Abstract
The cucurbit vegetable chieh-qua (Benincasa hispida var. chieh-qua How) is an important crop in South China and southeast Asian countries. Viral diseases cause substantial loss of chieh-qua yield. To identify the viruses that affect chieh-qua in China, ribosomal RNA-depleted total RNA sequencing was performed using chieh-qua leaf samples with typical viral symptoms. The virome of chieh-qua comprises four known viruses (melon yellow spot virus (MYSV), cucurbit chlorotic yellows virus (CCYV), papaya ringspot virus (PRSV) and watermelon silver mottle virus (WSMoV) and two novel viruses: cucurbit chlorotic virus (CuCV) in the genus Crinivirus and chieh-qua endornavirus (CqEV) in the genus Alphaendornavirus. The complete genomes of the two novel viruses in chieh-qua and three other isolates of CuCV in pumpkin, watermelon and cucumber were determined and the recombination signals of pumpkin and watermelon isolates of CuCV were detected. A reverse transcriptase PCR indicated that the dominant viruses of chieh-qua in Hainan are MYSV (66.67%) and CCYV (55.56%), followed by CuCV (27.41%), WSMoV (7.41%), cucumber mosaic virus (8.15%), zucchini yellow mosaic virus (6.67%), PRSV (6.67%) and CqEV (35.56%). Our findings support diagnostic and prevalence studies of viruses infecting chieh-qua in China, enabling sustainable control strategies for cucurbit viruses worldwide.
1. Introduction
Chieh-qua (Benincasa hispida Cogn. var. chieh-qua How), also known as fuzzy gourd, hairy melon or moa qua, is a variety of wax gourd (Benincasa hispida (Thunb.) Cogn.) in the Cucurbitaceae family. As an important vegetable crop, chieh-qua is widely cultivated throughout the world for its immature fruit, especially in South China and southeast Asian countries [1]. In China in 2021, the planting area of Benincasa hispida was about 6 million acres, with a yield of 58 million tons [2]. Chieh-qua is mainly planted in Guangdong, Guangxi and Hainan provinces in Southern China and the planting area has gradually spread from south to north China with increasing consumer demand [3,4,5].
However, with the expansion of cultivation, various diseases have arisen, especially viral diseases, which have been a major factor restricting the yield and quality of chieh-qua in China. Diverse virus-like symptoms, including mosaic, interveinal chlorosis, mottling and leaf deformation, are frequently observed in chieh-qua; however, there has been limited research on sporadic chieh-qua samples and only four viruses have been reported: zucchini tigre mosaic virus (ZTMV, genus Potyvirus), zucchini yellow mosaic virus (ZYMV, genus Potyvirus), papaya ringspot virus (PRSV, genus Potyvirus) and cucumber mosaic virus (CMV, genus Cucumovirus) [6,7,8]. ZYMV, PRSV and CMV have been frequently identified in cucurbit vegetables in China [9], whereas ZTMV has only been discovered in China in recent years, with a potential risk to infect more cucurbit crops in the field [9,10]. Recently, Nong [11] investigated both wax gourd and chieh-qua plants in Guangdong Province, China, and they discovered the above four viruses as well as six other viruses: cucurbit aphid-borne yellows virus (CABYV), melon aphid-borne yellows virus (MABYV), squash leaf curl China virus (SLCCNV), cucumber green mottle mosaic virus (CGMMV), watermelon silver mottle virus (WSMoV) and tobacco stripe virus (TSV). Among them, WSMoV was considered the dominant virus in the gourd population. Nevertheless, deep virome profiling and an extensive investigation of the virus diversity, occurrence and distribution on chieh-qua plants in China are still lacking.
The present study aimed to investigate in depth the viruses causing chieh-qua viral disease in China. For this, a ribosomal RNA (rRNA)-depleted total RNA method was used to construct the virome profiles of symptomatic chieh-qua samples. This approach identified four known viruses (MYSV, cucurbit chlorotic yellows virus (CCYV), PRSV and WSMoV) and two novel viruses in the genus Crinivirus and Endornavirus, whose complete genomes and natural hosts within gourd vegetables were also determined. Furthermore, an extensive virus screen was conducted on over two hundred samples collected from various plantations in Hainan province from 2021 to 2022. This study provided comprehensive information on the virus diversity of chieh-qua in China, the prevalence and geographical spread of viruses infecting chieh-qua and the dominant viruses in different plantations.
2. Materials and Methods
2.1. Plant Materials
A total of 135 chieh-qua samples showing virus-like symptoms were collected for the prevalence survey of viruses infecting chieh-qua. In total, 98 samples from other cucurbit vegetables showing virus-like symptoms, including 23 pumpkin (Cucurbita moschata) samples, 26 watermelon (Citrullus lanatus) samples and 49 cucumber (Cucumis sativus) samples, were collected to determine whether the potential new viruses occur in other cucurbit vegetables. The above 223 symptomatic samples and 28 asymptomatic chieh-qua samples were collected from 7 main cucurbit vegetable cultivation regions (Chengmai, Wenchang, Ding’an, Lingshui, Sanya, Dongfang and Danzhou) in Hainan province from 2021 to 2022 (Figure 1A).
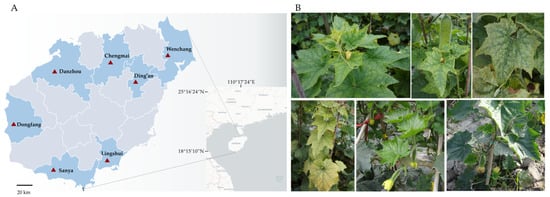
Figure 1.
The cucurbit vegetable sampling sites in Hainan Province, China, and various virus-like symptoms in chieh-qua. (A) The cucurbit vegetable sampling sites in Hainan Province, China. (B) Various virus-like symptoms in chieh-qua. (Upper left) interveinal chlorosis and deformation. (Upper middle), interveinal chlorosis and mottling. (Upper right), mottling, mosaic and interveinal chlorosis. (Lower left) interveinal chlorosis and yellowing. (Lower middle), mosaic and interveinal chlorosis, and (lower right), healthy plant.
2.2. Nucleic Acid Preparation and HTS
Leaves from five individual chieh-qua plants (numbered CS-1, CS-15, CS-16, CS-37 and CS-78) representing the distinct symptoms (Figure 1B) were pooled together in equal amounts to form a composite sample (PL-1) for downstream total RNA extraction and HTS. Total RNAs were extracted using the TRIzol Reagent (Invitrogen, Carlsbad, CA, USA) following the supplier’s guidelines. A Nanodrop ND-1000 spectrophotometer (Thermo Fisher Scientific Inc., Waltham, MA, USA) and gel electrophoresis were used to monitor the quality and quantity of the RNA. Its concentration was determined using a Qubit® 2.0 Fluorometer (Life Technologies, Carlsbad, CA, USA) and its integrity was evaluated using a 2100 Bioanalyzer (Agilent, Santa Clara, CA, USA). An Epicentre Ribo-Zero Gold Kit (Illumina, San Diego, CA, USA) was used to deplete ribosomal RNAs from the total RNA samples. An Illumina TruSeq® RNA Sample Prep Kit (Illumina, San Diego, CA, USA) was then used to construct a cDNA library from the ribosomal RNA-depleted RNA. The cDNA library was then subjected to HTS using the PE150 sequencing platform on an Illumina NovaSeq 6000 sequencer (Illumina, San Diego, CA, USA), carried out by LC-Bio Technology Co., Ltd. (Hangzhou, China).
2.3. Data Processing and Virus Annotation
The raw reads from rRNA-depleted total RNA sequencing were trimmed to remove low-quality and adapter sequences using fastp [12]. The genome of Benincasa hispida Cogn. var. chieh-qua How has not been published; therefore, the genome sequence of Wax gourd (Benincasa hispida) (https://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/009/727/055/GCF_009727055.1_ASM972705v1/, accessed on 13 January 2021) was used as the reference to eliminate host-derived sequences using HISAT v2.1.0 [13] for dataset size reduction. Trinity v2.13.3 [14] was used to de novo assemble the unmapped reads, which were then annotated according to BLASTn and BLASTx [15] searches against the nonredundant nucleotide (nt) and protein (nr) databases in GenBank [16], employing conservative cut-off e-values of 10−4 and 10−6, respectively.
2.4. RT-PCR Protocol
An RNAPrep Pure Plant Kit (TianGen, Beijing, China) was used to extract the total RNA from individual plants, which was reverse transcribed into cDNA using random hexamers and TransScript One-Step gDNA Removal and cDNA Synthesis SuperMix (TransGen, Beijing, China) following the supplier’s recommendations. The 2×Rapid Master Mix (Vazyme, Nanjing, China) was used to carry out subsequent PCR reactions using specific virus detection primers (Table S1). These primers were designed against the assembled contig sequences using Primer3 [17]. The reaction conditions for virus-specific PCR were: heating at 95 °C for 2 min; followed by 35 cycles of 95 °C for 45 s, 53 °C or 55 °C for 45 s and 72 °C for 45 s; followed by holding at 72 °C for 10 min and then holding at 4 °C until the analysis. A TIANgel Midi Purification kit (TianGen, Beijing, China) was used to purify the target amplicons, which were either sequenced directly or first ligated into vector pMD18-T (Takara, Kusatsu, Japan). The recombinant vectors were transformed into Escherichia coli DH5α competent cells (Takara, Kusatsu, Japan). We then selected three individual clones of each amplicon that had inserts of the expected size, which were sequenced in both directions using Sanger sequencing, carried out at BGI (Shenzhen, China).
2.5. Recovery of the Complete Genome and Characterization of the Newly Identified Viruses
Following confirmation of their virus status, the whole genomes of the newly discovered viruses were amplified using 2× Phanta Flash Master Mix (Vazyme, Nanjing, China) with various pairs of virus-specific primers (Table S2). The sequence of the novel crinivirus of Closteroviridae was obtained with complete 5′ and 3′ untranslated regions (UTRs). A SMARTer® RACE 5′/3′ kit with SeqAmp DNA Polymerase (Takara, San Jose, CA, USA) was used to determine the 5′ UTR sequence of the novel crinivirus. Poly(A) Polymerase (Takara, Kusatsu, Japan) was then used to add additional ATPs to the 3′ terminus of the viral genome lacking a poly(A) tail following the supplier’s guidelines. Subsequently, the 3′ terminal sequence was completed using an anchored oligo (dT) primer (5′-TGTGTTGGGTGTGTTTGGTTTTTTTTTTTTTTT-3′) to synthesize the cDNA from the poly(A)-tailed RNA virus, followed by amplification using the primer anchored-dt-rev (5′-TGTGTTGGGTGTGTTTGG-3′) together with a gene-specific primer.
For genomic characterization, the web tool ORFfinder (https://www.ncbi.nlm.nih.gov/orffinder/, accessed on 2 December 2022) was used to predict the open reading frames (ORFs) and the Conserved Domain Database (CDD, https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi, accessed on 2 December 2022) was used to analyze conserved motifs and domains. Transmembrane domains were predicted using DeepTMHMM [18]. Pairwise comparisons were calculated from CLUSTALW multiple alignments and MEGA 11 [19] was used to construct neighbor-joining (NJ) phylogenetic trees with 1000 bootstraps.
2.6. Recombination Analysis
We scanned the complete genome sequence alignment file for potential recombination events involving five cucurbit chlorotic virus (CuCV) isolates (cucumber-SY, pumpkin-DF, chieh-qua-CM, watermelon-LS and Thailand) and the CYSDV isolates available in the NCBI database using the recombination analysis software SimPlot version 3.5.1 [20] and the Recombination Detection Program v.4.43 (RDP4) [21]. Except for the selection of the linear genome option, each RDP4 analysis used the default settings. We considered only the events detected by a minimum of five of the seven detection methods (RDP, GENECONV, Bootscan, MaxChi, Chimaera, 3Seq and SiScan), which had at least three p-values < 10−6 and combination scores above 0.6. Events with a recombination score between 0.4 and 0.6 were considered fairly likely.
3. Results
3.1. HTS Output, Discovery of Viruses and HTS Validation
The rRNA-depleted total RNA sequencing of sample PL-1 produced 84,840,500 raw reads (81,484,200 clean reads), whose Q20 and Q30 values were >94%. A total of 77,465,294 (95.07%) of the reads mapped to the wax gourd reference genome. De novo assembly of the remaining 4,018,906 (4.93%) unmapped reads produced 23,111 contigs (lengths = 201–15,020 nt). BLASTn and BLASTx analyses identified 31 contigs corresponding to viruses of the Closteroviridae family (12 contigs, length ranging from 904 to 8886 nt), Tospoviridae (14 contigs, length ranging from 251 to 8902 nt), Potyviridae (3 contigs, length ranging from 270 to 10,320 nt) and Endornaviridae (2 contigs, with lengths of 14,990 and 15,020 nt) (Table 1). According to the generally low level of the amino acid sequence identity between the contigs and those of the known viruses in the NCBI database, we initially assumed that the chieh-qua composite sample (PL-1) comprised at least six viruses: MYSV, CCYV, WSMoV, PRSV, a novel crinivirus and a novel alphaendornavirus.

Table 1.
Statistics for rRNA-depleted total RNA sequencing output of chieh-qua sample PL-1 by BLASTx analysis.
To confirm that the putative viruses were not artefacts of HTS sequencing, seven primer pairs (Table S1) were designed based on the assembled contigs for individual RT-PCR detection on all five plant samples used for HTS and the resultant amplicons were sequenced. The results confirmed that the five samples tested positive for one to six of the six viruses (Table S3) and sequences of these amplicons showed 99–100% identity with the sequences obtained by HTS. CuCV, MYSV and chieh-qua endornavirus (CqEV) were identified in chieh-qua for the first time.
3.2. Recovery of the Complete Genomes and Characterization of a Potential New Crinivirus in the Family Closteroviridae
The entire genomes of the new crinivirus, CuCV, in chieh-qua were 9157 nt for RNA1 and 7788 nt for RNA2, whose poly(A) tails were of an undefined length (Figure 2A). To better understand the genomic sequence diversity of CuCV in different natural hosts, we also obtained the complete or near-complete genomes of CuCV from other cucurbit vegetables (pumpkin, watermelon and cucumber) (Table 2). The four CuCV isolates shared a high nt identity for the RNA1 (98.32–99.33%) and RNA2 (99.68–99.83%) genomic segments (Table 3).
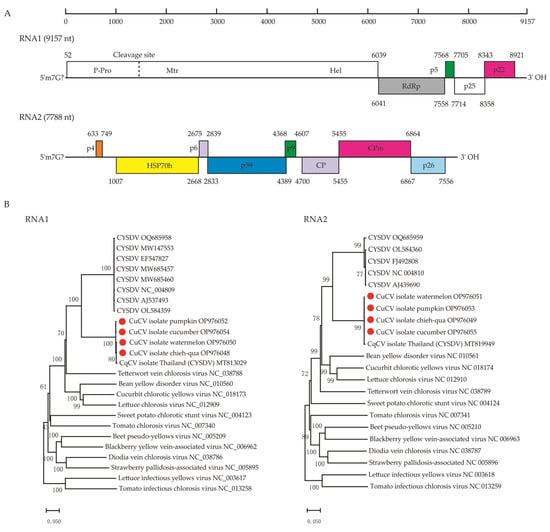
Figure 2.
Cucurbit chlorotic virus (CuCV) genomic structure and phylogeny. (A) Schematic diagram of the CuCV genomic organization. The numbers below diagrams are the genomic nucleotide (nt) positions. Boxes represent the predicted open reading frames (ORFs), with indicated nt coordinates. Different colors represent the conserved motifs, domains and viral proteins within the ORFs: a viral methyltransferase domain (Mtr, pfam01660), viral helicase domain (Hel, pfam 01443), an RNA-dependent RNA polymerase (RdRp), heat-shock-protein-70-like protein (HSP70h), coat protein (CP), minor coat protein (CPm). Predicted proteins with unknown functions are also shown: a 5 kDa protein (p5), a 25 kDa protein (p25), a 22 kDa protein (p22) in CuCV RNA1 and a 4 kDa protein (p4), a 6 kDa protein (p6), a 59 kDa protein (p59), a 9 kDa protein (p9), a 26 kDa protein (p26) in CuCV RNA2. (B), Neighbor-joining (NJ) phylogenetic trees based on the complete nucleotide sequences of RNA1 (Left) and RNA2 (Right) segments of the representative members of the genus Crinivirus, family Closteroviridae. CYSDV, cucurbit yellow stunting disorder virus. The newly identified viruses in this study are indicated by red dots.

Table 2.
The cucurbit chlorotic virus (CuCV) isolates used in this study.

Table 3.
Occurrence of viruses infecting chieh-qua in different plantations in Hainan Province, China.
BLASTn analyses revealed that CuCV and a so-called CYSDV Thailand isolate (MT813029 and MT819949) available in the GenBank database are molecularly identified as the same virus species. This conclusion is based on their complete genomes sharing more than 98% nucleotide identity (with 99% coverage). However, they exhibit significant divergence from other CYSDV isolates, with nucleotide identities ranging from 70.25% to 74.05% (coverage of 67% to 77%, e-value = 0.0) and a shared amino acid identity of 74.1% for the coat protein (Table S4). These findings challenge the existing nomenclature for the CYSDV Thailand isolate (MT813029 and MT819949), suggesting a misclassification. Through sequence comparison, phylogenetic analysis and literature research, we have determined that the CuCV identified in this study, along with the so-called CYSDV isolates from Thailand, potentially represent a distinct and divergent species separate from known CYSDV members (Figure 2B, Table S4).
The genomic organization of the characterized virus (Figure 2A) was very similar to those of members of the genus Crinivirus [22]. The RNA1 genomic segment contains five predicted ORFs, which encode a 1996 aa polyprotein (ORF1a, nt 52–6039, 229.83 kDa), RNA-dependent RNA polymerase (RdRp) (ORF1b, nt 6041–7558, 505 aa, 59.11 kDa; expressed from ORF1a via a +1 frame shift) and three proteins of unknown function (p5, p25 and p22). Based on similarities with other members of the genus Crinivirus, a putative P-Pro domain [23] (P-Pro, nt 52–1491) was predicted in the N-terminal of ORF1a, which has two conserved catalytic amino acids residues (Cys-412 and His-461), while the putative cleavage sites are between Gly-480 and Val-481. Viral methyltransferase (Mtr, nt 1705–2637, pfam 01660) domains, viral helicase (Hel, nt 5146–5937, pfam 01443) domains and two transmembrane domains between residues 1369 to 1379 and 1383 to 1394 were also predicted in ORF1a. The 5′- and 3′- UTRs of RNA1 are 51 and 237 nt, respectively.
RNA2 comprises eight predicted ORFs flanked by a 5′-UTR of 632 nt and a 3′-UTR of 231 nt, namely p4 (nt 633–749, 38aa, 4.5 kDa), a heat-shock-protein-70-like protein (HSP70h, nt 1007–2668, 553 aa, 62.4 kDa), p6 (nt 2675–2839, 54 aa, 6.62 kDa), p59 (nt 2833–4389, 518 aa, 59.67 kDa), p9 (nt 4368–4607, 79 aa, 9.44 kDa), coat protein (CP, nt 4700–5455, 251 aa, 28.80 kDa), minor CP (CPm, nt 5455–6864, 469 aa, 54.38 kDa) and p26 (nt 6867–7556, 229 aa, 26.89 kDa). Neither the p5 in RNA1 nor p4 in RNA2 have any homologs in the database. The seven nucleotides (ACATGGG) at the 5′-UTR of RNAs 1 and 2 are identical and although this feature is common in criniviruses [24], this sequence is otherwise unique.
3.3. The Genetic Variability and Recombination of CuCV Isolates from Cucurbit Vegetables
Phylogenetic relationships based on the RdRp and HSP70 protein sequences of representative members of Crinivirus in the Closteroviridae family revealed that four CuCV isolates (chieh-qua, cucumber, pumpkin and watermelon) in this study and the so-called CYSDV isolate from Thailand were consistently placed in a branch distinct from other CYSDV members (Figure S1). Although the RdRp and HSP70h proteins of CuCV shared the highest aa identities, of 90.10% and 83.91%, with those of CYSDV (Table S4), the CP shared only a 74.1% aa identity with CYSDV, which satisfies the accepted molecular criteria for species demarcation in the Closteroviridae family of <75% aa identity for the RdRp, HSP70h and CP genes [25].
The phylogenetic reconstruction of the CP genes of 51 available CYSDV isolates, 5 CuCV isolates (cucumber-SY, Pumpkin-DF, watermelon-LS, chieh-qua-CM and Thailand) and the formally identified criniviruses showed that the CuCV isolates were located in a distinct cluster away from the CYSDV clusters, which, similar to the results of previous studies [26], was separated into two groups (Figure 3A). The two groups of CYSDV were located in (I) the Mediterranean Basin (South Europe, North Africa and the Near East) and North America and (II) the Middle East (Iran, Sudan and Arabia). No significant evidence of geographical association or host structure was observed in the population.
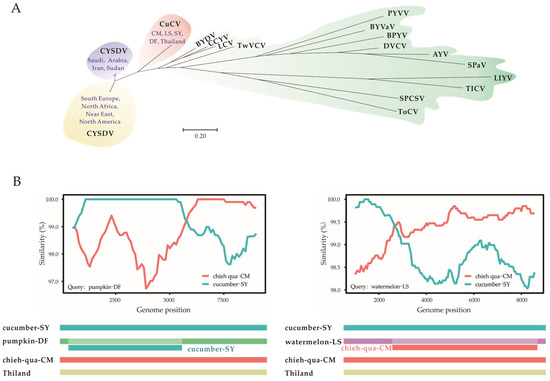
Figure 3.
(A) The neighbor-joining (NJ) phylogenetic trees constructed based on the coat protein gene nucleotide sequences of cucurbit chlorotic virus (CuCV), cucurbit yellow stunting disorder virus (CYSDV) and representative members of the genus Crinivirus, family Closteroviridae. (B) Recombination analyses of five CuCV isolates using the recombination detection program Simplot and RDP4. Abbreviations and accession numbers for the viruses are AYV (abutilon yellows virus, AY422070), BPYV (beet pseudoyellows virus, NC_005210), BYDV (bean yellow disorder virus, NC_010561), BYVaV (blackberry yellow vein-associated virus, NC_006963), CCYV (cucurbit chlorotic yellows virus, NC_018174), CuCV (OP976049, OP976051, OP976053, OP976055, MT819949), CYSDV (AJ439690, AJ243000, AY730779, DQ903105-DQ903111, EF210558, EF210560, EF210561, EF210559, FJ492808, HG939523, KC677625, KC677626, KC469990-KC470000, KX768875, LT992903-LT992905, JF340435, JN083790, LT992890-LT992902, OL584360, OQ685959, NC_004810, MW685458, MW685459), DVCV (diodia vein chlorosis virus, NC_038787), LCV (lettuce chlorosis virus, NC_012910), LIYV (lettuce infectious yellows virus, NC_003618), PYVV (potato yellow vein virus, YP_054421), SPaV (strawberry pallidosis-associated virus, NC_005896), SPCSV (sweet potato chlorotic stunt virus, NC_004124), TICV (tomato infectious chlorosis virus, NC_013259), ToCV (tomato chlorosis virus, NC_007341), TwVCV (tetterwort vein chlorosis virus, NC_038789).
The Simplot and RDP software detected the two recombinants of CuCV isolate pumpkin-DF and watermelon-LS, the potential parents of which were CuCV isolate chieh-qua-CM and cucumber-SY (Figure 3B). The recombination break points in the CuCV isolate pumpkin-DF started at nt 370 and terminated at nt 5334, with the highest probability value of 10−32 according to the 3Seq method and a high recombination score of 0.815, which targeted the region encoding the replication-associated protein. The break points of the watermelon-LS isolate started at nt 2170 and terminated at nt 8686, with the highest probability value of 10−12 according to the SiScan method and a recombination score of 0.526 (Figure 3B). No significant recombination signals were detected between CuCV local isolates and the Thailand isolate or between any of the CuCV isolates and CYSDV isolates.
3.4. A New Alphaendornavirus in Family Endornaviridae
The novel alphaendornavirus, provisionally named chieh-qua endornavirus (CqEV) has a monopartite genome of ~15,020 nt based on the long scaffolds that were assembled from the rRNA-depleted total RNA sequencing, which lacks 5′- and 3′-terminal sequences (GenBank accession number OQ851472). Similar to other endornaviruses, the CqEV genome has only a single ORF encoding a 4981 aa polyprotein (estimated molecular mass = 573.13 kDa; encoded by nt 34–14,979) (Figure 4A). A conserved domain search detected a putative viral helicase 1 domain (Hel-1, aa 1438–1639, pfam01443), two capsular polysaccharide synthesis protein domains (CPS, aa 2772–2955, aa 3193–3321, pfam05704), a UDP-glycosyltransferases domain (UGT, aa 3709–3843, cd03784) and an RdRp domain (aa 4612–4848, cd23255). As expected, BlastN and BlastX analyses showed that cucumis melo endornavirus (CmEV) was most closely related to the endornaviral sequences.
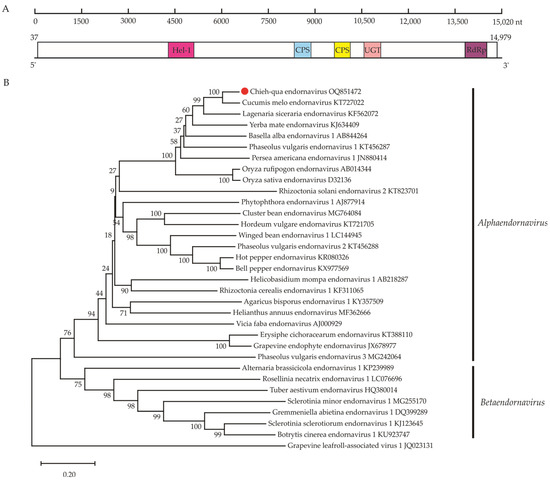
Figure 4.
Genome organization and phylogeny of chieh-qua endornavirus (CqEV). (A) Schematic diagram showing the CqEV genomic organization: The numbers under the diagram are the nucleotide (nt) positions in the genome. Boxes indicate the predicted open reading frames (ORFs), with nt coordinates indicated below. Different colors indicate conserved domains: viral helicase 1 domain (Hel-1, pfam01443), two capsular polysaccharide synthesis protein domains (CPS, pfam05704), a UDP-glycosyltransferases domain (UGT, cd03784) and an RNA-dependent RNA polymerase domain (RdRp, cd23255). (B) The neighbor-joining (NJ) phylogenetic trees constructed based on the amino acid sequences of the RNA-dependent RNA polymerase domains of the representative Endornaviridae members with grapevine-leafroll-associated virus 1 as an outgroup member. The newly identified viruse in this study is indicated by red dots.
To ascertain the relationship between CqEV and other members of the Alphaendornavirus, phylogenetic analysis of RdRP protein sequences from different endornaviruses (Figure 3B) revealed that CqEV consistently clustered with viruses in the genus Alphaendornavirus and formed a sister branch to CmEV (KT727022). Furthermore, the genome sequence of CqEV was compared with those of the 24 reported formal species in the genus Alphaendornavirus. The results indicated that CqEV is most closely related to CmEV (KT727022), sharing a 65.41% nt identity (Table S5). This fulfills the species demarcation criteria of overall nucleotide sequence identity below 75% [27]; therefore, the new virus belongs to a new species of the genus Alphaendornavirus in the family Endornaviridae.
3.5. The Prevalence of Viruses Infecting Chieh-Qua
We examined 135 chieh-qua leaf samples from 7 counties (Table 3) to ascertain the distribution and incidence of the 6 viruses discovered by HTS and 3 viruses (i.e., CMV, ZYMV and ZTMV) reported for chieh-qua. RT-PCR and electrophoretic analyses indicated that MYSV, CCYV, CqEV and CuCV could be detected in all sampling sites. MYSV had the highest prevalence of 66.67% (90/135) and its incidence rate was 100% in samples from 4 sampled counties (Ding’an, Lingshui, Sanya, Dongfang). CCYV had a high prevalence of 55.56% (75/135). The new alphaendornavirus, CqEV, had a prevalence of 35.56% (48/135), with a high incidence of 92.86% and 100% in the Ding’an and Donfang regions, respectively. The new crinivirus, CuCV, had a prevalence of 27.41% (37/135) and was detected in samples from Chengmai, Wenchang, Ding’an, Lingshui and Sanya. The incidence of WSMoV, PRSV, ZYMV and CMV was generally low in most of the sites sampled (Table 3). MYSV and CCYV might be the main viruses infecting chieh-qua in Hainan province. Among the 135 samples, CqEV, WSMoV and PRSV were never detected to singly infect chieh-qua. A total of 78 (57.78%) samples were co-infected by multiple viruses with 8 types of co-infection, and 57 samples were detected as positive for 1 specific virus (Figure 5). Among the 28 asymptomatic chieh-qua samples collected from Chengmai in Hainan province in 2022, 9 were positive for CqEV according to RT-PCR, while all the samples were negative for CuCV.
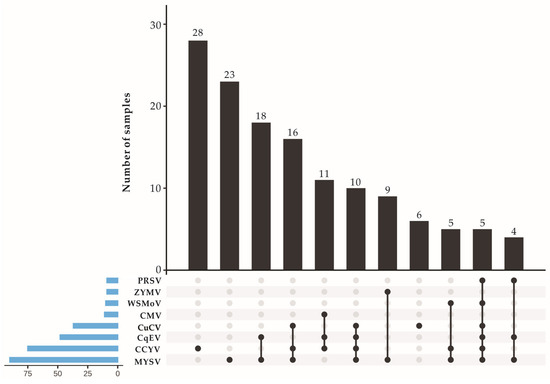
Figure 5.
Virus infection types and the number of samples corresponding to each infection type in chieh-qua (this figure was created using EVenn [28]). Abbreviations for the viruses are PRSV (papaya ringspot virus), ZYMV (zucchini yellow mosaic virus), WSMoV (watermelon silver mottle virus), CMV (cucumber mosaic virus), CuCV (cucurbit chlorotic virus), CqEV (chieh-qua endornavirus), CCYV (cucurbit chlorotic yellows virus) and MYSV (melon yellow spot virus).
To further explore the occurrence of the potential new crinivirus (CuCV) and the new endogenousvirus (CqEV) in different cucurbit vegetables, 98 symptomatic samples of 4 cucurbit vegetables were collected from 7 counties of Hainan. A total of 16 samples were CuCV positive by RT-PCR, including samples from pumpkin (4/23), watermelon (4/26) and cucumber (8/49) (Table S6). No CqEV was found in any of these 98 samples (Table S6), which indicated that CqEV is an endogenousvirus unique to chieh-qua.
4. Discussion
In this study, a putative novel crinivirus was identified and molecularly characterized. The closest relative of CuCV appears to be CYSDV, a Bemisia-tabaci-transmitted crinivirus causing extensive infections of cucurbit crops in many warm and temperate production areas [29,30,31,32,33]. The taxonomy of CuCV was first determined based on the low complete genome identity (65–69%) with CYSDV isolates compared to the 99% intraspecific similarity of CYSDV isolates (Tables S7 and S8), despite the extensive and discontinuous geographical distribution and different years of collection. However, CuCV and CYSDV share a high identity for the conserved RdRp and HSP70 proteins, which is frequently observed for other criniviruses, for example, amino acid similarities of 76.63–85.74% were discovered between diodia vein chlorosis virus (DVCV) and strawberry-pallidosis-associated virus (SPaV), bean yellow disorder virus (BYDV) and lettuce chlorosis virus (LCV) and tetterwort vein chlorosis virus (TwVCV) and BYDV (Table S9). In addition, the CP and all the other predicted proteins shared similarities of <75% at the AA level and at nt level with those of CYSDV (Table S10), which is far lower than the CYSDV intraspecific protein similarity. CuCV and CYSDV have a slightly different genomic arrangement. 5′- UTR of RNA1 and RNA2 of CuCV are approximately 1/2 of those of CYSDV in length and shared no significant nucleotide identities (Table S6). The 5′- end of CYSDV RNA 2 encodes a putative p4.9 protein that overlaps with HSP70h; however, the corresponding region of the CuCV genome encodes a unique p4 protein located distally from HSP70h. This evidence indicates the taxonomy of CuCV, a putative novel crinivirus in the family Closteroviridae.
Five CuCV isolates (four from this study and one from Thailand) from different natural hosts (chieh-qua, pumpkin, watermelon, cucumber and melon) showed generally limited spatial and temporal sequence variability, which is also found in some other viruses in the family Closteroviridae [33,34,35,36]. Although we identified significant recombination signals on the isolates from pumpkin and watermelon that appeared to be derived from chieh-qua and cucumber-hosted isolates, further validation is needed because the current biological data of the hosts and the geographical information cannot completely explain these recombination events.
In this study, a strong association between CuCV and specific symptoms was suggested because CuCV was found exclusively in symptomatic cucurbit samples but not in any of the 28 asymptomatic samples, which aligns with the results reported by Krishnan et al. [37] where CuCV (referred to as CYSDV in this article) was detected in all 16 symptomatic cucurbits samples from India but not in any of the asymptomatic samples. The statistical analysis results indicated that the specific symptoms caused by CuCV on cucurbit plants are potentially interveinal chlorosis, which is also in line with the study reported in India that CuCV causes interveinal chlorosis followed by bright yellowing in cucurbits (bitter gourd, cucumber and watermelon) [37].
CuCV is the fourth crinivirus reported to infect cucurbit plants in addition to CYSDV, CCYV and beet pseudoyellows virus (BPYV) [35,36]. So far, CuCV has been discovered in various cucurbits (melons, cucumbers, bitter melons, pumpkin, chieh-qua and watermelons) in Thailand, India and China, which indicates the potential broad host range of CuCV and the risk of a widespread outbreak of this virus. In addition, it has been suggested that CuCV is transmitted by whiteflies, like other criniviruses [38], thus further research is required to determine its biological, pathogenic and phytosanitary risks.
CqEV is the first endornavirus reported to infect chieh-qua. No CqEV was found as a single infection in symptomatic samples and 9 out of 28 asymptomatic samples were positive for CqEV, which indicated that CqEV is in a symbiotic relationship with chieh-qua causing no severe symptoms, similar to most plant endornaviruses, with the exception of Vicia faba endornavirus [39,40,41]. CqEV was only found in chieh-qua and not in cucumber, pumpkin and watermelon, which indicated that CqEV has a narrow host range. Except for cucumis melo endornavirus, which has the ability to infect three distinct genera in its family [42], the majority of endornaviruses have been identified in only one host species or, in some cases, in a limited number of closely related species within the same genus. Certain endornaviruses display an even narrower host range, only associating with specific genotypes or varieties of a particular host species, as is the case with phaseolus vulgaris endornavirus 1 (PvEV-1) and phaseolus vulgaris endornavirus 2 (PvEV-2) [43]. Although the percentage of plants containing endornavirus sequences is high (~8%) [44], the nature of their relationship with their host plants remains unclear.
In this study, chieh-qua is for the first time reported as the natural host of CuCV, MYSV and CqEV. Complex mix infection was commonly found in chieh-qua, though 57 samples were “single infected”. The virus-induced symptoms cannot differentiate between single or multiple infections, since the single-infected samples might have been infected with other unknown viruses besides the tested ones. Among the identified viruses, MYSV and CCYV were the most prevalent in this study and they were also reported to cause great losses in cucurbit vegetable production in many countries [45,46,47,48,49]. Further large-scale and in-depth investigations are needed concerning the virus diversity, epidemiology and evolution for the effective prevention and control of disease outbreaks in cucurbit and other plants.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/v15061396/s1, Figure S1: The neighbor-joining (NJ) phylogenetic trees constructed on the RNA-dependent RNA polymerase (RdRp) and heat shock protein 70 (HSP70) gene nucleotide sequences of cucurbit chlorotic virus (CuCV), cucurbit yellow stunting disorder virus (CYSDV) and the representative members of the genus Crinivirus, family Closteroviridae; Table S1: Specific primers used to confirm the presence of the virus in the original plants and to survey virus prevalence; Table S2: Primers used to complete the genomes of the new viruses; Table S3: RT-PCR confirmation of the presence of the virus in individual plants; Table S4: The percent amino acid identity RdRP, HSP70, and CP between the cucurbit chlorotic virus (CuCV) and other unclassified and formal members in the genus Crinivirus of the family Closteroviridae; Table S5: Overall nucleotide sequence identity (%) between chieh-qua endornavirus (CqEV) and members of different species in the genus Alphaendornavirus of the family Endornaviridae; Table S6: Occurrence of cucurbit chlorotic virus (CuCV) in different hosts; Table S7: The pairwise nucleotide identities (%) of genome RNA1 among cucurbit chlorotic virus (CuCV) virus and cucurbit yellow stunting disorder virus (CYSDV) isolates available in the NCBI database; Table S8: The pairwise nucleotide identities (%) of genome RNA2 among cucurbit chlorotic virus (CuCV) virus and cucurbit yellow stunting disorder virus (CYSDV) isolates available in the NCBI database; Table S9: The percent amino acid identity of RdRp (lower left) and HSP70h (upper right) among the formal species in the genus Crinivirus of the family Closteroviridae; Table S10. Percentages of the nucleotide (Nt) and deduced protein sequence (AA) identities between CuCV (RNA1: OP976048, OP976050, OP976052, OP976054, MT813029; RNA2: OP976049, OP976051, OP976053, OP976055 and MT819949) and CYSDV (RNA1: MW147553, EF547827, MW685457, MW685460, AY242077, AJ537493 and OL584359; RNA2: FJ492808, AY242078, MW685458, MW685459, OL584360 and AJ439690) available in the NCBI database.
Author Contributions
Conceptualization, H.C., Y.M. and H.L.; methodology, H.C. and Y.M.; software, Y.M. and H.C.; validation, H.C. and Y.L.; formal analysis, H.C.; investigation, H.C., Y.L., T.F., D.L. and H.L.; resources, H.C.; data curation, H.C. and Y.M.; writing—original draft preparation, H.C.; writing—review and editing, Y.M. and H.L.; visualization, H.C. and T.F.; supervision, H.C.; project administration, H.C. and H.L.; funding acquisition, H.C., Y.M. and D.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Hainan Provincial Natural Science Foundation of China, grant Number 2019RC284; the Central Public-Interest Scientific Institution Basal Research Fund for Chinese Academy of Tropical Agricultural Sciences, grant Number No. 1630042019001; the National Natural Science Foundation of China, grant number 32102182.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available from the corresponding author upon reasonable request.
Acknowledgments
The authors thank Xueren Cao (Environment and Plant Protection Institute, Chinese Academy of Tropical Agricultural Sciences) for his help with sampling.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Mao, Y.Z.; Jiang, B.; Peng, Q.W.; Liu, W.R.; Lin, Y.; Xie, D.S.; He, X.M.; Li, S.S. Cloning and characterization of WRKY gene homologs in Chieh-qua (Benincasa hispida Cogn. var. Chieh-qua How) and their expression in response to fusaric acid treatment. 3 Biotech 2017, 7, 86–95. [Google Scholar] [CrossRef] [PubMed]
- Department of Agriculture and Rural Affairs of Guangdong Province. Available online: http://dara.gd.gov.cn/nyyw/content/post_3327295.html (accessed on 15 April 2023).
- He, X.M.; Xie, D.S.; Chen, Q.H.; Peng, Q.W. Chieh-qua biotechnology, progress and prospects. Asian Australas. J. Plant Sci. Biotechnol. 2007, 1, 19–22. [Google Scholar]
- Zhou, S.J.; Chen, X.J.; Zhu, Y.Q.; Chen, L.P.; Zhang, P. Research progress and suggestion on germplasms of ash gourd and chieh-qua. J. Plant Genet. Resour. 2014, 15, 211–214. [Google Scholar]
- He, X.M.; Peng, Q.W.; Wang, M.; Yan, J.Q. Research Progresses in genetic breeding of chieh-qua in China. Guangdong Agri. Sci. 2021, 48, 1–11. [Google Scholar]
- Lu, C.G.; Li, H.F.; Fan, Z.F. Identification and cloning of coat protein gene of zucchini yellow mosaic virus infecting hairy squash. In Proceedings of the Annual Meeting of Chinese Society for Plant Pathology 2004, Ningbo, China, 1–4 September 2004; pp. 224–226. [Google Scholar]
- Zhou, C.J.; Liang, Z.R.; Zhang, J.B.; Huang, B.; He, G.Z.; Zhong, C.N.; Ouyang, T.X. First report of zucchini tigre mosaic virus infecting four cucurbit crops in China. Plant Dis. 2023, 107, 1247. [Google Scholar] [CrossRef]
- Zhao, Q.; Li, H.P.; Xie, D.S.; Luo, S.B.; Peng, Q.W. Development of a multiplex RT-PCR protocol for the detection of three viruses on Benincasa hispida. var. chieh-qua. Acta Hortic. Sin. 2011, 38, 2215–2222. [Google Scholar]
- Liu, Y.; Li, F.; Li, Y.Y.; Zhang, S.B.; Gao, X.W.; Xie, Y.; Yan, F.; Zhang, A.S.; Dai, L.Y.; Cheng, Z.B.; et al. Identification, distribution and occurrence of viruses in the main vegetables of China. Sci. Agri. Sin. 2019, 52, 239–261. [Google Scholar]
- Xiao, L.; Li, Y.Y.; Tan, G.L.; Lan, P.X.; Zhong, L.; Liu, Y.; Li, R.; Li, F. First report of zucchini tigre mosaic virus infecting several cucurbit plants in China. Plant Dis. 2016, 100, 1253. [Google Scholar] [CrossRef]
- Nong, Y. Identification of Viruses Infecting Cucurbit Crops in Guangdong Province and Analysis of Important Virus Characteristics; South China Agricultural University: Guangzhou, China, 2020; pp. 25–26. [Google Scholar]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Sayers, E.W.; Cavanaugh, M.; Clark, K.; Ostell, J.; Pruitt, K.D.; Karsch-Mizrachi, I. GenBank. Nucleic Acids Res. 2020, 48, D84–D86. [Google Scholar] [PubMed]
- Koressaar, T.; Lepamets, M.; Kaplinski, L.; Raime, K.; Andreson, R.; Remm, M. Primer3_masker: Integrating masking of template sequence with primer design software. Bioinformatics 2018, 34, 1937–1938. [Google Scholar] [CrossRef]
- Hallgren, J.; Tsirigos, K.D.; Pedersen, M.D.; Armenteros, J.J.A.; Marcatili, P.; Nielsen, H.; Krogh, A.; Winther, O. DeepTMHMM predicts alpha and beta transmembrane proteins using deep neural networks. BioRxiv 2022. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Lole, K.S.; Bollinger, R.C.; Paranjape, R.S.; Gadkari, D.; Kulkarni, S.S.; Novak, N.G.; Ingersoll, R.; Sheppard, H.W.; Ray, S.C. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 1999, 73, 152–160. [Google Scholar] [CrossRef]
- Martin, D.P.; Varsani, A.; Roumagnac, P.; Botha, G.; Maslamoney, S.; Schwab, T.; Kelz, Z.; Kumar, V.; Murrell, B. RDP5: A computer program for analyzing recombination in, and removing signals of recombination from, nucleotide sequence datasets. Virus Evol. 2021, 7, veaa087. [Google Scholar] [CrossRef]
- Fuchs, M.; Bar-Joseph, M.; Candresse, T.; Maree, H.J.; Martelli, G.P.; Melzer, M.J.; Menzel, W.; Minafra, A.; Sabanadzovic, S. ICTV Report Consortium. ICTV Virus Taxonomy Profile: Closteroviridae. J. Gen. Virol. 2020, 101, 364–365. [Google Scholar] [CrossRef]
- Peng, C.W.; Peremyslov, V.V.; Mushegian, A.R.; Dawson, W.O.; Dolja, V.V. Functional specialization and evolution of leader proteinases in the family Closteroviridae. J. Virol. 2001, 75, 12153–12160. [Google Scholar] [CrossRef]
- Wintermantel, W.M.; Wisler, G.C.; Anchieta, A.G.; Liu, H.Y.; Karasev, A.V.; Tzanetakis, I.E. The complete nucleotide sequence and genome organization of tomato chlorosis virus. Arch. Virol. 2005, 150, 2287–2298. [Google Scholar] [CrossRef] [PubMed]
- International Committee on Taxonomy of Viruses. Available online: https://ictv.global/report/chapter/closteroviridae/closteroviridae/crinivirus (accessed on 16 April 2023).
- Rubio, L.; Guerri, J.; Moreno, P. Genetic variability and evolutionary dynamics of viruses of the family Closteroviridae. Front. Microbiol. 2013, 4, 151. [Google Scholar] [CrossRef]
- International Committee on Taxonomy of Viruses. Available online: https://ictv.global/report/chapter/endornaviridae/endornaviridae/alphaendornavirus (accessed on 16 April 2023).
- Chen, T.; Zhang, H.Y.; Liu, Y.; Liu, Y.X.; Huang, L.Q. EVenn: Easy to create repeatable and editable Venn diagrams and Venn networks online. J. Genet. Genom. 2021, 48, 863–866. [Google Scholar] [CrossRef]
- Hassan, A.A.; Duffus, J.E. A review of a yellowing and stunting disorder of cucurbits in the United Arab Emirates. Emir. J. Food Agric. 1991, 2, 1–16. [Google Scholar] [CrossRef]
- Coutts, R.H.A.; Livieratos, I.C. Nucleotide sequence and genome organization of cucurbit yellow stunting disorder virus RNA1. Arch. Virol. 2003, 148, 2055–2062. [Google Scholar] [CrossRef]
- Jailani, A.A.K.; Iriarte, F.; Hochmuth, B.; Willis, S.M.; Warren, M.; Dey, K.; Velez-Climentet, M.; McVay, J.; Bag, S.; Paret, M.L. First report of cucurbit chlorotic yellows virus affecting watermelon in USA. Plant Dis. 2022, 106, 774. [Google Scholar] [CrossRef]
- Mondal, S.; Hladky, L.J.; Wintermantel, W.M. Differential seasonal prevalence of yellowing viruses infecting melon crops in southern California and Arizona determined by multiplex RT-PCR and RT-qPCR. Plant Dis. 2023; in press. [Google Scholar] [CrossRef] [PubMed]
- Rubio, L.; Abou-Jawdah, Y.; Lin, H.X.; Falk, B.W. Geographically distant isolates of the crinivirus cucurbit yellow stunting disorder virus show very low genetic diversity in the coat protein gene. J. Gen. Virol. 2001, 82, 929–933. [Google Scholar] [CrossRef] [PubMed]
- Marco, C.F.; Aranda, M.A. Genetic diversity of a natural population of cucurbit yellow stunting disorder virus. J. Gen. Virol. 2005, 86, 815–822. [Google Scholar] [CrossRef]
- Rubio, L.; Soong, J.; Kao, J.; Falk, B.W. Geographic distribution and molecular variation of isolates of three whitefly-borne closteroviruses of cucurbits: Lettuce infectious yellows virus, cucurbit yellow stunting disorder virus, and beet pseudo-yellows virus. Phytopathology 1999, 89, 707–711. [Google Scholar] [CrossRef]
- Boubourakas, I.N.; Avgelis, A.D.; Kyriakopoulou, P.E.; Katis, N.I. Occurrence of yellowing viruses (Beet pseudo-yellows virus, cucurbit yellow stunting disorder virus and cucurbit aphid-borne yellows virus) affecting cucurbits in Greece. Plant Pathol. 2006, 55, 276–283. [Google Scholar] [CrossRef]
- Krishnan, N.; Kumari, S.; Pandey, S.; Ram, D.; Behera, T.K.; Gandhi, K. Occurrence of cucurbit yellow stunting disorder virus causing yellowing disease of cucurbits in India. Crop Protect. 2022, 158, 106013. [Google Scholar] [CrossRef]
- Fuchs, M. Closteroviruses (Closteroviridae). In Encyclopedia of Virology, 4th ed.; Bamford, D.H., Zuckerman, M., Eds.; Academic Press: New York, NY, USA, 2021; pp. 336–347. ISBN 9780128145166. [Google Scholar]
- Grill, L.K.; Garger, S.J. Identification and characterization of double-stranded RNA associated with cytoplasmic male sterility in Vicia faba. Proc. Natl. Acad. Sci. USA 1981, 78, 7043–7046. [Google Scholar] [CrossRef]
- Fukuhara, T. Endornaviruses: Persistent dsRNA viruses with symbiotic properties in diverse eukaryotes. Virus Genes 2019, 55, 165–173. [Google Scholar] [CrossRef] [PubMed]
- Fukuhara, T. Endornaviruses (Endornaviridae). In Encyclopedia of Virology, 4th ed.; Bamford, D.H., Zuckerman, M., Eds.; Academic Press: New York, NY, USA, 2021; pp. 388–395. ISBN 9780128145166. [Google Scholar]
- Sabanadzovic, S.; Wintermantel, W.M.; Valverde, R.A.; McCreight, J.D.; Aboughanem-Sabanadzovic, N. Cucumis melo endornavirus: Genome organization, host range and co-divergence with the host. Virus Res. 2016, 214, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Khankhum, S.; Valverde, R.A.; Pastor-Corrales, M.A.; Osorno, J.M.; Sabanadzovic, S. Two endornaviruses show differential infection patterns between gene pools of Phaseolus vulgaris. Arch. Virol. 2015, 160, 1131–1137. [Google Scholar] [CrossRef]
- Roossinck, M.J.; Saha, P.; Wiley, G.B.; Quan, J.; White, J.D.; Lai, H.; Chavarria, F.; Shen, G.; Roe, B. Ecogenomics: Using massively parallel pyrosequencing to understand virus ecology. Mol. Ecol. 2010, 19, 81–88. [Google Scholar] [CrossRef]
- Kato, K.; Hanada, K.; Kameya-Iwaki, M. Transmissions mode, host range and electron microscopy of a pathogen causing a new disease of melon (Cucumis melo) in Japan. Ann. Phytopathol. Soc. Jpn. 1999, 65, 624–627. [Google Scholar] [CrossRef]
- Quito-Avila, D.F.; Peralta, E.L.; Martin, R.R.; Ibarra, M.A.; Alvarez, R.A.; Mendoza, A.; Insuasti, M.; Ochoa, J. Detection and occurrence of melon yellow spot virus in Ecuador: An emerging threat to cucurbit production in the region. Eur. J. Plant Pathol. 2014, 140, 193–197. [Google Scholar] [CrossRef]
- Che, H.Y.; Cao, X.R.; He, Y.H.; Luo, D.Q. Distribution and identification of watermelon viruses in Hainan Island. Acta Phytopathol. Sin. 2020, 50, 632–636. [Google Scholar]
- Orfanidou, C.G.; Baltzi, A.; Dimou, N.A.; Katis, N.I.; Maliogka, V.I. Cucurbit chlorotic yellows virus: Insights into its natural host range, genetic variability, and transmission parameters. Plant Dis. 2017, 101, 2053–2058. [Google Scholar] [CrossRef] [PubMed]
- Abrahamian, P.; Sobh, H.; Seblani, R.; Abou-Jawdah, Y. Co-infection of two criniviruses and a begomovirus enhances the disease severity in cucumber. Eur. J. Plant Pathol. 2015, 142, 521–530. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).