Antiproliferative Effects of Naja anchietae and Naja senegalensis Venom Peptides on Glioblastoma Cell Lines
Abstract
1. Introduction
2. Results and Discussion
2.1. Physicochemical Characterization
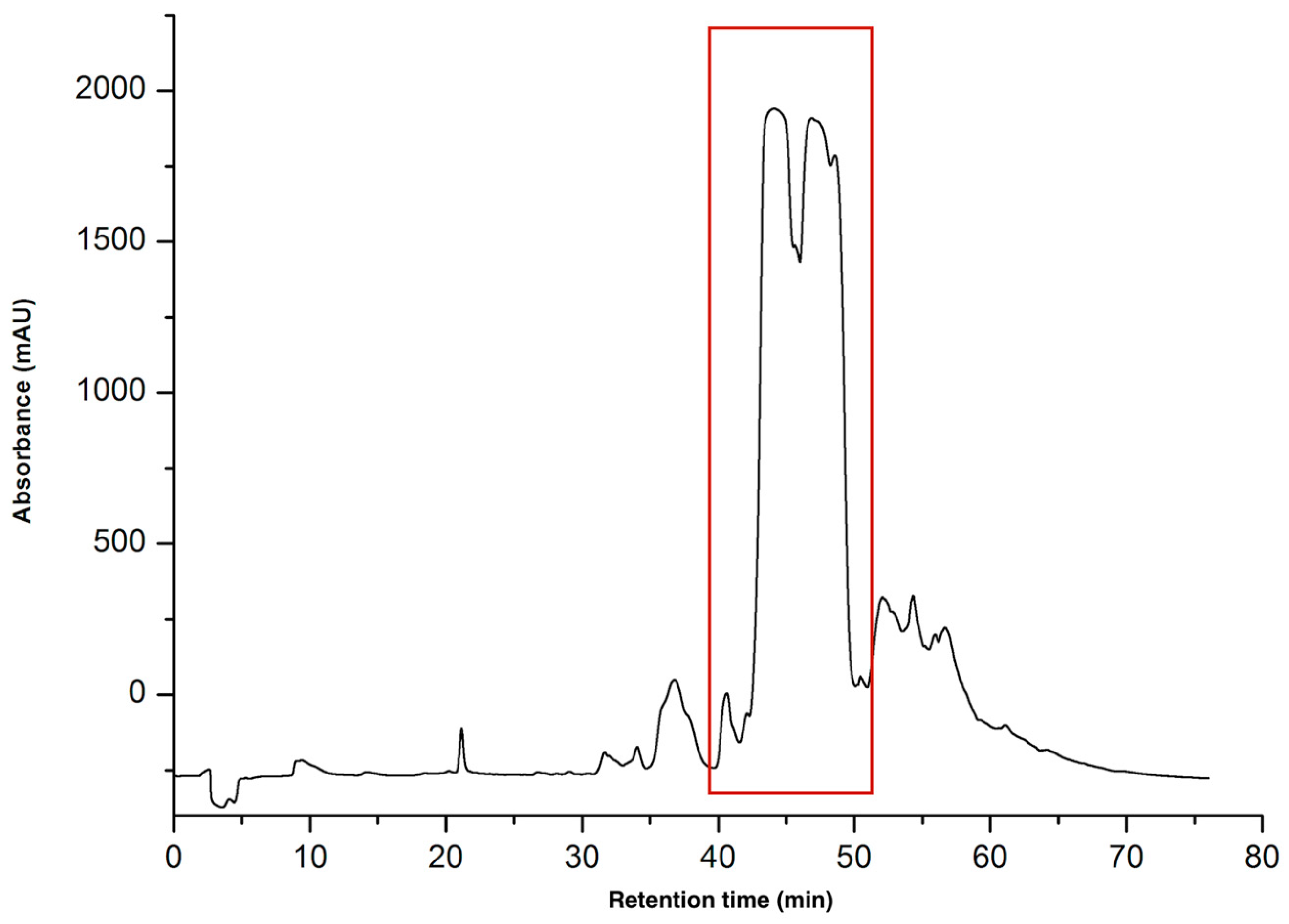
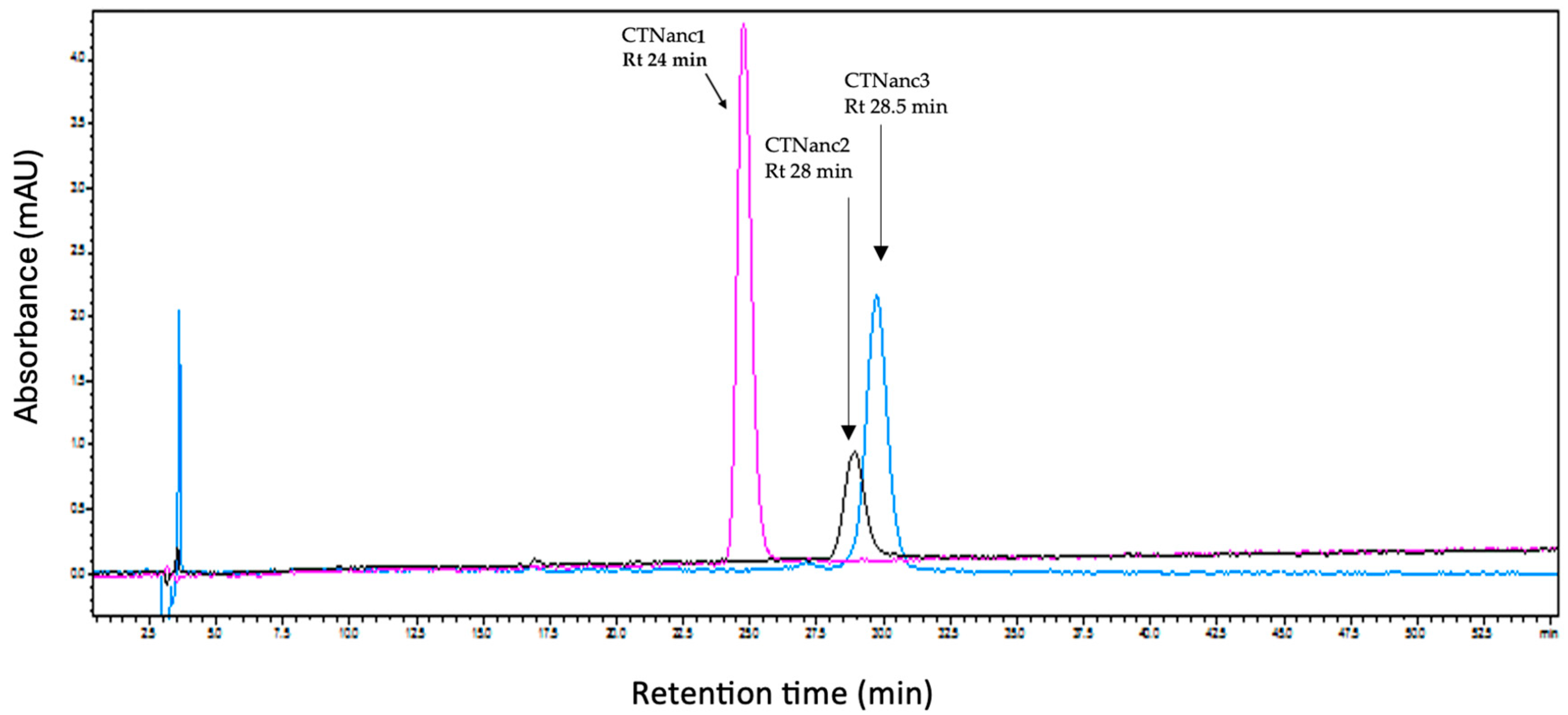
2.1.1. Purification of CTNs
2.1.2. MALDI-TOF Mass Spectrometry
2.1.3. N-Terminal Edman Sequencing
2.2. Pharmacological Characterization
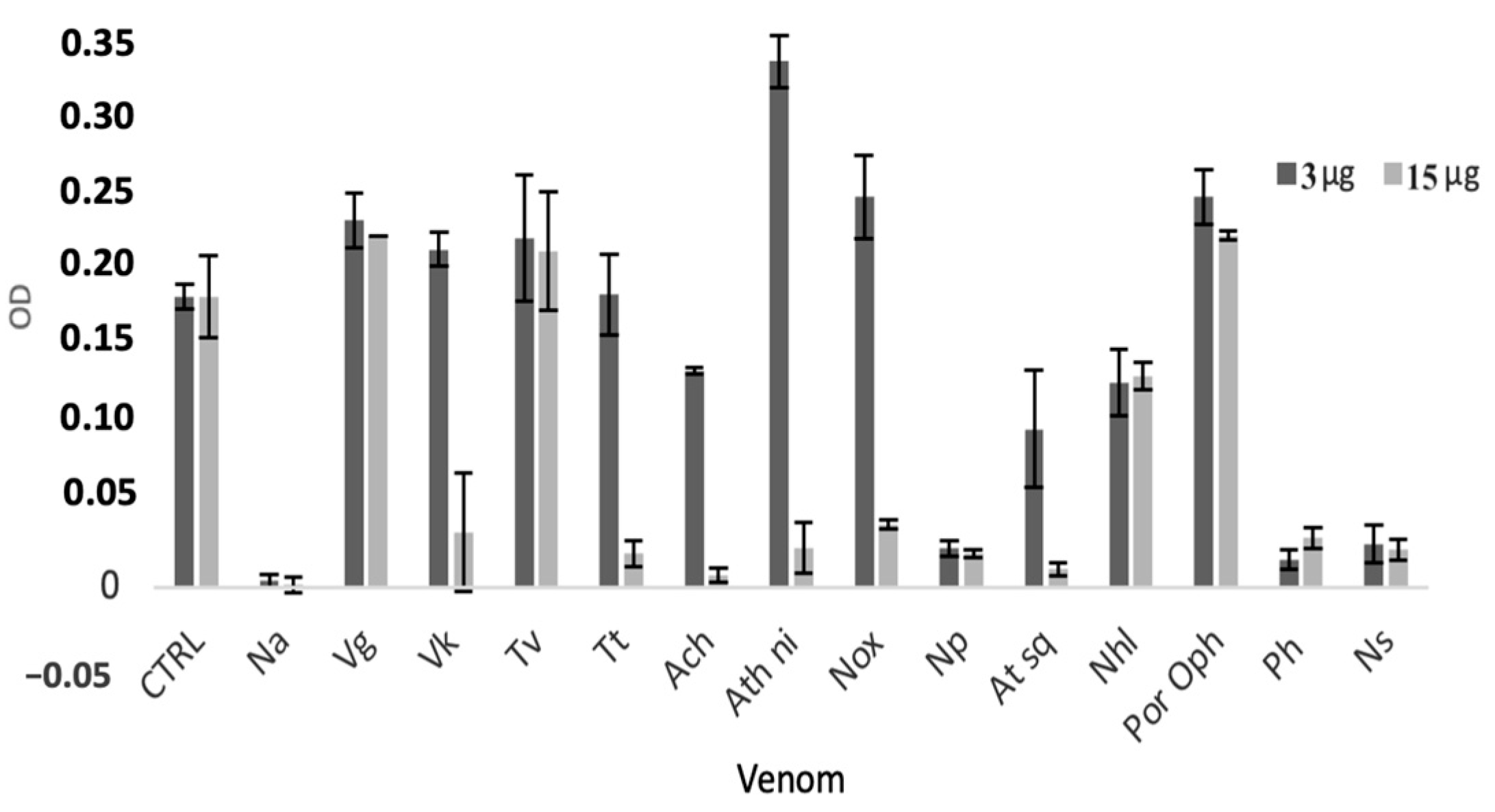
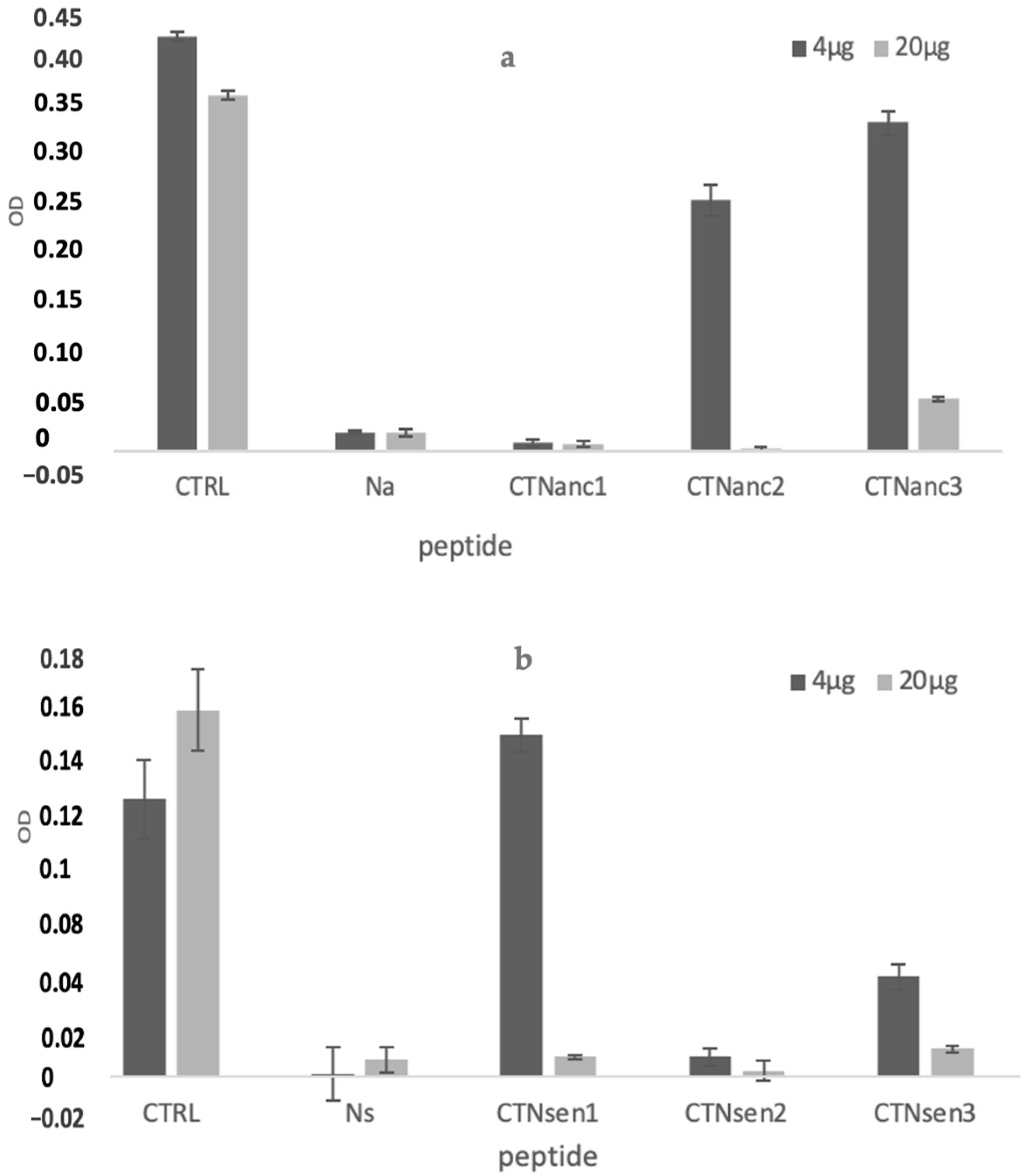
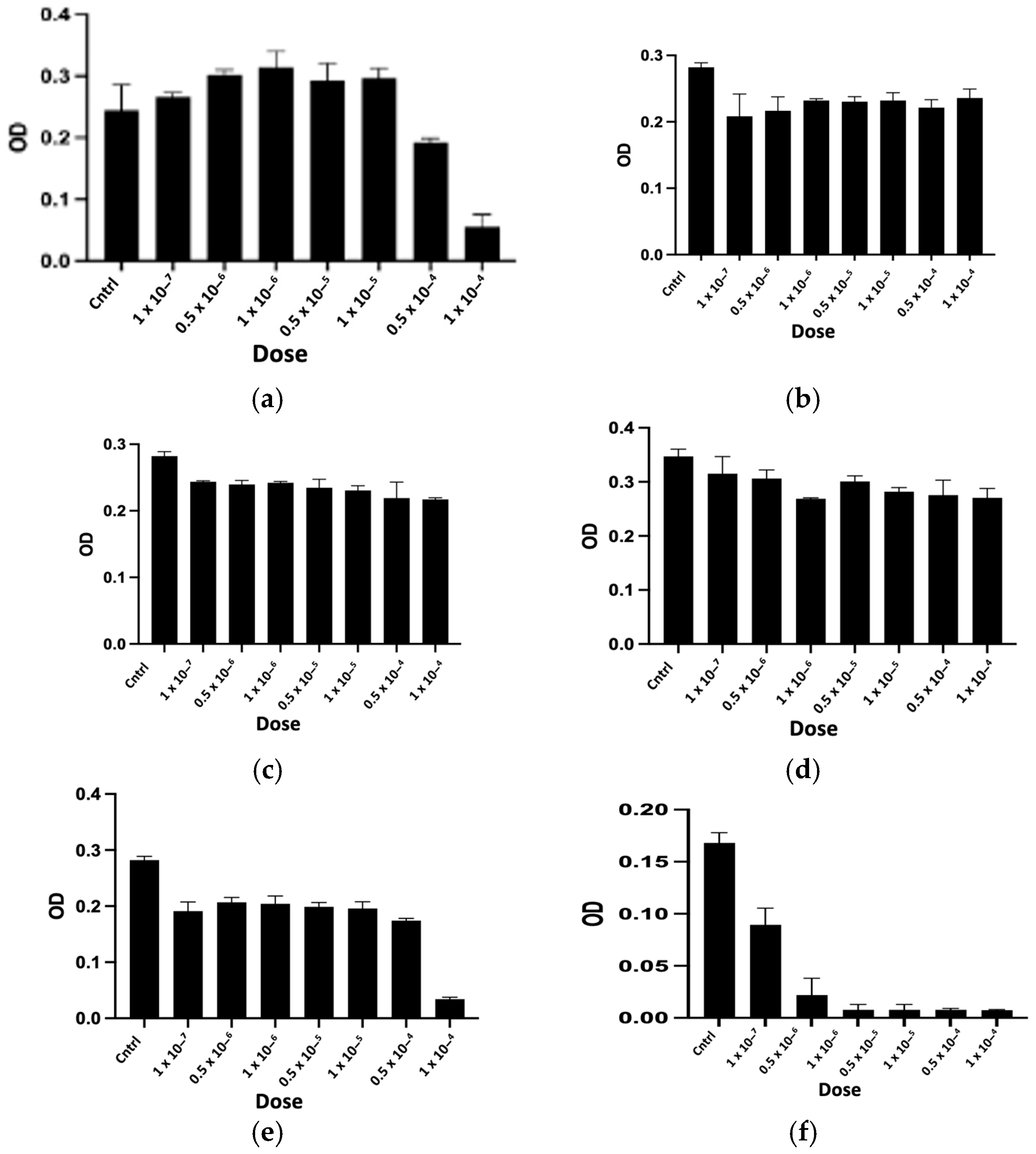
2.2.1. In Vitro Cytotoxicity Assay
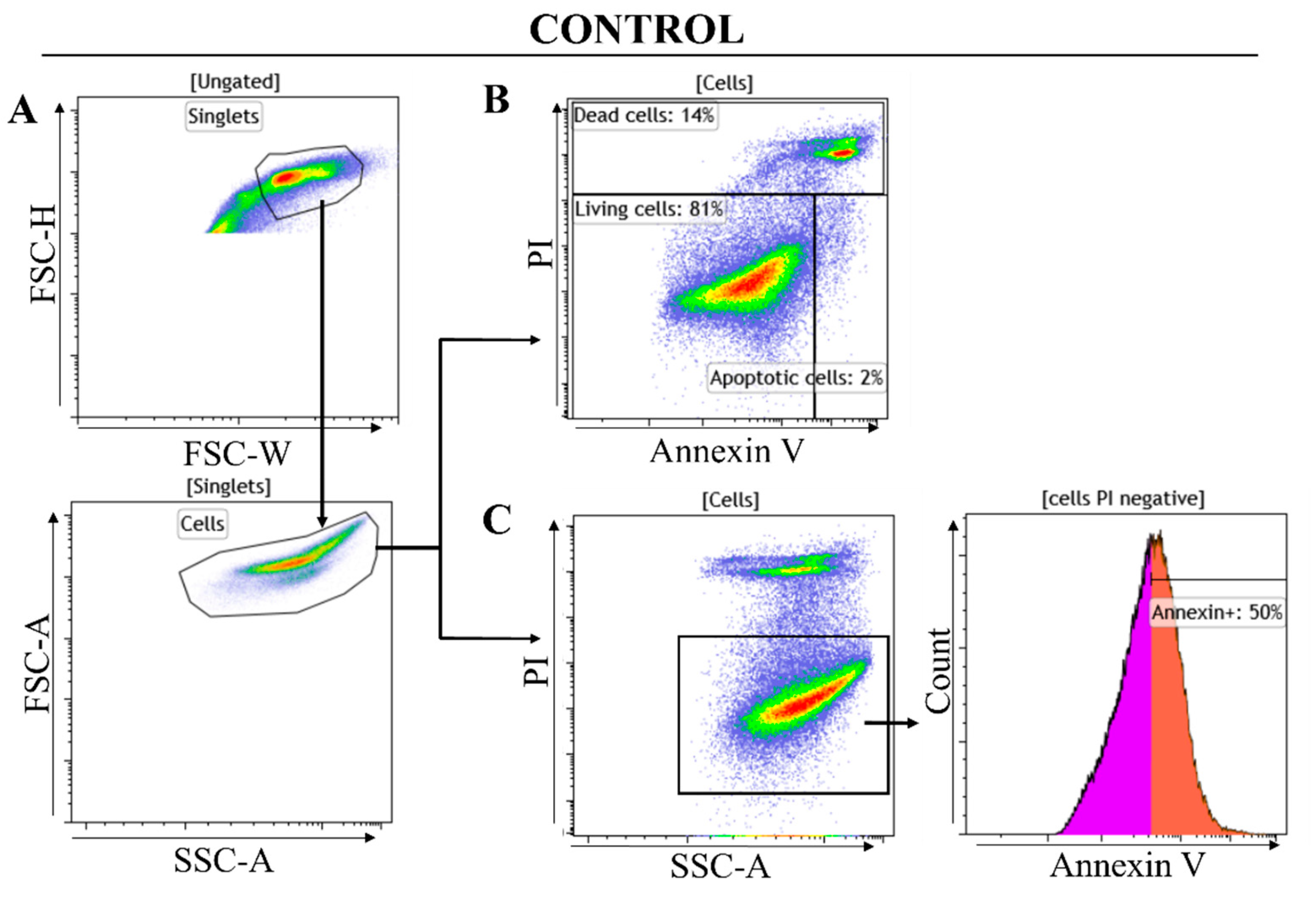
2.2.2. Flow Cytometry Analysis Results
- a.
- Gating strategy
- b.
- Cytotoxicity analysis by flow cytometry
3. Conclusions
4. Materials and Methods
4.1. RP-HPLC Purification
4.2. Mass Spectrometry Analysis
4.3. Reduction and Alkylation of Peptides
4.4. N-Terminal Sequencing
4.5. Peptide Mass Fingerprintings
4.6. MS/MS Analysis
4.7. Cell Culture
4.8. Cell Viability Assay
4.9. Flow Cytometry Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
References
- Dubovskii, P.V.; Utkin, Y.N. Specific Amino Acid Residues in the Three Loops of Snake Cytotoxins Determine Their Membrane Activity and Provide a Rationale for a New Classification of These Toxins. Toxins 2024, 16, 262. [Google Scholar] [CrossRef] [PubMed]
- Chong, H.P.; Tan, K.Y.; Tan, C.H. Cytotoxicity of snake venoms and cytotoxins from two Southeast Asian cobras (Naja sumatrana, Naja kaouthia): Exploration of anticancer potential, selectivity, and cell death mechanism. Front. Mol. Biosci. 2020, 7, 58358. [Google Scholar] [CrossRef]
- Kalita, B.; Utkin, Y.N.; Mukherjee, A.K. Current insights in the mechanisms of cobra venom cytotoxins and their complexes in inducing toxicity: Implications in antivenom therapy. Toxins 2022, 14, 839. [Google Scholar] [CrossRef] [PubMed]
- Hiu, J.J.; Yap, M.K.K. The myth of cobra venom cytotoxin: More than just direct cytolytic actions. Toxicon X 2022, 14, 100123. [Google Scholar] [CrossRef] [PubMed]
- Dubovskii, P.V.; Utkin, Y.N. Antiproliferative activity of cobra venom cytotoxins. Curr. Top. Med. Chem. 2015, 15, 638–648. [Google Scholar] [CrossRef]
- Kumar, T.K.S.; Jayaraman, G.; Lee, C.S.; Arunkumar, A.I.; Sivaraman, T.; Samuel, D.; Yu, C. Snake venom cardiotoxins-structure, dynamics, function and folding. J. Biomol. Struct. Dyn. 1997, 15, 431–463. [Google Scholar] [CrossRef]
- GKonshina, A.; VDubovskii, P.; GEfremov, R. Structure and dynamics of cardiotoxins. Curr. Protein Pept. Sci. 2012, 13, 570–584. [Google Scholar] [CrossRef]
- Chiou, J.T.; Wang, L.J.; Lee, Y.C.; Chang, L.S. Naja atra cardiotoxin 1 induces the FasL/Fas death pathway in human leukemia cells. Cells 2021, 10, 2073. [Google Scholar] [CrossRef] [PubMed]
- Boughanmi, Y.; Aboudou, S.; Boyadjian, K.; Charouandi, A.; Bouzid, S.; Barnetche, T.; Mabrouk, K. One-Step Chromatographic Approach for Purifying Peptides and Proteins from Venoms. Separations 2024, 11, 179. [Google Scholar] [CrossRef]
- Galat, A.; Gross, G.; Drevet, P.; Sato, A.; Ménez, A. Conserved structural determinants in three-fingered protein domains. FEBS J. 2008, 275, 3207–3225. [Google Scholar] [CrossRef]
- Suzuki-Matsubara, M.; Athauda, S.B.; Suzuki, Y.; Matsubara, K.; Moriyama, A. Comparison of the primary structures, cytotoxicities, and affinities to phospholipids of five kinds of cytotoxins from the venom of Indian cobra, Naja naja. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2016, 179, 158–164. [Google Scholar] [CrossRef] [PubMed]
- Dubovskii, P.V.; Lesovoy, D.M.; Dubinnyi, M.A.; Konshina, A.G.; Utkin, Y.N.; Efremov, R.G.; Arseniev, A.S. Interaction of three-finger toxins with phospholipid membranes: Comparison of S-and P-type cytotoxins. Biochem. J. 2005, 387, 807–815. [Google Scholar] [CrossRef]
- Efremov, R.G.; Volynsky, P.E.; Nolde, D.E.; Dubovskii, P.V.; Arseniev, A.S. Interaction of cardiotoxins with membranes: A molecular modeling study. Biophys. J. 2002, 83, 144–153. [Google Scholar] [CrossRef] [PubMed]
- Conlon, J.M.; Attoub, S.; Musale, V.; Leprince, J.; Casewell, N.R.; Sanz, L.; Calvete, J.J. Isolation and characterization of cytotoxic and insulin-releasing components from the venom of the black-necked spitting cobra Naja nigricollis (Elapidae). Toxicon X 2020, 6, 100030. [Google Scholar] [CrossRef] [PubMed]
- Ebrahim, K.; Shirazi, F.H.; Vatanpour, H.; Kobarfard, F.; Rabiei, H. Anticancer activity of cobra venom polypeptide, cytotoxin-II, against human breast adenocarcinoma cell line (MCF-7) via the induction of apoptosis. J. Breast Cancer 2014, 17, 314–322. [Google Scholar] [CrossRef]
- Liu, Y.; Ming, W.; Wang, Y.; Liu, S.; Qiu, Y.; Xiang, Y.; Guo, Q. Cytotoxin 1 from Naja atra Cantor venom induced necroptosis of leukemia cells. Toxicon 2019, 165, 110–115. [Google Scholar] [CrossRef]
- Attarde, S.S.; Pandit, S.V. Cytotoxic activity of NN-32 toxin from Indian spectacled cobra venom on human breast cancer cell lines. BMC Complement. Altern. Med. 2017, 17, 1–9. [Google Scholar] [CrossRef]
- Feofanov, A.V.; Sharonov, G.V.; Dubinnyi, M.A.; Astapova, M.V.; Kudelina, I.A.; Dubovskii, P.V.; Rodionov, D.I.; Utkin, Y.N.; Arseniev, A.S. Comparative study of structure and activity of cytotoxins from venom of the cobras Naja oxiana, Naja kaouthia, and Naja haje. Biochemistry 2004, 69, 1148–1157. [Google Scholar] [CrossRef]
- Chen, K.C.; Chiou, Y.L.; Kao, P.H.; Lin, S.R.; Chang, L.S. Taiwan cobra cardiotoxins induce apoptotic death of human neuroblastoma SK-N-SH cells mediated by reactive oxygen species generation and mitochondrial depolarization. Toxicon 2008, 51, 624–634. [Google Scholar] [CrossRef]
- Lin, K.L.; Su, J.C.; Chien, C.M.; Chuang, P.W.; Chang, L.S.; Lin, S.R. Down-regulation of the JAK2/PI3K-mediated signaling activation is involved in Taiwan cobra cardiotoxin III-induced apoptosis of human breast MDA-MB-231 cancer cells. Toxicon 2010, 55, 1263–1273. [Google Scholar] [CrossRef]
- Vogel, T.W.; Zhuang, Z.; Li, J.; Okamoto, H.; Furuta, M.; Lee, Y.S.; Weil, R.J. Proteins and protein pattern differences between glioma cell lines and glioblastoma multiforme. Clin. Cancer Res. 2005, 11, 3624–3632. [Google Scholar] [CrossRef] [PubMed]
- Patil, V.; Pal, J.; Somasundaram, K. Elucidating the cancer-specific genetic alteration spectrum of glioblastoma derived cell lines from whole exome and RNA sequencing. Oncotarget 2015, 6, 43452. [Google Scholar] [CrossRef] [PubMed]
- Lafnoune, A.; Lee, S.Y.; Heo, J.Y.; Gourja, I.; Darkaoui, B.; Abdelkafi-Koubaa, Z.; Oukkache, N. Anti-cancer effect of moroccan cobra Naja haje venom and its fractions against hepatocellular carcinoma in 3d cell culture. Toxins 2021, 13, 402. [Google Scholar] [CrossRef] [PubMed]
- Waghule, T.; Rapalli, V.K.; Singhvi, G.; Gorantla, S.; Khosa, A.; Dubey, S.K.; Saha, R.N. Design of temozolomide-loaded proliposomes and lipid crystal nanoparticles with industrial feasible approaches: Comparative assessment of drug loading, entrapment efficiency, and stability at plasma pH. J. Liposome Res. 2020, 31, 158–168. [Google Scholar] [CrossRef]
- Kumthekar, P.; Tang, S.C.; Brenner, A.J.; Kesari, S.; Piccioni, D.E.; Anders, C.; Ibrahim, N. ANG1005, a brain-penetrating peptide–drug conjugate, shows activity in patients with breast cancer with leptomeningeal carcinomatosis and recurrent brain metastases. Clin. Cancer Res. 2020, 26, 2789–2799. [Google Scholar] [CrossRef]
- Taal, W.; Oosterkamp, H.M.; Walenkamp, A.M.; Dubbink, H.J.; Beerepoot, L.V.; Hanse, M.C.; van den Bent, M.J. Single-agent bevacizumab or lomustine versus a combination of bevacizumab plus lomustine in patients with recurrent glioblastoma (BELOB trial): A randomised controlled phase 2 trial. Lancet Oncol. 2014, 15, 943–953. [Google Scholar] [CrossRef]
- Chen, D.; Yu, J.; Zhang, L. Necroptosis: An alternative cell death program defending against cancer. Biochim. Biophys. Acta (BBA)-Rev. Cancer 2016, 1865, 228–236. [Google Scholar] [CrossRef]
- Yuan, J.; Najafov, A.; Py, B.F. Roles of caspases in necrotic cell death. Cell 2016, 167, 1693–1704. [Google Scholar] [CrossRef]
- Suckau, D.; Resemann, A.; Schuerenberg, M.; Hufnagel, P.; Franzen, J.; Holle, A. A novel MALDI LIFT-TOF/TOF mass spectrometer for proteomics. Anal. Bioanal. Chem. 2003, 376, 952–965. [Google Scholar] [CrossRef]
- Ritz, C.; Baty, F.; Streibig, J.C.; Gerhard, D. Dose-Response Analysis Using R. PLoS ONE 2015, 10, e0146021. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing R; Foundation for Statistical Computing: Vienna, Austria, 2023; Available online: https://www.R-project.org/ (accessed on 31 August 2024).
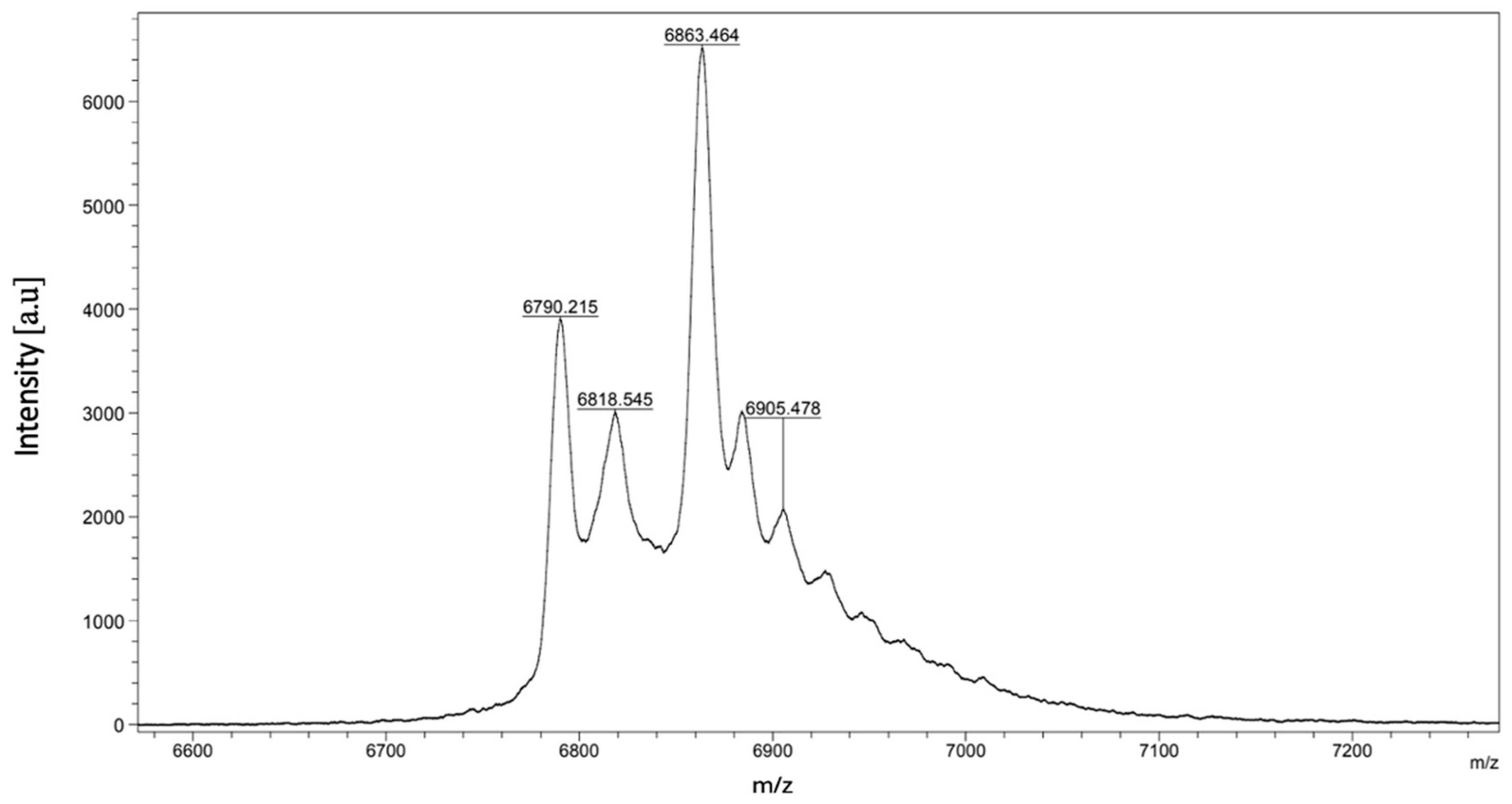
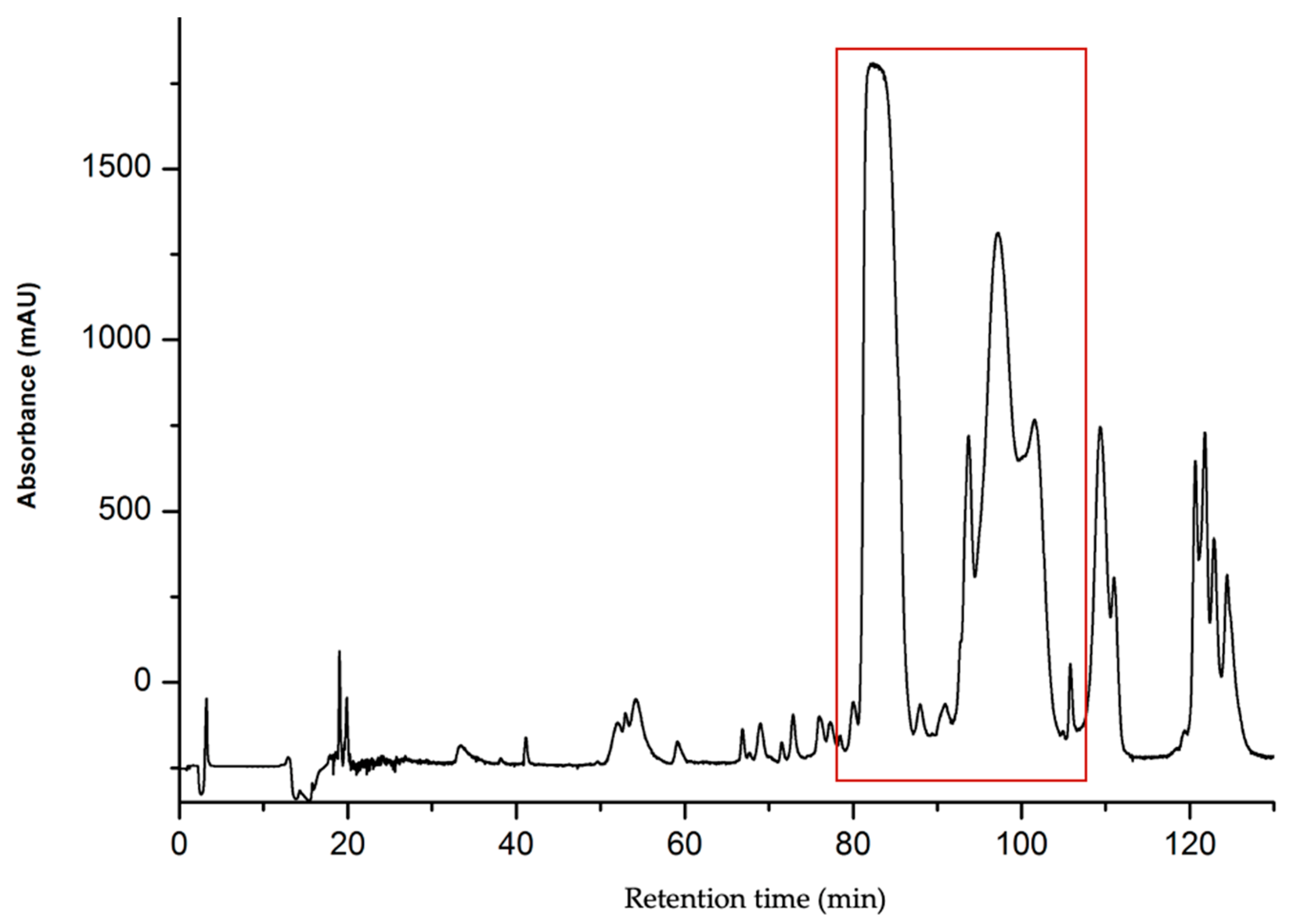
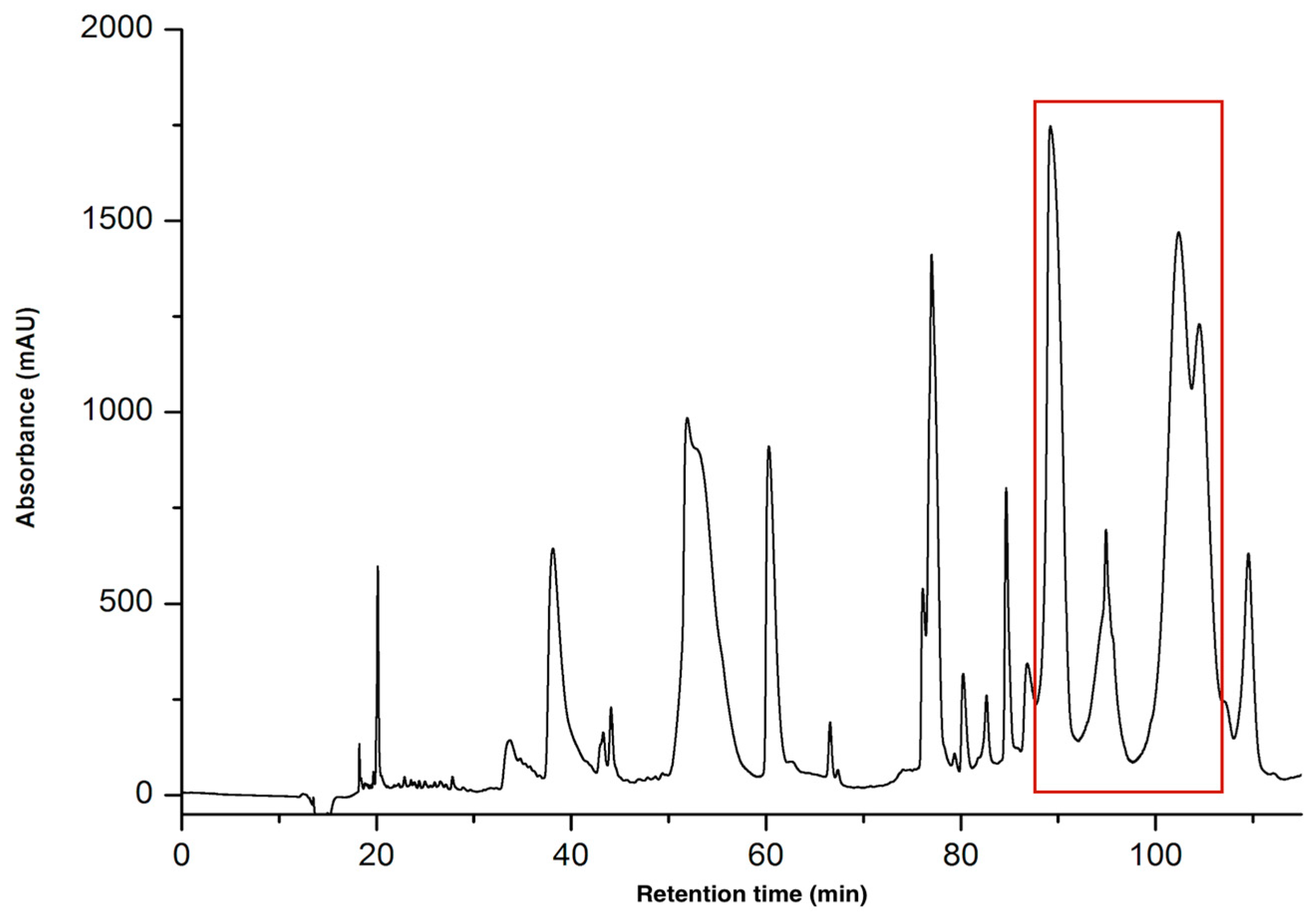
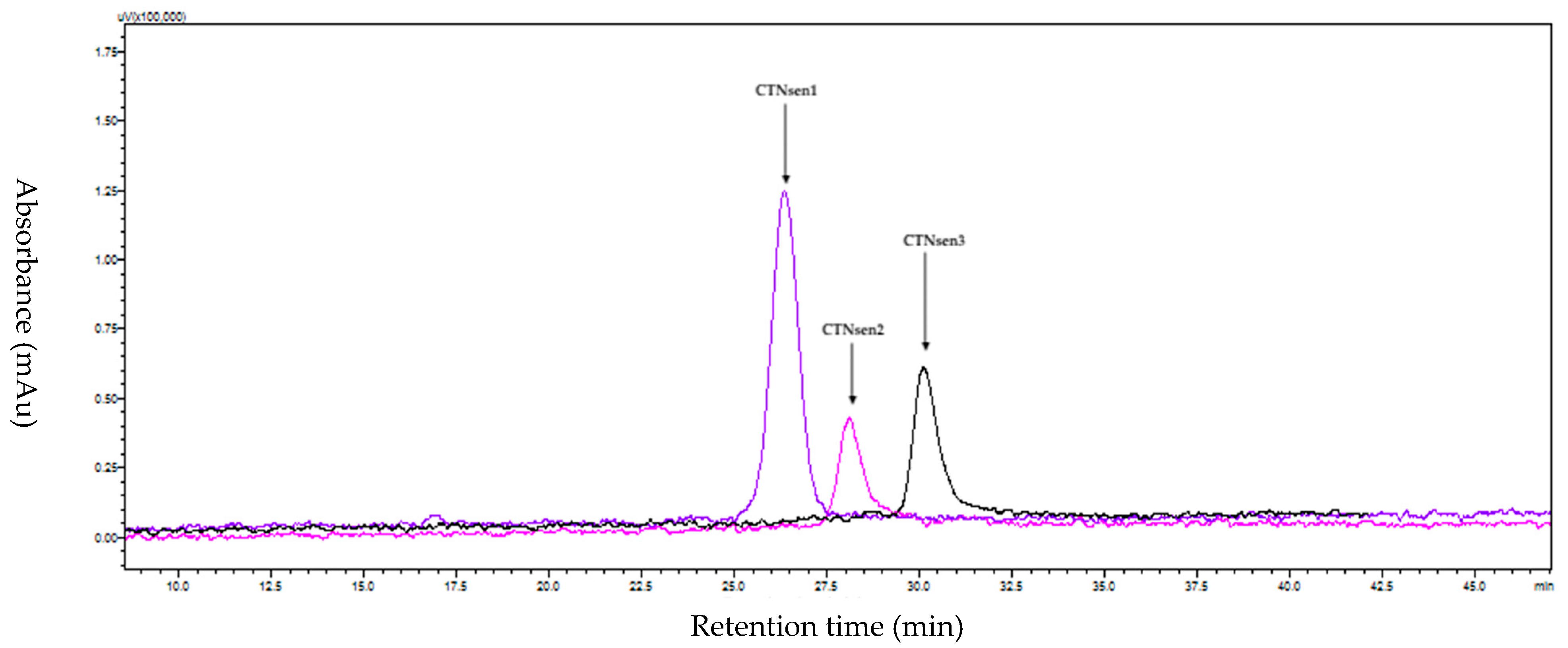
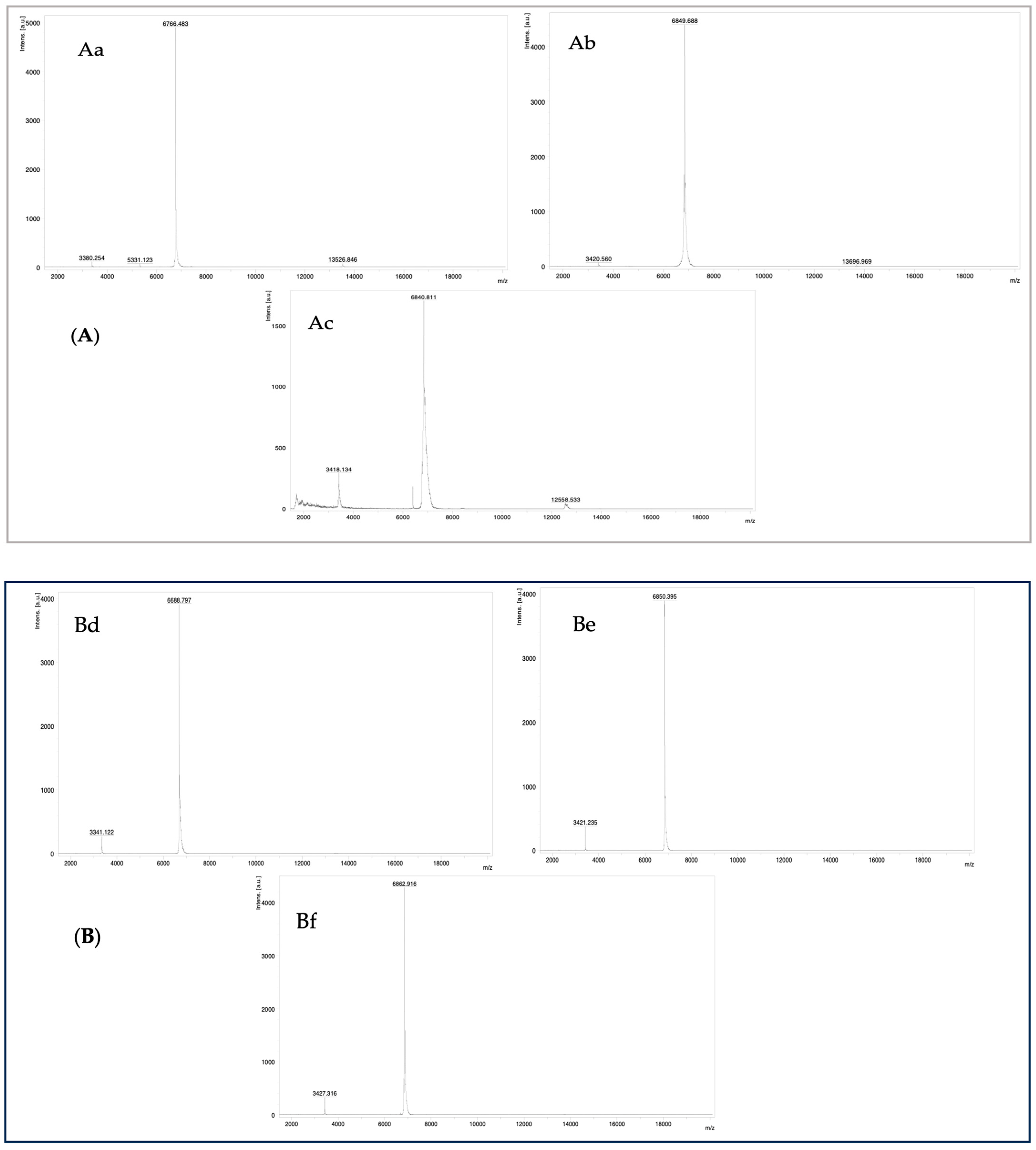
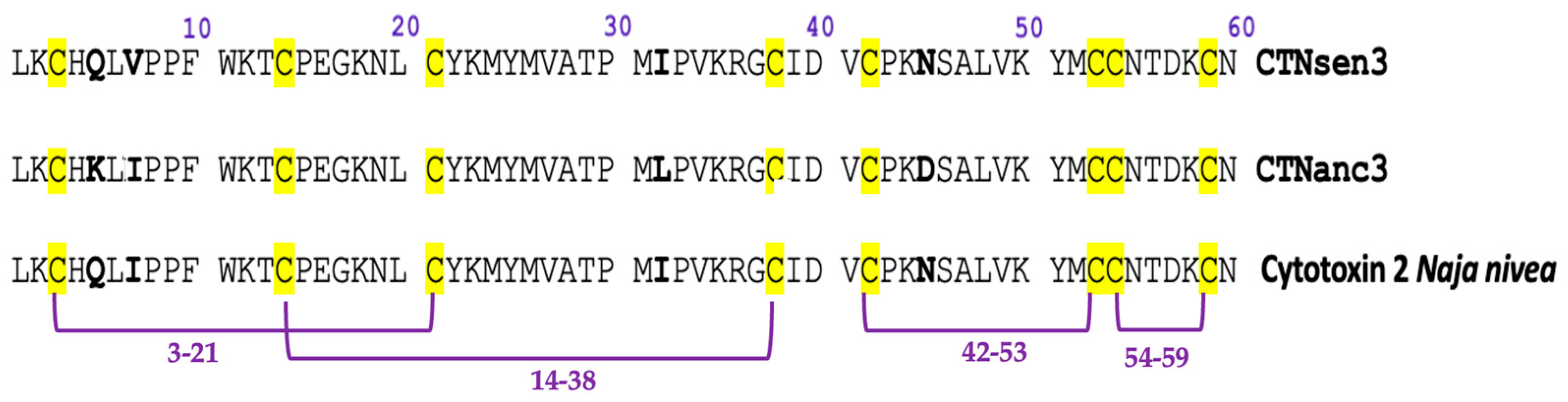

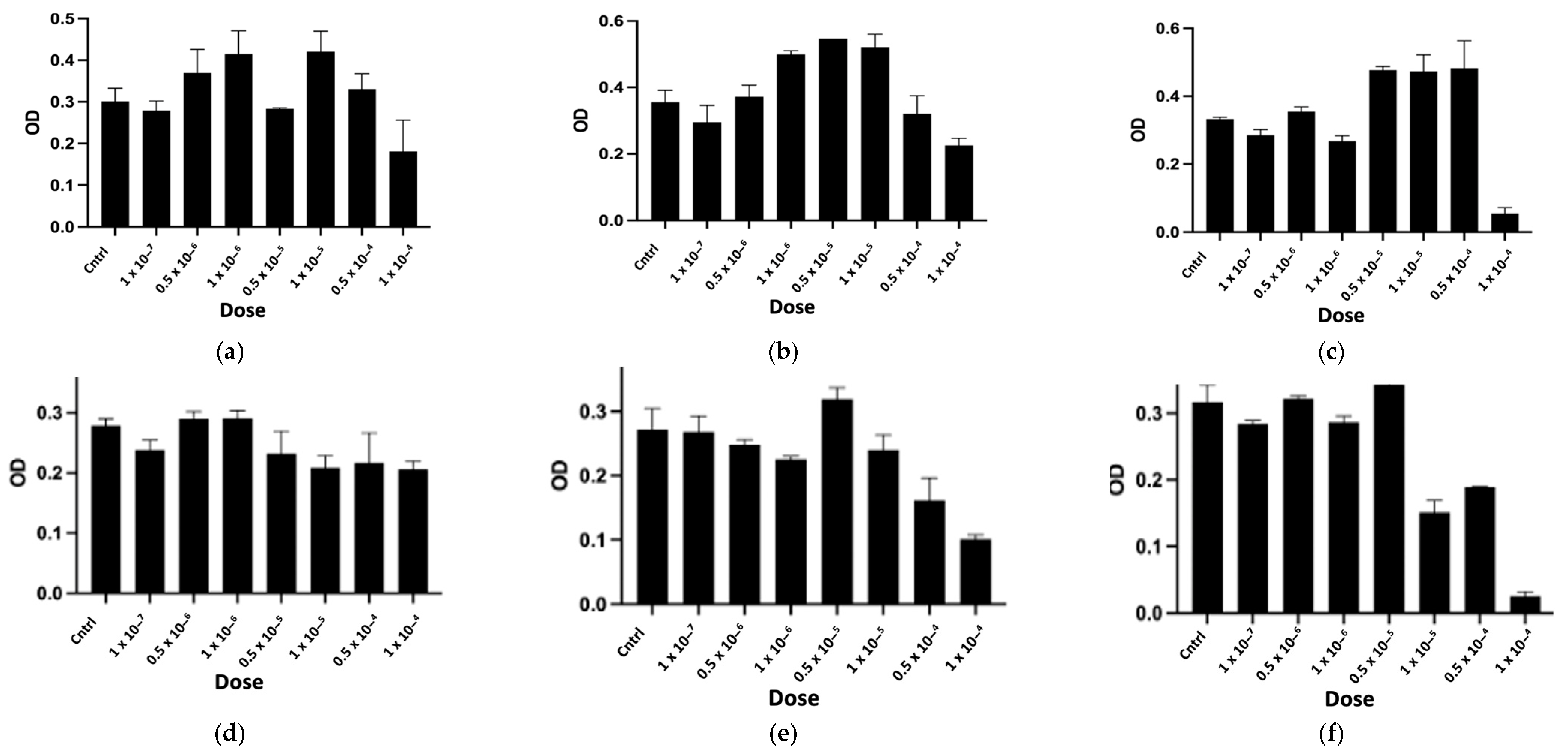
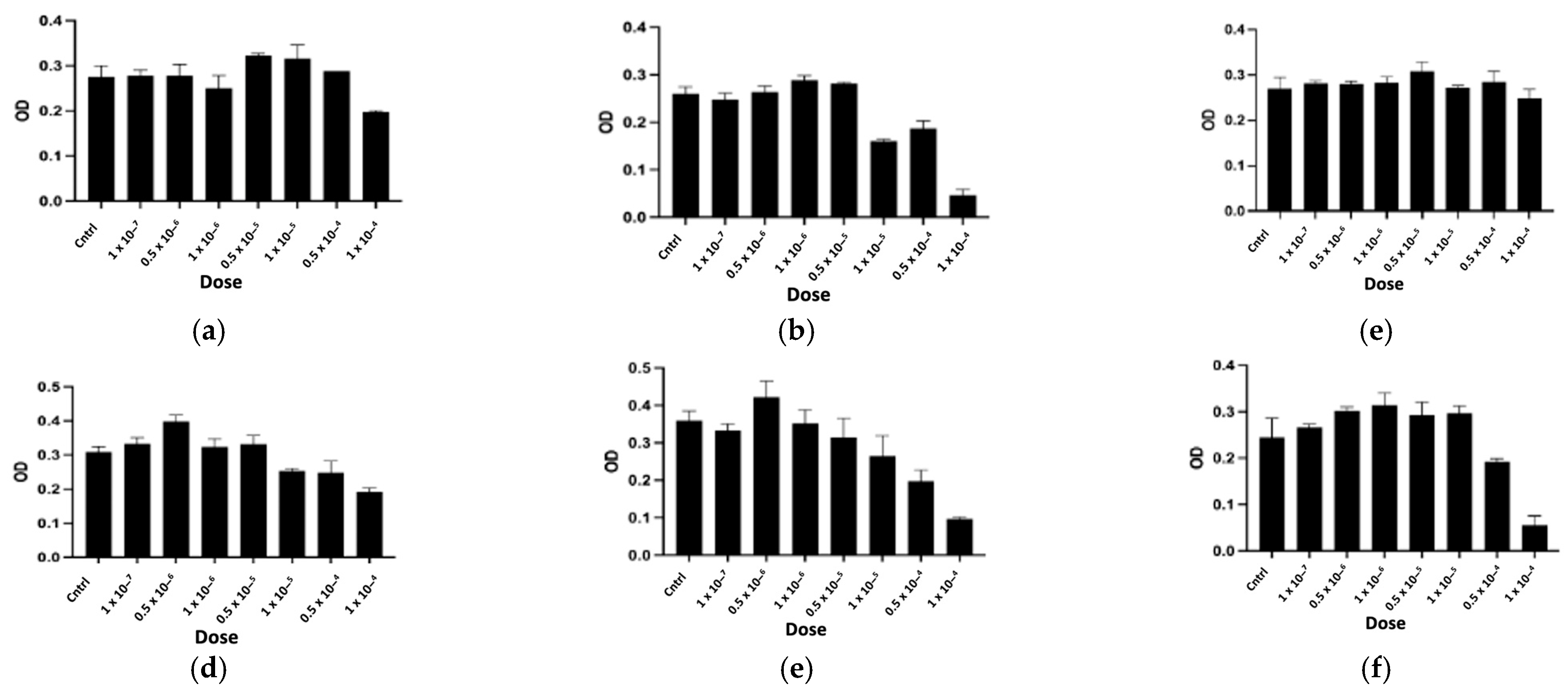


| Peptide | Determined Sequence | Homologous Sequence Identified Using BLAST/UniProtKB/Swiss-Prot-% of Identity |
|---|---|---|
| CTNsen1 | LKCHQLVPPFWKTCPEGKNLCYKMYMVSSSTVPVKRGCIDVCPKNSALVKYVCCNTDKCN | P01457.1 Cytotoxin 5 Naja haje haje 100% |
| CTNsen2 | LKCHKLVPPFWKTCPEGKNLCYKMYMVATPMIPVKRGCIDVCPKNSALVKYVCCNTNKCN | P01464.1 Cytotoxin 5 Naja haje annulifera 98.33% |
| CTNsen3 | LKCHQLVPPFWKTCPEGKNLCYKMYMVATPMIPVKRGCIDVCPKNSALVKYMCCNTDKCN | P01463.1 Cytotoxin 2 Naja nivea 98.33% |
| CTNanc1 | LKCHKLVPPVWKTCPEGKNLCYKMFMVSTSTVPVKRGCIDVCPKDSALVKYVCCSTDKCN | P01456.1 Cytotoxin 1 Naja nivea 100.00% |
| CTNanc2 | LKCHKLVPPFWKTCPEGKNLCYKMYMVATPMLPVKRGCIDVCPKDSALVKYMCCNTDK | P01462.1 Cytotoxin 2 Naja annulifera 100.00% |
| CTNanc3 | LKCHKLIPPFWKTCPEGKNLCYKMYMVATPMIPVKRGCIDVCPKDSALVKYMCCNTDKCN | P01463.1 Cytotoxin 2 Naja nivea 98.33% |
| Cytotoxin | Type | Active Against/Dose | Venom Source | Reference |
|---|---|---|---|---|
| CTX-2N | Type S | 0.8 μM for A549 non-small cell lung adenocarcinoma | Naja nigricollis | Conlon et al., 2020 [14] |
| NK-CTX/Cytotoxin 3 | Type S | Lung cancer (A549) A549 = 1.22 μg/mL Prostate cancer PC-3 = 4.46 μg/m Breast cancer MCF-7 = 12.23 μg/mL | Naja kaouthia | Chong et al., 2020 [2] |
| CTX-II | Type P | Breast cancer (MCF-7) | Naja oxiana | Ebrahim et al., 2014 [15] |
| Cytotoxin 2a/NS-CTX | Type P | Lung cancer (A549) 0.88 μg/mL Prostate cancer PC-3 = 3.13 μg/m Breast cancer MCF-7 = 9.10 μg/mL | Naja sumatrana | Chong et al., 2020 [2] |
| Cytotoxin 1 | Type S | Acute myeloid leukemia (KG-1a) 3.31 μg/mL | Naja atra | Liu et al. (2019) [16] |
| Cytotoxin 3/NN-32 | Type S | Breast cancer MCF-7 = 2.5 μg/mL | Naja naja | Attarde and Pandit (2017) [17] |
| CT1No | Type S | Leukemia WEHI-3 = 340 nM Myelogenous leukemia K562 = 650 nM Acute promyelocytic leukemia HL-60 = 1400 nM | Naja oxiana | Feofanov et al. (2004) [18] |
| CT2Nh | Type P | Leukemia WEHI-3 = 1190 nM Acute promyelocytic leukemia HL-60 = 3700 nM | Naja haje | Feofanov et al. (2004) [18] |
| Cytotoxin 3 | Type P | Human neuroblastoma SK-N-SH = 0.8 µM Breast adenocarcinoma MDA-MB-231 = 0.09–0.15 mM | Naja atra | Chen et al. (2008) [19], Lin et al. (2010) [20] |
| Cytotoxin 4 | Type S | Human neuroblastoma SK-N-SH = 1.2 µM | Naja atra | Chen et al. (2008) [19] |
| Tissue Origin | Cell Line | IC50 (μg/mL) | |||||
|---|---|---|---|---|---|---|---|
| CTNanc1 | CTNanc2 | CTNanc3 | CTNsen1 | CTNsen2 | CTNsen3 | ||
| Brain Glioblastoma | U87 | 36.41 | 38.34 | 5.33 | 54.69 | Inactive | 45.38 |
| 95% confidence interval | 32.87–39.96 | 34.63–42.04 | 4.65–6.02 | 46.11–63.28 | 36.80–53.97 | ||
| Brain Glioblastoma | U251 | Inactive | 475.8 | 416.37 | Inactive | Inactive | Inactive |
| 95% confidence interval | 395.04–556.56 | 338.15–494.59 | |||||
| Brain Glioblastoma | T98G | Inactive | 389.59 | 420.01 | Inactive | 462.18 | Inactive |
| 95% confidence interval | 296–483.18 | 384.96–455.06 | 386.35–538.01 | ||||
| Endothelial | HUVEC | Inactive * | 467.21 | 3.58 | 225.65 | Inactive * | Inactive * |
| 95% confidence interval | 423.44–510.97 | 3.42–3.74 | 167.03–284.28 | ||||
| Sample Tested | % Living Cells Gated | % Apoptotic Cells Gated | % Dead Cells Gated |
|---|---|---|---|
| Control U87 cells | 81 | 2 | 14 |
| CTNanc1-U87 | 7 | 2 | 90 |
| CTNanc2-U87 | 11 | 5 | 84 |
| CTNanc3-U87 | 83 | 3 | 12 |
| CTNsen2-U87 | 33 | 17 | 43 |
| CTNsen3-U87 | 85 | 2 | 10 |
| Sample Tested | [Cells PI Negative Cells] Number | [Cells PI Negative Cells] Annexin FITC MFI | [AnnexinV+] % Gated from Histograms |
|---|---|---|---|
| Control U87 cells | 71,033 | 29,764 | 50 |
| CTNanc1-U87 | 312 | 59,862 | 76 |
| CTNanc2-U87 | 2737 | 164,227 | 93 |
| CTNanc3-U87 | 68,626 | 46,465 | 69 |
| CTNsen2-U87 | 3689 | 200,997 | 96 |
| CTNsen3-U87 | 65,486 | 29,041 | 49 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Boughanmi, Y.; Berenguer-Daizé, C.; Balzano, M.; Mosrati, H.; Moulard, M.; Mansuelle, P.; Fourquet, P.; Torre, F.; de Pomyers, H.; Gigmes, D.; et al. Antiproliferative Effects of Naja anchietae and Naja senegalensis Venom Peptides on Glioblastoma Cell Lines. Toxins 2024, 16, 433. https://doi.org/10.3390/toxins16100433
Boughanmi Y, Berenguer-Daizé C, Balzano M, Mosrati H, Moulard M, Mansuelle P, Fourquet P, Torre F, de Pomyers H, Gigmes D, et al. Antiproliferative Effects of Naja anchietae and Naja senegalensis Venom Peptides on Glioblastoma Cell Lines. Toxins. 2024; 16(10):433. https://doi.org/10.3390/toxins16100433
Chicago/Turabian StyleBoughanmi, Yasmine, Caroline Berenguer-Daizé, Marielle Balzano, Hend Mosrati, Maxime Moulard, Pascal Mansuelle, Patrick Fourquet, Franck Torre, Harold de Pomyers, Didier Gigmes, and et al. 2024. "Antiproliferative Effects of Naja anchietae and Naja senegalensis Venom Peptides on Glioblastoma Cell Lines" Toxins 16, no. 10: 433. https://doi.org/10.3390/toxins16100433
APA StyleBoughanmi, Y., Berenguer-Daizé, C., Balzano, M., Mosrati, H., Moulard, M., Mansuelle, P., Fourquet, P., Torre, F., de Pomyers, H., Gigmes, D., Ouafik, L., & Mabrouk, K. (2024). Antiproliferative Effects of Naja anchietae and Naja senegalensis Venom Peptides on Glioblastoma Cell Lines. Toxins, 16(10), 433. https://doi.org/10.3390/toxins16100433





