Investigating Common Pathogenic Mechanisms between Homo sapiens and Different Strains of Candida albicans for Drug Design: Systems Biology Approach via Two-Sided NGS Data Identification
Abstract
1. Introduction
2. Results
2.1. The Identified GEINs under the Infection of C. albicans SC5314 and C. albicans WO-1
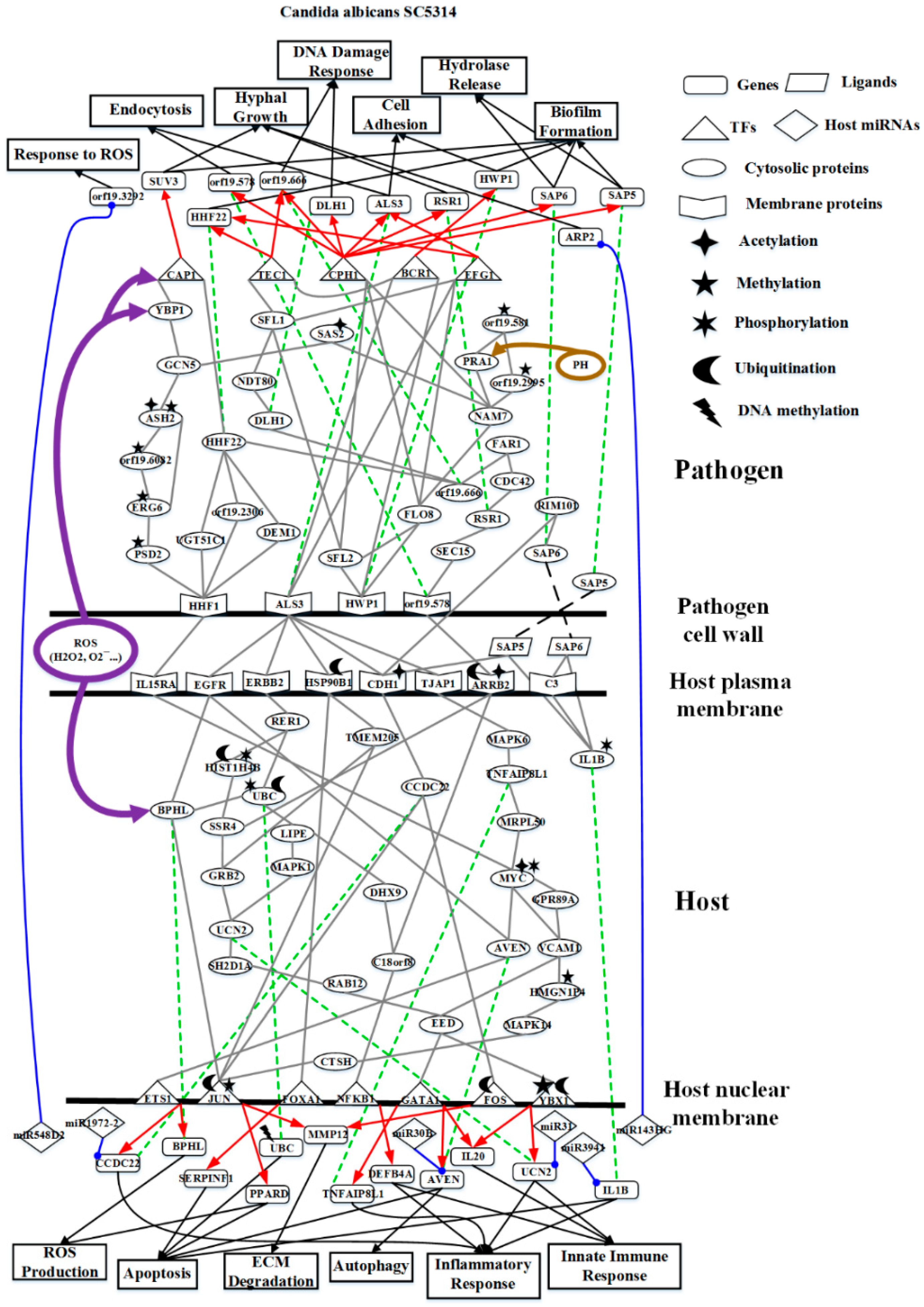
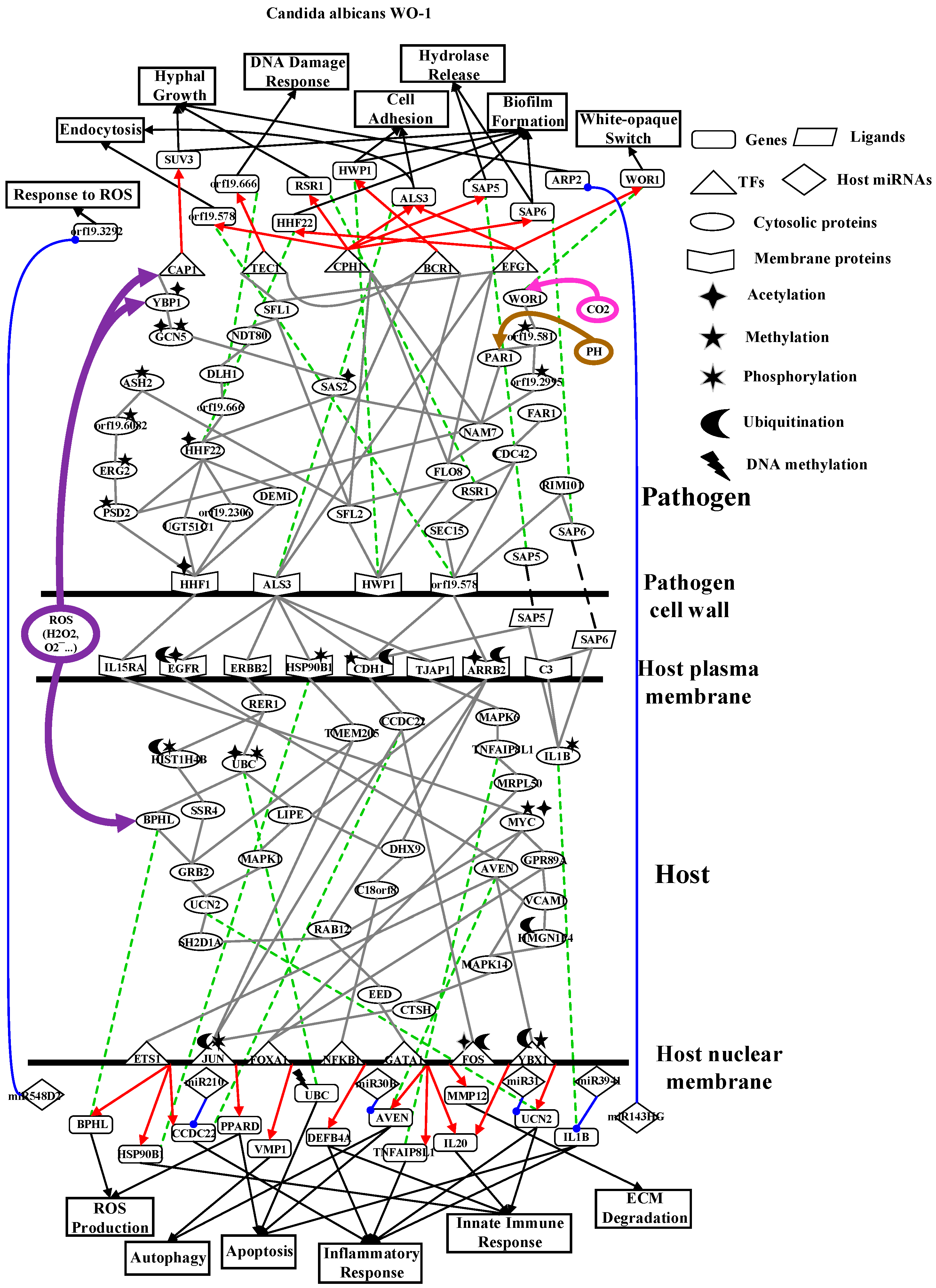
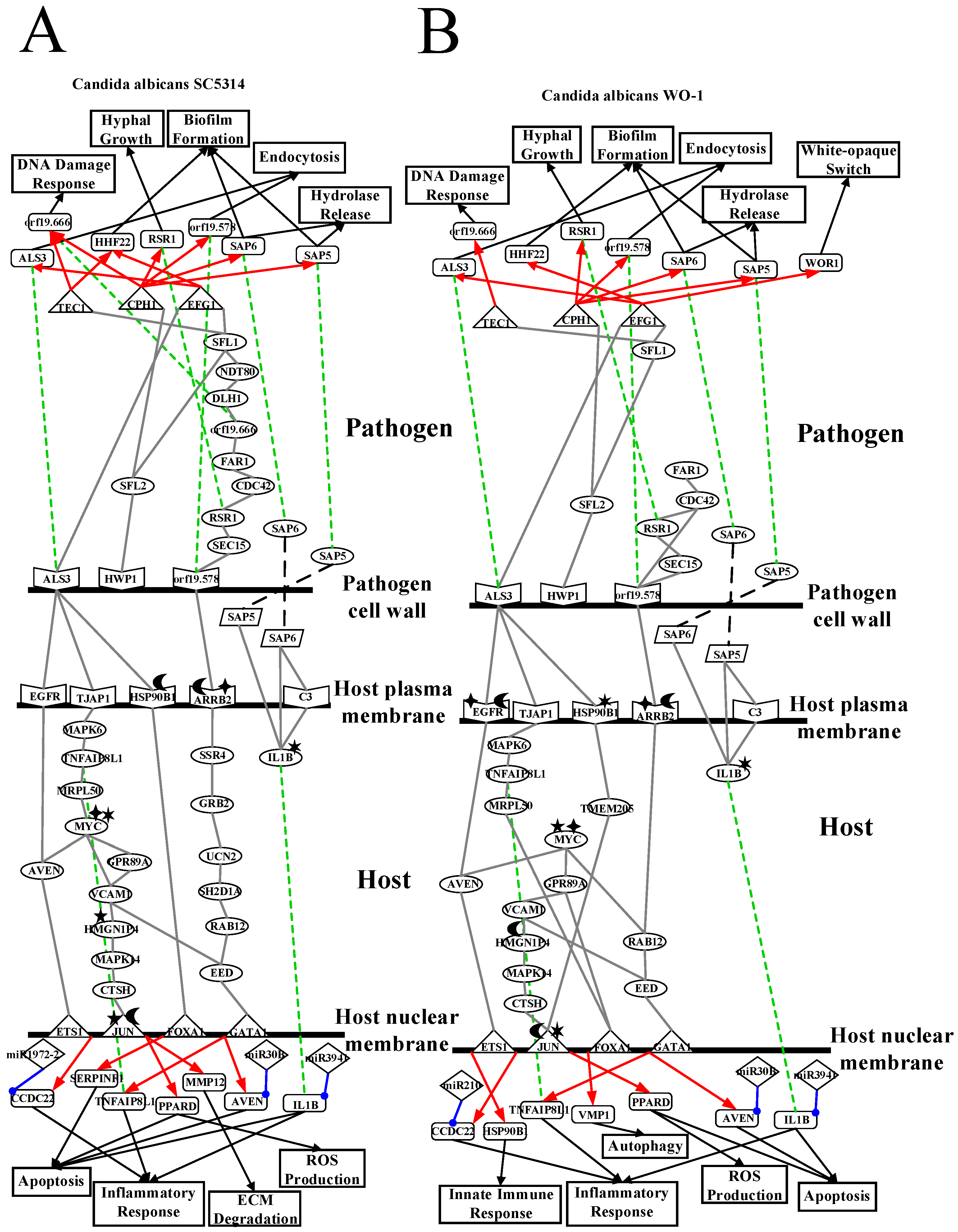
2.2. The Core Host-pathogen Cross-talk Networks (HPCNs) during the Infection of C. albicans SC5314 and C. albicans WO-1
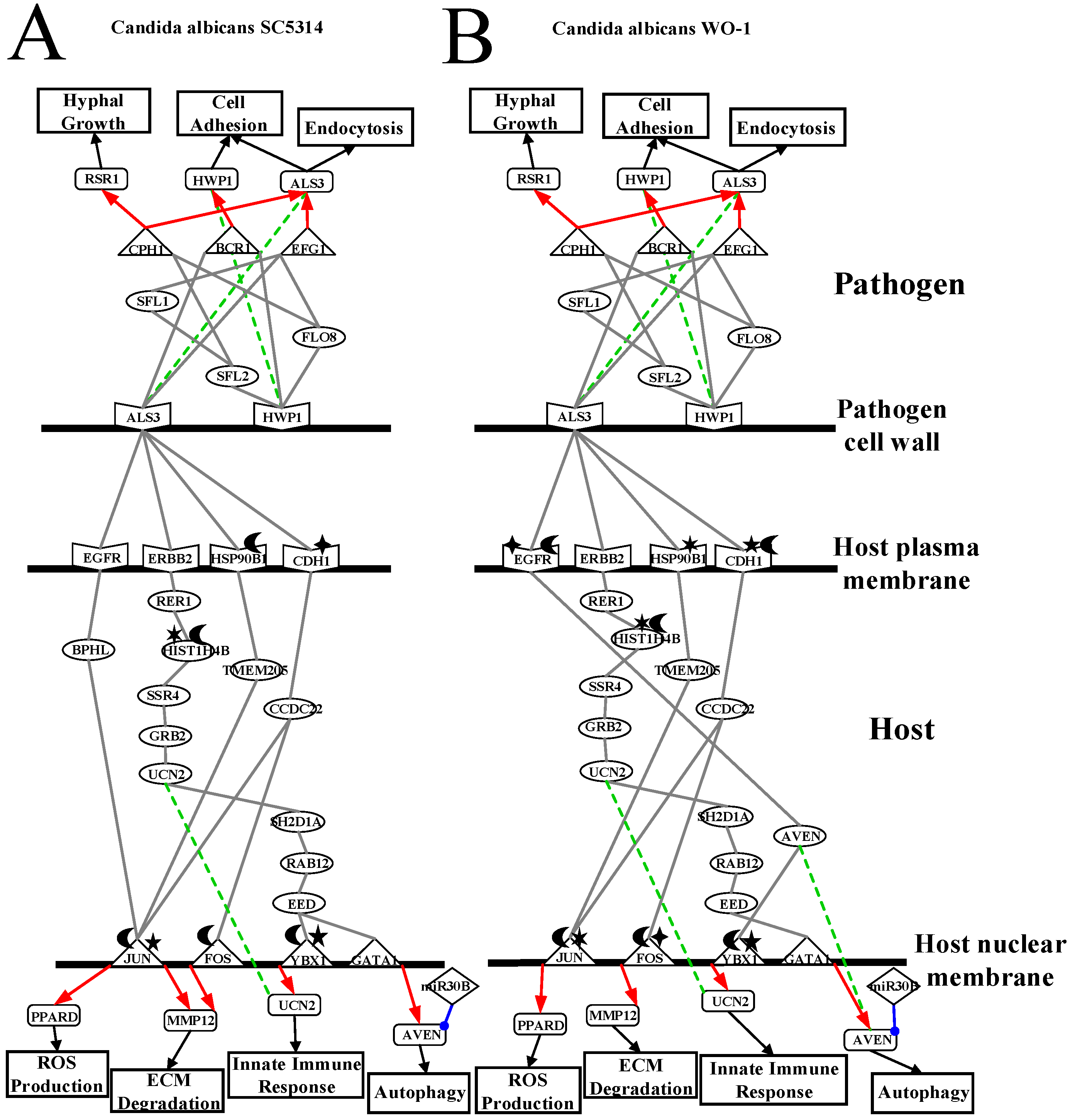
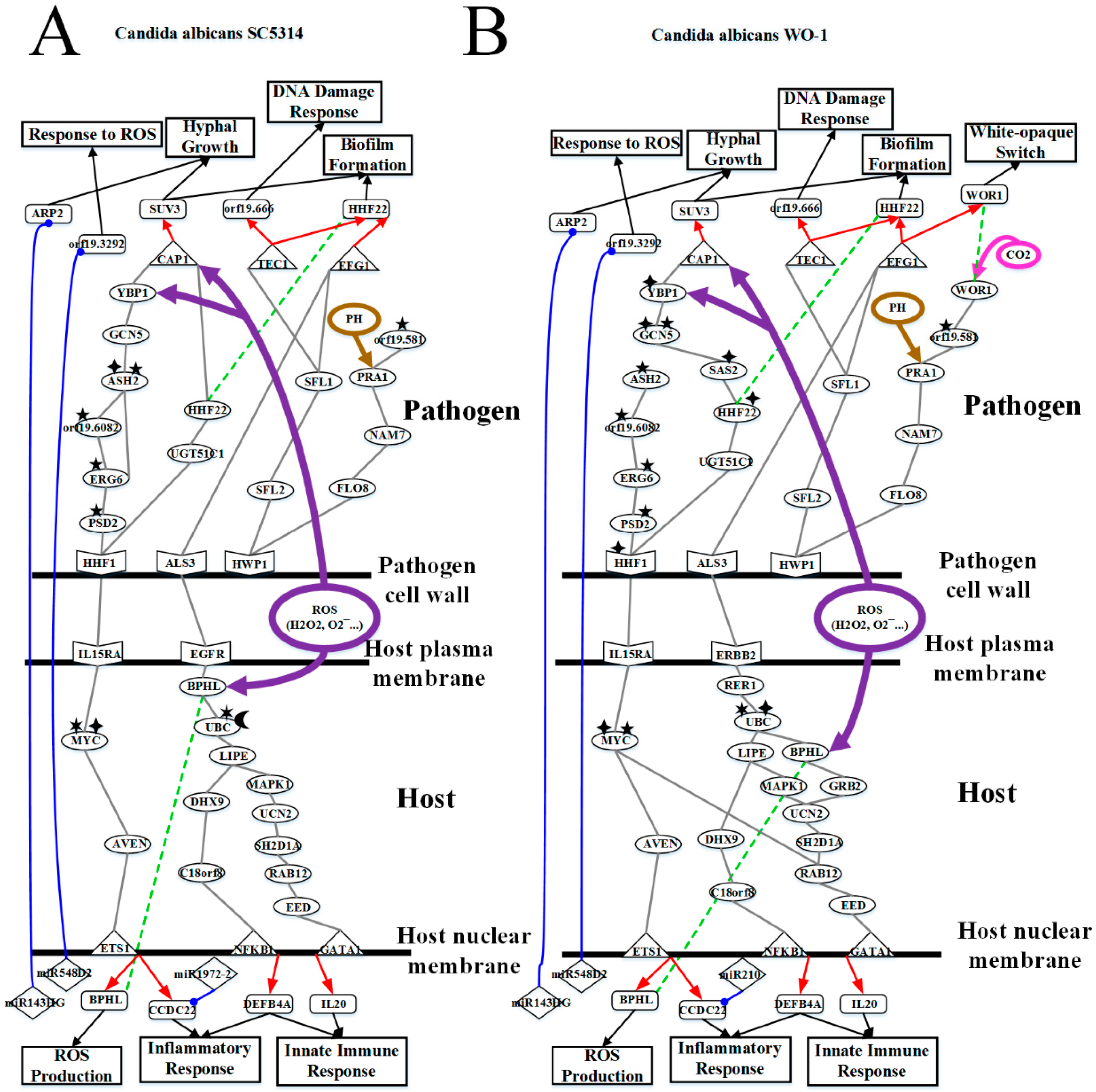
2.3. Analysis of Core Interspecies Pathways to Investigate Host/Pathogen Cross-Talk Common and Specific Pathogenic Progression Mechanisms during C. albicans SC5314 Infection
2.4. Analysis of Core Interspecies Pathways to Investigate Host/Pathogen Cross-talk Common and Specific Pathogenic Mechanisms during C. albicans WO-1 Infection
3. Discussion
3.1. Defensive Mechanism of OKF6/TERT-2 Cell and the Offensive Mechanism of Different Strains of C. albicans at Host Cell Surface
3.2. OKF6/TERT-2 Cell Confronts Different Strains of C. albicans by Strong ROS and Microenvironment Response
3.3. Released Pathogenic Factor and Accumulated Cellular Response Result in Apoptosis and Inflammatory Response Further Leading to Necrosis
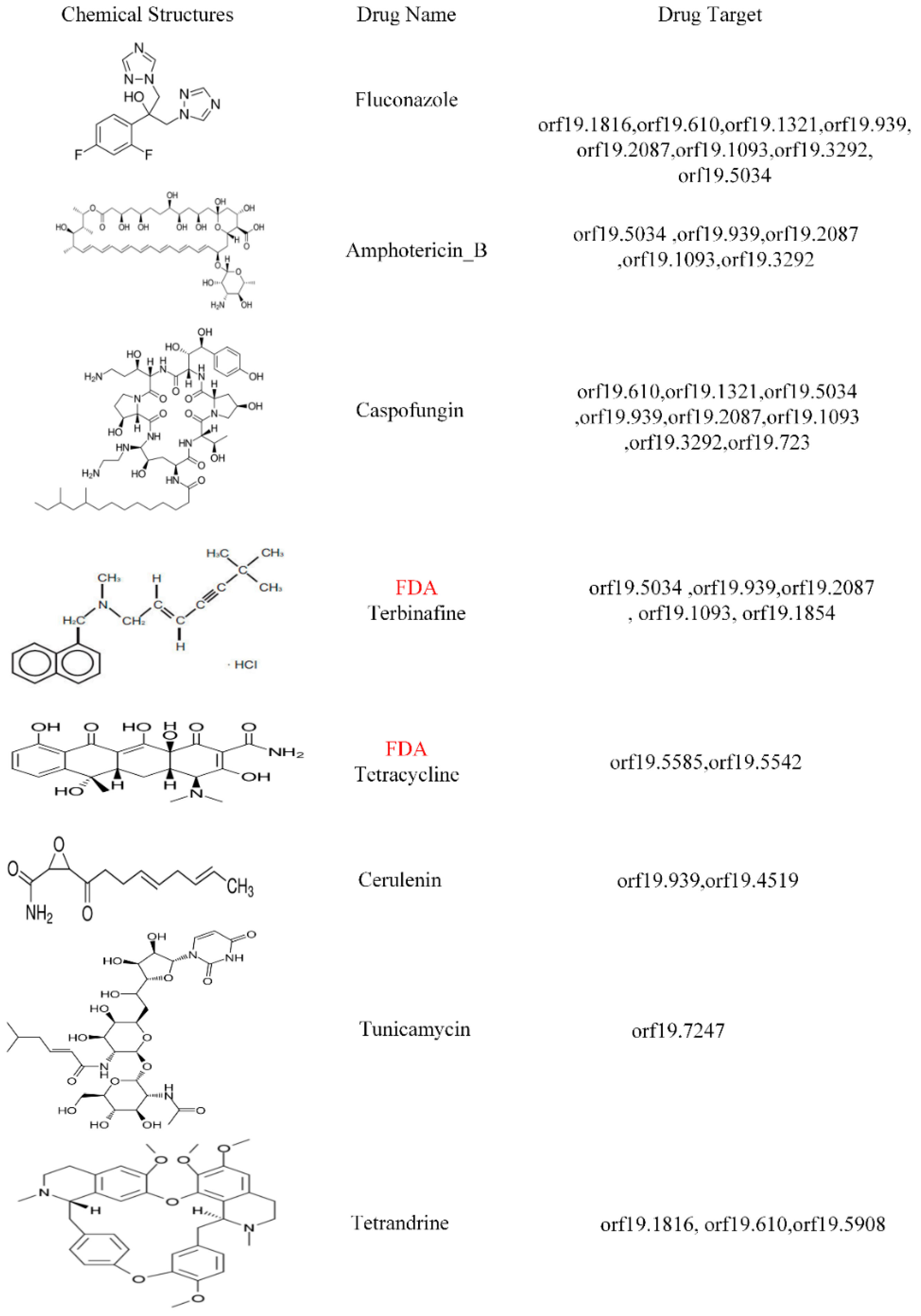
3.4. Prediction of Drug Target Proteins and the Multiple-Molecule Drug Design for the Infection of Different Strains of C. albicans
4. Conclusions
5. Materials and Methods
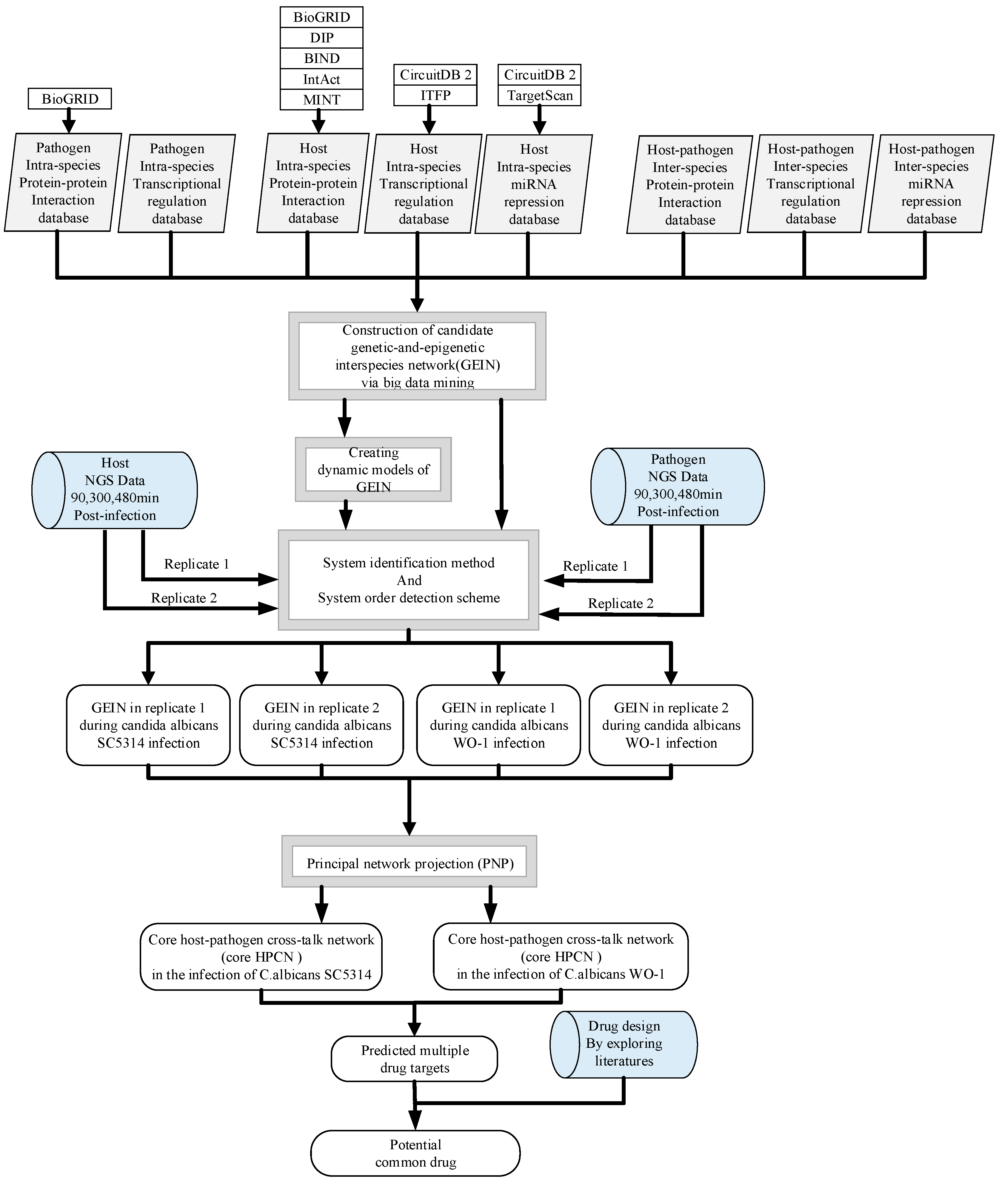
5.1. Overview of the Construction of GEINs and Core HPCNs in OKF6/TERT-2 Cells Line during the Infection of C. albicans SC5314 and C. albicans WO-1
5.2. NGS Data Preprocessing for Human and Pathogen
5.3. Construction of Candidate GEINs by the Inference of Putative Interspecies and Intraspecies PPINs and GRNs for C. albicans SC5314 and WO-1
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Hebecker, B.; Naglik, J.B.; Hube, B.; Jacobsen, I.D. Pathogenicity mechanisms and host response during oral Candida albicans infections. Expert Rev. Anti-Infect. Ther. 2014, 12, 867–879. [Google Scholar] [CrossRef] [PubMed]
- Williams, D.W.; Jordan, R.P.; Wei, X.Q.; Alves, C.T.; Wise, M.P.; Wilson, M.J.; Lewis, M.A. Interactions of Candida albicans with host epithelial surfaces. J. Oral Microbiol. 2013, 5. [Google Scholar] [CrossRef] [PubMed]
- Slutsky, B.; Staebell, M.; Anderson, J.; Risen, L.; Pfaller, M.; Soll, D.R. “White-opaque transition”: A second high-frequency switching system in Candida albicans. J. Bacteriol. 1987, 169, 189–197. [Google Scholar] [CrossRef] [PubMed]
- Arnaud, M.B.; Costanzo, M.C.; Marek, S.S.; Sherlock, G. The candida genome database (CGD), a community resource for Candida albicans gene and protein information. Nucleic Acids Res. 2005, 33, D358–D363. [Google Scholar] [CrossRef] [PubMed]
- Mishra, P.K.; Baum, M.; Carbon, J. DNA methylation regulates phenotype-dependent transcriptional activity in Candida albicans. Proc. Natl. Acad. Sci. USA 2011, 108, 11965–11970. [Google Scholar] [CrossRef] [PubMed]
- Shirtliff, M.E.; Peters, B.M.; Jabra-Rizk, M.A. Cross-kingdom interactions: Candida albicans and bacteria. FEMS Microbiol. Lett. 2009, 299, 1–8. [Google Scholar] [CrossRef]
- Dongari-Bagtzoglou, A.; Kashleva, H. Development of a highly reproducible three-dimensional organotypic model of the oral mucosa. Nat. Protoc. 2006, 1, 2012–2018. [Google Scholar] [CrossRef]
- Mayer, F.L.; Wilson, D.; Hube, B. Candida albicans pathogenicity mechanisms. Virulence 2013, 4, 119–128. [Google Scholar] [CrossRef]
- Goyer, M.; Loiselet, A.; Bon, F.; L’Ollivier, C.; Laue, M.; Holland, G.; Bonnin, A.; Dalle, F. Intestinal cell tight junctions limit invasion of Candida albicans through active penetration and endocytosis in the early stages of the interaction of the fungus with the intestinal barrier. PLoS ONE 2016, 11, e0149159. [Google Scholar] [CrossRef]
- Phan, Q.T.; Myers, C.l.; Fu, Y.; Sheppard, D.C.; Yeaman, M.R.; Welch, W.H.; Ibrahim, A.S.; Edwards, J.E., Jr.; Filler, S.G. Als3 is a Candida albicans invasin that binds to cadherins and induces endocytosis by host cells. PLoS Biol. 2007, 5, e64. [Google Scholar] [CrossRef]
- Zhu, W.; Phan, Q.T.; Boontheung, P.; Solis, N.V.; Loo, J.A.; Filler, S.G. EGFR and HER2 receptor kinase signaling mediate epithelial cell invasion by Candida albicans during oropharyngeal infection. Proc. Natl. Acad. Sci. USA 2012, 109, 14194–14199. [Google Scholar] [CrossRef] [PubMed]
- Edwards, J.E., Jr.; Gaither, T.A.; O’Shea, J.J.; Rotrosen, D.; Lawley, T.J.; Wright, S.A.; Frank, M.M.; Green, I. Expression of specific binding sites on candida with functional and antigenic characteristics of human complement receptors. J. Immunol. 1986, 137, 3577–3583. [Google Scholar] [PubMed]
- Kumamoto, C.A.; Vinces, M.D. Contributions of hyphae and hypha-co-regulated genes to Candida albicans virulence. Cell Microbiol. 2005, 7, 1546–1554. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-C.; Lan, C.-Y.; Hsieh, W.-P.; Murillo, L.A.; Agabian, N.; Chen, B.-S. Global screening of potential Candida albicans biofilm-related transcription factors via network comparison. BMC Bioinf. 2010, 11, 53. [Google Scholar] [CrossRef] [PubMed]
- Sohn, K.; Urban, C.; Brunner, H.; Rupp, S. EFG1 is a major regulator of cell wall dynamics in Candida albicans as revealed by DNA microarrays. J. Bacteriol. 2001, 183, 4090–4093. [Google Scholar] [CrossRef]
- Wu, C.-C.; Chen, B.-S. A systems biology approach to the coordination of defensive and offensive molecular mechanisms in the innate and adaptive host–pathogen interaction networks. PLoS ONE 2016, 11, e0149303. [Google Scholar] [CrossRef] [PubMed]
- Bonazzi, M.; Cossart, P. Impenetrable barriers or entry portals? The role of cell–cell adhesion during infection. J. Cell Biol. 2011, 195, 349–358. [Google Scholar] [CrossRef]
- Nobile, C.J.; Andes, D.R.; Nett, J.E.; Smith, F.J.; Yue, F.; Phan, Q.-T.; Edwards, J.E.; Filler, S.G.; Mitchell, A.P. Critical role of bcr1-dependent adhesins in C. albicans biofilm formation in vitro and in vivo. PLoS Pathogens. 2006, 2, e63. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Da Huang, W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using david bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Moazeni, M.; Khorramizadeh, M.R.; Teimoori-Toolabi, L.; Noorbakhsh, F.; Fallahi, A.A.; Rezaie, S. Down-regulation of the als3 gene as a consequent effect of rna-mediated silencing of the efg1 gene in Candida albicans. Iran. Biomed. J. 2012, 16, 172–178. [Google Scholar] [PubMed]
- Ryan, O.; Shapiro, R.S.; Kurat, C.F.; Mayhew, D.; Baryshnikova, A.; Chin, B.; Lin, Z.-Y.; Cox, M.J.; Vizeacoumar, F.; Cheung, D.; et al. Global gene deletion analysis exploring yeast filamentous growth. Science 2012, 337, 1353–1356. [Google Scholar] [CrossRef] [PubMed]
- Fanning, S.; Xu, W.; Solis, N.; Woolford, C.A.; Filler, S.G.; Mitchell, A.P. Divergent targets of Candida albicans biofilm regulator Bcr1 in vitro and in vivo. Eukaryotic. Cell 2012, 11, 896–904. [Google Scholar] [CrossRef] [PubMed]
- Da Silva Dantas, A.; Patterson, M.J.; Smith, D.A.; Maccallum, D.M.; Erwig, L.P.; Morgan, B.A.; Quinn, J. Thioredoxin regulates multiple hydrogen peroxide-induced signaling pathways in Candida albicans. Acta Pharmacol. Sin. 2010, 31, 616–628. [Google Scholar] [CrossRef] [PubMed]
- Patterson, M.J.; McKenzie, C.G.; Smith, D.A.; da Silva Dantas, A.; Sherston, S.; Veal, E.A.; Morgan, B.A.; MacCallum, D.M.; Erwig, L.-P.; Quinn, J. Ybp1 and Gpx3 signaling in Candida albicans govern hydrogen peroxide-induced oxidation of the cap1 transcription factor and macrophage escape. Antioxid. Redox Signal. 2013, 19, 2244–2260. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, A.; Diefenbacher, M.E.; Mylona, A.; Kassel, O.; Behrens, A. The E3 ubiquitin ligase Trim7 mediates c-Jun/AP-1 activation by ras signalling. Nat. Commun. 2015, 6, 6782. [Google Scholar] [CrossRef] [PubMed]
- Bhogaraju, S.; Kalayil, S.; Liu, Y.; Bonn, F.; Colby, T.; Matic, I.; Dikic, I. Phosphoribosylation of ubiquitin promotes serine ubiquitination and impairs conventional ubiquitination. Cell 2016, 167, 1636–1649.e13. [Google Scholar] [CrossRef] [PubMed]
- Chung, W.O.; Dale, B.A. Innate immune response of oral and foreskin keratinocytes: Utilization of different signaling pathways by various bacterial species. Infect. Immun. 2004, 72, 352–358. [Google Scholar] [CrossRef]
- Schulz, A.; Kluver, E.; Schulz-Maronde, S.; Adermann, K. Engineering disulfide bonds of the novel human beta-defensins hbd-27 and hbd-28: Differences in disulfide formation and biological activity among human beta-defensins. Biopolymers 2005, 80, 34–49. [Google Scholar] [CrossRef]
- Buck, I.; Morceau, F.; Cristofanon, S.; Reuter, S.; Dicato, M.; Diederich, M. The inhibitory effect of the proinflammatory cytokine TNFalpha on erythroid differentiation involves erythroid transcription factor modulation. Int. J. Oncol. 2009, 34, 853–860. [Google Scholar]
- Jin, S.; Song, Y.C.; Emili, A.; Sherman, P.M.; Chan, V.L. JlpA of campylobacter jejuni interacts with surface-exposed heat shock protein 90alpha and triggers signalling pathways leading to the activation of nf-kappab and p38 MAP kinase in epithelial cells. Cell Microbiol. 2003, 5, 165–174. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Chen, S. ER regulates an evolutionarily conserved apoptosis pathway. Biochem. Biophys. Res. Commun. 2010, 400, 34–38. [Google Scholar] [CrossRef] [PubMed]
- Geng, F.; Zhu, W.; Anderson, R.A.; Leber, B.; Andrews, D.W. Multiple post-translational modifications regulate e-cadherin transport during apoptosis. Virulence 2012, 125 Pt 11, 2615–2625. [Google Scholar] [CrossRef]
- Jariel-Encontre, I.; Salvat, C.; Steff, A.M.; Pariat, M.; Acquaviva, C.; Furstoss, O.; Piechaczyk, M. Complex mechanisms for c-fos and c-jun degradation. Mol. Biol. Rep. 1997, 24, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.; Shi, Y.; Xiang, B.; Qu, B.; Su, W.; Zhu, M.; Zhang, M.; Bao, G.; Wang, F.; Zhang, X.; et al. A nuclear function of beta-arrestin1 in GPCR signaling: Regulation of histone acetylation and gene transcription. Cell 2005, 123, 833–847. [Google Scholar] [CrossRef] [PubMed]
- Tsoni, S.V.; Marakalala, M.J.; Srinivasan, N.; Duffield, M.; Taylor, P.R.; Botto, M.; Steele, C.; Brown, G.D. Complement C3 plays an essential role in the control of opportunistic fungal infections. Infect. Immun. 2009, 77, 3679–3685. [Google Scholar] [CrossRef] [PubMed]
- Yano, S.; Banno, T.; Walsh, R.; Blumenberg, M. Transcriptional responses of human epidermal keratinocytes to cytokine interleukin-1. J. Cell Physiol. 2008, 214, 1–13. [Google Scholar] [CrossRef]
- Pietrella, D.; Rachini, A.; Pandey, N.; Schild, L.; Netea, M.; Bistoni, F.; Hube, B.; Vecchiarelli, A. The inflammatory response induced by aspartic proteases of Candida albicans is independent of proteolytic activity. Infect. Immun. 2010, 78, 4754–4762. [Google Scholar] [CrossRef]
- Chen, Z.; Jin, T.; Lu, Y. AntimiR-30b inhibits TNF-alpha mediated apoptosis and attenuated cartilage degradation through enhancing autophagy. Cell Physiol. Biochem. 2016, 40, 883–894. [Google Scholar] [CrossRef]
- Verhelst, K.; Verstrepen, L.; Carpentier, I.; Beyaert, R. Linear ubiquitination in NF-kappab signaling and inflammation: What we do understand and what we do not. Biochem. Pharmacol. 2011, 82, 1057–1065. [Google Scholar] [CrossRef]
- Sonneborn, A.; Tebarth, B.; Ernst, J.F. Control of white-opaque phenotypic switching in Candida albicans by the Efg1p morphogenetic regulator. Infect. Immun. 1999, 67, 4655–4660. [Google Scholar]
- Hirakawa, M.P.; Martinez, D.A.; Sakthikumar, S.; Anderson, M.Z.; Berlin, A.; Gujja, S.; Zeng, Q.; Zisson, E.; Wang, J.M.; Greenberg, J.M.; et al. Genetic and phenotypic intra-species variation in Candida albicans. Genome Res. 2015, 25, 413–425. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Huang, G. Environmental pH adaption and morphological transitions in Candida albicans. Curr Genet. 2016, 62, 283–286. [Google Scholar] [CrossRef] [PubMed]
- Farah, C.S.; Lynch, N.; McCullough, M.J. Oral fungal infections: An update for the general practitioner. Aust. Dent. J. 2010, 55 (Suppl. 1), 48–54. [Google Scholar] [CrossRef] [PubMed]
- Dühring, S.; Germerodt, S.; Skerka, C.; Zipfel, P.F.; Dandekar, T.; Schuster, S. Host-pathogen interactions between the human innate immune system and Candida albicans—Understanding and modeling defense and evasion strategies. Front. Microbiol. 2015, 6, 625. [Google Scholar] [CrossRef] [PubMed]
- Sandai, D.; Tabana, Y.M.; Ouweini, A.E.; Ayodeji, I.O. Resistance of Candida albicans biofilms to drugs and the host immune system. Jundishapur J. Microbiol. 2016, 9, e37385. [Google Scholar] [CrossRef]
- Xie, Z.; Thompson, A.; Sobue, T.; Kashleva, H.; Xu, H.; Vasilakos, J.; Dongari-Bagtzoglou, A. Candida albicans biofilms do not trigger reactive oxygen species and evade neutrophil killing. J. Infect. Dis. 2012, 206, 1936–1945. [Google Scholar] [CrossRef]
- Pappas, P.G.; Kauffman, C.A.; Andes, D.R.; Clancy, C.J.; Marr, K.A.; Ostrosky-Zeichner, L.; Reboli, A.C.; Schuster, M.G.; Vazquez, J.A.; Walsh, T.J.; et al. Clinical practice guideline for the management of candidiasis: 2016 update by the infectious diseases society of america. Clin. Infect. Dis. 2016, 62, e1–e50. [Google Scholar] [CrossRef]
- Villar, C.C.; Kashleva, H.; Nobile, C.J.; Mitchell, A.P.; Dongari-Bagtzoglou, A. Mucosal tissue invasion by Candida albicans is associated with E-cadherin degradation, mediated by transcription factor Rim101p and protease Sap5p. Infect. Immun. 2007, 75, 2126–2135. [Google Scholar] [CrossRef]
- Xu, D.; Jiang, B.; Ketela, T.; Lemieux, S.; Veillette, K.; Martel, N.; Davison, J.; Sillaots, S.; Trosok, S.; Bachewich, C.; et al. Genome-wide fitness test and mechanism-of-action studies of inhibitory compounds in Candida albicans. PLoS Pathog. 2007, 3, e92. [Google Scholar] [CrossRef]
- Staib, P.; Lermann, U.; Blass-Warmuth, J.; Degel, B.; Wurzner, R.; Monod, M.; Schirmeister, T.; Morschhauser, J. Tetracycline-inducible expression of individual secreted aspartic proteases in Candida albicans allows isoenzyme-specific inhibitor screening. Antimicrob. Agents Chemother. 2008, 52, 146–156. [Google Scholar] [CrossRef] [PubMed]
- Braga-Silva, L.A.; Santos, A.L. Aspartic protease inhibitors as potential anti-Candida albicans drugs: Impacts on fungal biology, virulence and pathogenesis. Curr. Med. Chem. 2011, 18, 2401–2419. [Google Scholar] [CrossRef] [PubMed]
- Niewerth, M.; Kunze, D.; Seibold, M.; Schaller, M.; Korting, H.C.; Hube, B. Ciclopirox olamine treatment affects the expression pattern of Candida albicans genes encoding virulence factors, iron metabolism proteins, and drug resistance factors. Antimicrob. Agents Chemother. 2003, 47, 1805–1817. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.X.; Li, D.D.; Hu, D.-D.; Hu, G.-H.; Yan, L.; Wang, Y.; Jiang, Y.Y. Effect of tetrandrine against Candida albicans biofilms. PLoS ONE 2013, 8, e79671. [Google Scholar] [CrossRef] [PubMed]
- Soysa, N.S.; Samaranayake, L.P.; Ellepola, A.N. Antimicrobials as a contributory factor in oral candidosis—A brief overview. Oral Dis. 2008, 14, 138–143. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Lee, J.E.; Lee, J.S. Histone deacetylase-mediated morphological transition in Candida albicans. J. Microbiol. 2015, 53, 805–811. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Shetty, A.C.; Schwartz, J.A.; Bradford, L.L.; Xu, W.; Phan, Q.T.; Kumari, P.; Mahurkar, A.; Mitchell, A.P.; Ravel, J.; et al. New signaling pathways govern the host response to C. albicans infection in various niches. Genome Res. 2015, 25, 679–689. [Google Scholar] [CrossRef] [PubMed]
- Licata, L.; Briganti, L.; Peluso, D.; Perfetto, L.; Iannuccelli, M.; Galeota, E.; Sacco, F.; Palma, A.; Nardozza, A.P.; Santonico, E.; et al. Mint, the molecular interaction database: 2012 update. Nucleic Acids Res. 2012, 40, D857–D861. [Google Scholar] [CrossRef]
- Orchard, S.; Ammari, M.; Aranda, B.; Breuza, L.; Briganti, L.; Broackes-Carter, F.; Campbell, N.H.; Chavali, G.; Chen, C.; del-Toro, N.; et al. The mintact project—intact as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res. 2014, 42, D358–D363. [Google Scholar] [CrossRef]
- Salwinski, L.; Miller, C.S.; Smith, A.J.; Pettit, F.K.; Bowie, J.U.; Eisenberg, D. The database of interacting proteins: 2004 update. Nucleic Acids Res. 2004, 32, D449–D451. [Google Scholar] [CrossRef]
- Bader, G.D.; Betel, D.; Hogue, C.W.V. Bind: The biomolecular interaction network database. Nucleic Acids Res. 2003, 31, 248–250. [Google Scholar] [CrossRef] [PubMed]
- Chatr-Aryamontri, A.; Breitkreutz, B.J.; Oughtred, R.; Boucher, L.; Heinicke, S.; Chen, D.; Stark, C.; Breitkreutz, A.; Kolas, N.; O’Donnell, L.; et al. The biogrid interaction database: 2015 update. Nucleic Acids Res. 2015, 43, D470–D478. [Google Scholar] [CrossRef]
- Friard, O.; Re, A.; Taverna, D.; De Bortoli, M.; Corá, D. Circuitsdb: A database of mixed microrna/transcription factor feed-forward regulatory circuits in human and mouse. BMC Bioinf. 2010, 11, 435. [Google Scholar] [CrossRef]
- Zheng, G.; Tu, K.; Yang, Q.; Xiong, Y.; Wei, C.; Xie, L.; Zhu, Y.; Li, Y. Itfp: An integrated platform of mammalian transcription factors. Bioinformatics 2008, 24, 2416–2417. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-C.; Lin, C.; Chuang, M.-T.; Hsieh, W.-P.; Lan, C.-Y.; Chuang, Y.-J.; Chen, B.-S. Interspecies protein-protein interaction network construction for characterization of host-pathogen interactions: A Candida albicans-zebrafish interaction study. BMC Syst. Biol. 2013, 7, 79. [Google Scholar] [CrossRef] [PubMed]
- Cherry, J.M.; Adler, C.; Ball, C.; Chervitz, S.A.; Dwight, S.S.; Hester, E.T.; Jia, Y.; Juvik, G.; Roe, T.; Schroeder, M.; et al. SGD: Saccharomyces genome database. Nucleic Acids Res. 1998, 26, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The string database in 2017: Quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. String v10: Protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef]
- Bird, A.J.; Blankman, E.; Stillman, D.J.; Eide, D.J.; Winge, D.R. The Zap1 transcriptional activator also acts as a repressor by binding downstream of the TATA box in ZET2. EMBO J. 2004, 23, 1123–1132. [Google Scholar] [CrossRef]
- Chen, H.-F.; Lan, C.-Y. Role of sfp1 in the regulation of Candida albicans biofilm formation. PLoS ONE 2015, 10, e0129903. [Google Scholar] [CrossRef]
- Colman-Lerner, A.; Chin, T.E.; Brent, R. Yeast Cbk1 and Mob2 activate daughter-specific genetic programs to induce asymmetric cell fates. Cell 2014, 107, 739–750. [Google Scholar] [CrossRef]
- Murciano, C.; Moyes, D.L.; Runglall, M.; Tobouti, P.; Islam, A.; Hoyer, L.L.; Naglik, J.R. Evaluation of the role of Candida albicans agglutinin-like sequence (Als) proteins in human oral epithelial cell interactions. PLoS ONE 2012, 7, e33362. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.N.; Solis, N.V.; Phan, Q.T.; Bajwa, J.S.; Kashleva, H.; Thompson, A.; Liu, Y.; Dongari-Bagtzoglou, A.; Edgerton, M.; Filler, S.G. Host cell invasion and virulence mediated by Candida albicans Ssa1. PLoS Pathogens. 2010, 6, e1001181. [Google Scholar] [CrossRef] [PubMed]
- Taylor, D.M.; Tradewell, M.L.; Minotti, S.; Durham, H.D. Characterizing the role of Hsp90 in production of heat shock proteins in motor neurons reveals a suppressive effect of wild-type Hsf1. Cell Stress Chaperones. 2007, 12, 151–162. [Google Scholar] [CrossRef] [PubMed]
- Argimon, S.; Wishart, J.A.; Leng, R.; Macaskill, S.; Mavor, A.; Alexandris, T.; Nicholls, S.; Knight, A.W.; Enjalbert, B.; Walmsley, R.; et al. Developmental regulation of an adhesin gene during cellular morphogenesis in the fungal pathogen Candida albicans. Eukaryot. Cell 2007, 6, 682–692. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, M.; Uppuluri, P.; Zhao, X.R.; Carlisle, P.L.; Vipulanandan, G.; Villar, C.C.; López-Ribot, J.L.; Kadosh, D. Expression of UME6, a key regulator of Candida albicans hyphal development, enhances biofilm formation via Hgc1- and Sun41-dependent mechanisms. Eukaryot. Cell 2013, 12, 224–232. [Google Scholar] [CrossRef] [PubMed]
- Bastidas, R.J.; Heitman, J.; Cardenas, M.E. The protein kinase Tor1 regulates adhesin gene expression in Candida albicans. PLoS Pathog. 2009, 5, e1000294. [Google Scholar] [CrossRef]
- Braun, B.R.; Kadosh, D.; Johnson, A.D. NRG1, a repressor of filamentous growth in C. albicans, is down-regulated during filament induction. EMBO J. 2001, 20, 4753–4761. [Google Scholar] [CrossRef]

| Nodes | Candidates_SC5314 | SC5314_R1 | SC5314_R2 | Candidates_WO-1 | WO-1_R1 | WO-1_R2 |
|---|---|---|---|---|---|---|
| HP | 23,217 | 15,817 | 9979 | 20,694 | 14,682 | 15,621 |
| HR | 3025 | 1960 | 1141 | 2488 | 1719 | 1743 |
| HT | 5649 | 855 | 357 | 4696 | 384 | 427 |
| HM | 607 | 385 | 370 | 412 | 331 | 342 |
| HL | 333 | 166 | 101 | 258 | 131 | 129 |
| PP | 3766 | 2414 | 2195 | 3736 | 3413 | 3411 |
| PT | 487 | 254 | 255 | 483 | 253 | 280 |
| Total nodes | 37,229 | 21,851 | 14,398 | 32,767 | 20,913 | 21,953 |
| Edges | Candidates_SC5314 | SC5314R1 | SC5314R2 | Candidates_WO-1 | WO-1_R1 | WO-1_R2 |
|---|---|---|---|---|---|---|
| HT→HG | 152,491 | 8036 | 5039 | 141,001 | 7963 | 7638 |
| HT→HM | 1347 | 154 | 21 | 901 | 62 | 42 |
| HT→HL | 271 | 162 | 98 | 200 | 129 | 124 |
| HL→HG | 37 | 1 | 0 | 35 | 0 | 0 |
| HM→HG | 170,671 | 2401 | 2408 | 118,017 | 2799 | 3744 |
| HM→HL | 130 | 4 | 3 | 78 | 2 | 5 |
| HM→HM | 45 | 6 | 1 | 21 | 0 | 4 |
| HT→PG | 21,328 | 374 | 242 | 20,177 | 586 | 489 |
| HM→PG | 23,730 | 394 | 328 | 16,639 | 543 | 771 |
| HL→PG | 1 | 0 | 0 | 1 | 0 | 0 |
| PT→HM | 48 | 9 | 1 | 29 | 0 | 1 |
| PT→HG | 8910 | 208 | 169 | 8726 | 220 | 248 |
| PT→PG | 86,491 | 7225 | 5112 | 85,211 | 5723 | 7167 |
| HG—HG | 6,449,171 | 30,892 | 24,665 | 5,521,448 | 119,623 | 127,687 |
| HG—PG | 1,615,845 | 10,248 | 7540 | 1,453,353 | 45,214 | 47,146 |
| PG—PG | 132,600 | 4830 | 3988 | 131,485 | 33,643 | 34,707 |
| SC5314 | ||
| Category | Term | p-Value |
| GOTERM_MF_DIRECT | GO:0016712~oxidoreductase activity (0.47%) | 0.00298310389230338 |
| GOTERM_MF_DIRECT | GO:0003676~nucleic acid binding (5.5%) | 0.0129970927095585 |
| UP_SEQ_FEATURE | Metal ion-binding (0.43%) | 0.0176534099242106 |
| GOTERM_BP_DIRECT | GO:0016338~cell adhesion (0.3%) | 0.0181497359081671 |
| KEGG_PATHWAY | hsa03030:DNA replication (0.39%) | 0.034840174 |
| WO-1 | ||
| Category | Term | p-Value |
| GOTERM_MF_FAT | GO:0008135~nucleic acid binding (0.89%) | 0.00700213339377146 |
| UP_SEQ_FEATURE | Metal ion-binding (0.45%) | 0.00288724333819777 |
| KEGG_PATHWAY | hsa03030:DNA replication (0.45%) | 0.015170596995848 |
| SP_PIR_KEYWORDS | cell adhesion (2.64%) | 0.0332759579717729 |
| GOTERM_MF_FAT | GO:0016651~oxidoreductase activity (0.7%) | 0.0337336128643766 |
| SC5314 | ||
| GOID | GO_Term | p-Value |
| 5198 | structural molecule activity (6.8%) | 3.18 × 10−19 |
| 5515 | protein binding (12.2%) | 8.85 × 10−15 |
| 16740 | transferase activity (15.6%) | 4.38 × 10−10 |
| 98772 | molecular function regulator (5.7%) | 2.53 × 10−8 |
| 16787 | hydrolase activity (16.3%) | 5.32 × 10−5 |
| WO-1 | ||
| Category | GO_Term | p-Value |
| 5515 | protein binding (12.2%) | 2.14 × 10−17 |
| 16787 | hydrolase activity (17.7%) | 1.13 × 10−14 |
| 16740 | transferase activity (15.8%) | 6.16 × 10−13 |
| 98772 | molecular function regulator (5.7%) | 7.02 × 10−10 |
| 5198 | structural molecule activity (5.4%) | 4.98 × 10−6 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yeh, S.-J.; Yeh, C.-C.; Lan, C.-Y.; Chen, B.-S. Investigating Common Pathogenic Mechanisms between Homo sapiens and Different Strains of Candida albicans for Drug Design: Systems Biology Approach via Two-Sided NGS Data Identification. Toxins 2019, 11, 119. https://doi.org/10.3390/toxins11020119
Yeh S-J, Yeh C-C, Lan C-Y, Chen B-S. Investigating Common Pathogenic Mechanisms between Homo sapiens and Different Strains of Candida albicans for Drug Design: Systems Biology Approach via Two-Sided NGS Data Identification. Toxins. 2019; 11(2):119. https://doi.org/10.3390/toxins11020119
Chicago/Turabian StyleYeh, Shan-Ju, Chun-Chieh Yeh, Chung-Yu Lan, and Bor-Sen Chen. 2019. "Investigating Common Pathogenic Mechanisms between Homo sapiens and Different Strains of Candida albicans for Drug Design: Systems Biology Approach via Two-Sided NGS Data Identification" Toxins 11, no. 2: 119. https://doi.org/10.3390/toxins11020119
APA StyleYeh, S.-J., Yeh, C.-C., Lan, C.-Y., & Chen, B.-S. (2019). Investigating Common Pathogenic Mechanisms between Homo sapiens and Different Strains of Candida albicans for Drug Design: Systems Biology Approach via Two-Sided NGS Data Identification. Toxins, 11(2), 119. https://doi.org/10.3390/toxins11020119




