Abstract
Genetic testing for BRCA1 and BRCA2 genes has led to the identification of many unique variants of uncertain significance (VUS). Multifactorial likelihood models that predict the odds ratio for VUS in favor or against cancer causality, have been developed, but their use is conditioned by the amount of necessary data, which are difficult to obtain if a variant is rare. As an alternative, variants mapping to the coding regions can be examined using in vitro functional assays. BRCA1 and BRCA2 proteins promote genome protection by interacting with different proteins. In this study, we assessed the functional effect of two sets of variants in BRCA genes by exploiting the green fluorescent protein (GFP)-reassembly in vitro assay, which was set-up to test the BRCA1/BARD1, BRCA1/UbcH5a, and BRCA2/DSS1 interactions. Based on the findings observed for the validation panels of previously classified variants, BRCA1/UbcH5a and BRCA2/DSS1 binding assays showed 100% sensitivity and specificity in identifying pathogenic and non-pathogenic variants. While the actual efficiency of these assays in assessing the clinical significance of BRCA VUS has to be verified using larger validation panels, our results suggest that the GFP-reassembly assay is a robust method to identify variants affecting normal protein functioning and contributes to the classification of VUS.
1. Introduction
Genetic testing for the breast cancer susceptibility genes, BRCA1 (MIM# 113705) and BRCA2 (MIM# 600185), has become part of routine clinical care for patients with a personal or family history indicative of Hereditary Breast/Ovarian Cancer Syndrome (HBOC). A recent prospective study estimates the average cumulative risks by age 70 years of BRCA1 carriers to be 60% and 59% for breast and ovarian cancer, respectively. For BRCA2 carriers, the corresponding values are 55% and 16.5%, respectively [1]. Most pathogenic variants identified in the BRCA1 and BRCA2 genes are nonsense or frameshift that, by introducing a premature termination codon, lead to non-functional proteins. Splice site variants with a similar effect on the protein have also been identified [2]. In addition to these clearly loss-of-function alterations, a growing number of variants of uncertain significance (VUS) are currently being identified. These include missense substitutions, small in-frame deletions or insertions, alterations of both exonic and intronic sites potentially affecting splicing, and variants in regulatory sequences. The inability to classify the VUS either as disease causing or as rare non-pathogenic (benign) variation represents a challenge for genetic counselling, risk evaluation, and adoption of preventive and risk-reduction measures.
A five-tier classification scheme has been developed for the assessment of the clinical relevance of VUS by the International Agency for Research on Cancer (IARC) Working Group [3]. According to this scheme, Class 1 (posterior probability of pathogenicity < 0.001) and Class 2 (0.001 < posterior probability < 0.049) are considered non-pathogenic and likely non-pathogenic variants, Class 3 (0.05 < posterior probability < 0.949) VUS remain unclassified because of a lack of sufficient information for classification, Class 4 (0.95 < posterior probability < 0.99) and Class 5 (posterior probability > 0.99) are likely pathogenic and pathogenic variants, respectively. Posterior probability may be estimated using a multi-factorial model. For missense variants, this model combines a prior probability of causality, based on the amino acid evolutionary conservation model (A-GVGD, URL: http://agvgd.iarc.fr/) [4], with a likelihood ratio for causality derived from genetic and family-based data and histopathological tumor features [5,6,7,8]. However, since most BRCA1 and BRCA2 VUS are detected in a single or a few families only, this model is not able to classify the majority of them. Therefore, functional assays have been developed to independently classify VUS based on their effect on established functions of the BRCA1 and BRCA2 proteins [9,10].
Mounting evidence implicates both BRCA1 and BRCA2 proteins in all phases of the cell cycle and in the regulation of its orderly events [11] through their association with different proteins [12,13]. As a consequence, a defective protein-protein interaction may lead to uncontrolled cell replication and genomic instability, which are both hallmarks of tumor formation and progression. Thus, biochemical in vitro assays analyzing the formation of protein complexes provide a context in which to assess the functional effect of the BRCA1 [14,15] and BRCA2 VUS [16,17,18].
One of the most important BRCA1-interacting proteins is BARD1 (MIM# 601593). Under physiological conditions, BRCA1 is prevalently found to be associated with BARD1. This interaction is mediated by the N-terminal RING-finger domains of both proteins [19], which suggests that the heterodimer is the physiological relevant form of BRCA1 and the two proteins require each other for most of their cellular functions. Several lines of evidence support this view: BRCA1-BARD1 binding appears to stabilize BRCA1 protein expression [20] and to increase its nuclear accumulation by masking the nuclear export sequence (NES) flanking the RING-finger domain [21]. The two proteins co-localize in nuclear dots during the S-phase of the cell cycle and in nuclear foci at sites of DNA damage [22,23,24,25]. Most importantly, the binding with the E2 ubiquitin-conjugating enzyme UbcH5a (MIM# 602961) enables the BRCA1-BARD1 complex to acquire an ubiquitin ligase activity, linked to BRCA1 tumor suppression function by regulating transcription and double stand break (DSB) repair [26,27,28]. This is consistent with the reported hypersensitivity to Ionizing Radiation (IR) and to agent crosslinking the DNA, such as Cisplatin or Mitomycin C (MMC), of cells carrying BRCA1 RING pathogenic variants [29,30,31].
Concerning BRCA2, the strong physical interaction with the small (70-residues) DSS1 peptide (MIM# 601285), through its highly conserved C-terminal DSS1/ssDNA Binding Domain (DBD), appears to be crucial for the maintenance of its stability and correct conformation [32,33], for the repair of DNA DSBs by homologous recombination (HR) and the formation of DNA damage induced RAD51 foci [34]. Recently, it was shown that the nuclear retention of BRCA2 is regulated by DSS1 interaction through the masking of a NES, such that variation impairing BRCA2-DSS1 binding mis-localizes BRCA2 to the cytoplasm [35].
In this study, we analyzed the effect of a group of VUS mapped to the BRCA1 and BRCA2 gene regions encoding the RING-finger domain of BRCA1 and the DBD of BRCA2 on the binding with the interacting proteins BARD1, UbcH5a, and DSS1, using the previously established GFP-fragment reassembly screening [15,36]. In this assay, the GFP is dissected into two fragments (NfrGFP and CfrGFP) that, when expressed together in E. coli bacterial cells, do not spontaneously reassemble into a fluorescence protein. However, if each of the two fragments of the GFP are individually fused to two interacting protein domains, this interaction can mediate the reassembly of the GFP in co-transformed bacteria with consequent cellular fluorescence. We evaluated the performance of these approaches in correctly discriminating between pathogenic and non-pathogenic analysis of a panel of variants classified according to the IARC 5-class model (Leiden Open Variation Database, URL: http://hci-exlovd.hci.utah.edu/variants.php). In addition, we verified the consistency of the VUS classification with the results of other published functional analyses investigating the same variants [14,30,37,38,39,40,41,42,43,44,45,46,47,48,49,50].
2. Results
2.1. Selection of the BRCA1 and BRCA2 Variants
We selected a total of 58 BRCA1 (n = 36) and BRCA2 (n = 22) variants mapping to the gene regions coding for the conserved motifs encompassing the RING-finger domain of BRCA1 and the DBD of BRCA2 [13] (Figure 1). Of these, 28 BRCA1 (n = 13) and BRCA2 (n = 15) variants, classified by the IARC-5 class system as pathogenic or non-pathogenic, were used for assay validation (Table 1). Thirty BRCA1 (n = 23) and BRCA2 (n = 7) variants were VUS (Table 2) of which 21 were identified among families referred for BRCA gene testing. Of the remaining nine variants, seven were extracted from the ClinVar database (https://www.ncbi.nlm.nih.gov/clinvar/), while two, p.Cys27Ala and p.His41Ala, were synthetic variants not yet observed in HBOC patients and were included so that at least one variant was examined for each of the critical cystein and histidine residues of the RING domain.
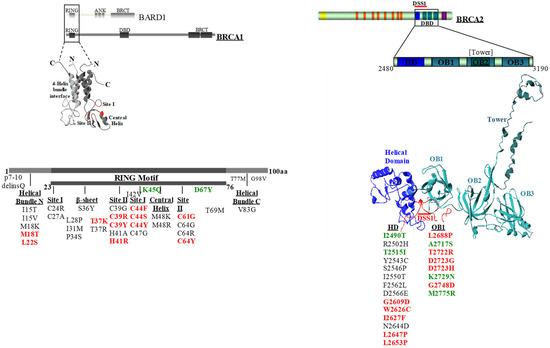
Figure 1.
Diagram showing the location of specific motifs of the BRCA1 and BRCA2 proteins and the variants analyzed. Non-pathogenic variants (class 1 and class 2) are shown in green, pathogenic variants (class 4 and class 5) are shown in red, and VUS are shown in black. DBD, DNA binding domain. HD, helical domain. OB1, oligonucleotide/oligosaccharide binding fold 1. VUS, variants of uncertain significance.

Table 1.
Variants of validation panels.

Table 2.
List of selected BRCA1 and BRCA2 VUS.
2.2. Evaluation of the Effect of BRCA1 Variants on the Interaction with the BARD1 and the UbcH5a Proteins by the GFP-Reassembly Assay
The N-terminal RING domain (aa 1–109) mediates the E3-ubiquitin ligase activity of BRCA1. It consists of a RING finger motif (aa 23–76) and two flanking α helices. It is characterized by a conserved pattern of seven cysteine and one histidine residues, which form two Zn2+-binding sites (termed Site I and Site II) stabilizing the RING structure. The RING finger motif assembles in a globular structure with three strand β-sheet and a central helix.
All 36 mutant constructs were co-transformed in bacterial cells with the pET11a-NfrGFP-BARD1 and pET11a-NfrGFP-UbcH5a constructs expressing the wild-type BARD1 (BDN) and UbcH5a (UbN) proteins, which is in frame with the N-terminal fragment of the GFP protein, respectively. In addition, pET11a-NfrGFP-BARD1 and pET11a-NfrGFP-UbcH5a were co-transformed with pMRBAD-BRCA1-CfrGFP expressing wild-type BRCA1 (BR1C) and pMRBAD-Z-CfrGFP expressing the leucine zipper (Z) peptide (ZC), both in a frame with the C-terminal fragment of the GFP protein, as positive and non-cognate negative controls. The results are listed in Table 3.

Table 3.
BRCA1 variants analyzed with different functional approaches.
Under inducing conditions, a bright fluorescence was observed in bacterial cells co-expressing the strong interacting anti-parallel leucine zipper NfrGFP-Z/Z-CfrGFP (ZN/ZC) fusion peptides and the positive controls (BDN/BR1C, UbN/BR1C). No fluorescence was observed in bacterial cells co-expressing the non-cognate negative controls (BDN/ZC, UbN/ZC) (Figure 2 and Figure 3). The following variants of the validation panel showed a fluorescence resembling that of the positive controls: p.Lys45Gln and p.Asp67Tyr (Class 1), p.Met18Thr (Class 4), p.Thr37Lys and p.His41Arg (Class 5), while no fluorescent signal was observed for the validation panel variants p.Leu22Ser, p.Cys39Arg, p.Cys39Tyr, p.Cys44Phe, p.Cys44Ser, p.Cys44Tyr, p.Cys61Gly and p.Cys64Tyr (all class 5). Among the analyzed VUS, p.Ile15Thr, p.Leu28Pro, p.Ile31Met, p.Ser36Tyr, p.Ile42Val, p.Met48Lys, p.Cys64Gly, p.Cys64Arg, p.Thr77Met, and p.Gly98Val showed bright fluorescence. Conversely, no fluorescence was observed in bacteria expressing the variants p.7-10delinsGln, p.Ile15Val, p.Met18Lys, p.Cys24Arg, p.Cys27Ala, p.Pro34Ser, p.Thr37Arg, p.Cys39Gly, p.His41Ala, p.Cys47Gly, p.Met48Arg, p.Cys64Tyr, p.Thr69Met., and p.Val83Gly (Figure 2).
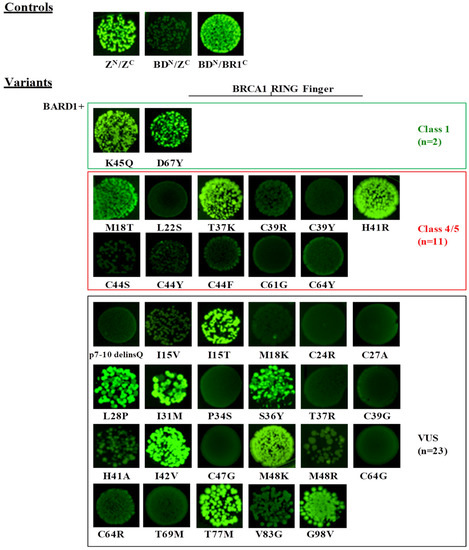
Figure 2.
Detection of the BARD1/BRCA1 interaction by the GFP-fragment reassembly assay. Fluorescence was assessed after 24 h of growth at 30 °C followed by 2 days of incubation at RT. All pictures were taken with the same setting of digital camera (long-wave UV light, 365 nm). [ZN, H6-NfrGFPZ, ZC, ZCfrGFP, BDN, H6-NfrGFPBARD1, BR1C, BRCA1CfrGFP].
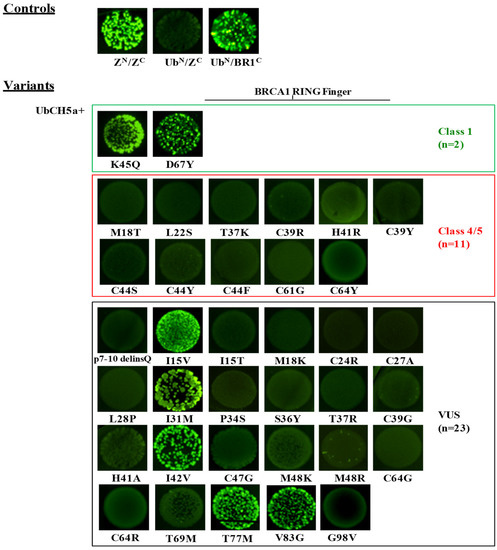
Figure 3.
Detection of the UbcH5a/BRCA1 interaction by the GFP-fragment reassembly assay. Fluorescence was assessed after 24 h of growth at 30 °C followed by 2 days of incubation at RT. All pictures were taken with the same setting of digital camera (long-wave UV light, 365 nm). [ZN, H6-NfrGFPZ, ZC, ZCfrGFP, UbN, H6-NfrGFPUbcH5a, BR1C, BRCA1CfrGFP].
The screening assessing the effect of BRCA1 variants on the binding to UbcH5a revealed bright fluorescence for bacterial cells co-expressing NfrGFP-UbcH5a together with BRCA1-CfrGFP bearing the class1 variants p.Lys45Gln and p.Asp67Tyr and the VUS p.Ile31Met, p.Ile42Val, p.Thr77Met, p.Ile15Val, and Val83Gly in the helical bundle interface (Figure 3). Conversely, all tested class 4 and class 5 variants were not able to bind to UbcH5a. These results indicate 100% sensitivity and specificity of this assay. All remaining tested VUS, including some of those that maintained the interaction with BARD1, namely p.Ile15Thr, p.Leu28Pro, p.Ser36Tyr, and p.Met48Lys, abrogated the binding with UbcH5a.
To confirm that the loss of fluorescence was attributable to the lack of binding to BARD1 and/or UbcH5a and not to poor expression of the BRCA1 mutants, the cell lysates were visualized by Western blotting against the GFP tag. The results showed that all fusion peptides were expressed to a similar extent (Figure 4A).

Figure 4.
Expression of BRCA1-CfrGFP (A) and NfrGFP-BRCA2HD/OB1 (B) wild-type (wt) and mutant constructs. The molecular masses are indicated on the left.
2.3. Evaluation of the Effect of the BRCA2 Variants on the Interaction with the DSS1 Protein by the GFP-Reassembly Assay
The C-terminal region of BRCA2 contains a DSS1/ssDNA Binding Domain (DBD) consisting of five components. The first is described as the helical domain (HD) due to its 190 amino acids consisting mainly of α-helices. This is followed by three oligonucleotide/oligosaccharide binding folds (OB1, OB2 and OB3). In addition, within OB2, a 130 amino acid insertion exists, which was named the tower domain due to its tower-like structure protruding from the OB fold. The crystallographic study of BRCA2-DBD revealed a stoichiometric complex with DSS1, which is almost entirely mediated by highly conserved residues spanning the HD and OB1 motifs. Solely, one DSS1 residue interacts with the first portion of the OB2 motif [32].
We replaced the leucine zipper peptides in the original constructs (pET11a-NfrGFP-Z and pMRBAD-Z-CfrGFP) with HD, the three OB motifs, each of which is in a distinct construct and the protein DSS1 is in all the possible orientations (i.e., pET11a-NfrGFP-HD/OB1/OB2/OB3 pET11a-NfrGFP-DSS1, pMRBAD-HD/OB1/OB2/OB3-CfrGFP, and pMRBAD-DSS1-CfrGFP). Upon co-transformation in bacterial cells, bright fluorescence was observed only when the HD and OB1 were fused to the NfrGFP and DSS1 was fused to the CfrGFP while a less pronounced fluorescence was observed in bacterial cells co-expressing BRCA2-OB2 and DSS1 inserted in the same orientation (Figure 5). No fluorescence was detected when the above inserts were fused in the opposite orientation and co-transfected likely due to a conformational restraint of GFP-reassembly under these conditions. Consistently with the BRCA2 DBD-DSS1 crystal structure, no fluorescence was observed in bacteria co-expressing BRCA2-OB3 and DSS1 in either orientation (Figure 5 and Supplemental Figure S1).
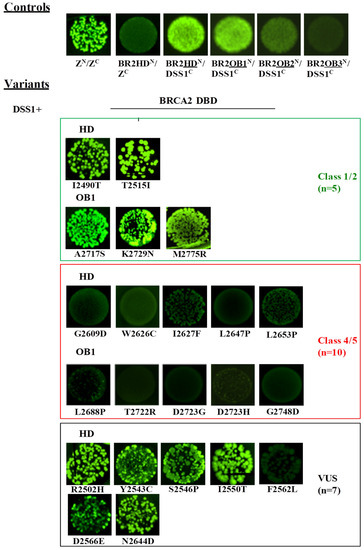
Figure 5.
Detection of the DSS1/BRCA2 interaction by GFP-fragment reassembly assay. Fluorescence was assessed after 24 h of growth at 30 °C followed by 2 days of incubation at RT. All pictures were taken with the same setting of digital camera (long-wave UV light, 365 nm). [ZN, H6-NfrGFPZ, ZC, ZCfrGFP, BR2HDN, H6-NfrGFPBRCA2HD, BR2OB1N, H6-NfrGFPBRCA2OB1, BR2OB2N, H6-NfrGFPBRCA2OB2, BR2OB3N, H6-NfrGFPBRCA2OB3, DSS1C, DSS1CfrGFP].
Based on the above results, we decided to evaluate the effect of the BRCA2 variants mapped to the gene regions encoding for the HD and OB1 motifs only. All 22 mutants and the corresponding wild-type forms were co-transformed with the pMRBAD-DSS1-CfrGFP expressing the wild-type DSS1. In addition, the pET11a-NfrGFP-HD was co-transformed with the pMRBAD-Z-CfrGFP expressing the leucine zipper peptide as a non-cognate negative control.
As reported in Figure 5, no fluorescence was observed in bacterial cells co-expressing non-cognate negative control NfrGFP-HD/Z-CfrGFP (HDN/ZC) while bright fluorescence was detected in bacteria co-expressing the positive controls ZN/ZC, NfrGFP-HD/DSS1-CfrGFP (HDN/DSS1C), and NfrGFP-OB1/DSS1-CfrGFP (OB1N/DSS1C). Comparable fluorescence was observed in bacterial cells co-expressing DSS1-CfrGFP with NfrGFP fused to the BRCA2 peptides bearing all the variants of Class 1/2 and the following VUS: p.Arg2502His, p.Tyr2543Cys, p.Ser2546Pro, p.Ile2550Thr, p.Asp2566Glu, and p.Asn2644Asp (HD). No fluorescence was observed in all the bacteria expressing BRCA2 peptides carrying the Class 4/5 mutations and the VUS p.Phe2562Leu (HD). The expression level of all the NfrGFP-HD/OB1 peptides was verified by Western blotting analysis using an anti-GFP antibody and no significant differences were observed (Figure 4B).
2.4. Validation of the GFP-Reassembly Screenings
The re-assembled complexes were purified by Immobilized Metal Affinity Chromatography (IMAC) from the soluble fraction of co-transformed bacterial cell lysates exploiting the hexahistidine tag (H6) fused to the N-terminus of the NGFP fragments [14,15]. Equal aliquots of the cleared lysates of co-transformed E. coli BL21 (DE3) cells were incubated with Ni-NTA agarose resin and the affinity-captured material was analyzed by Western blotting using a polyclonal anti-GFP antibody (Supplemental Figures S2–S4). The detection of two bands corresponding to the complex components confirmed interactions with the binding proteins for all BRCA1 and BRCA2 constructs that tested positive in the fluorescence reassembly assay. No bands, or bands of very low intensity, were observed in case BRCA1 and BRCA2 variants for whom no fluorescence was detected, which confirms the lack of interaction. Assay specificity was supported by the absence of signals detected in cell lysates co-expressing the NfrGFP-BARD1, NfrGFP-UbcH5a, or the NfrGFP-HD with the CfrGFP-Z peptide (non-cognate negative controls).
2.5. Evaluation of the Effect of VUS on mRNA Splicing
Exonic variants may affect protein functions not only by altering the amino acid sequence, but also abolishing naturally-occurring splice sites or introducing/activating new or cryptic splice sites. To account for this possibility, we performed an in silico analysis on all 30 VUS included in the study, from which only three were predicted to be spliceogenic by ≥three algorithms of the Alamut software. These included BRCA1 c.110C>G and c.115T>G, predicted to introduce novel acceptor splice sites, and BRCA1 c.190T>G, predicted to reinforce the strength of an alternatively used donor splice site in exon 5. Existing mRNA assay data reports no effect on mRNA splicing for BRCA1 c.110C>G [57], while BRCA1 c.190T>G has been shown to lead to increased levels of a non-productive naturally-occurring isoform (Δ5q) [58], which carries the out of frame deletion of the distal portion of exon 5 [59]. To the best of our knowledge, no experimental data has been reported as for the c.115T>G. No spliceogenic effect was predicted for all remaining VUS.
3. Discussion
The reporting of VUS can cause uncertainty and anxiety regarding cancer risk and clinical management. In addition, appropriate medical care may be delayed for individuals carrying VUS that are later determined to be pathogenic. In the near future, the relevance of these questions is likely to rise, as the relative frequency of VUS on the total number of variants identified in HBOC individuals is expected to increase dramatically with the development of next generation sequencing (NGS) analyses and the possibility to screen gene panels containing several verified or putative breast/ovarian cancer predisposition genes at minimal additional costs [60]. Furthermore, a study evaluating the coding regions and exon-intron boundaries of a 42-cancer gene sequencing panel (including BRCA1 and BRCA2) identified one or more VUS in approximately 90% of the patients [61]. This underlines the need to develop accurate methods to discriminate VUS that are actually related to cancer risk (pathogenic) from those without clinical relevance (non-pathogenic or benign).
In this study, we evaluated the use of an in vitro protein-fragment complementation assay and the GFP-fragment reassembly screening for the functional characterization of BRCA1 and BRCA2 VUS. We had previously applied this method to functionally characterize an Italian founder variant (p.Cys64Arg) mapped to the RING-finger domain of BRCA1 [15]. This variant was shown to abolish the interaction of BRCA1 with the partner protein BARD1, in a manner similar to the known cancer predisposing variant p.Cys61Gly [62]. In this case, we applied the same approach to analyze the effect of 25 BRCA1 VUS mapped to the RING-finger domain on the binding with BARD1 and UbcH5a proteins and seven BRCA2 VUS mapped to the DBD domain on the binding with the DSS1 protein.
To evaluate the accuracy of the assays in discriminating pathogenic from non-pathogenic variants, we tested the effect of a panel of 13 BRCA1 and 15 BRCA2 variants (validation panels) previously classified according to the IARC 5-class system. The assays analyzing the BRCA1/UbcH5a and BRCA2/DSS1 interactions show 100% sensitivity (percentage of correctly classified pathogenic variants) and 100% specificity (percentage of correctly classified non-pathogenic variants). In fact, all constructs carrying the Class 1 and 2 variants included in the validation panels displayed complex formation with the selected binding partners, while all constructs carrying the Class 4 and 5 variants did not. Based on these findings, our investigation shows that 19 (18 BRCA1 and 1 BRCA2) VUS behaved as pathogenic and 11 (5 BRCA1 and 6 BRCA2) as neutral variants (Figure 3 and Figure 5, Table 3 and Table 4).

Table 4.
BRCA2 variants analyzed with different functional approaches.
Two issues need to be considered when evaluating the performance of the GFP-fragment reassembly assay. First, since the assays described are based on an artificial experimental model, where functional domains of human proteins are expressed in prokaryotic cells, the behavior of mutant constructs that we observed might not reflect the properties of full-length BRCA proteins expressed in their ‘natural’ context. However, it must be considered that, although our observations might actually provide clues for the molecular mechanisms mediating the tumor suppressor activity of BRCA proteins (see below), the aim of this study was essentially to collect evidence for the clinical relevance of BRCA VUS by the use of functional assays that efficiently dichotomize variants into pathogenic or non-pathogenic. Second, some exonic non-synonymous changes can affect gene functionality by altering mRNA splicing. In a few cases, i.e., when the spliceogenic allele retains the ability to produce a full-length protein, this occurrence may co-exist with a pathogenic effect of the missense substitution such as in the case of the BRCA2 c.8168A>G (p.Asp2723Gly) [47,56]. Previous data and in silico analyses revealed for two of the BRCA1 VUS we tested, an actual (c.190T>G) or potential (c.115T>G) spliceogenic effect. Based on the outcomes of the BRCA/UbcH5a interaction assay, all the above variants were predicted to be pathogenic. However, no data are presently available to ascertain if full-length proteins carrying such variants are actually expressed, and, therefore, no data are present to hypothesize dual mRNA/protein defects.
The assay analyzing the BRCA1/BARD1 binding reveals high specificity (100%), but lower sensitivity (62.5%), compared to the assay exploiting BRCA1/UbcH5a interaction, which indicates that the analyses of different protein-protein interactions may have a different level of accuracy in predicting the pathogenicity of amino acid changes in binding domains. In fact, the class 4 p.Met18Thr and the class 5 p.Tyr37Lys and p.His41Arg variants preserved the interaction with BARD1 as the wild-type protein and the proteins carrying the Class1 variants p.Lys45Gln and p.Asp67Glu.
Similar results were observed using alternative approaches in independent studies (Table 3). Morris and co-workers reported a comprehensive analysis of missense mutations mapped to the RING-finger domain of BRCA1 and, using yeast 2-hybrid (Y2H) and in vitro ubiquitin ligase assays, they showed that, in the presence of the p.Met18Thr variant, BRCA1: (a) retains the interaction with BARD1, (b) is no longer able to bind UbcH5a, and (c) loses the E3 ubiquitin ligase activity [37]. Ransburgh and co-workers exploiting the co-immunoprecipitation approach showed that the variant p.Met18Thr only modestly affects the interaction between BRCA1 and BARD1 [38]. Moreover, a recent analysis measuring the effect of approximately 2000 BRCA1 missense substitutions, both on BARD1 binding and E3 ubiquitin ligase activity, reported that the BRCA1 proteins carrying the variants p.Met18Thr, p.Thr37Lys, and p.His41Arg are able to interact with BARD1 and lose the E3 ligase activity [43]. All these observations, together with our results, suggest that, in the absence of the BRCA1-UbcH5a complex, the maintenance of the binding with BARD1 is dispensable for the BRCA1 tumor suppressor activity. Thus, we speculate that the reduced or abrogated E3 ubiquitin ligase activity, through loss of binding with UbcH5a, is correlated with disease susceptibility and the prediction of pathogenicity. The observations of Drost and co-workers support this hypothesis, who reported that mice carrying the pathogenic p.Cys61Gly variant, inactivating BRCA1 E3 ligase activity [37], are embryonically lethal because of a developmental delay and that, in addition, the latency and nature of mammary tumors with one Brca1 allele carrying the p.Cys61Gly variant and one Brca1 conditional null allele were similar to those carrying two Brca1 null alleles [63]. Consistently with these findings, two of the BRCA1 VUS that, in our analysis, did not inhibit the BARD1 interaction, but lose the UbcH5a, and for which data on enzymatic properties are available (p.Ile15Thr, p.Leu28Pro) and lack ubiquitin ligase activity [37] (Table 3). However, it has to be mentioned that, in another study, the missense substitution p.Ile26Ala, abrogating E2 binding and thus ubiquitin-ligase activity, was found to prevent tumor formation to the same degree as wild-type Brca1 in three conditional genetic models. Furthermore, the DNA damage response remained intact with no changes in chromosome stability or sensitivity to genotoxic stress in mouse embryonic fibroblasts [64]. Given the discrepancy of the results from the two studies, the implication of BRCA1 E3 ligase activity in tumor suppression remains to be elucidated.
In the presence of p.Thr77Met, BRCA1 is able to bind both BARD1 and UbcH5a proteins. However, as reported by Morris and co-workers, the proteins lose the ubiquitin ligase activity [37]. A possible explanation is that a weak interaction between BRCA1 and UbcH5a is not sufficient to promote the ubiquitin ligase activity. Actually, the GFP-fragment reassembly approach can detect very weak and transient interactions and the fluorescence intensity observed does not necessarily correlate quantitatively to the interaction affinity of the proteins, which represents a limitation for this assay. Therefore, hypomorphic variants (i.e., variants weakening, but not completely disrupting, the interaction of BRCA1 and BRCA2 with partner binding proteins, which retains partial protein activity) are likely to produce a positive fluorescent signal and can be categorized as non-pathogenic with this approach. In effect, using the Y2H assay, Morris and co-workers showed a reduction in binding to the UbcH5a of BRCA1 carrying the p.Thr77Met variant. These observations suggest that the p.Thr77Met may be a hypomorphic variant associated with moderate-to-low risk of breast and ovarian cancers. Furthermore, this variant was observed in trans with a pathogenic variant in an individual without clinical signs of Fanconi anemia (FA) or FA-like disease, which is consistent with the suggestion that the p.Thr77Met is hypomorphic and is not associated with a high risk of cancer. Further analyses, using additional approaches such as the Y2H assay and the split biosensor assays measuring protein interaction kinetics (the tripartite split-GFP association assay [65]), are required to define the thresholds of binding activity that discriminate between pathogenic and neutral variants and assess potential intermediate effects.
A series of important considerations have to be applied to functional assays evaluating the BRCA1 and BRCA2 VUS [10,11]. In particular, a panel of different validated assays, representative of different protein functions, should be used, in order to avoid the misinterpretation of the clinical relevance of BRCA gene variants. We observed good correlation of results obtained from the GFP-reassembly screenings with those of other reported functional assays, including homology-directed repair (HDR), mouse embryo stem cells (mESCs) viability, and drug resistance assays (Table 3 and Table 4). In fact, BRCA1 and BRCA2 VUS that were proficient in these assays, retain binding activity with UbCH5a and DSS1, respectively. All variants that were not proficient in other reported assays showed a loss of the binding activities. These included the BRCA2 p.Phe2562Leu, which is the only VUS that, in our assay, abolishes the ability of BRCA2 DBD to bind DSS1, which is reported to be deficient in HDR activity [48].
To date, the most accurate medium throughput functional approaches in evaluating BRCA1 and BRCA2 VUS are the yeast-cell based colony size phenotype (SCP) assay [40,66] and the HDR assay in mammalian cells [47,48], respectively (Table 3 and Table 4). Compared to the GFP-reassembly screening, the SCP assay shows lower sensitivity (96%) and specificity (93%). Importantly, contrary to the SCP assay, the GFP-reassembly screening analyzing the effect of BRCA1 variants on UbcH5a binding correctly classifies the p.Cys61Gly variant as pathogenic (Table 3). While the GFP-reassembly assay and the HDR functional assay show the same level of accuracy, the latter is more technically and experimentally demanding and time consuming.
While this manuscript was in preparation, Findlay and co-workers published the results of a massive functional characterization of BRCA1 single nucleotide variants, (SNVs) based on a highly sensitive and specific saturation genome editing (SGE) approach [44]. Findlay et al. measured the effect on the survival of the near-haploid HAP1 cell line of almost 4,000 SNVs corresponding to 96.5% of all possible base-pair changes in 13 exons that encode the RING and BRCT domains. The assay was based on the observation that the maintenance of BRCA1-mediated HDR is necessary for the viability of HAP1 cells. Notably, we observed a 100% correlation between the functional classification based on SGE scores and the outcomes of our GFP-reassembly assay evaluating the BRCA1/UbCH5a interaction (Table 3). This strict consistency suggests that BRCA1/UbCH5a interaction, which mediates BRCA1 ligase activity, is essential for proficient HDR. In addition, since HDR deficiency is targeted by drugs exploiting synthetic lethality, such as Poly (ADP-ribose) polymerase 1 (PARP1) inhibitors [67], our assay could be used to predict a response to therapeutic treatments.
SGE has demonstrated to be a viable and highly productive strategy for functionally classifying thousands of variants in clinically actionable genes such as BRCA1, including a large number of variants that, to date, have not been observed in humans. In fact, a major strength of this approach is represented by the possibility of examining variants located throughout the entire gene sequence, while, on the contrary, the GFP-reassembly assay may be applied only to gene regions coding for protein-interaction domains. However, it must be noted that the vast majority of pathogenic BRCA gene variants lie in these domains [6,51]. On the other hand, SGE analyses are limited by the extreme difficulty of generating all possible gene variants that may occur in patients. In particular, substitutions and insertions/deletions involve more than one nucleotide. For example, in our study, we could analyze three variants identified in families referred for BRCA gene testing, namely the BRCA1 c.20_29del9 (p.7-10delinsGln) and c.206_207delCCinsTG (p.Thr69Met) in frame deletions, and the c.107C>A (p.Ser36Tyr) SNV, that were not evaluated in the high-throughput study of Findlay and co-workers.
This represents an important and attractive advantage of the split-GFP reassembly in vitro method, which, together with its easiness of use and robustness, makes it suitable for routine analyses of naturally occurring genetic variants. Moreover, this assay could be applied to large-scale studies that are required to achieve robust estimates of the accuracy of functional approaches for VUS classification and clinical applicability.
4. Materials and Methods
4.1. Variant Selection
All examined variants were observed in high risk HBOC families fulfilling the ascertainment criteria for BRCA1/2 testing in use at the collaborating institutions, including the Fondazione IRCCS Istituto Nazionale dei Tumori (Milan, Italy) [68], the Catalan Institute of Oncologia (Hospitalet de Llobregat, Barcelona, Spain) [69], and the Rigshospitalet (Copenhagen, Denmark) [70].
4.2. Plasmid Construction
The pET11a-NfrGFP-Z, pMRBAD-Z-CfrGFP, pET11a-NfrGFP-BARD1, and pMRBAD-BRCA1-CfrGFP expression vectors were obtained and generated as previously described [14]. The BRCA2 fragments encoding the Helical Domain (HD, amino acids 2480–2667), three oligonucleotide-binding folds (OB1, amino acids 2668–2807, OB2, amino acids 2808–3048, OB3, amino acids 3050–3185), the full-length coding sequences of the DSS1, and the UbcH5a genes were obtained by PCR amplification of cDNA prepared from 293T cells using a RevertAid H Minus First Strand cDNA Synthesis Kit (Thermo Scientific, Waltham, MA, USA), and cloned into pET11a-NfrGFP-Z between XhoI and BamHI restriction sites (HD, OB1, OB2, OB3, DSS1, and UbcH5a), and pMRBAD-Z-CfrGFP between NcoI and AatII restriction sites (HD, OB1, OB2, OB3, and DSS1) after removing the leucine zipper motifs (Z). All of the BRCA1 and BRCA2 variants were obtained by direct mutagenesis of pMRBAD-BRCA1-CfrGFP and of pET11a-NfrGFP-BRCA2HD/OB1 using the QuickChange XL Site-directed Mutagenesis Kit (Stratagene, La Jolla, CA, USA), according to the manufacturer’s instruction. DNA sequencing (Eurofins Genomics, Ebersberg, Germany) technology checked the recombinant clones.
4.3. Expression of Recombinant Peptides
E. coli BL21 (DE3) cells were transformed with pET11a-NfrGFP vector series or pMRBAD-CfrGFP vector series. Single colonies were picked and used to inoculate 20 mL of LB broth medium containing 100 µg/mL of ampicillin (pET11a-NfrGFP vector series, Addgene, Watertown, MA, USA) or 35 µg/mL of kanamycin (pMRBAD-CfrGFP vector series, Addgene, Watertown, MA, USA). The colonies were then incubated at 37 °C until the Optical Density measured at a wavelength of 600 nm (OD600) reached 0.6. Then, induction of recombinant peptides was carried out with the addition of IPTG to a final concentration of 20 µM (pET11a-NfrGFP vector series) and 0.2% of L-arabinose (pMRBAD-CfrGFP vector series). After growing for 24 h at 30 °C followed by two days at room temperature (RT), the cells were harvested by centrifugation. Each pellet was resuspended in 1 mL of lysis buffer [50 mM Tris-HCl, 300 mM NaCl, 0.1% v/v Triton X-100, 100 µM EDTA pH8.0, 0.5 mg/mL lysozime, 20 mM Imidazole, protease inhibitors (0.5 mM PMSF, 0.4 µg/mL leupeptin, 0.5 µg/mL aprotinin), and 5 µg/mL DNase and RNase]. The resuspended cells were lysed by sonication. The cell debris were removed by centrifugation at 4 °C for 20 min at 13,000 rpm. The protein concentration of the whole cell extracts was determined by the Bradford method [71], using the Bio-Rad protein assay kit (Bio-Rad Laboratories, Hercules, CA, USA). Equal amounts of protein (20 μg) were subjected to 13% SDS-PAGE and visualized by Western blotting using an anti-GFP antibody (# 600-101-215, Rockland, Limerik, PA, USA).
4.4. GFP-Fragment Reassembly Screening
E. coli BL21 (DE3) cells were co-transformed with the following compatible pairs of plasmids: pET11a-NfrGFP-Z and pMRBAD-Z-CfrGFP, pET11a-NfrGFP-BARD1, or pET11a-NfrGFP-UbcH5a and pMRBAD-BRCA1-CfrGFP (both as wild-type or mutant forms), pET11a-NfrGFP-BRCA2HD/OB1/OB2/OB3, or pMRBAD-BRCA2HD/OB1/OB2/OB3-CfrGFP (both as wild-type or mutant forms) and pMRBAD-DSS1-CfrGFP or pET11a-NfrGFP-DSS1, and screened for the occurrence of GFP-fragment reassembly, as previously described [14]. Fluorescence was observed after excitation with long-wave (365 nm) UV light combined with the short pass (SP) emission filter using a Syngene image capture system (SYNGENE, Cambridge, UK), as specified by the manufacturer.
4.5. Purification of the Reassembled GFP Complexes
The H6-NfrGFP-BARD1(or UbcH5a)/BRCA1-CfrGFP (both as wild-type or mutant forms) and H6-NfrGFP-BRCA2HD/OB1 (both as wild-type or mutant forms)/DSS1-CfrGFP complexes were purified from the soluble fraction of co-transformed E. coli strain BL21 (DE3) by IMAC using nickel nitrilotriacetic (Ni-NTA) agarose resin (QIAGEN, Hilden, Germany), according to the manufacturer’s instruction. The protein complexes were subjected to 13% SDS-PAGE and visualized by Western blotting using a polyclonal anti-GFP antibody (# 600-101-215, Rockland).
4.6. Protein Structures
The 3D crystallographic structures of the BRCA1-BARD1 heterodimeric complex and of BRCA2 in complex with the protein DSS1 were represented by the Swiss-PdbViewer tool (SPDBV, URL: http://www.expasy.org/spdbv/, accessed on 7 May 2015) [72] using the pdb as a template (protein data bank, URL: http://www.pdb.org, accessed on 7 May 2015) [73] files 1JM7 and 1MJE, respectively.
4.7. In Silico Analyses
The Alamut Visual version 2.10 (Interactive Biosoftware, Rouen, France) was used to evaluate the possible effect of examined VUS on mRNA transcript splicing.
5. Conclusions
The concordance between genetic data and the functional assays analyzing the BRCA1/UbcH5a and the BRCA2/DSS1 interactions, observed for the validation panels of previously classified variants, strongly suggests that the approaches described in the present study are able to differentiate between VUS that either inactivate or have no effect on BRCA1 and BRCA2 functions and support their utility in the assessment of the clinical relevance of VUS. Caution is warranted when the interpretation of VUS is based only on results from an assay focusing on a single specific biochemical activity. Therefore, these assays should be used in combination with other evidence when assessing variant pathogenicity, and the efficacy will be further verified using larger validation panels.
Supplementary Materials
The following are available online at http://www.mdpi.com/2072-6694/11/2/151/s1, Figure S1: Fluorescence complementation through GFP-reassembly assay depends on fusion orientation of DSS1 and BRCA2 fragments, Figure S2: Purification of the BARD1/BRCA1 reassembled complexes by the IMAC Method, Figure S3: Purification of the UbCH5a/BRCA1 reassembled complexes by the IMAC Method, Figure S4: Purification of the DSS1/BRCA2 reassembled complexes by the IMAC method.
Author Contributions
L.C. conceived the study, performed experimental analyses, analyzed the data, and wrote the manuscript. M.C. performed in silico analyses. T.v.O.H., C.L., S.M., M.T.P., and A.B.S. provided genetic data. P.R. conceived and supervised the study and contributed to the drafting of the manuscript. All authors critically revised and approved the manuscript.
Funding
Partially supported by grants from the Italian Association for Cancer Research (AIRC) (Investigator grant 15547) to P.R. and from the Carlos III National Health Institute and Ministerio de Educación y Ciencia funded by FEDER funds—a way to build Europe—[PI16/00563 and CIBERONC] and the Government of Catalonia [Pla estratègic de recerca i innovació en salut (PERIS) 2017SGR496] to C.L.
Acknowledgments
We thank Thomas J Magliery from the Ohio State University in Columbus (OH, USA) for kindly providing the pET11a-NfrGFP-Z and pMRBAD-Z-CfrGFP plasmids necessary for the GFP-reassembly assay and Maria Teresa Radice of the Fondazione IRCCS Istituto Nazionale dei Tumori, Milan, for technical assistance with plasmid construction.
Conflicts of Interest
P.R. was scientific coordinator of a course on the classification of BRCA gene allele variants sponsored by AstraZeneca (Milan, June 2018).
References
- Mavaddat, N.; Peock, S.; Frost, D.; Ellis, S.; Platte, R.; Fineberg, E.; Evans, D.G.; Izatt, L.; Eeles, R.A.; Adlard, J.; et al. Cancer risks for BRCA1 and BRCA2 mutation carriers: Results from prospective analysis of EMBRACE. J. Natl. Cancer Inst. 2013, 105, 812–822. [Google Scholar] [CrossRef]
- Rebbeck, T.R.; Friebel, T.M.; Friedman, E.; Hamann, U.; Huo, D.; Kwong, A.; Olah, E.; Olopade, O.I.; Solano, A.R.; et al. Mutational spectrum in a worldwide study of 29,700 families with BRCA1 or BRCA2 mutations. Hum. Mutat. 2018, 39, 593–620. [Google Scholar] [CrossRef] [PubMed]
- Plon, S.E.; Eccles, D.M.; Easton, D.; Foulkes, W.D.; Genuardi, M.; Greenblatt, M.S.; Hogervorst, F.B.; Hoogerbrugge, N.; Spurdle, A.B.; Tavtigian, S.V.; et al. Sequence variant classification and reporting: Recommendations for improving the interpretation of cancer susceptibility genetic test results. Hum. Mutat. 2008, 29, 1282–1291. [Google Scholar] [CrossRef] [PubMed]
- Tavtigian, S.V.; Byrnes, G.B.; Goldgar, D.E.; Thomas, A. Classification of rare missense substitutions, using risk surfaces, with genetic- and molecular-epidemiology applications. Hum. Mutat. 2008, 29, 1342–1354. [Google Scholar] [CrossRef] [PubMed]
- Goldgar, D.E.; Easton, D.F.; Deffenbaugh, A.M.; Monteiro, A.N.; Tavtigian, S.V.; Couch, F.J.; Breast Cancer Information Core (BIC) Steering Committee. Integrated Evaluation of DNA Sequence Variants of Unknown Clinical Significance: Application to BRCA1 and BRCA2. Am. J. Hum. Genet. 2004, 75, 535–544. [Google Scholar] [CrossRef] [PubMed]
- Easton, D.F.; Deffenbaugh, A.M.; Pruss, D.; Frye, C.; Wenstrup, R.J.; Allen-Brady, K.; Tavtigian, S.V.; Monteiro, A.N.; Iversen, E.S.; Couch, F.J.; et al. A systematic genetic assessment of 1433 sequence variants of unknown clinical significance in the BRCA1 and BRCA2 breast cancer-predisposition genes. Am. J. Hum. Genet. 2007, 81, 873–883. [Google Scholar] [CrossRef]
- Spurdle, A.B. Clinical relevance of rare germline sequence variants in cancer genes: Evolution and application of classification models. Curr. Opin. Genet. 2010, 20, 315–323. [Google Scholar] [CrossRef]
- Lindor, N.M.; Guidugli, L.; Wang, X.; Vallee, M.P.; Monteiro, A.N.; Tavtigian, S.; Goldgar, D.E.; Couch, F.J. A review of a multifactorial probability-based model for classification of BRCA1 and BRCA2 variants of uncertain significance (VUS). Hum. Mutat. 2012, 33, 8–21. [Google Scholar] [CrossRef]
- Millot, G.A.; Carvalho, M.A.; Caputo, S.M.; Vreeswijk, M.P.; Brown, M.A.; Webb, M.; Rouleau, E.; Neuhausen, S.L.; Hansen, T.V.; Galli, A.; et al. A guide for functional analysis of BRCA1 variants of uncertain significance. Hum. Mutat. 2012, 33, 1526–1537. [Google Scholar] [CrossRef]
- Guidugli, L.; Carreira, A.; Caputo, S.M.; Ehlen, A.; Galli, A.; Monteiro, A.N.; Neuhausen, S.L.; Hansen, T.V.; Couch, F.J.; Vreeswijk, M.P.; et al. ENIGMA consortium. Functional assays for analysis of variants of uncertain significance in BRCA2. Hum. Mutat. 2014, 35, 151–164. [Google Scholar] [CrossRef]
- Venkitaraman, A.R. Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell 2002, 108, 171–182. [Google Scholar] [CrossRef]
- Gudmundsdottir, K.; Ashworth, A. The roles of BRCA1 and BRCA2 and associated proteins in themaintenance of genomic stability. Oncogene 2006, 25, 5864–5874. [Google Scholar] [CrossRef] [PubMed]
- Roy, R.; Chun, J.; Powell, S.N. BRCA1 and BRCA2: Different roles in a common pathway of genome protection. Nat. Rev. Cancer 2011, 12, 68–78. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, M.; Magliery, T.J. Re-engineering a split-GFP reassembly screen to examine RING-domain interactions between BARD1 and BRCA1 mutants observed in cancer patients. Mol. Biosyst. 2008, 4, 599–605. [Google Scholar] [CrossRef] [PubMed]
- Caleca, L.; Putignano, A.L.; Colombo, M.; Congregati, C.; Sarkar, M.; Magliery, T.J.; Ripamonti, C.B.; Foglia, C.; Peissel, B.; Zaffaroni, D.; et al. Characterization of an Italian Founder Mutation in the RING-Finger Domain of BRCA1. PLoS ONE 2014, 9, e86924. [Google Scholar] [CrossRef] [PubMed]
- Xia, B.; Sheng, Q.; Nakanishi, K.; Ohashi, A.; Wu, J.; Christ, N.; Liu, X.; Jasin, M.; Couch, F.J.; Livingston, D.M. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol. Cell 2006, 22, 719–729. [Google Scholar] [CrossRef] [PubMed]
- Biswas, K.; Das, R.; Alter, B.P.; Kuznetsov, S.G.; Stauffer, S.M.; North, S.L.; Burkett, S.; Brody, L.C.; Meyer, S.; Byrd, R.A.; et al. A comprehensive functional characterization of BRCA2 variants associated with Fanconi anemia using mouse ES cell based assay. Blood 2011, 118, 2430–2442. [Google Scholar] [CrossRef]
- Brough, R.; Bajrami, I.; Vatcheva, R.; Natrajan, R.; Reis-Filho, J.S.; Lord, C.J.; Ashworth, A. APRIN is a cell cycle specific BRCA2-interacting protein required for genome integrity and a predictor of outcome after chemotherapy in breast cancer. EMBO J. 2012, 31, 1160–1176. [Google Scholar] [CrossRef]
- Brzovic, P.S.; Rajagopal, P.; Hoyt, D.W.; King, M.C.; Klevit, R.E. Structure of a BRCA1-BARD1 heterodimeric RING-RING complex. Nat. Struct. Biol. 2001, 8, 833–837. [Google Scholar] [CrossRef]
- Joukov, V.; Chen, J.; Fox, E.A.; Green, J.B.; Livingston, D.M. Functional communication between endogenous BRCA1 and its partner, BARD1, during Xenopus laevis development. Proc. Natl. Acad. Sci. USA 2001, 98, 12078–12083. [Google Scholar] [CrossRef]
- Thompson, M.E.; Robinson-Benion, C.L.; Holt, J.T. An amino-terminal motif functions as a second nuclear export sequence in BRCA1. J. Biol. Chem. 2005, 280, 21854–21857. [Google Scholar] [CrossRef]
- Wu, L.C.; Wang, Z.W.; Tsan, J.T.; Spillman, M.A.; Phung, A.; Xu, X.L.; Yang, M.C.; Hwang, L.Y.; Bowcock, A.M.; Baer, R. Identification of a RING protein that can interact in vivo with the BRCA1 gene product. Nat. Genet. 1996, 14, 430–440. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.; Kleiman, F.E.; Manley, J.L.; Ouchi, T.; Pan, Z.Q. Autoubiquitination of the BRCA1*BARD1 RING ubiquitin ligase. J. Biol. Chem. 2002, 2772, 2085–2092. [Google Scholar] [CrossRef]
- Mallery, D.L.; Vandenberg, C.J.; Hiom, K. Activation of the E3 ligase function of the BRCA1/BARD1 complex by polyubiquitin chains. EMBO J. 2002, 21, 6755–6762. [Google Scholar] [CrossRef] [PubMed]
- Wu-Baer, F.; Lagrazon, K.; Yuan, W.; Baer, R. The BRCA1/BARD1 heterodimer assembles polyubiquitin chains through an unconventional linkage involving lysine residue K6 of ubiquitin. J. Biol. Chem. 2003, 278, 34743–34746. [Google Scholar] [CrossRef] [PubMed]
- Hongtae, K.; Junjie, C.; Xiaochun, Y. Ubiquitin-Binding Protein RAP80 Mediates BRCA1-Dependent DNA Damage Response. Science 2007, 316, 1202–1205. [Google Scholar] [CrossRef]
- Horwitz, A.A.; Affar, E.B.; Heine, G.F.; Shi, Y.; Parvin, D.J. A mechanism for transcriptional repression dependent on the BRCA1 E3 ubiquitin ligase. PNAS 2007, 104, 6614–6619. [Google Scholar] [CrossRef]
- Wu, W.; Koike, A.; Takeshita, T.; Ohta, T. The ubiquitin E3 ligase activity of BRCA1 and its biological functions. Cell. Div. 2008, 3, 1. [Google Scholar] [CrossRef] [PubMed]
- Moynahan, M.E.; Cui, T.Y.; Jasin, M. Homology-directed dna repair, mitomycin-c resistance, and chromosome stability is restored with correction of a Brca1 mutation. Cancer Res. 2001, 61, 4842–4850. [Google Scholar]
- Ruffner, H.; Joazeiro, C.A.; Hemmati, D.; Hunter, T.; Verma, I.M. Cancer-predisposing mutations within the RING domain of BRCA1: Loss of ubiquitin protein ligase activity and protection from radiation hypersensitivity. Proc. Natl. Acad. Sci. USA 2001, 98, 5134–5139. [Google Scholar] [CrossRef]
- Nelson, A.C.; Holt, J.T. Impact of RING and BRCT domain mutations on BRCA1 protein stability, localization and recruitment to DNA damage. Radiat. Res. 2010, 174, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Jeffrey, P.D.; Miller, J.; Kinnucan, E.; Sun, Y.; Thoma, N.H.; Zheng, N.; Chen, P.L.; Lee, W.H.; Pavletich, N.P. BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure. Science 2002, 297, 1837–1848. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zou, C.; Bai, Y.; Wazer, D.E.; Band, V.; Gao, Q. DSS1 is required for the stability of BRCA2. Oncogene 2006, 25, 1186–1194. [Google Scholar] [CrossRef] [PubMed]
- Gudmundsdottir, K.; Lord, C.J.; Witt, E.; Tutt, A.N.; Ashworth, A. DSS1 is required for RAD51 focus formation and genomic stability in mammalian cells. EMBO Rep. 2004, 5, 989–993. [Google Scholar] [CrossRef] [PubMed]
- Jeyasekharan, A.D.; Liu, Y.; Hattori, H.; Pisupati, V.; Jonsdottir, A.B.; Rajendra, E.; Lee, M.; Sundaramoorthy, E.; Schlachter, S.; Kaminski, C.F.; et al. A cancer-associated BRCA2 mutation reveals masked nuclear export signals controlling localization. Nat. Struct. Mol. Biol. 2013, 20, 1191–1198. [Google Scholar] [CrossRef] [PubMed]
- Wilson, C.G.; Magliery, T.J.; Regan, L. Detecting protein-protein interactions with GFP-fragment reassembly. Nat. Methods. 2004, 1, 255–262. [Google Scholar] [CrossRef] [PubMed]
- Morris, J.R.; Pangon, L.; Boutell, C.; Katagiri, T.; Keep, N.H.; Solomon, E. Genetic analysis of BRCA1 ubiquitin ligase activity and its relationship to breast cancer susceptibility. Hum. Mol. Genet. 2006, 15, 599–606. [Google Scholar] [CrossRef] [PubMed]
- Ransburgh, D.J.; Chiba, N.; Ishioka, C.; Toland, A.E.; Parvin, J.D. Identification of breast tumor mutations in BRCA1 that abolish its function in homologous DNA recombination. Cancer Res. 2010, 70, 988–995. [Google Scholar] [CrossRef] [PubMed]
- Bouwman, P.; van der Gulden, H.; van der Heijden, I.; Drost, R.; Klijn, C.N.; Prasetyanti, P.; Pieterse, M.; Wientjens, E.; Seibler, J.; Hogervorst, F.B.; et al. A high-throughput functional complementation assay for classification of BRCA1 missense variants. Cancer Discov. 2013, 3, 1142–1155. [Google Scholar] [CrossRef] [PubMed]
- Millot, G.A.; Berger, A.; Lejour, V.; Boulé, J.B.; Bobo, C.; Cullin, C.; Lopes, J.; Stoppa-Lyonnet, D.; Nicolas, A. Assessment of human Nter and Cter BRCA1 mutations using growth and localization assays in yeast. Hum. Mutat. 2011, 32, 1470–1480. [Google Scholar] [CrossRef] [PubMed]
- Towler, W.I.; Zhang, J.; Ransburgh, D.J.; Toland, A.E.; Ishioka, C.; Chiba, N.; Parvin, J.D. Analysis of BRCA1 variants in double-strand break repair by homologous recombination and single-strand annealing. Hum. Mutat. 2013, 34, 439–445. [Google Scholar] [CrossRef] [PubMed]
- Kais, Z.; Chiba, N.; Ishioka, C.; Parvin, J.D. Functional differences among BRCA1 missense mutations in the control of centrosome duplication. Oncogene 2012, 31, 799–804. [Google Scholar] [CrossRef]
- Starita, L.M.; Young, D.L.; Islam, M.; Kitzman, J.O.; Gullingsrud, J.; Hause, R.J.; Fowler, D.M.; Parvin, J.D.; Shendure, J.; Fields, S.; et al. Massively Parallel Functional Analysis of BRCA1 RING Domain Variants. Genetics 2015, 200, 413–422. [Google Scholar] [CrossRef] [PubMed]
- Findlay, G.M.; Daza, R.M.; Martin, B.; Zhang, M.D.; Leith, A.P.; Gasperini, M.; Janizek, J.D.; Huang, X.; Starita, L.M.; Shendure, J. Accurate classification of BRCA1 variants with saturation genome editing. Nature 2018. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.; Hinson, S.R.; Ohashi, A.; Farrugia, D.; Wendt, P.; Tavtigian, S.V.; Deffenbaugh, A.; Goldgar, D.; Couch, F.J. Functional evaluation and cancer risk assessment of BRCA2 unclassified variants. Cancer Res. 2005, 65, 417–426. [Google Scholar]
- Farrugia, D.J.; Agarwal, M.K.; Pankratz, V.S.; Deffenbaugh, A.M.; Pruss, D.; Frye, C.; Wadum, L.; Johnson, K.; Mentlick, J.; Tavtigian, S.V.; et al. Functional assays for classification of BRCA2 variants of uncertain significance. Cancer Res. 2008, 68, 3523–3531. [Google Scholar] [CrossRef] [PubMed]
- Guidugli, L.; Pankratz, V.S.; Singh, N.; Thompson, J.; Erding, C.A.; Engel, C.; Schmutzler, R.; Domchek, S.; Nathanson, K.; Radice, P.; et al. A classification model for BRCA2 DNA binding domain missense variants based on homology-directed repair activity. Cancer Res. 2013, 73, 265–275. [Google Scholar] [CrossRef] [PubMed]
- Guidugli, L.; Shimelis, H.; Masica, D.L.; Pankratz, V.S.; Lipton, G.B.; Singh, N.; Hu, C.; Monteiro, A.N.A.; Lindor, N.M.; Goldgar, D.E.; et al. Assessment of the Clinical Relevance of BRCA2 Missense Variants by Functional and Computational Approaches. Am. J. Hum. Genet. 2018, 102, 233–248. [Google Scholar] [CrossRef]
- Hendriks, G.; Morolli, B.; Calléja, F.M.; Plomp, A.; Mesman, R.L.; Meijers, M.; Sharan, S.K.; Vreeswijk, M.P.; Vrieling, H. An efficient pipeline for the generation and functional analysis of human BRCA2 variants of uncertain significance. Hum. Mutat. 2014, 35, 1382–1391. [Google Scholar] [CrossRef]
- Mesman, R.L.S.; Calléja, F.M.G.R.; Hendriks, G.; Morolli, B.; Misovic, B.; Devilee, P.; van Asperen, C.J.; Vrieling, H.; Vreeswijk, M.P.G. The functional impact of variants of uncertain significance in BRCA2. Genet. Med. 2018. [Google Scholar] [CrossRef] [PubMed]
- Sweet, K.; Senter, L.; Pilarski, R.; Wei, L.; Toland, A.E. Characterization of BRCA1 ring finger variants of uncertain significance. Breast Cancer Res. Treat. 2010, 119, 737–743. [Google Scholar] [CrossRef] [PubMed]
- Parsons, M.; Tudini, E.; Li, H.; Hahnen, E.; Wappenschmidt, B.; Feliubadalo, L.; Aalfs, C.M.; Agata, S.; Aittomäki, K.; Alducci, E.; et al. Large scale multifactorial likelihood analysis of BRCA1 and BRCA2 variants: An ENIGMA resource to support clinical variant classification. Hum. Mutat. 2019. submitted. [Google Scholar]
- Whiley, P.J.; Parsons, M.T.; Leary, J.; Tucker, K.; Warwick, L.; Dopita, B.; Thorne, H.; Lakhani, S.R.; Goldgar, D.E.; Brown, M.A.; et al. Multifactorial likelihood assessment of BRCA1 and BRCA2 missense variants confirms that BRCA1:c.122A>G(p.His41Arg) is a pathogenic mutation. PLoS ONE 2014, 9, e86836. [Google Scholar] [CrossRef] [PubMed]
- Spearman, A.D.; Sweet, K.; Zhou, X.P.; McLennan, J.; Couch, F.J.; Toland, A.E. Clinically applicable models to characterize BRCA1 and BRCA2 variants of uncertain significance. J. Clin. Oncol. 2008, 26, 5393–5400. [Google Scholar] [CrossRef] [PubMed]
- Spurdle, A.B.; Lakhani, S.R.; Healey, S.; Parry, S.; Da Silva, L.M.; Brinkworth, R.; Hopper, J.L.; Brown, M.A.; Babikyan, D.; Chenevix-Trench, G.; et al. Clinical classification of BRCA1 and BRCA2 DNA sequence variants: The value of cytokeratin profiles and evolutionary analysis-a report from the kConFab Investigators. J. Clin. Oncol. 2008, 26, 1657–1663. [Google Scholar] [CrossRef] [PubMed]
- Walker, L.C.; Whiley, P.J.; Couch, F.J.; Farrugia, D.J.; Healey, S.; Eccles, D.M.; Lin, F.; Butler, S.A.; Goff, S.A.; Thompson, B.A.; et al. Detection of splicing aberrations caused by BRCA1 and BRCA2 sequence variants encoding missense substitutions: Implications for prediction of pathogenicity. Hum. Mutat. 2010, 31, 1484–1505. [Google Scholar] [CrossRef]
- Houdayer, C.; Caux-Moncoutier, V.; Krieger, S.; Barrois, M.; Bonnet, F.; Bourdon, V.; Bronner, M.; Buisson, M.; Coulet, F.; Gaildrat, P.; et al. Guidelines for splicing analysis in molecular diagnosis derived from a set of 327 combined in silico/in vitro studies on BRCA1 and BRCA2 variants. Hum. Mutat. 2012, 33, 1228–1238. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Swaminathan, S.; Martin, B.K.; Sharan, S.K. Aberrant splicing induced by missense mutations in BRCA1: Clues from a humanized mouse model. Hum. Mol. Genet. 2003, 12, 2121–2131. [Google Scholar] [CrossRef] [PubMed]
- Colombo, M.; Blok, M.J.; Whiley, P.; Santamariña, M.; Gutiérrez-Enríquez, S.; Romero, A.; Garre, P.; Becker, A.; Smith, L.D.; De Vecchi, G.; et al. Comprehensive annotation of splice junctions supports pervasive alternative splicing at the BRCA1 locus: A report from the ENIGMA consortium. Hum. Mol. Genet. 2014, 23, 3666–3680. [Google Scholar] [CrossRef] [PubMed]
- Kurian, A.W.; Hare, E.E.; Mills, M.A.; Kingham, K.E.; McPherson, L.; Whittemore, A.S.; McGuire, V.; Ladabaum, U.; Kobayashi, Y.; et al. Clinical evaluation of a multiple-gene sequencing panel for hereditary cancer risk assessment. J. Clin. Oncol. 2014, 32, 2001–2009. [Google Scholar] [CrossRef] [PubMed]
- Hilbers, F.S.; Vreeswijk, M.P.; van Asperen, C.J.; Devilee, P. The impact of next generation sequencing on the analysis of breast cancer susceptibility: A role for extremely rare genetic variation? Clin. Genet. 2013, 84, 407–414. [Google Scholar] [CrossRef] [PubMed]
- Brzovic, P.S.; Meza, J.; King, M.C.; Klevit, R.E. The cancer-predisposing mutation C61G disrupts homodimer formation in the NH2-terminal BRCA1 RING finger domain. J. Biol. Chem. 1998, 273, 7795–7799. [Google Scholar] [CrossRef]
- Drost, T.; Bouwman, P.; Rottemberg, S.; Boon, U.; Schut, S.; Klarenbeek, S.; Klijn, C.; van der Heijden, I.; van der Gulden, H.; Wientjens, E.; et al. BRCA1 RING Function Is Essential for Tumor Suppression but Dispensable for Therapy Resistance. Cancer Cell 2011, 20, 797–809. [Google Scholar] [CrossRef] [PubMed]
- Shakya, R.; Reid, L.J.; Reczek, C.R.; Cole, F.; Egli, D.; Lin, C.S.; de Rooij, D.G.; Hirsch, S.; Ravi, K.; Hicks, J.B.; et al. BRCA1 Tumor Suppression Depends on BRCT Phosphoprotein Binding, But not Its E3 Ligase Activity. Science 2011, 334, 525–528. [Google Scholar] [CrossRef] [PubMed]
- Cabantous, S.; Nguyen, H.B.; Pedelacq, J.D.; Koraïchi, F.; Chaudhary, A.; Ganguly, K.; Lockard, M.A.; Favre, G.; Terwilliger, T.C.; Waldo, G.S.; et al. A new protein-protein interaction sensor based on tripartite split-GFP association. Sci. Rep. 2013, 3, 2854. [Google Scholar] [CrossRef] [PubMed]
- Thouvenot, P.; Ben, Y.B.; Fourrière, L.; Lescure, A.; Boudier, T.; Del Nery, E.; Chauchereau, A.; Goldgar, D.E.; Houdayer, C.; Stoppa-Lyonnet, D.; et al. Functional Assessment of Genetic Variants with Outcomes Adapted to Clinical Decision-Making. PLoS Genet. 2016, 12, e1006096. [Google Scholar] [CrossRef] [PubMed]
- De Lorenzo, S.B.; Patel, A.G.; Hurley, R.M.; Kaufmann, S.H. The Elephant and the Blind Men: Making Sense of PARP Inhibitors in Homologous Recombination Deficient Tumor. Cells Front. Oncol. 2013, 3, 228. [Google Scholar] [CrossRef]
- Azzollini, J.; Scuvera, G.; Bruno, E.; Pasanisi, P.; Zaffaroni, D.; Calvello, M.; Pasini, B.; Ripamonti, C.B.; Colombo, M.; Pensotti, V.; et al. Mutation detection rates associated with specific selection criteria for BRCA1/2 testing in 1854 high-risk families: A monocentric Italian study. Eur. J. Int. Med. 2016, 32, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Stradella, A.; Del Valle, J.; Rofes, P.; Feliubadaló, L.; Grau Garces, È.; Velasco, À.; González, S.; Vargas, G.; Izquierdo, Á.; Campos, O.; et al. Does multilocus inherited neoplasia alleles syndrome have severe clinical expression? J. Med. Genet. 2018. [Google Scholar] [CrossRef]
- Hansen, T.V.; Jønson, L.; Steffensen, A.Y.; Andersen, M.K.; Kjaergaard, S.; Gerdes, A.M.; Ejlertsen, B.; Nielsen, F.C. Screening of 1331 Danish breast and/or ovarian cancer families identified 40 novel BRCA1 and BRCA2 mutations. Fam. Cancer 2011, 2, 207–212. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Guex, N.; Peitsch, M.C. SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis 1997, 18, 2714–2723. [Google Scholar] [CrossRef] [PubMed]
- Sussman, J.L.; Lin, D.; Jiang, J.; Manning, N.O.; Prilusky, J.; Ritter, O.; Abola, E.E. Protein Data Bank (PDB): Database of three-dimensional structural information of biological macromolecules. Acta Crystallogr. D Biol. Crystallogr. 1998, 54, 1078–1084. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).