [112]. In this connection, the expression of the Rac1b splice variant in intestinal epithelial cells from transgenic mice is associated with an increase in the number of β-catenin nuclear positive cells at the bottom of the crypts [14]. Rac1b also exacerbates the cellular production of ROS leading to increased expression of the Snail transcription factor, and induction of EMT and invasiveness [27,113,114]. ROS act as signaling molecules through protein oxidation, e.g., phosphatase inactivation, but also causes oxidative damage and induces genomic instability stimulating carcinogenesis. It has been recently proposed that Rac1b might be involved in the Hutchinson–Gilford progeria syndrome, a genetic disease wherein an aging phenotype manifests in childhood. Rac1b phosphorylation at Ser71 by ROCK1 facilitates its interaction with cytochrome c, and increases mitochondrial ROS production, leading to cellular senescence [27].
Interestingly, Rac1b antagonizes TGF-β-induced EMT, suggesting that Rac1b promotes a selective response to EMT inducers [15,115]. This differential effect might be related to a mutual negative regulation of Rac1/Rac1b [116].
Experimental studies revealed that Rac1b increases G1/S progression and survival of NIH3T3 cells, and is sufficient to induce the transformation of these mouse fibroblasts [117,118]. Rac1b might also be involved in later stages of carcinogenesis. Although Rac1b overexpression alone is not sufficient to drive intestinal neoplasia in transgenic mice, it enhances Apc-dependent intestinal tumorigenesis, and promotes cecum and proximal colon carcinogenesis upon chronic inflammation [14]. Thus, it is likely that Rac1b overexpression provides a proliferative advantage to cancer cells. In this concern it has been proposed that Rac1b might alleviate the oncogenic senescence induced by BRAF mutation allowing neoplastic progression [119].
In human colorectal cancers, Rac1b overexpression is associated with BRAF mutation, which in advanced stage is characterized by a bad prognosis [120]. Likewise, Rac1b overexpression is associated with a poor outcome of patients with wild-type KRAS/BRAF metastatic colorectal cancer treated with FOLFOX/XELOX chemotherapy [121]. Rac1b proves also to facilitate chemo-resistance of colon cancer cell lines to oxaliplatin through activation of NFκB signaling [122].
Selectively targeting Rac1b and/or its interaction with molecular partners, e.g., by the use of nanobodies directed against the extra 19-amino acid sequence or by the use of the corresponding cell-permeable competitor peptides, would constitute a powerful therapeutic approach for the treatment of human malignancies with Rac1b ectopic expression.
3.4. Rac1 Regulators & Effectors
A second level of dysregulation of Rac1 signaling pathway in CRC might involve Rac1 upstream regulators or downstream effector systems (Table 2 and Table 3).

Table 2.
Upstream effectors; RhoGAsP, RhoGDIs, GEFs.

Table 3.
Interacting partners and functional consequences.
3.4.1. Guanine nucleotide Exchange Factors (GEFs)
Two classes of exchange factors were described for Rho GTPases: the classical Dbl-related exchange factors and more recently the atypical Dock family exchange factors (Table 2).
Many of these Rac1-GEFs encompass a PH domain and are activated and/or recruited to the plasma membrane leading to Rac1 activation in response to the activation of the PI3K pathway. Activation of this pathway is frequently observed in CRC, and might account for overexpression or dysregulation of receptor tyrosine kinases (e.g., EGFR, MET), activation of the nonreceptor tyrosine kinase Src, activating mutations in RAS or in PIK3CA, or downregulation of the tumor suppressor PTEN [4,17,273].
Among the Dbl-related exchange factors, as stated above, Tiam1 is generally associated under physiological conditions with Rac1-induced E-cadherin junctional complex stabilization and inversely linked with invasive potential [188]. Nevertheless, Tiam1 might be involved at different steps of colorectal carcinogenesis.
Tiam1 is a target gene of the Wnt signaling pathway [53], and the accumulation of Tiam1 transcripts is associated with the invasiveness of colorectal carcinoma cell lines in vitro [191]. Conversely, Tiam1 knockdown inhibits cell growth and invasive properties of the human colon cancer SW480 cell line in vitro, and reduces the growth of subcutaneous xenograft and the development of metastasis after intracaecal orthotopic xenograft in nude mice [192].
Tiam1 transcripts are the target of a series of microRNAs, including miR-29b and miR-21-5p. The accumulation of miR-29b is decreased in human colorectal cancers and cell lines. The ectopic expression of miR-29b and Tiam1 downregulation in human colonic cell lines inhibits epithelial–mesenchymal transition and suppresses tumor growth and metastasis in athymic nude mice [193]. Circular RNAs (CircRNAs) contain miRNA binding sites and can function as miRNA sponges. The circRNA-ACAP2 targets miR-21-5p which exhausts Tiam1. In human CRC and in the colon cancer SW480 cell line, CircRNA-ACAP2 and Tiam1 are expressed at a high level. The depletion of circRNA-ACAP2 or Tiam1, or overexpression of miR-21, inhibits SW480 cell migration and invasiveness [194].
Tiam1 transgenic mice display more invasive tumors with metastatic potential than wild-type mice, after chemical induction by 1,2-dimethylhydrazine [195]. These tumors are characterized by a marked EMT phenotype (nuclear β-catenin, decreased E-cadherin, vimentin enrichment).
Tiam1 is also overexpressed in human CRC [53,58,196], and this significantly correlated with poor prognosis and the absence of response to chemotherapy [197]. This overexpression is associated with EGFR level [198]. Accordingly, Tiam1 phosphorylation by the EGFR/Akt pathway triggers its interaction and stabilization by the 14-3-3 proteins, leading to an increased accumulation of Rac1-GTP and expression of the Wnt responsive genes cyclin-D1 and c-Myc involved in cell proliferation, [198]. Mechanistically, Tiam1 and Rac1 are components of transcriptionally active β-catenin/TCF complexes at Wnt-responsive promoters, and serve to enhance Wnt target gene transcription [213]. The interplay of Rac1/Tiam1 and the Wnt pathway is illustrated by the decrease in the number and the size of intestinal tumors following knockout of Tiam1 in APCMin/+ mice [53]. It is worth noting that, although Tiam1 effects on cellular migration are mediated by Rac1, Tiam1-induced resistance of colon cancer cells to anoikis is independent of the GTPase [196].
On the other hand, Tiam1 proved also to be a critical antagonist of CRC progression through inhibiting TAZ/YAP. These effectors of Wnt and Hippo signaling pathways are co-activators of the transcriptional enhanced associate domain (TEAD) proteins that regulate cell proliferation and stem cell functions. Tiam1 antagonizes TAZ/YAP (1) in the cytoplasm by promoting TAZ degradation consecutively to the interaction with βTrCP E3-ligase, (2) in the nucleus by suppressing TAZ/YAP interaction with TEADs, and the transactivation of target genes implicated in epithelial-mesenchymal transition and cell migration [274].
Other Rac1 GEFs have been involved in colorectal carcinogenesis. Vav1 overexpression is associated human colorectal tumor at advanced stage and with lymph node metastasis [200]. Vav2 mutations are identified in 4.2% of CRC (COSMIC), and have been associated with high risk of recurrence for patients with stages II and III CRC [201].
Asef1 (APC-Stimulated Guanine Nucleotide Exchange Factor 1)/Rho Guanine Nucleotide Exchange Factor 4 (ARHGEF4) is a Rac1- exchange factor stimulated by APC. Asef1 GEF stimulation by APC leads to Rac1 activation, lamellipodia formation, and increased cell migration [200]. Kawasaki et al. have identified a second Asef, termed Asef2, that shows significant structural and functional similarities to Asef1. Asef2 increases the levels of the active forms of Rac1 when co-transfected with truncated mutant APC expressed in colorectal tumor cells [275].
Consistent with previous reports [276], the levels of cytoplasmic APC in colorectal tumor cells harboring an APC mutation (SW480, SW620, HT29, Caco-2 and LoVo cells) were significantly higher than those in MDCK II and colorectal tumor cells with wild-type APC (HCT116 and SW48 cells). Similarly, the levels of Asef in the cytoplasm were also increased in colorectal tumor cells bearing APC mutations. The different subcellular localizations of Asef may determine whether Asef stimulates E-cadherin-mediated cell–cell adhesion or cell migration. Interestingly, cells coinfected with full-length Asef1 and the armadillo repeat region migrate more rapidly than cells coinfected with Asef1 and full-length APC [148]. Asef2 activated by truncated mutant APC is required for aberrant migration of colorectal tumor cells [150,275] (see Table 2). It has therefore been proposed that truncated forms of APC often found in colorectal cancer are not only deleterious due to unregulated β-catenin accumulation but may also enhance cellular metastasis due to constitutive Asef activation.
Immunostaining of ARHGEF7 (beta-PIX) is markedly increased in CRC compared with control tissues. This expression correlates with colorectal adenocarcinoma metastasis, and is associated with a shorter disease-free survival and a shorter overall survival [145]. The overexpression of ARHGEF7 in human colon cancer HCT116 and LoVo cells significantly enhanced cell migration and invasion, whereas the knockdown of ARHGEF7 in colorectal adenocarcinoma cells significantly decreased cell migration and invasion. Recently whole genome studies of colorectal metastases vs. matched primary tumors extended multistage colorectal cancer progression model with new specific for metastases components ARHGEF7 and ARHGEF33 [146]. Recurrent mutations in ARHGEF genes and the distribution of mutations in several ARHGEF genes clustered toward the RhoGEF and Plekstrin homology (PH) domains. ARHGEF7 mutations associate with worse disease-free survival [146]. These mutations were mutually exclusive with KRAS or NRAS mutations, suggesting that these may play a similar role to RAS activation. This is of special importance as patients with RAS activation do not respond well to EGFR inhibitors panitumumab and cetuximab, which may mean that metastasis with EGFR amplification and carrying an ARHGEF7 or ARHGEF33 mutation, may not respond to EGFR targeted therapy. On the other hand, ARHGEF7 prove to be determinants for irinotecan sensitivity of colorectal cancer cell lines [147].
The Dock (dedicator of cytokinesis) family proteins constitute another evolutionary conserved exchange factor (GEFs) for the Rho GTPases Rac1 and Cdc42, encompassing 11 related proteins gathered in four groups (A-D). Because Docks lack a Dbl domain, they are often referred to as ‘‘atypical GEFs”.
The DHR1 domain of Dock GEFs favors their recruitment to the membrane in response to PI3-kinase activation by directly binding to PIP3. The ELMO (engulfment and cell motility) scaffolding molecules binds Dock A/B group of proteins (Docks 1-5) relieve Docks autoinhibition and orchestrate their recruitment to discrete cell areas to allow polarized Rac1 activation and cytoskeleton remodeling [151].
In this context, ELMO3 transcript and protein levels are upregulated in CRC, and this overexpression is associated with tumor size, tumor differentiation, lymph node metastasis, and distant metastasis. Silencing of ELMO3 in the human colon cancer HCT116 cell line inhibits proliferation, invasion, and F-actin polymerization [277].
The cortactin-driven (CTTN) invasion by CRC cells is dependent of the activation of DOCK1-Rac1 [160]. CTTN expression in CRC correlates with depth of invasion, lymph node metastasis, and Tumor-Node-Metastasis stage [161]. Mechanistically, CTTN increases accumulation of DOCK1 and Rac1, and DOCK1 silencing partially abolishes the migration and invasion capacity induced by CTTN in SW480 colon cancer cells [160]. This observation emphasizes the potential role of DOCK1 in CTTN-mediated colorectal cell migration and invasion.
A recent, whole genome sequencing of CRC identified non-silent DOCK2 mutations at frequencies of >7%. DOCK2 is more frequently mutated in hypermutated (MSI or POLE/POLD1 driver mutations) CRC (38.0%) than in non-hyper-mutated CRC (3.9%) [130]. As regards DOCK2 expression, strong immunostaining in CRC was associated with good clinical outcome (tumor size, TNM stage, metastasis), and a longer overall survival [163]. These observations might be related to the recruitment CD8+ T lymphocytes in tumors expressing DOCK2.
Several recent studies have reported that the gut microbiome influences the efficacy of anti-PD-1/L1 immunotherapy. Accordingly, DOCK2 may participate in the immune response initiated by gut microbes, and its deficiency is most likely involved in an immune evasion mechanism of high-risk hypermutated CRC.
Other mediator of the canonical Wnt/β-catenin signaling pathway, implicated in Rac1 activity regulation, and important player in colorectal tumorigenesis and progression, is HEF1 (Human Enhancer of Filamentation 1), also known as NEDD9 or Cas-L (Cas-L adaptor molecule) [179]. NEDD9 is a member of the p130Cas family, a multi-domain skeleton protein that serves an important role in the cell signaling process via modulating invasion, metastasis, proliferation, and apoptosis, as well as mediating the hypoxia-induced migration of colorectal carcinoma cells [177,278]. NEDD9 complexes with DOCK3 to regulate Rac1 activity [9,165].
DOCK3, also referred to as modifier of cell adhesion (MOCA), was also shown to be an inhibitor of Wnt/ β-catenin signaling [279]. Moreover, multiple studies reported DOCK3 to be implicated in cancer cell invasion and migration [280]. Genome-wide association studies (GWAS) of advanced colorectal adenomas and control tissues identify a significant association of the intronic variant rs17659990 of DOCK3 with colorectal cancer risk [166].
In addition, NEDD9 is positively correlated with the expression of mesenchymal-type marker proteins such as vimentin and Zeb, while E-cadherin is opposite [178,281]. The elevated expression in colorectal cancer significantly correlated with high TNM stage and associated with poor prognosis of CRC patients [179]. Downregulation of NEDD9 by apigenin suppresses the migration, invasion, and metastasis of colorectal cancer cells [178].
The Rac1-GEF FARP2 contributes to the collective invasion of colorectal cancer cells as glandular structure maintaining apico-basolateral cell polarity. Decreased ROCK2 activity in a restricted number of cells within a cohort leads to leader cell formation. This process involves the recruitment of FARP2 to the apical junctional complex and F-actin polymerization through polarized Rac1 activation, and F-actin contractility through the Myosin II inhibition [173]. This contrasts with the ROCK pro-invasive function in other cancers, illustrating that the molecular mechanism of metastatic spread likely depends on tumor types and invasion mode.
In contrast, although the Rac1-GEFs P-REX1/2 stimulate cell migration, to our knowledge, no study has reported the implication of these GEFs in CRC.
3.4.2. GTPase-Activating Proteins (GAP)/Rho GDP Dissociation Inhibitor (RhoGDI)
Conversely, Rac1 activation in CRC might result from the dysregulation of RhoGAPs (Table 2). In this connection, ARHGAP15 mRNA and protein levels are obviously lower in CRC as compared to control mucosa, and are significantly correlated with clinical stage, tumor size metastasis, vital status, and overall survival of CRC patients. The downregulation of this Rac1-specific GAP may account for the inactivation of the FOXO1 transcription factor by Akt. Overexpression of ARHGAP15 inhibits the growth, migration, and invasive properties of the human colonic HT-29 cell line, whereas opposite effects are observed in ARHGAP15-silenced LoVo cells [124].
RacGAP1 is overexpressed in CRC at both mRNA and protein levels [135]. RacGAP1 is present in the nucleus, but a diffuse distribution in the cytoplasm also occurs. Colorectal cancer patients had opposite prognoses depending on the subcellular accumulation of RacGAP1 expression. Patients with high nuclear RacGAP1 expression had poor outcomes, whereas those with high cytoplasmic RacGAP1 expression had favorable prognosis [136].
RCC2 (Regulator of Chromosome Condensation 2) is a guanine exchange factor for the RalA small GTPase and is implicated in kinetochore-microtubule function mitosis. RCC2 has been identified as a biomarker for colorectal cancer. RCC2 is a transcriptional target of TP53 -frequently mutated in MSS CRC- that proved to physically interacts and deactivates Rac1 and inhibits migration of colon cancer cells [185]. Decreased expression of RCC2 as a result of mutation in the 5’ UTR of RCC2 gene is associated with improved outcome in CRC with microsatellite instability (MSI). In contrast, weak protein RCC2 accumulation in microsatellite stable (MSS) CRC is associated with a poor prognosis [186].
Diacylglycerol kinase ζ (DGKζ) act as a critical regulator of both Rac1 and RhoA activity. DGKζ-derived phosphatidic acid activates PAK1, which phosphorylates RhoGDI, allowing for the release and subsequent Rac1 activation [282]. DGKζ protein levels were elevated approximately 3-fold in the colonic SW620 carcinoma cells compared to SW480 cells. The SW480 and SW620 cell lines are derived from primary and secondary tumors resected from a single patient. Knockdown of DGKζ expression in SW620 cells significantly reduced Rac1 activity and attenuated the invasiveness of SW620 cells in vitro [283].
3.4.3. Rac1 Effectors and Signaling Pathways
Activated Rac1 interacts with a wide range of effectors that mediate signaling pathways converging to various biological functions including cytoskeleton remodeling, junctional complexes activity, cell spreading, cell motility, cell proliferation and differentiation (Figure 2, Table 3).
IQGAP1 (IQ motif-containing GTPase-activating protein 1) is a scaffolding molecule that interacts with a multitude of molecular partners [284] among them, as stated above, some components of E-cadherin junctional complexes and the guanine exchange factor Tiam1. IQGAP1 is an effector of Rac1/Cdc42 GTPases in the regulation of actin cytoskeleton dynamics and cell-cell adhesion. On the other hand, IQGAP1 inhibits intrinsic Rac1 GTPase activity, increasing Rac1-GTP pools. In human colorectal cancers, IQGAP1 is overexpressed in tumor tissues as compared with control mucosa. IQGAP1 expression is preferentially expressed at the invasion front, and especially in advanced carcinomas that invaded into the subserosa, suggesting a role of IQGAP1 in CRC invasion [231]. SUMOylation might be responsible of IQGAP1 stabilization in CRC, and in the increased proliferation and migration of colon cancer cells in vitro and xenografted in nude mice, through activating ERK, MEK, and Akt signaling pathways [285].
Although the causal link has not been investigated, IQGAP2 expression in CRC correlated positively with patient survival, while on the contrary, IQGAP3 expression levels correlated inversely with survival [233]. Up-regulation of IQGAP3 was identified in the subgroup of CRC with MSS-TP53 mutant phenotypes associated with a poor CRC-specific survival [234].
As stated above, Rac1 stimulates the Wnt/β-catenin signaling pathway, which exerts a critical role in the stemness of normal and neoplastic intestinal epithelial cells. The ROS produced by the NOX1 complex at the membrane or by the mitochondria through the Rac1/Bcl2 or Rac1/cytochrome c interactions and the downstream activation of JNK and NFkB pathways and their downstream target genes are clearly involved in EMT and cell invasiveness. Genetic deletion of Rac1 in mouse intestinal epithelial cells demonstrates the requirement of the GTPase in Wnt-induced transformation and colorectal cancer tumor initiation. The pro-tumorigenic effect was independent of β-catenin nuclear translocation and was mediated by NOX1 -induced ROS production [58].
Besides the level of ROS intracellular accumulation, the source of production and their subcellular accumulation might trigger opposing cellular responses. In this connection, Cheung et al. have demonstrated that, following APC loss, ROS produced by the NOX holoenzyme trigger intestinal epithelial cell proliferation, whereas mitochondrial ROS accumulation as a result of depletion of TIGAR—a protein that control glucose metabolism—decreases cell proliferation [286]. Interestingly, TIGAR and Rac1 expression are both controlled by the Wnt -induced Myc transactivation, suggesting that Wnt pathway integrates ROS signals to promotes cell proliferation.
Activated Rac1 also directly interacts with STAT3 to promote STAT3 phosphorylation, thus triggering EMT of CRC cells [24]. Interestingly, STAT3 proved to be overexpressed in CRC and to promote the transcription of the Nck1 adaptor molecule [216]. Nck1 serves as a scaffold for the recruitment of PAK1, Cdc42 and Rac1. Nck1 levels in CRC correlates with serosal invasion and lymph node metastasis, and cause invasiveness of CRC cell lines in vitro and enhance their metastatic potential in vivo through the activation of PAK1/Erk1 pathway.
As regards the p21-activated kinases (PAKs), they act as downstream effectors of Cdc42 and Rac1 GTPases. All PAKs are characterized by an N-terminal p21-GTPase binding domain and a highly conserved C-terminal kinase domain. These Ser/Thr protein kinases are composed of 6 members gathered in two groups. Groups I PAKs (PAKs 1-3) includes several domains that are absent in Group II PAKs (PAKs 4–6), i.e., autoinhibitory domain that overlaps with the GTPase binding domain (except for PAK5), proline rich domain, binding site for the ARHGEF7/β-PIX GEF and for the Nck adapter protein [218,219]. These proteins exert a critical role in many cellular processes, including cell morphology, survival, transcription, cell cycle progression, motility, apoptosis. PAKs are activated by both GTPase-dependent and -independent mechanisms, and mediate their biological activity through phosphorylation or acting as molecular scaffolds.
PAK1 immunostaining gradually increases during colonic carcinogenesis from normal mucosa to adenomas and adenocarcinomas [287]. PAK1 is overexpressed in primary CRC, and high PAK1 immunoexpression is associated with disease recurrence. However, there was no association with most clinicopathological parameters [288]. Song et al. reported that PAK1 and PAK4 expression are associated with CRC metastasis and infiltration, and that high PAK1 expression may indicate poor prognosis [220]. PAK1 is also overexpressed in inflammatory bowel disease and colitis associated carcinomas [289].
The cross-talk of PAK and the Wnt signaling pathway is highlighted by the inhibition of endogenous PAKs that impedes the transition of adenoma to carcinoma in an Apc-driven mouse model of colorectal cancer [290]. Similarly, PAK4 phosphorylates and stabilizes β-catenin by inhibiting its degradation. Moreover, PAK4 nuclear accumulation enhances β-catenin nuclear import and increases TCF/LEF transcriptional activity [223].
PAK5 proved also to be overexpressed in CRC. Immunostaining of PAK5 barely detectable in normal mucosa, gradually increases along the CRC progression from adenoma to adenocarcinoma and liver metastases, and correlates with tumor stage and dedifferentiation [224]. Gain and loss of function experiments of PAK5 in the SW480 colon cancer cell line revealed that PAK5 reduced cell adhesion but promoted their migration [224]. The mitochondrial localization of PAK5 and the phosphorylation of Bad protein (Bcl2-associated agonist of cell death) plays an essential in cell protection from apoptosis. PAK5 inhibits camptothecin-induced apoptosis by suppressing the activity of caspase-8 and the phosphorylation of Bad in CRC cells [291].
PAK1 might also be related to resistance to chemotherapies. Accordingly, PAK1 overexpression partially overcomes the 5-fluoro-uracile (5-FU) -induced growth inhibition of human colon cancer cells xenografted in scid mice, whereas PAK1 inhibition acts in synergy with 5-FU treatment [292]. Such PAK1-induced resistance to chemotherapies might be related to the involvement of PAK1 in DNA damage repair [218,293].
The activation of the Rac1 and Cdc42 signaling results in the formation of actin stress fibers, membrane ruffles, lamellipodia, and filopodia respectively and in cortical actin assembly. Pathways through which Rho GTPases elicit these effects are through direct interaction with members of the Wiskott–Alrich syndrome protein (WASP) family which stimulates structures such as lamellipodia and filopodia.
Rac1 -induced lamellipodia formation involves the WAVE regulatory complex and Arp2/3 complex that binds to the sides of existing actin filaments and initiates growth of a new filament. In CRC, Arp2 and WAVE2 proteins are frequently colocalized in the same cancer cells, whereas they are not detected in the normal colonic epithelial cells. Arp2 and WAVE2 were scattered throughout the cancer tissue and frequently appeared at the invasive front, where budding processes are formed. The colocalization of WAVE2 and Arp2 prove to be an independent risk factor for liver metastasis of colorectal carcinoma [291]. High mRNA levels of Arp2, is significantly associated with liver metastasis. In lines with these results, the accumulation of Wave2 and Arp2 was lower in human colonic SW480 cell line as compared to the corresponding SW620 cell line derived from the metastatic lymph node of the same primary CRC [294].
Epidemiological evidence suggests type 2 diabetes mellitus as a risk factor for cancer. In this connection, high glucose levels promote the proliferation of breast cancer cell lines through Rac1-mediated EGFR activation [295]. The search for common susceptibility genes for diabetes mellitus target organ damage and CRC pointed out several pathways, including inflammation and Wnt/β-catenin pathways [296]. Interestingly, Myc—a Wnt target gene—increases glucose transporters and glycolytic enzymes expression and mediates the Warburg effect, but also stimulates mitochondrial biogenesis [297]. Tigar transactivation by Myc might also alleviate the deleterious mitochondrial oxidative stress and ROS overproduction, consecutive to hyperglycemia-induced Rac1 activation [209,286]. Rac1 and Bcl-2 crosstalk in mitochondria to activate STAT3, promoting a permissive redox milieu for cell survival [298]. STAT3 dependent gene transcription and its nontranscriptional mitochondrial function converge to promote cell survival and likely contribute in the neoplastic progression of colorectal epithelial cells [299,300].
4. Conclusions, Perspectives
This review illustrates the pleiotropic activity of Rac1 under physiopathological conditions and during colorectal carcinogenesis. The Rac1 signaling pathway exerts a critical role in intestinal homeostasis from crypt formation during ontogeny, to cell differentiation, polarization, and cell–cell cohesion, to maintain intestinal barrier integrity on the one hand, cell proliferation and motility to allow mucosa repair following injuries on the other. These processes are obviously reactivated and hijacked during neoplastic progression. Although Rac1 mutation proves to be a rare event in CRC, this Review unveils the different levels of dysregulation of this signaling pathway during colorectal carcinogenesis (Figure 4). The increased accumulation and activity of Rac1, its altered subcellular distribution, and the ectopic expression of the Rac1b splice variant are clearly related with increased proliferative and invasive properties of colon cancer cell in vitro, and associated to invasiveness, metastatic spread, and refractoriness to chemotherapies of human CRC. The critical role of Rac1 in colorectal carcinogenesis is further highlighted by the unbalance of Rac1 regulators (GAP, GEF, RHOGDI) and upstream and downstream effectors systems. Targeting the many pathways controlled by Rac1 has so far been a holy grail in cancer drug discovery. Promising strategies are actually devoted to target Rac1 itself, and several leads are under investigation, including nucleotide binding inhibitors, prenylation inhibitors, SUMOylation inhibitors, GEF interaction inhibitors, RhoGDI and GAP modulators, as well as PAK1 inhibitors [301,302]. In conjunction with these strategies, the ongoing development of organoid technology opens up new perspectives in precision medicine to assess individual tumor responses to these targeted therapeutic agents.
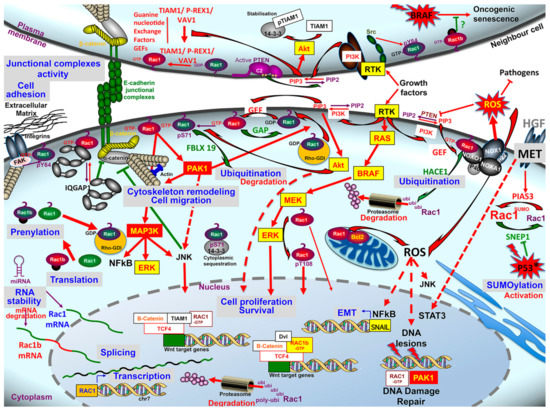
Figure 4.
Schematic overview of Rac1 signaling dysregulation in colorectal cancer, at the levels of Rac1 itself (transcripts stability, splice variant, post-translational modifications), of its upstream regulators and interacting partners, of its downstream effectors systems, and the biological impact of these dysregulations. For details see the text.
Funding
This research received no external funding.
Acknowledgments
This work was supported by French minister of higher education and research, INSERM and Sorbonne Université.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ait Ouakrim, D.; Pizot, C.; Boniol, M.; Malvezzi, M.; Boniol, M.; Negri, E.; Bota, M.; Jenkins, M.A.; Bleiberg, H.; Autier, P. Trends in colorectal cancer mortality in Europe: Retrospective analysis of the WHO mortality database. BMJ 2015, 351, h4970. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef]
- Fearon, E.R.; Vogelstein, B. A genetic model for colorectal tumorigenesis. Cell 1990, 61, 759–767. [Google Scholar] [CrossRef]
- Kotelevets, L.; Chastre, E.; Desmaële, D.; Couvreur, P. Nanotechnologies for the treatment of colon cancer: From old drugs to new hope. Int. J. Pharm. 2016, 514, 24–40. [Google Scholar] [CrossRef] [PubMed]
- Svensmark, J.H.; Brakebusch, C. Rho GTPases in cancer: Friend or foe? Oncogene 2019, 38, 7447–7456. [Google Scholar] [CrossRef] [PubMed]
- De, P.; Aske, J.C.; Dey, N. RAC1 Takes the Lead in Solid Tumors. Cells 2019, 8, 382. [Google Scholar] [CrossRef]
- Marei, H.; Carpy, A.; Woroniuk, A.; Vennin, C.; White, G.; Timpson, P.; Macek, B.; Malliri, A. Differential Rac1 signalling by guanine nucleotide exchange factors implicates FLII in regulating Rac1-driven cell migration. Nat. Commun. 2016, 7, 10664. [Google Scholar] [CrossRef]
- Marei, H.; Carpy, A.; Macek, B.; Malliri, A. Proteomic analysis of Rac1 signaling regulation by guanine nucleotide exchange factors. Cell Cycle 2016, 15, 1961–1974. [Google Scholar] [CrossRef]
- Sanz-Moreno, V.; Gadea, G.; Ahn, J.; Paterson, H.; Marra, P.; Pinner, S.; Sahai, E.; Marshall, C.J. Rac activation and inactivation control plasticity of tumor cell movement. Cell 2008, 135, 510–523. [Google Scholar] [CrossRef]
- Friedl, P.; Alexander, S. Cancer Invasion and the Microenvironment: Plasticity and Reciprocity. Cell 2011, 147, 992–1009. [Google Scholar] [CrossRef]
- Zhang, B.; Gao, Y.; Moon, S.Y.; Zhang, Y.; Zheng, Y. Oligomerization of Rac1 gtpase mediated by the carboxyl-terminal polybasic domain. J. Biol. Chem. 2001, 276, 8958–8967. [Google Scholar] [CrossRef] [PubMed]
- Casado-Medrano, V.; Baker, M.J.; Lopez-Haber, C.; Cooke, M.; Wang, S.; Caloca, M.J.; Kazanietz, M.G. The role of Rac in tumor susceptibility and disease progression: From biochemistry to the clinic. Biochem. Soc. Trans. 2018, 46, 1003–1012. [Google Scholar] [CrossRef] [PubMed]
- Jordan, P.; Brazåo, R.; Boavida, M.G.; Gespach, C.; Chastre, E. Cloning of a novel human Rac1b splice variant with increased expression in colorectal tumors. Oncogene 1999, 18, 6835–6839. [Google Scholar] [CrossRef] [PubMed]
- Kotelevets, L.; Walker, F.; Mamadou, G.; Lehy, T.; Jordan, P.; Chastre, E. The Rac1 splice form Rac1b favors mouse colonic mucosa regeneration and contributes to intestinal cancer progression. Oncogene 2018, 37, 6054–6068. [Google Scholar] [CrossRef] [PubMed]
- Melzer, C.; Hass, R.; Lehnert, H.; Ungefroren, H. RAC1B: A Rho GTPase with Versatile Functions in Malignant Transformation and Tumor Progression. Cells 2019, 8, 21. [Google Scholar] [CrossRef]
- Kwon, T.; Kwon, D.Y.; Chun, J.; Kim, J.H.; Kang, S.S. Akt protein kinase inhibits Rac1-GTP binding through phosphorylation at serine 71 of Rac1. J. Biol. Chem. 2000, 275, 423–428. [Google Scholar] [CrossRef]
- Kotelevets, L.; Scott, M.G.H.; Chastre, E. Targeting PTEN in Colorectal Cancers. Adv. Exp. Med. Biol. 2018, 1110, 55–73. [Google Scholar]
- Wang, F.-S.; Wu, W.-H.; Hsiu, W.-S.; Liu, Y.-J.; Chuang, K.-W. Genome-Scale Metabolic Modeling with Protein Expressions of Normal and Cancerous Colorectal Tissues for Oncogene Inference. Metabolites 2020, 10, 16. [Google Scholar] [CrossRef]
- Tong, J.; Li, L.; Ballermann, B.; Wang, Z. Phosphorylation of Rac1 T108 by extracellular signal-regulated kinase in response to epidermal growth factor: A novel mechanism to regulate Rac1 function. Mol. Cell. Biol. 2013, 33, 4538–4551. [Google Scholar] [CrossRef]
- Fang, J.Y.; Richardson, B.C. The MAPK signalling pathways and colorectal cancer. Lancet Oncol. 2005, 6, 322–327. [Google Scholar] [CrossRef]
- García-Aranda, M.; Redondo, M. Targeting Receptor Kinases in Colorectal Cancer. Cancers 2019, 11, 433. [Google Scholar] [CrossRef] [PubMed]
- Chang, F.; Lemmon, C.; Lietha, D.; Eck, M.; Romer, L. Tyrosine Phosphorylation of Rac1: A Role in Regulation of Cell Spreading. PLoS ONE 2011, 6, e28587. [Google Scholar] [CrossRef] [PubMed]
- Frame, M.C. Newest findings on the oldest oncogene how activated src does it. J. Cell Sci. 2004, 117, 989–998. [Google Scholar] [CrossRef] [PubMed]
- Aljagthmi, A.A.; Hill, N.T.; Cooke, M.; Kazanietz, M.G.; Abba, M.C.; Long, W.; Kadakia, M.P. ΔNp63α suppresses cells invasion by downregulating PKCγ/Rac1 signaling through miR-320a. Cell Death Dis. 2019, 10, 680. [Google Scholar] [CrossRef]
- Parsons, M.; Adams, J.C. Rac regulates the interaction of fascin with protein kinase C in cell migration. J. Cell Sci. 2008, 121, 2805–2813. [Google Scholar] [CrossRef]
- Islam, S.M.A.; Patel, R.; Acevedo-Duncan, M. Protein Kinase C-ζ stimulates colorectal cancer cell carcinogenesis via PKC-ζ/Rac1/Pak1/β-Catenin signaling cascade. Cell Res. 2018, 1865, 650–664. [Google Scholar]
- Kang, H.T.; Park, J.T.; Choi, K.; Choi, H.J.C.; Jung, C.W.; Kim, G.R.; Lee, Y.-S.; Park, S.C. Chemical screening identifies ROCK as a target for recovering mitochondrial function in Hutchinson-Gilford progeria syndrome. Aging Cell 2017, 16, 541–550. [Google Scholar] [CrossRef]
- Cai, S.-D.; Chen, J.-S.; Xi, Z.-W.; Zhang, L.-J.; Niu, M.-L.; Gao, Z.-Y. MicroRNA-144 inhibits migration and proliferation in rectal cancer by downregulating ROCK-1. Mol. Med. Rep. 2015, 12, 7396–7402. [Google Scholar] [CrossRef]
- Guo, Y.; Kenney, S.R.; Muller, C.Y.; Adams, S.; Rutledge, T.; Romero, E.; Murray-Krezan, C.; Prekeris, R.; Sklar, L.A.; Hudson, L.G.; et al. R-Ketorolac Targets Cdc42 and Rac1 and Alters Ovarian Cancer Cell Behaviors Critical for Invasion and Metastasis. Mol. Cancer Ther. 2015, 14, 2215–2227. [Google Scholar] [CrossRef]
- Castillo-Lluva, S.; Tatham, M.H.; Jones, R.C.; Jaffray, E.G.; Edmondson, R.D.; Hay, R.T.; Malliri, A. SUMOylation of the GTPase Rac1 is required for optimal cell migration. Nat. Cell Biol. 2010, 12, 1078–1085. [Google Scholar] [CrossRef]
- Ma, J.; Yang, Y.; Fu, Y.; Guo, F.; Zhang, X.; Xiao, S.; Zhu, W.; Huang, Z.; Zhang, J.; Chen, J. PIAS3-mediated feedback loops promote chronic colitis-associated malignant transformation. Theranostics 2018, 8, 3022–3037. [Google Scholar] [CrossRef] [PubMed]
- Yue, X.; Zhang, C.; Zhao, Y.; Liu, J.; Lin, A.W.; Tan, V.M.; Drake, J.M.; Liu, L.; Boateng, M.N.; Li, J.; et al. Gain-of-function mutant p53 activates small GTPase Rac1 through SUMOylation to promote tumor progression. Genes Dev. 2017, 31, 1641–1654. [Google Scholar] [CrossRef] [PubMed]
- Castillo-Lluva, S.; Tan, C.-T.; Daugaard, M.; Sorensen, P.H.B.; Malliri, A. The tumour suppressor HACE1 controls cell migration by regulating Rac1 degradation. Oncogene 2013, 32, 1735–1742. [Google Scholar] [CrossRef] [PubMed]
- Daugaard, M.; Nitsch, R.; Razaghi, B.; McDonald, L.; Jarrar, A.; Torrino, S.; Castillo-Lluva, S.; Rotblat, B.; Li, L.; Malliri, A.; et al. Hace1 controls ROS generation of vertebrate Rac1-dependent NADPH oxidase complexes. Nat. Commun. 2013, 4, 2180. [Google Scholar] [CrossRef]
- Acosta, M.I.; Urbach, S.; Doye, A.; Ng, Y.-W.; Boudeau, J.X.R.X.M.; Mettouchi, A.; Debant, A.; Manser, E.; Visvikis, O.; Lemichez, E. Group-I PAKs-mediated phosphorylation of HACE1 at serine 385 regulates its oligomerization state and Rac1 ubiquitination. Sci. Rep. 2018, 8, 1410. [Google Scholar] [CrossRef]
- Torrino, S.; Visvikis, O.; Doye, A.; Boyer, L.; Stefani, C.; Munro, P.; Bertoglio, J.; Gacon, G.; Mettouchi, A.; Lemichez, E. The E3 ubiquitin-ligase HACE1 catalyzes the ubiquitylation of active Rac1. Dev. Cell 2011, 21, 959–965. [Google Scholar] [CrossRef] [PubMed]
- Andrio, E.; Lotte, R.; Hamaoui, D.; Cherfils, J.; Doye, A.; Daugaard, M.; Sorensen, P.H.; Bost, F.; Ruimy, R.; Mettouchi, A.; et al. Identification of cancer-associated missense mutations in in hace1 that impair cell growth control and Rac1 ubiquitylation. Sci. Rep. 2017, 7, 44779. [Google Scholar] [CrossRef]
- Zhang, L.; Anglesio, M.S.; O’Sullivan, M.; Zhang, F.; Yang, G.; Sarao, R.; Nghiem, M.P.; Cronin, S.; Hara, H.; Melnyk, N.; et al. The E3 ligase HACE1 is a critical chromosome 6q21 tumor suppressor involved in multiple cancers. Nat. Med. 2007, 13, 1060–1069. [Google Scholar] [CrossRef]
- Hibi, K.; Sakata, M.; Sakuraba, K.; Shirahata, A.; Goto, T.; Mizukami, H.; Saito, M.; Ishibashi, K.; Kigawa, G.; Nemoto, H.; et al. Aberrant methylation of the HACE1 gene is frequently detected in advanced colorectal cancer. Anticancer Res. 2008, 28, 1581–1584. [Google Scholar]
- Zhao, J.; Mialki, R.K.; Wei, J.; Coon, T.A.; Zou, C.; Chen, B.B.; Mallampalli, R.K.; Zhao, Y. SCF E3 ligase F-box protein complex SCF FBXL19 regulates cell migration by mediating Rac1 ubiquitination and degradation. FASEB J. 2013, 27, 2611–2619. [Google Scholar] [CrossRef]
- Oberoi-Khanuja, T.K.; Rajalingam, K. IAPs as E3 ligases of Rac1: Shaping the move. Small GTPases 2012, 3, 131–136. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Oberoi, T.K.; Dogan, T.; Hocking, J.C.; Scholz, R.-P.; Mooz, J.; Anderson, C.L.; Karreman, C.; Meyer zu Heringdorf, D.; Schmidt, G.; Ruonala, M.; et al. IAPs regulate the plasticity of cell migration by directly targeting Rac1 for degradation. EMBO J. 2012, 31, 14–28. [Google Scholar] [CrossRef] [PubMed]
- Miura, K.; Fujibuchi, W.; Ishida, K.; Naitoh, T.; Ogawa, H.; Ando, T.; Yazaki, N.; Watanabe, K.; Haneda, S.; Shibata, C.; et al. Inhibitor of apoptosis protein family as diagnostic markers and therapeutic targets of colorectal cancer. Surg. Today 2011, 41, 175–182. [Google Scholar] [CrossRef] [PubMed]
- Voth, D.E.; Ballard, J.D. Clostridium difficile Toxins: Mechanism of Action and Role in Disease. Clin. Microbiol. Rev. 2005, 18, 247–263. [Google Scholar] [CrossRef]
- Yarbrough, M.L.; Li, Y.; Kinch, L.N.; Grishin, N.V.; Ball, H.L.; Orth, K. AMPylation of Rho GTPases by Vibrio VopS disrupts effector binding and downstream signaling. Science 2009, 323, 269–272. [Google Scholar] [CrossRef]
- Shao, F.; Dixon, J.E. YopT is a cysteine protease cleaving Rho family GTPases. Adv. Exp. Med. Biol. 2003, 529, 79–84. [Google Scholar]
- Okada, R.; Zhou, X.; Hiyoshi, H.; Matsuda, S.; Chen, X.; Akeda, Y.; Kashimoto, T.; Davis, B.M.; Iida, T.; Waldor, M.K.; et al. The Vibrio parahaemolyticuseffector VopC mediates Cdc42-dependent invasion of cultured cells but is not required for pathogenicity in an animal model of infection. Cell Microbiol. 2014, 16, 938–947. [Google Scholar] [CrossRef]
- Masuda, M.; Betancourt, L.; Matsuzawa, T.; Kashimoto, T.; Takao, T.; Shimonishi, Y.; Horiguchi, Y. Activation of rho through a cross-link with polyamines catalyzed by Bordetella dermonecrotizing toxin. EMBO J. 2000, 19, 521–530. [Google Scholar] [CrossRef]
- Rocha, C.L.; Rucks, E.A.; Vincent, D.M.; Olson, J.C. Examination of the Coordinate Effects of Pseudomonas aeruginosa ExoS on Rac1. Infect. Immun. 2005, 73, 5458–5467. [Google Scholar] [CrossRef]
- Sumigray, K.D.; Terwilliger, M.; Lechler, T. Morphogenesis and Compartmentalization of the Intestinal Crypt. Dev. Cell 2018, 45, 183–197. [Google Scholar] [CrossRef]
- Beaulieu, J.-F. Integrin α6β4 in Colorectal Cancer: Expression, Regulation, Functional Alterations and Use as a Biomarker. Cancers 2020, 12, 41. [Google Scholar] [CrossRef] [PubMed]
- Chono, E.; Kurokawa, T.; Oda, C.; Kawasaki, K.; Yamamoto, T.; Ishibashi, S. Expression of rac1 protein in the crypt-villus axis of rat small intestine: In reference to insulin action. Biochem. Biophys. Res. Commun. 1997, 233, 455–458. [Google Scholar] [CrossRef] [PubMed]
- Malliri, A.; Rygiel, T.P.; van der Kammen, R.A.; Song, J.-Y.; Engers, R.; Hurlstone, A.F.L.; Clevers, H.; Collard, J.G. The rac activator Tiam1 is a Wnt-responsive gene that modifies intestinal tumor development. J. Biol. Chem. 2006, 281, 543–548. [Google Scholar] [CrossRef] [PubMed]
- Johnsson, A.-K.E.; Dai, Y.; Nobis, M.; Baker, M.J.; McGhee, E.J.; Walker, S.; Schwarz, J.P.; Kadir, S.; Morton, J.P.; Myant, K.B.; et al. The Rac-FRET Mouse Reveals Tight Spatiotemporal Control of Rac Activity in Primary Cells and Tissues. Cell Rep. 2014, 6, 1153–1164. [Google Scholar] [CrossRef]
- Carmon, K.S.; Gong, X.; Yi, J.; Wu, L.; Thomas, A.; Moore, C.M.; Masuho, I.; Timson, D.J.; Martemyanov, K.A.; Liu, Q.J. LGR5 receptor promotes cell–cell adhesion in stem cells and colon cancer cells via the IQGAP1–Rac1 pathway. J. Biol. Chem. 2017, 292, 14989–15001. [Google Scholar] [CrossRef] [PubMed]
- Jamieson, C.; Lui, C.; Brocardo, M.G.; Martino-Echarri, E.; Henderson, B.R. Rac1 augments Wnt signaling by stimulating β-catenin-lymphoid enhancer factor-1 complex assembly independent of β-catenin nuclear import. J. Cell Sci. 2015, 128, 3933–3946. [Google Scholar] [CrossRef] [PubMed]
- van de Wetering, M.; Sancho, E.; Verweij, C.; de Lau, W.; Oving, I.; Hurlstone, A.; van der Horn, K.; Batlle, E.; Coudreuse, D.; Haramis, A.P.; et al. The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell 2002, 111, 241–250. [Google Scholar] [CrossRef]
- Myant, K.B.; Cammareri, P.; McGhee, E.J.; Ridgway, R.A.; Huels, D.J.; Cordero, J.B.; Schwitalla, S.; Kalna, G.; Ogg, E.-L.; Athineos, D.; et al. ROS production and NF-κB activation triggered by RAC1 facilitate WNT-driven intestinal stem cell proliferation and colorectal cancer initiation. Cell Stem Cell 2013, 12, 761–773. [Google Scholar] [CrossRef]
- Myant, K.B.; Scopelliti, A.; Haque, S.; Vidal, M.; Sansom, O.J.; Cordero, J.B. Rac1 drives intestinal stem cell proliferation and regeneration. Cell Cycle 2013, 12, 2973–2977. [Google Scholar] [CrossRef]
- Singh, S.R.; Zeng, X.; Zhao, J.; Liu, Y.; Hou, G.; Liu, H.; Hou, S.X. The lipolysis pathway sustains normal and transformed stem cells in adult Drosophila. Nature 2016, 538, 109–113. [Google Scholar] [CrossRef]
- Sato, T.; van Es, J.H.; Snippert, H.J.; Stange, D.E.; Vries, R.G.; van den Born, M.; Barker, N.; Shroyer, N.F.; van de Wetering, M.; Clevers, H. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature 2011, 469, 415–418. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Colman, M.J.; Schewe, M.; Meerlo, M.; Stigter, E.; Gerrits, J.; Pras-Raves, M.; Sacchetti, A.; Hornsveld, M.; Oost, K.C.; Snippert, H.J.; et al. Interplay between metabolic identities in the intestinal crypt supports stem cell function. Nature 2017, 543, 424–427. [Google Scholar] [CrossRef] [PubMed]
- Stappenbeck, T.S.; Gordon, J.I. Rac1 mutations produce aberrant epithelial differentiation in the developing and adult mouse small intestine. Development 2000, 127, 2629–2642. [Google Scholar] [PubMed]
- Wang, S.; Walton, K.D.; Gumucio, D.L. Signals and forces shaping organogenesis of the small intestine. Curr. Top. Dev. Biol. 2019, 132, 31–65. [Google Scholar] [PubMed]
- Gehart, H.; Clevers, H. Tales from the crypt: New insights into intestinal stem cells. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 19–34. [Google Scholar] [CrossRef]
- Garcia, M.A.; Nelson, W.J.; Chavez, N. Cell-Cell Junctions Organize Structural and Signaling Networks. Cold Spring Harb. Perspect Biol. 2018, 10, a029181. [Google Scholar] [CrossRef]
- Schlegel, N.; Meir, M.; Spindler, V.; Germer, C.-T.; Waschke, J. Differential role of Rho GTPases in intestinal epithelial barrier regulation in vitro. J. Cell. Physiol. 2011, 226, 1196–1203. [Google Scholar] [CrossRef]
- Garcia-Cattaneo, A.; Braga, V.M.M. Hold on tightly: How to keep the local activation of small GTPases. Cell Adh. Migr. 2013, 7, 283–287. [Google Scholar] [CrossRef][Green Version]
- Citi, S.; Guerrera, D.; Spadaro, D.; Shah, J. Epithelial junctions and Rho family GTPases: The zonular signalosome. Small GTPases 2014, 5, e973760. [Google Scholar] [CrossRef]
- Arnold, T.R.; Stephenson, R.E.; Miller, A.L. Rho GTPases and actomyosin: Partners in regulating epithelial cell-cell junction structure and function. Exp. Cell Res. 2017, 358, 20–30. [Google Scholar] [CrossRef]
- Marei, H.; Malliri, A. GEFs: Dual regulation of Rac1 signaling. Small GTPases 2017, 8, 90–99. [Google Scholar] [CrossRef] [PubMed]
- Mayor, R.; Etienne-Manneville, S. The front and rear of collective cell migration. Nat. Rev. Mol. Cell Biol. 2016, 17, 97–109. [Google Scholar] [CrossRef]
- Zheng, X.-B.; Liu, H.-S.; Zhang, L.-J.; Liu, X.-H.; Zhong, X.-L.; Zhou, C.; Hu, T.; Wu, X.-R.; Hu, J.-C.; Lian, L.; et al. Engulfment and Cell Motility Protein 1 Protects Against DSS-induced Colonic Injury in Mice via Rac1 Activation. J. Crohns Colitis 2019, 13, 100–114. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.-J.; Leoni, G.; Neumann, P.A.; Chun, J.; Nusrat, A.; Yun, C.C. Distinct Phospholipase C-β Isozymes Mediate Lysophosphatidic Acid Receptor 1 Effects on Intestinal Epithelial Homeostasis and Wound Closure. Mol. Cell. Biol. 2013, 33, 2016–2028. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Qi, Z.; Li, Y.; Wang, C.; Fu, W.; Chen, Y.-G. The non-muscle-myosin-II heavy chain Myh9 mediates colitis-induced epithelium injury by restricting Lgr5+ stem cells. Nat. Commun. 2015, 6, 7166. [Google Scholar] [CrossRef] [PubMed]
- Seidelin, J.B.; Larsen, S.; Linnemann, D.; Vainer, B.; Coskun, M.; Troelsen, J.T.; Nielsen, O.H. Cellular inhibitor of apoptosis protein 2 controls human colonic epithelial restitution, migration, and Rac1 activation. Am. J. Physiol. Gastrointest. Liver Physiol. 2015, 308, G92–G99. [Google Scholar] [CrossRef]
- Birkl, D.; Quiros, M.; Garcia-Hernandez, V.; Zhou, D.W.; Brazil, J.C.; Hilgarth, R.; Keeney, J.; Yulis, M.; Bruewer, M.; García, A.J.; et al. TNFα promotes mucosal wound repair through enhanced platelet activating factor receptor signaling in the epithelium. Mucosal Immunol. 2019, 12, 909–918. [Google Scholar] [CrossRef]
- Power, K.A.; Lu, J.T.; Monk, J.M.; Lepp, D.; Wu, W.; Zhang, C.; Liu, R.; Tsao, R.; Robinson, L.E.; Wood, G.A.; et al. Purified rutin and rutin-rich asparagus attenuates disease severity and tissue damage following dextran sodium sulfate-induced colitis. Mol. Nutr. Food Res. 2016, 60, 2396–2412. [Google Scholar] [CrossRef]
- Monk, J.M.; Wu, W.; Hutchinson, A.L.; Pauls, P.; Robinson, L.E.; Power, K.A. Navy and black bean supplementation attenuates colitis-associated inflammation and colonic epithelial damage. J. Nutr. Biochem. 2018, 56, 215–223. [Google Scholar] [CrossRef]
- Terzić, J.; Grivennikov, S.; Karin, E.; Karin, M. Inflammation and colon cancer. Gastroenterology 2010, 138, 2101–2114. [Google Scholar] [CrossRef]
- den Hartog, G.; Chattopadhyay, R.; Ablack, A.; Hall, E.H.; Butcher, L.D.; Bhattacharyya, A.; Eckmann, L.; Harris, P.R.; Das, S.; Ernst, P.B.; et al. Regulation of Rac1 and Reactive Oxygen Species Production in Response to Infection of Gastrointestinal Epithelia. PLoS Pathog. 2016, 12, e1005382. [Google Scholar] [CrossRef] [PubMed]
- Porter, A.P.; Papaioannou, A.; Malliri, A. Deregulation of Rho GTPases in cancer. Small GTPases 2016, 7, 123–138. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Xu, W.; Lu, T.; Zhou, J.; Ge, X.; Hua, D. MicroRNA-142-3p Promotes Cellular Invasion of Colorectal Cancer Cells by Activation of RAC1. Technol. Cancer Res. Treat. 2018, 17, 153303381879050. [Google Scholar] [CrossRef] [PubMed]
- Bao, Y.; Guo, H.; Lu, Y.; Feng, W.; Sun, X.; Tang, C.; Wang, X.; Shen, M. Blocking hepatic metastases of colon cancer cells using an shRNA against Rac1 delivered by activatable cell-penetrating peptide. Oncotarget 2016, 7, 77183–77195. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Xia, L.; Lin, J.; Su, J.; Oyang, L.; Wang, H.; Tan, S.; Tang, Y.; Chen, X.; Liu, W.; Luo, X.; et al. Diallyl disulfide inhibits colon cancer metastasis by suppressing Rac1-mediated epithelial-mesenchymal transition. OncoTargets Ther. 2019, 12, 5713–5728. [Google Scholar] [CrossRef]
- Lou, S.; Wang, P.; Yang, J.; Ma, J.; Liu, C.; Zhou, M. Prognostic and Clinicopathological Value of Rac1 in Cancer Survival: Evidence from a Meta-Analysis. J. Cancer 2018, 9, 2571–2579. [Google Scholar] [CrossRef]
- Zhao, H.; Dong, T.; Zhou, H.; Wang, L.; Huang, A.; Feng, B.; Quan, Y.; Jin, R.; Zhang, W.; Sun, J.; et al. miR-320a suppresses colorectal cancer progression by targeting Rac1. Carcinogenesis 2014, 35, 886–895. [Google Scholar] [CrossRef]
- Zheng, L.; Zhang, Y.; Lin, S.; Sun, A.; Chen, R.; Ding, Y.; Ding, Y. Down-regualtion of miR-106b induces epithelial-mesenchymal transition but suppresses metastatic colonization by targeting Prrx1 in colorectal cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 10534–10544. [Google Scholar]
- Zhou, K.; Rao, J.; Zhou, Z.-H.; Yao, X.-H.; Wu, F.; Yang, J.; Yang, L.; Zhang, X.; Cui, Y.-H.; Bian, X.-W.; et al. RAC1-GTP promotes epithelial-mesenchymal transition and invasion of colorectal cancer by activation of STAT3. Lab. Investig. 2018, 98, 989–998. [Google Scholar] [CrossRef]
- Payapilly, A.; Malliri, A. Compartmentalisation of RAC1 signalling. Curr. Opin. Cell Biol. 2018, 54, 50–56. [Google Scholar] [CrossRef]
- Hu, L.; Hittelman, W.; Lu, T.; Ji, P.; Arlinghaus, R.; Shmulevich, I.; Hamilton, S.R.; Zhang, W. NGAL decreases E-cadherin-mediated cell-cell adhesion and increases cell motility and invasion through Rac1 in colon carcinoma cells. Lab. Investig. 2009, 89, 531–548. [Google Scholar] [CrossRef] [PubMed]
- Lanning, C.C.; Daddona, J.L.; Ruiz-Velasco, R.; Shafer, S.H.; Williams, C.L. The Rac1 C-terminal Polybasic Region Regulates the Nuclear Localization and Protein Degradation of Rac1. J. Biol. Chem. 2004, 279, 44197–44210. [Google Scholar] [CrossRef]
- Abdrabou, A.; Brandwein, D.; Liu, C.; Wang, Z. Rac1 S71 Mediates the Interaction between Rac1 and 14-3-3 Proteins. Cells 2019, 8, 1006. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, J.; Proff, J.; Hävemeier, A.; Ladwein, M.; Rottner, K.; Barlag, B.; Pich, A.; Tatge, H.; Just, I.; Gerhard, R. Serine-71 phosphorylation of Rac1 modulates downstream signaling. PLoS ONE 2012, 7, e44358. [Google Scholar] [CrossRef]
- Schnelzer, A.; Prechtel, D.; Knaus, U.; Dehne, K.; Gerhard, M.; Graeff, H.; Harbeck, N.; Schmitt, M.; Lengyel, E. Rac1 in human breast cancer: Overexpression, mutation analysis, and characterization of a new isoform, Rac1b. Oncogene 2000, 19, 3013–3020. [Google Scholar] [CrossRef] [PubMed]
- Silva, A.L.; Carmo, F.; Bugalho, M.J. RAC1b overexpression in papillary thyroid carcinoma: A role to unravel. Eur. J. Endocrinol. 2013, 168, 795–804. [Google Scholar] [CrossRef]
- Ungefroren, H.; Sebens, S.; Giehl, K.; Helm, O.; Groth, S.; Fandrich, F.; Rocken, C.; Sipos, B.; Lehnert, H.; Gieseler, F. Rac1b negatively regulates TGF-β1-induced cell motility in pancreatic ductal epithelial cells by suppressing Smad signalling. Oncotarget 2014, 5, 277–290. [Google Scholar] [CrossRef]
- Mehner, C.; Miller, E.; Nassar, A.; Bamlet, W.R.; Radisky, E.S.; Radisky, D.C. Tumor cell expression of MMP3 as a prognostic factor for poor survival in pancreatic, pulmonary, and mammary carcinoma. Genes Cancer 2015, 6, 480–489. [Google Scholar]
- Stallings-Mann, M.L.; Waldmann, J.; Zhang, Y.; Miller, E.; Gauthier, M.L.; Visscher, D.W.; Downey, G.P.; Radisky, E.S.; Fields, A.P.; Radisky, D.C. Matrix metalloproteinase induction of Rac1b, a key effector of lung cancer progression. Sci. Transl. Med. 2012, 4, 142ra95. [Google Scholar] [CrossRef]
- Zhou, C.; Licciulli, S.; Avila, J.L.; Cho, M.; Troutman, S.; Jiang, P.; Kossenkov, A.V.; Showe, L.C.; Liu, Q.; Vachani, A.; et al. The Rac1 splice form Rac1b promotes K-ras-induced lung tumorigenesis. Oncogene 2013, 32, 903–909. [Google Scholar] [CrossRef]
- Matos, P.; Kotelevets, L.; Goncalves, V.; Henriques, A.F.A.; Henriques, A.; Zerbib, P.; Moyer, M.P.; Chastre, E.; Jordan, P. Ibuprofen inhibits colitis-induced overexpression of tumor-related Rac1b. Neoplasia 2013, 15, 102–111. [Google Scholar] [CrossRef] [PubMed]
- Goncalves, V.; Matos, P.; Jordan, P. Antagonistic SR proteins regulate alternative splicing of tumor-related Rac1b downstream of the PI3-kinase and Wnt pathways. Hum. Mol. Genet. 2009, 18, 3696–3707. [Google Scholar] [CrossRef] [PubMed]
- Goncalves, V.; Henriques, A.F.A.; Henriques, A.; Pereira, J.F.S.; Pereira, J.; Neves Costa, A.; Moyer, M.P.; Moita, L.F.; Gama-Carvalho, M.; Matos, P.; et al. Phosphorylation of SRSF1 by SRPK1 regulates alternative splicing of tumor-related Rac1b in colorectal cells. RNA 2014, 20, 474–482. [Google Scholar] [CrossRef] [PubMed]
- Pelisch, F.; Khauv, D.; Risso, G.; Stallings-Mann, M.; Blaustein, M.; Quadrana, L.; Radisky, D.C.; Srebrow, A. Involvement of hnRNP A1 in the matrix metalloprotease-3-dependent regulation of Rac1 pre-mRNA splicing. J. Cell. Biochem. 2012, 113, 2319–2329. [Google Scholar] [CrossRef] [PubMed]
- Ishii, H.; Saitoh, M.; Sakamoto, K.; Kondo, T.; Katoh, R.; Tanaka, S.; Motizuki, M.; Masuyama, K.; Miyazawa, K. Epithelial splicing regulatory proteins 1 (ESRP1) and 2 (ESRP2) suppress cancer cell motility via different mechanisms. J. Biol. Chem. 2014, 289, 27386–27399. [Google Scholar] [CrossRef]
- Wang, F.; Fu, X.; Chen, P.; Wu, P.; Fan, X.; Li, N.; Zhu, H.; Jia, T.-T.; Ji, H.; Wang, Z.; et al. SPSB1-mediated HnRNP A1 ubiquitylation regulates alternative splicing and cell migration in EGF signaling. Cell Res. 2017, 27, 540–558. [Google Scholar] [CrossRef]
- Matos, P.; Collard, J.G.; Jordan, P. Tumor-related alternatively spliced Rac1b is not regulated by Rho-GDP dissociation inhibitors and exhibits selective downstream signaling. J. Biol. Chem. 2003, 278, 50442–50448. [Google Scholar] [CrossRef]
- Matos, P.; Jordan, P. Increased Rac1b expression sustains colorectal tumor cell survival. Mol. Cancer Res. 2008, 6, 1178–1184. [Google Scholar] [CrossRef]
- Matos, P.; Jordan, P. Rac1, but not Rac1B, stimulates RelB-mediated gene transcription in colorectal cancer cells. J. Biol. Chem. 2006, 281, 13724–13732. [Google Scholar] [CrossRef]
- Lozano, E.; Frasa, M.A.M.; Smolarczyk, K.; Knaus, U.G.; Braga, V.M.M. PAK is required for the disruption of E-cadherin adhesion by the small GTPase Rac. J. Cell Sci. 2008, 121, 933–938. [Google Scholar] [CrossRef]
- Orlichenko, L.; Geyer, R.; Yanagisawa, M.; Khauv, D.; Radisky, E.S.; Anastasiadis, P.Z.; Radisky, D.C. The 19-amino acid insertion in the tumor-associated splice isoform Rac1b confers specific binding to p120 catenin. J. Biol. Chem. 2010, 285, 19153–19161. [Google Scholar] [CrossRef]
- Esufali, S.; Charames, G.S.; Pethe, V.V.; Buongiorno, P.; Bapat, B. Activation of tumor-specific splice variant Rac1b by dishevelled promotes canonical Wnt signaling and decreased adhesion of colorectal cancer cells. Cancer Res. 2007, 67, 2469–2479. [Google Scholar] [CrossRef]
- Radisky, D.C.; Levy, D.D.; Littlepage, L.E.; Liu, H.; Nelson, C.M.; Fata, J.E.; Leake, D.; Godden, E.L.; Albertson, D.G.; Angela Nieto, M.; et al. Rac1b and reactive oxygen species mediate MMP-3-induced EMT and genomic instability. Nature 2005, 436, 123–127. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Chen, Q.K.; Lui, C.; Cichon, M.A.; Radisky, D.C.; Nelson, C.M. Matrix compliance regulates Rac1b localization, NADPH oxidase assembly, and epithelial-mesenchymal transition. Mol. Biol. Cell 2012, 23, 4097–4108. [Google Scholar] [CrossRef] [PubMed]
- Witte, D.; Otterbein, H.; Rster, M.F.X.; Giehl, K.; Zeiser, R.; Lehnert, H.; Ungefroren, H. Negative regulation of TGF-β1-induced MKK6-p38 and MEK-ERK signalling and epithelial-mesenchymal transition by Rac1b. Sci. Rep. 2017, 7, 17313. [Google Scholar] [CrossRef] [PubMed]
- Melzer, C.; Hass, R.; Ohe von der, J.; Lehnert, H.; Ungefroren, H. The role of TGF-β and its crosstalk with RAC1/RAC1b signaling in breast and pancreas carcinoma. Cell Commun. Signal. 2017, 15, 19. [Google Scholar] [CrossRef]
- Matos, P.; Jordan, P. Expression of Rac1b stimulates NF-kappaB-mediated cell survival and G1/S progression. Exp. Cell Res. 2005, 305, 292–299. [Google Scholar] [CrossRef]
- Singh, A.; Karnoub, A.E.; Palmby, T.R.; Lengyel, E.; Sondek, J.; Der, C.J. Rac1b, a tumor associated, constitutively active Rac1 splice variant, promotes cellular transformation. Oncogene 2004, 23, 9369–9380. [Google Scholar] [CrossRef]
- Henriques, A.F.A.; Barros, P.; Moyer, M.P.; Matos, P.; Jordan, P. Expression of tumor-related Rac1b antagonizes B-Raf-induced senescence in colorectal cells. Cancer Lett. 2015, 369, 368–375. [Google Scholar] [CrossRef]
- Matos, P.; Oliveira, C.; Velho, S.; Goncalves, V.; da Costa, L.T.; Moyer, M.P.; Seruca, R.; Jordan, P. B-Raf(V600E) cooperates with alternative spliced Rac1b to sustain colorectal cancer cell survival. Gastroenterology 2008, 135, 899–906. [Google Scholar] [CrossRef]
- Alonso-Espinaco, V.; Cuatrecasas, M.; Alonso, V.; Escudero, P.; Marmol, M.; Horndler, C.; Ortego, J.; Gallego, R.; Codony-Servat, J.; Garcia-Albeniz, X.; et al. RAC1b overexpression correlates with poor prognosis in KRAS/BRAF WT metastatic colorectal cancer patients treated with first-line FOLFOX/XELOX chemotherapy. Eur. J. Cancer 2014, 50, 1973–1981. [Google Scholar] [CrossRef] [PubMed]
- Goka, E.T.; Chaturvedi, P.; Lopez, D.T.M.; Garza, A.D.L.; Lippman, M.E. RAC1b Overexpression Confers Resistance to Chemotherapy Treatment in Colorectal Cancer. Mol. Cancer Ther. 2019, 18, 957–968. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Chernoff, J.; Zheng, Y. Interaction of Rac1 with GTPase-activating proteins and putative effectors. A comparison with Cdc42 and RhoA. J. Biol. Chem. 1998, 273, 8776–8782. [Google Scholar] [CrossRef] [PubMed]
- Pan, S.; Deng, Y.; Fu, J.; Zhang, Y.; Zhang, Z.; Ru, X.; Qin, X. Decreased expression of ARHGAP15 promotes the development of colorectal cancer through PTEN/AKT/FOXO1 axis. Cell Death Dis. 2018, 9, 673. [Google Scholar] [CrossRef]
- Akilesh, S.; Suleiman, H.; Yu, H.; Stander, M.C.; Lavin, P.; Gbadegesin, R.; Antignac, C.; Pollak, M.; Kopp, J.B.; Winn, M.P.; et al. Arhgap24 inactivates Rac1 in mouse podocytes, and a mutant form is associated with familial focal segmental glomerulosclerosis. J. Clin. Investig. 2011, 121, 4127–4137. [Google Scholar] [CrossRef]
- Zhang, S.; Sui, L.; Zhuang, J.; He, S.; Song, Y.; Ye, Y.; Xia, W. ARHGAP24 regulates cell ability and apoptosis of colorectal cancer cells via the regulation of P53. Oncol. Lett. 2018, 16, 3517–3524. [Google Scholar]
- Nakamura, T.; Komiya, M.; Sone, K.; Hirose, E.; Gotoh, N.; Morii, H.; Ohta, Y.; Mori, N. Grit, a GTPase-activating protein for the Rho family, regulates neurite extension through association with the TrkA receptor and N-Shc and CrkL/Crk adapter molecules. Mol. Cell. Biol. 2002, 22, 8721–8734. [Google Scholar] [CrossRef][Green Version]
- Zhao, C.; Ma, H.; Bossy-Wetzel, E.; Lipton, S.A.; Zhang, Z.; Feng, G.-S. GC-GAP, a Rho family GTPase-activating protein that interacts with signaling adapters Gab1 and Gab2. J. Biol. Chem. 2003, 278, 34641–34653. [Google Scholar] [CrossRef]
- Moon, S.Y.; Zang, H.; Zheng, Y. Characterization of a brain-specific Rho GTPase-activating protein, p200RhoGAP. J. Biol. Chem. 2003, 278, 4151–4159. [Google Scholar] [CrossRef]
- Ge, W.; Cai, W.; Bai, R.; Hu, W.; Wu, D.; Zheng, S.; Hu, H. A novel 4-gene prognostic signature for hypermutated colorectal cancer. Cancer Manag. Res. 2019, 11, 1985–1996. [Google Scholar] [CrossRef]
- Mogensen, M.B.; Rossing, M.; Østrup, O.; Larsen, P.N.; Heiberg Engel, P.J.; Jørgensen, L.N.; Hogdall, E.V.; Eriksen, J.; Ibsen, P.; Jess, P.; et al. Genomic alterations accompanying tumour evolution in colorectal cancer: Tracking the differences between primary tumours and synchronous liver metastases by whole-exome sequencing. BMC Cancer 2018, 18, 752. [Google Scholar] [CrossRef]
- Parrini, M.C.; Sadou-Dubourgnoux, A.; Aoki, K.; Kunida, K.; Biondini, M.; Hatzoglou, A.; Poullet, P.; Formstecher, E.; Yeaman, C.; Matsuda, M.; et al. SH3BP1, an exocyst-associated RhoGAP, inactivates Rac1 at the front to drive cell motility. Mol. Cell 2011, 42, 650–661. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Feng, Y.; Chen, X.; Du, Z.; Jiang, S.; Ma, S.; Zou, W. SH3BP1-induced Rac-Wave2 pathway activation regulates cervical cancer cell migration, invasion, and chemoresistance to cisplatin. J. Cell. Biochem. 2018, 119, 1733–1745. [Google Scholar] [CrossRef] [PubMed]
- Zago, G.; Biondini, M.; Camonis, J.; Parrini, M.C. A family affair: A Ral-exocyst-centered network links Ras, Rac, Rho signaling to control cell migration. Small GTPases 2019, 10, 323–330. [Google Scholar] [CrossRef] [PubMed]
- Imaoka, H.; Toiyama, Y.; Saigusa, S.; Kawamura, M.; Kawamoto, A.; Okugawa, Y.; Hiro, J.; Tanaka, K.; Inoue, Y.; Mohri, Y.; et al. RacGAP1 expression, increasing tumor malignant potential, as a predictive biomarker for lymph node metastasis and poor prognosis in colorectal cancer. Carcinogenesis 2015, 36, 346–354. [Google Scholar] [CrossRef]
- Yeh, C.-M.; Sung, W.-W.; Lai, H.-W.; Hsieh, M.-J.; Yen, H.-H.; Su, T.-C.; Chang, W.-H.; Chen, C.-Y.; Ko, J.-L.; Chen, C.-J. Opposing prognostic roles of nuclear and cytoplasmic RACGAP1 expression in colorectal cancer patients. Hum. Pathol. 2016, 47, 45–51. [Google Scholar] [CrossRef]
- Lian, L.Y.; Barsukov, I.; Golovanov, A.P.; Hawkins, D.I.; Badii, R.; Sze, K.H.; Keep, N.H.; Bokoch, G.M.; Roberts, G.C. Mapping the binding site for the GTP-binding protein Rac-1 on its inhibitor RhoGDI-1. Structure 2000, 8, 47–55. [Google Scholar] [CrossRef]
- Fauré, J.; Dagher, M.C. Interactions between Rho GTPases and Rho GDP dissociation inhibitor (Rho-GDI). Biochimie 2001, 83, 409–414. [Google Scholar] [CrossRef]
- Del Pozo, M.A.; Kiosses, W.B.; Alderson, N.B.; Meller, N.; Hahn, K.M.; Schwartz, M.A. Integrins regulate GTP-Rac localized effector interactions through dissociation of Rho-GDI. Nat. Cell Biol. 2002, 4, 232–239. [Google Scholar] [CrossRef]
- Cho, H.J.; Kim, J.-T.; Baek, K.E.; Kim, B.-Y.; Lee, H.G. Regulation of Rho GTPases by RhoGDIs in Human Cancers. Cells 2019, 8, 1037. [Google Scholar] [CrossRef]
- Huang, D.; Lu, W.; Zou, S.; Wang, H.; Jiang, Y.; Zhang, X.; Li, P.; Songyang, Z.; Wang, L.; Wang, J.; et al. Rho GDP-dissociation inhibitor α is a potential prognostic biomarker and controls telomere regulation in colorectal cancer. Cancer Sci. 2017, 108, 1293–1302. [Google Scholar] [CrossRef] [PubMed]
- Feng, Q.; Baird, D.; Cerione, R.A. Novel regulatory mechanisms for the Dbl family guanine nucleotide exchange factor Cool-2/alpha-Pix. EMBO J. 2004, 23, 3492–3504. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zheng, T. Screening of Hub Genes and Pathways in Colorectal Cancer with Microarray Technology. Pathol. Oncol. Res. 2014, 20, 611–618. [Google Scholar] [CrossRef] [PubMed]
- Klooster ten, J.P.; Jaffer, Z.M.; Chernoff, J.; Hordijk, P.L. Targeting and activation of Rac1 are mediated by the exchange factor beta-Pix. J. Cell Biol. 2006, 172, 759–769. [Google Scholar] [CrossRef] [PubMed]
- Lei, X.; Deng, L.; Liu, D.; Liao, S.; Dai, H.; Li, J.; Rong, J.; Wang, Z.; Huang, G.; Tang, C.; et al. ARHGEF7 promotes metastasis of colorectal adenocarcinoma by regulating the motility of cancer cells. Int. J. Oncol. 2018, 53, 1980–1996. [Google Scholar] [CrossRef] [PubMed]
- Ishaque, N.; Abba, M.L.; Hauser, C.; Patil, N.; Paramasivam, N.; Huebschmann, D.; Leupold, J.H.; Balasubramanian, G.P.; Kleinheinz, K.; Toprak, U.H.; et al. Whole genome sequencing puts forward hypotheses on metastasis evolution and therapy in colorectal cancer. Nat. Commun. 2018, 9, 4782. [Google Scholar] [CrossRef]
- Li, X.-X.; Zheng, H.-T.; Peng, J.-J.; Huang, L.-Y.; Shi, D.-B.; Liang, L.; Cai, S.-J. RNA-seq reveals determinants for irinotecan sensitivity/resistance in colorectal cancer cell lines. Int. J. Clin. Exp. Pathol. 2014, 7, 2729–2736. [Google Scholar]
- Kawasaki, Y.; Sato, R.; Akiyama, T. Mutated APC and Asef are involved in the migration of colorectal tumour cells. Nat. Cell Biol. 2003, 5, 211–215. [Google Scholar] [CrossRef]
- Kawasaki, Y.; Tsuji, S.; Muroya, K.; Furukawa, S.; Shibata, Y.; Okuno, M.; Ohwada, S.; Akiyama, T. The adenomatous polyposis coli-associated exchange factors Asef and Asef2 are required for adenoma formation in Apc(Min/+)mice. EMBO Rep. 2009, 10, 1355–1362. [Google Scholar] [CrossRef]
- Kawasaki, Y.; Furukawa, S.; Sato, R.; Akiyama, T. Differences in the localization of the adenomatous polyposis coli-Asef/Asef2 complex between adenomatous polyposis coli wild-type and mutant cells. Cancer Sci. 2013, 104, 1135–1138. [Google Scholar] [CrossRef]
- Komander, D.; Patel, M.; Laurin, M.; Fradet, N.; Pelletier, A.; Barford, D.; Côté, J.-F. An alpha-helical extension of the ELMO1 pleckstrin homology domain mediates direct interaction to DOCK180 and is critical in Rac signaling. Mol. Biol. Cell 2008, 19, 4837–4851. [Google Scholar] [CrossRef] [PubMed]
- Kiyokawa, E.; Hashimoto, Y.; Kobayashi, S.; Sugimura, H.; Kurata, T.; Matsuda, M. Activation of Rac1 by a Crk SH3-binding protein, DOCK180. Genes Dev. 1998, 12, 3331–3336. [Google Scholar] [CrossRef] [PubMed]
- Gumienny, T.L.; Brugnera, E.; Tosello-Trampont, A.C.; Kinchen, J.M.; Haney, L.B.; Nishiwaki, K.; Walk, S.F.; Nemergut, M.E.; Macara, I.G.; Francis, R.; et al. CED-12/ELMO, a novel member of the CrkII/Dock180/Rac pathway, is required for phagocytosis and cell migration. Cell 2001, 107, 27–41. [Google Scholar] [CrossRef]
- Makino, Y.; Tsuda, M.; Ichihara, S.; Watanabe, T.; Sakai, M.; Sawa, H.; Nagashima, K.; Hatakeyama, S.; Tanaka, S. Elmo1 inhibits ubiquitylation of Dock180. J. Cell Sci. 2006, 119, 923–932. [Google Scholar] [CrossRef] [PubMed]
- White, D.T.; McShea, K.M.; Attar, M.A.; Santy, L.C. GRASP and IPCEF promote ARF-to-Rac signaling and cell migration by coordinating the association of ARNO/cytohesin 2 with Dock180. Mol. Biol. Cell 2010, 21, 562–571. [Google Scholar] [CrossRef] [PubMed]
- Patel, M.; Chiang, T.-C.; Tran, V.; Lee, F.-J.S.; Côté, J.-F. The Arf family GTPase Arl4A complexes with ELMO proteins to promote actin cytoskeleton remodeling and reveals a versatile Ras-binding domain in the ELMO proteins family. J. Biol. Chem. 2011, 286, 38969–38979. [Google Scholar] [CrossRef] [PubMed]
- Feng, H.; Li, Y.; Yin, Y.; Zhang, W.; Hou, Y.; Zhang, L.; Li, Z.; Xie, B.; Gao, W.-Q.; Sarkaria, J.N.; et al. Protein kinase A-dependent phosphorylation of Dock180 at serine residue 1250 is important for glioma growth and invasion stimulated by platelet derived-growth factor receptor α. NeuroOncology 2015, 17, 832–842. [Google Scholar] [CrossRef]
- Brugnera, E.; Haney, L.; Grimsley, C.; Lu, M.; Walk, S.F.; Tosello-Trampont, A.-C.; Macara, I.G.; Madhani, H.; Fink, G.R.; Ravichandran, K.S. Unconventional Rac-GEF activity is mediated through the Dock180-ELMO complex. Nat. Cell Biol. 2002, 4, 574–582. [Google Scholar] [CrossRef]
- Lu, M.; Kinchen, J.M.; Rossman, K.L.; Grimsley, C.; Hall, M.; Sondek, J.; Hengartner, M.O.; Yajnik, V.; Ravichandran, K.S. A Steric-inhibition model for regulation of nucleotide exchange via the Dock180 family of GEFs. Curr. Biol. 2005, 15, 371–377. [Google Scholar] [CrossRef]
- Jing, X.; Wu, H.; Ji, X.; Wu, H.; Shi, M.; Zhao, R. Cortactin promotes cell migration and invasion through upregulation of the dedicator of cytokinesis 1 expression in human colorectal cancer. Oncol. Rep. 2016, 36, 1946–1952. [Google Scholar] [CrossRef]
- Hirakawa, H.; Shibata, K.; Nakayama, T. Localization of cortactin is associated with colorectal cancer development. Int. J. Oncol. 2009, 35, 1271–1276. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nishihara, H.; Kobayashi, S.; Hashimoto, Y.; Ohba, F.; Mochizuki, N.; Kurata, T.; Nagashima, K.; Matsuda, M. Non-adherent cell-specific expression of DOCK2, a member of the human CDM-family proteins. Biochim. Biophys. Acta 1999, 1452, 179–187. [Google Scholar] [CrossRef]
- Miao, S.; Zhang, R.-Y.; Wang, W.; Wang, H.-B.; Meng, L.-L.; Zu, L.-D.; Fu, G.-H. Overexpression of dedicator of cytokinesis 2 correlates with good prognosis in colorectal cancer associated with more prominent CD8+ lymphocytes infiltration: A colorectal cancer analysis. J. Cell. Biochem. 2018, 119, 8962–8970. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Wu, W.K.K.; Li, X.; He, J.; Li, X.-X.; Ng, S.S.M.; Yu, C.; Gao, Z.; Yang, J.; Li, M.; et al. Novel recurrently mutated genes and a prognostic mutation signature in colorectal cancer. Gut 2015, 64, 636–645. [Google Scholar] [CrossRef]
- Namekata, K.; Enokido, Y.; Iwasawa, K.; Kimura, H. MOCA induces membrane spreading by activating Rac1. J. Biol. Chem. 2004, 279, 14331–14337. [Google Scholar] [CrossRef]
- Hofer, P.; Hagmann, M.; Brezina, S.; Dolejsi, E.; Mach, K.; Leeb, G.; Baierl, A.; Buch, S.; Sutterlüty-Fall, H.; Karner-Hanusch, J.; et al. Bayesian and frequentist analysis of an Austrian genome-wide association study of colorectal cancer and advanced adenomas. Oncotarget 2017, 8, 98623–98634. [Google Scholar] [CrossRef]
- Upadhyay, G.; Goessling, W.; North, T.E.; Xavier, R.; Zon, L.I.; Yajnik, V. Molecular association between β-catenin degradation complex and Rac guanine exchange factor DOCK4 is essential for Wnt/β-catenin signaling. Oncogene 2008, 27, 5845–5855. [Google Scholar] [CrossRef]
- Zheng, Y.; Zhou, J.; Tong, Y. Gene signatures of drug resistance predict patient survival in colorectal cancer. Pharmacogenomics 2015, 15, 135–143. [Google Scholar] [CrossRef]
- Ashraf, S.Q.; Nicholls, A.M.; Wilding, J.L.; Ntouroupi, T.G.; Mortensen, N.J.; Bodmer, W.F. Direct and immune mediated antibody targeting of ERBB receptors in a colorectal cancer cell-line panel. Proc. Natl. Acad. Sci. USA 2012, 109, 21046–21051. [Google Scholar] [CrossRef]
- Yamauchi, J.; Miyamoto, Y.; Chan, J.R.; Tanoue, A. ErbB2 directly activates the exchange factor Dock7 to promote Schwann cell migration. J. Cell Biol. 2008, 181, 351–365. [Google Scholar] [CrossRef]
- Watabe-Uchida, M.; John, K.A.; Janas, J.A.; Newey, S.E.; Van Aelst, L. The Rac activator DOCK7 regulates neuronal polarity through local phosphorylation of stathmin/Op18. Neuron 2006, 51, 727–739. [Google Scholar] [CrossRef] [PubMed]
- Arandjelovic, S.; Perry, J.S.A.; Lucas, C.D.; Penberthy, K.K.; Kim, T.-H.; Zhou, M.; Rosen, D.A.; Chuang, T.-Y.; Bettina, A.M.; Shankman, L.S.; et al. A noncanonical role for the engulfment gene ELMO1 in neutrophils that promotes inflammatory arthritis. Nat. Immunol. 2019, 20, 141–151. [Google Scholar] [CrossRef] [PubMed]
- Libanje, F.; Raingeaud, J.; Luan, R.; Thomas, Z.; Zajac, O.; Veiga, J.; Marisa, L.; Adam, J.; Boige, V.; Malka, D.; et al. ROCK2 inhibition triggers the collective invasion of colorectal adenocarcinomas. EMBO J. 2019, 38, e99299. [Google Scholar] [CrossRef] [PubMed]
- Alam, M.R.; Johnson, R.C.; Darlington, D.N.; Hand, T.A.; Mains, R.E.; Eipper, B.A. Kalirin, a cytosolic protein with spectrin-like and GDP/GTP exchange factor-like domains that interacts with peptidylglycine alpha-amidating monooxygenase, an integral membrane peptide-processing enzyme. J. Biol. Chem. 1997, 272, 12667–12675. [Google Scholar] [CrossRef] [PubMed]
- Rabiner, C.A.; Mains, R.E.; Eipper, B.A. Kalirin: A dual Rho guanine nucleotide exchange factor that is so much more than the sum of its many parts. Neuroscientist 2005, 11, 148–160. [Google Scholar] [CrossRef]
- Wu, J.-H.; Fanaroff, A.C.; Sharma, K.C.; Smith, L.S.; Brian, L.; Eipper, B.A.; Mains, R.E.; Freedman, N.J.; Zhang, L. Kalirin promotes neointimal hyperplasia by activating Rac in smooth muscle cells. Arterioscler. Thromb. Vasc. Biol. 2013, 33, 702–708. [Google Scholar] [CrossRef]
- Meng, H.; Wu, J.; Huang, Q.; Yang, X.; Yang, K.; Qiu, Y.; Ren, J.; Shen, R.; Qi, H. NEDD9 promotes invasion and migration of colorectal cancer cell line HCT116 via JNK/EMT. Oncol. Lett. 2019, 18, 4022–4029. [Google Scholar] [CrossRef]
- Dai, J.; Van Wie, P.G.; Fai, L.Y.; Kim, D.; Wang, L.; Poyil, P.; Luo, J.; Zhang, Z. Downregulation of NEDD9 by apigenin suppresses migration, invasion, and metastasis of colorectal cancer cells. Toxicol. Appl. Pharmacol. 2016, 311, 106–112. [Google Scholar] [CrossRef]
- Li, Y.; Bavarva, J.H.; Wang, Z.; Guo, J.; Qian, C.; Thibodeau, S.N.; Golemis, E.A.; Liu, W. HEF1, a novel target of Wnt signaling, promotes colonic cell migration and cancer progression. Oncogene 2011, 30, 2633–2643. [Google Scholar] [CrossRef]
- Li, P.; Zhou, H.; Zhu, X.; Ma, G.; Liu, C.; Lin, B.; Mao, W. High expression of NEDD9 predicts adverse outcomes of colorectal cancer patients. Int. J. Clin. Exp. Pathol. 2014, 7, 2565–2570. [Google Scholar]
- Barrows, D.; He, J.Z.; Parsons, R. PREX1 Protein Function Is Negatively Regulated Downstream of Receptor Tyrosine Kinase Activation by p21-activated Kinases (PAKs). J. Biol. Chem. 2016, 291, 20042–20054. [Google Scholar] [CrossRef] [PubMed]
- Cash, J.N.; Davis, E.M.; Tesmer, J.J.G. Structural and Biochemical Characterization of the Catalytic Core of the Metastatic Factor P-Rex1 and Its Regulation by PtdIns(3,4,5)P3. Structure 2016, 24, 730–740. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Wang, Q.; Wang, Y.; Chen, X.; Wang, Z. PLC-gamma1 and Rac1 coregulate EGF-induced cytoskeleton remodeling and cell migration. Mol. Endocrinol. 2009, 23, 901–913. [Google Scholar] [CrossRef] [PubMed]
- Williamson, R.C.; Cowell, C.A.M.; Hammond, C.L.; Bergen, D.J.M.; Roper, J.A.; Feng, Y.; Rendall, T.C.S.; Race, P.R.; Bass, M.D. Coronin-1C and RCC2 guide mesenchymal migration by trafficking Rac1 and controlling GEF exposure. J. Cell Sci. 2014, 127, 4292–4307. [Google Scholar] [CrossRef]
- Song, C.; Liang, L.; Jin, Y.; Li, Y.; Liu, Y.; Guo, L.; Wu, C.; Yun, C.-H.; Yin, Y. RCC2 is a novel p53 target in suppressing metastasis. Oncogene 2018, 37, 8–17. [Google Scholar] [CrossRef]
- Bruun, J.; Kolberg, M.; Ahlquist, T.C.; Røyrvik, E.C.; Nome, T.; Leithe, E.; Lind, G.E.; Merok, M.A.; Rognum, T.O.; Bjørkøy, G.; et al. Regulator of Chromosome Condensation 2 Identifies High-Risk Patients within Both Major Phenotypes of Colorectal Cancer. Clin. Cancer Res. 2015, 21, 3759–3770. [Google Scholar] [CrossRef]
- Nimnual, A.S.; Yatsula, B.A.; Bar-Sagi, D. Coupling of Ras and Rac guanosine triphosphatases through the Ras exchanger Sos. Science 1998, 279, 560–563. [Google Scholar] [CrossRef]
- Malliri, A.; van Es, S.; Huveneers, S.; Collard, J.G. The Rac Exchange Factor Tiam1 Is Required for the Establishment and Maintenance of Cadherin-based Adhesions. J. Biol. Chem. 2004, 279, 30092–30098. [Google Scholar] [CrossRef]
- Michiels, F.; Habets, G.G.; Stam, J.C.; van der Kammen, R.A.; Collard, J.G. A role for Rac in Tiam1-induced membrane ruffling and invasion. Nature 1995, 375, 338–340. [Google Scholar] [CrossRef]
- Ehler, E.; van Leeuwen, F.; Collard, J.G.; Salinas, P.C. Expression of Tiam-1 in the developing brain suggests a role for the Tiam-1-Rac signaling pathway in cell migration and neurite outgrowth. Mol. Cell. Neurosci. 1997, 9, 1–12. [Google Scholar] [CrossRef]
- Liu, L.; Wu, D.-H.; Ding, Y.-Q. Tiam1 gene expression and its significance in colorectal carcinoma. World J. Gastroenterol. 2005, 11, 705–707. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Zhang, Q.; Zhang, Y.; Wang, S.; Ding, Y. Lentivirus-Mediated Silencing of Tiam1 Gene Influences Multiple Functions of a Human Colorectal Cancer Cell Line. Neoplasia 2006, 8, 917–924. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Li, W.; Liu, H.; Yang, L.; Liao, Q.; Cui, S.; Wang, H.; Zhao, L. miR-29b suppresses tumor growth and metastasis in colorectal cancer via downregulating Tiam1 expression and inhibiting epithelial-mesenchymal transition. Cell Death Dis. 2014, 5, e1335. [Google Scholar] [CrossRef] [PubMed]
- He, J.-H.; Li, Y.-G.; Han, Z.-P.; Zhou, J.-B.; Chen, W.-M.; Lv, Y.-B.; He, M.-L.; Zuo, J.-D.; Zheng, L. The CircRNA-ACAP2/Hsa-miR-21-5p/ Tiam1 Regulatory Feedback Circuit Affects the Proliferation, Migration, and Invasion of Colon Cancer SW480 Cells. Cell Physiol. Biochem. 2018, 49, 1539–1550. [Google Scholar] [CrossRef]
- Yu, L.-N.; Zhang, Q.-L.; Li, X.; Hua, X.; Cui, Y.-M.; Zhang, N.-J.; Liao, W.-T.; Ding, Y.-Q. Tiam1 Transgenic Mice Display Increased Tumor Invasive and Metastatic Potential of Colorectal Cancer after 1,2-Dimethylhydrazine Treatment. PLoS ONE 2013, 8, e73077. [Google Scholar] [CrossRef]
- Minard, M.E.; Ellis, L.M.; Gallick, G.E. Tiam1 regulates cell adhesion, migration and apoptosis in colon tumor cells. Clin. Exp. Metastasis 2006, 23, 301–313. [Google Scholar] [CrossRef]
- Izumi, D.; Toden, S.; Ureta, E.; Ishimoto, T.; Baba, H.; Goel, A. TIAM1 promotes chemoresistance and tumor invasiveness in colorectal cancer. Cell Death Dis. 2019, 10, 267. [Google Scholar] [CrossRef]
- Zhu, G.; Fan, Z.; Ding, M.; Zhang, H.; Mu, L.; Ding, Y.; Zhang, Y.; Jia, B.; Chen, L.; Chang, Z.; et al. An EGFR/PI3K/AKT axis promotes accumulation of the Rac1-GEF Tiam1 that is critical in EGFR-driven tumorigenesis. Oncogene 2015, 34, 5971–5982. [Google Scholar] [CrossRef]
- Hornstein, I.; Alcover, A.; Katzav, S. Vav proteins, masters of the world of cytoskeleton organization. Cell. Signal. 2004, 16, 1–11. [Google Scholar] [CrossRef]
- Huang, M.-Y.; Wang, J.-Y.; Chang, H.-J.; Kuo, C.-W.; Tok, T.-S.; Lin, S.-R. CDC25A, VAV1, TP73, BRCA1 and ZAP70 gene overexpression correlates with radiation response in colorectal cancer. Oncol. Rep. 2011, 25, 1297–1306. [Google Scholar]
- Sho, S.; Court, C.M.; Winograd, P.; Russell, M.M.; Tomlinson, J.S. A prognostic mutation panel for predicting cancer recurrence in stages II and III colorectal cancer. J. Surg. Oncol. 2017, 116, 996–1004. [Google Scholar] [CrossRef] [PubMed]
- Kroviarski, Y.; Debbabi, M.; Bachoual, R.; Périanin, A.; Gougerot-Pocidalo, M.-A.; El-Benna, J.; Dang, P.M.-C. Phosphorylation of NADPH oxidase activator 1 (NOXA1) on serine 282 by MAP kinases and on serine 172 by protein kinase C and protein kinase A prevents NOX1 hyperactivation. FASEB J. 2010, 24, 2077–2092. [Google Scholar] [CrossRef] [PubMed]
- Diekmann, D.; Abo, A.; Johnston, C.; Segal, A.W.; Hall, A. Interaction of Rac with p67phox and regulation of phagocytic NADPH oxidase activity. Science 1994, 265, 531–533. [Google Scholar] [CrossRef] [PubMed]
- Gorzalczany, Y.; Alloul, N.; Sigal, N.; Weinbaum, C.; Pick, E. A prenylated p67phox-Rac1 chimera elicits NADPH-dependent superoxide production by phagocyte membranes in the absence of an activator and of p47phox: Conversion of a pagan NADPH oxidase to monotheism. J. Biol. Chem. 2002, 277, 18605–18610. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.; Prigmore, E.; Govind, S.; Veryard, C.; Kozma, R.; Wientjes, F.B.; Segal, A.W.; Lim, L. Cryptic Rac-binding and p21(Cdc42Hs/Rac)-activated kinase phosphorylation sites of NADPH oxidase component p67(phox). J. Biol. Chem. 1998, 273, 15693–15701. [Google Scholar] [CrossRef]
- Prigmore, E.; Ahmed, S.; Best, A.; Kozma, R.; Manser, E.; Segal, A.W.; Lim, L. A 68-kDa kinase and NADPH oxidase component p67phox are targets for Cdc42Hs and Rac1 in neutrophils. J. Biol. Chem. 1995, 270, 10717–10722. [Google Scholar] [CrossRef]
- Lapouge, K.; Smith, S.J.; Walker, P.A.; Gamblin, S.J.; Smerdon, S.J.; Rittinger, K. Structure of the TPR domain of p67phox in complex with Rac.GTP. Mol. Cell 2000, 6, 899–907. [Google Scholar] [CrossRef]
- Rolland, T.; Taşan, M.; Charloteaux, B.; Pevzner, S.J.; Zhong, Q.; Sahni, N.; Yi, S.; Lemmens, I.; Fontanillo, C.; Mosca, R.; et al. A proteome-scale map of the human interactome network. Cell 2014, 159, 1212–1226. [Google Scholar] [CrossRef]
- Pan, Y.; Wang, N.; Xia, P.; Wang, E.; Guo, Q.; Ye, Z. Inhibition of Rac1 ameliorates neuronal oxidative stress damage via reducing Bcl-2/Rac1 complex formation in mitochondria through PI3K/Akt/mTOR pathway. Exp. Neurol. 2018, 300, 149–166. [Google Scholar] [CrossRef]
- Huang, Q.; Li, S.; Cheng, P.; Deng, M.; He, X.; Wang, Z.; Yang, C.-H.; Zhao, X.-Y.; Huang, J. High expression of anti-apoptotic protein Bcl-2 is a good prognostic factor in colorectal cancer: Result of a meta-analysis. World J. Gastroenterol. 2017, 23, 5018–5033. [Google Scholar] [CrossRef]
- Osborn-Heaford, H.L.; Ryan, A.J.; Murthy, S.; Racila, A.-M.; He, C.; Sieren, J.C.; Spitz, D.R.; Carter, A.B. Mitochondrial Rac1 GTPase import and electron transfer from cytochrome c are required for pulmonary fibrosis. J. Biol. Chem. 2012, 287, 3301–3312. [Google Scholar] [CrossRef] [PubMed]
- Kuncewicz, T.; Balakrishnan, P.; Snuggs, M.B.; Kone, B.C. Specific association of nitric oxide synthase-2 with Rac isoforms in activated murine macrophages. Am. J. Physiol. Renal Physiol. 2001, 281, F326–F336. [Google Scholar] [CrossRef] [PubMed]
- Buongiorno, P.; Pethe, V.V.; Charames, G.S.; Esufali, S.; Bapat, B. Rac1 GTPase and the Rac1 exchange factor Tiam1 associate with Wnt-responsive promoters to enhance beta-catenin/TCF-dependent transcription in colorectal cancer cells. Mol. Cancer 2008, 7, 73. [Google Scholar] [CrossRef]
- Esufali, S.; Bapat, B. Cross-talk between Rac1 GTPase and dysregulated Wnt signaling pathway leads to cellular redistribution of β-catenin and TCF/LEF-mediated transcriptional activation. Oncogene 2004, 23, 8260–8271. [Google Scholar] [CrossRef] [PubMed]
- Simon, A.R.; Vikis, H.G.; Stewart, S.; Fanburg, B.L.; Cochran, B.H.; Guan, K.L. Regulation of STAT3 by direct binding to the Rac1 GTPase. Science 2000, 290, 144–147. [Google Scholar] [CrossRef]
- Zhang, F.; Lu, Y.-X.; Chen, Q.; Zou, H.-M.; Zhang, J.-M.; Hu, Y.-H.; Li, X.-M.; Zhang, W.-J.; Zhang, W.; Lin, C.; et al. Identification of NCK1 as a novel downstream effector of STAT3 in colorectal cancer metastasis and angiogenesis. Cell. Signal. 2017, 36, 67–78. [Google Scholar] [CrossRef]
- Lorès, P.; Visvikis, O.; Luna, R.; Lemichez, E.; Gacon, G. The SWI/SNF protein BAF60b is ubiquitinated through a signalling process involving Rac GTPase and the RING finger protein Unkempt. FEBS J. 2010, 277, 1453–1464. [Google Scholar] [CrossRef]
- Pérez-Yépez, E.A.; Saldívar-Cerón, H.I.; Villamar-Cruz, O.; Pérez-Plasencia, C.; Arias-Romero, L.E. p21 Activated kinase 1: Nuclear activity and its role during DNA damage repair. DNA Repair 2018, 65, 42–46. [Google Scholar] [CrossRef]
- Rane, C.K.; Minden, A. P21 activated kinase signaling in cancer. Semin. Cancer Biol. 2019, 54, 40–49. [Google Scholar] [CrossRef]
- Song, B.; Wang, W.; Zheng, Y.; Yang, J.; Xu, Z. P21-activated kinase 1 and 4 were associated with colorectal cancer metastasis and infiltration. J. Surg. Res. 2015, 196, 130–135. [Google Scholar] [CrossRef]
- Wang, S.E.; Shin, I.; Wu, F.Y.; Friedman, D.B.; Arteaga, C.L. HER2/Neu (ErbB2) signaling to Rac1-Pak1 is temporally and spatially modulated by transforming growth factor beta. Cancer Res. 2006, 66, 9591–9600. [Google Scholar] [CrossRef] [PubMed]
- King, H.; Nicholas, N.S.; Wells, C.M. Role of p-21-activated kinases in cancer progression. Int. Rev. Cell Mol. Biol. 2014, 309, 347–387. [Google Scholar] [PubMed]
- Li, Y.; Shao, Y.; Tong, Y.; Shen, T.; Zhang, J.; Li, Y.; Gu, H.; Li, F. Nucleo-cytoplasmic shuttling of PAK4 modulates β-catenin intracellular translocation and signaling. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2012, 1823, 465–475. [Google Scholar] [CrossRef] [PubMed]
- Gong, W.; An, Z.; Wang, Y.; Pan, X.; Fang, W.; Jiang, B.; Zhang, H. P21-activated kinase 5 is overexpressed during colorectal cancer progression and regulates colorectal carcinoma cell adhesion and migration. Int. J. Cancer 2009, 125, 548–555. [Google Scholar] [CrossRef]
- Fanger, G.R.; Johnson, N.L.; Johnson, G.L. MEK kinases are regulated by EGF and selectively interact with Rac/Cdc42. EMBO J. 1997, 16, 4961–4972. [Google Scholar] [CrossRef]
- Nagata, K.I.; Puls, A.; Futter, C.; Aspenstrom, P.; Schaefer, E.; Nakata, T.; Hirokawa, N.; Hall, A. The MAP kinase kinase kinase MLK2 co-localizes with activated JNK along microtubules and associates with kinesin superfamily motor KIF3. EMBO J. 1998, 17, 149–158. [Google Scholar] [CrossRef]
- Gerwins, P.; Blank, J.L.; Johnson, G.L. Cloning of a novel mitogen-activated protein kinase kinase kinase, MEKK4, that selectively regulates the c-Jun amino terminal kinase pathway. J. Biol. Chem. 1997, 272, 8288–8295. [Google Scholar] [CrossRef]
- Kuroda, S.; Fukata, M.; Kobayashi, K.; Nakafuku, M.; Nomura, N.; Iwamatsu, A.; Kaibuchi, K. Identification of IQGAP as a putative target for the small GTPases, Cdc42 and Rac1. J. Biol. Chem. 1996, 271, 23363–23367. [Google Scholar] [CrossRef]
- Hart, M.J.; Callow, M.G.; Souza, B.; Polakis, P. IQGAP1, a calmodulin-binding protein with a rasGAP-related domain, is a potential effector for cdc42Hs. EMBO J. 1996, 15, 2997–3005. [Google Scholar] [CrossRef]
- Fukata, M.; Watanabe, T.; Noritake, J.; Nakagawa, M.; Yamaga, M.; Kuroda, S.; Matsuura, Y.; Iwamatsu, A.; Perez, F.; Kaibuchi, K. Rac1 and Cdc42 capture microtubules through IQGAP1 and CLIP-170. Cell 2002, 109, 873–885. [Google Scholar] [CrossRef]
- Nabeshima, K.; Shimao, Y.; Inoue, T.; Koono, M. Immunohistochemical analysis of IQGAP1 expression in human colorectal carcinomas: Its overexpression in carcinomas and association with invasion fronts. Cancer Lett. 2002, 176, 101–109. [Google Scholar] [CrossRef]
- Brill, S.; Li, S.; Lyman, C.W.; Church, D.M.; Wasmuth, J.J.; Weissbach, L.; Bernards, A.; Snijders, A.J. The Ras GTPase-activating-protein-related human protein IQGAP2 harbors a potential actin binding domain and interacts with calmodulin and Rho family GTPases. Mol. Cell. Biol. 1996, 16, 4869–4878. [Google Scholar] [CrossRef] [PubMed]
- Kumar, D.; Hassan, M.K.; Pattnaik, N.; Mohapatra, N.; Dixit, M. Reduced expression of IQGAP2 and higher expression of IQGAP3 correlates with poor prognosis in cancers. PLoS ONE 2017, 12, e0186977. [Google Scholar] [CrossRef] [PubMed]
- Katkoori, V.R.; Shanmugam, C.; Jia, X.; Vitta, S.P.; Sthanam, M.; Callens, T.; Messiaen, L.; Chen, D.; Zhang, B.; Bumpers, H.L.; et al. Prognostic Significance and Gene Expression Profiles of p53 Mutations in Microsatellite-Stable Stage III Colorectal Adenocarcinomas. PLoS ONE 2012, 7, e30020. [Google Scholar] [CrossRef]
- Tapon, N.; Nagata, K.; Lamarche, N.; Hall, A. A new rac target POSH is an SH3-containing scaffold protein involved in the JNK and NF-kappaB signalling pathways. EMBO J. 1998, 17, 1395–1404. [Google Scholar] [CrossRef]
- Xu, Z.; Kukekov, N.V.; Greene, L.A. POSH acts as a scaffold for a multiprotein complex that mediates JNK activation in apoptosis. EMBO J. 2003, 22, 252–261. [Google Scholar] [CrossRef]
- Kärkkäinen, S.; van der Linden, M.; Renkema, G.H. POSH2 is a RING finger E3 ligase with Rac1 binding activity through a partial CRIB domain. FEBS Lett. 2010, 584, 3867–3872. [Google Scholar] [CrossRef]
- Uhlik, M.T.; Abell, A.N.; Johnson, N.L.; Sun, W.; Cuevas, B.D.; Lobel-Rice, K.E.; Horne, E.A.; Dell’Acqua, M.L.; Johnson, G.L. Rac–MEKK3–MKK3 scaffolding for p38 MAPK activation during hyperosmotic shock. Nat. Cell Biol. 2003, 5, 1104–1110. [Google Scholar] [CrossRef]
- Sandrock, K.; Bielek, H.; Schradi, K.; Schmidt, G.; Klugbauer, N. The nuclear import of the small GTPase Rac1 is mediated by the direct interaction with karyopherin alpha2. Traffic 2010, 11, 198–209. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, M.; Yu, F.; Lu, S.; Sun, H.; Tang, H.; Peng, Z. Karyopherin alpha 2 is a novel prognostic marker and a potential therapeutic target for colon cancer. J. Exp. Clin. Cancer Res. 2015, 34, 145. [Google Scholar] [CrossRef]
- Perathoner, A.; Pirkebner, D.; Brandacher, G.; Spizzo, G.; Stadlmann, S.; Obrist, P.; Margreiter, R.; Amberger, A. 14-3-3sigma expression is an independent prognostic parameter for poor survival in colorectal carcinoma patients. Clin. Cancer Res. 2005, 11, 3274–3279. [Google Scholar] [CrossRef] [PubMed]
- Hamel, B.; Monaghan-Benson, E.; Rojas, R.J.; Temple, B.R.S.; Marston, D.J.; Burridge, K.; Sondek, J. SmgGDS is a guanine nucleotide exchange factor that specifically activates RhoA and RhoC. J. Biol. Chem. 2011, 286, 12141–12148. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, S.-I.; Fukumoto, Y.; Nochioka, K.; Minami, T.; Kudo, S.; Shiba, N.; Takai, Y.; Williams, C.L.; Liao, J.K.; Shimokawa, H. Statins exert the pleiotropic effects through small GTP-binding protein dissociation stimulator upregulation with a resultant Rac1 degradation. Arterioscler. Thromb. Vasc. Biol. 2013, 33, 1591–1600. [Google Scholar] [CrossRef] [PubMed]
- Berg, T.J.; Gastonguay, A.J.; Lorimer, E.L.; Kuhnmuench, J.R.; Li, R.; Fields, A.P.; Williams, C.L. Splice variants of SmgGDS control small GTPase prenylation and membrane localization. J. Biol. Chem. 2010, 285, 35255–35266. [Google Scholar] [CrossRef]
- Van Aelst, L.; Joneson, T.; Bar-Sagi, D. Identification of a novel Rac1-interacting protein involved in membrane ruffling. EMBO J. 1996, 15, 3778–3786. [Google Scholar] [CrossRef]
- D’Souza-Schorey, C.; Boshans, R.L.; McDonough, M.; Stahl, P.D.; Van Aelst, L. A role for POR1, a Rac1-interacting protein, in ARF6-mediated cytoskeletal rearrangements. EMBO J. 1997, 16, 5445–5454. [Google Scholar] [CrossRef]
- Shin, O.H.; Exton, J.H. Differential binding of arfaptin 2/POR1 to ADP-ribosylation factors and Rac1. Biochem. Biophys. Res. Commun. 2001, 285, 1267–1273. [Google Scholar] [CrossRef]
- Miki, H.; Yamaguchi, H.; Suetsugu, S.; Takenawa, T. IRSp53 is an essential intermediate between Rac and WAVE in the regulation of membrane ruffling. Nature 2000, 408, 732–735. [Google Scholar] [CrossRef]
- Williamson, R.C.; Cowell, C.A.M.; Reville, T.; Roper, J.A.; Rendall, T.C.S.; Bass, M.D. Coronin-1C Protein and Caveolin Protein Provide Constitutive and Inducible Mechanisms of Rac1 Protein Trafficking. J. Biol. Chem. 2015, 290, 15437–15449. [Google Scholar] [CrossRef]
- Kobayashi, K.; Kuroda, S.; Fukata, M.; Nakamura, T.; Nagase, T.; Nomura, N.; Matsuura, Y.; Yoshida-Kubomura, N.; Iwamatsu, A.; Kaibuchi, K. p140Sra-1 (specifically Rac1-associated protein) is a novel specific target for Rac1 small GTPase. J. Biol. Chem. 1998, 273, 291–295. [Google Scholar] [CrossRef]
- Teng, Y.; Qin, H.; Bahassan, A.; Bendzunas, N.G.; Kennedy, E.J.; Cowell, J.K. The WASF3-NCKAP1-CYFIP1 Complex Is Essential for Breast Cancer Metastasis. Cancer Res. 2016, 76, 5133–5142. [Google Scholar] [CrossRef] [PubMed]
- Shifrin, Y.; Pinto, V.I.; Hassanali, A.; Arora, P.D.; McCulloch, C.A. Force-induced apoptosis mediated by the Rac/Pak/p38 signalling pathway is regulated by filamin A. Biochem. J. 2012, 445, 57–67. [Google Scholar] [CrossRef] [PubMed]
- Tian, Z.-Q.; Shi, J.-W.; Wang, X.-R.; Li, Z.; Wang, G.-Y. New cancer suppressor gene for colorectal adenocarcinoma: Filamin A. World J. Gastroenterol. 2015, 21, 2199–2205. [Google Scholar] [CrossRef] [PubMed]
- Jeon, Y.J.; Choi, J.S.; Lee, J.Y.; Yu, K.R.; Ka, S.H.; Cho, Y.; Choi, E.-J.; Baek, S.H.; Seol, J.H.; Park, D.; et al. Filamin B serves as a molecular scaffold for type I interferon-induced c-Jun NH2-terminal kinase signaling pathway. Mol. Biol. Cell 2008, 19, 5116–5130. [Google Scholar] [CrossRef][Green Version]
- Jeon, Y.J.; Choi, J.S.; Lee, J.Y.; Yu, K.R.; Kim, S.M.; Ka, S.H.; Oh, K.H.; Kim, I.K.; Zhang, D.-E.; Bang, O.S.; et al. ISG15 modification of filamin B negatively regulates the type I interferon-induced JNK signalling pathway. EMBO Rep. 2009, 10, 374–380. [Google Scholar] [CrossRef]
- Yayoshi-Yamamoto, S.; Taniuchi, I.; Watanabe, T. FRL, a novel formin-related protein, binds to Rac and regulates cell motility and survival of macrophages. Mol. Cell. Biol. 2000, 20, 6872–6881. [Google Scholar] [CrossRef]
- Grikscheit, K.; Frank, T.; Wang, Y.; Grosse, R. Junctional actin assembly is mediated by Formin-like 2 downstream of Rac1. J. Cell Biol. 2015, 209, 367–376. [Google Scholar] [CrossRef]
- Zhu, X.-L.; Liang, L.; Ding, Y.-Q. Overexpression of FMNL2 is closely related to metastasis of colorectal cancer. Int. J. Colorectal Dis. 2008, 23, 1041–1047. [Google Scholar] [CrossRef]
- Zhu, X.-L.; Zeng, Y.-F.; Guan, J.; Li, Y.-F.; Deng, Y.-J.; Bian, X.-W.; Ding, Y.-Q.; Liang, L. FMNL2 is a positive regulator of cell motility and metastasis in colorectal carcinoma. J. Pathol. 2011, 224, 377–388. [Google Scholar] [CrossRef]
- Chan, D.; Citro, A.; Cordy, J.M.; Shen, G.C.; Wolozin, B. Rac1 protein rescues neurite retraction caused by G2019S leucine-rich repeat kinase 2 (LRRK2). J. Biol. Chem. 2011, 286, 16140–16149. [Google Scholar] [CrossRef]
- Schreij, A.M.A.; Chaineau, M.; Ruan, W.; Lin, S.; Barker, P.A.; Fon, E.A.; McPherson, P.S. LRRK2 localizes to endosomes and interacts with clathrin-light chains to limit Rac1 activation. EMBO Rep. 2015, 16, 79–86. [Google Scholar] [CrossRef]
- Nethe, M.; de Kreuk, B.J.; Tauriello, D.V.F.; Anthony, E.C.; Snoek, B.; Stumpel, T.; Salinas, P.C.; Maurice, M.M.; Geerts, D.; Deelder, A.M.; et al. Rac1 acts in conjunction with Nedd4 and dishevelled-1 to promote maturation of cell-cell contacts. J. Cell Sci. 2012, 125, 3430–3442. [Google Scholar] [CrossRef]
- Eide, P.W.; Cekaite, L.; Danielsen, S.A.; Eilertsen, I.A.; Kjenseth, A.; Fykerud, T.A.; Ågesen, T.H.; Bruun, J.; Rivedal, E.; Lothe, R.A.; et al. NEDD4 is overexpressed in colorectal cancer and promotes colonic cell growth independently of the PI3K/PTEN/AKT pathway. Cell. Signal. 2013, 25, 12–18. [Google Scholar] [CrossRef]
- Noda, Y.; Takeya, R.; Ohno, S.; Naito, S.; Ito, T.; Sumimoto, H. Human homologues of the Caenorhabditis elegans cell polarity protein PAR6 as an adaptor that links the small GTPases Rac and Cdc42 to atypical protein kinase C. Genes Cells 2001, 6, 107–119. [Google Scholar] [CrossRef]
- Cheung, L.W.T.; Yu, S.; Zhang, D.; Li, J.; Ng, P.K.S.; Panupinthu, N.; Mitra, S.; Ju, Z.; Yu, Q.; Liang, H.; et al. Naturally Occurring Neomorphic PIK3R1 Mutations Activate the MAPK Pathway, Dictating Therapeutic Response to MAPK Pathway Inhibitors. Cancer Cell 2014, 26, 479–494. [Google Scholar] [CrossRef]
- Liu, J.; Liu, F.; Li, X.; Song, X.; Zhou, L.; Jie, J. Screening key genes and miRNAs in early-stage colon adenocarcinoma by RNA-sequencing. Tumor Biol. 2017, 39, 1010428317714899. [Google Scholar] [CrossRef]
- Jezyk, M.R.; Snyder, J.T.; Gershberg, S.; Worthylake, D.K.; Harden, T.K.; Sondek, J. Crystal structure of Rac1 bound to its effector phospholipase C-beta2. Nat. Struct. Mol. Biol. 2006, 13, 1135–1140. [Google Scholar] [CrossRef]
- Klooster ten, J.P.; Leeuwen, I.V.; Scheres, N.; Anthony, E.C.; Hordijk, P.L. Rac1-induced cell migration requires membrane recruitment of the nuclear oncogene SET. EMBO J. 2007, 26, 336–345. [Google Scholar] [CrossRef]
- Wang, H.; Qiu, P.; Zhu, S.; Zhang, M.; Li, Y.; Zhang, M.; Wang, X.; Shang, J.; Qu, B.; Liu, J.; et al. SET nuclear proto-oncogene gene expression is associated with microsatellite instability in human colorectal cancer identified by co-expression analysis. Dig. Liver Dis. 2019. [Google Scholar] [CrossRef]
- Cristóbal, I.; Torrejón, B.; Rubio, J.; Santos, A.; Pedregal, M.; Caramés, C.; Zazo, S.; Luque, M.; Sanz-Alvarez, M.; Madoz-Gúrpide, J.; et al. Deregulation of SET is Associated with Tumor Progression and Predicts Adverse Outcome in Patients with Early-Stage Colorectal Cancer. J Clin Med 2019, 8, 346. [Google Scholar] [CrossRef]
- Huelsenbeck, S.C.; Schorr, A.; Roos, W.P.; Huelsenbeck, J.; Henninger, C.; Kaina, B.; Fritz, G. Rac1 protein signaling is required for DNA damage response stimulated by topoisomerase II poisons. J. Biol. Chem. 2012, 287, 38590–38599. [Google Scholar] [CrossRef]
- Zhang, R.; Xu, J.; Zhao, J.; Bai, J.H. Proliferation and invasion of colon cancer cells are suppressed by knockdown of TOP2A. J. Cell. Biochem. 2018, 119, 7256–7263. [Google Scholar] [CrossRef]
- Kotelevets, L.; Trifault, B.; Chastree, E.; Scott, M.G.H. Posttranslational Regulation and Conformational Plasticity of PTEN. Cold Spring Harb. Perspect Med. 2020. [Google Scholar] [CrossRef]
- Diamantopoulou, Z.; White, G.; Fadlullah, M.Z.H.; Dreger, M.; Pickering, K.; Maltas, J.; Ashton, G.; MacLeod, R.; Baillie, G.S.; Kouskoff, V.; et al. TIAM1 Antagonizes TAZ/YAP Both in the Destruction Complex in the Cytoplasm and in the Nucleus to Inhibit Invasion of Intestinal Epithelial Cells. Cancer Cell 2017, 31, 621–634. [Google Scholar] [CrossRef]
- Kawasaki, Y.; Sagara, M.; Shibata, Y.; Shirouzu, M.; Yokoyama, S.; Akiyama, T. Identification and characterization of Asef2, a guanine-nucleotide exchange factor specific for Rac1 and Cdc42. Oncogene 2007, 26, 7620–7627. [Google Scholar] [CrossRef]
- Rosin-Arbesfeld, R.; Ihrke, G.; Bienz, M. Actin-dependent membrane association of the APC tumour suppressor in polarized mammalian epithelial cells. EMBO J. 2001, 20, 5929–5939. [Google Scholar] [CrossRef]
- Peng, H.-Y.; Yu, Q.-F.; Shen, W.; Guo, C.-M.; Li, Z.; Zhou, X.-Y.; Zhou, N.-J.; Min, W.-P.; Gao, D. Knockdown of ELMO3 Suppresses Growth, Invasion and Metastasis of Colorectal Cancer. Int. J. Mol. Sci. 2016, 17, 2119. [Google Scholar] [CrossRef]
- Kim, S.-H.; Xia, D.; Kim, S.-W.; Holla, V.; Menter, D.G.; Dubois, R.N. Human enhancer of filamentation 1 Is a mediator of hypoxia-inducible factor-1alpha-mediated migration in colorectal carcinoma cells. Cancer Res. 2010, 70, 4054–4063. [Google Scholar] [CrossRef]
- Caspi, E.; Rosin-Arbesfeld, R. A novel functional screen in human cells identifies MOCA as a negative regulator of Wnt signaling. Mol. Biol. Cell 2008, 19, 4660–4674. [Google Scholar] [CrossRef][Green Version]
- Gadea, G.; Blangy, A. Dock-family exchange factors in cell migration and disease. Eur. J. Cell Biol. 2014, 93, 466–477. [Google Scholar] [CrossRef]
- Feng, J.; Zhao, J.; Xie, H.; Yin, Y.; Luo, G.; Zhang, J.; Feng, Y.; Li, Z. Involvement of NEDD9 in the invasion and migration of gastric cancer. Tumor Biol. 2015, 36, 3621–3628. [Google Scholar] [CrossRef]
- Abramovici, H.; Mojtabaie, P.; Parks, R.J.; Zhong, X.-P.; Koretzky, G.A.; Topham, M.K.; Gee, S.H. Diacylglycerol kinase zeta regulates actin cytoskeleton reorganization through dissociation of Rac1 from RhoGDI. Mol. Biol. Cell 2009, 20, 2049–2059. [Google Scholar] [CrossRef]
- Cai, K.; Mulatz, K.; Ard, R.; Nguyen, T.; Gee, S.H. Increased diacylglycerol kinase ζ expression in human metastatic colon cancer cells augments Rho GTPase activity and contributes to enhanced invasion. BMC Cancer 2014, 14, 208. [Google Scholar] [CrossRef]
- Hedman, A.C.; Smith, J.M.; Sacks, D.B. The biology of IQGAPproteins: Beyond the cytoskeleton. EMBO Rep. 2015, 16, 427–446. [Google Scholar] [CrossRef]
- Liang, Z.; Yang, Y.; He, Y.; Yang, P.; Wang, X.; He, G.; Zhang, P.; Zhu, H.; Xu, N.; Zhao, X.; et al. Cancer Letters. Cancer Lett. 2017, 411, 90–99. [Google Scholar] [CrossRef]
- Cheung, E.C.; Lee, P.; Ceteci, F.; Nixon, C.; Blyth, K.; Sansom, O.J.; Vousden, K.H. Opposing effects of TIGAR- and RAC1-derived ROS on Wnt-driven proliferation in the mouse intestine. Genes Dev. 2016, 30, 52–63. [Google Scholar] [CrossRef]
- Carter, J.H.; Douglass, L.E.; Deddens, J.A.; Colligan, B.M.; Bhatt, T.R.; Pemberton, J.O.; Konicek, S.; Hom, J.; Marshall, M.; Graff, J.R. Pak-1 expression increases with progression of colorectal carcinomas to metastasis. Clin. Cancer Res. 2004, 10, 3448–3456. [Google Scholar] [CrossRef]
- Al-Maghrabi, J.; Emam, E.; Gomaa, W.; Al-Qaydy, D.; Al-Maghrabi, B.; Buhmeida, A.; Abuzenadah, A.; Al-Qahtani, M.; Al-Ahwal, M. Overexpression of PAK-1 is an independent predictor of disease recurrence in colorectal carcinoma. Int. J. Clin. Exp. Pathol. 2015, 8, 15895–15902. [Google Scholar]
- Gerçeker, E.; Boyacıoglu, S.O.; Kasap, E.; Baykan, A.; Yuceyar, H.; Yıldırım, H.; Ayhan, S.; Ellidokuz, E.; Korkmaz, M. Never in mitosis gene A-related kinase 6 and aurora kinase A: New gene biomarkers in the conversion from ulcerative colitis to colorectal cancer. Oncol. Rep. 2015, 34, 1905–1914. [Google Scholar] [CrossRef]
- Chow, H.Y.; Dong, B.; Valencia, C.A.; Zeng, C.T.; Koch, J.N.; Prudnikova, T.Y.; Chernoff, J. Group I Paks are essential for epithelial- mesenchymal transition in an Apc-driven model of colorectal cancer. Nat. Commun. 2018, 9, 3473. [Google Scholar] [CrossRef]
- Wang, X.; Gong, W.; Qing, H.; Geng, Y.; Wang, X.; Zhang, Y.; Peng, L.; Zhang, H.; Jiang, B. p21-activated kinase 5 inhibits camptothecin-induced apoptosis in colorectal carcinoma cells. Tumor Biol. 2010, 31, 575–582. [Google Scholar] [CrossRef] [PubMed]
- Huynh, N.; Shulkes, A.; Baldwin, G.; He, H. Up-regulation of stem cell markers by P21- activated kinase 1 contributes to 5-fluorouracil resistance of colorectal cancer. Cancer Biol. Ther. 2016, 17, 813–823. [Google Scholar] [CrossRef] [PubMed]
- Fritz, G.; Henninger, C. Rho GTPases: Novel Players in the Regulation of the DNA Damage Response? Biomolecules 2015, 5, 2417–2434. [Google Scholar] [CrossRef] [PubMed]
- Iwaya, K.; Oikawa, K.; Semba, S.; Tsuchiya, B.; Mukai, Y.; Otsubo, T.; Nagao, T.; Izumi, M.; Kuroda, M.; Domoto, H.; et al. Correlation between liver metastasis of the colocalization of actin-related protein 2 and 3 complex and WAVE2 in colorectal carcinoma. Cancer Sci. 2007, 98, 992–999. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Zhou, M.; Xie, J.; Chao, P.; Feng, Q.; Wu, J. High glucose levels promote the proliferation of breast cancer cells through GTPases. Breast Cancer 2017, 9, 429–436. [Google Scholar] [CrossRef] [PubMed]
- González, N.; Prieto, I.; Del Puerto-Nevado, L.; Portal-Nuñez, S.; Ardura, J.A.; Corton, M.; Fernández-Fernández, B.; Aguilera, O.; Gomez-Guerrero, C.; Mas, S.; et al. Diabetes Cancer Connect Consortium 2017 update on the relationship between diabetes and colorectal cancer: Epidemiology, potential molecular mechanisms and therapeutic implications. Oncotarget 2017, 8, 18456–18485. [Google Scholar] [CrossRef]
- Cannino, G.; Ciscato, F.; Masgras, I.; Sánchez-Martín, C.; Rasola, A. Metabolic Plasticity of Tumor Cell Mitochondria. Front. Oncol. 2018, 8, 333. [Google Scholar] [CrossRef]
- Kang, J.; Chong, S.J.F.; Ooi, V.Z.Q.; Vali, S.; Kumar, A.; Kapoor, S.; Abbasi, T.; Hirpara, J.L.; Loh, T.; Goh, B.-C.; et al. Overexpression of Bcl-2 induces STAT-3 activation via an increase in mitochondrial superoxide. Oncotarget 2015, 6, 34191–34205. [Google Scholar] [CrossRef]
- Han, J.; Theiss, A.L. Stat3: Friend or foe in colitis and colitis-associated cancer? Inflamm. Bowel Dis. 2014, 20, 2405–2411. [Google Scholar] [CrossRef]
- Valle-Mendiola, A.; Soto-Cruz, I. Energy Metabolism in Cancer: The Roles of STAT3 and STAT5 in the Regulation of Metabolism-Related Genes. Cancers 2020, 12, 124. [Google Scholar] [CrossRef]
- Marei, H.; Malliri, A. Rac1 in human diseases: The therapeutic potential of targeting Rac1 signaling regulatory mechanisms. Small GTPases 2017, 8, 139–163. [Google Scholar] [CrossRef] [PubMed]
- Maldonado, M.D.M.; Dharmawardhane, S. Targeting Rac and Cdc42 GTPases in Cancer. Cancer Res. 2018, 78, 3101–3111. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).