Abstract
In the United States, several land use and land cover (LULC) data sets are available based on satellite data, but these data sets often fail to accurately represent features on the ground. Alternatively, detailed mapping of heterogeneous landscapes for informed decision-making is possible using high spatial resolution orthoimagery from the National Agricultural Imagery Program (NAIP). However, large-area mapping at this resolution remains challenging due to radiometric differences among scenes, landscape heterogeneity, and computational limitations. Various machine learning (ML) techniques have shown promise in improving LULC maps. The primary purposes of this study were to evaluate bagging (Random Forest, RF), boosting (Gradient Boosting Machines [GBM] and extreme gradient boosting [XGB]), and stacking ensemble ML models. We used these techniques on a time series of Sentinel 2A data and NAIP orthoimagery to create a LULC map of a portion of Irion and Tom Green counties in Texas (USA). We created several spectral indices, structural variables, and geometry-based variables, reducing the dimensionality of features generated on Sentinel and NAIP data. We then compared accuracy based on random cross-validation without accounting for spatial autocorrelation and target-oriented cross-validation accounting for spatial structures of the training data set. Comparison of random and target-oriented cross-validation results showed that autocorrelation in the training data offered overestimation ranging from 2% to 3.5%. The XGB-boosted stacking ensemble on-base learners (RF, XGB, and GBM) improved model performance over individual base learners. We show that meta-learners are just as sensitive to overfitting as base models, as these algorithms are not designed to account for spatial information. Finally, we show that the fusion of Sentinel 2A data with NAIP data improves land use/land cover classification using geographic object-based image analysis.
Keywords:
bagging; boosting; stacking; GEOBIA; autocorrelation; target-oriented cross-validation; data fusion 1. Introduction
Accurate extraction of information from remote sensing data is of utmost importance for environmental monitoring, land-use planning, and the management of natural resources. Unlike field-based data, remote sensing offers cost-effective, time-efficient, and frequent data delivery over a broad geographical extent. In the recent past, high-resolution land use/land cover (LULC) map production was hindered by the geographical coverage of an area by satellite sensors and the cost of commercial data [1,2]. In the last three decades, however, these limitations have largely been resolved, with several mapping efforts accomplished in the United States using Landsat data and, more recently, Sentinel [3,4]. Satellite images are still the dominant data used in LULC mapping; however, these maps are regional with a relatively moderate spatial resolution (10–60 m) that often do not capture small-scale resources present on the ground. In contrast to the relatively coarse scale of such data, the National Agricultural Imagery Program (NAIP) in the conterminous United States captures high spatial resolution (0.6 m) data every 2–3 years. Although NAIP data are acquired chiefly for agricultural purposes, they have been rarely used for general mapping of land cover in relatively large (county to state)-scale LULC classification efforts. Such large-area mapping at a fine resolution remains challenging due to radiometric differences among scenes, landscape heterogeneity, and computational limitations. Various classification approaches have been developed to address these challenges.
With the launch of Landsat in 1972, pixel-based classification soon became the dominant paradigm [5]. However, with the advent of high-resolution satellite sensing, another paradigm challenged the pixel-based approach, focusing on grouping high-resolution pixels into one based on several criteria, a method called geographic object-based image analysis (GEOBIA). Several studies have employed pixel-based [6,7,8] or object-based [9] approaches to NAIP data. Comparative studies have shown that GEOBIA yields higher accuracy than traditional pixel-based classification [10]. Additionally, in recent decades, remote sensing data analysis has benefited from exploiting the power of machine learning (ML) algorithms in LULC classification, such as support vector machines (SVM) [11,12], Decision Trees (DT) [13], Random Forest (RF) [9,14,15], Nearest Neighbor [16,17], and Deep Learning (DL) [18].
Because each approach has its own biases and advantages, typically, a variety of ensemble models are subjectively selected to decrease variance (bagging), reduce bias (boosting), and improve the predictive power of the classifier (stacking) [19,20]. Specifically, ensemble ML methods have gained much recent attention in various fields, especially in applied remote sensing [20,21,22,23,24], because they can handle high-resolution images to capture fine structural information. After building such an ensemble model, current state-of-the-art practices involve model evaluation using held-out data (a random subset of training data, i.e., random cross-validation). The data collection process is not usually random; therefore, a random selection of observations to create the testing data set does not guarantee independence from training observations when a spatial dependency is inherent in the data. Spatial dependency in the sampling data arises from spatial sampling biases, as observers tend to collect data from a clustered hotspot of their activity. This phenomenon of spatial dependency [25] is called spatial autocorrelation (SAC) [26]. When spatial data are autocorrelated, the considered model fails to account for a critical determinant that is spatially structured and thus causes spatial structure in the response. Another SAC issue in the input data set can result in unreliable results through validation approaches because an observation point cannot act as spatially independent validation of proximity data points [27,28]. Using such cross-validation approaches without accounting for spatial structures of input data provides overoptimistic results, making accuracy assessment unreliable [9]. We compared accuracy based on random cross-validation without accounting for spatial autocorrelation and on target-oriented cross-validation accounting for spatial structures of the training data set.
This study took a GEOBIA approach to Sentinel 2A and NAIP data to evaluate the effect of the stacking ensemble on classification accuracy. We compared random cross-validation to target-oriented cross-validation strategies (i.e., leave-location-out cross-validation) in reducing the effects of spatial bias in training data in LULC classification. We demonstrated a reduction in overinflation of overall accuracy and documented the effect of accounting for spatial autocorrelation.
2. Materials and Methods
2.1. Study Area
The 5381.73 km2 study area was a heterogeneous landscape in central Texas in Irion and Tom Green counties (Figure 1). The mean annual precipitation is 508 mm, and the minimum and maximum temperatures range from 0 °C to 35 °C. Most of the study area is situated on the Semiarid Edwards Plateau with scattered woody vegetation [29]. The study area contains cropland [29] as well as forests of live oak (Quercus fusiformis), Texas oak (Q. buckleyi), Ashe juniper (Juniperus ashei), and mixed grass prairie with scatted honey mesquite (Prosopis glandulosa) [29].
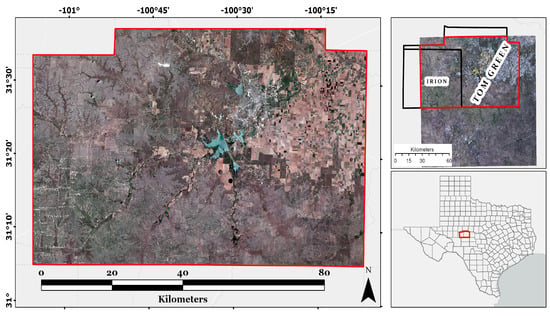
Figure 1.
Study area in Irion and Tom Green counties in Texas, with a red, green, and blue composite of a Sentinel 2A image from June 2018.
2.2. Imagery Data Set
2.2.1. NAIP Orthoimagery
The National Agriculture Imagery Program acquires digital orthophoto quarter quads (DOQQ) every 2–3 years. The NAIP DOQQs used in this study were acquired in 2020 between August 23 and October 18 through ten flight missions. Each DOQQ contains four working bands, including red (465–676 nm), green (604–664 nm), blue (533–587 nm), and near-infrared (NIR) (420–492 nm). The spatial resolution of the entire data set is 0.6 m, and the radiometric resolution is 8-bit (digital number values range from 0 to 255).
2.2.2. Sentinel Multispectral Imagery
The Sentinel-2 multispectral instrument is managed by the European Space Agency, which consists of a constellation of two polar-orbiting satellites in the same Sun-synchronous orbit covering areas from 56° to 84° latitude [30,31]. Sentinel-2 has 13 spectral bands between visible and short-wave infrared, with three bands at 60 m, six at 20 m, and four at 10 m spatial resolution. The radiometric resolution of 13 bands is 12-bit (digital number values range from 0 to 4095). The temporal resolution of Sentinel 2B is five days, whereas Sentinel 2A has a slightly coarser temporal resolution of ten days.
This study used 11 Sentinel 2A images for the year 2020, with one image per month except for February. The criterion for image selection was that the images must have little or no contamination from clouds or haze. All Sentinel 2A scenes were downloaded in level-1C processing format from the Copernicus Open Access Hub (https://scihub.copernicus.eu/) accessed on 7 June 2021. This study only used four bands (red, green, blue, and near-infrared) available at 10 m spatial resolution.
2.3. Layer Generation and Dicing
In addition to original visible and NIR bands from NAIP and Sentinel, we generated byproducts and indices following methods by Subedi et al. [32,33]. We first calculated the soil-adjusted vegetation index (SAVI) on each NAIP and Sentinel image. Then, for Sentinel imagery, we extracted all near-infrared bands and produced principal component axes (PCA) [9,34,35]. In addition, we also ran PCA on the SAVI layers. Finally, we calculated textures on the first PCA axis on SAVI layers [32,33].
To harness the high spatial resolution of NAIP imagery and the radiometric and temporal resolution of Sentinel imagery, we applied multi-resolution image segmentation on NAIP imagery using eCognition software (Version 10.5) to derive quality objects [36]. Due to the large extent of the study area, all necessary images were first generated and then diced (clipped) into manageable sizes. Next, we generated several features based on NAIP and Sentinel images on each object/polygon in eCognition. We used multi-resolution segmentation in NAIP, NIR and PCA axis (bands) at a scale value of 50 pixels with shape and compactness weights of 0.04 and 0.98, respectively. Multi-resolution segmentation was followed by spectral difference segmentation and region growth at a spatial scale of 75 pixels. The detailed segmentation workflow for each diced tile is described in our previous work on land use/land cover mapping using the GEOBIA approach [9]. Finally, all objects with generated features were exported to merge into the final data set.
2.4. Selection of Features
Combining geometry, spectral bands, spectral indices, principal component axes on NAIP data, spectral indices, and PCAs on near-infrared and spectral indices resulted in several feature variables. However, since these features were produced on the original NAIP and Sentinel bands, the information held by these features became redundant. Therefore, we performed recursive feature elimination (RFE) in the scikit-learn package in Python [37] to retain the most critical features. A RFE relies on variable importance scores calculated using training data [38].
2.5. Machine Learning Algorithms
2.5.1. Random Forest
RF is an ensemble learning technique proposed by Breiman [39]. A RF uses several classification and regression trees to make a prediction. Initial data are split into in-bag (about 2/3rds of the data) and out-of-bag (OOB) (the remaining 3rd) samples. Trees are created using in-bag samples, whereas OOB samples are used to estimate the OOB error [39]. The central idea of RF is that several bootstrap aggregated classifiers perform better than a single classifier [39]. Each tree is created by bootstrapping in-bag samples. The resulting classification is based on the majority votes of all trees for the most frequent input feature , where is the class predictin of the bth Random Forest tree. Each decision tree is independently produced, and each node is split using a randomly selected defined number of features (mtry). RF has been widely applied in LULC classification [9,40,41].
2.5.2. Gradient Boosting
Gradient Boosting Machines (GBM) [42] build the boosting procedure as an optimization task over a differential loss function in an iterative fashion. A classical GBM trains a series of base learners that minimizes some loss function iteratively in a sequence [43]. In classification, GBM uses multi-class logistic likelihood algorithms for fitting criteria. Extreme gradient boosting (XGB) implements a traditional GBM that builds additive models while optimizing the loss function [44].
2.6. Stacking Ensemble Machine Learning
Before stacking models, all base learning algorithms (level 0) were first developed. Stacking, also called stack generalization (level 1), uses meta-learning to integrate different algorithms. Stacking ensemble machine learning algorithms (in this case, classification trees and boosted trees) estimate base learner generalization biases and minimize these biases while stacking [45]. The essence of stacking is to use the level 1 model to learn from the predictions of the base models (level 0). Generally, a stacked generalization framework improves prediction performance compared to the best level 0 model. We tested RF, GBM, XGB, and logistic regression and selected the model that produced better results than the base learner. Figure 2 shows the basic workflow of the stack generalization used in this study.
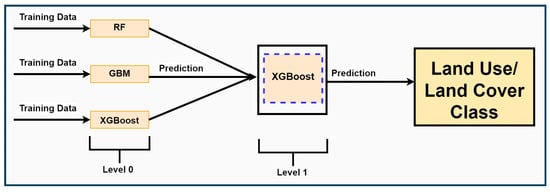
Figure 2.
A schematic overview of stacking ensemble machine learning using bagging and boosting algorithms.
2.7. Accuracy Assessment
The performances of the base and stack models were measured using a 20% holdout of the total data in random cross-validation. In the target-oriented cross-validation (leave-location-out (LLO-CV)), the holdout data varied at each iteration (Appendix A). Five performance criteria derived from the confusion matrix were evaluated to evaluate the model: (a) overall accuracy (OA), (b) Precision (User’s Accuracy), (c) Recall (Producer’s Accuracy), (d) Kappa [46], and (e) Mathews correlation coefficient (MCC) [47]. In general, the higher the values of these performance metrics, the better the performance of the classifiers in discriminating land use/land cover classes.
3. Results
3.1. Level 0 and Level 1 Classification Comparison
OA, Kappa, and MCC of the three base learners were relatively similar (Table 1) when random cross-validation was used on holdout data. All metrics were derived based on the confusion matrix (Appendix B). In classification, XGB produced the highest OA (0.942), followed by GBM and Random Forest. Kappa and MCC followed the same pattern: XGB produced the highest Kappa and MCC statistics. GBM, followed by XGB, yielded the highest producer’s accuracy on cropland.

Table 1.
Performance metrics produced by base learners and meta-learners using random cross-validation. These statistics were calculated using a confusion matrix in the holdout data set (Figure 3).
The stacking model (level 1) produced the highest OA, Kappa, and MCC statistics. At the land use/land cover class level, however, the stacking model did not produce the best results, although it was consistent with other competitive models (Figure 3).
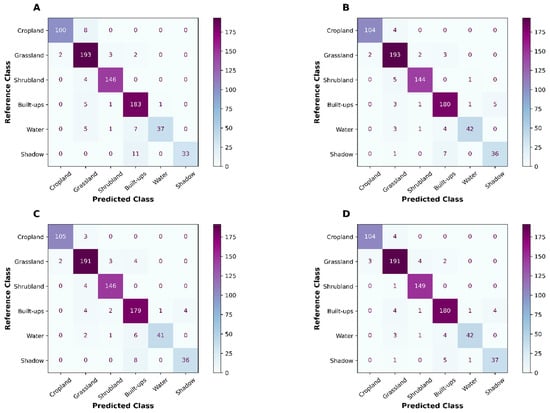
Figure 3.
Confusion matrices produced on holdout (20%) of the total training data using RF (A), GBM (B), XGB (C), and stacking (XGB) (D) classifiers.
3.2. Identifying and Reducing Overfitting
Both cross-validation approaches produced competitive overall accuracy statistics (Figure 4). Selected base learners and stacked models produced higher accuracy statistics when repeated with random cross-validation (10 folds, 5 repetitions). However, the overall accuracy values were reduced when predicted at unknown locations, hinting at spatial overfitting of the models.
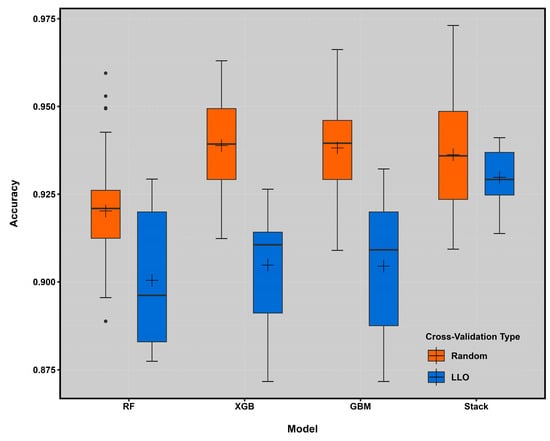
Figure 4.
Box plot of overall accuracy across folds in the base-learner model and meta-learner model using random cross-validation (random, in red) and target-oriented cross-validation (LLO, in blue). The horizontal black line in each box plot indicates the median and the crosshairs indicate the mean.
A random validation data set that was not part of the training or testing data set showed slightly different results about the performance metrics (Table 2). In general, leave-location-out cross-validation (LLO-CV) resulted in similar statistics and three accuracy metrics that were lower than those produced on holdout data. This indicates that random cross-validation could predict better when the testing data set was closer to the training data set. On the other hand, LLO-CV showed similar results to the training data set. The Mathews correlation coefficient suggested that all the base models and stack models were consistent and produced better spatial prediction abilities (random cross-validation < LLO-CV). The class-level prediction ability of these models varied significantly in both cross-validation strategies (Appendix B).

Table 2.
Accuracy metrics produced in the independent validation data set. These metrics were produced on the confusion matrix.
3.3. Contribution of Features
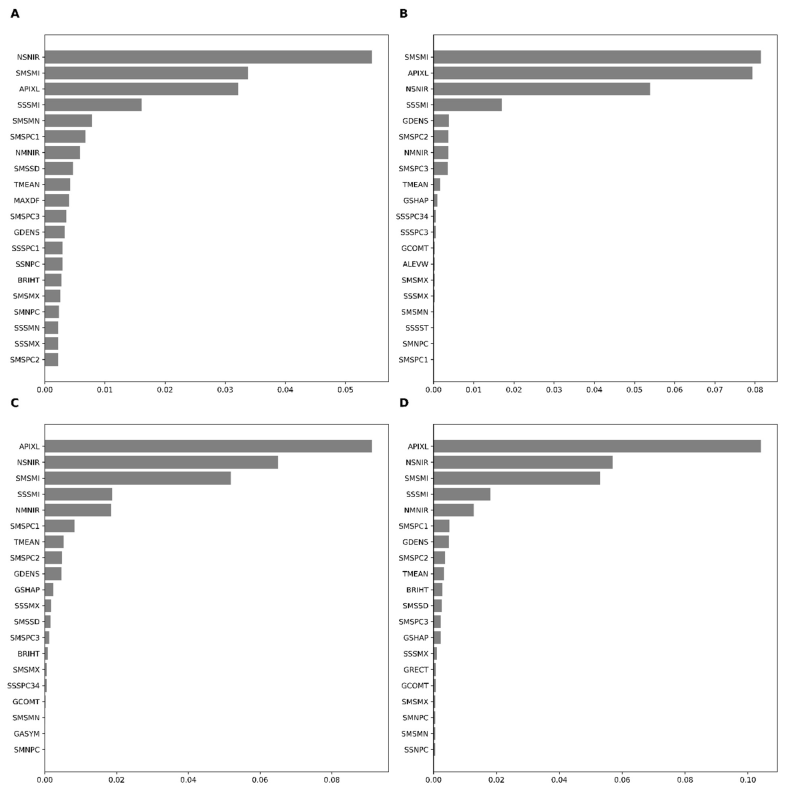
The 20 highest contributing features across the base learners (RF, XGB, and GBM) and the meta-learner indicated that SAVI minimum (SMSMI), SAVI standard deviation (SSSMI), the density of an object (GDENS), NAIP’s near-infrared band standard deviation (NSNIR), and area of the object (AIPXL) were among the essential variables in model performance (Figure 5). Permutation-based variable importance demonstrated similar features as important features of boosting (GBM and XGB) save for a few exceptions in the top ten features. For example, PCA1 on SAVI was indicated as one of the top ten important features in RF but not in XGB. The stacking algorithm (meta-learner) found the area of objects (AIPXL) to be the most important feature, followed by SMSMI, NSNIR, and SMSSD.
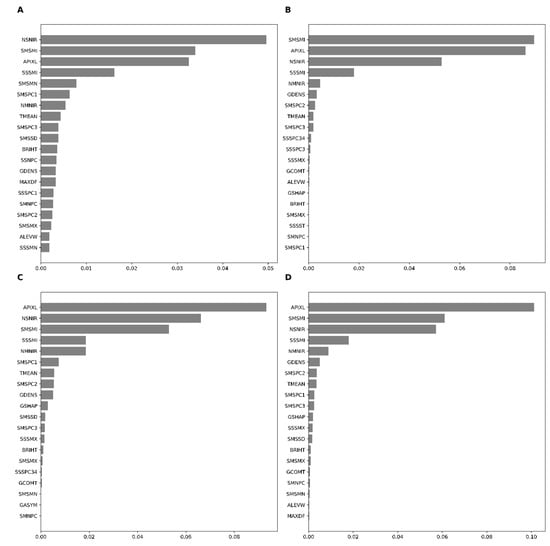
Figure 5.
Permutation-based feature importance for RF (A), GBM (B), XGB (C), and stacked (D) models in target-oriented cross-validation.
In general, the area of objects, density mean, and standard deviation of SAVI in the Sentinel data and the standard deviation on the near-infrared band of NAIP data were among the most significant. In contrast, the standard deviation from the PCA bands extracted in time-series SAVI layers of the Sentinel images was not so crucial in the LLO cross-validations (Figure 5). Random cross-validation demonstrated patterns somewhat similar to those of LLO-CV. The XGB and stacking algorithms precisely identified the same variables as the most critical features in random cross-validation (Appendix C). The final LULC map based on stacking and LLO-CV is presented in Figure 6. Several subsets in the study area using level 0 and level 1 are presented in Appendix D. Selected variables used in classification are defined in Appendix E.
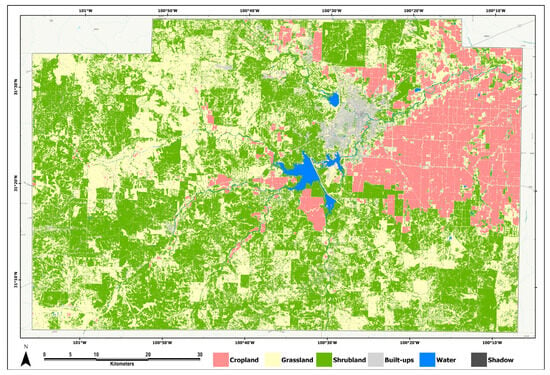
Figure 6.
Classified map of the study area based on the stacking model (meta-learner) using target-oriented cross-validation, and geographic object-based image analysis (GEOBIA) approach.
4. Discussion
Machine learning on remotely sensed data has become widespread and promises to improve LULC classification results over traditional supervised methods [48]. Our study evaluated geographic object-based implementation of bagging (Random Forest), boosting (GBM and XGB), and stacking ensemble machine learning in land use/land cover mapping and found that machine learning enhanced the discrimination ability of base learners in multi-class classification problems. The improvement was apparent in the target-oriented cross-validation approach with consistency and reduced overfitting problems, in contrast to base learners and meta-learners in random cross-validation. A stacking ensemble does not necessarily guarantee improvement over its base models. Results from random cross-validation often inflate accuracy estimates, which may not hold when validated on independent data sets or when there is a spatial structure in the data [9]. Here, we evaluated the fusion of Sentinel data with NAIP and compared image accuracy using random cross-validation and target-oriented validation. Although the accuracy on the holdout data set from random cross-validation appeared similar, the stacking ensemble performed best under the target-oriented LLO-CV (Figure 4). The independent data set suggested (Table 2) that the stacking ensemble is the best approach because the overall accuracy obtained from random cross-validation was overoptimistic. Our results suggest that meta-learners outperform base learners by training weak-performing learners into super-learners. Although the meta-learner showed improved accuracy over base learners, our validation results do not warrant indicating that meta-learner ML models consistently improve discrimination ability among classes. The LLO-CV demonstrated that the meta-learner was weak in discriminating among grassland, shrubland, and built-up LULC classes. In our case, we used two boosting models based on our initial classification results with different hyperparameters. We did not find significant performance enhancement in one of the widely used ensembles of machine learning models, such as SVM. In stacking ensemble models, each base learner should be carefully selected as a candidate base learner.
Our evaluation showed that the random cross-validation method introduces spatial overfitting when the spatial structure of the data is not considered. All models performed better, producing better evaluation metrics but resulting in smaller accuracy metrics when used with target-oriented cross-validation (LLO-CV). This highlights that the discrimination ability of the models increased when the training and testing samples were in spatial proximity. However, when tested with the independent validation data set, the accuracy metrics performed poorly compared to the usual training/testing data splitting approach. Unlike random cross-validation, the consistent performance of learners in target-oriented cross-validation showed that avoidance of spatial structure in the data could overcome spatial overfitting issues [28,49]. Despite advocation to account for spatial structure in remote sensing data [50] and the implications of failing to do so, the debate is ongoing on whether spatial validation approaches should be used to improve model performance [28,51,52].
Our findings indicate that spatial structures in the data can inflate overall model performances if the training data are spatially structured. Mannel et al. [53] reported that the random cross-validation of training data resulted in 5–8% inflation of overall accuracy when training data were autocorrelated. In our case, the overall accuracy overestimation based on random cross-validation ranged from 2% to 3.4%. Moreover, our approach showed that the spatial structure in the data still exists even after feature selection. A possible limitation of variable selection is based on the fact that different algorithms calculate the importance of each variable differently. For example, RFE depends on the importance of initial model training. RFE removes the least important features until the best subset is determined [38]; however, the algorithm ranks the features without considering their importance and prediction ability in spatial space [49].
Our assessment of random and target-oriented cross-validation in Sentinel 2A time series and NAIP data showed that spatial resolution of NAIP, variation in the near-infrared band of NAIP, variations of SAVI, and sizes of objects were among the most important factors for the assessment of land use/land cover in a heterogeneous landscape of Texas. Using time-series Sentinel data helped discriminate vegetation from other LULC classes. NAIP DOQQS were captured in autumn, which hindered the generation of meaningful comparable high-resolution information from NAIP data. Moreover, the noise in the data may have cumulative effects due to the use of multi-temporal Sentinel data by increasing dimensionality and redundancy. As described above, RFE failed to remove such variables effectively. Feature selection combined with a target-oriented cross-validation approach reduced the overestimation of accuracy.
5. Conclusions
High-resolution LULC maps are essential for land-use planning, yield estimation, examining the effects of habitat fragmentation, and many other applications. Therefore, it is axiomatic that more accurate and reliable high-resolution maps can provide a critical decision base for land managers, researchers, and wildlife biologists. However, accurate high-resolution LULC map generation is prohibited by the high cost associated with high-resolution remotely sensed data. The effective use of high-resolution NAIP data is marred by heterogeneity in sensors, multiple acquisitions, higher volume, and low radiometric resolution. Sentinel spectral bands are slightly coarser but have high temporal, spectral, and radiometric resolutions, so they can provide more temporal depth and the capability of capturing differences in moisture and vegetation phenology. The approach we used can be extended to other geographic locations; however, the specific band combinations and initial pre-processing steps may vary depending on the characteristics and objectives of the project.
This study demonstrated the efficacy of fusing Sentinel and NAIP data and the effect of a target-oriented cross-validation approach in reducing spatial overfitting. In multi-class LULC mapping, we demonstrated a 2–3.5% accuracy inflation using random cross-validation. Furthermore, independent validation data sets suggested that target-oriented cross-validation produced consistent accuracy metrics, avoiding the overinflation of accuracy.
This study applied widely used bagging algorithms (Random Forest) and boosting (GBM and XGB) in remote sensing. In addition, we stacked these models to improve the final classification. The main contribution of this research is that meta-learners are just as sensitive to overfitting as base models that are not designed to consider spatial autocorrelation. We found that classification performance could be improved by harvesting spatial resolution from NAIP and time-series information from Sentinel data. Furthermore, the boosting algorithms were more proficient in discriminating LULC classes compared to bagging. The LULC map generated from stacking diverse machine learning models coupled with feature selection and target-oriented cross-validation presents a practical method for improved land use management. This approach can assist decision-makers in providing cost-effective ways to accurately quantify resources on the grounds. Testing different base learners with target-oriented cross-validation in different landscapes on a relatively large scale would offer broader insight into improving overall land use/land cover mapping.
Author Contributions
Conceptualization, M.R.S.; methodology, M.R.S.; software, M.R.S.; validation, M.R.S., C.P.-Q., R.D.C., N.E.M., S.S.K. and G.P.; formal analysis, M.R.S.; writing—original draft preparation, M.R.S.; writing—review and editing, M.R.S., C.P.-Q., R.D.C., N.E.M., S.S.K., G.P. and X.S.; visualization, M.R.S.; funding acquisition, C.P.-Q., N.E.M., S.S.K. and G.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Texas Comptroller’s Office.
Data Availability Statement
Data sets used in this study are available in registries that are freely accessible to the public. The National Agricultural Imagery Program (NAIP) Digital Ortho Quadrangle Quads are accessible through EarthExplorer (https://earthexplorer.usgs.gov/; accessed on 6 February 2021) and Sentinel data collected by the European Space Agency can be accessed through https://dataspace.copernicus.eu/ (previously https://scihub.copernicus.eu/dhus/#/home; accessed on 10 February 2021).
Acknowledgments
We thank the Texas Comptroller’s Office for the financial assistance of this study. In addition, we extend our thanks to the Urbanovsky Foundation for the Research Assistantship to the first author. Comments from two anonymous reviewers improved the manuscript. Finally, thanks are due to the National Agricultural Imagery Program (NAIP) for Digital Ortho Quadrangle Quads (DOQQS) data and the European Space Agency for Sentinel data.
Conflicts of Interest
The authors declare no conflicts of interest.
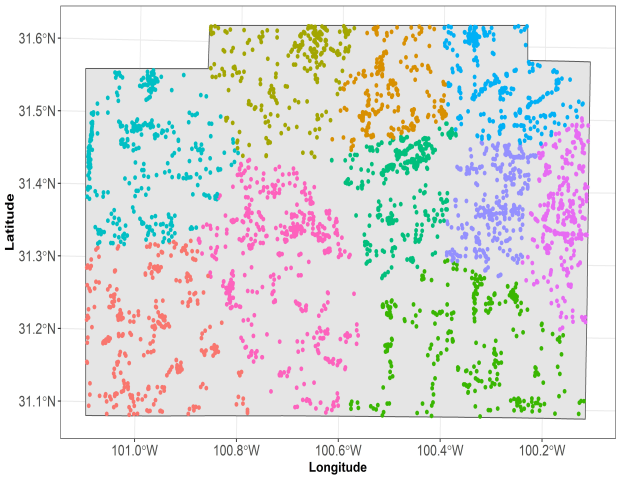
Appendix A
Clustering of training data points. Each color represents the fold in machine learning. We used k-means on spatial coordinates of training data to group these data points.
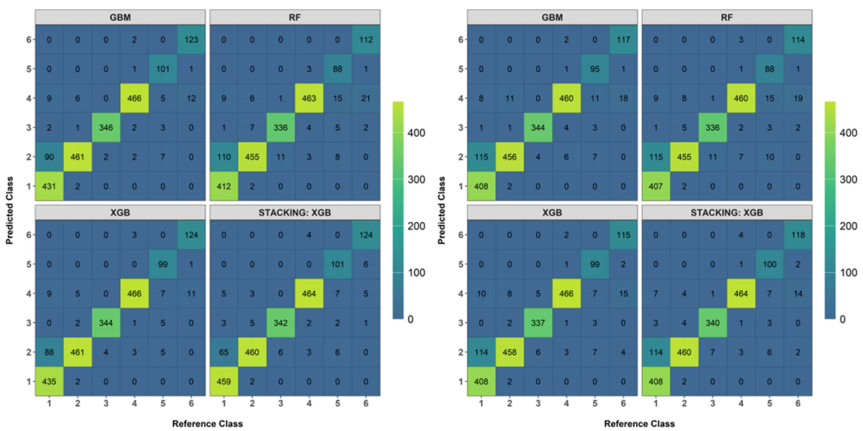
Appendix B
Confusion matrices produced on independent validation data points: left (target-oriented cross-validation; LLO-CV), right (random cross-validation; R-CV). Validation data points were generated using random sampling, and labeling was performed using Google Earth Pro, version 7.3.
Appendix C
Permutation-based variable importance of base learners (RF [A], GBM [B], XGB [C]) and stack learner (Stack-XGB [D]) in random cross-validation.
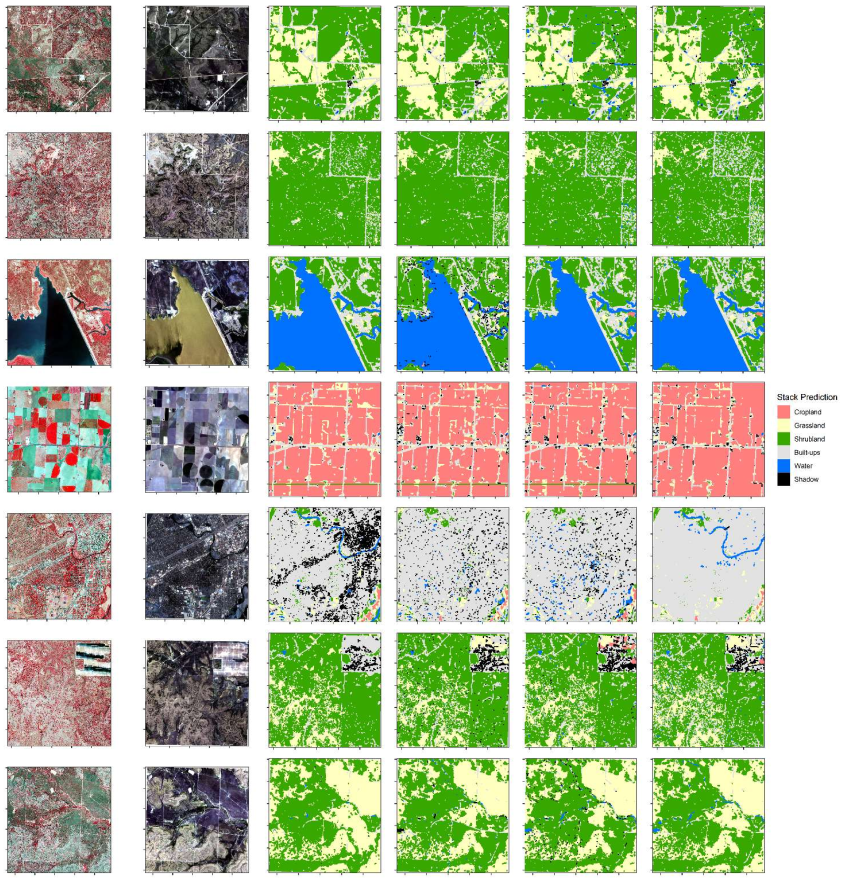
Appendix D
Land cover classification result. From left to right (NIR, green, and red): NAIP, natural color composites of Sentinel 2A, base learner (RF, GBM, XGB), and stacking (extreme-right). The produced maps were based on target-oriented validations.
Appendix E
Imagery data source, extracted variables and their definitions
| Source/Type | Variable Name | Definition |
| NAIP | NMNIR | Mean of Near Infrared (NIR) |
| NSNIR | Standard deviation of NIR | |
| Sentinel | SMNPC | Mean of PC of NIR |
| SMSMI | Mean SAVI Minimum | |
| SMSMN | Mean SAVI Mean | |
| SMSMX | Mean SAVI Maximum | |
| SMSPC1 | Mean SAVI PC1 | |
| SMSPC2 | Mean SAVI PC2 | |
| SMSPC3 | Mean SAVI PC3 | |
| SMSSD | Mean SAVI Standard Deviation | |
| SSNPC | Standard Deviation of PC of NIR | |
| SSSMI | Standard Deviation of SAVI Minimum | |
| SSSMN | Standard Deviation of SAVI Mean | |
| SSSMX | Standard Deviation of Maximum | |
| SSSPC1 | Standard Deviation of SAVI PCA1 | |
| SSSPC2 | Standard Deviation of SAVI PCA2 | |
| SSSPC3 | Standard Deviation of SAVI PCA3 | |
| SSSST | Standard Deviation of SAVI | |
| TMEAN | GLCM Mean of SAVI PCA1[All Direction] | |
| ALEVW | Ratio of Area to Width | |
| APIXL | Area in Pixels | |
| BRIHT | Brightness | |
| Geometry | GASYM | Asymmetry |
| GCOMT | Compactness | |
| GDENS | Density | |
| GRECT | Rectangular Fit | |
| GROND | Roundness | |
| GSHAP | Shape Index | |
| MAXDF | Maximum Difference |
References
- Hirayama, H.; Sharma, R.C.; Tomita, M.; Hara, K. Evaluating Multiple Classifier System for the Reduction of Salt-and-Pepper Noise in the Classification of Very-High-Resolution Satellite Images. Int. J. Remote Sens. 2019, 40, 2542–2557. [Google Scholar] [CrossRef]
- Maxwell, A.E.; Strager, M.P.; Warner, T.A.; Zégre, N.P.; Yuill, C.B. Comparison of NAIP Orthophotography and Rapideye Satellite Imagery for Mapping of Mining and Mine Reclamation. GISci. Remote Sens. 2014, 51, 301–320. [Google Scholar] [CrossRef]
- Homer, C.G.C.; Dewitz, J.A.J.; Yang, L.; Jin, S.; Danielson, P.; Xian, G.; Coulston, J.; Herold, N.D.N.; Wickham, J.D.J.; Megown, K. Completion of the 2011 National Land Cover Database for the Conterminous United States-Representing a Decade of Land Cover Change Information. Photogramm. Eng. Remote Sens. 2015, 81, 345–354. [Google Scholar] [CrossRef]
- Fry, J.A.; Xian, G.; Jin, S.; Dewitz, J.A.; Homer, C.G.; Yang, L.; Barnes, C.A.; Herold, N.D.; Wickham, J.D. Completion of the 2006 National Land Cover Database for the Conterminous United States. Photogramm. Eng. Remote Sens. 2011, 77, 858–864. [Google Scholar]
- Hay, G.J.; Castilla, G. Geographic Object-Based Image Analysis (GEOBIA): A New Name for a New Discipline. In Object-Based Image Analysis: Spatial Concepts for Knowledge-Driven Remote Sensing Applications; Lecture Notes in Geoinformation and Cartography; Blaschke, T., Lang, S., Hay, G., Eds.; Springer: Berlin/Heidelberg, Germany, 2008; pp. 75–89. [Google Scholar]
- Hayes, M.M.; Miller, S.N.; Murphy, M.A. High-Resolution Landcover Classification Using Random Forest. Remote Sens. Lett. 2014, 5, 112–121. [Google Scholar] [CrossRef]
- Knight, J.F.; Tolcser, B.P.; Corcoran, J.M.; Rampi, L.P. The Effects of Data Selection and Thematic Detail on the Accuracy of High Spatial Resolution Wetland Classifications. Photogramm. Eng. Remote Sens. 2013, 79, 613–623. [Google Scholar] [CrossRef]
- Zurqani, H.A.; Post, C.J.; Mikhailova, E.A.; Cope, M.P.; Allen, J.S.; Lytle, B.A. Evaluating the Integrity of Forested Riparian Buffers over a Large Area Using LiDAR Data and Google Earth Engine. Sci. Rep. 2020, 10, 14096. [Google Scholar] [CrossRef] [PubMed]
- Subedi, M.R.; Portillo-Quintero, C.; Kahl, S.S.; McIntyre, N.E.; Cox, R.D.; Perry, G. Leveraging NAIP Imagery for Accurate Large-Area Land Use/Land Cover Mapping: A Case Study in Central Texas. Photogramm. Eng. Remote Sens. 2023, 89, 547–560. [Google Scholar] [CrossRef]
- Li, X.; Shao, G. Object-Based Land-Cover Mapping with High Resolution Aerial Photography at a County Scale in Midwestern USA. Remote Sens. 2014, 6, 11372–11390. [Google Scholar] [CrossRef]
- Zylshal; Sulma, S.; Yulianto, F.; Nugroho, J.T.; Sofan, P. A Support Vector Machine Object Based Image Analysis Approach on Urban Green Space Extraction Using Pleiades-1A Imagery. Model. Earth Syst. Environ. 2016, 2, 54. [Google Scholar] [CrossRef]
- Tzotsos, A.; Argialas, D. Support Vector Machine Classification for Object-Based Image Analysis. In Object-Based Image Analysis: Spatial Concepts for Knowledge-Driven Remote Sensing Applications; Lecture Notes in Geoinformation and Cartography; Blaschke, T., Lang, S., Hay, G., Eds.; Springer: Berlin/Heidelberg, Germany, 2008; pp. 663–677. [Google Scholar]
- Ruiz, L.Á.; Recio, J.A.; Crespo-Peremarch, P.; Sapena, M. An Object-Based Approach for Mapping Forest Structural Types Based on Low-Density LiDAR and Multispectral Imagery. Geocarto Int. 2018, 33, 443–457. [Google Scholar] [CrossRef]
- Amini, S.; Homayouni, S.; Safari, A.; Darvishsefat, A.A. Object-Based Classification of Hyperspectral Data Using Random Forest Algorithm. Geo-Spat. Inf. Sci. 2018, 21, 127–138. [Google Scholar] [CrossRef]
- van Leeuwen, B.; Tobak, Z.; Kovács, F. Machine Learning Techniques for Land Use/Land Cover Classification of Medium Resolution Optical Satellite Imagery Focusing on Temporary Inundated Areas. J. Environ. Geogr. 2020, 13, 43–52. [Google Scholar] [CrossRef]
- Myint, S.W.; Gober, P.; Brazel, A.; Grossman-Clarke, S.; Weng, Q. Per-Pixel vs. Object-Based Classification of Urban Land Cover Extraction Using High Spatial Resolution Imagery. Remote Sens. Environ. 2011, 115, 1145–1161. [Google Scholar] [CrossRef]
- Yu, Q.; Gong, P.; Clinton, N.; Biging, G.; Kelly, M.; Schirokauer, D. Object-Based Detailed Vegetation Classification with Airborne High Spatial Resolution Remote Sensing Imagery. Photogramm. Eng. Remote Sens. 2006, 72, 799–811. [Google Scholar] [CrossRef]
- Yuan, Q.; Shen, H.; Li, T.; Li, Z.; Li, S.; Jiang, Y.; Xu, H.; Tan, W.; Yang, Q.; Wang, J.; et al. Deep Learning in Environmental Remote Sensing: Achievements and Challenges. Remote Sens. Environ. 2020, 241, 111716. [Google Scholar] [CrossRef]
- Taghizadeh-Mehrjardi, R.; Schmidt, K.; Amirian-Chakan, A.; Rentschler, T.; Zeraatpisheh, M.; Sarmadian, F.; Valavi, R.; Davatgar, N.; Behrens, T.; Scholten, T. Improving the Spatial Prediction of Soil Organic Carbon Content in Two Contrasting Climatic Regions by Stacking Machine Learning Models and Rescanning Covariate Space. Remote Sens. 2020, 12, 1095. [Google Scholar] [CrossRef]
- Das, B.; Rathore, P.; Roy, D.; Chakraborty, D.; Jatav, R.S.; Sethi, D.; Kumar, P. Comparison of Bagging, Boosting and Stacking Algorithms for Surface Soil Moisture Mapping Using Optical-Thermal-Microwave Remote Sensing Synergies. Catena 2022, 217, 106485. [Google Scholar] [CrossRef]
- Jafarzadeh, H.; Mahdianpari, M.; Gill, E.; Mohammadimanesh, F.; Homayouni, S. Bagging and Boosting Ensemble Classifiers for Classification of Multispectral, Hyperspectral and PolSAR Data: A Comparative Evaluation. Remote Sens. 2021, 13, 4405. [Google Scholar] [CrossRef]
- Wu, X.; Wang, J. Application of Bagging, Boosting and Stacking Ensemble and EasyEnsemble Methods for Landslide Susceptibility Mapping in the Three Gorges Reservoir Area of China. Int. J. Environ. Res. Public Health 2023, 20, 4977. [Google Scholar] [CrossRef]
- Helber, P.; Bischke, B.; Dengel, A.; Borth, D. Eurosat: A Novel Dataset and Deep Learning Benchmark for Land Use and Land Cover Classification. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2019, 12, 2217–2226. [Google Scholar] [CrossRef]
- Kattenborn, T.; Leitloff, J.; Schiefer, F.; Hinz, S. Review on Convolutional Neural Networks(CNN) in Vegetation Remote Sensing. ISPRS J. Photogramm. Remote Sens. 2021, 173, 24–49. [Google Scholar] [CrossRef]
- Legendre, P.; Dale, M.R.T.; Fortin, M.J.; Gurevitch, J.; Hohn, M.; Myers, D. The Consequences of Spatial Structure for the Design and Analysis of Ecological Field Surveys. Ecography 2002, 25, 601–615. [Google Scholar] [CrossRef]
- Getis, A. A History of the Concept of Spatial Autocorrelation: A Geographer’s Perspective. Geogr. Anal. 2008, 40, 297–309. [Google Scholar] [CrossRef]
- Stehman, S.V.; Foody, G.M. Key Issues in Rigorous Accuracy Assessment of Land Cover Products. Remote Sens. Environ. 2019, 231, 111199. [Google Scholar] [CrossRef]
- Roberts, D.R.; Bahn, V.; Ciuti, S.; Boyce, M.S.; Elith, J.; Guillera-Arroita, G.; Hauenstein, S.; Lahoz-Monfort, J.J.; Schröder, B.; Thuiller, W.; et al. Cross-Validation Strategies for Data with Temporal, Spatial, Hierarchical, or Phylogenetic Structure. Ecography 2017, 40, 913–929. [Google Scholar] [CrossRef]
- Griffith, G.E.; Bryce, S.; Omernik, J.; Rogers, A. Ecoregions of Texas. U.S. Environmental Protection Agency: Corvallis, OR, USA, 2004. [Google Scholar]
- Drusch, M.; Del Bello, U.; Carlier, S.; Colin, O.; Fernandez, V.; Gascon, F.; Hoersch, B.; Isola, C.; Laberinti, P.; Martimort, P.; et al. Sentinel-2: ESA’s Optical High-Resolution Mission for GMES Operational Services. Remote Sens. Environ. 2012, 120, 25–36. [Google Scholar] [CrossRef]
- Hagolle, O.; Sylvander, S.; Huc, M.; Claverie, M.; Clesse, D.; Dechoz, C.; Lonjou, V.; Poulain, V. SPOT-4(Take 5): Simulation of Sentinel-2 Time Series on 45 Large Sites. Remote Sens. 2015, 7, 12242–12264. [Google Scholar] [CrossRef]
- Franklin, S.E.; Wulder, M.A.; Gerylo, G.R. Texture Analysis of IKONOS Panchromatic Data for Douglas-Fir Forest Age Class Separability in British Columbia. Int. J. Remote Sens. 2001, 22, 2627–2632. [Google Scholar] [CrossRef]
- Haralick, R.M.; Dinstein, I.; Shanmugam, K. Textural Features for Image Classification. IEEE Trans. Syst. Man Cybern. 1973, 6, 610–621. [Google Scholar] [CrossRef]
- Legendre, P.; Legendre, L. Numerical Ecology, 3rd ed.; Elsevier: Amsterdam, The Netherlands; Oxford, UK, 2012. [Google Scholar]
- Good, E.J.; Kong, X.; Embury, O.; Merchant, C.J.; Remedios, J.J. An Infrared Desert Dust Index for the Along-Track Scanning Radiometers. Remote Sens. Environ. 2012, 116, 159–176. [Google Scholar] [CrossRef]
- ECognition Developer, version 9; Trimble: Sunnyvale, CA, USA, 2020.
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-Learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Kuhn, M.; Johnson, K. Classification Trees and Rule-Based Models BT. In Applied Predictive Modeling; Kuhn, M., Johnson, K., Eds.; Springer: New York, NY, USA, 2013; pp. 369–413. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Silveyra Gonzalez, R.; Latifi, H.; Weinacker, H.; Dees, M.; Koch, B.; Heurich, M. Integrating LiDAR and High-Resolution Imagery for Object-Based Mapping of Forest Habitats in a Heterogeneous Temperate Forest Landscape. Int. J. Remote Sens. 2018, 39, 8859–8884. [Google Scholar] [CrossRef]
- Guo, L.; Chehata, N.; Mallet, C.; Boukir, S. Relevance of Airborne Lidar and Multispectral Image Data for Urban Scene Classification Using Random Forests. ISPRS J. Photogramm. Remote Sens. 2011, 66, 56–66. [Google Scholar] [CrossRef]
- Friedman, J.H. Greedy function approximation: A gradient boosting machine. Ann. Stat. 2001, 29, 1189–1232. [Google Scholar] [CrossRef]
- Feng, J.; Xu, Y.-X.; Jiang, Y.; Zhou, Z.-H. Soft Gradient Boosting Machine. arXiv 2020, arXiv:2006.04059. [Google Scholar]
- Chen, T.; Guestrin, C. XGBoost: A Scalable Tree Boosting System. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13 August 2016; pp. 785–794. [Google Scholar]
- Wolpert, D. Stacked Generalization. Neural Netw. 1992, 5, 241–259. [Google Scholar] [CrossRef]
- Congalton, R.G. A Review of Assessing the Accuracy of Classifications of Remotely Sensed Data. Remote Sens. Environ. 1991, 37, 35–46. [Google Scholar] [CrossRef]
- Matthews, B.W. Comparison of the Predicted and Observed Secondary Structure of T4 Phage Lysozyme. Biochim. Biophys. Acta-Protein Struct. 1975, 405, 442–451. [Google Scholar] [CrossRef]
- Dou, J.; Yunus, A.P.; Bui, D.T.; Merghadi, A.; Sahana, M.; Zhu, Z.; Chen, C.W.; Han, Z.; Pham, B.T. Improved Landslide Assessment Using Support Vector Machine with Bagging, Boosting, and Stacking Ensemble Machine Learning Framework in a Mountainous Watershed, Japan. Landslides 2020, 17, 641–658. [Google Scholar] [CrossRef]
- Meyer, H.; Reudenbach, C.; Hengl, T.; Katurji, M.; Nauss, T. Improving Performance of Spatio-Temporal Machine Learning Models Using Forward Feature Selection and Target-Oriented Validation. Environ. Model. Softw. 2018, 101, 1–9. [Google Scholar] [CrossRef]
- Congalton, R.G. A Comparison of Sampling Schemes Used in Generating Error Matrices for Assessing the Accuracy of Maps Generated from Remotely Sensed Data. Photogramm. Eng. Remote Sens. 1998, 54, 593–600. [Google Scholar]
- Wadoux, A.M.J.C.; Heuvelink, G.B.M.; de Bruin, S.; Brus, D.J. Spatial Cross-Validation Is Not the Right Way to Evaluate Map Accuracy. Ecol. Model. 2021, 457, 109692. [Google Scholar] [CrossRef]
- Karasiak, N.; Dejoux, J.F.; Monteil, C.; Sheeren, D. Spatial Dependence between Training and Test Sets: Another Pitfall of Classification Accuracy Assessment in Remote Sensing. Mach. Learn. 2021, 111, 2715–2740. [Google Scholar] [CrossRef]
- Mannel, S.; Price, M.; Hua, D. Impact of Reference Datasets and Autocorrelation on Classification Accuracy. Int. J. Remote Sens. 2011, 32, 5321–5330. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).