A Review of Advanced Technologies and Development for Hyperspectral-Based Plant Disease Detection in the Past Three Decades
Abstract
1. Background
2. Complex Presence of Pathogens and Plant–Pathogen Interactions Make Hyperspectral Technologies Indispensable
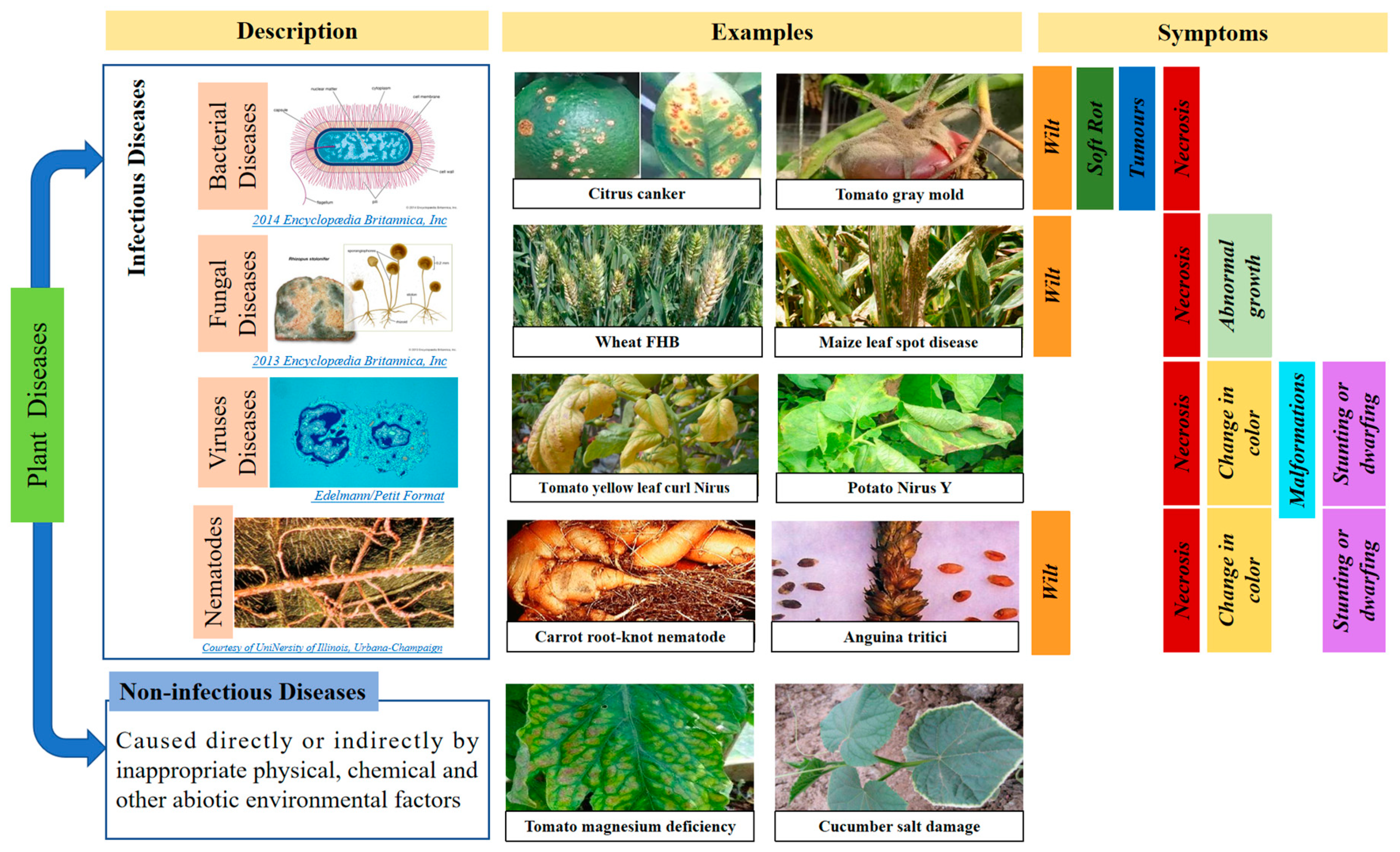
2.1. Plant Diseases May Be Caused by More Than One Causal Agent and Different Agents May Have the Same Symptoms
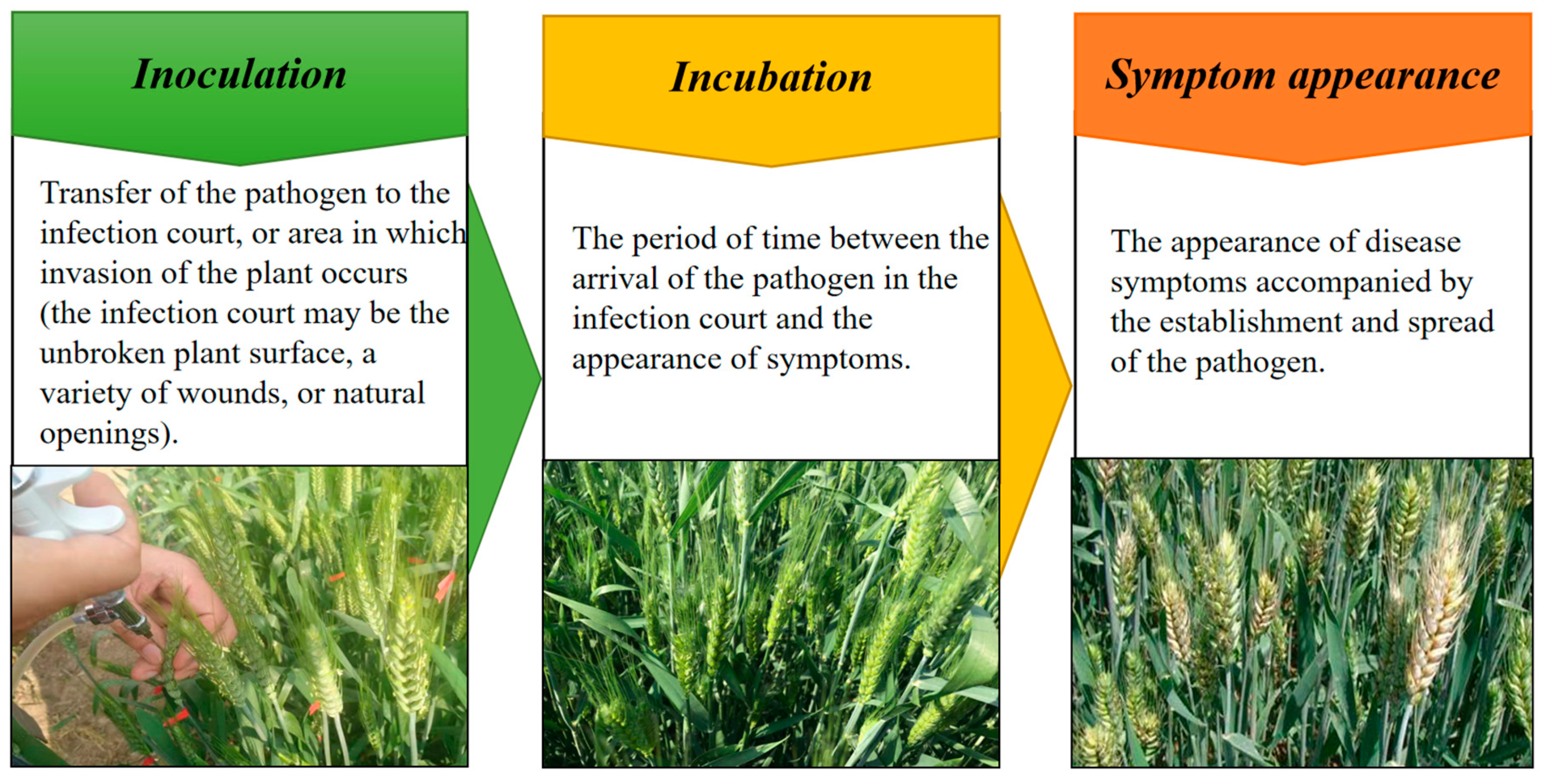
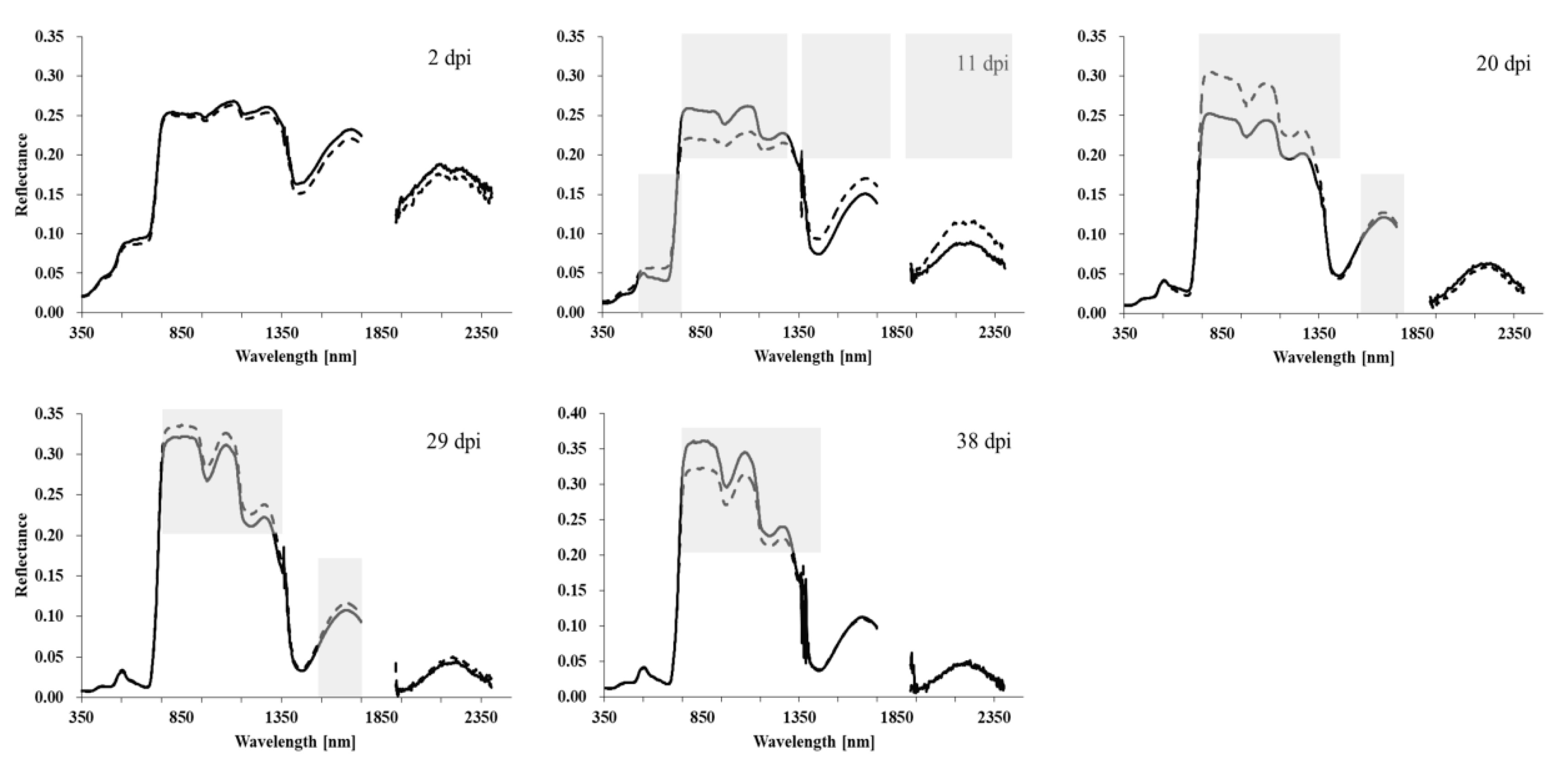
2.2. Host Plant–Pathogen Interaction Is a Complex Dynamic Process with Changes of Various Physiological and Biochemical Parameters
2.3. Hyperspectral Technology Has Its Specific Necessity in Plant Disease Detection
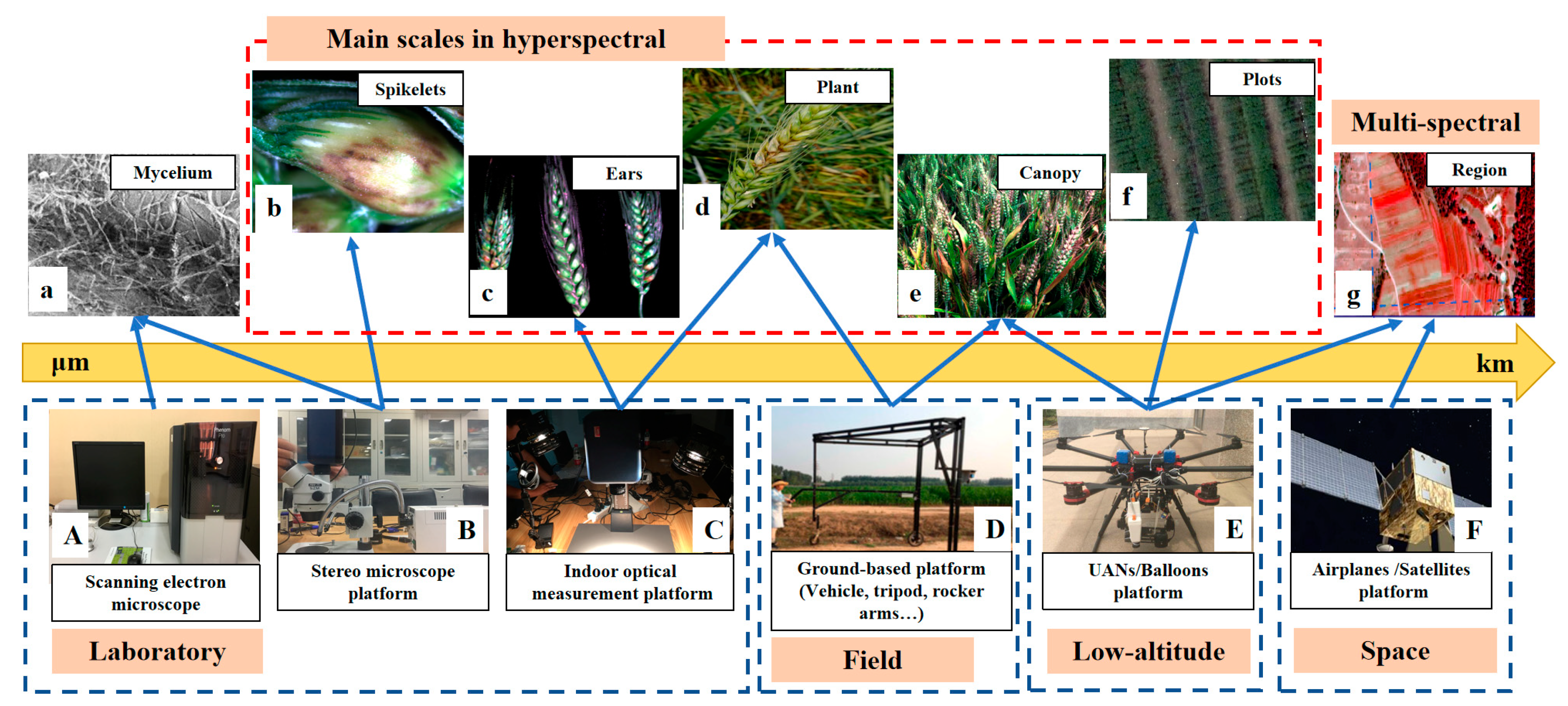
2.4. Applicable Hyperspectral Sensors and Platforms Are Different for Different Pathogens with Different Symptoms
3. Main Hyperspectral Technologies for Plant Disease Analysis: Choosing Suitable Methods to Achieve Target Details
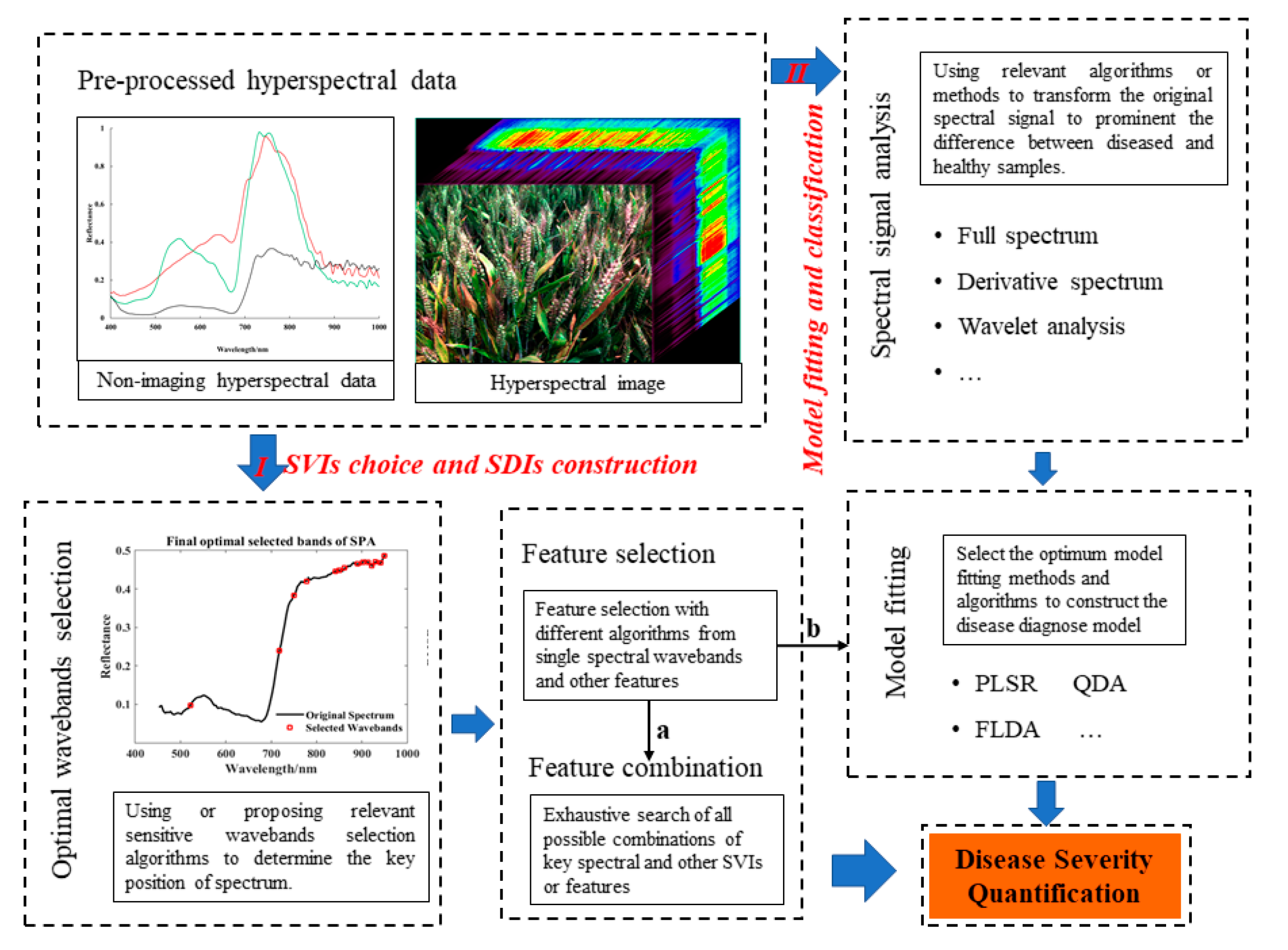
3.1. Choosing a Suitable Data Measurement System Is a Prerequisite for Obtaining Accurate Results
3.2. Complete and Appropriate Pre-Processing Guarantees Accurate Results
3.3. Special Hyperspectral Technologies and Frameworks for Different Plant Disease Analysis Directions
3.3.1. Detection Is One of the Earliest, Basic but Important Applications in Hyperspectral-Based Plant Disease
3.3.2. Diseases Classification Is the Attempt to Identify and Label the Pathogens Affecting the Plant Simultaneously
3.3.3. Quantitative Diagnosis of Plant Disease Severity is the Main Direction of Hyperspectral Disease Analysis
4. Discussion and Prospect
4.1. Identification of Different Pathogens and Discrimination of Biotic and Abiotic Stresses Are Always the Primary Challenges in the Disease Research Field
- Extension to smaller scales and higher spatial resolutions for pathogen identification. Nowadays, the spectral resolution of hyperspectral technologies can reach 1 nm or higher, which makes hyperspectral data more sensitive to the subtle differences caused by different pathogens. However, the mixed pixel problems of lower spatial resolution data are more complex with improved spectral resolution. The effects of the atmosphere, light source, background, etc. are also relatively complex under lower spatial resolution. Thus, increasingly, many researchers have extended the spatial resolution to the sub-cell level by microscopy. In the smallest scale case, the characteristics of the spectral signature of each pathogen are exactly matched, which makes it easy to accurately determine the spectral difference between pathogens on the same plant. Furthermore, the development of UAV and other aviation facilities effectively improves the flexibility of data acquisition. From another perspective, it is possible to rely upon the pathogen changes in the plant–pathogen interaction process and the bioecological characteristics of different pathogens to acquire hyperspectral data at different times to achieve effective pathogen division.
- Accounting for auxiliary data to realize discrimination of biotic and abiotic stresses. Almost all infectious plant diseases (biotic stresses) only appear on individual plants in the early stage, and they usually present point distributions before large-scale outbreaks. However, abiotic stresses, including both nutrient stresses and meteorological disasters, occur across wide ranges, and there is no extension process. Biotic stresses show the inhomogeneity of spectral characteristics, correlation indices, and features in hyperspectral images, while abiotic stresses have relatively even distributions. Thus, biotic and abiotic stresses can be discriminated based on their symptom distributions. Nevertheless, the combination with meteorological, soil, and field management data in the early stage and at the field or relatively large scales is necessary for the implementation of discrimination. Once an infectious disease breaks out, it is necessary to coordinate changes in meteorological data in the process of disease development with spatial distribution analysis.
4.2. Plant Disease Early Warning Is the Key Point of Applying RS Technologies to Field Work
- Early-stage detection of plant diseases with multi-source data at the field scale. In recent years, many websites and mobile applications related to agricultural consultation and assistance have increasingly provided crop disease detection and pesticide application guidance. The detection and identification of each pathogen are mainly based on image information recognition by big data analysis; thus, these analyses are always performed after the symptoms have appeared. However, starting from the actual situation of agricultural production, the most important and useful detection should be in the incubation or sporadic occurrence period. Various RS systems are available that could potentially be applied to detect and monitor plant diseases such as VIS-SWIR spectral systems, fluorescence and thermal systems, synthetic aperture radar (SAR), light detection and ranging (LIDAR) systems, and even gamma rays, X-rays, and ultraviolet rays. Each system has its advantages and disadvantages in plant disease detection. Zhang et al. [37] reviewed the characteristics and potential of each system in plant disease detection. The VIS-SWIR system has stable performance with respect to pigment changes and is always used after symptom occurrence but performs poorly in early stage detection. Fluorescence and thermal systems have considerable potential to capture pre-symptom physiological changes but are not suitable for large-scale analysis. The SAR and LIDAR systems are more suitable for structural change analysis. It is not difficult to find that if these different RS systems can be used together, they can achieve complementary advantages, then achieve a plant disease incubation period detection at the field scale.
- Early warning of plant diseases on the regional or larger scale. The research conducted by the National Center for Atmospheric Research (NCAR) of America has shown that the land surface temperature exhibits a rising trend because of the increase in greenhouse gas emissions and destruction of the ozone sphere, and this rising trend has intensified since the 1970s [140]. Globalization and human activities promote the rapid spread and distribution of plant pathogens, and the globalization climate changes indirectly influence disease occurrence and plant distributions. However, although there has been some research on plant disease warning under climate changes, most studies are focused on niche simulations based on independent time points at small scales [141,142]. For instance, the Intergovernmental Panel on Climate Change provides regular scientific assessments on climate change, implications, and potential future risks for policymakers, as well as putting forward adaptation and mitigation options; hence, plant disease early warning should also be developed in this direction. Therefore, long time-series climate changes and plant disease parameters cannot be ignored. On this premise, the occurrence and development regularities of each disease can be summarized and founded in more detailed and accurate. With large-scale RS image data, expert or prognosis systems based on regional weather data and epidemiological parameters of plant diseases can be utilized to forecast the temporal and spatial spread of diseases in specific growing regions.
4.3. Spaceborne Hyperspectral Technology Requires Synchronous Development of Basic Research and Joint Spaceborne–Airborne–Ground Applications
- Joint application of existing mature technologies. The first airborne-based hyperspectral imaging sensor AIS was developed in 1983 by JPL/NASA. Since then, numerous airborne hyperspectral imaging technologies have been developed successively, including AVIRIS (JPL), the Fluorescence Line Imager (Moniteq Ltd. and Itres Research Ltd. for Canadian Department of Fisheries and Oceans), the Compact Airborne Spectrographic Imager (Itres Research Ltd. of Calgary, Alberta, Canada), the Hyperspectral Mapper (Australian Integrated Spectronics Ltd.), and many others. After the success of airborne hyperspectral technologies, satellite-based hyperspectral technology was continuously developed in the late 1990s. Although the first HSI on the Lewis satellite of NASA failed to work properly after it was put into orbit on 23 August 1997, it has also become the beginning of satellite-based hyperspectral technology. Since then, the Fourier Transform Hyperspectral Imager on MightiSat Ⅱ, Hyperion on EO-1, HJ-1A/HSI on HJ-1A, and AHSI on GF-5 have succeeded. All of these types of airborne and spaceborne hyperspectral images have been widely but separately applied in plant RS monitoring, but they are rare in crop disease detection. Nowadays, ground- and UAV-based hyperspectral images form the relevant perfect systems for plant disease detection on small scales. However, these must be combined with airborne or spaceborne hyperspectral technologies to extend the application range and scale. Ground-based hyperspectral images have the advantages of unmixed pixels, flexible and high spatial resolution. Thus, it greatly improves the accuracy of hyperspectral analysis of specific diseases. These characteristics are complementary to those of airborne and spaceborne data, which have lower spatial and time resolutions but higher widths. Thus, the joint application of ground-, airborne-, and spaceborne-based hyperspectral technologies in plant disease analysis is the development trend of hyperspectral technology practices.
- Establishment of a comprehensive spectrum library of plant diseases. Through the above analysis of different pathogens, advantages and limitations of different scales, and hyperspectral technologies for different plant disease analyses, it can easily be seen that the most significant aspect of plant disease detection by RS is the accuracy of the hyperspectral signature of each pathogen. JPL/NASA has established abundant spectrum datasets for plants, minerals, snow, ice, and other objects. These spectrum libraries contain three sub-libraries: laboratory spectrum library, ground spectrum library, and the hyperspectral remote sensing spectrum library. However, there is no unified standard spectral library for crop diseases. Considering the actual application requirements, the establishment of a comprehensive spectrum library of the global main crop disease is anticipated. To meet the needs of integrated spaceborne–airborne–ground analysis, the spectrum library should include at least three scales: ground, airborne, and spaceborne. Perfect spectrum libraries can provide significant references in practical applications and provide the basis for new and targeted hyperspectral technology.
- Implementation of targeted spaceborne hyperspectral missions and expansion of its scope of commercialization. Although there are some existing spaceborne hyperspectral sensors, and most of them can be used to monitoring vegetation changes, fewer are clearly focused on vegetation. The HyperSpectral Imager on the IMS-1 satellite of India, which operates in the VNIR spectral range from 450 to 950 nm with a total of 64 spectral bands at a spectral resolution of 8 nm, is specific to the vegetation type measurement and resource characterization. Meanwhile, the HSI of HJ-1A in China is focused on environment and disaster monitoring with 115 bands from 450 to 950 nm. Even so, most spaceborne hyperspectral sensors are non-commercial, significantly limiting their large-scale industrial applications. In recent years, facing the frequent global climate change and disasters, increasingly more countries and organizations have proposed the special hyperspectral RS missions and speed up these trends. The 5 m optical service satellite (ZY-1 02D) equipped with one hyperspectral sensor and one multispectral sensor was put into the predetermined orbit in 2019 and can effectively obtain nine-band multispectral data of 115 km width and 166-band hyperspectral data of 60 km width. This is the first civil hyperspectral service satellite in China and could provide services for precision agriculture in the future. Furthermore, the European Space Agency selected the Fluorescence EXplorer mission proposed for the global monitoring of steady-state chlorophyll fluorescence in terrestrial vegetation, which will operate in a three-instrument array for measurement of the interrelated features of fluorescence, hyperspectral reflectance, and canopy temperature. The HyspIRI mission, which is being developed by JPL/NASA, USA, is planned to be launched in 2021. The equipped VIR-SWIR and thermal infrared sensors will be utilized to study ecosystems worldwide; provide critical information on natural disasters such as volcanoes, wildfires, and drought; and may be useful for plant disease early warning. Furthermore, the successful launch of the Environmental Mapping and Analysis Program (EnMAP) in Germany, Hyperspectral Imager Suite (HISUI) in Japan, and Hyperspectral Precursor and Application Mission (PRISMA) of the Italian Space Agency can be used for vegetation status detection, product development for agricultural areas, and the management and monitoring of natural and induced hazards in the future.
5. Conclusions
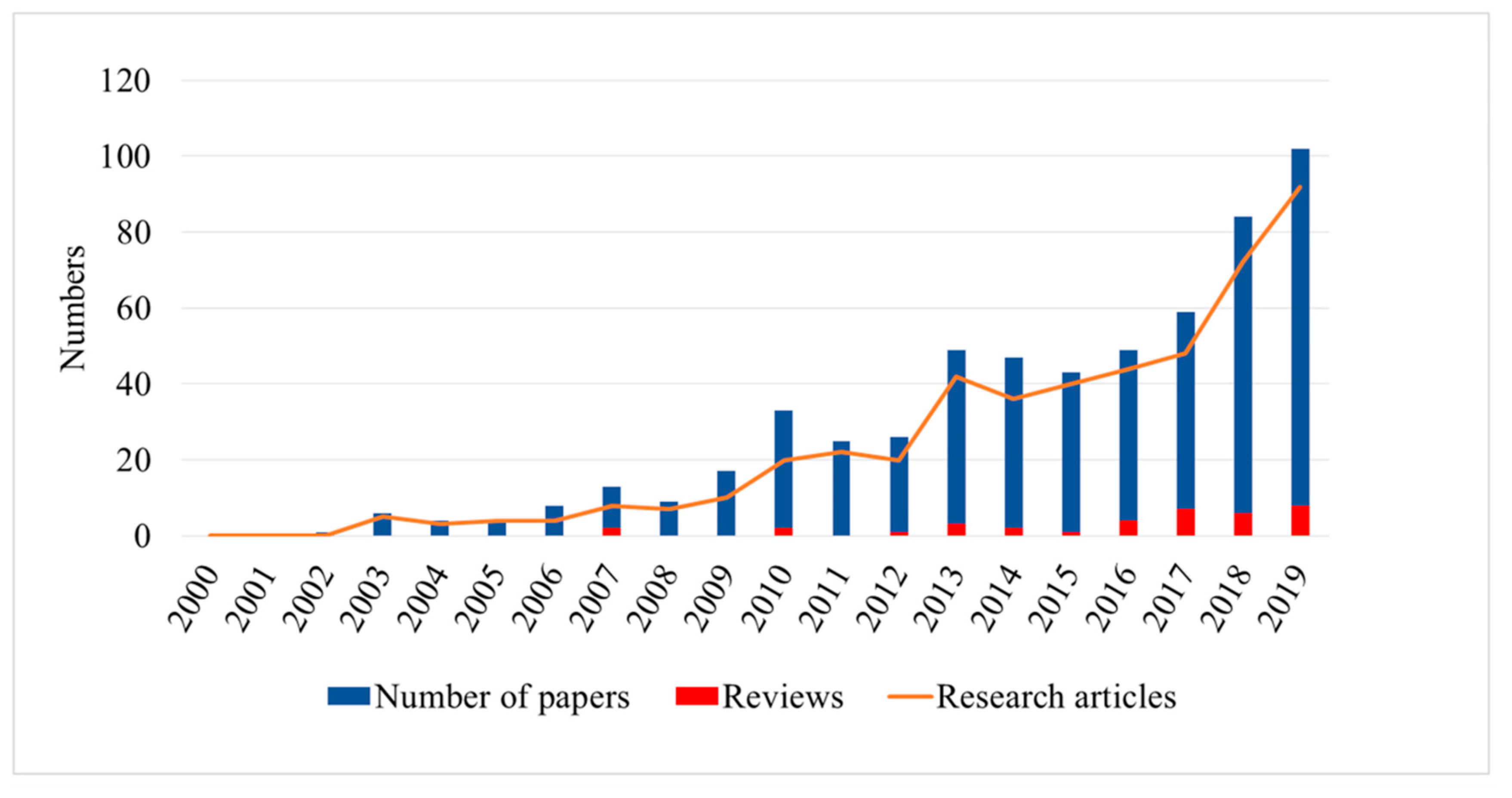
- Hyperspectral technology-based plant disease detection is drawing increasing attention. As shown in Figure 1, hyperspectral-based plant disease analysis technology emerged in 2002 and has been developing rapidly in the following 10 years. It has been developing continuously with the maturity of related technologies in the past 10 years. These developments provide many methods and ideas for future research and analysis, as well as reliable support for plant protection.
- The mainstream technologies are focused on small scales, and satellite payloads require further development and attention. In the past three decades, almost 86% of hyperspectral imaging research has been focused on field and laboratory environments and more concern has been placed on the leaf and canopy scales. However, large-scale accurate analysis is necessary for practical applications. Thus, scale transformation methods for both the spatial and spectral scales require more attention. Although the algorithms for hyperspectral data analysis on small scales can provide technical support for regional or larger scales, it is difficult to achieve large-scale monitoring without the assistance of satellite payload.
- Close attention should be paid to the information integration analysis of satellite scales. After the implementation of targeted hyperspectral satellite missions, big data collection, pre-processing, and analysis will be the priorities. The real-time dynamic monitoring of plant disease at the regional, national, and global scales can be realized only if large-scale data integration analysis is achieved. With the development of multi-source RS data, the fusion of multi-source data may be a development trend in the future.
Author Contributions
Funding
Conflicts of Interest
References
- Moshou, D.; Pantazi, X.E.; Kateris, D.; Gravalos, I. Water stress detection based on optical multisensor fusion with a least squares support vector machine classifier. Biosyst. Eng. 2014, 117, 15–22. [Google Scholar] [CrossRef]
- FAO. New Standards to Curb the Global Spread of Plant Pests and Diseases. Available online: http://www.fao.org/news/story/en/item/1187738/icode/ (accessed on 3 April 2019).
- FAO. Declaration of the World Summit on Food Security; Food and Agriculture Organization: Rome, Italy, 2009. [Google Scholar]
- Carvajal-Yepes, M.; Cardwell, K.; Nelson, A.; Garrett, K.A.; Giovani, B.; Saunders, D.G.O.; Kamoun, S.; Legg, J.P.; Verdier, V.; Lessel, J.; et al. A global surveillance system for crop diseases. Science 2019, 364, 1237–1239. [Google Scholar] [CrossRef]
- Nganje, W.E.; Bangsund, D.A.; Leistritz, F.L.; Wilson, W.W.; Tiapo, N.M. Regional economic impacts of Fusarium head blight in wheat and barley. Rev. Agric. Econ. 2004, 26, 332–347. [Google Scholar] [CrossRef]
- Barbedo, J.G.A. A review on the main challenges in automatic plant disease identification based on visible range images. Biosyst. Eng. 2016, 144, 52–60. [Google Scholar] [CrossRef]
- Ruiz-Ruiz, S.; Ambrós, S.; Vives, M.D.C.; Navarro, L.; Moreno, P.; Guerri, J. Detection and quantitation of Citrus leaf blotch virus by TaqMan real-time RT-PCR. J. Virol. Methods 2009, 160, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Sankaran, S.; Mishra, A.; Ehsani, R.; Davis, C. A review of advanced techniques for detecting plant diseases. Comput. Electron. Agric. 2010, 72, 1–12. [Google Scholar] [CrossRef]
- Yvon, M.; Thébaud, G.; Alary, R.; Labonne, G. Specific detection and quantification of the phytopathogenic agent ‘Candidatus Phytoplasma prunorum’. Mol. Cell. Probes 2009, 23, 227–234. [Google Scholar] [CrossRef]
- Ali, M.M.; Bachik, N.A.; Muhadi, N.A.; Yusof, T.N.T.; Gomes, C. Non-destructive techniques of detecting plant diseases: A review. Physiol. Mol. Plant Pathol. 2019, 108, 101426–101437. [Google Scholar] [CrossRef]
- Golhani, K.; Balasundram, S.K.; Vadamalai, G.; Pradhan, B. A review of neural networks in plant disease detection using hyperspectral data. Inf. Process. Agric. 2018, 5, 354–371. [Google Scholar] [CrossRef]
- Purcell, D.E.; O’Shea, M.G.; Johnson, R.A.; Kokot, S. Near-infrared spectroscopy for the prediction of disease ratings for Fiji leaf gall in sugarcane clones. Appl. Spectrosc. 2009, 63, 450–457. [Google Scholar] [CrossRef]
- Shrestha, S.; Deleuran, L.C.; Gislum, R. Classification of different tomato seed cultivars by multispectral visible-near infrared spectroscopy and chemometrics. J. Spectr. Imaging 2016, 5, 1–8. [Google Scholar] [CrossRef]
- Borges, E.; Matos, A.P.; Cardoso, J.M.; Correia, C.; Vasconcelos, T.; Gomes, N. Early detection and monitoring of plant diseases by Bioelectric Impedance Spectroscopy. In Proceedings of the 2012 IEEE 2nd Portuguese Meeting in Bioengineering (ENBENG), Coimbra, Portugal, 23–25 February 2012; pp. 1–4. [Google Scholar]
- Belasque, J., Jr.; Gasparoto, M.C.G.; Marcassa, L.G. Detection of mechanical and disease stresses in citrus plants by fluorescence spectroscopy. Appl. Opt. 2008, 47, 1922–1926. [Google Scholar] [CrossRef] [PubMed]
- Dhingra, G.; Kumar, V.; Joshi, H.D. Study of digital image processing techniques for leaf disease detection and classification. Multimed. Tools Appl. 2017, 77, 19951–20000. [Google Scholar] [CrossRef]
- Barbedo, J.G.A. Digital image processing techniques for detecting, quantifying and classifying plant diseases. Springerplus 2013, 2, 660–671. [Google Scholar] [CrossRef]
- Cen, H.; Weng, H.; Yao, J.; He, M.; Lv, J.; Hua, S.; Li, H.; He, Y. Chlorophyll fluorescence imaging uncovers photosynthetic fingerprint of citrus Huanglongbing. Front. Plant Sci. 2017, 8, 1509–1519. [Google Scholar] [CrossRef]
- Raza, S.-e.-A.; Prince, G.; Clarkson, J.P.; Rajpoot, N.M. Automatic detection of diseased tomato plants using thermal and stereo visible light images. PLoS ONE 2015, 10, e0123262–e0123281. [Google Scholar] [CrossRef]
- Sankaran, S.; Maja, J.M.; Buchanon, S.; Ehsani, R. Huanglongbing (citrus greening) detection using visible, near infrared and thermal imaging techniques. Sensors 2013, 13, 2117–2130. [Google Scholar] [CrossRef]
- Söderström, M.; Börjesson, T.; Roland, B.; Stadig, H. Modelling within-field variations in deoxynivalenol (DON) content in oats using proximal and remote sensing. Precis. Agric. 2014, 16, 1–14. [Google Scholar] [CrossRef]
- Wahabzada, M.; Mahlein, A.-K.; Bauckhage, C.; Steiner, U.; Oerke, E.-C.; Kersting, K. Plant phenotyping using probabilistic topic models: Uncovering the hyperspectral language of plants. Sci. Rep. 2016, 6, 22482–22492. [Google Scholar] [CrossRef]
- López-López, M.; Calderón, R.; González-Dugo, V.; Zarco-Tejada, P.; Fereres, E. Early detection and quantification of almond red leaf blotch using high-resolution hyperspectral and thermal imagery. Remote Sens. 2016, 8, 276. [Google Scholar] [CrossRef]
- Mahlein, A.-K.; Hammersley, S.; Oerke, E.-C.; Dehne, H.-W.; Goldbach, H.; Grieve, B. Supplemental blue LED lighting array to improve the signal quality in hyperspectral imaging of plants. Sensors 2015, 15, 12834–12840. [Google Scholar] [CrossRef]
- Al-Saddik, H.; Simon, J.-C.; Cointault, F. Development of spectral disease indices for ‘Flavescence Doree’ grapevine disease identification. Sensors 2017, 17, 2772. [Google Scholar] [CrossRef] [PubMed]
- Ghamisi, P.; Plaza, J.; Chen, Y.; Li, J.; Plaza, A. Advanced Supervised Spectral Classifiers for Hyperspectral Images A review. IEEE Geosci. Remote Sens. Mag. 2017, 5, 8–32. [Google Scholar] [CrossRef]
- Zhou, W.; Zhang, J.; Zou, M.; Liu, X.; Du, X.; Wang, Q.; Liu, Y.; Liu, Y.; Li, J. Prediction of cadmium concentration in brown rice before harvest by hyperspectral remote sensing. Environ. Sci. Pollut. Res. Int. 2019, 26, 1848–1856. [Google Scholar] [CrossRef] [PubMed]
- Thomas, S.; Kuska, M.T.; Bohnenkamp, D.; Brugger, A.; Alisaac, E.; Wahabzada, M.; Behmann, J.; Mahlein, A.-K. Benefits of hyperspectral imaging for plant disease detection and plant protection: A technical perspective. J. Plant Dis. Prot. 2017, 125, 5–20. [Google Scholar] [CrossRef]
- Virlet, N.; Sabermanesh, K.; Sadeghi-Tehran, P.; Hawkesford, M.J. Field Scanalyzer: An automated robotic field phenotyping platform for detailed crop monitoring. Funct. Plant Biol. 2017, 44, 143–153. [Google Scholar] [CrossRef] [PubMed]
- Bock, C.H.; Poole, G.H.; Parker, P.E.; Gottwald, T.R. Plant disease severity estimated visually, by digital photography and image analysis, and by hyperspectral imaging. Crit. Rev. Plant Sci. 2010, 29, 59–107. [Google Scholar] [CrossRef]
- Lee, W.S.; Alchanatis, V.; Yang, C.; Hirafuji, M.; Moshou, D.; Li, C. Sensing technologies for precision specialty crop production. Comput. Electron. Agric. 2010, 74, 2–33. [Google Scholar] [CrossRef]
- West, J.S.; Bravo, C.; Oberti, R.; Lemaire, D.; Moshou, D.; McCartney, H.A. The potential of optical canopy measurement for targeted control of field crop diseases. Annu. Rev. Phytopathol. 2003, 41, 593–614. [Google Scholar] [CrossRef]
- Bauriegel, E.; Herppich, W. Hyperspectral and chlorophyll fluorescence imaging for early detection of plant diseases, with special reference to Fusarium spec. infections on wheat. Agriculture 2014, 4, 32–57. [Google Scholar] [CrossRef]
- Baranowski, P.; Jedryczka, M.; Mazurek, W.; Babula-Skowronska, D.; Siedliska, A.; Kaczmarek, J. Hyperspectral and thermal imaging of oilseed rape (Brassica napus) response to fungal species of the genus Alternaria. PLoS ONE 2015, 10, e0012313–e0122913. [Google Scholar] [CrossRef] [PubMed]
- Graeff, S.; Link, J.; Claupein, W. Identification of powdery mildew (Erysiphe graminis sp. tritici) and take-all disease (Gaeumannomyces graminis sp. tritici) in wheat (Triticum aestivum L.) by means of leaf reflectance measurements. Open Life Sci. 2006. [Google Scholar] [CrossRef]
- Shurtleff, M.C.; Pelczar, M.J.; Kelman, A.; Pelczar, R.M. Plant disease. In Plant Pathology; Encyclopedia Britannica: Chicago, IL, USA, 2020; Available online: https://www.britannica.com/science/plant-disease (accessed on 1 April 2020).
- Zhang, J.; Huang, Y.; Pu, R.; Gonzalez-Moreno, P.; Yuan, L.; Wu, K.; Huang, W. Monitoring plant diseases and pests through remote sensing technology: A review. Comput. Electron. Agric. 2019, 165, 104943–104956. [Google Scholar] [CrossRef]
- Boyd, L.A.; Christopher, R.; O’Sullivan, D.M.; Leach, J.E.; Hei, L. Plant-pathogen interactions: Disease resistance in modern agriculture. Trends Genet. 2013, 29, 233–240. [Google Scholar] [CrossRef] [PubMed]
- Mahlein, A.-K.; Kuska, M.T.; Behmann, J.; Polder, G.; Walter, A. Hyperspectral sensors and imaging technologies in phytopathology: State of the art. Annu. Rev. Phytopathol. 2018, 56, 535–558. [Google Scholar] [CrossRef]
- Pinzón, A.; Barreto, E.; Bernal, A.; Achenie, L.; González Barrios, A.F.; Isea, R.; Restrepo, S. Computational models in plant-pathogen interactions: The case of Phytophthora infestans. Theor. Biol. Med. Model. 2009, 6, 24–34. [Google Scholar] [CrossRef]
- Jiang, Z. Large-Scale Transcriptional Data Analyses of Plant Immune Responses. Ph.D. Thesis, China Agricultural University, Beijing, China, 2017. [Google Scholar]
- Mahlein, A.-K.; Alisaac, E.; Masri, A.A.; Behmann, J.; Dehne, H.-W.; Oerke, E.-C. Comparison and combination of thermal, fluorescence and hyperspectral imaging for monitoring Fusarium head blight of wheat on spikelet scale. Sensors 2019, 19, 2281. [Google Scholar] [CrossRef]
- Yuan, L.; Huang, Y.; Loraamm, R.W.; Nie, C.; Wang, J.; Zhang, J. Spectral analysis of winter wheat leaves for detection and differentiation of diseases and insects. Field Crop. Res. 2014, 156, 199–207. [Google Scholar] [CrossRef]
- Oerke, E.-C.; Herzog, K.; Toepfer, R. Hyperspectral phenotyping of the reaction of grapevine genotypes to Plasmopara viticola. J. Exp. Bot. 2016, 67, 5529–5543. [Google Scholar] [CrossRef]
- Polder, G.; Blok, P.M.; de Villiers, H.A.C.; van der Wolf, J.M.; Kamp, J. Potato virus Y detection in seed potatoes using deep learning on hyperspectral images. Front. Plant Sci. 2019, 10, 209–221. [Google Scholar] [CrossRef]
- Jin, X.; Jie, L.; Wang, S.; Qi, H.; Li, S. Classifying wheat hyperspectral pixels of healthy heads and Fusarium head blight disease using a deep neural network in the wild field. Remote Sens. 2018, 10, 395. [Google Scholar] [CrossRef]
- Chen, D.; Shi, Y.; Huang, W.; Zhang, J.; Wu, K. Mapping wheat rust based on high spatial resolution satellite imagery. Comput. Electron. Agric. 2018, 152, 109–116. [Google Scholar] [CrossRef]
- Alisaac, E.; Behmann, J.; Kuska, M.T.; Dehne, H.-W.; Mahlein, A.-K. Hyperspectral quantification of wheat resistance to Fusarium head blight: Comparison of two Fusarium species. Eur. J. Plant Pathol. 2018, 152, 869–884. [Google Scholar] [CrossRef]
- Ashourloo, D.; Aghighi, H.; Matkan, A.A.; Mobasheri, M.R.; Rad, A.M. An Investigation Into Machine Learning Regression Techniques for the Leaf Rust Disease Detection Using Hyperspectral Measurement. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2016, 9, 4344–4351. [Google Scholar] [CrossRef]
- Ashourloo, D.; Mobasheri, M.; Huete, A. Developing Two Spectral Disease Indices for Detection of Wheat Leaf Rust (Pucciniatriticina). Remote Sens. 2014, 6, 4723–4740. [Google Scholar] [CrossRef]
- Huang, W.; Lu, J.; Ye, H.; Kong, W.; Mortimer, A.H.; Shi, Y. Quantitative identification of crop disease and nitrogen-water stress in winter wheat using continuous wavelet analysis. Int. J. Agric. Biol. Eng. 2018, 11, 145–152. [Google Scholar] [CrossRef]
- Zhu, M.; Yang, H.; Li, Z. Early Detection and Identification of Rice Sheath Blight Disease Based on Hyperspectral Image and Chlorophyll Content. Spectrosc Spectr. Anal. 2019, 39, 1898–1904. [Google Scholar] [CrossRef]
- Huang, S.; Qi, L.; Ma, X.; Xue, K.; Wang, W.; Zhu, X. Hyperspectral image analysis based on BoSW model for rice panicle blast grading. Comput. Electron. Agric. 2015, 118, 167–178. [Google Scholar] [CrossRef]
- Dhau, I.; Adam, E.; Mutanga, O.; Ayisi, K.; Abdel-Rahman, E.M.; Odindi, J.; Masocha, M. Testing the capability of spectral resolution of the new multispectral sensors on detecting the severity of grey leaf spot disease in maize crop. Geocarto Int. 2017, 33, 1223–1236. [Google Scholar] [CrossRef]
- Adam, E.; Deng, H.; Odindi, J.; Abdel-Rahman, E.M.; Mutanga, O. Detecting the early stage of Phaeosphaeria leaf spot infestations in maize crop using in situ hyperspectral data and guided regularized random forest algorithm. J. Spectrosc. 2017, 2017, 1–8. [Google Scholar] [CrossRef]
- Williams, P.J.; Bezuidenhout, C.; Rose, L.J. Differentiation of maize ear rot pathogens, on growth media, with near infrared hyperspectral imaging. Food Anal. Methods 2019, 12, 1556–1570. [Google Scholar] [CrossRef]
- Nagasubramanian, K.; Jones, S.; Sarkar, S.; Singh, A.K.; Singh, A.; Ganapathysubramanian, B. Hyperspectral band selection using genetic algorithm and support vector machines for early identification of charcoal rot disease in soybean stems. Plant Methods 2018, 14, 86–92. [Google Scholar] [CrossRef] [PubMed]
- Gazala, I.F.S.; Sahoo, R.N.; Pandey, R.; Mandal, B.; Gupta, V.K.; Singh, R.; Sinha, P. Spectral reflectance pattern in soybean for assessing yellow mosaic disease. Indian J. Virol. 2013, 24, 242–249. [Google Scholar] [CrossRef] [PubMed]
- Franceschini, M.H.D.; Bartholomeus, H.; van Apeldoorn, D.F.; Suomalainen, J.; Kooistra, L. Feasibility of unmanned aerial vehicle optical imagery for early detection and severity assessment of late blight in potato. Remote Sens. 2019, 11, 224. [Google Scholar] [CrossRef]
- Mahlein, A.-K.; Steiner, U.; Dehne, H.-W.; Oerke, E.-C. Spectral signatures of sugar beet leaves for the detection and differentiation of diseases. Precis. Agric. 2010, 11, 413–431. [Google Scholar] [CrossRef]
- Mahlein, A.-K.; Steiner, U.; Hillnhutter, C.; Dehne, H.-W.; Oerke, E.-C. Hyperspectral imaging for small-scale analysis of symptoms caused by different sugar beet diseases. Plant Methods 2012, 8, 3–15. [Google Scholar] [CrossRef]
- Rumpf, T.; Mahlein, A.-K.; Steiner, U.; Oerke, E.-C.; Dehne, H.W.; Plümer, L. Early detection and classification of plant diseases with Support Vector Machines based on hyperspectral reflectance. Comput. Electron. Agric. 2010, 74, 91–99. [Google Scholar] [CrossRef]
- Reynolds, G.J.; Windels, C.E.; MacRae, I.V.; Laguette, S. Remote sensing for assessing Rhizoctonia crown and root rot severity in sugar beet. Plant Dis. 2012, 96, 497–505. [Google Scholar] [CrossRef]
- Xie, C.; Yang, C.; He, Y. Hyperspectral imaging for classification of healthy and gray mold diseased tomato leaves with different infection severities. Comput. Electron. Agric. 2017, 135, 154–162. [Google Scholar] [CrossRef]
- Lu, J.; Zhou, M.; Gao, Y.; Jiang, H. Using hyperspectral imaging to discriminate yellow leaf curl disease in tomato leaves. Precis. Agric. 2017, 19, 379–394. [Google Scholar] [CrossRef]
- Abdulridha, J.; Batuman, O.; Ampatzidis, Y. UAV-based remote sensing technique to detect citrus canker disease utilizing hyperspectral imaging and machine learning. Remote Sens. 2019, 11, 1373. [Google Scholar] [CrossRef]
- Li, X.; Lee, W.S.; Li, M.; Ehsani, R.; Mishra, A.R.; Yang, C.; Mangan, R.L. Spectral difference analysis and airborne imaging classification for citrus greening infected trees. Comput. Electron. Agric. 2012, 83, 32–46. [Google Scholar] [CrossRef]
- Couture, J.J.; Singh, A.; Charkowski, A.O.; Groves, R.L.; Gray, S.M.; Bethke, P.C.; Townsend, P.A. Integrating spectroscopy with potato disease management. Plant Dis. 2018, 102, 2233–2240. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, N.; Yuan, L.; Chen, F.; Wu, K. Discrimination of winter wheat disease and insect stresses using continuous wavelet features extracted from foliar spectral measurements. Biosyst. Eng. 2017, 162, 20–29. [Google Scholar] [CrossRef]
- Kuska, M.T.; Wahabzada, M.; Leucker, M.; Dehne, H.-W.; Kersting, K.; Oerke, E.-C.; Steiner, U.; Mahlein, A.-K. Hyperspectral phenotyping on the microscopic scale: Towards automated characterization of plant-pathogen interactions. Plant Methods 2015, 11, 28–41. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Burks, T.F.; Ritenour, M.A.; Bonn, W.G. Detection of citrus canker using hyperspectral reflectance imaging with spectral information divergence. J. Food Eng. 2009, 93, 183–191. [Google Scholar] [CrossRef]
- Abdulridha, J.; Ampatzidis, Y.; Ehsani, R.; de Castro, A.I. Evaluating the performance of spectral features and multivariate analysis tools to detect laurel wilt disease and nutritional deficiency in avocado. Comput. Electron. Agric. 2018, 155, 203–211. [Google Scholar] [CrossRef]
- Shi, Y.; Huang, W.; González-Moreno, P.; Luke, B.; Dong, Y.; Zheng, Q.; Ma, H.; Liu, L. Wavelet-based rust spectral feature set (WRSFs): A novel spectral feature set based on continuous wavelet transformation for tracking progressive host–pathogen interaction of yellow rust on wheat. Remote Sens. 2018, 10, 525. [Google Scholar] [CrossRef]
- Pinto, F.; Damm, A.; Schickling, A.; Panigada, C.; Cogliati, S.; Muller-Linow, M.; Balvora, A.; Rascher, U. Sun-induced chlorophyll fluorescence from high-resolution imaging spectroscopy data to quantify spatio-temporal patterns of photosynthetic function in crop canopies. Plant Cell Environ. 2016, 39, 1500–1512. [Google Scholar] [CrossRef]
- Bravo, C.; Moshou, D.; West, J.; McCartney, A.; Ramon, H. Early disease detection in wheat fields using spectral reflectance. Biosyst. Eng. 2003, 84, 137–145. [Google Scholar] [CrossRef]
- Huang, L.; Zhao, J.; Zhang, D.; Yuan, L.; Dong, Y.; Zhang, J. Identifying and mapping stripe rust in winter wheat using multi-temporal airborne hyperspectral images. Int. J. Agric. Biol. 2012, 14, 697–704. [Google Scholar]
- Vanegas, F.; Bratanov, D.; Powell, K.; Weiss, J.; Gonzalez, F. A novel methodology for improving plant pest surveillance in vineyards and crops using UAV-based hyperspectral and spatial data. Sensors 2018, 18, 260. [Google Scholar] [CrossRef] [PubMed]
- Hillnhütter, C.; Mahlein, A.-K.; Sikora, R.A.; Oerke, E.-C. Remote sensing to detect plant stress induced by Heterodera schachtii and Rhizoctonia solani in sugar beet fields. Field Crop. Res. 2011, 122, 70–77. [Google Scholar] [CrossRef]
- Zhang, M.; Qin, Z.; Liu, X.; Ustin, S.L. Detection of stress in tomatoes induced by late blight disease in California, USA, using hyperspectral remote sensing. Int. J. Appl. Earth Obs. Geoinf. 2003, 4, 295–310. [Google Scholar] [CrossRef]
- Mirik, M.; Ansley, R.J.; Price, J.A.; Workneh, F.; Rush, C.M. Remote monitoring of wheat streak mosaic progression using sub-pixel classification of Landsat 5 TM imagery for site specific disease management in winter wheat. Adv. Remote Sens. 2013, 2, 16–28. [Google Scholar] [CrossRef]
- Suomalainen, J.; Anders, N.; Iqbal, S.; Roerink, G.; Franke, J.; Wenting, P.; Hünniger, D.; Bartholomeus, H.; Becker, R.; Kooistra, L. A Lightweight hyperspectral mapping system and photogrammetric processing chain for unmanned aerial vehicles. Remote Sens. 2014, 6, 11013–11030. [Google Scholar] [CrossRef]
- Jay, S.; Bendoula, R.; Hadoux, X.; Féret, J.-B.; Gorretta, N. A physically-based model for retrieving foliar biochemistry and leaf orientation using close-range imaging spectroscopy. Remote Sens. Environ. 2016, 177, 220–236. [Google Scholar] [CrossRef]
- Zhang, N.; Pan, Y.; Feng, H.; Zhao, X.; Yang, X.; Ding, C.; Yang, G. Development of Fusarium head blight classification index using hyperspectral microscopy images of winter wheat spikelets. Biosyst. Eng. 2019, 186, 83–99. [Google Scholar] [CrossRef]
- Moshou, D.; Bravo, C.; West, J.; Wahlen, S.; McCartney, A.; Ramon, H. Automatic detection of ‘yellow rust’ in wheat using reflectance measurements and neural networks. Comput. Electron. Agric. 2004, 44, 173–188. [Google Scholar] [CrossRef]
- Singh, C.B.; Jayas, D.S.; Paliwal, J.; White, N.D.G. Detection of midge-damaged wheat kernels using short-wave near-infrared hyperspectral and digital colour imaging. Biosyst. Eng. 2010, 105, 380–387. [Google Scholar] [CrossRef]
- Shahin, M.A.; Symons, S.J. Detection of Fusarium damaged kernels in Canada Western Red Spring wheat using visible/near-infrared hyperspectral imaging and principal component analysis. Comput. Electron. Agric. 2011, 75, 107–112. [Google Scholar] [CrossRef]
- Mirik, M.; Jones, D.C.; Price, J.A.; Workneh, F.; Ansley, R.J.; Rush, C.M. Satellite remote sensing of wheat infected by wheat streak mosaic virus. Plant Dis. 2011, 95, 4–12. [Google Scholar] [CrossRef]
- Nagasubramanian, K.; Jones, S.; Singh, A.K.; Singh, A.; Ganapathysubramanian, B.; Sarkar, S. Explaining hyperspectral imaging based plant disease identification: 3D CNN and saliency maps. In Proceedings of the 31st Conference on Neural Information Processing Systems (NIPS 2017), Long Beach, CA, USA, 4–9 December 2017. [Google Scholar]
- Mishra, P.; Asaari, M.S.M.; Herrero-Langreo, A.; Lohumi, S.; Diezma, B.; Scheunders, P. Close range hyperspectral imaging of plants: A review. Biosyst. Eng. 2017, 164, 49–67. [Google Scholar] [CrossRef]
- Pandey, P.; Ge, Y.; Stoerger, V.; Schnable, J.C. High throughput In vivo analysis of plant leaf chemical properties using hyperspectral imaging. Front. Plant Sci. 2017, 8, 1348–1359. [Google Scholar] [CrossRef]
- Tekle, S.; Mage, I.; Segtnan, V.H.; Bjørnstad, A. Near-infrared hyperspectral imaging of Fusarium-damaged oats (Avena sativa L.). Cereal Chem. 2015, 92, 73–80. [Google Scholar] [CrossRef]
- Li, J.; Zhang, R.; Li, J.; Wang, Z.; Zhang, H.; Zhan, B.; Jiang, Y. Detection of early decayed oranges based on multispectral principal component image combining both bi-dimensional empirical mode decomposition and watershed segmentation method. Postharvest Biol. Technol. 2019, 158, 110986–110996. [Google Scholar] [CrossRef]
- Liu, J.; Chiang, C.; Chen, S. Image-based plant recognition by fusion of multimodal information. In Proceedings of the 10th International Conference on Innovative Mobile and Internet Services in Ubiquitous Computing (IMIS), Fukuoka, Japan, 6–8 July 2016; pp. 5–11. [Google Scholar]
- Liu, J.; Lin, T. Location and image-based plant recognition and recording system. J. Inf. Hiding Multimed. Signal Process. 2015, 6, 898–910. [Google Scholar]
- Williams, D.; Britten, A.; McCallum, S.; Jones, H.; Aitkenhead, M.; Karley, A.; Loades, K.; Prashar, A.; Graham, J. A method for automatic segmentation and splitting of hyperspectral images of raspberry plants collected in field conditions. Plant Methods 2017, 13, 74–85. [Google Scholar] [CrossRef]
- Sun, G.; Zhang, A.; Ren, J.; Ma, J.; Wang, P.; Zhang, Y.; Jia, X. Gravitation-based edge detection in hyperspectral images. Remote Sens. 2017, 9, 592. [Google Scholar] [CrossRef]
- Perez-Sanz, F.; Navarro, P.J.; Egea-Cortines, M. Plant phenomics: An overview of image acquisition technologies and image data analysis algorithms. Gigascience 2017, 6, 1–18. [Google Scholar] [CrossRef]
- Xie, C.; He, Y. Spectrum and image texture features analysis for early blight disease detection on eggplant leaves. Sensors 2016, 16, 676. [Google Scholar] [CrossRef] [PubMed]
- Wei, F. Research on Feature Extraction and Feature Selection for Hyperspectral Remote Sensing Data; Northwestern Polytechnical University: Xi’an, China, 2015. [Google Scholar]
- Knauer, U.; Matros, A.; Petrovic, T.; Zanker, T.; Scott, E.S.; Seiffert, U. Improved classification accuracy of powdery mildew infection levels of wine grapes by spatial-spectral analysis of hyperspectral images. Plant Methods 2017, 13, 31–47. [Google Scholar] [CrossRef] [PubMed]
- Lowe, A.; Harrison, N.; French, A.P. Hyperspectral image analysis techniques for the detection and classification of the early onset of plant disease and stress. Plant Methods 2017, 13, 80–91. [Google Scholar] [CrossRef] [PubMed]
- Yeh, Y.F.; Chung, W.; Liao, J.; Chung, C.; Kuo, Y.; Lin, T. A Comparison of Machine Learning Methods on Hyperspectral Plant Disease Assessments. IFAC Proc. Vol. 2013, 46, 361–365. [Google Scholar] [CrossRef]
- Moshou, D.; Bravo, C.; Oberti, R.; West, J.; Bodria, L.; McCartney, A.; Ramon, H. Plant disease detection based on data fusion of hyper-spectral and multi-spectral fluorescence imaging using Kohonen maps. Real-Time Imaging 2005, 11, 75–83. [Google Scholar] [CrossRef]
- Golhani, K.; Balasundram, S.K.; Vadamalai, G.; Pradhan, B. Estimating chlorophyll content at leaf scale in viroid-inoculated oil palm seedlings (Elaeis guineensis Jacq.) using reflectance spectra (400 nm–1050 nm). Int. J. Remote Sens. 2019, 40, 7647–7662. [Google Scholar] [CrossRef]
- Joalland, S.; Screpanti, C.; Liebisch, F.; Varella, H.V.; Gaume, A.; Walter, A. Comparison of visible imaging, thermography and spectrometry methods to evaluate the effect of Heterodera schachtii inoculation on sugar beets. Plant Methods 2017, 13, 73–86. [Google Scholar] [CrossRef]
- Prabhakar, M.; Prasad, Y.G.; Thirupathi, M.; Sreedevi, G.; Dharajothi, B.; Venkateswarlu, B. Use of ground based hyperspectral remote sensing for detection of stress in cotton caused by leafhopper (Hemiptera: Cicadellidae). Comput. Electron. Agric. 2011, 79, 189–198. [Google Scholar] [CrossRef]
- Geospatial, L.H. EO-1 Hyperion Vegetation Indices Tutorial. Available online: http://www.harrisgeospatial.com/docs/narrowbandgreenness.html (accessed on 3 April 2019).
- Calderón, R.; Navas-Cortés, J.A.; Lucena, C.; Zarco-Tejada, P.J. High-resolution airborne hyperspectral and thermal imagery for early detection of Verticillium wilt of olive using fluorescence, temperature and narrow-band spectral indices. Remote Sens. Environ. 2013, 139, 231–245. [Google Scholar] [CrossRef]
- Devadas, R.; Lamb, D.W.; Simpfendorfer, S.; Backhouse, D. Evaluating ten spectral vegetation indices for identifying rust infection in individual wheat leaves. Precis. Agric. 2008, 10, 459–470. [Google Scholar] [CrossRef]
- Berdugo, C.A.; Zito, R.; Paulus, S.; Mahlein, A.-K. Fusion of sensor data for the detection and differentiation of plant diseases in cucumber. Plant Pathol. 2014, 63, 1344–1356. [Google Scholar] [CrossRef]
- Mahlein, A.-K.; Rumpf, T.; Welke, P.; Oerke, E.-C.; Plümer, L.; Steiner, U.; Oerke, E.-C. Development of spectral indices for detecting and identifying plant diseases. Remote Sens. Environ. 2013, 128, 21–30. [Google Scholar] [CrossRef]
- Kononenko, I. Analysis and extensions of relief. In Proceedings of the European Conference on Machine Learning; Springer: Secaucus, NJ, USA; pp. 171–182.
- Cao, X.; Luo, Y.; Zhou, Y.; Fan, J.; Xu, X.; West, J.S.; Duan, X.; Cheng, D. Detection of powdery mildew in two winter wheat plant densities and prediction of grain yield using canopy hyperspectral reflectance. PLoS ONE 2015, 10, e0121462–e0121479. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Qin, F.; Ruan, L.I.; Wang, R.; Liu, Q.; Ma, Z.; Li, X.; Cheng, P.; Wang, H. Identification and severity determination of wheat stripe rust and wheat leaf rust based on hyperspectral data acquired using a black-paper-based measuring method. PLoS ONE 2016, 11, e0154648–e0154674. [Google Scholar] [CrossRef] [PubMed]
- Susič, N.; Žibrat, U.; Širca, S.; Strajnar, P.; Razinger, J.; Knapič, M.; Vončina, A.; Urek, G.; Gerič Stare, B. Discrimination between abiotic and biotic drought stress in tomatoes using hyperspectral imaging. Sens. Actuators B Chem. 2018, 273, 842–852. [Google Scholar] [CrossRef]
- Zhang, N.; Zhang, X.; Yang, G.; Zhu, C.; Huo, L.; Feng, H. Assessment of defoliation during the Dendrolimus tabulaeformis Tsai et Liu disaster outbreak using UAV-based hyperspectral images. Remote Sens. Environ. 2018, 217, 323–339. [Google Scholar] [CrossRef]
- Menesatti, P.; Antonucci, F.; Pallottino, F.; Giorgi, S.; Matere, A.; Nocente, F.; Pasquini, M.; D’Egidio, M.G.; Costa, C. Laboratory vs. in-field spectral proximal sensing for early detection of Fusarium head blight infection in durum wheat. Biosyst. Eng. 2013, 114, 289–293. [Google Scholar] [CrossRef]
- Cheng, T.; Rivard, B.; Sánchez-Azofeifa, G.A.; Feng, J.; Calvo-Polanco, M. Continuous wavelet analysis for the detection of green attack damage due to mountain pine beetle infestation. Remote Sens. Environ. 2010, 114, 899–910. [Google Scholar] [CrossRef]
- Li, G.; Wang, C.; Feng, M.; Yang, W.; Li, F.; Feng, R. Hyperspectral prediction of leaf area index of winter wheat in irrigated and rainfed fields. PLoS ONE 2017, 12, e0183338–e0183352. [Google Scholar] [CrossRef]
- Zhang, J.; Rivard, B.; Rogge, D.M. The successive projection algorithm (SPA), an algorithm with a spatial constraint for the automatic search of endmembers in hyperspectral data. Sensors 2008, 8, 1321–1342. [Google Scholar] [CrossRef]
- Wang, F.; Li, Y.; Peng, Y.; Yang, B.; Li, L.; Liu, Y. Multi-Parameter Potato Quality Non-Destructive Rapid Detection by Visible/Near-Infrared Spectra. Spectrosc Spect. Anal. 2018, 38, 3736–3742. [Google Scholar] [CrossRef]
- Li, H.; Liang, Y.; Xu, Q.; Cao, D. Key wavelengths screening using competitive adaptive reweighted sampling method for multivariate calibration. Anal. Chim. Acta 2009, 648, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Ye, S.; Wang, D.; Min, S. Successive projections algorithm combined with uninformative variable elimination for spectral variable selection. Chemom. Intell. Lab. Syst. 2008, 91, 194–199. [Google Scholar] [CrossRef]
- Rouse, J.W.; Haas, R.H.; Schell, J.A.; Deering, D.W. Monitoring vegetation systems in the great plains with ERTS. NASA Spec. Publ. 1973, 351, 309–317. [Google Scholar]
- Gitelson, A.A.; Gritz, Y.; Merzlyak, M.N. Relationships between leaf chlorophyll content and spectral reflectance and algorithms for non-destructive chlorophyll assessment in higher plant leaves. J. Plant Physiol. 2003, 160, 271–282. [Google Scholar] [CrossRef]
- Haboudane, D.; Miller, J.R.; Pattey, E.; Zarco-Tejada, P.J.; Strachan, I.B. Hyperspectral vegetation indices and novel algorithms for predicting green LAI of crop canopies: Modeling and validation in the context of precision agriculture. Remote Sens. Environ. 2004, 90, 337–352. [Google Scholar] [CrossRef]
- Gamon, J.A.; Serrano, L.; Surfus, J.S. The photochemical reflectance index: An optical indicator of photosynthetic radiation use efficiency across species, functional types, and nutrient levels. Oecologia 1997, 112, 492–501. [Google Scholar] [CrossRef]
- Penuelas, J.; Baret, F.; Filella, I. Semi-empirical indices to assess carotenoids/chlorophyll-a ratio from leaf spectral reflectance. Photosynth. Res. 1995, 43, 67–74. [Google Scholar]
- Gamon, J.A.; Surfus, J.S. Assessing leaf pigment content and activity with a reflectometer. New Phytol. 1999, 143, 105–117. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Merzlyak, M.N.; Chivkunova, O.B. Optical properties and nondestructive estimation of anthocyanin content in plant leaves. Photochem. Photobiol. 2010, 74, 38–45. [Google Scholar] [CrossRef]
- Gitelson, A.; Zur, Y.; Chivkunova, O.B.; Merzlyak, M.N. Assessing carotenoid content in plant leaves with reflectance spectroscopy. Photochem. Photobiol. 2010, 75, 272–281. [Google Scholar] [CrossRef]
- Gitelson, A.; Merzlyak, M.N. Spectral reflectance changes associated with autumn senescence of Aesculus Hippocastanum L. and Acer Platanoides L. Leaves. Spectral features and relation to chlorophyII estimation. J. Plant Physiol. 1994, 143, 286–292. [Google Scholar] [CrossRef]
- Chen, J.M. Evaluation of Vegetation Indices and a Modified Simple Ratio for Boreal Applications. Can. J. Remote Sens. 1996, 22, 229–242. [Google Scholar] [CrossRef]
- Ceccatoa, P.; Flasseb, S.P.; Tarantolac, S.; Jacquemoudd, S.P.; Gre’goirea, J.-M. Detecting vegetation leaf water content using reflectance in the optical domain. Remote Sens. Environ. 2001, 77, 22–33. [Google Scholar] [CrossRef]
- Hardisky, M.; Klemas, V.; Smart, R.M. The influences of soil salinity, growth form, and leaf moisture on the spectral reflectance of spartina alterniflora canopies. Photogramm. Eng. Remote Sens. 1983, 49, 77–83. [Google Scholar]
- Serrano, L.; Penuelas, J.; Ustin, S.L. Remote sensing of nitrogen and lignin in Mediterranean vegetation from AVIRIS data: Decomposing biochemical from structural signals. Remote Sens. Environ. 2002, 81, 355–364. [Google Scholar] [CrossRef]
- Heim, R.H.J.; Wright, I.J.; Allen, A.P.; Geedicke, I.; Oldeland, J. Developing a spectral disease index for myrtle rust (Austropuccinia psidii). Plant Pathol. 2019, 68, 738–745. [Google Scholar] [CrossRef]
- Jones, C.D.; Jones, J.B.; Lee, W.S. Diagnosis of bacterial spot of tomato using spectral signatures. Comput. Electron. Agric. 2010, 74, 329–335. [Google Scholar] [CrossRef]
- Muir, A.Y.; Porteous, R.L.; Wastie, R.L. Experiments in the detection of incipient diseases in potato tubers by optical methods. J. Agric. Eng. Res. 1982, 27, 131–138. [Google Scholar] [CrossRef]
- IPCC. 2019 Refinement to the 2006 IPCC Guidelines for National Greenhouse Gas Inventorie; Task Force on National Greenhouse Gas Inventories (TFI): Kyoto, Japan, 2019. [Google Scholar]
- Rizzetto, S.; Belyazid, S.; Gegout, J.C.; Nicolas, M.; Alard, D.; Corcket, E.; Gaudio, N.; Sverdrup, H.; Probst, A. Modelling the impact of climate change and atmospheric N deposition on French forests biodiversity. Environ. Pollut. 2016, 213, 1016–1027. [Google Scholar] [CrossRef]
- Ma, H.; Jing, Y.; Huang, W.; Shi, Y.; Dong, Y.; Zhang, J.; Liu, L. Integrating early growth information to monitor winter wheat powdery mildew using multi-temporal landsat-8 imagery. Sensors 2018, 18, 3290. [Google Scholar] [CrossRef] [PubMed]

| Crop Types | Crop Names | Disease Names | Disease Types | Main Infected Sites | Sensors/Platforms/Scales | Analysis Approach | References |
|---|---|---|---|---|---|---|---|
| Cereal Crops | Wheat | Wheat stripe rust | Fungal diseases | Leaves * | Analytical spectral device (ASD)/handheld/leaves and canopy | Partial least squares discriminant analysis (PLSR), support vector regression (SVR), and Gaussian process regression (GPR) | [49] |
| Wheat leaf rust | Fungal diseases | Leaves * | ASD and digital camera/handheld/leaves | Linear spectral mixture analysis, Fisher function | [50] | ||
| Wheat powdery mildew | Fungal diseases | Leaves * | ASD/handheld/canopy | Continuous wavelet analysis, Fisher’s linear discrimination analysis (FLDA) and support vector machine (SVM) | [51] | ||
| Fusarium head blight | Fungal diseases | Ears * and stalks | ImSpector V10E and ImSpector N25E/indoor measurement platform/spikelets | Linear model fitting, spectral vegetation indices (SVIs) | [42] | ||
| Rice | Rice sheath blight | Fungal diseases | Leaves * | ImSpector V10E/indoor measurement platform/single plant | Linear discriminant analysis (LDA) and SVM | [52] | |
| Rice blast | Fungal diseases | Leaves *, stems and ears | ORCA-05G/darkroom/panicle | “Bag of spectra words” (BoSW) model and chi-square support vector machine (chi-SVM) | [53] | ||
| Maize | Grey leaf spot disease | Fungal diseases | Leaves * | ASD and three multi-spectral satellite Resampled/handheld/satellite/leaves and canopy | Random forest algorithm (RF) | [54] | |
| Leaf spot disease | Fungal diseases | Leaves * and bracks | ASD/handheld/leaves | Guided regularized random forest (GRRF) and RF | [55] | ||
| Ear rot | Fungal diseases | Ears and kernels * | SisuChema/HgCdTe detector/fungal isolates | Principal component analysis (PCA) and PLSR | [56] | ||
| Legume Crops | Soybean | Soybean anthracnose | Fungal diseases | Stems *, pods and leaves * | Pika XC/mounting tower/stems | Genetic algorithm as an optimizer and SVM as a classifier | [57] |
| Yellow mosaic virus | Viral disease | Leaves * | ASD/handheld/leaves | Spectral derivative and red edge analysis | [58] | ||
| Tuber Crops | Potato | Late blight disease | Fungal diseases | Leaves * and fruits | Rikola/unmanned aerial vehicle (UAV)/plots | Simplex volume maximization (SiVM) and pixel-wise log-likelihood ratio (LLR) calculation | [59] |
| Potato virus Y | Viral disease | Leaves * | Specim FX10/tractor/canopy | Deep learning, fully convolutional neural network | [45] | ||
| Sugar Crops | Sugar Beet | Cercospora leaf spot | Fungal diseases | Leaves * | ASD/handheld/leaves | Spectral signature analysis and vegetation indices | [60] |
| Beet rust | Fungal diseases | Leaves * | ImSpector V10E/ microscope/tissue | Spectral angle mapper (SAM) | [61] | ||
| Beet powdery mildew | Fungal diseases | Leaves * | ASD/leaf clip/leaves | SVIs and SVM | [62] | ||
| Root rot | Fungal diseases | Roots (leaves) 1 | ASD/handheld/canopy | SVIs and nonlinear regressions | [63] | ||
| Vegetables | Tomato | Gray mold | Bacterial diseases | Fruits, leaves * and stems | ImSpector V10E/indoor measurement platform/leaves | K-nearest neighbor (KNN), C5.0 models and feature rank | [64] |
| Tomato yellow leaf curl virus | Viral disease | Leaves * | Imspector V10E-QE/indoor measurement platform/leaves | Grey level co-occurrence matrix (GLCM) | [65] | ||
| Fruits | Citrus | Citrus canker | Bacterial diseases | Fruits * and leaves * | Pika L 2.4/mounting tower and UAV/leaves, fruits and single plant | Radial basis function (RBF) and KNN | [66] |
| Huanglongbing (Citrus greening) | Bacterial diseases | Fruits *, leaves * and roots | AISA Eagle/airborne/canopy | SVM | [67] |
| Plant and Diseases | Targets * | Scales | Methods and Algorithms | Classification Accuracy | Reference |
|---|---|---|---|---|---|
| Sugar beet and Cercospora leaf spot/powdery mildew/sugar beet rust | Disease identification | Leaf | Spectral angle mapper (SAM) | 98.9% for Cercospora leaf spot at 8 dai; 97.23% for powdery mildew at 14 dai; 61.70% for sugar beet rust at 20 dai. | [61] |
| Wheat and Fusarium head blight | Disease identification | Spike | Support Vector Machine (SVM) with reflectance and spectral vegetation indices (SVIs) | 95.0% and 99.0% for two classes classification using SVIs and reflectance; 76.0% and 77.0% for multiclass classification using SVIs and reflectance; | [48] |
| Wheat and yellow rust (Puccinia striiformis) | Disease detection | Leaf | Quadratic discriminant analysis (QDA)/self-organizing map (SOM) NN | 94.5% by using QDA; Around 99% by using SOM NN | [103] |
| Citrus and citrus bacterial canker | Disease severity classification (asymptomatic, early, and late symptoms) | Leaf/fruit/plant | Neural network radial basis function (RBF); KNN with SVIs. | 94%, 96%, and 100% by RBF and 94%, 95%, and 96% by KNN for three levels at leaf scale; 92% canker detection at fruit scale and 100% and plant scale | [66] |
| Soybean and charcoal rot | Disease identification | Stem | Three dimensional convolutional neural network (3D CNN) | 95.73% | [88] |
| Wheat and stripe rust | Disease identification and mapping | Canopy/plot | Linear regression model | —— | [76] |
| Index | Formula | Definition and Description | Possible Symptoms | Possible Diseases | References |
|---|---|---|---|---|---|
| Normalized Difference Vegetation Index (NDVI) | Used to analyze healthy and green vegetation. It is robust over a wide range of conditions. | All | Almost all of green plants’ disease * | [124] | |
| Green Chlorophyll Index (GCI) | Used to estimate the leaf chlorophyll content of a plant. | Pigment | Myrtle rust Powdery mildew Stripe rust Flavescence Dorée Leaf spot | [125] | |
| Transformed Chlorophyll Absorption Reflectance Index (TCARI) | Indicates the relative abundance of chlorophyll. | [126] | |||
| Photochemical Reflectance Index (PRI) | Sensitive to the changes in carotenoid pigments (particularly xanthophyll pigments). | [127] | |||
| Structure Insensitive Pigment Index (SIPI) | Maximizes the sensitivity of the index to the ratio of bulk carotenoids to chlorophyll. | [128] | |||
| Red Green Ratio Index (RGRI) | It is an indicator of leaf production and stress, used to estimate the course of foliage development in canopies. | [129] | |||
| Anthocyanin Reflectance Index 1 (ARI1) | Weakening vegetation contains higher concentrations of anthocyanins, so this index is one measure of stressed vegetation. | [130] | |||
| Carotenoid Reflectance Index 1 (CRI1) | Weakening vegetation contains higher concentrations of carotenoids, so this index is one measure of stressed vegetation. | [131] | |||
| Red Edge Normalized Difference Vegetation Index (RENDVI) | Modification of the NDVI, using red edge instead of the absorption and reflectance peaks to enhance the sensitivity to small changes in canopy foliage content, gap fraction, and senescence. | Structure Pigment | Apple scab | [132] | |
| Modified Simple Ratio (MSR) | Used to increase the sensitivity of vegetation biophysical parameters. | [133] | |||
| Moisture Stress Index (MSI) | A reflectance measurement that is sensitive to increasing leaf water content. | Water | Root rot | [134] | |
| Normalized Difference Infrared Index (NDII) | A reflectance measurement that is sensitive to changes in the water content of plant canopies. | [135] | |||
| Normalized Difference Nitrogen Index (NDNI) | Estimates the relative amounts of nitrogen contained in vegetation canopies. | Nutrient | Yellow mosaic disease | [136] |
| Plant and Disease | Formula * | Sensors | Scales | Methods and Algorithms | Reference |
|---|---|---|---|---|---|
| Grapevine and Flavescence Dorée | FieldSpec 3 ASD | Leaf | D.A.: Genetic algorithm (GA) for feature selection | [25] | |
| Lemon Myrtle and Myrtle Rust | Spectral Evolution PSR+ 3500 | Leaf | D.A.: Random-forest-based for feature selection | [137] | |
| Sugar Beet and Cercospora Leaf Spot | ImSpector V10E | Leaf | D.A.: RELIEF-F for feature selection | [111] | |
| Sugar Beet and Sugar Beet Rust | ImSpector V10E | Leaf | D.A.: RELIEF-F for feature selection | [111] | |
| Sugar Beet and Powdery Mildew | ImSpector V10E | Leaf | D.A.: RELIEF-F for feature selection | [111] | |
| Winter Wheat and Fusarium Head Blight | ] | UHD 185 | Kernel | D.A.: Instability index-spectral angle mapper (ISI-SAM) for feature selection | [83] |
| Chinese Pine and Dendrolimus tabulaeformis Tsai et Liu | UHD 185 | Plant | D.A. and D.B.: Instability index between classes-successive projection algorithm (ISIC-SPA) for feature selection and PLSR for model fitting | [116] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, N.; Yang, G.; Pan, Y.; Yang, X.; Chen, L.; Zhao, C. A Review of Advanced Technologies and Development for Hyperspectral-Based Plant Disease Detection in the Past Three Decades. Remote Sens. 2020, 12, 3188. https://doi.org/10.3390/rs12193188
Zhang N, Yang G, Pan Y, Yang X, Chen L, Zhao C. A Review of Advanced Technologies and Development for Hyperspectral-Based Plant Disease Detection in the Past Three Decades. Remote Sensing. 2020; 12(19):3188. https://doi.org/10.3390/rs12193188
Chicago/Turabian StyleZhang, Ning, Guijun Yang, Yuchun Pan, Xiaodong Yang, Liping Chen, and Chunjiang Zhao. 2020. "A Review of Advanced Technologies and Development for Hyperspectral-Based Plant Disease Detection in the Past Three Decades" Remote Sensing 12, no. 19: 3188. https://doi.org/10.3390/rs12193188
APA StyleZhang, N., Yang, G., Pan, Y., Yang, X., Chen, L., & Zhao, C. (2020). A Review of Advanced Technologies and Development for Hyperspectral-Based Plant Disease Detection in the Past Three Decades. Remote Sensing, 12(19), 3188. https://doi.org/10.3390/rs12193188






