Nutritional Factors Modulating Alu Methylation in an Italian Sample from The Mark-Age Study Including Offspring of Healthy Nonagenarians
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population and Blood sample Collection
2.2. NMR Analysis of Lipoprotein Subclasses
2.3. Metal Trace Element Determination in Plasma Samples
2.4. Systemic Inflammation Parameters
2.5. Determination of Total Glutathione and Total Free Cysteine in Whole Blood
2.6. Determination of Ascorbic Acid and Uric Acid in Plasma
2.7. Determination of Total Carotenoid Plasma Levels
2.8. DNA Extraction and Bisulfite Treatment
2.9. Bisulfite Pyrosequencing Analysis of Alu DNA Methylation
2.10. Statistical Analysis
3. Results
3.1. Characteristics of Subjects
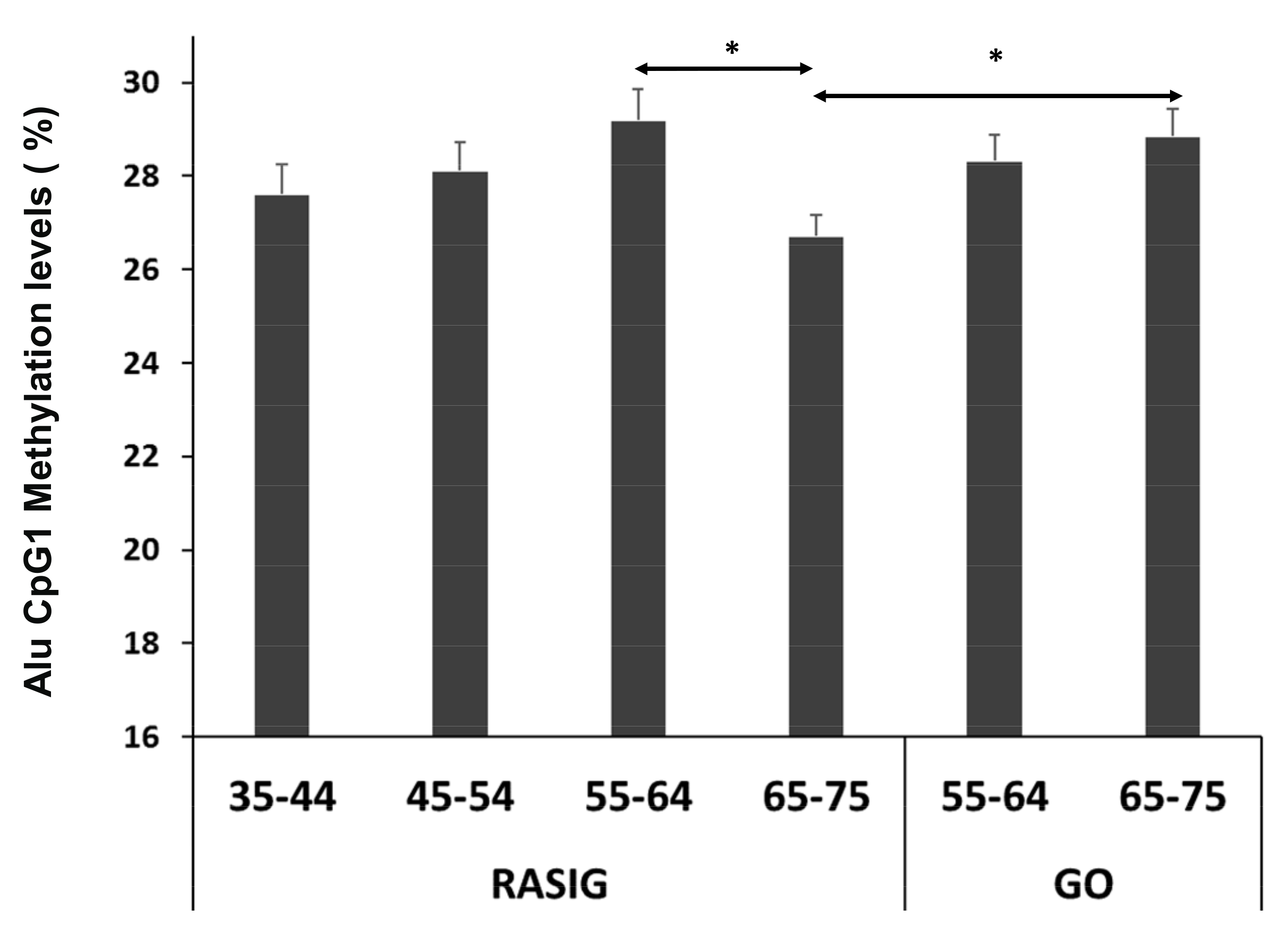
3.2. Alu Methylation
3.3. The Association Between Alu CpG1 Methylation Levels and Nutritional, Metabolic and Inflammatory Factors
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sadakierska-Chudy, A.; Kostrzewa, R.M.; Filip, M. A comprehensive view of the epigenetic landscape part I: DNA methylation, passive and active DNA demethylation pathways and histone variants. Neurotox. Res. 2015, 27, 84–97. [Google Scholar] [CrossRef] [PubMed]
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; et al. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921. [Google Scholar] [PubMed]
- Cardelli, M. The epigenetic alterations of endogenous retroelements in aging. Mech. Ageing Dev. 2018, 174, 30–46. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Lu, X.; Xie, H. Dynamic Alu methylation during normal development, aging, and tumorigenesis. BioMed Res. Int. 2014, 2014, 784706. [Google Scholar] [CrossRef] [PubMed]
- Weisenberger, D.J.; Campan, M.; Long, T.I.; Kim, M.; Woods, C.; Fiala, E.; Ehrlich, M.; Laird, P.W. Analysis of repetitive element DNA methylation by MethyLight. Nucleic Acids Res. 2005, 33, 6823–6836. [Google Scholar] [CrossRef]
- Deininger, P.L.; Moran, J.V.; Batzer, M.A.; Kazazian, H.H. Mobile elements and mammalian genome evolution. Curr. Opin. Genet. Dev. 2003, 13, 651–658. [Google Scholar] [CrossRef]
- Goodier, J.L. Retrotransposition in tumors and brains. Mob. DNA 2014, 5, 11. [Google Scholar] [CrossRef]
- Tubio, J.M.C.; Li, Y.; Ju, Y.S.; Martincorena, I.; Cooke, S.L.; Tojo, M.; Gundem, G.; Pipinikas, C.P.; Zamora, J.; Raine, K.; et al. Mobile DNA in cancer. Extensive transduction of nonrepetitive DNA mediated by L1 retrotransposition in cancer genomes. Science 2014, 345, 1251343. [Google Scholar] [CrossRef]
- Kochanek, S.; Renz, D.; Doerfler, W. DNA methylation in the Alu sequences of diploid and haploid primary human cells. EMBO J. 1993, 12, 1141–1151. [Google Scholar] [CrossRef]
- Meissner, A.; Mikkelsen, T.S.; Gu, H.; Wernig, M.; Hanna, J.; Sivachenko, A.; Zhang, X.; Bernstein, B.E.; Nusbaum, C.; Jaffe, D.B.; et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 2008, 454, 766–770. [Google Scholar] [CrossRef]
- Patchsung, M.; Settayanon, S.; Pongpanich, M.; Mutirangura, D.; Jintarith, P.; Mutirangura, A. Alu siRNA to increase Alu element methylation and prevent DNA damage. Epigenomics 2018, 10, 175–185. [Google Scholar] [CrossRef] [PubMed]
- Daskalos, A.; Nikolaidis, G.; Xinarianos, G.; Savvari, P.; Cassidy, A.; Zakopoulou, R.; Kotsinas, A.; Gorgoulis, V.; Field, J.K.; Liloglou, T. Hypomethylation of retrotransposable elements correlates with genomic instability in non-small cell lung cancer. Int. J. Cancer 2009, 124, 81–87. [Google Scholar] [CrossRef] [PubMed]
- Bollati, V.; Schwartz, J.; Wright, R.; Litonjua, A.; Tarantini, L.; Suh, H.; Sparrow, D.; Vokonas, P.; Baccarelli, A. Decline in genomic DNA methylation through aging in a cohort of elderly subjects. Mech. Ageing Dev. 2009, 130, 234–239. [Google Scholar] [CrossRef] [PubMed]
- Thongsroy, J.; Patchsung, M.; Mutirangura, A. The association between Alu hypomethylation and severity of type 2 diabetes mellitus. Clin. Epigenet. 2017, 9, 93. [Google Scholar] [CrossRef] [PubMed]
- Jordà, M.; Díez-Villanueva, A.; Mallona, I.; Martín, B.; Lois, S.; Barrera, V.; Esteller, M.; Vavouri, T.; Peinado, M.A. The epigenetic landscape of Alu repeats delineates the structural and functional genomic architecture of colon cancer cells. Genome Res. 2017, 27, 118–132. [Google Scholar] [CrossRef] [PubMed]
- Jintaridth, P.; Tungtrongchitr, R.; Preutthipan, S.; Mutirangura, A. Hypomethylation of Alu elements in post-menopausal women with osteoporosis. PLoS ONE 2013, 8, e70386. [Google Scholar] [CrossRef] [PubMed]
- Muka, T.; Koromani, F.; Portilla, E.; O’Connor, A.; Bramer, W.M.; Troup, J.; Chowdhury, R.; Dehghan, A.; Franco, O.H. The role of epigenetic modifications in cardiovascular disease: A systematic review. Int. J. Cardiol. 2016, 212, 174–183. [Google Scholar] [CrossRef]
- Gentilini, D.; Mari, D.; Castaldi, D.; Remondini, D.; Ogliari, G.; Ostan, R.; Bucci, L.; Sirchia, S.M.; Tabano, S.; Cavagnini, F.; et al. Role of epigenetics in human aging and longevity: Genome-wide DNA methylation profile in centenarians and centenarians’ offspring. Age 2013, 35, 1961–1973. [Google Scholar] [CrossRef]
- Horvath, S.; Pirazzini, C.; Bacalini, M.G.; Gentilini, D.; Di Blasio, A.M.; Delledonne, M.; Mari, D.; Arosio, B.; Monti, D.; Passarino, G.; et al. Decreased epigenetic age of PBMCs from Italian semi-supercentenarians and their offspring. Aging 2015, 7, 1159–1170. [Google Scholar] [CrossRef]
- Tedone, E.; Arosio, B.; Gussago, C.; Casati, M.; Ferri, E.; Ogliari, G.; Ronchetti, F.; Porta, A.; Massariello, F.; Nicolini, P.; et al. Leukocyte telomere length and prevalence of age-related diseases in semisupercentenarians, centenarians and centenarians’ offspring. Exp. Gerontol. 2014, 58, 90–95. [Google Scholar] [CrossRef]
- Zhu, Z.Z.; Hou, L.; Bollati, V.; Tarantini, L.; Marinelli, B.; Cantone, L.; Yang, A.S.; Vokonas, P.; Lissowska, J.; Fustinoni, S.; et al. Predictors of global methylation levels in blood DNA of healthy subjects: A combined analysis. Int. J. Epidemiol. 2012, 41, 126–139. [Google Scholar] [CrossRef]
- Perng, W.; Villamor, E.; Shroff, M.R.; Nettleton, J.A.; Pilsner, J.R.; Liu, Y.; Diez-Roux, A.V. Dietary intake, plasma homocysteine, and repetitive element DNA methylation in the Multi-Ethnic Study of Atherosclerosis (MESA). Nutr. Metab. Cardiovasc. Dis. 2014, 24, 614–622. [Google Scholar] [CrossRef] [PubMed]
- Ono, H.; Iwasaki, M.; Kuchiba, A.; Kasuga, Y.; Yokoyama, S.; Onuma, H.; Nishimura, H.; Kusama, R.; Ohnami, S.; Sakamoto, H.; et al. Association of dietary and genetic factors related to one-carbon metabolism with global methylation level of leukocyte DNA. Cancer Sci. 2012, 103, 2159–2164. [Google Scholar] [CrossRef] [PubMed]
- Sae-Lee, C.; Corsi, S.; Barrow, T.M.; Kuhnle, G.G.C.; Bollati, V.; Mathers, J.C.; Byun, H.M. Dietary Intervention Modifies DNA Methylation Age Assessed by the Epigenetic Clock. Mol. Nutr. Food Res. 2018, 62, 1800092. [Google Scholar] [CrossRef]
- Tremblay, B.L.; Guénard, F.; Lamarche, B.; Pérusse, L.; Vohl, M.C. Network Analysis of the Potential Role of DNA Methylation in the Relationship between Plasma Carotenoids and Lipid Profile. Nutrients 2019, 11, 1265. [Google Scholar] [CrossRef]
- Speckmann, B.; Schulz, S.; Hiller, F.; Hesse, D.; Schumacher, F.; Kleuser, B.; Geisel, J.; Obeid, R.; Grune, T.; Kipp, A.P. Selenium increases hepatic DNA methylation and modulates one-carbon metabolism in the liver of mice. J. Nutr. Biochem. 2017, 48, 112–119. [Google Scholar] [CrossRef]
- Perng, W.; Rozek, L.S.; Mora-Plazas, M.; Duchin, O.; Marin, C.; Forero, Y.; Baylin, A.; Villamor, E. Micronutrient status and global DNA methylation in school-age children. Epigenetics 2012, 7, 1133–1141. [Google Scholar] [CrossRef]
- Zappe, K.; Pointner, A.; Switzeny, O.J.; Magnet, U.; Tomeva, E.; Heller, J.; Mare, G.; Wagner, K.H.; Knasmueller, S.; Haslberger, A.G. Counteraction of Oxidative Stress by Vitamin E Affects Epigenetic Regulation by Increasing Global Methylation and Gene Expression of MLH1 and DNMT1 Dose Dependently in Caco-2 Cells. Oxidative Med. Cell. Longev. 2018, 2018, 3734250. [Google Scholar] [CrossRef]
- Niedzwiecki, M.M.; Hall, M.N.; Liu, X.; Oka, J.; Harper, K.N.; Slavkovich, V.; Ilievski, V.; Levy, D.; van Geen, A.; Mey, J.L.; et al. Blood glutathione redox status and global methylation of peripheral blood mononuclear cell DNA in Bangladeshi adults. Epigenetics 2013, 8, 730–738. [Google Scholar] [CrossRef]
- Bürkle, A.; Moreno-Villanueva, M.; Bernhard, J.; Blasco, M.; Zondag, G.; Hoeijmakers, J.H.J.; Toussaint, O.; Grubeck-Loebenstein, B.; Mocchegiani, E.; Collino, S.; et al. MARK-AGE biomarkers of ageing. Mech. Ageing Dev. 2015, 151, 2–12. [Google Scholar] [CrossRef]
- Moreno-Villanueva, M.; Kötter, T.; Sindlinger, T.; Baur, J.; Oehlke, S.; Bürkle, A.; Berthold, M.R. The MARK-AGE phenotypic database: Structure and strategy. Mech. Ageing Dev. 2015, 151, 26–30. [Google Scholar] [CrossRef]
- Moreno-Villanueva, M.; Capri, M.; Breusing, N.; Siepelmeyer, A.; Sevini, F.; Ghezzo, A.; de Craen, A.J.M.; Hervonen, A.; Hurme, M.; Schön, C.; et al. MARK-AGE standard operating procedures (SOPs): A successful effort. Mech. Ageing Dev. 2015, 151, 18–25. [Google Scholar] [CrossRef]
- Heijmans, B.T.; Beekman, M.; Houwing-Duistermaat, J.J.; Cobain, M.R.; Powell, J.; Blauw, G.J.; van der Ouderaa, F.; Westendorp, R.G.J.; Slagboom, P.E. Lipoprotein particle profiles mark familial and sporadic human longevity. PLoS Med. 2006, 3, e495. [Google Scholar] [CrossRef] [PubMed]
- Giacconi, R.; Muti, E.; Malavolta, M.; Cipriano, C.; Costarelli, L.; Bernardini, G.; Gasparini, N.; Mariani, E.; Saba, V.; Boccoli, G.; et al. The +838 C/G MT2A polymorphism, metals, and the inflammatory/immune response in carotid artery stenosis in elderly people. Mol. Med. 2007, 13, 388–395. [Google Scholar] [CrossRef] [PubMed]
- Malavolta, M.; Giacconi, R.; Piacenza, F.; Santarelli, L.; Cipriano, C.; Costarelli, L.; Tesei, S.; Pierpaoli, S.; Basso, A.; Galeazzi, R.; et al. Plasma copper/zinc ratio: An inflammatory/nutritional biomarker as predictor of all-cause mortality in elderly population. Biogerontology 2010, 11, 309–319. [Google Scholar] [CrossRef]
- Jansen, E.; Beekhof, P.; Cremers, J.; Weinberger, B.; Fiegl, S.; Toussaint, O.; Bernhard, J.; Gonos, E.; Capri, M.; Franceschi, C.; et al. Quality control data of physiological and immunological biomarkers measured in serum and plasma. Mech. Ageing Dev. 2015, 151, 54–59. [Google Scholar] [CrossRef] [PubMed]
- Weber, D.; Stuetz, W.; Toussaint, O.; Debacq-Chainiaux, F.; Dollé, M.E.T.; Jansen, E.; Gonos, E.S.; Franceschi, C.; Sikora, E.; Hervonen, A.; et al. Associations between Specific Redox Biomarkers and Age in a Large European Cohort: The MARK-AGE Project. Oxidative Med. Cell. Longev. 2017, 2017, 1401452. [Google Scholar] [CrossRef] [PubMed]
- Stuetz, W.; Weber, D.; Dollé, M.E.T.; Jansen, E.; Grubeck-Loebenstein, B.; Fiegl, S.; Toussaint, O.; Bernhardt, J.; Gonos, E.S.; Franceschi, C.; et al. Plasma Carotenoids, Tocopherols, and Retinol in the Age-Stratified (35–74 Years) General Population: A Cross-Sectional Study in Six European Countries. Nutrients 2016, 8, 614. [Google Scholar] [CrossRef]
- Bollati, V.; Baccarelli, A.; Hou, L.; Bonzini, M.; Fustinoni, S.; Cavallo, D.; Byun, H.M.; Jiang, J.; Marinelli, B.; Pesatori, A.C.; et al. Changes in DNA methylation patterns in subjects exposed to low-dose benzene. Cancer Res. 2007, 67, 876–880. [Google Scholar] [CrossRef]
- Johanning, K.; Stevenson, C.A.; Oyeniran, O.O.; Gozal, Y.M.; Roy-Engel, A.M.; Jurka, J.; Deininger, P.L. Potential for retroposition by old Alu subfamilies. J. Mol. Evol. 2003, 56, 658–664. [Google Scholar] [CrossRef]
- Yang, A.S.; Estécio, M.R.H.; Doshi, K.; Kondo, Y.; Tajara, E.H.; Issa, J.P.J. A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res. 2004, 32, e38. [Google Scholar] [CrossRef] [PubMed]
- Erichsen, L.; Beermann, A.; Arauzo-Bravo, M.J.; Hassan, M.; Dkhil, M.A.; Al-Quraishy, S.; Hafiz, T.A.; Fischer, J.C.; Santourlidis, S. Genome-wide hypomethylation of LINE-1 and Alu retroelements in cell-free DNA of blood is an epigenetic biomarker of human aging. Saudi J. Biol. Sci. 2018, 25, 1220–1226. [Google Scholar] [CrossRef] [PubMed]
- Pellicano, M.; Buffa, S.; Goldeck, D.; Bulati, M.; Martorana, A.; Caruso, C.; Colonna-Romano, G.; Pawelec, G. Evidence for Less Marked Potential Signs of T-Cell Immunosenescence in Centenarian Offspring Than in the General Age-Matched Population. J. Gerontol. Ser. A Biol. Sci. Med. Sci. 2014, 69, 495–504. [Google Scholar] [CrossRef] [PubMed]
- Van Leeuwen, N.; Beekman, M.; Deelen, J.; van den Akker, E.B.; de Craen, A.J.M.; Slagboom, P.E.; ’t Hart, L.M. Low mitochondrial DNA content associates with familial longevity: The Leiden Longevity Study. Age 2014, 36, 9629. [Google Scholar] [CrossRef]
- Cui, F.; Sirotin, M.V.; Zhurkin, V.B. Impact of Alu repeats on the evolution of human p53 binding sites. Biol. Direct 2011, 6, 2. [Google Scholar] [CrossRef]
- Petrovich, M.; Veprintsev, D.B. Effects of CpG methylation on recognition of DNA by the tumour suppressor p53. J. Mol. Biol. 2009, 386, 72–80. [Google Scholar] [CrossRef]
- Deininger, P. Alu elements: Know the SINEs. Genome Biol. 2011, 12, 236. [Google Scholar] [CrossRef]
- Ciccarone, F.; Malavolta, M.; Calabrese, R.; Guastafierro, T.; Bacalini, M.G.; Reale, A.; Franceschi, C.; Capri, M.; Hervonen, A.; Hurme, M.; et al. Age-dependent expression of DNMT1 and DNMT3B in PBMCs from a large European population enrolled in the MARK-AGE study. Aging Cell 2016, 15, 755–765. [Google Scholar] [CrossRef]
- Valentini, E.; Zampieri, M.; Malavolta, M.; Bacalini, M.G.; Calabrese, R.; Guastafierro, T.; Reale, A.; Franceschi, C.; Hervonen, A.; Koller, B.; et al. Analysis of the machinery and intermediates of the 5hmC-mediated DNA demethylation pathway in aging on samples from the MARK-AGE Study. Aging 2016, 8, 1896–1922. [Google Scholar] [CrossRef]
- Tiffon, C. The Impact of Nutrition and Environmental Epigenetics on Human Health and Disease. Int. J. Mol. Sci. 2018, 19, 3425. [Google Scholar] [CrossRef]
- Gadecka, A.; Bielak-Zmijewska, A. Slowing Down Ageing: The Role of Nutrients and Microbiota in Modulation of the Epigenome. Nutrients 2019, 11, 1251. [Google Scholar] [CrossRef] [PubMed]
- Xavier, A.A.O.; Pérez-Gálvez, A. Carotenoids as a Source of Antioxidants in the Diet. Carotenoids Nat. 2016, 79, 359–375. [Google Scholar]
- Agodi, A.; Barchitta, M.; Quattrocchi, A.; Maugeri, A.; Canto, C.; Marchese, A.E.; Vinciguerra, M. Low fruit consumption and folate deficiency are associated with LINE-1 hypomethylation in women of a cancer-free population. Genes Nutr. 2015, 10, 30. [Google Scholar] [CrossRef] [PubMed]
- Hou, L.; Wang, H.; Sartori, S.; Gawron, A.; Lissowska, J.; Bollati, V.; Tarantini, L.; Zhang, F.F.; Zatonski, W.; Chow, W.H.; et al. Blood leukocyte DNA hypomethylation and gastric cancer risk in a high-risk Polish population. Int. J. Cancer 2010, 127, 1866–1874. [Google Scholar] [CrossRef]
- Chong, Y.S.; Mai, C.W.; Leong, C.O.; Wong, L.C. Lutein improves cell viability and reduces Alu RNA accumulation in hydrogen peroxide challenged retinal pigment epithelial cells. Cutan. Ocul. Toxicol. 2018, 37, 52–60. [Google Scholar] [CrossRef]
- Bakken, T.; Braaten, T.; Olsen, A.; Kyrø, C.; Lund, E.; Skeie, G. Consumption of Whole-Grain Bread and Risk of Colorectal Cancer among Norwegian Women (the NOWAC Study). Nutrients 2016, 8, 40. [Google Scholar] [CrossRef]
- Zhu, Y.; Wang, P.; Sha, W.; Sang, S. Urinary Biomarkers of Whole Grain Wheat Intake Identified by Non-targeted and Targeted Metabolomics Approaches. Sci. Rep. 2016, 6, 36278. [Google Scholar] [CrossRef]
- Rangel-Salazar, R.; Wickström-Lindholm, M.; Aguilar-Salinas, C.A.; Alvarado-Caudillo, Y.; Døssing, K.B.V.; Esteller, M.; Labourier, E.; Lund, G.; Nielsen, F.C.; Rodríguez-Ríos, D.; et al. Human native lipoprotein-induced de novo DNA methylation is associated with repression of inflammatory genes in THP-1 macrophages. BMC Genom. 2011, 12, 582. [Google Scholar] [CrossRef]
- Zhang, P.; Chu, T.; Dedousis, N.; Mantell, B.S.; Sipula, I.; Li, L.; Bunce, K.D.; Shaw, P.A.; Katz, L.S.; Zhu, J.; et al. DNA methylation alters transcriptional rates of differentially expressed genes and contributes to pathophysiology in mice fed a high fat diet. Mol. Metab. 2017, 6, 327–339. [Google Scholar] [CrossRef]
- Antonini, J.M.; Kodali, V.; Meighan, T.G.; Roach, K.A.; Roberts, J.R.; Salmen, R.; Boyce, G.R.; Zeidler-Erdely, P.C.; Kashon, M.; Erdely, A.; et al. Effect of Age, High-Fat Diet, and Rat Strain on Serum Biomarkers and Telomere Length and Global DNA Methylation in Peripheral Blood Mononuclear Cells. Sci. Rep. 2019, 9, 1996. [Google Scholar] [CrossRef]
- Ciccarone, F.; Castelli, S.; Ioannilli, L.; Ciriolo, M.R. High Dietary Fat Intake Affects DNA Methylation/Hydroxymethylation in Mouse Heart: Epigenetic Hints for Obesity-Related Cardiac Dysfunction. Mol. Nutr. Food Res. 2019, 63, e1800970. [Google Scholar] [CrossRef] [PubMed]
- Ford, E.S. Serum Copper Concentration and Coronary Heart Disease among US Adults. Am. J. Epidemiol. 2000, 151, 1182–1188. [Google Scholar] [CrossRef] [PubMed]
- Zowczak, M.; Iskra, M.; Torliński, L.; Cofta, S. Analysis of serum copper and zinc concentrations in cancer patients. Biol. Trace Elem. Res. 2001, 82, 1–8. [Google Scholar] [CrossRef]
- Malavolta, M.; Piacenza, F.; Basso, A.; Giacconi, R.; Costarelli, L.; Mocchegiani, E. Serum copper to zinc ratio: Relationship with aging and health status. Mech. Ageing Dev. 2015, 151, 93–100. [Google Scholar] [CrossRef]
- Quinn, J.F.; Harris, C.; Kaye, J.A.; Lind, B.; Carter, R.; Anekonda, T.; Ralle, M. Gender effects on plasma and brain copper. Int. J. Alzheimers Dis. 2011, 2011, 150916. [Google Scholar] [CrossRef]
- Battaglia Richi, E.; Baumer, B.; Conrad, B.; Darioli, R.; Schmid, A.; Keller, U. Health Risks Associated with Meat Consumption: A Review of Epidemiological Studies. Int. J. Vitam. Nutr. Res. 2015, 85, 70–78. [Google Scholar] [CrossRef]
- Wang, W.; Wang, W.H.; Azadzoi, K.M.; Dai, P.; Wang, Q.; Sun, J.B.; Zhang, W.T.; Shu, Y.; Yang, J.H.; Yan, Z. Alu RNA accumulation in hyperglycemia augments oxidative stress and impairs eNOS and SOD2 expression in endothelial cells. Mol. Cell. Endocrinol. 2016, 426, 91–100. [Google Scholar] [CrossRef]

| RASIG (n = 60) | GO (n = 32) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Age Classes (years) | 35–44 (n = 12) | 45–54 (n = 14) | 55–64 (n = 11) | 65–75 (n = 23) | p Value a | 55–64 (n = 17) | p Value b | 65–75 (n = 15) | p Value b | p Value c |
| Females (%) | 4 (33.3%) | 9 (64.3%) | 6 (54.5%) | 11 (47.8%) | NS | 6 (35.3%) | NS | 6 (40.0%) | NS | |
| RBC (×106/μL) | 5.01 ± 0.12 | 4.87 ± 0.08 | 4.77 ± 0.11 | 4.90 ± 0.10 | NS | 4.96 ± 0.10 | NS | 4.77 ± 0.11 | NS | NS |
| Hemoglobine (g/dl) | 14.30 ± 0.38 | 13.64 ± 0.41 | 14.51 ± 0.28 | 14.49 ± 0.30 | NS | 14.74 ± 0.34 | NS | 14.12 ± 0.30 | NS | NS |
| WBC (×103/μL) | 6.16 ± 0.38 | 6.40 ± 0.48 | 6.08 ± 0.33 | 5.83 ± 0.27 | NS | 6.28 ± 0.44 | NS | 5.78 ± 0.25 | NS | NS |
| Neutrophils (×103/μL) | 3.51 ± 0.26 | 3.61 ± 0.38 | 3.45 ± 0.29 | 3.46 ± 0.25 | NS | 3.79 ± 0.31 | NS | 3.28 ± 0.18 | NS | NS |
| Lymphocytes (×103/μL) | 1929 ± 148 | 2008 ± 118 | 1892 ± 186 | 1764 ± 96 | NS | 1790 ± 151 | NS | 1776 ± 90 | NS | NS |
| Monocytes (×103/μL) | 364 ± 38 | 401 ± 30 | 371 ± 30 | 366 ± 19 | NS | 357 ± 31 | NS | 334 ± 24 | NS | NS |
| Platelets (×103/μL) | 241 ± 50 | 289 ± 67 | 278 ± 74 | 226 ± 70 | 0.031 | 238 ± 44 | NS | 248 ± 52 | NS | NS |
| CRP (μg/L) | 1.23 ± 0.39 | 1.13 ± 0.32 | 1.25 ± 0.40 | 1.87 ± 0.35 | NS | 1.65 ± 0.24 | NS | 3.18 ± 0.97 | NS | NS |
| TC (mmol/L) | 5.02 ± 0.23 | 5.22 ± 0.23 | 6.28 ± 0.31 | 5.75 ± 0.30 | 0.045 | 5.54 ± 0.18 | 0.041 | 5.71 ± 0.19 | NS | NS |
| HDL (mmol/L) | 1.31 ± 0.10 | 1.38 ± 0.12 | 1.51 ± 0.08 | 1.49 ± 0.12 | NS | 1.37 ± 0.10 | NS | 1.47 ± 0.11 | NS | NS |
| LDL (mmol/L) | 2.92 ± 0.16 | 3.18 ± 0.23 | 4.07 ± 0.22 | 3.47 ± 0.24 | 0.035 | 3.44 ± 0.15 | 0.030 | 3.52 ± 0.18 | NS | NS |
| TG (mmol/L) | 2.20 ± 0.90 | 1.47 ± 0.40 | 1.30 ± 0.13 | 1.49 ± 0.20 | NS | 1.14 ± 0.53 | NS | 1.15 ± 0.46 | NS | NS |
| FG (mmol/L) | 5.15 ± 0.29 | 5.30 ± 0.16 | 5.43 ± 0.11 | 5.82 ± 0.16 | NS | 5.82 ± 0.34 | NS | 5.88 ± 0.31 | NS | NS |
| Creatinine (μmol/L) | 73.2 ± 5.4 | 65.7 ± 4.1 | 72.3 ± 5.0 | 75.4 ± 3.1 | NS | 73.3 ± 3.8 | NS | 69.9 ± 3.0 | NS | NS |
| BMI | 25.7 ± 1.3 | 24.8 ± 1.4 | 26.3 ± 1.1 | 28.9 ± 0.9 | NS | 25.7 ± 1.6 | NS | 27.1 ± 0.9 | NS | NS |
| Smoking Never | 7 (58.3%) | 7 (50.0%) | 4 (36.4%) | 14 (60.9%) | NS | 9 (52.9%) | NS | 9 (60.0%) | NS | NS |
| Former | 2 (16.7%) | 5 (35.7%) | 5 (45.5%) | 8 (34.8%) | 6 (35.3%) | 3 (20.0%) | ||||
| current | 3 (25.0%) | 2 (14.3%) | 2 (18.2%) | 1 (4.3%) | 2 (11.8%) | 3 (20.0%) | ||||
| Alchol consumption < 1 serv./day | 10 (83.3%) | 10 (71.4%) | 5 (45.5%) | 17 (73.9%) | NS | 10 (58.8%) | NS | 7(46.7%) | NS | NS |
| =1 serv./day | 1 (8.3%) | 2 (14.3%) | 2 (18.2%) | 1 (4.3%) | 1 (5.9%) | 4 (26.7%) | ||||
| >1 serv./day | 1 (8.3%) | 2 (14.3%) | 4 (36.4%) | 5 (21.7%) | 6 (35.3%) | 4 (26.7%) | ||||
| Predictors | Coefficient | Std. Error | Importance | Sig |
|---|---|---|---|---|
| Subject group a | 2.219 | 0.542 | 0.200 | 0.0001 |
| Plasma Cu | 0.0005 | 0.001 | 0.156 | 0.001 |
| LDL2-C b | 0.043 | 0.015 | 0.104 | 0.004 |
| Ascorbic acid | −0.218 | 0.080 | 0.088 | 0.008 |
| Total Glutathione | −0.002 | 0.001 | 0.073 | 0.015 |
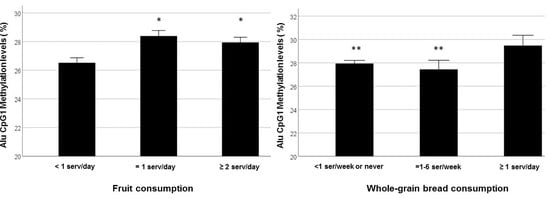
| Whole-grain bread consumption c | −1.906 | 0.804 | 0.067 | 0.020 |
| Age | −0.054 | 0.024 | 0.063 | 0.024 |
| HDL2-C | 0.081 | 0.037 | 0.057 | 0.032 |
| Plasma Zn | −0.005 | 0.002 | 0.054 | 0.036 |
| Fruit consumption c | −1.687 | 0.806 | 0.052 | 0.040 |
| Predictors | Coefficient | Std. Error | Sig |
|---|---|---|---|
| Subject group a | 0.070 | 0.022 | 0.002 |
| Plasma Cu | 0.153 | 0.052 | 0.005 |
| LDL2-C b | 0.078 | 0.039 | 0.048 |
| Whole-grain bread consumption (1-6 serv/week) c | −0.086 | 0.037 | 0.024 |
| Whole-grain bread consumption (≥ 1 serv/day) c | −0.118 | 0.042 | 0.007 |
| Fruit consumption (≥ 1 serv/day) d | −0.070 | 0.035 | 0.046 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Giacconi, R.; Malavolta, M.; Bürkle, A.; Moreno-Villanueva, M.; Franceschi, C.; Capri, M.; Slagboom, P.E.; Jansen, E.H.J.M.; Dollé, M.E.T.; Grune, T.; et al. Nutritional Factors Modulating Alu Methylation in an Italian Sample from The Mark-Age Study Including Offspring of Healthy Nonagenarians. Nutrients 2019, 11, 2986. https://doi.org/10.3390/nu11122986
Giacconi R, Malavolta M, Bürkle A, Moreno-Villanueva M, Franceschi C, Capri M, Slagboom PE, Jansen EHJM, Dollé MET, Grune T, et al. Nutritional Factors Modulating Alu Methylation in an Italian Sample from The Mark-Age Study Including Offspring of Healthy Nonagenarians. Nutrients. 2019; 11(12):2986. https://doi.org/10.3390/nu11122986
Chicago/Turabian StyleGiacconi, Robertina, Marco Malavolta, Alexander Bürkle, María Moreno-Villanueva, Claudio Franceschi, Miriam Capri, P. Eline Slagboom, Eugène H. J. M. Jansen, Martijn E. T. Dollé, Tilman Grune, and et al. 2019. "Nutritional Factors Modulating Alu Methylation in an Italian Sample from The Mark-Age Study Including Offspring of Healthy Nonagenarians" Nutrients 11, no. 12: 2986. https://doi.org/10.3390/nu11122986
APA StyleGiacconi, R., Malavolta, M., Bürkle, A., Moreno-Villanueva, M., Franceschi, C., Capri, M., Slagboom, P. E., Jansen, E. H. J. M., Dollé, M. E. T., Grune, T., Weber, D., Hervonen, A., Stuetz, W., Breusing, N., Ciccarone, F., Zampieri, M., Aversano, V., Caiafa, P., Formentini, L., ... Cardelli, M. (2019). Nutritional Factors Modulating Alu Methylation in an Italian Sample from The Mark-Age Study Including Offspring of Healthy Nonagenarians. Nutrients, 11(12), 2986. https://doi.org/10.3390/nu11122986