Abstract
Gene function analysis, molecular breeding, and the introduction of new traits in crop plants all require the development of a high-performance genetic transformation system. In numerous crops, including tomatoes, Agrobacterium-mediated genetic transformation is the preferred method. As one of our ongoing research efforts, we are in the process of mapping a broad-spectrum nematode resistance gene (Me1) in pepper. We work to transform tomato plants with candidate genes to confer resistance to nematodes in Solanaceae members. The transformation technology development is designed to produce a reproducible, rapid, and highly effective Agrobacterium-mediated genetic transformation system of Micro-Tom. In our system, a transformation efficiency of over 90% was achieved. The entire procedure, starting from the germination of seeds to the establishment of transformed plants in soil, was completed in 53 days. We confirmed the presence of the NeoR/KanR and DsRed genes in the transformed roots by polymerase chain reaction. The hairy root plants were infected with nematodes, and after 3 months, the presence of DsRed and NeoR/KanR genes was detected in the transformant roots to confirm the long-term effectiveness of the method. The presented study may facilitate root-related research and exploration of root–pathogen interactions.
1. Introduction
With an estimated yield of 186 million tons in 2020 [], the tomato (Solanum lycopersicum L.) is a widely cultivated vegetable crop of global significance. The tomato is important for scientific research as a model plant of the Solanaceae family, which is often used as an experimental plant for fruit-related studies. For this reason, tomato may offer new opportunities for investigating biological mechanisms that are not possible in other popular model plants such as Arabidopsis. In recent years, the use of tomato as a model crop has increased due to its readily available resources, such as high-quality assembled reference genomes [,], as well as the research efforts to improve the efficiency of Agrobacterium-mediated transformation and the recovery time of transgenic lines [,,,]. The tomato genome was published in 2012 [], and as a result, tomatoes have become an increasingly favored research material over the years.
Among the many known tomato varieties, the “Micro-Tom” has become a widely used model plant. Micro-Tom is a tomato mutant with a short life cycle that allows harvesting of the ripe fruit within 70–90 days after sowing. Seedlings grow with ease under ordinary fluorescent lighting. It can tolerate close planting, it is easily transformed, and it has the same hereditary information as the common tomato except for two main genes (dwarf gene and miniature gene). Thus, it is most useful for studying the gene functions of model plants such as Arabidopsis [,]. Due to the rapid development of genome editing technologies, the availability of efficient transformation methods is especially important. When root biology research is the primary focus, Agrobacterium rhizogenes-mediated transformation provides a quick and reliable alternative method to stable transformation. A. rhizogenes contains root loci (rol) genes, which, upon infection, trigger the formation of genetically transformed roots (hairy root phenotype), referring to the striking abundance of adventitious roots present [,]. Composite plants with healthy, untransformed shoots and transgenic hairy roots can be obtained as a result of A. rhizogenes transformation. Protocols were proposed for the production of composite plants in order to efficiently identify root–pathogen resistance genes in plants. Furthermore, several molecular studies were carried out in vitro with tomato hairy root crops, e.g., Fusarium tolerance [], transgenic nematode resistance [,], salt stress [], and rhizobial symbiosis study [].
Almost 40 years ago, McCormick et al. [] published the first report of Agrobacterium-mediated tomato (S. lycopersicum) transformation. Transformations of various species and explant types, such as leaves, cotyledons, and hypocotyls, have been reported in recent years [,,]. In recent years, great progress has been made in the development of efficient Agrobacterium-mediated transformation protocols for Micro-Tom [,,]. In most published papers, the transformation rates of A. rhizogenes-mediated Micro-Tom transformation were around 40–60%. In the majority of cases, no essential details (medium components, transplantation frequency, and infection method) were provided on the transformation protocol, which would be essential for the reproducibility of Micro-Tom’s transformation procedures. It was also unclear whether the transformation frequency was determined by the number of independent transgenic events or the number of transgenic plants.
In this paper, we present a high-throughput transformation system in detail developed in S. lycopersicum cv. Micro-Tom, using radicles and hypocotyls as starting explants and applying A. rhizogenes ARqua1 in order to solve the problem of long transformation periods and low-efficiency transformation rates in tomato plant. We set out to evaluate two different inoculation methods and compare their effectiveness. In an effort to make it easier to control the transformation process and to distinguish between non-transformed and transgenic roots, DsRed was used as a visual marker that produced red fluorescence, and its use as a marker was tested for the reliable identification and selection of transformed hairy roots of tomato during in vitro culture. One of the main aims of the present study is to show that the expression of transgenes persists in the transformant roots for several months []. We intended to demonstrate that this alternative transformation method by means of nematode infection is suitable for long-term studies since, in the majority of cases, 2–3 months are required for the development of the corresponding infection phenotype in Meloidogyne infection. Our final objective was to establish a highly efficient, reliable, and reproducible A. rhizogenes transformation system for S. lycopersicum cv. Micro-Tom to validate and describe candidate genes (Me1) of pepper (Capsicum annuum, PI201234) [] that could confer resistance against root-knot nematode species (Meloidogyne spp.).
We hypothesized that A. rhizogenes is suitable for transforming S. lycopersicum cv. Micro-Tom with obtaining high efficiency. Furthermore based on our preliminary experiments in C. annuum [], we expected that rhizogenes infection by puncturing the hypocotyl will show the greatest efficiency in the case of tomato.
2. Materials and Methods
2.1. Plant Material
The seeds of tomato (S. lycopersicum) cv. Micro-Tom were soaked in water for 3 h, surface sterilized with filtered 10% solution of CaClO2 and a drop of Tween20 for 20 min, and rinsed three times with sterile distilled water. MS20 (Murashige and Skoog supplied with 20 g sucrose) media were used to germinate the sterilized seeds. A total of 360 seeds were germinated this way in 3 separate rounds, each round with 120 seeds.
2.2. Agrobacterium Strain and Vector
We used the A. rhizogenes strain ARqua1 harboring our desired plasmid vector for the transformation. For the experiments, we used the plasmid pKWGFS7-RR (VIB-Ugent Center for Plant Systems Biology modified by Á. Domonkos, Gödöllő, Hungary) [] containing an aminoglycoside adenylyltransferase gene for the bacterial selection, an aminoglycoside phosphotransferase gene for the plant selection, and a DsRed reporter gene. The control line was transformed with the same plasmid pKGWFS7 [] construction but without the DsRed reporter.
2.3. Agrobacterium Culture Preparation and Infection Medium
For the infection procedure to be carried out, Agrobacterium strains were cultured in plastic Petri dishes at 28 °C on solid Luria Broth (LB) medium containing 50 mg/L spectinomycin and 50 mg/L rifampicin for 48 h.
2.4. Preparation of Explants, Agrobacterium Infection and Co-Cultivation
The plant roots were cut horizontally at the desired parts (radicles and hypocotyl) using a sterile sharp scalpel. In this study, we used 3 transformation methods to compare their transformation efficiency and studied a total of 40 plants/method (30 + 10 control) in each round: (1) coating the wound surface of radicles in bacterial mass; (2) coating the wound surface hypocotyl in bacterial mass; (3) puncturing the hypocotyl with tungsten needle dipped in bacterial mass. For Methods (1) and (2), we crossed the wounded surface over the bacterial mass in the Petri dishes. For Method (3) we gently dipped the tungsten needle in the bacterial mass, then punctured the hypocotyl vertically numerous times. The co-cultivation with the bacteria was carried out on MS20 media. To observe the effect of co-cultivation length, the first round of germinated seeds (120 plantlets) was co-cultivated for 6 days, while the other two rounds (240 plantlets) were co-cultivated for 4 days. The media were poured obliquely into sterile glass cultivating tubes 24 mm in diameter and 15 cm in height in order to be able to observe the plants individually during the experiment. Figure 1 and Figure 2 highlight the key steps in the transformation process.
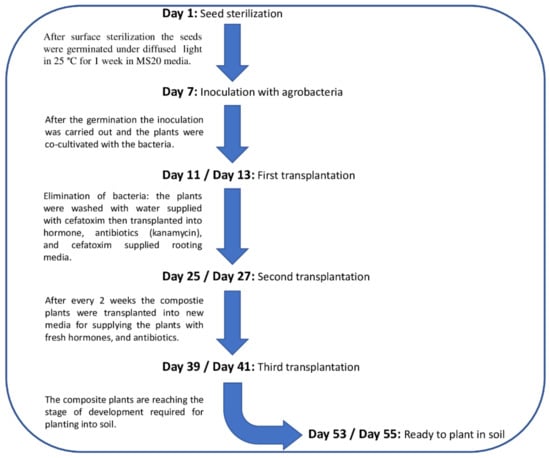
Figure 1.
Schematic representation of the timeline for the production of composite tomato plants.

Figure 2.
Essential steps in the production of tomato composite plants. Seed germination (a) and seeds placed in sterilized glass storage jars with MS20 media (b). Vigorous plants were removed from the MS20 medium (c). A. rhizogenes bacterial culture and observed transformation methods (d–f). Resulting vigorous composite tomato plantlets growing in sterile glass cultivating tubes 24 mm in diameter and 15 cm in height (g,h).
2.5. Tissue Culture and Transplantation
The plants were washed in sterile distilled water containing 500 mg/L cefotaxime after co-cultivation and then transferred to a rooting medium containing 400 mg/L cefotaxime, 60 mg/L kanamycin, and 0.5 mg/L IAA, to eliminate the bacteria and apply selection pressure to the roots. The plants were transferred every 2 weeks to a new rooting medium to supply them with fresh hormones, nutrients, and antibiotics.
2.6. Fluorescence Observation and Imaging
The red fluorescence in the seedling roots was observed using a Leica MZ10 F stereomicroscope (Leica, Wetzlar, Germany) with an external Leica EL6000 light source for enhanced fluorescence imaging, visualized using the manufacturer’s own computer software (Leica Application Suite). The images were processed using Adobe Photoshop software ver. 23.2.2 (Adobe Inc., San Jose, CA, USA).
2.7. DNA Extraction and PCR Verification
Total DNA was isolated from fresh root tissue (100 mg/sample) using a Zenogene DNA purification kit (Zenon Bio, Szeged, Hungary) according to the manufacturer’s instructions. The PCR reactions were carried out in 12 µL reaction mixtures containing 1.2 µL of 10× PCR buffer, 0.3 U Taq polymerase, 0.2 mM dNTP, 0.5 µM of each primer, and 10 ng template DNA. Thermal cycling conditions consisted of one cycle of initial denaturation for 4 min at 94 °C, followed by 30 cycles of 94 °C for 60 s, 58 °C for 60 s, 72 °C for 60 s, with a final extension step of 7 min at 72 °C. The amplified products were electrophoresed on a 1.0% (w/v) agarose gel stained with 0.5 mg/L ethidium bromide solution and visualized on a UV transilluminator. pKWGFS7-RR vector-specific NeoR/KanR primers (F-5′-GTTCTTTTTGTCAAGACCGACCT-3′; R-5′-CTCTTCAGCAATATCACGGGTAG-3′) and DsRed primers (F-5′-TGCCCTGCGCGCTCCTCCA-3′; R-5′-CTACAGGAACAGGTGGTGG-3′) were used for PCR identification of positive plants.
2.8. Determination of Agrobacterium Infection and Transformation Efficiency
DsRed fluorescence and PCR screening of NeoR/KanR and DsRed genes from roots of four-week-old in vitro putative T0 plants were used to calculate transformation efficiency. The statistical calculation is based on the following equation: the transformation efficiency (%) = (Number of positive transgenic plants/number of infected explants) × 100.
2.9. Nematode Susceptibility Test
The culture of Meloidogyne incognita was maintained in the susceptible tomato line. Before infection with nematodes, the two-month-old plantlets were transplanted individually into 250 mL plastic pots filled with steam-sterilized sandy soil and grown to the fourth/fifth true leaf stage. The tomato plants were inoculated with egg masses and J2 juveniles collected from infected roots and soil. Plants were grown in a 25 °C growth chamber, and after 8–10 weeks, the roots were evaluated, and transgenes in the root samples were tested.
3. Results
3.1. Rhizogenes Inoculation Method
The infiltration method and the length of co-cultivation play a key role in achieving a highly efficient Agrobacterium transformation. To identify the most appropriate method for tomato transformation, we applied three transformation methods to compare their transformation efficiency. S. lycopersicum plants were cut horizontally at the desired parts (radicles and hypocotyl) using a sterile sharp scalpel and infected by coating the wound surface in bacterial mass or puncturing the hypocotyl with a tungsten needle dipped in bacterial mass. The typical phenotype of the infection after one week was characterized by cell proliferation that could be observed at the site of infection and on the stem, as well as by brown spots, which were probably the signs of local cell death. The transformed roots typically grew from the cell proliferation zone. As in our previous experiments in peppers, both normal and hairy roots grew on the plant, and the formation of transformant roots did not depend on the development of the hairy root phenotype. Since the newly formed roots were highly fragile and easily damaged, during the transplantation, extra care had to be taken. The formation of the first roots started on the rooting medium after two weeks of cultivation. The plants infected with the tungsten needle (Method (3)) were the fastest in rooting while the coating of hypocotyl with bacterial mass (Method (2)) showed the slowest rooting (usually 4 weeks).
3.2. Elimination of A. rhizogenes
Upon each transplantation, we washed the plants with sterile water supplemented with 500 mg/L cefotaxime. The length of co-cultivation greatly influenced the survival rates of plants. After 6 days of co-cultivation, the bacteria typically overgrew the medium and were impossible to eliminate during transplantation despite the washing step. Because of the overgrown bacteria, we observed an 85% mortality rate among the plants. Method (1) had the highest survival rates with 11 living plants, while Method (3) only had 1, and Method (2) produced no surviving plants. The results showed that the plants infected with Method (2) were the least tolerant to the overgrowing bacteria, while plants infected with Method (1) showed significantly higher survival rates. Therefore, the infection with Method (1) showed the highest tolerance against the aggressive growth of bacteria. The dead plants were characterized by whitening (a typical symptom of kanamycin, most likely because the plants failed to develop roots due to the high amount of bacteria present in the rooting area), lack of cell proliferation, stem watering, and bacterial mass that penetrated the medium even after multiple elimination attempts. The surviving plants successfully developed roots and the elimination of the bacteria was successful. After 4 days of co-cultivation, the bacteria were eliminated after the first transplantation; therefore, we could not observe the high mortality rates that occurred during the 6-day-long co-cultivation period (Table 1). These results supported the finding that the length of co-cultivation played a key role in the survival rate of plants and the success of the rhizogenes transformation of S. lycopersicum cv. Micro-Tom. The 4-day-long co-cultivation turned out to be the most efficient way of infection in regard to transformation and survival rates.

Table 1.
Mortality rate of different organs inoculated with binary vector inserted in the A. rhizogenes strain ARqua1 after 6 days of co-cultivation. Plants transformed with pKWGFS7-RR and pKWGFS7 (mock) are represented together in this table.
3.3. Agrobacterium Transformation of Explants
Hypocotyl explants of the Micro-Tom genotypes were inoculated with pKWGFS7-RR by using the puncturing and coating inoculation method. Two weeks after the first transplantation approximately 60% of the puncture-inoculated plants succeeded in developing intact roots, while only 20% of the coating-inoculated plants had roots during the same period (Figure 3). Four weeks after the inoculation, this number was 90% in the case of Method (3) and 80% for Method (2). During this time, the puncture-inoculated plants rapidly developed multiple roots from shared nodes, which appeared to be the cell proliferation zones on the surface of the hypocotyl. Concerning transformation, the punctured plants showed higher efficiency, with 95% of the inoculated plants developing transformant roots, while in the case of Method (2), this number was 92%. Six weeks after the inoculation, the puncture-inoculated plants showed a substantially more extensive root system compared to the other methods. The hypocotyl-coating-inoculated plants showed the highest sensitivity to the overgrowing bacteria of the three inoculation methods. Our results suggested that if the study focused on the rhizogenes transformation of hypocotyl explants, the puncturing method offered a more efficient way to generate composite plants.
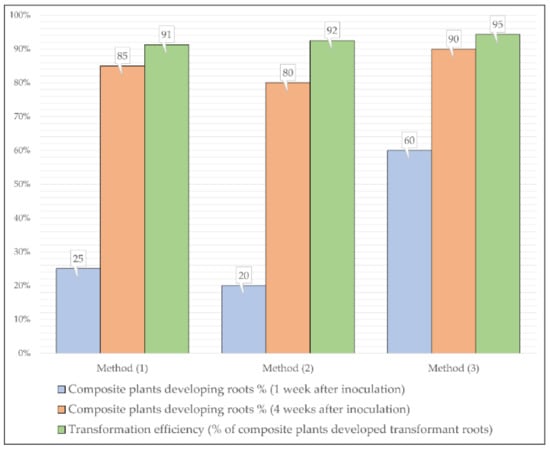
Figure 3.
The effectiveness of different methods for the production of tomato composite plants and the development of roots on the composite plants transformed by different methods.
Radicle explants from Micro-Tom genotypes were inoculated with pKWGFS7-RR by using the coating infection method. The growing of roots was similar to the hypocotyl explants transformed by the coating of the wound surface. Two weeks after the first transplantation, only 25% of the plants succeeded in root generation, but 4 weeks after the first transplantation, 85% of all the radicle-transformed plants were able to grow intact roots. The transformation efficiency was similar to the methods involving the puncturing of hypocotyl, with 91% of the transformed plants growing transformant roots (Table 2).

Table 2.
Transformation efficiency of different organs inoculated with binary vector inserted in the A. rhizogenes strain ARqua1 (pKWGFS7-RR) and mock inoculated (pKWGFS7). The table represents only the 4-day-long co-cultivation experiments.
In comparison with the composite plants of Method (3), 6 weeks after the first transplantation, 99% of the plants with both transformation methods were ready to be transplanted into soil, but the radicle-transformed samples showed a less extensive root system compared to the plants obtained by Method (3). The radicle-transformed plants showed the highest tolerance to the aggressively overgrowing bacteria of all three methods; therefore, using radicle explants for the transformation can be advantageous if the plant type used is sensitive to the growing bacteria.
3.4. DsRed Allows for Quick and Easy Identification of Positive Plants
We decided to use the DsRed gene (pKWGFS7-RR) driven by the Ubiquitin promoter as a molecular marker to identify transformed hairy roots and to quantify transformation efficiency. The resultant red fluorescence in transgenic tissue could be observed through a fluorescence microscope because DsRed could be expressed in all tissues at every developmental stage. As a result, the red fluorescence could be used to screen positive roots at various stages of genetic transformation and quickly distinguish between positive and negative roots. Furthermore, DsRed provides an opportunity to eliminate non-transformant roots during the experiment. The wavelengths of excitation and emission for DsRed range from 554 to 563 nm for excitation and from 582 to 592 nm for emission. As we expected, strong red fluorescence appeared in the positive roots, while the transgene-negative roots showed almost no signal or weak autofluorescence (Figure 4).
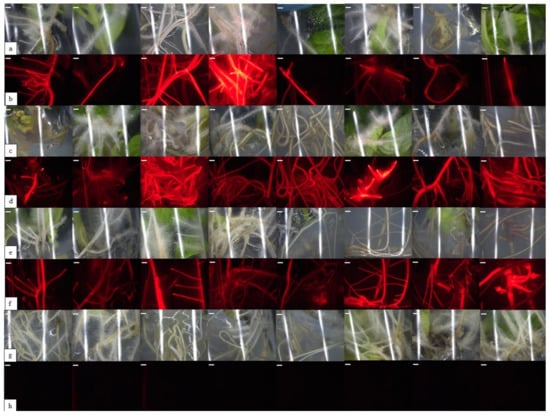
Figure 4.
Selection of transformed hairy roots expressing the DsRed gene in tomato plants. The roots were visualized with a stereomicroscope under bright light (lines a,c,e) and fluorescence (lines b,d,f). Control roots were visualized with stereomicroscope under bright light (line g) and under fluorescence (line h). Each bar represents 8 mm.
We showed that the applied vector pKWGFS7-RR was highly suitable for A. rhizogenes transformation in the case of S. lycopersicum cv. Micro-Tom. In the roots, which showed a higher level of expression of the DsRed gene than the average, the presence of the red fluorescent protein was visible under normal light in the form of pale pink colorations on the roots of the composite plants.
3.5. Confirmation of Genetic Co-Transformation of the Roots by PCR
To test whether the desired target DNA fragments were integrated into hairy roots that emitted red fluorescence and showed kanamycin resistance, all transgenic roots were selected by PCR amplification. DNA was isolated from the roots of all independent Micro-Tom plantlets and a non-transformed Micro-Tom plant, and by using DsRed and NeoR/KanR-specific primers, PCR analysis was performed. In line with our expectations, the DsRed and NeoR/KanR amplification products appeared as 678 bp and 558 bp long PCR products, respectively (Figure 5). PCR amplifications demonstrated the simultaneous presence of the NeoR/KanR and DsRed genes in the roots that were previously evaluated as DsRed positive under the stereomicroscope. In contrast, only the NeoR/KanR gene was amplified in the DNA sample deriving from control hairy roots. In the DsRed-negative roots (based on microscopy), none of the desired fragments appeared in the agarose gel. The results support the fact that upon transformation, the whole T-DNA sequence is integrated into the roots.
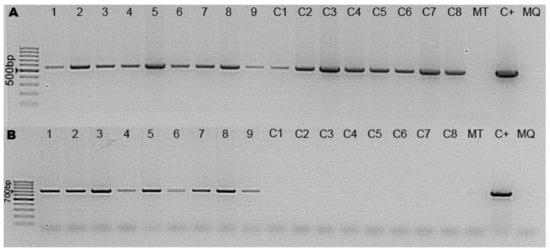
Figure 5.
PCR screening for NeoR/KanR and Dsred genes. (A) PCR results of NeoR/KanR primers: Samples 1–9 represent transformant plants which were selected based on microscopy; Samples C1-C8 represent the control transformant line without the DsRed reporter; MT is a non-transformant Micro-Tom sample; C+ is the vector-positive control; MQ is a MilliQ water sample. (B) PCR results of DsRed-specific primers with the same samples in the same order.
3.6. Long-Term Evaluation of Transformed Roots after Nematode Infection and Analysis of the Presence of Transgenes
To confirm that the expression of transgenes persists for several months in transformed roots, we evaluated plants infected with Meloidogyne after 4 months. Several months after infection, transformed roots infected with M. incognita developed a variety of gall symptoms. We proved that the nematode could complete its life cycle in transformed roots. We observed the presence of DsRed in the hairy roots of each infected plant through a fluorescence microscope. The study confirmed that the strong red fluorescence signal was detected in the roots of all adult plants without exception (Figure 6). It can be concluded that the method we have developed is suitable for testing the long-term function of root-specific genes in S. lycopersicum cv. Micro-Tom plants.
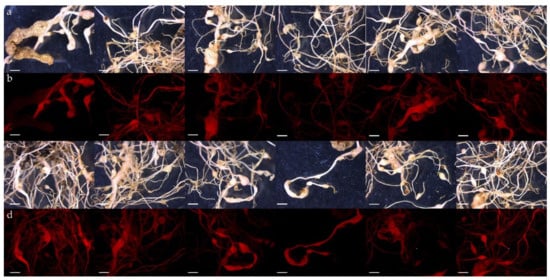
Figure 6.
Selection of transformed hairy roots expressing the DsRed gene in tomato plants infected with Meloidogyne incognita after 4 months. The roots of adult plants were visualized with a stereomicroscope under bright light (line a,c) and under fluorescence (line b,d). Rounded or irregular galls are clearly observed on the roots as a phenotype of typical Meloidogyne spp. infection. Each bar represents 8 mm.
4. Discussion
In comparison with A. tumefaciens-mediated transformation, A. rhizogenes-mediated transformation has a range of benefits [], including the ability to induce genetic transformation in hairy roots deriving from explants quickly and efficiently. Furthermore, individual transformed clones can be identified by using selection marker genes, which enable this technique to be applied in studying gene functions [], plant transformation events [], and secondary metabolism []. In tomato, the cotyledon, hypocotyl, and leaf discs [] were used as materials for transformation []. The most common inoculation method used for the production of composite plants involves stab inoculation []. Recently, several authors reported that the growth rates of some species could substantially be improved when A. rhizogenes was inoculated directly onto the radicles or hypocotyls of freshly sliced seedlings [,]. Similar to other studies, hypocotyl explants produced more hairy roots than leaf discs or cotyledon segments in tomato when inoculated with A. rhizogenes strains [].
In the present study, we used radicles and hypocotyls for inoculation with A. rhizogenes strain ARqua1, and 120 hairy root clones per method were obtained after 7 weeks of cultivation (Figure 1). The DsRed gene was also used as a selection marker, with its expression pattern monitored at various developmental stages, followed by the confirmation of its integration through PCR analysis. The number of re-generate explants was increased by establishing a durable and efficient transformation method. Using roots transformed by A. rhizogenes made the functional validation of candidate genes involved in pathogen–plant root interactions possible [,]. Since most of the studies only presented short-term investigations, it was necessary to confirm that the plants transformed could maintain the expression level of the gene of interest for several months until the infection phenotype of Meloidogyne species appeared. An exception is the study in which Kátia et al. [] verified by PCR and enzyme assays that the nptII gene remained active for several months in S. lycopersicum Mill. cv. Ailsa Craig explants co-cultivated with A. rhizogenes strain R1601. It was also an important goal to investigate whether DsRed could possibly be used to screen transformant roots from the early stages of the experiment because a number of tissues could provide strong autofluoroescent signals at certain wavelengths.
In brief, our results verified that DsRed could be used as a visual screening marker to reduce the difficulties and improve the efficiency of the screening process for positive transgenic plants at different developmental stages and that the red fluorescent signals remained visible throughout the whole cycle of root-knot nematode infection. We established a method for producing A. rhizogenes-dependent transformed roots from S. lycopersicum cv. Micro-Tom that was efficient, convenient, and fast. Three important factors were investigated in our experiments: first, the part of the plant to be transformed; second, the difference between inoculation methods; and third, the quality and efficiency of the transformation method. We found that even though both radicle and hypocotyl segments could be infected by the same ARqua1, the highest transformation efficiency was obtained with hypocotyls of Micro-Tom infected by Method (3) and the lowest with radicles of Micro-Tom infected by Method (1). Our results showed that the puncturing inoculation method proved to be highly efficient in producing composite plants harboring our genes of interest in the roots. Presumably, the puncturing process allowed the bacteria to penetrate deeper into the stem, while the effect of small individual wounds promoted enhanced rooting on the wound sites. In the past, numerous tools were used for puncturing such as sterile toothpicks, stainless steel needles, and sterile scalpels. The tungsten needle we used might be effective since, due to its thin and smooth surface, the needle was able to inflict minimal damage, making it easier for the plant to survive.
In this experiment, hairy roots with red fluorescence were observed in the transformed radicles and hypocotyls using a stereo fluorescence microscope and UV light. Over 90% of the hairy roots showing strong DsRed expression were derived from hypocotyls and radicles. Seven to ten days after the infection, a large number of adventitious roots appeared on the surface of each hypocotyl, which took place much faster than in the radicle segments, which took 15–20 days to develop the roots. In addition, our results showed that the hypocotyl was the most suitable material for A. rhizogenes-mediated transformation of tomato and that the hypocotyl explants of S. lycopersicum cv. Micro-Tom and the A. rhizogenes strain ARqua1 were an appropriate combination for tomato transformation. The microscopy results showed the possibility of extended visual screening with DsRed for Meloidogyne studies, as well as the opportunity to efficiently screen the positive transformation events in the early stages of root formation. Similarly, the PCR validation results confirmed the presence of DsRed in the plants previously evaluated as DsRed positive under the microscope.
The improved A. rhizogenes-mediated transformation method had numerous benefits. Firstly, DsRed served as an observable marker for identifying transformed plants, making the process easier and faster and shortening the transformed plant selection period. Furthermore, the highest transformation efficiency was found to be 95% in tomato plants.
The co-transformation efficiency (number of transformed explants/number of infected explants) of S. lycopersicum cv. Micro-Tom varied from 90% to 95% according to the type of organ (hypocotyl or radicles) and the transformation method used (Figure 3). In a previous study with S. lycopersicum cv. Moneymaker, Plágaro et al. [] achieved a similar high transformation efficiency of almost 90% using A. rhizogenes strain MSU440. The transformation method presented in this study provided a fast and reliable means of studying root-specific genes, promoters, and host–pathogen interactions. The A. rhizogenes-mediated transformation can be used in species that appear to be recalcitrant to plant regeneration or A. tumefaciens transformation such as Capsicum species [], since the method only involves plant rooting.
Several studies reported the possibility of generating stable transformant plants (whole-plant level) by regenerating rhizogenes-transformed root tissues, although the publications mostly focused on ornamental plants [,,]. In the future, we would like to explore this novel approach to generating a high number of transformant plants from transformed root tissues since, in theory, this procedure excludes the generation of mosaic transformation. This method can also aid research work in the field of mycorrhizal studies, as well as be applied in the creation of transgenic plants producing secondary metabolites.
5. Conclusions
In this study, we developed a novel and highly efficient protocol for A. rhizogenes-mediated transformation of S. lycopersicum Micro-Tom cultivars. Furthermore, using the fluorescent molecular marker DsRed, the selection of transformed tomato hairy roots could be effectively performed. Hairy root culture has a number of advantages, including intensive growth, repeatability, and vegetative propagation, which allows the transformed roots to be permanently maintained. This method improves the ability to study root-specific genes and could be used for molecular studies of root–pathogen interactions as well.
As a result of composite plants with well-developed roots that were acclimatized, transformed rootstocks were produced in 2 months, which could be directly used for nematode resistance validation and functional testing. This method had the significant advantage of allowing functional analysis studies on root genes at the whole-plant level. In comparison, in most cases, it takes 1 year to produce well-developed transformed seedlings from primary explants, such as leaf cuttings, using A. tumefaciens-mediated transformation.
The presented study can assist root-specific experiments by offering an effective in vitro protocol to introduce genes into plant roots and enabling the gene products to be screened and localized when the reporter is used in a fusion-protein construction. The method we present can facilitate the advancement of root-related research in the future.
Author Contributions
Conceptualization, M.T. and Z.T.; Data curation, M.T. and Z.T.; Formal analysis, M.T., Z.G.T., Z.S. and Z.T.; Investigation, M.T. and Z.T.; Methodology, M.T. and Z.T.; Project administration, Z.T.; Resources, Z.T.; Supervision, S.F. and Z.T.; Validation, M.T., Z.G.T. and Z.T.; Visualization, Z.T.; Writing—original draft, M.T. and Z.T.; Writing—review & editing, M.T., Z.S. and Z.T. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Food and Agriculture Organization (FAO). FAOSTAT. 2020. Available online: http://www.fao.org/faostat/en/#home (accessed on 15 April 2020).
- Hosmani, P.S.; Flores-Gonzalez, M.; van de Geest, H.; Maumus, F.; Bakker, L.V.; Schijlen, E.; van Haarst, J.; Cordewener, J.; Sanchez-Perez, G.; Peters, S.; et al. An improved de novo assembly and annotation of the tomato reference genome using single-molecule sequencing, Hi-C proximity ligation and optical maps. bioRxiv 2019. [Google Scholar] [CrossRef]
- Su, X.; Wang, B.; Geng, X.; Du, Y.; Yang, Q.; Liang, B.; Meng, G.; Gao, Q.; Yang, W.; Zhu, Y.; et al. A high-continuity and annotated tomato reference genome. BMC Genom. 2021, 22, 898. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.-J.; Uchii, S.; Watanabe, S.; Ezura, H. A Highly Efficient Transformation Protocol for Micro-Tom, a Model Cultivar for Tomato Functional Genomics. Plant Cell Physiol. 2006, 47, 426–431. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Mendívil, A.; Rivera-López, J.; Germán-Báez, L.J.; Lopez-Meyer, M.; Hernández-Verdugo, S.; López-Valenzuela, J.A.; Reyes-Moreno, C.; Valdez-Ortiz, A. A Simple and Efficient Protocol for Plant Regeneration and Genetic Transformation of Tomato cv. Micro-Tom from Leaf Explants. HortScience 2011, 46, 1655–1660. [Google Scholar] [CrossRef] [Green Version]
- Wu, Z.; Sun, S.; Wang, F.; Guo, D. Establishment of Regeneration and Transformation System of Lycopersicon esculentum MicroTom. Br. Biotechnol. J. Int. 2011, 1, 53–60. [Google Scholar] [CrossRef]
- Van Eck, J.; Keen, P.; Tjahjadi, M. Agrobacterium tumefaciens-Mediated Transformation of Tomato. Methods Mol. Biol. 2019, 1864, 225–234. [Google Scholar] [CrossRef]
- Tomato Genome Consortium. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 2012, 485, 635–641. [Google Scholar] [CrossRef] [Green Version]
- Sun, W.; Dunning, F.M.; Pfund, C.; Weingarten, R.; Bent, A.F. Within-species flagellin polymorphism in Xanthomonas campestris pv campestris and its impact on elicitation of Arabidopsis FLAGELLIN SENSING2–dependent defenses. Plant Cell 2006, 18, 764–779. [Google Scholar] [CrossRef] [Green Version]
- Chopra, R.; Johnson, E.B.; Daniels, E.; McGinn, M.; Dorn, K.M.; Esfahanian, M.; Folstad, N.; Amundson, K.; Altendorf, K.; Betts, K.; et al. Translational genomics using Arabidopsis as a model enables the characterization of pennycress genes through forward and reverse genetics. Plant J. 2018, 96, 1093–1105. [Google Scholar] [CrossRef]
- Van Altvorst, A.C.; Bino, R.J.; Van Dijk, A.J.; Lamers AM, J.; Lindhout, W.H.; Van der Mark, F.; Dons, J.J.M. Effects of the introduction of Agrobacterium rhizogenes rol genes on tomato plant and flower development. Plant Sci. 1992, 83, 77–85. [Google Scholar] [CrossRef]
- Bettini, P.P.; Marvasi, M.; Fani, F.; Lazzara, L.; Cosi, E.; Melani, L.; Mauro, M.L. Agrobacterium rhizogenes rolB gene affects photosynthesis and chlorophyll content in transgenic tomato (Solanum lycopersicum L.) plants. J. Plant Physiol. 2016, 204, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Xue, R.F.; Wu, X.B.; Wang, Y.; Zhuang, Y.; Chen, J.; Wu, J.; Ge, W.; Wang, L.; Wang, S.; Blair, M.W. Hairy root transgene expression analysis of a secretory peroxidase (PvPOX1) from common bean infected by Fusarium wilt. Plant Sci. 2017, 260, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plovie, E.; De Buck, S.; Goeleven, E.; Tanghe, M.; Vercauteren, I.; Gheysen, G. Hairy roots to test for transgenic nematode resistance: Think twice. Nematology 2003, 5, 831–841. [Google Scholar] [CrossRef]
- Wiśniewska, A.; Dabrowska-Bronk, J.; Szafrański, K.; Fudali, S.; Święcicka, M.; Czarny, M.; Wilkowska, A.; Morgiewicz, K.; Matusiak, J.; Sobczak, M.; et al. Analysis of tomato gene promoters activated in syncytia induced in tomato and potato hairy roots by Globodera rostochiensis. Transgenic Res. 2013, 22, 557–569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Talano, M.A.; Agostini, E.; Medina, M.I.; Forchetti, D.; Milrad, S.; Tigier, H.A. Tomato (Lycopersicon esculentum cv. Pera) Hairy Root Cultures: Characterization and Changes in Peroxidase Activity under NaCl Treatment. Vitr. Cell. Dev. Biol. Plant 2003, 39, 354–359. [Google Scholar] [CrossRef]
- Rudaya, E.S.; Dolgikh, E.A. Production and analysis of composite tomato plants Solanum lycopersicum L. carrying pea genes encoding the receptors to rhizobial signal molecules. Agric. Biol. 2021, 56, 465–474. [Google Scholar] [CrossRef]
- McCormick, S.; Niedermeyer, J.; Fry, J.; Barnason, A.; Horsch, R.; Fraley, R. Leaf disc transformation of cultivated tomato (L. esculentum) using Agrobacterium tumefaciens. Plant Cell Rep. 1986, 5, 81–84. [Google Scholar] [CrossRef]
- Moghaieb, R.E.; Saneoka, H.; Fujita, K. Shoot regeneration from GUS-transformed tomato (Lycopersicon esculentum) hairy root. Cell. Mol. Biol. Lett. 2004, 9, 439–450. [Google Scholar]
- Karmakar, S.; Molla, K.A.; Gayen, D.; Karmakar, A.; Das, K.; Sarkar, S.N.; Datta, K.; Datta, S.K. Development of a rapid and highly efficient Agrobacterium-mediated transformation system for pigeon pea [Cajanus cajan (L.) Millsp]. GM Crop. Food 2019, 10, 115–138. [Google Scholar] [CrossRef] [Green Version]
- Xiao, Y.; Zhang, S.; Liu, Y.; Chen, Y.; Zhai, R.; Yang, C.; Wang, Z.; Ma, F.; Xu, L. Efficient Agrobacterium-mediated genetic transformation using cotyledons, hypocotyls and roots of ‘Duli’ (Pyrus betulifolia Bunge). Sci. Hortic. 2022, 296, 110906. [Google Scholar] [CrossRef]
- Dan, Y.; Yan, H.; Munyikwa, T.; Dong, J.; Zhang, Y.; Armstrong, C.L. MicroTom—A high-throughput model transformation system for functional genomics. Plant Cell Rep. 2006, 25, 432–441. [Google Scholar] [CrossRef] [PubMed]
- Peres, L.E.P.; Morgante, P.G.; Vecchi, C.; Kraus, J.E.; van Sluys, M.-A. Shoot regeneration capacity from roots and transgenic hairy roots of tomato cultivars and wild related species. Plant Cell Tissue Organ Cult. 2001, 65, 37–44. [Google Scholar] [CrossRef]
- Shikata, M.; Ezura, H. Micro-Tom Tomato as an Alternative Plant Model System: Mutant Collection and Efficient Transformation. Methods Mol Biol. 2016, 1363, 47–55. [Google Scholar] [CrossRef]
- Joäo KH, L.; Brown, T.A. Long-term stability of root cultures of tomato transformed with Agrobacterium rhizogenes R1601. J. Exp. Bot. 1994, 45, 641–647. [Google Scholar] [CrossRef]
- Tóth, Z.; Szabó, Z.; Földi, T.; Szabadi, N.; Hajnik, L.; Jeney, A.; Kiss, G.; Kaló, P. Genetic mapping and identification of the Me1 gene conferring resistance to root-knot nematodes in pepper (Capsicum annuum L.). In Proceedings of the XVIth EUCARPIA, Kecskemét, Hungary, 12–14 September 2016; pp. 542–545. [Google Scholar]
- Mate, T.; Zoltan, S.; Zoltan, T. Alternative method for the transformation of Capsicum species. J. Plant Sci. Phytopathol. 2021, 5, 1–3. [Google Scholar] [CrossRef]
- Horváth, B.; Domonkos, Á.; Kereszt, A.; Szűcs, A.; Ábrahám, E.; Ayaydin, F.; Kaló, P. Loss of the nodule-specific cysteine rich peptide, NCR169, abolishes symbiotic nitrogen fixation in the Medicago truncatula dnf7 mutant. Proc. Natl. Acad. Sci. USA 2015, 112, 15232–15237. [Google Scholar] [CrossRef] [Green Version]
- Domonkos, A.; Kovács, S.; Gombár, A.; Kiss, E.; Horváth, B.; Kováts, G.Z.; Farkas, A.; Tóth, M.T.; Ayaydin, F.; Bóka, K.; et al. NAD1 Controls Defense-Like Responses in Medicago truncatula Symbiotic Nitrogen Fixing Nodules Following Rhizobial Colonization in a BacA-Independent Manner. Genes 2017, 8, 387. [Google Scholar] [CrossRef] [Green Version]
- Sharma, P.K.; Goud, V.V.; Yamamoto, Y.; Sahoo, L. Efficient Agrobacterium tumefaciens-mediated stable genetic transformation of green microalgae, Chlorella sorokiniana. 3 Biotech 2021, 11, 196. [Google Scholar] [CrossRef]
- Ho-Plágaro, T.; Huertas, R.; Tamayo-Navarrete, M.I.; Ocampo, J.A.; García-Garrido, J.M. An improved method for Agrobacterium rhizogenes-mediated transformation of tomato suitable for the study of arbuscular mycorrhizal symbiosis. Plant Methods 2018, 14, 34. [Google Scholar] [CrossRef] [Green Version]
- Aggarwal, P.R.; Nag, P.; Choudhary, P.; Chakraborty, N.; Chakraborty, S. Genotype-independent Agrobacterium rhizogenes-mediated root transformation of chickpea: A rapid and efficient method for reverse genetics studies. Plant Methods 2018, 14, 55. [Google Scholar] [CrossRef] [Green Version]
- Cui, M.-L.; Liu, C.; Piao, C.-L.; Liu, C.-L. A Stable Agrobacterium rhizogenes-Mediated Transformation of Cotton (Gossypium hirsutum L.) and Plant Regeneration from Transformed Hairy Root via Embryogenesis. Front. Plant Sci. 2020, 11, 604255. [Google Scholar] [CrossRef]
- Shahin, E.A.; Sukhapinda, K.; Simpson, R.B.; Spivey, R. Transformation of cultivated tomato by a binary vector in Agrobacterium rhizogenes: Transgenic plants with normal phenotypes harbor binary vector T-DNA, but no Ri-plasmid T-DNA. Theor. Appl. Genet. 1986, 72, 770–777. [Google Scholar] [CrossRef] [PubMed]
- Quandt, H.J.; Pühler, A.; Broer, I.N.G.E. Transgenic root nodules of Vicia hirsuta: A fast and efficient system for the study of gene expression in indeterminate-type nodules. Mol. Plant Microbe Interact. 1993, 6, 699–706. [Google Scholar] [CrossRef]
- Boisson-Dernier, A.; Chabaud, M.; Garcia, F.; Bécard, G.; Rosenberg, C.; Barker, D.G. Agrobacterium rhizogenes-transformed roots of Medicago truncatula for the study of nitrogen-fixing and endomycorrhizal symbiotic associations. Mol. Plant Microbe Interact. 2001, 14, 695–700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tomilov, A.; Tomilova, N.; Yoder, J.I. Agrobacterium tumefaciens and Agrobacterium rhizogenes transformed roots of the parasitic plant Triphysaria versicolor retain parasitic competence. Planta 2007, 225, 1059–1071. [Google Scholar] [CrossRef]
- Kifle, S.; Shao, M.; Jung, C.; Cai, D. An improved transformation protocol for studying gene expression in hairy roots of sugar beet (Beta vulgaris L.). Plant Cell Rep. 1999, 18, 514–519. [Google Scholar] [CrossRef]
- Hwang, C.-F.; Bhakta, A.V.; Truesdell, G.M.; Pudlo, W.M.; Williamson, V.M. Evidence for a Role of the N Terminus and Leucine-Rich Repeat Region of the Mi Gene Product in Regulation of Localized Cell Death. Plant Cell 2000, 12, 1319–1329. [Google Scholar] [CrossRef] [Green Version]
- Li, D.; Zhao, K.; Xie, B.; Zhang, B.; Luo, K. Establishment of a highly efficient transformation system for pepper (Capsicum annuum L.). Plant Cell Rep. 2003, 21, 785–788. [Google Scholar] [CrossRef]
- Bhat, S.R.; Chitralekha, P.; Chandel, K.P.S. Regeneration of plants from long-term root culture of lime, Citrus aurantifolia (Christm.) Swing. Plant Cell Tissue Organ Cult. 1992, 29, 19–25. [Google Scholar] [CrossRef]
- Giri, A.; Narasu, M.L. Transgenic hairy roots: Recent trends and applications. Biotechnol. Adv. 2000, 18, 1–22. [Google Scholar] [CrossRef]
- Choi, P.S.; Kim, Y.D.; Choi, K.M.; Chung, H.J.; Choi, D.W.; Liu, J.R. Plant regeneration from hairy-root cultures transformed by infection with Agrobacterium rhizogenes in Catharanthus roseus. Plant Cell Rep. 2004, 22, 828–831. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).