Abstract
Pneumothorax is a potentially life-threatening condition defined as the collapse of the lung due to air leakage into the chest cavity. Delays in the diagnosis of pneumothorax can lead to severe complications and even mortality. A significant challenge in pneumothorax diagnosis is the shortage of radiologists, resulting in the absence of written reports in plain X-rays and, consequently, impacting patient care. In this paper, we propose an automatic triage system for pneumothorax detection in X-ray images based on deep learning. We address this problem from the perspective of multi-source domain adaptation where different datasets available on the Internet are used for training and testing. In particular, we use datasets which contain chest X-ray images corresponding to different conditions (including pneumothorax). A convolutional neural network (CNN) with an EfficientNet architecture is trained and optimized to identify radiographic signs of pneumothorax using those public datasets. We present the results using cross-dataset validation, demonstrating the robustness and generalization capabilities of our multi-source solution across different datasets. The experimental results demonstrate the model’s potential to assist clinicians in prioritizing and correctly detecting urgent cases of pneumothorax using different integrated deployment strategies.
1. Introduction
In recent years, advancements in machine learning and deep learning have significantly enhanced the development of different areas in the healthcare domain, providing powerful tools for improving diagnostic accuracy and workflow efficiency. Radiology, in particular, has been one of the medical subspecialties more affected by this recent development [1], with many different use cases regarding medical imaging [2,3,4] such as disease classification (disease vs. no disease), localization (using saliency maps or bounding boxes), segmentation (which can be from both normal and pathologic areas), and image registration (aligning images from different techniques such as computed tomography, CT, and magnetic resonance imaging, MRI), but also more clinically related ones such as worklist prioritization [5], test scheduling and protocoling, image quality assessment [6], structured reporting, monitoring (temporal tracking of diseases) [7], or even multimodality (a combination of images plus clinical EMR data) [8]. Most of the medical devices and algorithms approved by the FDA in 2020, 72%, were related to radiology [9].
Nevertheless, radiology suffers from an inherent lack of resources, due to the shortage of radiologists [10] and growing demand for medical imaging procedures; according to the WHO [11], in 2016, “an estimated 3.6 billion diagnostic medical examinations, such as X-rays, are performed every year”, a number that “continues to grow as more people access medical care”.
Despite the number of recent techniques that has emerged in the daily workflow of radiology departments, such as ultrasound, computed tomography (CT), or magnetic resonance imaging (MRI), conventional radiology (X-ray) continues to be the most frequently performed imaging procedure [12], up to 75% in a radiology department [13], due to their lower cost, higher availability and accessibility, and great usefulness in the diagnosis of a broad range of conditions Figure 1.
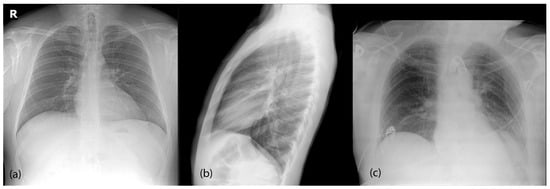
Figure 1.
Most common radiological projections for chest X-rays. (a) Posteroanterior (PA) is the standard frontal projection. (b) Lateral (L) projection is generally taken in conjunction with the PA view. Both are taken in standing position and full inspiration. (c) Anteroposterior (AP) view is generally performed for patients with more severe diseases, as it can be performed using portable devices and in bed, outside of the radiology department.
This has increased the pressure on radiology departments, generally without enough resources to focus on X-rays, which results in delays in report generation, clinical errors due to excessive workload, or even a direct interpretation from the referring physician of X-ray studies, without a proper radiological report [13], something widely acknowledged as leading to legal issues, as the formal reporting of radiological procedures is a professional duty mandated by the current regulations.
There is not a simple solution to these challenges, as they are expected to continue rising, so it is essential that we explore strategies to mitigate them, and the implementation of artificial intelligence in radiology departments can provide the enhanced efficiency required to reduce this impact [14], for instance, by enabling early triage and the preliminary diagnosis of critical cases.
In these situations, triaging using machine-learning tools can be highly effective in reducing diagnostic delays and avoiding a missed diagnosis. Triaging in medicine refers to the process of sorting and prioritizing patients based on the severity of their conditions. The ultimate goal is to optimize resource allocation and save lives by ensuring critical cases receive prompt attention.
Several studies have focused recently on the application of deep learning to chest radiographs in the emergency department.
Annaruma et al. [15] developed an AI system for the real-time triaging of adult chest radiographs, classifying them in critical, urgent, nonurgent, or normal, with a high negative predictive value (94%) and specificity (95%), and a significant reduction in the delay for critical and urgent image findings.
Hwuang et al. [16] evaluated a commercially available deep-learning algorithm designed to classify four thoracic diseases (malignancy, active tuberculosis, pneumonia, and pneumothorax) and retrospectively compared its performance to that of the radiology residents, showing a lower specificity (69.6 vs. 98.1%) but higher sensitivity (88.7% vs. 65.6%), making it feasible for helping to reveal clinical abnormalities in an emergency scenario.
Khader et al. [17] trained a neural-network-based model to identify several disease patterns (cardiomegaly, pulmonary congestion, pleural effusion, pulmonary opacities, and atelectasis) using images from intensive care unit patients, with the aim of helping non-radiologist physicians to improve their interpretation in their clinical routine.
Yun et al. [18] developed an algorithm for the longitudinal follow-up of patients (with a focus on intensive care units), showing that the algorithm could effectively detect the presence or absence of changes given a pair of radiographs from the same patients, effectively detecting urgent findings and reducing radiologists’ workload.
Kolossvâry et al. [19] trained an algorithm for the initial evaluation of patients with acute chest pain (ACP), in order to predict, using chest radiographs, the need of further cardiovascular or pulmonary tests, and showing that, at a highly sensitive threshold (99%), 14% of the patients could be deferred from further testing.
Pneumothorax is a good example of this specific problem. It is an acute condition where air enters the pleural space (a virtual cavity between the lungs and the thoracic wall—ribs), happening spontaneously (with or without underlying lung disease) or after chest trauma [20] Figure 2. The air that fills the pleural space comprises the lung, causing partial or complete lung collapse and preventing lung expansion during breathing, ultimately preventing the correct gas exchange and leading to shortness of breath. If this situation is maintained over time, it can lead to severe complications, with the ultimate consequence of death.
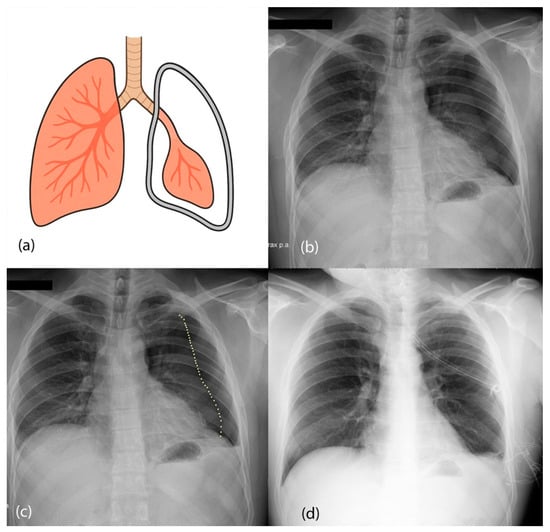
Figure 2.
(a) Diagram of the pneumothorax presentation. Air is leaked to the space between the pleura and the lungs, collapsing the left lung. (b) Chest X-ray image from a posteroanterior view of a patient with left pneumothorax. (c) Dotted line showing the collapsed lung (pneumothorax line). (d) Treated pneumothorax from the same patient as (b) after placement of a chest tube.
A plain chest radiograph is typically used for diagnosis confirmation, although a small pneumothorax may need a CT [21]. The treatment of acute pneumothorax generally needs immediate needle decompression (to manually extract the air from the pleural cavity) or insertion of a chest tube. Small pneumothoraces may be managed conservatively, as they can resolve spontaneously [20].
In this context, delays in the detection of pneumothorax are critical as they can lead to increased morbimortality. Automatic systems that detect pneumothorax can help in early detection and treatment, and, considering the severity of the condition, also help to avoid complications and potentially save lives.
Machine-learning algorithms are considered Class II devices by the U.S. Food and Drug Administration (FDA) [22,23] and Class IIa or IIb under the European Union Medical Device Regulation (EU MDR), depending on their intended use and associated risk [24]. That categorization means that, for clinical implementation, these algorithms must undergo robust evaluation to demonstrate their clinical benefit and safety.
In recent years, the release of several datasets of chest X-rays (Indiana [25], NIH ChestX-ray14 [26], CheXpert [27], MIMIC-CXR [28], PadChest [29], VinDr-CXR [30], etc.), publicly available on the Internet and with thousands of images of different pathologic conditions, has enabled the research and development of machine-learning techniques to improve the detection of several conditions, providing us with powerful Internet-based multi-source data, with pneumothorax being one of the labels present in many of those datasets. But there some challenges with the usage of these datasets for pneumothorax detection [31]:
First, many datasets use NLP techniques for extracting labels out of radiology reports (instead of using expert labelling from radiologists directly), which has led to the inconsistent assignment of labels due to NLP limitations and implicit knowledge in the reports. Some of the datasets, such as Indiana, MIMIC-CXR, PadChest, and, recently, CheXpert Plus [32] (an extension of the original CheXpert dataset), also release the radiology reports along with the images, in an attempt to allow research on the information extraction from the reports.
Second, some of the datasets suffer confounding biases due to treated pneumothoraces with the presence of chest tubes, labelled as pneumothorax-positive and sometimes even without visible pneumothorax in the chest X-ray images, a fact that limits the power of generalization if not mitigated properly [33].
Third, many of the datasets do not include the size or localization of the pneumothorax (small vs. large, and left vs. right lung), and few of them include information that would allow object detection or segmentation, information that would be important for evaluating the performance of difficult cases (small pneumothoraces).
There have been recent attempts to overcome those limitations: ref. [34] showed that expert-labeled models outperformed NLP-labeled models and released enhanced labels for four diseases in a subset of the NIH dataset (pneumothorax amongst them). Ref. [35] released new manually curated labels for all pathologies for a subset of the NIH dataset. Ref. [36] trained a model to annotate chest tubes and applied it to the NIH and PadChest datasets, publicly releasing their annotations [37].
Ref. [38] performed a review for the entire NIH using the expert radiologists’ dataset, regenerating the entire pneumothorax label, and showing that 2231/5302 were false positive labels (no actual pneumothorax), and 2067/106,818 were false negative (labeled as no-pneumothorax but actually pathological), releasing their new labels for public use. Ref. [39] trained Mask R-CNN models for pneumothorax detection and released the segmentation masks they used for the training.
All this evidence shows that, despite pneumothorax being an important condition that needs to be ruled out in the emergency department, the use of machine learning for detection and triaging has some difficulties that need to be addressed in order to achieve clinical implementation.
In this paper, we present a solution based on multi-source data publicly available on the Internet. Our main insight is that, by unifying and harmonizing all available datasets, we can obtain a more powerful set of data that has the potential to improve the results of deep-learning methods. In this sense, we create a multi-source dataset that demonstrates its advantages over individual datasets when training CNN-based models.
Our proposed model has the potential to be used in a general emergency department in any hospital since the model includes datasets from different hospitals and countries. The generalization capabilities of this new model are demonstrated in the resulting experiments in this paper.
Once the CNN model is trained with the multi-source data, it could be used to analyze and automatically detect pneumothorax using chest X-ray images in real time, allowing a rapid triage of emergency patients, as it is compact enough for cloud storage and deployment across multiple departments: it could run on a local server receiving images from X-ray machines, PACS systems, other computers, or mobile devices, and also could be deployed to be used online by hospitals globally, as it contains no personal patient information.
In summary, the main contributions of our paper are as follows:
- -
- We present a new unified and harmonized chest X-ray dataset framework to facilitate multi-source deep learning for pneumothorax detection. We modified and harmonized the structure from individual datasets (including label standardization) to make data unification possible.
- -
- We perform a set of experiments with models trained on multi-source data, trying to address the biases that pneumothorax models commonly suffer (chest tubes as confounder biases, label mismatch, etc.) with the aim of creating a model that is useful for clinical usage.
- -
- We evaluated the performance of the trained models on unseen data from other public datasets in a pseudo-clinical scenario. Our results show that multi-source datasets, in general, lead to models with better performance than single-source datasets.
2. Materials and Methods
This retrospective study was approved by the Ethical Committee of the hospital because of the retrospective nature of the study and the usage of external datasets only.
2.1. Datasets
We performed a review of all public chest X-ray datasets available, to include as much relevant information as possible, with focus on datasets containing pneumothorax labels. We discarded datasets containing only pediatric population (VinDr-PCXR [40,41]), datasets that we were unable to download or that were currently unavailable (CXR-AL14 [42] and CANDID-II [43]) or other datasets with labels not fully relevant to our analysis (PLCO, NLST, VinDr-RibXR, datasets focusing only on tuberculosis detection, etc.).
We finally included 11 datasets in our work:
- ChestX-ray14 (‘NIH’) [26]: This was the first dataset with a massive amount of images, released in 2016 by the US National Institute of Health (NIH). It contains 112,120 frontal images from 30,805 individual patients, containing 14 different labels (amongst them, 5302 labelled with pneumothorax (4.73%) and 60,361 with no findings) labelled mainly via NLP techniques. A small subset of the dataset also contains bounding boxes for disease localization. Posterior research and expert labelling of the dataset changed the final pneumothorax count to 5138 (4.58%).
- CheXpert [27,44]: This was released in 2019 by Stanford University (US), containing 224,316 frontal and lateral images from 65,240 patients, using also 14 labels (most of them overlapping with the ones from NIH dataset). It has 19,466 pneumothorax (8.68%) and 22,528 normal images (10.04%). It also contains a label ‘Support Devices’ for external devices such as pacemakers, endotracheal tubes, valves, catheters, etc., and introduces the possibility of uncertainty of a label (‘−1’ value). Labels were extracted via NLP, and they have recently released an extension with radiology reports included [32].
- MIMIC-CXR [28,45]: This was released in 2019 by the Massachusetts Institute of Technology (MIT, US); it contains 377,110 multi-view images from 65,379 patients, using the same labels and NLP labeler as the previous dataset, but also releasing the radiology reports and original images in DICOM format. It contains 14,239 pneumothorax images (3.78%) and 143,363 normal images (38.02%).
- PadChest [29]: This was made public in 2019 by University of Alicante (Spain); it comprises 160,868 multi-view images from 67,625 unique patients, containing the Spanish radiology report and the extracted hierarchical labels using Unified Medical Language System (UMLS) terminology. A subset of the dataset is also manually labeled. It contains 851 pneumothorax images (0.52%, 411 of them manually labeled) and 50,616 normal images (31.47%).
- VinDr-CXR [30,46]: This was released by VinBigData in 2022; it contains 18,000 frontal images from two Vietnamese hospitals, manually annotated from radiologists, using both global and local labels. It contains 12,657 normal images (70.3%) and 76 pneumothorax images (0.004%). The dataset was released in DICOM format.
- SIIM-ACR Pneumothorax Detection Challenge [47]: This dataset was released in 2019 by the Society of Imaging Informatics in Medicine (SIIM) and American College of Radiology (ACR) for a competition focused on detecting pneumothorax in chest radiographs using segmentation. It contains 12,047 frontal images—9378 (78%) without pneumothorax and 2669 with pneumothorax (22%)—and their corresponding segmentation masks.
- BRAX [48,49]: This was released in 2022, and contains 40,967 frontal and lateral images from 18,442 patients from the Hospital Israelita Albert Einstein (Brazil). They adapted the NLP labeler used in CheXpert and MIMIC-CXR to Portuguese, thus extracting the same 14 disease labels. Original DICOM files were also publicly released. It contains 214 pneumothorax images (0.52%) and 29,009 images without findings (71%).
- Indiana [25,50]: This was the first multilabel chest X-ray dataset, released in 2012 by Indiana University (US), and contains 7470 images (frontal and lateral) from 3851 studies coming from two hospitals from the Indiana region, along with the radiology reports and MeSH codification. It contains 54 images with pneumothorax (0.72%) and 2696 normal images (36.09%).
- CANDID-PTX [51,52]: This was released in 2021, and contains 19,237 frontal images from Dunedin Hospital (New Zealand), labelled for pneumothorax (3196 samples, 16.61%) and also for acute rib fracture and chest tube presence.
- PTX-498 [53,54]: This was released in 2021, and contains 498 frontal images from three different hospitals in Shanghai, China. All of them correspond to pneumothorax (100%, no other labels), along with manually labelled segmentation masks.
- CRADI [55,56]: This was another dataset released in 2021 with images from several institutions in Shanghai, China. It contains 25 different labels annotated using NLP. The training set contains 74,082 frontal images, but it is not publicly available without prior request due to PII reasons; the external test set contains 10,440 images (one per patient), 201 with pneumothorax (1.92%), and 2737 without findings (26.22%).
We divided those datasets into 6 training datasets (the first ones) and 5 external test set datasets (the latter ones). This division was made to balance the number of pneumothorax images in train and test datasets and also to allow having bigger datasets in the training set, to capture a wider range of pathologies and radiological features to create the model, while still having a considerable number of images in testing datasets.
Images from test datasets are not used at all for training or hyperparameter tuning and are just used for model evaluation purposes. Object detection labels or segmentation masks from available datasets were not used in our analysis.
We took the decision of not integrating other popular public datasets (JSRT [57], PLCO [58], NLST [59], TBX11K [60], Montgomery-Shenzhen [61], etc.) because pneumothorax was not included in their label sets. Although unlikely, this could result in the presence of some pneumothorax cases being incorrectly treated as false positives, thereby introducing label noise and biasing the evaluation. Additionally, the absence of positive cases limits their value for assessing some of the performance metrics (sensitivity and PPV), which are critical in the clinical scenario of pneumothorax detection.
Table 1 shows information about all datasets used in our work.

Table 1.
Details of the individual datasets used in our work. The symbol ‘#’ means “number of”.
2.2. Dataset Harmonization
Publicly available datasets employ different directory structures, metadata schemas (csv, text files, and JSON files), file formats (image formats vs. DICOM radiology format), label encodings, etc. To be able to use them for our work, we had to develop a unified ingest pipeline that included several functions:
- -
- Metadata organization: We cataloged everything into a single master table/CSV file, where each row corresponds to one image and includes all important metadata related to it (patient ID, study ID, image ID, view position, date, patient sex and age, and image path and image labels, amongst other values).
- -
- Label harmonization: We mapped each label to a common convention (e.g., “Pneumothorax” and “PTX” were mapped to “pneumothorax”, whereas “normal” or “pathological” were mapped to “no_finding”). We performed one-hot encoding on all labels. For images with uncertainty labels (labeled as ‘−1’ in CheXpert and MIMIC datasets), we relabeled them as disease-negative (‘0’). For datasets not containing the ‘no_finding’ column, it was set to positive if none of the other pathological columns was positive, and negative otherwise.
- -
- Split identifiers: To prevent data leakage, we assigned each patient study a unique cross-dataset identifier in order to allow train–validation splits at a patient level, ensuring no patient’s images appeared in both sets.
- -
- Projections: We correctly handled data projections and corrected some labelling errors for specific images, especially in pneumothorax-positive images.
- -
- DICOM conversion: Datasets that used DICOM (Digital Imaging and Communications in Medicine) format required additional processing to extract metadata out of the DICOM files, as well as the raw images, which were converted to 8-bit PNG images and saved to disk prior to training.
- -
- Original splits: If present in the dataset, original training, validation, and test splits were not used, as the ratio was sometimes different to the one we used, and our target was evaluation in external datasets.
- -
- Enhanced annotations: Whenever enhanced annotations or labels were available, they were used in replacement of the original labels from the dataset—in the NIH dataset, labels from experts [34,38], and, for NIH and PadChest datasets, annotation from chest tubes [36].
- -
- Segmentation information: Binary labels of interest such as ‘pneumothorax’ or ‘tubes’ were inferred if there were segmentation masks available. Segmentation masks themselves were not used for training.
2.3. Preprocessing
Before reaching the model, all images (both from training and from test datasets) were filtered to include only frontal images (PA or AP), removing the remaining projections (including lateral projection). We also filtered out all pediatric images (<18 years old) from the datasets containing age.
Regardless of their original resolution or bit depth, all images were rescaled to a fixed size of 456 × 456 pixels with three channels. Grayscale images were replicated across channels to create RGB representations, enabling compatibility with models pretrained on natural images. Pixel intensities were normalized to [0, 1] range based on their original bit depth.
Training time augmentation was used by applying random transformations to the input images at training time, as it is known to improve model performance [62]. We used horizontal flipping, random rotations (up to ±10°), brightness adjustments (80–120%), horizontal and vertical translations (up to 3%), and shearing (up to 3°).
To create training and validation datasets, each individual dataset was applied a function to split individual patients into training (95%) and validation (5%) sets preserving the label distribution of the dataset for the pneumothorax label. Then, to partially mitigate the label imbalance of pneumothorax across all datasets, we applied 2× oversampling on pneumothorax images and random undersampling to non-pneumothorax images to obtain a ratio of 4:1 (pneumothorax to no pneumothorax) in the training set.
Table 2 shows the final number of unique images used in the training set, where pneumothorax images are oversampled, so each image is used twice as input for each epoch. Table 3 shows the number of images in the test set.

Table 2.
Training datasets used in our work.

Table 3.
Test datasets used in our work.
2.4. Model Architecture
We chose EfficientNet B5 as the base architecture of our analysis. EfficientNet [63] is a family of CNN (Convolutional Neural Network) architectures released in 2019 that achieved state-of-the-art (SOTA) metrics on large-scale image classification benchmarks, with focus on reducing model size and computational cost. This was achieved by using compound scaling, which applies a set of constants to uniformly scale model depth, width, and input resolution simultaneously (instead of only increasing one dimension of network capacity). This balanced approach yielded several models (from B0 to B7) that progressively increased parameter count, image resolution, and computational cost, with the benefit of better classification performance.
There are several reasons for the selection of this architecture. First, some recent studies suggest EfficientNet architecture (in particular, EfficientNet B3) may be superior to DenseNet121 [64] (previously the most common and best-performing amongst other architectures) in the setting of chest X-ray classification [38,65].
Second, because some papers reflect the importance of the image size in the detection of pneumothorax (particularly in small pneumothorax). While 224 × 224 was the standard for some architectures (ResNet50 and DenseNet121) due to the usage of ImageNet pretraining, pneumothorax detection seems to benefit from larger image sizes, even up to 1024 × 1024 [66,67,68]. Thus, we selected an architecture that we could train using a local GPU, has enough inference speed to enable triaging, and could benefit from an already available ImageNet-pretrained model.
We adopted a two-stage training pipeline to leverage both general and radiological image features. First, we used the EfficientNet B5-pretrained model (on ImageNet general-purpose images), where we applied domain-specific intermediate pretraining on a binary task (frontal vs. lateral radiographic view) using a subset of all datasets combined, by retraining the last three convolutional blocks (blocks 5, 6, and 7 from the original architecture). This helps the model to learn fine features specific to the radiology domain [69]. After that, we froze all the weights from the base architecture and replaced the classification head of the model to suit the final classification task for pneumothorax classification, with gradual unfreezing of blocks 7, 6, and 5 after several epochs.
Instead of training a plain binary pneumothorax classification model, we designed a multi-label model with three classes: pneumothorax, no_finding (if images contained any pathology or disease), and support_devices (if images contained external devices such as pacemakers, chest tubes, endotracheal tubes, etc.). While pneumothorax was the primary classification goal, two auxiliary outputs were included to mitigate the potential effect of confounder biases, rather than as targets of clinical interest. That allowed the model to learn to differentiate different devices from chest tubes, and also to detect features of pathologies that may be correlated with pneumothorax but are independent.
Training was performed for 20 epochs with batch size 8, using weighted binary cross entropy loss (which allows us to leverage the importance of each label to give more weight to pneumothorax detection itself, using a weighted value of 5) and Adam optimizer with a starting learning rate of 1 × 10−4, reducing the learning rate automatically by a factor of two if the pneumothorax validation AUROC did not improve after two epochs.
2.5. Evaluation
Model performance was assessed on each test cohort, both globally and per-dataset, using the following metrics: Sensitivity (Recall), Specificity, Positive Predictive Value (PPV or Precision), Negative Predictive Value (NPV), and F1-score.
We also calculated the confusion matrices and generated the ROC (receiver operating characteristic) and PR (precision–recall) curves, along with calculating the area under the curve (ROC–AUC or AUROC, and PR–AUC or AP, average precision) in order to visualize performance at different classification thresholds. PR–AUC is useful given the imbalanced scenario of pneumothorax in the different datasets [70].
We estimated 95% confidence intervals for all performance metrics by nonparametric bootstrapping with 1000 resamples.
2.6. Experiments
In addition to the main goal, training a model using multi-source training data (from different datasets) and evaluating the results across independent test datasets, we conducted two supplementary studies:
- Single-source vs. multi-source training: We retrained the same network architecture on each dataset independently (NIH, CheXpert, MIMIC.-CXR, PadChest, SIIM-ACR, and VinDR-CXR), and compared test-set metrics from single-source models to those of the multi-source model.
- Threshold optimization: To maximize clinical utility, we evaluated threshold selection strategies on the validation set. Choosing an optimal threshold is crucial, as a suboptimal choice can lead to a model that can obtain high performance metrics but no clinical utility. Our aim was to balance false negatives (which risk missed pneumothoraces and can be potentially life-threatening) and false positives (which can lead to lack of clinical confidence).
All training was performed using Tensorflow framework on a single GPU (NVIDIA RTX2080ti). All training, evaluation, and inference code is available in a public repository, something that can help containerization and potential integration in real world.
3. Results
3.1. Evaluation of Multi-Source Model on External Datasets
The primary objective of our work was to train a model using multi-source data (including data from all training datasets: NIH, CheXpert, MIMIC-CXR, PadChest, SIIM-ACR, and VinDR-CXR), and evaluate the model performance on unseen data using external test sets (BRAX, CANDID-PTX, CRADI, PTX-498, and Indiana).
The results are shown in Table 4, and are calculated both per-dataset and aggregated in two ways:

Table 4.
Evaluation of the model trained on multi-source data on external datasets.
- -
- Mean ± SD (macro-averaged): Taking the arithmetic mean and standard deviation of each metric across individual datasets, each dataset is treated equally (irrespective of the number of samples), which is useful for evaluating model robustness to different dataset sources.
- -
- Overall (micro-averaged): By combining all test samples into a single dataset and calculating their metrics, so that each image has equal weight irrespective of its source, this is useful for reflecting real-world performance but can hide biases in smaller test datasets.
While we trained a multi-label model (‘pneumothorax’, ‘support devices’, and ‘no finding’ labels), the evaluation focuses on pneumothorax, as it is the primary target of the study. The performance on the remaining labels will be only shown in the www.github.com/santibacat (accessed on 26 June 2025).
The PTX-498 dataset lacks any negative samples (only contains pneumothorax images), and, therefore, some metrics cannot be calculated. Those metrics will be excluded, and the ROC and PR curves will not be shown for this dataset.
The results for the overall external test set were as follows: ROC–AUC 0.961 (0.957–0.964), PR–AUC 0.825 (0.814–0.834), sensitivity 0.862 (0.852–0.872), specificity 0.945 (0.943–0.947), PPV (precision) 0.566 (0.554–0.577), and NPV 0.988 (0.987–0.989).
The metrics are similar across individual datasets except for the BRAX dataset, which shows a much worse performance than the other datasets. The Indiana dataset also shows a lower performance but that may be secondary to the low number of simples and the pneumothorax label coming directly from reports (which includes confounders like ‘hydropneumothorax’).
Figure 3 shows the receiver operating characteristic (ROC) and precision–recall curves, with their respective areas under the curve (AUROC or ROC–AUC, and AP or PR–AUC). They contain information both from the overall performance (micro-average, thick line) and from the individual datasets.
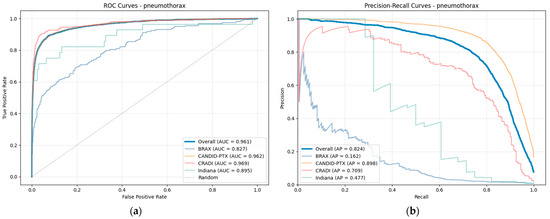
Figure 3.
(a) ROC curve and (b) precision–recall curve for pneumothorax. The thick blue line represents the overall curve (micro-average), and the thin lines are the curves for individual datasets. PTX-498 is not shown due to lack of negative samples.
The curves from the individual datasets are shown with the exception of the PTX-498 dataset (which lacks positive samples), and are similar to the curve from the aggregated dataset with the exception of the BRAX and Indiana datasets.
The distribution of real and predicted values is shown in Figure 4 as confusion matrices. Percentages are row-normalized, with the number of samples in parentheses. Individual confusion matrices per-dataset are shown along with the confusion matrix for the overall test dataset.
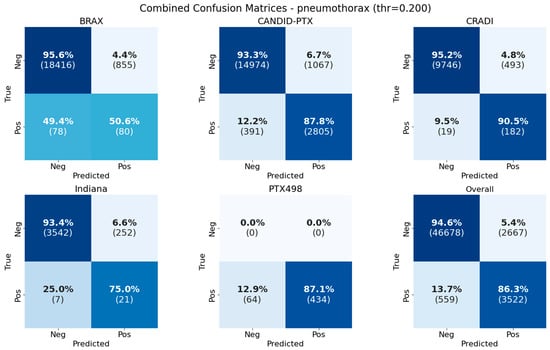
Figure 4.
Confusion matrices for pneumothorax classification in the different test datasets and the overall performance in all test datasets (individually and combined). Percentages are row-normalized.
Table 5 shows the results of the model evaluation on the other two labels the model was trained on along with pneumothorax: no_finding (which defines normal or pathological images, including many pathologies, pneumothorax amongst them), and support_devices (which include several devices such as tubes, pacemakers, catheters, etc.). Although those labels have no clinical use in our work, they are useful for training because they help the model learn other patterns of disease that can become confounders if they are present along with pneumothorax.

Table 5.
Evaluation of the model trained on for labels.
3.2. Single-Source vs. Multi-Source Training
We wanted to investigate whether the use of multi-source data allows trained models to have a better performance and higher generalization power in comparison with training with single-source data.
Therefore, in addition to the model trained on multi-source data, we trained another set of models using the same architecture and training pipeline but using only data from each training dataset individually.
The results from the training are shown in Table 6, Table 7, Table 8, Table 9, Table 10 and Table 11. Each table shows a matrix of train and test datasets (x and y axes) with the values of a specific metric, so that each metric corresponds to the model trained on the specified training dataset and evaluated on the specified test dataset.

Table 6.
Average precision (PR–AUC) results for external test sets using single-source and multi-source training datasets. Best results in bold.

Table 7.
Area under the ROC curve (ROC–AUC) results for external test sets using single-source and multi-source training datasets. Best results in bold.

Table 8.
Sensitivity (Recall) results for external test sets using single-source and multi-source training datasets. Best results in bold.

Table 9.
Specificity results for external test sets using single-source and multi-source training datasets. Best results in bold.

Table 10.
Positive Predictive Value (PPV–Precision) results for external test sets using single-source and multi-source training datasets. Best results in bold.

Table 11.
F1-score results for external test sets using single-source and multi-source training datasets. Best results in bold.
A visual comparison of all the evaluation metrics (for the overall training set) for the multi-source and single-source models is shown in Figure 5.
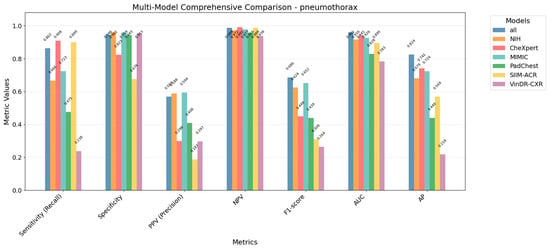
Figure 5.
Comparison of evaluation metrics for multi-source model and each single-source trained model, showing that multi-source model outperforms in general metrics to single-source models.
ROC and PR curves were also calculated for each single-source training dataset independently, as shown in Figure 6 and Figure 7.
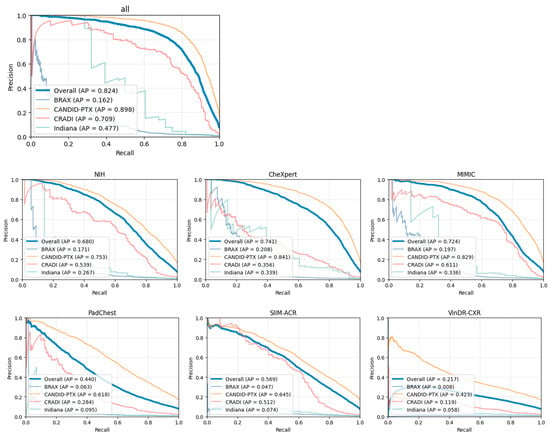
Figure 6.
Precision–recall curves for each independent single-source dataset. The plot entitled “all” represents the multi-source dataset.
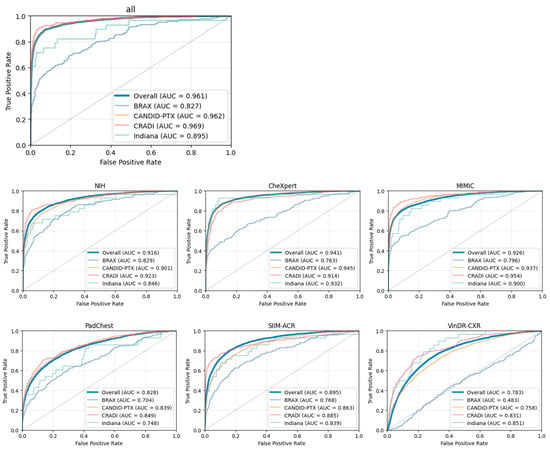
Figure 7.
ROC curves for each independent single-source dataset. The plot entilted “all” represents the multi-source dataset.
The results show that the model trained with multi-source data is better balanced and has higher generalization capabilities to unseen data compared to the models trained with single-source data. For every metric, the multi-source model usually exceeds the individual models, while individual models can exceed in one specific metric while generally dropping the performance in the others.
That shows using multi-source data helps to mitigate the ‘domain shift’, where metrics drop significantly in external datasets compared to training–validation datasets due to a lack of generalization, which can be attributed to different factors (equipment, projection, labeling strategies and quality, etc.).
We calculated the confusion matrices for all trained models (using multi-source and single-source training datasets), both using a combined test set and also on a per-dataset basis. All results are shown in Figure 8.

Figure 8.
Confusion matrices for all combinations of training datasets in vertical axis (individual single-source and also multi-source as all) and all test datasets in the horizontal axis (including micro-average as Overall).
3.3. Threshold Optimization
A small split (5%) of the data from the training datasets was used as a held-out (validation) set, in order to tune the model hyperparameters; amongst them, we used the validation set to choose the optimal threshold value for the predictions.
Binary and multilabel models use sigmoid activation functions in the last layer, which means it outputs a continuous probability score in the range of [0, 1] for each label. ROC and PR curves are able to show the overall performance at multiple classification thresholds, offering a global interpretation of the model ability to linearly separate two given classes in a binary classification, which is reflected in the value of their metrics in the area under the curve (AUC).
In a real-world scenario, a fixed threshold needs to be set to consider a prediction as positive or negative. A 0.5 value for the classification threshold is considered the standard, although the actual value can be modified to be more suitable for a given task.
In the context of disease triage, and, even more, in the specific case of pneumothorax (where minimizing the number of false negatives is a priority, given the severity and potential mortality of the disease), lowering the classification threshold is generally beneficial, although some careful assessment needs to be made to avoid increasing the false positives too much (which would increase the untrustworthiness of the model and would make it unsuitable and unusable in a real-world scenario).
Table 12 shows the different metrics for the validation set at different threshold values, as well as the number of true positives, false positives, false negatives, and true negatives. Figure 9 shows the F1 values at different classification thresholds, showing the value where the F1-score is maximized (0.641). That threshold maximizes F1 but does not lead to a useful model due to its inability to linearly separate the classes, as shown in Figure 10, which shows the confusion matrices at different specific threshold scores.

Table 12.
Metrics for the validation set at different threshold values.
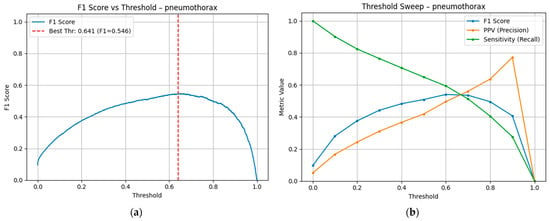
Figure 9.
(a) F1-score at different threshold values. The red line indicates the threshold value where the F1-score is maximized. (b) Sensitivity (Recall) and PPV (Precision), both components of the F1-score, are shown independently.
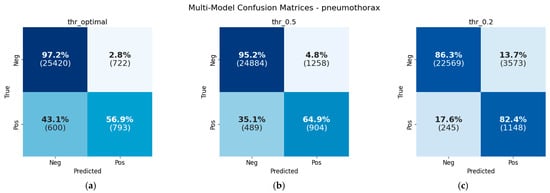
Figure 10.
Confusion matrices at different threshold values: (a) 0.641, which maximizes F1-score; (b) 0.5, standard classification threshold; and (c) 0.2, low classification threshold for reducing false negatives.
Our aim was to minimize the number of false negatives (missed pneumothoraces) by pushing the recall as high as possible while maintaining the precision (lowering the number of false positives to a reasonable number). We estimated 0.8 as the minimum recall for achieving our target and, thus, evaluated the different precision values with this objective. Table 13 shows the calculated precision values at fixed recall values over 0.8, showing that precision was not as high as would be advisable.

Table 13.
Calculated precision values at different fixed recall values. We estimated a minimum of 80% of recall for minimizing the number of missed pneumothoraces; thus, we calculated precision values at different recall values over 0.8.
In a real-world triaging scenario and considering the severity of the disease, we decided that, in order to maintain the recall and minimize the number of false negatives, we would use a threshold of 0.2 in our work. That threshold would mean, considering the estimated prevalence of the disease, around three false positives per true positive (3:1 ratio). That ratio, although higher than desirable, is low enough to still be clinically useful at the benefit of reducing the number of missed pneumothoraces.
4. Discussion
In this study, we wanted to investigate the viability of training a model suitable for pneumothorax detection in an emergency scenario, by using public chest X-ray datasets for training (creating a multi-source dataset) and evaluation (in order to assess the generalizability and robustness in unseen data).
We selected the EfficientNet-B5 model and trained it using a multi-source training strategy with 322,569 pooled chest X-rays from six public datasets, showing an overall AUROC of 0.961 and PR–AUC of 0.825 in the external test set (which include five unseen public datasets not used for training or validation), where the pneumothorax prevalence was 7.6% (4081 out of 53,426 samples).
Those results show robust discrimination on heterogeneous unseen data, which was the main objective of our work, in order to assess the potential usage in a clinical scenario. Nevertheless, threshold tuning remains an important task that can substantially modify the clinical application of trained models. In our case, we chose to lower the threshold to 0.2, to sacrifice specificity for safety, so we minimize the number of missed pneumothoraces at the cost of more false alarms. Our results in the validation set showed that, at a recall of >80%, we obtained a ratio of 3:1 for false positives versus true positives. If we selected a threshold to increase the recall to >90%, we would have obtained a ratio larger than 6:1, which would have decreased the PPV too much and given too many false alarms to make the model clinically useful.
The choice of this threshold yields a recall of 0.862 in the external test set, which still gives 13.7% false negatives, probably still too high in a real-world scenario given the potential severity of the disease. In a real scenario, missed pneumothoraces are generally small pneumothoraces or in anteroposterior radiographic projections, which are more difficult to detect; but, in triage systems, we should minimize false negatives as low as technically feasible (approaching 0%) without a high number of false positives.
Although the PPV in the external test set is not too high (0.569), given the low prevalence of the disease, it gives a ratio of false positives versus true positives of ~0.8:1, which corresponds to nearly a false alarm for every real pneumothorax, something that is acceptable in a clinical scenario given the triage setting and the severity of the condition.
The results are similar across individual test datasets except for the BRAX dataset, which show worse results compared to the rest of the datasets. An error analysis (shown in Figure 11) suggests images from pediatric patients and lateral images in the false positives (suggesting potential labelling issues), and images without pneumothorax in the false negatives, which suggest potential labelling issues or treated pneumothoraces; all those issues should be investigated further in order to improve the evaluation of models trained or evaluated using this dataset.
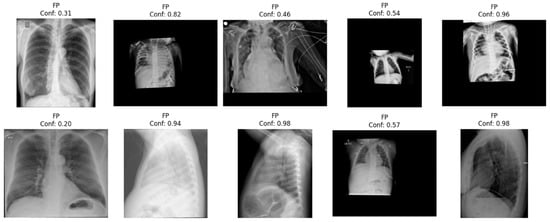
Figure 11.
Error analysis of false positives from the BRAX dataset, showing several images that would not have to be used as input, such as pediatric images (three on the first row) or images from lateral views (three in the second row), suggesting potential limitations in the evaluation of the performance in this dataset.
The Indiana dataset also showed decreased metrics compared to other test datasets, but an error analysis does not show clear problems in false negatives (missed pneumothoraces).
We also performed an analysis of the contribution of the individual datasets by training single-source models and comparing their performance with the multi-source dataset, showing that the multi-source model outperforms the single-source counterparts’ global metrics (AUROC, PR–AUC, and F1-score) and almost in all specific metrics.
For instance, specificity is higher in models trained using the VinDR-CXR dataset, but that is probably due to the low prevalence of pneumothorax in that dataset, producing very few false positives but a very low recall. The MIMIC-CXR-trained model has a higher PPV probably due to their data distribution (a moderate pneumothorax prevalence, and high number of anteroposterior views). The model trained using the CheXpert dataset has the largest recall (0.908) probably due to the largest prevalence of pneumothorax (8.7%), which, in addition to the class-balance oversampling, effectively lowers its internal decision boundary at the cost of extra false positives.
But, even if specific single-source models outperform the multi-source model in a specific metric, it always comes at the cost of worse overall performance. Training using multi-source heterogeneous data improves generalizability and model robustness and mitigates the ‘domain shift’ where models trained with one dataset significantly decrease their performance when tested against external datasets that resemble real-world performance [71].
This effect is something that has been extensively discussed before for the pneumothorax scenario and is something that heavily affects some of the used datasets. For example, the NIH dataset suffered from incorrect labelling coming from NLP techniques used in radiology report extraction, and also from confounder biases coming from the correlation between treated pneumothoraces (labelled with ‘pneumothorax’) and chest tubes (that did not contain any label), creating spurious correlations [31,72]. Using expert labelling and chest tube information can help mitigate these issues, which also happens (to a lesser extent) in other datasets [73,74,75,76]. With the objective of mitigating this domain shift, we incorporated auxiliary tasks to the model, by also predicting no disease and support devices, in order to prevent shortcut learning and enforce the model to learn real pneumothorax features.
The CheXNet [77] was the first paper which attracted wide interest from the global scientific community in chest X-ray classification (although many of the papers that were published in that period used only the NIH dataset [78,79], which, as we discussed, had problems that needed to be mitigated). The release of many public datasets in 2019 (including CheXpert, MIMIC-CXR, and PadChest), which featured more and better labels, and more radiologic projections, and included radiology reports, had enabled richer scientific research that identified and resolved obstacles to developing models suited for real-world scenarios.
Recent studies using multicenter datasets have achieved pneumothorax AUROCs in the 0.94–0.98 range [65,80,81,82,83]. Our off-the-shelf EfficientNet-B5 model matches the upper end of that spectrum (AUROC 0.961 and PR–AUC 0.825) while being trained on a broad, carefully curated pool of six public datasets and validated on five additional public datasets (with >53,000 radiographs). These results demonstrate that data harmonization, label cleaning, and domain-aware adjustments are more critical for performance and generalizability than specific and elaborate network architectures.
Despite these findings, comparing our metrics to those of other studies requires caution for several reasons. First, some papers do not show the threshold tuning (that might be different than the one used in this paper, where we focused on diminishing the number of false negatives). Second, we do not use official splits for the datasets, even though some validation or test splits have manual annotations versus automatic annotations for the training split. Third, we must prefer using PR–AUC to interpret the results, as shown in [70], given the severe imbalance of the dataset and the expected low prevalence of pneumothorax in a real-world setting. Last, even though we use the ‘no finding’ label to help the model learn features from other pathologies, we do not train the model to identify other pathologies different than pneumothorax, which is something that could further help the model (even more with pathologies with free air such as pneumoperitoneum, subcutaneous emphysema, etc.)
Our work also has some limitations. First, other training strategies could have improved the results, such as the use of more negative samples from already available datasets (using batch dynamic sampling instead of choosing a fixed number of negative samples) or freezing more layers from the pretrained base model. Second, we also could have chosen a model architecture (an ensemble of models, custom models, etc., such as [84]) or image size (bigger image size) that yielded better results, but we preferred to use simpler models that could have ImageNet-pretrained weights and that could be retrained in a local server with an accessible GPU. Third, we do not carry out a subgroup analysis on the test datasets to dissect the model performance by pneumothorax size (small or large), location, or radiographic projection (posteroanterior or anteroposterior). That would show the specific needs to obtain better metrics on difficult samples.
Although the usage of saliency maps has been shown to be useful for clinical interpretability [85], it is not something that can be trusted to always show the localization of a disease [86]. We believe object detection or segmentation models are much more useful in clinical use, both due to their capacity to detect and localize the pathology correctly, and also due to their interpretability capabilities [53,87].
Further work should analyze the utility of segmentation in real-time pneumothorax triaging, as well as a prospective real workflow with a clinician-in-the-loop or radiologist-in-the-loop trial to assess the real model impact, false-alarm fatigue, and potential utility in terms of the detected pathology and reduced time-to-diagnosis.
Given the current metrics, many models (including ours) are best positioned as a worklist-prioritization tool, flagging high-risk cases for expedited review, rather than a full autonomous diagnosis, which would require almost 100% recall with less false alarms. Appropriate site-specific threshold calibration and continuous performance monitoring is needed in order to use these models in a clinical scenario.
5. Deployment and Integrability
The healthcare integration of technological advancements, and, specifically, deep-learning systems, retain its own challenges due to the presence of inherent PHI/PII (personal health information/personal identification information), both in the headers of the DICOM files (that holds up radiological information), and, sometimes, also burned into the image itself. That implies the removal of PHI/PII for all data that leaves the hospital/clinic network, a fact that partially limits the available deployment options.
That was what motivated the decision to use a model from the EfficientNet family, which not only is already compatible with popular frameworks (Tensorflow and PyTorch), but also allows a decent inference speed using local resources while retaining high metrics.
Below are shown the different integration options that could be implemented:
5.1. In-Device Integration
Local integration embedded directly in the X-ray acquisition device, where the DICOM object is directly present in-device after its generation and inference, is performed automatically using local resources, returning a pneumothorax probability and triggering an alarm for the technician even before the patient leaves the X-ray acquisition room. The technician could alert the referring physician or the radiologist, reducing the diagnosis time and providing faster care.
As the data is managed inside the modality itself, it does not leave the hospital/clinic network. The drawback is that the model must be embedded inside the modality itself, something that is currently not supported out of the box for most of X-ray vendors.
5.2. Server Integration with PACS-RIS Service
Local integration in a server connected within the hospital/clinic network allows seamless PII/PHI management (as the data does not leave the hospital network) while requiring minimum hardware upgrades (only the GPU requirements according to the number of acquired chest X-rays).
The model would be exposed as a stateless micro-service that accepts studies via standard DICOM protocols (C-STORE), processes them, and returns the result, either as a DICOM Structured Report to the referring DICOM node, or as an HL7-FHIR DiagnosticReport resource. This design allows any PACS to dispatch studies and receive triage scores without modifying the existing radiological or clinical workflow.
This approach, along with containerization, has the benefit of allowing multiple parallel models for different clinical use in the same server (e.g., model detection, segmentation, x-ray quality assurance, other pathologies, etc.).
The disadvantage of this implementation is the lack of automatic prioritization for the high-probability pneumothorax studies, as their score would be sent to PACS directly.
5.3. Local Web Dashboard
A lightweight web front-end can enable clinicians to drag-and-drop single images (mainly JPG/PNG, but DICOM could also be accepted) on an intranet page. Inference would run on the local computer, entirely on the CPU, using Tensorflow-Lite [88] or similar web-deployment machine-learning frameworks.
Although much slower than inference on GPU or cloud services, this approach provides an alternative workflow in resource-constrained scenarios where PII/PHI cannot be transmitted to external services.
5.4. Cloud Dashboard
A cloud-based dashboard is another option for institutions that prefer a fully managed cloud-accessible solution. A micro-service would run in a virtual private cloud (VPC) behind a DICOM-web gateway that enforces automatic de-identification. All patient identifiers, birth dates, and free-text (burned into the image or in DICOM headers) would be stripped in transit, ensuring that only anonymized data reaches the cloud, while reverse-mapping to the original study would be retained on-premises.
This has the clear advantage of cloud accessibility from outside the hospital/clinic network, allowing for remote or mobile access (for instance, for consultation with senior physicians/radiologists), and dashboard hooks (push notifications to mobile devices when a new positive case is predicted), with the drawback of higher costs (cloud computation, storage, and anonymization) and potential data leaks.
5.5. Mobile Integration
Apart from the two previous options (which would also allow access through mobile devices), deployment in mobile devices could be an alternative for constrained resources (remote or rural locations), as it does not need specific hardware or a connection to the clinic network. The model would be directly deployed to the phone after being adapted to mobile deployment (using frameworks like Tensorflow-Lite [88], and using 8-bit integer quantization or other techniques to limit the computer processing needed).
The main drawbacks for this method are the lack of integration with the rest of the network, as well as the drop in speed and accuracy versus the full-precision pipeline.
All described methods would need to comply with local regulations in terms of PHI/PII (EU GDPR/US HIPAA alignment), especially when the data leave the hospital network, and also enforce security measures such as HTTPS/TLS in transit, AES-256 encryption at rest, and role-based access control.
6. Conclusions
We demonstrate that, by using multi-source data from available chest X-ray datasets, we are able to create a deep-learning model for pneumothorax detection that can generalize well to unseen data. Nevertheless, caution is warranted before deploying these models in clinical practice; even after threshold optimization, they still miss more than 10% of pneumothoraces and, because of the condition’s low prevalence, generate roughly one false alarm per true case detected, underscoring the need for continued refinement and for clinician review to remain integral to the diagnostic workflow.
Author Contributions
Conceptualization (S.I.C. and O.M.M.), data curation (S.I.C.), formal analysis (S.I.C. and O.M.M.), funding acquisition (O.M.M.), investigation (S.I.C., J.d.D.B.M. and O.M.M.), methodology (S.I.C. and O.M.M.), project administration (O.M.M. and J.d.D.B.M.), resources (S.I.C. and O.M.M.), software (S.I.C.), supervision (J.d.D.B.M. and O.M.M.), validation (S.I.C. and O.M.M.), visualization (S.I.C. and O.M.M.), writing—original draft (S.I.C. and O.M.M.), writing—review and editing (S.I.C., J.d.D.B.M. and O.M.M.). All authors have read and agreed to the published version of the manuscript.
Funding
This research has received funding from the project Robotic-Based Well-Being Monitoring and Coaching for Elderly People during Daily Life Activities (RobWell) funded by the Spanish Ministerio de Ciencia, Innovación y Universidades with ID: RTI2018-095599-A-C22. and from the Wallenberg AI, Autonomous Systems and Software Program (WASP) through the Knut and Alice Wallenberg Foundation.
Data Availability Statement
Data used for the generation of this manuscript will be available in www.github.com/santibacat (accessed on 26 June 2025). Public datasets are available in their respective sources.
Acknowledgments
We would like to thank the authors of the public datasets used in this work.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| Abbreviation | Meaning |
| ACP | Acute Chest Pain |
| ACR | American College of Radiology |
| AES | Advanced Encryption Standard |
| AI | Artificial Intelligence |
| AUC | Area Under the Curve |
| AP | Anteroposterior (radiographic projection) Average Precision |
| AUROC | Area Under the Receiver Operating Characteristic Curve |
| CNN | Convolutional Neural Network |
| CSV | Comma Separated Values |
| C-STORE | DICOM Composite Object Store Service Class |
| CPU | Central Processing Unit |
| CT | Computed Tomography |
| DICOM | Digital Imaging and Communications in Medicine |
| EMR | Electronic Medical Record |
| EU | European Union |
| FDA | Food and Drug Administration (U.S.) |
| FHIR | Fast Healthcare Interoperability Resources |
| GPU | Graphics Processing Unit |
| HL7 | Health Level 7 |
| HTTPS | Hypertext Transfer Protocol Secure |
| IMIB | Instituto Murciano de Investigación Biosanitaria |
| JPG | Joint Photographic Experts Group |
| JSON | Javascript Object Notation |
| MDR | Medical Device Regulation |
| MeSH | Medical Subject Headings |
| MDPI | Multidisciplinary Digital Publishing Institute |
| MRI | Magnetic Resonance Imaging |
| NIH | National Institute of Health (U.S.) |
| NLP | Natural Language Processing |
| NPV | Negative Predictive Value |
| PA | Posteroanterior (radiographic projection) |
| PACS | Picture Archiving and Communication System |
| PHI | Personal Health Information |
| PII | Personal Identification Information |
| PNG | Portable Network Graphics |
| PPV | Positive Predictive Value |
| PR | Precision-Recall |
| PTX | Pneumothorax |
| RIS | Radiology Information System |
| RGB | Red Green Blue |
| ROC | Receiver Operating Characteristic |
| SIIM | Society of Imaging Informatics in Medicine |
| SOTA | State Of The Art |
| TLS | Transport Layer Security |
| UMLS | Unified Medical Language System |
| US | United States |
| VPC | Virtual Private Cloud |
| WHO | World Health Organization |
References
- Wenderott, K.; Krups, J.; Zaruchas, F.; Weigl, M. Effects of Artificial Intelligence Implementation on Efficiency in Medical Imaging—A Systematic Literature Review and Meta-Analysis. npj Digit. Med. 2024, 7, 265. [Google Scholar] [CrossRef]
- Chen, X.; Wang, X.; Zhang, K.; Fung, K.-M.; Thai, T.C.; Moore, K.; Mannel, R.S.; Liu, H.; Zheng, B.; Qiu, Y. Recent Advances and Clinical Applications of Deep Learning in Medical Image Analysis. Med. Image Anal. 2022, 79, 102444. [Google Scholar] [CrossRef] [PubMed]
- Mazurowski, M.A.; Buda, M.; Saha, A.; Bashir, M.R. Deep Learning in Radiology: An Overview of the Concepts and a Survey of the State of the Art with Focus on MRI. J. Magn. Reson. Imaging JMRI 2019, 49, 939–954. [Google Scholar] [CrossRef]
- Choy, G.; Khalilzadeh, O.; Michalski, M.; Do, S.; Samir, A.E.; Pianykh, O.S.; Geis, J.R.; Pandharipande, P.V.; Brink, J.A.; Dreyer, K.J. Current Applications and Future Impact of Machine Learning in Radiology. Radiology 2018, 288, 318–328. [Google Scholar] [CrossRef] [PubMed]
- Kapoor, N.; Lacson, R.; Khorasani, R. Workflow Applications of Artificial Intelligence in Radiology and an Overview of Available Tools. J. Am. Coll. Radiol. 2020, 17, 1363–1370. [Google Scholar] [CrossRef]
- Selby, I.A.; González Solares, E.; Breger, A.; Roberts, M.; Escudero Sánchez, L.; Babar, J.; Rudd, J.H.F.; Walton, N.A.; Sala, E.; Schönlieb, C.-B.; et al. A Pipeline for Automated Quality Control of Chest Radiographs. Radiol. Artif. Intell. 2025, 7, e240003. [Google Scholar] [CrossRef] [PubMed]
- Hosny, A.; Parmar, C.; Quackenbush, J.; Schwartz, L.H.; Aerts, H.J.W.L. Artificial Intelligence in Radiology. Nat. Rev. Cancer 2018, 18, 500–510. [Google Scholar] [CrossRef]
- Hwang, E.J.; Goo, J.M.; Park, C.M. AI Applications for Thoracic Imaging: Considerations for Best Practice. Radiology 2025, 314, e240650. [Google Scholar] [CrossRef]
- Benjamens, S.; Dhunnoo, P.; Meskó, B. The State of Artificial Intelligence-Based FDA-Approved Medical Devices and Algorithms: An Online Database. npj Digit. Med. 2020, 3, 118. [Google Scholar] [CrossRef]
- Afshari Mirak, S.; Tirumani, S.H.; Ramaiya, N.; Mohamed, I. The Growing Nationwide Radiologist Shortage: Current Opportunities and Ongoing Challenges for International Medical Graduate Radiologists. Radiology 2025, 314, e232625. [Google Scholar] [CrossRef]
- World Health Organization (WHO). To X-Ray or Not to X-Ray? Available online: https://www.who.int/news-room/feature-stories/detail/to-x-ray-or-not-to-x-ray- (accessed on 27 April 2025).
- NHS England. Diagnostic Imaging Dataset Statistical Release: 24 April 2025; NHS England: London, UK, 2025. Available online: https://www.england.nhs.uk/statistics/wp-content/uploads/sites/2/2025/04/Statistical-Release-24th-April-2025.pdf (accessed on 27 April 2025).
- Sociedad Española de Radiología Médica (SERAM). Informe sobre la Radiografía Convencional. Versión 3; SERAM: Madrid, Spain, 2021; Available online: https://seram.es/wp-content/uploads/2021/09/informe_rx_simple_v3.pdf (accessed on 27 April 2025).
- Tajmir, S.H.; Alkasab, T.K. Toward Augmented Radiologists: Changes in Radiology Education in the Era of Machine Learning and Artificial Intelligence. Acad. Radiol. 2018, 25, 747–750. [Google Scholar] [CrossRef]
- Annarumma, M.; Withey, S.J.; Bakewell, R.J.; Pesce, E.; Goh, V.; Montana, G. Automated Triaging of Adult Chest Radiographs with Deep Artificial Neural Networks. Radiology 2019, 291, 196–202. [Google Scholar] [CrossRef] [PubMed]
- Hwang, E.J.; Nam, J.G.; Lim, W.H.; Park, S.J.; Jeong, Y.S.; Kang, J.H.; Hong, E.K.; Kim, T.M.; Goo, J.M.; Park, S.; et al. Deep Learning for Chest Radiograph Diagnosis in the Emergency Department. Radiology 2019, 293, 573–580. [Google Scholar] [CrossRef]
- Khader, F.; Han, T.; Müller-Franzes, G.; Huck, L.; Schad, P.; Keil, S.; Barzakova, E.; Schulze-Hagen, M.; Pedersoli, F.; Schulz, V.; et al. Artificial Intelligence for Clinical Interpretation of Bedside Chest Radiographs. Radiology 2022, 307, 220510. [Google Scholar] [CrossRef] [PubMed]
- Yun, J.; Ahn, Y.; Cho, K.; Oh, S.Y.; Lee, S.M.; Kim, N.; Seo, J.B. Deep Learning for Automated Triaging of Stable Chest Radiographs in a Follow-up Setting. Radiology 2023, 309, e230606. [Google Scholar] [CrossRef]
- Kolossváry, M.; Raghu, V.K.; Nagurney, J.T.; Hoffmann, U.; Lu, M.T. Deep Learning Analysis of Chest Radiographs to Triage Patients with Acute Chest Pain Syndrome. Radiology 2023, 306, e221926. [Google Scholar] [CrossRef] [PubMed]
- Bintcliffe, O.; Maskell, N. Spontaneous pneumothorax. BMJ 2014, 348, g2928. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, A.R.; Morgan, W.E. Radiological Review of Pneumothorax. BMJ 2005, 330, 1493–1497. [Google Scholar] [CrossRef]
- Medical Devices; Radiology Devices; Classification of the Radiological Computer Aided Triage and Notification Software. Available online: https://www.federalregister.gov/documents/2020/01/22/2020-00496/medical-devices-radiology-devices-classification-of-the-radiological-computer-aided-triage (accessed on 3 May 2025).
- Electronic Code of Federal Regulations. U.S. Government Publishing Office. Code of Federal Regulations—21 CFR 892.2080—Radiological Computer Aided Triage and Notification Software. Available online: https://www.ecfr.gov/current/title-21/part-892/section-892.2080 (accessed on 3 May 2025).
- European Commission MDCG 2021-24—Guidance on Classification of Medical Devices—Annex VIII Rule 11. Available online: https://health.ec.europa.eu/latest-updates/mdcg-2021-24-guidance-classification-medical-devices-2021-10-04_en (accessed on 3 May 2025).
- Demner-Fushman, D.; Kohli, M.D.; Rosenman, M.B.; Shooshan, S.E.; Rodriguez, L.; Antani, S.; Thoma, G.R.; McDonald, C.J. OpenI Indiana Dataset: Preparing a Collection of Radiology Examinations for Distribution and Retrieval. J. Am. Med. Inform. Assoc. 2016, 23, 304–310. [Google Scholar] [CrossRef]
- Wang, X.; Peng, Y.; Lu, L.; Lu, Z.; Bagheri, M.; Summers, R.M. ChestX-Ray8: Hospital-Scale Chest X-Ray Database and Benchmarks on Weakly-Supervised Classification and Localization of Common Thorax Diseases. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition, CVPR 2017, Honolulu, HI, USA, 21–26 July 2017; pp. 3462–3471. [Google Scholar] [CrossRef]
- Irvin, J.; Rajpurkar, P.; Ko, M.; Yu, Y.; Ciurea-Ilcus, S.; Chute, C.; Marklund, H.; Haghgoo, B.; Ball, R.; Shpanskaya, K.; et al. CheXpert: A Large Chest Radiograph Dataset with Uncertainty Labels and Expert Comparison. arXiv 2019, arXiv:1901.07031. [Google Scholar] [CrossRef]
- Johnson, A.E.W.; Pollard, T.J.; Berkowitz, S.J.; Greenbaum, N.R.; Lungren, M.P.; Deng, C.; Mark, R.G.; Horng, S. MIMIC-CXR: A Large Publicly Available Database of Labeled Chest Radiographs. arXiv 2019, arXiv:1901.07042. [Google Scholar] [CrossRef]
- Bustos, A.; Pertusa, A.; Salinas, J.-M.; de la Iglesia-Vayá, M. PadChest: A Large Chest x-Ray Image Dataset with Multi-Label Annotated Reports. arXiv 2019, arXiv:1901.07441. [Google Scholar] [CrossRef]
- Nguyen, H.Q.; Lam, K.; Le, L.T.; Pham, H.H.; Tran, D.Q.; Nguyen, D.B.; Le, D.D.; Pham, C.M.; Tong, H.T.T.; Dinh, D.H.; et al. VinDr-CXR: An Open Dataset of Chest X-Rays with Radiologist’s Annotations. Sci. Data 2022, 9, 429. [Google Scholar] [CrossRef]
- Oakden-Rayner, L. Exploring Large Scale Public Medical Image Datasets. Acad. Radiol. 2019, 27, 106–112. [Google Scholar] [CrossRef] [PubMed]
- Chambon, P.; Delbrouck, J.-B.; Sounack, T.; Huang, S.-C.; Chen, Z.; Varma, M.; Truong, S.Q.; Chuong, C.T.; Langlotz, C.P. CheXpert Plus: Augmenting a Large Chest X-Ray Dataset with Text Radiology Reports, Patient Demographics and Additional Image Formats. arXiv 2024, arXiv:2405.19538. [Google Scholar] [CrossRef]
- Oakden-Rayner, L.; Dunnmon, J.; Carneiro, G.; Ré, C. Hidden Stratification Causes Clinically Meaningful Failures in Machine Learning for Medical Imaging. arXiv 2019, arXiv:1909.12475. [Google Scholar] [CrossRef]
- Majkowska, A.; Mittal, S.; Steiner, D.F.; Reicher, J.J.; McKinney, S.M.; Duggan, G.E.; Eswaran, K.; Cameron Chen, P.-H.; Liu, Y.; Kalidindi, S.R.; et al. Chest Radiograph Interpretation with Deep Learning Models: Assessment with Radiologist-Adjudicated Reference Standards and Population-Adjusted Evaluation. Radiology 2019, 294, 421–431. [Google Scholar] [CrossRef] [PubMed]
- Nabulsi, Z.; Sellergren, A.; Jamshy, S.; Lau, C.; Santos, E.; Ye, W.; Yang, J.; Pilgrim, R.; Kazemzadeh, S.; Yu, J.; et al. Deep Learning for Distinguishing Normal versus Abnormal Chest Radiographs and Generalization to Unseen Diseases. Nat. Sci. Rep. 2021. [Google Scholar] [CrossRef]
- Damgaard, C.; Eriksen, T.N.; Juodelyte, D.; Cheplygina, V.; Jiménez-Sánchez, A. Augmenting Chest X-Ray Datasets with Non-Expert Annotations. arXiv 2023, arXiv:2309.02244. [Google Scholar] [CrossRef]
- Cheplygina, V.; Cathrine, D.; Eriksen, T.N.; Jiménez-Sánchez, A. NEATX: Non-Expert Annotations of Tubes in X-Rays. Zenodo 2025. [Google Scholar] [CrossRef]
- Hallinan, J.T.P.D.; Feng, M.; Ng, D.; Sia, S.Y.; Tiong, V.T.Y.; Jagmohan, P.; Makmur, A.; Thian, Y.L. Detection of Pneumothorax with Deep Learning Models: Learning From Radiologist Labels vs Natural Language Processing Model Generated Labels. Acad. Radiol. 2022, 29, 1350–1358. [Google Scholar] [CrossRef] [PubMed]
- Filice, R.W.; Stein, A.; Wu, C.C.; Arteaga, V.A.; Borstelmann, S.; Gaddikeri, R.; Galperin-Aizenberg, M.; Gill, R.R.; Godoy, M.C.; Hobbs, S.B.; et al. Crowdsourcing Pneumothorax Annotations Using Machine Learning Annotations on the NIH Chest X-Ray Dataset. J. Digit. Imaging 2020, 33, 490–496. [Google Scholar] [CrossRef]
- Goldberger, A.L.; Amaral, L.A.; Glass, L.; Hausdorff, J.M.; Ivanov, P.C.; Mark, R.G.; Mietus, J.E.; Moody, G.B.; Peng, C.K.; Stanley, H.E. PhysioBank, PhysioToolkit, and PhysioNet: Components of a New Research Resource for Complex Physiologic Signals. Circulation 2000, 101, E215–E220. [Google Scholar] [CrossRef] [PubMed]
- Pham, H.H.; Tran, T.T.; Nguyen, H.Q. VinDr-PCXR: An Open, Large-Scale Pediatric Chest X-Ray Dataset for Interpretation of Common Thoracic Diseases (Ver-sion 1.0.0). PhysioNet. RRID:SCR_007345. 2022. Available online: https://doi.org/10.13026/k8qc-na36 (accessed on 26 June 2025).
- Fan, W.; Yang, Y.; Qi, J.; Zhang, Q.; Liao, C.; Wen, L.; Wang, S.; Wang, G.; Xia, Y.; Wu, Q.; et al. A Deep-Learning-Based Framework for Identifying and Localizing Multiple Abnormalities and Assessing Cardiomegaly in Chest X-Ray. Nat. Commun. 2024, 15, 1347. [Google Scholar] [CrossRef]
- Feng, S. CANDID-II Dataset. 2022. Available online: https://doi.org/10.17608/k6.auckland.19606921.v1 (accessed on 26 June 2025).
- Stanford Center for AI in Medicine & Imaging. CheXpert Dataset. Available online: https://doi.org/10.71718/y7pj-4v93 (accessed on 28 May 2025).
- Johnson, A.; Pollard, T.; Mark, R.; Berkowitz, S.; Horng, S. MIMIC-CXR Database (Version 2.1.0). PhysioNet. RRID:SCR_007345. 2024. Available online: https://doi.org/10.13026/4jqj-jw95 (accessed on 26 June 2025).
- Nguyen, H.Q.; Pham, H.H.; Tuan Linh, L.; Dao, M.; Khanh, L. VinDr-CXR: An Open Dataset of Chest X-Rays with Radiologist Annotations (Version 1.0.0). PhysioNet. RRID:SCR_007345. 2021. Available online: https://doi.org/10.13026/3akn-b287 (accessed on 26 June 2025).
- SIIM-ACR Pneumothorax Segmentation. Available online: https://kaggle.com/competitions/siim-acr-pneumothorax-segmentation (accessed on 5 March 2023).
- Reis, E.P.; de Paiva, J.P.Q.; da Silva, M.C.B.; Ribeiro, G.A.S.; Paiva, V.F.; Bulgarelli, L.; Lee, H.M.H.; Santos, P.V.; Brito, V.M.; Amaral, L.T.W.; et al. BRAX, Brazilian Labeled Chest x-Ray Dataset. Sci. Data 2022, 9, 487. [Google Scholar] [CrossRef]
- Reis, E.P.; Paiva, J.; Bueno da Silva, M.C.; Sousa Ribeiro, G.A.; Fornasiero Paiva, V.; Bulgarelli, L.; Lee, H.; dos Santos, P.V.; brito v Amaral, L.; Beraldo, G.; et al. BRAX, a Brazilian Labeled Chest X-Ray Dataset (Version 1.1.0). PhysioNet. RRID:SCR_007345. 2022. Available online: https://doi.org/10.13026/grwk-yh18 (accessed on 26 June 2025).
- Indiana University; U.S. National Library of Medicine. Open-I: Open Access Biomedical Image Search Engine. Available online: https://openi.nlm.nih.gov/faq (accessed on 28 May 2025).
- Feng, S.; Azzollini, D.; Kim, J.S.; Jin, C.-K.; Gordon, S.P.; Yeoh, J.; Kim, E.; Han, M.; Lee, A.; Patel, A.; et al. Curation of the CANDID-PTX Dataset with Free-Text Reports. Radiol. Artif. Intell. 2021, 3, e210136. [Google Scholar] [CrossRef] [PubMed]
- CANDID-PTX Dataset. 2021. Available online: https://doi.org/10.17608/k6.auckland.14173982 (accessed on 28 May 2025).
- Wang, Y.; Wang, K.; Peng, X.; Shi, L.; Sun, J.; Zheng, S.; Shan, F.; Shi, W.; Liu, L. DeepSDM: Boundary-Aware Pneumothorax Segmentation in Chest X-Ray Images. Neurocomputing 2021, 454, 201–211. [Google Scholar] [CrossRef]
- Wang, Y. PTX-498: A Multi-Center Pneumothorax Segmentation Chest X-Ray Image Dataset. Zenodo 2021. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, M.; Hu, S.; Shen, Y.; Lan, J.; Jiang, B.; de Bock, G.H.; Vliegenthart, R.; Chen, X.; Xie, X. Development and Multicenter Validation of Chest X-Ray Radiography Interpretations Based on Natural Language Processing. Commun. Med. 2021, 1, 1–12. [Google Scholar] [CrossRef]
- Liu, M.; Xie, X. Chest Radiograph at Diverse Institutes (CRADI) Dataset. Zenodo 2021. [Google Scholar] [CrossRef]
- Development of a Digital Image Database for Chest Radiographs With and Without a Lung Nodule. Available online: https://www.ajronline.org/doi/epdf/10.2214/ajr.174.1.1740071 (accessed on 28 May 2025).
- Gohagan, J.K.; Prorok, P.C.; Greenwald, P.; Kramer, B.S. The PLCO Cancer Screening Trial: Background, Goals, Organization, Operations, Results. Rev. Recent Clin. Trials 2015, 10, 173–180. [Google Scholar] [CrossRef] [PubMed]
- National Lung Screening Trial Research Team Data from the National Lung Screening Trial (NLST). 2013. Available online: https://doi.org/10.7937/TCIA.HMQ8-J677 (accessed on 26 June 2025).
- Liu, Y.; Wu, Y.-H.; Ban, Y.; Wang, H.; Cheng, M.-M. TBX11K: Rethinking Computer-Aided Tuberculosis Diagnosis. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 13–19 June 2020; pp. 2646–2655. [Google Scholar] [CrossRef]
- Jaeger, S.; Candemir, S.; Antani, S.; Wáng, Y.-X.J.; Lu, P.-X.; Thoma, G. Two Public Chest X-Ray Datasets for Computer-Aided Screening of Pulmonary Diseases. Quant. Imaging Med. Surg. 2014, 4, 475–477. [Google Scholar] [CrossRef]
- Ogawa, R.; Kido, T.; Kido, T.; Mochizuki, T. Effect of Augmented Datasets on Deep Convolutional Neural Networks Applied to Chest Radiographs. Clin. Radiol. 2019, 74, 697–701. [Google Scholar] [CrossRef] [PubMed]
- Tan, M.; Le, Q.V. EfficientNet: Rethinking Model Scaling for Convolutional Neural Networks. arXiv 2020, arXiv:1905.11946. [Google Scholar] [CrossRef]
- Huang, G.; Liu, Z.; van der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. arXiv 2016, arXiv:1608.06993. [Google Scholar] [CrossRef]
- Thian, Y.L.; Ng, D.; Hallinan, J.T.P.D.; Jagmohan, P.; Sia, S.Y.; Tan, C.H.; Ting, Y.H.; Kei, P.L.; Pulickal, G.G.; Tiong, V.T.Y.; et al. Deep Learning Systems for Pneumothorax Detection on Chest Radiographs: A Multicenter External Validation Study. Radiol. Artif. Intell. 2021, 3, e200190. [Google Scholar] [CrossRef] [PubMed]
- Haque, M.I.U.; Dubey, A.K.; Danciu, I.; Justice, A.C.; Ovchinnikova, O.S.; Hinkle, J.D. Effect of Image Resolution on Automated Classification of Chest X-Rays. J. Med. Imaging 2023, 10, 044503. [Google Scholar] [CrossRef]
- Pereira, S.C.; Rocha, J.; Campilho, A.; Sousa, P.; Mendonça, A.M. Lightweight Multi-Scale Classification of Chest Radiographs via Size-Specific Batch Normalization. Comput. Methods Programs Biomed. 2023, 236, 107558. [Google Scholar] [CrossRef]
- Wollek, A.; Hyska, S.; Sabel, B.; Ingrisch, M.; Lasser, T. Higher Chest X-Ray Resolution Improves Classification Performance. arXiv 2023, arXiv:2306.06051. [Google Scholar] [CrossRef]
- Comparison of Fine-Tuning Strategies for Transfer Learning in Medical Image Classification. Available online: https://arxiv.org/html/2406.10050v1 (accessed on 2 April 2025).
- Mosquera, C.; Ferrer, L.; Milone, D.H.; Luna, D.; Ferrante, E. Class Imbalance on Medical Image Classification: Towards Better Evaluation Practices for Discrimination and Calibration Performance. Eur. Radiol. 2024, 34, 7895–7903. [Google Scholar] [CrossRef]
- Cohen, J.P.; Hashir, M.; Brooks, R.; Bertrand, H. On the Limits of Cross-Domain Generalization in Automated X-Ray Prediction. arXiv 2020, arXiv:2002.02497. [Google Scholar] [CrossRef]
- Rueckel, J.; Huemmer, C.; Fieselmann, A.; Ghesu, F.-C.; Mansoor, A.; Schachtner, B.; Wesp, P.; Trappmann, L.; Munawwar, B.; Ricke, J.; et al. Pneumothorax Detection in Chest Radiographs: Optimizing Artificial Intelligence System for Accuracy and Confounding Bias Reduction Using in-Image Annotations in Algorithm Training. Eur. Radiol. 2021, 31, 7888–7900. [Google Scholar] [CrossRef] [PubMed]
- Pooch, E.H.P.; Ballester, P.L.; Barros, R.C. Can We Trust Deep Learning Models Diagnosis? The Impact of Domain Shift in Chest Radiograph Classification. arXiv 2019, arXiv:1909.01940. [Google Scholar] [CrossRef]
- Bercean, B.; Buburuzan, A.; Birhala, A.; Avramescu, C.; Tenescu, A.; Marcu, M. Breaking Down Covariate Shift on Pneumothorax Chest X-Ray Classification. In Proceedings of the Uncertainty for Safe Utilization of Machine Learning in Medical Imaging; Sudre, C.H., Baumgartner, C.F., Dalca, A., Mehta, R., Qin, C., Wells, W.M., Eds.; Springer Nature: Cham, Switzerland, 2023; pp. 157–166. [Google Scholar]
- Jiménez-Sánchez, A.; Juodelyte, D.; Chamberlain, B.; Cheplygina, V. Detecting Shortcuts in Medical Images—A Case Study in Chest X-Rays. In Proceedings of the 2023 IEEE 20th International Symposium on Biomedical Imaging (ISBI), Cartagena, Colombia, 18–21 April 2023; pp. 1–5. [Google Scholar]
- Seah, J.; Tang, C.; Buchlak, Q.D.; Milne, M.R.; Holt, X.; Ahmad, H.; Lambert, J.; Esmaili, N.; Oakden-Rayner, L.; Brotchie, P.; et al. Do Comprehensive Deep Learning Algorithms Suffer from Hidden Stratification? A Retrospective Study on Pneumothorax Detection in Chest Radiography. BMJ Open 2021, 11, e053024. [Google Scholar] [CrossRef] [PubMed]
- Rajpurkar, P.; Irvin, J.; Zhu, K.; Yang, B.; Mehta, H.; Duan, T.; Ding, D.; Bagul, A.; Langlotz, C.; Shpanskaya, K.; et al. CheXNet: Radiologist-Level Pneumonia Detection on Chest X-Rays with Deep Learning. arXiv 2017, arXiv:1711.05225. [Google Scholar]
- Taylor, A.G.; Mielke, C.; Mongan, J. Automated Detection of Moderate and Large Pneumothorax on Frontal Chest X-Rays Using Deep Convolutional Neural Networks: A Retrospective Study. PLOS Med. 2018, 15, e1002697. [Google Scholar] [CrossRef]
- Gündel, S.; Grbic, S.; Georgescu, B.; Liu, S.; Maier, A.; Comaniciu, D. Learning to Recognize Abnormalities in Chest X-Rays with Location-Aware Dense Networks. In Progress in Pattern Recognition, Image Analysis, Computer Vision, and Applications: CIARP 2018; Lecture Notes in Computer Science (LNIP, Volume 11401); Springer: Cham, Switzerland; pp. 757–765. [CrossRef]
- Cid, Y.D.; Macpherson, M.; Gervais-Andre, L.; Zhu, Y.; Franco, G.; Santeramo, R.; Lim, C.; Selby, I.; Muthuswamy, K.; Amlani, A.; et al. Development and Validation of Open-Source Deep Neural Networks for Comprehensive Chest x-Ray Reading: A Retrospective, Multicentre Study. Lancet Digit. Health 2024, 6, e44–e57. [Google Scholar] [CrossRef]
- Wang, C.-H.; Lin, T.; Chen, G.; Lee, M.-R.; Tay, J.; Wu, C.-Y.; Wu, M.-C.; Roth, H.R.; Yang, D.; Zhao, C.; et al. Deep Learning-Based Diagnosis and Localization of Pneumothorax on Portable Supine Chest X-Ray in Intensive and Emergency Medicine: A Retrospective Study. J. Med. Syst. 2023, 48, 1. [Google Scholar] [CrossRef]
- Hillis, J.M.; Bizzo, B.C.; Mercaldo, S.; Chin, J.K.; Newbury-Chaet, I.; Digumarthy, S.R.; Gilman, M.D.; Muse, V.V.; Bottrell, G.; Seah, J.C.Y.; et al. Evaluation of an Artificial Intelligence Model for Detection of Pneumothorax and Tension Pneumothorax in Chest Radiographs. JAMA Netw. Open 2022, 5, e2247172. [Google Scholar] [CrossRef]
- Feng, S.; Liu, Q.; Patel, A.; Bazai, S.U.; Jin, C.-K.; Kim, J.S.; Sarrafzadeh, M.; Azzollini, D.; Yeoh, J.; Kim, E.; et al. Automated Pneumothorax Triaging in Chest X-Rays in the New Zealand Population Using Deep-Learning Algorithms. J. Med. Imaging Radiat. Oncol. 2022, 66, 1035–1043. [Google Scholar] [CrossRef]
- Sze-To, A.; Riasatian, A.; Tizhoosh, H.R. Searching for Pneumothorax in X-Ray Images Using Autoencoded Deep Features. Sci. Rep. 2021, 11, 9817. [Google Scholar] [CrossRef] [PubMed]
- Wollek, A.; Graf, R.; Čečatka, S.; Fink, N.; Willem, T.; Sabel, B.O.; Lasser, T. Attention-Based Saliency Maps Improve Interpretability of Pneumothorax Classification. Radiol. Artif. Intell. 2023, 5, e220187. [Google Scholar] [CrossRef] [PubMed]
- Arun, N.; Gaw, N.; Singh, P.; Chang, K.; Aggarwal, M.; Chen, B.; Hoebel, K.; Gupta, S.; Patel, J.; Gidwani, M.; et al. Assessing the Trustworthiness of Saliency Maps for Localizing Abnormalities in Medical Imaging. Radiol. Artif. Intell. 2021, 3, e200267. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Yin, X.; Zhang, T.; Feng, Y.; Zhao, Y.; Jin, M.; Peng, M.; Xing, C.; Li, F.; Wang, Z.; et al. Detection and Semiquantitative Analysis of Cardiomegaly, Pneumothorax, and Pleural Effusion on Chest Radiographs. Radiol. Artif. Intell. 2021, 3, e200172. [Google Scholar] [CrossRef]
- David, R.; Duke, J.; Jain, A.; Reddi, V.J.; Jeffries, N.; Li, J.; Kreeger, N.; Nappier, I.; Natraj, M.; Wang, T.; et al. TensorFlow Lite Micro: Embedded Machine Learning for TinyML Systems. Proc. Mach. Learn. Syst. 2021, 3, 800–811. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).