Comparison of a Genotype 1 and a Genotype 2 Macaque Foamy Virus env Gene Indicates Distinct Infectivity and Cell-Cell Fusion but Similar Tropism and Restriction of Cell Entry by Interferon-Induced Transmembrane Proteins
Abstract
1. Introduction
2. Materials and Methods
2.1. Construction of the SFVmmu-DPZ9524 Infectious Clone
2.2. Construction of SFV Reporter Viruses
2.3. Design and Visualization
2.4. Cell Culture
2.5. Western Blot
2.6. Fusion Assay
2.7. Retroviral Vectors, Foamy Viruses and Pseudotyped Lentiviral Particles
2.8. Neutralization Assay
2.9. Infection Experiments
2.10. Quantitative Reverse Transcription Realtime-PCR (RT-qPCR)
2.11. Statistical Analysis
3. Results
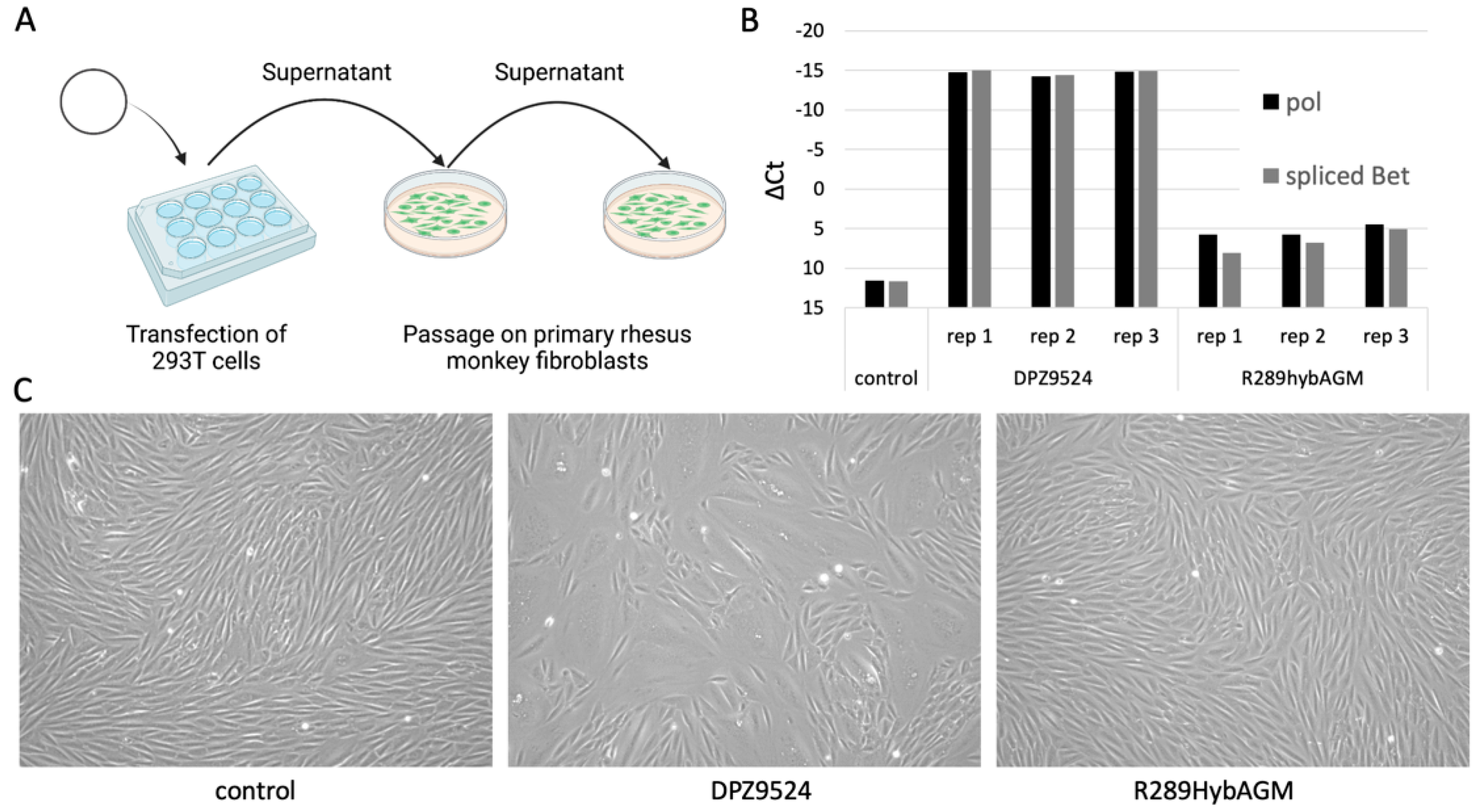
3.1. Generation of an Infectious Clone of SFVmmu-DPZ9524
3.2. Generation of a SFV Reporter Virus with Genotype 1 and Genotype 2 env
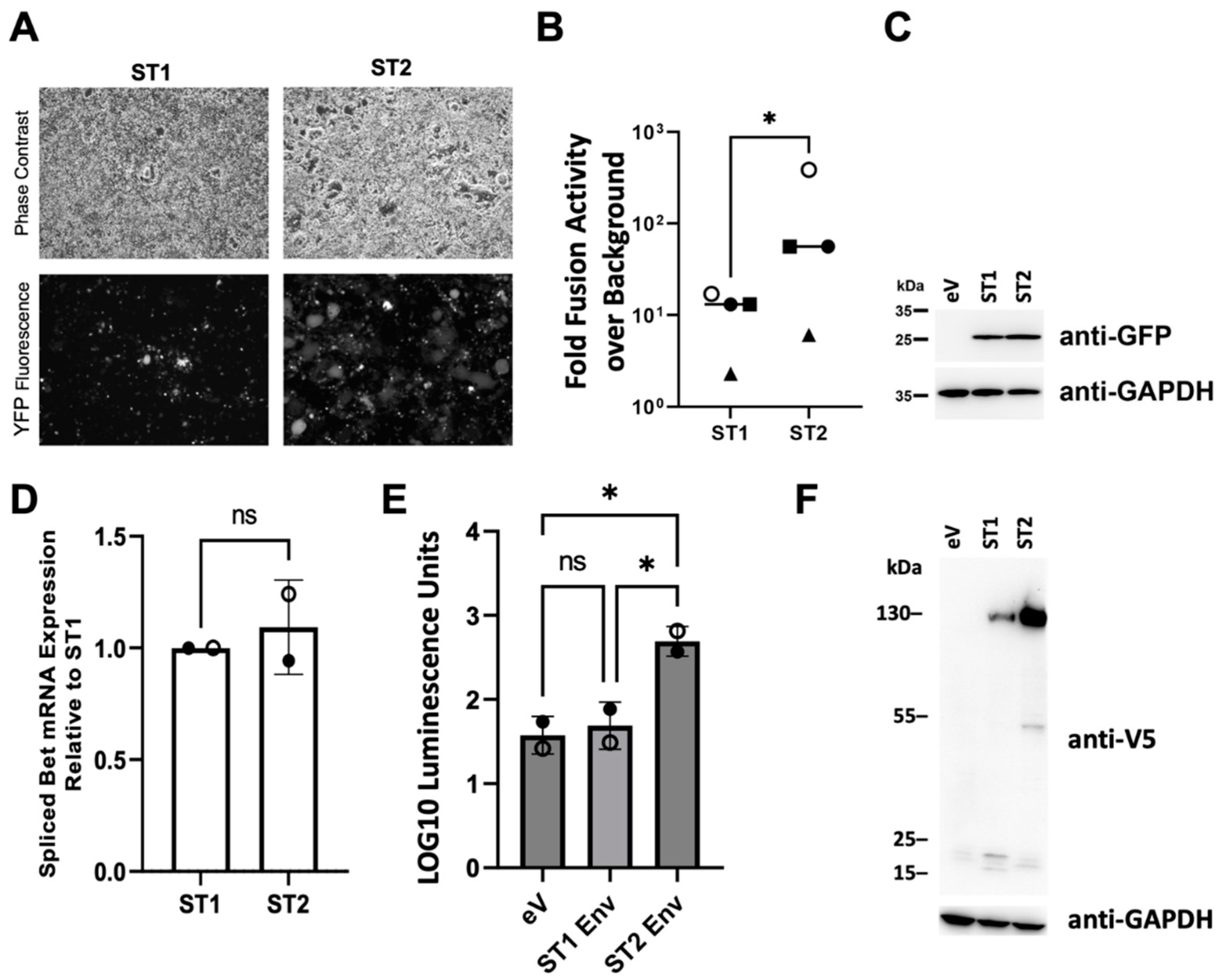
3.3. The SFVmmu_R289hybAGM env Gene Is Associated with Significantly Higher Cell-Cell Fusion Than That of SFVmmu-DPZ9524
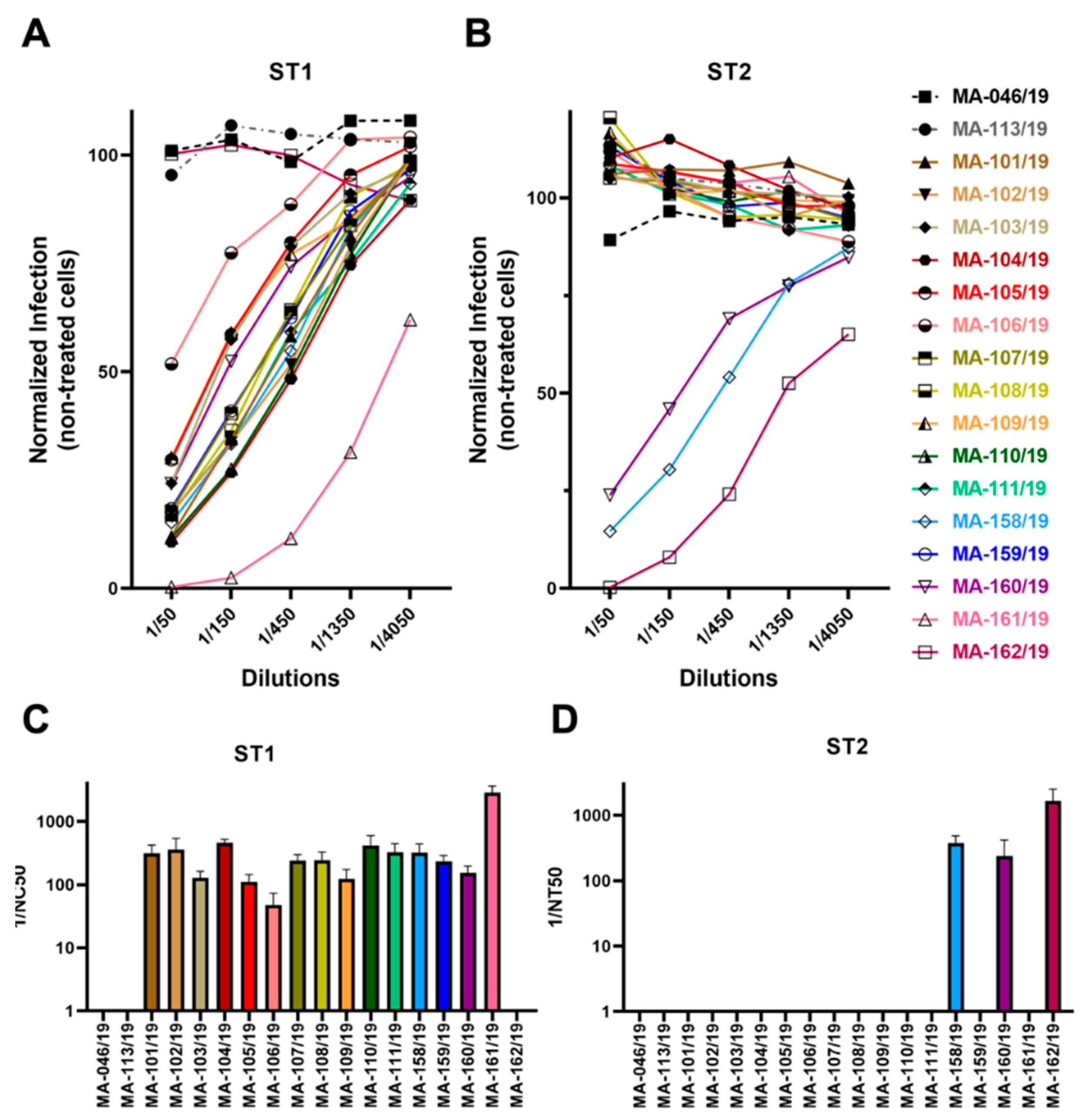
3.4. Low Frequency of Cross-Neutralization between Sera Neutralizing Genotype 1 or Genotype 2
3.5. No Evidence for env-Associated Differences in Cell Tropism
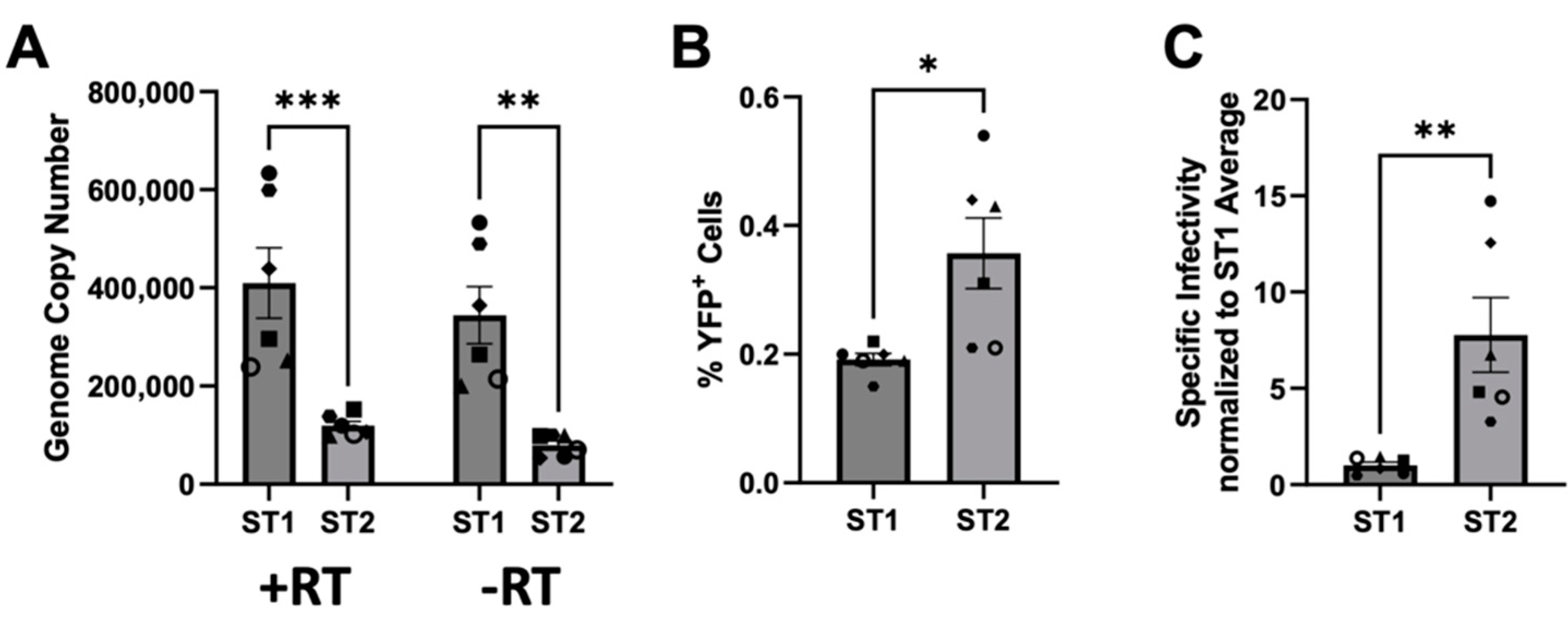
3.6. SFVmmu_R289hybAGM env Effects Lower Particle Release and Higher Specific Infectivity
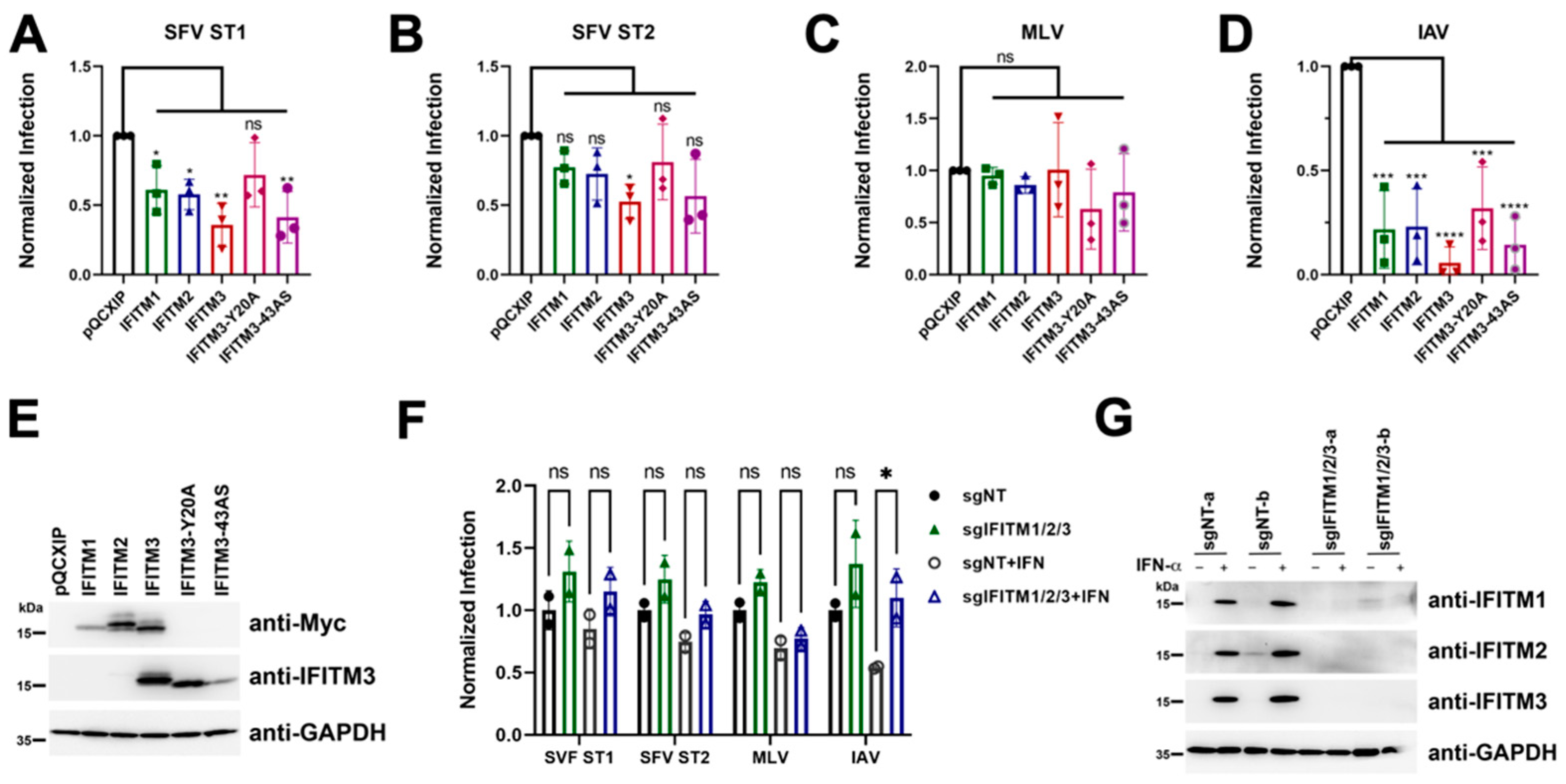
3.7. No Evidence for env-Associated Differences in Susceptibility to IFITM-Mediated Restriction at the Cell Entry Step
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Switzer, W.M.; Salemi, M.; Shanmugam, V.; Gao, F.; Cong, M.-E.; Kuiken, C.; Bhullar, V.; Beer, B.E.; Vallet, D.; Gautier-Hion, A.; et al. Ancient Co-Speciation of Simian Foamy Viruses and Primates. Nature 2005, 434, 376–380. [Google Scholar] [CrossRef]
- Katzourakis, A.; Gifford, R.J.; Tristem, M.; Gilbert, M.T.P.; Pybus, O.G. Macroevolution of Complex Retroviruses. Science 2009, 325, 1512. [Google Scholar] [CrossRef]
- Han, G.-Z.; Worobey, M. Endogenous Viral Sequences from the Cape Golden Mole (Chrysochloris asiatica) Reveal the Presence of Foamy Viruses in All Major Placental Mammal Clades. PLoS ONE 2014, 9, e97931. [Google Scholar] [CrossRef] [PubMed]
- Meiering, C.D.; Linial, M.L. Historical Perspective of Foamy Virus Epidemiology and Infection. Clin. Microbiol. Rev. 2001, 14, 165–176. [Google Scholar] [CrossRef]
- Han, G.-Z.; Worobey, M. An Endogenous Foamy-Like Viral Element in the Coelacanth Genome. PLoS Pathog. 2012, 8, e1002790. [Google Scholar] [CrossRef]
- Chen, Y.; Zhang, Y.-Y.; Wei, X.; Cui, J. Multiple Infiltration and Cross-Species Transmission of Foamy Viruses across the Paleozoic to the Cenozoic Era. J Virol. 2021, 95, e0048421. [Google Scholar] [CrossRef] [PubMed]
- Naville, M.; Volff, J.-N. Endogenous Retroviruses in Fish Genomes: From Relics of Past Infections to Evolutionary Innovations? Front. Microbiol. 2016, 7, 1197. [Google Scholar] [CrossRef]
- Aiewsakun, P. Avian and Serpentine Endogenous Foamy Viruses, and New Insights into the Macroevolutionary History of Foamy Viruses. Virus Evol. 2020, 6, vez057. [Google Scholar] [CrossRef]
- Buseyne, F.; Betsem, E.; Montange, T.; Njouom, R.; Bilounga Ndongo, C.; Hermine, O.; Gessain, A. Clinical Signs and Blood Test Results among Humans Infected with Zoonotic Simian Foamy Virus: A Case-Control Study. J. Infect. Dis. 2018, 218, 144–151. [Google Scholar] [CrossRef] [PubMed]
- Gessain, A.; Montange, T.; Betsem, E.; Bilounga Ndongo, C.; Njouom, R.; Buseyne, F. Case-Control Study of the Immune Status of Humans Infected with Zoonotic Gorilla Simian Foamy Viruses. J. Infect. Dis. 2020, 221, 1724–1733. [Google Scholar] [CrossRef]
- Lambert, C.; Couteaudier, M.; Gouzil, J.; Richard, L.; Montange, T.; Betsem, E.; Rua, R.; Tobaly-Tapiero, J.; Lindemann, D.; Njouom, R.; et al. Potent Neutralizing Antibodies in Humans Infected with Zoonotic Simian Foamy Viruses Target Conserved Epitopes Located in the Dimorphic Domain of the Surface Envelope Protein. PLoS Pathog. 2018, 14, e1007293. [Google Scholar] [CrossRef] [PubMed]
- Engel, G.A.; Small, C.T.; Soliven, K.; Feeroz, M.M.; Wang, X.; Kamrul Hasan, M.; Oh, G.; Rabiul Alam, S.M.; Craig, K.L.; Jackson, D.L.; et al. Zoonotic Simian Foamy Virus in Bangladesh Reflects Diverse Patterns of Transmission and Co-Infection. Emerg. Microbes Infect. 2013, 2, e58. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Wang, H.; Jing, S.; Zeng, W. Simian Foamy Virus Prevalence in Macaca Mulatta and Zookeepers. AIDS Res. Hum. Retroviruses 2012, 28, 591–593. [Google Scholar] [CrossRef]
- Olszko, M.E.; Trobridge, G.D. Foamy Virus Vectors for HIV Gene Therapy. Viruses 2013, 5, 2585–2600. [Google Scholar] [CrossRef]
- Rajawat, Y.S.; Humbert, O.; Kiem, H.-P. In-Vivo Gene Therapy with Foamy Virus Vectors. Viruses 2019, 11, 1091. [Google Scholar] [CrossRef]
- Trobridge, G.D. Foamy Virus Vectors for Gene Transfer. Expert Opin. Biol. Ther. 2009, 9, 1427–1436. [Google Scholar] [CrossRef] [PubMed]
- Williams, D.A. Foamy Virus Vectors Come of Age. Mol. Ther. 2008, 16, 635. [Google Scholar] [CrossRef]
- Budzik, K.M.; Nace, R.A.; Ikeda, Y.; Russell, S.J. Evaluation of the Stability and Intratumoral Delivery of Foreign Transgenes Encoded by an Oncolytic Foamy Virus Vector. Cancer Gene Ther. 2022, 29, 1240–1251. [Google Scholar] [CrossRef] [PubMed]
- Budzik, K.M.; Nace, R.A.; Ikeda, Y.; Russell, S.J. Oncolytic Foamy Virus—Generation and Properties of a Nonpathogenic Replicating Retroviral Vector System That Targets Chronically Proliferating Cancer Cells. J. Virol. 2021, 95, 10. [Google Scholar] [CrossRef]
- Ensser, A.; Großkopf, A.K.; Mätz-Rensing, K.; Roos, C.; Hahn, A.S. Isolation and Sequence Analysis of a Novel Rhesus Macaque Foamy Virus Isolate with a Serotype-1-like Env. Arch. Virol. 2018, 163, 2507–2512. [Google Scholar] [CrossRef] [PubMed]
- Nandakumar, S.; Bae, E.H.; Khan, A.S. Complete Genome Sequence of a Naturally Occurring Simian Foamy Virus Isolate from Rhesus Macaque (SFVmmu_K3T). Genome Announc. 2017, 5, 33. [Google Scholar] [CrossRef]
- Aiewsakun, P.; Richard, L.; Gessain, A.; Mouinga-Ondémé, A.; Vicente Afonso, P.; Katzourakis, A. Modular Nature of Simian Foamy Virus Genomes and Their Evolutionary History. Virus Evol. 2019, 5, vez032. [Google Scholar] [CrossRef] [PubMed]
- Duda, A.; Lüftenegger, D.; Pietschmann, T.; Lindemann, D. Characterization of the Prototype Foamy Virus Envelope Glycoprotein Receptor-Binding Domain. J. Virol. 2006, 80, 8158–8167. [Google Scholar] [CrossRef]
- Flower, R.L.; Wilcox, G.E.; Cook, R.D.; Ellis, T.M. Detection and Prevalence of Serotypes of Feline Syncytial Spumaviruses. Arch. Virol. 1985, 83, 53–63. [Google Scholar] [CrossRef] [PubMed]
- Winkler, I.G.; Flügel, R.M.; Löchelt, M.; Flower, R.L. Detection and Molecular Characterisation of Feline Foamy Virus Serotypes in Naturally Infected Cats. Virology 1998, 247, 144–151. [Google Scholar] [CrossRef] [PubMed]
- Zemba, M.; Alke, A.; Bodem, J.; Winkler, I.G.; Flower, R.L.; Pfrepper, K.; Delius, H.; Flügel, R.M.; Löchelt, M. Construction of Infectious Feline Foamy Virus Genomes: Cat Antisera Do Not Cross-Neutralize Feline Foamy Virus Chimera with Serotype-Specific Env Sequences. Virology 2000, 266, 150–156. [Google Scholar] [CrossRef]
- Blochmann, R.; Curths, C.; Coulibaly, C.; Cichutek, K.; Kurth, R.; Norley, S.; Bannert, N.; Fiebig, U. A Novel Small Animal Model to Study the Replication of Simian Foamy Virus in Vivo. Virology 2014, 448, 65–73. [Google Scholar] [CrossRef]
- Alkhatib, G.; Combadiere, C.; Broder, C.C.; Feng, Y.; Kennedy, P.E.; Murphy, P.M.; Berger, E.A. CC CKR5: A RANTES, MIP-1alpha, MIP-1beta Receptor as a Fusion Cofactor for Macrophage-Tropic HIV-1. Science 1996, 272, 1955–1958. [Google Scholar] [CrossRef]
- Feng, Y.; Broder, C.C.; Kennedy, P.E.; Berger, E.A. HIV-1 Entry Cofactor: Functional CDNA Cloning of a Seven-Transmembrane, G Protein-Coupled Receptor. Science 1996, 272, 872–877. [Google Scholar] [CrossRef]
- Bailey, C.C.; Zhong, G.; Huang, I.-C.; Farzan, M. IFITM-Family Proteins: The Cell’s First Line of Antiviral Defense. Annu. Rev. Virol. 2014, 1, 261–283. [Google Scholar] [CrossRef]
- Huang, I.-C.; Bailey, C.C.; Weyer, J.L.; Radoshitzky, S.R.; Becker, M.M.; Chiang, J.J.; Brass, A.L.; Ahmed, A.A.; Chi, X.; Dong, L.; et al. Distinct Patterns of IFITM-Mediated Restriction of Filoviruses, SARS Coronavirus, and Influenza a Virus. PLoS Pathog. 2011, 7, e1001258. [Google Scholar] [CrossRef]
- Li, K.; Markosyan, R.M.; Zheng, Y.-M.; Golfetto, O.; Bungart, B.; Li, M.; Ding, S.; He, Y.; Liang, C.; Lee, J.C.; et al. IFITM Proteins Restrict Viral Membrane Hemifusion. PLoS Pathog. 2013, 9, e1003124. [Google Scholar] [CrossRef]
- Hörnich, B.F.; Großkopf, A.K.; Dcosta, C.J.; Schlagowski, S.; Hahn, A.S. Interferon-Induced Transmembrane Proteins Inhibit Infection by the Kaposi’s Sarcoma-Associated Herpesvirus and the Related Rhesus Monkey Rhadinovirus in a Cell-Specific Manner. mBio 2021, 12, e0211321. [Google Scholar] [CrossRef]
- Kim, J.; Shin, C.-G. IFITM Proteins Inhibit the Late Step of Feline Foamy Virus Replication. Anim. Cells Syst. 2020, 24, 282–288. [Google Scholar] [CrossRef]
- Feeroz, M.M.; Soliven, K.; Small, C.T.; Engel, G.A.; Andreina Pacheco, M.; Yee, J.L.; Wang, X.; Kamrul Hasan, M.; Oh, G.; Levine, K.L.; et al. Population Dynamics of Rhesus Macaques and Associated Foamy Virus in Bangladesh. Emerg. Microbes Infect. 2013, 2, e29. [Google Scholar] [CrossRef]
- Hahn, A.S.; Desrosiers, R.C. Rhesus Monkey Rhadinovirus Uses Eph Family Receptors for Entry into B Cells and Endothelial Cells but not Fibroblasts. PLoS Pathog. 2013, 9, e1003360. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Campeau, E.; Ruhl, V.E.; Rodier, F.; Smith, C.L.; Rahmberg, B.L.; Fuss, J.O.; Campisi, J.; Yaswen, P.; Cooper, P.K.; Kaufman, P.D. A Versatile Viral System for Expression and Depletion of Proteins in Mammalian Cells. PLoS ONE 2009, 4, e6529. [Google Scholar] [CrossRef] [PubMed]
- Hörnich, B.F.; Großkopf, A.K.; Schlagowski, S.; Tenbusch, M.; Kleine-Weber, H.; Neipel, F.; Stahl-Hennig, C.; Hahn, A.S. SARS-CoV-2 and SARS-CoV Spike-Mediated Cell-Cell Fusion Differ in Their Requirements for Receptor Expression and Proteolytic Activation. J. Virol. 2021, 95, 9. [Google Scholar] [CrossRef] [PubMed]
- Großkopf, A.K.; Schlagowski, S.; Fricke, T.; Ensser, A.; Desrosiers, R.C.; Hahn, A.S. Plxdc Family Members Are Novel Receptors for the Rhesus Monkey Rhadinovirus (RRV). PLoS Pathog. 2021, 17, e1008979. [Google Scholar] [CrossRef]
- Sanjana, N.E.; Shalem, O.; Zhang, F. Improved Vectors and Genome-Wide Libraries for CRISPR Screening. Nat. Meth. 2014, 11, 783–784. [Google Scholar] [CrossRef]
- Wrensch, F.; Karsten, C.B.; Gnirß, K.; Hoffmann, M.; Lu, K.; Takada, A.; Winkler, M.; Simmons, G.; Pöhlmann, S. Interferon-Induced Transmembrane Protein-Mediated Inhibition of Host Cell Entry of Ebolaviruses. J. Infect. Dis. 2015, 212, S210–S218. [Google Scholar] [CrossRef] [PubMed]
- Großkopf, A.K.; Ensser, A.; Neipel, F.; Jungnickl, D.; Schlagowski, S.; Desrosiers, R.C.; Hahn, A.S. A Conserved Eph Family Receptor-Binding Motif on the GH/GL Complex of Kaposi’s Sarcoma-Associated Herpesvirus and Rhesus Monkey Rhadinovirus. PLoS Pathog. 2018, 14, e1006912. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing Real-Time PCR Data by the Comparative C(T) Method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Bastone, P.; Löchelt, M. Kinetics and Characteristics of Replication-Competent Revertants Derived from Self-Inactivating Foamy Virus Vectors. Gene Ther. 2004, 11, 465–473. [Google Scholar] [CrossRef] [PubMed]
- Meiering, C.D.; Linial, M.L. Reactivation of a Complex Retrovirus Is Controlled by a Molecular Switch and Is Inhibited by a Viral Protein. Proc. Natl. Acad. Sci. USA 2002, 99, 15130–15135. [Google Scholar] [CrossRef] [PubMed]
- Kaul, A.; Schönmann, U.; Pöhlmann, S. Seroprevalence of Viral Infections in Captive Rhesus and Cynomolgus Macaques. Primate Biol. 2019, 6, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Plochmann, K.; Horn, A.; Gschmack, E.; Armbruster, N.; Krieg, J.; Wiktorowicz, T.; Weber, C.; Stirnnagel, K.; Lindemann, D.; Rethwilm, A.; et al. Heparan Sulfate Is an Attachment Factor for Foamy Virus Entry. J. Virol. 2012, 86, 10028–10035. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.F.; Sullivan, M.D.; Linial, M.L. Evidence That the Human Foamy Virus Genome Is DNA. J. Virol. 1999, 73, 1565–1572. [Google Scholar] [CrossRef]
- John, S.P.; Chin, C.R.; Perreira, J.M.; Feeley, E.M.; Aker, A.M.; Savidis, G.; Smith, S.E.; Elia, A.E.H.; Everitt, A.R.; Vora, M.; et al. The CD225 Domain of IFITM3 Is Required for Both IFITM Protein Association and Inhibition of Influenza a Virus and Dengue Virus Replication. J. Virol. 2013, 87, 7837–7852. [Google Scholar] [CrossRef] [PubMed]
- Williams, D.E.J.; Wu, W.-L.; Grotefend, C.R.; Radic, V.; Chung, C.; Chung, Y.-H.; Farzan, M.; Huang, I.-C. IFITM3 Polymorphism Rs12252-C Restricts Influenza a Viruses. PLoS ONE 2014, 9, e110096. [Google Scholar] [CrossRef] [PubMed]
- Couteaudier, M.; Montange, T.; Njouom, R.; Bilounga-Ndongo, C.; Gessain, A.; Buseyne, F. Plasma Antibodies from Humans Infected with Zoonotic Simian Foamy Virus Do Not Inhibit Cell-to-Cell Transmission of the Virus despite Binding to the Surface of Infected Cells. PLoS Pathog. 2022, 18, e1010470. [Google Scholar] [CrossRef]
- Stirnnagel, K.; Schupp, D.; Dupont, A.; Kudryavtsev, V.; Reh, J.; Müllers, E.; Lamb, D.C.; Lindemann, D. Differential PH-Dependent Cellular Uptake Pathways among Foamy Viruses Elucidated Using Dual-Colored Fluorescent Particles. Retrovirology 2012, 9, 71. [Google Scholar] [CrossRef]
- Gärtner, K.; Wiktorowicz, T.; Park, J.; Mergia, A.; Rethwilm, A.; Scheller, C. Accuracy Estimation of Foamy Virus Genome Copying. Retrovirology 2009, 6, 32. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, T.C.; Albrecht, P.; Hannah, J.E.; Tauraso, N.M.; Robbins, B.; Trimmer, R.W. Foamy Virus Serotypes 1 and 2 in Rhesus Monkey Tissues. Arch. Gesamte Virusforsch. 1972, 38, 216–224. [Google Scholar] [CrossRef] [PubMed]
- Berka, U.; Hamann, M.V.; Lindemann, D. Early Events in Foamy Virus-Host Interaction and Intracellular Trafficking. Viruses 2013, 5, 1055–1074. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Tuo, X.; Zhang, J.; Chai, K.; Tan, J.; Qiao, W. Antiviral Role of IFITM3 in Prototype Foamy Virus Infection. Virol. J. 2022, 19, 195. [Google Scholar] [CrossRef]


| Plasmid | Source | Reference/Identifier |
|---|---|---|
| psPAX2 | Addgene (kind gift from Didier Trono) | Addgene #12260 |
| VSV-G (pMD2.G) | Addgene Addgene (kind gift from Didier Trono) | Addgene #12259 |
| gag/pol | Addgene (kind gift from Tannishtha Reya) | Addgene #14887 |
| pLenti CMV GFP Neo (657-2) | Lenti CMV GFP Neo (657-2) was a gift from Eric Campeau and Paul Kaufman | Addgene #17447 [38] |
| Gal4-TurboGFP-Luc (Gal4- driven TurboGFP-luciferase reporter plasmid) | Laboratory of Alexander Hahn | [39] |
| AX526 (Gal4-driven TurboGFP-luciferase reporter lentivirus, pLenti-Gal4-TurboGFP-Luc) | Laboratory of Alexander Hahn | [40] |
| plentiCRISPRv2 encoding sgRNAs targeting IFITM1-3 (sgIFITM1/2/3-a, sgIFITM1/2/3-b) or non-targeting sgRNAs (sgNT-a, sgNT-b) | lentiCRISRPv2 backbone from Addgene (kind gift from Feng Zhang), inserts from the laboratory of Alexander Hahn | [33,41] |
| Vp16-Gal4 | Laboratory of Alexander Hahn | [39] |
| pCAGGS IAV_WSN-HA | Laboratory of Stefan Pöhlmann | [42] |
| pCAGGS IAV_WSN-NA | Laboratory of Stefan Pöhlmann | [42] |
| paMLV_env | Laboratory of Michael Farzan | [31] |
| pQXCIP | Laboratory of Stefan Pöhlmann | [42] |
| pQCXIP-IFITM1 | Laboratory of Stefan Pöhlmann | [42] |
| pQCXIP-IFITM2 | Laboratory of Stefan Pöhlmann | [42] |
| pQCXIP-IFITM3 | Laboratory of Stefan Pöhlmann | [42] |
| pQCXIP-IFITM3-Y20A | Laboratory of Stefan Pöhlmann | [33] |
| pQCXIP-IFITM3-43AS | Laboratory of Stefan Pöhlmann | [33] |
| Cell Line/Cells | Cell Line Origin |
|---|---|
| HEK 293T | RRID:CVCL_0063, a kind gift from Vladan Rankovic, Göttingen, and originally purchased from the ATCC |
| A549 | A kind gift from the laboratory of Stefan Pöhlmann, German Primate Center-Leibniz Institute for Primate Research, Göttingen, Germany |
| SLK | RRID:CVCL_9569, NIH AIDS Research and Reference Reagent program |
| Human foreskin fibroblasts (HFF) | A kind gift from the laboratory of Klaus Korn, Universitätsklinikum Erlangen, Institute for Clinical and Molecular Virology, Erlangen, Germany |
| Human vascular endothelial cells (HUVEC) | PromoCell |
| HaCaT | RRID:CVCL_0038, CLS Cell Lines Service GmbH |
| MDA-MB-231 | ATCC HTB-26, a kind gift from the laboratory of Felix Engel |
| BJAB | Leibniz-Institute DSMZ (Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH) |
| MFB5487 | Macaca fuscata B cell line, established from PBMC through immortalization with Herpesvirus papio. A kind gift from Ulrike Sauermann [40] |
| Primary rhesus monkey fibroblasts | A kind gift from the laboratory of Rüdiger Behr. |
| Target | Details |
|---|---|
| IFITM1 | Goat, R&D (AF4827), 1:500 (secondary: Proteintech rabbit anti-goat 1:10,000) |
| IFITM2 | Mouse, Proteintech (66137-1-lg), 1:1000 (secondary: Dianova donkey anti-mouse 1:10,000) |
| IFITM3 | Rabbit, Cell Signal Technology (D8E8G) 1:1000 (secondary: Life Technologies goat anti-rabbit 1:10,000) |
| c-Myc | Mouse, Santa Cruz Biotechnology (9E10) 1:1000 (secondary: Dianova donkey anti-mouse 1:10,000) |
| V5 | Mouse, BioRad (MCA1360), 1:1000 (secondary: Dianova donkey anti-mouse 1:10,000) |
| GAPDH | Mouse, GenScript, 1:5000 (secondary: Dianova donkey anti-mouse 1:10,000) |
| GFP | Rabbit, GenScript, 1:1000 (secondary: Life Technologies goat anti-rabbit 1:10,000) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fricke, T.; Schlagowski, S.; Liu, S.; Yang, X.; Fiebig, U.; Kaul, A.; Ensser, A.; Hahn, A.S. Comparison of a Genotype 1 and a Genotype 2 Macaque Foamy Virus env Gene Indicates Distinct Infectivity and Cell-Cell Fusion but Similar Tropism and Restriction of Cell Entry by Interferon-Induced Transmembrane Proteins. Viruses 2023, 15, 262. https://doi.org/10.3390/v15020262
Fricke T, Schlagowski S, Liu S, Yang X, Fiebig U, Kaul A, Ensser A, Hahn AS. Comparison of a Genotype 1 and a Genotype 2 Macaque Foamy Virus env Gene Indicates Distinct Infectivity and Cell-Cell Fusion but Similar Tropism and Restriction of Cell Entry by Interferon-Induced Transmembrane Proteins. Viruses. 2023; 15(2):262. https://doi.org/10.3390/v15020262
Chicago/Turabian StyleFricke, Thomas, Sarah Schlagowski, Shanchuan Liu, Xiaoliang Yang, Uwe Fiebig, Artur Kaul, Armin Ensser, and Alexander S. Hahn. 2023. "Comparison of a Genotype 1 and a Genotype 2 Macaque Foamy Virus env Gene Indicates Distinct Infectivity and Cell-Cell Fusion but Similar Tropism and Restriction of Cell Entry by Interferon-Induced Transmembrane Proteins" Viruses 15, no. 2: 262. https://doi.org/10.3390/v15020262
APA StyleFricke, T., Schlagowski, S., Liu, S., Yang, X., Fiebig, U., Kaul, A., Ensser, A., & Hahn, A. S. (2023). Comparison of a Genotype 1 and a Genotype 2 Macaque Foamy Virus env Gene Indicates Distinct Infectivity and Cell-Cell Fusion but Similar Tropism and Restriction of Cell Entry by Interferon-Induced Transmembrane Proteins. Viruses, 15(2), 262. https://doi.org/10.3390/v15020262






