Abstract
Human endogenous retrovirus group E (HERV-E) elements are stably integrated into the human genome, transmitted vertically in a Mendelian manner, and are endowed with transcriptional activity as alternative promoters or enhancers. Such effects are under the control of the proviral long terminal repeats (LTR) that are organized into three HERV-E phylogenetic subgroups, namely LTR2, LTR2B, and LTR2C. Moreover, HERV-E expression is tissue-specific, and silenced by epigenetic constraints that may be disrupted in cancer, autoimmunity, and human placentation. Interest in HERV-E with regard to these conditions has been stimulated further by concerns regarding the capacity of HERV-E elements to modify the expression of neighboring genes and/or to produce retroviral proteins, including immunosuppressive env peptides, which in turn may induce (auto)-antibody (Ab) production. Finally, better understanding of HERV-E elements may have clinical applications for prevention, diagnosis, prognosis, and therapy.
1. Introduction
Human endogenous retroviruses (HERV) are descendants of occasional germline invasion by exogenous retroviruses. HERVs occupy as much as 8% of the ~3 billion base pairs (bp) of the human genome [1,2]. Such elements, distributed in ~400,000 loci and transmitted vertically in a Mendelian manner, are categorized into 30–40 families depending on the method of classification [3,4]. Each family encompasses tens to thousands of loci [5], and several criteria are used for HERV classification, including (i) the single letter amino acid tRNA primer binding site used by the provirus; (ii) the similarity with animal retroviruses, which divides them into three classes (class I: gammaretrovirus; class II: betaretrovirus; and class III: spuma(retro)virus); and (iii) the molecular genetic means. Needless to say, a systematic nomenclature is necessary for naming all the retroviral elements present in the genome, but such a consensus nomenclature has not yet been established [6,7].
In their original form, HERVs possess two unique (U)3-repeat(R)-U5 long terminal repeats (LTR) present at the extremities, and flanking the three HERV genes: the group-specific antigen (gag), reverse transcriptase/polymerase (pol) and envelope (env) genes. The HERV gag, pol and env genes, when present, have the particularity of being disrupted by frame shift mutations, stop codons and/or deletions that affect their retroviral replicative capacity as well as their ability to transpose and to create de novo insertional mutations. Similarly, the regulatory sequences present in the LTRs have accumulated mutations, and most of them are transcriptionally silenced by epigenetic constraints [8].
This review will focus on the HERV group E family (HERV-E), which uses tRNAGlu and which is related to class I gammaretrovirus based on homology with the Moloney Murine Leukaemia Virus (MoMuLV) pol, gag and env genes [9]. Interest in HERV-E elements has been stimulated further by concerns regarding their occurrence and implication in malignancies, autoimmune diseases, and human placentation.
2. The HERV-E Family
2.1. HERV-E Phylogeny
More than 1,300 HERV-E elements have been described in the human genome (DFAM 1.3 release December 2014, [7,10], and these elements are subdivided into three distinct subgroups referred to as LTR2 (n = 811, 60%), LTR2B (n = 302, 23%) and LTR2C (n = 223, 17%) based on divergences observed between sequences of the 5’LTR part of the HERV-E [11]. Similarities between the three subgroups range from 75% (LTR2B/LTR2C), to 81% (LTR2/LTR2C), and 83% (LTR2/LTR2B). To go further in the exploration of these three subgroups, we have updated our initial 5’LTR HERV-E phylogenic tree [12] with new sequences in order to build a new phylogenic tree containing 16/46 (34.8%) LTR2 sequences, 21/46 (45.6%) LTR2B sequences, and 9/46 (19.6%) LTR2C sequences (Table 1 and Figure 1).

Table 1.
Description of the 46 HERV-E elements explored (see Experimental Section).
| Locus | HERV Name | Subgroup | 5’LTR Genomic Location | 3’LTR Genomic Location | Size (bp) | Structure | 5’ and 3’LTR Divergences | Molecular Clock Dating | EPO Alignment Dating | Genomic Amplification |
|---|---|---|---|---|---|---|---|---|---|---|
| 19q13.32 | HERV-E.ApoC1 | LTR2 | 19:44913964-44914947 | - | 987 | solo LTR | - | - | >15 My | >15 My [ 65] |
| 3q22.2 | - | LTR2 | 3:134027135-134027667 | 3:134019829-134020359 | 7839 | gag, pol truncated | 9.5% | 32 My | >15 My | |
| 8q12.1 | - | LTR2 | 8:58690826-58691341 | 8:58698386-58698895 | 8069 | gag, pol truncated | 7.0% | 23 My | >15 My | |
| 13q22.3 | HERV-E.EDNRB | LTR2 | 13:77975613-77976074 | 13:77975589-77976108 | 5777 | gag, pol, env truncated | 49.7% | >50 My | >25 My | >25 My [ 65] |
| 17q21.31 | HERV-E.BRCA1 | LTR2 | 17:43160221-43160752 | 17:43167223-43167735 | 7514 | gag, pol truncated | 9.4% | 31 My | >15 My | |
| 7p22.1 | - | LTR2 | 7:100919594-100920108 | - | 514 | solo LTR | - | - | >15 My | |
| 3p21.33 | LTR2 | 3:43678285-34678772 | - | 487 | solo LTR | - | - | >15 My | ||
| 6q22.31 | HERV-E.FABP7 | LTR2 | 6:122748781-122749289 | 6:122741492-122741993 | 7752 | pol truncated | 9.0% | 30 My | >25 My | |
| 11q12.2 | HERV-E.CD5 | LTR2 | 11:61093564-61094073 | 11:61098368-61098863 | 5254 | pol, env truncated | 7.8% | 26 My | >25 My | > 25 My [ 11] |
| 4q35.1 | - | LTR2 | 4:183377788-183378289 | - | 501 | solo LTR | - | - | >25 My | |
| 2q11.2 | - | LTR2 | 2:101186575-101186124 | - | 451 | solo LTR | - | - | - | |
| 17q24.3 | - | LTR2 | 17:71020856-71021358 | - | 502 | solo LTR | - | - | >15 My | |
| 4q28.2 | - | LTR2 | 4:128703926-128704424 | - | 498 | solo LTR | - | - | >25 My | |
| 1q44 | - | LTR2 | 1:247127397-247127939 | - | 542 | Solo LTR | 12.3% | 41 My | >15 My | |
| 19q13.43 | - | LTR2 | 19:57815303-57815707 | - | 1163 | solo LTR | - | - | >25 My | |
| 12p12.2 | - | LTR2 | 12:20944374-20944873 | - | 499 | solo LTR | - | - | >15 My | |
| 10q23.1 | - | LTR2B | 10:84172465-84171987 | - | 997 | solo LTR | - | - | >15 My | |
| 6q21.33 | LTR2B | 6:31186291-31186769 | - | 478 | solo LTR | - | - | >8 My | ||
| 5q31.2 | HERV-E.UBE2D2 | LTR2B | 5:139525866-139526346 | 5:139531087-139531583 | 5644 | gag, pol truncated | 5.2% | 17 My | >15 My | |
| 1p21 | HERV-E-AMY1B | LTR2B | 1:103696448-103696893 | 1:103703817-103704285 | 7837 | full length | 11.9% | 40 My | >6 My | |
| 2p22.3 | - | LTR2B | 2:34677243-34677662 | - | 888 | solo LTR | - | - | >15 My | |
| 1q24.2 | HERV-E.IQWD1 (DCAF6, PC326) | LTR2B | 1:167863096-167863572 | 1:167869427-167869928 | 6832 | gag, pol truncated | 6.9% | 23 My | >15 My | |
| 11q13.4 | - | LTR2B | 11:73271302-73271710 | - | 408 | solo LTR | - | - | >15 My | |
| 15q14 | - | LTR2B | 15:34812862-34813351 | - | 489 | solo LTR | - | - | >15 My | |
| 6q23.1 | - | LTR2B | 6:130260127-130260610 | 6:130267508-130267970 | 7843 | gag, pol truncated | 9.7% | 32 My | >15 My | |
| 7p12.3 | - | LTR2B | 7:46031148-46031644 | - | 496 | solo LTR | - | - | >15 My | |
| 6p21.31 | - | LTR2B | 6:35004307-35003824 | - | 483 | solo LTR | - | - | >15 My | |
| Xp22.22 | HERV-E.MID1 | LTR2B | X:10585282-10584793 | X:10590339-10589853 | 5,546 | gag, pol, env truncated | 6.4% | 20 My | >15 My | > 25 My [ 55] |
| 16q24.3 | - | LTR2B | 16:90056524-90057012 | - | 488 | solo LTR | - | - | >15 My | |
| 21q22.3 | - | LTR2B | 21:42807398-42807919 | - | 521 | solo LTR | - | - | >15 My | |
| 3q11.2 | - | LTR2B | 3:97856881-97857316 | - | 435 | solo LTR | - | - | - | |
| 7q33 | HERV-E.PTN | LTR2B | 7:136947291-136947711 | 7:137262486-137263044 | 6,360 | gag, pol, env truncated | 7.9% | 26 My | >15 My | >15 My [ 11,20,40] |
| 18q21.33 | - | LTR2B | 18:61617742-61618212 | - | 470 | solo LTR | - | - | - | |
| 1q24.2 | LTR2B | 1:167869414-167869892 | - | 478 | solo LTR | - | - | >15 My | ||
| 5p13.1 | - | LTR2B | 5:40872315-40872812 | - | 497 | solo LTR | - | - | > 15 My | |
| Yq11.21 | - | LTR2B | Y:12245468-12245973 | Y:12251519-12252038 | 6571 | gag truncated | 14.4% | 48 My | - | |
| 3p25.3 | - | LTR2B | 3:9590155-9590642 | - | 487 | solo LTR | - | - | > 25 My | |
| 6q15 | CT-RCC HERV-E | LTR2C | 6:88662137-88662613 | 6:88670478-88670972 | 8792 | full length | 5.8% | 19 My | > 15 My | |
| 2q37.1 | - | LTR2C | 2:231400694-231401230 | 2:231408328-231408867 | 8173 | full length | 2.9% | 10 My | > 15 My | |
| 2q23.1 | - | LTR2C | 2:149035320-149035863 | - | 543 | solo LTR | - | - | > 8 My | |
| 12p13.31 | - | LTR2C | 12:7703612-7704158 | - | 546 | solo LTR | - | - | > 8 My | |
| 11q13.31 | - | LTR2C | 11:66000626-66001172 | - | 546 | solo LTR | - | - | > 8 My | |
| 2q14.3 | - | LTR2C | 2:127683400-127683948 | - | 548 | solo LTR | - | - | > 8 My | |
| 19p12 | HERV-E clone 4-1 | LTR2C | 19:20755113-20755654 | 19:20746795-20747340 | 8,806 | full length | 4.5% | 15 My | > 8 My | |
| 8p12 | - | LTR2C | 8:30728558-30729101 | - | 5,121 | env, 3’LTR truncated | - | - | > 8 My | |
| 7p14.3 | - | LTR2C | 7:32762447-32762988 | - | 541 | solo LTR | - | - | > 6 My |
EPO: Enredo-Pecan-Ortheus whole genome primate alignment tool from ensembl.
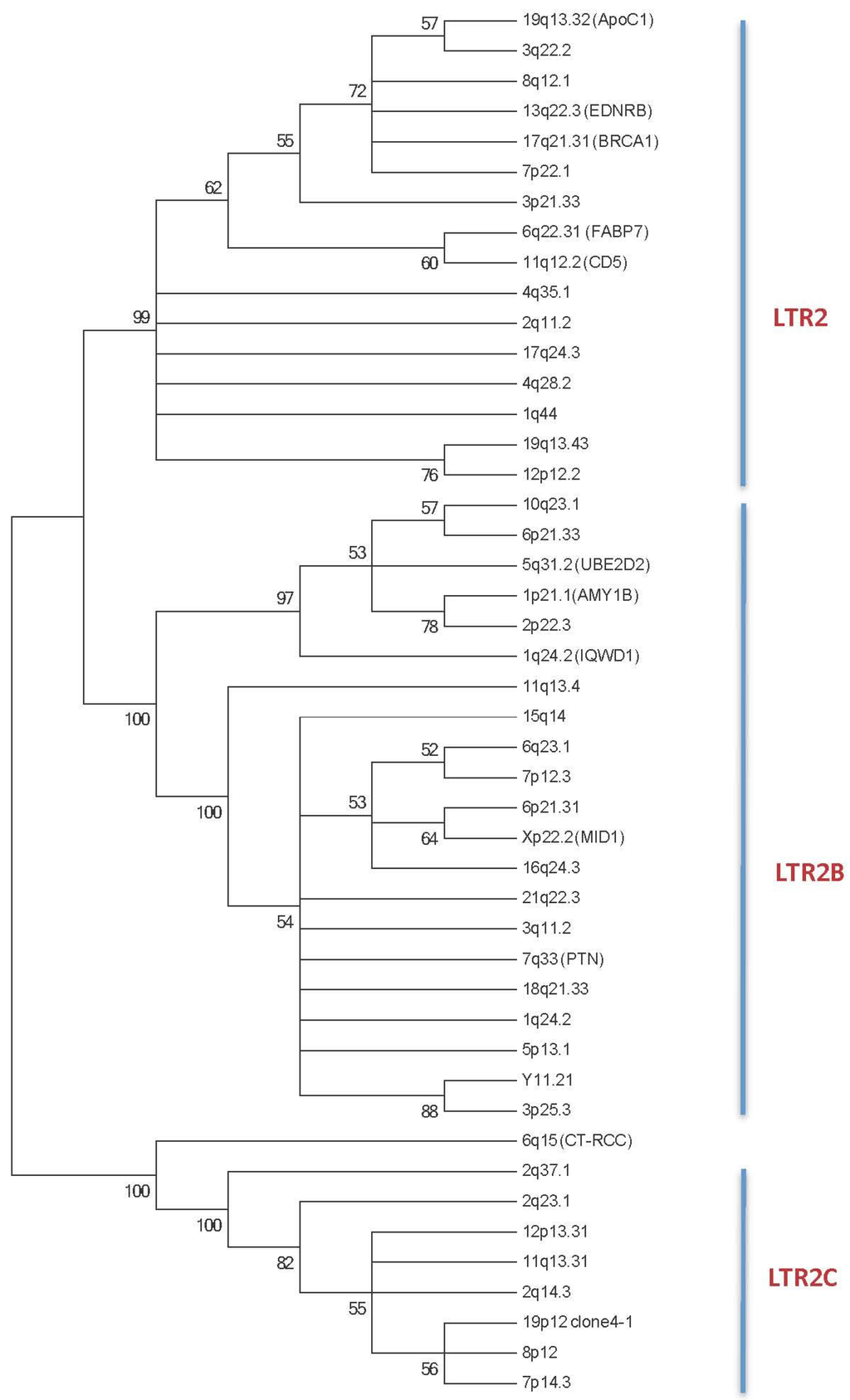
Figure 1.
The phylogenetic tree of the 5’ LTR HERV-E sequences reveals three main subgroups: LTR2, LTR2B and LTR2C. Phylogenetic analyses were inferred using the Maximum Parsimony Methods on the ClustalW Multiple alignment of 46 HERV-E 5’LTR nucleotide sequences. The most parsimonious tree is shown; it shows that the 46 HERV-E LTRs are segregated into 3 distinct subgroups. Bootstrap values greater than 50% are shown. The 5’ HERV-E LTR sequences are identified with their respective human chromosomal locus location (See Experimental Section plus Table 1). When known, the gene used for alternative transcript usage is reported.
2.1.1. HERV-E Subgroups and Integration Time
Using both the phylogenic approach by genomic amplification with different primates, from New World monkeys to humans, and the molecular clock approach, Li et al. have established two waves in HERV-E lineage integration [13]. The molecular clock approach is used to date the age of a HERV-E element insertion by applying an evolutionary mutation rate of ~0.3% mutation per million years (My) [13,14]. The first insertion occurred after the divergence of the New World primates from the Old World primates ~45 My ago, and a second insertion started ~25 My ago with the beginning of hominoid evolution.
Applied to our selection of HERV-E elements, we can complete Li’s observations and establish that HERV-E integration has occurred in several waves according to the LTR2 subgroup (for more information, see the Experimental Section). As summarized in Figure 2 and reported in Table 1, integration for LTR2 occurred predominantly before the divergence of hominoids from Old World monkeys, for LTR2B at the time of hominoid divergence, and in the case of LTR2C elements throughout hominoid evolution.
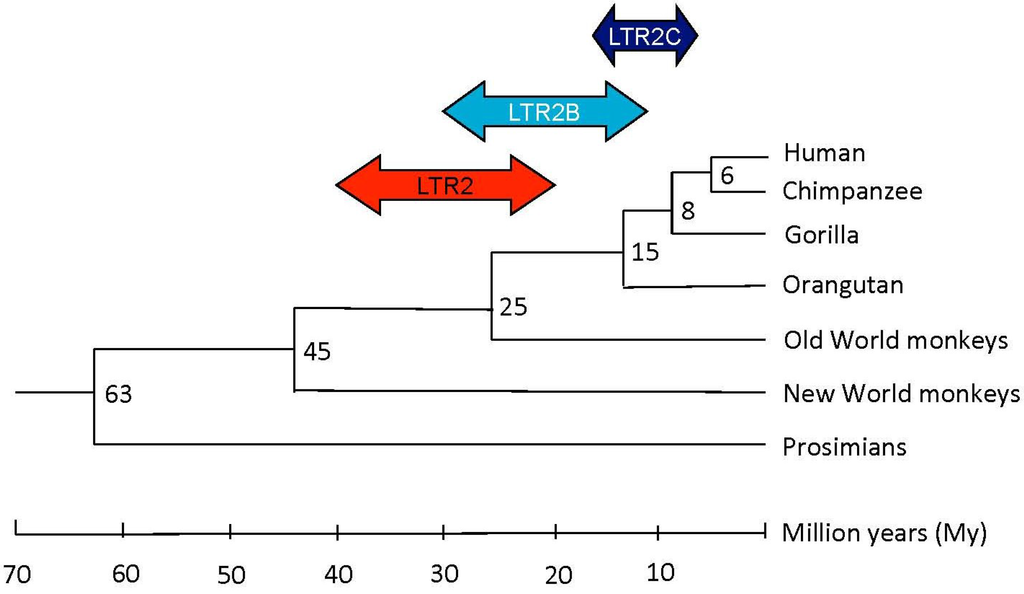
Figure 2.
Putative integration times of the LTR2, LTR2B, LTR2C HERV-E familly members during primate evolution. The evolutionary tree was adapted from Yi et al. [12,13].
2.1.2. HERV-E Subgroups and Structure
We and others have established that HERV-E elements are disseminated within the genome as solitary LTR elements, HERV-E gag, pol, env and/or 3’LTR deleted provirus, while full length and mutated HERV-E elements were detected in a few cases (e.g., HERV-E clone 4-1) [12,15,16]. With regards to our selection of HERV-E elements (Table 1), solitary LTRs (n = 29/46, 63.0%) represent the main group, followed by truncated provirus (n = 13/46, 28.3%), and full length HERV-Es (n = 4/46, 8.7%).
2.1.3. HERV-E Subgroups and Promoter Activity
Basically, the consensus sequences of LTR2 (463 bp, DF0000449), LTR2B (483 bp, DF0000484) and LTR2C (501 bp, DF0000485) [10] are composed of three parts: a transcriptional control U3 region, a transcription initiation site which defines the U3/R boundary at positions 298 to 337, and a poly-adenylation signal which defines the R/U5 boundary at positions 399 to 435.
The alignment analysis of the U3, R and U5 parts reveals important variations between LTR2, LTR2B and LTR2C with similarities ranging from 57.0% to 71.8% in U3; 73.5% to 87.8% in R; and 84.4% to 93.7% in U5. As a consequence, the differences observed in the regulatory U3 region between the three subgroups suggest distinct transcription binding sites and promoter activity between LTR2, LTR2B and LTR2C elements. This assertion is reinforced by the observation that placentally expressed LTR2B (HERV-E.PTN, HERV-E.MID1, and HERV-E PC326/IQWD1) shared >88.0% homology in U3, and that B-cell expressing LTR2 provirus (HERV-E.CD5 and HERV-E.FABP7) shared 94.0% homology in U3. Further analyses are required to characterize consensus transcription factor binding sites in each subgroup.
2.1.4. Chromosomal Integration and Consequences
Analysis of the DFAM 1.3 database for the presence of HERV-E in the human genome reveals HERV-E sequence integrations on all chromosomes with a preferential distal position. Furthermore, recent technological advances in sequencing and bioinformatics have further established a predominant HERV intergenic integration and an antisense orientation to the host gene containing the HERV with a ratio of 3:1 for HERV-E elements [17]. Such observations suggest that HERVs, when integrated in the same orientation and/or present in the introns, are more likely to be deleted to prevent a negative effect on the neighbouring or integrated gene.
Analysis of the human Expressed Sequence Tag (EST) database reveals an important proportion of HERVs and LTR elements in the transcriptome and, among them, HERV-Es are overrepresented and initiate 9.3% of the chimeric LTR transcripts when leukemic B cells from diffuse large B cell lymphoma are analyzed [18]. In general, when the HERV element is in the sense strand, upstream of the host promoter (exon 1) or in the first intron, the HERV element can act as an alternative promoter, splice with exon 2, and generate a fusion mRNA with a longer 5’ untranslated region (UTR). The splicing of the HERV element to a splice acceptor site in the host gene sequence could shift the open reading frame (ORF), yielding a transcript that cannot be translated into the functional host gene product as observed with HERV-E.CD5 (Figure 3). In contrast, when HERV elements are in the anti-sense strand relative to neighbouring host genes, the active HERV element could either disrupt expression of the host genes, dampening the host gene activity or act as a promoter (e.g., HERV-E.FABP7) or an enhancer (e.g., HERV-E.AMY1B [19,20]) to promote an alternative tissue expression. When the HERV element is in the body of a host gene, the HERV can provide additional splice donor site(s), acceptor site(s), or premature polyadenylation signals resulting in creation of incorrectly spliced and/or truncated variants.
Last but not least, HERV-E transcripts can also arise from the HERV-E element, and transcripts containing gag, pol and/or env genes may be expressed as observed with the HERV-E clone 4-1 [16]. The translated products from HERV-E genes could have antigenic potential (gag and env peptides), and/or immunosuppressive functions (env peptides).
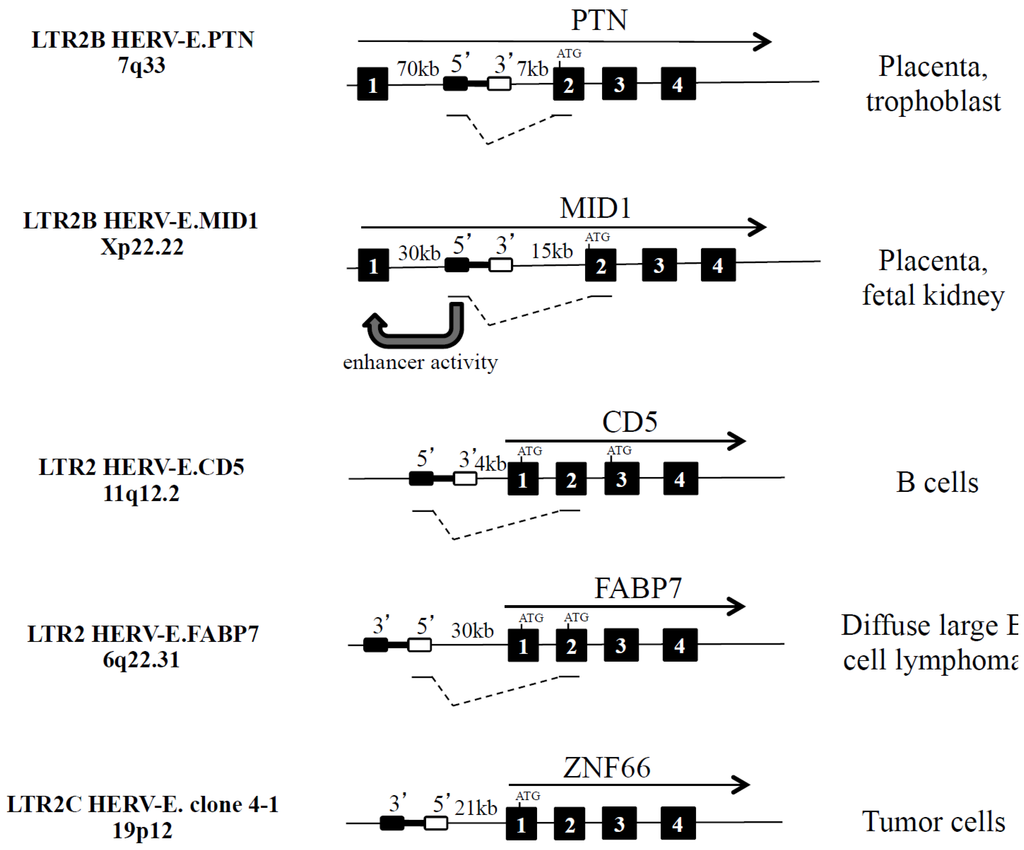
Figure 3.
Examples of five integrated HERV-Es in the vicinity of PTN, MID1, CD5, FABP7 and ZNF66 human genes. HERV-E is depicted as a black rectangle (5’LTR) linked to a white rectangle (3’LTR). The direction of gene transcription is indicated (black arrow). Black boxes refer to exons, and fusion transcripts are indicated.
2.2. Examples
2.2.1. Alternative Promoter: LTR2B HERV-E.PTN and LTR2B HERV-E.MID1
As indicated in Figure 3 and Table 2, both HERV-E.PTN and HERV-E.MID1 are integrated in the same orientation and upstream from the second exon of the secreted heparin-binding cytokine pleiotrophin (PTN) on one hand [21,22] and the microtubule associated protein Mid1 that targets the phosphatase 2A protein for degradation on the other hand [23]. Mid1 is involved in the pathogenesis of the X-linked form of Opitz syndrome when mutated [24]. For these two HERV-E elements, the presence of HERV-E pseudogene deletions (gag, pol and env) and an insertion of a truncated RTLV-Ib element (pol and env) were reported [21,22].
For the LTR-PTN and the LTR-Mid1 transcripts, the ORF, present in exon 2, is conserved, and as a consequence, differences are related to the acquisition of a cell-specific expression in the placenta. Furthermore, it was also observed that the 5’ LTR2B part of HERV-E.MID1 participates in the regulation of the non-retroviral Mid1 promoter as an enhancer in placental cells [24].

Table 2.
HERV-E integration influences human genes.
| Locus | Gene Name (Function) | Subgroup | Gene Location | Orientation | Transcripts | Chimeric Transcript | Protein Variant | Reference |
|---|---|---|---|---|---|---|---|---|
| 1p21 | AMY1B (salivary amylase) | LTR2 | Upstream | antisense | salivary | No (enhancer) | No | [18,19] |
| 1q24.2 | IQWD1/DCAF6 (nuclear receptor) | LTR2B | Upstream | sense | placenta | Yes | unknown | [11,40] |
| 7q33 | PTN (secreted heparin-binding cytokine) | LTR2B | Intron 1 | sense | Placenta, trophoblast | Yes | No | [11,20,40] |
| 5q31.2 | UBE2D2 (ubiquitin conjugating enzyme) | LTR2B | Upstream | sense | unknown | Yes | No | [11] |
| 11q12.2 | CD5 (T cell and B cell subset antigen) | LTR2 | Upstream | sense | B cells | Yes | Yes | [11,66] |
| 13q22.3 | EDNRB (G protein-coupled receptor of endothelin) | LTR2 | Upstream | sense | placenta | Yes | No | [11,40,65,67] |
| 19q13.32 | ApoC1 (lipoprotein metabolism) | LTR2 | Upstream | Liver, macrophage | Two variants | No | [65] | |
| 6q22.31 | FABP7 (brain lipid binding protein) | LTR2 | Upstream | antisense | B cell lymphoma | Yes | Yes | [17] |
| Xp22.22 | MID1 (microtubule-associated protein) | LTR2B | Intron 1 | sense | Placenta, fetal kidney | Yes | No | [11,23,40,67] |
2.2.2. Alternative Variants: HERV-E.CD5 and HERV-E.FABP7
The 5254 bp retroviral sequence HERV-E.CD5 is integrated into chromosome 11 at position 11q12.2 within the cd5 gene locus and 5 kilobases (kb) upstream of the host CD5 gene exon 1, in the same transcriptional direction, and downstream of the cd6 gene [25]. Within the two LTRs, the gag gene is present while the pol gene and the env gene, in part, are lacking. Using the molecular clock approach (5’ and 3’ LTR2 identity is 92.2%), and a panel of hominoids, Old World monkeys, and New World monkeys, we have established that HERV-E.CD5 integration occurred just prior to the divergence of hominoids from Old World monkeys ~25 My ago [12].
In human B cells, HERV-E insertion introduces another promoter for the cd5 gene, and enables transcription of a fusion transcript that splices in CD5 exon 2 with the consequence of an ORF shift to exon 3, leading to an intracellular variant of CD5 [26]. The LTR2-CD5 fusion transcripts are found in lymphoid organs, and such expression is confined to B cells with the higher expression observed in the B1 cell subset [27,28]. We have not detected proviral HERV-E.CD5 transcripts containing gag and/or env genes.
As recently described, the HERV-E.FABP7 element was observed to drive a new isoform in diffuse large B cell lymphoma [18]. Inserted 30 kb upstream of the fabp7 gene locus in the antisense orientation, the HERV-E.FABP7 element creates an alternative promoter which produces an ectopic LTR2-driven chimeric transcript. The LTR2-FABP7 transcript skips the normal ORF site from exon 1 to exon 2 (Figure 3). The contribution from the three conserved residues of the nuclear localisation signal (NLS) present in exon 1 is lost. As a consequence, the chimeric FABP7 protein cannot translocate to the nucleus and cannot exert its repressive action on leukemic B cell proliferation.
2.3. DNA Methylation Controls Transcription
HERV elements are tightly controlled by DNA methylation [32,33,34]. As a consequence, several authors have used HERV-E elements as a sensor of the DNA methylation state in response to external stimuli and/or in pathological situations to test for possible epigenetic deregulation. Epigenetics is defined as mechanisms by which stable and heritable changes in DNA methylation and packaging of genes can control transcription without affecting the underlying DNA sequences (reviewed in [35,36]). The epigenetic machinery includes the control of DNA methylation, which involves the addition of a methyl group at position 5 of the cytosine pyrimidine ring to form 5-methylcytosine (5mC) within CpG sites. The DNA methylation reaction is controlled by DNA methyltransferases (DNMTs) which use S-adenosylmethionine (SAM) as the methyl donor.
Analysis of the CpG sites in the 5’ LTR2 of HERV-E.CD5 using methylation-sensitive endonuclease assays followed by polymerase chain reaction and bisulfite sequencing revealed that U3 CpG motifs are hypomethylated in B cells expressing LTR2-CD5 fusion transcripts [37]. In addition to these observations, our studies have revealed that cytokines, such as IL-6, may be involved in influencing the HERV-E.CD5 U3 DNA methylation status, and, in turn, LTR2-CD5 expression. Interestingly, LTR2-CD5 overexpression is effective in inducing cytokine expression, such as IL-5, IL-10 and IL-13, and in controlling the cell surface expression of the host gene which controls the negative selection of autoreactive B cells, raising the possibility that HERV-E.CD5 participates in an autoimmune response process [38,39,40].
Cell-specific LTR-promoter DNA demethylation leading to expression of the HERV-E transcripts is not restricted to HERV-E.CD5. It has also been described for HERV-E.FABP7 in leukemic B cells [18]; HERV-E.PTN, HERV-E.EDNRB, and HERV-E.MID1 in placenta but not in blood cells [41]; and HERV-E clone 4-1 in peripheral blood lymphocytes (CD4+, CD8+ and B cells) but not in neutrophils from patients with systemic lupus erythematosus (SLE) [42,43,44]. SLE is regarded as the prototype of autoimmune diseases.
3. HERV-E and Diseases
HERV-Es have attracted significant attention due to their association with malignancies, autoimmune diseases, and human placentation. Interest has been stimulated further by concerns regarding (i) their capacity to produce retroviral protein and in turn to induce (auto)-antibody (Ab) production; (ii) the immunosuppressive functions of the retroviral env proteins; (iii) their capacity to act as alternative promoters or enhancers; (iv) and their DNA methylation dependence.
3.1. Cancer
HERV-E expression has been reported in several tumour cells, such as those in human mammary glands and the prostate, ovary, colon, germinal cells, and uterus [45,46]. Regarding the HERV-E clone 4-1, its env transcript was reported in prostate, ovary and uterus cancer but not in healthy controls [31]. As a consequence, Abs directed against the p15env clone 4-1 peptide were further detected by ELISA in 40% of the sera obtained from women with ovarian cancer but not from normal controls [31]. Anti-p15env Abs are effective in inhibiting the immunosuppressive activity of the p15env clone 4-1 peptide [47]. The immunosuppressive effect of retroviral env peptides is also observed with other HERVs (e.g., HERV-K/HML-2 and HERV-R/ERV3), and that effect is related to the presence of a conserved immunosuppressive retroviral domain located within the plasma-membrane insertion part of the env protein [48].
In renal cell carcinoma (RCC), but not in normal tissues, the LTR2C full length CT-RCC HERV-E element located on chromosome 6 at position 6q15 has been shown to encode a highly immunogenic 10 amino acid env peptide (CT-RCC-1) that can be considered to be a tumour-specific antigen [49]. The CT-RCC-1 env peptide was found to promote RCC reactive CD8+ T cells and RCC cytotoxic T cells as observed in one patient with RCC. Expression of this proviral env peptide is controlled by DNA methylation at LTR sequences, and transcription is under the positive control of the HIF-2α transcription factor, which is itself repressed by the von Hippel-Lindau tumor-suppressor gene [50].
3.2. Autoimmune Diseases
Enhanced expression of mRNA from HERV-E clone 4-1 (M10976) was reported in T cells from patients with SLE [51], and in salivary glands isolated from patients with Sjögren’s syndrome [52,53]. A repressive effect of UVB and steroid treatment on the expression of HERV-E clone 4-1 was also observed in cultured human keratinocytes and in CD4+ T cells from SLE patients, respectively [54,55]. Interestingly, a positive correlation was reported between the 5’ LTR2C clone 4-1 demethylation status in CD4+ T cells with SLE activity, leukopenia and lymphopenia [42].
Abs directed against clone 4-1 p30gag protein were detected in 48% of SLE patients, 35.0% of patients with Sjögren’s syndrome, 33.3% of patients with mixed connective disease, and none of the healthy controls tested [56]. Anti-p30gag Abs also cross-react with two nuclear autoantigens used in the diagnosis of autoimmune diseases, U1-RNP and Sm autoantigens [57]. As a consequence, anti-p30gag Abs are believed to participate in the formation of immune complexes that, in turn, contribute to the activation in the complement pathway in the target organs. The effects of the clone 4-1 p15env protein were further tested in vitro, revealing, on one hand, its capacity to induce T cell activation and anergy, and, on the other hand, its capacity to induce IL-6 and IL-16 cytokine production [58].
Another SLE susceptible endogenous gammaretrovirus is the non-HERV-E human T cell leukaemia-related endogenous retrovirus (HRES-1) that is inserted in the long arm of chromosome 1 at position 1q42 [59]. HRES-1 possesses several similarities with HERV-E clone 4-1: (i) epigenetic control by DNA methylation [60]; (ii) the capacity to produce a p38gag protein that can, in turn, induce the development of Abs as observed in 52% of patients with SLE and in contrast to 3.6% in healthy donors [61]; and (iii) a cross-reactivity of the anti-p38gag HRES-1 Ab with the nuclear autoantigen U1-RNP.
3.3. Human Placentation
In placenta, several studies have highlighted the contribution of various HERV proviruses in normal placenta development and in maintenance of foetomaternal tolerance. The placenta, and particularly the syncytiotrophoblasts, shows expression of several HERVs, such as the ERVW-1 and ERVFRD-1 loci (syncytin-1 and syncytin-2) which are located on chromosomes 7 and 6, respectively [62], and HERV-E proviruses that function as tissue-specific and alternative gene promoters for PTN (growth function), Mid1 (placental development), ApoC1 (lipid metabolism) and EDNRB (anti-apoptotic activity) and appear essential to placental development and function [21,22,23]. HERV-E provirus expression results from a lower DNA methylation state in the placenta, ranging from 4% to 91% when analysing the HERV-E LTRs [41]. Other factors, such as transcription factors, are also involved for cellular specificity. This assertion is reinforced by the characterization of an Sp1 binding site present in the 5’ LTR of HERV-E.PTN, HERV-E.ApoC1 and HERV-E.EDNRB that was found to be critical for strong HERV-E placental transcriptional activity [22,63].
4. Experimental Section
Starting from the HERV-E elements characterized in our previous study [12], we have (i) performed a new selection of HERV-E elements using the online basic local alignment search tool (human BLASTN) provided by ensembl [64] ; (ii) retrieved the 5’LTR and 3’LTR proviral chromosomal locations and HERV-E sizes for the 46 selected HERV-Es using the DFAM 1.3 database [10]; (iii) performed an analysis of HERV-Es relative to sequences from HERV-E clone 4-1 in order to characterize gag, pol and/or env deletions using the dot plot matrix tool [65]; (iv) performed an alignment analysis to test the divergences among U3, R and U5 parts between LTR2, LTR2B and LTR2C subgroups [66]; and (v) used the ensembl genome browser for positioning the HERV-E elements in the human genome. When known, the gene used for alternative transcript usage was used to name the HERV-E element.
Phylogenetic reconstructions were conducted in MEGA 6 [67] with the Maximum Parsimony (MP) approach with a bootstrap resampling of 1000; they were inferred on 46 HERV-E 5’LTR nucleotide sequences aligned using the ClustalW Multiple Alignment method of BioEdit version 7.2.5. There were a total of 637 positions in the final dataset. The MP tree was obtained using the Subtree-Pruning-Regrafting (SPR) algorithm [68] with search level 1, in which the initial trees were obtained by the random addition of sequences.
To date HERV-E integration, three approaches were selected: (1) molecular clock using divergences between 5’LTR and 3’LTR [66] and an evolutionary rate of 0.3% mutation per My [13,14]; (2) whole-genome alignments using the eight-primate Enredo-Pecan-Ortheus (EPO) alignment tool generated by ensembl to determine the integration time during primate evolution (>6 My: Chimpanzee, >8 My: Gorilla, >15 My: Orangutan, >25 My: Old World monkeys, i.e. vervet-AGM, macaque and olive baboon; and >45 My: Marmoset as New World monkeys); and (3) genomic amplification when reported in the literature.
5. Conclusions
The widespread distribution of HERV elements near human gene promoters in the human genome implies a large panel of putative biological activities. The results that may occur can include truncation of host gene products; improper localization of host gene products due to missing localization signals; and ectopic increased or decreased expression of the host gene. The HERV elements can also, from their own sequence, generate antigenic material, and produce immunosuppressive env peptides. Almost all these effects have been reported for HERV-E elements in pathological conditions and/or during placental formation, supporting an important contribution for this family to particular disease states and human evolution. The HERV-E family is also characterized by its heterogeneity, which is in part related to the presence of three main subgroups.
To date, most of the studies testing HERV-E elements in human diseases consider the HERV-E family a minor group with few members, and the HERV-E clone 4-1 as the prototype for HERV-E. However, such assertions need to be revised based on the observations that the HERV-E family possesses at least 1300 members and that HERV-E clone 4-1 belongs to the LTR2C HERV-E subgroup, which is distinct from the two other subgroups in terms of regulation and integration time in the human genome. As a consequence, we recommend, for future studies, using a panel of HERV-Es selected from the three subgroups.
According to the growing importance of HERV-E elements in human diseases, the next challenges are related to acquiring more details about the characterization of HERV-E regulation, HERV-E insertion sites, HERV-E (fusion) transcripts, and HERV-E peptide production, specifically focusing on the antigenic gag peptides and the immunosuppressive env peptides. Altogether, a better understanding of the HERV-E elements and their impact on cellular biology will have applications in the development of new biomarkers for prevention, diagnosis, prognosis, and therapy.
Acknowledgments
The authors are grateful to Simone Forest and Genevieve Michel for their secretarial assistance. This work is supported by the Région Bretagne, the Ligue Contre le Cancer, the Association Française du Gougerot Sjögren et des Syndrome Secs (AFGS), and the Association Dentaire Française (ADF). The research leading to these results has received support from the Innovative Medicines Initiative Joint Undertaking under grant agreement n°115565, resources of which are composed of financial contribution from the European Unin’s Seventh Framework Programme (FP7/2007-2013) and EFPIA companies in kind contribution.
Author Contributions
Analyzed the data: CLD, SV, YR. Wrote the paper: WB, YR. All authors agree to be listed and approve the submitted version of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Belshaw, R.; Pereira, V.; Katzourakis, A.; Talbot, G.; Paces, J.; Burt, A.; Tristem, M. Long-term reinfection of the human genome by endogenous retroviruses. Proc. Natl. Acad. Sci. USA 2004, 101, 4894–4899. [Google Scholar] [CrossRef]
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; et al. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921. [Google Scholar] [CrossRef] [PubMed]
- Jurka, J. Repbase update: a database and an electronic journal of repetitive elements. Trends Genet. 2000, 16, 418–420. [Google Scholar] [CrossRef] [PubMed]
- Jern, P.; Sperber, G.O.; Ahlsen, G.; Blomberg, J. Sequence variability, gene structure, and expression of full-length human endogenous retrovirus H. J. Virol. 2005, 79, 6325–6337. [Google Scholar] [CrossRef] [PubMed]
- Katzourakis, A.; Rambaut, A.; Pybus, O.G. The evolutionary dynamics of endogenous retroviruses. Trends Microbiol 2005, 13, 463–468. [Google Scholar] [CrossRef] [PubMed]
- Mayer, J.; Blomberg, J.; Seal, R.L. A revised nomenclature for transcribed human endogenous retroviral loci. Mob. DNA 2011, 2, e7. [Google Scholar] [CrossRef]
- Wheeler, T.J.; Clements, J.; Eddy, S.R.; Hubley, R.; Jones, T.A.; Jurka, J.; Smit, A.F.; Finn, R.D. Dfam: a database of repetitive DNA based on profile hidden Markov models. Nucl. Acids Res. 2013, 41, D70–D82. [Google Scholar] [CrossRef] [PubMed]
- Belshaw, R.; Watson, J.; Katzourakis, A.; Howe, A.; Woolven-Allen, J.; Burt, A.; Tristem, M. Rate of recombinational deletion among human endogenous retroviruses. J. Virol. 2007, 81, 9437–9442. [Google Scholar] [CrossRef] [PubMed]
- Hayward, A.; Grabherr, M.; Jern, P. Broad-scale phylogenomics provides insights into retrovirus-host evolution. Proc. Natl. Acad. Sci. USA 2013, 110, 20146–20151. [Google Scholar] [CrossRef] [PubMed]
- DFAM. Available online: http://www.dfam.org (accessed on 1 December 2014).
- Steele, P.E.; Rabson, A.B.; Bryan, T.; Martin, M.A. Distinctive termini characterize two families of human endogenous retroviral sequences. Science 1984, 225, 943–947. [Google Scholar] [CrossRef] [PubMed]
- Renaudineau, Y.; Vallet, S.; Le Dantec, C.; Hillion, S.; Saraux, A.; Youinou, P. Characterization of the human CD5 endogenous retrovirus-E in B lymphocytes. Genes Immun. 2005, 6, 663–671. [Google Scholar] [PubMed]
- Yi, J.M.; Kim, H.S. Molecular evolution of the HERV-E family in primates. Arch. Virol. 2006, 151, 1107–1116. [Google Scholar] [CrossRef] [PubMed]
- Li, W.H.; Tanimura, M. The molecular clock runs more slowly in man than in apes and monkeys. Nature 1987, 326, 93–96. [Google Scholar] [CrossRef]
- Tristem, M. Identification and characterization of novel human endogenous retrovirus families by phylogenetic screening of the human genome mapping project database. J. Virol. 2000, 74, 3715–3730. [Google Scholar] [CrossRef] [PubMed]
- Prusty, B.K.; zur Hausen, H.; Schmidt, R.; Kimmel, R.; de Villiers, E.M. Transcription of HERV-E and HERV-E-related sequences in malignant and non-malignant human haematopoietic cells. Virology 2008, 382, 37–45. [Google Scholar] [CrossRef] [PubMed]
- van de Lagemaat, L.N.; Medstrand, P.; Mager, D.L. Multiple effects govern endogenous retrovirus survival patterns in human gene introns. Genome Biol. 2006, 7, eR86. [Google Scholar]
- Lock, F.E.; Rebollo, R.; Miceli-Royer, K.; Gagnier, L.; Kuah, S.; Babaian, A.; Sistiaga-Poveda, M.; Lai, C.B.; Nemirovsky, O.; Serrano, I.; et al. Distinct isoform of FABP7 revealed by screening for retroelement-activated genes in diffuse large B-cell lymphoma. Proc. Natl. Acad. Sci. USA 2014, 111, E3534–E3543. [Google Scholar] [CrossRef] [PubMed]
- Samuelson, L.C.; Wiebauer, K.; Snow, C.M.; Meisler, M.H. Retroviral and pseudogene insertion sites reveal the lineage of human salivary and pancreatic amylase genes from a single gene during primate evolution. Mol. Cell. Biol. 1990, 10, 2513–2520. [Google Scholar] [PubMed]
- Ting, C.N.; Rosenberg, M.P.; Snow, C.M.; Samuelson, L.C.; Meisler, M.H. Endogenous retroviral sequences are required for tissue-specific expression of a human salivary amylase gene. Genes Dev 1992, 6, 1457–1465. [Google Scholar] [CrossRef] [PubMed]
- Schulte, A.M.; Wellstein, A. Structure and phylogenetic analysis of an endogenous retrovirus inserted into the human growth factor gene pleiotrophin. J. Virol. 1998, 72, 6065–6072. [Google Scholar] [PubMed]
- Schulte, A.M.; Malerczyk, C.; Cabal-Manzano, R.; Gajarsa, J.J.; List, H.J.; Riegel, A.T.; Wellstein, A. Influence of the human endogenous retrovirus-like element HERV-E.PTN on the expression of growth factor pleiotrophin: a critical role of a retroviral Sp1-binding site. Oncogene 2000, 19, 3988–3998. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Wu, K.; Didoronkute, A.; Levy, M.V.; Todi, N.; Shchelokova, A.; Massiah, M.A. MID1 catalyzes the ubiquitination of protein phosphatase 2A and mutations within its Bbox1 domain disrupt polyubiquitination of alpha4 but not of PP2Ac. PLoS One 2014, 9, e107428. [Google Scholar] [CrossRef] [PubMed]
- Landry, J.R.; Rouhi, A.; Medstrand, P.; Mager, D.L. The Opitz syndrome gene Mid1 is transcribed from a human endogenous retroviral promoter. Mol. Biol. Evol. 2002, 19, 1934–1942. [Google Scholar] [CrossRef] [PubMed]
- Alonso-Ramirez, R.; Loisel, S.; Buors, C.; Pers, J.O.; Montero, E.; Youinou, P.; Renaudineau, Y. Rationale for Targeting CD6 as a Treatment for Autoimmune Diseases. Arthritis 2010, 2010, 130646. [Google Scholar] [CrossRef] [PubMed]
- Renaudineau, Y.; Hillion, S.; Saraux, A.; Mageed, R.A.; Youinou, P. An alternative exon 1 of the CD5 gene regulates CD5 expression in human B lymphocytes. Blood 2005, 106, 2781–2789. [Google Scholar] [CrossRef] [PubMed]
- Garaud, S.; Le Dantec, C.; Berthou, C.; Lydyard, P.M.; Youinou, P.; Renaudineau, Y. Selection of the alternative exon 1 from the cd5 gene down-regulates membrane level of the protein in B lymphocytes. J. Immunol. 2008, 181, 2010–2018. [Google Scholar] [CrossRef] [PubMed]
- Renaudineau, Y.; Bariller, E.; Pers, J.Q. B1 and CD5-positive B cells. eLS 2014. [Google Scholar] [CrossRef]
- Martin, M.A.; Bryan, T.; Rasheed, S.; Khan, A.S. Identification and cloning of endogenous retroviral sequences present in human DNA. Proc. Natl. Acad. Sci. USA 1981, 78, 4892–4896. [Google Scholar] [CrossRef] [PubMed]
- Repaske, R.; Steele, P.E.; O'Neill, R.R.; Rabson, A.B.; Martin, M.A. Nucleotide sequence of a full-length human endogenous retroviral segment. J. Virol. 1985, 54, 764–772. [Google Scholar] [PubMed]
- Wang-Johanning, F.; Frost, A.R.; Jian, B.; Azerou, R.; Lu, D.W.; Chen, D.T.; Johanning, G.L. Detecting the expression of human endogenous retrovirus E envelope transcripts in human prostate adenocarcinoma. Cancer 2003, 98, 187–197. [Google Scholar] [CrossRef] [PubMed]
- Deplus, R.; Blanchon, L.; Rajavelu, A.; Boukaba, A.; Defrance, M.; Luciani, J.; Rothe, F.; Dedeurwaerder, S.; Denis, H.; Brinkman, A.B.; et al. Regulation of DNA methylation patterns by CK2-mediated phosphorylation of Dnmt3a. Cell. Rep. 2014, 8, 743–753. [Google Scholar] [CrossRef] [PubMed]
- Rowe, H.M.; Trono, D. Dynamic control of endogenous retroviruses during development. Virology 2011, 411, 273–287. [Google Scholar] [CrossRef] [PubMed]
- Macfarlan, T.S.; Gifford, W.D.; Driscoll, S.; Lettieri, K.; Rowe, H.M.; Bonanomi, D.; Firth, A.; Singer, O.; Trono, D.; Pfaff, S.L. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature 2012, 487, 57–63. [Google Scholar] [PubMed]
- Brooks, W.H.; Le Dantec, C.; Pers, J.O.; Youinou, P.; Renaudineau, Y. Epigenetics and autoimmunity. J. Autoimmun. 2010, 34, J207–J219. [Google Scholar] [CrossRef] [PubMed]
- Renaudineau, Y.; Youinou, P. Epigenetics and autoimmunity, with special emphasis on methylation. Keio J. Med. 2011, 60, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Garaud, S.; Le Dantec, C.; Jousse-Joulin, S.; Hanrotel-Saliou, C.; Saraux, A.; Mageed, R.A.; Youinou, P.; Renaudineau, Y. IL-6 modulates CD5 expression in B cells from patients with lupus by regulating DNA methylation. J. Immunol. 2009, 182, 5623–5632. [Google Scholar] [CrossRef] [PubMed]
- Garaud, S.; Le Dantec, C.; de Mendoza, A.R.; Mageed, R.A.; Youinou, P.; Renaudineau, Y. IL-10 production by B cells expressing CD5 with the alternative exon 1B. Ann. N.Y. Acad. Sci. 2009, 1173, 280–285. [Google Scholar] [CrossRef] [PubMed]
- Garaud, S.; Morva, A.; Lemoine, S.; Hillion, S.; Bordron, A.; Pers, J.O.; Berthou, C.; Mageed, R.A.; Renaudineau, Y.; Youinou, P. CD5 promotes IL-10 production in chronic lymphocytic leukemia B cells through STAT3 and NFAT2 activation. J. Immunol. 2011, 186, 4835–4844. [Google Scholar] [CrossRef]
- Mageed, R.A.; Garaud, S.; Taher, T.E.; Parikh, K.; Pers, J.O.; Jamin, C.; Renaudineau, Y.; Youinou, P. CD5 expression promotes multiple intracellular signaling pathways in B lymphocyte. Autoimmun Rev 2012, 11, 795–798. [Google Scholar] [CrossRef] [PubMed]
- Reiss, D.; Zhang, Y.; Mager, D.L. Widely variable endogenous retroviral methylation levels in human placenta. Nucl. Acids Res. 2007, 35, 4743–4754. [Google Scholar] [CrossRef] [PubMed]
- Sukapan, P.; Promnarate, P.; Avihingsanon, Y.; Mutirangura, A.; Hirankarn, N. Types of DNA methylation status of the interspersed repetitive sequences for LINE-1, Alu, HERV-E and HERV-K in the neutrophils from systemic lupus erythematosus patients and healthy controls. J. Hum. Genet. 2014, 59, 178–188. [Google Scholar] [CrossRef] [PubMed]
- Nakkuntod, J.; Sukkapan, P.; Avihingsanon, Y.; Mutirangura, A.; Hirankarn, N. DNA methylation of human endogenous retrovirus in systemic lupus erythematosus. J. Hum. Genet. 2013, 58, 241–249. [Google Scholar] [CrossRef] [PubMed]
- Garaud, S.; Youinou, P.; Renaudineau, Y. DNA methylation and B-cell autoreactivity. Adv Exp Med Biol 2011, 711, 50–60. [Google Scholar] [PubMed]
- Frank, O.; Verbeke, C.; Schwarz, N.; Mayer, J.; Fabarius, A.; Hehlmann, R.; Leib-Mosch, C.; Seifarth, W. Variable transcriptional activity of endogenous retroviruses in human breast cancer. J Virol 2008, 82, 1808–1818. [Google Scholar] [CrossRef] [PubMed]
- Gimenez, J.; Montgiraud, C.; Pichon, J.P.; Bonnaud, B.; Arsac, M.; Ruel, K.; Bouton, O.; Mallet, F. Custom human endogenous retroviruses dedicated microarray identifies self-induced HERV-W family elements reactivated in testicular cancer upon methylation control. Nucl. Acids Res. 2010, 38, 2229–2246. [Google Scholar] [CrossRef] [PubMed]
- Turbeville, M.A.; Rhodes, J.C.; Hyams, D.M.; Distler, C.M.; Steele, P.E. Expression of a putative immunosuppressive protein in human tumors and tissues. Pathobiology 1996, 64, 233–238. [Google Scholar] [CrossRef] [PubMed]
- Lv, H.; Han, J.; Liu, J.; Zheng, J.; Zhong, D.; Liu, R. ISDTool 2.0: a computational model for predicting immunosuppressive domain of retroviruses. J. Theor. Biol. 2014, 360, 78–82. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; Harashima, N.; Kajigaya, S.; Yokoyama, H.; Cherkasova, E.; McCoy, J.P.; Hanada, K.; Mena, O.; Kurlander, R.; Tawab, A.; et al. Regression of human kidney cancer following allogeneic stem cell transplantation is associated with recognition of an HERV-E antigen by T cells. J. Clin. Invest. 2008, 118, 1099–1109. [Google Scholar] [CrossRef] [PubMed]
- Cherkasova, E.; Malinzak, E.; Rao, S.; Takahashi, Y.; Senchenko, V.N.; Kudryavtseva, A.V.; Nickerson, M.L.; Merino, M.; Hong, J.A.; Schrump, D.S.; et al. Inactivation of the von Hippel-Lindau tumor suppressor leads to selective expression of a human endogenous retrovirus in kidney cancer. Oncogene 2011, 30, 4697–4706. [Google Scholar] [CrossRef] [PubMed]
- Ogasawara, H.; Okada, M.; Kaneko, H.; Hishikawa, T.; Sekigawa, I.; Iida, N.; Maruyama, N.; Yamamoto, N.; Hashimoto, H. Quantitative comparison of human endogenous retrovirus mRNA between SLE and rheumatoid arthritis. Lupus 2001, 10, 517–518. [Google Scholar] [CrossRef] [PubMed]
- Le Dantec, C.; Varin, M.M.; Brooks, W.H.; Pers, J.O.; Youinou, P.; Renaudineau, Y. Epigenetics and Sjogren's syndrome. Cur. Pharm. Biotechnol. 2012, 13, 2046–2053. [Google Scholar]
- Konsta, O.D.; Thabet, Y.; Le Dantec, C.; Brooks, W.H.; Tzioufas, A.G.; Pers, J.O.; Renaudineau, Y. The contribution of epigenetics in Sjogren's Syndrome. Front. Genet. 2014, 5, e71. [Google Scholar] [CrossRef]
- Ogasawara, H.; Naito, T.; Kaneko, H.; Hishikawa, T.; Sekigawa, I.; Hashimoto, H.; Kaneko, Y.; Yamamoto, N.; Maruyama, N. Quantitative analyses of messenger RNA of human endogenous retrovirus in patients with systemic lupus erythematosus. J. Rheumatol. 2001, 28, 533–538. [Google Scholar] [PubMed]
- Bessis, D.; Moles, J.P.; Basset-Seguin, N.; Tesniere, A.; Arpin, C.; Guilhou, J.J. Differential expression of a human endogenous retrovirus E transmembrane envelope glycoprotein in normal, psoriatic and atopic dermatitis human skin. Br. J. Dermatol. 2004, 151, 737–745. [Google Scholar] [CrossRef] [PubMed]
- Hishikawa, T.; Ogasawara, H.; Kaneko, H.; Shirasawa, T.; Matsuura, Y.; Sekigawa, I.; Takasaki, Y.; Hashimoto, H.; Hirose, S.; Handa, S.; et al. Detection of antibodies to a recombinant gag protein derived from human endogenous retrovirus clone 4-1 in autoimmune diseases. Viral. Immunol. 1997, 10, 137–147. [Google Scholar] [CrossRef] [PubMed]
- Piotrowski, P.C.; Duriagin, S.; Jagodzinski, P.P. Expression of human endogenous retrovirus clone 4-1 may correlate with blood plasma concentration of anti-U1 RNP and anti-Sm nuclear antibodies. Clin. Rheumatol. 2005, 24, 620–624. [Google Scholar] [CrossRef] [PubMed]
- Naito, T.; Ogasawara, H.; Kaneko, H.; Hishikawa, T.; Sekigawa, I.; Hashimoto, H.; Maruyama, N. Immune abnormalities induced by human endogenous retroviral peptides: with reference to the pathogenesis of systemic lupus erythematosus. J. Clin. Immunol. 2003, 23, 371–376. [Google Scholar] [CrossRef] [PubMed]
- Pullmann, R., Jr.; Bonilla, E.; Phillips, P.E.; Middleton, F.A.; Perl, A. Haplotypes of the HRES-1 endogenous retrovirus are associated with development and disease manifestations of systemic lupus erythematosus. Arthritis Rheum. 2008, 58, 532–540. [Google Scholar] [CrossRef] [PubMed]
- Fali, T.; Le Dantec, C.; Thabet, Y.; Jousse, S.; Hanrotel, C.; Youinou, P.; Brooks, W.H.; Perl, A.; Renaudineau, Y. DNA methylation modulates HRES1/p28 expression in B cells from patients with Lupus. Autoimmunity 2014, 47, 265–271. [Google Scholar] [CrossRef] [PubMed]
- Perl, A.; Colombo, E.; Dai, H.; Agarwal, R.; Mark, K.A.; Banki, K.; Poiesz, B.J.; Phillips, P.E.; Hoch, S.O.; Reveille, J.D.; et al. Antibody reactivity to the HRES-1 endogenous retroviral element identifies a subset of patients with systemic lupus erythematosus and overlap syndromes. Correlation with antinuclear antibodies and HLA class II alleles. Arthritis Rheum. 1995, 38, 1660–1671. [Google Scholar] [CrossRef] [PubMed]
- Lokossou, A.G.; Toudic, C.; Barbeau, B. Implication of human endogenous retrovirus envelope proteins in placental functions. Viruses 2014, 6, 4609–4627. [Google Scholar] [CrossRef] [PubMed]
- Landry, J.R.; Mager, D.L. Functional analysis of the endogenous retroviral promoter of the human endothelin B receptor gene. J Virol 2003, 77, 7459–7466. [Google Scholar] [CrossRef] [PubMed]
- E!Ensembl. Available online: http://www.ensembl.org (accessed on 1 December 2014).
- NCBI DCODE Tool zPicture. Available online: http://zpicture.dcode.org (accessed on 1 December 2014).
- ClustalW2. Available online: http://www.ebi.ac.uk/Tools/msa/clustalw2 (accessed on December 2014).
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Nei, M.; Kumar, S. Molecular Evolution and Phylogenetics; Oxford University Press: New York, NY, USA, 2000; p. 333. [Google Scholar]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).