Abstract
Background: Cryptosporidium spp. cause opportunistic infections in immunosuppressed individuals, such as people living with HIV (PLWH). However, the association between giardiasis and HIV infection remains uncertain. This study assessed co-infections in Ghanaian PLWH and HIV-negative individuals, analyzing socio-economic, clinical, and immunological implications, including the Giardia duodenalis assemblage and Cryptosporidium spp. sub-family levels. Methods: Stool samples from Ghanaian PLWH were tested using several real-time PCR assays targeting G. duodenalis at the species level and assemblages A and B to optimize diagnostic accuracy. GD60 gene-based Sanger sequencing was used for Cryptosporidium spp. subtyping. Results were correlated with anonymized patient data to evaluate interactions with HIV infection. Results: In PLWH, C. hominis Ib, C. hominis Ie, and C. parvum IIc were detected at similar frequencies, followed by C. hominis Ia, C. hominis Id, and C. parvum IIe in decreasing order. Only C. parvum IIc was repeatedly observed in individuals with CD4+ T cell counts above 200/µL, while other sub-families occurred preferentially in those with lower counts. C. hominis Ia and Ib were associated with PLWH not receiving antiretroviral therapy; C. hominis Ia was linked to recently diagnosed HIV infections. No relevant associations between G. duodenalis assemblages and HIV infection were found. Conclusions: Sub-families Ia and Ib of C. hominis preferentially occur in individuals with severe immunosuppression, while C. parvum IIc is also detectable in individuals with better immune function. The prevalence of giardiasis in Ghana appears to be influenced by factors other than HIV-induced immunosuppression.
1. Introduction
Cryptosporidium spp. are protozoan parasites that cause opportunistic enteric infections in humans and various non-human vertebrates, depending on host susceptibility [1]. Besides immunosuppressed individuals, such as people living with HIV (PLWH), children under two years of age are particularly at risk of infection in endemic areas, often due to poor hygiene conditions [2]. Infection typically occurs through contaminated water sources [3], and the parasite’s oocysts are highly resistant to environmental inactivation [4]. Recent evidence also suggests associations with sexually active populations and potential sexual transmission routes, especially among men who have sex with men (MSM) [5]. Therapeutic options remain limited, particularly when the patient’s immune status cannot be improved. Although drugs like paromomycin and nitazoxanide have been used, their efficacy remains unconvincing [4].
In contrast to Cryptosporidium spp., the relationship between HIV infection and enteric infection or colonization with the protozoan parasite Giardia duodenalis remains controversial and uncertain [6]. Studies from Sub-Saharan Africa report a likely association between HIV and increased G. duodenalis co-infection rates in children, with a meta-analysis suggesting a pooled prevalence of 25.7% in this subgroup [7]. Similar trends have been observed in Asia among PLWH [8]. Historically, HIV enteropathy and AIDS-related diarrhea have been linked to G. duodenalis among other pathogens [9,10]. However, a global review on giardiasis in HIV patients found only a slightly to moderately increased odds ratio (OR) of 1.7 for giardiasis in co-infected individuals compared to HIV-negative persons, with a pooled prevalence close to 5% [11]. Importantly, factors such as diarrhea showed a stronger association, increasing the likelihood of G. duodenalis carriage by 3.8 times. The review also indicated a negligible influence of immune competence on giardiasis prevalence, with little difference in the OR between HIV patients on anti-retroviral therapy (ART) compared to HIV patients not receiving this treatment [11]. Potential explanations for a link between giardiasis and HIV include sexual routes shared by both infections [12]. However, this does not explain the high co-infection rates in young African children unlikely to engage in sexual activity [7]. A small US study suggested that HIV patients co-infected with G. duodenalis exhibited higher leukocyte counts and a trend toward lower CD4+ T-lymphocytes. In 24.1% of co-infected individuals, initial metronidazole treatment failed, with increased blood hemoglobin levels uniquely associated with treatment failure [13]. Generally, low CD4+ T-lymphocyte counts are risk factors for severe, disseminated, and atypical protozoan infections, including giardiasis [14]. Impaired antibody responses to G. duodenalis are also considered a risk for poor immunological control [15]. Nevertheless, G. duodenalis infection in HIV patients is usually acute and painful but rarely chronic due to effective antimicrobial treatment [15]. Despite ongoing debate on HIV’s role in giardiasis, there is limited information on G. duodenalis assemblages below the species level [15,16,17,18], especially regarding assemblages A and B, in particular, which are linked to zoonotic and human infections but lack data on HIV associations [16,19].
The quality of prevalence data for protozoan infections like cryptosporidiosis and giardiasis also depends heavily on diagnostic accuracy. Various diagnostic methods exist, including light and fluorescence microscopy, as well as molecular tools [3]. Cryptosporidium spp. show high genetic diversity, with dozens of species and over 100 genotypes described [20]. Sanger sequencing of the GD60 gene is a traditional typing method developed over 20 years ago [21,22] and remains in use in recent Ghanaian studies [23]. Stool microscopy is less sensitive than real-time PCR for detecting G. duodenalis [24], but PCR-based assays also face challenges with suboptimal sensitivity and specificity, including those targeting specific G. duodenalis assemblages [25]. Thus, these limitations should be considered when planning studies on HIV and parasitic co-infections.
In Ghana, PLWH and children are among the groups most susceptible to cryptosporidiosis, with prevalence estimates ranging from 5% to 10% in cross-sectional studies [23,26]. GD60 gene typing in Ghanian children has identified sub-families IIc, Ib, and Ia as the most frequent for C. hominis (Ib, Ia) and C. parvum (IIc) [23]. The present study aims to explore associations between giardiasis overall, G. duodenalis assemblages A and B in particular, and sequence-confirmed Cryptosporidium spp. sub-families with socio-economic, clinical, and immunological factors in Ghanaian PWLH. Cryptosporidium spp. are included as a well-established parameter linked to HIV infection, while such associations are controversial for giardiasis. This exploratory approach seeks to clarify both established and uncertain interactions. To address issues of diagnostic accuracy, multiple Giardia-specific real-time PCR assays were employed alongside GD60 gene Sanger sequencing of Cryptosporidium-positive samples, as detailed in the Methods. Ultimately, this study aims to contribute valuable epidemiological insights into cryptosporidiosis and giardiasis within Ghanaian PLWH as an example of a lower-income setting.
2. Materials and Methods
2.1. Study Design and Sample Materials
PLWH attending the HIV outpatient department at the Komfo Anokye Teaching Hospital (Kumasi, Ghana) were invited to participate in this cross-sectional study, which focused on associations between gastrointestinal and other pathogens and socio-demographic, clinical, and immunological parameters. For comparison, HIV-negative adults from the same region were also included [27,28]. Equal numbers of PLWH receiving and not receiving antiretroviral therapy ensured the assessment of possible therapy effects. The entire study population, comprising both HIV-positive and HIV-negative individuals, was investigated over a 12-month period. Demographic, socio-economic, immunological, and clinical data were collected using standardized questionnaires administered by trained investigators.
2.2. Laboratory Diagnostics
Venous blood samples were used for CD4+ T lymphocytes counts, performed with a FACSCalibur flow cytometer (Becton Dickinson, Mountain View, CA, USA) in Ghana. HIV-1 viral loads were measured with a Real-Time HIV-1 PCR system (Abbott Diagnostics, Wiesbaden, Germany).
Peripheral blood mononuclear cells (PBMCs) were isolated from heparinized venous blood by Ficoll/Hypaque-based density gradient centrifugation (Biocoll Separating Solution, Biochrom AG, Berlin, Germany), washed with phosphate-buffered saline, and resuspended in Roswell Park Memorial Institute 1640 medium (Gibco Invitrogen, Carlsbad, CA, USA) supplemented with heat-inactivated fetal calf serum (Biochrom AG, Berlin, Germany). Cryopreserved PBMCs were shipped in liquid nitrogen to Germany for cell surface marker staining, as previously reported [28]. Cytometric results were obtained using an LSRII flow cytometer (BD Biosciences, Heidelberg, Germany) and analyzed with FlowJo (version 9.6.2, Tree Star, San Carlos, CA, USA).
Native stool aliquots were stored at −80 °C until DNA extraction, which was performed using the QIAamp stool DNA mini kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. In a previous investigation, stool samples had been assessed with three different real-time PCR assays for Cryptosporidium spp., which showed sensitivities of 88.8–100% and specificities of 96.9–99.6% [29]. Samples with at least one positive signal in this prior assessment were included and reanalyzed with a nested PCR on a LightCycler Pro device (Roche, Basel, Switzerland), as detailed in Appendix A, Table A1 and Table A2. Each run included a plasmid-based positive control (sequence inserts in a pEX A128 vector backbone (Eurofins Genomics, Luxembourg)) and a PCR-grade water-based negative control for quality control (Appendix A, Table A1). Amplicons were visualized using a Lonza FlashGel system (Lonza Group, Basel, Switzerland). Gel-positive samples were submitted for nucleic acid extraction and Sanger sequencing to Microsynth Seqlab GmbH (Göttingen, Germany). The same primers used for the secondary PCR reaction (see Appendix A, Table A1, for details) were applied for sequencing. Forward and reverse strand sequences were manually aligned, and quality control was performed using the Finch TV software (Version 1.4, Geospiza Inc., 2004–2012, Seattle, WA, USA). Because sequencing was solely diagnostically performed and not associated with sample-associated characteristics, no sequence files were deposited at international databases. Instead, aligned sequence raw reads are provided in the Appendix A section of this article (please also see the Results for details). Cryptosporidium spp. sub-families were identified using the NCBI GenBank database [30].
For molecular diagnosis of giardiasis, laboratory-developed real-time PCR assays from the literature [25,31,32,33,34,35] were applied to detect G. duodenalis generally, as well as assemblages A and B. These targeted the following: a 75-base pair sequence of the beta giardin (bg) gene (referred to as G. duodenalis PCR 1), a 63-base pair sequence of the 18S rRNA gene (referred to as G. duodenalsis PCR 2), a 98-base pair sequence of the glutamate dehydrogenase (gdh) gene (referred to as G. duodenalis PCR 3) of G. duodenalis, a 75-base pair sequence of the bg gene amplified by assays with (referred to as G. duodenalis assemblage A PCR 1) and without (referred to as G. duodenalis assemblage A PCR 2) the use of hybridization probes containing locked nucleic acids, and a 76-base pair sequence of the triosephosphate isomerase (tpi) gene of of G. duodenalis assemblage A, as well as a 75-base pair sequence of the bg gene amplified by assays with (referred to as G. duodenalis assemblage B PCR 1) and without (referred to as G. duodenalis assemblage B PCR 2) the use of hybridization probes containing locked nucleic acids, and a 82-base pair sequence of the triosephosphate isomerase (tpi) gene of G. duodenalis assemblage B. Assay performance estimates included sensitivities of 17.5–100%, specificities of 84–100%, and limits of detection ranging from <10 to 83 copies per µL eluate, with kappa between 0.155 and 0.908 [25]. Each PCR run included negative (PCR-grade water) and positive (plasmid target sequence in a pEX-A128 vector) controls. Appendix A, Table A3, details oligonucleotide sequences and assay limits of detection. Real-time PCR assays were conducted on Corbett Q cyclers (Qiagen, Hilden, Germany), using master mix compositions and cycling conditions provided in Appendix A, Table A4. To control for sample inhibition, a Phocid herpes virus (PhHV) DNA-specific real-time PCR was used [36].
2.3. Case Definitions and Exclusion and Inclusion Criteria
For Cryptosporidium spp. sub-family identification, only samples with sequences of sufficient quality for sub-family-level assignment were considered positive. Given limitations in giardiasis diagnostic accuracy [25], samples were only considered truly positive if at least two out of three G. duodenalis-specific assays yielded positive results; single positive signals were excluded as likely false positives. Only confirmed G. duodenalis-positive samples were further tested for assemblages A and B using additional real-time PCR assays. Again, positive results in at least 2 out of 3 assemblage-specific assays defined true positives; a lone positive was considered a likely false positive. Samples positive for G. duodenalis but unassignable to assemblage A or B were classified as non-A-non-B assemblages. Duplicate samples from the same patient were excluded unless a new positive result was detected.
Cycle threshold (Ct) values for G. duodenalis-specific real-time PCR were categorized into three semi-quantitative ranges: “High target DNA amount” (Ct < 25), “intermediate target DNA amount” (Ct 25–35), and “low target DNA amount” (Ct > 35). Assignment to these categories was based on the lowest Ct value recorded for each sample. Semi-quantitative assessment was limited to giardiasis, as nested PCR for Cryptosporidium typing did not yield Ct values.
2.4. Statistics
Continuous variables were summarized as median and interquartile range (IQR) and compared using the Kruskal–Wallis test. Categorical variables were analyzed with the generalized Fisher’s exact test. Pairwise post hoc tests with false discovery rate (FDR) correction were conducted for variables with significant results. Multiple linear regression analysis was performed using the R package (Version 4.4.3, R Foundation for Statistical Computing, Vienna, Austria) “forestmodel”. Associations between ordinal and continuous variables were assessed with Kendall’s rank correlation tau. Two-sided p-values were provided, with statistical significance set at α = 5%. All statistical analyses were conducted using R (version 4.4.3, R Foundation for Statistical Computing, Vienna, Austria). Notably, no sample size calculations were performed for this hypothesis-generating, exploratory analysis, which did not include any predefined effect size assumptions.
2.5. Ethics
The study was conducted in accordance with the Declaration of Helsinki and its amendments. Sample collection and analysis followed protocols approved by the Committee on Human Research of the Kwame Nkrumah University of Science and Technology in Kumasi, Ghana, CHRPE/AP/12/11 (approved on 8 September 2011), and the ethics committee of the Medical Council in Hamburg, Germany, PV3771 (approved 13 May 2011). Written informed consent was obtained from all participants.
3. Results
3.1. Application of the Exclusion and Inclusion Criteria to the Samples Tested
From 114 samples that had shown at least one positive Cryptosporidium spp.-specific real-time PCR signal in the previous assessment [29], amplification of a Cryptosporidium spp.-specific sequence allowing for discrimination at the sub-family level was achieved in 47 samples (Appendix A Table A5), which corresponded to 41 patients. Specifically, C. hominis sequences were assigned to 30 patient clusters: Ia (n = 4), Ib (n = 15), Id (n = 3), and Ie (n = 8). Among the remaining 11 patients, sequences were assigned to C. parvum clusters IIc (n = 8) and IIe (n = 3). Detailed sequence results and assignments are presented in Appendix A, Table A5.
Of those tested for molecular evidence of giardiasis, 26 samples were classified as true positives for G. duodenalis DNA, as they yielded positive results in at least two out of three G. duodenalis-specific real-time PCR assays. Signals regarded as likely false positives included 33 from G. duodenalis PCR 1, 64 from G. duodenalis PCR 2, and none from G. duodenalis PCR 3. Among the 26 true-positive samples for G. duodenalis, assemblage-specific PCR assessment identified 6 as assemblage A, 12 as assemblage B, and 8 as non-A-non-B assemblages. Assemblage-specific signals excluded as likely false positives comprised two for assemblage B PCR 2 and one each for assemblage A PCR 2 and assemblage B PCR 3; no such likely false positives were recorded for the remaining assays.
Table 1 presents a visualization of these findings. Notably, co-occurrence of G. duodenalis and Cryptosporidium spp. in the same sample was exceptionally rare. Specifically, G. duodenalis assemblage B and C. hominis Ib were detected together in one sample, and G. duodenalis assemblage B and C. parvum IIc in another. No clear evidence of clustering was observed.

Table 1.
Assignment of samples to Cryptosporidium spp. sub-families and G. duodenalis assemblages. The assessment included 905 stool samples from 730 Ghanaian patients with and without HIV infection.
3.2. Prevalence of DNA of the Various Cryptosporidium spp. Sub-Families as Well as G. duodenalis and Assemblages A and B Within the Stool Samples of the Study Population
Among 730 stool samples analyzed, G. duodenalis DNA was detected in 26 cases (3.6%). Detection rates differed by HIV status: 6 out of 83 HIV-negative individuals (7.2%) tested positive, compared to 20 out of 647 HIV-positive individuals (3.1%, p = 0.105; Figure 1). Assemblage A was identified in six samples (0.8%), comprising one from an HIV-negative subject (1.2%) and five from HIV-positive subjects (0.8%; p = 0.517). Assemblage B was detected in twelve samples (1.6%), including three HIV-negative (3.6%) and nine HIV-positive individuals (1.4%, p = 0.147). Non-A-non-B assemblages accounted for eight cases (1.1%), with two detected among HIV-negative (2.4%) and six among HIV-positive participants (0.9%, p = 0.147).
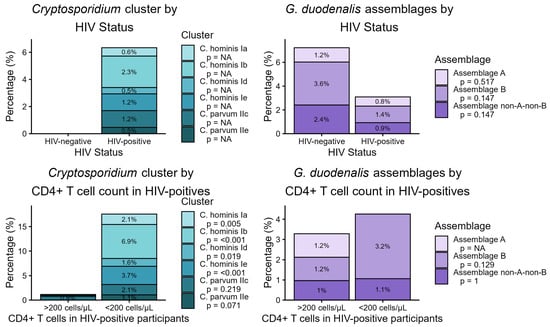
Figure 1.
Prevalence of Cryptosporidium spp. sub-families, as well as G. duodenalis and assemblages A and B, according to the HIV status and stratified by CD4+ T-lymphocyte count. The letter p describes the significance level.
Cryptosporidium spp. DNA was found exclusively in HIV-positive individuals, with 41 out of 647 participants (6.3%) testing positive. Analyzing sub-family prevalence by CD4+ T cell count showed significantly higher detection rates in those with counts below 200/µL. Specifically, C. hominis Ia was detected in 4 out of 188 (2.1%) with low CD4+ T cell counts and in none of the 518 with higher counts (p = 0.005); C. hominis Ib was observed in 13 out of 188 (6.9%) with low CD4+ counts versus 1 out of 518 (0.2%; p < 0.001); C. hominis Id was found in 3 out of 188 (1.6%) compared to none in those with higher counts (0.0%; p = 0.019). C. hominis Ie was present in 7 out of 188 (3.7%) versus 1 out of 518 (0.2%; p < 0.001). For C. parvum IIc, 4 out of 188 (2.1%) samples from individuals with low CD4+ T cell counts and 4 out of 518 (0.8%) samples from those with higher counts were positive (p = 0.219). A non-significant trend was noted for C. parvum IIe, with 2 out of 188 (1.1%) positive in the low CD4+ group and none found among participants with higher counts (p = 0.071).
3.3. Comparison of Demographic, Socio-Economic, and Clinical Characteristics of the HIV-Positive Cohort According to the Presence or Absence of DNA of the Various Cryptosporidium spp. Sub-Families, as Well as G. duodenalis and Assemblages A and B, in Stool Samples
Demographic, socio-economic, medical-treatment-related, and clinical characteristics of HIV-positive participants were evaluated according to the detection of Cryptosporidium spp. sub-families (Table 2). The median age was similar across groups, with no significant differences observed (p = 0.889). Female participants comprised 73.5% of those without detectable Cryptosporidium spp. DNA and varied among sub-families, though without statistically significant differences (p = 0.121). Access to tap water, household electricity, and refrigerator ownership showed no significant variation between groups (p = 0.341, 0.325, and 0.524, respectively). Intake of combination antiretroviral therapy (cART) differed significantly across sub-families (p = 0.001), notably with none of the participants in the Ia and Ib sub-groups receiving cART. Use of trimethoprim/sulfamethoxazole (TMP/SMX) prophylaxis showed a trend toward significance (p = 0.067), with higher proportions in certain sub-families. Body mass index did not differ significantly (p = 0.527). Duration since HIV diagnosis varied significantly (p = 0.005), with the shortest duration observed in the Ia sub-family group. Self-reported clinical symptoms within the previous six months revealed notable findings: cough was more frequently reported in some sub-families (p = 0.006), while diarrhea (p = 0.007), fever (p = 0.005), skin rash (p = 0.001), and weight loss (p = 0.001) also showed variation. Nonetheless, pairwise comparisons with FDR correction did not indicate statistically significant differences.

Table 2.
Demographics, socio-economic parameters, medical treatment, and clinical parameters in HIV-positive participants according to the detection of Cryptosporidium spp. sub-families.
The demographic, socio-economic, medical treatment-related, and clinical characteristics of HIV-positive participants were also analyzed according to the detection of G. duodenalis assemblages (Table 3). The median age across groups showed no statistically significant difference, ranging from 34 (assemblage B) to 46 years (assemblage non-A-non-B) (p = 0.153). Females constituted approximately 73% of the group without detectable G. duodenalis DNA and ranged from 50% to 100% across the assemblage groups, without significant variation (p = 0.124). Access to tap water differed significantly between groups (p = 0.025); however, FDR-corrected pairwise comparisons did not reveal a significant pairwise difference. No significant differences were observed for household electricity access or refrigerator ownership (p = 1.000 and p = 0.324, respectively). The proportion of participants receiving cART and TMP/SMX prophylaxis did not differ significantly across assemblage groups (p = 0.756 and p = 0.241, respectively). Body mass index and time since HIV diagnosis also showed no significant differences (p = 0.678 and p = 0.497, respectively). Self-reported clinical symptoms during the preceding six months, including cough, diarrhea, fever, weight loss, and skin rash, were similar among groups. Although skin rash showed a significant difference in the overall comparison (p = 0.012), FDR-adjusted pairwise comparisons did not detect significant individual differences.

Table 3.
Demographics, socio-economic, medical-treatment-related, and clinical parameters in HIV-positive participants according to the detection of G. duodenalis assemblages.
3.4. Comparison of Virological and Immunological Characteristics of HIV-Positive Participants Depending on the Abundance or Absence of DNA of the Various Cryptosporidium spp. Sub-Families, as Well as G. duodenalis and Assemblages A and B, in Their Stool Samples
Analysis of virological and immunological parameters revealed significant differences among HIV-positive participants, as stratified by the detection of Cryptosporidium spp. sub-families (Table 4). Median viral loads (log_10 copies/mL) were markedly higher in all Cryptosporidium spp.-positive groups compared to participants without detectable Cryptosporidium spp. DNA, ranging from 5.2 to 6.6 versus 4.1, respectively (p < 0.001). Pairwise comparisons adjusted for false discovery rate indicated statistically significant differences between the control group and sub-families Ia, Ib, Id, Ie, and IIc. Similarly, CD4+ T cell counts were significantly lower in all Cryptosporidium spp.-positive sub-groups, with medians ranging between 24 and 155 cells/µL compared to 359 cells/µL in participants without proof of Cryptosporidium spp. DNA (p < 0.001). Significant pairwise differences were observed for all sub-families except IIe. CD8+ T cell counts did not differ significantly among groups (p = 0.307). The CD4+/CD8+ T cell ratio showed pronounced reductions in Cryptosporidium spp.-infected participants, with median ratios between 0.02 and 0.12 versus 0.39 in the control group (p < 0.001). Significant pairwise contrasts were noted for all sub-families (Ia, Ib, Id, Ie, IIc, and IIe).

Table 4.
Virological and immunological parameters according to Cryptosporidium spp. sub-families.
The distribution of virological and immunological parameters stratified by G. duodenalis assemblages in HIV-positive participants did not show statistically significant differences (Table 5). Median viral loads (log_10 copies/mL) were comparable across groups, ranging from 1.6 in the assemblage A group to 4.3 in participants without detectable G. duodenalis DNA (p = 0.556). CD4+ T-lymphocyte counts also did not differ significantly, with medians of 345 cells/µL for assemblage A, 116 for assemblage B, 265 for non-A-non-B assemblages, and 348 for participants without detectable G. duodenalis DNA (p = 0.153). Similarly, CD8+ T cell counts were consistent across groups (p = 0.915). The CD4+/CD8+ T cell ratio showed a non-significant trend toward variation (p = 0.067), with the highest median ratio observed in the assemblage A group (0.7) and the lowest in the group positive for assemblage B (0.1).

Table 5.
Virological and immunological parameters according to G. duodenalis assemblages.
3.5. Factors Associated with Co-Infection with the Various Cryptosporidium spp. Sub-Families as Well as G. duodenalis and Assemblages A and B in the HIV-Positive Cohort
A multivariable linear regression model was employed to examine associations between Cryptosporidium spp. sub-families and G. duodenalis assemblages and CD4+ T cell counts among HIV-positive participants (Figure 2). Using individuals without detectable Cryptosporidium spp. or G. duodenalis as reference groups, the model demonstrated that infection with several Cryptosporidium spp. sub-families was significantly associated with lower CD4+ T cell counts. Specifically, the presence of C. hominis sub-families Ia, Ib, Id, and Ie corresponded to reductions in CD4+ T cell counts ranging from approximately 333 to 384 cells/µL compared to the reference group (all p < 0.05). C. parvum sub-families IIc and IIe were associated with non-significant reductions. None of the G. duodenalis assemblages showed a significant association with CD4+ T cell counts in this model.
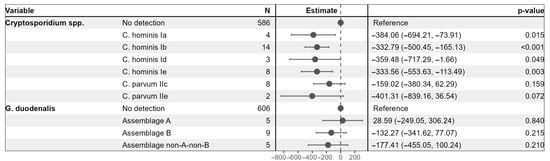
Figure 2.
Multivariable linear regression model to examine the association of Cryptosporidium spp. sub-families and G. duodenalis assemblages with CD4+ T cell counts among HIV-positive participants.
3.6. Correlations of Cycle Threshold (Ct) Values for Specific Sequences of the Various Cryptosporidium spp. Sub-Families as Well as G. duodenalis and Assemblages A and B with CD4+ T Cell Count, CD4+/CD8+ T Cell Ratio, and HIV Viral Load
Correlation analysis between Ct values from real-time PCR targeting G. duodenalis and its assemblages and immunological and virological parameters showed no statistically significant associations in the overall cohort (Table 6). Kendall’s tau coefficients indicated weak positive correlations between Ct values and CD4+ T cell counts (tau = 0.27 for overall G. duodenalis Ct values) and the CD4+/CD8+ T cell ratio (tau = 0.32), although these did not reach significance (p > 0.10). Viral load exhibited a weak negative correlation with Ct values (tau = −0.25), which was also non-significant (p = 0.227). Subgroup analyses of assemblages A and B demonstrated similar non-significant correlations across all immunological and virological parameters.

Table 6.
Correlation of cycle threshold (Ct) values of the real-time PCR targeting G. duodenalis and its assemblages with immunological and virological parameters in the entire cohort.
4. Discussion
The study aimed to provide epidemiological data on Cryptosporidium spp. sub-families and Giardia duodenalis assemblage in Ghanaian PLWH. Several key findings emerged.
Focusing on Cryptosporidium spp. sub-families, stool samples from Ghanaian PLWH harbored DNA of C. hominis Ib, C. hominis Ie, and C. parvum IIc at similar frequencies, followed by C. hominis Ia, C. hominis Id, and C. parvum IIe at lower frequencies. Notably, C. parvum IIc exhibited the highest detection rate among participants with CD4+ T cell counts >200/µL and was the only sub-family detected more than once in this subgroup.
Compared to a previous study in Ghanaian children without known HIV infection during a similar period, the most frequently observed sub-families (C. parvum IIc, C. hominis Ia, and C. hominis Ib) were consistent [23]. The other detected sub-families, C. hominis Id, C. hominis Ie, and C. parvum IIc, also matched previous findings [23]. These observations suggest a relatively stable distribution of these six Cryptospordium spp. sub-families in Ghana regardless of the immunological status or population studied. Even though the order of prevalence differed, C. hominis Ib and C. parvum IIc were among the top three in both Ghanaian PLWH and children [23].
Despite similarities, notable peculiarities were observed in the PLWH population. Specifically, while most Cryptosporidium spp. detections occurred in individuals with low CD4+ T cell counts, C. parvum IIc was most frequently detected among PLWH with CD4+ T cell counts above 200/µL. This parallel with findings in Ghanaian children [23] suggests that C. parvum IIc may be less dependent on host immunosuppression compared to other Cryptosporidium spp. sub-families. This hypothesis aligns with the multivariable regression results, which revealed significant associations between C. hominis sub-families and reduced CD4+ T cell counts, but not for C. parvum IIc or IIe. Additionally, similarly high levels of anthroponotic C. hominis IIc circulation, irrespective of HIV status, have been reported in West African Nigeria [37]. In severely immunocompromised individuals, C. parvum IIc infection has been linked to severe cryptosporidiosis [38], suggesting a potential for increased virulence. Conversely, C. hominis Ia and Ib sub-families were more frequent among HIV-positive individuals not receiving anti-retroviral therapy, with C. hominis Ia especially common in recently diagnosed cases. Together with their significant associations with reduced CD4+ counts, these findings support the hypothesis that these sub-families thrive under severe immunosuppression. Future studies should evaluate whether cluster-specific detection of C. parvum IIc warrants intensified infection prevention and control measures, given its persistence at higher CD4+ levels and possible altered virulence or transmission dynamics.
The distribution of Cryptosporidium spp. sub-families among PLWH appears remarkably consistent across West African countries, with similar profiles reported in Nigeria and Equatorial Guinea [39,40]. Even in East African Ethiopia, the sub-family distribution was comparable [41].
Regardless of sub-family, Cryptosporidium spp. preferentially occurred in newly diagnosed PLWH with CD4+ T cell counts <200/µL, a population at particular risk. This emphasizes the need to explore low-level diagnostic screening for Cryptosporidium spp. in such immunocompromised patients.
Epidemiological data on Cryptosporidium sub-family distributions may prove valuable for future therapeutic development. Current comprehensive reviews [42,43] highlight ongoing efforts to expand treatment beyond immune restoration [4]. While sub-family-specific therapies remain distant [42,43], differential drug effects could influence future therapeutic strategies.
Regarding Giardia duodenalis, no significant associations with HIV infection or any specific assemblage were detected in this study. The observed trend linking lack of tap water access to giardiasis reaffirms established fecal–oral transmission routes [44]. A non-significant tendency associating assemblage B and non-A-non-B assemblages with skin rash was a likely statistical artifact. These results support the critical view questioning a strong HIV–giardiasis link [11]. Notably, non-significant trends suggested a reduced CD4+/CD8+ ratio and increased G. duodenalis quantity (by Ct value) associated with low CD4+ cell counts and high HIV viral loads, paralleling a previous US study indicating trends toward reduced CD4+ T-lymphocytes in HIV patients co-infected with G. duodenalis [13]. However, the lack of statistical significance tempers interpretation.
This study has a number of limitations. First and most importantly, the retrospective design precluded sample size adjustments based on expected effect sizes, potentially missing small associations. The associated very small subgroup sizes reduced the statistical power of the study. Second, limited epidemiologic data constrained analytical options. Third, interpretable Cryptospodium spp. sequencing was possible in only a minority of PCR-positive samples, with some sequences short and low quality, necessitating cautious interpretation. Therefore, phylogenetic analysis beyond the sub-family level was avoided to prevent erroneous conclusions.
5. Conclusions
This study confirmed a strong association between cryptosporidiosis and HIV infection in the Ghanaian cohort examined. DNA of C. hominis Ib, C. hominis Ie, and C. parvum IIc were detected at similar frequencies, followed by C. hominis Ia and C. hominis Id, and C. parvum IIe at lower frequencies. Notably, only C. parvum IIc was repeatedly detected in individuals with CD4+ T cell counts greater than 200/µL. C. hominis Ia and Ib were associated with PLWH not receiving ART, with C. hominis Ia additionally linked to recently diagnosed HIV infections. These findings suggest the preferential occurrence of certain sub-families in individuals with severe immunosuppression. In contrast, no significant associations were found between G. duodenalis assemblages and HIV infection, indicating that other factors likely influence G. duodenalis epidemiology in Ghana. Future research should focus on longitudinal studies to track infection dynamics relative to cART initiation and evaluate the integration of both traditional and molecular parasitic diagnostics into routine HIV care in endemic settings. Particularly for resource-limited areas, there is a critical need to develop and implement affordable, field-adapted diagnostic tools.
Author Contributions
Conceptualization, H.F. and K.A.E.; methodology, H.F., L.G., F.W., A.E. and V.D.C.; software, H.F., F.W., L.G. and K.A.E.; validation, H.F., F.W., L.G. and K.A.E.; formal analysis, F.W., L.G. and K.A.E.; investigation, H.F., F.S.S., B.R.N., A.D., S.O.A., R.B., E.O.K., E.A.-A., F.W., L.G., M.K.A., V.D.C., T.F. and K.A.E.; resources, H.F. and K.A.E.; data curation, L.G. and K.A.E.; writing—original draft preparation, K.A.E., H.F. and L.G.; writing—review and editing, H.F., F.S.S., B.R.N., A.D., S.O.A., R.B., E.O.K., F.W., S.P., M.K.A., T.B.T., L.G., E.A.-A., A.E., V.D.C., T.F. and K.A.E.; visualization, K.A.E.; supervision, H.F. and K.A.E.; project administration, H.F. and K.A.E.; funding acquisition, H.F. and K.A.E. All authors have read and agreed to the published version of the manuscript.
Funding
The provided study was financially supported by the ESTHER Alliance for Global Health Partnerships, the German Federal Ministry of Education and Research (Project No. 01KA1102), and the German Ministry of Defense (grant 36K2-S-45 1922). In addition, we acknowledge the generous support of the Heinz Ansmann Foundation for AIDS Research.
Institutional Review Board Statement
This study was performed in compliance with the Declaration of Helsinki and all its amendments. The assessed samples were collected and analyzed based on protocols ethically cleared by the Committee on Human Research of the Kwame Nkrumah University of Science and Technology in Kumasi, Ghana, CHRPE/AP/12/11, approved on was 8 September 2011, and the ethics committee of the Medical Council in Hamburg, Germany, PV3771, approved on 13 May 2011.
Informed Consent Statement
Participants were only included in the study if written informed consent was either provided by themselves or by their next-of-kin if applicable.
Data Availability Statement
All relevant data are provided in the manuscript or its Appendix A tables. Raw data can be made available upon reasonable request.
Acknowledgments
Annett Michel and Simone Priesnitz are gratefully acknowledged for excellent technical assistance. In addition, we acknowledge the generous support of the Heinz Ansmann Foundation for AIDS Research.
Conflicts of Interest
The authors declare that this research was conducted in the absence of any commercial or financial relationships that could be construed as potential conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of the data; in the writing of the manuscript; or in the decision to publish the results.
Abbreviations
The following abbreviations are used in this manuscript:
| 95% CI | 95% confidence interval |
| AIDS | acquired immune deficiency syndrome |
| cART | combined anti-retroviral therapy |
| Ct | cycle threshold |
| DNA | deoxyribonucleic acid |
| FDR | false discovery rate |
| HIV | human immunodeficiency virus |
| IQR | interquartile range |
| Min. | minute |
| MSM | men who have sex with men |
| N | number |
| n.e. | not estimable |
| OR | odds ratio |
| PBMC | peripheral blood mononuclear cells |
| PCR | polymerase chain reaction |
| PLWH | people living with HIV |
| SD | standard deviation |
| Sec. | second |
| TMP/SMX | trimethoprim/sulfamethoxazole |
Appendix A

Table A1.
Oligonucleotides used for the Cryptosporidium spp.-specific nested PCR and for the subsequent Sanger sequencing. Hyphens in the oligonucleotide sequences have been inserted to increase the readability, not to delineate codon triplets.
Table A1.
Oligonucleotides used for the Cryptosporidium spp.-specific nested PCR and for the subsequent Sanger sequencing. Hyphens in the oligonucleotide sequences have been inserted to increase the readability, not to delineate codon triplets.
| PCR target | Cryptosporidium spp. (primary PCR) |
| Target gene | GP60 gene |
| Forward primer | 5′-ATA-GTC-TCC-GCT-GTA-TTC-3′ |
| Reverse primer | 5′-GGA-AGG-AAC-GAT-GTA-TCT-3′ |
| Positive control plasmid insert | 5′-ACT-CTC-CGT-TAT-AGT-CTC-CGC-TGT-ATT-CTC-AGC-CCC-AGC-CGT-TCC-ACT-CAG-AGG-AAC-TTT-AAA-GGA-TGT-TCC-TGT-TGA-GGG-CTC-ATC-ATC-GTC-ATC-GTC-ATC-GTC-ATC-GTC-ATC-ATC-ATC-ATC-ATC-ATC-ATC-ATC-ATC-ATC-ATC-AAC-ATC-AAC-CGT-CGC-ACC-AGC-AAA-TAA-GGC-AAG-AAC-TGG-AGA-AGA-CGC-AGA-AGG-CAG-TCA-AGA-TTC-TAG-TGG-TAC-TGA-AGC-TTC-TGG-TAG-CCA-GGG-TTC-TGA-AGA-GGA-AGG-TAG-TGA-AGA-CGA-TGG-CCA-AAC-TAG-TGC-TGC-TTC-CCA-ACC-CAC-TAC-TCC-AGC-TCA-AAG-TGA-AGG-CGC-AAC-TAC-CGA-AAC-CAT-AGA-AGC-TAC-TCC-AAA-AGA-AGA-ATG-CGG-CAC-TTC-ATT-TGT-AAT-GTG-GTT-CGG-AGA-AGG-TAC-CCC-AGC-TGC-GAC-ATT-GAA-GTG-TGG-TGC-CTA-CAC-TAT-CGT-CTA-TGC-ACC-TAT-AAA-AGA-CCA-AAC-AGA-TCC-CGC-ACC-AAG-ATA-TAT-CTC-TGG-TGA-AGT-TAC-ATC-TGT-AAC-CTT-TGA-AAA-GAG-TGA-TAA-TAC-AGT-TAA-AAT-CAA-GGT-TAA-CGG-TCA-GGA-TTT-CAG-CAC-TCT-CTC-TGC-TAA-TTC-AAG-TAG-TCC-AAC-TGA-AAA-TGG-CGG-ATC-TGC-GGG-TCA-GGC-TTC-ATC-AAG-ATC-AAG-AAG-ATC-ACT-CTC-AGA-GGA-AAC-CAG-TGA-AGC-TGC-TGC-AAC-CGT-CGA-TTT-GTT-TGC-CTT-TAC-CCT-TGA-TGG-TGG-TAA-AAG-AAT-TGA-AGT-GGC-TGT-ACC-AAA-CGT-CGA-AGA-TGC-ATC-TAA-AAG-AGA-CAA-GTA-CAG-TTT-GGT-TGC-AGA-CGA-TAA-ACC-TTT-CTA-TAC-CGG-CGC-AAA-CAG-CGG-CAC-TAC-CAA-TGG-TGT-CTA-CAG-GTT-GAA-TGA-GAA-CGG-AGA-CTT-GGT-TGA-TAA-GGA-CAA-CAC-AGT-TCT-TTT-GAA-GGA-TGC-TGG-TTC-CTC-TGC-TTT-TGG-ACT-CAG-ATA-CAT-CGT-TCC-TTC-CGT-TTT-TGC-AA-3′ |
| GenBank accession number used for the insert | MK034696.1 |
| Reference | [21,22] |
| PCR target | Cryptosporidium spp. (secondary PCR) |
| Target gene | GP60 gene |
| Forward primer | 5′-TCC-GCT-GTA-TTC-TCA-GCC-3′ |
| Reverse primer | 5′-GCA-GAG-GAA-CCA-GCA-TC-3′ |
| Reference | [21,22] |
| Sequencing target | Cryptosporidium spp. (Sanger sequencing) |
| Target gene | GP60 gene |
| Forward primer | 5′-TCC-GCT-GTA-TTC-TCA-GCC-3′ |
| Reverse primer | 5′-GCA-GAG-GAA-CCA-GCA-TC-3′ |
| Reference | [21,22] |

Table A2.
Reaction mixes and run conditions for PCR assays targeting Cryptosporidium spp.
Table A2.
Reaction mixes and run conditions for PCR assays targeting Cryptosporidium spp.
| Cryptosporidium spp. (Primary PCR) | Cryptosporidium spp. (Secondary PCR) | |
|---|---|---|
| Reaction chemistry | ||
| Master Mix | HotStar master mix (Qiagen, Hilden, Germany) | HotStar master mix (Qiagen, Hilden, Germany) |
| Reaction volume (µL) | 25 | 50 |
| Forward primer concentration (pmol/µL) | 40 | 20 |
| Reverse primer concentration (pmol/µL) | 40 | 20 |
| Final Mg2+ concentration (mM) | 5 | 3 |
| Eluate volume (µL) | 2 | 10 |
| Run conditions | ||
| Initial denaturation | 10 min. 94 °C | 10 min. 94 °C |
| Cycle numbers | 35 | 35 |
| Denaturation | 45 s. 94 °C | 45 s. 94 °C |
| Annealing | 45 s. 55 °C | 45 s. 55 °C |
| Amplification | 60 s. 72 °C | 60 s. 72 °C |
| Hold Cooling | 7 min. 72 °C 30 s. 40 °C | 7 min. 72 °C 30 s. 40 °C |
min. = minute; s. = second.

Table A3.
Target genes, calculated detection limits, and oligonucleotides used for the applied real-time PCR screening assays for Giardia duodenalis, as well as assemblages A and B. Hyphens in the oligonucleotide sequences have been inserted to increase the readability, not to delineate codon triplets.
Table A3.
Target genes, calculated detection limits, and oligonucleotides used for the applied real-time PCR screening assays for Giardia duodenalis, as well as assemblages A and B. Hyphens in the oligonucleotide sequences have been inserted to increase the readability, not to delineate codon triplets.
| PCR target | Giardia duodenalis PCR 1 |
| Target gene | bg |
| Detection limit | <10 copies/µL |
| Forward primer | 5′-CAT-CCG-CGA-GGA-GGT-CAA-3′ |
| Reverse primer | 5′-GCA-GCC-ATG-GTG-TCG-ATC-T-3′ |
| Probe and modifications | 5′-Cy5-AAG-TCC-GCC-GAC-AAC-ATG-TAC-CTA-ACG-A-BHQ2-3′ |
| Positive control plasmid insert | 5′-CGT-TCG-AGG-ACA-TCC-GCG-AGG-AGG-TCA-AGA-AGT-CCG-CCG-ACA-ACA-TGT-ACC-TAA-CGA-TCA-AGG-AGG-AGA-TCG-ACA-CCA-TGG-CTG-CAA-ACT-TCC-GC-3′ |
| GenBank accession number used for the insert | KT728533.1 |
| Reference | [25,31] |
| PCR target | Giardia duodenalis PCR 2 |
| Target gene | 18S rRNA gene sequence |
| Detection limit | <10 copies/µL |
| Forward primer | 5′-GAC-GGC-TCA-GGA-CAA-CGG-TT-3′ |
| Reverse primer | 5′-TTG-CCA-GCG-GTG-TCC-G-3′ |
| Probe and modifications | 5′-FAM-CCC-GCG-GCG-GTC-CCT-GCT-AG-BHQ1-3′ |
| Positive control plasmid insert | 5′-GGA-CGC-GGC-GGA-CGG-CTC-AGG-ACA-ACG-GTT-GCA-CCC-CCC-GCG-GCG-GTC-CCT-GCT-AGC-CGG-ACA-CCG-CTG-GCA-ACC-CGG-CGC-CA-3′ |
| GenBank accession number used for the insert | MT484081.1 |
| Reference | [25,32] |
| PCR target | Giardia duodenalis PCR 3 |
| Target gene | gdh |
| Detection limit | <10 copies/µL |
| Forward primer | 5′-CTG-AAG-AAC-TCC-CTC-ACC-AC-3′ |
| Reverse primer | 5′-CAG-AAG-CGC-ATG-ACC-TCG-TTG-3′ |
| Probe and modifications | 5′-HEX-CAA-GGG-CGG-CTC-CGA-CTT-TGA-CCC-AA-BHQ1-3′ |
| Positive control plasmid insert | 5′-TCG-AGC-AGA-TCC-TGA-AGA-ACT-CCC-TCA-CCA-CGC-TCC-CGA-TGG-GCG-GCG-GCA-AGG-GCG-GCT-CCG-ACT-TTG-ACC-CAA-AGG-GCA-AGT-CCG-ACA-ACG-AGG-TCA-TGC-GCT-TCT-GCC-AGT-CCT-TC-3′ |
| GenBank accession number used for the insert | MH311031.1 |
| Reference | [25,33] |
| PCR target | Giardia duodenalis assemblage A PCR 1 |
| Target gene | Bg |
| Detection limit | <10 copies/µL |
| Forward primer | 5′-CCT-CAA-GAG-CCT-GAA-CGA-TCT-C-3′ |
| Reverse primer | 5′-AGC-TGG-TCG-TAC-ATC-TTC-TTC-CTT-3′ |
| Probe and modifications | 5′-FAM-TTC-TCC-GTG-GCA-ATG-CCC-GTC-T-BHQ1-3′ |
| Positive control plasmid insert | 5′-GGA-AGG-AGG-CCC-TCA-AGA-GCC-TGA-ACG-ATC-TCG-AGA-CGG-GCA-TTG-CCA-CGG-AGA-ACG-CAG-AAA-GGA-AGA-AGA-TGT-ACG-ACC-AGC-TCA-ACG-AGA-AG-3′ |
| GenBank accession number used for the insert | KT728533.1 |
| Reference | [25,31] |
| PCR target | Giardia duodenalis assemblage A PCR 2 |
| Target gene | bg |
| Detection limit | 83 copies/µL |
| Forward primer | 5′-CCT-CAA-GAG-CCT-GAA-CGA-TCT-C-3′ |
| Reverse primer | 5′-AGC-TGG-TCG-TAC-ATC-TTC-TTC-CTT-3′ |
| Probe and modifications | 5′-6-FAM-TGG-C+A+A-TGC-C+CG-+TCT-BHQ1-3′ |
| Positive control plasmid insert | 5′-GGA-AGG-AGG-CCC-TCA-AGA-GCC-TGA-ACG-ATC-TCG-AGA-CGG-GCA-TTG-CCA-CGG-AGA-ACG-CAG-AAA-GGA-AGA-AGA-TGT-ACG-ACC-AGC-TCA-ACG-AGA-AG-3′ |
| GenBank accession number used for the insert | KT728533.1 |
| Reference | [25,34] |
| PCR target | Giardia duodenalis assemblage A PCR 3 |
| Target gene | tpi |
| Detection limit | <10 copies/µL |
| Forward primer | 5′-CAT-TGC-CCC-TTC-CGC-C-3′ |
| Reverse primer | 5′-CTG-CGC-TGC-TAT-CCT-CAA-CTG-3′ |
| Probe and modifications | 5′-VIC-CCA-TTG-CGG-CAA-ACA-MGB-NFQ-3′ |
| Positive control plasmid insert | 5′-TGG-ACG-TCG-TCA-TTG-CCC-CTT-CCG-CCG-TAC-ACC-TGT-CAA-CAG-CCA-TTG-CGG-CAA-ACA-CGT-CAA-AAC-AGT-TGA-GGA-TAG-CAG-CGC-AGA-ATG-TGT-ACC-3′ |
| GenBank accession number used for the insert | MZ822181.1 |
| Reference | [25,35] |
| PCR target | Giardia duodenalis assemblage B PCR 1 |
| Target gene | bg |
| Detection limit | <10 copies/µL |
| Forward primer | 5′-CCT-CAA-GAG-CCT-GAA-CGA-CCT-C-3′ |
| Reverse primer | 5′-AGC-TGG-TCA-TAC-ATC-TTC-TTC-CTC-3′ |
| Probe and modifications | 5′-Cy5-TTC-TCC-GTG-GCG-ATG-CCT-GTC-T-BHQ2-3′ |
| Positive control plasmid insert | 5′-GGA-AGG-AGG-CCC-TCA-AGA-GCC-TGA-ACG-ACC-TCG-AGA-CAG-GCA-TCG-CCA-CGG-AGA-ACG-CCG-AGA-GGA-AGA-AGA-TGT-ATG-ACC-AGC-TCA-ACG-AGA-AA-3′ |
| GenBank accession number used for the insert | PP566783.1 |
| Reference | [25,31] |
| PCR target | Giardia duodenalis assemblage B PCR 2 |
| Target gene | bg |
| Detection limit | 83 copies/µL |
| Forward primer | 5′-CCT-CAA-GAG-CCT-GAA-CGA-CCT-C-3′ |
| Reverse primer | 5′-AGC-TGG-TCA-TAC-ATC-TTC-TTC-CTC-3′ |
| Probe and modifications | 5′-Cy5-TGG-CG+A-TGC-+C+T+G-TCT-BHQ2-3′ |
| Positive control plasmid insert | 5′-GGA-AGG-AGG-CCC-TCA-AGA-GCC-TGA-ACG-ACC-TCG-AGA-CAG-GCA-TCG-CCA-CGG-AGA-ACG-CCG-AGA-GGA-AGA-AGA-TGT-ATG-ACC-AGC-TCA-ACG-AGA-AA-3′ |
| GenBank accession number used for the insert | PP566783.1 |
| Reference | [25,34] |
| PCR target | Giardia duodenalis assemblage B PCR 3 |
| Target gene | tpi |
| Detection limit | <10 copies/µL |
| Forward primer | 5′-GAT-GAA-CGC-AAG-GCC-AAT-AA-3′ |
| Reverse primer | 5′-TCT-TTG-ATT-CTC-CAA-TCT-CCT-TCT-T-3′ |
| Probe and modifications | 5′-FAM-AAT-ATT-GCT-CAG-CTC-GAG-MGB-NFQ-3′ |
| Positive control plasmid insert | 5′-AGA-GAC-CCT-GGA-TGA-ACG-CAA-GGC-CAA-TAA-CAC-TAT-GGA-GGT-GAA-TAT-TGC-TCA-GCT-CGA-GGC-TCT-TAA-GAA-GGA-GAT-TGG-AGA-ATC-AAA-GAA-GTT-ATG-GGA-3′ |
| GenBank accession number used for the insert | KU311953.1 |
| Reference | [25,35] |
+ = following base is LCA (locked nucleic acid).

Table A4.
Reaction mixes and run conditions for Giardia duodenalis, as well as assemblages A and B.
Table A4.
Reaction mixes and run conditions for Giardia duodenalis, as well as assemblages A and B.
| G. duodenalis PCR 1-3 | G. duodenalis Assemblage A PCR 1, G. duodenalis Assemblage B PCR 1 | G. duodenalis Assemblage A PCR 2, G. duodenalis Assemblage B PCR 2 | G. duodenalis Assemblage A PCR 3, G. duodenalis Assemblage B PCR 3 | |
|---|---|---|---|---|
| Reaction chemistry | ||||
| Master Mix | HotStar master mix (Qiagen, Hilden, Germany) | HotStar master mix (Qiagen, Hilden, Germany) | HotStar master mix (Qiagen, Hilden, Germany) | HotStar master mix (Qiagen, Hilden, Germany) |
| Reaction volume (µL) | 20.0 | 20.0 | 20.0 | 20.0 |
| Forward primer concentration (nM) | 300.0 (PCR 1), 125.0 (PCR 2), 200.0 (PCR 3) | 300.0 | 300.0 | 300.0 |
| Reverse primer concentration (nM) | 300.0 (PCR 1), 125.0 (PCR 2), 200.0 (PCR 3) | 300.0 | 300.0 | 900.0 |
| Probe concentration (nM) | 6.0 (PCR 1), 10.0 (PCR 2), 200.0 (PCR 3) | 200.0 | 200.0 | 100.0 |
| Final Mg2+ concentration (mM) | 1.5 | 4.0 | 3.0 | 3.0 |
| Bovine serum albumin (mg/mL) | 2.0 | 2.0 | 2.0 | 2.0 |
| Eluate volume (µL) | 2.0 | 2.0 | 2.0 | 2.0 |
| Run conditions | ||||
| Initial denaturation | 15 min. at 95 °C | 15 min. at 95 °C | 15 min. at 95 °C | 15 min. at 95 °C |
| Cycle numbers | 40 | 40 | 50 | 50 |
| Denaturation | 15 s. at 95 °C | 15 s. at 95 °C | 15 s. at 95 °C | 15 s. at 95 °C |
| Annealing | 60 s. at 60 °C | 60 s. at 60 °C | 8 s. at 58 °C | 60 s. at 60 °C |
| Amplification | same as annealing | same as annealing | 3 s. at 72 °C | same as annealing |
| Hold | 10 s. at 40 °C | 10 s. at 40 °C | 10 s. at 40 °C | 10 s. at 40 °C |
min. = minute; s. = second.

Table A5.
Sequences of the obtained amplicons. Dashes within the sequences are included to improve readability, not to delineate amino acid–coding base triplets.
Table A5.
Sequences of the obtained amplicons. Dashes within the sequences are included to improve readability, not to delineate amino acid–coding base triplets.
| Anonymized Sample-ID | Sequence | NCBI GenBank Accession Number of Best Matching Sequence | Species Level Assignment | Sub-Family Level Assignment |
|---|---|---|---|---|
| 1 | 5′-TTT-TTC-TCA-GCC-CCA-GCC-GTT-CCA-CTC-AGA-GGC-ACT-TTA-AAG-GAT-GTT-TCT-GTT-GAG-AGC-TCA-TCG-TCA-TCA-TCG-TCA-TCG-TCA-ACA-ACC-CCC-GCA-CCA-GCT-CCA-AAG-AAG-GTA-AGA-GAA-AGC-GAA-GAA-GGG-AAG-AAC-AGT-GAA-GAT-AGT-CAA-ACT-CCC-GCT-AGT-CCT-GGA-AGT-GAT-TCT-CAG-GAT-AGC-TCT-AAA-GGA-GAC-GAA-GTT-GTA-GAT-GGA-GGC-GCT-TCC-GGA-TCT-AGT-ACC-CCA-ACT-CAA-GCT-GCT-GAA-AAG-GAG-CCC-GAA-ACT-CCA-GAA-TCT-ACT-CCA-AAG-GAA-GAA-TGT-GGT-ACT-TCA-TTT-ATA-ATG-TGG-TTC-GGA-GAA-GGT-ACT-CCA-GCC-ACA-ACT-TTG-AAG-TGC-GGT-GGC-TAC-ACT-ATC-GTC-TAT-GCA-CCA-GAA-AAG-GAT-AAT-AAA-GAA-CCC-GCA-CCA-AGA-TAC-ATC-TCT-GGT-GAT-GTT-AAG-GCT-GTA-ACC-TTT-GAA-AAG-GGA-GAA-GAT-AAT-ACA-GTT-AAA-ATC-AAG-GTT-GAT-GGT-AAG-GAG-TTC-AGT-ACT-CTC-TCT-TCT-AGC-TCA-AGC-AGT-CCA-ACT-GAA-AAT-AAC-GGA-TCT-ACG-GGC-CAG-GTT-GCA-TCA-AGA-TCA-AGA-AGA-TCG-CTC-TCA-GAG-GAA-AAT-AGT-GAA-ACT-GCT-GCA-ACC-GTC-GAT-TTG-TTT-GCC-TTC-ACC-CTT-CAA-GGT-GGT-AAA-AGA-ATC-GAA-GTC-GCT-GTG-CCA-AGT-GAC-AAA-GAT-GTA-TCC-AAG-AGA-AAC-AAG-TAC-AGT-TTG-GTT-GCA-GGC-GAT-AAG-ACT-TTC-TAT-ACC-GGC-GCA-AAT-AGC-GGT-AAT-ACC-GAC-GGT-ATCT-ACA-GGT-TGA-ATG-ATG-ATG-GAG-ACT-TGG-TGG-ACA-AGA-ACA-ACA-ACG-TTC-TTT-TGA-AGG-ATG-TG-3′ | KU670812 | C. parvum | IIc |
| 2 | 5′-KTT-TTT-TTT-YAC-CCA-GCC-GTT-CCA-CTC-AGA-GGC-ACT-TTA-AAG-GAT-GCT-TCT-GTT-GAG-GGC-TCA-TCA-TCA-TCA-TCA-TCA-TCA-TCA-TCA-TCA-TCG-ACC-ACC-GTC-GCA-CCA-GCT-CCA-AAG-AAA-GAA-AGA-ACT-GGA-GAG-GGC-GTA-GAT-GGA-AAG-GAC-CAA-GTA-GAT-AGT-ACA-GGT-TCT-GAT-CAG-AAC-AGT-AAA-GGA-GAC-ACT-AAA-GGA-ACC-ACA-GAA-GAT-GGT-AAA-GAG-ACC-GAA-GGT-ACT-GTT-TCC-AAA-CCC-ACT-ACT-CCA-GAT-CAA-GGT-GAG-AGC-GCA-ACT-CCC-GGA-TCC-ACG-GAA-ACT-ACT-CCA-AAG-GAA-GAA-TGC-GGT-ACT-TCA-TTT-GTA-ATG-TGG-TTC-GGA-GAA-GGT-ACC-CCA-GTT-GCG-ACC-TTG-AAG-TGT-GGT-GGT-TAC-ACT-ATC-GTC-TAT-GCA-CCT-GTA-AAG-GAA-CAA-ACA-AAT-CCC-GCA-CCA-AGA-TAT-ATC-TCT-GGT-GAG-GTA-AAA-AAT-GTA-TCC-TTC-CAA-AAA-GAA-AGT-GAT-GGT-ACA-ATT-AAAA-TCA-AGA-TTG-ACG-AAA-AGG-AGT-TCA-GTT-CTC-TCT-CTA-CTG-ACT-CAA-GCA-CTC-CAA-CTG-CAA-ATA-GCG-GAT-CCG-CGG-AAC-AGG-TTC-AAT-CAA-GAT-CAA-GAA-GAT-CAC-TCA-CAG-AGG-GAA-GTG-AAA-CAC-CTG-CAA-CCG-TCG-ATT-TGT-TTG-CCT-TCA-CCC-TTG-ATG-GTG-GTA-AAA-GAA-TTG-AAG-TGG-CTG-TAC-CAA-ACA-ACG-AGG-ATG-CAT-CCA-AAA-GAA-CCG-AGT-ACA-GTT-TGG-TTG-CAA-ACG-ATA-AGC-CTT-TCT-ATA-CCG-GCG-CAA-ATA-GCG-GCA-CCG-AAA-ATG-GTG-TCT-ACA-AGT-TGA-ATG-AGA-ACG-GAG-ACT-TGG-TTG-ACA-AGG-ACA-ATA-AAG-TTC-TTT-TGA-ARS-GG-3′ | MW480843 | C. parvum | IIe |
| 3 | 5′-TTT-TTT-TTY-ACC-CAC-CGT-CCC-ACT-CAG-AGG-CAC-CTT-GAA-GGA-TGT-TTC-TGT-TGA-GGG-CTC-ATC-ATC-ATC-ATC-TTC-ATC-ATC-GTC-TTC-ATC-TTC-ATC-ATC-ATC-ATC-ATC-GTC-AAC-AAC-CCC-AGC-ACC-AGC-TTC-AAA-GAA-GGT-AAG-AGA-AGC-AGA-AGG-CAG-TGT-AGA-AAA-GGG-CAG-TGA-AGA-AAA-GGA-CAG-TGA-AGA-AAA-GGG-CAG-TGA-AGA-AAA-GGG-CAG-TGA-AGA-AGA-TAG-CCA-AAC-TCC-CGC-TAG-TCC-TGG-AGG-TGG-AGG-GGT-GAG-TGA-AGG-AGA-TAC-TCA-AGG-TGA-CTC-TAA-AGG-AGA-CGG-AGT-TAG-TGA-AGA-TGA-GAA-CCA-AAG-TCA-AGG-TGG-GGA-CGC-TAC-TTC-CGA-ATC-TAG-CAC-CCA-AAC-TCA-AGC-TAC-TGA-AAA-AGA-ACC-CGG-ATC-TTC-AGA-AGC-TAC-TCC-AAA-GGA-AGA-GTG-CGG-TAC-TTC-ATT-TGT-AAT-GTG-GTT-CGG-ACA-GGG-TGT-TCC-AGT-TGT-AAC-TTT-GAA-GTG-TGG-TGG-CTA-TAC-TAT-GGT-CTA-TGC-ACC-AGA-AAA-TGG-CAA-AAC-AGA-TCC-CGC-ACC-AAG-ATA-TAT-CTC-TGG-TGA-AGT-TTC-AAC-CGT-AAA-CTT-TGA-AAA-ACA-AGA-TAG-TAC-AGT-TAA-AAT-CAA-GGT-TAA-TGA-TGT-GGA-GTT-CAG-CAC-TCT-CTC-TAC-TAG-CTC-AAG-TAA-TCC-AAC-TGA-AAA-TAG-CGG-ATC-TGA-GAG-CCA-GGC-TCA-ATC-AAG-ATC-AAG-AAG-ATC-ACT-CGC-AGA-GGA-TGG-TAC-TGA-GAC-TGC-TGC-AAC-CGT-CGA-TTT-GAT-TGC-CTT-CAC-CCT-TCA-AGG-TGG-TAA-AAG-AAT-CGA-AGT-CGC-TGT-GCC-AAG-TGA-CGA-AGA-TGC-AGA-CAA-AAG-AAG-CAA-GTA-CAG-TTT-GGT-TGC-AGA-CGA-TAA-GCC-TTT-CTA-TAC-CGG-TGC-AAA-CAG-TGG-AGC-CAC-TGA-TGG-TGT-CTA-CAA-ATT-GGA-TGA-TAA-TGG-AAA-CTT-GGT-AGA-TAA-GGA-CAA-CAA-CGT-TCT-TTG-AGW-GGT-3′ | JF727754 | C. hominis | Ie |
| 4 | 5′-CTG-TTG-AGG-RAT-CAT-CAT-CAT-CAT-CAT-CAT-CAT-CAT-CAT-CAT-CAT-CAT-CAT-CAT-CAT-CAT-CAT-CAT-CAT-CAA-CAT-CGA-CCG-TCG-CAC-CAG-CTC-CAA-AGA-AAG-AAA-GAA-CTG-TAG-AGG-GCG-GCA-CGG-AAG-GAA-AGA-ACG-AAG-AAA-GCA-GTC-CAG-GTT-CTG-AAG-AAC-AAG-ACG-GTG-GTA-AGG-AGG-ACG-GTG-GTA-AGG-AAA-ACG-GTG-AAG-GAG-ACA-CAG-TAG-ACG-GGG-AAC-AAA-CCG-GGA-GTG-GTT-CTC-AAG-TTA-CTC-CAT-CTG-GAA-GTG-CCG-GCA-CAG-CTA-CCG-AGT-CCA-CAG-CAA-CTA-CTA-CTC-CAA-AGG-AAG-AAT-GTG-GTA-CTT-CAT-TTG-TCA-TGT-GGT-TCG-AGA-AAG-GCA-CCC-CGG-TTG-CGA-CCT-TGA-AGT-GTG-GTG-ATT-ACA-CTA-TCG-TCT-ATG-CAC-CTA-TAA-AAG-ATC-AAA-CAG-ATC-CCG-CAC-CAA-GAT-ATA-TCT-CTG-GTG-AAG-TTA-CAT-CTG-TAT-CCT-TTG-AAA-AGA-GTG-AAA-GTA-CAG-TTA-CAA-TCA-AGG-TTA-ATG-GTA-AAG-AGT-TCA-GCA-CTC-TCT-CTG-CTA-ACT-CAA-GTA-GTC-CAA-CTA-AAG-ATA-ACG-GTG-AAT-CTA-GTG-ACA-GTC-AGG-TTC-AAT-CAA-GAT-CAA-GAA-GAT-CAC-TCG-CAG-AGG-AGA-ATG-GTG-AAA-CAG-TTG-CAA-CAG-TTG-ATT-TGT-TTG-CCT-TTA-CTC-TTG-ATG-GTG-GTA-GAA-GAA-TTG-AAG-TGG-CTG-TGC-CAA-AGG-ACG-AAA-ATG-CAG-ACA-AAA-GAA-GCG-AGT-ACA-GTT-TGG-TTG-CAG-ACG-ATA-AGC-CTT-TCT-ATA-CCG-GCG-CAA-ACA-GTG-GCA-TCA-CCA-ATG-GTG-TCT-ACA-AAT-TGG-ATG-AGA-ATG-GAA-ACT-TGG-TG-3′ | KU727289 | C. hominis | Ia |
| 5 | 5′-TTT-CTC-AGC-CCC-AGC-CCG-TTC-CAC-TCA-GAG-GCA-CTT-TAA-AGG-ATG-TTT-CTG-TTG-AGA-GCT-CAT-CGT-CAT-CAT-CGT-CAT-CGT-CAA-CAA-CCC-CCG-CAC-CAG-CTC-CAA-AGA-AGG-TAA-GAG-AAA-GCG-AAG-AAG-GGA-AGA-ACA-GTG-AAG-ATA-GTC-AAA-CTC-CCG-CTA-GTC-CTG-GAA-GTG-GTT-CTC-AGG-ATA-GCT-CTA-AAG-GAG-ACG-AAG-TTG-TAG-ATG-GAG-GCGT-TTC-CGG-ATC-TAG-TAC-CCC-AAC-TCA-AGC-TGC-TGA-AAA-GGA-GCC-CGA-AAC-TCC-AGA-ATC-TAC-TCC-AAA-GGA-AGA-ATG-TGG-TAC-TTC-ATT-TAT-AAT-GTG-GTT-CGG-AGA-AGG-TAC-TCC-AGC-CAC-AAC-TTT-GAA-GTG-CGG-TGG-CTA-CAC-TAT-CGT-CTA-TGC-ACC-AGA-AAA-GGA-TAA-TAA-AGA-ACC-CGC-ACC-AAG-ATA-CAT-CTC-TGG-TGA-TGT-TAA-GGC-TGT-AAC-CTT-TGA-AAA-GGG-AGA-AGA-TAA-TAC-AGT-TAA-AAT-CAA-GGT-TGA-TGG-TAG-GGA-GTT-CAG-TAC-TCT-CTC-TTC-TAG-CTC-AAG-CAG-TCC-AAC-TGA-AAA-TAG-CGG-ATC-TGA-GAG-CCA-GGC-TCA-ATC-AAG-ATC-AAG-AAG-ATC-ACT-CAC-AGA-GGG-AAG-TGA-AAC-ACC-TGC-AAC-CGT-CGA-TTT-GTT-TGC-CTT-CAC-CCT-TCA-AGG-TGG-TAA-AAG-AAT-CGA-AGT-CGC-TGT-GCC-AAG-TGA-CGT-AGA-TGC-ATC-CAA-GAG-AAA-CAA-GTA-CAG-TTT-GGT-TGC-AGG-CGA-TAA-GAC-TTT-CTA-TAC-CGG-CGC-AAA-TAG-CGG-TAA-TAC-CGA-CGG-TAT-CTA-CAG-GTT-GAA-TGA-TGA-TGG-AGA-CTT-GGT-GGA-CAA-GAA-CAA-CAA-CGT-TCT-TTT-GAA-GGW-GGK-G-3′ | JF802124 | C. parvum | IIc |
| 6 | 5′-TTC-CGC-TGY-AMT-CTC-AGC-CCC-AGC-CGT-TCC-ACT-CAG-AGG-CAC-TTT-AAA-GGA-TGT-TTC-TGT-TGA-GAG-CTC-ATC-GTC-ATC-ATY-GTC-ATC-GTC-AAC-AAC-AAC-CCC-CGC-ACC-AGC-TCC-AAA-GAA-GGT-AAG-AGA-AAG-CGA-AGA-AGG-GAA-GAA-CAG-TGA-AGA-TAG-TCA-GAC-TCC-CGC-TAG-TCC-TGG-AAG-TGA-TTC-TCA-GGA-TAG-CTC-TAA-AGG-AGA-CGA-AGC-TGT-AGA-TGG-ATC-CGC-TTC-CGG-ATC-TAG-TAC-CCC-AAC-TCA-AGC-TGC-TGA-AAA-GGA-GCC-CGA-AAC-TCC-AGA-ATC-TAC-TCC-AAA-GGA-AGA-ATG-TGG-TAC-TTC-ATT-TAT-AAT-GTG-GTT-CGG-AGA-AGG-TAC-TCC-AGC-CAC-AAC-TTT-GAA-GTG-CGG-TGG-CTA-CAC-TAT-CGT-CTA-TGC-ACC-AGA-AAA-GGA-TAA-TAA-AGA-ACC-CGC-ACC-AAG-ATA-CAT-CTC-TGG-TGA-TGT-TAA-GGC-TGT-AAC-CTT-TGA-AAA-GGG-AGA-AGA-TAA-TAC-AGT-TAA-AAT-CAA-GGT-TGA-TGG-TAA-GGA-GTT-CAG-TAC-TCT-CTC-TTC-TAG-CTC-AAG-CAG-TCC-AAC-TGA-AAA-TAA-CGG-ATC-TAC-GGG-CCA-GGT-TGC-ATC-AAG-ATC-AAG-AAG-ATC-GCT-CTC-AGA-GGA-AAA-TAG-TGA-AAC-TGC-TGC-AAC-CGT-CGA-TTT-GTT-TGC-CTT-CAC-CCT-TGA-TGG-TGG-CCG-AAG-AAT-TGA-AGT-TGC-TGT-ACC-CAG-CGT-CGA-AGA-TGC-AAC-CAA-AAG-AGA-CAA-GTA-CAG-TTT-GGT-TGC-AAA-CGG-TAA-GCC-TTT-CTA-TAC-CGG-CGC-AAA-CAG-CGG-CAC-TAC-CAA-TGG-TGT-CTA-CAG-GTT-GAA-TGA-GAA-CGG-AGA-CTT-GGT-TGA-CAA-GGA-CAA-CAC-AGT-TCT-CTT-GAA-GGA-TGC-TGG-TTC-CTC-TGC-AAT-T-3′ | GU214366 | C. parvum | IIc |
| 7 | 5′-KWW-TTT-TCA-GCC-CCA-GCC-GTC-CCA-CTC-AGA-GGC-ACC-TTG-AAG-GAT-GTT-TCT-GTT-GAG-GGC-TCA-TCA-TCA-TCT-TCA-TCA-TCG-TCT-TCA-TCT-TCA-TCA-TCA-TCA-TCG-TCG-TCA-ACA-ACC-CCA-GCA-CCA-GCT-TCA-AAG-AAG-GTA-AGA-GAA-GCA-GAA-GGC-AGT-GAA-GAA-AAG-GAC-AGC-GAA-GAA-AAG-GAC-AGT-GAA-GAA-AAG-GGC-AGT-GAA-GAA-GGT-AGC-CAA-ACT-CCC-GCT-AGT-CCT-GGA-GGT-GGA-GGG-GTG-AGT-GAA-GGA-GAT-ACT-CAA-GGT-GAC-TCT-AAA-GGA-GAC-GGA-GTT-AGT-TCA-GAT-GAG-AAC-CAA-AGT-CAA-GGT-GGG-GAC-GCT-ACT-CCC-GGA-TCT-AGC-ACC-CAA-ACT-CAA-GCT-ACT-GAA-AAA-GAA-CCC-GGA-TCT-TCA-GAA-GCT-ACT-CCA-AAG-GAA-GAG-TGC-GGT-ACT-TCA-TTT-GTA-ATG-TGG-TTC-GGA-CAG-GGT-GTT-CCA-GTT-GTA-ACT-TTG-AAG-TGT-GGT-GGT-TAT-ACT-ATG-GTC-TAT-GCA-CCA-GAA-AAT-GGC-AAA-ACA-GAT-CCC-GCA-CCA-AGA-TAT-ATC-TCT-GGT-AAA-GTT-TCA-ACC-GTA-GAC-TTT-GAA-AAA-CAA-GAT-AGT-ACA-GTT-AAA-ATC-AAG-GTT-AAT-GGT-GTG-GAG-TTC-AGC-ACT-CTC-TCT-ACT-AGC-TCA-AGT-AAT-CCA-ACT-GAA-AAT-AGC-GGA-TCT-GAG-AGC-CAG-GCT-CAA-TCA-AGA-TCA-AGA-AGA-TCA-CTC-GCA-GAG-GAT-GGT-ACT-GAG-ACT-GCT-GCA-ACC-GTC-GAT-TTG-ATT-GCC-TTC-ACC-CTT-CAA-GGT-GGT-AAA-AGA-ATC-GAA-GTC-GCT-GTG-CCA-AGT-GAC-GAA-GAT-GTA-TCC-AAG-AGA-AAC-AAG-TAC-AGT-TTG-GTT-GCA-GGC-GAT-AAG-ACT-TTC-TAT-ACC-GGC-GCA-AAT-AGC-GGT-AAT-ACC-GAC-GGT-ATC-TAC-AGG-TTG-AAT-GAT-GAT-GGA-GAC-TTG-GTG-GAC-AAG-AAC-AAC-AAC-GTT-CTT-TTG-AAG-GAT-GTG-KGK-T-3′ | ON863593 | C. hominis | Ie |
| 8 | 5′-TTT-TTT-ACC-CCA-GCC-GTT-CCA-CTC-AGA-GGC-ACT-TTA-AAG-GAT-GTT-TCT-GTT-GAG-AGC-TCA-TCG-TCA-TCA-TCG-TCA-TCG-TCA-ACA-ACA-ACC-CCC-GCA-CCA-GCT-CCA-AAG-AAG-GTA-AGA-GAA-AGC-GAA-GAA-GGG-AAG-AAC-AGT-GAA-GAT-AGT-CAG-ACT-CCC-GCT-AGT-CCT-GGA-AGT-GAT-TCT-CAG-GAT-AGC-TCT-AAA-GGA-GAC-GAA-GCT-GTA-GAT-GGA-TCC-GCT-TCC-GGA-TCT-AGT-ACC-CCA-ACT-CAA-GCT-GCT-GAA-AAG-GAG-CCC-GAA-ACT-CCA-GAA-TCT-ACT-CCA-AAG-GAA-GAA-TGT-GGT-ACT-TCA-TTT-ATA-ATG-TGG-TTC-GGA-GAA-GGT-ACT-CCA-GCC-ACA-ACT-TTG-AAG-TGC-GGT-GGC-TAC-ACT-ATC-GTC-TAT-GCA-CCA-GAA-AAG-GAT-AAT-AAA-GAA-CCC-GCA-CCA-AGA-TAC-ATC-TCT-GGT-GAT-GTT-AAG-GCT-GTA-ACC-TTT-GAA-AAG-GGA-GAA-GAT-AAT-ACA-GTT-AAA-ATC-AAG-GTT-GAT-GGT-AAG-GAG-TTC-AGT-ACT-CTC-TCT-TCT-AGC-TCA-AGC-AGT-CCA-ACT-GAA-AAT-AAC-GGA-TCT-ACG-GGC-CAG-GTT-GCA-TCA-AGA-TCA-AGA-AGA-TCG-CTC-TCA-GAG-GAA-AAT-AGT-GAA-ACT-GCT-GCA-ACC-GTC-GAT-TTG-TTT-GCC-TTC-ACC-CTT-GAT-GGT-GGC-CGA-AGA-ATT-GAA-GTT-GCT-GTA-CCC-AGC-GTC-GAA-GAT-GCA-ACC-AAA-AGA-GAC-AAG-TAC-AGT-TTG-GTT-GCA-AAC-GGT-AAG-CCT-TTC-TAT-ACC-GGC-GCA-AAC-AGC-GGC-ACT-ACC-AAT-GGT-GTC-TAC-AGG-TTG-AAT-GAG-AAC-GGA-GAC-TTG-GTT-GAC-AAG-GAC-AAC-ACA-GTT-CTC-TTG-AAG-GWW-Y-3′ | MW480840 | C. parvum | IIc |
| 9 | 5′-TTT-YYA-GCC-CCA-GCC-GTT-CCA-CTC-AGA-GGC-ACC-TTG-AAA-GAT-GTT-TCT-GTT-GAG-AGC-TCA-TCA-TCA-TCA-TCA-TCA-TCA-TCG-TCA-TCA-TCA-TCG-TCA-TCA-TCG-TCA-ACA-ACA-ACC-CCC-GCA-CCA-GCT-CCA-AAG-AAG-GCA-AGA-GAA-GCA-GAT-GGC-GGA-GAA-GAA-AAG-AAC-AAT-GAA-GAA-AGC-CAA-ACT-CCC-GCT-AGT-CCT-GGA-AGT-GGT-GGG-GTG-AGT-GAA-GGA-CAA-GAT-ACT-CAA-GGT-GGC-TCC-AAA-GGA-GAC-GCT-GAG-GAA-GGC-ACT-GAA-GAC-AAT-GAA-CAA-GCC-GAT-GAG-AGT-GCT-ACC-CAA-CCT-TCT-ACC-CCA-GGT-CAA-GGC-TCC-GTT-AAA-ACC-GAA-TCC-ACA-GAA-ACT-ACT-CCA-AAG-GAG-AAG-TGC-GGT-ACT-TCA-TTT-GTT-ATG-TGG-TTC-GGA-CAG-GGT-GTT-CCA-GTC-GCA-ACT-TTG-AAG-TGC-GGT-GAC-TAT-ACT-ATG-GTC-TAT-GCA-CCA-GAA-AAG-GAC-AAA-ACA-GAT-CCC-GCA-CCA-AGA-TAT-ATC-TCT-GGT-GAA-GTT-ACA-ACC-GTA-ACC-TTT-GAT-AAA-CAA-GAG-AGT-ACA-GTT-ACA-ATC-AAG-GTT-AAT-AAT-GTA-GAG-TTC-GGC-ACT-CTC-TCT-ACT-AGC-TCA-AGT-AAA-CCA-ACT-GAA-AAT-AAA-GGT-GAG-TCT-AGC-GAT-CAG-GTT-GGG-TCA-AGA-TCA-AGA-AGA-TCA-CTC-ACA-GAG-GAA-ACT-AGT-GAA-ACT-GCA-ACC-GTC-GAT-TTG-TTT-GCC-TTT-ACC-CTT-GAT-GGT-GGT-AAA-AGA-ATT-GAA-GTG-GCT-GTA-CCA-AGT-GAC-GAA-GAT-GTA-TCC-AAG-AGA-AAC-AAG-TAC-AGT-TTG-GTT-GCA-AAC-GAT-AAG-ACT-TTC-TAT-ACC-GGC-GCA-AAT-AGC-GGT-AAT-ACC-GAC-GGT-ATC-TAC-AGG-TTG-AAT-GAT-AAT-GGA-GAC-TTG-GTG-GAC-AAG-AAC-AAC-AAC-GTT-CTT-TTG-AAG-W-3′ | MW480830 | C. hominis | Ib |
| 10 | 5′-YTT-TTT-TTT-TCA-CCC-AGC-CGT-TCC-ACT-CAG-AGG-CAC-CTT-GAA-AGA-TGT-TTC-TGT-TGA-GAG-CTC-ATC-ATC-ATC-ATC-ATC-ATC-ATC-GTC-ATC-ATC-ATC-GTC-ATC-ATC-GTC-AAC-AAC-AAC-CCC-CGC-ACC-AGC-TCC-AAA-GAA-GGC-AAG-AGA-AGC-AGA-TGG-CGG-AGA-AGA-AAA-GAA-CAA-TGA-AGA-AAG-CCA-AAC-TCC-CGC-TAG-TCC-TGG-AAG-TGG-TGG-GGT-GAG-TGA-AGG-ACA-AGA-TAC-TCA-AGG-TGG-CTC-CAA-AGG-AGA-CGC-TGA-GGA-AGG-CAC-TGA-AGA-CAA-TGA-ACA-AGC-CGA-TGA-GAG-TGC-TGC-CCA-ACC-TTC-TAC-CCC-AGG-TCA-AGG-CTC-CGT-TAA-AAC-CGA-ATC-CAC-AGA-AAC-TAC-TCC-AAA-GGA-GAA-GTG-CGG-TAC-TTC-ATT-TGT-TAT-GTG-GTT-CGG-ACA-GGG-TGT-TCC-AGT-CGC-AAC-TTT-GAA-GTG-CGG-TGA-CTA-TAC-TAT-GGT-CTA-TGC-ACC-AGA-AAA-GGA-CAA-AAC-AGA-TCC-CGC-ACC-AAG-ATA-TAT-CTC-TGG-TGA-AGT-TAC-AAC-CGT-AAC-CTT-TGA-TAA-ACA-AGA-GAG-TAC-AGT-TAC-AAT-CAA-GGT-TAA-TAA-TGT-AGA-GTT-CGG-CAC-TCT-CTC-TAC-TAG-CTC-AAG-TAA-ACC-AAC-TGA-AAA-TAA-AGG-TGA-GTC-TAG-CGA-TCA-GGT-TGG-GTC-AAG-ATC-AAG-AAG-ATC-ACT-CAC-AGA-GGA-AAC-TAG-TGA-AAC-TGC-AAC-CGT-CGA-TTT-GTT-TGC-CTT-TAC-CCT-TGA-TGG-TGG-TAA-AAG-AAT-TGA-AGT-GGC-TGT-ACC-AAG-TGA-CGA-AGA-TGT-ATC-CAA-GAG-AAA-CAA-GTA-CAG-TTT-GGT-TGC-AAA-CGA-TAA-GAC-TTT-CTA-TAC-CGG-CGC-AAA-TAG-CGG-TAA-TAC-CGA-CGG-TAT-CTA-CAG-GTT-GAA-TGA-TAA-TGG-AGA-CTT-GGT-GGA-CAA-GAA-CAA-CAA-CGT-TCT-TTT-GAA-GWG-KT-3′ | MK105902 | C. hominis | Ib |
| 11 | 5′-TTC-TCA-GCC-CCA-GCC-GTT-CCA-CTC-AGA-GGC-ACC-TTG-AAA-GAT-GTT-TCT-GTT-GAG-AGC-TCA-TCA-TCA-TCA-TCA-TCA-TCA-TCG-TCA-TCA-TCA-TCG-TCA-TCA-TCG-TCA-ACA-ACA-ACC-CCC-GCA-CCA-GCT-CCA-AAG-AAG-GCA-AGA-GAA-GCA-GAT-GGC-GGA-GAA-GAA-AAG-AAC-AAT-GAA-GAA-AGC-CAA-ACT-CCC-GCT-AGT-CCT-GGA-AGT-GGT-GGG-GTG-AGT-GAA-GGA-CAA-GAT-ACT-CAA-GGT-GGC-TCC-AAA-GGA-GAC-GCT-GAG-GAA-GGC-ACT-GAA-GAC-AAT-GAA-CAA-GCC-GAT-GAG-AGT-GCT-ACC-CAA-CCT-TCT-ACC-CCA-GGT-CAA-GGC-TCC-GTT-AAA-ACC-GAA-TCC-ACA-GAA-ACT-ACT-CCA-AAG-GAG-AAG-TGC-GGT-ACT-TCA-TTT-GTT-ATG-TGG-TTC-GGA-CAG-GGT-GTT-CCA-GTC-GCA-ACT-TTG-AAG-TGC-GGT-GAC-TAT-ACT-ATG-GTC-TAT-GCA-CCA-GAA-AAG-GAC-AAA-ACA-GAT-CCC-GCA-CCA-AGA-TAT-ATC-TCT-GGT-GAA-GTT-ACA-ACC-GTA-ACC-TTT-GAT-AAA-CAA-GAG-AGT-ACA-GTT-ACA-ATC-AAG-GTT-AAT-AAT-GTA-GAG-TTC-GGC-ACT-CTC-TCT-ACT-AGC-TCA-AGT-AAA-CCA-ACT-GAA-AAT-AAA-GGT-GAG-TCT-AGC-GAT-CAG-GTT-GGG-TCA-AGA-TCA-AGA-AGA-TCA-CTC-ACA-GAG-GAA-ACT-AGT-GAA-ACT-GCA-ACC-GTC-GAT-TTG-TTT-GCC-TTT-ACC-CTT-GAT-GGT-GGT-AAA-AGA-ATT-GAA-GTG-GCT-GTA-CCA-AGT-GAC-GAAG-ATGT-ATC-CAA-GAG-AAA-CAA-GTA-CAG-TTT-GGT-TGC-AAA-CGA-TAA-GAC-TTT-CTA-TAC-CGG-CGC-AAA-TAG-CGG-TAA-TAC-CGA-CGG-TAT-CTA-CAG-GTT-GAA-TGA-TAA-TGG-AGA-CTT-GGT-GGA-CAA-GAA-CAA-CAA-CGT-TCT-TTT-GAA-GGA-TGT-GGG-3′ | MT053132 | C. hominis | Ib |
| 12 | 5′-TWA-TTT-TCA-GCC-CCA-GCC-GTC-CCA-CTC-AGA-GGC-ACC-TTG-AAG-GAT-GTT-TCT-GTT-GAG-GGC-TCA-TCA-TCA-TCT-TCA-TCA-TCG-TCT-TCA-TCT-TCA-TCA-TCA-TCA-TCG-TCG-TCA-ACA-ACC-CCA-GCA-CCA-GCT-TCA-AAG-AAG-GTA-AGA-GAA-GCA-GAA-GGC-AGT-GAA-GAA-AAG-GAC-AGC-GAA-GAA-AAG-GAC-AGT-GAA-GAA-AAG-GGC-AGT-GAA-GAA-GGT-AGC-CAA-ACT-CCC-GCT-AGT-CCT-GGA-GGT-GGA-GGG-GTG-AGT-GAA-GGA-GAT-ACT-CAA-GGT-GAC-TCT-AAA-GGA-GAC-GGA-GTT-AGT-TCA-GAT-GAG-AAC-CAA-AGT-CAA-GGT-GGG-GAC-GCT-ACT-CCC-GGA-TCT-AGC-ACC-CAA-ACT-CAA-GCT-ACT-GAA-AAA-GAA-CCC-GGA-TCT-TCA-GAA-GCT-ACT-CCA-AAG-GAA-GAG-TGC-GGT-ACT-TCA-TTT-GTA-ATG-TGG-TTC-GGA-CAG-GGT-GTT-CCA-GTT-GTA-ACT-TTG-AAG-TGT-GGT-GGT-TAT-ACT-ATG-GTC-TAT-GCA-CCA-GAA-AAT-GGC-AAA-ACA-GAT-CCC-GCA-CCA-AGA-TAT-ATC-TCT-GGT-AAA-GTT-TCA-ACC-GTA-GAC-TTT-GAA-AAA-CAA-GAT-AGT-ACA-GTT-AAA-ATC-AAG-GTT-AAT-GGT-GTG-GAG-TTC-AGC-ACT-CTC-TCT-ACT-AGC-TCA-AGT-AAT-CCA-ACT-GAA-AAT-AGC-GGA-TCT-GAG-AGC-CAG-GCT-CAA-TCA-AGA-TCA-AGA-AGA-TCA-CTC-GCA-GAG-GAT-GGT-ACT-GAG-ACT-GCT-GCA-ACC-GTC-GAT-TTG-ATT-GCC-TTC-ACC-CTT-CAA-GGT-GGT-AAA-AGA-ATC-GAA-GTC-GCT-GTG-CCA-AGT-GAC-GAA-GAT-GTA-TCC-AAG-AGA-AAC-AAG-TAC-AGT-TTG-GTT-GCA-GGC-GAT-AAG-ACT-TTC-TAT-ACC-GGC-GCA-AAT-AGC-GGT-AAT-ACC-GAC-GGT-ATC-TAC-AGG-TTG-AAT-GAT-GAT-GGA-GAC-TTG-GTG-GAC-AAG-AAC-AAC-AAC-GTT-CTT-TTG-AAG-GAT-GTG-GTT-3′ | GU214354 | C. hominis | Ie |
| 13 | 5′-CGC-TGT-WAT-TCT-CAG-CCC-CAG-CCG-TTC-CAC-TCA-GAG-GCA-CCT-TGA-AAG-ATG-TTT-CTG-TTG-AGA-GCT-CAT-CAT-CAT-CAT-CAT-CAT-CAT-CGT-CAT-CAT-CAT-CGT-CAT-CAT-CGT-CAA-CAA-CAA-CCC-CCG-CAC-CAG-CTC-CAA-AGA-AGG-CAA-GAG-AAG-CAG-ATG-GCG-GAG-AAG-AAA-AGA-ACA-ATG-AAG-AAA-GCC-AAA-CTC-CCG-CTA-GTC-CTG-GAA-GTG-GTG-GGG-TGA-GTG-AAG-GAC-AAG-ATA-CTC-AAG-GTG-GCT-CCA-AAG-GAG-ACG-CTG-AGG-AAG-GCA-CTG-AAG-ACA-ATG-AAC-AAG-CCG-ATG-AGA-GTG-CTA-CCC-AAC-CTT-CTA-CCC-CAG-GTC-AAG-GCT-CCG-TTA-AAA-CCG-AAT-CCA-CAG-AAA-CTA-CTC-CAA-AGG-AGA-AGT-GCG-GTA-CTT-CAT-TTG-TTA-TGT-GGT-TCG-GAC-AGG-GTG-TTC-CAG-TCG-CAA-CTT-TGA-AGT-GCG-GTG-ACT-ATA-CTA-TGG-TCT-ATG-CAC-CAG-AAA-AGG-ACA-AAA-CAG-ATC-CCG-CAC-CAA-GAT-ATA-TCT-CTG-GTG-AAG-TTA-CAA-CCG-TAA-CCT-TTG-ATA-AAC-AAG-AGA-GTA-CAG-TTA-CAA-TCA-AGG-TTA-ATA-ATG-TAG-AGT-TCG-GCA-CTC-TCT-CTA-CTA-GCT-CAA-GTA-AAC-CAA-CTG-AAA-ATA-AAG-GTG-AGT-CTA-GCG-ATC-AGG-TTG-GGT-CAA-GAT-CAA-GAA-GAT-CAC-TCA-CAG-AGG-AAA-CTA-GTG-AAA-CTG-CAA-CCG-TCG-ATT-TGT-TTG-CCT-TTA-CCC-TTG-ATG-GTG-GTA-AAA-GAA-TTG-AAG-TGG-CTG-TAC-CAA-GTG-ACG-AAG-ATG-TAT-CCA-AGA-GAA-ACA-AGT-ACA-GTT-TGG-TTG-CAA-ACG-ATA-AGA-CTT-TCT-ATA-CCG-GCG-CAA-ATA-GCG-GTA-ATA-CCG-ACG-GTA-TCT-ACA-GGT-TGA-ATG-ATA-ATG-GAG-ACT-TGG-TGG-ACA-AGA-ACA-ACA-ACG-TTC-TTT-TGA-AGG-ATG-CTG-TTT-YCC-TCT-GC-3′ | MT053132 | C. hominis | Ib |
| 14 | 5′-TTT-TTC-AGC-CCC-AGC-CGT-TCC-ACT-CAG-AGG-CAC-CTT-GAA-AGA-TGT-TTC-TGT-TGA-GAG-CTC-ATC-ATC-ATC-ATC-ATC-ATC-ATC-GTC-ATC-ATC-ATC-GTC-ATC-ATC-GTC-AAC-AAC-AAC-CCC-CGC-ACC-AGC-TCC-AAA-GAA-GGC-AAG-AGA-AGC-AGA-TGG-CGG-AGA-AGA-AAA-GAA-CAA-TGA-AGA-AAG-CCA-AAC-TCC-CGC-TAG-TCC-TGG-AAG-TGG-TGG-GGT-GAG-TGA-AGG-ACA-AGA-TAC-TCA-AGG-TGG-CTC-CAA-AGG-AGA-CGC-TGA-GGA-AGG-CAC-TGA-AGA-CAA-TGA-ACA-AGC-CGA-TGA-GAG-TGC-TAC-CCA-ACC-TTC-TAC-CCC-AGG-TCA-AGG-CTC-CGT-TAA-AAC-CGA-ATC-CAC-AGA-AAC-TAC-TCC-AAA-GGA-GAA-GTG-CGG-TAC-TTC-ATT-TGT-TAT-GTG-GTT-CGG-ACA-GGG-TGT-TCC-AGT-CGC-AAC-TTT-GAA-GTG-CGG-TGA-CTA-TAC-TAT-GGT-CTA-TGC-ACC-AGA-AAA-GGA-CAA-AAC-AGA-TCC-CGC-ACC-AAG-ATA-TAT-CTC-TGG-TGA-AGT-TAC-AAC-CGT-AAC-CTT-TGA-TAA-ACA-AGA-GAG-TAC-AGT-TAC-AAT-CAA-GGT-TAA-TAA-TGT-AGA-GTT-CGG-CAC-TCT-CTC-TAC-TAG-CTC-AAG-TAA-ACC-AAC-TGA-AAA-TAA-AGG-TGA-GTC-TAG-CGA-TCA-GGT-TGG-GTC-AAG-ATC-AAG-AAG-ATC-ACT-CAC-AGA-GGA-AAC-TAG-TGA-AAC-TGC-AAC-CGT-CGA-TTT-GTT-TGC-CTT-TAC-CCT-TGA-TGG-TGG-TAA-AAG-AAT-TGA-AGT-GGC-TGT-ACC-AAG-TGA-CGA-AGA-TGT-ATC-CAA-GAG-AAA-CAA-GTA-CAG-TTT-GGT-TGC-AAA-CGA-TAA-GAC-TTT-CTA-TAC-CGG-CGC-AAA-TAG-CGG-TAA-TAC-CGA-CGG-TAT-CTA-CAG-GTT-GAA-TGA-TAA-TGG-AGA-CTT-GGT-GGA-CAA-GAA-CAA-CAA-CGT-TCT-TTT-GAA-GWW-G-3′ | MT053132 | C. hominis | Ib |
| 15 | 5′-TTT-TTT-TYA-CCC-CAG-CCG-TTC-CAC-TCA-GAG-GCA-CCT-TGA-AAG-ATG-TTT-CTG-TTG-AGA-GCT-CAT-CAT-CAT-CAT-CAT-CAT-CAT-CGT-CAT-CAT-CAT-CGT-CAT-CAT-CGT-CAA-CAA-CAA-CCC-CCG-CAC-CAG-CTC-CAA-AGA-AGG-CAA-GAG-AAG-CAG-ATG-GCG-GAG-AAG-AAA-AGA-ACA-ATG-AAG-AAA-GCC-AAA-CTC-CCG-CTA-GTC-CTG-GAA-GTG-GTG-GGG-TGA-GTG-AAG-GAC-AAG-ATA-CTC-AAG-GTG-GCT-CCA-AAG-GAG-ACG-CTG-AGG-AAG-GCA-CTG-AAG-ACA-ATG-AAC-AAG-CCG-ATG-AGA-GTG-CTA-CCC-AAC-CTT-CTA-CCC-CAG-GTC-AAG-GCT-CCG-TTA-AAA-CCG-AAT-CCA-CAG-AAA-CTA-CTC-CAA-AGG-AGA-AGT-GCG-GTA-CTT-CAT-TTG-TTA-TGT-GGT-TCG-GAC-AGG-GTG-TTC-CAG-TCG-CAA-CTT-TGA-AGT-GCG-GTG-ACT-ATA-CTA-TGG-TCT-ATG-CAC-CAG-AAA-AGG-ACA-AAA-CAG-ATC-CCG-CAC-CAA-GAT-ATA-TCT-CTG-GTG-AAG-TTA-CAA-CCG-TAA-CCT-TTG-ATA-AAC-AAG-AGA-GTA-CAG-TTA-CAA-TCA-AGG-TTA-ATA-ATG-TAG-AGT-TCG-GCA-CTC-TCT-CTA-CTA-GCT-CAA-GTA-AAC-CAA-CTG-AAA-ATA-AAG-GTG-AGT-CTA-GCG-ATC-AGG-TTG-GGT-CAA-GAT-CAA-GAA-GAT-CAC-TCA-CAG-AGG-AAA-CTA-GTG-AAA-CTG-CAA-CCG-TCG-ATT-TGT-TTG-CCT-TTA-CCC-TTG-ATG-GTG-GTA-AAA-GAA-TTG-AAG-TGG-CTG-TAC-CAA-GTG-ACG-AAG-ATG-TAT-CCA-AGA-GAA-ACA-AGT-ACA-GTT-TGG-TTG-CAA-ACG-ATA-AGA-CTT-TCT-ATA-CCG-GCG-CAA-ATA-GCG-GTA-ATA-CCG-ACG-GTA-TCT-ACA-GGT-TGA-ATG-ATA-ATG-GAG-ACT-TGG-TGG-ACA-AGA-ACA-ACA-ACG-TTC-TTT-GAG-WGG-3′ | MK105902 | C. hominis | Ib |
| 16 | 5′-CAG-CCC-CGG-CCG-TTS-CWC-TCA-GAG-GCA-CCT-YGA-AGG-ATG-TTT-CTG-TTG-AGG-GAT-CAT-CAT-CAT-CAT-CAT-CAT-CAT-CAT-CAT-CAT-CAT-CAT-CAT-CAT-CAT-CAA-CAT-CGA-CCG-TCG-CAC-CAG-CTC-CAA-AGA-AAG-AAA-GAA-CTG-TAG-AGG-GCG-GCA-CGG-AAG-GAA-AGA-ACG-AAG-AAA-GCA-GTC-CAG-GTT-CTG-AAG-AAC-AAG-ACG-GTG-GTA-AGG-AAG-ACG-GTG-GTA-AGG-AAA-ACG-GTG-AAG-GAG-ACA-CAG-TAG-ACG-GGG-AAC-AAA-CCG-GGA-GTG-GTT-CTC-AAG-TTA-CTC-CAT-CTG-GAA-GTG-CCG-GCA-CAG-CTA-CCG-AGT-CCA-CAG-CAA-CTA-CTA-CTC-CAA-AGG-AAG-AAT-GTG-GTA-CTT-CAT-TTG-TCA-TGT-GGT-TCG-AGA-AAG-GCA-CCC-CGG-TTG-CGA-CTT-TGA-AGT-GTG-GTG-ATT-ACA-CTA-TCG-TCT-ATG-CAC-CTA-TAA-AAG-ATC-AAA-CAG-ATC-CCG-CAC-CAA-GAT-ATA-TCT-CTG-GTG-AAG-TTA-CAT-CTG-TAT-CCT-TTG-AAA-AGA-GTG-AAA-GTA-CAG-TTA-CAA-TCA-AGG-TTA-ATG-GTA-AAG-AGT-TCA-GCA-CTC-TCT-CTG-CTA-ACT-CAA-GTA-GTC-CAA-CTA-AAG-ATA-ACG-GTG-AAT-CTA-GTG-ACA-GTC-AGG-TTC-AAT-CAA-GAT-CAA-GAA-GAT-CAC-TCG-CAG-AGG-AGA-ATG-GTG-AAA-CAG-TTG-CAA-CAG-TTG-ATT-TGT-TTG-CCT-TTA-CTC-TTG-ATG-GTG-GTA-GAA-GAA-TTG-AAG-TGG-CTG-TGC-CAA-AGG-ACG-AAA-ATG-CAG-ACA-AAA-GAA-GTG-AGT-ACA-GTT-TGG-TTG-CAG-ACG-ATA-AGC-CTT-TCT-ATA-CCG-GCG-CAA-ACA-GTG-GCA-TCA-CCA-ATG-GTG-TCT-ACA-AAT-TGG-ATG-AGA-ATG-GAA-ACT-TGG-WTG-ACR-AGG-ACA-ACA-AAG-TTC-TCT-TGA-AAG-ATG-3′ | KY990897 | C. hominis | Ia |
| 17 | 5′-TAC-CGC-TAT-TTG-CGC-CGG-TAT-AGA-AAG-TCT-TAT-CGT-TTG-CAA-CCA-ARC-WGT-ACT-TGT-TTC-TCT-GGA-KAC-ATC-TTC-GTC-ACW-TGG-TAC-AGC-CAY-TTC-AAT-TCT-TTT-WCC-ACC-ATC-AAG-GGT-AAA-GGC-AAR-CAA-ATM-GAC-GGT-TGC-AGT-TTC-AST-AGT-TTC-CTC-TGT-GAG-TGA-TCT-TCT-TGA-TCW-TGA-CCC-AAC-CTG-ATC-GCT-AGA-CTC-ACC-TTT-ATT-TTC-AGT-TGG-TKT-ACW-TGA-GCT-AGT-AGA-GAG-AGT-GCC-GAM-CTC-TAC-ATT-ATT-AAC-CTT-GAT-KGT-ARC-TGT-ACT-CTC-TTG-TTW-ATC-AAA-GGT-TAC-GGT-TGK-AAC-3′ | OP778252 | C. hominis | Ib |
| 18 | 5′-ACA-ACC-CCC-ACA-CCA-GCT-CCA-AAG-AAG-GCA-AGA-GAA-GCA-GAT-GGC-GGA-GAA-GAA-AAG-ATC-AAC-GAA-GAA-AGC-CAA-ACT-CCC-GCT-AGT-CCT-GGA-AGT-GGT-GGG-GTG-AGT-GAA-GGA-CAA-GAT-ACT-CAA-GGT-GGC-TCC-AAA-GGA-GAC-GCT-GAG-GAA-GGC-ACT-GAA-GTC-AAT-GAA-CAA-CCC-GAT-GAG-AGT-GCT-ACC-CAA-CCT-TCT-ACC-CCA-GGT-CAA-GGC-TCC-GTT-AAA-ACC-GAA-TCC-ACA-GAA-ACT-ACT-CCA-AAG-GAG-AAG-TGC-GGT-ACT-TCA-TTT-GTT-ATG-TGG-TTC-GGA-CAG-GGT-GTT-CCA-GTC-GCA-ACT-TTG-AAG-TGC-GGT-GAC-TAT-ACT-ATG-GTC-TAT-GCA-CCA-GAA-AAG-GAC-AAA-ACA-GAT-CCC-GCA-CCA-AGA-TAT-ATC-TCT-GGT-GAA-GTT-ACA-ACC-GTA-ACY-TTC-VAA-ARV-MAC-SAC-MMY-MCA-RDC-CCC-CCC-GCM-WCT-ATA-GTC-TCC-GCT-GTA-TTY-TTT-GTA-ACG-CTG-AAT-GAA-TCG-ATC-AGC-TCA-CTG-TMT-GGT-GCA-TCG-CTC-TCT-YCC-GAA-AGA-TTA-TAC-ACA-TTT-ACA-GAA-ATC-TTT-TGA-TCT-GTC-ACA-TCA-AAA-ATA-GCA-TAA-CTT-GGA-CTA-TAC-TCC-TGA-TTC-CAC-TCT-TGA-TTT-CCC-ACA-GGA-AGA-KTT-CCT-TCC-AGA-TAG-GAT-GCA-GAA-CCA-CTC-TCT-CCA-TTT-GCT-AAA-ATC-KTT-SC-3′ | OP778252 | C. hominis | Ib |
| 19 | 5′-KKT-TTT-TTC-AGC-CCC-AGC-CGT-TCC-ACT-CAG-AGG-CAC-CTT-GAA-AGA-TGT-TTC-TGT-TGA-GAG-CTC-ATC-ATC-ATC-ATC-ATC-ATC-ATC-GTC-ATC-ATC-ATC-GTC-ATC-ATC-GTC-AAC-AAC-AAC-CCC-CGC-ACC-AGC-TCC-AAA-GAA-GGC-AAG-AGA-AGC-AGA-TGG-CGG-AGA-AGA-AAA-GAA-CAA-TGA-AGA-AAG-CCA-AAC-TCC-CGC-TAG-TCC-TGG-AAG-TGG-TGG-GGT-GAG-TGA-AGG-ACA-AGA-TAC-TCA-AGG-TGG-CTC-CAA-AGG-AGA-CGC-TGA-GGA-AGG-CAC-TGA-AGA-CAA-TGA-ACA-AGC-CGA-TGA-GAG-TGC-TAC-CCA-ACC-TTC-TAC-CCC-AGG-TCA-AGG-CTC-CGT-TAA-AAC-CGA-ATC-CAC-AGA-AAC-TAC-TCC-AAA-GGA-GAA-GTG-CGG-TAC-TTC-ATT-TGT-TAT-GTG-GTT-CGG-ACA-GGG-TGT-TCC-AGT-CGC-AAC-TTT-GAA-GTG-CGG-TGA-CTA-TAC-TAT-GGT-CTA-TGC-ACC-AGA-AAA-GGA-CAA-AAC-AGA-TCC-CGC-ACC-AAG-ATA-TAT-CTC-TGG-TGA-AGT-TAC-AAC-CGT-AAC-CTT-TGA-TAA-ACA-AGA-GAG-TAC-AGT-TAC-AAT-CAA-GGT-TAA-TAA-TGT-AGA-GTT-CGG-CAC-TCT-CTC-TAC-TAG-CTC-AAG-TAA-ACC-AAC-TGA-AAA-TAA-AGG-TGA-GTC-TAG-CGA-TCA-GGT-TGG-GTC-AAG-ATC-AAG-AAG-ATC-ACT-CAC-AGA-GGA-AAC-TAG-TGA-AAC-TGC-AAC-CGT-CGA-TTT-GTT-TGC-CTT-TAC-CCT-TGA-TGG-TGG-TAA-AAG-AAT-TGA-AGT-GGC-TGT-ACC-AAG-TGA-CGA-AGA-TGT-ATC-CAA-GAG-AAA-CAA-GTA-CAG-TTT-GGT-TGC-AAA-CGA-TAA-GAC-TTT-CTA-TAC-CGG-CGC-AAA-TAG-CGG-TAA-TAC-CGA-CGG-TAT-CTA-CAG-GTT-GAA-TGA-TAA-TGG-AGA-CTT-GGT-GGA-CAA-GAAC-AAC-AAC-GTT-CTT-TTG-AAG-GAT-GTG-GGK-3′ | MT053132 | C. hominis | Ib |
| 20 | 5′-TTT-TTT-TTY-ACC-CAG-CCG-TTC-CAC-TCA-GAG-GCA-CCT-TGA-AAG-ATG-TTT-CTG-TTG-AGA-GCT-CAT-CAT-CAT-CAT-CAT-CAT-CAT-CGT-CAT-CAT-CAT-CGT-CAT-CAT-CGT-CAA-CAA-CAA-CCC-CCG-CAC-CAG-CTC-CAA-AGA-AGG-CAA-GAG-AAG-CAG-ATG-GCG-GAG-AAG-AAA-AGA-ACA-ATG-AAG-AAA-GCC-AAA-CTC-CCG-CTA-GTC-CTG-GAA-GTG-GTG-GGG-TGA-GTG-AAG-GAC-AAG-ATA-CTC-AAG-GTG-GCT-CCA-AAG-GAG-ACG-CTG-AGG-AAG-GCA-CTG-AAG-ACA-ATG-AAC-AAG-CCG-ATG-AGA-GTG-CTA-CCC-AAC-CTT-CTA-CCC-CAG-GTC-AAG-GCT-CCG-TTA-AAA-CCG-AAT-CCA-CAG-AAA-CTA-CTC-CAA-AGG-AGA-AGT-GCG-GTA-CTT-CAT-TTG-TTA-TGT-GGT-TCG-GAC-AGG-GTG-TTC-CAG-TCG-CAA-CTT-TGA-AGT-GCG-GTG-ACT-ATA-CTA-TGG-TCT-ATG-CAC-CAG-AAA-AGG-ACA-AAA-CAG-ATC-CCG-CAC-CAA-GAT-ATA-TCT-CTG-GTG-AAG-TTA-CAA-CCG-TAA-CCT-TTG-ATA-AAC-AAG-AGA-GTA-CAG-TTA-CAA-TCA-AGG-TTA-ATA-ATG-TAG-AGT-TCG-GCA-CTC-TCT-CTA-CTA-GCT-CAA-GTA-AAC-CAA-CTG-AAA-ATA-AAG-GTG-AGT-CTA-GCG-ATC-AGG-TTG-GGT-CAA-GAT-CAA-GAA-GAT-CAC-TCA-CAG-AGG-AAA-CTA-GTG-AAA-CTG-CAA-CCG-TCG-ATT-TGT-TTG-CCT-TTA-CCC-TTG-ATG-GTG-GTA-AAA-GAA-TTG-AAG-TGG-CTG-TAC-CAA-GTG-ACG-AAG-ATG-TAT-CCA-AGA-GAA-ACA-AGT-ACA-GTT-TGG-TTG-CAA-ACG-ATA-AGA-CTT-TCT-ATA-CCG-GCG-CAA-ATA-GCG-GTA-ATA-CCG-ACG-GTA-TCT-ACA-GGT-TGA-ATG-ATA-ATG-GAG-ACT-TGG-TGG-ACA-AGA-ACA-ACA-ACG-TTC-TTT-GAG-RGG-K-3′ | MK105902 | C. hominis | Ib |
| 21 | 5′-ACC-CAG-CCG-TCC-CAC-TCA-GAG-GCA-CCT-TGA-AGG-ATG-TTT-CTG-TTG-AGG-GCT-CAT-CAT-CAT-CTT-CAT-CAT-CGT-CTT-CAT-CTT-CAT-CAT-CAT-CAT-CGT-CGT-CAA-CAA-CCC-CAG-CAC-CAG-CTT-CAA-AGA-AGG-TAA-GAG-AAG-CAG-AAG-GCA-GTG-AAG-AAA-AGG-ACA-GCG-AAG-AAA-AGG-ACA-GTG-AAG-AAA-AGG-GCA-GTG-AAG-AAG-GTA-GCC-AAA-CTC-CCG-CTA-GTC-CTG-GAG-GTG-GAG-GGG-TGA-GTG-AAG-GAG-ATA-CTC-AAG-GTG-ACT-CTA-AAG-GAG-ACG-GAG-TTA-GTT-CAG-ATG-AGA-ACC-AAA-GTC-AAG-GTG-GGG-ACG-CTA-CTC-CCG-GAT-CTA-GCA-CCC-AAA-CTC-AAG-CTA-CTG-AAA-AAG-AAC-CCG-GAT-CTT-CAG-AAG-CTA-CTC-CAA-AGG-AAG-AGT-GCG-GTA-CTT-CAT-TTG-TAA-TGT-GGT-TCG-GAC-AGG-GTG-TTC-CAG-TTG-TAA-CTT-TGA-AGT-GTG-GTG-GTT-ATA-CTA-TGG-TCT-ATG-CAC-CAG-AAA-ATG-GCA-AAA-CAG-ATC-CCG-CAC-CAA-GAT-ATA-TCT-CTG-GTA-AAG-TTT-CAA-CCG-TAG-ACT-TTG-AAA-AAC-AAG-ATA-GTA-CAG-TTA-AAA-TCA-AGG-TTA-ATG-GTG-TGG-AGT-TCA-GCA-CTC-TCT-CTA-CTA-GCT-CAA-GTA-ATC-CAA-CTG-AAA-ATA-GCG-GAT-CTG-AGA-GCC-AGG-CTC-AAT-CAA-GAT-CAA-GAA-GAT-CAC-TCG-CAG-AGG-ATG-GTA-CTG-AGA-CTG-CTG-CAA-CCG-TCG-ATT-TGA-TTG-CCT-TCA-CCC-TTC-AAG-GTG-GTA-AAA-GAA-TCG-AAG-TCG-CTG-TGC-CAA-GTG-ACG-AAG-ATG-TAT-CCA-AGA-GAA-ACA-AGT-ACA-GTT-TGG-TTG-CAG-GCG-ATA-AGA-CTT-TCT-ATA-CCG-GCG-CAA-ATA-GCG-GTA-ATA-CCG-ACG-GTA-TCT-ACA-GGT-TGA-ATG-ATG-ATG-GAG-ACT-TGG-TGG-ACA-AGA-ACA-ACA-ACG-TTC-TTT-GAA-GWS-KKK-3′ | ON863660 | C. hominis | Ie |
| 22 | 5′-CTT-TTT-TTY-ACC-CAG-CCG-TTC-CAC-TCA-GAG-GCA-CTT-TAA-AGG-ATG-TTT-CTG-TTG-AGA-GCT-CAT-CGT-CAT-CAT-CGT-CAT-CGT-CAA-CAA-CAA-CCC-CCG-CAC-CAG-CTC-CAA-AGA-AGG-TAA-GAG-AAA-GCG-AAG-AAG-GGA-AGA-ACA-GTG-AAG-ATA-GTC-AGA-CTC-CCG-CTA-GTC-CTG-GAA-GTG-ATT-CTC-AGG-ATA-GCT-CTA-AAG-GAG-ACG-AAG-CTG-TAG-ATG-GAT-CCG-CTT-CCG-GAT-CTA-GTA-CCC-CAA-CTC-AAG-CTG-CTG-AAA-AGG-AGC-CCG-AAA-CTC-CAG-AAT-CTA-CTC-CAA-AGG-AAG-AAT-GTG-GTA-CTT-CAT-TTA-TAA-TGT-GGT-TCG-GAG-AAG-GTA-CTC-CAG-CCA-CAA-CTT-TGA-AGT-GCG-GTG-GCT-ACA-CTA-TCG-TCT-ATG-CAC-CAG-AAA-AGG-ATA-ATA-AAG-AAC-CCG-CAC-CAA-GAT-ACA-TCT-CTG-GTG-ATG-TTA-AGG-CTG-TAA-CCT-TTG-AAA-AGG-GAG-AAG-ATA-ATA-CAG-TTA-AAA-TCA-AGG-TTG-ATG-GTA-AGG-AGT-TCA-GTA-CTC-TCT-CTT-CTA-GCT-CAA-GCA-GTC-CAA-CTG-AAA-ATA-ACG-GAT-CTA-CGG-GCC-AGG-TTG-CAT-CAA-GAT-CAA-GAA-GAT-CGC-TCT-CAG-AGG-AAA-ATA-GTG-AAA-CTG-CTG-CAA-CCG-TCG-ATT-TGT-TTG-CCT-TCA-CCC-TTG-ATG-GTG-GCC-GAA-GAA-TTG-AAG-TTG-CTG-TAC-CCA-GCG-TCG-AAG-ATG-CAA-CCA-AAA-GAG-ACA-AGT-ACA-GTT-TGG-TTG-CAA-ACG-GTA-AGC-CTT-TCT-ATA-CCG-GCG-CAA-ACA-GCG-GCA-CTA-CCA-ATG-GTG-TCT-ACA-GGT-TGA-ATG-AGA-ACG-GAG-ACT-TGG-TTG-ACA-AGG-ACA-ACA-CAG-TTC-TCT-TGA-AGA-GKG-3′ | KU670811 | C. parvum | IIc |
| 23 | 5′-TCY-CTC-AGA-GCA-CCT-TGA-AGG-ATG-TTT-CTG-TTG-AGG-RCT-CAT-CAT-CAT-CAT-CAT-CAT-CAT-CAT-CAT-CAT-CAT-CAT-CAT-CAT-CAA-CGA-CCG-TCG-CAC-CAG-CTT-CAA-ATA-AGG-CAA-GAA-CTG-GAG-AGG-ACA-CAG-GAC-GAA-GCG-AAG-GAA-GTC-AAG-GTT-CTG-AAG-AAC-ACC-AAG-ACG-GAG-AGG-ACG-ATA-GTT-CAG-ATT-CTA-GTG-GAG-GCA-GTG-TAG-GAG-GCA-CAG-AGA-GCG-GAA-GTG-CAG-GAG-GAA-AGA-ACG-AAG-AAG-ATA-GTT-CAA-GTT-CTG-GAG-GTG-CTC-AGG-ACG-GCA-GTG-GAG-GCA-CTG-CAG-AAG-GCG-CTA-CTC-AGT-CCG-AGG-CTA-CTG-CTT-CTC-AAG-GTG-CTC-CAT-CTC-AAG-GTT-CTG-ACA-AAA-CTA-CCG-AGT-CCA-CAC-AAA-CTA-CTC-CAA-AGG-AAG-AGT-GCG-GTA-CTT-CGT-TTG-TAA-TGT-GGT-TCG-GTG-AAG-GTA-CCC-CGG-TTG-CGA-CCT-TGA-AGT-GTG-GTG-GTT-ACA-CTA-TCG-TCT-ATG-CAC-CTG-TAA-AGG-ATC-AAG-CAA-ATC-CCG-CAC-CAA-GAT-ATA-TCT-CTG-GTG-AAG-TAA-AGA-ATG-TAT-CCT-TCC-AAA-AAG-AAA-GTG-ATA-ATA-CAA-TTA-AAA-TCA-AGG-TTG-ACG-GTC-AGG-ATT-TCA-GCA-CTC-TCT-CTG-CTA-GCT-CAA-GTA-GTC-CAA-CCG-AAA-ATA-AAG-GTG-AGT-CTG-GCA-ATC-AGG-TTG-AGT-CAA-GAT-CAA-GAA-GAT-CAC-TCA-CAG-AGG-AAA-CTA-GTG-AAA-CTG-CAA-CCG-TCG-ATT-TGT-TTG-CCT-TTA-CCC-TTA-ATG-GTG-GTA-AGA-GAA-TTG-AAG-TGG-CTG-TGC-CAA-ACG-CCG-AAG-AAA-CAT-CGA-AAA-GAG-ACA-AGT-ACA-GTT-TGG-TTG-CAG-ACG-ATA-GTG-CTT-TCT-ATA-CCG-GCA-AAA-ATA-GCG-GCA-GCA-CCG-ATG-GTG-TCT-ACA-AGT-TGA-ATG-AGA-ACG-GAG-ACT-TAG-T-3′ | MK982538 | C. hominis | Id |
| 24 | 5′-KTT-TTT-CTC-AGC-CCC-AGC-CGT-TCC-ACT-CAG-AGG-CAC-CTT-GAA-AGA-TGT-TTC-TGT-TGA-GAG-CTC-ATC-ATC-ATC-ATC-ATC-ATC-ATC-GTC-ATC-ATC-ATC-GTC-ATC-ATC-GTC-AAC-AAC-AAC-CCC-CGC-ACC-AGC-TCC-AAA-GAA-GGC-AAG-AGA-AGC-AGA-TGG-CGG-AGA-AGA-AAA-GAA-CAA-TGA-AGA-AAG-CCA-AAC-TCC-CGC-TAG-TCC-TGG-AAG-TGG-TGG-GGT-GAG-TGA-AGG-ACA-AGA-TAC-TCA-AGG-TGG-CTC-CAA-AGG-AGA-CGC-TGA-GGA-AGG-CAC-TGA-AGA-CAA-TGA-ACA-AGC-CGA-TGA-GAG-TGC-TAC-CCA-ACC-TTC-TAC-CCC-AGG-TCA-AGG-CTC-CGT-TAA-AAC-CGA-ATC-CAC-AGA-AAC-TAC-TCC-AAA-GGA-GAA-GTG-CGG-TAC-TTC-ATT-TGT-TAT-GTG-GTT-CGG-ACA-GGG-TGT-TCC-AGT-CGC-AAC-TTT-GAA-GTG-CGG-TGA-CTA-TAC-TAT-GGT-CTA-TGC-ACC-AGA-AAA-GGA-CAA-AAC-AGA-TCC-CGC-ACC-AAG-ATA-TAT-CTC-TGG-TGA-AGT-TAC-AAC-CGT-AAC-CTT-TGA-TAA-ACA-AGA-GAG-TAC-AGT-TAC-AAT-CAA-GGT-TAA-TAA-TGT-AGA-GTT-CGG-CAC-TCT-CTC-TAC-TAG-CTC-AAG-TAA-ACC-AAC-TGA-AAA-TAA-AGG-TGA-GTC-TAG-CGA-TCA-GGT-TGG-GTC-AAG-ATC-AAG-AAG-ATC-ACT-CAC-AGA-GGA-AAC-TAG-TGA-AAC-TGC-AAC-CGT-CGA-TTT-GTT-TGC-CTT-TAC-CCT-TGA-TGG-TGG-TAA-AAG-AAT-TGA-AGT-GGC-TGT-ACC-AAG-TGA-CGA-AGA-TGT-ATC-CAA-GAG-AAA-CAA-GTA-CAG-TTT-GGT-TGC-AAA-CGA-TAA-GAC-TTT-CTA-TAC-CGG-CGC-AAA-TAG-CGG-TAA-TAC-CGA-CGG-TAT-CTA-CAG-GTT-GAA-TGA-TAA-TGG-AGA-CTT-GGT-GGA-CAA-GAA-CAA-CAA-CGT-TCT-TTT-GAA-GGW-GTG-G-3′ | MT053132 | C. hominis | Ib |
| 25 | 5′-TTT-TTT-MYA-CCC-CGG-CCG-TTC-CAC-TCA-GAG-GCA-CCT-TGA-AGG-ATG-TTT-CTG-TTG-AGG-GAT-CAT-CGT-CAT-CGT-CAT-CAT-CAT-CAT-CAT-CAT-CAT-CAT-CAT-CAT-CAT-CAA-CAT-CGA-CCG-TCG-CAC-CAG-CTC-CAA-AGA-AAG-AAA-GAA-CTG-TAG-AGG-GCA-CGG-AAG-GAA-AGA-ACG-AAG-AAA-GCA-GTC-CAG-GTT-CTG-AAG-AAC-AAG-ACG-GTG-GTA-AGG-AAG-ACG-GTG-GTA-AGG-AAG-ACG-GTG-GTA-AGG-AAG-ACG-GTG-GTA-AGG-AAG-ACG-CTG-AAG-AAG-GAG-ACA-CAG-TAG-ACG-GGG-AAC-AAA-CCG-GGA-GTG-GTT-CTC-AAG-TTA-CTC-CAT-CTG-AAA-GTG-CCG-GCA-CAG-CTA-CCG-AGT-CCA-CAG-CAA-CTA-CTA-CTC-CAA-AGG-AAG-AAT-GTG-GTA-CTT-CAT-TTG-TCA-TGT-GGT-TCG-AGC-AAG-GCA-CCC-CGG-TTG-CGA-CCT-TGA-AGT-GTG-GTG-ATT-ACA-CTA-TCG-TCT-ATG-CAC-CTA-TAA-AAG-ATC-AAA-CAG-ATC-CCG-CAC-CAA-GAT-ATA-TCT-CTG-GTG-AAG-TTA-CAT-CTG-TAT-CCT-TTG-AAA-AGA-GTG-AAA-GTA-CAG-TTA-CAA-TCA-AGG-TTA-ATG-GTA-AAG-AGT-TCA-GCA-CTC-TCT-CTG-CTA-ACT-CAA-GTA-GTC-CAA-CTA-AAG-ATA-ACG-GTG-AAT-CTA-GTG-ACA-GTC-GGG-TTC-AAT-CVA-GAT-CAA-GAA-GAT-CAC-TCG-CAG-AGG-AGA-ATG-GTG-AAA-CAG-YTG-CAA-CCG-TTG-ATT-TGT-TTG-CCT-TTA-CTC-TTG-ATG-GTG-GTA-AAA-KAA-TTG-ARG-TGG-CTG-TGC-CAR-AGG-ACG-AAA-ATG-MMG-ACA-AAA-GAA-GCG-AGT-ACA-GTT-TGG-TTG-CAG-ACG-ATA-AGC-CTT-TCT-ATA-CCG-GCG-CAA-ACA-GTG-GCA-CCA-CCR-ATG-GTG-TCT-ACA-AAT-TGG-ATG-AKA-ATG-GAA-ACT-TGG-TTG-ACR-AGG-ACA-ACA-AAG-TTC-TCT-TGA-AAT-GTG-K-3′ | MN661180 | C. hominis | Ia |
| 26 | 5′-KKT-TTT-TTC-AGC-CCC-AGC-CGT-TCC-ACT-CAG-AGG-CAC-CTT-GAA-AGA-TGT-TTC-TGT-TGA-GAG-CTC-ATC-ATC-ATC-ATC-ATC-ATC-ATC-GTC-ATC-ATC-ATC-GTC-ATC-ATC-GTC-AAC-AAC-AAC-CCC-CGC-ACC-AGC-TCC-AAA-GAA-GGC-AAG-AGA-AGC-AGA-TGG-CGG-AGA-AGA-AAA-GAA-CAA-TGA-AGA-AAG-CCA-AAC-TCC-CGC-TAG-TCC-TGG-AAG-TGG-TGG-GGT-GAG-TGA-AGG-ACA-AGA-TAC-TCA-AGG-TGG-CTC-CAA-AGG-AGA-CGC-TGA-GGA-AGG-CAC-TGA-AGA-CAA-TGA-ACA-AGC-CGA-TGA-GAG-TGC-TAC-CCA-ACC-TTC-TAC-CCC-AGG-TCA-AGG-CTC-CGT-TAA-AAC-CGA-ATC-CAC-AGA-AAC-TAC-TCC-AAA-GGA-GAA-GTG-CGG-TAC-TTC-ATT-TGT-TAT-GTG-GTT-CGG-ACA-GGG-TGT-TCC-AGT-CGC-AAC-TTT-GAA-GTG-CGG-TGA-CTA-TAC-TAT-GGT-CTA-TGC-ACC-AGA-AAA-GGA-CAA-AAC-AGA-TCC-CGC-ACC-AAG-ATA-TAT-CTC-TGG-TGA-AGT-TAC-AAC-CGT-AAC-CTT-TGA-TAA-ACA-AGA-GAG-TAC-AGT-TAC-AAT-CAA-GGT-TAA-TAA-TGT-AGA-GTT-CGG-CAC-TCT-CTC-TAC-TAG-CTC-AAG-TAA-ACC-AAC-TGA-AAA-TAA-AGG-TGA-GTC-TAG-CGA-TCA-GGT-TGG-GTC-AAG-ATC-AAG-AAG-ATC-ACT-CAC-AGA-GGA-AAC-TAG-TGA-AAC-TGC-AAC-CGT-CGA-TTT-GTT-TGC-CTT-TAC-CCT-TGA-TGG-TGG-TAA-AAG-AAT-TGA-AGT-GGC-TGT-ACC-AAG-TGA-CGA-AGA-TGT-ATC-CAA-GAG-AAA-CAA-GTA-CAG-TTT-GGT-TGC-AAA-CGA-TAA-GAC-TTT-CTA-TAC-CGG-CGC-AAA-TAG-CGG-TAA-TAC-CGA-CGG-TAT-CTA-CAG-GTT-GAA-TGA-TAA-TGG-AGA-CTT-GGT-GGA-CAA-GAA-CAA-CAA-CGT-TCT-TTT-GAA-GRW-GT-3′ | MT053132 | C. hominis | Ib |
| 27 | 5′-TTT-TTC-AGC-CCC-AGC-CGT-TCC-ACT-CAG-AGG-CAC-CTT-GAA-AGA-TGT-TTC-TGT-TGA-GAG-CTC-ATC-ATC-ATC-ATC-ATC-ATC-ATC-GTC-ATC-ATC-ATC-GTC-ATC-ATC-GTC-AAC-AAC-AAC-CCC-CGC-ACC-AGC-TCC-AAA-GAA-GGC-AAG-AGA-AGC-AGA-TGG-CGG-AGA-AGA-AAA-GAA-CAA-TGA-AGA-AAG-CCA-AAC-TCC-CGC-TAG-TCC-TGG-AAG-TGG-TGG-GGT-GAG-TGA-AGG-ACA-AGA-TAC-TCA-AGG-TGG-CTC-CAA-AGG-AGA-CGC-TGA-GGA-AGG-CAC-TGA-AGA-CAA-TGA-ACA-AGC-CGA-TGA-GAG-TGC-TAC-CCA-ACC-TTC-TAC-CCC-AGG-TCA-AGG-CTC-CGT-TAA-AAC-CGA-ATC-CAC-AGA-AAC-TAC-TCC-AAA-GGA-GAA-GTG-CGG-TAC-TTC-ATT-TGT-TAT-GTG-GTT-CGG-ACA-GGG-TGT-TCC-AGT-CGC-AAC-TTT-GAA-GTG-CGG-TGA-CTA-TAC-TAT-GGT-CTA-TGC-ACC-AGA-AAA-GGA-CAA-AAC-AGA-TCC-CGC-ACC-AAG-ATA-TAT-CTC-TGG-TGA-AGT-TAC-AAC-CGT-AAC-CTT-TGA-TAA-ACA-AGA-GAG-TAC-AGT-TAC-AAT-CAA-GGT-TAA-TAA-TGT-AGA-GTT-CGG-CAC-TCT-CTC-TAC-TAG-CTC-AAG-TAA-ACC-AAC-TGA-AAA-TAA-AGG-TGA-GTC-TAG-CGA-TCA-GGT-TGG-GTC-AAG-ATC-AAG-AAG-ATC-ACT-CAC-AGA-GGA-AAC-TAG-TGA-AAC-TGC-AAC-CGT-CGA-TTT-GTT-TGC-CTT-TAC-CCT-TGA-TGG-TGG-TAA-AAG-AAT-TGA-AGT-GGC-TGT-ACC-AAG-TGA-CGA-AGA-TGT-ATC-CAA-GAG-AAA-CAA-GTA-CAG-TTT-GGT-TGC-AAA-CGA-TAA-GAC-TTT-CTA-TAC-CGG-CGC-AAA-TAG-CGG-TAA-TAC-CGA-CGG-TAT-CTA-CAG-GTT-GAA-TGA-TAA-TGG-AGA-CTT-GGT-GGA-CAA-GAA-CAA-CAA-CGT-TCT-TTT-GAA-GWG-GGG-3′ | MT053132 | C. hominis | Ib |
| 28 | 5′-TYT-GGT-GAG-GTA-AAA-AAT-GTA-TCC-TTC-CAA-AAA-GAA-AGY-GAT-GGT-ACA-WTT-AAA-ATC-AAG-ATT-GAS-GAA-AAG-GAG-TTC-AGT-TCT-CTY-TAR-YYG-AST-CAA-GCA-CTC-CRA-MYK-CAW-ATA-GYG-GAT-YCS-CGG-AAC-AGG-TKC-AWT-CAA-GAK-CAA-GAA-GAT-CAC-TCA-CAG-AGG-GAA-GTG-AAA-CWC-SKG-CTS-CCG-TMG-ATT-TGT-YYG-CCT-TCA-CCC-TTG-ATG-GTG-GTA-AAA-GAA-TTG-AAG-TGA-MTT-TCC-GCT-GTA-TTC-TCA-GCC-AGT-ATC-TCG-TAC-CCT-GCC-CTG-CCG-TTG-TAT-TCC-TGA-AGC-CAG-AAA-TCT-TCA-CTG-TAA-GCA-GTT-CCG-TGC-TCC-TGC-TGG-ATT-TGA-ATC-TTT-GTG-TCA-TAC-TCG-ACC-GAC-TTC-ATG-CAG-TCA-AGA-TAT-TCT-TCT-CTG-CTG-ATG-CTG-GTT-CCT-CTG-CAA-SGY-3′ | JF268649 | C. parvum | IIe |
| 29 | 5′-TTT-TTT-YAC-CCA-GCC-GTT-CCA-CTC-AGA-GGC-ACT-TTA-AAG-GAT-GCT-TCT-GTT-GAG-GGC-TCA-TCA-TCA-TCA-TCA-TCA-TCA-TCA-TCA-TCA-TCG-ACC-ACC-GTC-GCA-CCA-GCT-CCA-AAG-AAA-GAA-AGA-ACT-GGA-GAG-GGC-GTA-GAT-GGA-AAG-GAC-CAA-GTA-GAT-AGT-ACA-GGT-TCT-GAT-CAG-AAC-AGT-AAA-GGA-GAC-ACT-AAA-GGA-ACC-ACA-GAA-GAT-GGT-AAA-GAG-ACC-GAA-GGT-ACT-GTT-TCC-AAA-CCC-ACT-ACT-CCA-GAT-CAA-GGT-GAG-AGC-GCA-ACT-CCC-GGA-TCC-ACG-GAA-ACT-ACT-CCA-AAG-GAA-GAA-TGC-GGT-ACT-TCA-TTT-GTA-ATG-TGG-TTC-GGA-GAA-GGT-ACC-CCA-GTT-GCG-ACC-TTG-AAG-TGT-GGT-GGT-TAC-ACT-ATC-GTC-TAT-GCA-CCT-GTA-AAG-GAA-CAA-ACA-AAT-CCC-GCA-CCA-AGA-TAT-ATC-TCT-GGT-GAG-GTA-AAA-AAT-GTA-TCC-TTC-CAA-AAA-GAA-AGT-GAT-GGT-ACA-ATT-AAA-ATC-AAG-ATT-GAC-GAA-AAG-GAG-TTC-AGT-TCT-CTC-TCT-ACT-GAC-TCA-AGC-ACT-CCA-ACT-GCA-AAT-AGC-GGA-TCC-GCG-GAA-CAG-GTT-CAA-TCA-AGA-TCA-AGA-AGA-TCA-CTC-ACA-GAG-GGA-AGT-GAA-ACA-CCT-GCA-ACC-GTC-GAT-TTG-TTT-GCC-TTC-ACC-CTT-GAT-GGT-GGT-AAA-AGA-ATT-GAA-GTG-GCT-GTA-CCA-AAC-AAC-GAG-GAT-GCA-TCC-AAA-AGA-ACC-GAG-TAC-AGT-TTG-GTT-GCA-AAC-GAT-AAG-CCT-TTC-TAT-ACC-GGC-GCA-AAT-AGC-GGC-ACC-GAA-AAT-GGT-GTC-TAC-AAG-TTG-AAT-GAG-AAC-GGA-GAC-TTG-GTN-GAC-AAG-GAC-AAT-AAA-GTT-CTT-TTG-AGR-SGT-3′ | MW480843 | C. parvum | IIe |
| 30 | 5′-TGW-TGA-GGG-ATC-ATC-ATC-ATC-ATC-ATC-ATC-ATC-ATC-ATC-ATC-ATC-ATC-ATC-ATC-ATC-ATC-ATC-AAC-ATC-GAC-CGT-CGC-ACC-AGC-TCC-AAA-GAA-AGA-AAG-AAC-TGT-AGA-GGG-CGG-CAC-GGA-AGG-AAA-GAA-CGA-AGA-AAG-CAG-TCC-AGG-TTC-TGA-AGA-ACA-AGA-CGG-TGG-TAA-GGA-GGA-CGG-TGG-TAA-GGA-AAA-CGG-TGA-AGG-AGA-CAC-AGT-AGA-CGG-GGA-ACA-AAC-CGG-GAG-TGG-TTC-TCA-AGT-TAC-TCC-ATC-TGG-AAG-TGC-CGG-CAC-AGC-TAC-CGA-GTC-CAC-AGC-AAC-TAC-TAC-TCC-AAA-GGA-AGA-ATG-TGG-TAC-TTC-ATT-TGT-CAT-GTG-GTT-CGA-GAA-AGG-CAC-CCC-GGT-TGC-GAC-CTT-GAA-GTG-TGG-TGA-TTA-CAC-TAT-CGT-CTA-TGC-ACC-TAT-AAA-AGA-TCA-AAC-AGA-TCC-CGC-ACC-AAG-ATA-TAT-CTC-TGG-TGA-AGT-TAC-ATC-TGT-ATC-CTT-TGA-AAA-GAG-TGA-AAG-TAC-AGT-TAC-AAT-CAA-GGT-TAA-TGG-TAA-AGA-GTT-CAG-CAC-TCT-CTC-TGC-TAA-CTC-AAG-TAG-TCC-AAC-TAA-AGA-TAA-CGG-TGA-ATC-TAG-TGA-CAG-TCA-GGT-TCA-ATC-AAG-ATC-AAG-AAG-ATC-ACT-CGC-AGA-GGA-GAA-TGG-TGA-AAC-AGT-TGC-AAC-AGT-TGA-TTT-GTT-TGC-CTT-TAC-TCT-TGA-TGG-TGG-TAG-AAG-AAT-TGA-AGT-GGC-TGT-GCC-AAA-GGA-CGA-AAA-TGC-AGA-CAA-AAG-AAG-CGA-GTA-CAG-TTT-GGT-TGC-AGA-CGA-TAA-GCC-TTT-CTA-TAC-CGG-CGC-AAA-CAG-TGG-CAT-CAC-CAA-TGG-TGT-CTA-CAA-ATT-GGA-TGA-GAA-TGG-AAA-CTT-GGK-3′ | GU214345 | C. hominis | Ia |
| 31 | 5′-TTT-TTT-CYA-CCC-CAG-CCG-TTC-CAC-TCA-GAG-GCA-CTT-TAA-AGG-ATG-CTT-CTG-TTG-AGG-GCT-CAT-CAT-CAT-CAT-CAT-CAT-CAT-CAT-CAT-CAT-CGA-CCA-CCG-TCG-CAC-CAG-CTC-CAA-AGA-AAG-AAA-GAA-CTG-GAG-AGG-GCG-TAG-ATG-GAA-AGG-ACC-AAG-TAG-ATA-GTA-CAG-GTT-CTG-ATC-AGA-ACA-GTA-AAG-GAG-ACA-CTA-AAG-GAA-CCA-CAG-AAG-ATG-GTA-AAG-AGA-CCG-AAG-GTA-CTG-TTT-CCA-AAC-CCA-CTA-CTC-CAG-ATC-AAG-GTG-AGA-GCG-CAA-CTC-CCG-GAT-CCA-CGG-AAA-CTA-CTC-CAA-AGG-AAG-AAT-GCG-GTA-CTT-CAT-TTG-TAA-TGT-GGT-TCG-GAG-AAG-GTA-CCC-CAG-TTG-CGA-CCT-TGA-AGT-GTG-GTG-GTT-ACA-CTA-TCG-TCT-ATG-CAC-CTG-TAA-AGG-AAC-AAA-CAA-ATC-CCG-CAC-CAA-GAT-ATA-TCT-CTG-GTG-AGG-TAA-AAA-ATG-TAT-CCT-TCC-AAA-AAG-AAA-GTG-ATG-GTA-CAA-TTA-AAA-TCA-AGA-TTG-ACG-AAR-AGG-AGT-TCA-GTT-CTC-TCT-CTA-CTG-ACT-CAA-GCA-CTC-CAA-CTG-CAA-ATA-GCG-GAT-CCG-CGG-AAC-AGG-TTC-AAT-CAA-GAT-CAA-GAA-GAT-CAC-TCA-CAG-AGG-GAA-GTG-AAA-CAC-CTG-CAA-CCG-TCG-ATT-TGT-TTG-CCT-TCA-CCC-TTG-ATG-GTG-GTA-AAA-GAA-TTG-AAG-TGG-CTG-TAC-CAA-ACA-ACG-AGG-ATG-CAT-CCA-AAA-GAA-CCG-AGT-ACA-GTT-TGG-TTG-CAA-ACG-ATA-AGC-CTT-TCT-ATA-CCG-GCG-CAA-ATA-GCG-GCA-CCG-AAR-ATG-GTG-TCT-ACA-AGT-TGA-ATG-AGA-ACG-GAG-ACT-TGG-TTG-ACA-AGG-ACA-ATA-AAG-TTC-TTT-TGA-AGG-ATG-YGG-GG-3′ | MW480843 | C. parvum | IIe |
| 32 | 5′-GAG-GAC-GAT-AGT-TCA-GAT-TCT-AGT-GGA-GGC-AGT-GTA-GGA-GGC-ACA-GAG-AGC-GGA-AGT-GCA-GGA-GGA-AAG-AAC-GAA-GAA-GAT-AGT-TCA-AGT-TCY-GGA-GGT-GCT-CAG-GAC-GGC-AGT-GGA-GGC-ACT-GCA-GAA-GGC-GCT-ACT-CAG-TCC-GAG-GST-ACT-GCT-TYT-CAA-GRT-GCT-CCA-TCT-CAA-GGT-TCT-GAC-AAA-ACT-ACC-GAG-TCC-ACA-CAA-ACT-ACT-CCA-AAG-GAA-GAG-TGC-GGT-ACT-TCG-TTT-GTA-ATG-TGG-TTC-GGT-GAA-GGT-ACC-CCG-GTY-GSG-ACC-TTG-AAG-TGT-GGT-GGT-TAC-ACT-ATC-GTY-TAT-GCA-CCT-GTA-AAG-GAT-CAA-GCA-AAT-CCC-GCA-CCA-AGA-TAT-ATC-TCT-GGT-GAA-GTA-AAG-AAT-GTA-TCC-TTC-CAA-AAA-GAA-AGT-GAT-AAT-ACA-ATT-AAA-ATC-AAG-GTT-GAC-GGT-CAG-GAT-TTC-AGC-ACT-CTC-TCT-GCT-AGC-TCA-AGT-AGT-CCA-ACC-GAA-AAT-AAA-GGT-GAG-TMT-GGC-AAT-CAG-GTT-GAG-TCA-AGA-TCA-AGA-AGA-TCA-CTC-ACA-GAG-GAA-ACT-AGT-GAA-ACT-GCA-ACC-GTC-GAT-TTG-TTT-GCC-TTT-ACC-CTT-AAT-GGT-GGT-AAG-AGA-ATT-GAA-GTG-GCT-GTG-CCA-AAC-GCC-GAA-GAA-ACA-TCG-AAA-AGA-GAC-AAG-TAC-AGT-TTG-GTT-GCA-GAC-GAT-AGT-GCT-TTC-3′ | PV261913 | C. hominis | Id |
| 33 | 5′-TTT-TYA-CCC-CAG-CCG-TCC-CAC-TCA-GAG-GCA-CCT-TGA-AGG-ATG-TTT-CTG-TTG-AGG-GCT-CAT-CAT-CAT-CTT-CAT-CAT-CGT-CTT-CAT-CTT-CAT-CAT-CAT-CAT-CGT-CGT-CAA-CAA-CCC-CAG-CAC-CAG-CTT-CAA-AGA-AGG-TAA-GAG-AAG-CAG-AAG-GCA-GTG-AAG-AAA-AGG-ACA-GCG-AAG-AAA-AGG-ACA-GTG-AAG-AAA-AGG-GCA-GTG-AAG-AAG-GTA-GCC-AAA-CTC-CCG-CTA-GTC-CTG-GAG-GTG-GAG-GGG-TGA-GTG-AAG-GAG-ATA-CTC-AAG-GTG-ACT-CTA-AAG-GAG-ACG-GAG-TTA-GTT-CAG-ATG-AGA-ACC-AAA-GTC-AAG-GTG-GGG-ACG-CTA-CTC-CCG-GAT-CTA-GCA-CCC-AAA-CTC-AAG-CTA-CTG-AAA-AAG-AAC-CCG-GAT-CTT-CAG-AAG-CTA-CTC-CAA-AGG-AAG-AGT-GCG-GTA-CTT-CAT-TTG-TAA-TGT-GGT-TCG-GAC-AGG-GTG-TTC-CAG-TTG-TAA-CTT-TGA-AGT-GTG-GTG-GTT-ATA-CTA-TGG-TCT-ATG-CAC-CAG-AAA-ATG-GCA-AAA-CAG-ATC-CCG-CAC-CAA-GAT-ATA-TCT-CTG-GTA-AAG-TTT-CAA-CCG-TAG-ACT-TTG-AAA-AAC-AAG-ATA-GTA-CAG-TTA-AAA-TCA-AGG-TTA-ATG-GTG-TGG-AGT-TCA-GCA-CTC-TCT-CTA-CTA-GCT-CAA-GTA-ATC-CAA-CTG-AAA-ATA-GCG-GAT-CTG-AGA-GCC-AGG-CTC-AAT-CAA-GAT-CAA-GAA-GAT-CAC-TCG-CAG-AGG-ATG-GTA-CTG-AGA-CTG-CTG-CAA-CCG-TCG-ATT-TGA-TTG-CCT-TCA-CCC-TTC-AAG-GTG-GTA-AAA-GAA-TCG-AAG-TCG-CTG-TGC-CAA-GTG-ACG-AAG-ATG-TAT-CCA-AGA-GAA-ACA-AGT-ACA-GTT-TGG-TTG-CAG-GCG-ATA-AGA-CTT-TCT-ATA-CCG-GCG-CAA-ATA-GCG-GTA-ATA-CCG-ACG-GTA-TCT-ACA-GGT-TGA-ATG-ATG-ATG-GAG-ACT-TGG-TGG-ACA-AGA-ACA-ACA-ACG-TTC-TTT-TGA-AGA-BG-3′ | ON863660 | C. hominis | Ie |
| 34 | 5′-CCC-CAG-CCG-TTC-CAC-TCA-GAG-GCA-CCT-TGA-AGG-ATG-TTT-CTG-TTG-AGG-GCT-CAT-CAT-CAT-CTT-CAT-CAT-CGT-CTT-CAT-CTT-CAT-CAT-CAT-CAT-CGT-CGT-CAA-CAA-CCC-CAG-CAC-CAG-CTT-CAA-AGA-AGG-TAA-GAG-AAG-CAG-AAG-GCA-GTG-AAG-AAA-AGG-ACA-GCG-AAG-AAA-AGG-ACA-GTG-AAG-AAA-AGG-GCA-GTG-AAG-AAG-GTA-GCC-AAA-CTC-CCG-CTA-GTC-CTG-GAG-GTG-GAG-GGG-TGA-GTG-AAG-GAG-ATA-CTC-AAG-GTG-ACT-CTA-AAG-GAG-ACG-GAG-TTA-GTT-CAG-ATG-AGA-ACC-AAA-GTC-AAG-GTG-GGG-ACG-CTA-CTC-CCG-GAT-CTA-GCA-CCC-AAA-CTC-AAG-CTA-CTG-AAA-AAG-AAC-CCG-GAT-CTT-CAG-AAG-CTA-CTC-CAA-AGG-AAG-AGT-GCG-GTA-CTT-CAT-TTG-TAA-TGT-GGT-TCG-GAC-AGG-GTG-TTC-CAG-TTG-TAA-CTT-TGA-AGT-GTG-GTG-GTT-ATA-CTA-TGG-TCT-ATG-CAC-CAG-AAA-ATG-GCA-AAA-CAG-ATC-CCG-CAC-CAA-GAT-ATA-TCT-CTG-GTA-AAG-TTT-CAA-CCG-TAG-ACT-TTG-AAA-AAC-AAG-ATA-GTA-CAG-TTA-AAA-TCA-AGG-TTA-ATG-GTG-TGG-AGT-TCA-GCA-CTC-TCT-CTA-CTA-GCT-CAA-GTA-ATC-CAA-CTG-AAA-ATA-GCG-GAT-CTG-AGA-GCC-AGG-CTC-AAT-CAA-GAT-CAA-GAA-GAT-CAC-TCG-CAG-AGG-ATG-GTA-CTG-AGA-CTG-CTG-CAA-CCG-TCG-ATT-TGA-TTG-CCT-TCA-CCC-TTC-AAG-GTG-GTA-AAA-GAA-TCG-AAG-TCG-CTG-TGC-CAA-GTG-ACG-AAG-ATG-TAT-CCA-AGA-GAA-ACA-AGT-ACAG-TTT-GGT-TGC-AGG-CGA-TAA-GAC-TTT-CTA-TAC-CGG-CGC-AAA-TAG-CGG-TAA-TAC-CGA-CGG-TAT-CTA-CAG-GTT-GAA-TGA-TGA-TGG-AGA-CTT-GGT-GGA-CAA-GAA-CAA-CAA-CGT-TCT-TTG-AAG-WSG-GGG-3′ | ON863660 | C. hominis | Ie |
| 35 | 5′-CAC-AGC-CGT-TCY-KCT-CAG-AGG-CAC-CTT-GAA-GGA-TGT-TTC-TGT-TGA-GGR-CTC-ATC-ATC-ATC-ATC-ATC-ATC-ATC-ATC-ATC-ATC-ATC-ATC-ATC-ATC-AAC-GAC-CGT-CGC-ACC-AGC-TTC-AAA-TAA-GGC-AAG-AAC-TGG-AGA-GGA-CAC-AGG-ACG-AAG-CGA-AGG-AAG-TCA-AGG-TTC-TGA-AGA-ACA-CCA-AGA-CGG-AGA-GGA-CGA-TAG-TTC-AGA-TTC-TAG-TGG-AGG-CAG-TGT-AGG-AGG-CAC-AGA-GAG-CGG-AAG-TGC-AGG-AGG-AAA-GAA-CGA-AGA-AGA-TAG-TTC-AAG-TTC-TGG-AGG-TGC-TCA-GGA-CGG-CAG-TGG-AGG-CAC-TGC-AGA-AGG-CGC-TAC-TCA-GTC-CGA-GGC-TAC-TGC-TTC-TCA-AGG-TGC-TCC-ATC-TCA-AGG-TTC-TGA-CAA-AAC-TAC-CGA-GTC-CAC-ACA-AAC-TAC-TCC-AAA-GGA-AGA-GTG-CGG-TAC-TTC-GTT-TGT-AAT-GTG-GTT-CGG-TGA-AGG-TAC-CCC-GGT-TGC-GAC-CTT-GAA-GTG-TGG-TGG-TTA-CAC-TAT-CGT-CTA-TGC-ACC-TGT-AAA-GGA-TCA-AGC-AAA-TCC-CGC-ACC-AAG-ATA-TAT-CTC-TGG-TGA-AGT-AAA-GAA-TGT-ATC-CTT-CCA-AAA-AGA-AAG-TGA-TAA-TAC-AAT-TAA-AAT-CAA-GGT-TGA-CGG-TCA-GGA-TTT-CAG-CAC-TCT-CTC-TGC-TAG-CTC-AAG-TAG-TCC-AAC-CGA-AAA-TAA-AGG-TGA-GTC-TGG-CAA-TCA-GGT-TGA-GTC-AAG-ATC-AAG-AAG-ATC-ACT-CAC-AGA-GGA-AAC-TAG-TGA-AAC-TGC-AAC-CGT-CGA-TTT-GTT-TGC-CTT-TAC-CCT-TAA-TGG-TGG-TAA-GAG-AAT-TGA-AGT-GGC-TGT-GCC-AAA-CGC-CGA-AGA-AAC-ATC-GAA-AAG-AGA-CAA-GTA-CAG-TTT-GGT-TGC-AGA-CGA-TAG-TGC-TTT-CTA-TAC-CGG-CAA-AAA-TAG-CGG-CAG-CAC-CGA-TGG-TGT-CTA-CAA-GTT-GAA-TGA-GAA-CGG-AGA-CTT-AGT-GA-3′ | MK982538 | C. hominis | Id |
| 36 | 5′-KKT-TTT-TCT-CAG-CCC-CAG-CCG-TTC-CAC-TCA-GAG-GCA-CTT-TAA-AGG-ATG-TTT-CTG-TTG-AGA-GCT-CAT-CGT-CAT-CAT-CGT-CAT-CGT-CAA-CAA-CCC-CCG-CAC-CAG-CTC-CAA-AGA-AGG-TAA-GAG-AAA-GCG-AAG-AAG-GGA-AGA-ACA-GTG-AAG-ATA-GTC-AAA-CTC-CCG-CTA-GTC-CTG-GAA-GTG-GTT-CTC-AGG-ATA-GCT-CTA-AAG-GAG-ACG-AAG-TTG-TAG-ATG-GAG-GCG-TTT-CCG-GAT-CTA-GTA-CCC-CAA-CTC-AAG-CTG-CTG-AAA-AGG-AGC-CCG-AAA-CTC-CAG-AAT-CTA-CTC-CAA-AGG-AAG-AAT-GTG-GTA-CTT-CAT-TTA-TAA-TGT-GGT-TCG-GAG-AAG-GTA-CTC-CAG-CCA-CAA-CTT-TGA-AGT-GCG-GTG-GCT-ACA-CTA-TCG-TCT-ATG-CAC-CAG-AAA-AGG-ATA-ATA-AAG-AAC-CCG-CAC-CAA-GAT-ACA-TCT-CTG-GTG-ATG-TTA-AGG-CTG-TAA-CCT-TTG-AAA-AGG-GAG-AAG-ATA-ATA-CAG-TTA-AAA-TCA-AGG-TTG-ATG-GTA-GGG-AGT-TCA-GTA-CTC-TCT-CTT-CTA-GCT-CAA-GCA-GTC-CAA-CTG-AAA-ATA-GCG-GAT-CTG-AGA-GCC-AGG-CTC-AAT-CAA-GAT-CAA-GAA-GAT-CAC-TCA-CAG-AGG-GAA-GTG-AAA-CAC-CTG-CAA-CCG-TCG-ATT-TGT-TTG-CCT-TCA-CCC-TTC-AAG-GTG-GTA-AAA-GAA-TCG-AAG-TCG-CTG-TGC-CAA-GTG-ACG-TAG-ATG-CAT-CCA-AGA-GAA-ACA-AGT-ACA-GTT-TGG-TTG-CAG-GCG-ATA-AGA-CTT-TCT-ATA-CCG-GCG-CAA-ATA-GCG-GTA-ATA-CCG-ACG-GTA-TCT-ACA-GGT-TGA-ATG-ATG-ATG-GAG-ACT-TGG-TGG-ACA-AGA-ACA-ACA-ACG-TTC-TTT-TGA-AGG-ATK-3′ | JF802124 | C. parvum | IIc |
| 37 | 5′-TCA-GCC-CCA-GCC-GTT-CCA-CTC-AGA-GGC-ACC-TTG-AAA-GAT-GTT-TCT-GTT-GAG-AGC-TCA-TCA-TCA-TCA-TCA-TCA-TCA-TCG-TCA-TCA-TCA-TCG-TCA-TCA-TCG-TCA-ACA-ACA-ACC-CCC-GCA-CCA-GCT-CCA-AAG-AAG-GCA-AGA-GAA-GCA-GAT-GGC-GGA-GAA-GAA-AAG-AAC-AAT-GAA-GAA-AGC-CAA-ACT-CCC-GCT-AGT-CCT-GGA-AGT-GGT-GGG-GTG-AGT-GAA-GGA-CAA-GAT-ACT-CAA-GGT-GGC-TCC-AAA-GGA-GAC-GCT-GAG-GAA-GGC-ACT-GAA-GAC-AAT-GAA-CAA-GCC-GAT-GAG-AGT-GCT-ACC-CAA-CCT-TCT-ACC-CCA-GGT-CAA-GGC-TCC-GTT-AAA-ACC-GAA-TCC-ACA-GAA-ACT-ACT-CCA-AAG-GAG-AAG-TGC-GGT-ACT-TCA-TTT-GTT-ATG-TGG-TTC-GGA-CAG-GGT-GTT-CCA-GTC-GCA-ACT-TTG-AAG-TGC-GGT-GAC-TAT-ACT-ATG-GTC-TAT-GCA-CCA-GAA-AAG-GAC-AAA-ACA-GAT-CCC-GCA-CCA-AGA-TAT-ATC-TCT-GGT-GAA-GTT-ACA-ACC-GTA-ACC-TTT-GAT-AAA-CAA-GAG-AGT-ACA-GTT-ACA-ATC-AAG-GTT-AAT-AAT-GTA-GAG-TTC-GGC-ACT-CTC-TCT-ACT-AGC-TCA-AGT-AAA-CCA-ACT-GAA-AAT-AAA-GGT-GAG-TCT-AGC-GAT-CAG-GTT-GGG-TCA-AGA-TCA-AGA-AGA-TCA-CTC-ACA-GAG-GAA-ACT-AGT-GAA-ACT-GCA-ACC-GTC-GAT-TTG-TTT-GCC-TTT-ACC-CTT-GAT-GGT-GGT-AAA-AGA-ATT-GAA-GTG-GCT-GTA-CCA-AGT-GAC-GAA-GAT-GTA-TCC-AAG-AGA-AAC-AAG-TAC-AGT-TTG-GTT-GCA-AAC-GAT-AAG-ACT-TTC-TAT-ACC-GGC-GCA-AAT-AGC-GGT-AAT-ACC-GAC-GGT-ATC-TAC-AGG-TTG-AAT-GAT-AAT-GGA-GAC-TTG-GTG-GAC-AAG-AAC-AAC-AAC-GTT-CTT-TTG-AAG-GAT-G-3′ | MT053132 | C. hominis | Ib |
| 38 | 5′-YTT-TTT-TTY-ACC-CAG-CCG-TTC-CAC-TCA-GAG-GCA-CTT-TAA-AGG-ATG-TTT-CTG-TTG-AGA-GCT-CAT-CGT-CAT-CAT-CGT-CAT-CGT-CAA-CAA-CCC-CCG-CAC-CAG-CTC-CAA-AGA-AGG-TAA-GAG-AAA-GCG-AAG-AAG-GGA-AGA-ACA-GTG-AAG-ATA-GTC-AAA-CTC-CCG-CTA-GTC-CTG-GAA-GTG-GTT-CTC-AGG-ATA-GCT-CTA-AAG-GAG-ACG-AAG-TTG-TAG-ATG-GAG-GCG-TTT-CCG-GAT-CTA-GTA-CCC-CAA-CTC-AAG-CTG-CTG-AAA-AGG-AGC-CCG-AAA-CTC-CAG-AAT-CTA-CTC-CAA-AGG-AAG-AAT-GTG-GTA-CTT-CAT-TTA-TAA-TGT-GGT-TCG-GAG-AAG-GTA-CTC-CAG-CCA-CAA-CTT-TGA-AGT-GCG-GTG-GCT-ACA-CTA-TCG-TCT-ATG-CAC-CAG-AAA-AGG-ATA-ATA-AAG-AAC-CCG-CAC-CAA-GAT-ACA-TCT-CTG-GTG-ATG-TTA-AGG-CTG-TAA-CCT-TTG-AAA-AGG-GAG-AAG-ATA-ATA-CAG-TTA-AAA-TCA-AGG-TTG-ATG-GTA-GGG-AGT-TCA-GTA-CTC-TCT-CTT-CTA-GCT-CAA-GCA-GTC-CAA-CTG-AAA-ATA-GCG-GAT-CTG-AGA-GCC-AGG-CTC-AAT-CAA-GAT-CAA-GAA-GAT-CAC-TCA-CAG-AGG-GAA-GTG-AAA-CAC-CTG-CAA-CCG-TCG-ATT-TGT-TTG-CCT-TCA-CCC-TTC-AAG-GTG-GTA-AAA-GAA-TCG-AAG-TCG-CTG-TGC-CAA-GTG-ACG-TAG-ATG-CAT-CCA-AGA-GAA-ACA-AGT-ACA-GTT-TGG-TTG-CAG-GCG-ATA-AGA-CTT-TCT-ATA-CCG-GCG-CAA-ATA-GCG-GTA-ATA-CCG-ACG-GTA-TCT-ACA-GGT-TGA-ATG-ATG-ATG-GAG-ACT-TGG-TGG-ACA-AGA-ACA-ACA-ACG-TTC-TTT-TGA-GAT-S-3′ | JF802124 | C. parvum | IIc |
| 39 | 5′-TTT-TTY-ACC-CCA-GCC-GTT-CCM-YTC-AGA-GGC-ACC-TTG-AAA-GAT-GTT-TCT-GTT-GAG-AGC-TCA-TCA-TCA-TCA-TCA-TCA-TCA-TCG-TCA-TCA-TCA-TCG-TCA-TCA-TCG-TCA-ACA-ACA-ACC-CCC-GCA-CCA-GCT-CCA-AAG-AAG-GCA-AGA-GAA-GCA-GAT-GGC-GGA-GAA-GAA-AAG-AAC-AAT-GAA-GAA-AGC-CAA-ACT-CCC-GCT-AGT-CCT-GGA-AGT-GGT-GGG-GTG-AGT-GAA-GGA-CAA-GAT-ACT-CAA-GGT-GGC-TCC-AAA-GGA-GAC-GCT-GAG-GAA-GGC-ACT-GAA-GAC-AAT-GAA-CAA-GCC-GAT-GAG-AGT-GCT-ACC-CAA-CCT-TCT-ACC-CCA-GGT-CAA-GGC-TCC-GTT-AAA-ACC-GAA-TCC-ACA-GAA-ACT-ACT-CCA-AAG-GAG-AAG-TGC-GGT-ACT-TCA-TTT-GTT-ATG-TGG-TTC-GGA-CAG-GGT-GTT-CCA-GTC-GCA-ACT-TTG-AAG-TGC-GGT-GAC-TAT-ACT-ATG-GTC-TAT-GCA-CCA-GAA-AAG-GAC-AAA-ACA-GAT-CCC-GCA-CCA-AGA-TAT-ATC-TCT-GGT-GAA-GTT-ACA-ACC-GTA-ACC-TTT-GAT-AAA-CAA-GAG-AGT-ACA-GTT-ACA-ATC-AAG-GTT-AAT-AAT-GTA-GAG-TTC-GGC-ACT-CTC-TCT-ACT-AGC-TCA-AGT-AAA-CCA-ACT-GAA-AAT-AAA-GGT-GAG-TCT-AGC-GAT-CAG-GTT-GGG-TCA-AGA-TCA-AGA-AGA-TCA-CTC-ACA-GAG-GAA-ACT-AGT-GAA-ACT-GCA-ACC-GTC-GAT-TTG-TTT-GCC-TTT-ACC-CTT-GAT-GGT-GGT-AAA-AGA-ATT-GAA-GTG-GCT-GTA-CCA-AGT-GAC-GAA-GAT-GTA-TCC-AAG-AGA-AAC-AAG-TAC-AGT-TTG-GTT-GCA-AAC-GAT-AAG-ACT-TTC-TAT-ACC-GGC-GCA-AAT-AGC-GGT-AAT-ACC-GAC-GGT-ATC-TAC-AGG-TTG-AAT-GAT-AAT-GGA-GAC-TTG-GTG-GAC-AAG-AAC-AAC-AAC-GTT-CTT-TTG-AAG-WWB-3′ | MK105902 | C. hominis | Ib |
| 40 | 5′-TTY-ACC-CAG-CCG-TCC-CAC-TCA-GAG-GCA-CCT-TGA-AGG-ATG-TTT-CTG-TTG-AGG-GCT-CAT-CAT-CAT-CTT-CAT-CAT-CGT-CTT-CAT-CTT-CAT-CAT-CAT-CAT-CGT-CGT-CAA-CAA-CCC-CAG-CAC-CAG-CTT-CAA-AGA-AGG-TAA-GAG-AAG-CAG-AAG-GCA-GTG-AAG-AAA-AGG-ACA-GCG-AAG-AAA-AGG-ACA-GTG-AAG-AAA-AGG-GCA-GTG-AAG-AAG-GTA-GCC-AAA-CTC-CCG-CTA-GTC-CTG-GAG-GTG-GAG-GGG-TGA-GTG-AAG-GAG-ATA-CTC-AAG-GTG-ACT-CTA-AAG-GAG-ACG-GAG-TTA-GTT-CAG-ATG-AGA-ACC-AAA-GTC-AAG-GTG-GGG-ACG-CTA-CTC-CCG-GAT-CTA-GCA-CCC-AAA-CTC-AAG-CTA-CTG-AAA-AAG-AAC-CCG-GAT-CTT-CAG-AAG-CTA-CTC-CAA-AGG-AAG-AGT-GCG-GTA-CTT-CAT-TTG-TAA-TGT-GGT-TCG-GAC-AGG-GTG-TTC-CAG-TTG-TAA-CTT-TGA-AGT-GTG-GTG-GTT-ATA-CTA-TGG-TCT-ATG-CAC-CAG-AAA-ATG-GCA-AAA-CAG-ATC-CCG-CAC-CAA-GAT-ATA-TCT-CTG-GTA-AAG-TTT-CAA-CCG-TAG-ACT-TTG-AAA-AAC-AAG-ATA-GTA-CAG-TTA-AAA-TCA-AGG-TTA-ATG-GTG-TGG-AGT-TCA-GCA-CTC-TCT-CTA-CTA-GCT-CAA-GTA-ATC-CAA-CTG-AAA-ATA-GCG-GAT-CTG-AGA-GCC-AGG-CTC-AAT-CAA-GAT-CAA-GAA-GAT-CAC-TCG-CAG-AGG-ATG-GTA-CTG-AGA-CTG-CTG-CAA-CCG-TCG-ATT-TGA-TTG-CCT-TCA-CCC-TTC-AAG-GTG-GTA-AAA-GAA-TCG-AAG-TCG-CTG-TGC-CAA-GTG-ACG-AAG-ATG-TAT-CCA-AGA-GAA-ACA-AGT-ACA-GTT-TGG-TTG-CAG-GCG-ATA-AGA-CTT-TCT-ATA-CCG-GCG-CAA-ATA-GCG-GTA-ATA-CCG-ACG-GTA-TCT-ACA-GGT-TGA-ATG-ATG-ATG-GAG-ACT-TGG-TGG-ACA-AGA-ACA-ACA-ACG-TTC-TTT-GAG-RGK-3′ | ON863660 | C. hominis | Ie |
| 41 | 5′-AGG-AGC-CAG-CAT-CTT-TCA-AGA-GAA-CTT-TGT-TGT-CST-TGT-CAA-CCA-AGT-TTC-CAT-TMT-CAT-CCA-ATT-TGT-AGA-CAC-CAT-TGG-TGA-TGC-CAC-TGT-TTG-CGC-CGG-TAT-AGA-AAG-GCT-TAT-CGT-CTG-CAA-CCA-AAC-TGT-ACT-CGC-TTC-TTT-TGT-CTG-CAT-YTT-CGT-CCT-TTG-GCA-CAG-CCA-CTT-CAA-TTC-TTC-TAC-CAC-CAT-CAA-GAG-TAA-AGG-CAA-ACA-AAT-CAA-CTG-TTG-CAA-CTG-TTT-CAC-CAT-TCT-CCT-CTG-CGA-GTG-ATC-TTC-TTG-ATC-TTG-ATT-GAA-CCT-GAC-TGT-CAC-TAG-ATT-CAC-CGT-TAT-CTT-TAG-TTG-GAC-TAC-TTG-AGT-TAG-CAG-AGA-GAG-TGC-TGA-ACT-CTT-TAC-CAT-TAA-CCT-TGA-TTG-TAA-CTG-TAC-TTT-CAC-TCT-TTT-CAA-AGG-ATA-CAG-ATG-TAA-CTT-CAC-CAG-AGA-TAT-ATC-TTG-GTG-CGG-GAT-CTG-TTT-GAT-CTT-TTA-TAG-GTG-CAT-AGA-CGA-TAG-TGT-AAT-CAC-CAC-ACT-TCA-AGG-TCG-CAA-CCG-GGG-TGC-CTT-TCT-CGA-ACC-ACA-TGA-CAA-ATG-AAG-TAC-CAC-ATT-CTT-CCT-TTG-GAG-TAG-TAG-TTG-CTG-TGG-ACT-CGG-TAG-CTG-TGC-CGG-CAC-TTC-CAG-ATG-GAG-TAA-CTT-GAG-AAC-CAC-TCC-CGG-TTT-GTT-CCC-CGT-CTA-CTG-TGT-CTC-CTT-CAC-CGT-TTT-CCT-TAC-CAC-CGT-CTT-CCT-TAC-CAC-CGT-CTT-GTT-CTT-CAG-AAC-CTG-GAC-TGC-TTT-CTT-CGT-TCT-TTC-CTT-CCG-TGC-CGC-CCT-CTA-CAG-TTC-TTT-CTT-TCT-TTG-GAG-CTG-GTG-CGA-CGG-TCG-ATG-TTG-ATG-ATG-ATG-ATG-ATG-ATG-ATG-ATG-ATG-ATG-ATG-ATG-ATG-ATG-ATC-CCT-CAR-CAG-3′ | KY990897 | C. hominis | Ia |
| 42 | 5′-CTC-AKC-CGT-TCC-ACT-CAG-AGG-CAC-CTT-GAT-AGA-TGT-TTC-TGT-TGA-GAG-CTC-ATC-ATC-ATC-ATC-ATC-ATC-ATC-GTC-ATC-ATC-ATC-GTC-ATC-ATC-GTC-AAC-AAC-AAC-CCC-CGC-ACC-AGC-TCC-AAA-GAA-GGC-AAG-AGA-AGC-AGA-TGG-CGG-AGA-AGA-AAA-GAA-CAA-TGA-AGA-AAG-CCA-AAC-TCC-CGC-TAG-TCC-TGG-AAG-TGG-TGG-GGT-GAG-TGA-AGG-ACA-AGA-TAC-TCA-AGG-TGG-CTC-CAA-AGG-AGA-CGC-TGA-GGA-AGG-CAC-TGA-AGA-CAA-TGA-ACA-AGC-CGA-TGA-GAG-TGC-TAC-CCA-ACC-TTC-TAC-CCC-AGG-TCA-AGG-CTC-CGT-TAA-AAC-CGA-ATC-CAC-AGA-AAC-TAC-TCC-AAA-GGA-GAA-GTG-CGG-TAC-TTC-ATT-TGT-TAT-GTG-GTT-CGG-ACA-GGG-TGT-TCC-AGT-CGC-AAC-TTT-GAA-GTG-CGG-TGA-CTA-TAC-TAT-GGT-CTA-TGC-ACC-AGA-AAA-GGA-CAA-AAC-AGA-TCC-CGC-ACC-AAG-ATA-TAT-CTC-TGG-TGA-AGT-TAC-AAC-CGT-AAC-CTT-TGA-TAA-ACA-AGA-GAG-TAC-AGT-TAC-AAT-CAA-GGT-TAA-TAA-TGT-AGA-GTT-CGG-CAC-TCT-CTC-TAC-TAG-CTC-AAG-TAA-ACC-AAC-TGA-AAA-TAA-AGG-TGA-GTC-TAG-CGA-TCA-GGT-TGG-GTC-AAG-ATC-AAG-AAG-ATC-ACT-CAC-AGA-GGA-AAC-TAG-TGA-AAC-TGC-AAC-CGT-CGA-TTT-GTT-TGC-CTT-TAC-CCT-TGA-TGG-TGG-TAA-AAG-AAT-TGA-AGT-GGC-TGT-ACC-AAG-TGA-CGA-AGA-TGT-ATC-CAA-GAG-AAA-CAA-GTA-CAG-TTT-GGT-TGC-AAA-CGA-TAA-GAC-TTT-CTA-TAC-CGG-CGC-AAA-TAG-CGG-TAA-TAC-CGA-CGG-TAT-CTA-CAG-GTT-GAA-TGA-TAA-TGG-AGA-CTT-GGT-GGA-CAA-GAA-CAA-CAA-CGT-TCT-TTT-GAA-GGA-TGC-TGT-CT-3′ | MT053132 | C. hominis | Ib |
| 43 | 5′-CTT-CCG-CTG-TAW-TCT-CAG-CCC-CAG-CCG-TTC-CAC-TCA-GAG-GCA-CTT-TAA-AGG-ATG-TTT-CTG-TTG-AGA-GCT-CAT-CGT-CAT-CAT-CGT-CAT-CGT-CAA-CAA-CAA-CCC-CCG-CAC-CAG-CTC-CAA-AGA-AGG-TAA-GAG-AAA-GCG-AAG-AAG-GGA-AGA-ACA-GTG-AAG-ATA-GTC-AGA-CTC-CCG-CTA-GTC-CTG-GAA-GTG-ATT-CTC-AGG-ATA-GCT-CTA-AAG-GAG-ACG-AAG-CTG-TAG-ATG-GAT-CCG-CTT-CCG-GAT-CTA-GTA-CCC-CAA-CTC-AAG-CTG-CTG-AAA-AGG-AGC-CCG-AAA-CTC-CAG-AAT-CTA-CTC-CAA-AGG-AAG-AAT-GTG-GTA-CTT-CAT-TTA-TAA-TGT-GGT-TCG-GAG-AAG-GTA-CTC-CAG-CCA-CAA-CTT-TGA-AGT-GCG-GTG-GCT-ACA-CTA-TCG-TCT-ATG-CAC-CAG-AAA-AGG-ATA-ATA-AAG-AAC-CCG-CAC-CAA-GAT-ACA-TCT-CTG-GTG-ATG-TTA-AGG-CTG-TAA-CCT-TTG-AAA-AGG-GAG-AAG-ATA-ATA-CAG-TTA-AAA-TCA-AGG-TTG-ATG-GTA-AGG-AGT-TCA-GTA-CTC-TCT-CTT-CTA-GCT-CAA-GCA-GTC-CAA-CTG-AAA-ATA-ACG-GAT-CTA-CGG-GCC-AGG-TTG-CAT-CAA-GAT-CAA-GAA-GAT-CGC-TCT-CAG-AGG-AAA-ATA-GTG-AAA-CTG-CTG-CAA-CCG-TCG-ATT-TGT-TTG-CCT-TCA-CCC-TTG-ATG-GTG-GCC-GAA-GAA-TTG-AAG-TTG-CTG-TAC-CCA-GCG-TCG-AAG-ATG-CAA-CCA-AAA-GAG-ACA-AGT-ACA-GTT-TGG-TTG-CAA-ACG-GTA-AGC-CTT-TCT-ATA-CCG-GCG-CAA-ACA-GCG-GCA-CTA-CCA-ATG-GTG-TCT-ACA-GGT-TGA-ATG-AGA-ACG-GAG-ACT-TGG-TTG-ACA-AGG-ACA-ACA-CAG-TTC-TCT-TGA-AGG-ATG-CTG-TTT-YCY-TCT-G-3′ | GU214366 | C. parvum | IIc |
| 44 | 5′-AGG-ACG-GTG-GTA-ATG-AAA-TCG-GTG-AAG-GAG-ACA-CAG-TAG-ACG-GCG-AAC-AAA-CCG-GGA-GTG-GTT-CTC-AAG-TTA-CTC-CAT-CTG-GAA-GTG-CCG-GCA-CAG-CTA-CCG-AGT-CCA-CAG-CAA-CTA-CTA-CTC-CAA-AGG-AAG-AAT-GTG-GTA-CTT-CAT-TTG-TCA-TGT-GGT-TCG-AGA-AAG-GCA-CCC-CGG-TTG-CGA-CCT-TGA-AGT-GTG-GTG-ATT-ACA-CTA-TCG-TCT-ATG-CAC-CTA-TAA-AAG-ATC-AAA-CAG-ATC-CCG-CAC-CAA-GAT-ATA-TCT-CTG-GTG-AAG-TTA-CAT-CTG-TAT-CCT-TTG-AAA-AGA-GTG-AAA-GTA-CAG-TTA-CAA-TCA-AGG-TTA-ATG-GTA-AAG-AGT-TCA-GCA-CTC-TCT-CTG-CTA-ACT-CAA-GTA-GTC-CAA-CTA-AAG-ATA-ACG-GTG-AAT-CTA-GTG-ACA-GTC-AGG-TTC-AAT-CAA-GAT-CAA-GAA-GAT-CAC-TCG-CAG-AGG-AGA-ATG-GTG-AAA-CAG-TTG-CAA-CAG-TTG-ATT-TGT-TTG-CCT-TTA-CTC-TTG-ATG-GTG-GTA-GAA-GAA-TTG-AAG-TGG-CTG-TGC-CAA-AGG-ACG-AAA-ATG-CAG-ACA-AAA-GAA-GCG-AGT-ACA-GTT-TGG-TTG-CAG-ACG-ATA-AGC-CTT-TCT-ATA-CCG-GCG-CAA-ACA-GTG-GCA-TCA-CCA-ATG-GTG-TCT-ACA-AAT-T-3′ | OL598576 | C. hominis | Ia |
| 45 | 5′-GCT-TCC-GCT-GTA-TTC-TCA-GCC-CCA-GCC-GTT-CCA-CTC-AGA-GGC-ACC-TTG-AAA-GAT-GTT-TCT-GTT-GAG-AGC-TCA-TCA-TCA-TCA-TCA-TCA-TCA-TCG-TCA-TCA-TCA-TCG-TCA-TCA-TCG-TCA-ACA-ACA-ACC-CCC-GCA-CCA-GCT-CCA-AAG-AAG-GCA-AGA-GAA-GCA-GAT-GGC-GGA-GAA-GAA-AAG-AAC-AAT-GAA-GAA-AGC-CAA-ACT-CCC-GCT-AGT-CCT-GGA-AGT-GGT-GGG-GTG-AGT-GAA-GGA-CAA-GAT-ACT-CAA-GGT-GGC-TCC-AAA-GGA-GAC-GCT-GAG-GAA-GGC-ACT-GAA-GAC-AAT-GAA-CAA-GCC-GAT-GAG-AGT-GCT-ACC-CAA-CCT-TCT-ACC-CCA-GGT-CAA-GGC-TCC-GTT-AAA-ACC-GAA-TCC-ACA-GAA-ACT-ACT-CCA-AAG-GAG-AAG-TGC-GGT-ACT-TCA-TTT-GTT-ATG-TGG-TTC-GGA-CAG-GGT-GTT-CCA-GTC-GCA-ACT-TTG-AAG-TGC-GGT-GAC-TAT-ACT-ATG-GTC-TAT-GCA-CCA-GAA-AAG-GAC-AAA-ACA-GAT-CCC-GCA-CCA-AGA-TAT-ATC-TCT-GGT-GAA-GTT-ACA-ACC-GTA-ACC-TTT-GAT-AAA-CAA-GAG-AGT-ACA-GTT-ACA-ATC-AAG-GTT-AAT-AAT-GTA-GAG-TTC-GGC-ACT-CTC-TCT-ACT-AGC-TCA-AGT-AAA-CCA-ACT-GAA-AAT-AAA-GGT-GAG-TCT-AGC-GAT-CAG-GTT-GGG-TCA-AGA-TCA-AGA-AGA-TCA-CTC-ACA-GAG-GAA-ACT-AGT-GAA-ACT-GCA-ACC-GTC-GAT-TTG-TTT-GCC-TTT-ACC-CTT-GAT-GGT-GGT-AAA-AGA-ATT-GAA-GTG-GCT-GTA-CCA-AGT-GAC-GAA-GAT-GTA-TCC-AAG-AGA-AAC-AAG-TAC-AGT-TTG-GTT-GCA-AAC-GAT-AAG-ACT-TTC-TAT-ACC-GGC-GCA-AAT-AGC-GGT-AAT-ACC-GAC-GGT-ATC-TAC-AGG-TTG-AAT-GAT-AAT-GGA-GAC-TTG-GTG-GAC-AAG-AAC-AAC-AAC-GTT-CTT-TTG-AAG-GAT-GYT-GYT-TYC-YTC-TGC-AAT-AGG-ATT-CAG-ATA-CAT-CGG-TTC-CTT-CCA-G-3′ | GU214349 | C. hominis | Ib |
| 46 | 5′-CGC-TGT-ATT-CTC-AGC-CCC-AGC-CGT-CCC-ACT-CAG-AGG-CAC-CTT-GAA-GGA-TGT-TTC-TGT-TGA-GGG-CTC-ATC-ATC-ATC-TTC-ATC-ATC-GTC-TTC-ATC-TTC-ATC-ATC-ATC-ATC-GTC-GTC-AAC-AAC-CCC-AGC-ACC-AGC-TTC-AAA-GAA-GGT-AAG-AGA-AGC-AGA-AGG-CAG-TGA-AGA-AAA-GGA-CAG-CGA-AGA-AAA-GGA-CAG-TGA-AGA-AAA-GGG-CAG-TGA-AGA-AGG-TAG-CCA-AAC-TCC-CGC-TAG-TCC-TGG-AGG-TGG-AGG-GGT-GAG-TGA-AGG-AGA-TAC-TCA-AGG-TGA-CTC-TAA-AGG-AGA-CGG-AGT-TAG-TTC-AGA-TGA-GAA-CCA-AAG-TCA-AGG-TGG-GGA-CGC-TAC-TCC-CGG-ATC-TAG-CAC-CCA-AAC-TCA-AGC-TAC-TGA-AAA-AGA-ACC-CGG-ATC-TTC-AGA-AGC-TAC-TCC-AAA-GGA-AGA-GTG-CGG-TAC-TTC-ATT-TGT-AAT-GTG-GTT-CGG-ACA-GGG-TGT-TCC-AGT-TGT-AAC-TTT-GAA-GTG-TGG-TGG-TTA-TAC-TAT-GGT-CTA-TGC-ACC-AGA-AAA-TGG-CAA-AAC-AGA-TCC-CGC-ACC-AAG-ATA-TAT-CTC-TGG-TAA-AGT-TTC-AAC-CGT-AGA-CTT-TGA-AAA-ACA-AGA-TAG-TAC-AGT-TAA-AAT-CAA-GGT-TAA-TGG-TGT-GGA-GTT-CAG-CAC-TCT-CTC-TAC-TAG-CTC-AAG-TAA-TCC-AAC-TGA-AAA-TAG-CGG-ATC-TGA-GAG-CCA-GGC-TCA-ATC-AAG-ATC-AAG-AAG-ATC-ACT-CGC-AGA-GGA-TGG-TAC-TGA-GAC-TGC-TGC-AAC-CGT-CGA-TTT-GAT-TGC-CTT-CAC-CCT-TCA-AGG-TGG-TAA-AAG-AAT-CGA-AGT-CGC-TGT-GCC-AAG-TGA-CGA-AGA-TGT-ATC-CAA-GAG-AAA-CAA-GTA-CAG-TTT-GGT-TGC-AGG-CGA-TAA-GAC-TTT-CTA-TAC-CGG-CGC-AAA-TAG-CGG-TAA-TAC-CGA-CGG-TAT-CTA-CAG-GTT-GAA-TGA-TGA-TGG-AGA-CTT-GGT-GGA-CAA-GAA-CAA-CAA-CGT-TCT-TTT-GAA-GGA-TGC-TGG-TTY-CCT-CTG-CAA-3′ | GU214354 | C. hominis | Ie |
| 47 | 5′-TTT-TTT-CTC-CGC-TGT-ATT-CTC-AGC-CAC-AGC-CGT-TCC-ACT-CAG-AGG-CAC-TTT-AAA-GGA-TGT-TTC-TGT-TGA-GAG-CTC-ATC-GTC-ATC-ATC-GTC-ATC-GTC-AAC-AAC-AAC-CCC-CGC-ACC-AGC-TCC-AAA-GAA-GGT-AAG-AGA-AAG-CGA-AGA-AGG-GAA-GAA-CAG-TGA-AGA-TAG-TCA-GAC-TCC-CGC-TAG-TCC-TGG-AAG-TGA-TTC-TCA-GGA-TAG-CTC-TAA-AGG-AGA-CGA-AGC-TGT-AGA-TGG-ATC-CGC-TTC-CGG-ATC-TAG-TAC-CCC-AAC-TCA-AGC-TGC-TGA-AAA-GGA-GCC-CGA-AAC-TCC-AGA-ATC-TAC-TCC-AAA-GGA-AGA-ATG-TGG-TAC-TTC-ATT-TAT-AAT-GTG-GTT-CGG-AGA-AGG-TAC-TCC-AGC-CAC-AAC-TTT-GAA-GTG-CGG-TGG-CTA-CAC-TAT-CGT-CTA-TGC-ACC-AGA-AAA-GGA-TAA-TAA-AGA-ACC-CGC-ACC-AAG-ATA-CAT-CTC-TGG-TGA-TGT-TAA-GGC-TGT-AAC-CTT-TGA-AAA-GGG-AGA-AGA-TAA-TAC-AGT-TAA-AAT-CAA-GGT-TGA-TGG-TAA-GGA-GTT-CAG-TAC-TCT-CTC-TTC-TAG-CTC-AAG-CAG-TCC-AAC-TGA-AAA-TAA-CGG-ATC-TAC-GGG-CCA-GGT-TGC-ATC-AAG-ATC-AAG-AAG-ATC-GCT-CTC-AGA-GGA-AAA-TAG-TGA-AAC-TGC-TGC-AAC-CGT-CGA-TTT-GTT-TGC-CTT-CAC-CCT-TGA-TGG-TGG-CCG-AAG-AAT-TGA-AGT-TGC-TGT-ACC-CAG-CGT-CGA-AGA-TGC-AAC-CAA-AAG-AGA-CAA-GTA-CAG-TTT-GGT-TGC-AAA-CGG-TAA-GCC-TTT-CTA-TAC-CGG-CGC-AAA-CAG-CGG-CAC-TAC-CAA-TGG-TGT-CTA-CAG-GTT-GAA-TGA-GAA-CGG-AGA-CTT-GGT-TGA-CAA-GGA-CAA-CAC-AGT-TCT-CTT-GAA-GGA-TGC-TGG-TTT-CCT-CTG-CAT-TTG-GAC-TCA-GAT-ACA-TCG-TT-3′ | GU214366 | C. parvum | IIc |
References
- Bouzid, M.; Hunter, P.R.; Chalmers, R.M.; Tyler, K.M. Cryptosporidium pathogenicity and virulence. Clin. Microbiol. Rev. 2013, 26, 115–134. [Google Scholar] [CrossRef]
- Mmbaga, B.T.; Houpt, E.R. Cryptosporidium and Giardia Infections in Children: A Review. Pediatr. Clin. N. Am. 2017, 64, 837–850. [Google Scholar] [CrossRef] [PubMed]
- Luka, G.; Samiei, E.; Tasnim, N.; Dalili, A.; Najjaran, H.; Hoorfar, M. Comprehensive review of conventional and state-of-the-art detection methods of Cryptosporidium. J. Hazard. Mater. 2022, 421, 126714. [Google Scholar] [CrossRef]
- Ali, M.; Xu, C.; Wang, J.; Kulyar, M.F.; Li, K. Emerging therapeutic avenues against Cryptosporidium: A comprehensive review. Vet. Parasitol. 2024, 331, 110279. [Google Scholar] [CrossRef]
- To, B.; Chai, N.; Fitzpatrick, C.; Richardson, D. Factors associated with Cryptosporidium in men who have sex with men: A systematic review. Int. J. STD AIDS 2024, 35, 668–674. [Google Scholar] [CrossRef]
- Tian, L.G.; Zhou, X.N. The neglected intestinal parasite co-infection with AIDS. Zhongguo Ji Sheng Chong Xue Yu Ji Sheng Chong Bing Za Zhi 2008, 26, 376–381. [Google Scholar]
- Kalavani, S.; Matin, S.; Rahmanian, V.; Meshkin, A.; Bahadori Mazidi, B.; Taghipour, A.; Abdoli, A. Prevalence of Giardia duodenalis among African children: A systematic review and meta-analysis. Parasite Epidemiol. Control 2024, 26, e00365. [Google Scholar] [CrossRef]
- Dib, H.H.; Lu, S.Q.; Wen, S.F. Prevalence of Giardia lamblia with or without diarrhea in South East, South East Asia and the Far East. Parasitol. Res. 2008, 103, 239–251. [Google Scholar] [CrossRef]
- Bhaijee, F.; Subramony, C.; Tang, S.J.; Pepper, D.J. Human immunodeficiency virus-associated gastrointestinal disease: Common endoscopic biopsy diagnoses. Patholog. Res. Int. 2011, 2011, 247923. [Google Scholar] [CrossRef]
- Chui, D.W.; Owen, R.L. AIDS and the gut. J. Gastroenterol. Hepatol. 1994, 9, 291–303. [Google Scholar] [CrossRef]
- Mahdavi, F.; Shams, M.; Sadrebazzaz, A.; Shamsi, L.; Omidian, M.; Asghari, A.; Hassanipour, S.; Salemi, A.M. Global prevalence and associated risk factors of diarrheagenic Giardia duodenalis in HIV/AIDS patients: A systematic review and meta-analysis. Microb. Pathog. 2021, 160, 105202. [Google Scholar] [CrossRef]
- Estambale, B.B.; Knight, R. Protozoan infections and HIV-1 infection: A review. East Afr. Med. J. 1992, 69, 373–377. [Google Scholar] [PubMed]
- Debnath, A.; Reed, S.L.; Morris, S.R. Predictors of Failure from Primary Therapy for Giardiasis in San Diego: A Single Institution Retrospective Review. Pathogens 2019, 8, 165. [Google Scholar] [CrossRef] [PubMed]
- Stark, D.; Barratt, J.L.; van Hal, S.; Marriott, D.; Harkness, J.; Ellis, J.T. Clinical significance of enteric protozoa in the immunosuppressed human population. Clin. Microbiol. Rev. 2009, 22, 634–650. [Google Scholar] [CrossRef]
- Janoff, E.N.; Smith, P.D. Perspectives on gastrointestinal infections in AIDS. Gastroenterol. Clin. N. Am. 1988, 17, 451–463. [Google Scholar] [CrossRef]
- Heyworth, M.F. Giardia duodenalis genetic assemblages and hosts. Parasite 2016, 23, 13. [Google Scholar] [CrossRef]
- Lasek-Nesselquist, E.; Welch, D.M.; Sogin, M.L. The identification of a new Giardia duodenalis assemblage in marine vertebrates and a preliminary analysis of G. duodenalis population biology in marine systems. Int. J. Parasitol. 2010, 40, 1063–1074. [Google Scholar] [CrossRef]
- Feng, Y.; Xiao, L. Zoonotic potential and molecular epidemiology of Giardia species and giardiasis. Clin. Microbiol. Rev. 2011, 24, 110–140. [Google Scholar] [CrossRef]
- Argüello-García, R.; Ortega-Pierres, M.G. Giardia duodenalis Virulence—“To Be, or Not To Be”. Curr. Trop. Med. Rep. 2021, 8, 246–256. [Google Scholar] [CrossRef]
- Ryan, U.M.; Feng, Y.; Fayer, R.; Xiao, L. Taxonomy and molecular epidemiology of Cryptosporidium and Giardia—A 50 year perspective (1971–2021). Int. J. Parasitol. 2021, 51, 1099–1119. [Google Scholar] [CrossRef]
- Alves, M.; Xiao, L.; Sulaiman, I.; Lal, A.A.; Matos, O.; Antunes, F. Subgenotype analysis of Cryptosporidium isolates from humans, cattle, and zoo ruminants in Portugal. J. Clin. Microbiol. 2003, 41, 2744–2747. [Google Scholar] [CrossRef] [PubMed]
- Peng, M.M.; Matos, O.; Gatei, W.; Das, P.; Stantic-Pavlinic, M.; Bern, C.; Sulaiman, I.M.; Glaberman, S.; Lal, A.A.; Xiao, L. A comparison of Cryptosporidium subgenotypes from several geographic regions. J. Eukaryot. Microbiol. 2001, 48, 28S–31S. [Google Scholar] [CrossRef] [PubMed]
- Eibach, D.; Krumkamp, R.; Al-Emran, H.M.; Sarpong, N.; Hagen, R.M.; Adu-Sarkodie, Y.; Tannich, E.; May, J. Molecular characterization of Cryptosporidium spp. among children in rural Ghana. PLoS Negl. Trop. Dis. 2015, 9, e0003551. [Google Scholar] [CrossRef]
- Frickmann, H.; Hoffmann, T.; Köller, T.; Hahn, A.; Podbielski, A.; Landt, O.; Loderstädt, U.; Tannich, E. Comparison of five commercial real-time PCRs for in-vitro diagnosis of Entamoeba histolytica, Giardia duodenalis, Cryptosporidium spp., Cyclospora cayetanensis, and Dientamoeba fragilis in human stool samples. Travel Med. Infect. Dis. 2021, 41, 102042. [Google Scholar] [CrossRef]
- Weinreich, F.; Hahn, A.; Eberhardt, K.A.; Kann, S.; Feldt, T.; Sarfo, F.S.; Di Cristanziano, V.; Frickmann, H.; Loderstädt, U. Comparative Evaluation of Real-Time Screening PCR Assays for Giardia duodenalis and of Assays Discriminating the Assemblages A and B. Microorganisms 2022, 10, 1310. [Google Scholar] [CrossRef]
- Sarfo, F.S.; Frickmann, H.; Dompreh, A.; Osei Asibey, S.; Boateng, R.; Weinreich, F.; Osei Kuffour, E.; Norman, B.R.; Di Cristanziano, V.; Feldt, T.; et al. High Clinical Burden of Cryptosporidium spp. in Adult Patients with Acquired Immunodeficiency in Ghana. Microorganisms 2024, 12, 2151. [Google Scholar] [CrossRef]
- Sarfo, F.S.; Eberhardt, K.A.; Dompreh, A.; Kuffour, E.O.; Soltau, M.; Schachscheider, M.; Drexler, J.F.; Eis-Hübinger, A.M.; Häussinger, D.; Oteng-Seifah, E.E.; et al. Helicobacter pylori Infection Is Associated with Higher CD4 T Cell Counts and Lower HIV-1 Viral Loads in ART-Naïve HIV-Positive Patients in Ghana. PLoS ONE 2015, 10, e0143388. [Google Scholar] [CrossRef]
- Eberhardt, K.A.; Sarfo, F.S.; Dompreh, A.; Kuffour, E.O.; Geldmacher, C.; Soltau, M.; Schachscheider, M.; Drexler, J.F.; Eis-Hübinger, A.M.; Häussinger, D.; et al. Helicobacter pylori Coinfection Is Associated with Decreased Markers of Immune Activation in ART-Naive HIV-Positive and in HIV-Negative Individuals in Ghana. Clin. Infect. Dis. 2015, 61, 1615–1623. [Google Scholar] [CrossRef]
- Weinreich, F.; Hahn, A.; Eberhardt, K.A.; Feldt, T.; Sarfo, F.S.; Di Cristanziano, V.; Frickmann, H.; Loderstädt, U. Comparison of Three Real-Time PCR Assays Targeting the SSU rRNA Gene, the COWP Gene and the DnaJ-Like Protein Gene for the Diagnosis of Cryptosporidium spp. in Stool Samples. Pathogens 2021, 10, 1131. [Google Scholar] [CrossRef]
- Sayers, E.W.; Cavanaugh, M.; Clark, K.; Pruitt, K.D.; Sherry, S.T.; Yankie, L.; Karsch-Mizrachi, I. GenBank 2024 Update. Nucleic Acids Res. 2024, 52, D134–D137. [Google Scholar] [CrossRef]
- Guy, R.A.; Payment, P.; Krull, U.J.; Horgen, P.A. Real-time PCR for quantification of Giardia and Cryptosporidium in environmental water samples and sewage. Appl. Environ. Microbiol. 2003, 69, 5178–5185. [Google Scholar] [CrossRef]
- Verweij, J.J.; Blangé, R.A.; Templeton, K.; Schinkel, J.; Brienen, E.A.; van Rooyen, M.A.; van Lieshout, L.; Polderman, A.M. Simultaneous detection of Entamoeba histolytica, Giardia lamblia, and Cryptosporidium parvum in fecal samples by using multiplex real-time PCR. J. Clin. Microbiol. 2004, 42, 1220–1223. [Google Scholar] [CrossRef]
- Shin, J.H.; Lee, S.E.; Kim, T.S.; Ma, D.W.; Cho, S.H.; Chai, J.Y.; Shin, E.H. Development of Molecular Diagnosis Using Multiplex Real-Time PCR and T4 Phage Internal Control to Simultaneously Detect Cryptosporidium parvum, Giardia lamblia, and Cyclospora cayetanensis from Human Stool Samples. Korean J. Parasitol. 2018, 56, 419–427. [Google Scholar] [CrossRef] [PubMed]
- Alonso, J.L.; Amorós, I.; Cuesta, G. LNA probes in a real-time TaqMan PCR assay for genotyping of Giardia duodenalis in wastewaters. J. Appl. Microbiol. 2010, 108, 1594–1601. [Google Scholar] [CrossRef] [PubMed]
- Elwin, K.; Fairclough, H.V.; Hadfield, S.J.; Chalmers, R.M. Giardia duodenalis typing from stools: A comparison of three approaches to extracting DNA, and validation of a probe-based real-time PCR typing assay. J. Med. Microbiol. 2014, 63, 38–44. [Google Scholar] [CrossRef]
- Niesters, H.G. Quantitation of viral load using real-time amplification techniques. Methods 2001, 25, 419–429. [Google Scholar] [CrossRef]
- Odeniran, P.O.; Ademola, I.O. Epidemiology of Cryptosporidium infection in different hosts in Nigeria: A meta-analysis. Parasitol. Int. 2019, 71, 194–206. [Google Scholar] [CrossRef]
- Del Chierico, F.; Onori, M.; Di Bella, S.; Bordi, E.; Petrosillo, N.; Menichella, D.; Cacciò, S.M.; Callea, F.; Putignani, L. Cases of cryptosporidiosis co-infections in AIDS patients: A correlation between clinical presentation and GP60 subgenotype lineages from aged formalin-fixed stool samples. Ann. Trop. Med. Parasitol. 2011, 105, 339–349. [Google Scholar] [CrossRef]
- Ukwah, B.N.; Ezeonu, I.M.; Ezeonu, C.T.; Roellig, D.; Xiao, L. Cryptosporidium species and subtypes in diarrheal children and HIV-infected persons in Ebonyi and Nsukka, Nigeria. J. Infect. Dev. Ctries. 2017, 11, 173–179. [Google Scholar] [CrossRef]
- Blanco, M.A.; Montoya, A.; Iborra, A.; Fuentes, I. Identification of Cryptosporidium subtype isolates from HIV-seropositive patients in Equatorial Guinea. Trans. R. Soc. Trop. Med. Hyg. 2014, 108, 594–596. [Google Scholar] [CrossRef]
- Adamu, H.; Petros, B.; Zhang, G.; Kassa, H.; Amer, S.; Ye, J.; Feng, Y.; Xiao, L. Distribution and clinical manifestations of Cryptosporidium species and subtypes in HIV/AIDS patients in Ethiopia. PLoS Negl. Trop. Dis. 2014, 8, e2831. [Google Scholar] [CrossRef] [PubMed]
- Dhal, A.K.; Panda, C.; Yun, S.I.; Mahapatra, R.K. An update on Cryptosporidium biology and therapeutic avenues. J. Parasit. Dis. 2022, 46, 923–939. [Google Scholar] [CrossRef]
- Lu, Y.; Zhang, X.; Guan, Z.; Ji, R.; Peng, F.; Zhao, C.; Gao, W.; Gao, F. Molecular pathogenesis of Cryptosporidium and advancements in therapeutic interventions. Parasite 2025, 32, 7. [Google Scholar] [CrossRef]
- Nhambirre, O.; Lobo, M.L.; Cossa-Moiane, I.; Bauhofer, A.; Deus, N.; Matos, O. Risk Factors and Circulating Subtypes of Cryptosporidium spp. and Giardia duodenalis in Hospitalized Children in Mozambique. Microorganisms 2025, 13, 196. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).