Abstract
In the ongoing arms race between virus and host, fine-tuned gene expression plays a critical role in antiviral signaling. However, viruses have evolved to disrupt this process and promote their own replication by targeting host restriction factors. Polymerase-associated factor 1 complex (PAF1C) is a key player in this relationship, recruiting other host factors to regulate transcription and modulate innate immune gene expression. Consequently, PAF1C is consistently targeted by a diverse range of viruses, either to suppress its antiviral functions or co-opt them for their own benefit. In this review, we delve into the current mechanisms through which PAF1C restricts viruses by activating interferon and inflammatory responses at the transcriptional level. We also highlight how the ubiquity of these mechanisms makes PAF1C especially vulnerable to viral hijacking and antagonism. Indeed, as often as PAF1C is revealed to be a restriction factor, viruses are found to have targeted the complex in reply.
1. Introduction
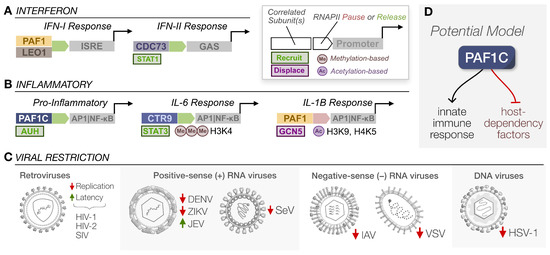
As the first line of host defense, pathogen sensing by the innate immune response culminates in the reconfiguration of host gene expression to restrict virus replication. Polymerase-associated factor 1 complex (PAF1C) plays a critical role in this response. Its diverse molecular functions may bring about death by a thousand cuts for viruses, stemming from PAF1C’s ability to regulate transcription elongation, chromatin remodeling, histone modification, and more.
PAF1C was initially characterized in yeast [1], and the highly conserved complex is composed of PAF1, LEO1, CTR9, RTF1, and CDC73 [2,3]. Concentrated in the nucleus, PAF1C acts as a scaffold to modulate transcription by recruiting histone- and RNA polymerase II (RNAPII)-modifying enzymes to transcription start sites (TSS). The complex coordinates the activity of multiple partners, including elongation factor TFIIS/SI [4], members of the chromatin transcription complex FACT (SPT5, SPT16, POB3) [5], the DRB sensitivity-inducing factor (DSIF) [6], cleavage and polyadenylation specificity factor (CPSF) [7,8], and MYC [9,10]. Moreover, complex members can interact with other transcription factors independently, such as free RTF1 facilitating elongation exclusively [11,12,13]. The diversity of scaffolding partners associated with PAF1C has produced several distinct models for RNAPII pausing, release, and eventual transcript elongation.
PAF1C has been classically viewed as a positive regulator of gene expression, wherein its occupancy at a TSS creates a transcriptional environment conducive to RNAPII release and productive transcription elongation [4,13,14,15,16]. Mechanistically, the SPT6 recruitment of PAF1C likely displaces negative elongation factor (NELF) to mediate RNAPII release and then maintain transcript elongation [17]. However, this model has become more complex in the last decade, with the discovery that RNAPII release is negatively regulated by PAF1C for highly paused genes [10,18,19]. This could be attributed to upstream super-enhancers, where PAF1C occupation potentially blocks RNAPII function, driving the negative regulation of gene expression [18]. Additionally, PAF1C may recruit transcriptional repressors, such as Integrator-PP2A (INTAC) [20] and NELF/SPT5 [21], with a growing number of studies detecting PAF1C-mediated negative regulation [22,23,24,25]. Of course, these distinct mechanisms for transcription are not mutually exclusive, and others have attributed this difference to the modulation of RNAPII velocity and processivity instead of direct suppression [26]. Various molecular switches may also exist to actively reconfigure PAF1C from a transcription attenuator to an elongation factor [27]. Ultimately, the model remains fluid, yet PAF1C remains a key player in modulating gene expression as an RNAPII scaffold.
Beyond the direct modulation of transcription, PAF1C can affect gene expression indirectly via epigenetic and structural modifications to chromatin, altering accessibility and activity at various TSS. As with transcription, PAF1C relies on several interacting partners to alter histone methylation and ubiquitination [28,29,30,31]. Although this early work was conducted in yeast only, PAF1C interacts with the homologous mammalian complex of proteins associated with Set1 (COMPASS)—composed of SET methyltransferases, myeloid leukemia factors (MLL), and MLL fusion partners—to modulate gene expression [32,33,34,35]. Based on functions in yeast, PAF1C likely modulates DNA replication and damage response, telomere regulation, genome and nucleosome stability, and RNA export [16]. In other words, PAF1C is far from limited to its roles in transcriptional regulation, and its multiplicity of functions makes the complex a keystone of regulating stress responses (Figure 1).
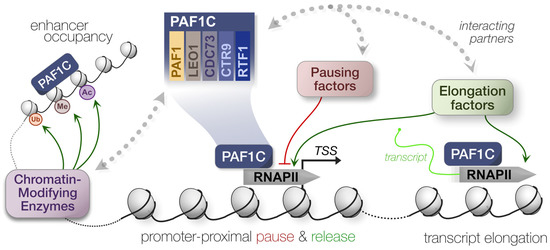
Figure 1.
Summary of PAF1C-mediated transcriptional regulation. Polymerase-associated factor 1 complex (PAF1C) engages with several histone- and RNAPII-modifying enzymes to modulate transcription. This can alter the pausing, release, and elongation dynamics of RNAPII.
Since the innate immune recognition of viral infections often culminates in an antiviral gene expression program [36], in this review, we will discuss how PAF1C is inherently involved in this process. PAF1C can effectively mount an antiviral response by modulating both interferon and inflammatory gene expression, restricting several distinct virus families. Of course, viruses are notorious for their ability to hinder or hijack host proteins, including the disruption of antiviral signaling [37,38,39]. As methods to identify virus–host interactions are more accessible and widespread [40], we often find that PAF1C is both the archer and the prey when it comes to viruses and innate immunity.
3. Karma: PAF1C Is Vulnerable to Viral Antagonism
PAF1C can facilitate an antiviral response across several axes, sometimes leveraging multiple functions at once. As a positive regulator of transcription elongation, PAF1C can recruit various transcription factors and histone-modifying enzymes to modulate RNAPII pausing, release, and elongation, resulting in the increase or decrease of interferon or inflammatory gene expression, depending on the genetic landscape. All these processes drive both canonical and non-canonical pathways that restrict virus replication (Figure 3A). PAF1C’s antiviral function, in other words, is a unique product of the stimulus, the complex’s interactions with other host factors, and the genomic sites it occupies. It is pleiotropic and virtually ubiquitous in the nucleus. However, here we argue that PAF1C performs its functions all too well, making it a prime target in the great war between virus and host. Unsurprisingly, much of the research we have discussed regarding PAF1C’s role in the innate immune response is a byproduct of studying why viruses target PAF1C at all.
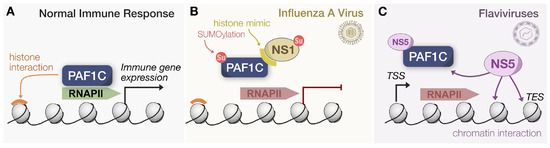
Figure 3.
Influenza and Flaviviruses target PAF1C for displacement. (A) Under normal conditions, PAF1C relies on chromatin interactions via histones to associate with RNAPII and recruit additional factors for immune gene expression. (B) During IAV infection, NS1 translocates to the nucleus. Relying on a histone mimic tail, NS1 can displace PAF1C from RNAPII by competitively binding the complex. Additionally, global SUMOylation increased during IAV infection, and NS1 relies on SUMOylation for functionality during virus replication. (C) Flavivirus NS5 localizes to the nucleus and relies on its MTase domain to interact with chromatin. Such an interaction effectively displaces PAF1C from RNAPII during transcript elongation, causing the premature termination of transcription.
3.1. Influenza Virus: Histone Mimic and Reprogramming SUMOylation
Though not the first study to associate PAF1C with viruses, antagonism by IAV has brought the complex to the forefront of innate immunity in the virus–host arms race. In a seminal study, the NS1 protein of IAV H3N2 was found to possess a histone mimic “tail” that interacted with the PAF1 subunit in a virus strain-specific manner [42]. This interaction was facilitated by the similarity of the NS1 tail to histone H3 and was not observed in H5N1 or H3N2 with truncated versions of NS1. Wild-type IAV exhibited decreased PAF1 and RNAPII levels at transcription end sites (TES) compared to a mutant virus lacking the PAF1 binding sequence. Additionally, more TSS-associated short transcripts were detected, antiviral gene expression was suppressed, and viral titers were higher relative to the mutant virus. These observations collectively confirmed that NS1 prevented PAF1C from facilitating transcription elongation to engage antiviral signaling (Figure 3B). The production of a full transcript for both wild-type and mutant NS1 was measured in vitro to validate that NS1 inhibits elongation. Furthermore, infection by IAV mutants unable to antagonize PAF1 resulted in stronger ISG expression, and these IAV mutants had improved replication in PAF1-depleted cells.
Interestingly, recent studies have correlated PAF1C activity with IAV-induced SUMOylation, an epigenetic regulator of transcription. It has been previously demonstrated that IAV infection increases the rate of SUMOylation [60]. A subsequent study implemented a proteomics-based approach to uncover novel SUMO1/2 substrates during IAV infection [61]. Every subunit of PAF1C was an IAV-induced target, and the shRNA knockdowns of several subunits led to an expected increase in IAV replication. Importantly, the CDC73 subunit relied on SUMOylation to properly stimulate ISG expression, so how does this coincide with PAF1C antagonism? Notably, these trends were proven with a virus incapable of directly targeting PAF1 via a histone mimic. Thus, the mechanism by which IAV antagonizes PAF1C has the potential to be strain specific. Future studies should investigate the two potential modes of antagonism. First, whether the degree of SUMOylation across IAV strains impacts PAF1C-mediated antiviral signaling. Second, whether this is correlated with the likelihood of antagonizing PAF1C through the evolution of a histone mimic on NS1.
3.2. Flaviviruses: NS5 Antagonism via Chromatin Interaction
The NS5 protein of flaviviruses is also well-characterized in its ability to interfere with PAF1C-mediated immune responses. Global virus–host proteomics for DENV and ZIKV first revealed that NS5 interacted with several PAF1C subunits [52], with the interaction being subsequently validated for all four DENV serotypes, ZIKV, and West Nile Virus (WNV). Eventually, this interaction was found to be conserved across mosquito- and tick-borne flaviviruses [43,53], suggesting that it plays a consequential role in flavivirus replication. As established previously, the knockdown of various subunits led to increased virus replication (except for JEV). Yet, more importantly, NS5 reduced LEO1 occupancy at DENV-induced ISGs, abrogating the expression of those same genes [52]. It was readily apparent that flavivirus NS5 disrupted PAF1C recruitment to ISGs, but the exact mechanism behind this interference remained unresolved.
Recent work by us and others has addressed this need to characterize the NS5–PAF1C interaction. Our own work established that DENV NS5 relied on nuclear localization to facilitate an interaction with PAF1C [43]. Additionally, the C-terminus region of the NS5 methyltransferase (MTase) domain, referred to as the pivot region, was required for interaction with PAF1C. Upon mutating these regions and eliminating interaction with PAF1C, immune gene expression was significantly rescued, establishing the causal link between this virus–host interaction and the antagonism of antiviral signaling. Another group focusing on ZIKV NS5 expanded our understanding of localization dynamics by discovering NS5’s ability to colocalize and associate with chromatin [62]. Intriguingly, the MTase domain facilitated this chromatin binding, allowing NS5 to repress transcription via the direct occupation of certain TSS, including that of neurodevelopment and immune genes. This mechanism directly resulted in the displacement of PAF1C from several TSS because the strength of NS5-PAF1C and NS5-chromatin interactions exceeds that of PAF1C and chromatin (Figure 3C).
Connecting these two studies would require discerning whether disruption of the pivot region we isolated may be sufficient to break the competitive binding between NS5, PAF1C, and chromatin. It is also relevant to elucidate whether NS5 antagonism is sufficient to displace PAF1C from paused sites of expression, as current work has focused predominantly on active transcript elongation. Regardless, both studies effectively highlight how flavivirus NS5 quite literally blocks antiviral signaling at PAF1C-mediated sites of expression, boosting virus replication and exacerbating pathogenesis.
3.3. Retroviruses: Exile from the Provirus
Aside from the antiviral signaling outcomes of PAF1C transcriptional regulation, the actual, direct functionality of the complex is especially relevant for the expression of integrated viruses. Following the integration of HIV-1, the transactivator protein, Tat, binds the viral promoter and phosphorylates RNAPII and associated factors to drive the productive elongation of viral transcripts [63]. P-TEFb—part of the super elongation complex (SEC)—is well implicated in this mechanism, so it is unsurprising that every PAF1C member was identified upon the pulldown of HIV-1 Tat protein [64,65]. In a knockdown model, PAF1C and MLL-fusion partners—known as Tatcom1—were required for the transactivation of HIV-1 transcription and basal long terminal repeat (LTR) activity [64] (Figure 4A). Based on these results, Tat actively recruits PAF1 to the viral promoter, and the subsequent formation of Tatcom1 allows for the productive elongation of viral transcripts. Herein, HIV appears to leverage PAF1C function directly, rather than through the antagonism of its antiviral roles.
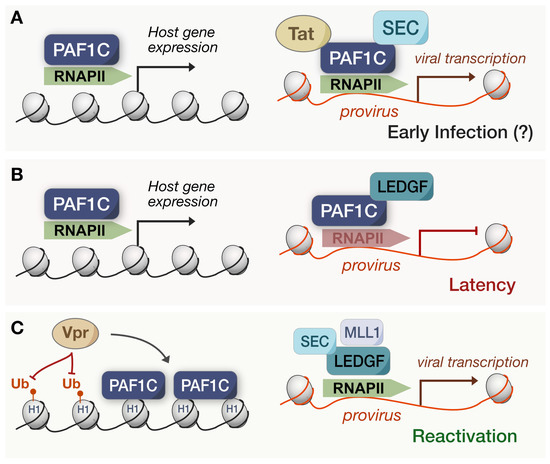
Figure 4.
Suggested models of PAF1C in HIV infection. (A) In early models, HIV-1 Tat was associated with PAF1C to facilitate proviral expression despite PAF1C maintaining an antiviral state via host gene expression. This early infection model remains questionable. (B) During latency, PAF1C partners with LEDGF/p75 to pause RNAPII at the provirus, upholding the latency of the viral infection. (C) Upon HIV-1 reactivation, MLL1 displaces PAF1C from LEDGF/p75, allowing for the SEC to mediate the resumed transcription of the provirus. Additionally, HIV-1 Vpr has been associated with reduced H1.2 ubiquitination and promotes the association of PAF1C with these sites.
However, this model for PAF1C-mediated HIV transcription does not readily align with the maintenance of latency and viral restriction correlated with PAF1C subunits [49,50,51]. How does HIV effectively antagonize and restructure PAF1C-mediated restriction to exit latency? Recent work has uncovered a potential mechanism for reconciling this disparity. Lens epithelium-derived growth factor (LEDGF)/p75—a chromatin-associated factor—facilitates HIV integration by interacting with the viral integrase while repressing proviral transcription during latency [66,67]. In the recent study, LEDGF/p75 interacted with several PAF1C subunits in latent HIV infected cells [32], and the depletion of PAF1—surprisingly—increased the reactivation of HIV transcription. Upon intentionally stimulating the reversal of latency, PAF1 depletion did not impact proviral transcription. Such a connection suggests that LEDGF/p75 associates with PAF1C to maintain latency (Figure 4B), yet PAF1C does not revert to its previous roles in Tatcom1 upon reactivation. Perhaps the early PAF1C-mediated inhibition of HIV is then distinct from its role during latency or is dependent on the genomic context of integrated proviral DNA. To that end, the same study found that RNAPII occupancy increased at HIV genomic sites after PAF1 and LEDGF/p75 depletions, and PAF1 was unable to associate with the LTR without LEDGF/p75 [32] (Figure 4C). Thus, the PAF1C pausing of RNAPII—orchestrated by LEDGF/p75—seems to be the culprit for maintaining HIV latency, highlighting the importance of PAF1C-mediated pausing for viral antagonism.
The same study articulated how PAF1C is possibly displaced following HIV reactivation. A cascade underlies proviral transcriptional activation during HIV latency reversal: LEDGF/p75 associates and recruits the MLL1 branch of COMPASS to catalyze H3K4 trimethylation [32]. This epigenetic mark promotes SEC binding at the proviral LTR, stimulating proviral initiation and elongation via RNAPII. MLL1 and PAF1 share the same binding site on LEDGF/p75, wherein the forced expression of MLL1 dissociates PAF1 from LEDGF/p75 and vice versa. Although this model recapitulates PAF1C as a restriction factor for HIV, its shortcomings lie in its inability to explain how the HIV provirus hijacks PAF1C during latency. For one, it does not necessarily dispute past work on PAF1C being associated with Tat, for MLL-fusion partners were also necessary for transactivation [64]. Moreover, others have resolved the overall structure of the SEC and found it to be based on a highly flexible and unstructured scaffold [68]. Such flexibility inherently allows for changes in binding affinities, including the dynamic reconfiguration of the binding partners and transcriptional landscape. Therefore, both models of PAF1C and HIV proviral transcription may be distinct manifestations of the same complex.
Tat may contribute to PAF1C displacement in this way, yet other HIV proteins are implicated as well. Recent work found that Vpr expression reduced histone H1.2 ubiquitination, and when H1.2 was affinity purified, proteomics showed Vpr expression enhanced PAF1C binding to H1.2 [69] (Figure 4C). Histone H1 can be integral to chromatin accessibility and the resulting differential gene expression, so Vpr may indirectly antagonize PAF1C via this process to sequester its antiviral functions. Either way, future studies should focus on whether Vpr and Tat are targeting the immune or proviral transcription roles in which PAF1C is involved. It also begs the question as to whether PAF1C displacement and viral activation is a byproduct of host functions over time or a process initiated by the virus during latency. Our convoluted model exemplifies how the relationship between PAF1C and viruses is highly complex, and the functional trade-offs may create a delicate balance between viral hijacking and host restriction.
Being able to reconfigure this balance to benefit the virus or host could have powerful implications, and the contemporary identification of a small molecule inhibitor, referred to as iPAF1C, makes this a current reality. iPAF1C targets a key binding interface between CTR9 and PAF1 subunits, and the drug disrupted PAF1C chromatin localization and induced RNAPII release [70]. Moreover, the drug’s effect mimicked that of PAF1 depletion across the transcriptomic landscape. Thus, iPAF1C became a candidate for the “kick and kill” HIV therapeutic strategy. This approach forces viral reactivation with latency reversal agents (LRAs) to purportedly clear reactivated cells via cytopathic effects or the host immune response [71,72]. iPAF1C enhanced the activity of existing LRAs in CD4+ T cells because the subsequent release of RNAPII advanced the transcriptional elongation of the provirus [70]. Importantly, iPAF1C alone could reverse latency in human peripheral blood mononuclear cells (PBMCs) derived from persons undergoing ART. Advances such as these reinforce PAF1C as a strong candidate for epigenetic-targeted therapies against viruses [73]. Whether disrupting the complex intentionally or displacing viral antagonists to restore native functions, the pharmacological targeting of PAF1C could modulate gene expression to benefit human health. It also means understanding the complex mechanisms behind the viral hijacking of PAF1C is all the more pertinent.
3.4. DNA Viruses: Another Case of Hijacking
Similar complexities extend to DNA viruses, such as HSV-1. During HSV-1 infection, the transcription of immediate early (IE) genes is a critical rate-limiting step. This step relies on the SEC, including pTEFb, as the inhibition of SEC led to the reduced occupancy of viral IE promoters, less viral transcription, and suppressed viral reactivation as a result [74]. As with HIV Tat, the HSV-1 protein HCF-1—a component of the IE regulatory complex—associates with the SEC, pTEFb, and PAF1C [75]. HCF-1 interactions are likely dynamic, and while interactions with SEC and pTEFb are understood to facilitate RNAPII release, it is unknown whether PAF1C is fulfilling roles in elongation as well or is being displaced from potential pausing status.
Another IE gene, ICP22, further complicates PAF1C antagonism. As an inhibitor of pTEFb activity [76], ICP22 inhibited RNAPII elongation at the TSS and TES of a model cellular gene [77]. Following immunoprecipitation, such as HCF-1, ICP22 interacted with several transcription elongation factors, including PAF1C and the related FACT complex [77,78]. Considering ICP22 and HCF-1 are predominantly associated with host gene expression [79,80,81] and viral transcription [82], respectively, PAF1C antagonism is potentially doubling: HCF-1 hijacks PAF1C at the provirus while ICP22 stalls it at sites of host gene expression. This may be especially relevant since recent proteomics revealed that PAF1C specifically associates with oligomerized interferon inducible factor 16 (IFI16)—an immune DNA sensor—and colocalizes with ICP4 at HSV-1 genome compartments throughout multiple stages of infection [52]. Although the connection between the IFI16 interaction and ICP4 colocalization is yet to be established as causative, it did correlate with reduced HSV-1 late gene expression. Upon PAF1 knockdown, IE gene expression was unaffected while late gene expression was increased. Therefore, it will be valuable to discern whether the ICP22 targeting of PAF1C attenuates this restriction, affects antiviral sites of expression, or is simply a byproduct of colocalization with viral compartments. Recognizing how these dynamics are shaped by the timeline of DNA virus replication could provide further insights as well. Human adenovirus protein E1A also similarly recruits PAF1C for proviral transcription [83], so fully understanding these recurrent mechanisms is key to discerning how viruses are both similar and distinct in their hijacking of PAF1C.
3.5. SARS-CoV-2: Nucleocapsid as a New Candidate for Study
PAF1C continues to appear in virus–host interaction studies. Most recently, SARS-CoV-2 nucleocapsid (N) has been suggested to interact with PAF1C subunits, yet these interactions were weak and inconsistent across several proteomics-based studies [84,85,86,87]. However, this interaction became more apparent in a concurrent study with the expression and purification of a truncated N (N*) protein expressed differentially across SARS-CoV-2 variants [88]. Upon affinity purification and subsequent mass spectrometry on N*, all PAF1C subunits were clearly identified to be interactors. Additionally, PAF1C was previously implicated in the host regulation of the response to SARS-CoV-2 infection [89,90]. Considering the Alpha, Gamma, and Omicron variants specifically express N* during infection compared to the original strain [39], enrichment for the interaction with PAF1C raises the possibility of a variant-specific mechanism for immune antagonism which evolved over the course of the pandemic. Such a rapid selection for the expression of distinct viral proteins to target PAF1C would underscore how relevant the complex is to antiviral immunity. Further study on N* and its effects on gene expression will allow us to confirm immune antagonism and add SARS-CoV-2 variants to the list of viruses that target PAF1C rather than tolerate it.
4. Conclusions
Despite being able to activate several distinct antiviral regulatory programs through transcriptional and epigenetic controls, PAF1C is the anti-hero of its own story, typically being antagonized or hijacked by the same viruses it restricts. Considering the broad scope of the studies we have discussed, and the sheer evolutionary diversity of viruses implicated, repeated association with PAF1C is far from a coincidence. It is more likely to be an invisible string tying together viruses that evolutionarily converged on targeting such a pleiotropic conserved host factor. Then again, our understanding of the context-based effects of PAF1C, especially as it relates to viruses, is lacking, and new virus–PAF1C interactions continue to be identified contemporarily. Since PAF1C fulfills several functions simultaneously, recognizing which viruses hijack which functions will be crucial to navigating the labyrinth of PAF1C and antiviral immunity.
Author Contributions
M.W.K. and P.S.S. conceived of the work and conducted the literature review. M.W.K. wrote the manuscript and designed the figures. M.W.K. and P.S.S. revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded through a grant to P.S.S. from the NIH, grant number R21AI168716.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We would like to thank members of the Shah Lab for their critical review of the manuscript and constructive feedback. This work is dedicated to Taylor A. Swift, a constant source of inspiration.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Shi, X.; Finkelstein, A.; Wolf, A.J.; Wade, P.A.; Burton, Z.F.; Jaehning, J.A. Paf1p, an RNA Polymerase II-Associated Factor in Saccharomyces Cerevisiae, May Have Both Positive and Negative Roles in Transcription. Mol. Cell. Biol. 1996, 16, 669–676. [Google Scholar] [CrossRef]
- Mueller, C.L.; Jaehning, J.A. Ctr9, Rtf1, and Leo1 Are Components of the Paf1/RNA Polymerase II Complex. Mol. Cell. Biol. 2002, 22, 1971–1980. [Google Scholar] [CrossRef]
- Xu, Y.; Bernecky, C.; Lee, C.-T.; Maier, K.C.; Schwalb, B.; Tegunov, D.; Plitzko, J.M.; Urlaub, H.; Cramer, P. Architecture of the RNA Polymerase II-Paf1C-TFIIS Transcription Elongation Complex. Nat. Commun. 2017, 8, 15741. [Google Scholar] [CrossRef]
- Kim, J.; Guermah, M.; Roeder, R.G. The Human PAF1 Complex Acts in Chromatin Transcription Elongation Both Independently and Cooperatively with SII/TFIIS. Cell 2010, 140, 491–503. [Google Scholar] [CrossRef]
- Squazzo, S.L.; Costa, P.J.; Lindstrom, D.L.; Kumer, K.E.; Simic, R.; Jennings, J.L.; Link, A.J.; Arndt, K.M.; Hartzog, G.A. The Paf1 Complex Physically and Functionally Associates with Transcription Elongation Factors in Vivo. EMBO J. 2002, 21, 1764–1774. [Google Scholar] [CrossRef]
- Wier, A.D.; Mayekar, M.K.; Héroux, A.; Arndt, K.M.; VanDemark, A.P. Structural Basis for Spt5-Mediated Recruitment of the Paf1 Complex to Chromatin. Proc. Natl. Acad. Sci. USA 2013, 110, 17290–17295. [Google Scholar] [CrossRef]
- Arnold, M.; Bressin, A.; Jasnovidova, O.; Meierhofer, D.; Mayer, A. A BRD4-Mediated Elongation Control Point Primes Transcribing RNA Polymerase II for 3’-Processing and Termination. Mol. Cell 2021, 81, 3589–3603.e13. [Google Scholar] [CrossRef]
- Nordick, K.; Hoffman, M.G.; Betz, J.L.; Jaehning, J.A. Direct Interactions between the Paf1 Complex and a Cleavage and Polyadenylation Factor Are Revealed by Dissociation of Paf1 from RNA Polymerase II. Eukaryot. Cell 2008, 7, 1158–1167. [Google Scholar] [CrossRef]
- Gerlach, J.M.; Furrer, M.; Gallant, M.; Birkel, D.; Baluapuri, A.; Wolf, E.; Gallant, P. PAF1 Complex Component Leo1 Helps Recruit Drosophila Myc to Promoters. Proc. Natl. Acad. Sci. USA 2017, 114, E9224–E9232. [Google Scholar] [CrossRef]
- Jaenicke, L.A.; von Eyss, B.; Carstensen, A.; Wolf, E.; Xu, W.; Greifenberg, A.K.; Geyer, M.; Eilers, M.; Popov, N. Ubiquitin-Dependent Turnover of MYC Antagonizes MYC/PAF1C Complex Accumulation to Drive Transcriptional Elongation. Mol. Cell 2016, 61, 54–67. [Google Scholar] [CrossRef]
- Cao, Q.-F.; Yamamoto, J.; Isobe, T.; Tateno, S.; Murase, Y.; Chen, Y.; Handa, H.; Yamaguchi, Y. Characterization of the Human Transcription Elongation Factor Rtf1: Evidence for Nonoverlapping Functions of Rtf1 and the Paf1 Complex. Mol. Cell. Biol. 2015, 35, 3459–3470. [Google Scholar] [CrossRef]
- Mbogning, J.; Nagy, S.; Pagé, V.; Schwer, B.; Shuman, S.; Fisher, R.P.; Tanny, J.C. The PAF Complex and Prf1/Rtf1 Delineate Distinct Cdk9-Dependent Pathways Regulating Transcription Elongation in Fission Yeast. PLoS Genet. 2013, 9, e1004029. [Google Scholar] [CrossRef]
- Žumer, K.; Maier, K.C.; Farnung, L.; Jaeger, M.G.; Rus, P.; Winter, G.; Cramer, P. Two Distinct Mechanisms of RNA Polymerase II Elongation Stimulation in Vivo. Mol. Cell 2021, 81, 3096–3109.e8. [Google Scholar] [CrossRef]
- Yu, M.; Yang, W.; Ni, T.; Tang, Z.; Nakadai, T.; Zhu, J.; Roeder, R.G. RNA Polymerase II-Associated Factor 1 Regulates the Release and Phosphorylation of Paused RNA Polymerase II. Science 2015, 350, 1383–1386. [Google Scholar] [CrossRef] [PubMed]
- van den Heuvel, D.; Spruijt, C.G.; González-Prieto, R.; Kragten, A.; Paulsen, M.T.; Zhou, D.; Wu, H.; Apelt, K.; van der Weegen, Y.; Yang, K.; et al. A CSB-PAF1C Axis Restores Processive Transcription Elongation after DNA Damage Repair. Nat. Commun. 2021, 12, 1342. [Google Scholar] [CrossRef]
- Francette, A.M.; Tripplehorn, S.A.; Arndt, K.M. The Paf1 Complex: A Keystone of Nuclear Regulation Operating at the Interface of Transcription and Chromatin. J. Mol. Biol. 2021, 433, 166979. [Google Scholar] [CrossRef]
- Aoi, Y.; Shah, A.P.; Ganesan, S.; Soliman, S.H.A.; Cho, B.-K.; Goo, Y.A.; Kelleher, N.L.; Shilatifard, A. SPT6 Functions in Transcriptional Pause/Release via PAF1C Recruitment. Mol. Cell 2022, 82, 3412–3423.e5. [Google Scholar] [CrossRef]
- Chen, F.X.; Xie, P.; Collings, C.K.; Cao, K.; Aoi, Y.; Marshall, S.A.; Rendleman, E.J.; Ugarenko, M.; Ozark, P.A.; Zhang, A.; et al. PAF1 Regulation of Promoter-Proximal Pause Release via Enhancer Activation. Science 2017, 357, 1294–1298. [Google Scholar] [CrossRef]
- Chen, F.X.; Woodfin, A.R.; Gardini, A.; Rickels, R.A.; Marshall, S.A.; Smith, E.R.; Shiekhattar, R.; Shilatifard, A. PAF1, a Molecular Regulator of Promoter-Proximal Pausing by RNA Polymerase II. Cell 2015, 162, 1003–1015. [Google Scholar] [CrossRef]
- Wang, Z.; Song, A.; Xu, H.; Hu, S.; Tao, B.; Peng, L.; Wang, J.; Li, J.; Yu, J.; Wang, L.; et al. Coordinated Regulation of RNA Polymerase II Pausing and Elongation Progression by PAF1. Sci. Adv. 2022, 8, eabm5504. [Google Scholar] [CrossRef]
- Yoo, H.-S.; Seo, J.-H.; Yoo, J.-Y. CTR9, a Component of PAF Complex, Controls Elongation Block at the c-Fos Locus via Signal-Dependent Regulation of Chromatin-Bound NELF Dissociation. PLoS ONE 2013, 8, e61055. [Google Scholar] [CrossRef]
- Bai, X.; Kim, J.; Yang, Z.; Jurynec, M.J.; Akie, T.E.; Lee, J.; LeBlanc, J.; Sessa, A.; Jiang, H.; DiBiase, A.; et al. TIF1gamma Controls Erythroid Cell Fate by Regulating Transcription Elongation. Cell 2010, 142, 133–143. [Google Scholar] [CrossRef] [PubMed]
- Kenaston, M.W.; Pham, O.H.; Petit, M.J.; Shah, P.S. Transcriptomic Profiling Implicates PAF1 in Both Active and Repressive Immune Regulatory Networks. BMC Genom. 2022, 23, 787. [Google Scholar] [CrossRef]
- Yoo, H.-S.; Choi, Y.; Ahn, N.; Lee, S.; Kim, W.-U.; Jang, M.S.; Jang, M.H.; Kim, Y.S.; Yoo, J.-Y. Transcriptional Regulator CTR9 Inhibits Th17 Differentiation via Repression of IL-17 Expression. J. Immunol. 2014, 192, 1440–1448. [Google Scholar] [CrossRef] [PubMed]
- Van Oss, S.B.; Cucinotta, C.E.; Arndt, K.M. Emerging Insights into the Roles of the Paf1 Complex in Gene Regulation. Trends Biochem. Sci. 2017, 42, 788–798. [Google Scholar] [CrossRef]
- Hou, L.; Wang, Y.; Liu, Y.; Zhang, N.; Shamovsky, I.; Nudler, E.; Tian, B.; Dynlacht, B.D. Paf1C Regulates RNA Polymerase II Progression by Modulating Elongation Rate. Proc. Natl. Acad. Sci. USA 2019, 116, 14583–14592. [Google Scholar] [CrossRef]
- Liu, X.; Guo, Z.; Han, J.; Peng, B.; Zhang, B.; Li, H.; Hu, X.; David, C.J.; Chen, M. The PAF1 Complex Promotes 3′ Processing of Pervasive Transcripts. Cell Rep. 2022, 38, 110519. [Google Scholar] [CrossRef] [PubMed]
- Pavri, R.; Zhu, B.; Li, G.; Trojer, P.; Mandal, S.; Shilatifard, A.; Reinberg, D. Histone H2B Monoubiquitination Functions Cooperatively with FACT to Regulate Elongation by RNA Polymerase II. Cell 2006, 125, 703–717. [Google Scholar] [CrossRef]
- Wood, A.; Schneider, J.; Dover, J.; Johnston, M.; Shilatifard, A. The Paf1 Complex Is Essential for Histone Monoubiquitination by the Rad6-Bre1 Complex, Which Signals for Histone Methylation by COMPASS and Dot1p. J. Biol. Chem. 2003, 278, 34739–34742. [Google Scholar] [CrossRef]
- Krogan, N.J.; Dover, J.; Wood, A.; Schneider, J.; Heidt, J.; Boateng, M.A.; Dean, K.; Ryan, O.W.; Golshani, A.; Johnston, M.; et al. The Paf1 Complex Is Required for Histone H3 Methylation by COMPASS and Dot1p: Linking Transcriptional Elongation to Histone Methylation. Mol. Cell 2003, 11, 721–729. [Google Scholar] [CrossRef]
- Mulder, K.W.; Brenkman, A.B.; Inagaki, A.; van den Broek, N.J.F.; Timmers, H.T.M. Regulation of Histone H3K4 Tri-Methylation and PAF Complex Recruitment by the Ccr4-Not Complex. Nucleic Acids Res. 2007, 35, 2428–2439. [Google Scholar] [CrossRef] [PubMed]
- Gao, R.; Bao, J.; Yan, H.; Xie, L.; Qin, W.; Ning, H.; Huang, S.; Cheng, J.; Zhi, R.; Li, Z.; et al. Competition between PAF1 and MLL1/COMPASS Confers the Opposing Function of LEDGF/P75 in HIV Latency and Proviral Reactivation. Sci. Adv. 2020, 6, eaaz8411. [Google Scholar] [CrossRef]
- Crump, N.T.; Smith, A.; Godfrey, L.; Jackson, N.; Rice, S.; Kim, J.; Basrur, V.; Fermin, D.; Elenitoba-Johnson, K.; Roeder, R.G.; et al. PAF1 and FACT Cooperate with MLL-AF4 to Drive Enhancer Activity in Leukemia. bioRxiv 2022. [Google Scholar] [CrossRef]
- Muntean, A.G.; Tan, J.; Sitwala, K.; Huang, Y.; Bronstein, J.; Connelly, J.A.; Basrur, V.; Elenitoba-Johnson, K.S.J.; Hess, J.L. The PAF Complex Synergizes with MLL Fusion Proteins at HOX Loci to Promote Leukemogenesis. Cancer Cell 2010, 17, 609–621. [Google Scholar] [CrossRef] [PubMed]
- Hetzner, K.; Garcia-Cuellar, M.-P.; Büttner, C.; Slany, R.K. The Interaction of ENL with PAF1 Mitigates Polycomb Silencing and Facilitates Murine Leukemogenesis. Blood 2018, 131, 662–673. [Google Scholar] [CrossRef] [PubMed]
- Kawai, T.; Akira, S. Innate Immune Recognition of Viral Infection. Nat. Immunol. 2006, 7, 131–137. [Google Scholar] [CrossRef]
- Fishburn, A.T.; Pham, O.H.; Kenaston, M.W.; Beesabathuni, N.S.; Shah, P.S. Let us Get Physical: Flavivirus–host Protein–Protein Interactions in Replication and Pathogenesis. Front. Microbiol. 2022, 13, 847588. [Google Scholar] [CrossRef]
- Weber-Gerlach, M.; Weber, F. To Conquer the Host, Influenza Virus Is Packing It In: Interferon-Antagonistic Strategies beyond NS1. J. Virol. 2016, 90, 8389–8394. [Google Scholar] [CrossRef]
- Thorne, L.G.; Bouhaddou, M.; Reuschl, A.-K.; Zuliani-Alvarez, L.; Polacco, B.; Pelin, A.; Batra, J.; Whelan, M.V.X.; Hosmillo, M.; Fossati, A.; et al. Evolution of Enhanced Innate Immune Evasion by SARS-CoV-2. Nature 2022, 602, 487–495. [Google Scholar] [CrossRef]
- Shah, P.S.; Beesabathuni, N.S.; Fishburn, A.T.; Kenaston, M.W.; Minami, S.A.; Pham, O.H.; Tucker, I. Systems Biology of Virus–host Protein Interactions: From Hypothesis Generation to Mechanisms of Replication and Pathogenesis. Annu. Rev. Virol. 2022, 9, 397–415. [Google Scholar] [CrossRef]
- Murira, A.; Lamarre, A. Type-I Interferon Responses: From Friend to Foe in the Battle against Chronic Viral Infection. Front. Immunol. 2016, 7, 609. [Google Scholar] [CrossRef] [PubMed]
- Marazzi, I.; Ho, J.S.Y.; Kim, J.; Manicassamy, B.; Dewell, S.; Albrecht, R.A.; Seibert, C.W.; Schaefer, U.; Jeffrey, K.L.; Prinjha, R.K.; et al. Suppression of the Antiviral Response by an Influenza Histone Mimic. Nature 2012, 483, 428–433. [Google Scholar] [CrossRef]
- Petit, M.J.; Kenaston, M.W.; Pham, O.H.; Nagainis, A.A.; Fishburn, A.T.; Shah, P.S. Nuclear Dengue Virus NS5 Antagonizes Expression of PAF1-Dependent Immune Response Genes. PLoS Pathog. 2021, 17, e1010100. [Google Scholar] [CrossRef]
- Kim, N.; Sun, H.-Y.; Youn, M.-Y.; Yoo, J.-Y. IL-1β-Specific Recruitment of GCN5 Histone Acetyltransferase Induces the Release of PAF1 from Chromatin for the de-Repression of Inflammatory Response Genes. Nucleic Acids Res. 2013, 41, 4495–4506. [Google Scholar] [CrossRef]
- Wei, J.; Lian, H.; Zhong, B.; Shu, H.-B. Parafibromin Is a Component of IFN-γ–Triggered Signaling Pathways That Facilitates JAK1/2-Mediated Tyrosine Phosphorylation of STAT1. J. Immunol. 2015, 195, 2870–2878. [Google Scholar] [CrossRef]
- Parnas, O.; Jovanovic, M.; Eisenhaure, T.M.; Herbst, R.H.; Dixit, A.; Ye, C.J.; Przybylski, D.; Platt, R.J.; Tirosh, I.; Sanjana, N.E.; et al. A Genome-Wide CRISPR Screen in Primary Immune Cells to Dissect Regulatory Networks. Cell 2015, 162, 675–686. [Google Scholar] [CrossRef]
- Youn, M.-Y.; Yoo, H.-S.; Kim, M.-J.; Hwang, S.-Y.; Choi, Y.; Desiderio, S.V.; Yoo, J.-Y. HCTR9, a Component of Paf1 Complex, Participates in the Transcription of Interleukin 6-Responsive Genes through Regulation of STAT3-DNA Interactions. J. Biol. Chem. 2007, 282, 34727–34734. [Google Scholar] [CrossRef]
- Liu, L.; Oliveira, N.M.; Cheney, K.M.; Pade, C.; Dreja, H.; Bergin, A.-M.H.; Borgdorff, V.; Beach, D.H.; Bishop, C.L.; Dittmar, M.T.; et al. A Whole Genome Screen for HIV Restriction Factors. Retrovirology 2011, 8, 94. [Google Scholar] [CrossRef] [PubMed]
- Hecke, C.V.; Trypsteen, W.; Malatinkova, E.; Spiegelaere, W.D.; Vervisch, K.; Rutsaert, S.; Loes, S.K.; Sips, M.; Vandekerckhove, L. Early Treated HIV-1 Positive Individuals Demonstrate Similar Restriction Factor Expression Profile as Long-Term Non-Progressors. eBioMedicine 2019, 41, 443–454. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Mohsen, M.; Wang, C.; Strain, M.C.; Lada, S.M.; Deng, X.; Cockerham, L.R.; Pilcher, C.D.; Hecht, F.M.; Liegler, T.; Richman, D.D.; et al. Select Host Restriction Factors Are Associated with HIV Persistence During Antiretroviral Therapy. AIDS Lond. Engl. 2015, 29, 411–420. [Google Scholar] [CrossRef] [PubMed]
- Raposo, R.A.S.; Abdel-Mohsen, M.; Bilska, M.; Montefiori, D.C.; Nixon, D.F.; Pillai, S.K. Effects of Cellular Activation on Anti-HIV-1 Restriction Factor Expression Profile in Primary Cells. J. Virol. 2013, 87, 11924–11929. [Google Scholar] [CrossRef]
- Lum, K.K.; Howard, T.R.; Pan, C.; Cristea, I.M. Charge-Mediated Pyrin Oligomerization Nucleates Antiviral IFI16 Sensing of Herpesvirus DNA. mBio 2019, 10, e01428-19. [Google Scholar] [CrossRef] [PubMed]
- Shah, P.S.; Link, N.; Jang, G.M.; Sharp, P.P.; Zhu, T.; Swaney, D.L.; Johnson, J.R.; Von Dollen, J.; Ramage, H.R.; Satkamp, L.; et al. Comparative Flavivirus–host Protein Interaction Mapping Reveals Mechanisms of Dengue and Zika Virus Pathogenesis. Cell 2018, 175, 1931–1945.e18. [Google Scholar] [CrossRef] [PubMed]
- Kovanich, D.; Saisawang, C.; Sittipaisankul, P.; Ramphan, S.; Kalpongnukul, N.; Somparn, P.; Pisitkun, T.; Smith, D.R. Analysis of the Zika and Japanese Encephalitis Virus NS5 Interactomes. J. Proteome Res. 2019, 18, 3203–3218. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Shu, W.-J.; Li, Y.-M.; Mari, M.; Yan, C.; Wang, D.; Yin, Z.-H.; Jiang, W.; Zhou, Y.; Okamoto, K.; et al. The Paf1 Complex Transcriptionally Regulates the Mitochondrial-Anchored Protein Atg32 Leading to Activation of Mitophagy. Autophagy 2020, 16, 1366–1379. [Google Scholar] [CrossRef]
- Li, Y.; Wu, K.; Zeng, S.; Zou, L.; Li, X.; Xu, C.; Li, B.; Liu, X.; Li, Z.; Zhu, W.; et al. The Role of Mitophagy in Viral Infection. Cells 2022, 11, 711. [Google Scholar] [CrossRef] [PubMed]
- Ponia, S.S.; Robertson, S.J.; McNally, K.L.; Subramanian, G.; Sturdevant, G.L.; Lewis, M.; Jessop, F.; Kendall, C.; Gallegos, D.; Hay, A.; et al. Mitophagy Antagonism by ZIKV Reveals Ajuba as a Regulator of PINK1 Signaling, PKR-Dependent Inflammation, and Viral Invasion of Tissues. Cell Rep. 2021, 37, 109888. [Google Scholar] [CrossRef]
- Wang, H.; Zheng, Y.; Huang, J.; Li, J. Mitophagy in Antiviral Immunity. Front. Cell Dev. Biol. 2021, 9, 723108. [Google Scholar] [CrossRef]
- Ke, P.-Y. The Multifaceted Roles of Autophagy in Flavivirus–host Interactions. Int. J. Mol. Sci. 2018, 19, 3940. [Google Scholar] [CrossRef]
- Pal, S.; Santos, A.; Rosas, J.M.; Ortiz-Guzman, J.; Rosas-Acosta, G. Influenza A Virus Interacts Extensively with the Cellular SUMOylation System during Infection. Virus Res. 2011, 158, 12–27. [Google Scholar] [CrossRef]
- Domingues, P.; Golebiowski, F.; Tatham, M.H.; Lopes, A.M.; Taggart, A.; Hay, R.T.; Hale, B.G. Global Reprogramming of Host SUMOylation during Influenza Virus Infection. Cell Rep. 2015, 13, 1467–1480. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Wu, J.; Liu, S.; Lu, R.; Jiang, H.; Wang, N.; Luo, M.; Guo, L.; Xiao, J.; Bu, L.; et al. The RNA Polymerase of Cytoplasmically Replicating Zika Virus Binds with Chromatin DNA in Nuclei and Regulates Host Gene Transcription. Proc. Natl. Acad. Sci. USA 2022, 119, e2205013119. [Google Scholar] [CrossRef] [PubMed]
- Van Lint, C.; Bouchat, S.; Marcello, A. HIV-1 Transcription and Latency: An Update. Retrovirology 2013, 10, 67. [Google Scholar] [CrossRef]
- Sobhian, B.; Laguette, N.; Yatim, A.; Nakamura, M.; Levy, Y.; Kiernan, R.; Benkirane, M. HIV-1 Tat Assembles a Multifunctional Transcription Elongation Complex and Stably Associates with the 7SK SnRNP. Mol. Cell 2010, 38, 439–451. [Google Scholar] [CrossRef]
- He, N.; Liu, M.; Hsu, J.; Xue, Y.; Chou, S.; Burlingame, A.; Krogan, N.J.; Alber, T.; Zhou, Q. HIV-1 Tat and Host AFF4 Recruit Two Transcription Elongation Factors into a Bifunctional Complex for Coordinated Activation of HIV-1 Transcription. Mol. Cell 2010, 38, 428–438. [Google Scholar] [CrossRef]
- Ciuffi, A.; Llano, M.; Poeschla, E.; Hoffmann, C.; Leipzig, J.; Shinn, P.; Ecker, J.R.; Bushman, F. A Role for LEDGF/P75 in Targeting HIV DNA Integration. Nat. Med. 2005, 11, 1287–1289. [Google Scholar] [CrossRef] [PubMed]
- Gérard, A.; Ségéral, E.; Naughtin, M.; Abdouni, A.; Charmeteau, B.; Cheynier, R.; Rain, J.-C.; Emiliani, S. The Integrase Cofactor LEDGF/P75 Associates with Iws1 and Spt6 for Postintegration Silencing of HIV-1 Gene Expression in Latently Infected Cells. Cell Host Microbe 2015, 17, 107–117. [Google Scholar] [CrossRef]
- Chou, S.; Upton, H.; Bao, K.; Schulze-Gahmen, U.; Samelson, A.J.; He, N.; Nowak, A.; Lu, H.; Krogan, N.J.; Zhou, Q.; et al. HIV-1 Tat Recruits Transcription Elongation Factors Dispersed along a Flexible AFF4 Scaffold. Proc. Natl. Acad. Sci. USA 2013, 110, E123–E131. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.R.; Crosby, D.C.; Hultquist, J.F.; Kurland, A.P.; Adhikary, P.; Li, D.; Marlett, J.; Swann, J.; Hüttenhain, R.; Verschueren, E.; et al. Global Post-Translational Modification Profiling of HIV-1-Infected Cells Reveals Mechanisms of Host Cellular Pathway Remodeling. Cell Rep. 2022, 39, 110690. [Google Scholar] [CrossRef]
- Soliman, S.H.A.; Cisneros, W.J.; Iwanaszko, M.; Aoi, Y.; Ganesan, S.; Walter, M.; Zeidner, J.M.; Mishra, R.K.; Kim, E.-Y.; Wolinsky, S.M.; et al. Enhancing HIV-1 Latency Reversal through Regulating the Elongating RNA Pol II Pause-Release by a Small-Molecule Disruptor of PAF1C. Sci. Adv. 2023, 9, eadf2468. [Google Scholar] [CrossRef]
- Lewin, S.R.; Rasmussen, T.A. Kick and Kill for HIV Latency. Lancet 2020, 395, 844–846. [Google Scholar] [CrossRef] [PubMed]
- Thorlund, K.; Horwitz, M.S.; Fife, B.T.; Lester, R.; Cameron, D.W. Landscape Review of Current HIV ‘Kick and Kill’ Cure Research—Some Kicking, Not Enough Killing. BMC Infect. Dis. 2017, 17, 595. [Google Scholar] [CrossRef]
- Nehme, Z.; Pasquereau, S.; Herbein, G. Control of Viral Infections by Epigenetic-Targeted Therapy. Clin. Epigenetics 2019, 11, 55. [Google Scholar] [CrossRef] [PubMed]
- Alfonso-Dunn, R.; Arbuckle, J.H.; Vogel, J.L.; Kristie, T.M. Inhibition of the Super Elongation Complex Suppresses Herpes Simplex Virus Immediate Early Gene Expression, Lytic Infection, and Reactivation from Latency. mBio 2020, 11, e01216-20. [Google Scholar] [CrossRef]
- Alfonso-Dunn, R.; Turner, A.-M.W.; Jean Beltran, P.M.; Arbuckle, J.H.; Budayeva, H.G.; Cristea, I.M.; Kristie, T.M. Transcriptional Elongation of HSV Immediate Early Genes by the Super Elongation Complex Drives Lytic Infection and Reactivation from Latency. Cell Host Microbe 2017, 21, 507–517.e5. [Google Scholar] [CrossRef] [PubMed]
- Zaborowska, J.; Baumli, S.; Laitem, C.; O’Reilly, D.; Thomas, P.H.; O’Hare, P.; Murphy, S. Herpes Simplex Virus 1 (HSV-1) ICP22 Protein Directly Interacts with Cyclin-Dependent Kinase (CDK)9 to Inhibit RNA Polymerase II Transcription Elongation. PLoS ONE 2014, 9, e107654. [Google Scholar] [CrossRef]
- Isa, N.F.; Bensaude, O.; Aziz, N.C.; Murphy, S. HSV-1 ICP22 Is a Selective Viral Repressor of Cellular RNA Polymerase II-Mediated Transcription Elongation. Vaccines 2021, 9, 1054. [Google Scholar] [CrossRef]
- Fox, H.L.; Dembowski, J.A.; DeLuca, N.A. A Herpesviral Immediate Early Protein Promotes Transcription Elongation of Viral Transcripts. mBio 2017, 8, e00745-17. [Google Scholar] [CrossRef]
- He, Q.; Wu, Y.; Wang, M.; Chen, S.; Jia, R.; Yang, Q.; Zhu, D.; Liu, M.; Zhao, X.; Zhang, S.; et al. ICP22/IE63 Mediated Transcriptional Regulation and Immune Evasion: Two Important Survival Strategies for Alphaherpesviruses. Front. Immunol. 2021, 12, 743466. [Google Scholar] [CrossRef]
- Rice, S.A.; Davido, D.J. HSV-1 ICP22: Hijacking Host Nuclear Functions to Enhance Viral Infection. Future Microbiol. 2013, 8, 311–321. [Google Scholar] [CrossRef]
- Matundan, H.H.; Jaggi, U.; Wang, S.; Ghiasi, H. Loss of ICP22 in HSV-1 Elicits Immune Infiltration and Maintains Stromal Keratitis Despite Reduced Primary and Latent Virus Infectivity. Investig. Ophthalmol. Vis. Sci. 2019, 60, 3398–3406. [Google Scholar] [CrossRef]
- Vogel, J.L.; Kristie, T.M. The Dynamics of HCF-1 Modulation of Herpes Simplex Virus Chromatin during Initiation of Infection. Viruses 2013, 5, 1272–1291. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, G.J.; Cohen, M.J.; Mymryk, J.S. Adenovirus E1A Recruits the Human Paf1 Complex To Enhance Transcriptional Elongation. J. Virol. 2014, 88, 5630–5637. [Google Scholar] [CrossRef]
- Zheng, X.; Sun, Z.; Yu, L.; Shi, D.; Zhu, M.; Yao, H.; Li, L. Interactome Analysis of the Nucleocapsid Protein of SARS-CoV-2 Virus. Pathogens 2021, 10, 1155. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Liu, Y.; Gupta, S.; Paramo, M.I.; Hou, Y.; Mao, C.; Luo, Y.; Judd, J.; Wierbowski, S.; Bertolotti, M.; et al. A Comprehensive SARS-CoV-2–Human Protein–Protein Interactome Reveals COVID-19 Pathobiology and Potential Host Therapeutic Targets. Nat. Biotechnol. 2022, 41, 128–139. [Google Scholar] [CrossRef] [PubMed]
- Gordon, D.E.; Jang, G.M.; Bouhaddou, M.; Xu, J.; Obernier, K.; White, K.M.; O’Meara, M.J.; Rezelj, V.V.; Guo, J.Z.; Swaney, D.L.; et al. A SARS-CoV-2 Protein Interaction Map Reveals Targets for Drug Repurposing. Nature 2020, 583, 459–468. [Google Scholar] [CrossRef] [PubMed]
- Gordon, D.E.; Hiatt, J.; Bouhaddou, M.; Rezelj, V.V.; Ulferts, S.; Braberg, H.; Jureka, A.S.; Obernier, K.; Guo, J.Z.; Batra, J.; et al. Comparative Host-Coronavirus Protein Interaction Networks Reveal Pan-Viral Disease Mechanisms. Science 2020, 370, eabe9403. [Google Scholar] [CrossRef] [PubMed]
- Bouhaddou, M.; Reuschl, A.-K.; Polacco, B.J.; Thorne, L.G.; Ummadi, M.R.; Ye, C.; Rosales, R.; Pelin, A.; Batra, J.; Jang, G.M.; et al. Global Landscape of the Host Response to SARS-CoV-2 Variants Reveals Viral Evolutionary Trajectories. bioRxiv 2022. [Google Scholar] [CrossRef]
- Shaath, H.; Vishnubalaji, R.; Elkord, E.; Alajez, N.M. Single-Cell Transcriptome Analysis Highlights a Role for Neutrophils and Inflammatory Macrophages in the Pathogenesis of Severe COVID-19. Cells 2020, 9, 2374. [Google Scholar] [CrossRef]
- Vishnubalaji, R.; Shaath, H.; Alajez, N.M. Protein Coding and Long Noncoding RNA (LncRNA) Transcriptional Landscape in SARS-CoV-2 Infected Bronchial Epithelial Cells Highlight a Role for Interferon and Inflammatory Response. Genes 2020, 11, 760. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).