Abstract
The current status of multi-mycotoxin contamination in edible and medicinal plants demands urgent development of high-throughput analytical methods for mycotoxin detection. In this study, a reliable and sensitive method for the simultaneous analysis of 73 mycotoxins was established and successfully applied to detect mycotoxins in 260 samples of four dual-purpose plants (lotus seed, coix seed, licorice root, and dried tangerine peel). Sample preparation involved optimized QuEChERS (Quick, Easy, Cheap, Effective, Rugged, and Safe) extraction combined with liquid–liquid extraction purification, and an enhanced ion pair library was established to reduce matrix interference and improve the method’s universality. Method validation demonstrated recovery rates ranging from 61.6% to 118.6% for all compounds, with relative standard deviations (RSDs) below 15%. The limits of detection (LODs) and quantification (LOQs) ranged from 0.25–12.25 μg/kg and 0.5–25 μg/kg, respectively. Based on the contamination analysis and health risk assessment using Margin of Exposure (MOE) and Hazard Index (HI) methods, we found that multi-mycotoxin contamination is highly prevalent in edible and medicinal plants, with different components being susceptible to invasion by distinct fungal genera. Seed-type plants showed high susceptibility to Aspergillus (53.3%) and Fusarium (22.2%) contamination, with MOE values below 10,000 for aflatoxins indicating potential health risks. Physical state and good storage conditions significantly influenced contamination levels, with fragmented samples showing substantially higher mycotoxin levels. Additionally, mycotoxins with associated biosynthetic metabolic pathways were frequently detected simultaneously in highly contaminated samples. Based on these findings, we recommend implementing strict moisture control during storage, maintaining intact product form where possible, and establishing comprehensive supplier qualification systems. This study provides valuable reference for monitoring mycotoxin contamination in similar plants.
Keywords:
mycotoxins; edible and medicinal plants; QuEChERS; UHPLC-MS/MS; contamination status; health risk assessment Key Contribution:
This study developed an optimized QuEChERS method and effectively applied it to four edible and medicinal plants, encompassing a comprehensive range of monitored and regulated mycotoxins, totaling 73 different types under surveillance. Multiple mycotoxin contamination is widespread in dual-purpose plants, and mycotoxins with associated biosynthetic metabolic pathways were frequently co-detected in highly contaminated samples. Seeds demonstrated higher health risks, showing susceptibility to aflatoxins and zearalenone contamination. The exceptionally high contamination rate of fumonisins in coix seeds suggests the need for continuous monitoring.
1. Introduction
Mycotoxins, as secondary metabolites produced by fungi, readily contaminate various food matrices, including foodstuffs, oils, and traditional Chinese medicines (TCMs) [1,2,3]. To date, over 400 mycotoxins have been identified and isolated, with several, such as aflatoxins and ochratoxins, being demonstrated to possess severe toxic effects, including the induction of hepatocellular carcinoma and various diseases affecting the urinary and gastrointestinal systems [1,4,5]. Furthermore, co-contamination by multiple mycotoxins may result in synergistic toxic effects. For instance, the combined toxicity of aflatoxin B1, zearalenone, and deoxynivalenol mixture exhibits enhanced hepatotoxicity in rat hepatocytes compared to their individual effects. Furthermore, certain matrices are susceptible to contamination by both masked and emerging mycotoxins [6,7,8]. These circumstances pose significant threats to public health [9,10,11]. Consequently, numerous countries and organizations have established limits for mycotoxins. For example, the European Commission recently issued Commission Regulation (EU) No 2023/915 of 25 April 2023 on the maximum levels for certain contaminants in food, repealing Regulation (EC) No 1881/2006, which now stipulates maximum levels for sixteen mycotoxins (including three newly added ones) in foodstuffs.
Globally, over two billion people rely on TCMs for their health benefits and, with the increasing emphasis on health and wellness, the consumption of edible TCMs has risen substantially [12,13,14,15]. However, regulatory standards for mycotoxin limits in plants lag behind those for food products, primarily due to insufficient detection methods, contamination data, and related risk assessments. Current high-throughput mycotoxin detection methods predominantly focus on food matrices, typically employing a QuEChERS or “dilute and shoot” method for sample preparation. For instance, Michael Sulyok et al. developed a method combining direct extraction with LC-MS/MS to determine 39 mycotoxins in wheat and corn [16,17]. Similarly, Ádám Tölgyesi et al. developed a novel LC-MS/MS multi-method for the simultaneous determination of 295 food contaminants in cereals, including 266 pesticides, 12 mycotoxins, 14 alkaloid toxins, and three Alternaria toxins [18,19,20].
Compared to food matrices, edible and medicinal plants present unique analytical challenges as dried materials containing numerous metabolites, some structurally similar to mycotoxins. This complexity creates significant matrix interference for trace mycotoxin detection, affecting methods’ sensitivity and accuracy. When applying food-based high-throughput detection methods to plants, several limitations emerge. For example, Zhao et al. observed significant matrix interference affecting mycotoxin recovery rates in nutmeg, galangal, and coix seeds using a “dilute and shoot” method [21]. When applying the QuEChERS methodology, certain aminopropyl (NH2), primary secondary amine (PSA) cleanup sorbents, while effective at removing fatty acids and organic acids, can inadvertently adsorb acid-sensitive toxins containing carboxyl groups, resulting in reduced recovery rates [21,22]. To minimize matrix interference and achieve higher recovery rates and sensitivity, combining different sample preparation methods to leverage their respective advantages presents a viable solution [23]. For instance, Nouri and Sereshti developed a rapid method combining SPE with DLLME for determining aflatoxins in soybeans [24].
Currently, research on mycotoxin contamination distribution patterns primarily focuses on food matrices and typically examines only a few specific mycotoxins or mycotoxin classes. For example, Abirami Ramu Ganesan et al. investigated the distribution patterns of Ochratoxin A and deoxynivalenol in agricultural products and related foods, while Sun et al. studied the contamination profiles of aflatoxins, ochratoxins, and fumonisins in Chinese rice [25,26]. However, there is limited research exploring the potential correlations between edible and medicinal plants and their specific mycotoxin contamination.
Chemical compounds with interactive effects may exhibit lower or higher toxic effects compared to individual substances, necessitating cumulative exposure risk assessment for multiple chemical compounds [27]. Consequently, cumulative exposure assessment methods are more appropriate for evaluating multi-mycotoxin contamination in matrices. The main cumulative exposure assessment methods include the Margin of Exposure (MOE), Hazard Index (HI), Relative Potency Factor (RPF), and Point of Departure (POD). The RPF requires similar toxicological targets, exposure routes, and duration among components in chemical mixtures, making it unsuitable for assessing contamination by diverse mycotoxin types. Additionally, there is no internationally standardized evaluation method for the POD approach. Therefore, the MOE and HI are currently the primary methods employed for cumulative mycotoxin exposure assessment. The European Food Safety Authority (EFSA) has specifically identified the MOE as the most suitable approach for evaluating genotoxic carcinogens [28]. For example, Zhang et al. applied both MOE and HI methods to assess different mycotoxins in dual-purpose plants such as coix seed and lotus seed based on their toxicity profiles [29]. Similarly, Lu et al. utilized the HI method to evaluate 31 mycotoxins in six edible and medicinal plants [30].
This study encompasses 73 mycotoxins produced by major toxigenic fungi, including Fusarium, Claviceps, Alternaria, and Penicillium species [2] (Table 1). The coverage extends to regulated mycotoxins, their associated masked forms, and emerging mycotoxins such as Enniatins and Beauvericin, aiming to provide comprehensive contamination data. Considering exposure levels, four commonly used edible and medicinal plants were selected as research subjects: lotus seed (LS), coix seed (CS) [14,31,32], licorice root (LR), commonly used as a sweetener [33], and dried tangerine peel, named “chenpi” in China (CP), often preserved as candied fruit [34]. CP can be stored for decades and its source material (tangerines) is particularly susceptible to fungal contamination.

Table 1.
Chemical Information of 73 mycotoxins.
This study developed a robust, high-throughput analytical method for these 73 mycotoxins by combining optimized QuEChERS with liquid–liquid extraction and establishing a more comprehensive ion pair library. This method was successfully applied to four edible and medicinal plants, enabling a detailed analysis of their contamination levels and characteristics. A risk assessment for Chinese populations was conducted using both the MOE and HI approaches. The findings provide valuable reference data for mycotoxin risk assessment in edible and medicinal plants and the development of relevant regulatory standards.
2. Results and Discussion
2.1. Method Optimization
2.1.1. Optimization of UHPLC-MS/MS Conditions
At the beginning of this study, mass spectrometric conditions from previous literature were referenced, including the detection of 191 mycotoxins reported by Elisabeth Varga et al. and 41 mycotoxins reported by Ann-Kristin Rausch et al. [35,36]. When applied to herbal medicine matrices, significant matrix interference was observed near some target peaks. However, this issue could be effectively resolved by modifying the MRM transitions. This demonstrates that differences in matrices require the consideration of ion pair specificity rather than merely ion response intensity. Subsequently, standard solutions of 73 mycotoxins (dissolved in 50% methanol at 500 ng/mL) were individually injected into the MS/MS system at a constant flow rate of 5 μL/min. The Analyst 1.5.1 software was used to compare and select the optimal precursor and product ions. For each mycotoxin, 3–5 ion pairs were optimized to enhance method applicability (Table 2). As shown in Figure 1, for AFB1 quantification in licorice ([M+H]+), the optimal MRM transitions were 313.0 > 241.0 and 313.0 > 269.0, while for AFB1 in tangerine peel, they were 313.0 > 241.0 and 313.0 > 285.1. Notably, although the product ion transition 313.0 > 285.1 exhibited higher intensity, undesirable interference peaks were observed near the AFB1 peak (m/z 313.0 > 285.1) in the LR. The establishment of a more comprehensive MRM transition library significantly improved the method’s versatility. To our knowledge, such an extensive ion pair spectral library for more than 70 mycotoxins has not been previously reported.

Table 2.
Optimized MS/MS parameters for the analytes studied.
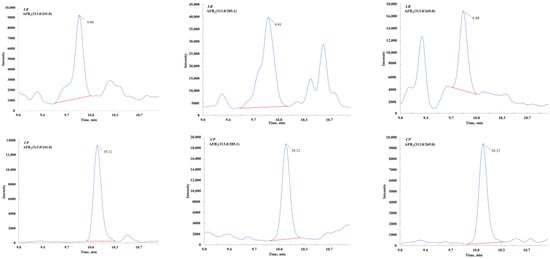
Figure 1.
The optimal ion pairs in different plants for certain mycotoxin.
Due to the significant matrix effects in Chinese herbal medicines and the large number of target analytes, the optimized chromatographic conditions were designed to achieve maximum response intensity and optimal resolution for all analytes. Following Elisabeth Varga’s approach, chromatographic separation was performed in both positive and negative ionization modes [36]. Since more mycotoxins were separated in the positive mode, methanol (MeOH) was selected as the organic phase due to its relatively weaker elution strength, enabling better separation. Various modifiers, including formic acid, acetic acid, ammonium formate, and ammonium acetate, were evaluated to enhance ionization efficiency. The addition of 0.4% formic acid improved the response of many mycotoxins, particularly fumonisins and ochratoxins. Ammonium formate supported better peak shapes through the formation of [M+NH4]+ adducts. The optimal concentration was determined to be 2 mM, as higher concentrations (5 mM) led to ionization suppression (e.g., for ochratoxin A). In negative mode, with only 9 mycotoxins being detected, switching the organic phase from methanol to acetonitrile improved peak shapes and enhanced sensitivity without requiring modifiers. Additionally, the liquid chromatographic gradient, column temperature, and flow rate were optimized. The final mobile phases consisted of water–acetonitrile (A/B) with 0.4% formic acid and 2 mM ammonium formate for the positive mode and water–acetonitrile (A/B) for the negative mode. Based on previous research, a core-shell column (Poroshell EC-C18) was selected for its low column pressure and superior separation performance [37]. Although separating the positive and negative ionization modes sacrificed some analytical efficiency, this approach provided a better resolution when analyzing edible and medicinal plant samples, avoiding interference from matrix components and achieving a higher sensitivity. This method demonstrates broader applicability across similar matrix types.
2.1.2. Optimization of Sample Preparation
In 2006, Sulyok et al. first developed an LC-MS/MS method for multi-mycotoxin determination, using direct dilution to analyze 39 mycotoxins in cereals [17]. However, the applicability of this simplified method to plants remained uncertain. We selected LR as the model sample for preparation optimization due to its significant matrix interference. Accuracy evaluation of the “dilute and shoot” method was performed using spiked LR samples (mixed standard solution added to blank LR samples, left overnight at room temperature in a fume hood to better simulate actual mycotoxin contamination). Results indicated that the extraction solvent (acetonitrile/water/acetic acid, 79:20:1, v/v/v) was not compatible with all mycotoxins and matrix interference affected accurate quantification. Alternative extraction solvents were explored to enhance the selectivity and reduce interference, comparing extraction systems composed of formic acid, acetic acid, or citric acid buffer–acetonitrile. As no significant differences were observed among these systems, we maintained the “dilute and shoot” method’s extract solvent system for operational simplicity.
The salting-out step in QuEChERS is commonly used to remove some polar impurities, organic compounds, and proteins. To address the complexity of edible and medicinal plants, we introduced a simplified salting-out step to reduce matrix interference which proved effective in three tested plant matrices (Figure 2). Further comparison of sodium chloride, sodium acetate, and sodium citrate salt packets revealed that anhydrous sodium citrate stabilized solution pH, improving recovery rates of acid-sensitive mycotoxins by 5–8%, consistent with expectations (Figure 3). Conversely, sodium acetate decreased acidity, causing some losses of these mycotoxins.
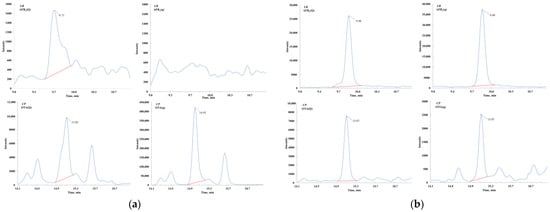
Figure 2.
Comparation of chromatograms for the salt-out procedure. (a) Chromatography of quantitative (Q) and qualitative (q) ion pairs for sample without salt-out procedure; (b) chromatography of quantitative (Q) and qualitative (q) ion pairs for sample with salt-out procedure.
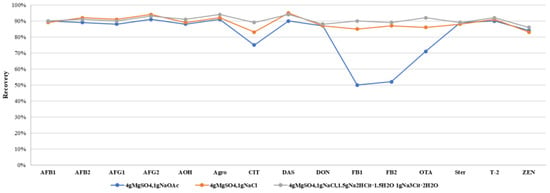
Figure 3.
Recoveries of representative mycotoxins in different salt package.
Innovatively, unlike conventional QuEChERS, we separated the extract from the matrix before adding the aqueous solution for salting-out to minimize the co-extraction of interferents. The effects of water and 5% formic acid solution on mycotoxin recovery were investigated, with a 5% formic acid solution yielding satisfactory recovery rates (70–120%) for most of the mycotoxins.
A challenging issue arose with LS samples, which formed white precipitates during 4 °C storage after processing, affecting measurement accuracy and necessitating effective cleanup. Given the high content of starch, protein, and lipids in lotus seeds, various dispersive solid-phase extraction (d-SPE) sorbents were evaluated, including graphitized carbon black (GCB), enhanced matrix removal-lipid (EMR-Lipid), octadecyl silane (C18), aminopropyl (NH2), primary secondary amine (PSA), silica (Si), neutral aluminum oxide (Al-N), carboxymethyl (CBA), diethylaminopropyl (DEA), and cyanopropyl (CN-U). These sorbents were combined with MgSO4 (100 mg:900 mg), but none met the requirements due to their poor recovery of important mycotoxins or failure to resolve precipitation issues.
Inspired by Hyun-Deok Cho et al.’s work using hexane for preliminary lipid removal before immunoaffinity column cleanup [22], we modified the approach using cyclohexane. Adding 12.0 mL cyclohexane to 6.0 mL extract significantly improved the precipitation issue while minimally affecting mycotoxin recovery, with only 1–2 mycotoxins showing losses around 7.8% and others below 1.8%. Ultimately, liquid–liquid extraction with cyclohexane was adopted as the cleanup method.
2.2. Method Validation
Method validation was performed on three different edible and medicinal plants (LS, LR, and CP), evaluating key analytical parameters including the linearity, accuracy, limits of detection (LOD), limits of quantification (LOQ), and precision. The comprehensive validation data are summarized in Table A1, Table A2 and Table A3.
2.2.1. Linearity
Due to matrix effects exceeding ±20% for most mycotoxins, matrix-matched calibration curves were necessary for accurate quantification. Blank sample extracts after nitrogen evaporation were reconstituted with 0.5 mL acetonitrile, followed by the addition of varying amounts of mixed standard stock solutions. The solutions were then made up to 2 mL with solvent (acetonitrile/water/acetic acid, 20:79:1). Three concentration ranges were prepared: G1 (0.1, 0.5, 1, 5, 10, 20, and 50 ng/mL), G2 (0.5, 2.5, 5, 25, 50, 100, and 250 ng/mL), and G3 (2.5, 12.5, 25, 125, 250, 500, and 1250 ng/mL). Calibration curves were constructed using peak area versus concentration relationships. All mycotoxins demonstrated good linearity with correlation coefficients (r) greater than 0.998.
2.2.2. Method Limit of Quantification (LOQ) and Limit of Detection (LOD)
Spiking experiments were conducted to determine the method’s quantification limits (LOQs) for each matrix. At spiking levels of 0.5 μg/kg (calculated as AFB1), LS and CP samples met requirements for signal-to-noise ratio, recovery, and precision. However, LR samples required a higher LOQ of 1.0 μg/kg (calculated as AFB1), which better reflected actual sample conditions. The LOQs for the three matrices ranged from 0.5 to 25.0 µg/kg, as shown in Table A1, Table A2 and Table A3. Despite using generic extraction and cleanup procedures, the method achieved lower LOQs for several mycotoxins compared to existing reports [29,38]. The LOQs were significantly below the maximum residue limits (MRLs) set by Commission Regulation (EU) No 2023/915. For example, the LOQ for FB1 and FB2 was 2.5 µg/kg, well below the MRL of 200 µg/kg, demonstrating the method’s suitability for regulatory monitoring of these edible and medicinal plants. Limits of detection (LODs) were determined at spiking levels of 0.25 μg/kg (calculated as AFB1) for LS and 0.5 μg/kg (calculated as AFB1) for CP and LR.
During the method’s development, matrix interference for certain mycotoxins in several plants remained unresolved. Consequently, some mycotoxins, such as tenuazonic acid, were excluded from the final method and require further optimization.
2.2.3. Method Accuracy and Precision
In the absence of certified reference materials, the method’s accuracy was evaluated using recovery rates (obtained by spiking known amounts of analytes into blank matrices). Recovery studies were performed at three concentration levels in three blank matrices (n = 6): 1.0 μg/kg (Level 1), 5.0 μg/kg (Level 2), and 10.0 μg/kg (Level 3) (calculated as AFB1). The recovery rates for the 73 target analytes ranged from 61.6% to 116.4%, with RSDs less than 14.9%. These results largely comply with European Commission Regulation (EC) No 401/2006, indicating the satisfactory accuracy and precision of the method.
2.3. Mycotoxin Contamination of Edible and Medicinal Plants
The established analytical method was applied to analyze 260 batches of four different edible and medicinal plants to characterize their mycotoxin contamination patterns and summarize the distinct contamination characteristics across different plants.
2.3.1. Lotus Seed (LS)
Lotus seeds have a 7000-year history as a vegetable, functional food, and medicinal herb. China is the world’s largest lotus root cultivator and consumer, with a cultivation area of 200,000 hectares [31]. By 2017, Fujian Province’s annual lotus seed production reached 12,205 tons, contributing approximately 1.8 billion RMB to the country’s GDP [39].
This study analyzed twenty-nine LS samples, including nine special samples (LS28-36): fresh powder (LS31), moldy powder (LS35), discolored powder (LS36), three farm-cultivated powders (LS32-34), and three commercial medicinal samples (LS28-30). In total, 17 mycotoxins were detected in the samples (Table A4), with an 86.2% detection rate, primarily produced by Aspergillus species (Figure 4 and Figure 5). Notably, LS showed the highest aflatoxin contamination rate (41.4%) among the four studied plants, at 34.5%, exceeding the Chinese Pharmacopoeia (Ch.P) limits. Three samples—the moldy, discolored, and one farm-cultivated sample—contained AFB1 levels up to 4000 μg/kg (Figure 6), indicating rapid aflatoxin accumulation in deteriorated lotus seeds to alarming levels.
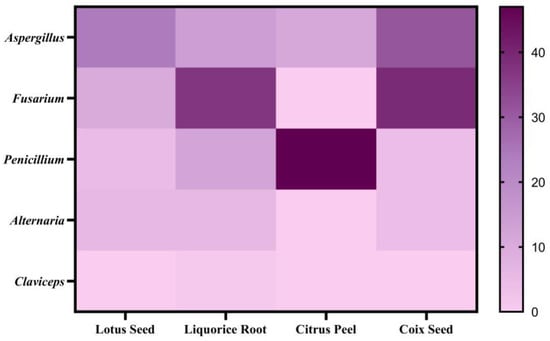
Figure 4.
Distribution of susceptible fungi types of four plants.
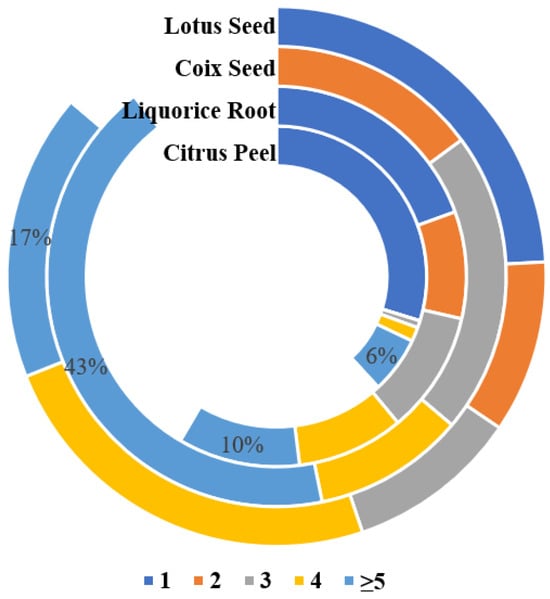
Figure 5.
Content percentage of co-occurrence number of mycotoxins in four matrices.
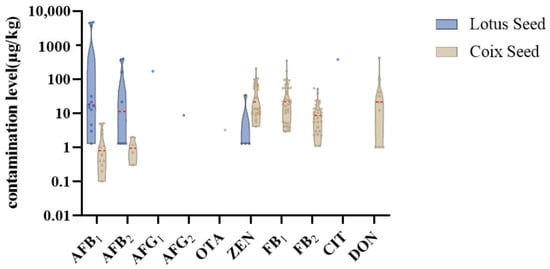
Figure 6.
Comparison of the concentration of the regulated mycotoxins in lotus seed and coix seed.
The data revealed that highly aflatoxin-contaminated samples frequently contained related metabolites such as AFM1, AFM2, Ster, and O-m-ster. Interestingly, AFM1 and AFM2, previously reported only in milk as AFB1 metabolites in animals, had never been detected in herbs and spices [40], suggesting possible non-animal AFB1 metabolism pathways worthy of further investigation. Additionally, CPA levels exceeded 10,000 μg/kg in these samples, confirming previous reports of AF-CPA co-occurrence and increased toxicity risks [41,42]. Research suggests CPA may serve as a fungal colonization signal molecule, related to its calcium ion-inhibitory activity [43]. Our finding of CPA as the sole mycotoxin in fresh lotus seeds partially supports this hypothesis.
The significance analysis of mycotoxin contamination in LS of different forms was conducted using the Mann–Whitney U test. Statistical analysis revealed significantly higher detection rates of AFB1, AFB2, AFM1, AFM2, CIT, CPA, and O-m-Ster in powder form compared to the original form (p < 0.05). No significant differences were observed in the contamination levels of other mycotoxins between the two forms (Figure 7 and Figure 8). The average increase in detection rates was calculated to be 24.7%. Additionally, only discolored lotus seeds contained CIT, a nephrotoxic mycotoxin produced by Aspergillus, Penicillium, or related fungi, possibly explaining the color change. Therefore, appearance may serve as an important quality indicator for lotus seeds.
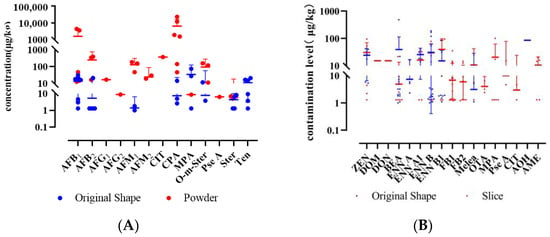
Figure 7.
Comparison of mycotoxin levels (mean with 95% CI) between intact and processed forms of LS (A) and LR (B).
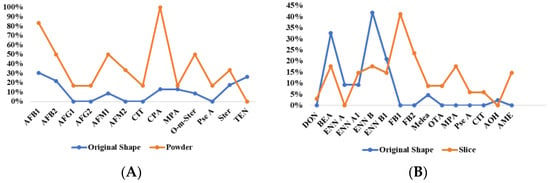
Figure 8.
Comparison of mycotoxin detection rates between intact and processed forms of LS (A) and LR (B).
2.3.2. Licorice Root (LR)
LR, derived from the dried roots and rhizomes of Glycyrrhiza uralensis Fisch, G. glabra L., or G. inflata Bat., appears in approximately 60% of TCM prescriptions due to its complementary properties [37]. As one of China’s most widely used herbs, LR is included in multiple pharmacopeias, including Chinese, Korean, European, and United States Pharmacopeias, due to its widespread global use for its sweet taste.
Among 77 samples, 18 mycotoxins were detected, with an overall detection rate of 59.7% (Table A5). The concerning OTA showed a low detection rate of 3.9%, with no samples exceeding the European Pharmacopoeia 11.0 limit (20 μg/kg), while ZEN was detected in 15.6% of samples, indicating potential risks. Contrary to previous reports of high OTA occurrence in licorice [44], 47 samples (LR31-77) from five Chinese regions (Xinjiang, Inner Mongolia, Gansu, Jilin, and Ningxia) showed no OTA contamination, suggesting a possible geographical difference between European and Chinese cultivation regions. Using the same Mann–Whitney U test, sliced LR showed higher contamination levels and detection rates of regulated mycotoxins (FB1, FB2, OTA, MPA, Pse A, CIT, and AME) compared to raw materials (p < 0.05), with increases of 3.7% to 42.1% (Figure 7 and Figure 8).
ENNs and BEA were the predominant contaminants in LR, typically co-occurring due to their similar chemical structures produced by Fusarium species. The four enniatins consistently showed a concentration pattern of ENN B > ENN B1 > ENN A1 > ENN A. Research suggests that different Fusarium species preferentially incorporate specific amino acids to biosynthesize certain ENNs, explaining why some ENNs can only be isolated from specific fungal strains [45]. The consistent concentration pattern observed in this study suggests contamination by a single Fusarium species.
This study analyzed 47 batches of GR samples (GR31-77) that were collected and processed between 2015 and 2020. All samples were maintained in a temperature-controlled storage facility (≤20 °C). Statistical analysis using the Kruskal–Wallis test (α = 0.05) revealed no significant temporal differences in mycotoxin levels, with the exception of beauvericin (BEA), enniatin A (ENN A), and enniatin A1 (ENN A1). This indicates that strict collection and storage management with controlled environmental conditions effectively reduces mycotoxin contamination in LR.
2.3.3. Dried Tangerine Peel (CP)
CP, derived from mature fruit peels of Citrus reticulata Blanco and its cultivars, is not only one of the most renowned TCMs but also serves as an ingredient in fermented foods [34]. Studies suggest that the quality improves with storage duration [46]. Given its extended storage requirements–typically over three years before use as a medicinal herb–CP faces potential mycotoxin contamination risks. However, multi-mycotoxin contamination in CP has not been extensively documented.
Surprisingly, CP showed the lowest contamination risk among the four matrices studied. Only five mycotoxins were detected in 131 samples, with a detection rate of 38.2%, primarily from Penicillium species (Figure 4) (Table A6). Consistent with previous research, MPA showed the highest detection rate (35.9%), mainly produced by Penicillium [47], confirming citrus fruits’ susceptibility to Penicillium contamination [48]. Similar to LR, BEA-positive samples showed concurrent ENN detection, following the same contamination pattern of ENN B > ENN B1 > ENN A1, suggesting possible contamination by the same Fusarium species. However, our findings differ from previous studies on fresh citrus peel fungal communities, which identified Erythrobasidium, Penicillium, Aspergillus, Rhodotorula, and Mycosphaerella as the dominant genera, with the rare detection of Fusarium [49]. The low levels of BEA and ENNs in CP suggest initial field contamination by multiple fungi including Fusarium, Penicillium, and Aspergillus, with Fusarium gradually being replaced by other dominant fungi.
2.3.4. Coix Seed (CS)
CS is a widely used medicinal and edible grain that has gained popularity as a health food, especially among women, for its properties in eliminating dampness and reducing swelling. It is increasingly consumed as a daily beverage alternative to coffee.
Among 47 samples, 27 mycotoxins were detected, with a total detection rate of 89.4% (Table A7). The analysis of mycotoxin-producing fungi revealed that CS was most susceptible to Aspergillus and Fusarium contamination (Figure 4). Fusarium mycotoxins showed the highest detection rates, with FBs (2.9–430.7 μg/kg) at 74.4% and ZEN (4.1–206.9 μg/kg) at 59.6%. Additionally, AFs showed a significant detection rate of 27.7%, validating the necessity of aflatoxin and zearalenone limits in coix seeds as specified in the Ch.P.
Consistent with previous literature reports, CS showed significant multi-mycotoxin contamination, including both parent mycotoxins and their modified forms [12]. The most severely contaminated sample contained 17 different mycotoxins, with over 50% of samples containing at least four mycotoxins (Figure 5). Samples with high levels of parent mycotoxins often showed a concurrent detection of their modified forms. For example, sample CS3 contained ZEN along with ZAN, α-ZEL, β-ZEL, and α-ZAL, while sample CS6 showed both DON and 3-Ac-DON. Modified mycotoxins showed lower detection rates and contamination levels compared to their parent compounds.
Interestingly, ENNs, which were common contaminants in the other three matrices and are produced by Fusarium, were not detected in CS samples. This might be related to differences in the dominant fungal species colonizing CS, suggesting possible competitive relationships among fungi.
2.4. Health Risk Assessment
A health risk assessment was performed according to the guidelines established by the International Agency for Research on Cancer (IARC) and the Joint FAO/WHO Expert Committee on Food Additives (JECFA), evaluating mycotoxins that showed detection frequencies above 20% in the studied matrices.
2.4.1. Estimation of Exposure
The exposure to mycotoxins was calculated using the average contamination levels from 260 batches of four matrices, average body weight, and daily intake doses. Daily intake doses were based on the maximum recommended dosages specified in the 2020 edition of the Ch.P, with maximum daily intakes set at 15 g for LS, 10 g for LR, 10 g for CP, and 30 g for CS. The average body weight of 64.4 kg was derived from the “Report on Nutrition and Chronic Diseases of Chinese Residents (2020)”, accounting for male-to-female population ratios. Mycotoxin exposure was calculated using the following formula, with detailed exposure levels presented in Table 3.
Exposure = (C × IR)/BW

Table 3.
Exposure of detected mycotoxins in four matrices for Chinese people.
C represents the average mycotoxin contamination level in medicinal materials (ng/g); IR represents the daily intake rate (g·day−1); and BW represents the average body weight (kg). Following the principles for handling non-detect data from the WHO Global Environment Monitoring System/Food Contamination Monitoring and Assessment Programme (GEMS/Food) Second Workshop on “Reliable Evaluation of Low-Level Contamination of Food” and the standards proposed in the European Commission’s Scientific Cooperation Task 3.2.10 (SCOOP) [50], the contamination level was calculated as the mean of all samples, with non-detect samples assigned a value of 1/2 LOD.
2.4.2. Risk Assessment of Mycotoxins Based on Margin of Exposure Margin of Exposure (MOE)
For non-threshold carcinogenic chemical hazards such as aflatoxins, risk assessment was conducted using the MOE approach, based on the Benchmark Dose Lower confidence limit of 10% extra risk (BMDL10) parameters published by EFSA (see Table 3) [51]. An MOE value greater than 10,000 indicates an acceptable health risk, while values below this threshold suggest potential health concerns.
MOE = BMDL10/Exposure
MOE values were calculated for seven mycotoxins (AFB1, AFB2, AFG1, AFG2, AFM1, AFM2, and Ster), as shown in Figure 9. CP and LR were excluded from the analysis due to the non-detection of the relevant mycotoxins. The analysis revealed that Ster posed minimal risks in both LS and CS for its lower toxicity. However, the MOE values for the remaining six aflatoxins were all below 10,000, indicating potential health concerns. While AFM2 showed values approaching 10,000, the actual risk might be higher considering that LS, as both medicinal and food items, may be consumed in larger quantities than the calculated dose. Additionally, children with lower body weights may face elevated risks. These findings highlight the necessity for monitoring aflatoxin risks in LS and CS.
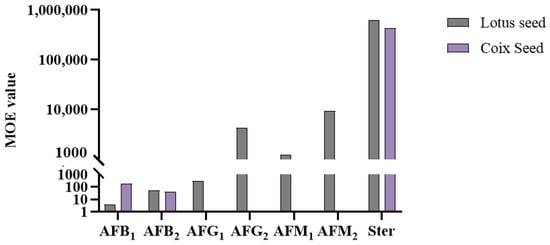
Figure 9.
MOE values from lotus seed and coix seed consumption for Chinese people.
2.4.3. Risk Assessment of Mycotoxins Based on Hazard Index (HI)
For threshold hazardous compounds such as DON and ZEN, a risk assessment was conducted using the HI method, based on the Provisional Maximum Tolerable Daily Intake (PMTDI) [52]. The HI is calculated as the sum of Hazard Quotients (HQ) for individual chemical compounds. An HI value less than 1 indicates an acceptable exposure risk level, while values exceeding 1 suggest potential adverse effects on human health. For certain mycotoxins with insufficient toxicological data and no official PMTDI values, a reference value of 2000 ng·kg−1 b.w.·day−1 was adopted (including BEA, CPA, ENNs, and MPA), based on fumonisin toxicity data.
HQ = Exposure/PMTDI
HI = ∑HQ
As shown in Figure 10, the overall HI values for all four matrices were relatively low, with ZEN and CIT being the primary risk contributors. Among these, CPA was the largest contributor to the HI in LS, with a value of 0.23. Although CS showed low overall risk, the high detection rate of fumonisins (74.4%) suggests potential exposure risks, indicating the need for expanded data collection to comprehensively evaluate the necessity of including these compounds in regulatory standards.
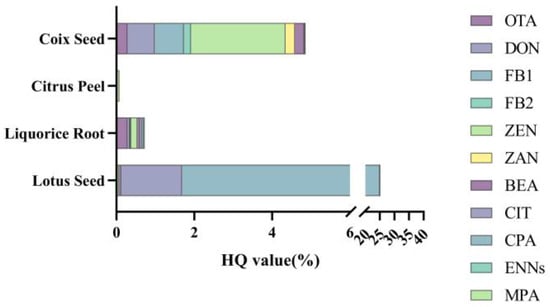
Figure 10.
HQ values from lotus seed and coix seed consumption for Chinese people.
3. Conclusions
This study systematically revealed the mycotoxin contamination characteristics and potential risks in four dual-use medicinal and edible plants through contamination analysis and health risk assessment.
Contamination analysis demonstrated that different types of plants exhibited unique contamination profiles: seed-type plants (LS and CS) were susceptible to Aspergillus and Fusarium contamination, with detection rates of 53.3% and 22.2%, respectively. Despite low detection rates, the MOE values of six aflatoxins remained below 10,000 due to their high toxicity, indicating potential health risks. Peel-type materials (citrus peel) were primarily contaminated with MPA produced by Penicillium (35.9% detection rate), while root-type materials (licorice) were mainly affected by Fusarium species (52.9% detection rate).
Significant differences were observed in contamination levels between samples of different physical states, with fragmented samples showing more severe mycotoxin contamination. Powdered LS showed a 52.9% higher detection rate of AFB1 compared to whole seeds, while sliced LR demonstrated a 41.2% higher detection rate of FB1 than intact samples. Three highly positive lotus samples, including moldy samples, discolored samples, and farm-cultivated samples, although limited in number, suggested the importance of timely drying and standardized sourcing in preventing mycotoxin contamination. GR samples collected between 2015 and 2020 and stored under cool conditions showed no significant differences in mycotoxin contamination.
Furthermore, highly contaminated samples revealed the co-occurrence of metabolically related toxins, such as the simultaneous detection of AFM1 (2.1 μg/kg) and AFM2 (1.8 μg/kg) in lotus seeds, providing new directions for studies on the metabolic mechanisms of mycotoxins in plants. Interestingly, the CP samples included in this study showed very low exposure risk (HI = 0.07%), possibly attributed to their specific processing techniques (e.g., low-temperature drying) or natural antimicrobial components.
The multi-toxin detection method established in this study can be extended to other plants with similar compositions, providing important technical support and data references for establishing scientific quality control systems. Based on our findings, we propose the following recommendations for industrial applications: (1) Emphasize supplier qualification verification during raw material procurement and strictly control moisture content; (2) Process and store materials in a non-fragmented state when possible and use sealed packaging after drying to prevent moisture absorption; (3) Maintain storage temperatures below 20 °C during storage and transportation and utilize dehumidification equipment when possible.
4. Materials and Methods
4.1. Sample Collection
A total of 260 samples of four edible and medicinal plants were collected, including LR (77 batches), LS (29 batches), CS (47 batches), and CP (131 batches). The majority of samples were obtained through national or local quality surveillance programs from various provinces in China, with a few collected directly from local farmers. All samples were authenticated by Chief Pharmacist Yang Xinhua from the Traditional Chinese Medicine/Natural Medicine and Health Food Institute, Shanghai Institute of Food and Drug Control. For each matrix, 200 g of sample was collected, ground into fine powder, passed through a 50-mesh sieve, and stored at −20 °C.
4.2. Chemicals and Reagents
Acetonitrile and methanol were purchased from Merck (Darmstadt, Germany). LC-MS grade acetic acid and formic acid were supplied by Fisher Scientific (Somerville, USA). Ammonium formate and ammonium acetate were obtained from Sigma Aldrich (Zwijndrecht, The Netherlands). Analytically pure acetic acid, formic acid, ammonium formate, and ammonium acetate were purchased from Merck (Darmstadt, Germany). Deionized water was obtained using a Milli-Q Gradient Water System (Millipore, Bedford, MA, USA).
Anhydrous magnesium sulfate, sodium chloride, trisodium citrate dihydrate, disodium citrate hydrate, anhydrous sodium acetate, dispersed solid-phase extraction (d-SPE) sorbent octadecylsilane (C18), primary secondary amine (PSA), silica gel (Si), and propane sulfonic acid (PRS) were obtained from Bonna-Agela Technologies (Tianjin, China). Solid reagent anhydrous sodium acetate prepared for buffer solution was obtained from Sinopharm Chemical Reagent Co. Ltd. (Shanghai, China). All other reagents were of analytical grade.
4.3. Preparation of Standard Solution
Stock solutions of mycotoxins were prepared in acetonitrile at concentrations ranging from 10 to 250 μg/mL and stored in brown glass vials at −20 °C, respectively. Based on their mass spectrometric response intensities, the 73 mycotoxins were divided into three groups. Group 1 (G1) included the following mycotoxins: 7-dechloro griseofulvin, aflatoxin B1, B2, G1, G2, and M1, agroclavine, anisomycin, beauvericin, diacetoxyscirpenol, dihydrolysergamide, enniatin A, enniatin A1, enniatin B, enniatin B1, ergocornine, ergocorninine, ergocristine, ergocristinine, ergocryptine, ergocryptinine, ergosine, griseofulvin, lysergamide, meleagrin, mycophenolic acid, ochratoxin A, ochratoxin B, ochratoxin C, oxaline, puromycin, roquefortine C, and sterigmatocystin. Group 2 (G2) included 15-acetoxyscirpenol, aflatoxin M2, aflatoxin P1, apicidin, chaetocin, citrinin, cyclopiazonic acid, equisetin, fumonisin B1, fumonisin B2, fumonisin B3, gliotoxin, monocerin, neosolaniol, o-methylsterigmatocystin, pseurotin A, secalonic acid D, T-2 toxin, α-zearalanol, α-zearalenol, β-zearalanol, β-zearalenol, and zearalenone. Group 3 (G3) included 15-acetyldeoxynivalenol, 3-acetyldeoxynivalenol, chetomin, citreoviridin, deepoxy-deoxynivalenol, deoxynivalenol, fumagillin, fusarenon X, HT-2 toxin, Ostreogrycin A, T-2-triol, tentoxin, wortmannin, patulin, alternariol-methylether, and alternariol.
Mixed standard stock solutions were prepared by combining individual stock solutions from each group to achieve the following concentrations: 100 ppb for G1, 500 ppb for G2, and 2500 ppb for G3. Working standard solutions at various concentration levels were subsequently prepared by appropriate dilution with suitable solvents as required by the analytical method.
4.4. Sample Preparation
An accurately weighed 2.0 g portion of homogenized sample was transferred into a 50 mL polypropylene centrifuge tube. Extraction was carried out with 20 mL of acetonitrile–water–acetic acid (80:19:1, v/v/v) using an orbital shaker (IKA, Guangzhou, China) for 90 min, followed by centrifugation (Eppendorf, Hamburg, Germany) at 3900 rpm for 5 min. Subsequently, 10 mL of the supernatant was transferred and combined with 10 mL of 5% formic acid solution. A QuEChERS salt mixture (sodium chloride, anhydrous magnesium sulfate, sodium citrate, and sodium citrate sesquihydrate; 1 g:4 g:1 g:0.5 g) was added immediately, followed by high-speed vortexing for 5 min (SPEX, New York, NY, USA) and centrifugation at 3900 rpm for 5 min. For further purification, 6.0 mL of the supernatant was subjected to liquid–liquid partitioning with 12 mL cyclohexane, followed by centrifugation at 3900 rpm. The lower phase (4 mL) was collected and concentrated to near dryness under a gentle nitrogen stream at 40 °C. The residue was reconstituted in 0.5 mL acetonitrile and diluted to 2 mL with water. The final extract was filtered through a 0.22 μm PTFE membrane filter (Agilent, Shanghai, China) prior to UHPLC-MS/MS analysis, with an injection volume of 5 μL.
4.5. UHPLC-MS/MS Analysis
Chromatographic separation was performed on a 1290 UHPLC system equipped with a quaternary solvent delivery system, degasser, autosampler, and column thermostat, coupled to a 5500 triple quadrupole mass spectrometer (AB SCIEX, Framingham, MA, USA) with an electrospray ionization (ESI) source operating in both positive and negative modes.
Chromatographic separation of the 73 mycotoxins was achieved on a Poroshell EC-C18 column (150 × 3.0 mm, 2.7 μm) (Agilent, Wilmington, DE, USA) at a flow rate of 450 μL/min. For the 64 mycotoxins analyzed in the positive mode (ESI+), mobile phase A consisted of 0.4% formic acid and 2.0 mM ammonium formate in water and mobile phase B consisted of 0.4% formic acid and 2.0 mM ammonium formate in methanol. The gradient program was 0–2 min, 20% B; 2–6 min, 20–50% B; 6–11 min, 50–55% B; 11–15 min, 55–100% B; 15–19 min, 100% B; 19–21 min, 100–20% B; and 21–25 min, 100% B. For the remaining mycotoxins analyzed in negative mode (ESI−), mobile phase A was water and mobile phase B was acetonitrile, with the following gradient: 0–2 min, 10% B; 2–8 min, 10–50% B; 8–13 min, 50–60% B; 13–15 min, 60–100% B; 15–16 min, 100% B; 16–18 min, 100–10% B; and 18–20 min, 10% B. The injection volume was 1.0 μL, the column temperature was maintained at 35 °C, and the sample tray temperature was set at 15 °C to enhance sample stability.
Mass spectrometric detection was performed under the following conditions: for the positive mode, the ion spray voltage was 5.5 kV, curtain gas was 30 psi, ion source gas 1 and gas 2 were both 50 psi, and source temperature was 450 °C; for the negative mode, the ion spray voltage was 4.5 kV, curtain gas was 30 psi, ion source gas 1 and gas 2 were both 50 psi, and source temperature was 400 °C.
A multiple reaction monitoring (MRM) mode was employed, with at least one precursor ion and two product ions monitored for each mycotoxin. The two most intense product ions free from matrix interference were selected for quantification and qualification, respectively. Declustering potentials (DPs) and collision energies (CEs) were optimized individually using standard solutions for each analyte.
Data acquisition and processing were performed using Analyst 1.5.1 and MultiQuant™ 2.1.1 software (AB SCIEX). Detailed information regarding the retention times (RTs), monitored precursor and product ions, and optimized DPs and CEs for each mycotoxin is presented in Table 1.
4.6. Statistical Analysis
All statistical analyses were performed using R software (version 4.2.0; R Core Team, 2023). Due to the non-normal distribution of data and presence of numerous null values, non-parametric analyses were conducted using the “stats” package. The wilcox.test() function was employed for Mann–Whitney U tests to evaluate differences in mycotoxin contamination between different forms of samples, while the kruskal.test() function was used for Kruskal–Wallis tests to analyze differences among different harvest years. Data visualization was accomplished using the “ggplot2” package. Statistical significance was defined as p < 0.05.
Author Contributions
Conceptualization, Investigation, Writing—Original draft preparation: X.H.; Validation, Data Curation: R.F.; Visualization: Q.H.; Reviewing and Editing: X.M.; Supervision, Project administration: H.Z.; All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Natural Science Foundation of Shanghai (23ZR1457200), Shanghai Science and Technology Commission R&D Platform Program (21DZ2290200), and Shanghai Municipal Administration of Market Regulation Technical Support Project (2024BZ20).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in this study are included in the article. Further inquiries can be directed to the corresponding author(s).
Conflicts of Interest
The authors declare no conflict of interest.
Appendix A

Table A1.
Overview of the extraction recovery (R), repeatability (RSD), limit of detection (LOD), and limit of quantification (LOQ) for each mycotoxin in LS.
Table A1.
Overview of the extraction recovery (R), repeatability (RSD), limit of detection (LOD), and limit of quantification (LOQ) for each mycotoxin in LS.
| Mycotoxin | Spiked Levels (μg/kg) | LOD (μg/kg) | LOQ (μg/kg) | Linear Range (μg/kg) | Coefficient (r) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Level 1 | Level 2 | Level 3 | ||||||||
| R (%) | RSD (%) | R (%) | RSD (%) | R (%) | RSD (%) | |||||
| 15-Asp | 88.8 | 3.1 | 86.4 | 2.3 | 83.1 | 3.5 | 1.25 | 2.5 | 2~1000 | 0.99973 |
| 15-ADON | 81.9 | 5.2 | 82.2 | 4.2 | 79.8 | 4.1 | 6.25 | 12.5 | 10~5000 | 0.99990 |
| 3-ADON | 90.8 | 2.7 | 89.1 | 2.2 | 88.0 | 3.5 | 6.25 | 12.5 | 10~5000 | 0.99973 |
| 7-D-G | 94.9 | 1.0 | 97.0 | 2.2 | 94.9 | 3.7 | 0.25 | 0.5 | 0.4~200 | 0.99959 |
| AFB1 | 91.5 | 2.0 | 92.3 | 2.8 | 86.1 | 3.4 | 0.25 | 0.5 | 0.4~200 | 0.99967 |
| AFB2 | 92.4 | 1.4 | 92.5 | 2.2 | 88.3 | 2.2 | 0.25 | 0.5 | 0.4~200 | 0.99995 |
| AFG1 | 91.1 | 1.4 | 90.5 | 4.2 | 83.8 | 0.9 | 0.25 | 0.5 | 0.4~200 | 0.99993 |
| AFG2 | 87.2 | 1.9 | 83.4 | 3.8 | 82.2 | 4.8 | 0.25 | 0.5 | 0.4~200 | 0.99985 |
| AFM1 | 86.8 | 3.3 | 89.1 | 3.0 | 87.4 | 2.2 | 0.25 | 0.5 | 0.4~200 | 0.99925 |
| AFM2 | 89.2 | 2.1 | 89.8 | 3.9 | 85.6 | 3.6 | 1.25 | 2.5 | 2~1000 | 0.99929 |
| AFP1 | 90.0 | 2.4 | 82.1 | 2.0 | 84.9 | 7.5 | 1.25 | 2.5 | 2~1000 | 0.99993 |
| Agro | 82.8 | 2.5 | 87.6 | 3.9 | 86.2 | 2.4 | 0.25 | 0.5 | 0.4~200 | 0.99964 |
| Anis | 96.4 | 2.7 | 102.5 | 2.7 | 103.3 | 1.9 | 0.25 | 0.5 | 0.4~200 | 0.99936 |
| Apici | 80.4 | 3.2 | 81.9 | 2.8 | 79.2 | 3.5 | 1.25 | 2.5 | 2~1000 | 0.99988 |
| BEA | 63.5 | 5.9 | 69.5 | 6.7 | 69.4 | 11.4 | 0.25 | 0.5 | 0.4~200 | 0.99985 |
| Chae | 82.9 | 9.6 | 91.3 | 6.8 | 88.9 | 8.4 | 6.25 | 12.5 | 10~5000 | 0.99928 |
| Che | 85.9 | 9.0 | 85.7 | 3.7 | 81.3 | 3.9 | 1.25 | 2.5 | 2~1000 | 0.99986 |
| CIT | 75.6 | 2.6 | 72.9 | 3.0 | 73.4 | 4.4 | 1.25 | 2.5 | 2~1000 | 0.99991 |
| CVD | 81.3 | 4.7 | 84.2 | 2.8 | 85.0 | 4.2 | 6.25 | 12.5 | 10~5000 | 0.99987 |
| CPA | 69.1 | 5.0 | 73.1 | 3.1 | 77.1 | 8.9 | 1.25 | 2.5 | 2~1000 | 0.99964 |
| DAS | 84.3 | 4.6 | 88.2 | 3.0 | 89.9 | 3.4 | 0.25 | 0.5 | 0.4~200 | 0.99928 |
| DiLy | 72.7 | 6.5 | 85.0 | 1.6 | 88.2 | 3.7 | 0.25 | 0.5 | 0.4~200 | 0.99942 |
| DOM | 92.5 | 2.4 | 89.6 | 8.2 | 87.0 | 6.5 | 6.25 | 12.5 | 10~5000 | 0.99977 |
| DON | 89.0 | 1.4 | 89.3 | 3.6 | 89.3 | 2.5 | 6.25 | 12.5 | 10~5000 | 0.99978 |
| ENN A | 76.0 | 3.5 | 71.8 | 5.0 | 72.1 | 8.7 | 0.25 | 0.5 | 0.4~200 | 0.99985 |
| ENN A1 | 73.6 | 4.6 | 78.1 | 3.4 | 78.0 | 6.3 | 0.25 | 0.5 | 0.4~200 | 0.99984 |
| ENN B | 80.1 | 3.5 | 82.3 | 2.4 | 83.1 | 5.1 | 0.25 | 0.5 | 0.4~200 | 0.99964 |
| ENN B1 | 81.3 | 2.0 | 83.9 | 3.2 | 88.0 | 5.0 | 0.25 | 0.5 | 0.4~200 | 0.99990 |
| Equi | 71.4 | 2.0 | 71.5 | 1.7 | 77.2 | 6.9 | 1.25 | 2.5 | 2~1000 | 0.99957 |
| EGCN | 82.0 | 2.8 | 87.9 | 1.8 | 85.0 | 3.0 | 0.25 | 0.5 | 0.4~200 | 0.99957 |
| EGCNN | 93.5 | 4.0 | 100.0 | 3.2 | 91.8 | 3.0 | 0.25 | 0.5 | 0.4~200 | 0.99984 |
| EGST | 101.4 | 1.9 | 103.7 | 3.9 | 103.2 | 4.9 | 0.25 | 0.5 | 0.4~200 | 0.99977 |
| EGSTN | 81.4 | 2.2 | 82.7 | 3.4 | 87.2 | 3.0 | 0.25 | 0.5 | 0.4~200 | 0.99983 |
| EGPT | 79.5 | 1.8 | 79.7 | 3.6 | 79.9 | 3.9 | 0.25 | 0.5 | 0.4~200 | 0.99955 |
| EGPTN | 90.6 | 2.1 | 91.6 | 2.6 | 89.6 | 3.3 | 0.25 | 0.5 | 0.4~200 | 0.99979 |
| EGSN | 116.4 | 3.1 | 113.2 | 3.2 | 106.1 | 4.7 | 0.25 | 0.5 | 0.4~200 | 0.99928 |
| FB1 | 76.4 | 2.6 | 77.4 | 1.7 | 75.4 | 3.0 | 1.25 | 2.5 | 2~1000 | 0.99938 |
| FB2 | 73.4 | 3.5 | 76.5 | 3.5 | 74.7 | 5.0 | 1.25 | 2.5 | 2~1000 | 0.99905 |
| FB3 | 74.3 | 7.1 | 81.0 | 5.4 | 77.0 | 5.1 | 1.25 | 2.5 | 2~1000 | 0.99951 |
| Fum | 84.0 | 12.8 | 75.6 | 5.8 | 79.1 | 2.6 | 6.25 | 12.5 | 10~5000 | 0.99933 |
| FuX | 87.8 | 2.3 | 86.3 | 5.1 | 82.7 | 2.8 | 6.25 | 12.5 | 10~5000 | 0.99978 |
| Glio | 83.9 | 1.6 | 82.3 | 3.9 | 81.9 | 3.4 | 1.25 | 2.5 | 2~1000 | 0.99920 |
| Grise | 90.5 | 1.2 | 90.0 | 2.3 | 85.3 | 3.6 | 0.25 | 0.5 | 0.4~200 | 0.99950 |
| HT-2 | 88.3 | 1.7 | 84.9 | 2.5 | 82.5 | 3.4 | 6.25 | 12.5 | 10~5000 | 0.99978 |
| Lyser | 76.3 | 2.2 | 80.2 | 2.0 | 78.5 | 3.6 | 0.25 | 0.5 | 0.4~200 | 0.99959 |
| Melea | 83.8 | 1.9 | 83.7 | 3.9 | 80.6 | 2.8 | 0.25 | 0.5 | 0.4~200 | 0.99945 |
| Mono | 95.1 | 0.7 | 98.8 | 3.2 | 95.3 | 3.5 | 1.25 | 2.5 | 2~1000 | 0.99997 |
| MPA | 114.7 | 3.0 | 109.3 | 2.7 | 97.9 | 3.2 | 0.25 | 0.5 | 0.4~200 | 0.99997 |
| NEO | 78.5 | 3.8 | 77.5 | 2.5 | 76.1 | 3.9 | 1.25 | 2.5 | 2~1000 | 0.99979 |
| O-m-Ster | 92.3 | 0.9 | 99.9 | 3.3 | 93.6 | 3.3 | 1.25 | 2.5 | 2~1000 | 0.99961 |
| Ostre A | 95.5 | 1.8 | 103.9 | 2.3 | 98.3 | 2.8 | 6.25 | 12.5 | 10~5000 | 0.99941 |
| OTA | 82.5 | 4.7 | 85.2 | 4.4 | 83.3 | 10.2 | 0.25 | 0.5 | 0.4~200 | 0.99927 |
| OTB | 88.2 | 1.9 | 95.0 | 1.7 | 90.9 | 4.0 | 0.25 | 0.5 | 0.4~200 | 0.99972 |
| OTC | 80.6 | 6.2 | 79.4 | 2.1 | 79.8 | 2.6 | 0.25 | 0.5 | 0.4~200 | 0.99981 |
| Oxa | 83.3 | 4.7 | 88.4 | 4.2 | 83.6 | 3.3 | 0.25 | 0.5 | 0.4~200 | 0.99944 |
| Pse A | 95.5 | 4.2 | 101.0 | 3.6 | 97.7 | 2.1 | 1.25 | 2.5 | 2~1000 | 0.99927 |
| Puro | 72.8 | 2.6 | 78.9 | 4.9 | 80.9 | 5.5 | 0.25 | 0.5 | 0.4~200 | 0.99959 |
| Rq C | 78.8 | 2.1 | 74.5 | 2.5 | 78.0 | 3.8 | 0.25 | 0.5 | 0.4~200 | 0.99966 |
| Secal A | 75.4 | 5.3 | 88.7 | 3.6 | 78.2 | 6.2 | 1.25 | 2.5 | 2~1000 | 0.99944 |
| Ster | 90.0 | 6.3 | 88.7 | 0.4 | 81.0 | 4.1 | 0.25 | 0.5 | 0.4~200 | 0.99963 |
| T2-triol | 83.5 | 2.8 | 82.9 | 2.8 | 79.3 | 3.1 | 6.25 | 12.5 | 10~5000 | 0.99946 |
| T-2 | 93.9 | 3.9 | 94.7 | 3.7 | 92.7 | 3.1 | 1.25 | 2.5 | 2~1000 | 0.99930 |
| Ten | 96.7 | 2.1 | 110.5 | 2.1 | 109.2 | 3.1 | 6.25 | 12.5 | 10~5000 | 0.99913 |
| Wor-man | 92.9 | 3.8 | 99.6 | 3.5 | 93.0 | 3.7 | 6.25 | 12.5 | 10~5000 | 0.99999 |
| α-ZAL | 88.8 | 2.7 | 87.0 | 2.2 | 88.0 | 3.0 | 1.25 | 2.5 | 2~400 | 0.99939 |
| α-ZEL | 89.2 | 3.5 | 83.7 | 1.2 | 81.8 | 1.9 | 1.25 | 2.5 | 2~400 | 0.99955 |
| β-ZAL | 94.0 | 4.2 | 88.2 | 2.7 | 79.6 | 3.1 | 1.25 | 2.5 | 2~1000 | 0.99978 |
| β-ZEL | 87.2 | 4.6 | 84.5 | 2.8 | 83.4 | 2.6 | 1.25 | 2.5 | 2~1000 | 0.99970 |
| PAT | 85.0 | 2.7 | 80.5 | 3.6 | 78.0 | 2.4 | 6.25 | 12.5 | 10~5000 | 0.99946 |
| ZAN | 92.6 | 3.2 | 83.6 | 4.4 | 72.3 | 2.5 | 1.25 | 2.5 | 2~1000 | 0.99948 |
| ZEN | 94.9 | 4.8 | 80.8 | 2.1 | 83.5 | 4.2 | 1.25 | 2.5 | 2~1000 | 0.99985 |
| AME | 91.9 | 1.7 | 94.3 | 2.4 | 94.5 | 2.9 | 6.25 | 12.5 | 10~2000 | 0.99952 |
| AOH | 87.0 | 5.7 | 77.7 | 4.7 | 81.6 | 3.7 | 6.25 | 12.5 | 10~2000 | 0.99931 |

Table A2.
Overview of the extraction recovery (R), repeatability (RSD), limit of detection (LOD), and limit of quantification (LOQ) for each mycotoxin in GR.
Table A2.
Overview of the extraction recovery (R), repeatability (RSD), limit of detection (LOD), and limit of quantification (LOQ) for each mycotoxin in GR.
| Mycotoxin | Spiked Levels (μg/kg) | LOD (μg/kg) | LOQ (μg/kg) | Linear Range (μg/kg) | Coefficient (r) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Level 1 | Level 2 | Level 3 | ||||||||
| R (%) | RSD (%) | R (%) | RSD (%) | R (%) | RSD (%) | |||||
| 15-Asp | 84.6 | 5.2 | 79.7 | 4.5 | 79.6 | 4.1 | 2.5 | 5.0 | 2~1000 | 0.99950 |
| 15-ADON | 83.6 | 6.1 | 83.6 | 2.4 | 81.7 | 6.9 | 12.5 | 25.0 | 10~5000 | 0.99956 |
| 3-ADON | 88.5 | 5.3 | 81.5 | 3.2 | 78.3 | 3.1 | 12.5 | 25.0 | 10~5000 | 0.99987 |
| 7-D-G | 84.0 | 2.7 | 81.2 | 3.2 | 79.9 | 2.4 | 0.5 | 1.0 | 0.4~200 | 0.99916 |
| AFB1 | 95.9 | 8.0 | 83.3 | 9.9 | 83.6 | 6.4 | 0.5 | 1.0 | 0.4~200 | 0.99924 |
| AFB2 | 109.6 | 7.2 | 82.5 | 4.2 | 79.7 | 4.5 | 0.5 | 1.0 | 0.4~200 | 0.99957 |
| AFG1 | 93.7 | 5.9 | 75.8 | 4.5 | 77.3 | 7.6 | 0.5 | 1.0 | 0.4~200 | 0.99946 |
| AFG2 | 87.5 | 13.2 | 88.3 | 9.0 | 88.8 | 7.7 | 0.5 | 1.0 | 0.4~200 | 0.99990 |
| AFM1 | 94.4 | 6.1 | 84.3 | 5.5 | 87.0 | 3.5 | 0.5 | 1.0 | 0.4~200 | 0.99969 |
| AFM2 | 92.2 | 3.5 | 80.9 | 4.3 | 84.8 | 4.3 | 2.5 | 5.0 | 2~1000 | 0.99924 |
| AFP1 | 94.7 | 7.1 | 74.0 | 4.5 | 72.7 | 6.1 | 2.5 | 5.0 | 2~1000 | 0.99959 |
| Agro | 78.9 | 7.3 | 72.9 | 6.2 | 73.3 | 5.0 | 0.5 | 1.0 | 0.4~200 | 0.99901 |
| Anis | 86.0 | 4.4 | 87.4 | 4.7 | 87.6 | 3.3 | 0.5 | 1.0 | 0.4~200 | 0.99962 |
| Apici | 84.9 | 7.8 | 75.2 | 5.7 | 77.1 | 6.6 | 2.5 | 5.0 | 2~1000 | 0.99919 |
| BEA | 91.2 | 5.4 | 84.3 | 1.7 | 81.6 | 4.3 | 0.5 | 1.0 | 0.4~200 | 0.99920 |
| Chae | 83.3 | 10.2 | 75.2 | 9.4 | 78.1 | 6.5 | 12.5 | 25.0 | 10~5000 | 0.99959 |
| Che | 83.2 | 10.6 | 79.5 | 6.4 | 79.5 | 8.5 | 2.5 | 5.0 | 2~1000 | 0.99899 |
| CIT | 70.6 | 3.5 | 72.4 | 4.3 | 74.1 | 5.3 | 2.5 | 5.0 | 2~1000 | 0.99963 |
| CVD | 86.1 | 14.2 | 78.4 | 7.8 | 76.5 | 8.0 | 12.5 | 25.0 | 10~5000 | 0.99989 |
| CPA | 73.6 | 11.8 | 67.6 | 8.8 | 64.3 | 4.7 | 2.5 | 5.0 | 2~1000 | 0.99982 |
| DAS | 83.3 | 3.0 | 79.8 | 3.1 | 78.0 | 4.1 | 0.5 | 1.0 | 0.4~200 | 0.99968 |
| DiLy | 62.7 | 2.5 | 67.4 | 3.5 | 65.9 | 4.9 | 0.5 | 1.0 | 0.4~200 | 0.99939 |
| DOM | 85.2 | 2.6 | 82.1 | 3.2 | 76.8 | 2.3 | 12.5 | 25.0 | 10~5000 | 0.99948 |
| DON | 75.4 | 4.6 | 76.7 | 3.4 | 74.9 | 3.4 | 12.5 | 25.0 | 10~5000 | 0.99981 |
| ENN A | 86.3 | 5.0 | 82.0 | 2.7 | 79.3 | 2.7 | 0.5 | 1.0 | 0.4~200 | 0.99920 |
| ENN A1 | 87.5 | 3.6 | 82.9 | 3.8 | 79.3 | 4.1 | 0.5 | 1.0 | 0.4~200 | 0.99943 |
| ENN B | 92.1 | 4.9 | 83.6 | 3.1 | 80.3 | 4.3 | 0.5 | 1.0 | 0.4~200 | 0.99976 |
| ENN B1 | 85.9 | 4.1 | 81.5 | 4.3 | 79.9 | 2.8 | 0.5 | 1.0 | 0.4~200 | 0.99930 |
| Equi | 80.4 | 7.5 | 73.1 | 4.3 | 69.2 | 4.1 | 2.5 | 5.0 | 2~1000 | 0.99892 |
| EGCN | 94.2 | 4.6 | 73.6 | 5.2 | 78.0 | 4.5 | 0.5 | 1.0 | 0.4~200 | 0.99928 |
| EGCNN | 88.4 | 3.2 | 80.6 | 3.1 | 77.4 | 5.2 | 0.5 | 1.0 | 0.4~200 | 0.99980 |
| EGST | 102.8 | 6.8 | 86.0 | 4.1 | 80.6 | 3.4 | 0.5 | 1.0 | 0.4~200 | 0.99946 |
| EGSTN | 87.5 | 4.6 | 75.1 | 3.0 | 71.0 | 3.2 | 0.5 | 1.0 | 0.4~200 | 0.99912 |
| EGPT | 95.8 | 6.3 | 77.5 | 3.9 | 70.6 | 2.9 | 0.5 | 1.0 | 0.4~200 | 0.99936 |
| EGPTN | 85.2 | 6.5 | 76.8 | 4.4 | 79.8 | 6.1 | 0.5 | 1.0 | 0.4~200 | 0.99911 |
| EGSN | 94.7 | 7.6 | 91.3 | 4.2 | 94.0 | 6.2 | 0.5 | 1.0 | 0.4~200 | 0.99927 |
| FB1 | 74.5 | 7.5 | 76.6 | 7.9 | 79.6 | 8.6 | 2.5 | 5.0 | 2~1000 | 0.99951 |
| FB2 | 71.7 | 11.4 | 77.9 | 9.4 | 87.4 | 11.1 | 2.5 | 5.0 | 2~1000 | 0.99935 |
| FB3 | 79.2 | 11.1 | 86.6 | 9.2 | 92.2 | 4.8 | 2.5 | 5.0 | 2~1000 | 0.99945 |
| Fum | 81.6 | 7.9 | 82.2 | 9.0 | 105.4 | 10.3 | 12.5 | 25.0 | 10~5000 | 0.99945 |
| FuX | 84.6 | 4.8 | 80.3 | 2.7 | 77.0 | 2.7 | 12.5 | 25.0 | 10~5000 | 0.99958 |
| Glio | 80.7 | 3.0 | 82.6 | 3.4 | 78.4 | 4.2 | 2.5 | 5.0 | 2~1000 | 0.99907 |
| Grise | 69.8 | 7.0 | 72.2 | 9.2 | 80.5 | 4.4 | 0.5 | 1.0 | 0.4~200 | 0.99917 |
| HT-2 | 87.8 | 4.9 | 83.7 | 3.9 | 82.1 | 4.2 | 12.5 | 25.0 | 10~5000 | 0.99949 |
| Lyser | 71.3 | 4.7 | 67.1 | 5.0 | 68.4 | 3.8 | 0.5 | 1.0 | 0.4~200 | 0.99998 |
| Melea | 87.9 | 2.2 | 80.8 | 3.8 | 76.1 | 2.9 | 0.5 | 1.0 | 0.4~200 | 0.99923 |
| Mono | 83.7 | 3.7 | 79.4 | 4.4 | 76.7 | 1.6 | 2.5 | 5.0 | 2~1000 | 0.99972 |
| MPA | 82.7 | 5.4 | 87.4 | 6.6 | 84.2 | 3.1 | 0.5 | 1.0 | 0.4~200 | 0.99894 |
| NEO | 87.1 | 1.9 | 82.8 | 5.4 | 77.2 | 2.5 | 2.5 | 5.0 | 2~1000 | 0.99909 |
| O-m-Ster | 77.9 | 4.8 | 70.3 | 7.4 | 72.9 | 14.4 | 2.5 | 5.0 | 2~1000 | 0.99947 |
| Ostre A | 86.7 | 5.5 | 75.9 | 6.1 | 76.6 | 9.1 | 12.5 | 25.0 | 10~5000 | 0.99906 |
| OTA | 83.2 | 6.8 | 77.7 | 6.3 | 80.5 | 7.3 | 0.5 | 1.0 | 0.4~200 | 0.99904 |
| OTB | 87.2 | 4.5 | 85.8 | 3.4 | 82.4 | 2.0 | 0.5 | 1.0 | 0.4~200 | 0.99940 |
| OTC | 98.5 | 6.1 | 80.5 | 4.2 | 77.5 | 5.2 | 0.5 | 1.0 | 0.4~200 | 0.99984 |
| Oxa | 85.1 | 2.8 | 81.8 | 4.5 | 78.8 | 3.5 | 0.5 | 1.0 | 0.4~200 | 0.99952 |
| Pse A | 86.8 | 5.4 | 83.8 | 4.2 | 82.4 | 5.0 | 2.5 | 5.0 | 2~1000 | 0.99925 |
| Puro | 72.2 | 9.1 | 69.9 | 7.7 | 75.6 | 4.1 | 0.5 | 1.0 | 0.4~200 | 0.99917 |
| Rq C | 78.3 | 7.4 | 73.3 | 12.7 | 73.5 | 14.9 | 0.5 | 1.0 | 0.4~200 | 0.99989 |
| Secal A | 86.7 | 13.2 | 78.6 | 9.4 | 74.2 | 7.5 | 2.5 | 5.0 | 2~1000 | 0.99928 |
| Ster | 80.9 | 10.8 | 70.8 | 4.8 | 70.5 | 14.6 | 0.5 | 1.0 | 0.4~200 | 0.99966 |
| T2-triol | 93.8 | 2.6 | 85.7 | 4.6 | 80.3 | 3.3 | 12.5 | 25.0 | 10~5000 | 0.99914 |
| T-2 | 91.6 | 5.0 | 86.0 | 1.6 | 81.1 | 3.5 | 2.5 | 5.0 | 2~1000 | 0.99975 |
| Ten | 80.8 | 7.2 | 86.0 | 4.0 | 88.1 | 2.1 | 12.5 | 25.0 | 10~5000 | 0.99995 |
| Wor-man | 88.7 | 2.8 | 82.9 | 2.8 | 80.8 | 4.1 | 12.5 | 25.0 | 10~5000 | 0.99984 |
| α-ZAL | 85.5 | 10.2 | 72.0 | 6.4 | 75.2 | 7.6 | 2.5 | 5.0 | 2~400 | 0.99973 |
| α-ZEL | 83.9 | 4.3 | 72.9 | 5.5 | 76.8 | 4.8 | 2.5 | 5.0 | 2~400 | 0.99908 |
| β-ZAL | 76.5 | 8.0 | 78.4 | 10.0 | 82.8 | 5.5 | 2.5 | 5.0 | 2~1000 | 0.99960 |
| β-ZEL | 82.5 | 10.2 | 94.8 | 4.1 | 98.6 | 13.3 | 2.5 | 5.0 | 2~1000 | 0.99897 |
| PAT | 77.9 | 8.3 | 76.7 | 7.6 | 78.9 | 9.8 | 12.5 | 25.0 | 10~5000 | 0.99957 |
| ZAN | 83.4 | 7.6 | 78.3 | 4.6 | 77.6 | 2.5 | 2.5 | 5.0 | 2~1000 | 0.99998 |
| ZEN | 90.2 | 6.9 | 80.9 | 3.4 | 76.5 | 4.7 | 2.5 | 5.0 | 2~1000 | 0.99965 |
| AME | 98.8 | 4.1 | 93.4 | 2.1 | 89.7 | 2.5 | 12.5 | 25.0 | 10~1000 | 0.99869 |
| AOH | 106.9 | 6.7 | 93.1 | 2.5 | 81.6 | 4.7 | 12.5 | 25.0 | 10~1000 | 0.99849 |

Table A3.
Overview of the extraction recovery (R), repeatability (RSD), limit of detection (LOD), and limit of quantification (LOQ) for each mycotoxin in CP.
Table A3.
Overview of the extraction recovery (R), repeatability (RSD), limit of detection (LOD), and limit of quantification (LOQ) for each mycotoxin in CP.
| Mycotoxin | Spiked Levels (μg/kg) | LOD (μg/kg) | LOQ (μg/kg) | Linear Range (μg/kg) | Coefficient (r) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Level 1 | Level 2 | Level 3 | ||||||||
| R (%) | RSD (%) | R (%) | RSD (%) | R (%) | RSD (%) | |||||
| 15-Asp | 82.6 | 6.3 | 82.9 | 1.7 | 78.5 | 3.0 | 1.25 | 2.5 | 2~1000 | 0.99917 |
| 15-ADON | 87.8 | 4.4 | 79.5 | 3.8 | 73.8 | 5.3 | 6.25 | 12.5 | 10~5000 | 0.99981 |
| 3-ADON | 87.7 | 4.7 | 83.5 | 3.2 | 79.9 | 2.5 | 6.25 | 12.5 | 10~5000 | 0.99934 |
| 7-D-G | 79.0 | 6.0 | 83.8 | 3.4 | 80.6 | 2.5 | 0.25 | 0.5 | 0.4~200 | 0.99981 |
| AFB1 | 81.6 | 5.4 | 84.2 | 3.2 | 78.5 | 2.6 | 0.25 | 0.5 | 0.4~200 | 0.99911 |
| AFB2 | 78.5 | 5.5 | 83.7 | 2.7 | 78.4 | 3.4 | 0.25 | 0.5 | 0.4~200 | 0.99923 |
| AFG1 | 81.7 | 6.6 | 83.6 | 3.0 | 80.6 | 3.0 | 0.25 | 0.5 | 0.4~200 | 0.99980 |
| AFG2 | 83.1 | 6.4 | 83.6 | 2.0 | 83.3 | 3.7 | 0.25 | 0.5 | 0.4~200 | 0.99929 |
| AFM1 | 81.9 | 4.0 | 83.4 | 4.5 | 78.9 | 3.7 | 0.25 | 0.5 | 0.4~200 | 0.99979 |
| AFM2 | 87.3 | 8.6 | 82.6 | 5.9 | 75.2 | 4.8 | 1.25 | 2.5 | 2~1000 | 0.99975 |
| AFP1 | 80.3 | 6.5 | 81.9 | 4.7 | 77.4 | 4.0 | 1.25 | 2.5 | 2~1000 | 0.99912 |
| Agro | 80.3 | 4.8 | 80.2 | 4.2 | 75.3 | 4.6 | 0.25 | 0.5 | 0.4~200 | 0.99985 |
| Anis | 82.3 | 7.0 | 81.3 | 4.0 | 77.6 | 4.6 | 0.25 | 0.5 | 0.4~200 | 0.99902 |
| Apici | 82.3 | 4.7 | 83.9 | 2.0 | 79.0 | 3.6 | 1.25 | 2.5 | 2~1000 | 0.99904 |
| BEA | 79.2 | 6.8 | 81.7 | 2.4 | 79.3 | 2.9 | 0.25 | 0.5 | 0.4~200 | 0.99926 |
| Chae | 86.3 | 9.8 | 83.7 | 6.6 | 79.9 | 6.1 | 6.25 | 12.5 | 10~5000 | 0.99957 |
| Che | 82.0 | 8.6 | 82.3 | 4.6 | 80.4 | 4.8 | 1.25 | 2.5 | 2~1000 | 0.99928 |
| CIT | 77.3 | 6.4 | 79.2 | 7.5 | 74.3 | 8.0 | 1.25 | 2.5 | 2~1000 | 0.99902 |
| CVD | 81.9 | 6.3 | 92.5 | 5.0 | 82.8 | 3.6 | 6.25 | 12.5 | 10~5000 | 0.99919 |
| CPA | 81.2 | 7.9 | 77.3 | 5.1 | 77.6 | 5.0 | 1.25 | 2.5 | 2~1000 | 0.99903 |
| DAS | 87.8 | 3.7 | 81.8 | 2.8 | 78.3 | 2.7 | 0.25 | 0.5 | 0.4~200 | 0.99904 |
| DiLy | 67.0 | 3.6 | 64.0 | 2.3 | 61.6 | 2.5 | 0.25 | 0.5 | 0.4~200 | 0.99966 |
| DOM | 84.8 | 3.6 | 71.6 | 4.8 | 78.2 | 7.4 | 6.25 | 12.5 | 10~5000 | 0.99960 |
| DON | 77.7 | 4.5 | 76.7 | 2.5 | 72.1 | 3.2 | 6.25 | 12.5 | 10~5000 | 0.99983 |
| ENN A | 74.1 | 3.9 | 79.7 | 1.4 | 77.0 | 2.7 | 0.25 | 0.5 | 0.4~200 | 0.99978 |
| ENN A1 | 76.0 | 3.3 | 82.7 | 3.3 | 82.2 | 3.3 | 0.25 | 0.5 | 0.4~200 | 0.99990 |
| ENN B | 82.0 | 3.7 | 84.2 | 3.8 | 82.7 | 3.8 | 0.25 | 0.5 | 0.4~200 | 0.99996 |
| ENN B1 | 80.1 | 4.3 | 84.2 | 4.2 | 83.7 | 2.9 | 0.25 | 0.5 | 0.4~200 | 0.99981 |
| Equi | 75.0 | 4.2 | 72.2 | 2.9 | 77.4 | 2.3 | 1.25 | 2.5 | 2~1000 | 0.99908 |
| EGCN | 88.9 | 3.9 | 85.1 | 1.5 | 78.9 | 4.2 | 0.25 | 0.5 | 0.4~200 | 0.99940 |
| EGCNN | 78.7 | 5.5 | 80.7 | 4.0 | 76.4 | 3.8 | 0.25 | 0.5 | 0.4~200 | 0.99916 |
| EGST | 86.1 | 4.3 | 84.1 | 1.8 | 82.9 | 2.5 | 0.25 | 0.5 | 0.4~200 | 0.99990 |
| EGSTN | 80.9 | 4.4 | 81.0 | 3.0 | 76.0 | 3.0 | 0.25 | 0.5 | 0.4~200 | 0.99936 |
| EGPT | 86.8 | 3.8 | 81.7 | 4.7 | 78.8 | 3.0 | 0.25 | 0.5 | 0.4~200 | 0.99919 |
| EGPTN | 85.8 | 3.9 | 81.1 | 2.7 | 75.8 | 3.2 | 0.25 | 0.5 | 0.4~200 | 0.99979 |
| EGSN | 87.6 | 13.6 | 88.9 | 11.6 | 78.0 | 4.5 | 0.25 | 0.5 | 0.4~200 | 0.99933 |
| FB1 | 78.0 | 7.3 | 70.7 | 5.1 | 77.0 | 2.6 | 1.25 | 2.5 | 2~1000 | 0.99999 |
| FB2 | 79.1 | 7.3 | 76.1 | 2.8 | 73.1 | 5.5 | 1.25 | 2.5 | 2~1000 | 0.99983 |
| FB3 | 63.6 | 8.4 | 73.2 | 2.9 | 72.0 | 2.2 | 1.25 | 2.5 | 2~1000 | 0.99997 |
| Fum | 71.2 | 6.2 | 81.8 | 3.8 | 76.9 | 6.7 | 6.25 | 12.5 | 10~5000 | 0.99963 |
| FuX | 89.3 | 4.4 | 81.1 | 3.5 | 76.2 | 2.6 | 6.25 | 12.5 | 10~5000 | 0.99948 |
| Glio | 80.4 | 5.4 | 81.7 | 4.1 | 79.7 | 4.3 | 1.25 | 2.5 | 2~1000 | 0.99983 |
| Grise | 82.7 | 5.4 | 83.1 | 3.6 | 79.3 | 2.5 | 0.25 | 0.5 | 0.4~200 | 0.99960 |
| HT-2 | 78.9 | 7.5 | 81.5 | 7.3 | 80.4 | 3.4 | 6.25 | 12.5 | 10~5000 | 0.99936 |
| Lyser | 68.4 | 2.2 | 69.7 | 3.5 | 66.9 | 2.4 | 0.25 | 0.5 | 0.4~200 | 0.99923 |
| Melea | 82.0 | 4.3 | 80.0 | 3.9 | 78.2 | 3.2 | 0.25 | 0.5 | 0.4~200 | 0.99926 |
| Mono | 75.6 | 7.1 | 82.1 | 2.8 | 79.5 | 3.2 | 1.25 | 2.5 | 2~1000 | 0.99979 |
| MPA | 79.1 | 10.5 | 87.7 | 4.4 | 84.7 | 2.1 | 0.25 | 0.5 | 0.4~200 | 0.99907 |
| NEO | 91.4 | 2.9 | 81.3 | 4.3 | 75.5 | 3.0 | 1.25 | 2.5 | 2~1000 | 0.99972 |
| O-m-Ster | 81.0 | 4.4 | 83.3 | 2.5 | 81.1 | 2.8 | 1.25 | 2.5 | 2~1000 | 0.99904 |
| Ostre A | 90.0 | 5.0 | 89.0 | 4.1 | 81.3 | 6.9 | 6.25 | 12.5 | 10~5000 | 0.99941 |
| OTA | 73.6 | 7.0 | 83.4 | 10.3 | 81.0 | 6.4 | 0.25 | 0.5 | 0.4~200 | 0.99926 |
| OTB | 70.4 | 7.0 | 85.8 | 3.1 | 88.5 | 3.6 | 0.25 | 0.5 | 0.4~80 | 0.99953 |
| OTC | 77.2 | 5.6 | 79.6 | 3.3 | 78.7 | 1.9 | 0.25 | 0.5 | 0.4~200 | 0.99980 |
| Oxa | 85.3 | 3.6 | 79.9 | 3.4 | 76.6 | 4.3 | 0.25 | 0.5 | 0.4~200 | 0.99956 |
| Pse A | 89.5 | 4.0 | 82.5 | 4.3 | 79.3 | 4.3 | 1.25 | 2.5 | 2~1000 | 0.99977 |
| Puro | 71.1 | 3.5 | 62.9 | 3.2 | 67.1 | 4.8 | 0.25 | 0.5 | 0.4~200 | 0.99963 |
| Rq C | 87.0 | 2.7 | 78.5 | 3.0 | 74.5 | 3.2 | 0.25 | 0.5 | 0.4~200 | 0.99931 |
| Secal A | 73.5 | 5.3 | 79.3 | 3.4 | 75.4 | 1.0 | 1.25 | 2.5 | 2~1000 | 0.99928 |
| Ster | 77.8 | 10.6 | 82.3 | 4.0 | 80.5 | 2.9 | 0.25 | 0.5 | 0.4~200 | 0.99910 |
| T2-triol | 91.2 | 2.7 | 82.1 | 4.0 | 77.9 | 3.2 | 6.25 | 12.5 | 10~5000 | 0.99984 |
| T-2 | 82.1 | 3.3 | 83.7 | 2.3 | 82.3 | 4.0 | 1.25 | 2.5 | 2~1000 | 0.99971 |
| Ten | 81.7 | 3.3 | 83.0 | 2.9 | 82.5 | 3.1 | 6.25 | 12.5 | 10~5000 | 0.99926 |
| Wor-man | 90.9 | 3.4 | 83.2 | 3.7 | 79.7 | 6.7 | 6.25 | 12.5 | 10~5000 | 0.99907 |
| α-ZAL | 86.0 | 9.8 | 92.6 | 5.6 | 75.9 | 5.1 | 1.25 | 2.5 | 2~1000 | 0.99934 |
| α-ZEL | 106.5 | 11.1 | 85.2 | 4.5 | 84.3 | 5.6 | 1.25 | 2.5 | 2~1000 | 0.99921 |
| β-ZAL | 92.4 | 9.3 | 105.3 | 3.9 | 89.6 | 5.8 | 1.25 | 2.5 | 2~1000 | 0.99906 |
| β-ZEL | 80.1 | 8.3 | 97.4 | 4.4 | 86.6 | 6.7 | 1.25 | 2.5 | 2~1000 | 0.99906 |
| PAT | 88.7 | 3.5 | 75.9 | 7.6 | 73.0 | 9.9 | 6.25 | 12.5 | 10~5000 | 0.99995 |
| ZAN | 88.7 | 7.5 | 92.3 | 4.0 | 80.6 | 3.5 | 1.25 | 2.5 | 2~1000 | 0.99900 |
| ZEN | 91.7 | 11.5 | 81.5 | 3.6 | 78.4 | 3.5 | 1.25 | 2.5 | 2~1000 | 0.99961 |
| AME | 96.2 | 10.7 | 77.3 | 3.6 | 76.0 | 3.8 | 6.25 | 12.5 | 10~1000 | 0.99912 |
| AOH | 84.4 | 9.5 | 90.2 | 7.0 | 83.1 | 4.5 | 6.25 | 12.5 | 10~1000 | 0.99868 |

Table A4.
The muti-mycotoxin contamination situation of LS.
Table A4.
The muti-mycotoxin contamination situation of LS.
| Sample | Comment | Contamination Level (μg/kg) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| AFB1 | AFB2 | AFG1 | AFG2 | AFM1 | AFM2 | CIT | CPA | MPA | ||
| LS1 | original shape | 3.0 | 21.4 | ND | ND | 1.0 | ND | ND | ND | ND |
| LS2 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LS3 | original shape | ND | ND | ND | ND | 1.8 | ND | ND | ND | ND |
| LS4 | original shape | 4.6 | 1.3 | ND | ND | ND | ND | ND | ND | ND |
| LS5 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | 1.3 |
| LS6 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LS7 | original shape | 1.3 | ND | ND | ND | ND | ND | ND | ND | ND |
| LS8 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LS9 | original shape | 18.4 | 1.3 | ND | ND | ND | ND | ND | ND | ND |
| LS10 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LS11 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LS12 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LS13 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LS14 | original shape | 31.6 | 1.3 | ND | ND | ND | ND | ND | ND | ND |
| LS15 | original shape | 21.5 | ND | ND | ND | ND | ND | ND | ND | 27.8 |
| LS16 | original shape | 16.4 | 1.3 | ND | ND | ND | ND | ND | ND | 75.0 |
| LS17 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LS18 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LS19 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LS20 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LS21 | powder | ND | ND | ND | ND | ND | ND | ND | 406.9 | ND |
| LS22 | powder | 12.6 | ND | ND | ND | ND | ND | ND | 1043.7 | 8.6 |
| LS23 | powder | 14.9 | ND | ND | ND | ND | ND | ND | 1563.7 | ND |
| LS24 | powder, fresh | 3638.6 | 160.8 | ND | ND | 47.5 | ND | ND | 12199.8 | ND |
| LS25 | powder, farm-grown | 4445.4 | 351.9 | 17ND | 8.8 | 189.6 | 19.5 | 376.8 | 19520.6 | ND |
| LS26 | powder, farm-grown | 4700.6 | 396.5 | ND | ND | 155.1 | 29.8 | ND | 23574.6 | ND |
| LS27 | powder, farm-grown | ND | ND | ND | ND | ND | ND | ND | 14.6 | ND |
| LS28 | Powder, moldy | ND | ND | ND | ND | ND | ND | ND | 5.0 | ND |
| LS29 | Powder, discolored | ND | ND | ND | ND | ND | ND | ND | 2.6 | ND |
| Contamination rate (%) | 41.4 | 27.6 | 3.4 | 3.4 | 17.2 | 6.9 | 3.4 | 31.0 | 13.8 | |
| Sample | Comment | Contamination Level (μg/kg) | ||||||||
| ZEN | BEA | ENN A1 | Glio | O-m-Ster | Pse A | Ster | Ten | |||
| LS1 | original shape | ND | ND | ND | ND | ND | ND | ND | 1.3 | |
| LS2 | original shape | ND | ND | ND | ND | ND | ND | 6.5 | ND | |
| LS3 | original shape | ND | ND | ND | ND | 11.7 | ND | ND | ND | |
| LS4 | original shape | 1.3 | ND | ND | ND | ND | ND | 1.3 | ND | |
| LS5 | original shape | ND | 11.1 | 1.3 | ND | ND | ND | 1.3 | ND | |
| LS6 | original shape | ND | ND | 1.3 | ND | 3.8 | ND | ND | ND | |
| LS7 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | |
| LS8 | original shape | ND | ND | ND | ND | ND | ND | 8.2 | ND | |
| LS9 | original shape | ND | 14.0 | 1.8 | ND | ND | ND | ND | 9.3 | |
| LS10 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | |
| LS11 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | |
| LS12 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | |
| LS13 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | |
| LS14 | original shape | ND | 10.5 | ND | ND | ND | ND | ND | 16.6 | |
| LS15 | original shape | ND | 14.3 | ND | ND | ND | ND | ND | 15.1 | |
| LS16 | original shape | 34.1 | 35.5 | 16.2 | ND | ND | ND | ND | 21.1 | |
| LS17 | original shape | ND | 11.6 | ND | 70.7 | ND | ND | ND | 3.7 | |
| LS18 | original shape | 1.3 | 10.6 | 30.4 | 12.5 | ND | ND | ND | ND | |
| LS19 | original shape | 1.3 | 27.3 | 3.3 | 8.6 | ND | ND | ND | ND | |
| LS20 | original shape | ND | 15.9 | 1.5 | ND | ND | ND | ND | ND | |
| LS21 | powder | ND | ND | ND | ND | ND | ND | ND | ND | |
| LS22 | powder | ND | ND | ND | ND | ND | ND | ND | ND | |
| LS23 | powder | ND | ND | ND | ND | ND | 6.4 | ND | ND | |
| LS24 | powder, fresh | ND | ND | ND | ND | 11.4 | ND | 7.0 | ND | |
| LS25 | powder, farm-grown | ND | ND | ND | ND | 158.8 | ND | 5.0 | ND | |
| LS26 | powder, farm-grown | ND | ND | ND | ND | 118.8 | ND | ND | ND | |
| LS27 | powder, farm-grown | ND | ND | ND | ND | ND | ND | ND | ND | |
| LS28 | Powder, moldy | ND | ND | ND | ND | ND | ND | ND | ND | |
| LS29 | Powder, discolored | ND | ND | ND | ND | ND | ND | ND | ND | |
| Contamination rate (%) | 13.8 | 31.0 | 24.1 | 10.3 | 17.2 | 3.4 | 20.7 | 20.7 | ||

Table A5.
The muti-mycotoxin contamination situation of LR.
Table A5.
The muti-mycotoxin contamination situation of LR.
| Sample | Comment | Vintage Year | Contamination Level (μg/kg) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ZEN | DOM | DON | BEA | ENN A | ENN A1 | ENN B | ENN B1 | FB1 | |||
| LR1 | slice | / | ND | ND | ND | ND | ND | ND | 1.3 | 1.3 | 8.3 |
| LR2 | slice | / | ND | ND | ND | ND | ND | ND | 13.8 | 1.3 | 13 |
| LR3 | slice | / | 2.2 | ND | ND | ND | ND | ND | ND | ND | 1.3 |
| LR4 | slice | / | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR5 | slice | / | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR6 | slice | / | 26.9 | 15.1 | 15.1 | 1.3 | ND | 32.3 | 77.2 | 104.8 | ND |
| LR7 | slice | / | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR8 | slice | / | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR9 | slice | / | ND | ND | ND | 1.3 | ND | 1.3 | 1.3 | ND | 4.8 |
| LR10 | slice | / | ND | ND | ND | ND | ND | ND | ND | ND | 1.3 |
| LR11 | slice | / | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR12 | slice | / | ND | ND | ND | 2.5 | ND | 1.3 | ND | ND | 14.6 |
| LR13 | slice | / | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR14 | slice | / | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR15 | slice | / | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR16 | slice | / | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR17 | slice | / | ND | ND | ND | 1.3 | ND | 17.7 | 61.2 | 85.4 | 34.9 |
| LR18 | slice | / | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR19 | slice | / | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR20 | slice | / | 1.3 | ND | ND | ND | ND | ND | ND | ND | 1.3 |
| LR21 | slice | / | 94.6 | ND | ND | 21.4 | ND | 27.2 | 23.2 | 94.8 | 10.2 |
| LR22 | slice | / | 53.1 | ND | ND | ND | ND | ND | ND | ND | 1.3 |
| LR23 | slice | / | ND | ND | ND | ND | ND | ND | ND | ND | 1.3 |
| LR24 | slice | / | ND | ND | ND | ND | ND | ND | ND | ND | 20.7 |
| LR25 | slice | / | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR26 | slice | / | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR27 | slice | / | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR28 | slice | / | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR29 | slice | / | ND | ND | ND | ND | ND | ND | ND | ND | 1.3 |
| LR30 | slice | / | ND | ND | ND | 1.3 | ND | ND | ND | ND | 8 |
| LR31 | original shape, a * | 2018 | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR32 | original shape, b | 2018 | 39.5 | ND | ND | ND | ND | ND | ND | ND | ND |
| LR33 | original shape, c | 2018 | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR34 | original shape, d | 2018 | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR35 | original shape, d | 2018 | 42 | ND | ND | ND | ND | ND | ND | ND | ND |
| LR36 | original shape, d | 2018 | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR37 | original shape, d | 2018 | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR38 | original shape | 2019 | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR39 | original shape, d | 2019 | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR40 | original shape, d | 2019 | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR41 | original shape, d | 2019 | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR42 | original shape, d | 2019 | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR43 | original shape, d | 2019 | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR44 | original shape, d | 2019 | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR45 | original shape, d | 2019 | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR46 | slice, d | 2019 | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR47 | slice, d | 2019 | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR48 | slice, d | 2019 | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR49 | slice, d | 2019 | 40.4 | ND | ND | ND | ND | ND | ND | ND | ND |
| LR50 | original shape, d | 2016 | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR51 | original shape, d | 2016 | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR52 | original shape, d | 2018 | ND | ND | ND | ND | ND | ND | 1.6 | ND | ND |
| LR53 | original shape, d | 2018 | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR54 | original shape, d | 2015 | ND | ND | ND | ND | ND | ND | 1.1 | ND | ND |
| LR55 | original shape, d | 2015 | ND | ND | ND | 1.1 | ND | ND | ND | ND | ND |
| LR56 | original shape, d | 2015 | ND | ND | ND | 2 | ND | ND | ND | ND | ND |
| LR57 | original shape, d | 2015 | ND | ND | ND | 1 | ND | ND | ND | ND | ND |
| LR58 | original shape, d | 2017 | ND | ND | ND | ND | ND | ND | 1.7 | ND | ND |
| LR59 | original shape, d | 2017 | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR60 | original shape, c | 2019 | 34.1 | ND | ND | 1.8 | ND | ND | 1.3 | ND | ND |
| LR61 | original shape, c | 2019 | ND | ND | ND | 5.9 | ND | ND | 1.4 | ND | ND |
| LR62 | original shape, c | 2015 | 5.3 | ND | ND | 90.2 | 15.9 | 38.7 | 119.7 | 90.6 | ND |
| LR63 | original shape, e | 2018 | ND | ND | ND | 7.2 | 5.2 | 14 | 53.2 | 31.0 | ND |
| LR64 | original shape, e | 2019 | ND | ND | ND | 483.2 | 2.3 | 18.2 | 166.8 | 84.5 | ND |
| LR65 | original shape, e | 2020 | ND | ND | ND | ND | 5.9 | 31.2 | 188.2 | 100.8 | ND |
| LR66 | original shape, a | 2018 | ND | ND | ND | 21.8 | ND | ND | 3.2 | 1.9 | ND |
| LR67 | original shape, a | 2019 | ND | ND | ND | ND | ND | ND | 3.5 | 1.9 | ND |
| LR68 | original shape, a | 2020 | 5.8 | ND | ND | ND | ND | ND | 1.1 | ND | ND |
| LR69 | original shape, c | 2019 | ND | ND | ND | 3.2 | ND | ND | ND | ND | ND |
| LR70 | original shape, c | 2020 | ND | ND | ND | 3.2 | ND | ND | 3.1 | 1.8 | ND |
| LR71 | original shape, c | 2019 | ND | ND | ND | ND | ND | ND | 1.4 | ND | ND |
| LR72 | original shape, c | 2019 | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR73 | original shape, c | 2020 | ND | ND | ND | ND | ND | ND | 1.7 | 1.1 | ND |
| LR74 | original shape, b | 2017 | 17.4 | ND | ND | 4.9 | ND | ND | 1 | ND | ND |
| LR75 | original shape, b | 2020 | ND | ND | ND | 10.8 | ND | ND | 2.1 | 1.5 | ND |
| LR76 | original shape, b | 2018 | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR77 | original shape, b | 2019 | ND | ND | ND | 2.1 | ND | ND | 1.9 | ND | ND |
| Contamination rate (%) | 15.6 | 1.3 | 1.3 | 26.0 | 5.2 | 11.7 | 31.2 | 18.2 | 18.2 | ||
| Sample | Comment | Vintage year | Contamination level (μg/kg) | ||||||||
| FB2 | Melea | OTA | MPA | Pse A | CIT | AOH | AME | DiLy | |||
| LR1 | slice | / | ND | ND | 3.2 | ND | 15.3 | 4.6 | ND | ND | ND |
| LR2 | slice | / | ND | ND | ND | ND | ND | 1.3 | ND | ND | ND |
| LR3 | slice | / | ND | ND | ND | 1.3 | ND | ND | ND | 1.3 | ND |
| LR4 | slice | / | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR5 | slice | / | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR6 | slice | / | ND | ND | ND | 1.3 | ND | ND | ND | ND | ND |
| LR7 | slice | / | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR8 | slice | / | ND | ND | ND | ND | ND | ND | ND | 14.5 | ND |
| LR9 | slice | / | 14.4 | ND | ND | ND | ND | ND | ND | ND | ND |
| LR10 | slice | / | ND | ND | 2.4 | ND | 4.6 | ND | ND | ND | ND |
| LR11 | slice | / | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR12 | slice | / | 6.3 | ND | ND | ND | ND | ND | ND | ND | ND |
| LR13 | slice | / | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR14 | slice | / | ND | ND | ND | ND | ND | ND | ND | 19.2 | ND |
| LR15 | slice | / | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR16 | slice | / | ND | ND | ND | 107.0 | ND | ND | ND | ND | ND |
| LR17 | slice | / | 10.2 | ND | ND | 99.8 | ND | ND | ND | ND | ND |
| LR18 | slice | / | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR19 | slice | / | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR20 | slice | / | 12.4 | ND | ND | ND | ND | ND | ND | 1.3 | ND |
| LR21 | slice | / | ND | ND | ND | 1.3 | ND | ND | ND | ND | ND |
| LR22 | slice | / | ND | ND | 6.4 | ND | ND | ND | ND | ND | ND |
| LR23 | slice | / | 1.3 | ND | ND | ND | ND | ND | ND | ND | ND |
| LR24 | slice | / | 8.9 | 5.7 | ND | 1.3 | ND | ND | ND | ND | ND |
| LR25 | slice | / | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR26 | slice | / | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR27 | slice | / | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR28 | slice | / | 1.3 | ND | ND | ND | ND | ND | ND | ND | ND |
| LR29 | slice | / | ND | 24.8 | ND | ND | ND | ND | ND | 15.9 | ND |
| LR30 | slice | / | 1.3 | 1.3 | ND | ND | ND | ND | ND | ND | ND |
| LR31 | original shape, a * | 2018 | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR32 | original shape, b | 2018 | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR33 | original shape, c | 2018 | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR34 | original shape, d | 2018 | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR35 | original shape, d | 2018 | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR36 | original shape, d | 2018 | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR37 | original shape, d | 2018 | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR38 | original shape | 2019 | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR39 | original shape, d | 2019 | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR40 | original shape, d | 2019 | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR41 | original shape, d | 2019 | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR42 | original shape, d | 2019 | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR43 | original shape, d | 2019 | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR44 | original shape, d | 2019 | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR45 | original shape, d | 2019 | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR46 | slice, d | 2019 | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR47 | slice, d | 2019 | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR48 | slice, d | 2019 | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR49 | slice, d | 2019 | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR50 | original shape, d | 2016 | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR51 | original shape, d | 2016 | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR52 | original shape, d | 2018 | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR53 | original shape, d | 2018 | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR54 | original shape, d | 2015 | ND | ND | ND | ND | ND | ND | ND | ND | 2.5 |
| LR55 | original shape, d | 2015 | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR56 | original shape, d | 2015 | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR57 | original shape, d | 2015 | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR58 | original shape, d | 2017 | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR59 | original shape, d | 2017 | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR60 | original shape, c | 2019 | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR61 | original shape, c | 2019 | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR62 | original shape, c | 2015 | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR63 | original shape, e | 2018 | ND | 5.1 | ND | ND | ND | ND | ND | ND | ND |
| LR64 | original shape, e | 2019 | ND | ND | ND | ND | ND | ND | 85.5 | ND | ND |
| LR65 | original shape, e | 2020 | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR66 | original shape, a | 2018 | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR67 | original shape, a | 2019 | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR68 | original shape, a | 2020 | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR69 | original shape, c | 2019 | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR70 | original shape, c | 2020 | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR71 | original shape, c | 2019 | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR72 | original shape, c | 2019 | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR73 | original shape, c | 2020 | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR74 | original shape, b | 2017 | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR75 | original shape, b | 2020 | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| LR76 | original shape, b | 2018 | ND | 1.1 | ND | ND | ND | ND | ND | ND | ND |
| LR77 | original shape, b | 2019 | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| Contamination rate (%) | 10.4 | 6.5 | 3.9 | 7.8 | 2.6 | 2.6 | 1.3 | 6.5 | 1.3 | ||
*: a: Jilin, b: Ningxia, c: Neimenggu, d: Xinjiang, e: Gansu.

Table A6.
The muti-mycotoxin contamination situation of CP.
Table A6.
The muti-mycotoxin contamination situation of CP.
| Sample | Comment | Contamination Level (μg/kg) | ||||
|---|---|---|---|---|---|---|
| BEA | ENN A1 | ENN B | ENN B1 | MPA | ||
| CP1 | original shape | ND | ND | ND | ND | 6.7 |
| CP2 | original shape | ND | ND | ND | ND | ND |
| CP3 | original shape | ND | ND | ND | ND | 9.8 |
| CP4 | original shape | ND | ND | ND | ND | 14 |
| CP5 | original shape | ND | ND | ND | ND | ND |
| CP6 | original shape | ND | ND | ND | ND | ND |
| CP7 | original shape | ND | ND | ND | ND | 25.7 |
| CP8 | original shape | ND | ND | ND | ND | ND |
| CP9 | original shape | ND | ND | ND | ND | 24.1 |
| CP10 | original shape | ND | ND | ND | ND | ND |
| CP11 | original shape | ND | ND | ND | ND | ND |
| CP12 | original shape | ND | ND | ND | ND | ND |
| CP13 | original shape | ND | ND | ND | ND | 15.2 |
| CP14 | original shape | ND | ND | ND | ND | 16 |
| CP15 | original shape | 6.9 | 3 | 19.3 | 6.1 | ND |
| CP16 | original shape | ND | ND | ND | ND | 17.9 |
| CP17 | original shape | ND | ND | ND | ND | ND |
| CP18 | original shape | ND | ND | ND | ND | 14.2 |
| CP19 | original shape | ND | ND | ND | ND | ND |
| CP20 | original shape | 14 | 2.7 | 15.8 | 4.7 | 11.4 |
| CP21 | original shape | ND | ND | ND | ND | ND |
| CP22 | original shape | ND | ND | ND | ND | 15.8 |
| CP23 | original shape | ND | ND | ND | ND | ND |
| CP24 | original shape | ND | ND | ND | ND | ND |
| CP25 | original shape | ND | ND | ND | ND | ND |
| CP26 | original shape | ND | ND | ND | ND | 20.5 |
| CP27 | original shape | ND | ND | ND | ND | 18.3 |
| CP28 | original shape | ND | ND | ND | ND | ND |
| CP29 | original shape | ND | ND | ND | ND | 16 |
| CP30 | original shape | ND | ND | ND | ND | ND |
| CP31 | original shape | ND | ND | ND | ND | 22.3 |
| CP32 | original shape | ND | ND | ND | ND | ND |
| CP33 | original shape | ND | ND | ND | ND | 12 |
| CP34 | original shape | 8.7 | 2.7 | 30.5 | 13.2 | 22.8 |
| CP35 | original shape | ND | ND | ND | ND | ND |
| CP36 | original shape | ND | ND | ND | ND | ND |
| CP37 | original shape | ND | ND | ND | ND | 16.8 |
| CP38 | original shape | ND | ND | ND | ND | 34.4 |
| CP39 | original shape | ND | ND | ND | ND | ND |
| CP40 | original shape | ND | ND | ND | ND | 14.2 |
| CP41 | original shape | ND | ND | ND | ND | ND |
| CP42 | original shape | ND | ND | ND | ND | ND |
| CP43 | original shape | ND | ND | ND | ND | 21 |
| CP44 | original shape | ND | ND | ND | ND | ND |
| CP45 | original shape | ND | ND | ND | ND | 15.8 |
| CP46 | original shape | ND | ND | ND | ND | ND |
| CP47 | original shape | ND | ND | ND | ND | 15 |
| CP48 | original shape | 12.1 | 1.2 | 16.4 | 3.7 | ND |
| CP49 | original shape | 10.5 | 1.6 | 22.7 | 5.1 | 22.0 |
| CP50 | original shape | ND | ND | ND | ND | ND |
| CP51 | original shape | ND | ND | ND | ND | 16.3 |
| CP52 | original shape | ND | ND | ND | ND | ND |
| CP53 | original shape | ND | ND | ND | ND | 10.5 |
| CP54 | original shape | ND | ND | ND | ND | ND |
| CP55 | original shape | ND | ND | ND | ND | 11.9 |
| CP56 | original shape | ND | ND | ND | ND | 21.4 |
| CP57 | original shape | ND | ND | ND | ND | 16.5 |
| CP58 | original shape | ND | ND | ND | ND | ND |
| CP59 | original shape | ND | ND | ND | ND | ND |
| CP60 | original shape | ND | ND | ND | ND | ND |
| CP61 | original shape | ND | ND | ND | ND | ND |
| CP62 | original shape | ND | ND | ND | ND | 18.3 |
| CP63 | original shape | ND | ND | ND | ND | ND |
| CP64 | original shape | 5.5 | 7.3 | 65.8 | 20.0 | 16.8 |
| CP65 | original shape | ND | ND | ND | ND | ND |
| CP66 | original shape | ND | ND | ND | ND | 22.8 |
| CP67 | original shape | ND | ND | ND | ND | ND |
| CP68 | original shape | ND | ND | ND | ND | 12.1 |
| CP69 | original shape | ND | ND | ND | ND | 22.4 |
| CP70 | original shape | ND | ND | ND | ND | ND |
| CP71 | original shape | ND | ND | ND | ND | ND |
| CP72 | original shape | ND | ND | ND | ND | 16.7 |
| CP73 | original shape | ND | ND | ND | ND | ND |
| CP74 | original shape | ND | ND | ND | ND | 16.8 |
| CP75 | original shape | ND | ND | ND | ND | ND |
| CP76 | original shape | ND | ND | ND | ND | ND |
| CP77 | original shape | ND | ND | ND | ND | 20.4 |
| CP78 | original shape | ND | ND | ND | ND | ND |
| CP79 | original shape | ND | ND | ND | ND | 15.6 |
| CP80 | original shape | ND | ND | ND | ND | ND |
| CP81 | original shape | ND | ND | ND | ND | 19.9 |
| CP82 | original shape | ND | ND | ND | ND | ND |
| CP83 | original shape | ND | ND | ND | ND | ND |
| CP84 | original shape | ND | ND | ND | ND | 26.7 |
| CP85 | original shape | ND | ND | ND | ND | ND |
| CP86 | original shape | ND | ND | ND | ND | ND |
| CP87 | original shape | ND | ND | ND | ND | ND |
| CP88 | original shape | ND | ND | ND | ND | ND |
| CP89 | original shape | ND | ND | ND | ND | 32.3 |
| CP90 | original shape | ND | ND | ND | ND | ND |
| CP91 | original shape | ND | ND | ND | ND | ND |
| CP92 | original shape | ND | ND | ND | ND | 11.8 |
| CP93 | original shape | 6.2 | 15.4 | 167.2 | 34.6 | 12.4 |
| CP94 | original shape | 5.1 | 7.3 | 81.3 | 20.2 | 14.5 |
| CP95 | original shape | 8.8 | 3.7 | 35.9 | 11 | 21.8 |
| CP96 | original shape | ND | ND | ND | ND | ND |
| CP97 | original shape | ND | ND | ND | ND | ND |
| CP98 | original shape | ND | ND | ND | ND | ND |
| CP99 | original shape | ND | ND | ND | ND | 33 |
| CP100 | original shape | 11.5 | 11.7 | 100.4 | 25.6 | 21.7 |
| CP101 | original shape | ND | ND | ND | ND | ND |
| CP102 | original shape | ND | ND | ND | ND | ND |
| CP103 | original shape | ND | ND | ND | ND | ND |
| CP104 | original shape | ND | ND | ND | ND | ND |
| CP105 | original shape | ND | ND | ND | ND | ND |
| CP106 | original shape | ND | ND | ND | ND | ND |
| CP107 | original shape | ND | ND | ND | ND | ND |
| CP108 | original shape | ND | ND | ND | ND | ND |
| CP109 | original shape | ND | ND | ND | ND | ND |
| CP110 | original shape | ND | ND | ND | ND | ND |
| CP111 | original shape | ND | ND | ND | ND | ND |
| CP112 | original shape | 10.3 | ND | 10.7 | 3.3 | ND |
| CP113 | powder | ND | ND | ND | ND | ND |
| CP114 | powder | ND | ND | ND | ND | ND |
| CP115 | powder | ND | ND | ND | ND | ND |
| CP116 | original shape | ND | ND | ND | ND | ND |
| CP117 | original shape | ND | ND | ND | ND | ND |
| CP118 | original shape | ND | ND | ND | ND | ND |
| CP119 | original shape | ND | ND | ND | ND | ND |
| CP120 | original shape | ND | ND | ND | ND | ND |
| CP121 | original shape | ND | ND | ND | ND | ND |
| CP122 | original shape | ND | ND | ND | ND | ND |
| CP123 | original shape | ND | ND | ND | ND | ND |
| CP124 | original shape | ND | ND | ND | ND | ND |
| CP125 | original shape | ND | ND | ND | ND | ND |
| CP126 | original shape | ND | ND | ND | ND | ND |
| CP127 | original shape | ND | ND | ND | ND | ND |
| CP128 | original shape | ND | ND | ND | ND | ND |
| CP129 | original shape | ND | ND | ND | ND | ND |
| CP130 | original shape | ND | ND | ND | ND | ND |
| CP131 | original shape | ND | ND | ND | ND | ND |
| Contamination rate (%) | 8.4 | 7.6 | 8.4 | 8.4 | 35.9 | |

Table A7.
The muti-mycotoxin contamination situation of CS.
Table A7.
The muti-mycotoxin contamination situation of CS.
| Sample | Comment | Contamination Level (μg/kg) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 15-Asp | 3-ADON | Apici | BEA | DON | DAS | Equi | FB1 | FB2 | ||
| CS1 | original shape | 21.3 | ND | ND | ND | ND | 37.1 | ND | 24.3 | 10.5 |
| CS2 | original shape | 2.2 | ND | ND | ND | ND | 2.3 | ND | 13.2 | 3.0 |
| CS3 | original shape | 6.8 | ND | ND | 0.2 | 100.4 | 7.3 | ND | 353.0 | 50.5 |
| CS4 | original shape | 1.0 | ND | ND | ND | ND | 1.9 | ND | 91.5 | 9.0 |
| CS5 | original shape | 1.0 | ND | ND | ND | 41.7 | 2.6 | ND | 98.0 | 5.7 |
| CS6 | original shape | 1.6 | 20.6 | ND | ND | 32.3 | 2.8 | ND | ND | ND |
| CS7 | original shape | 1.0 | ND | ND | 2.0 | 1.0 | 2.7 | 1.0 | ND | ND |
| CS8 | original shape | 1.0 | ND | ND | ND | 1.0 | 1.5 | ND | 5.0 | ND |
| CS9 | original shape | 1.0 | ND | ND | ND | 1.0 | 0.7 | ND | 96.3 | 5.5 |
| CS10 | original shape | 5.2 | ND | 0.6 | ND | 12.3 | 3.8 | ND | 36.4 | ND |
| CS11 | original shape | 4.6 | ND | ND | ND | 422.3 | 3.0 | ND | ND | ND |
| CS12 | original shape | 2.1 | ND | ND | ND | ND | 9.4 | ND | ND | ND |
| CS13 | original shape | ND | ND | ND | ND | 21.5 | 1.2 | ND | 18.3 | 2.3 |
| CS14 | original shape | 8.8 | ND | ND | ND | ND | 4.4 | ND | 4.1 | ND |
| CS15 | original shape | 10.5 | ND | ND | ND | ND | 9.6 | ND | 56.4 | 10.7 |
| CS16 | original shape | ND | ND | ND | ND | ND | ND | ND | 5.6 | ND |
| CS17 | original shape | ND | ND | ND | 3.1 | ND | ND | ND | ND | 7.1 |
| CS18 | original shape | ND | ND | ND | 11.1 | ND | ND | ND | 18.1 | 19.6 |
| CS19 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | 3.9 |
| CS20 | original shape | ND | ND | ND | ND | ND | ND | ND | 11.1 | 8.4 |
| CS21 | original shape | ND | ND | ND | 5.1 | ND | ND | ND | 3.2 | 1.1 |
| CS22 | original shape | ND | ND | ND | 46.2 | ND | ND | ND | 2.9 | ND |
| CS23 | original shape | ND | ND | ND | 4.5 | ND | ND | ND | 23.5 | 2.4 |
| CS24 | original shape | ND | ND | ND | 18.6 | ND | ND | ND | 10.1 | 1.1 |
| CS25 | original shape | ND | ND | ND | 13.7 | ND | ND | ND | 54.6 | 23.4 |
| CS26 | original shape | ND | ND | ND | 13.9 | ND | ND | ND | 26.6 | 23.0 |
| CS27 | original shape | ND | ND | ND | ND | ND | ND | ND | 3.9 | ND |
| CS28 | original shape | ND | ND | ND | 14.2 | ND | ND | ND | 172.8 | 38.1 |
| CS29 | original shape | ND | ND | ND | 50.0 | ND | ND | ND | 20.4 | 3.0 |
| CS30 | original shape | ND | ND | ND | 5.8 | ND | ND | ND | 5.2 | 7.4 |
| CS31 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| CS32 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| CS33 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| CS34 | original shape | ND | ND | ND | 11.7 | ND | ND | ND | 21.9 | 10.3 |
| CS35 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| CS36 | original shape | ND | ND | ND | 2.1 | ND | ND | ND | 73.4 | 13.9 |
| CS37 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| CS38 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| CS39 | original shape | ND | ND | ND | ND | ND | ND | ND | 4.1 | ND |
| CS40 | original shape | ND | ND | ND | ND | ND | ND | ND | 16.2 | 5.5 |
| CS41 | original shape | ND | ND | ND | 4.4 | ND | ND | ND | 43.0 | 12.6 |
| CS42 | original shape | ND | ND | ND | 109.9 | ND | ND | ND | 69.8 | 17.5 |
| CS43 | original shape | ND | ND | ND | 56.2 | ND | ND | ND | 55.7 | 53.9 |
| CS44 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| CS45 | original shape | ND | ND | ND | ND | ND | ND | ND | 12.4 | 2.2 |
| CS46 | original shape | ND | ND | ND | 111.1 | ND | ND | ND | 54.4 | 13.2 |
| CS47 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| Contamination rate (%) | 29.8 | 2.1 | 2.1 | 40.4 | 19.1 | 31.9 | 2.1 | 70.2 | 59.6 | |
| Sample | comment | Contamination level (μg/kg) | ||||||||
| FB3 | FuX | MPA | ZEN | ZAN | α-ZEL | β-ZAL | β-ZEL | CVD | ||
| CS1 | original shape | 2.4 | ND | ND | ND | ND | ND | ND | ND | ND |
| CS2 | original shape | 6.4 | ND | ND | ND | ND | ND | ND | ND | ND |
| CS3 | original shape | 27.2 | 50.8 | ND | 59.1 | 23.0 | 21.7 | 22.5 | 22.5 | ND |
| CS4 | original shape | ND | ND | ND | 53.4 | 37.2 | 35.9 | 35.8 | 35.6 | ND |
| CS5 | original shape | 8.3 | ND | 24.5 | 62.0 | 27.1 | 51.2 | 71.8 | 27.0 | ND |
| CS6 | original shape | ND | 1.0 | ND | 10.0 | ND | ND | ND | ND | ND |
| CS7 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| CS8 | original shape | ND | ND | ND | 8.5 | ND | ND | ND | ND | ND |
| CS9 | original shape | ND | 36.7 | ND | 9.6 | ND | ND | ND | ND | ND |
| CS10 | original shape | ND | 21.0 | ND | 11.2 | ND | ND | ND | ND | ND |
| CS11 | original shape | ND | 52.2 | ND | 9.4 | ND | ND | ND | ND | ND |
| CS12 | original shape | ND | 29.3 | ND | 96.7 | ND | ND | ND | ND | ND |
| CS13 | original shape | ND | 13.6 | ND | 9.8 | ND | ND | ND | ND | ND |
| CS14 | original shape | ND | ND | ND | 10.8 | ND | ND | ND | ND | ND |
| CS15 | original shape | 10.0 | ND | ND | 29.1 | ND | ND | ND | ND | 25.7 |
| CS16 | original shape | ND | ND | ND | 12.9 | ND | ND | ND | ND | ND |
| CS17 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| CS18 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| CS19 | original shape | ND | ND | ND | 69.1 | 3.8 | ND | ND | ND | ND |
| CS20 | original shape | ND | ND | ND | 96.8 | 2.3 | ND | ND | ND | ND |
| CS21 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| CS22 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| CS23 | original shape | ND | ND | ND | 5.6 | ND | ND | ND | ND | ND |
| CS24 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| CS25 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| CS26 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| CS27 | original shape | ND | ND | ND | 14.5 | ND | ND | ND | ND | ND |
| CS28 | original shape | ND | ND | ND | 77.5 | 2.0 | ND | ND | ND | ND |
| CS29 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| CS30 | original shape | ND | ND | ND | 6.6 | ND | ND | ND | ND | ND |
| CS31 | original shape | ND | ND | ND | 105.1 | 1.2 | ND | ND | ND | ND |
| CS32 | original shape | ND | ND | ND | 28.8 | 1.0 | ND | ND | ND | ND |
| CS33 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| CS34 | original shape | ND | ND | ND | 81.4 | 3.8 | ND | ND | ND | ND |
| CS35 | original shape | ND | ND | ND | 65.4 | 2.1 | ND | ND | ND | ND |
| CS36 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| CS37 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| CS38 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| CS39 | original shape | ND | ND | ND | 13.7 | ND | ND | ND | ND | ND |
| CS40 | original shape | ND | ND | ND | 49.5 | ND | ND | ND | ND | ND |
| CS41 | original shape | ND | ND | ND | 206.9 | 6.0 | ND | ND | ND | ND |
| CS42 | original shape | ND | ND | ND | 4.1 | ND | ND | ND | ND | ND |
| CS43 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| CS44 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| CS45 | original shape | ND | ND | ND | 13.2 | ND | ND | ND | ND | ND |
| CS46 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| CS47 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| Contamination rate (%) | 10.6 | 14.9 | 2.1 | 59.6 | 23.4 | 6.4 | 6.4 | 6.4 | 2.1 | |
| Sample | comment | Contamination level (μg/kg) | ||||||||
| Grise | CPA | 7-D-G | AFB1 | AFB2 | OTA | Ster | Glio | Ten | ||
| CS1 | original shape | ND | 13.3 | ND | 0.2 | ND | ND | ND | ND | 1.7 |
| CS2 | original shape | ND | 3.1 | ND | ND | ND | ND | ND | ND | ND |
| CS3 | original shape | ND | ND | ND | 0.6 | ND | 3.2 | ND | ND | 0.5 |
| CS4 | original shape | ND | ND | ND | 0.4 | ND | ND | ND | ND | 0.2 |
| CS5 | original shape | ND | ND | ND | 5.1 | 2.0 | ND | 0.8 | ND | 0.2 |
| CS6 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| CS7 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| CS8 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| CS9 | original shape | ND | ND | ND | 0.4 | ND | ND | ND | 1.0 | ND |
| CS10 | original shape | ND | ND | 54.4 | ND | ND | ND | ND | 57.8 | ND |
| CS11 | original shape | 1.8 | ND | ND | 4.9 | 0.3 | ND | ND | ND | ND |
| CS12 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| CS13 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| CS14 | original shape | ND | ND | ND | 0.1 | ND | ND | ND | ND | ND |
| CS15 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| CS16 | original shape | ND | ND | ND | 0.3 | ND | ND | ND | ND | ND |
| CS17 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| CS18 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| CS19 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| CS20 | original shape | ND | ND | ND | 1.0 | ND | ND | ND | ND | ND |
| CS21 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| CS22 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| CS23 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| CS24 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| CS25 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| CS26 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| CS27 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| CS28 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| CS29 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| CS30 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| CS31 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| CS32 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| CS33 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| CS34 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| CS35 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| CS36 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| CS37 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| CS38 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| CS39 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| CS40 | original shape | ND | ND | ND | 3.3 | 1.2 | ND | ND | ND | ND |
| CS41 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| CS42 | original shape | ND | ND | ND | 2.4 | ND | ND | ND | ND | ND |
| CS43 | original shape | ND | ND | ND | 1.1 | ND | ND | ND | ND | ND |
| CS44 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| CS45 | original shape | ND | ND | ND | ND | 0.7 | ND | ND | ND | ND |
| CS46 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| CS47 | original shape | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| Contamination rate (%) | 2.1 | 4.3 | 2.1 | 25.5 | 8.5 | 2.1 | 2.1 | 4.3 | 8.5 | |

Table A8.
Source and purity of 73 reference standards.
Table A8.
Source and purity of 73 reference standards.
| No. | Mycotoxin | Lot. | Purity | Company |
|---|---|---|---|---|
| 1 | 15-Acetoxyscirpenol | AS466216 | 99% | Apollo Scientific, Stockport, UK |
| 2 | 15-Acetyldeoxynivalenol | L14261A | 99.9% | Romer Labs, Tullin, Austria |
| 3 | 3-Acetyldeoxynivalenol | SZBD036XV | 98.3% | Sigma-Aldrich, Laramie, USA |
| 4 | 7-Dechloro Griseofulvin | 10-KPA-158-4 | 98.85% | Toronto Research Chemicals, Toronto, Canada |
| 5 | Aflatoxin B1 | LKB0P75 | 98% | J&K Scientific, Shanghai, China |
| 6 | Aflatoxin B2 | L260Q63 | 98% | J&K Scientific, Shanghai, China |
| 7 | Aflatoxin G1 | 282095 | 98% | J&K Scientific, Shanghai, China |
| 8 | Aflatoxin G2 | LQ10Q76 | 98.72% | J&K Scientific, Shanghai, China |
| 9 | Aflatoxin M1 | 2-AJK-95-1 | 95% | Toronto Research Chemicals, Toronto, Canada |
| 10 | Aflatoxin M2 | LB40Q56 | 95% | J&K Scientific, Shanghai, China |
| 11 | Aflatoxin P1 | 2-BSR-92-3 | 98% | Toronto Research Chemicals, Toronto, Canada |
| 12 | Agroclavine | L13212A | 98.2% | Chiron AS, Trondheim, Norway |
| 13 | Anisomycin | LDC0Q85 | 99.04% | J&K Scientific, Shanghai, China |
| 14 | Apicidin | 3-PQY-12-1 | 96% | Toronto Research Chemicals, Toronto, Canada |
| 15 | Beauvericin | LHB0P07 | 99% | J&K Scientific, Shanghai, China |
| 16 | Chaetocin | 124M4012V | 98% | Sigma-Aldrich, Laramie, USA |
| 17 | Chetomin | 367820 | 97.17% | International Laboratory USA, San Bruno, USA |
| 18 | Citrinin | LH40O33 | 98.5 | J&K Scientific, Shanghai, China |
| 19 | Citreoviridin | L280Q47 | 97.17% | J&K Scientific, Shanghai, China |
| 20 | Cyclopiazonic acid | 085M4081V | 98% | Sigma-Aldrich, Laramie, USA |
| 21 | Diacetoxyscirpenol | L280Q35 | 99% | J&K Scientific, Shanghai, China |
| 22 | Dihydrolysergamide | 3-NAV-62-1 | 98% | Toronto Research Chemicals, Toronto, Canada |
| 23 | Deepoxy-deoxynivalenol | L16103A | 98% | Chiron AS, Trondheim, Norway |
| 24 | Deoxynivalenol | LR10Q103 | 99.83% | J&K Scientific, Shanghai, China |
| 25 | Enniatin A | AL26.129 | 99% | BioAustralis, New South Wales, Australia |
| 26 | Enniatin A1 | AL26.21 | 99% | BioAustralis, New South Wales, Australia |
| 27 | Enniatin B | AL26.135 | 99% | BioAustralis, New South Wales, Australia |
| 28 | Enniatin B1 | AL23.115 | 99% | BioAustralis, New South Wales, Australia |
| 29 | Equisetin | 2-LWJ-95-1 | 93.06% | Toronto Research Chemicals, Toronto, Canada |
| 30 | Ergocornine | L14331F | 97.8% | Chiron AS, Trondheim, Norway |
| 31 | Ergocorninine | L15212A | 97.8% | Chiron AS, Trondheim, Norway |
| 32 | Ergocristine | LK70P83 | 99% | Chiron AS, Trondheim, Norway |
| 33 | Ergocristinine | L15071E | 99% | Chiron AS, Trondheim, Norway |
| 34 | Ergocryptine | L14331E | 97.6% | Chiron AS, Trondheim, Norway |
| 35 | Ergocryptinine | L15071D | 97.6% | Chiron AS, Trondheim, Norway |
| 36 | Ergosine | L15282E | 99.9% | Chiron AS, Trondheim, Norway |
| 37 | Fumonisin B1 | SZBF083XV | 98.5% | Sigma-Aldrich, Laramie, USA |
| 38 | Fumonisin B2 | SZBF089XV | 98.5% | Sigma-Aldrich, Laramie, USA |
| 39 | Fumonisin B3 | SZBE295XV | 98.5% | Sigma-Aldrich, Laramie, USA |
| 40 | Fumagillin | LK70P67 | 98% | J&K Scientific, Shanghai, China |
| 41 | Fusarenon X | L14141F | 99.9% | Romer Labs, Tullin, Austria |
| 42 | Gliotoxin | LJ50Q77 | 99% | J&K Scientific, Shanghai, China |
| 43 | Griseofulvin | L5C0O27 | 99.6% | J&K Scientific, Shanghai, China |
| 44 | HT-2 toxin | SZBA287X | 99.9% | Sigma-Aldrich, Laramie, USA |
| 45 | Lysergamide | 1-GAC-130-2 | 99.6% | Toronto Research Chemicals, Toronto, Canada |
| 46 | Meleagrin | 2-LXM-37-1 | 97% | Toronto Research Chemicals, Toronto, Canada |
| 47 | Monocerin | AS467363 | 99.9% | Apollo Scientific, Stockport, UK |
| 48 | Mycophenolic Acid | LJ20O39 | 98% | J&K Scientific, Shanghai, China |
| 49 | Neosolaniol | SZBD144XV | 99.3% | Sigma-Aldrich, Laramie, USA |
| 50 | O-methylsterigmatocystin | 00013645-201 | 99% | ChromaDex Standards, Irvine, USA |
| 51 | Ostreogrycin A | 361047 | 99% | International Laboratory USA, San Bruno, USA |
| 52 | Ochratoxin A | 5-JKS-45-3 | 98% | Toronto Research Chemicals, Toronto, Canada |
| 53 | Ochratoxin B | LAC0P30 | 99% | J&K Scientific, Shanghai, China |
| 54 | Ochratoxin C | 5-UKS-34-1 | 98% | Toronto Research Chemicals, Toronto, Canada |
| 55 | Oxaline | AS463714 | 92.87% | Apollo Scientific, Stockport, UK |
| 56 | Pseurotin A | 3-PQY-14-1 | 95% | Toronto Research Chemicals, Toronto, Canada |
| 57 | Puromycin | 00016435-939 | 99% | ChromaDex Standards, Irvine, USA |
| 58 | Roquefortine C | AS467985 | 98% | Apollo Scientific, Stockport, UK |
| 59 | Secalonic acid D | L15471S | 99% | Chiron AS, Trondheim, Norway |
| 60 | Sterigmatocystin | SZBF020XV | 99.3% | Sigma-Aldrich, Laramie, USA |
| 61 | T-2-triol | L15354A | 98.2% | Chiron AS, Trondheim, Norway |
| 62 | T-2 toxin | 2J0A02 | 99% | Pribolab, Qingdao, China |
| 63 | Tentoxin | 1I1J28 | 99% | Pribolab, Qingdao, China |
| 64 | Wortmannin | L6B0P68 | 98% | J&K Scientific, Shanghai, China |
| 65 | α-zearalanol | 086K4024 | 97% | Sigma-Aldrich, Laramie, USA |
| 66 | α-zearalenol | 056W4021V | 97% | Sigma-Aldrich, Laramie, USA |
| 67 | β-zearalanol | 115K4037 | 98% | Sigma-Aldrich, Laramie, USA |
| 68 | β-zearalenol | 115M4019V | 98% | Sigma-Aldrich, Laramie, USA |
| 69 | Patulin | LAS467986 | 99% | Apollo Scientific, Stockport, UK |
| 70 | Zearalanone | 4-RNP-61-1 | 96% | Toronto Research Chemicals, Toronto, Canada |
| 71 | Zearalenone | SZBC355XV | 99.3% | Sigma-Aldrich, Laramie, USA |
| 72 | Alternariol-methylether | 045M4017V | 96% | Sigma-Aldrich, Laramie, USA |
| 73 | Alternariol | 084M4167V | 96% | Sigma-Aldrich, Laramie, USA |
References
- Alassane-Kpembi, I.; Schatzmayr, G.; Taranu, I.; Marin, D.; Puel, O.; Oswald, I.P. Mycotoxins co-contamination: Methodological aspects and biological relevance of combined toxicity studies. Crit. Rev. Food Sci. Nutr. 2017, 57, 3489–3507. [Google Scholar] [CrossRef] [PubMed]
- Bennett, J.W.; Klich, M. Mycotoxins. Clin. Microbiol. Rev. 2003, 16, 497–516. [Google Scholar] [CrossRef] [PubMed]
- Pleadin, J.; Frece, J.; Markov, K. Chapter Eight-Mycotoxins in food and feed. In Advances in Food and Nutrition Research; Toldrá, F., Ed.; Academic Press: Cambridge, MA, USA, 2019; Volume 89, pp. 297–345. [Google Scholar]
- Alshannaq, A.; Yu, J.H. Occurrence, Toxicity, and Analysis of Major Mycotoxins in Food. Int. J. Environ. Res. Public Health 2017, 14, 632. [Google Scholar] [CrossRef]
- Awuchi, C.G.; Ondari, E.N.; Nwozo, S.; Odongo, G.A.; Eseoghene, I.J.; Twinomuhwezi, H.; Ogbonna, C.U.; Upadhyay, A.K.; Adeleye, A.O.; Okpala, C.O.R. Mycotoxins’ Toxicological Mechanisms Involving Humans, Livestock and Their Associated Health Concerns: A Review. Toxins 2022, 14, 167. [Google Scholar] [CrossRef] [PubMed]
- Kabak, B.; Dobson, A.D. Mycotoxins in spices and herbs-An update. Crit. Rev. Food Sci. Nutr. 2017, 57, 18–34. [Google Scholar] [CrossRef]
- Lu, Q.; Qin, J.-A.; Fu, Y.-W.; Luo, J.-Y.; Lu, J.-H.; Logrieco, A.F.; Yang, M.-H. Modified mycotoxins in foodstuffs, animal feed, and herbal medicine: A systematic review on global occurrence, transformation mechanism and analysis methods. TrAC Trends Anal. Chem. 2020, 133, 116088. [Google Scholar] [CrossRef]
- Sun, L.H.; Lei, M.Y.; Zhang, N.Y.; Gao, X.; Li, C.; Krumm, C.S.; Qi, D.S. Individual and combined cytotoxic effects of aflatoxin B1, zearalenone, deoxynivalenol and fumonisin B1 on BRL 3A rat liver cells. Toxicon 2015, 95, 6–12. [Google Scholar] [CrossRef]
- Clarke, R.; Connolly, L.; Frizzell, C.; Elliott, C.T. Cytotoxic assessment of the regulated, co-existing mycotoxins aflatoxin B1, fumonisin B1 and ochratoxin, in single, binary and tertiary mixtures. Toxicon 2014, 90, 70–81. [Google Scholar] [CrossRef]
- Karsauliya, K.; Yahavi, C.; Pandey, A.; Bhateria, M.; Sonker, A.K.; Pandey, H.; Sharma, M.; Singh, S.P. Co-occurrence of mycotoxins: A review on bioanalytical methods for simultaneous analysis in human biological samples, mixture toxicity and risk assessment strategies. Toxicon 2022, 218, 25–39. [Google Scholar] [CrossRef]
- Šegvić Klarić, M. Adverse effects of combined mycotoxins. Arch. Ind. Hyg. Toxicol. 2012, 63, 519–530. [Google Scholar]
- Liu, H.; Zhu, L.; Chen, L.; Li, L. Therapeutic potential of traditional Chinese medicine in atherosclerosis: A review. Phytother. Res. 2022, 36, 4080–4100. [Google Scholar] [CrossRef] [PubMed]
- Shang, A.; Gan, R.-Y.; Xu, X.-Y.; Mao, Q.-Q.; Zhang, P.-Z.; Li, H.-B. Effects and mechanisms of edible and medicinal plants on obesity: An updated review. Crit. Rev. Food Sci. Nutr. 2021, 61, 2061–2077. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Li, Y.; Zhou, S.; Guo, C.; Dong, X.; Li, Q.; Guo, J.; Wang, Y.; Huang, L. The Differences of Nutrient Components in Edible and Feeding Coix Seed at Different Developmental Stages Based on a Combined Analysis of Metabolomics. Molecules 2023, 28, 3759. [Google Scholar] [CrossRef]
- Zhao, D.T.; Gao, Y.J.; Zhang, W.J.; Bi, T.C.; Wang, X.; Ma, C.X.; Rong, R. Development a multi-immunoaffinity column LC-MS-MS method for comprehensive investigation of mycotoxins contamination and co-occurrence in traditional Chinese medicinal materials. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2021, 1178, 122730. [Google Scholar] [CrossRef]
- Spanjer, M.C.; Rensen, P.M.; Scholten, J.M. LC–MS/MS multi-method for mycotoxins after single extraction, with validation data for peanut, pistachio, wheat, maize, cornflakes, raisins and figs. Food Addit. Contam. Part A 2008, 25, 472–489. [Google Scholar] [CrossRef]
- Sulyok, M.; Berthiller, F.; Krska, R.; Schuhmacher, R. Development and validation of a liquid chromatography/tandem mass spectrometric method for the determination of 39 mycotoxins in wheat and maize. Rapid Commun. Mass Spectrom. 2006, 20, 2649–2659. [Google Scholar] [CrossRef]
- Chau, S.L.; Zhao, A.; Jia, W.; Wang, L. Simultaneous Determination of Pesticide Residues and Mycotoxins in Storage Pu-erh Tea Using Ultra-High-Performance Liquid Chromatography Coupled with Tandem Mass Spectrometry. Molecules 2023, 28, 6883. [Google Scholar] [CrossRef]
- Mbisana, M.; Rebagamang, T.; Mogopodi, D.; Chibua, I. Development and validation of a QuEChERS-LC-MS/MS method for determination of multiple mycotoxins in maize and sorghum from Botswana. Front. Fungal Biol. 2023, 4, 1141427. [Google Scholar] [CrossRef]
- Tölgyesi, Á.; Cseh, A.; Simon, A.; Sharma, V.K. Development of a Novel LC-MS/MS Multi-Method for the Determination of Regulated and Emerging Food Contaminants Including Tenuazonic Acid, a Chromatographically Challenging Alternaria Toxin. Molecules 2023, 28, 1468. [Google Scholar] [CrossRef]
- Zhao, X.; Liu, D.; Yang, X.; Zhang, L.; Yang, M. Detection of seven Alternaria toxins in edible and medicinal herbs using ultra-high performance liquid chromatography-tandem mass spectrometry. Food Chem. X 2022, 13, 100186. [Google Scholar] [CrossRef]
- Cho, H.-D.; Suh, J.H.; Feng, S.; Eom, T.; Kim, J.; Hyun, S.M.; Kim, J.; Wang, Y.; Han, S.B. Comprehensive analysis of multi-class mycotoxins in twenty different species of functional and medicinal herbs using liquid chromatography–tandem mass spectrometry. Food Control 2019, 96, 517–526. [Google Scholar] [CrossRef]
- Bian, Y.; Zhang, Y.; Zhou, Y.; Wei, B.; Feng, X. Recent Insights into Sample Pretreatment Methods for Mycotoxins in Different Food Matrices: A Critical Review on Novel Materials. Toxins 2023, 15, 215. [Google Scholar] [CrossRef] [PubMed]
- Nouri, N.; Sereshti, H. Electrospun polymer composite nanofiber-based in-syringe solid phase extraction in tandem with dispersive liquid-liquid microextraction coupled with HPLC-FD for determination of aflatoxins in soybean. Food Chem. 2019, 289, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Ganesan, A.R.; Mohan, K.; Karthick Rajan, D.; Pillay, A.A.; Palanisami, T.; Sathishkumar, P.; Conterno, L. Distribution, toxicity, interactive effects, and detection of ochratoxin and deoxynivalenol in food: A review. Food Chem. 2022, 378, 131978. [Google Scholar] [CrossRef]
- Sun, X.D.; Su, P.; Shan, H. Mycotoxin Contamination of Rice in China. J. Food Sci. 2017, 82, 573–584. [Google Scholar] [CrossRef]
- USEPA. Guidelines for the Health Risk Assessment of Chemical Mixtures; USEPA: Washington, DC, USA, 1986. [Google Scholar]
- EFSA. Statement on the applicability of the Margin of Exposure approach for the safety assessment of impurities which are both genotoxic and carcinogenic in substances added to food/feed. EFSA J. 2012, 10, 2578. [Google Scholar]
- Zhang, Y.; Kuang, F.; Liu, C.; Ma, K.; Liu, T.; Zhao, M.; Lv, G.; Huang, H. Contamination and Health Risk Assessment of Multiple Mycotoxins in Edible and Medicinal Plants. Toxins 2023, 15, 209. [Google Scholar] [CrossRef]
- Lu, Q.; Ruan, H.-N.; Sun, X.-Q.; Luo, J.-Y.; Yang, M.-H. Contamination status and health risk assessment of 31 mycotoxins in six edible and medicinal plants using a novel green defatting and depigmenting pretreatment coupled with LC-MS/MS. LWT 2022, 161, 113401. [Google Scholar] [CrossRef]
- Arooj, M.; Imran, S.; Inam-Ur-Raheem, M.; Rajoka, M.S.R.; Sameen, A.; Siddique, R.; Sahar, A.; Tariq, S.; Riaz, A.; Hussain, A.; et al. Lotus seeds (Nelumbinis semen) as an emerging therapeutic seed: A comprehensive review. Food Sci. Nutr. 2021, 9, 3971–3987. [Google Scholar] [CrossRef]
- Du, J.; Yin, G.; Hu, Y.; Shi, S.; Jiang, J.; Song, X.; Zhang, Z.; Wei, Z.; Tang, C.; Lyu, H. Coicis semen protects against focal cerebral ischemia-reperfusion injury by inhibiting oxidative stress and promoting angiogenesis via the TGFβ/ALK1/Smad1/5 signaling pathway. Aging 2020, 13, 877–893. [Google Scholar] [CrossRef]
- Morita, A.; Omoya, Y.; Ito, R.; Ishibashi, Y.; Hiramoto, K.; Ohnishi, S.; Yoshikawa, N.; Kawanishi, S. Glycyrrhizin and its derivatives promote hepatic differentiation via sweet receptor, Wnt, and Notch signaling. Biochem. Biophys. Rep. 2021, 28, 101181. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Jiang, Z.; Wen, M.; Wu, Z.; Zha, M.; Xu, W.; Zhang, L. Chemical Variation of Chenpi (Citrus Peels) and Corresponding Correlated Bioactive Compounds by LC-MS Metabolomics and Multibioassay Analysis. Front. Nutr. 2022, 9, 825381. [Google Scholar] [CrossRef]
- Rausch, A.K.; Brockmeyer, R.; Schwerdtle, T. Development and validation of a liquid chromatography tandem mass spectrometry multi-method for the determination of 41 free and modified mycotoxins in beer. Food Chem. 2021, 338, 127801. [Google Scholar] [CrossRef] [PubMed]
- Varga, E.; Glauner, T.; Berthiller, F.; Krska, R.; Schuhmacher, R.; Sulyok, M. Development and validation of a (semi-)quantitative UHPLC-MS/MS method for the determination of 191 mycotoxins and other fungal metabolites in almonds, hazelnuts, peanuts and pistachios. Anal. Bioanal. Chem. 2013, 405, 5087–5104. [Google Scholar] [CrossRef]
- Huang, X.; Wang, S.; Mao, D.; Miao, S.; Hu, Q.; Ji, S. Optimized QuEChERS Method Combined with UHPLC-MS/MS for the Simultaneous Determination of 15 Mycotoxins in Liquorice. J. AOAC Int. 2018, 101, 633–642. [Google Scholar] [CrossRef]
- Liao, X.; Li, Y.; Long, N.; Xu, Q.; Li, P.; Wang, J.; Zhou, L.; Kong, W. Multi-mycotoxin detection and human exposure risk assessment in medicinal foods. Food Res. Int. 2023, 164, 112456. [Google Scholar] [CrossRef]
- He, J.; Tao, Z.; Liang, S.; Ye, D. Compression and shearing force on kernel rupture in shelling fresh lotus seeds. Int. J. Agric. Biol. Eng. 2021, 14, 237–242. [Google Scholar] [CrossRef]
- Zhao, X.; Liu, D.; Zhang, L.; Zhou, Y.; Yang, M. Development and optimization of a method based on QuEChERS-dSPE followed by UPLC-MS/MS for the simultaneous determination of 21 mycotoxins in nutmeg and related products. Microchem. J. 2021, 168, 106499. [Google Scholar] [CrossRef]
- Fernández Pinto, V.; Patriarca, A.; Locani, O.; Vaamonde, G. Natural co-occurrence of aflatoxin and cyclopiazonic acid in peanuts grown in Argentina. Food Addit. Contam. 2001, 18, 1017–1020. [Google Scholar] [CrossRef]
- Gqaleni, N.; Smith, J.E.; Lacey, J. Co-production of aflatoxins and cyclopiazonic acid in isolates of Aspergillus flavus. Food Addit. Contam. 1996, 13, 677–685. [Google Scholar] [CrossRef]
- Duran, R.M.; Cary, J.W.; Calvo, A.M. Production of cyclopiazonic acid, aflatrem, and aflatoxin by Aspergillus flavus is regulated by veA, a gene necessary for sclerotial formation. Appl. Microbiol. Biotechnol. 2007, 73, 1158–1168. [Google Scholar] [CrossRef] [PubMed]
- Sheikh-Zeinoddin, M.; Khalesi, M. Biological detoxification of ochratoxin A in plants and plant products. Toxin Rev. 2019, 38, 187–199. [Google Scholar] [CrossRef]
- Sy-Cordero, A.A.; Pearce, C.J.; Oberlies, N.H. Revisiting the enniatins: A review of their isolation, biosynthesis, structure determination and biological activities. J. Antibiot. 2012, 65, 541–549. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Jiang, Y.; Wen, L.; Yang, B. Characterization of polysaccharide structure in Citrus reticulate ’Chachi’ peel during storage and their bioactivity. Carbohydr. Res. 2021, 508, 108398. [Google Scholar] [CrossRef] [PubMed]
- Kirk, P.; Cannon, P.; Stalpers, J.; Minter, D.W. Dictionary of the Fungi, 10th ed.; CABI Publishing: Wallingford, UK, 2008. [Google Scholar]
- Yang, H.; Wang, W.; Zeng, L.; Liang, R.; Xiao, Q.; Zhou, Y.; Wu, W.; Deng, F. Development and optimization of a method based on dispersive solid phase extraction followed by ultra-high-performance liquid chromatography–quadrupole/orbitrap high-resolution mass spectrometry for simultaneous determination of 30 mycotoxins in Citrus products. J. Sep. Sci. 2022, 45, 4158–4166. [Google Scholar]
- Xin, Z.; Liu, S.-J.; Wang, Z.-L.; Fu, W.; Lin, C.; Chen, H.-P.; Liu, Y.-P. Community structure analysis of fungi isolated from citrus surfaces. Microbiol. China 2017, 44, 1089–1098. [Google Scholar]
- Schothorst, R.C.; van Egmond, H.P. Report from SCOOP task 3.2.10 “collection of occurrence data of Fusarium toxins in food and assessment of dietary intake by the population of EU member states”: Subtask: Trichothecenes. Toxicol. Lett. 2004, 153, 133–143. [Google Scholar] [CrossRef]
- Schrenk, D.; Bignami, M.; Bodin, L.; Chipman, J.K.; del Mazo, J.; Grasl-Kraupp, B.; Hogstrand, C.; Hoogenboom, L.; Leblanc, J.-C.; Nebbia, C.S.; et al. Risk assessment of aflatoxins in food. EFSA J. 2020, 18, e06040. [Google Scholar]
- EFSA. Scientific Opinion on the risks for public and animal health related to the presence of citrinin in food and feed. EFSA J. 2012, 10, 2605. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).