Abstract
Hyperspectral imaging has many applications. However, the high device costs and low hyperspectral image resolution are major obstacles limiting its wider application in agriculture and other fields. Hyperspectral image reconstruction from a single RGB image fully addresses these two problems. The robust HSCNN-R model with mean relative absolute error loss function and evaluated by the Mean Relative Absolute Error metric was selected through permutation tests from models with combinations of loss functions and evaluation metrics, using tomato as a case study. Hyperspectral images were subsequently reconstructed from single tomato RGB images taken by a smartphone camera. The reconstructed images were used to predict tomato quality properties such as the ratio of soluble solid content to total titratable acidity and normalized anthocyanin index. Both predicted parameters showed very good agreement with corresponding “ground truth” values and high significance in an F test. This study showed the suitability of hyperspectral image reconstruction from single RGB images for fruit quality control purposes, underpinning the potential of the technology—recovering hyperspectral properties in high resolution—for real-world, real time monitoring applications in agriculture any beyond.
Keywords:
hyperspectral image reconstruction; RGB image; deep learning; HSCNN-R; SSC; TTA; STR; lycopene; tomato 1. Introduction
Hyperspectral imaging (HSI) combines spectroscopy and optical imaging and provides information about the chemical properties of a material and its spatial distribution [1]. HSI is a form of non-invasive imaging that applies visible and near-infrared radiation (wavelengths 400 nm to 2500 nm) to chemicals or biological substances to measure differential reflection [2]. Due to the vast amount of information to be obtained from hyperspectral images—compared to images in the RGB (red, green, blue) color model—HSI has been widely applied in research and industry. Applications include rapid, environmentally friendly, and noninvasive analysis in remote sensing [3,4], biodiversity monitoring [5], health care [6], wood characterization [7], and the food industry [8,9].
However, despite the potential benefits, the wide application of HSI is restrained due to the considerable costs of high-quality imaging devices compared to conventional RGB sensors. Moreover, most of these HSI devices are scanning-based—using either push broom or filter scanning approaches—making them less portable and time consuming to operate, which seriously limits the broader application of HSI technology [10]. In addition, snapshot hyperspectral cameras—able to take images quickly—often feature a rather low spatial resolution [11]. High-resolution hyperspectral information is appealing as it not only provides spectral signatures of chemical elements but also spatial details [12].
Deep learning approaches are increasingly applied in many areas of research and industry [13,14,15], and recently allowed the development of hyperspectral image reconstruction approaches [16]. The reconstruction of hyperspectral information from RGB images is envisioned to provide a promising way of overcoming current limitations of both scanner- and snapshot camera-based hyperspectral imaging devices, providing image with both high spatial and spectral resolution, and being affordable, user friendly, and highly portable [17]. In particular, smartphone camera sensors could easily capture images in high spatial resolution, e.g., twelve million pixels per image, providing a sound basis for reconstructing high-resolution hyperspectral images. While reconstruction approaches were initially very rigid and complex [18]—limiting their usability for practical application—recent progress, in particular the application of deep learning approaches, enabled easier, faster, and more accurate hyperspectral image reconstruction pipelines [16,19]. Several contrasting approaches based on deep learning have been proposed recently [20,21,22].
While hyperspectral recovery from a single RGB image has seen a great improvement with the development of deep learning, it is still limited for several reasons. For example, hyperspectral images used during method development were previously restricted to the visual spectral range (VIS, 400–700 nm) with 31 wavebands and a spectral resolution of 10 nm [18]. Compared to the near-infrared range (NIR; 800 to 2500 nm), images in the visual range miss information important for many applications [23]. In addition, considerable uncertainty exists on criteria for model performance evaluation. Currently three major evaluation metrics are widely used in performance assessment: Mean Relative Absolute Error (MRAEEM), Root Mean Square Error (RMSE), and Spectral Angle Mapper (SAM) [17,19,21,24,25]. However, there is no general agreement over which criterion is most robust for indicating a better model.
A key application of HSI is food quality evaluation [26]. Tomato is one of the most important fruits for daily consumption, and the fast and non-destructive evaluation of its quality is of great interest both in research and industry—rendering it a suitable object for a case study [27,28]. Taste of different tomato varieties and qualities is mainly affected by sugar content, acidity, and the ratio between them [29]. Previous studies used diverse instruments such as a Raman spectrometer [30], near-infrared spectrophotometers [31,32] and a multichannel hyperspectral imaging instrument [33] for quantifying those parameters. The normalized anthocyanin index (NAI) has been shown to be very effective in predicting lycopene content [34,35]. Lycopene, a secondary plant compound of the carotenoid class, may reduce the risk of developing several cancer types and coronary heart diseases [36]. Making use of readily available RGB cameras, e.g., smartphone cameras, in combination with hyperspectral image reconstruction techniques would greatly facilitate the assessment of tomato quality parameters. In particular, it will promote the selection and sorting process of tomato fruits in industry [37] and might even support consumers in the choice of tomato qualities.
In this study, we demonstrate the use of a permutation test to select an appropriate state-of-the-art deep learning model for hyperspectral image reconstruction from a single RGB image. Subsequently, we show that the reconstructed images can be used to predict tomato quality properties through random forests (RF) regression at high accuracies—developing an efficient pipeline from automatic segmentation to quality assessment. Finally, the application potential of reconstructed hyperspectral image is discussed.
2. Materials and Methods
2.1. Plant Material, Growth Conditions, and Tomato Sampling
Ungrafted tomato plants (Solanum lycopersicum L., variety “Dometica” (Rijk Zwaan)) were used in this experiment. Seeds were sowed on 29th of July 2018 in a climate-controlled chamber; 39 days after sowing seedlings were transplanted to a Venlo-type greenhouse in southwestern Norway (58°42′49.2″ N 5°31′51.0″ E) and grown on rockwool slabs with drip irrigation according to common practice [38]. The plants were irrigated with a complete nutrient solution based on standardized recommendations: 17.81 mM NO3, 0.71 mM NH4, 1.74 mM P, 9.2 mM K, 4.73 mM Ca, 2.72 mM Mg, 2.74 mM S, 15 µM Fe, 10 µM Mn, 5 µM Zn, 30 µM B, 0.75 µM Cu, and 0.5 µM Mo. The electrical conductivity of the nutrient solution was maintained at around 3.2 mS cm−1 and the pH was 5.8. Average daily temperature, relative humidity, CO2 concentration, and natural solar radiation during the growing period were 22.4 ± 2.8°C, 74 ± 7.8%, 670 ± 192 ppm and 33 ± 77 W∙m−2, respectively. High-pressure sodium lamps (Philips GP Plus, Gavita Nordic AS, Norway) with an intensity of 300 W∙m−2 (1.5 m above the top of the canopy) were used in addition for ≤18 h per day (i.e., when solar radiation was <250 W∙m−2). Side shoots were pruned regularly, and the number of tomatoes in each truss was pruned to seven. Tomato fruits for the study were collected 210 days after sowing during the morning. Three undamaged tomatoes of similar size were selected from each of 12 color grades [39]. Color grades range from 1 to 12—where 1 is uniform green (e.g., mature green) and 12 is uniform dark red (i.e., red overripe).
2.2. Image Acquisition
Hyperspectral images of tomatoes were instantly taken by a portable hyperspectral camera, Specim IQ (Spectral Imaging Ltd., Oulu Finland; [40]), with a spatial resolution of 512 × 512 pixels, a spectral resolution of 7 nm, and 204 spectral bands from 397 to 1003 nm. Calibration was conducted following the user manual. When taking images, the camera was placed 100 cm above the table where 12 tomatoes were placed on a tripod. Two Arrilite 750 Plus halogen lamps (ARRI, Munich, Germany) were symmetrically placed beside the camera for illumination. A white panel (90% reflectance) was placed adjacent to the tomatoes as a reference target for reflectance transformation [40].
A set of RGB images was rendered directly from the hyperspectral images under CIE Standard Illuminant D65 with the CIE 1931 2° Standard Observer with gamma correction (γ = 1.4) (Figure 1). Cropped images of 21 × 31 pixels, excluding overexposed area, from both rendered RGB image and corresponding hyperspectral image area of each tomato were subsequently used to build the model (see below).
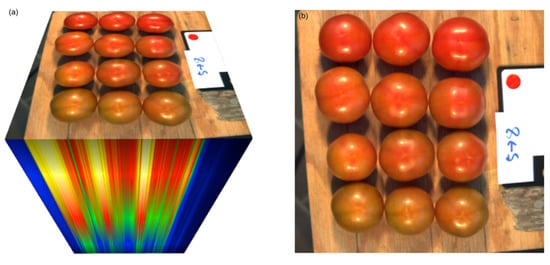
Figure 1.
(a) Illustration of the 204-band hyperspectral image captured with the hyperspectral camera Specim IQ, and (b) corresponding, rendered 3-band RGB image, rendered under Standard Illuminant D65 with the CIE 1931 2° Standard Observer with gamma correction. See text for details.
The built-in main camera of the smartphone Samsung Galaxy S9+, Android 9.0 (Samsung Corp., Seoul, South Korea), was used to take RGB images immediately after hyperspectral imaging. The main 12 Mp camera consists of a 1/2.55’’ sensor and f/1.5 to f/2.4 variable aperture lens; images were taken with a resolution of 3024 × 4032 pixels; the distance between the smartphone camera and the tomato was set to fit the fixed-size table inside the view finder—not using the optical zoom. The flashlight was turned off and autofocus mode was activated when shooting the RGB images. Images were saved in jpg format.
2.3. Tomato Quality Parameters (“Ground Truth”)
After the imaging campaign, each tomato was immediately and separately homogenized with a handheld blender. The fresh, uniform samples were used for estimation of soluble solid content (SSC, expressed as °Brix) and total titratable acidity (TTA, expressed as % of citric acid equivalents (CAE) per FW; [41,42]). SSC was measured with a digital refractometer PR-101α (ATAGO, Tokyo, Japan). TTA was determined using an automatic titrator 794 Basic Titrino (Metrohm, Herisau, Switzerland) by titrating with 0.1 M NaOH to pH 8.2. The ratio of SSC to TTA (STR) of each tomato was calculated.
Lycopene content was calculated with the normalized anthocyanin index (NAI) [35], using the reflectance at 570 nm and 780 nm as determined with the hyperspectral camera (see above). The NAI of each ith pixel was calculated through Equation (1).
are the reflectance of the ith pixel of the image at wavelengths 780 nm and 570 nm, respectively; is the NAI value of the ith pixel. The median value of all of each segmented tomato, excluding the overexposed areas [43] on tomato RGB images, was treated as the overall NAI of an individual tomato. These quality parameters, either determined according to laboratory measurements (SSC, TTA, STR) or calculated from reflectance measurements using the hyperspectral camera (NAI), were used as “ground truth” values in this study and subsequently compared with the predicted parameters (see below).
2.4. Model Selection, Training and Validation
A state-of-the-art deep learning model, i.e., a residual neural network model named HSCNN-R [17], was selected—showing very good performance for hyperspectral image reconstruction. In HSCNN-R, a modern residual block [44] was used to replace the plain CNN architecture of HSCNN [16] to improve the model performance. Six residual blocks, improving the time efficacy without harming performance during the validation procedure compared with the 16 residual blocks proposed originally (data not shown), were chosen for the HSCNN-R model with 64 filters in each residual block. Permutation test was applied during model selection and a total of 36 samples was randomly divided into a training set (24 samples), and a testing set (12 samples), for 5 times. The batch size, learning rate, learning rate decay, and optimizer weight initialization in different layers in HSCNN-R were set according to Shi et al. (2018).
Two different loss functions were used to compare their effectiveness in hyperspectral image reconstruction: mean square error (MSE, Equation (2)) [24,25], one of the most widely used loss functions, and mean relative absolute error (MRAELF, Equation (3)) which was suggested to reduce the bias from different illuminance levels [17].
and represent the ith pixel of the ground truth and reconstructed hyperspectral images, respectively.
Three evaluation metrics, MRAEEM (Equation (4)), RMSE (Equation (5)), and SAM (Equation (6)) were used to select models with their corresponding minimum values during validation; smaller values indicate less error on reconstructed hyperspectral images. RMSE was also used to evaluate the difference between ground truth and reconstructed hyperspectral images, and SAM quantifies the similarity of the original and reconstructed reflectance across the spectra through measuring the average angle between them [45].
, are the ith pixel in the ground truth and reconstructed hyperspectral images, respectively; T means transpose, and n is the total number of pixels of each image.
For model training, the batch size was set to 8 and the optimizer AdaMax [46] with settings of β1 = 0.9, β2 = 0.999, and eps = 10−8. The weights were initialized though HeNormal initialization [44] in each convolutional layer. The initial learning rate was set at 0.005 and the learning rate decreased by 10% every 100 epochs. The model performance was evaluated through the three evaluation metrics MRAEEM, RMSE, and SAM (see above). All models were trained until no further decrease in validation loss occurred.
During validation, the whole cropped RGB image (21 × 31 × 3) from the validation set for HSCNN-R was used as input to reconstruct the hyperspectral image with 204 spectral bands (21 × 31 × 204); MRAEEM, RMSE, and SAM values between reconstructed and ground truth hyperspectral images were calculated accordingly. Based on the 30 models selected, two loss functions × three evaluation metrics × 5 times’ random sampling, values from evaluation metrics were analyzed between and within two loss functions by permutation test using the EnvStats package [47] in R [48]. The model generating constant performance was selected and used to reconstruct hyperspectral images from single tomato RGB images.
2.5. Image Segmentation and Quality Parameter Prediction
For tomato quality parameter prediction, the overall spectral information of each tomato was considered. The RGB images from Samsung Galaxy S9+ and masks outlined manually through Labelme [49] were used for training. RetinaNet [50] was trained for 5 epochs with 10,000 iterations in each epoch to detect and segment individual tomato; learning rate was set at 2e−8 and kept constant during training.
The reconstructed hyperspectral reflectance of each tomato was extracted based on the segmented tomato mask from RGB images from smartphone, excluding the overexposed region, and then subjected to asymmetric least squares baseline correction of the logarithmic linearized reflectance log(1/R) [51]; the median values of spectral reflectance of different wavebands were extracted for each tomato for quality parameter prediction. The recursive feature elimination method from the Caret package with RF model and repeated 10-fold cross validation was applied to select important wavebands which were then used to build the prediction model; the optimal model was selected based on the prediction accuracy by tuning the parameter, mtry, in RF models. The parameter mtry sets the number of input variables randomly chosen at each node of the RF models. The predicted values of each sample were based on the model trained on the rest of the samples, R2 and p values in the F test were calculated based on the predictions and corresponding ground truth values (see above) of all tomato samples.
The free of charge cloud service Google Colaboratory (Colab) with Python 3 runtime served as major platform for model training and validation. Colab is equipped with a 2.3 GHz and 12.6 GB RAM Intel Xeon processor with two cores and a NVIDIA Tesla K80 GPU with 12 GB RAM.
Detailed implementation of the whole analysis pipeline is available upon request.
3. Results
3.1. Model Selection and Performance
The training and validation histories of the three evaluation metrics MRAEEM, RMSE, and SAM are displayed in Figure 2. MRAEEM, RMSE, and SAM of models with MRAELF loss function (Figure 2, left subpanels) decreased sharply at the beginning and reached minimum values of 0.0453, 6.223, and 1.676 at the 472nd, 476th, and 500th epoch, respectively. For all three evaluation metrics, values increased slightly with subsequent epochs and finally stabilized after approx. 1000 epochs. The minimum values of MRAEEM, RMSE, and SAM from models with MSE loss function (Figure 2, right subpanels) also decreased, possessing higher fluctuations, and showed higher minimum values of 0.0507, 6.778, and 1.735 at the 2663th, 2156th, and 2531st epoch, respectively (Figure 2, Table 1). Both the number of epochs and time consumed for reaching the minimum values of three criteria during validation were much less in models with the MRAELF loss function, i.e., less than 500 epochs and 90 seconds, compared with MSE ones.
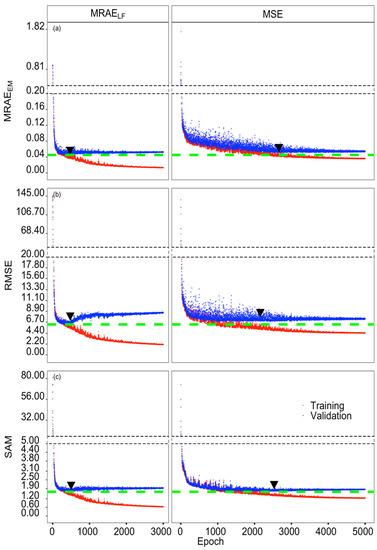
Figure 2.
Training and validation history of the HSCNN-R model reconstructing hyperspectral images from single RGB images. Mean Relative Absolute Error (MRAEEM) (a), Root Mean Square Error (RMSE) (b), and Spectral Angle Mapper (SAM) (c) change during model training (red dots), and testing (blue dots), with either MRAELF loss function (left; 3000 epochs) or MSE loss function (right; 5000 epochs). The green dashed lines indicate the minimum values obtained from each evaluation metric on validation data with both MSE and MRAELF loss functions considered, and the black triangles indicate the epochs where the minimum values of each evaluation metric with different loss functions are located. The dashed lines indicate breaks in the Y-axis to improve visualization.

Table 1.
Examples of minimum Mean Relative Absolute Error (MRAEEM), Root Mean Square Error (RMSE), and Spectral Angle Mapper (SAM) and the corresponding epoch and time consumed when reaching these minimum values of different evaluation metrics during validation with the HSCNN-R model with MRAELF and MSE loss functions, respectively.
The minimum values of MRAE (MRAEEMmin) models with MRAELF loss function were significantly lower than the ones from models with MSE loss function (Figure 3a). Minimum values of RMSE and SAM models were statistically the same using either of the two loss functions (Figure 3b,c). During the permutation test there was no significant difference in MRAEEMmin, RMSEmin, and SAMmin values between the three evaluation metrics within each loss function (Figure 3d–i). Thus, a model with MRAELF loss function evaluated by MRAEEM was used to reconstruct the hyperspectral images from single RGB images for non-destructive tomato quality parameter quantification.
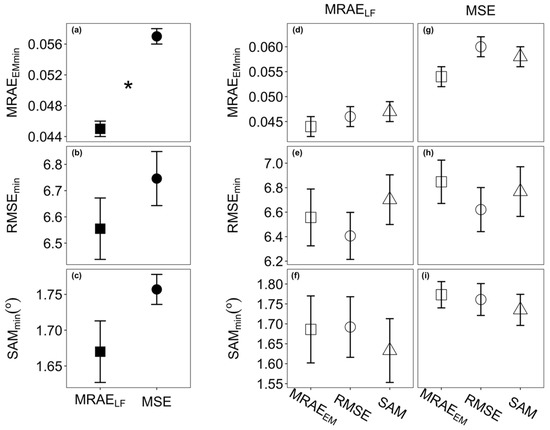
Figure 3.
Selection of models with a lower loss when training with the HSCNN-R model using different loss functions and evaluated with different evaluation metrics. Comparison of minimum values of MRAEEM (MRAEEMmin), RMSE (RMSEmin), and SAM (SAMmin) based on three evaluation metrics, and MRAEEM, RMSE, and SAM, from models trained with either MRAELF or MSE loss function during permutation test. (a–c) Comparisons between two loss functions, mean values with standard error, n = 15, * indicates a significant difference with p < 0.05; (d–f) comparisons within the loss functions MRAELF (d–f) and MSE (g–i), mean values with standard error, n = 5.
The reconstructed spectral reflectance from RGB images, either directly taken by a smartphone RGB sensor or rendered from hyperspectral images, were generally very similar to their corresponding spectral reflectance as determined with a hyperspectral camera (Figure 4a), and its 1st derivatives (Figure 4b). Largest deviations, with greater reconstructed reflectance in RGB images taken by the smartphone, occurred in the spectral range of approximately 380–740 nm. The reconstructed spectral reflectance of the central pixels of the validation set of tomatoes of color grades 1–12 are given in Supplementary Figure S1.
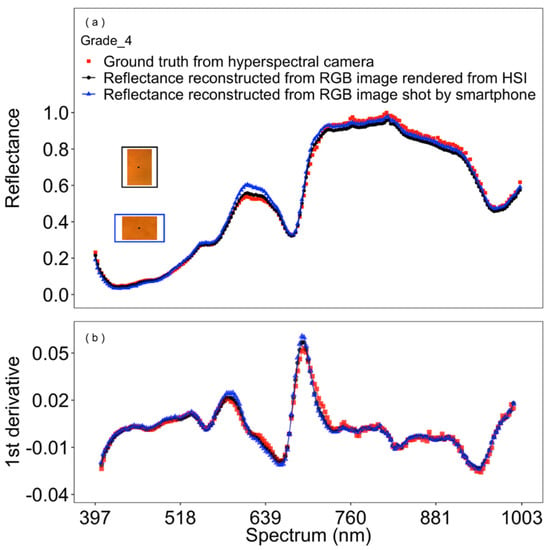
Figure 4.
Reconstructed spectral reflectance (a), and its first derivative (b) of tomato fruits along the visible to near-infrared spectra from 397 to 1003 nm— example of a tomato of color grade 4 (Grade_4; i.e., orange). The black dots at the center of the RGB image (inset in a)—images originating either from a hyperspectral camera (blue frame, triangles), or from an RGB smartphone camera (black frame, circles)—denote the analyzed region and respective reflectance along the spectra. Red squares denote the spectral reflectance from original hyperspectral images (i.e., the corresponding “ground truth”).
3.2. Tomato Quality Parameter Prediction
Tomato quality properties’ soluble solid content (SSC), total titratable acidity (TTA), and their ratio (STR) were predicted with good agreement to the corresponding laboratory measurements (Figure 5a–c), featuring R2 of 0.51, 0.61, and 0.78, respectively. Lycopene content, as indicated by NAI, was predicted with a high accuracy of R2 = 0.92 (Figure 5d). All four quality parameters were predicted significantly better than random guessing, denoted by corresponding p values of 4.02 × 10−6, 8.8 × 10−8, 1.37 × 10−11, and 2.54 × 10−18 in F test. The relationship between SSC, TTA, STR, and the NAI was found to be close to zero (Figure S2).
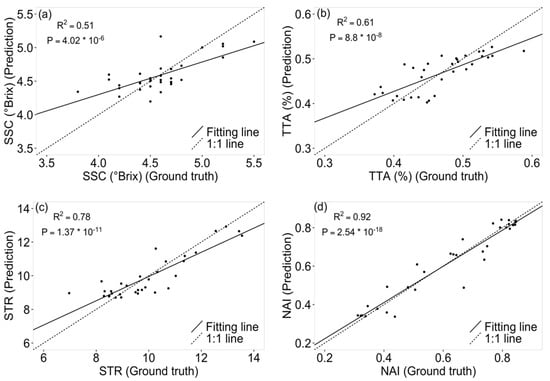
Figure 5.
Comparisons of predicted tomato quality parameters based on reconstructed hyperspectral images and “ground truth” measurements of soluble solid content (SSC) (a), total titratable acidity (TTA) (b), and the ratio of SSC to TTA (STR) (c) in the laboratory. Lycopene content as indicated by the normalized anthocyanin index (NAI) (d) is based on reconstructed hyperspectral images. R2 and corresponding p value from F test are given (n = 33).
4. Discussion
HSI reconstruction has become popular and opened a new field for low-cost methods of acquiring hyperspectral information in high resolution, both spatial and spectral. Even though there has been some research developing methods for reconstructing hyperspectral images [17,18,25], real world applications of these methods are still lacking [10]. This study has demonstrated the potential of using the HSCNN-R model for hyperspectral reconstruction in the visual near-infrared range to predict key quality parameters of tomato. Three models can be selected based on three evaluation metrics, MRAEEM, RMSE, and SAM, as their corresponding minimum values did not appear in the same model, as has been found previously [17,25,52]. A single lower value from any one of these three evaluation metrics thus cannot indicate a better model performance. As only the minimum errors reconstructed from models with MRAELF loss function and MRAEEM evaluation metric were found to be significantly lower than ones with MSE loss function, it can be concluded that these models were consistently superior in spectral reflectance reconstruction compared with models with other loss function and evaluation metric combinations. This was also found by Shi et al. [17] as MRAELF loss function was more robust to outliers and treated wavebands of the whole spectra with different illumination levels were more similar compared with the MSE loss function. As these loss functions can also be purpose specific, MRAELF loss function should thus be chosen if all wavebands are equally prioritized for better exploration of the whole spectra, while MSE should be preferred if the highly illuminated spectral reflectance is of greater interest.
Models trained with the MRAELF loss function were able to converge with fewer epochs at higher speed, and reached lower errors compared to the MSE loss function, which is beneficial for hyperspectral image reconstruction as model training is expected in practice to be implemented in real time prediction, e.g., sorting tomato based on lycopene content on the conveyor belt. The increase of validation error after reaching the minimum value, for either MRAEEM, RMSE, or SAM, was mainly due to overfitting on a small training dataset [53].
To further confirm the robustness of the selected model in reconstructing hyperspectral images, reconstructed spectral reflectance of RGB images either rendered from hyperspectral images or directly captured by smartphone camera, were compared with directly measured spectral reflectance from the hyperspectral camera (“ground truth”). The similarity of both the reflectance and corresponding 1st derivative demonstrated that the selected approach resulted in a reliable reconstruction of the spectral pattern. As the RGB image used for training was rendered from hyperspectral images by a standard CIE matching function while smartphone RGB sensors have different spectral sensitivity functions likely deviating from CIE [22], an increase of errors during the reconstruction of hyperspectral images from RGB smartphone sensors might be expected. However, although the RGB images from the smartphone were completely new to the trained model, the reconstruction results demonstrated the soundness of the model in recovering the spectral reflectance even from regular RGB images taken by a standard smartphone model.
The very high R2 value in NAI prediction showed that the reconstructed hyperspectral image was suitable for predicting tomato lycopene content non-destructively based on the RGB images of intact tomatoes. NAI is the indicator for lycopene which can be closely reflected by the color of tomato [54]. Tomato color change from green to red due to the degradation of chlorophyll while accumulating lycopene during development [55]. The high prediction accuracy for NAI via reconstructed hyperspectral image for lycopene is probably also related to the large range of color change in the corresponding tomato sample. Both TTA and SSC were less precisely predicted; however, their ratio STR was predicted with a high R2 value and very high significance in F test, which agrees with earlier findings [35]. The higher precision of reconstructed hyperspectral images in predicting STR values is fortunate as it is also more informative compared with either TTA or SSC values alone—tomato flavor is determined mainly by the ratio of sugar and acidity rather than the two separate properties [29]. Overall, the high accuracy in tomato quality prediction highlights the robustness and potential of hyperspectral image reconstruction.
The good to very good performance, both in reconstructing hyperspectral images from model unseen RGB images from smartphone camera and in predicting tomato quality parameters at moderate to high accuracies, makes the HSCNN-R model an important tool for future imaging applications. Hyperspectral reconstruction from a single RGB image makes HSI application mobile and low cost, and allows for easy implementation through either a cloud service or an app. With the selected model and trained weights, we can now generate hyperspectral images of tomatoes of the same variety at least with consumer level cameras and explore the other hyperspectral properties of interest—as the example on tomato quality predicted from smartphone images illustrates in this study (Figure 5). Specifically, this provides huge benefits for tomato research and industry, and potentially also for other fruit crops, such as cucumber and apple. Even though it is possible to predict the STR of each pixel of tomato image directly without reconstructing the whole spectrum from 400 to 1000 nm, the fully reconstructed spectral reflectance provides opportunities to explore other hyperspectral properties through different machine learning algorithms, offering much higher flexibility. Important bands can even be selected to reduce workload while improving prediction accuracy, as is commonly used in HSI analysis [56,57].
5. Conclusions
This study first demonstrates the use of hyperspectral image reconstruction from a single RGB image for a real-world application—using tomato fruits as an example. The capability of HSCNN-R for spectral reconstruction beyond the visual-range towards near-infrared was demonstrated. The reconstructed hyperspectral images from RGB images of tomato were able to estimate important tomato quality parameters with high accuracy.
Hyperspectral image reconstruction could be a promising approach for a range of other fields, thereby further developing its full potential. With HSCNN-R, we can potentially reconstruct hyperspectral images in both higher spatial and spectral resolution at much lower costs. However, the reconstructing model built on tomatoes can probably not be transferred easily to other categories, e.g., determination of chemical properties of other fruits, soils or rocks, or to different light conditions—which can be an obstacle for extending the application range. Thus, libraries containing hyperspectral images in different categories of interest (fruits of various varieties, and at different harvest stages and growing conditions (incl. stress), soil types, wood, skin etc.) should be built for training models to fit each category specifically or developing a general model that fits more categories.
Future advancement in this field should particularly focus on: (1) exploration of more robust models for hyperspectral image reconstruction in various illumination conditions, and (2) extending the application field using current state of the art models and building libraries for a wider range of objects as exemplified above. Thereby it can be expected that higher resolution hyperspectral images will be more accessible for a range of real-world applications in the future.
Supplementary Materials
The following are available online at https://www.mdpi.com/2072-4292/12/19/3258/s1, Figure S1: Reconstructed spectral reflectance (A–L) and their first derivatives (a–l) for tomato images in 12 grades based on color tile. Figure S2: The correlations between normalized anthocyanin index (NAI) and (a) soluble solid content (SSC), (b) total titratable acid (TTA), (c) the ratio of SSC to TTA (STR), respectively.
Author Contributions
J.Z. collected data, did data analysis and drafted the manuscript; D.K. performed the experiment and collected data; M.V. and J.L.C. designed the experiment; J.Z., D.K., B.R., G.B., M.V., N.C. and J.L.C. contributed substantially to the manuscript preparation and revision. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the European Union’s Horizon 2020 Research and Innovation program as part of the SiEUGreen project (Grant Agreement N 774233), by the Norwegian Ministry of Foreign Affairs as part of the Sinograin II project (Grant Agreement CHN 2152, 17/0019), by the Bionær program of the Research Council of Norway as part of the research project “Biofresh” (project no 255613/E50), and by the Research Council of Norway InnoLED project (project no: 297301).
Acknowledgments
We thank Anne Kvitvær for technical support during tomato harvesting. The comments of three anonymous reviewers helped improving a previous version of this work.
Conflicts of Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Dong, X.; Jakobi, M.; Wang, S.; Köhler, M.H.; Zhang, X.; Koch, A.W. A review of hyperspectral imaging for nanoscale materials research. Appl. Spectrosc. Rev. 2019, 54, 285–305. [Google Scholar] [CrossRef]
- Cozzolino, D.; Murray, I. Identification of animal meat muscles by visible and near infrared reflectance spectroscopy. LWT Food Sci. Technol. 2004, 37, 447–452. [Google Scholar] [CrossRef]
- Mahlein, A.K.; Kuska, M.T.; Thomas, S.; Bohnenkamp, D.; Alisaac, E.; Behmann, J.; Wahabzada, M.; Kersting, K. Plant disease detection by hyperspectral imaging: From the lab to the field. Adv. Anim. Biosci. 2017, 8, 238–243. [Google Scholar] [CrossRef]
- Qi, H.; Paz-Kagan, T.; Karnieli, A.; Jin, X.; Li, S. Evaluating calibration methods for predicting soil available nutrients using hyperspectral VNIR data. Soil Tillage Res. 2018, 175, 267–275. [Google Scholar] [CrossRef]
- Vance, C.K.; Tolleson, D.R.; Kinoshita, K.; Rodriguez, J.; Foley, W.J. Near infrared spectroscopy in wildlife and biodiversity. J. Near Infrared Spectrosc. 2016, 24, 1–25. [Google Scholar] [CrossRef]
- Afara, I.O.; Prasadam, I.; Arabshahi, Z.; Xiao, Y.; Oloyede, A. Monitoring osteoarthritis progression using near infrared (NIR) spectroscopy. Sci. Rep. 2017, 7, 11463. [Google Scholar] [CrossRef] [PubMed]
- Tsuchikawa, S.; Kobori, H. A review of recent application of near infrared spectroscopy to wood science and technology. J. Wood Sci. 2015, 61, 213–220. [Google Scholar] [CrossRef]
- Barbin, D.F.; ElMasry, G.; Sun, D.-W.; Allen, P.; Morsy, N. Non-destructive assessment of microbial contamination in porcine meat using NIR hyperspectral imaging. Innov. Food Sci. Emerg. Technol. 2013, 17, 180–191. [Google Scholar] [CrossRef]
- Menesatti, P.; Antonucci, F.; Pallottino, F.; Giorgi, S.; Matere, A.; Nocente, F.; Pasquini, M.; D’Egidio, M.G.; Costa, C. Laboratory vs. in-field spectral proximal sensing for early detection of Fusarium head blight infection in durum wheat. Biosyst. Eng. 2013, 114, 289–293. [Google Scholar] [CrossRef]
- Signoroni, A.; Savardi, M.; Baronio, A.; Benini, S. Deep learning meets hyperspectral image analysis: A multidisciplinary review. J. Imaging 2019, 5, 52. [Google Scholar] [CrossRef]
- Cao, X.; Du, H.; Tong, X.; Dai, Q.; Lin, S. A prism-mask system for multispectral video acquisition. IEEE Trans. Pattern Anal. Mach. Intell. 2011, 33, 2423–2435. [Google Scholar]
- Thomas, S.; Kuska, M.T.; Bohnenkamp, D.; Brugger, A.; Alisaac, E.; Wahabzada, M.; Behmann, J.; Mahlein, A.-K. Benefits of hyperspectral imaging for plant disease detection and plant protection: A technical perspective. J. Plant Dis. Prot. 2018, 125, 5–20. [Google Scholar] [CrossRef]
- Rahimy, E. Deep learning applications in ophthalmology. Curr. Opin. Ophthalmol. 2018, 29, 254–260. [Google Scholar] [CrossRef] [PubMed]
- Rao, Q.; Frtunikj, J. Deep learning for self-driving cars: Chances and challenges. In Proceedings of the 1st International Workshop on Software Engineering for AI in Autonomous Systems, Gothenburg, Sweden, 28 May 2018; pp. 35–38. [Google Scholar]
- Ghosal, S.; Zheng, B.; Chapman, S.C.; Potgieter, A.B.; Jordan, D.R.; Wang, X.; Singh, A.K.; Singh, A.; Hirafuji, M.; Ninomiya, S. A weakly supervised deep learning framework for sorghum head detection and counting. Plant Phenomics 2019, 2019, 1525874. [Google Scholar] [CrossRef]
- Xiong, Z.; Shi, Z.; Li, H.; Wang, L.; Liu, D.; Wu, F. HSCNN: CNN-based hyperspectral image recovery from spectrally undersampled projections. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 518–525. [Google Scholar]
- Shi, Z.; Chen, C.; Xiong, Z.; Liu, D.; Wu, F. HSCNN+: Advanced CNN-based hyperspectral recovery from rgb images. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops, Salt Lake City, UT, USA, 18–22 June 2018; pp. 939–947. [Google Scholar]
- Arad, B.; Ben-Shahar, O. Sparse recovery of hyperspectral signal from natural RGB images. In Proceedings of the European Conference on Computer Vision; Springer: New York, NY, USA, 2016; pp. 19–34. [Google Scholar]
- Galliani, S.; Lanaras, C.; Marmanis, D.; Baltsavias, E.; Schindler, K. Learned spectral super-resolution. arXiv 2017, arXiv:1703.09470. [Google Scholar]
- Tschannerl, J.; Ren, J.; Marshall, S. Low cost hyperspectral imaging using deep learning based spectral reconstruction. Lect. Notes Comput. Sci. (Including Subser. Lect. Notes Artif. Intell. Lect. Notes Bioinformatics) 2018, 11257 LNCS, s 206–s 217. [Google Scholar]
- Stiebel, T.; Koppers, S.; Seltsam, P.; Merhof, D. Reconstructing spectral images from rgb-images using a convolutional neural network. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops (CVPRW), Salt Lake City, UT, USA, 18–22 June 2018; pp. 948–953. [Google Scholar]
- Nie, S.; Gu, L.; Zheng, Y.; Lam, A.; Ono, N.; Sato, I. Deeply learned filter response functions for hyperspectral reconstruction. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 4767–4776. [Google Scholar]
- Nagasubramanian, K.; Jones, S.; Singh, A.K.; Sarkar, S.; Singh, A.; Ganapathysubramanian, B. Plant disease identification using explainable 3D deep learning on hyperspectral images. Plant Methods 2019, 15, 98. [Google Scholar] [CrossRef] [PubMed]
- Can, Y.B.; Timofte, R. An efficient CNN for spectral reconstruction from RGB images. arXiv 2018, arXiv:1804.04647. [Google Scholar]
- Yan, Y.; Zhang, L.; Li, J.; Wei, W.; Zhang, Y. Accurate Spectral Super-Resolution from Single RGB Image Using Multi-scale CNN. In Pattern Recognition and Computer Vision; Lai, J.H., Liu, C.L., Chen, X., Zhou, J., Tan, T., Zheng, N., Zha, H., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 206–217. [Google Scholar]
- Ma, J.; Sun, D.-W.; Pu, H.; Cheng, J.-H.; Wei, Q. Advanced techniques for hyperspectral imaging in the food industry: Principles and recent applications. Annu. Rev. Food Sci. Technol. 2019, 10, 197–220. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; Lopez, A.; Jeon, S.; de Freitas, S.T.; Yu, Q.; Wu, Z.; Labavitch, J.M.; Tian, S.; Powell, A.L.T.; Mitcham, E. Disassembly of the fruit cell wall by the ripening-associated polygalacturonase and expansin influences tomato cracking. Hortic. Res. 2019, 6, 17. [Google Scholar] [CrossRef]
- Polder, G.; Van Der Heijden, G.; Van der Voet, H.; Young, I.T. Measuring surface distribution of carotenes and chlorophyll in ripening tomatoes using imaging spectrometry. Postharvest Biol. Technol. 2004, 34, 117–129. [Google Scholar] [CrossRef]
- Simonne, A.H.; Fuzere, J.M.; Simonne, E.; Hochmuth, R.C.; Marshall, M.R. Effects of nitrogen rates on chemical composition of yellow grape tomato grown in a subtropical climate. J. Plant Nutr. 2007, 30, 927–935. [Google Scholar] [CrossRef]
- Qin, J.; Chao, K.; Kim, M.S. Investigation of Raman chemical imaging for detection of lycopene changes in tomatoes during postharvest ripening. J. Food Eng. 2011, 107, 277–288. [Google Scholar] [CrossRef]
- Clément, A.; Dorais, M.; Vernon, M. Nondestructive measurement of fresh tomato lycopene content and other physicochemical characteristics using visible—NIR spectroscopy. J. Agric. Food Chem. 2008, 56, 9813–9818. [Google Scholar] [CrossRef]
- Akinaga, T.; Tanaka, M.; Kawasaki, S. On-tree and after-harvesting evaluation of firmness, color and lycopene content of tomato fruit using portable NIR spectroscopy. J. Food Agric. Environ. 2008, 6, 327–332. [Google Scholar]
- Huang, Y.; Lu, R.; Chen, K. Assessment of tomato soluble solids content and pH by spatially-resolved and conventional Vis/NIR spectroscopy. J. Food Eng. 2018, 236, 19–28. [Google Scholar] [CrossRef]
- Ntagkas, N.; Min, Q.; Woltering, E.J.; Labrie, C.; Nicole, C.C.S.; Marcelis, L.F.M. Illuminating tomato fruit enhances fruit vitamin C content. In Proceedings of the VIII International Symposium on Light in Horticulture 1134, East Lansing, MI, USA, 22–26 May 2016; pp. 351–356. [Google Scholar]
- Farneti, B.; Schouten, R.E.; Woltering, E.J. Low temperature-induced lycopene degradation in red ripe tomato evaluated by remittance spectroscopy. Postharvest Biol. Technol. 2012, 73, 22–27. [Google Scholar] [CrossRef]
- Farinetti, A.; Zurlo, V.; Manenti, A.; Coppi, F.; Mattioli, A.V. Mediterranean diet and colorectal cancer: A systematic review. Nutrition 2017, 43, 83–88. [Google Scholar] [CrossRef]
- Chandrasekaran, I.; Panigrahi, S.S.; Ravikanth, L.; Singh, C.B. Potential of Near-Infrared (NIR) spectroscopy and hyperspectral imaging for quality and safety assessment of fruits: An overview. Food Anal. Methods 2019, 12, 2438–2458. [Google Scholar] [CrossRef]
- Paponov, M.; Kechasov, D.; Lacek, J.; Verheul, M.; Paponov, I.A. Supplemental LED inter-lighting increases tomato fruit growth through enhanced photosynthetic light use efficiency and modulated root activity. Front. Plant Sci. 2020, 10, 1656. [Google Scholar] [CrossRef]
- Cantwell, M. Optimum procedures for ripening tomatoes. Manag. Fruit Ripening Postharvest Hortic. Ser. 2000, 9, 80–88. [Google Scholar]
- Behmann, J.; Acebron, K.; Emin, D.; Bennertz, S.; Matsubara, S.; Thomas, S.; Bohnenkamp, D.; Kuska, M.; Jussila, J.; Salo, H. Specim IQ: Evaluation of a new, miniaturized handheld hyperspectral camera and its application for plant phenotyping and disease detection. Sensors 2018, 18, 441. [Google Scholar] [CrossRef]
- Mitcham, B.; Cantwell, M.; Kader, A. Methods for determining quality of fresh commodities. Perishables Handl. Newsl. 1996, 85, 1–5. [Google Scholar]
- Verheul, M.J.; Slimestad, R.; Tjøstheim, I.H. From producer to consumer: Greenhouse tomato quality as affected by variety, maturity stage at harvest, transport conditions, and supermarket storage. J. Agric. Food Chem. 2015, 63, 5026–5034. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, K.; Guo, W.; Yoshioka, Y.; Ninomiya, S. On plant detection of intact tomato fruits using image analysis and machine learning methods. Sensors 2014, 14, 12191–12206. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE conference on computer vision and pattern recognition, Las Vegas, NV, USA, 26 June–1 July 2016; pp. 770–778. [Google Scholar]
- Renza, D.; Martinez, E.; Molina, I. Unsupervised change detection in a particular vegetation land cover type using spectral angle mapper. Adv. Sp. Res. 2017, 59, 2019–2031. [Google Scholar] [CrossRef]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Millard, S.P.; Kowarik, M.A.; Imports, M. Package ‘EnvStats’. Packag. Environ. Stat. Version 2018, 2, 31–32. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013. [Google Scholar]
- Russell, B.C.; Torralba, A.; Murphy, K.P.; Freeman, W.T. LabelMe: A database and web-based tool for image annotation. Int. J. Comput. Vis. 2008, 77, 157–173. [Google Scholar] [CrossRef]
- Lin, T.-Y.; Goyal, P.; Girshick, R.; He, K.; Dollár, P. Focal loss for dense object detection. In Proceedings of the IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 2999–3007. [Google Scholar]
- Esquerre, C.; Gowen, A.A.; Burger, J.; Downey, G.; O’Donnell, C.P. Suppressing sample morphology effects in near infrared spectral imaging using chemometric data pre-treatments. Chemom. Intell. Lab. Syst. 2012, 117, 129–137. [Google Scholar] [CrossRef]
- Liu, W.; Lee, J. An Efficient Residual Learning Neural Network for Hyperspectral Image Superresolution. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2019, 12, 1240–1253. [Google Scholar] [CrossRef]
- Martin-Diaz, I.; Morinigo-Sotelo, D.; Duque-Perez, O.; de J. Romero-Troncoso, R. Early fault detection in induction motors using AdaBoost with imbalanced small data and optimized sampling. IEEE Trans. Ind. Appl. 2016, 53, 3066–3075. [Google Scholar] [CrossRef]
- Kozukue, N.; Friedman, M. Tomatine, chlorophyll, β-carotene and lycopene content in tomatoes during growth and maturation. J. Sci. Food Agric. 2003, 83, 195–200. [Google Scholar] [CrossRef]
- Schouten, R.E.; Farneti, B.; Tijskens, L.M.M.; Alarcón, A.A.; Woltering, E.J. Quantifying lycopene synthesis and chlorophyll breakdown in tomato fruit using remittance VIS spectroscopy. Postharvest Biol. Technol. 2014, 96, 53–63. [Google Scholar] [CrossRef]
- Singh, L.; Mutanga, O.; Mafongoya, P.; Peerbhay, K. Remote sensing of key grassland nutrients using hyperspectral techniques in KwaZulu-Natal, South Africa. J. Appl. Remote Sens. 2017, 11, 36005. [Google Scholar] [CrossRef]
- Kawamura, K.; Tsujimoto, Y.; Rabenarivo, M.; Asai, H.; Andriamananjara, A.; Rakotoson, T. Vis-NIR spectroscopy and PLS regression with waveband selection for estimating the total C and N of paddy soils in Madagascar. Remote Sens. 2017, 9, 1081. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).