Abstract
Pseudorabies virus (PRV) variants were discovered in immunized pigs in Northern China and have become the dominant strains since 2011, which caused huge economic losses. In this study, a classical PRV strain was successfully isolated in a PRV gE positive swine farm. The complete genome sequence was obtained using a high-throughput sequencing method and the virus was named JS-2020. The nucleotide homology analysis and phylogenetic tree based on complete genome sequences or gC gene showed that the JS-2020 strain was relatively close to the classical Ea strain in genotype II clade. However, a large number of amino acid variations occurred in the JS-2020 strain compared with the Ea strain, including multiple immunogenic and virulence-related genes. In particular, the gE protein of JS-2020 was similar to earlier Chinese PRV strains without Aspartate insertion. However, the amino acid variations analysis based on major immunogenic and virulence-related genes showed that the JS-2020 strain was not only homologous with earlier PRV strains, but also with strains isolated in recent years. Moreover, the JS-2020 strain was identified as a recombinant between the GXGG-2016 and HLJ-2013 strains. The pathogenicity analysis proved that the PRV JS-2020 strain has typical neurogenic infections and a strong pathogenicity in mice. Together, a novel recombinant classical strain was isolated and characterized in the context of the PRV variant pandemic in China. This study provided some valuable information for the study of the evolution of PRV in China.
1. Introduction
Pseudorabies virus (PRV) is a highly pathogenic and infectious pathogen in pigs, which causes pseudorabies (PR) or Aujeszky’s disease (AD) and is characterized by neurological symptoms, fever and itchiness. AD leads to abortion and stillbirth in swine, growth retardation in growing pigs, and high mortality in piglets, resulting in huge economic losses in pig production. PRV is a double-stranded linear DNA virus belonging to the Herpesviridae family, Alphaherpesvirinae subfamily and Varicellovirus genus. The genomes of herpesvirus were divided into six classes (A to F). The PRV genome belongs to the D class, which is similar to the varicella-zoster virus (VZV) genome [1]. The first complete DNA sequence of the PRV genome was obtained from several published incomplete sequences and multiple newly sequenced fragments derived from different strains [2]. The first full genome characterization of a single PRV strain (Bartha strain) was revealed using Illumina high-throughput sequencing [3]. These studies showed that PRV was organized into a unique long (UL) region, a unique short (US) region and two large inverted and terminal repeats (IR, TR) flanking the US region.
The earliest sporadic outbreaks of PRV were reported in the United States in 1813 and then PRV was spread around the world [1]. With the widespread use of the PRV Bartha-K61 strain, an attenuated live vaccine, PRV was effectively controlled in pigs [4]. Furthermore, it has been eradicated in the United States, Netherlands and various European countries [5]. In China, PRV was discovered in 1950s. With the use of the commercial Bartha-K61 vaccine since the 1970s, PRV was effectively controlled in Chinese swine farms. However, since 2011, the PRV variant strain was discovered in pigs immunized with the Bartha-K61 vaccine in Northern China, and then spread rapidly almost nationwide [6,7]. The epidemiological investigation revealed that the positive rate of wild-type PRV was 8.27% between 2012 and 2017, and it even reached 12% between 2012 and 2013 [8]. Although the Bartha-K61 vaccine was widely used in swine farms, more and more outbreaks of PR were reported in vaccinated swine farms since the variant strain was discovered [9,10,11]. Several research studies have shown that the Bartha-K61 vaccine could not provide full protection against the PRV variant strains [7,12,13]. Compared with classical PRV strains, the PRV variant showed stronger pathogenicity in pigs [10,14]. In recent years, several novel vaccines based on PRV variant strains have been developed and evaluated, including the gE/gI-deleted inactivated vaccine based on the PRV ZJ01 strain [15], the inactivated vaccine (gE-deleted) and the live attenuated vaccine (gE/gI/TK-deleted) based on the PRV HN1201 strain [16,17].
With the wide application of high-throughput sequencing technology, multiple complete genome sequences of PRV variant strains were reported and characterized. However, the novel classical strains were less reported on. In this study, a novel classical PRV strain was isolated in the context of the PRV variant pandemic in China. Its complete genome sequence was obtained using Illumina high-throughput sequencing. The genome characteristics, genetic evolution and amino acid variations were analyzed. Furthermore, the biological characteristics and pathogenicity were further revealed and tested.
2. Materials and Methods
2.1. Cells and Virus Infection
PK-15 cells were purchased from the American Type Culture Collection (ATCC) and cultured in Dulbecco’s modified Eagle medium (DMEM) (Solarbio, Shanghai, China) supplemented with 10% fetal bovine serum (FBS, Thermo Fisher Scientific, Waltham, MA, USA) at 37 °C in 5% CO2. Clinical brain tissue samples were collected from a case of PR-suspected aborted swine in Jiangsu Province of China in 2020. The fever and loss of appetite were observed from aborted swine. It was a PRV negative farm, and the pigs were not vaccinated with any PRV vaccines in recent years. Samples were homogenized and the PK-15 cells were infected with filtered supernatant for 1 h. Next, the infected cells were washed with phosphate-buffered saline (PBS) and incubated in DMEM supplemented with 1% FBS.
2.2. PCR Identification and Growth Curves
The supernatant samples from infected PK-15 cells which showed PRV-like cytopathic effects (CPEs), were purified using three rounds of plaque purification to obtain purified PRV. Total DNA strands of purified PRV samples were extracted using viral genomic DNA extraction kit according to the manufacturer’s instructions (TIANGEN, China). Next, purified DNA samples were identified with polymerase chain reaction (PCR) using PRV specific primers pairs for gD gene (F: 5′ CAG GAG GAC GAG CTG GGG CT -3′ and R: 5′ GTC CAC GCC CCG CTT GAA GCT -3′).
The purified PRV JS-2020 and Bartha strain were inoculated with PK-15 cells at 0.1 MOI in DMEM supplemented with 1% FBS, and the supernatants were harvested at 2, 6, 12, 24, 36 and 48 h post infection. The viral titers of supernatants were tested using PK-15 cells.
2.3. Immunofluorescence
PK-15 cells were infected with JS-2020 at 0.1 MOI in DMEM and supplemented with 1% FBS; cell samples were collected at 24 h post infection. For immunofluorescence, the infected PK-15 cells were fixed with 4% paraformaldehyde for 10 min and permeabilized with 0.5% Triton X-100 at room temperature. Cells were blocked with 3% bovine serum albumin (BSA) for 1 h at room temperature and incubated with PRV gB protein antibody (A general gift from Prof. Beibei Chu at Henan Agricultura University) overnight at 4 °C. Following three washes with PBS, cells were incubated with an Alexa fluor 555-conjugated anti-mouse IgG (Cell Signaling Technology, Danvers, MA, USA) for 1 h at the room temperature. The cells were visualized with an inverted fluorescence microscope (U-HGLGPS, OLYMPUS, Tokyo, Japan).
2.4. Complete Genome Sequencing and Analysis
Purified genomic DNA of PRV JS-2020 strain was sequenced through next-generation sequencing (NGS) technology using Illumina paired-end sequencing (Sangon Biotech, Shanghai, China). The complete genome sequence was annotated using SnapGene 6.0 software and submitted to the GenBank database (GenBank accession number: OR271601).
The complete sequences of JS-2020 strain were aligned with other 15 PRV strains (Table 1), the nucleotide homology analysis was performed using MegAlign module of DNASTAR Lasergene 7 software. Phylogenetic trees of genomic and gC sequences were constructed using maximum likelihood (ML) method of MEGA11 software (V11.0.13).

Table 1.
Information of pseudorabies virus strains for gC gene and complete genome analysis.
2.5. Amino Acid Variations Analysis
Amino acid sequence alignment of JS-2020 ORFs were performed using BLAST on the NCBI website (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastp&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome, accessed on 20 June 2023). The amino acid sequence homology and phylogenetic trees of PRV major immunogenic and virulence-related genes (including gB, gC, gD, gE, gI and TK) were analyzed using maximum likelihood (ML) method with MEGA11 software (V11.0.13).
2.6. Recombination Analysis
RDP4 software (V4.101) was used to detect the potential recombination signals in JS-2020 strain with Bootscan, 3seq, PhylPro, Maxchi, SiScan and Chimaera algorithms. Then, the major recombination events were further validated with SimPlot 3.5.1 software with a sliding window of 2000 nucleotides which moved every 200 nucleotide steps.
2.7. Animal Experiments
The purified PRV JS-2020 strain was diluted to 104.5 TCID50/mL in DMED. Six-week-old specific pathogen-free (SPF) BALB/c mice (Comparative Medicine Center of Yangzhou University) were randomly divided into two groups including DMEM group and PRV infected group. The 5 mice of PRV infected group were infected with 103.5 TCID50 PRV JS-2020 strain in 100 μL DMED by injecting intraperitoneally. Another 5 mice of DMEM group were intraperitoneally injected with 100 μL DMED. Mice were monitored daily, and the survival rates were recorded for 7 days.
2.8. Histopathology and Immunohistochemistry Staining
Brain tissues samples were collected from dead mice of PRV JS-2020 strain infected group and surviving mice of DMEM group. Samples were fixed with 10% formaldehyde, placed into paraffin blocks, and cut into sections. The hematoxylin and eosin staining were applied to sections for histopathological examination. PRV specific antibody (anti-gB protein) was used for immunohistochemistry staining as described previously [18].
3. Results
3.1. Isolation and Identification of PRV JS-2020 Strain
The PRV gE positive brain tissue samples from aborted piglets were identified with the real-time PCR method. Supernatants of the PRV gE-positive tissues were incubated in PK-15 cells and typical CPEs of PRV were observed within 24 h post infection (hpi) (Figure 1A). Moreover, the results of IFA showed that PRV gB protein was detected in infected PK-15 cells (Figure 1A). After three rounds of plaque purification and PCR identification (Figure 1B), the purified PRV was obtained and named JS-2020. Results of growth curves showed the highly efficient replication capability of JS-2020 in PK-15 cells (Figure 1C). The viral titers of JS-2020 in infected cells’ supernatants increased rapidly from 6 hpi to 12 hpi, and peaked with a viral copy number of more than 109 TCID50/mL after 36 hpi (Figure 1C). This result was similar to the Bartha-K61’s outcome. These results indicated that a field strain of PRV was successfully isolated from the clinical samples and grew well in PK-15 cells.
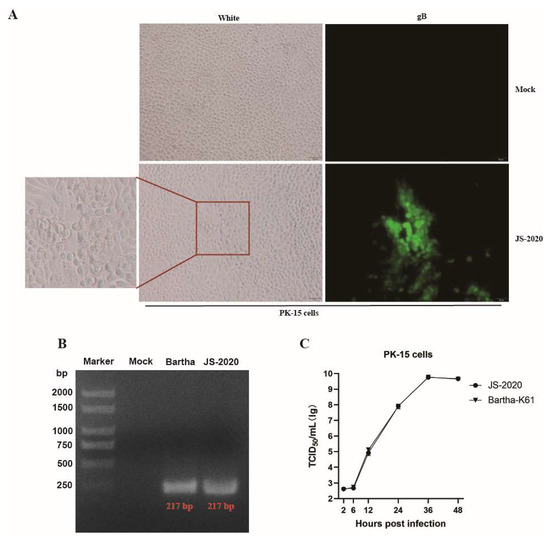
Figure 1.
Isolation and identification of PRV JS-2020 strain. (A) PRV-like CPEs were observed within 24 h post infection, and the gB protein of PRV was positive (IFA). (B) PCR identification of purified JS-2020 strain (gD gene, where Bartha was used as a positive control). (C) Growth curves of JS-2020 strain.
3.2. Genomic Characterization of the JS-2020 Strain
To identify the genetic characteristics of the JS-2020 strain, the complete genome sequences were obtained using the high-throughput sequencing method. The complete genome length of the JS-2020 strain was 143,246 bp, which encodes 69 open reading frames (ORFs) (Figure 2A). The genome sequence was divided into the following four parts: UL (101,287 bp), US (9183 bp), IR (16,388 bp) and TR (16,388 bp) (Table 2). GC content was 74%, which was similar to other published PRV strains [4].
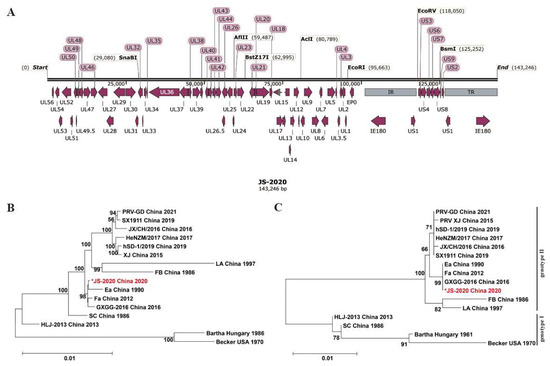
Figure 2.
Genomic characterization of PRV JS-2020 strain. Complete genome distribution of JS-2020 strain (A) and the phylogenetic trees based on both complete sequences (B) and gC (C). “*” indicates the target PRV strain in this study.

Table 2.
Genome organization and location of PRV JS-2020 strain.
Based on complete genome sequences, the nucleotide homology analysis revealed that JS-2020 shared 91.3–99% homology with other PRV strains. In addition, it had the highest homology with the Ea strain (99.8%), which is a Chinese classical PRV strain isolated in 1990 (Table 3). Moreover, the phylogenetic trees based on both genomic and gC sequences were constructed and analyzed (Figure 2B,C). The phylogenetic tree based on gC showed that the PRV strains were classified into genotype I and genotype II, and the JS-2020 strain was clustered within genotype II. The results based on the complete genome showed that the JS-2020 strain also clustered with Chinese PRV strains and had the closest genetic evolutionary relationship with the Ea strain, which was a Chinese classical PRV strain isolated in 1990. These results, which were based on the analysis of genome sequences, suggested that a classical PRV strain was isolated in the context of the PRV variant pandemic in China.

Table 3.
Nucleotide homology of 13 PRV strains compared with JS-2020 strain.
3.3. Amino Acid Variations Analysis of the JS-2020 Strain
Compared with the Ea strain, a total of 27 proteins of the JS-2020 strain were different, containing 86 mutations, 7 deletions and 20 insertions (Table 4). The major amino acid variations occurred in UL47 (7 aa), UL27 (6 aa), UL36 (34 aa), IE180 (7 aa), US1 (21 aa) and US3 (5 aa).

Table 4.
Protein coding variations of JS-2020 strain compared with Ea strain.
To assess the variations of major immunogenic and virulence-related genes (including gB, gC, gD, gE, gI and TK), the amino acid sequence of the JS-2020 strain was compared with 24 Chinese PRV strains and the Bartha strain. The homology analysis results showed that the gI and TK of JS-2020 was highly conserved (sharing 100% homology with most of the Chinese PRV strains) and gB, gC, gD and gE shared lower homology with other PRV strains (gB 96.5–99.7%, gC 92.4–100%, gD 97.3–100% and gE 83.9–100%). Moreover, the phylogenetic trees based on amino acid sequence of gB, gC, gD, gE, gI and TK were constructed and analyzed (Figure 3). The gB protein of the JS-2020 strain had the highest homology with the Fa, GXGG-2016 and SC strain (99.7%), but their evolutionary relationships belong to different branches (Figure 3A). The gC protein has 100% homology and the closest evolutionary relationship with the Ea, Fa and GXGG-2016 strain (Figure 3B). The gD protein has the highest homology (100%) and belongs to same branch of evolutionary relationship with the Fa, GXGG-2016, HLJ-2013 and SC strain. (Figure 3C). The gE protein has the highest homology (100%) and the closest evolutionary relationship with Ea, Fa, HLJ-2013 and SC strain (Figure 3D). The evolutionary relationships of highly conserved gI and TK proteins are similar to homology analysis results, which are also similar to most of the Chinese PRV strains (Figure 3E,F).
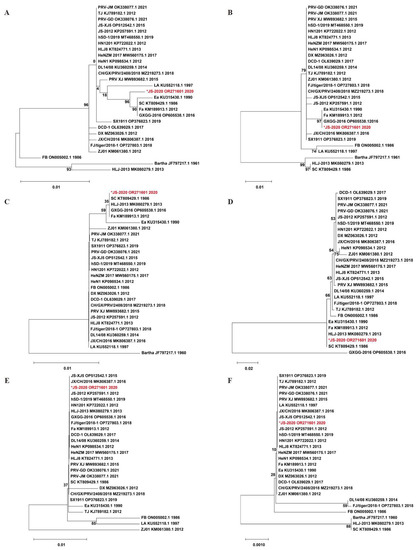
Figure 3.
Phylogenetic trees that are based on amino acid sequences of gB (A), gC (B), gD (C), gE (D), gI (E) and TK (F). “*” indicates the PRV isolate in this study.
The results of previous studies showed that the gE protein of the PRV variant strain contained two Aspartate (Asp, D) insertions when compared with earlier PRV strains that were isolated from China. Although Asp insertions were also observed in a few early PRV strains, the insertions in the variant strains were highly conserved [7,9]. The results of the amino acid sequence analysis showed that the gE protein of the JS-2020 strain is similar to the earlier Chinese PRV strains without the Asp insertion at the amino acid position 497 (Figure 4). These results further proved that a classical PRV strain was isolated, which is similar to earlier Chinese PRV strains.
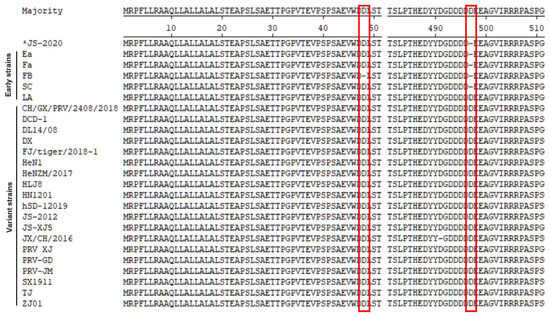
Figure 4.
Alignment of amino acid sequences of PRV gE protein. JS-2020 strain is similar to earlier Chinese PRV strains without Asp insertion at amino acid position 497. Red lines indicate two Asp insertions in the variant strains.
3.4. Recombination Analysis
Multiple studies have shown that recombinant events were found in a few isolated Chinese PRV strains [9,19,20]. The homology analysis of the JS-2020 results showed that partial genes of the JS-2020 have high homology and close evolutionary relationship not only with earlier PRV strain (Ea), but also with recent strains (GXGG-2016). These results suggested that the JS-2020 might be a recombinant strain. To test whether there are any recombination signals in the JS-2020 with other Chinese PRV strains (as shown in Table 5) and the Bartha strain, recombinant analysis was performed using RDP4 software (V4.101). The results showed that several major recombination events were detected in the JS-2020 strain with Bootscan, 3seq, PhylPro, Maxchi, SiScan and Chimaera algorithms. In addition, the major backbone of the JS-2020 was the GXGG-2016 strain, and the major recombination regions were obtained from the HLJ-2013 strain (minor backbone). There were no Ea and Fa strains associated with the recombination signals in the JS-2020 strain, although they shared high homology. Moreover, the recombination events were further verified using Simplot 3.5.1 software. The result showed that four potential major recombination regions which form the HLJ-2013 strain were detected and that they were located at 15,201 to 17201; 31,401 to 33,201; 109,401 to 119,401 and 126,401 to 132,001 (Figure 5). The major recombination regions include partial UL46, UL27, UL34, UL36 and US3 ORFs and complete UL35, US1 and US2 ORFs (Figure 5). The other regions of the JS-2020 strain were highly similar to the GXGG-2016 strain. These results indicated that the JS-2020 and the GXGG-2016 strain share a common parental strain and most of the fragments in the JS-2020 strain come from the GXGG-2016 strain. However, there is a continuous deletion of 69 amino acids in the TK gene of the GXGG-2016 strain [20], but the JS-2020 does not undergo deletion. Therefore, this indicates that the TK gene of the JS-2020 comes from other PRV strains.

Table 5.
Amino acid homology of major immunogenic and virulence-related genes.
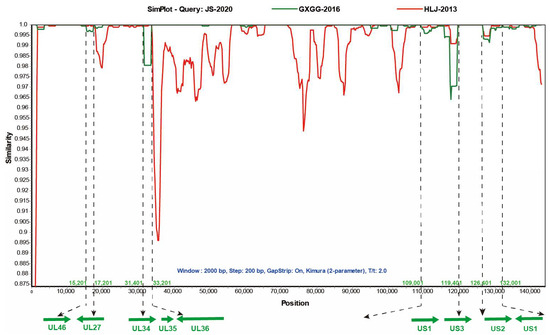
Figure 5.
Recombination analysis of JS-2020 strain using Simplot 3.5.1 software. The four recombination regions are shown with dotted lines. Parameters: Window 2000 bp, Step 200 bp, GapStrip On, Kimura (2-parameter), T/t 2.0.
3.5. Pathogenicity Analysis
Mice are commonly used as animal models since they show neurogenic infections of the central nervous systems (CNS) with high mortalities in a productive PRV infection [21,22]. To assess the pathogenicity of the JS-2020 strain, the six-week-old specific pathogen-free (SPF) BALB/c mice were infected with the PRV JS-2020 strain or the DMEM by injecting, intraperitoneally. The results showed that the mice infected with the PRV JS-2020 strain began to die on the third day post infection (Figure 6A). All PRV infected mice died on firth day post infection and the symptoms associated with the PRV infection (nervous symptoms and dead mice) were not observed in the DMEM group. In our previous study results, the higher dose of PRV Bartha -K61 (50 μL 1 × 105 TCID50/mL) infected mice started to die as early as the fifth day post infection. Therefore, the JS-2020 showed higher pathogenicity than Bartha-K61 in mice [23].
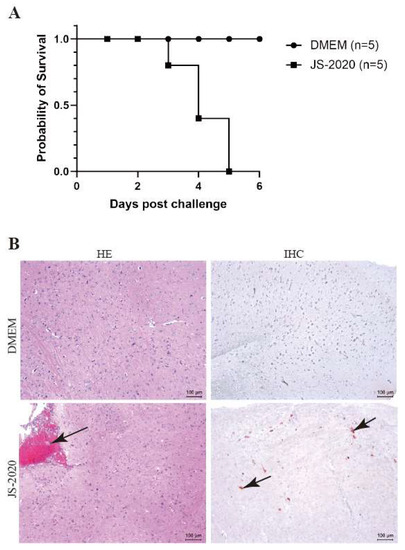
Figure 6.
Pathogenicity of JS-2020 strain in mice. (A) Six-week-old SPF BALB/c mice were infected with 103.5 TCID50 PRV JS-2020, survival of each group was recorded and the survival curves were generated. (B) The histopathological examination (HE) and immunohistochemistry staining (IHC) of brain tissues.
Next, histopathological examination and immunohistochemistry staining of brain tissues were performed to evaluate the neurovirulence of the JS-2020 strain. The hematoxylin-eosin (HE) staining results showed that the meningeal congestion was observed in mice infected with the PRV JS-2020 strain (Figure 6B, HE). Immunohistochemistry staining results revealed that PRV antigens were positive in brain tissues of the mice infected with the PRV JS-2020 strain (Figure 6B, IHC). No lesions or PRV antigens were observed in the brain tissues of mice in the DMEM group. These results indicated that the PRV JS-2020 strain has typical neurogenic infections and a strong pathogenicity in mice.
4. Discussion
Since the PRV variant strains were discovered in 2011 in China, it has spread to most areas of China and caused huge economic losses. Next, more and more PRV variant strains were isolated and identified. To control the spread of newly emerged PRV variants, several effective vaccines based on PRV variants have been developed and applied in recent years in China. However, PRV exhibits a definite neurotropism and results in acute infection in piglets or in the establishment of latent infection in trigeminal ganglion neuros [1,21]. Based on its characteristic of latent infection, the earlier PRV strains were able to maintain long-term infection in swine farms. These PRV strains can be reactivated and spread to healthy pigs. In this study, a classical PRV strain (JS-2020) was isolated and identified from PRV-infected pigs in 2020. It proved that the earlier PRV strains still persist in Chinese swine farms in the context of the PRV variant pandemic.
The complete genomic sequences of the PRV JS-2020 strain were obtained using a high-throughput sequencing method. It had similar genome organization, including a unique long (UL) region, a unique short (US) region and two inverted repeats (IR and TR) along with other published PRV strains. Based on complete genome sequences, the JS-2020 strain has high homology and a close evolutionary relationship with the Ea strain (an earlier classical PRV strain isolated in 1990 in China). In addition, it was classified into genotype II with most of the other Chinese PRV strains based on the gC phylogenetic tree. Compared with the Ea strain, a large number of amino acid variations occurred in the JS-2020 strain, including multiple immunogenic and virulence-related genes.
It was reported that the gB, gC, gD, gE, gI and TK genes are major immunogenic and virulence-related genes of PRV. The gB protein is one of envelope glycoproteins and major viral antigen in PRV, participates in the processes of viral entry and cell-to-cell spread [24,25,26]. The gC protein was regarded as a viral adhesion and immune response related glycoprotein [27]. The cell receptor nectin-1, an immunoglobulin-like cell adhesion molecule, is engaged by the gD protein at an early step of PRV infection. It was reported that the special antibody of gD protein was able to block PRV attachment to cell [28,29]. Both gE and gI are neurovirulence associated proteins of PRV. Deletion of gE or gI severely reduces anterograde spread capacity of PRV in processes of axonal transport [30,31]. The TK gene codes thymidine kinase of PRV. The pathogenicity of TK mutant PRV was highly attenuated in mice, rabbits and pigs [1]. The amino acid variations analysis based on these immunogenic and virulence-related genes showed that the gB, gC, gD and gE genes of the JS-2020 strain were not only homologous with earlier PRV strains (the Ea, Fa or SC strains), but also with strains isolated in recent years (the GXGG-2016 or HLJ-2013 strains). In particular, the gE gene of the JS-2020 strain was most similar to the earlier strains without an Asp insertion when compared to those with variant strains isolated in recent years. Both the gI and TK gene of the JS-2020 strain were highly conserved compared with most of the Chinese PRV strains.
It was reported that the GXGG-2016 and HLJ-2013 strains were classical PRV strains, and they were also homologous with earlier Chinese strains from previous studies [20,32]. The analysis of homology and variations based on nucleotide or amino acid sequences suggested that the JS-2020 strain might have evolved from the GXGG-2016 or HLJ-2013 strain. In recent studies, multiple recombinant PRV strains were identified, such as the JSY13, SC and FJ strains. The recombination regions of the JSY13 and SC strains were regarded as a Bartha strain [9,19]. The FJ strain was regarded as a recombination strain between the HLJ8 and Ea strains [33]. The evolutionary and recombination analysis based on a large number of PRV complete genome sequences indicated that novel PRV variants might evolve from classical PRV strains through recombination mechanisms [34]. In this study, the recombination events were further predicted using related software. The JS-2020 strain was identified as a recombinant, its major sequences was highly similar to GXGG-2016 strain and a fraction of recombination regions were from the HLJ-2013 strain. In summary, a recombinant classical PRV strain was isolated and characterized in this study. These results will provide some evidence for the PRV-evolution-related studies. Moreover, it indicated that the classical PRV strains were still spreading among Chinese swine farms in the context of the PRV variant pandemic and that the novel natural recombinant virus is constantly being produced.
Author Contributions
Conceptualization, K.T. and X.L.; methodology, Z.L. and Z.Z.; software, Z.L. and R.L.; validation, Z.L., Z.Z. and P.L.; formal analysis, Z.L.; investigation, Z.L., Z.S., X.Y. and J.D.; resources, X.L., Z.S., X.Y. and J.D.; data curation, Z.L.; writing—original draft preparation, Z.L.; writing—review and editing, Z.L. and X.L.; visualization, Z.L.; supervision, K.T. and X.L.; project administration, X.L.; funding acquisition, X.L. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the National Natural Science Foundation of China (No. 32172823 and No. 32102637), the Postgraduate Research and Practice Innovation Program of Jiangsu Province (Yangzhou University) (No. SJCX22_1805) and the Project of the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Institutional Review Board Statement
All the animal experiments were approved by the Jiangsu Administrative Committee for Laboratory Animals (Permission Number: 202304001) and complied with the Guidelines of Laboratory Animal Welfare and Ethics of Jiangsu Administrative Committee and Laboratory Animal Welfare and Ethics Committee of Yangzhou University for Laboratory Animals.
Data Availability Statement
All the data generated during this study are included in the manuscript. Additional data related to this article may be requested from the corresponding authors.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Pomeranz, L.E.; Reynolds, A.E.; Hengartner, C.J. Molecular biology of pseudorabies virus: Impact on neurovirology and veterinary medicine. Microbiol. Mol. Biol. Rev. 2005, 69, 462–500. [Google Scholar] [CrossRef]
- Klupp, B.G.; Hengartner, C.J.; Mettenleiter, T.C.; Enquist, L.W. Complete, annotated sequence of the pseudorabies virus genome. J. Virol. 2004, 78, 424–440. [Google Scholar] [CrossRef]
- Szpara, M.L.; Tafuri, Y.R.; Parsons, L.; Shamim, S.R.; Verstrepen, K.J.; Legendre, M.; Enquist, L.W. A wide extent of inter-strain diversity in virulent and vaccine strains of alphaherpesviruses. PLoS Pathog. 2011, 7, e1002282. [Google Scholar] [CrossRef]
- Zheng, H.H.; Fu, P.F.; Chen, H.Y.; Wang, Z.Y. Pseudorabies Virus: From Pathogenesis to Prevention Strategies. Viruses 2022, 14, 1638. [Google Scholar] [CrossRef]
- Zhou, J.; Li, S.; Wang, X.; Zou, M.; Gao, S. Bartha-k61 vaccine protects growing pigs against challenge with an emerging variant pseudorabies virus. Vaccine 2017, 35, 1161–1166. [Google Scholar] [CrossRef]
- Yu, X.; Zhou, Z.; Hu, D.; Zhang, Q.; Han, T.; Li, X.; Gu, X.; Yuan, L.; Zhang, S.; Wang, B.; et al. Pathogenic pseudorabies virus, China, 2012. Emerg. Infect. Dis. 2014, 20, 102–104. [Google Scholar] [CrossRef]
- An, T.Q.; Peng, J.M.; Tian, Z.J.; Zhao, H.Y.; Li, N.; Liu, Y.M.; Chen, J.Z.; Leng, C.L.; Sun, Y.; Chang, D.; et al. Pseudorabies virus variant in Bartha-K61-vaccinated pigs, China, 2012. Emerg. Infect. Dis. 2013, 19, 1749–1755. [Google Scholar] [CrossRef]
- Sun, Y.; Liang, W.; Liu, Q.; Zhao, T.; Zhu, H.; Hua, L.; Peng, Z.; Tang, X.; Stratton, C.W.; Zhou, D.; et al. Epidemiological and genetic characteristics of swine pseudorabies virus in mainland China between 2012 and 2017. PeerJ 2018, 6, e5785. [Google Scholar] [CrossRef]
- Bo, Z.; Miao, Y.; Xi, R.; Gao, X.; Miao, D.; Chen, H.; Jung, Y.S.; Qian, Y.; Dai, J. Emergence of a novel pathogenic recombinant virus from Bartha vaccine and variant pseudorabies virus in China. Transbound. Emerg. Dis. 2021, 68, 1454–1464. [Google Scholar] [CrossRef]
- Luo, Y.; Li, N.; Cong, X.; Wang, C.H.; Du, M.; Li, L.; Zhao, B.; Yuan, J.; Liu, D.D.; Li, S.; et al. Pathogenicity and genomic characterization of a pseudorabies virus variant isolated from Bartha-K61-vaccinated swine population in China. Vet. Microbiol. 2014, 174, 107–115. [Google Scholar] [CrossRef]
- Tan, L.; Yao, J.; Yang, Y.; Luo, W.; Yuan, X.; Yang, L.; Wang, A. Current Status and Challenge of Pseudorabies Virus Infection in China. Virol. Sin. 2021, 36, 588–607. [Google Scholar] [CrossRef] [PubMed]
- Tong, W.; Liu, F.; Zheng, H.; Liang, C.; Zhou, Y.J.; Jiang, Y.F.; Shan, T.L.; Gao, F.; Li, G.X.; Tong, G.Z. Emergence of a Pseudorabies virus variant with increased virulence to piglets. Vet. Microbiol. 2015, 181, 236–240. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.H.; Yuan, J.; Qin, H.Y.; Luo, Y.; Cong, X.; Li, Y.; Chen, J.; Li, S.; Sun, Y.; Qiu, H.J. A novel gE-deleted pseudorabies virus (PRV) provides rapid and complete protection from lethal challenge with the PRV variant emerging in Bartha-K61-vaccinated swine population in China. Vaccine 2014, 32, 3379–3385. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.Y.; Sun, Z.; Tan, F.F.; Guo, L.H.; Wang, Y.Z.; Wang, J.; Wang, Z.Y.; Wang, L.L.; Li, X.D.; Xiao, Y.; et al. Pathogenicity of a currently circulating Chinese variant pseudorabies virus in pigs. World J. Virol. 2016, 5, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.; Dong, J.; Wang, J.; Hou, C.; Sun, H.; Yang, W.; Bai, J.; Jiang, P. A novel inactivated gE/gI deleted pseudorabies virus (PRV) vaccine completely protects pigs from an emerged variant PRV challenge. Virus Res. 2015, 195, 57–63. [Google Scholar] [CrossRef]
- Wang, T.; Xiao, Y.; Yang, Q.; Wang, Y.; Sun, Z.; Zhang, C.; Yan, S.; Wang, J.; Guo, L.; Yan, H.; et al. Construction of a gE-Deleted Pseudorabies Virus and Its Efficacy to the New-Emerging Variant PRV Challenge in the Form of Killed Vaccine. Biomed. Res. Int. 2015, 2015, 684945. [Google Scholar] [CrossRef][Green Version]
- Zhang, C.; Guo, L.; Jia, X.; Wang, T.; Wang, J.; Sun, Z.; Wang, L.; Li, X.; Tan, F.; Tian, K. Construction of a triple gene-deleted Chinese Pseudorabies virus variant and its efficacy study as a vaccine candidate on suckling piglets. Vaccine 2015, 33, 2432–2437. [Google Scholar] [CrossRef]
- Yan, S.; Huang, B.; Bai, X.; Zhou, Y.; Guo, L.; Wang, T.; Shan, Y.; Wang, Y.; Tan, F.; Tian, K. Construction and Immunogenicity of a Recombinant Pseudorabies Virus Variant With TK/gI/gE/11k/28k Deletion. Front. Vet. Sci. 2021, 8, 797611. [Google Scholar] [CrossRef] [PubMed]
- Ye, C.; Guo, J.C.; Gao, J.C.; Wang, T.Y.; Zhao, K.; Chang, X.B.; Wang, Q.; Peng, J.M.; Tian, Z.J.; Cai, X.H.; et al. Genomic analyses reveal that partial sequence of an earlier pseudorabies virus in China is originated from a Bartha-vaccine-like strain. Virology 2016, 491, 56–63. [Google Scholar] [CrossRef]
- Qin, Y.; Qin, S.; Huang, X.; Xu, L.; Ouyang, K.; Chen, Y.; Wei, Z.; Huang, W. Isolation and identification of two novel pseudorabies viruses with natural recombination or TK gene deletion in China. Vet. Microbiol. 2023, 280, 109703. [Google Scholar] [CrossRef]
- Klopfleisch, R.; Klupp, B.G.; Fuchs, W.; Kopp, M.; Teifke, J.P.; Mettenleiter, T.C. Influence of pseudorabies virus proteins on neuroinvasion and neurovirulence in mice. J. Virol. 2006, 80, 5571–5576. [Google Scholar] [CrossRef]
- Klopfleisch, R.; Teifke, J.P.; Fuchs, W.; Kopp, M.; Klupp, B.G.; Mettenleiter, T.C. Influence of tegument proteins of pseudorabies virus on neuroinvasion and transneuronal spread in the nervous system of adult mice after intranasal inoculation. J. Virol. 2004, 78, 2956–2966. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yan, S.; Li, X.; Yang, Q.; Guo, L.; Wang, Y.; Xiao, Y.; Tan, F.; Li, X.; Tian, K. From mouse to pig: Is PRV vaccine safe across two species? Virus Res. 2017, 236, 44–49. [Google Scholar] [CrossRef] [PubMed]
- Peeters, B.; de Wind, N.; Hooisma, M.; Wagenaar, F.; Gielkens, A.; Moormann, R. Pseudorabies virus envelope glycoproteins gp50 and gII are essential for virus penetration, but only gII is involved in membrane fusion. J. Virol. 1992, 66, 894–905. [Google Scholar] [CrossRef]
- Rauh, I.; Mettenleiter, T.C. Pseudorabies virus glycoproteins gII and gp50 are essential for virus penetration. J. Virol. 1991, 65, 5348–5356. [Google Scholar] [CrossRef] [PubMed]
- Zaripov, M.M.; Morenkov, O.S.; Fodor, N.; Braun, A.; Schmatchenko, V.V.; Fodor, I.; Brown, A. Distribution of B-cell epitopes on the pseudorabies virus glycoprotein B. J. Gen. Virol. 1999, 80 Pt 3, 537–541. [Google Scholar] [CrossRef]
- Rue, C.A.; Ryan, P. Pseudorabies virus glycoprotein C attachment-proficient revertants isolated through a simple, targeted mutagenesis scheme. J. Virol. Methods 2008, 151, 101–106. [Google Scholar] [CrossRef]
- Li, A.; Lu, G.; Qi, J.; Wu, L.; Tian, K.; Luo, T.; Shi, Y.; Yan, J.; Gao, G.F. Structural basis of nectin-1 recognition by pseudorabies virus glycoprotein D. PLoS Pathog. 2017, 13, e1006314. [Google Scholar] [CrossRef]
- Zhang, T.; Liu, Y.; Chen, Y.; Wang, J.; Feng, H.; Wei, Q.; Zhao, S.; Yang, S.; Liu, D.; Zhang, G. A monoclonal antibody neutralizes pesudorabies virus by blocking gD binding to the receptor nectin-1. Int. J. Biol. Macromol. 2021, 188, 359–368. [Google Scholar] [CrossRef]
- Ch’ng, T.H.; Enquist, L.W. Neuron-to-cell spread of pseudorabies virus in a compartmented neuronal culture system. J. Virol. 2005, 79, 10875–10889. [Google Scholar] [CrossRef]
- Kratchmarov, R.; Kramer, T.; Greco, T.M.; Taylor, M.P.; Ch’ng, T.H.; Cristea, I.M.; Enquist, L.W. Glycoproteins gE and gI are required for efficient KIF1A-dependent anterograde axonal transport of alphaherpesvirus particles in neurons. J. Virol. 2013, 87, 9431–9440. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Shi, Z.; Liu, C.; Wang, P.; Wang, M.; Wang, S.; Liu, Z.; Wei, L.; Sun, Z.; He, X.; et al. Implication of the Identification of an Earlier Pseudorabies Virus (PRV) Strain HLJ-2013 to the Evolution of Chinese PRVs. Front. Microbiol. 2020, 11, 612474. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Tang, W.; Wang, X.; Zhao, J.; Peng, K.; Sun, X.; Li, S.; Kuang, S.; Zhu, L.; Zhou, Y.; et al. The Genetic Characterization of a Novel Natural Recombinant Pseudorabies Virus in China. Viruses 2022, 14, 978. [Google Scholar] [CrossRef] [PubMed]
- Hu, R.; Wang, L.; Liu, Q.; Hua, L.; Huang, X.; Zhang, Y.; Fan, J.; Chen, H.; Song, W.; Liang, W.; et al. Whole-Genome Sequence Analysis of Pseudorabies Virus Clinical Isolates from Pigs in China between 2012 and 2017 in China. Viruses 2021, 13, 1322. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).