Development of a Novel NGS Methodology for Ultrasensitive Circulating Tumor DNA Detection as a Tool for Early-Stage Breast Cancer Diagnosis
Abstract
1. Introduction
2. Results
2.1. The Genetic Landscape in Tumors and ctDNA of Localized BC Patients
2.2. Panel Utility for BC Detection Using a Non-Tumor Informed Pipeline and Association with Clinicopathological Variables
3. Discussion
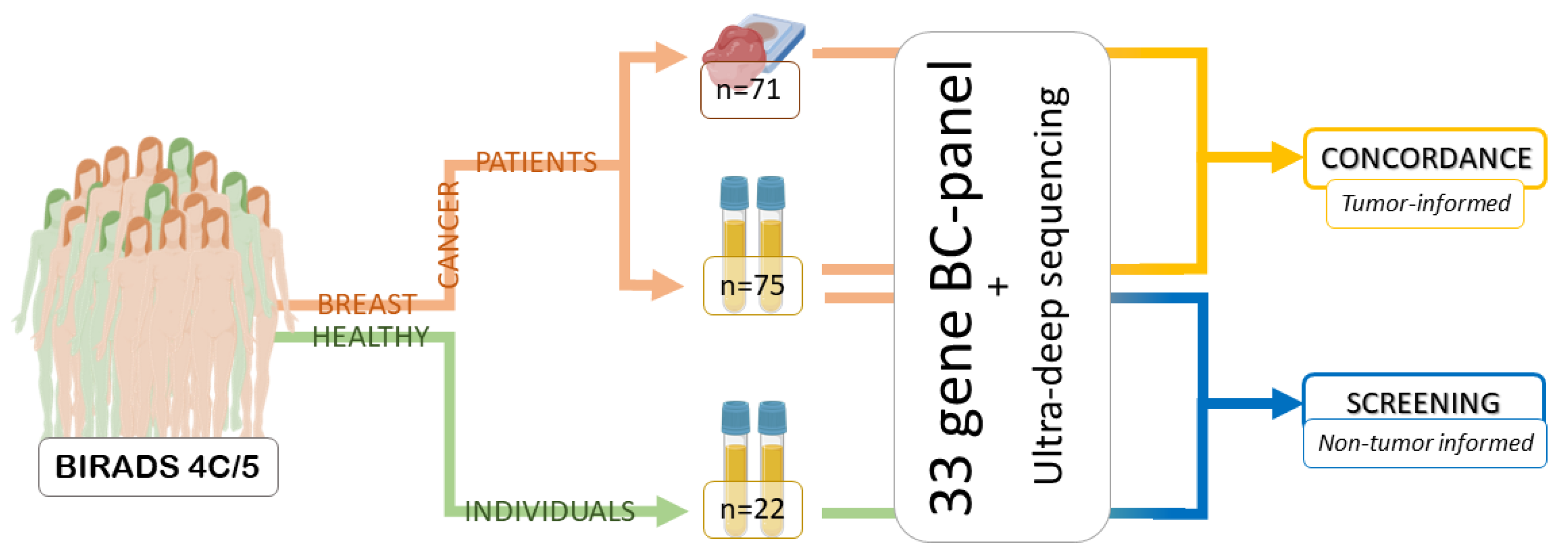
4. Materials and Methods
4.1. Patients and Women with Negative Biopsies
4.2. Blood Sample Processing
4.3. DNA Extraction and Quantification from Plasma and Solid Biopsies
4.4. Sequencing BC Panel Design
4.5. Sequencing Library Preparation
4.6. Sequencing Data Processing
4.7. Variant Filtration and Analysis
4.8. Statistical Analyses and Data Visualization
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gerlinger, M.; Rowan, A.J.; Horswell, S.; Math, M.; Larkin, J.; Endesfelder, D.; Gronroos, E.; Martinez, P.; Matthews, N.; Stewart, A.; et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 2012, 366, 883–892. [Google Scholar] [CrossRef] [PubMed]
- Tie, J.; Cohen, J.D.; Lahouel, K.; Lo, S.N.; Wang, Y.; Kosmider, S.; Wong, R.; Shapiro, J.; Lee, M.; Harris, S.; et al. Circulating Tumor DNA Analysis Guiding Adjuvant Therapy in Stage II Colon Cancer. N. Engl. J. Med. 2022, 386, 2261–2272. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Murillas, I.; Chopra, N.; Comino-Mendez, I.; Beaney, M.; Tovey, H.; Cutts, R.J.; Swift, C.; Kriplani, D.; Afentakis, M.; Hrebien, S.; et al. Assessment of Molecular Relapse Detection in Early-Stage Breast Cancer. JAMA Oncol. 2019, 5, 1473–1478. [Google Scholar] [CrossRef] [PubMed]
- Dawson, S.J.; Tsui, D.W.; Murtaza, M.; Biggs, H.; Rueda, O.M.; Chin, S.F.; Dunning, M.J.; Gale, D.; Forshew, T.; Mahler-Araujo, B.; et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N. Engl. J. Med. 2013, 368, 1199–1209. [Google Scholar] [CrossRef]
- Bettegowda, C.; Sausen, M.; Leary, R.J.; Kinde, I.; Wang, Y.; Agrawal, N.; Bartlett, B.R.; Wang, H.; Luber, B.; Alani, R.M.; et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 2014, 6, 224ra24. [Google Scholar] [CrossRef]
- Page, K.; Martinson, L.J.; Hastings, R.K.; Fernandez-Garcia, D.; Gleason, K.L.T.; Gray, M.C.; Rushton, A.J.; Goddard, K.; Guttery, D.S.; Stebbing, J.; et al. Prevalence of ctDNA in early screen-detected breast cancers using highly sensitive and specific dual molecular barcoded personalised mutation assays. Ann. Oncol. 2021, 32, 1057–1060. [Google Scholar] [CrossRef]
- McDonald, B.R.; Contente-Cuomo, T.; Sammut, S.J.; Odenheimer-Bergman, A.; Ernst, B.; Perdigones, N.; Chin, S.F.; Farooq, M.; Mejia, R.; Cronin, P.A.; et al. Personalized circulating tumor DNA analysis to detect residual disease after neoadjuvant therapy in breast cancer. Sci. Transl. Med. 2019, 11, eaax7392. [Google Scholar] [CrossRef]
- Cohen, J.D.; Li, L.; Wang, Y.; Thoburn, C.; Afsari, B.; Danilova, L.; Douville, C.; Javed, A.A.; Wong, F.; Mattox, A.; et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 2018, 359, 926–930. [Google Scholar] [CrossRef]
- Mansukhani, S.; Barber, L.J.; Kleftogiannis, D.; Moorcraft, S.Y.; Davidson, M.; Woolston, A.; Proszek, P.Z.; Griffiths, B.; Fenwick, K.; Herman, B.; et al. Ultra-Sensitive Mutation Detection and Genome-Wide DNA Copy Number Reconstruction by Error-Corrected Circulating Tumor DNA Sequencing. Clin. Chem. 2018, 64, 1626–1635. [Google Scholar] [CrossRef]
- Pereira, B.; Chin, S.F.; Rueda, O.M.; Vollan, H.K.; Provenzano, E.; Bardwell, H.A.; Pugh, M.; Jones, L.; Russell, R.; Sammut, S.J.; et al. The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat. Commun. 2016, 7, 11479. [Google Scholar] [CrossRef]
- Lang, G.T.; Jiang, Y.Z.; Shi, J.X.; Yang, F.; Li, X.G.; Pei, Y.C.; Zhang, C.H.; Ma, D.; Xiao, Y.; Hu, P.C.; et al. Characterization of the genomic landscape and actionable mutations in Chinese breast cancers by clinical sequencing. Nat. Commun. 2020, 11, 5679. [Google Scholar] [CrossRef]
- Kastenhuber, E.R.; Lowe, S.W. Putting p53 in Context. Cell 2017, 170, 1062–1078. [Google Scholar] [CrossRef] [PubMed]
- Dillon, R.L.; White, D.E.; Muller, W.J. The phosphatidyl inositol 3-kinase signaling network: Implications for human breast cancer. Oncogene 2007, 26, 1338–1345. [Google Scholar] [CrossRef] [PubMed]
- Kouros-Mehr, H.; Bechis, S.K.; Slorach, E.M.; Littlepage, L.E.; Egeblad, M.; Ewald, A.J.; Pai, S.Y.; Ho, I.C.; Werb, Z. GATA-3 links tumor differentiation and dissemination in a luminal breast cancer model. Cancer Cell 2008, 13, 141–152. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, B.J.; Cordoba, G.D.; Aranda, A.G.; Alvarez, M.; Vicioso, L.; Perez, C.L.; Hernando, C.; Bermejo, B.; Parreno, A.J.; Lluch, A.; et al. Detection of TP53 and PIK3CA Mutations in Circulating Tumor DNA Using Next-Generation Sequencing in the Screening Process for Early Breast Cancer Diagnosis. J. Clin. Med. 2019, 8, 1183. [Google Scholar] [CrossRef] [PubMed]
- Alba-Bernal, A.; Lavado-Valenzuela, R.; Dominguez-Recio, M.E.; Jimenez-Rodriguez, B.; Queipo-Ortuno, M.I.; Alba, E.; Comino-Mendez, I. Challenges and achievements of liquid biopsy technologies employed in early breast cancer. EBioMedicine 2020, 62, 103100. [Google Scholar] [CrossRef]
- Dang, D.K.; Park, B.H. Circulating tumor DNA: Current challenges for clinical utility. J Clin Invest 2022, 132, (12). [Google Scholar] [CrossRef] [PubMed]
- Garcia-Murillas, I.; Schiavon, G.; Weigelt, B.; Ng, C.; Hrebien, S.; Cutts, R.J.; Cheang, M.; Osin, P.; Nerurkar, A.; Kozarewa, I.; et al. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci. Transl. Med. 2015, 7, 302ra133. [Google Scholar] [CrossRef]
- Olsson, E.; Winter, C.; George, A.; Chen, Y.; Howlin, J.; Tang, M.H.; Dahlgren, M.; Schulz, R.; Grabau, D.; van Westen, D.; et al. Serial monitoring of circulating tumor DNA in patients with primary breast cancer for detection of occult metastatic disease. EMBO Mol. Med. 2015, 7, 1034–1047. [Google Scholar] [CrossRef]
- Klein, E.A.; Richards, D.; Cohn, A.; Tummala, M.; Lapham, R.; Cosgrove, D.; Chung, G.; Clement, J.; Gao, J.; Hunkapiller, N.; et al. Clinical validation of a targeted methylation-based multi-cancer early detection test using an independent validation set. Ann. Oncol. 2021, 32, 1167–1177. [Google Scholar] [CrossRef]
- Zhou, Y.; Xu, Y.; Gong, Y.; Zhang, Y.; Lu, Y.; Wang, C.; Yao, R.; Li, P.; Guan, Y.; Wang, J.; et al. Clinical factors associated with circulating tumor DNA (ctDNA) in primary breast cancer. Mol. Oncol. 2019, 13, 1033–1046. [Google Scholar] [CrossRef] [PubMed]
- Yi, Z.; Ma, F.; Rong, G.; Guan, Y.; Li, C.; Xu, B. Clinical spectrum and prognostic value of TP53 mutations in circulating tumor DNA from breast cancer patients in China. Cancer Commun. 2020, 40, 260–269. [Google Scholar] [CrossRef]
- Liu, B.; Yi, Z.; Guan, Y.; Ouyang, Q.; Li, C.; Guan, X.; Lv, D.; Li, L.; Zhai, J.; Qian, H.; et al. Molecular landscape of TP53 mutations in breast cancer and their utility for predicting the response to HER-targeted therapy in HER2 amplification-positive and HER2 mutation-positive amplification-negative patients. Cancer Med. 2022, 11, 2767–2778. [Google Scholar] [CrossRef] [PubMed]
- Wolff, A.C.; Hammond, M.E.; Schwartz, J.N.; Hagerty, K.L.; Allred, D.C.; Cote, R.J.; Dowsett, M.; Fitzgibbons, P.L.; Hanna, W.M.; Langer, A.; et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J. Clin. Oncol. 2007, 25, 118–145. [Google Scholar] [CrossRef] [PubMed]
- Giuliano, A.E.; Connolly, J.L.; Edge, S.B.; Mittendorf, E.A.; Rugo, H.S.; Solin, L.J.; Weaver, D.L.; Winchester, D.J.; Hortobagyi, G.N. Breast Cancer-Major changes in the American Joint Committee on Cancer eighth edition cancer staging manual. CA Cancer J. Clin. 2017, 67, 290–303. [Google Scholar] [CrossRef]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef]
- Grossman, R.L.; Heath, A.P.; Ferretti, V.; Varmus, H.E.; Lowy, D.R.; Kibbe, W.A.; Staudt, L.M. Toward a Shared Vision for Cancer Genomic Data. N. Engl. J. Med. 2016, 375, 1109–1112. [Google Scholar] [CrossRef]
- Robinson, J.T.; Thorvaldsdottir, H.; Wenger, A.M.; Zehir, A.; Mesirov, J.P. Variant Review with the Integrative Genomics Viewer. Cancer Res. 2017, 77, e31–e34. [Google Scholar] [CrossRef]
- Mayakonda, A.; Lin, D.C.; Assenov, Y.; Plass, C.; Koeffler, H.P. Maftools: Efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018, 28, 1747–1756. [Google Scholar] [CrossRef]

| Clinical Characteristics | n (%) |
|---|---|
| Diagnostic age (years) | |
| 30–50 | 13 (17.6) |
| >50 | 61 (82.4) |
| Tumor type | |
| IDC | 59 (79.7) |
| DCIS | 5 (6.8) |
| ILC | 3 (4.1) |
| PC | 1 (1.4) |
| TC | 3 (4.1) |
| MC | 3 (4.1) |
| Tumor size | |
| <2 cm | 32 (43.2) |
| 2–5 cm | 37 (50.0) |
| >5 cm | 5 (6.8) |
| Tumor grade | |
| I | 15 (20.3) |
| II | 37 (50.0) |
| III | 22 (29.7) |
| Axilar lymph node | |
| Positive | 29 (39.1) |
| Negative | 40 (54.0) |
| Unknown | 5 (6.7) |
| Disease stage | |
| Stage 1A | 19 (25.6) |
| Stage 1B | 3 (4.0) |
| Stage 2A | 28 (37.8) |
| Stage 2B | 10 (13.5) |
| Stage 3A | 5 (6.7) |
| Stage 3C | 4 (5.4) |
| Unknown | 5 (6.7) |
| Estrogen receptor | |
| Positive | 66 (89.2) |
| Negative | 7 (9.5) |
| Unknown | 1 (1.4) |
| Progesterone receptor | |
| Positive | 56 (75.7) |
| Negative | 17 (23) |
| Unknown | 1 (1.4) |
| HER2 status | |
| Positive | 6 (8.1) |
| Negative | 67 (90.5) |
| Unknown | 1 (1.4) |
| BIRADS category | |
| 4/B/C | 40 (54.1) |
| 5C | 33 (44.6) |
| Unknown | 1 (1.4) |
| Clinical relapse | |
| Yes | 8 (10.8) |
| No | 64 (86.5) |
| Unknown | 2 (2.7) |
| Tumor | Plasma | ||||||
|---|---|---|---|---|---|---|---|
| Sample | Gene | Nucleotide Change | Aa Change | VAF (%) | Caller Detected No (N)/Yes (Y) | Manually Detected No (N)/Yes (Y) | VAF (%) |
| 001MS | PIK3CA | c.G3145C | p.G1049R | 14.6 | N | N | - |
| 002MS | CDH1 | c.C2245T | p.R749W | 5.3 | N | N | - |
| TP53 | c.G524A | p.R175H | 54.5 | N | Y | 0.2 | |
| PIK3CA | c.G1252A | p.E418K | 35.0 | N | N | - | |
| PIK3CA | c.A3140T | p.H1047L | 34.4 | N | N | - | |
| 007MS | PIK3CA | c.A3140G | p.H1047R | 31.2 | Y | N | 0.4 |
| 009MS | TP53 | c.C637T | p.R213X | 45.2 | Y | N | 0.8 |
| 010MS | PIK3CA | c.A3140G | p.H1047R | 15.0 | N | N | - |
| 014MS | PIK3CA | c.A3140G | p.H1047R | 25.9 | N | N | - |
| 015MS | GATA3 | c.922-3_922-2delCA | p.X308_splice | 22.4 | N | Y | 0.08 |
| TP53 | c.A377G | p.Y126C | 58.2 | N | Y | 0.24 | |
| 016MS | TP53 | c.G743T | p.R248L | 37.9 | Y | N | 4 |
| SMAD4 | c.C725G | p.S242X | 23.7 | Y | N | 3.2 | |
| PIK3CA | c.A3140T | p.H1047L | 35.1 | Y | N | 4.6 | |
| 017MS | PIK3CA | c.A3140G | p.H1047R | 27.3 | N | N | - |
| 021MS | TP53 | c.376-2A>G | p.X126_splice | 33.4 | Y | N | 1.8 |
| 022MS | KDM6A | c.C1747T | p.Q583X | 12.5 | N | N | - |
| 023MS | TP53 | c.G524A | p.R175H | 63.0 | N | N | - |
| PIK3CA | c.G1633A | p.E545K | 39.3 | N | N | - | |
| 030MS | GATA3 | c.922-3_922-2delCA | p.X308_splice | 38.7 | N | N | - |
| PIK3CA | c.G1633A | p.E545K | 77.0 | N | N | - | |
| 031MS | PIK3CA | c.G1633A | p.E545K | 15.7 | N | N | - |
| 032MS | PIK3CA | c.G353A | p.G118D | 6.9 | N | N | - |
| PIK3CA | c.G2908A | p.E970K | 14.7 | N | N | - | |
| PIK3CA | c.A3140G | p.H1047R | 11.4 | N | N | - | |
| PIK3CA | c.A3140T | p.H1047L | 3.0 | N | N | - | |
| 033MS | TP53 | c.A503T | p.H168L | 37.6 | Y | N | 0.37 |
| 035MS | PIK3CA | c.G1633A | p.E545K | 34.8 | N | N | - |
| 036MS | NF1 | c.3478delG | p.G1160Vfs*6 | 5.5 | N | N | - |
| PIK3CA | c.A3140T | p.H1047L | 31.5 | N | N | - | |
| 039MS | AKT1 | c.G49A | p.E17K | 33.7 | N | N | - |
| NCOR1 | c.G6751T | p.G2251C | 10.3 | N | N | - | |
| 040MS | PIK3CA | c.G1093A | p.E365K | 21.7 | N | N | - |
| PIK3CA | c.G1624A | p.E542K | 40.0 | N | N | - | |
| 044MS | KRAS | c.G35C | p.G12A | 29.3 | Y | N | 0.97 |
| TP53 | c.G587C | p.R196P | 51.2 | Y | N | 1.2 | |
| 045MS | AKT1 | c.G49A | p.E17K | 6.9 | N | N | - |
| 047MS | PIK3CA | c.A3140G | p.H1047R | 7.2 | N | N | - |
| 052MS | PIK3CA | c.A1637G | p.Q546R | 19.8 | N | N | - |
| PIK3CA | c.A3073G | p.T1025A | 21.6 | N | N | - | |
| 056MS | PIK3CA | c.G1624A | p.E542K | 17.9 | N | N | - |
| 057MS | PIK3CA | c.G1633A | p.E545K | 32.1 | N | N | - |
| 060MS | MAP3K1 | c.813_814del | p.R273Sfs*27 | 11.6 | N | N | - |
| 064MS | PIK3CA | c.T1035A | p.N345K | 34.9 | N | N | - |
| 065MS | GATA3 | c.922-3_922-2delCA | p.X308_splice | 23.3 | N | N | - |
| PIK3CA | c.A3140G | p.H1047R | 25.9 | N | N | - | |
| 066MS | ERBB2 | c.G2305T | p.D769Y | 23.4 | N | N | - |
| PIK3CA | c.G1624A | p.E542K | 27.5 | N | N | - | |
| 067MS | TP53 | c.A842C | p.D281A | 51.8 | Y | N | 0.31 |
| PIK3CA | c.G3145C | p.G1049R | 86.2 | Y | N | 0.32 | |
| 079MS | TP53 | c.C742T | p.R248W | 9.1 | N | Y | 0.05 |
| PIK3CA | c.A1637G | p.Q546R | 11.9 | N | N | - | |
| 080MS | PIK3CA | c.G1633A | p.E545K | 26.4 | N | N | - |
| 081MS | PTEN | c.T406C | p.C136R | 54.0 | Y | N | 3.3 |
| TP53 | c.G743A | p.R248Q | 52.9 | Y | N | 1.5 | |
| 093MS | PIK3CA | c.A1634G | p.E545G | 29.1 | N | N | - |
| 095MS | PIK3CA | c.A3140T | p.H1047L | 52.0 | N | N | - |
| 099MS | PIK3CA | c.A3140T | p.H1047L | 28.6 | N | N | - |
| 101MS | SF3B1 | c.A2098G | p.K700E | 20.9 | N | N | - |
| 104MS | TP53 | c.A715G | p.N239D | 22.2 | N | N | - |
| 107MS | GATA3 | c.922-3_922-2delCA | p.X308_splice | 40.1 | N | Y | 0.03 |
| Sample | Gene | Nucleotide Change | Aa Change | VAF (%) | COSMIC + TCGA BC + TCGAall No (N)/Yes (Y) | Tumor Biopsy Sequenced No (N)/Yes (Y) | Any Mutation in Tumor No (N)/Yes (Y) |
|---|---|---|---|---|---|---|---|
| 011MS | MAP3K1 | c.C1292A | p.S431* | 0.238 | Y | Y | N |
| 030MS | SF3B1 | c.C1898T | p.A633V | 0.389 | Y | Y | Y |
| 049MS | NCOR1 | c.3022C>T | p.Q1008* | 0.361 | Y | Y | N |
| 050MS | SF3B1 | c.2098A>G | p.K700E | 1.4 | Y | Y | N |
| TP53 | c.733G>A | p.G245S | 0.321 | Y | Y | N | |
| 053MS | HRAS | c.34G>T | p.G12C | 0.894 | Y | Y | N |
| 056MS | USP9X | c.1795C>T | p.R599C | 0.481 | Y | Y | Y |
| 062MS | BAP1 | c.709C>T | p.R237C | 0.405 | Y | Y | N |
| 080MS | PIK3R1 | c.1669C>T | p.R557* | 0.318 | Y | Y | Y |
| 081MS | TP53 | c.637C>T | p.R213* | 0.643 | Y | Y | Y |
| 092MS | ARID1A | c.2879-1G>A | p.X960_splice | 0.329 | Y | Y | - |
| 094MS | TP53 | c.528C>A | p.C176* | 0.327 | Y | Y | N |
| 099MS | BRCA2 | c.1786G>A | p.D596N | 0.450 | Y | Y | Y |
| 101MS | USP9X | c.1795C>T | p.R599C | 0.209 | Y | Y | Y |
| CDH1 | c.220C>T | p.R74* | 0.279 | Y | Y | Y | |
| 105MS | GATA3 | c.914G>A | p.R305Q | 0.277 | Y | Y | N |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiménez-Rodríguez, B.; Alba-Bernal, A.; López-López, E.; Quirós-Ortega, M.E.; Carbajosa, G.; Garrido-Aranda, A.; Álvarez, M.; Godoy-Ortiz, A.; Queipo-Ortuño, M.I.; Vicioso, L.; et al. Development of a Novel NGS Methodology for Ultrasensitive Circulating Tumor DNA Detection as a Tool for Early-Stage Breast Cancer Diagnosis. Int. J. Mol. Sci. 2023, 24, 146. https://doi.org/10.3390/ijms24010146
Jiménez-Rodríguez B, Alba-Bernal A, López-López E, Quirós-Ortega ME, Carbajosa G, Garrido-Aranda A, Álvarez M, Godoy-Ortiz A, Queipo-Ortuño MI, Vicioso L, et al. Development of a Novel NGS Methodology for Ultrasensitive Circulating Tumor DNA Detection as a Tool for Early-Stage Breast Cancer Diagnosis. International Journal of Molecular Sciences. 2023; 24(1):146. https://doi.org/10.3390/ijms24010146
Chicago/Turabian StyleJiménez-Rodríguez, Begoña, Alfonso Alba-Bernal, Esperanza López-López, María Elena Quirós-Ortega, Guillermo Carbajosa, Alicia Garrido-Aranda, Martina Álvarez, Ana Godoy-Ortiz, María Isabel Queipo-Ortuño, Luis Vicioso, and et al. 2023. "Development of a Novel NGS Methodology for Ultrasensitive Circulating Tumor DNA Detection as a Tool for Early-Stage Breast Cancer Diagnosis" International Journal of Molecular Sciences 24, no. 1: 146. https://doi.org/10.3390/ijms24010146
APA StyleJiménez-Rodríguez, B., Alba-Bernal, A., López-López, E., Quirós-Ortega, M. E., Carbajosa, G., Garrido-Aranda, A., Álvarez, M., Godoy-Ortiz, A., Queipo-Ortuño, M. I., Vicioso, L., Díaz-Córdoba, G., Roldán-Díaz, M. D., Velasco-Suelto, J., Hernando, C., Bermejo, B., Julve-Parreño, A., Lluch, A., Pascual, J., Comino-Méndez, I., & Alba, E. (2023). Development of a Novel NGS Methodology for Ultrasensitive Circulating Tumor DNA Detection as a Tool for Early-Stage Breast Cancer Diagnosis. International Journal of Molecular Sciences, 24(1), 146. https://doi.org/10.3390/ijms24010146






