Broad-Spectrum Detection of HPV in Male Genital Samples Using Target-Enriched Whole-Genome Sequencing
Abstract
:1. Introduction
2. Materials and Methods
2.1. Samples
2.2. HPV Typing by Enriched Whole-Genome Sequencing (eWGS) Assay
2.3. Bioinformatics
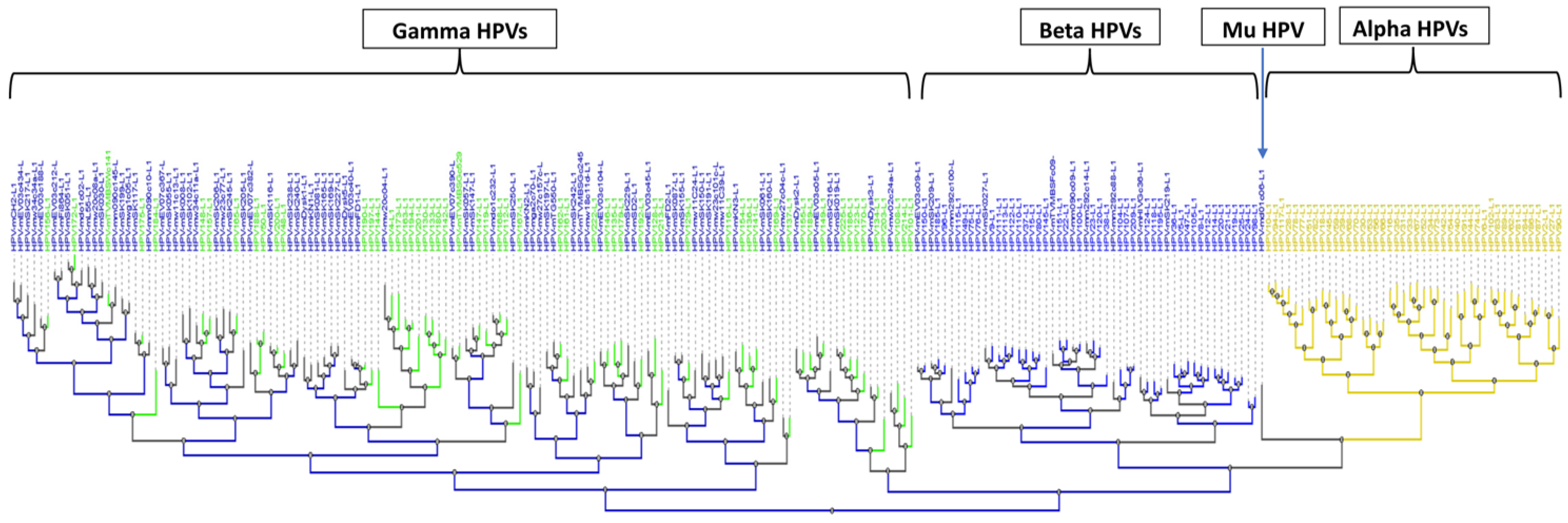
3. Results
3.1. HPV Identification in Male Genital Samples
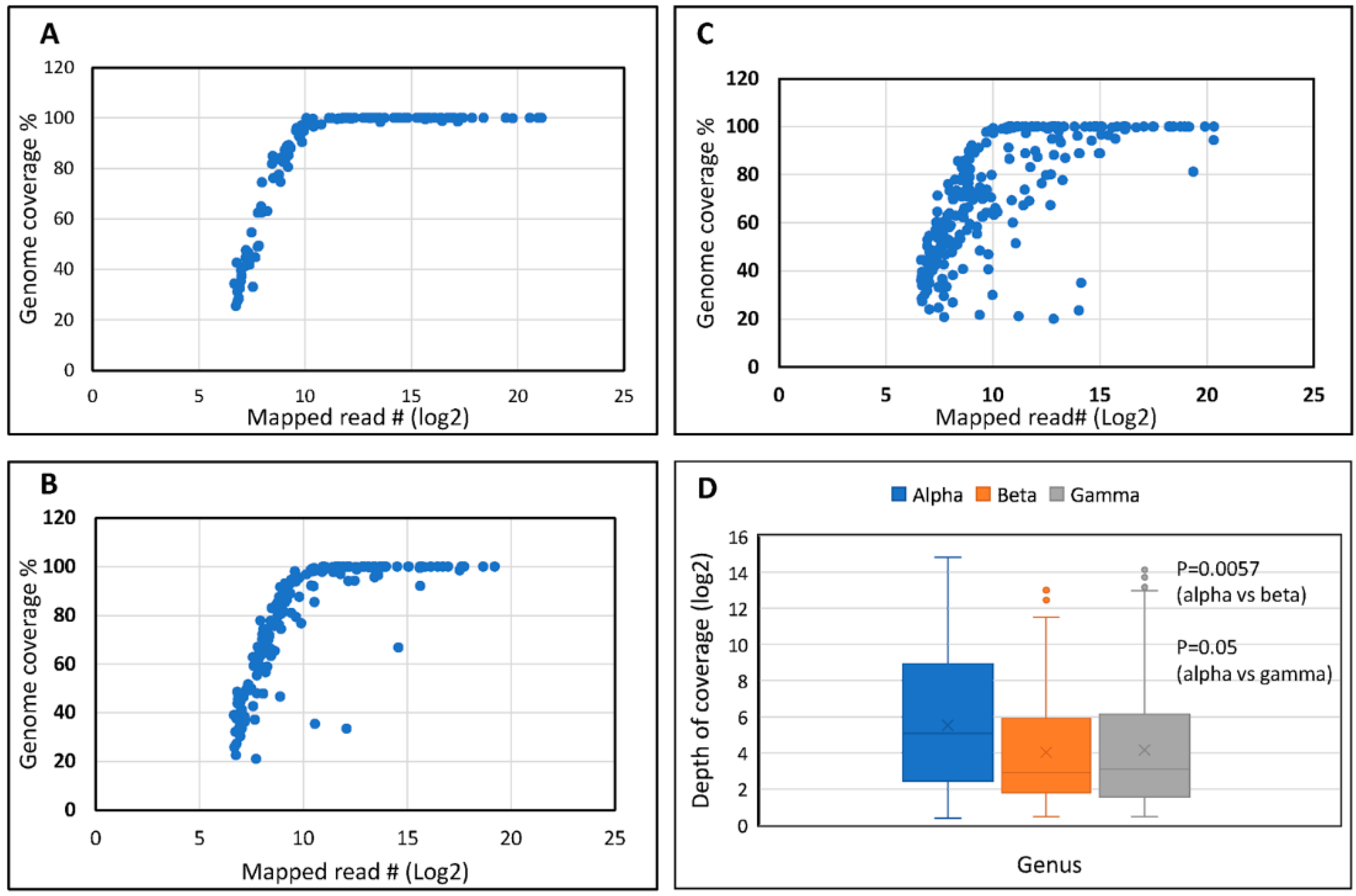
3.1.1. Read and Mapping Characteristics
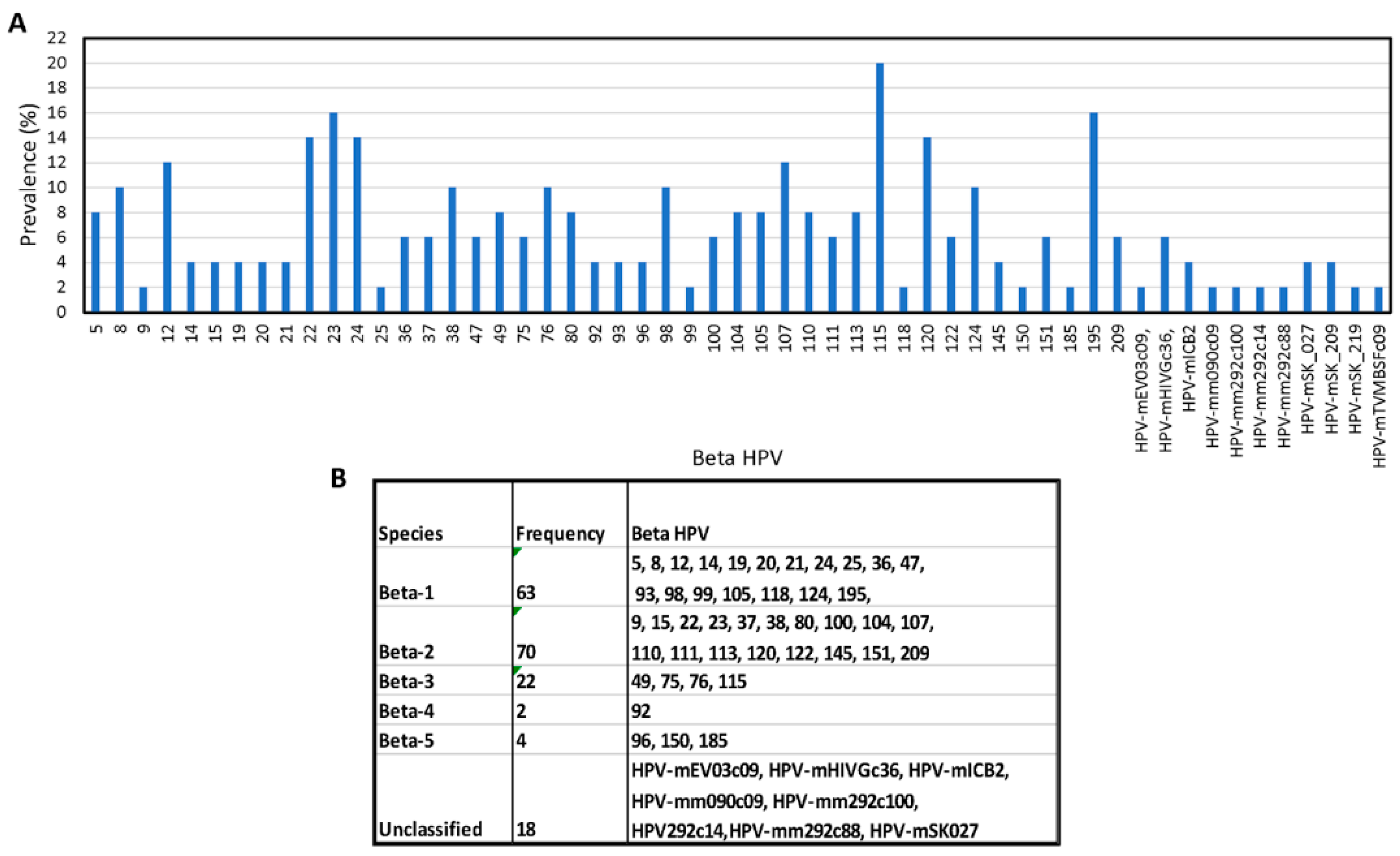
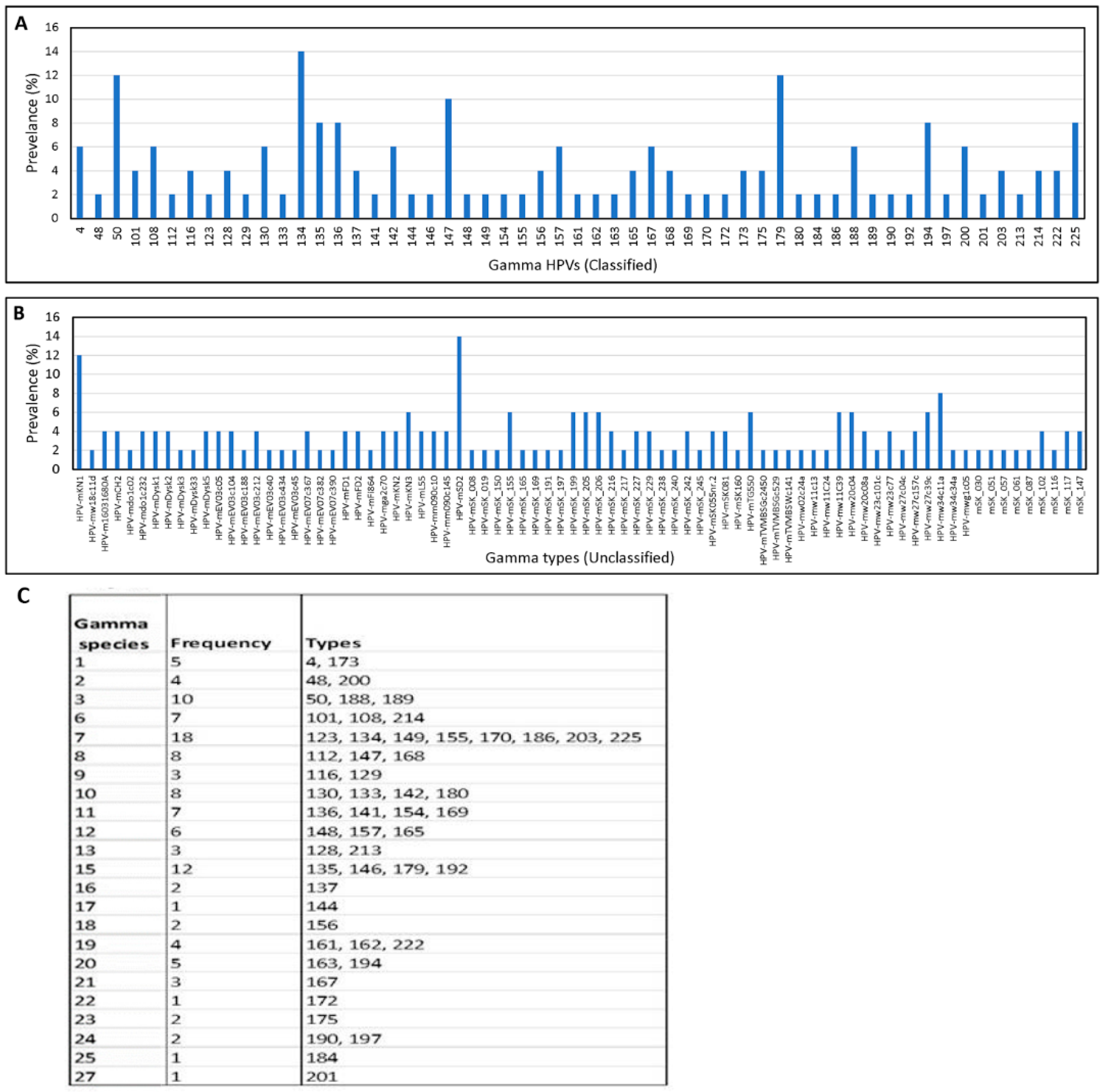
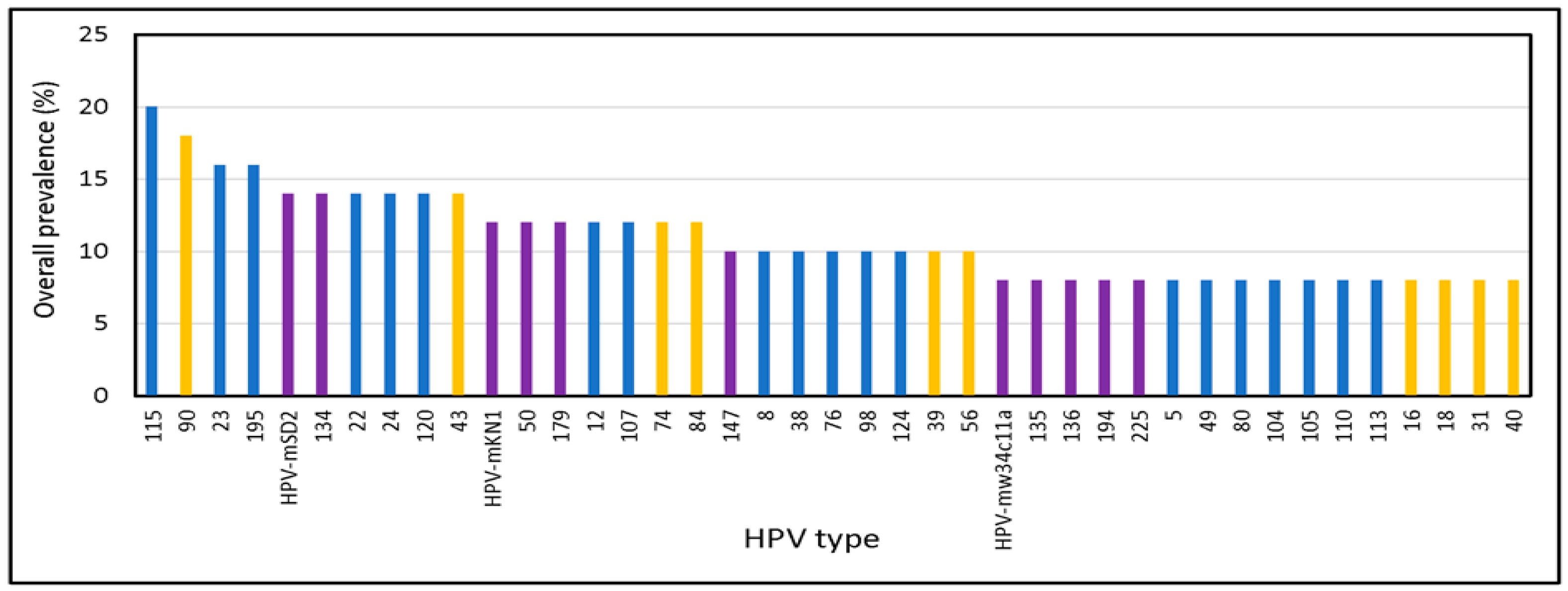
3.1.2. Broad Spectrum of HPV Genomes in Male Genital Samples
3.2. Identification of Novel and Unclassified Types
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Disclaimer
References
- Williams, J.; Kostiuk, M.; Biron, V.L. Molecular Detection Methods in HPV-Related Cancers. Front. Oncol. 2022, 12, 864820. [Google Scholar] [CrossRef] [PubMed]
- Gheit, T.; Rollo, F.; Brancaccio, R.N.; Robitaille, A.; Galati, L.; Giuliani, M.; Latini, A.; Pichi, B.; Benevolo, M.; Cuenin, C.; et al. Oral Infection by Mucosal and Cutaneous Human Papillomaviruses in the Men Who Have Sex with Men from the OHMAR Study. Viruses 2020, 12, 899. [Google Scholar] [CrossRef]
- Flores-Miramontes, M.G.; Olszewski, D.; Artaza-Irigaray, C.; Willemsen, A.; Bravo, I.G.; Vallejo-Ruiz, V.; Leal-Herrera, Y.A.; Pina-Sanchez, P.; Molina-Pineda, A.; Canton-Romero, J.C.; et al. Detection of Alpha, Beta, Gamma, and Unclassified Human Papillomaviruses in Cervical Cancer Samples from Mexican Women. Front. Cell Infect. Microbiol. 2020, 10, 234. [Google Scholar] [CrossRef] [PubMed]
- Agalliu, I.; Gapstur, S.; Chen, Z.; Wang, T.; Anderson, R.L.; Teras, L.; Kreimer, A.R.; Hayes, R.B.; Freedman, N.D.; Burk, R.D. Associations of Oral alpha-, beta-, and gamma-Human Papillomavirus Types with Risk of Incident Head and Neck Cancer. JAMA Oncol. 2016, 2, 599–606. [Google Scholar] [CrossRef] [PubMed]
- Smelov, V.; Muwonge, R.; Sokolova, O.; McKay-Chopin, S.; Eklund, C.; Komyakov, B.; Gheit, T. Beta and gamma human papillomaviruses in anal and genital sites among men: Prevalence and determinants. Sci. Rep. 2018, 8, 8241. [Google Scholar] [CrossRef] [PubMed]
- Bertinazzi, M.; Gheit, T.; Polesel, J.; McKay-Chopin, S.; Cutrone, C.; Sari, M.; Sbaraglia, M.; Tos, A.P.D.; Nicolai, P.; Tommasino, M.; et al. Clinical implications of alpha, beta, and gamma HPV infection in juvenile onset recurrent respiratory papillomatosis. Eur. Arch. Otorhinolaryngol. 2022, 279, 285–292. [Google Scholar] [CrossRef]
- Al-Soneidar, W.A.; Harper, S.; Alli, B.Y.; Nicolau, B. Interaction of HPV16 and Cutaneous HPV in Head and Neck Cancer. Cancers 2022, 14, 5197. [Google Scholar] [CrossRef]
- Meiring, T.L.; Mbulawa, Z.Z.A.; Lesosky, M.; Coetzee, D.; Williamson, A.L. High diversity of alpha, beta and gamma human papillomaviruses in genital samples from HIV-negative and HIV-positive heterosexual South African men. Papillomavirus Res. 2017, 3, 160–167. [Google Scholar] [CrossRef]
- Minoni, L.; Romero-Medina, M.C.; Venuti, A.; Sirand, C.; Robitaille, A.; Altamura, G.; Le Calvez-Kelm, F.; Viarisio, D.; Zanier, K.; Muller, M.; et al. Transforming Properties of Beta-3 Human Papillomavirus E6 and E7 Proteins. mSphere 2020, 5, e00398-20. [Google Scholar] [CrossRef]
- Bandolin, L.; Borsetto, D.; Fussey, J.; Da Mosto, M.C.; Nicolai, P.; Menegaldo, A.; Calabrese, L.; Tommasino, M.; Boscolo-Rizzo, P. Beta human papillomaviruses infection and skin carcinogenesis. Rev. Med. Virol. 2020, 30, e2104. [Google Scholar] [CrossRef]
- Venuti, A.; Lohse, S.; Tommasino, M.; Smola, S. Cross-talk of cutaneous beta human papillomaviruses and the immune system: Determinants of disease penetrance. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2019, 374, 20180287. [Google Scholar] [CrossRef] [PubMed]
- Brink, A.A.; Lloveras, B.; Nindl, I.; Heideman, D.A.; Kramer, D.; Pol, R.; Fuente, M.J.; Meijer, C.J.; Snijders, P.J. Development of a general-primer-PCR-reverse-line-blotting system for detection of beta and gamma cutaneous human papillomaviruses. J. Clin. Microbiol. 2005, 43, 5581–5587. [Google Scholar] [CrossRef] [PubMed]
- Chouhy, D.; Gorosito, M.; Sanchez, A.; Serra, E.C.; Bergero, A.; Fernandez Bussy, R.; Giri, A.A. New generic primer system targeting mucosal/genital and cutaneous human papillomaviruses leads to the characterization of HPV 115, a novel Beta-papillomavirus species 3. Virology 2010, 397, 205–216. [Google Scholar] [CrossRef] [PubMed]
- Giuliano, A.R.; Lazcano-Ponce, E.; Villa, L.L.; Flores, R.; Salmeron, J.; Lee, J.H.; Papenfuss, M.R.; Abrahamsen, M.; Jolles, E.; Nielson, C.M.; et al. The human papillomavirus infection in men study: Human papillomavirus prevalence and type distribution among men residing in Brazil, Mexico, and the United States. Cancer Epidemiol. Biomark. Prev. 2008, 17, 2036–2043. [Google Scholar] [CrossRef]
- Yin, L.; Yao, J.; Chang, K.; Gardner, B.P.; Yu, F.; Giuliano, A.R.; Goodenow, M.M. HPV Population Profiling in Healthy Men by Next-Generation Deep Sequencing Coupled with HPV-QUEST. Viruses 2016, 8, 28. [Google Scholar] [CrossRef] [PubMed]
- Bzhalava, D.; Muhr, L.S.; Lagheden, C.; Ekstrom, J.; Forslund, O.; Dillner, J.; Hultin, E. Deep sequencing extends the diversity of human papillomaviruses in human skin. Sci. Rep. 2014, 4, 5807. [Google Scholar] [CrossRef]
- Latsuzbaia, A.; Wienecke-Baldacchino, A.; Tapp, J.; Arbyn, M.; Karabegovic, I.; Chen, Z.; Fischer, M.; Muhlschlegel, F.; Weyers, S.; Pesch, P.; et al. Characterization and Diversity of 243 Complete Human Papillomavirus Genomes in Cervical Swabs Using Next Generation Sequencing. Viruses 2020, 12, 1437. [Google Scholar] [CrossRef]
- Li, T.; Unger, E.R.; Batra, D.; Sheth, M.; Steinau, M.; Jasinski, J.; Jones, J.; Rajeevan, M.S. Universal Human Papillomavirus Typing Assay: Whole-Genome Sequencing following Target Enrichment. J. Clin. Microbiol. 2017, 55, 811–823. [Google Scholar] [CrossRef]
- Muhr, L.S.A.; Guerendiain, D.; Cuschieri, K.; Sundstrom, K. Human Papillomavirus Detection by Whole-Genome Next-Generation Sequencing: Importance of Validation and Quality Assurance Procedures. Viruses 2021, 13, 1323. [Google Scholar] [CrossRef]
- Bruni, L.; Diaz, M.; Castellsague, X.; Ferrer, E.; Bosch, F.X.; de Sanjose, S. Cervical human papillomavirus prevalence in 5 continents: Meta-analysis of 1 million women with normal cytological findings. J. Infect. Dis. 2010, 202, 1789–1799. [Google Scholar] [CrossRef]
- Gargano, J.W.; Unger, E.R.; Liu, G.; Steinau, M.; Meites, E.; Dunne, E.; Markowitz, L.E. Prevalence of Genital Human Papillomavirus in Males, United States, 2013–2014. J. Infect. Dis. 2017, 215, 1070–1079. [Google Scholar] [CrossRef] [PubMed]
- Hariri, S.; Unger, E.R.; Sternberg, M.; Dunne, E.F.; Swan, D.; Patel, S.; Markowitz, L.E. Prevalence of genital human papillomavirus among females in the United States, the National Health And Nutrition Examination Survey, 2003–2006. J. Infect. Dis. 2011, 204, 566–573. [Google Scholar] [CrossRef] [PubMed]
- Han, J.J.; Beltran, T.H.; Song, J.W.; Klaric, J.; Choi, Y.S. Prevalence of Genital Human Papillomavirus Infection and Human Papillomavirus Vaccination Rates Among US Adult Men: National Health and Nutrition Examination Survey (NHANES) 2013–2014. JAMA Oncol. 2017, 3, 810–816. [Google Scholar] [CrossRef] [PubMed]
- Castle, P.E. Human papillomavirus (HPV) genotype 84 infection of the male genitalia: Further evidence for HPV tissue tropism? J. Infect. Dis. 2008, 197, 776–778. [Google Scholar] [CrossRef]
- Lomsadze, A.; Li, T.; Rajeevan, M.S.; Unger, E.R.; Borodovsky, M. Bioinformatics Pipeline for Human Papillomavirus Short Read Genomic Sequences Classification Using Support Vector Machine. Viruses 2020, 12, 710. [Google Scholar] [CrossRef]
- Sias, C.; Salichos, L.; Lapa, D.; Del Nonno, F.; Baiocchini, A.; Capobianchi, M.R.; Garbuglia, A.R. Alpha, Beta, gamma human PapillomaViruses (HPV) detection with a different sets of primers in oropharyngeal swabs, anal and cervical samples. Virol. J. 2019, 16, 27. [Google Scholar] [CrossRef]
- Bolatti, E.M.; Hosnjak, L.; Chouhy, D.; Casal, P.E.; Re-Louhau, M.F.; Bottai, H.; Komlos, K.F.; Poljak, M.; Giri, A.A. Assessing Gammapapillomavirus infections of mucosal epithelia with two broad-spectrum PCR protocols. BMC Infect. Dis. 2020, 20, 274. [Google Scholar] [CrossRef]
- Sichero, L.; Pierce Campbell, C.M.; Ferreira, S.; Sobrinho, J.S.; Luiza Baggio, M.; Galan, L.; Silva, R.C.; Lazcano-Ponce, E.; Giuliano, A.R.; Villa, L.L.; et al. Broad HPV distribution in the genital region of men from the HPV infection in men (HIM) study. Virology 2013, 443, 214–217. [Google Scholar] [CrossRef]
- Karimi, A.; Mohebbi, E.; McKay-Chopin, S.; Rashidian, H.; Hadji, M.; Peyghambari, V.; Marzban, M.; Naghibzadeh-Tahami, A.; Gholipour, M.; Kamangar, F.; et al. Human Papillomavirus and Risk of Head and Neck Squamous Cell Carcinoma in Iran. Microbiol. Spectr. 2022, 10, e0011722. [Google Scholar] [CrossRef]
- Li, T.; Unger, E.R.; Rajeevan, M.S. Universal human papillomavirus typing by whole genome sequencing following target enrichment: Evaluation of assay reproducibility and limit of detection. BMC Genom. 2019, 20, 231. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, T.; Unger, E.R.; Rajeevan, M.S. Broad-Spectrum Detection of HPV in Male Genital Samples Using Target-Enriched Whole-Genome Sequencing. Viruses 2023, 15, 1967. https://doi.org/10.3390/v15091967
Li T, Unger ER, Rajeevan MS. Broad-Spectrum Detection of HPV in Male Genital Samples Using Target-Enriched Whole-Genome Sequencing. Viruses. 2023; 15(9):1967. https://doi.org/10.3390/v15091967
Chicago/Turabian StyleLi, Tengguo, Elizabeth R. Unger, and Mangalathu S. Rajeevan. 2023. "Broad-Spectrum Detection of HPV in Male Genital Samples Using Target-Enriched Whole-Genome Sequencing" Viruses 15, no. 9: 1967. https://doi.org/10.3390/v15091967
APA StyleLi, T., Unger, E. R., & Rajeevan, M. S. (2023). Broad-Spectrum Detection of HPV in Male Genital Samples Using Target-Enriched Whole-Genome Sequencing. Viruses, 15(9), 1967. https://doi.org/10.3390/v15091967




