Nanogroove-Enhanced Hydrogel Scaffolds for 3D Neuronal Cell Culture: An Easy Access Brain-on-Chip Model
Abstract
:1. Introduction
2. Materials and Methods
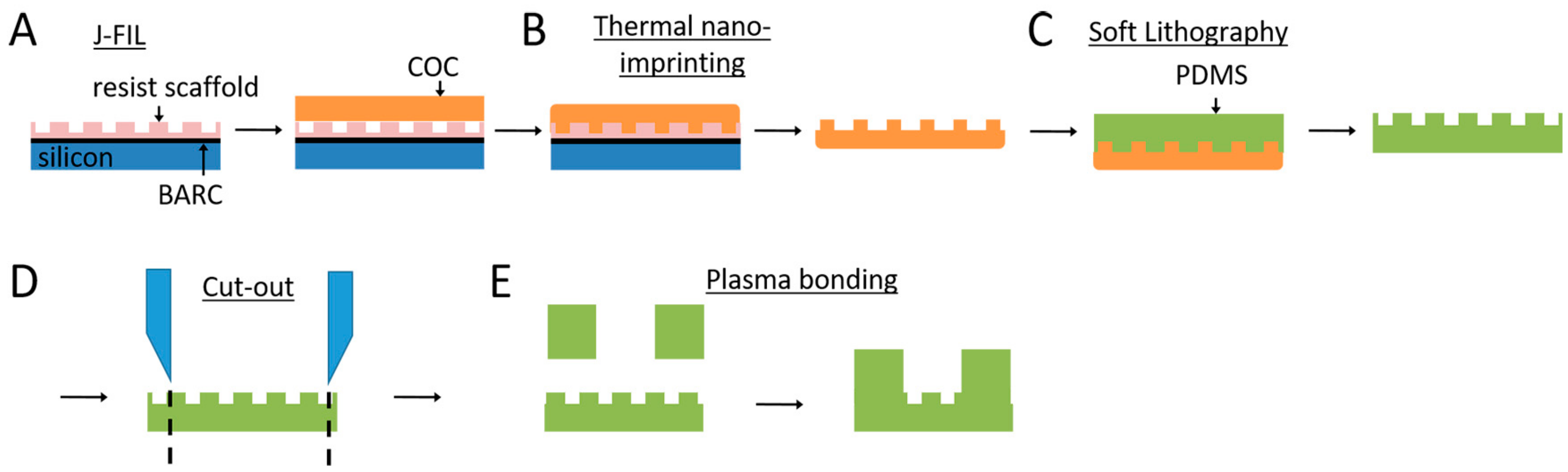
2.1. Nanogroove Substrate Fabrication
2.2. 3D Cell Culture on Nanogrooved Substrates
2.2.1. 3D SH-SY5Y Cells
2.2.2. 3D CTX Cells
2.3. Immunofluorescence Staining and Imaging
2.3.1. SH-SY5Y Cells
2.3.2. CTX Cells
2.4. Cell Culture Analysis: CTX Set and SH-SY5Y
3. Results
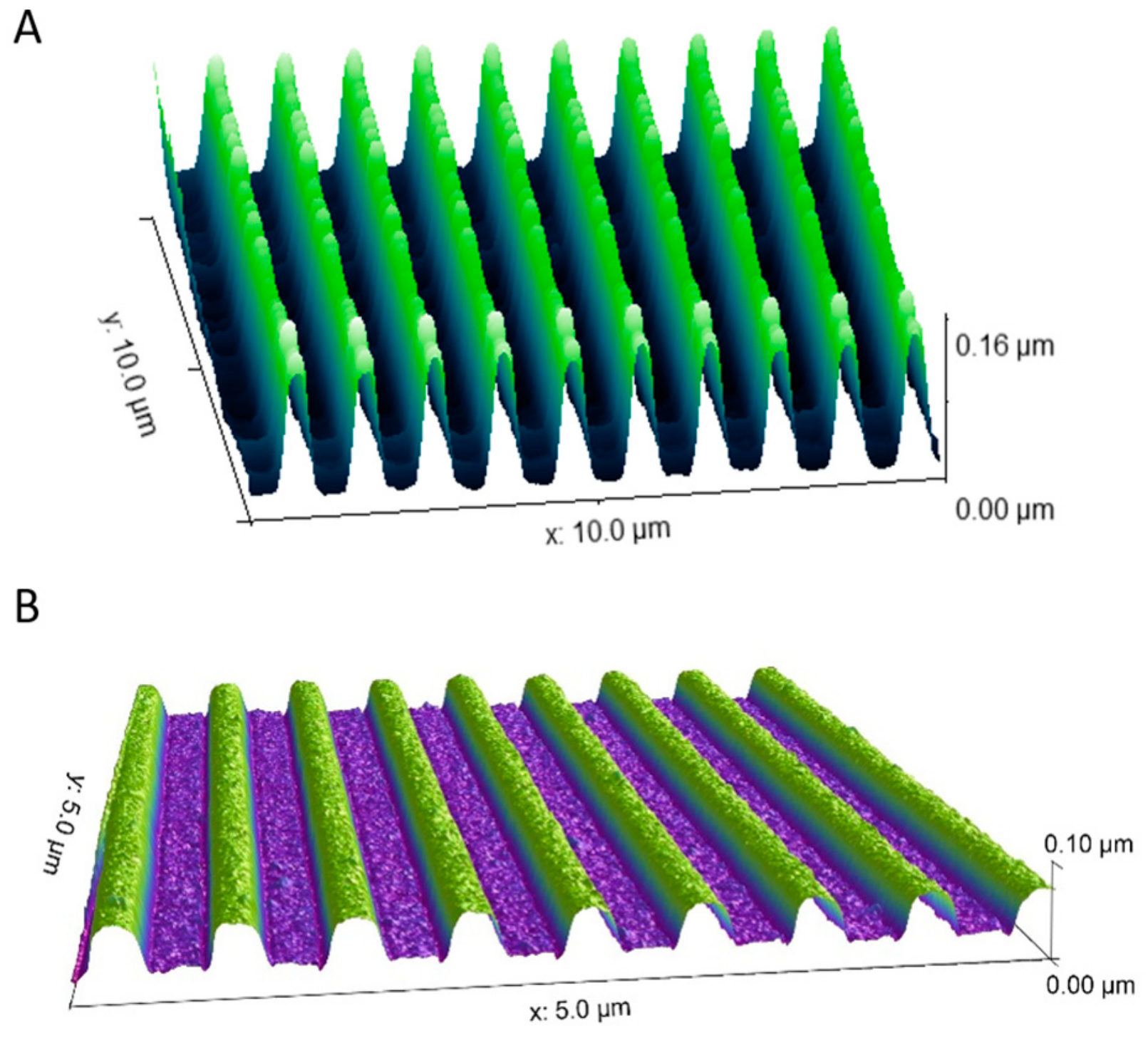
3.1. Nanogrooved Substrate Fidelity
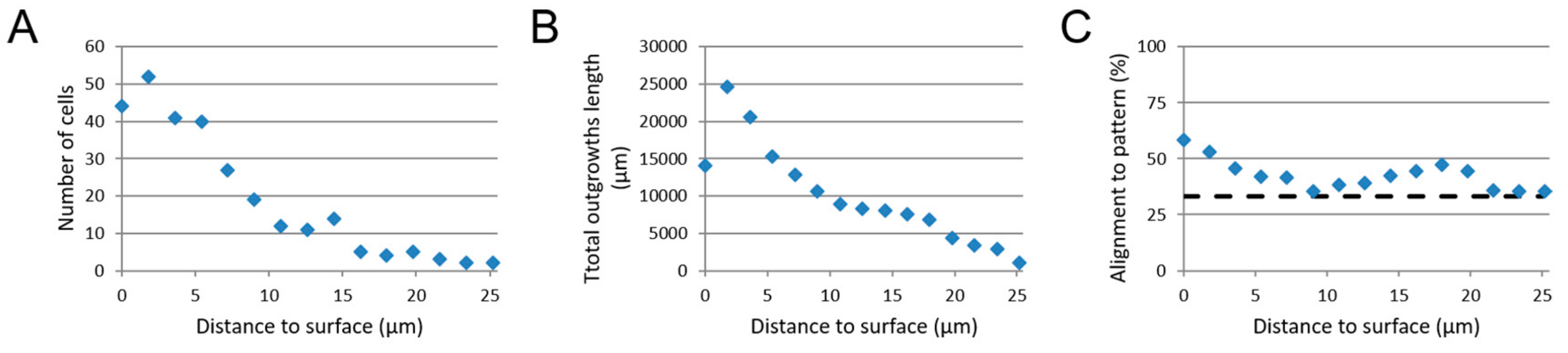
3.2. 3D SH-SY5Y Cell Culture
3.3. 3D CTX Cell Culture
3.3.1. Neuron-Astrocytes Alignment in 2D Culture
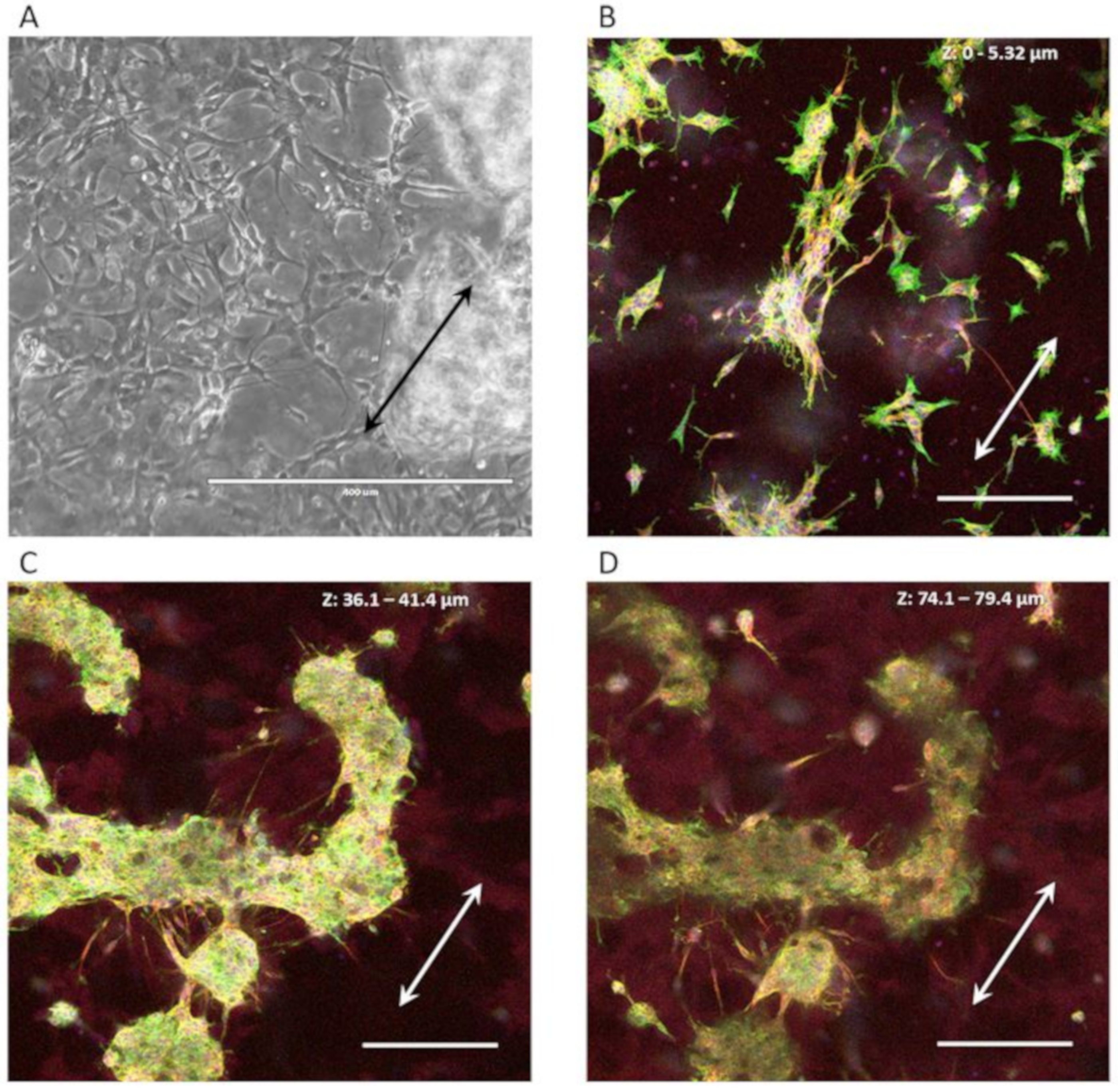
3.3.2. Neuron-Astrocyte Alignment in 3D Culture
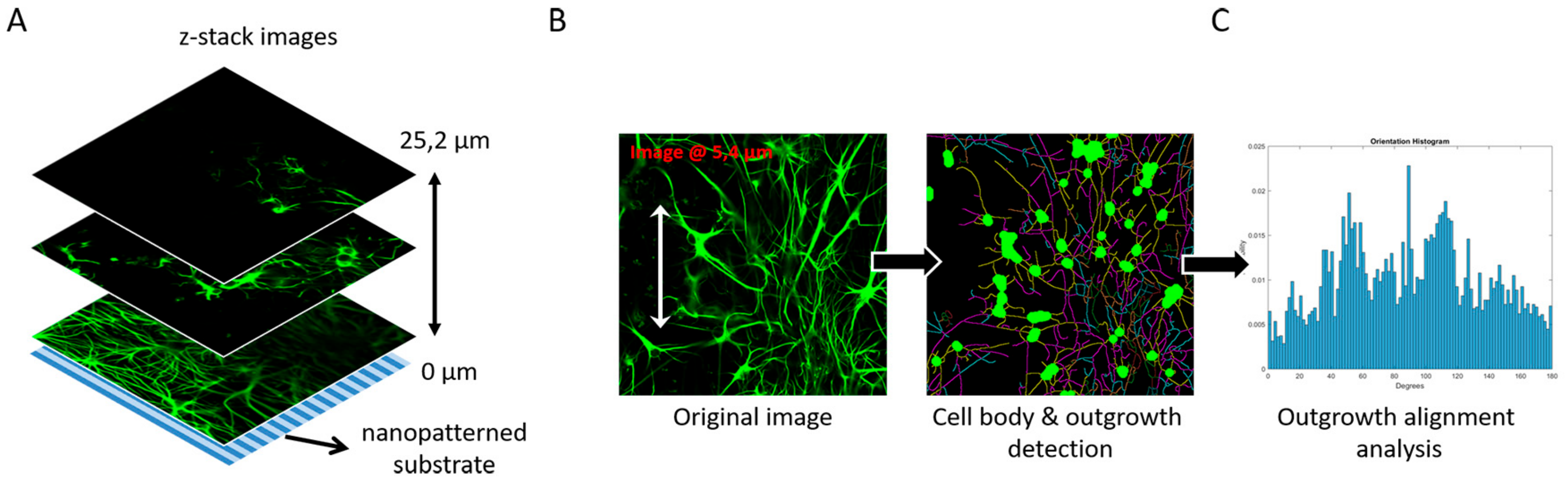
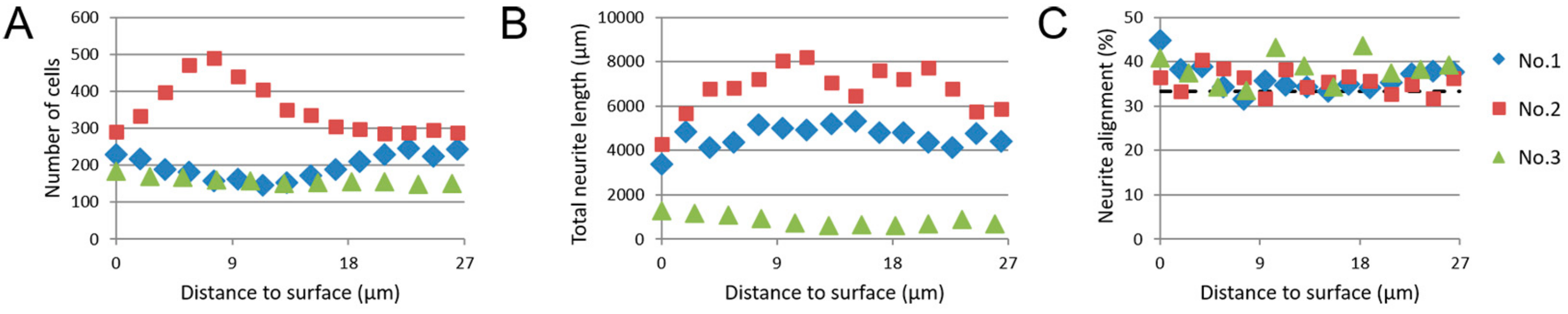
3.4. Image-Based Screening Method Analysis Using 3D CTX Data Set
3.5. Image-Based Screening of 3D SH-SY5Y Cell Cultures
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gribkoff, V.K.; Kaczmarek, L.K. The need for new approaches in CNS drug discovery: Why drugs have failed, and what can be done to improve outcomes. Neuropharmacology 2017, 120, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Finkbeiner, S. Bridging the Valley of Death of therapeutics for neurodegeneration. Nat. Med. 2010, 16, 1227–1232. [Google Scholar] [CrossRef] [PubMed]
- Cummings, J.; Reiber, C.; Kumar, P. The price of progress: Funding and financing Alzheimer’s disease drug development. Alzheimer’s Dement. Transl. Res. Clin. Interv. 2018, 4, 330–343. [Google Scholar] [CrossRef] [PubMed]
- Olanow, C.W.; Kieburtz, K.; Schapira, A.H.V. Why have we failed to achieve neuroprotection in Parkinson’s disease. Ann. Neurol. 2008, 64 (Suppl. 2), S101–S110. [Google Scholar] [CrossRef]
- Sosa-Hernández, J.E.; Villalba-Rodríguez, A.M.; Romero-Castillo, K.D.; Aguilar-Aguila-Isaías, M.A.; García-Reyes, I.E.; Hernández-Antonio, A.; Ahmed, I.; Sharma, A.; Parra-Saldívar, R.; Iqbal, H.M.N. Organs-on-a-Chip module: A review from the development and applications perspective. Micromachines 2018, 9, 536. [Google Scholar] [CrossRef] [PubMed]
- Moraes, C.; Mehta, G.; Lesher-Perez, S.C.; Takayama, S. Organs-on-a-Chip: A focus on compartmentalized microdevices. Ann. Biomed. Eng. 2012, 40, 1211–1227. [Google Scholar] [CrossRef] [PubMed]
- Frimat, J.-P.; Xie, S.; Bastiaens, A.; Schurink, B.; Wolbers, F.; den Toonder, J.; Luttge, R. Advances in 3D neuronal cell culture. J. Vac. Sci. Technol. B Nanotechnol. Microelectron. Mater. Process. Meas. Phenom. 2015, 33, 06F902. [Google Scholar] [CrossRef] [Green Version]
- Mountcastle, V. The columnar organization of the neocortex. Brain 1997, 120, 701–722. [Google Scholar] [CrossRef] [Green Version]
- Oberlaender, M.; De Kock, C.P.J.; Bruno, R.M.; Ramirez, A.; Meyer, H.S.; Dercksen, V.J.; Helmstaedter, M.; Sakmann, B. Cell type-specific three-dimensional structure of thalamocortical circuits in a column of rat vibrissal cortex. Cereb. Cortex 2012, 22, 2375–2391. [Google Scholar] [CrossRef]
- Shipp, S. Structure and function of the cerebral cortex. Curr. Biol. 2007, 17, R443–R449. [Google Scholar] [CrossRef] [Green Version]
- Lancaster, M.A.; Renner, M.; Martin, C.A.; Wenzel, D.; Bicknell, L.S.; Hurles, M.E.; Homfray, T.; Penninger, J.M.; Jackson, A.P.; Knoblich, J.A. Cerebral organoids model human brain development and microcephaly. Nature 2013, 501, 373–379. [Google Scholar] [CrossRef]
- Truskett, V.N.; Watts, M.P.C. Trends in imprint lithography for biological applications. Trends Biotechnol. 2006, 24, 312–317. [Google Scholar] [CrossRef]
- Bremus-Koebberling, E.A.; Beckemper, S.; Koch, B.; Gillner, A. Nano structures via laser interference patterning for guided cell growth of neuronal cells. J. Laser Appl. 2012, 24, 042013. [Google Scholar] [CrossRef] [Green Version]
- Johansson, F.; Carlberg, P.; Danielsen, N.; Montelius, L.; Kanje, M. Axonal outgrowth on nano-imprinted patterns. Biomaterials 2006, 27, 1251–1258. [Google Scholar] [CrossRef]
- Tonazzini, I.; Cecchini, A.; Elgersma, Y.; Cecchini, M. Interaction of SH-SY5Y cells with nanogratings during neuronal differentiation: Comparison with primary neurons. Adv. Healthc. Mater. 2014, 3, 581–587. [Google Scholar] [CrossRef]
- Miller, C.; Jeftinija, S.; Mallapragada, S. Synergistic effects of physical and chemical guidance cues on neurite alignment and outgrowth on biodegradable polymer substrates. Tissue Eng. 2002, 8, 367–378. [Google Scholar] [CrossRef]
- Kim, Y.; Meade, S.M.; Chen, K.; Feng, H.; Rayyan, J.; Hess-Dunning, A.; Ereifej, E.S. Nano-architectural approaches for improved intracortical interface technologies. Front. Neurosci. 2018, 12, 1–20. [Google Scholar] [CrossRef]
- Hoffman-Kim, D.; Mitchel, J.A.; Bellamkonda, R.V. Topography, cell response, and nerve regeneration. Annu. Rev. Biomed. Eng. 2010, 12, 203–231. [Google Scholar] [CrossRef]
- Kim, H.N.; Jiao, A.; Hwang, N.S.; Kim, M.S.; Kang, D.H.; Kim, D.-H.; Suh, K.-Y. Nanotopography-guided tissue engineering and regenerative medicine. Adv. Drug Deliv. Rev. 2013, 65, 536–558. [Google Scholar] [CrossRef]
- Xie, S.; Luttge, R. Imprint lithography provides topographical nanocues to guide cell growth in primary cortical cell culture. Microelectron. Eng. 2014, 124, 30–36. [Google Scholar] [CrossRef]
- Bastiaens, A.J.; Xie, S.; Luttge, R. Investigating the interplay of lateral and height dimensions influencing neuronal processes on nanogrooves. J. Vac. Sci. Technol. B Nanotechnol. Microelectron. Mater. Process. Meas. Phenom. 2018, 36, 06J801. [Google Scholar] [CrossRef] [Green Version]
- Bastiaens, A.J.; Xie, S.; Mustafa, D.A.M.; Frimat, J.-P.; den Toonder, J.M.J.; Luttge, R. Validation and optimization of an image-based screening method applied to the study of neuronal processes on nanogrooves. Front. Cell. Neurosci. 2018, 12, 1–14. [Google Scholar] [CrossRef]
- Xie, S.; Schurink, B.; Wolbers, F.; Luttge, R.; Hassink, G. Nanoscaffold’s stiffness affects primary cortical cell network formation. J. Vac. Sci. Technol. B Nanotechnol. Microelectron. Mater. Process. Meas. Phenom. 2014, 32, 06FD03. [Google Scholar] [CrossRef]
- Dwane, S.; Durack, E.; Kiely, P.A. Optimising parameters for the differentiation of SH-SY5Y cells to study cell adhesion and cell migration. BMC Res. Notes 2013, 6, 366. [Google Scholar] [CrossRef]
- Teppola, H.; Sarkanen, J.R.; Jalonen, T.O.; Linne, M.L. Morphological differentiation towards neuronal phenotype of SH-SY5Y neuroblastoma cells by estradiol, retinoic acid and cholesterol. Neurochem. Res. 2016, 41, 731–747. [Google Scholar] [CrossRef]
- Encinas, M.; Iglesias, M.; Liu, Y.; Wang, H.; Muhaisen, A.; Ceña, V.; Gallego, C.; Comella, J.X. Sequential treatment of SH-SY5Y cells with retinoic acid and brain-derived neurotrophic factor gives rise to fully differentiated, neurotrophic factor-dependent, human neuron-like cells. J. Neurochem. 2000, 75, 991–1003. [Google Scholar] [CrossRef]
- Wiertz, R. Regulation of In Vitro Cell-Cell and Cell-Substrate Adhesion. Ph.D. Thesis, University of Twente, Enschede, The Netherlands, 2010. [Google Scholar]
- Romijn, H.J.; van Huizen, F.; Wolters, P.S. Towards an improved serum-free, chemically defined medium for long-term culturing of cerebral cortex tissue. Neurosci. Biobehav. Rev. 1984, 8, 301–334. [Google Scholar] [CrossRef]
- Agholme, L.; Lindström, T.; Kågedal, K.; Marcusson, J.; Hallbeck, M. An In Vitro model for neuroscience: differentiation of SH-SY5Y cells into cells with morphological and biochemical characteristics of mature neurons. J. Alzheimer’s Dis. 2010, 20, 1069–1082. [Google Scholar] [CrossRef]
- Xie, S. Brain-On-A-Chip Integrated Neuronal Networks. Ph.D. Thesis, University of Twente, Enschede, The Netherlands, 2016. [Google Scholar]
- Chua, J.S.; Chng, C.P.; Moe, A.A.K.; Tann, J.Y.; Goh, E.L.K.; Chiam, K.H.; Yim, E.K.F. Extending neurites sense the depth of the underlying topography during neuronal differentiation and contact guidance. Biomaterials 2014, 35, 7750–7761. [Google Scholar] [CrossRef]
- Kang, K.; Park, Y.S.; Park, M.; Jang, M.J.; Kim, S.M.; Lee, J.; Choi, J.Y.; Jung, D.H.; Chang, Y.T.; Yoon, M.H.; et al. Axon-first neuritogenesis on vertical nanowires. Nano Lett. 2016, 16, 675–680. [Google Scholar] [CrossRef]
- Ferrari, A.; Cecchini, M.; Dhawan, A.; Micera, S.; Tonazzini, I.; Stabile, R.; Pisignano, D.; Beltram, F. Nanotopographic control of neuronal polarity. Nano Lett. 2011, 11, 505–511. [Google Scholar] [CrossRef]
- Boutin, M.E.; Hoffman-Kim, D. Application and assessment of optical clearing methods for imaging of tissue-engineered neural stem cell spheres. Tissue Eng. Part C Methods 2014, 21, 292–302. [Google Scholar] [CrossRef]
- Grist, S.M.; Nasseri, S.S.; Poon, T.; Roskelley, C.; Cheung, K.C. On-chip clearing of arrays of 3-D cell cultures and micro-tissues. Biomicrofluidics 2016, 10, 044107. [Google Scholar] [CrossRef] [Green Version]
- Biran, R.; Noble, M.D.; Tresco, P.A. Directed nerve outgrowth is enhanced by engineered glial substrates. Exp. Neurol. 2003, 184, 141–152. [Google Scholar] [CrossRef]
- Alexander, J.K.; Fuss, B.; Colello, R.J. Electric field-induced astrocyte alignment directs neurite outgrowth. Neuron Glia Biol. 2006, 2, 93–103. [Google Scholar] [CrossRef] [Green Version]
- Routh, B.N.; Johnston, D.; Harris, K.; Chitwood, R.A. Anatomical and electrophysiological comparison of CA1 pyramidal neurons of the rat and mouse. J. Neurophysiol. 2009, 102, 2288–2302. [Google Scholar] [CrossRef]
- Price, J.; Hynes, R.O. Astrocytes in culture synthesize and secrete a variant form of fibronectin. J. Neurosci. 1985, 5, 2205–2211. [Google Scholar] [CrossRef] [Green Version]
- Liesi, P.; Kirkwood, T.; Vaheri, A. Fibronectin is expressed by astrocytes cultured from embryonic and early postnatal rat brain. Exp. Cell Res. 1986, 163, 175–185. [Google Scholar] [CrossRef]
- Neugebauer, K.M.; Tomaselli, K.J.; Lilien, J.; Reichardt, L.F. N-cadherin, NCAM, and integrins promote retinal neurite outgrowth on astrocytes In Vitro. J. Cell Biol. 1988, 107, 1177–1187. [Google Scholar] [CrossRef]
- Lois, C.; García-Verdugo, J.M.; Alvarez-Buylla, A. Chain migration of neuronal precursors. Science 1996, 271, 978–981. [Google Scholar] [CrossRef]
- Snow, D.M.; Lemmon, V.; Carrino, D.A.; Caplan, A.I.; Silver, J. Sulfated proteoglycans in astroglial barriers inhibit neurite outgrowth in vitro. Exp. Neurol. 1990, 109, 111–130. [Google Scholar] [CrossRef]
- Schurink, B.; Luttge, R. Hydrogel/poly-dimethylsiloxane hybrid bioreactor facilitating 3D cell culturing. J. Vac. Sci. Technol. B Nanotechnol. Microelectron. Mater. Process. Meas. Phenom. 2013, 31, 06F903. [Google Scholar] [CrossRef]
- Bastiaens, A.J.; Frimat, J.-P.; van Nunen, T.; Schurink, B.; Homburg, E.F.G.A.; Luttge, R. Advancing a MEMS-based 3D cell culture system for in vitro neuro-electrophysiological recordings. Front. Mech. Eng. 2018, 4, 1–10. [Google Scholar] [CrossRef]
- Pirlo, R.K.; Sweeney, A.J.; Ringeisen, B.R.; Kindy, M.; Gao, B.Z. Biochip/laser cell deposition system to assess polarized axonal growth from single neurons and neuron/glia pairs in microchannels with novel asymmetrical geometries. Biomicrofluidics 2011, 5, 13408. [Google Scholar] [CrossRef] [Green Version]
- Choi, S.H.; Kim, Y.H.; Hebisch, M.; Sliwinski, C.; Lee, S.; D’Avanzo, C.; Chen, H.; Hooli, B.; Asselin, C.; Muffat, J.; et al. A three-dimensional human neural cell culture model of Alzheimer’s disease. Nature 2014, 515, 274–278. [Google Scholar] [CrossRef]
- Xu, T.; Gregory, C.A.; Molnar, P.; Cui, X.; Jalota, S.; Bhaduri, S.B.; Boland, T. Viability and electrophysiology of neural cell structures generated by the inkjet printing method. Biomaterials 2006, 27, 3580–3588. [Google Scholar] [CrossRef]
- Suri, S.; Han, L.H.; Zhang, W.; Singh, A.; Chen, S.; Schmidt, C.E. Solid freeform fabrication of designer scaffolds of hyaluronic acid for nerve tissue engineering. Biomed. Microdevices 2011, 13, 983–993. [Google Scholar] [CrossRef]
- Lozano, R.; Stevens, L.; Thompson, B.C.; Gilmore, K.J.; Gorkin, R.; Stewart, E.M.; in het Panhuis, M.; Romero-Ortega, M.; Wallace, G.G. 3D printing of layered brain-like structures using peptide modified gellan gum substrates. Biomaterials 2015, 67, 264–273. [Google Scholar] [CrossRef]
- Eiraku, M.; Watanabe, K.; Matsuo-Takasaki, M.; Kawada, M.; Yonemura, S.; Matsumura, M.; Wataya, T.; Nishiyama, A.; Muguruma, K.; Sasai, Y. Self-organized Formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell 2008, 3, 519–532. [Google Scholar] [CrossRef]
- Qian, X.; Song, H.; Ming, G.L. Brain organoids: Advances, applications and challenges. Development 2019. [Google Scholar] [CrossRef]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bastiaens, A.; Xie, S.; Luttge, R. Nanogroove-Enhanced Hydrogel Scaffolds for 3D Neuronal Cell Culture: An Easy Access Brain-on-Chip Model. Micromachines 2019, 10, 638. https://doi.org/10.3390/mi10100638
Bastiaens A, Xie S, Luttge R. Nanogroove-Enhanced Hydrogel Scaffolds for 3D Neuronal Cell Culture: An Easy Access Brain-on-Chip Model. Micromachines. 2019; 10(10):638. https://doi.org/10.3390/mi10100638
Chicago/Turabian StyleBastiaens, Alex, Sijia Xie, and Regina Luttge. 2019. "Nanogroove-Enhanced Hydrogel Scaffolds for 3D Neuronal Cell Culture: An Easy Access Brain-on-Chip Model" Micromachines 10, no. 10: 638. https://doi.org/10.3390/mi10100638




